Abstract
Abstract
The removal of antibiotics from the feeds used in the livestock industry has resulted in the use of a wide range of alternative antimicrobial products that aim to deliver the productivity and health benefits that have traditionally been associated with antibiotics. Amongst the most popular alternatives are phytogenic product-based extracts from herbs and spices with known antimicrobial properties. Despite embracing such alternatives, the industry is still largely unaware of modes of action, their overall effects on animal health, and interactions with other feed additives such as probiotics. To address some of these issues, three phytogenic products were selected and their interactions with caecal microbiota of layers, grown under six different production systems, were investigated in vitro. Caecal microbiotas were grown with and without phytogenic products, and the changes in microbiota composition were monitored by sequencing of 16S rRNA gene amplicons. Phytogenic products and production system both significantly influenced microbiota composition. The three phytogenic products all altered the relative abundance of species within the Lactobacillus genus, by promoting the growth of some and inhibiting other Lactobacillus species. There were also significant alterations in the Bacillus genus. This was further investigated by comparing the effects of the phytogenic products on the growth of a commercially used Bacillus-based probiotic. The phytogens affected the probiotic mix differently, with some promoting the growth of Bacillus sp. at lower phytogenic concentrations, and fully suppressing growth at higher concentrations, indicating the importance of finding an optimal concentration that can control pathogens while promoting beneficial bacteria.
Key points
• After removal of antibiotics from animal feed, urgent solutions for pathogen control were needed.
• Alternative products entered the market without much knowledge on their effects on animal health.
• Probiotic products are used in combination with phytogens despite the possible incompatibility.
Keywords: Microbiota, Chicken, In vitro, Phytobiotic, Bacillus
Introduction
Antibiotic growth promoters (AGPs) have been used in the livestock industry since the 1940s discovery that their addition can improve the growth of livestock, and they became a common practice in the 1950s. By 2001, approximately 70% of total antibiotics used in the USA were given to livestock (USA 2004; Tasho and Cho 2018). It became apparent that the growth-promoting effect of AGPs was related to intestinal microbiota when their administration failed to have any growth-promoting effects in germ-free animals; moreover, in germ-free animals, there were even negative effects instead of accelerated growth (Dibner and Richards 2005). It was speculated that the AGPs’ mode of action included pathogen control; this was especially beneficial for the proven ability of AGPs to control necrotic enteritis, a disease that imposes a significant economic burden on the poultry industry (Elwinger et al. 1998; Van Immerseel et al. 2004; Vissiennon et al. 2000) and Salmonella, a severe human pathogen, often transmitted from poultry (Wilson 1989). Despite the benefits of AGP use for the industry, in terms of lowered production cost and animal welfare via decreased mortality, very soon after the introduction of AGPs in livestock feed, the first reports of antimicrobial resistance (AMR) and calls for controlled usage started to appear (Starr and Reynolds 1951).
Although the initial rise of AMR is attributed to mutations, bacteria can exchange the AMR genes and hence AMR can spread rapidly and contribute to the reduced clinical efficacy of currently used antibiotics (Bywater 2005; O’Neill 2016). Extended exposure to antibiotics gives a competitive advantage to AMR carrying bacteria which results in AMR strains rapidly taking over the environment. Currently, there is AMR to all known antibiotics, including the last generation that was intended for use in worst-case scenarios (Prestinaci et al. 2015). The World Health Organization (WHO) declared AMR a priority research area with dismal predictions that the death toll due to AMR infections may be one person every 3 seconds, and 10 million per year by 2050. If this prediction came true, the cost to the world is expected to be over 100 trillion USD (O’Neill 2016) and the current coronavirus pandemics is an eye opener to all AMR doom scenarios.
After the evidence on AMR growing escalated, so did the media and public calls for control of AGPs in livestock. As a result of the growing concerns about the escalating occurrence of AMR pathogens, the first country to stop using AGPs was Sweden in 1986, followed by Denmark’s ban of avoparcin which was soon after endorsed by the European Union (EU). This started a chain reaction of AGP removal in EU countries and raised awareness in the rest of the world. A detailed history of the ban of AGPs has been documented in Dibner and Richards (2005). Further bans on AGP use, around the globe, have accelerated with public approval and often triggered by the media coverage of extreme cases of antibiotic abuse in some countries (Darwish et al. 2013; Davies and Walsh 2018; Li et al. 2013; Viswanathan 2014).
Following the implementation of bans on AGP use, there has been a burning need for alternative products that can be used for pathogen control. The alternatives to AGPs considered are many, including enzymes, bacteriocins and antimicrobial peptides, organic acids (such as fumaric, citric, formic, lactic, and sorbic acid), probiotics used continuously and at-hatch administered for early colonisation or in mix with other alternative products (Prakasita et al. 2019), prebiotics, natural antimicrobial products (bentonite, zeolite, charcoal, and biochar), and essential oils and their individual active antimicrobial phytochemicals such as carvacrol, thymol, cinnamaldehyde, eugenol, and eucalyptol (Anonye 2016; Huyghebaert et al. 2011; Tellez and Latorre 2017; Thacker 2013; Verstegen and Williams 2002). New products containing mixed phytochemicals, whole plant, or mushroom extracts are becoming widely accepted in the Australian and worldwide livestock industries and there are a variety of products available on the market, although little is known about their mode of action.
We investigated the validity of the hypothesis that the phytogenic products would influence beneficial and commercial probiotic species as well as pathogens, and that the range of both beneficial and pathogenic bacteria may have natural resistance to these products which could result in their use having the same AMR effects as the originally used banned antibiotics. We selected three commonly used products and investigated their effects on the complex microbial communities taken from the caeca of healthy layer birds.
Materials and methods
In vitro system
Enriched LYHBHI media was prepared by adding chicken feed extract (100 ml), bacterial ferment (100 ml), vitamin mix (1 ml), and trace mineral mix (1 ml) to 798 ml of LYHBHI media (brain heart infusion (37 g/l, BD Biosciences, San Jose, CA, USA), yeast extract (5 g/l, Alfa Aesar, Ward Hill, USA), cellobiose (1 g/l, BD), hemin (0.005 g/l, BD), L-cysteine (0.5 g/l, Alfa Aesar), resazurin sodium salt (5 mg/l, Alfa Aesar)) to make the final volume of 1 l. The LYHBHI media and the feed extract were sterilised by autoclaving while bacterial ferment was filter sterilised. The media was purged with nitrogen gas to remove the oxygen in the media.
Lactobacillus plantarum (ATCC® BAA-793™) and Lactobacillus rhamnosus (ATCC® 53103™) were cultured aerobically at 37 °C in 60 ml of LYHBHI. The culture was centrifuged and 50 ml of supernatant from each culture was filter sterilised to mix in the media. To prepare the feed extract, 100 g of chicken feed (Red Hen Chick premium micro starter crumbles antibiotic and anticoccidial free; Lauke Mills, Daveystone SA, Australia) was ground for 3 min (1500 W, Nutri Ninja Auto iQ Duo; SharkNinja, Needham, USA). Powdered feed was blended for an additional 3 min after adding 1 l of water. The feed extract was autoclaved and centrifuged (3220g, 5 min) and 100 ml of supernatant was added to LYHBHI.
One capsule of vitamin mix (Multivitamins and Minerals, Cenovis, Brisbane, Australia) and a capsule of vitamin K2 (Caruso’s Natural Health, Eastern Creek, Australia) were mixed in 10 ml of water and filter sterilised to use in the media. Similarly, 1 ml of trace elements mix (Youngevity, Chula Vista, USA) was also added in the media after filter sterilisation.
Caecum parent cultures
The caeca from 24 layer hens from the same commercial layer farm in Queensland, Australia, were collected, transported on dry ice, and stored at − 80 °C. The farm has its own hatchery and feed mill ensuring the same genetics and feed for the production systems used. The caeca were thawed inside an anaerobic (with N2/CO2 gas) workstation (A35, Whitley, Shipley, UK) at 37 °C. The caecal contents were squeezed into Erlenmeyer flasks containing 50 ml of enhanced LYHBHI media. The caeca were then opened and transferred into the flask together with the remaining content to ensure the inclusion of mucosa-associated microbes in the parent culture. The culture was incubated at 37 °C for 2 h in the anaerobic chamber with shaking at the rate of 90 rpm (orbital shaker) and aliquoted into cryotubes with 30% sterile glycerol and stored at − 80 °C. The Animal Ethics Committee of Central Queensland University approved this study under the approval number 0000020312.
Experimental design
Three phytogenic products were used in the study. Product 1 (Prod1) is a mix of phytochemicals including plant compounds such as carvacrol, cinnamaldehyde, and organic acids. Product 2 (Prod2) is a whole-plant extract containing caffeoylquinic acids, flavonoids, polyphenols, and pro-vitamins, and product 3 (Prod3) is another mix of phytochemicals containing 50% oregano essential oil and a food-grade silicic acid. Twenty-four parent cultures, 4 each from the six different production systems: cage, barn, organic, free-range, and 2 types of the open range, were prepared and grown in the control enriched LYHBHI media as well as in the same media with 0.1% v/v of the each of the 3 products. The cultures were grown for 24 h, collected, and immediately centrifuged and the microbial pellet was stored for DNA extraction.
DNA extraction
DNA from the microbial pellets of the anaerobic cultures was extracted using the method as previously described (Bauer et al. 2019; Gangadoo et al. 2019). Briefly, the microbial cells were lysed using a bead beater with 0.2 g of 0.1-mm glass beads and 500 μl of lysis buffer. DNA was purified using a DNA spin purification column (Enzymax LLC, Cat# EZC101, Kentucky, USA) and quantity and quality of DNA was measured with a NanoDrop Micro-UV/Vis Spectrophotometer (Thermo Fisher Scientific, Wilmington, USA).
16S rRNA gene sequencing
The V3–V4 regions of the 16S rRNA genes were amplified using the primers ACTCCTACGGGAGGCAGCAG (forward) and GGACTACHVGGGTWTCTAAT (reverse). The DNA sequencing library was prepared following the manufacturer’s protocol (Illumina Inc., San Diego, CA, USA) using barcodes, spacers, and Illumina sequencing linkers (Fadrosh et al. 2014). The DNA library was sequenced on Illumina MiSeq platform using 2 × 300 bp paired-end sequencing.
The upstream DNA sequence analysis was done using Quantitative Insights Into Microbial Ecology (QIIME v.1.9.1) (Caporaso et al. 2010) as this software package includes methods to deconvolute and process dual barcoded amplicons that are not available in more recent software packages. Fastq-Join algorithm was used to combine paired-end sequences allowing no mismatches within the overlapping region. The reads with Phred quality score of 20 or above were selected for the analysis. UCLUST (Edgar 2010) was used for OTU (operational taxonomic units) picking at 97% similarity while Pintail (Ashelford et al. 2005) was used for the inspection of chimeric sequences. The sequences were assigned with taxonomy against the GreenGenes database (v 2013_8) with QIIME default parameters (DeSantis et al. 2006). A rarefied OTU abundance table was used to obtain a UniFrac matrix. The OTU table was Hellinger transformed (Legendre and Gallagher 2001) for further downstream analysis and interpretation.
Downstream analysis of the data and visualisation of the result was performed using Calypso (Zakrzewski et al. 2017). The sequence data is publicly available at the MG-RAST database under a project ID mgp454253 and library ID mgl794100.
Bacillus inhibition growth curves
Bacillus probiotic product was a mix of 3 Bacillus species and will not be named. The probiotic product culture was grown at 37 °C overnight by inoculating the product powder into brain heart infusion (BHI) media. The primary culture was then subcultured and harvested in the exponential growth stage for use in the Bacillus inhibition growth curve analysis. The phytogenic product concentrations were optimised in a test run and the Bacillus growth curves (n = 7) were measured on a SpectroStar Omega microplate reader (BMG Labtech, Ortenberg, Germany), with ultrafast UV/Vis, absorbance spectrophotometer. Optical density was measured every hour for 20 h at 600 nm. Each data point for each sample was an average of 5 OD measurements, evenly covering all parts of the well. The 96-well plate reader was operating overnight at 37 °C and with 100 rpm plate orbital shaking. Each treatment was represented with n = 7 measurements (wells) and each product concentration had its own blank that was later deducted.
Results
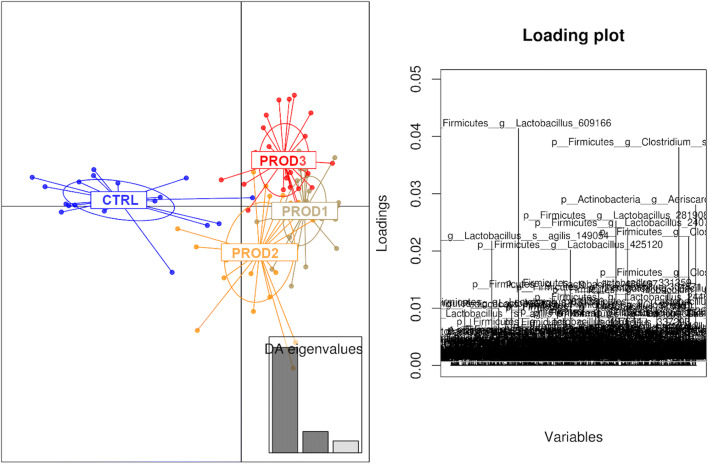
The influence of treatments and of the production systems on microbial communities in the in vitro system were both significant, with 2-way PERMANOVA (done using Primer 7e) P < 1E−5 and P = 3E−4, respectively. There was no significant interaction between the treatments and production systems P = 0.862. We tested a range of multivariate analyses for visualisation, and the control was consistently separated from more or less overlapping treatments. Discriminant analysis of principal components (DAPC plot) analysis showed the two more similar products (Prod1 and Prod3), both containing carvacrol and herbal phytogens, overlapped (DAPC plot, Fig. 1) while Prod2, a perennial edible plant extract, was slightly separated (Fig. 1). Control was distinctly separated from all three products.
Fig. 1.
Discriminant analysis of principal components (DAPC) showing the similarities between the treatments. Each dot represents a microbiota profile from a successfully sequenced sample
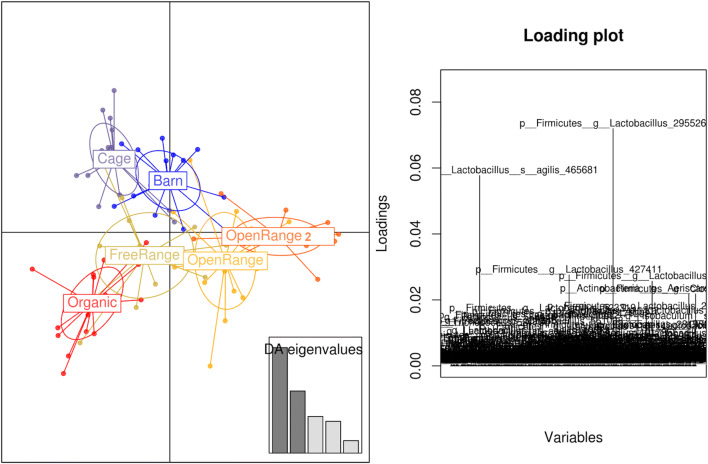
Similarly, the production system DAPC plot presented in Fig. 2 shows the separation of cage and barn originated microbiota and overlap of the organic, free-range, and open range, emphasising again that organic-fed birds are also grown in free-range sheds but fed organic feed mix while the open-range birds have a much bigger outdoor range like pasture or orchards access. Each production system was represented with 16 different replicates (4 biological replicates grown under 4 phytogenic conditions). Two different sheds under open-range production system showed high microbiota similarity. Cage production system seems the most unique compared to the others. The overlap of the physically closed cage and barn systems, where the birds have no access to outdoors, was relatively tight compared to the remaining systems with outdoor access that showed more variation.
Fig. 2.
Discriminant analysis of principal components (DAPC) showing the differences between the production systems. Each dot represents a microbiota from a successfully sequenced sample from that production system
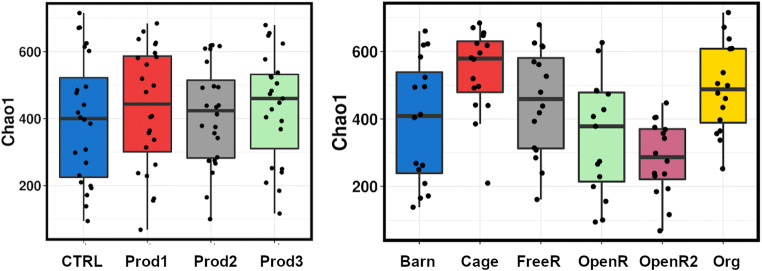
The treatments with different products did not have any major influence on the diversity measures inspected (Chao1 P = 0.86; Richness P = 0.98; Evenness P = 0.31; Shannon P = 0.41; and Simpson P = 0.14); however, the production system did show some significant alterations (Chao1 P = 3.5E−5; Richness P = 1.4E−4; Evenness P = 2.5E−3; Shannon P = 8.6E−4; and Simpson P = 4.8E−3) (Fig. 3). The barn-grown birds clustered into two groups with lower and higher Chao1-based diversity. Surprisingly cage bird samples showed relatively higher diversity than the other groups; however, it should be noted that these are in vitro culture diversity measures and that they should not be confused with original microbiota of the donor animal caecal diversity as that would be strongly influenced by the community supported by the microbiological media used.
Fig. 3.
Chao1 estimated richness between the treatments and production systems after in vitro experiment
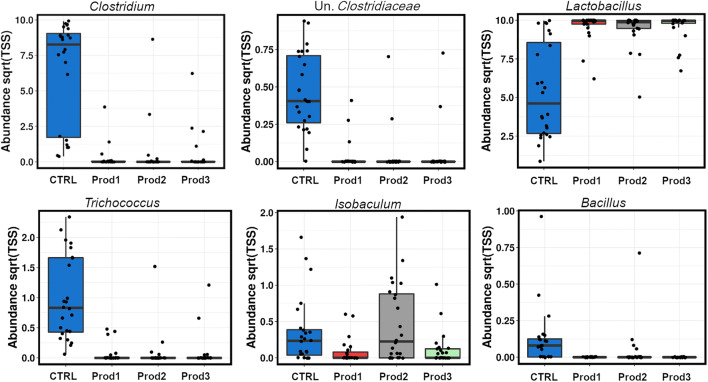
There were six genera significantly (ANOVA, P < 0.05) affected by the products (Fig. 4), with the overlapping effect of the three products in all genera except for Isobaculum, where Prod2 (whole-plant extract) had the opposite effect compared to herbal phytogenic mixes Prod1 and Prod3. In all other affected genera, the three products acted equally, by reducing Clostridium, unknown Clostridiaceae, Trichococcus and recently very popular poultry probiotic genus Bacillus. All three products increased the overall abundance of the genus Lactobacillus.
Fig. 4.
Genera significantly altered (ANOVA, P < 0.05) by the three treatments
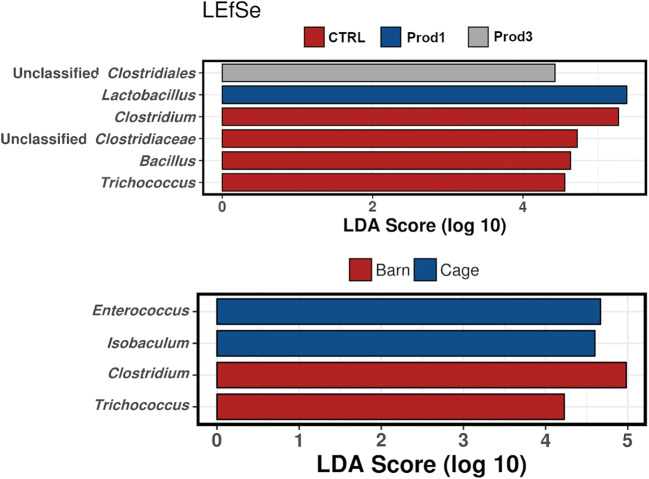
There were three genera significantly affected by production system: Pediococcus (P = 0.011, slightly higher in open range), Aeriscardovia (P = 1.2E−3), and Isobaculum (P = 5.9E−5) both slightly higher in the organic and in the cage. Organic and cage systems also showed more of the phylum Actinobacteria (P = 0.001). The linear discriminant analysis (LDA) effect size method (LEfSe, Fig. 5) selects the genera most likely to explain differences between the treatments and production systems microbiotas, via combining standard statistical tests with further tests encoding biological consistency and effect relevance. The analysis identifies Prod1 as the most Lactobacillus promoting and Prod3 as related to unclassified Clostridiales. Control birds microbiota was characterised with Clostridium, Bacillus, and Trichococcus. Similarly, the barn production system was associated with Clostridium and Trichococcus, whereas the cage system was an Enterococcus and Isobaculum represented environment (Fig. 5).
Fig. 5.
Linear discriminant analysis effect size method (LEfSe) showing the genera most likely to explain differences between the treatments (top image) and production systems (bottom image) microbiotas
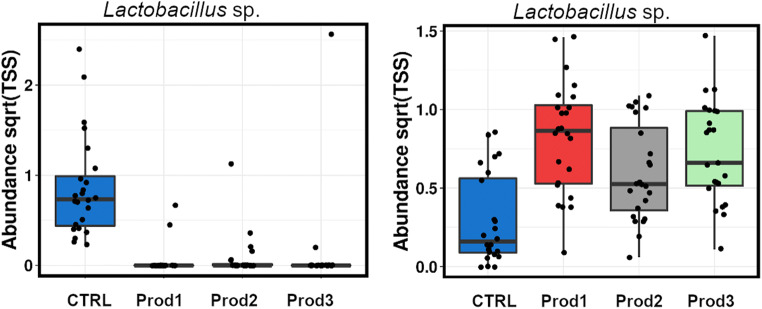
At the OTU level, there were 408 OTUs differentially abundant between the treatments (ANOVA, P < 0.05), 27 of those assigned to C. perfringens with blastn %ID up to 100%, all reduced in all three treatments as a major contributor to the reduction of genus Clostridium (Fig. 4). The overall increase in the Lactobacillus genus, by all three products, was a product of substantial rearrangement of the Lactobacillus species (Fig. 6), with 330 differential OTUs from this genus, 84 OTUs significantly reduced and 246 promoted by the products. This can be an indication of common multiple phytogenic antimicrobial resistance amongst some Lactobacillus species.
Fig. 6.
Two of 330 Lactobacillus OTUs significantly altered by the treatments
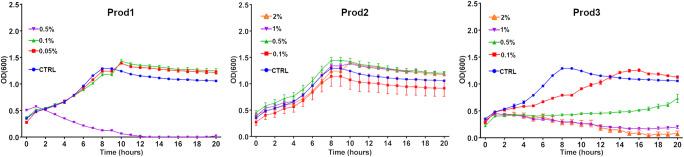
Unlike with genus Bacillus that contains a fewer probiotic and some pathogenic species, there are many unique species and strains of Lactobacillus used as probiotics in poultry production that, based on our data, can be either promoted or inhibited or even removed by the products we investigated. Contrastingly, the data in Fig. 4 shows the reproducible inhibition of Bacillus sp. by all three phytogenic products. We selected a widely used Bacillus commercial probiotic product that contains three species of Bacillus and performed a growth curve assay in the presence of the three products (Fig. 7). Figure 7 indicates that Prod1 and Prod3 at lower concentrations can stimulate the growth of the Bacillus probiotic but become inhibitory at higher concentrations. Prod3 was slightly inhibitory at the lowest concentration tested but became growth-promoting at all higher concentrations. It should be noted that the plate reader growth curves move out of the linear range after OD = 0.5 and since it is technically impossible to dilute the sample into liner range while in the plate reader, the growth curves at higher OD are “compressed” and the differences in the stationary phase would be more prominent if the growth curves were plotted using dilutions. Despite this, the differences between both 0.05 and 0.1% Prod1 and control were significant in all measurements after reaching stationary phase at 10 h (P < 0.01 using multiple t tests for each hour). There was no significant difference between the treatments and control for Prod2 and no difference between 0.1% Prod3 and control at the end of stationary phase measurements.
Fig. 7.
Growth curve of a commercial probiotic comprised of 3 Bacillus species grown under different concentrations of products in BHI media (n = 7)
Discussion
With the recent prohibition of the use of AGPs in livestock feed and an overall negative public perception of using AGPs in the livestock industry, there has been a flood of products promising effective pathogen control using organic-compatible, mostly plant-based products. Here we investigated the effects of three of these products on layer bird caecal microbiota using an in vitro system and microbial community analysis. We concluded that, while both the phytogenic products and the production systems did have a major influence on microbiota, there was no significant interaction between the two; thus, the effect of the phytogenic products should be more or less reproducible across the different layer production systems.
Interestingly, despite having a significant influence on microbiota, our results suggest that the phytogenic products did not reduce richness and diversity measures using in vitro model, as opposed to the production systems where differences in richness were quite substantial. This may come as a surprise because the role of these products is to control bacteria, specifically pathogen growth and overgrowth in the gut of livestock, and based on human data on the influence of prolonged use of antibiotics on richness and diversity (Fricke 2014), we could expect a significant reduction of diversity by antibiotic-alternative products. On the other hand, antibiotic growth promoters previously used in the industry also had no major influence on intestinal microbiota and no reduction in diversity with zinc bacitracin even increasing richness in broilers (Crisol-Martinez et al. 2017); thus, their mode of action is more accurately described as preventing microbial overgrowth rather than entirely removing a range of susceptible species. The reduction of some species/OTUs below detection limit can allow for growth of other rare microbiota above the detection limit, thus compensating diversity reductions.
Apart from numerous OTUs that were similar or identical to Clostridium perfringens, that were almost entirely removed by all 3 products, our samples had no major poultry pathogens; thus, the effect of the products on Salmonella and Campylobacter could not be investigated. However, the influence on the Bacillus commercial probiotic, as well as rearrangements of Lactobacillus species, is of industrial significance. High level of resistance to multiple phytogenic products by some of the Lactobacillus species indicates that some bacteria have natural resistance to a range of distinct phytogenic products. Mechanisms of this resistance are still unknown and the possibility of phytogenic products promoting AMR needs to be further investigated.
Lactobacillus and Bifidobacterium probiotic species have dominated the livestock probiotic industry for decades with other beneficial species of Enterococcus, Streptococcus, and Bacillus having a lesser role. However, in more recent years, Bacillus-based probiotics have become more common in the livestock market, to now become one of the most widely used. Bacillus probiotics are gaining attention in probiotics formulas for human use as detailed in a recent review (Elshaghabee et al. 2017), mostly due to their enhanced tolerance and survivability under the aggressive environments of probiotic production practices such as freeze-drying and bile and acid of the gastrointestinal tract. Bacillus probiotics have proven themselves as more stable during prolonged storage due to the spore-forming capability. Bacillus probiotic species possess the capability of pathogen exclusion, anti-oxidant, antimicrobial action, beneficial immuno-modulatory, and food fermentation abilities (Elshaghabee et al. 2017).
Despite the considerable range of Bacillus probiotic benefits, the recent rise in demand for Bacillus products in the livestock industry comes from their exceptional capacity to produce extracellular enzymes. Bacillus species are used for industrial production of food-grade amylase, glucoamylase, protease, pectinase, and cellulase (Elshaghabee et al. 2017; Kiran et al. 2018; Kuppers et al. 2014; Simair et al. 2017; Wiegand et al. 2013) as well as vitamins (Commichau et al. 2014; Mahdinia et al. 2018; Paracchini et al. 2017) and carotenoids (Xue et al. 2015; Yoshida et al. 2009) used for the production of numerous human health supplements. Bacillus production of digestive enzymes is significant for the poultry industry that uses hard to digest foods in animals that have very short food gut retention time. This is amplified during the current industry adjustment to ecological impacts of production and sustainability that involves experimenting with hard to digest unconventional foods and waste products that could be more readily available in future than poultry feedstuffs that have been traditionally used (Abudabos et al. 2017).
The recent rise in the use of Bacillus products coincides with the replacement of traditional antibiotics growth promoters with a range of phytogenic and other alternative products. It is interesting that, to date, no work has been published to demonstrate their mutual compatibility, despite the fact that the farmers commonly mix them in the same ration. Our data indicate that some products at lower concentrations can significantly promote Bacillus probiotic growth but suppress it at higher concentrations; thus, it is possible to find a “sweet spot” and adjust the phytogen concentration towards promoting probiotic growth while keeping pathogens under control. This requires careful on-farm optimisation. We must note that we performed growth curves on a mixed probiotic product containing 3 Bacillus species and that each species could be differently affected. It is, however, difficult to perform competitive exclusion curves with 3 species of the same genus without strain modification, but even individual species growth curves would be useful to answer this question. The results suggest that there is likely to be value in testing the compatibility of other probiotics with the phytogenic products in order to best guide their effective combinatorial use on farms.
This study has provided new insights into the action of phytogenic products on intestinal microbiota of layers, using in vitro experiments on the complex caecal microbial community. However, microbiota in the actual birds is more complex than in the in vitro model and it differs between the gut sections. In vitro methodology uses only subsection of the original bird microbiota that can grow on the specific media and can survive freezing of the inoculum for storage. Additionally, there is a robust variability of intestinal microbiota between and within the flocks (Stanley et al. 2013) further complicated by complex interactions between microbes and their metabolites in these communities, so that the effect of phytogenic products on the livestock cannot be easily predicted. Administration of both probiotics and alternative antibiotic products from early days post-hatch is likely to have a critical influence on the development of microbiota (Baldwin et al. 2018; Donaldson et al. 2017). More research is needed to better understand their impact on animal health, antimicrobial resistance on farms, pathogen control, and immunity.
Acknowledgements
The data was analysed using the Marie Curie High-Performance Computing System at Central Queensland University. We wish to acknowledge and appreciate help from Jason Bell provided in all aspects of High-Performance Computing.
Author contribution statement
YB: performed research, analysed data. JA, RM, TTHV: performed research. DS: conceived the study; analysed data, wrote the paper. All authors edited the manuscript and agreed with its final form.
Funding
This research did not receive any specific grant from funding agencies or the commercial sector. The project was funded internally by Central Queensland University Merit Grant fund.
Data availability
Sequencing data is publically available on MG-RAST Metagenomics Analysis Server Database (https://www.mg-rast.org/) with full sample annotation under project mgp454253 and a library ID mgl794100.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
Animal ethics approvals were obtained from the Animal Ethics Committee at Central Queensland University with the approval number 0000020312.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Abudabos AM, Al-Atiyat RM, Albatshan HA, Aljassim R, Aljumaah MR, Alkhulaifi MM, Stanley DM. Effects of concentration of corn distillers dried grains with solubles and enzyme supplementation on cecal microbiota and performance in broiler chickens. Appl Microbiol Biotechnol. 2017;101(18):7017–7026. doi: 10.1007/s00253-017-8448-5. [DOI] [PubMed] [Google Scholar]
- Anonye BO. General commentary on alternatives to antibiotic growth promoters in animals. Front Vet. 2016;3:74. doi: 10.3389/fvets.2016.00074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashelford KE, Chuzhanova NA, Fry JC, Jones AJ, Weightman AJ. At least 1 in 20 16S rRNA sequence records currently held in public repositories is estimated to contain substantial anomalies. Appl Environ Microbiol. 2005;71(12):7724–7736. doi: 10.1128/Aem.71.12.7724-7736.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldwin S, Hughes RJ, Hao Van TT, Moore RJ, Stanley D. At-hatch administration of probiotic to chickens can introduce beneficial changes in gut microbiota. PLoS One. 2018;13(3):e0194825. doi: 10.1371/journal.pone.0194825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer BW, Gangadoo S, Bajagai YS, Van TTH, Moore RJ, Stanley D. Oregano powder reduces Streptococcus and increases SCFA concentration in a mixed bacterial culture assay. PLoS One. 2019;14(12):e0216853. doi: 10.1371/journal.pone.0216853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bywater RJ. Identification and surveillance of antimicrobial resistance dissemination in animal production. Poult Sci. 2005;84(4):644–648. doi: 10.1093/ps/84.4.644. [DOI] [PubMed] [Google Scholar]
- Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Pena AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Tumbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7(5):335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Commichau FM, Alzinger A, Sande R, Bretzel W, Meyer FM, Chevreux B, Wyss M, Hohmann HP, Pragai Z. Overexpression of a non-native deoxyxylulose-dependent vitamin B6 pathway in Bacillus subtilis for the production of pyridoxine. Metab Eng. 2014;25:38–49. doi: 10.1016/j.ymben.2014.06.007. [DOI] [PubMed] [Google Scholar]
- Crisol-Martinez E, Stanley D, Geier MS, Hughes RJ, Moore RJ. Understanding the mechanisms of zinc bacitracin and avilamycin on animal production: linking gut microbiota and growth performance in chickens. Appl Microbiol Biotechnol. 2017;101(11):4547–4559. doi: 10.1007/s00253-017-8193-9. [DOI] [PubMed] [Google Scholar]
- Darwish WS, Eldaly EA, El-Abbasy MT, Ikenaka Y, Nakayama S, Ishizuka M. Antibiotic residues in food: the African scenario. Jpn J Vet Res. 2013;61(Suppl):S13–S22. [PubMed] [Google Scholar]
- Davies M, Walsh TR. A colistin crisis in India. Lancet Infect Dis. 2018;18(3):256–257. doi: 10.1016/S1473-3099(18)30072-0. [DOI] [PubMed] [Google Scholar]
- DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, Huber T, Dalevi D, Hu P, Andersen GL. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol. 2006;72(7):5069–5072. doi: 10.1128/Aem.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dibner JJ, Richards JD. Antibiotic growth promoters in agriculture: history and mode of action. Poult Sci. 2005;84(4):634–643. doi: 10.1093/ps/84.4.634. [DOI] [PubMed] [Google Scholar]
- Donaldson EE, Stanley D, Hughes RJ, Moore RJ. The time-course of broiler intestinal microbiota development after administration of cecal contents to incubating eggs. PeerJ. 2017;5:e3587. doi: 10.7717/peerj.3587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26(19):2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- Elshaghabee FMF, Rokana N, Gulhane RD, Sharma C, Panwar H. Bacillus as potential probiotics: status, concerns, and future perspectives. Front Microbiol. 2017;8:1490. doi: 10.3389/fmicb.2017.01490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elwinger K, Berndtson E, Engstrom B, Fossum O, Waldenstedt L. Effect of antibiotic growth promoters and anticoccidials on growth of Clostridium perfringens in the caeca and on performance of broiler chickens. Acta Vet Scand. 1998;39(4):433–441. doi: 10.1186/BF03547769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fadrosh DW, Ma B, Gajer P, Sengamalay N, Ott S, Brotman RM, Ravel J (2014) An improved dual-indexing approach for multiplexed 16S rRNA gene sequencing on the Illumina MiSeq platform. Microbiome:6. 10.1186/2049-2618-2-6 [DOI] [PMC free article] [PubMed]
- Fricke WF. The more the merrier? Reduced fecal microbiota diversity in preterm infants treated with antibiotics. J Pediatr. 2014;165(1):8–10. doi: 10.1016/j.jpeds.2014.03.022. [DOI] [PubMed] [Google Scholar]
- Gangadoo S, Bauer BW, Bajagai YS, Van TTH, Moore RJ, Stanley D. In vitro growth of gut microbiota with selenium nanoparticles. Anim Nutr. 2019;5(4):424–431. doi: 10.1016/j.aninu.2019.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huyghebaert G, Ducatelle R, Van Immerseel F. An update on alternatives to antimicrobial growth promoters for broilers. Vet J. 2011;188(2):182–188. doi: 10.1016/j.tvjl.2010.03.003. [DOI] [PubMed] [Google Scholar]
- Kiran T, Asad W, Ajaz M, Hanif M, Rasool SA. Industrially relevant cellulase production by indigenous thermophilic Bacillus licheniformis TLW-3 strain: isolation-molecular identification and enzyme yield optimisation. Pak J Pharm Sci. 2018;31(6):2333–2340. [PubMed] [Google Scholar]
- Kuppers T, Steffen V, Hellmuth H, O’Connell T, Bongaerts J, Maurer KH, Wiechert W. Developing a new production host from a blueprint: Bacillus pumilus as an industrial enzyme producer. Microb Cell Factories. 2014;13(1):46. doi: 10.1186/1475-2859-13-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legendre P, Gallagher ED. Ecologically meaningful transformations for ordination of species data. Oecologia. 2001;129(2):271–280. doi: 10.1007/s004420100716. [DOI] [PubMed] [Google Scholar]
- Li R, He L, Hao L, Wang Q, Zhou Y, Jiang H. Genotypic and phenotypic characterisation of antimicrobial-resistant Escherichia coli from farm-raised diarrheic sika deer in Northeastern China. PLoS One. 2013;8(9):e73342. doi: 10.1371/journal.pone.0073342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahdinia E, Demirci A, Berenjian A. Enhanced vitamin K (Menaquinone-7) production by Bacillus subtilis natto in biofilm reactors by optimisation of glucose-based medium. Curr Pharm Biotechnol. 2018;19(11):917–924. doi: 10.2174/1389201020666181126120401. [DOI] [PubMed] [Google Scholar]
- O’Neill J (2016) The review on antimicrobial resistance. Tackling drug-resistant infections globally: final report and recommendations Available at: http://amr-review.org/sites/default/files/160518_Final%20paper_with%20cover.pdf
- Paracchini V, Petrillo M, Reiting R, Angers-Loustau A, Wahler D, Stolz A, Schonig B, Matthies A, Bendiek J, Meinel DM, Pecoraro S, Busch U, Patak A, Kreysa J, Grohmann L. Molecular characterisation of an unauthorised genetically modified Bacillus subtilis production strain identified in a vitamin B2 feed additive. Food Chem. 2017;230:681–689. doi: 10.1016/j.foodchem.2017.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prakasita VC, Asmara W, Widyarini S, Wahyuni A. Combinations of herbs and probiotics as an alternative growth promoter: an in vitro study. Vet World. 2019;12(4):614–620. doi: 10.14202/vetworld.2019.614-620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prestinaci F, Pezzotti P, Pantosti A. Antimicrobial resistance: a global multifaceted phenomenon. Pathog Glob Health. 2015;109(7):309–318. doi: 10.1179/2047773215Y.0000000030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simair AA, Qureshi AS, Khushk I, Ali CH, Lashari S, Bhutto MA, Mangrio GS, Lu C. Production and partial characterisation of alpha-amylase enzyme from Bacillus sp. BCC 01-50 and potential applications. Biomed Res Int. 2017;2017:9173040. doi: 10.1155/2017/9173040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanley D, Geier MS, Hughes RJ, Denman SE, Moore RJ. Highly variable microbiota development in the chicken gastrointestinal tract. PLoS One. 2013;8(12):e84290. doi: 10.1371/journal.pone.0084290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starr MP, Reynolds DM. Streptomycin resistance of coliform bacteria from turkeys fed streptomycin. Am J Public Health Nations Health. 1951;41(11 Pt 1):1375–1380. doi: 10.2105/ajph.41.11_pt_1.1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tasho RP, Cho JY. Entry routes of veterinary antibiotics in the environment. In: Hashmi MZ, Strezov V, Varma A, editors. Antibiotics and antibiotics resistance genes in soils. Cham: Springer; 2018. pp. 55–71. [Google Scholar]
- Tellez G, Latorre JD. Editorial: alternatives to antimicrobial growth promoters and their impact in gut microbiota, health and disease. Front Vet. 2017;4:196. doi: 10.3389/fvets.2017.00196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thacker PA. Alternatives to antibiotics as growth promoters for use in swine production: a review. J Anim Sci Biotechnol. 2013;4(1):35. doi: 10.1186/2049-1891-4-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- USA UoCS (2004) Hogging It!: Estimates of antimicrobial abuse in livestock. https://www.ucsusa.org/resources/hogging-it-estimates-antimicrobial-abuse-livestock#.Wycey1MvzEY Accessed 28/01/2020 2020
- Van Immerseel F, De Buck J, Pasmans F, Huyghebaert G, Haesebrouck F, Ducatelle R. Clostridium perfringens in poultry: an emerging threat for animal and public health. Avian Pathol. 2004;33(6):537–549. doi: 10.1080/03079450400013162. [DOI] [PubMed] [Google Scholar]
- Verstegen MW, Williams BA. Alternatives to the use of antibiotics as growth promoters for monogastric animals. Anim Biotechnol. 2002;13(1):113–127. doi: 10.1081/ABIO-120005774. [DOI] [PubMed] [Google Scholar]
- Vissiennon T, Kroger H, Kohler T, Kliche R. Effect of avilamycin, tylosin and ionophore anticoccidials on Clostridium perfringens enterotoxaemia in chickens. Berl Munch Tierarztl Wochenschr. 2000;113(1):9–13. [PubMed] [Google Scholar]
- Viswanathan VK. Off-label abuse of antibiotics by bacteria. Gut Microbes. 2014;5(1):3–4. doi: 10.4161/gmic.28027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiegand S, Voigt B, Albrecht D, Bongaerts J, Evers S, Hecker M, Daniel R, Liesegang H. Fermentation stage-dependent adaptations of Bacillus licheniformis during enzyme production. Microb Cell Factories. 2013;12:120. doi: 10.1186/1475-2859-12-120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson S. Control of Salmonella enteritidis in poultry. Vet Rec. 1989;125(18):465–466. doi: 10.1136/vr.125.18.465. [DOI] [PubMed] [Google Scholar]
- Xue D, Abdallah II, de Haan IE, Sibbald MJ, Quax WJ. Enhanced C30 carotenoid production in Bacillus subtilis by systematic overexpression of MEP pathway genes. Appl Microbiol Biotechnol. 2015;99(14):5907–5915. doi: 10.1007/s00253-015-6531-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida K, Ueda S, Maeda I. Carotenoid production in Bacillus subtilis achieved by metabolic engineering. Biotechnol Lett. 2009;31(11):1789–1793. doi: 10.1007/s10529-009-0082-6. [DOI] [PubMed] [Google Scholar]
- Zakrzewski M, Proietti C, Ellis JJ, Hasan S, Brion MJ, Berger B, Krause L. Calypso: a user-friendly web-server for mining and visualising microbiome-environment interactions. Bioinformatics. 2017;33(5):782–783. doi: 10.1093/bioinformatics/btw725. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Sequencing data is publically available on MG-RAST Metagenomics Analysis Server Database (https://www.mg-rast.org/) with full sample annotation under project mgp454253 and a library ID mgl794100.