Abstract
The aim of the current study was to evaluate the relation between xeroderma pigmentosum complementation group C (XPC) polymorphisms and susceptibility to breast cancer (BC), the development and progression of disease, and response to different individualized drug treatments. We investigated two polymorphisms in XPC Ala499Val and Lys939Gln using PCR‐RFLP assays including 618 cases and 622 controls. The frequency of the TT genotype of Ala499Val (adjusted odds ratio = 1.575; 95% confidence interval, 1.104–2.245; P = 0.012) and the AC genotype of Lys939Gln (adjusted odds ratio = 1.330; 95% confidence interval, 1.045–1.694; P = 0.020) were found to significantly increase the risk of developing BC. The CT+TT genotypes of Ala499Val were associated with estrogen receptor positive, and Her‐2 and p53 negative status, and the AC+CC genotypes of Lys939Gln were associated with BRCA1 negative status. Moreover, a significantly increased chance of treatment with neoadjuvant anthracycline‐based chemotherapy response was found in women carrying TT genotype of Ala499Val, or CC and AC genotypes of Lys939Gln. In addition, a significantly longer overall survival after chemotherapy was observed in patients who had XPC Lys939Gln AC+CC genotypes with estrogen receptor positive (log–rank test, P = 0.086) and p53 negative (log–rank test, P = 0.020). The current data suggested that XPC Ala499Val and Lys939Gln polymorphisms may contribute to the identification of patients with increased risk for BC. Moreover, the polymorphisms were associated with the prognosis of BC patients. (Cancer Sci 2012; 103: 1207–1214)
Breast cancer is the most common neoplasm among women. Similar to other cancers, breast cancer is the ultimate outcome of multiple hereditary and environmental factors, most likely including genetic mutations that are particularly important in DNA repair genes.1 Polymorphisms in DNA repair genes have been reported that could lead to a deficiency in DNA repair capacity, subsequently contributing to genomic instability and an increase in an individual's susceptibility to cancer.2, 3, 4 Several epidemiologic studies have identified some of single nucleotide polymorphisms (SNPs) in DNA repair genes as risk factors for cancers.5, 6, 7
As an important DNA damage recognition protein, xeroderma pigmentosum complementation group C (XPC) is involved in global genome DNA repair, a subclass of nucleotide excision repair (NER).8 The XPC gene is localized at 3p25 and encodes a protein of 940 amino acids.9 In the XPC gene, two common polymorphisms have been widely studied: (i) a substitution of alanine for valine in codon 499 (Ala499Val), with a C‐to‐T transition in exon 8, in the interaction domain of XPC with hHRAD23; and (ii) an A‐to‐C transition in exon 15 resulting in a lysine‐to‐glutamine transition at position 939 (Lys939Gln), located in the interaction domain with TFIIH. Previous studies have reported on the associations between SNPs in the XPC gene with risk of breast cancer,4, 10, 11, 12, 13, 14 lung cancer,15, 16, 17, 18, 19, 20, 21 bladder cancer,22, 23, 24, 25 and other cancers,26, 27, 28, 29, 30, 31, 32 but the results were inconclusive. These findings suggested that the effects of these polymorphisms remained unclear, and further investigations were needed to resolve these discrepancies. Therefore, we investigated the association of the two SNPs with susceptibility to breast cancer (BC), the development and progression of disease, and patients’ responses to different individualized drug treatments.
In the present study, we carried out a case–control study of spontaneous BC patients and age/gender frequency‐matched healthy controls to assess whether the two SNPs XPC Ala499Val and XPC Lys939Gln contributed to increased BC susceptibility. We further explored the association between the polymorphisms, the clinicopathological profile of BC, and therapeutic outcomes after adjuvant anthracycline‐based chemotherapy.
Materials and Methods
Study subjects
The study was approved by the regional ethics committee at China Medical University (Shenyang, China). All breast cancer patients who participated in the study underwent surgery at the First Hospital of China Medical University between January 2005 and December 2010 and were of Han Chinese ethnicity, unrelated to each other, newly diagnosed, with pathologically confirmed disease, and previously untreated. The control subjects, women randomly selected from the same geographical region with no history of cancer, were frequency‐matched to the cases in terms of age (±5 years) and ethnicity.
A clinical oncologist collected clinical and pathological characteristics and therapeutic response after chemotherapy retrospectively from medical records. Clinical data were collected from medical records including age at the time of diagnosis, menopause status, tumor size, tumor stage, tumor type, lymph node status, and estrogen receptor (ER), progesterone receptor, human epidermal growth factor receptor‐2 (Her‐2), tumor protein 53 (p53), breast cancer type 1 susceptibility protein (BRCA1), and BRCA2 status. Although patients received different types of chemotherapy, hormones, radiotherapy, or biological treatment, therapeutic outcome was evaluated only in those patients who received anthracycline‐based (epirubicin [E] or doxorubicin [A]) chemotherapy. Anthracycline‐based chemotherapy consists of cyclophosphamide (C), the anthracycline agent (E or A), and/or 5‐fluorouracil (F), (CEF and CAF regimens) combined with radiotherapy.
According to Response Evaluation Criteria in Solid Tumors,33 for patients with four times of the CEF/CAF program, the degree of sensitivity to chemotherapeutic drugs is divided into: (i) complete response (CR), the disappearance of all target lesions and any pathological lymph nodes (whether target or non‐target) must have reduction in short axis to <10 mm; (ii) partial response (PR), at least a 30% decrease in the sum of diameters of target lesions, taking as reference the baseline sum diameters; (iii) progressive disease (PD), at least a 20% increase in the sum of diameters of target lesions, taking as reference the smallest sum on study. In addition to the relative increase of 20%, the sum must also show an absolute increase of at least 5 mm; and (iv) stable disease (SD), neither sufficient shrinkage to qualify for PR nor sufficient increase to qualify for PD, taking as reference the smallest sum diameters while on study.
Genotyping
DNA samples were obtained from stored blood samples using Qiagen standard protocols (Shanghai, China). Genotyping for XPC Ala499Val and Lys939Gln polymorphisms by PCR‐RFLP assay was done following a modified method of Guo et al.34 Primer sequences were: Ala499 Val, 5′‐TAAGGACCCAAGCTTGCCCG‐3′ (forward) and 5′‐CCCACTTTTCCTCCTGCTCACAG‐3′ (reverse); and Lys939Gln, 5′‐ACCAGCTCTCAAGCAGAAGC‐3′ (forward) and 5′‐CTGCCTCAGTTTGCCTTCTC‐3′ (reverse).
Polymerase chain reaction was carried out as follows: an initial melting step of 5 min at 94°C; 35 cycles of denaturation for 30 s at 94°C; annealing for 30 s at 63°C for Ala499Val, and 55°C for Lys939Gln; extension for 45 s at 72°C, followed by a 5 min final extension at 72°C. The PCR products were then digested with restriction endonucleases. For XPC Lys939Gln, the PCR products were digested with PvuII (New England BioLabs, Ipswich, MA, USA) overnight at 37°C. The variant C allele had a PvuII restriction site and after the digestion, two bands (150 and 131 bp) were generated; the wild‐type A allele lacked this restriction site and a single band with a size of 281 bp was obtained. For XPC Ala499Val, the wild‐type allele (C) produced two fragments of 131 and 21 bp and the polymorphic allele (T) produced a single 152 bp fragment.
For quality control, 10% of the two PCR‐RFLP assays were randomly selected for sequencing. These results of the quality control analysis confirmed 100% concordance.
Statistical analysis
In order to match the groups of case and control in terms of several putative confounding factors, Pearson's 2 × 2 χ2‐test (gender) and independent sample t‐tests (mean age) were used for analysis of the differences of several qualitative and quantitative traits. To evaluate deviation from the Hardy–Weinberg equilibrium, the discrepancies between observed and expected genotype frequencies in patients and controls were compared using a χ2‐test, with one degree of freedom. Genotypic risk of the two SNPs for BC, in terms of odds ratio (OR) and 95% confidence interval (CI), was derived from binary logistic regression analysis with SNP genotypes as the explainable variables. Overall survival (OS) was calculated as the time to progression or death without progression from the date of diagnosis. Survival distributions were estimated with the Kaplan–Meier method and compared with the log–rank test. Finally, the associations between individual epidemiologic risk factors, clinical characteristics, genotypes, and the time to the occurrence of BC were assessed using univariate and multivariate Cox regression analysis. In this study, the statistical analyses were carried out using spss version 13.0 statistical software (SPSS, Chicago, IL, USA). Statistical significance was set at P < 0.05 and all tests were two‐sided.
Results
Subject characteristics
There were no significant differences in the distributions of age and menopausal state between cases and controls (P = 0.819 and P = 0.574), and the mean age was also matched for cases (range, 21–83 years; median, 45 years) and controls (range, 23–79 years; median, 47 years). Before statistical analysis, we also tested the two SNPs loci with the Hardy–Weinberg equilibrium. The SNPs in control populations did not significantly deviate from the Hardy–Weinberg equilibrium (data not shown). The demographics, risk exposure, clinical variables, and therapeutic outcome for all 618 BC patients are summarized in Table 1.
Table 1.
Frequency distribution of selected variables in patients with breast cancer (n = 618)
| Characteristic | Cases (n = 618) | |
|---|---|---|
| No. | % | |
| Sex | ||
| Female | 618 | 100 |
| Age at diagnosis, years | 618 | |
| ≤45 | 325 | 52.6 |
| >45 | 293 | 47.4 |
| Menopausal status | 618 | |
| Premenopausal | 334 | 54.0 |
| Postmenopausal | 284 | 46.0 |
| First‐degree family history of breast cancer | 618 | |
| No | 472 | 76.4 |
| Yes | 146 | 23.6 |
| Tumor size (cm) | 605 | |
| ≤2.0 | 259 | 42.8 |
| 2.1–4.0 | 245 | 40.5 |
| 4.1+ | 101 | 16.7 |
| Lymph node metastasis | 410 | |
| Node‐negative | 216 | 52.7 |
| Node‐positive | 194 | 47.3 |
| Histology | 618 | |
| DIC | 530 | 85.8 |
| LIC | 66 | 10.7 |
| Others | 22 | 3.6 |
| Therapeutic regimen | ||
| Anthracycline‐based chemotherapy[Link] | 491 | 79.4 |
| Other chemotherapies or treatments[Link] | 127 | 20.6 |
†Contains cyclophosphamide, the anthracycline agent and/or 5‐fluorouracil. ‡Treatments include docetaxel plus pirarubicin, surgery only, total parenteral nutrition, or traditional Chinese treatments. DIC, ductal invasive carcinoma; LIC, lobular invasive carcinoma.
Genotypic association analysis of two SNPs between patients with BC and controls
Overall genotypes and allele frequencies for the XPC Ala499Val and Lys939Gln polymorphisms are shown in Table 2. We found a significant increase in the BC group (16.02%) compared with the control group (12.06%) in the frequency of the TT genotype of Ala499Val (P = 0.012). Significantly different frequency was also observed for the CT+TT genotypes (P = 0.030). For XPC Lys939Gln, the frequencies of the AC genotype were 51.94% and 46.92% for the BC and control group, respectively (P = 0.020). Moreover, the AC+CC genotypes in the BC group (65.53%) has a higher frequency than the control group (58.84%) (P = 0.015).
Table 2.
Frequency distribution of XPC genotypes and their associations with risk of developing breast cancer
| Genotype | Cases (n = 618) | Controls[Link] (n = 622) | P [Link] | Adjusted OR§ (95% CI) |
|---|---|---|---|---|
| No. (%) | No. (%) | |||
| XPC Ala499Val | ||||
| CC | 197 (31.88) | 235 (37.78) | 1.00 (Ref.) | |
| CT | 322 (52.10) | 312 (50.16) | 0.096 | 1.231 (0.963–1.573) |
| TT | 99 (16.02) | 75 (12.06) | 0.012 | 1.575 (1.104–2.245) |
| CT+TT | 421 (68.12) | 387 (62.22) | 0.030 | 1.298 (1.027–1.640) |
| XPC Lys939Gln | ||||
| AA | 213 (34.47) | 256 (41.2) | 1.00 (Ref.) | |
| AC | 321 (51.94) | 290 (46.6) | 0.020 | 1.330 (1.045–1.694) |
| CC | 84 (13.59) | 76 (12.2) | 0.121 | 1.328 (0.927–1.903) |
| AC+CC | 405 (65.53) | 366 (58.8) | 0.015 | 1.330 (1.056–1.674) |
†The observed genotype frequency among individuals in the control group was in agreement with Hardy–Weinberg equilibrium P = 0.064 for XPC Ala499Val; P = 0.659 for XPC Lys939Gln). ‡P‐values were calculated from two‐sided χ2‐tests for either genotype distribution or allele frequency. §Odds ratio (OR) and 95% confidence interval (CI) values were calculated by unconditional logistic regression adjusted for age and menopausal status.
Next, we used adjusted the OR values and 95% CI to predict risk of breast cancer using logistic regression analysis. The results showed that the CT+TT genotypes of XPC Ala499Val had a higher risk of BC (adjusted OR = 1.298; 95% CI, 1.027–1.640; P = 0.030). Patients with the TT genotype had a 1.58‐fold increased risk of developing BC (adjusted OR = 1.575; 95% CI, 1.104–2.245; P = 0.012). In addition, the AC genotype (adjusted OR = 1.330; 95% CI, 1.045–1.694; P = 0.020) and the AC+CC genotypes (adjusted OR = 1.330; 95% CI, 1.056–1.674; P = 0.015) of Lys939Gln both have increased the risk of BC (Table 2).
Relation between genotype distribution and clinicopathologic characteristics
To investigate the effects of the two SNPs on the clinicopathological features of BC patients, we used the χ2‐test and unconditional logistic regression adjusted by age and menopausal state. Clinical and pathological characteristics of the patients differentiated according to XPC genotypes are shown in Table 3.
Table 3.
Correlations between clinicopathologic parameters and XPC Ala499Val and Lys939Gln polymorphisms in patients with breast cancer (n = 618)
| Features |
Total no. |
XPC Ala499Val | XPC Lys939Gln | ||||||
|---|---|---|---|---|---|---|---|---|---|
|
CC No. (%) |
CT+TT No. (%) |
P [Link] | OR (95% CI)§ |
AA No. (%) |
AC+CC No. (%) |
P [Link] | OR (95% CI)§ | ||
| Age at diagnosis, years[Link] | 618 | 0.438 | 0.672 | ||||||
| ≤45 | 99 (16.0) | 226 (36.6) | 1.0 (ref.) | 115 (18.6) | 210 (34.0) | 1.0 (ref.) | |||
| >45 | 98 (15.9) | 195 (31.6) | 0.872 (0.621–1.223) | 98 (15.9) | 195 (31.6) | 1.090 (0.781–1.520) | |||
| Menopausal status | 618 | 0.603 | 0.309 | ||||||
| Premenopausal | 103 (16.7) | 231 (37.4) | 1.0 (ref.) | 109 (17.6) | 225 (36.4) | 1.00 (ref.) | |||
| Postmenopausal | 94 (15.2) | 190 (30.7) | 0.901 (0.642–1.265) | 104 (16.8) | 180 (29.1) | 0.838 (0.601–1.169) | |||
| First‐degree family history of breast cancer | 618 | 0.839 | 0.274 | ||||||
| No | 149 (24.1) | 323 (52.3) | 1.0 (ref.) | 157 (25.4) | 315 (51.0) | 1.00 (ref.) | |||
| Yes | 48 (7.8) | 98 (15.9) | 0.942 (0.634–1.400) | 56 (9.1) | 90 (14.6) | 0.801 (0.545–1.177) | |||
| Tumor size (cm) | 605 | 0.199 | 0.352 | ||||||
| ≤2.0 | 78 (12.9) | 181 (29.9) | NA | 83 (13.7) | 176 (29.1) | NA | |||
| 2.1–4.0 | 75 (12.4) | 170 (28.1) | NA | 93 (15.4) | 152 (25.1) | NA | |||
| ≥4.1 | 40 (6.6) | 61 (10.1) | NA | 33 (5.5) | 68 (11.2) | NA | |||
| Lymph node metastasis | 410 | 0.916 | 0.079 | ||||||
| Node‐negative | 69 (16.8) | 147 (35.8) | 1.0 (ref.) | 85 (20.7) | 131 (32.0) | 1.0 (ref.) | |||
| Node‐positive | 63 (15.4) | 131 (32.0) | 0.976 (0.645–1.478) | 60 (14.6) | 134 (32.7) | 1.449 (0.963–2.181) | |||
| Histology | 618 | 0.507 | 0.732 | ||||||
| DIC | 166 (26.9) | 364 (58.9) | NA | 185 (29.9) | 345 (55.8) | NA | |||
| LIC | 25 (4.0) | 41 (6.6) | NA | 22 (3.6) | 44 (7.1) | NA | |||
| Others | 6 (1.0) | 16 (2.6) | NA | 6 (1.0) | 16 (2.6) | NA | |||
| ER status | 610 | 0.011 | 0.438 | ||||||
| Negative | 98 (16.1) | 160 (26.2) | 1.0 (ref.) | 84 (13.8) | 174 (28.5) | 1.0 (ref.) | |||
| Positive | 99 (16.2) | 253 (41.5) | 1.565 (1.112–2.204) | 126 (20.7) | 226 (37.0) | 0.866 (0.617–1.216) | |||
| PR status | 609 | 0.078 | 0.795 | ||||||
| Negative | 90 (14.8) | 157 (25.8) | 1.0 (ref.) | 87 (14.3) | 160 (26.3) | 1.0 (ref.) | |||
| Positive | 107 (17.6) | 255 (41.9) | 1.366 (0.969–1.926) | 123 (20.2) | 239 (39.2) | 1.057 (0.752–1.484) | |||
| Her‐2 status | 600 | 0.019 | 0.384 | ||||||
| Negative | 41 (6.8) | 121 (20.2) | 1.0 (ref.) | 60 (10.0) | 102 (17.0) | 1.0 (ref.) | |||
| Positive | 155 (25.8) | 283 (47.2) | 0.619 (0.413–0.927) | 145 (24.2) | 293 (18.8) | 1.189 (0.816–1.731) | |||
| p53 status | 598 | 0.010 | 0.546 | ||||||
| Negative | 71 (11.9) | 197 (33.0) | 1.0 (ref.) | 97 (16.2) | 171 (28.6) | 1.0 (ref.) | |||
| Positive | 120 (20.1) | 209 (35.0) | 0.628 (0.441–0.893) | 111 (18.6) | 218 (36.5) | 1.114 (0.794–1.562) | |||
| BRCA1 status | 587 | 0.823 | <0.001 | ||||||
| Negative | 38 (6.5) | 77 (13.1) | 1.0 (ref.) | 19 (3.2) | 96 (16.4) | 1.0 (ref) | |||
| Positive | 149 (25.4) | 323 (55.0) | 1.070 (0.693–1.652) | 186 (31.7) | 286 (48.7) | 0.304 (0.180–0.515) | |||
| BRCA2 status | 574 | 0.235 | 0.789 | ||||||
| Negative | 79 (13.8) | 149 (26.0) | 1.0 (ref.) | 78 (13.6) | 150 (26.2) | 1.0 (ref.) | |||
| Positive | 103 (18.0) | 242 (42.2) | 1.246 (0.872–1.780) | 122 (21.3) | 223 (38.9) | 0.950 (0.669–1.351) | |||
†Median age was 45 years. ‡P‐ values were calculated from two‐sided χ2‐tests or Fisher's exact test. §Odds ratio (OR) and 95% confidence interval (CI) values were calculated by unconditional logistic regression adjusted for age and menopausal status. BRCA1, breast cancer type 1 susceptibility protein; BRCA2, breast cancer type 2 susceptibility protein; ER, estrogen receptor; DIC, ductal invasive carcinoma; LIC, lobular invasive carcinoma; Her‐2, human epidermal growth factor receptor‐2; NA, not applicable; p53, tumor protein 53; PR, progesterone receptor; ref., reference.
We observed that the distribution frequency of the CT+TT genotypes of XPC Ala499Val were associated with the patients who had ER positive status rather than the CC genotype (adjusted OR = 1.565; 95% CI, 1.112–2.204; P = 0.011). The CT+TT genotypes were associated with Her‐2 (adjusted OR = 0.619; 95% CI, 0.143–0.927; P = 0.019) and p53 negative status (adjusted OR = 0.628; 95% CI, 0.441–0.893; P = 0.010) (Table 3). For Lys939Gln, the AC+CC genotypes were associated with BRCA1 negative status (adjusted OR = 0.304; 95% CI, 0.180–0.515; P < 0.001). There were no statistically significant correlations between genotype distributions and age at diagnosis, menopausal state, first‐degree family history of breast cancer, tumor size, histology, clinical stage, or lymph node metastasis.
Association between genotypes and response to neoadjuvant chemotherapy
Neoadjuvant chemotherapy treatment is given to patients with the aim of reducing the size of the primary tumor to increase the likelihood of breast conservation and to abolish occult systemic metastases. In our study, only 72 patients received neoadjuvant anthracycline‐based chemotherapy before surgical operation and 36 of them were CR and PR. For XPC Ala499Val, the TT genotype had a significantly higher response rate (73.3%; 11 responders among 15 patients). Statistical analysis showed a significantly increased chance of treatment response in women carrying the TT genotype (OR = 0.218; 95% CI, 0.053–0.895; P = 0.029). In addition, there were statistically significant associations between polymorphism of Lys939Gln and successful treatment with neoadjuvant anthracycline‐based chemotherapy for the CC genotype (P = 0.013), the AC genotype (P = 0.049), and the AC+CC genotypes (P = 0.020) (Table 4). The results suggested that the polymorphisms of XPC may increase the sensitivity of neoadjuvant anthracycline‐based chemotherapy and improve the effects of treatment of BC patients.
Table 4.
Association of XPC polymorphisms with therapeutic response to neoadjuvant chemotherapy in breast cancer patients
| Variable | No. | RECIST | P [Link] | OR (95% CI)‡ | |
|---|---|---|---|---|---|
| CR and PR (%) | SD and PD (%) | ||||
| XPC Ala499Val | 72 | ||||
| CC | 24 | 9 (37.5) | 15 (62.5) | 1.00 (ref.) | |
| CT | 33 | 16 (48.5) | 17 (51.5) | 0.409 | 0.637 (0.218‐1.862) |
| TT | 15 | 11 (73.3) | 4 (26.7) | 0.029 | 0.218 (0.053‐0.895) |
| CT+TT | 48 | 27 (58.3) | 21 (41.7) | 0.137 | 0.467 (0.171‐1.274) |
| XPC Lys939Gln | 72 | ||||
| AA | 21 | 6 (28.6) | 15 (71.4) | 1.00 (ref.) | |
| AC | 42 | 23 (54.8) | 19 (45.2) | 0.049 | 0.330 (0.107‐1.018) |
| CC | 9 | 7 (77.8) | 2 (22.2) | 0.013 | 0.114 (0.018‐0.716) |
| AC+CC | 51 | 30 (58.8) | 21 (41.2) | 0.020 | 0.280 (0.093‐0.840) |
†P‐values were calculated from χ2‐tests or Fisher's exact test. ‡Odds ratio (OR) and 95% confidence interval (CI) values were calculated by unconditional logistic regression adjusted for age and menopausal status. CR, complete response; PD, progressive disease; PR, partial response; RECIST, Response Evaluation Criteria in Solid Tumors; ref., reference; SD, stable disease.
Genotypic difference and therapeutic regimen, prognosis
In our study, OS was evaluated using the Kaplan–Meier method in BC patients (n = 618). But we did not find an association between the polymorphisms of Ala499Val (Fig. 1A) or Lys939Gln (Fig. 1B) and OS.
Figure 1.
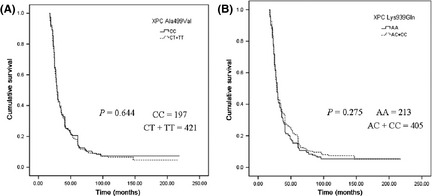
Kaplan–Meier curves illustrating the overall survival of breast cancer patients with XPC Ala499Val (A) and XPC Lys939Gln (B) polymorphisms.
It is well established that anthracycline‐based chemotherapy influences the outcome for BC patients, and it has been shown to be effective in the treatment of BC. Next, we further studied the hierarchical factors ER, progesterone receptor, Her‐2, and p53 status in patients with BC who received anthracycline‐based neoadjuvant and/or adjuvant chemotherapy. We found that the Lys939Gln polymorphism had the extended trend of OS for patients treated with anthracycline‐based chemotherapy (log–rank test, P = 0.187) (Fig. 2A). Moreover, this trend was also found in patients with ER positive status who received anthracycline‐based chemotherapy (log–rank test, P = 0.086) (Fig. 2B). The estimated median OS for patients with ER positive status who had the AA of XPC Lys939Gln genotype was 37.834 months (95%CI, 31.029–44.639) and for those with the AC+CC genotypes, 43.968 months (95% CI, 37.981–49.956). In addition, patients with p53 negative status with Lys939Gln AC+CC genotypes showed significantly longer OS (log–rank test, P = 0.020) (Fig. 2C). The estimated median OS for patients with p53 negative status who had the AA genotype and the AC+CC genotypes of XPC Lys939Gln was 34.883 months (95% CI, 29.748–40.018) and 42.569 months (95% CI, 37.844–47.294), respectively. However, for patients with p53 positive status, there was no statistically significant differences (log−rank test, P = 0.752) (Fig. 2D). And there was no statistically significant differences for Her‐2 negative (log–rank test, P = 0.213) (Fig. 2E) and Her‐2 positive status (log–rank test, P = 0.385) (Fig. 2F).
Figure 2.
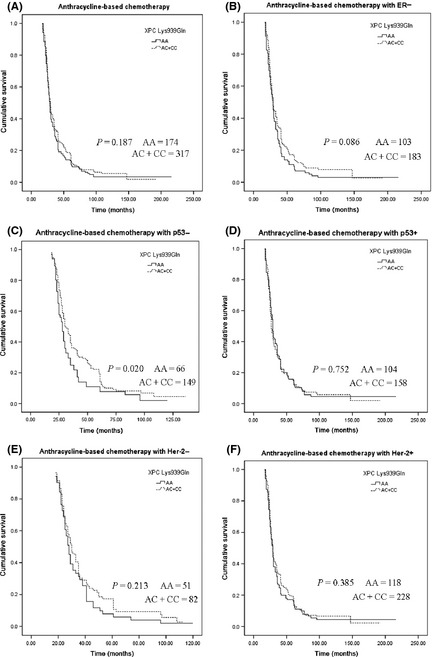
Kaplan–Meier curves illustrating the overall survival of breast cancer patients who received anthracycline‐based neoadjuvant and/or adjuvant chemotherapy (A), patients with estrogen receptor (ER) positive status (B), p53 negative status (C), p53 positive status (D), Her‐2 negative status (E), and Her‐2 positive status (F).
To estimate the independent impact of each variable on OS, a descriptive Cox proportional hazard model was carried out. Univariate analysis identified that the genotypes AC+CC of XPC Lys939Gln showed longer OS for the anthracycline‐based chemotherapy patients with p53 negative status. Finally, multivariate Cox regression analysis also identified that XPC Lys939Gln (OR = 0.620, 95% CI, 0.389–0.987; P = 0.044) was predictive of OS for patients (Table 5).
Table 5.
Hazard ratios for overall survival (OS) in the breast cancer patients with anthracycline‐based chemotherapy
| Variable | Univariate | P [Link] | Multivariate | P [Link]‡ |
|---|---|---|---|---|
| HR (95% CI)[Link] | HR (95% CI)[Link]‡ | |||
| Age (≥45 years vs <45 years) | 1.003 (0.764–1.316) | 0.983 | ||
| Menopausal status (postmenopausal vs premenopausal) | 0.870 (0.662–1.143) | 0.318 | ||
| XPC Ala499Val (CT+TT vs CC) | 1.227 (0.922–1.633) | 0.160 | ||
| XPC Lys939Gln (AC+CC vs AA) | 0.717 (0.536–0.960) | 0.025 | 0.620 (0.389–0.987) | 0.044 |
| First‐degree family history of breast cancer (yes vs no) | 1.064 (0.778–1.454) | 0.699 | ||
| Tumor size (>4 cm vs ≤4 cm) | 0.818 (0.667–1.002) | 0.053 | ||
| Histology (DIC/LIC vs others) | 1.064 (0.778–1.454) | 0.699 | ||
| Lymph node metastases (metastases vs non‐metastases) | 1.441 (1.013–2.049) | 0.042 | 1.585 (1.013–2.475) | 0.043 |
| ER status (positive vs negative) | 0.966 (0.734–1.272) | 0.805 | ||
| PR status (positive vs negative) | 1.074 (0.815–1.414) | 0.613 | ||
| Her‐2 status (positive vs negative) | 0.920 (0.692–1.221) | 0.562 | ||
| BRCA1 status (positive vs negative) | 0.997 (0.728–1.365) | 0.983 | ||
| BRCA2 status (positive vs negative) | 0.899 (0.674–1.199) | 0.469 |
†P‐values, odds ratios (OR), and 95% confidence intervals (CI) were assessed using univariate Cox regression analysis. ‡BRCA1, breast cancer type 1 susceptibility protein; BRCA2, breast cancer type 2 susceptibility protein; Her‐2, human epidermal growth factor receptor; DIC, ductal invasive carcinoma; ER, estrogen receptor; HR, hazards ratio; LIC, lobular invasive carcinoma; p53, tumor protein 53; PR, progesterone receptor.
Discussion
Recently, the associations between XPC polymorphisms and the risk of BC have been widely studied, but the results were inconclusive. Smith et al.,10 and Jorgensen et al.,4 did not find the genotypes distribution of XPC Ala499Val and Lys939Gln in the USA. Försti A et al.12 found that significant findings among Finnish were associations to XPC Lys939Gln, but these findings were not repeated in the Polish series. A meta‐analysis13 that involved a total of 11 studies including 2258 (44.36%) Caucasian, 1798 (35.32%) mixed, 814 (16.00%) African, and only 220 (4.32%) Asian BC cases (Zhang et al.14) reported that there was no significant association between the XPC Ala499Val or Lys939Gln polymorphisms and BC risk. However, they did not indicate an association between the polymorphisms and clinicopathological parameters or therapeutic outcome. Therefore, we investigated 618 BC cases to assess whether XPC Ala499Val and Lys939Gln polymorphisms contribute to increased susceptibility to BC, and we evaluated the association between the SNPs and the development and progression of disease, and response to different individualized drug treatments. We found that patients with the AC genotype of XPC Lys939Gln had a higher risk of BC (OR = 1.330), which agreed with the results of Zhang et al. and assessed many more BC cases in Asian women. Moreover, we found a significant increase in the frequency of the TT genotype of Ala499Val for the BC group, and the patients had increased risk of developing BC (OR = 1.575). It is suggested the XPC Ala499Val and Lys939Gln polymorphisms were both closely related to the development of BC in Chinese patients. It may be because the XPC protein binds with HR23B to form the heterodimeric complex, which is the first NER factor to detect a lesion and recruit the rest of the repair machinery to the damaged site.35, 36 The specific binding of XPC–HR23B is an important molecular process, based on detection by the NER machinery of a wide variety of lesions that vary in terms of chemical structure during DNA repair.8 The XPC gene polymorphisms, particularly nsSNP, may alter an individual's capacity to repair damaged DNA, possibly leading to genetic instability and carcinogenesis.
The XPC polymorphisms may cause instability in the DNA of tumor cells and promote tumor cell DNA damage, further increasing the efficacy of DNA‐damaging agents. Topoisomerase II is an important ribozyme involved in DNA repair and replication processes. Anthracycline‐based chemotherapy is currently one of the most commonly used chemotherapy regimens for BC patients and the main mechanism for it is the inhibition of DNA topoisomerase II. As an anticancer drug target, it combines to form stable drug–topoisomerase II–DNA complexes that cause the DNA breakage and lead to the death of tumor cells.37 Therefore, this study examined XPC gene polymorphisms on the clinical efficacy of a DNA damage agent. We first reported that XPC polymorphisms were associated with the prognosis of anthracycline‐based chemotherapy patients. Our study analyzed the therapeutic response to neoadjuvant anthracycline‐based chemotherapy in BC patients in relation to XPC gene polymorphisms and found that the TT genotype of XPC Ala499Val had a significantly higher response rate. Patients with the Lys939Gln polymorphism were also more likely to be successfully treated with neoadjuvant anthracycline‐based chemotherapy. In addition, we found that the Lys939Gln polymorphism has an extended trend of OS for anthracycline‐based chemotherapy patients, and anthracycline‐based chemotherapy patients with ER positive status. The result was consistent with Fleming et al.38 who indicated that SNPs in the XPC gene may represent novel markers of ovarian cancer response to platinum‐based chemotherapy. Notably, the patients with p53 negative status and Lys939Gln GC+CC genotypes showed significantly longer OS, suggesting that the GC+CC genotypes might increase the effects of anthracycline‐based chemotherapy. The tumor suppressor p53 mainly induced apoptosis in the DNA damage response and p53 negative expression could cause instability of genome function.39, 40 If p53 negative status was associated with the mutation of DNA repair gene XPC, it may decrease the ability to repair DNA, further increase the DNA double‐strand breakages, and induce more tumor cell to death by anthracycline. Therefore, the effects of anthracycline‐based chemotherapy could be more sensitive and have a longer OS in our study.
Moreover, our study analyzed the association between XPC polymorphisms and clinicopathological parameters. The results showed that the polymorphisms of XPC Ala499Val and Lys939Gln were associated with p53 and BRCA1 negative status, respectively. The results imply that XPC gene mutation can reduce the p53 and BRCA1 tumor suppressor role and further enhance the risk of cancer. In addition, the distribution frequencies of the XPC Ala499Val CT+TT genotypes were associated with patients who were ER positive. It showed us that XPC gene polymorphisms might be able to increase the ER level, and improve the efficiency of target and endocrine therapy, and the prognosis of BC patients further.
In conclusion, this hospital‐based case–control study showed that XPC Ala499Val and Lys939Gln are associated with BC risk in this Chinese population. The two SNPs may have an effect on preoperative diagnosis and postoperative treatment, and we hope these accompanying predictive and prognostic biomarker tests will help to improve the efficacy of treatment.
Disclosure Statement
The authors have no conflicts of interest.
Acknowledgments
This work was supported by grants from the Fund of the National Natural Science Foundation of the Republic of China (Grant Nos. 30873097 and 30973559), the study on the cooperation of major disease control techniques in Liaoning province (Grant No. 2010225001), and Colleges and Universities Doctoral Programs Specifically Research Fund (Grant No. 20092104110020).
These authors contributed equally to this work.
References
- 1. Jemal A, Siegel R, Xu J, Ward E. Cancer statistics, 2010. CA Cancer J Clin 2010; 60: 277–300. [DOI] [PubMed] [Google Scholar]
- 2. Berwick M, Vineis P. Markers of DNA repair and susceptibility to cancer in humans: an epidemiologic review. J Natl Cancer Inst 2000; 92: 874–97. [DOI] [PubMed] [Google Scholar]
- 3. Ramos JM, Ruiz A, Colen R, Lopez ID, Grossman L, Matta JL. DNA repair and breast carcinoma susceptibility in women. Cancer 2004; 100: 1352–7. [DOI] [PubMed] [Google Scholar]
- 4. Jorgensen TJ, Visvanathan K, Ruczinski I, Thuita L, Hoffman S, Helzlsouer KJ. Breast cancer risk is not associated with polymorphic forms of xeroderma pigmentosum genes in a cohort of women from Washington County, Maryland. Breast Cancer Res Treat 2007; 101: 65–71. [DOI] [PubMed] [Google Scholar]
- 5. Goode EL, Ulrich CM, Potter JD. Polymorphisms in DNA repair genes and associations with cancer risk. Cancer Epidemiol Biomarkers Prev 2002; 11: 1513–30. [PubMed] [Google Scholar]
- 6. Yang ZH, Liang WB, Jia J, Wei YS, Zhou B, Zhang L. The xeroderma pigmentosum group C gene polymorphisms and genetic susceptibility of nasopharyngeal carcinoma. Acta Oncol 2008; 47: 379–84. [DOI] [PubMed] [Google Scholar]
- 7. Lockett KL, Snowhite IV, Hu JJ. Nucleotide‐excision repair and prostate cancer risk. Cancer Lett 2005; 220: 125–35. [DOI] [PubMed] [Google Scholar]
- 8. Sugasawa K, Shimizu Y, Iwai S, Hanaoka F. A molecular mechanism for DNA damage recognition by the xeroderma pigmentosum group C protein complex. DNA Repair 2002; 1: 95–107. [DOI] [PubMed] [Google Scholar]
- 9. Araki M, Masutani C, Takemura M. Centrosome protein centrin 2/caltractin 1 is part of the xeroderma pigmentosum group C complex that initiates global genome nucleotide excision repair. J Biol Chem 2001; 276: 18665–72. [DOI] [PubMed] [Google Scholar]
- 10. Smith TR, Levine EA, Freimanis RI et al Polygenic model of DNA repair genetic polymorphisms in human breast cancer risk. Carcinogenesis 2008; 29: 2132–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Shen J, Desai M, Agrawal M et al Polymorphisms in nucleotide excision repair genes and DNA repair capacity phenotype in sisters discordant for breast cancer. Cancer Epidemiol Biomarkers Prev 2006; 15: 1614–9. [DOI] [PubMed] [Google Scholar]
- 12. Försti A, Angelini S, Festa F et al Single nucleotide polymorphisms in breast cancer. Oncol Rep 2004; 11: 917–22. [PubMed] [Google Scholar]
- 13. Zheng W, Cong XF, Cai WH, Yang S, Mao C, Zou HW. Current evidences on XPC polymorphisms and breast cancer susceptibility: a meta‐analysis. Breast Cancer Res Treat 2011; 128: 811–5. [DOI] [PubMed] [Google Scholar]
- 14. Zhang L, Zhang Z, Yan W. Single nucleotide polymorphisms for DNA repair genes in breast cancer patients. Clin Chim Acta 2005; 359: 150–5. [DOI] [PubMed] [Google Scholar]
- 15. Hu Z, Wang Y, Wang X et al DNA repair gene XPC genotypes/haplotypes and risk of lung cancer in a Chinese population. Int J Cancer 2005; 115: 478–83. [DOI] [PubMed] [Google Scholar]
- 16. Shen M, Berndt SI, Rothman N et al Polymorphisms in the DNA nucleotide excision repair genes and lung cancer risk in Xuan Wei, China. Int J Cancer 2005; 116: 768–73. [DOI] [PubMed] [Google Scholar]
- 17. Lee GY, Jang JS, Lee SY et al XPC polymorphisms and lung cancer risk. Int J Cancer 2005; 115: 807–13. [DOI] [PubMed] [Google Scholar]
- 18. Marin MS, Lopez‐Cima MF, Garcia‐Castro L, Pascual T, Marron MG, Tardon A. Poly (AT) polymorphism in intron 11 of the XPC DNA repair gene enhances the risk of lung cancer. Cancer Epidemiol Biomarkers Prev 2004; 13: 1788–93. [PubMed] [Google Scholar]
- 19. Vogel U, Overvad K, Wallin H, Tjonneland A, Nexo BA, Raaschou‐Nielsen O. Combinations of polymorphisms in XPD, XPC and XPA in relation to risk of lung cancer. Cancer Lett 2005; 222: 67–74. [DOI] [PubMed] [Google Scholar]
- 20. Yun Bai, Liang Xu, Xiaobo Yang et al Sequence variations in DNA repair gene XPC is associated with lung cancer risk in a Chinese population: a case‐control study. BMC Cancer 2007; 7: 81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Dong J, Hu Z, Shu Y et al Potentially functional polymorphisms in DNA repair genes and non‐small‐cell lung cancer survival: a pathway‐based analysis. Mol Carcinog 2011; doi: 10.1002/mc.20819. [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- 22. Sak SC, Barrett JH, Paul AB, Bishop DT, Kiltie AE. The polyAT, intronic IVS11‐6 and Lys939Gln XPC polymorphisms are not associated with transitional cell carcinoma of the bladder. Br J Cancer 2005; 92: 2262–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. García‐Closas M, Malats N, Real FX et al Genetic variation in the nucleotide excision repair pathway and bladder cancer risk. Cancer Epidemiol Biomarkers Prev 2006; 15: 536–42. [DOI] [PubMed] [Google Scholar]
- 24. Sak SC, Barrett JH, Paul AB, Bishop DT, Kiltie AE. Comprehensive analysis of 22 XPC polymorphisms and bladder cancer risk. Cancer Epidemiol Biomarkers Prev 2006; 15: 2537–41. [DOI] [PubMed] [Google Scholar]
- 25. Yimin Zhu, Maode Lai, Hushan Yang et al Genotypes, haplotypes and diplotypes of XPC and risk of bladder cancer. Carcinogenesis 2007; 28: 698–703. [DOI] [PubMed] [Google Scholar]
- 26. Yang M, Kang MJ, Choi Y et al Associations between XPC expression, genotype, and the risk of head and neck cancer. Environ Mol Mutagen 2005; 45: 374–9. [DOI] [PubMed] [Google Scholar]
- 27. Sugimura T, Kumimoto H, Tohnai I et al Gene‐environment interaction involved in oral carcinogenesis: molecular epidemiological study for metabolic and DNA repair gene polymorphisms. J Oral Pathol Med 2006; 35: 11–8. [DOI] [PubMed] [Google Scholar]
- 28. Kietthubthew S, Sriplung H, Au WW, Ishida T. Polymorphism in DNA repair genes and oral squamous cell carcinoma in Thailand. Int J Hyg Environ Health 2006; 209: 21–9. [DOI] [PubMed] [Google Scholar]
- 29. Ye W, Kumar R, Bacova G, Lagergren J, Hemminki K, Nyren O. The XPD 751Gln allele is associated with an increased risk for esophageal adenocarcinoma: a population‐based case‐control study in Sweden. Carcinogenesis 2006; 27: 1835–41. [DOI] [PubMed] [Google Scholar]
- 30. Weiss JM, Weiss NS, Ulrich CM, Doherty JA, Voigt LF, Chen C. Interindividual variation in nucleotide excision repair genes and risk of endometrial cancer. Cancer Epidemiol Biomarkers Prev 2005; 14: 2524–30. [DOI] [PubMed] [Google Scholar]
- 31. Huang WY, Berndt SI, Kang D et al Nucleotide excision repair gene polymorphisms and risk of advanced colorectal adenoma: XPC polymorphisms modify smoking‐related risk. Cancer Epidemiol Biomarkers Prev 2006; 15: 306–11. [DOI] [PubMed] [Google Scholar]
- 32. Hirata H, Hinoda Y, Tanaka Y et al Polymorphisms of DNA repair genes are risk factors for prostate cancer. Eur J Cancer 2007; 43: 231–7. [DOI] [PubMed] [Google Scholar]
- 33. Smith IC, Heys SD, Hutcheon AW et al Neoadjuvant chemotherapy in breast cancer: significantly enhanced response with docetaxel. J Clin Oncol 2002; 20: 1456–66. [DOI] [PubMed] [Google Scholar]
- 34. Guo W, Zhou RM, Wan LL et al Polymorphisms of the DNA repair gene xeroderma pigmentosum groups A and C and risk of esophageal squamous cell carcinoma in a population of high incidence region of North China. J Cancer Res Clin Oncol 2008; 134: 263–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kusumoto R, Masutani C, Sugasawa K et al Diversity of the damage recognition step in the global genomic nucleotide excision repair in vitro. Mutat Res 2001; 485: 219–27. [DOI] [PubMed] [Google Scholar]
- 36. Yokoi M, Masutani C, Maekawa T, Sugasawa K, Ohkuma Y, Hanaoka F. The xeroderma pigmentosum group C protein complex XPC‐HR23B plays an important role in the recruitment of transcription factor IIH to damaged DNA. J Biol Chem 2000; 275: 9870–5. [DOI] [PubMed] [Google Scholar]
- 37. Giaccone G, Pinedo HM. Drug resistance. Oncologist 1996; 1: 82–7. [PubMed] [Google Scholar]
- 38. Fleming ND, Agadjanian H, Nassanian H et al Xeroderma pigmentosum complementation group C single‐nucleotide polymorphisms in the nucleotide excision repair pathway correlate with prolonged progression‐free survival in advanced ovarian cancer. Cancer 2012; 118: 689–97. [DOI] [PubMed] [Google Scholar]
- 39. Chen F, Wang W, El‐Deiry WS. Current strategies to target p53 in cancer. Biochem Pharmacol 2010; 80: 724–30. [DOI] [PubMed] [Google Scholar]
- 40. Zhang XP, Liu F, Wang W. Two‐phase dynamics of p53 in the DNA damage response. Proc Natl Acad Sci USA 2011; 108: 8990–5. [DOI] [PMC free article] [PubMed] [Google Scholar]


