Graphical abstract
Keywords: Pandemic, COVID-19, Airborne transmission, Coronavirus, Virus concentration
Abstract
The COVID-19 disease spread at different rates in the different countries and in different regions of the same country, as happened in Italy. Transmission by contact or at close range due to large respiratory droplets is widely accepted, however, the role of airborne transmission due to small respiratory droplets emitted by infected individuals (also asymptomatic) is controversial. It was suggested that outdoor airborne transmission could play a role in determining the differences observed in the spread rate. Concentrations of virus-laden aerosol are still poorly known and contrasting results are reported, especially for outdoor environments. Here we investigated outdoor concentrations and size distributions of virus-laden aerosol simultaneously collected during the pandemic, in May 2020, in northern (Veneto) and southern (Apulia) regions of Italy. The two regions exhibited significantly different prevalence of COVID-19. Genetic material of SARS-CoV-2 (RNA) was determined, using both real time RT-PCR and ddPCR, in air samples collected using PM10 samplers and cascade impactors able to separate 12 size ranges from nanoparticles (diameter D < 0.056 µm) up to coarse particles (D > 18 µm). Air samples tested negative for the presence of SARS-CoV-2 at both sites, viral particles concentrations were <0.8 copies m−3 in PM10 and <0.4 copies m−3 in each size range investigated. Outdoor air in residential and urban areas was generally not infectious and safe for the public in both northern and southern Italy, with the possible exclusion of very crowded sites. Therefore, it is likely that outdoor airborne transmission does not explain the difference in the spread of COVID-19 observed in the two Italian regions.
1. Introduction
The pandemic of COVID-19 disease, due to the novel coronavirus SARS-CoV-2, was firstly reported in a cluster in Wuhan (China) in December 2019 and it rapidly spread all around the World. By June 27 (2020), infected cases reached 9,660,902 individuals and 491,195 deaths worldwide (https://covid19.who.int/). Starting from February 2020 it was clear that spread of the disease happened in specific outbreak areas and that significant differences were observed in COVID-19 prevalence and fatality rate in different countries and in different regions of the same country. In Europe, Italy was the first country severely hit, with the majority of cases observed in northern Italy and a much smaller number of cases observed in central and, especially, southern Italy. The rapid spread of SARS-CoV-2 and these spatial differences raised important questions regarding the mechanisms of transmission and the role potentially played by airborne transmission. It has also been suggested that airborne transmission could be responsible of the different COVID-19 prevalence observed in northern and southern Italy because of the different dispersion conditions in the two areas (Conticini et al., 2020, Setti et al., 2020). The Po Valley area in northern Italy is characterised, especially during winter period, by low wind speed accompanied to long periods of stable conditions with shallow mixing layers (Ferrero et al., 2010) and this limit both transport (because of limited ventilation) and dispersion of pollutants (because of limited turbulence) favouring large pollutant concentrations near the ground. Venice area, in the northeast of Italy is located near the sea and it has a typical circulation of air masses (Contini et al., 2015) with prevalent winds coming from NNE-NE directions (from the Alps mountains) mainly during the night and winds coming from SSE-SE during the day (from the Adriatic Sea). In contrast, Apulia region in southern Italy has a local meteorology characterised by greater solar radiation, compared to the Po Valley, increasing thermal turbulence and strong winds that favour transport and dilution of pollutants (Cesari et al., 2018). This difference in dispersion conditions could, in principle, influence the concentrations of virus-laden particles in outdoor air.
The spread of SARS-CoV-2 by contact (direct or indirect through contaminated surfaces) is widely accepted, however, the relative importance of airborne transmission is still controversial (Contini and Costabile, 2020, Domingo et al., 2020, Klompas et al., 2020, Morawska and Cao, 2020, Prather et al., 2020). Particles are emitted during sneezes, cough, respiration, speaking, singing, and shouting. In case of infected individuals, these particles could contain viable virus as happens for other respiratory viruses (Milton et al., 2013, Yan et al., 2018), including other coronaviruses such as NL63, OC43, 229E, and HKU1 (Leung et al., 2020). Sneezing and coughing are mainly associated with symptomatic individuals, however, emissions during respiration and speaking could happen also for asymptomatic individuals that have typically a viral load comparable to that of symptomatic patients (Lavezzo et al., 2020). Large respiratory droplets (conventionally with diameter D > 5 µm) settle faster than they evaporate, contaminating the immediate vicinity of the infected individuals. In contrast, small droplets (i.e. D < 5 µm) evaporate faster than they settle, leaving dry residuals (also called droplet nuclei) which might contain virus aggregates, proteins, and mineral salts (Asadi et al., 2020, Borouiba, 2020). Droplet nuclei can remain suspended in air for longer time compared to large droplets and potentially contribute to airborne transmission (Allen and Marr, 2020, Morawska and Cao, 2020, Martano, 2020).
The probability of airborne transmission is different in outdoor and indoor environments and depends on several parameters; the most important are: (i) concentration and size distribution of virus-laden particles in air; (ii) the fraction of viable viral particles; (iii) the minimum viral load necessary to transmit the infection by inhalation in susceptible individuals. The lifetime of aerosolized SARS-CoV-2 could be 3 h under laboratory controlled conditions (van Doremalen et al., 2020), but it could be less in outdoors depending on the degradation of the virus due to local meteorology conditions (Ratnesar-Shumate et al., 2020). The minimum infectious dose, expressed in viral RNA copies inhaled, is not defined for SARS-CoV-2 in current scientific literature. However, referring to the studies on SARS-CoV-1, the dose of airborne virus copies (i.e. the quantum) necessary to cause infection in 63% of susceptible individuals is variable between 10 and 100 and it is possible to assume an average of 20 for SARS-CoV-2 (Buonanno et al., 2020). Current knowledge of concentration and size distribution of virus-laden aerosol in air is extremely scarce and contrasting results have been observed. In outdoors, measurements in Wuhan (Hu et al., 2020, Liu et al., 2020) showed concentrations below the detection limit, except for crowded areas; while, a study conducted in Bergamo (north of Italy) identified traces of viral RNA in 23% of the analysed PM10 samples without quantifying the concentrations (Setti et al., 2020). Concentrations of viral particles in indoor environments (mainly measured in hospitals and quarantine areas) seems to be higher (Hu et al., 2020, Liu et al., 2020, Santarpia et al., 2020) than those observed in outdoors. However, other studies showed no detectable concentrations even in proximity of quarantined COVID-19 patients (Faridi et al., 2020, Ong et al., 2020).
This work tries to fill the gap in knowledge regarding atmospheric concentrations of virus-laden particles in Italy. It is focused on the analysis of concentration and size distribution of SARS-CoV-2 virus-laden aerosol in outdoor air, comparing data collected in the Veneto region (north Italy) and Apulia region (south Italy), to asses a possible role of outdoor airborne transmission in the difference of COVID-19 spread rate observed in north and south of Italy. Measurements were performed simultaneously in the two regions, collecting air samples using both PM10 samplers and cascade impactors. SARS-CoV-2 presence was determined looking for genetic material (RNA) using both real-time RT-PCR and droplet digital PCR (ddPCR) methods (Corman et al., 2020, Suo et al., 2020).
2. Methods
2.1. Samples collection
Aerosol sampling was simultaneously carried out from 13th to 27th of May 2020, in two different Italian regions: Veneto (in the northeaster Italy) and Apulia (in the southeaster Italy). In Veneto, samples were collected at the Scientific Campus of Ca’ Foscari University (45°28′47″N, 12°15′12″E, Mestre-Venice, Italy). The site is located in a working/residential area of Mestre, characterized by some major potential sources of particulate matter: high density residential areas; heavily trafficked roads; the industrial area of Porto Marghera, and an international airport (Squizzato et al., 2016). In Apulia, measurements were performed at the Lecce Environmental-Climate Observatory (ECO, 40.3°N 18.1°E; 36 m a.s.L.), located at the Institute of Atmospheric Sciences and Climate of the National Research Council (ISAC-CNR), inside the University Campus, at about 4 km (WSW) from the urban area of Lecce (Dinoi et al., 2020). The area, considered an urban background site, is affected by the integrated contribution of local anthropogenic sources (mainly road traffic and biomass burning) and by the long-range transport of natural and anthropogenic dust (Cesari et al., 2016, Cesari et al., 2018).
During both sampling campaigns, two different samplers were used. In Venice, PM10 samples were collected using a low volume aerosol sampler (Skypost PM-TCR Tecora) equipped with a sequential sampler (Charlie) that operates at flow rate of 38.3 L min−1. The sampling period for each sample was about 48 h, with a total average air volume of 110 m3 per sample. A second simultaneous sampling was performed using a model 110 MOUDI cascade impactor with an average flow of 30 L min−1. The inlet of the impactor has a nominal cut-off size of 18 μm, and the nominal cut-off sizes of the 10 impaction stages are: 10, 5.6, 3.2, 1.8, 1.0, 0.56, 0.32, 0.18, 0.10 and 0.056 μm. A back-up filter collected particles with aerodynamic diameter <0.056 μm. The sampling period for each impactor sample was about 6 days, with a total average air volume of about 250 m3 per sample. This setup for data collection with the impactor was already successfully used in other measurement campaigns (Cesari et al., 2020).
In Lecce, the 48-h PM10 samples were collected using a low volume (38 L min−1) sampler (SWAM 5a Dual Channel Monitor-FAI Instruments). Size-segregated samples were collected with a rotating model 120 MOUDI-II™ cascade impactor, operating at 30 L min−1 for about 6 days for each sample, to separate particles of different aerodynamic diameters in the same twelve intervals used in the Venice site.
At both sites, quartz fibre filters were used, after a decontamination process with a 4 h pre-combustion at 400 °C in a muffle furnace. In total, 12 PM10 filters (6 for each site) and 48 impactor filters (24 for each site) were collected. In addition, 4 field blank filters were obtained for each site, 2 for the PM10 sampler and 2 for the cascade impactor. All samples were vacuum packed in sealed sterile petri dishes and frozen at −25 °C immediately after sampling for conservation until the successive analysis. Laboratory analysis started within four days from the end of collection period.
It has been chosen to use both, PM10 samplers and cascade impactors, because it is important to know the size distribution of virus-laden particles to effectively understand the risk of airborne transmission. Sub-micrometric particles (in the accumulation mode <1 µm) could remained suspended in atmosphere for longer time compared to larger particles that have a greater deposition velocity. Therefore, particles in the accumulation mode could contribute to airborne transmission more than coarse particles and there are limited indication on this aspect in current studies. In Liu et al. (2020) in indoor environments in hospitals in China SARS-CoV-2 RNA was detected also in the size range 0.25–1 µm, instead, in the indoor measurements in Singapore hospitals (Chia et al., 2020) the smallest aerodynamic size fraction that contained detectable levels of SARS-CoV-2 was 1–4 μm.
2.2. Analytical method for RNA detection
RNA extraction for PCR experiments was achieved using Total RNA Purification Kit (Norgen Biotek Corp.) with a modified protocol to increase yield. Each filter was cut and placed inside a 2 mL centrifuge tube containing 1 mL of Phosphate Buffer Solution (PBS) pH 7.4. The tube was sealed and put in a sonicator water bath (Elmasonic S10H) for 30 min. Aerosol particles were separated from the quartz filter by centrifugation using a mini syringe placed in a collection tube. The obtained pellet, for each filter processed, was resuspended by 350 µL of supernatant, according to the manufacturer's protocol. The final eluted solutions (about 70 µL in total) were stored frozen at −80 ± 2 °C until PCR analysis that was performed within four days from extraction.
Molecular analysis for the detection of SARS-CoV-2 was carried out using real-time RT-PCR and Droplet Digital PCR (ddPCR) technologies. According to World Health Organization (WHO), real-time RT-PCR represents the gold standard for the diagnosis of SARS-CoV-2. Recently, ddPCR has demonstrated the best performance to detect SARS-CoV-2, because it reduces the false negatives (Suo et al., 2020).
Real-time RT-PCR for SARS-CoV-2 was carried out on a CFX96™ Real-Time system (Bio-Rad, Italy) using COVID-19 PCR DIATHEVA Detection kit (Diatheva, Cartoceto, PU, Italy) based on the WHO guideline (Corman et al., 2020). The COVID-19 PCR DIATHEVA Detection kit is a One-Step real-time reverse transcription (RT-PCR) multiplex assay based on fluorescent-labelled probe used to confirm the presence of SARS-CoV-2-RNA by amplification of RdRp and E gene. The kit provides all the reagents required for the analysis, PCR positive and PCR negative controls included. 5 µL of extracted RNA were added to 15 µL of Master mix for each sample and analysed according to kit instructions. Undiluted and 1:10 diluted samples were tested. In each run, two negative controls (molecular grade water) and a positive control were added. The interpretation of the sample results was done according to kit instructions. The limit of detection (LOD) of the COVID-19 PCR DIATHEVA Detection kit was previously defined through analysis of standard material RNA of SARS-CoV-2 and was equal to 10 copies/µL.
The ddPCR assays were performed using Bio-Rad SARS-CoV-2 ddPCR kit on QX200™ Droplet Digital™ PCR system (Bio-Rad, Italy). The Bio-Rad SARS-CoV-2 ddPCR is a reverse transcription (RT) droplet digital PCR (ddPCR) test designed for the qualitative detection of RNA from SARS-CoV-2. The assay includes the 2019-nCoV CDC ddPCR Triplex Probes and the One-Step RT-ddPCR Advanced Kit for Probes. The 2019-nCoV CDC ddPCR Triplex Probes contains specific oligonucleotide primers and probes for SARS-CoV-2 (N1 and N2), the same as those reported by Center for Disease Control and Prevention (CDC), mapping on regions of the virus nucleocapsid (N) gene into a single assay multiplex to enable a one-well reaction. The reaction mixtures were partitioned into approximately 20,000 droplets using a QX200 Droplet Generator™ (Bio-Rad, Italy) with the random dispersal of target nucleic acids into the droplets. The PCR assays were conducted in a C1000 Touch™ Thermal Cycler (Bio-Rad, Italy), according to kit instructions. After amplification, the droplets were individually assayed using the QX200™ Droplet Reader™. The fluorescence data were then analysed by the QuantaSoft v1.7 Software and QuantaSoft Analysis Pro v1.0 Software (Bio-Rad, Italy) to determine the presence of SARS-Cov-2 N1 and N2 in the specimen. The LOD of the Bio-Rad SARS CoV-2 ddPCR test was declared by the manufacturer in 0.625 copies/µL for targets N1 and N2.
The efficiency of the extraction procedure was evaluated through the recovery of a process control, a virus added prior to acid nucleic extraction. Mengo virus strain MC0, supplied by Istituto Superiore di Sanità (ISS, Rome, Italy), is a murine virus of the Picornaviridae family, a non-enveloped positive-sense ssRNA virus. The efficiency of the extraction method was evaluated comparing the Ct values obtained for Mengovirus on samples extracts. In detail, 10 µL of Mengovirus was added prior to extraction to (i) 1 mL of PBS (reference sample); (ii) 1 mL of PBS with a blank filter and (iii) 1 mL of PBS with an exposed environmental filter. Each condition was run in duplicate. The detection of Mengovirus was carried out on a CFX96™ Real-Time system (Bio-Rad, Italy) using amplification conditions, primers and probe and reagents RNA UltraSense™ One-Step Quantitative RT-PCR System (Life Technologies, Carlsbad, California, US) (Pintó et al., 2009). Results indicate an average recovery of 49% (±5%).
3. Results and discussion
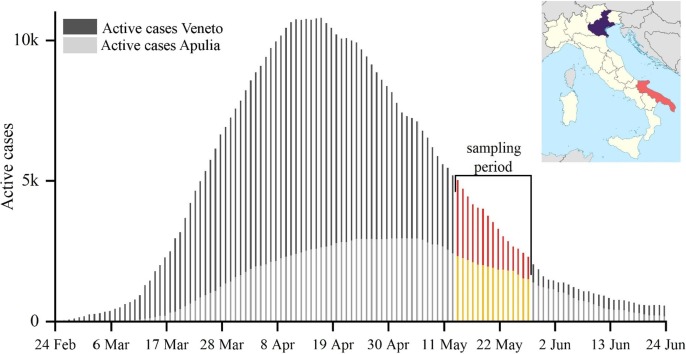
The outbreak of COVID-19 in Italy has resulted in 239,961 confirmed cases and 34,708 fatalities as of 27th June 2020. The transmission of SARS-CoV-2 was exceptionally severe in Veneto region (Fig. 1 ), with maximum active cases (i.e. currently infected individuals) of 10,800 on 16th April (about 10% of the overall Italian cases) over a population of 4.9 million people. Apulia region (southern Italy) reached the maximum of active cases on the 3rd of May with 2,955 cases (3% of the overall Italian cases) over a population of 4.0 million people. At the beginning of sampling period (13th May) Veneto and Apulia regions were affected by 5,020 and 2,322 active cases, respectively. These official numbers likely underestimate the real contagions. In Italy, cumulatively, 2.2–3.5 million individuals seem to have been infected as of May 4th, giving an attack rate of 3.6%-5.8% of the population (Flaxman et al., 2020).
Fig. 1.
Daily number of infected individuals observed in Veneto and Apulia regions during COVID-19 outbreak in Italy. The measurement sites are shown together with sampling period.
During sampling, the average temperature was 19.6 °C (±1.4 °C) in Venice and 21.0 °C (±1.9 °C) in Lecce; the average relative humidity was 69% (±9.5%) in Venice and 56% (±9.8%) in Lecce. No precipitations were observed at the two sites during the sampling period.
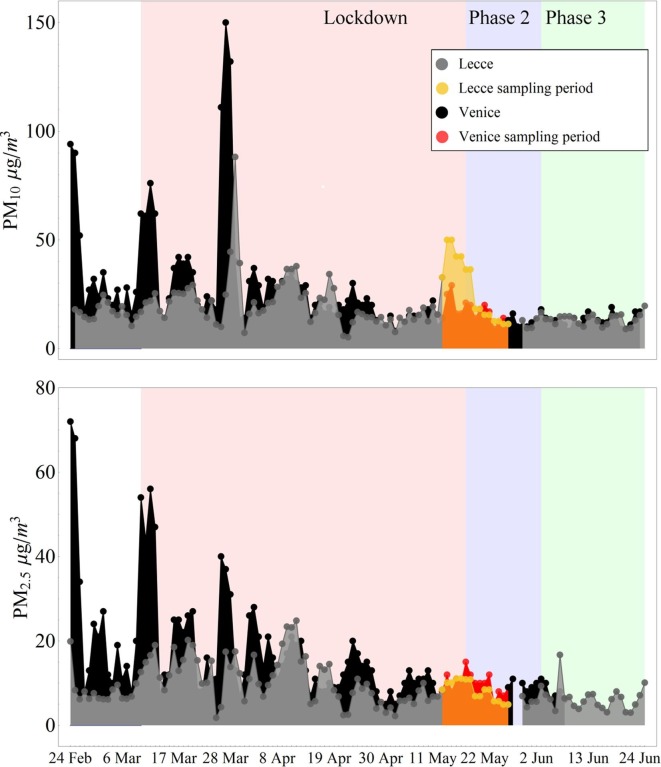
PM10 and PM2.5 concentration levels at the two sites are shown in Fig. 2 . The national lockdown in Italy (indicated as phase I) included the period between 10th March and 17th May, even if there were some differences of restrictive measures during this period. Successively, there was the post-lockdown divided in what we called phase II and phase III in Fig. 2. Phase II was from 18th May until 3rd June and it was a re-opening with several limitations (cinema and theatres closed, travels among different regions were interdict, employees of public administrations remained diffusely on smart working). During phase III it was removed the restriction of travels among different regions and a significant fraction of employees of public administrations started to work on offices again. Our measurements started near the end of the phase I and continued during phase II of the post-lockdown period. The concentrations of PM10, averaged over the whole measurement period, were 17.2 ± 5.2 µg m−3 (average ± standard deviation) in Venice, as provided by the Regional Agency of Environmental Prevention and Protection of Veneto (ARPAV) and 27.0 ± 14.8 µg m−3 in Lecce. The average concentrations of PM2.5 were 9.8 ± 2.5 µg m−3 in Venice and 8.3 ± 2.3 µg m−3 in Lecce. High PM10 concentration are determined in Lecce at the beginning of sampling period (13–18 May 2020), while the values of PM2.5 were comparable at the two sites (Fig. 2). This was due to a contribution of coarse particles due to African dust advection, that influenced only southern regions of Italy, contributing mainly to PM10 but not significantly to PM2.5.
Fig. 2.
Comparison of PM10 (top) and PM2.5 (bottom) concentrations at the two sites evidencing the sampling period. The lockdown period (phase I) and the post-lockdown (phase II and II) are reported.
The existence of SARS-CoV-2 in aerosol samples was determined through the detection of its genetic material (RNA) in collected samples. Air samples were extracted at the Istituto Zooprofilattico Sperimentale della Puglia e della Basilicata (IZSPB), COVID-19 laboratory for the Apulia region. All extracts were firstly analysed using real-time RT-PCR and were negative showing no detectable presence of SARS-CoV-2 RNA. The detection limit of the method, referred to the extracted solutions, was 10 genome copies µL−1. Successively, the same extracts were also analysed using the most sensitive technique available in the laboratory, the ddPCR, lowering the detection limit to 0.625 genome copies µL−1 and all samples tested negative for the presence of viral RNA.
The LODs (genome copies µL−1) were transformed in thresholds for atmospheric concentrations of viral particles (expressed in copies/m3) using the same approach employed for determination of concentration threshold of a chemical component of aerosol samples. Specifically, the LOD was transformed in a threshold of viral particles contained in a single filter (i.e. RNA copies per filter) considering the total volume of extraction solutions (about 70 µL) and the efficiency of the methodology (i.e. the recovery). These numbers were then normalised using the sampled volume to obtain the concentration threshold in copies m−3. The concentration of virus-laden aerosol in PM10 samples was <0.8 copies m−3 at both sites during the sampling period. The size-segregated concentrations from nanoparticles (D < 0.056 µm) up to coarse particles (D > 18 µm) were <0.4 copies m−3 at both sites.
These results are comparable with those found in outdoor residential area in Wuhan (China) during the pandemic (Liu et al., 2020). Liu et al. (2020) collected air samples suing both samplers and cascade impactors, between February and March 2020, in public areas in outdoor as well as in indoor in quarantine areas. Samples collected in outdoor residential areas tested negative (<3 copies m−3) with the exclusion of crowded zones in proximity of hospitals in which concentrations up to 11 copies m−3 were detected. Hu et al. (2020) found no viral RNA in air samples collected in residential community and an open public area (not crowded sites) in Wuhan (China). The analysis reported by Setti et al. (2020) shows that 23% of the 34 PM10 samples collected between February and March in outdoor in northern Italy (Bergamo) tested positive for SARS-CoV-2 RNA, however, concentrations of virus-laden particles were not evaluated. Results reported here suggest that in outdoor conditions, and excluding crowded areas, it is unlikely a role of airborne transmission of COVID-19.
The risk could be larger in community indoor environments where a certain number of infected individuals could be present in closed environments with limited ventilation. In this case, concentrations of virus-laden aerosol seems to be larger compared to outdoor, even if some contrasting results have been obtained. Liu et al. (2020) found SARS-CoV-2 RNA concentrations up to 42 copies m−3 in hospitals and quarantine areas in Wuhan (China) with a fraction of these viral particles in the fine size range (0.25–1 µm). Chia et al. (2020) found detectable SARS-CoV-2 genetic material in air in indoor COVID-19 patient care areas in Singapore in the size range >1 µm. Lednicky et al. (2020) found viable SARS-CoV-2 was isolated from air samples collected 2 to 4.8 m away from the patients in samples collected at the Student Health Care Center (SHCC, University of Florida, USA). Santarpia et al., 2020 found detectable concentrations of viral RNA in 63% of the samples collected in indoor at the Medical Center of the University of Nebraska (where COVID-19 patients were quarantined) with concentrations up to 2.86 copies L−1). Instead, Faridi et al. (2020) did not detect SARS-CoV-2 in ten air samples collected in patient rooms of the largest hospital in Iran. In Singapore, air samples collected in a quarantine area with three patients tested negative for the presence of SARS-CoV-2 RNA (Ong et al., 2020). The possible larger risk of community indoor environments compared to outdoors could be mitigated by the use of face masks and the ventilation of closed spaces with outdoor air.
4. Conclusions
The results found indicate that outdoor atmospheric concentrations of SARS-CoV-2 were very small (<0.8 copies m−3) in both northern and southern Italy. The same applies for each size range investigated with the impactor, which gave virus-laden aerosol concentrations <0.4 copies m−3. The measurements were taken in a period when the number of active cases (i.e. infected individuals) in the two regions were not at the maximum values (Fig. 1), thereby, it is possible to assume that higher concentrations (up to a factor 2 on average for Venice) were likely be present during the period of maximum spread of contagion. The average typical threshold of about 20 virus copies is necessary (Buonanno et al., 2020) to make a quantum of virus (i.e. the dose of airborne droplet nuclei that, if inhaled, is able to cause infection in 63% of susceptible persons). Considering a typical inhalation rate of about 1 m3/h, as average between rest and light exercise (Adams, 1993), the concentrations would be low to spread the contagion via airborne transmission even assuming the mentioned increase of a factor 2.
Therefore, it is possible to conclude that outdoor air in residential and urban areas was generally not infectious and safe for the public in both northern and southern Italy, with the possible exclusion of very crowded sites. In addition, outdoor airborne transmission of SARS-CoV-2 was likely not the main cause of the difference in diffusion rates of COVID-19 observed during outbreaks in north and south of Italy.
Author contributions
D. Contini, A. Gambaro, G. La Salandra conceptualized the study design; E. Barbaro, E. Gregoris, M. Feltracco collected samples in Venice and contributed to data post-processing; M. Conte, A. Dinoi collected samples in Lecce and contributed to data post-processing; D. Chirizzi, G. Ciccarese, G. La Salandra, and G. La Bella carried out the laboratory tests. All authors collaborated to interpretation of results, wrote, read, commented, and approved the final manuscript.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This study did not receive specific funding and was performed within the cooperation project AIR-CoV (Evaluation of the concentration and size distribution of SARS-CoV-2 in air in outdoor environments). Authors wish to thank Arpa Veneto for furnishing their meteorological and particulate matter measurements in Venice area.
Handling Editor: Frederic Coulon
References
- Adams, W.C., 1993. Measurement of Breathing Rate and Volume in Routinely Performed Daily Activities. Final Report. Human Performance Laboratory, Physical Education Department, University of California, Davis. Human Performance Laboratory, Physical Education Department, University of California, Davis. Prepared for the California Air Resources Board, Contract No. A033-205, April 1993.
- Allen J.G., Marr L.C. Recognizing and controlling airborne transmission of SARSCoV-2 in indoor environments. Indoor Air. 2020;30:557–558. doi: 10.1111/ina.12697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asadi S., Bouvier N., Wexler A.S., Ristenpart W.D. The coronavirus pandemic and aerosols: Does COVID-19 transmit via expiratory particles? Aerosol Sci. Tech. 2020;54(6):635–638. doi: 10.1080/02786826.2020.1749229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borouiba L. Turbulent gas clouds and respiratory pathogen emissions, potential implications for reducing transmission of COVID-19. JAMA. 2020;323(18):1837–1838. doi: 10.1001/jama.2020.4756. [DOI] [PubMed] [Google Scholar]
- Buonanno G., Stabile L., Morawska L. Estimation of airborne viral emission: quanta emission rate of SARS-CoV-2 for infection risk assessment. Environ. Int. 2020 doi: 10.1016/j.envint.2020.105794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cesari D., Amato F., Pandolfi M., Alastuey A., Querol X., Contini D. An intercomparison of PM10 source apportionment using PCA and PMF receptor models in three European sites. Environ. Sci. Pollut. Res. 2016;23(15):15133–15148. doi: 10.1007/s11356-016-6599-z. [DOI] [PubMed] [Google Scholar]
- Cesari D., De Benedetto G.E., Bonasoni P., Busetto M., Dinoi A., Merico E., Chirizzi D., Cristofanelli P., Donateo A., Grasso F.M., Marinoni A., Pennetta A., Contini D. Seasonal variability of PM2.5 and PM10 composition and sources in an urban background site in Southern Italy. Sci. Tot. Environ. 2018;612:202–213. doi: 10.1016/j.scitotenv.2017.08.230. [DOI] [PubMed] [Google Scholar]
- Cesari D., Merico E., Dinoi A., Gambaro A., Morabito E., Gregoris E., Barbarob E., Feltracco M., Alebić-Juretić A., Odorčić D., Kontošić D., Mifka B., Contini D. An inter-comparison of size segregated carbonaceous aerosol collected by low-volume impactor in the port-cities of Venice (Italy) and Rijeka (Croatia) Atmos. Pollut. Res. 2020 doi: 10.1016/j.apr.2020.06.027. [DOI] [Google Scholar]
- Chia P.Y., Coleman K.K., Tan Y.K., et al. Detection of air and surface contamination by SARS-CoV-2 in hospital rooms of infected patients. Nat. Commun. 2020;11:2800. doi: 10.1038/s41467-020-16670-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conticini E., Frediani B., Caro D. Can atmospheric pollution be considered a co-factor in extremely high level of SARS-CoV-2 lethality in Northern Italy? Environ. Pollut. 2020;261:114465. doi: 10.1016/j.envpol.2020.114465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Contini D., Gambaro A., Donateo A., Cescon P., Cesari D., Merico E., Belosi F., Citron M. Inter-annual trend of the primary contribution of ship emissions to PM2.5 concentrations in Venice (Italy): Efficiency of emissions mitigation strategies. Atmos. Environ. 2015;102:183–190. [Google Scholar]
- Contini D., Costabile F. Does air pollution influence COVID-19 outbreaks? Atmosphere. 2020;11:377. doi: 10.3390/atmos11040377. [DOI] [Google Scholar]
- Corman V., Landt O., Kaiser M., Molenkamp M., Meijer A., Chu D.K.W., et al. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. 2020;25(3) doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinoi A., Conte M., Grasso F.M., Contini D. Long-term characterization of submicron atmospheric particles in an urban background site in Southern Italy. Atmosphere. 2020;11:334. doi: 10.3390/atmos11040334. [DOI] [Google Scholar]
- Domingo J.L., Marquès M., Rovira J. Influence of airborne transmission of SARS-CoV-2 in COVID-19 pandemic. A review. Environ. Res. 2020;188:109861. doi: 10.1016/j.envres.2020.109861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faridi S., Niazi S., Sadeghi K., Naddafi K., Yavarian J., Shamsipour M., et al. A field indoor air measurement of SARS-CoV-2 in the patient rooms of the largest hospital in Iran. Sci. Total Environ. 2020;725:138401. doi: 10.1016/j.scitotenv.2020.138401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrero L., Perrone M.G., Petraccone S., Sangiorgi G., Ferrini B.S., Lo Porto C., Lazzati Z., Cocchi D., Bruno F., Greco F., Riccio A., Bolzacchini E. Vertically-resolved particle size distribution within and above the mixing layer over the Milan metropolitan area. Atmos. Chem. Phys. 2010;10:3915–3932. [Google Scholar]
- Flaxman S., Mishra S., Gandy A., Unwin H.J.T., Mellan T.A., Coupland H., et al. Estimating the effects of non-pharmaceutical interventions on COVID-19 in Europe. Nature. 2020 doi: 10.1038/s41586-020-2405-7. [DOI] [PubMed] [Google Scholar]
- Hu, J., Lei, C., Chen, Z., Liu, W., Hu, X., Pei, R., et al., 2020. Airborne SARS-CoV-2 and the Use of Masks for Protection against Its Spread in Wuhan, China. Preprint at https://www.preprints.org/manuscript/202005.0464/v1.
- Klompas M., Baker M.A., Rhee C. Airborne transmission of SARS-CoV-2. Theoretical considerations and available evidence. JAMA. 2020;324(5):441–442. doi: 10.1001/jama.2020.12458. [DOI] [PubMed] [Google Scholar]
- Lavezzo E., Franchin E., Ciavarella C., Cuomo-Dannenburg G., Barzon L., Del Vecchio C., et al. Suppression of a SARS-CoV-2 outbreak in the Italian municipality of Vo’. Nature. 2020 doi: 10.1038/s41586-020-2488-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lednicky J.A., Shankar S.N., Elbadry M.A., Gibson J.C., Alam Md.M., Stephenson C.J., Eiguren-Fernandez A., Morris J.G., Mavian C.N., Salemi M., Clugston J.R., Wu C. Collection of SARS-CoV-2 virus from the air of a clinic within a university student health care center and analyses of the viral genomic sequence. Aerosol Air Qual. Res. 2020;20:1167–1171. doi: 10.4209/aaqr.2020.02.0202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung N.H.L., Chu D.K.W., Shiu E.Y.C., Chan K.-H., McDevitt J.J., Hau B.J.P., et al. Respiratory virus shedding in exhaled breath and efficacy of face mask. Nat. Med. 2020;26:676–680. doi: 10.1038/s41591-020-0843-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Ning Z., Chen Y., Guo M., Liu Y., Gali N.K., et al. Aerodynamic analysis of SARS-CoV-2 in two Wuhan hospitals. Nature. 2020;582:557–560. doi: 10.1038/s41586-020-2271-3. [DOI] [PubMed] [Google Scholar]
- Martano P. Droplet fate in a cough puff. Atmosphere. 2020;11(8):841. doi: 10.3390/atmos11080841. [DOI] [Google Scholar]
- Milton D.K., Fabian M.P., Cowling B.J., Grantham M.L., McDevitt J.J. Influenza virus aerosols in human exhaled breath: Particle size, culturability, and effect of surgical masks. PLoS Pathog. 2013;9(3):e1003205. doi: 10.1371/journal.ppat.1003205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morawska L., Cao J. Airborne transmission of SARS-CoV-2: The world should face the reality. Environ. Int. 2020;139:105730. doi: 10.1016/j.envint.2020.105730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ong S.Y., Tan Y.K., Chia Y.K., Lee T.H., Ng O.T., Su M., et al. Air, surface environmental, and personal protective equipment contamination by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) from a symptomatic patient. JAMA. 2020;323(16):1610–1612. doi: 10.1001/jama.2020.3227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pintó R.M., Costafreda M.I., Bosch A. Risk assessment in shellfish-borne outbreaks of hepatitis A. Appl. Environ. Microbiol. 2009;75(23):7350–7355. doi: 10.1128/AEM.01177-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prather K.A., Wang C.C., Schooley R.T. Reducing transmission of SARS-COV-2. Science. 2020 doi: 10.1126/science.abc6197. [DOI] [PubMed] [Google Scholar]
- Ratnesar-Shumate S., Williams G., Green B., Krause M., Holland B., Wood S., et al. Simulated sunlight rapidly inactivates SARS-CoV-2 on surfaces. J. Infect. Dis. 2020;222:214–222. doi: 10.1093/infdis/jiaa274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santarpia, J.L., Rivera, D.N., Herrera, V., Morwitzer, M.J., Creager, H., Santarpia, G.W. et al., 2020. Transmission Potential of SARS-CoV-2 in Viral Shedding Observed at the University of Nebraska Medical Center. Available online: https://www.medrxiv.org/content/10.1101/2020.03.23.20039446v2.
- Setti L., Passarini F., De Gennaro G., Baribieri P., Perrone M.G., Borelli M., et al. SARS-Cov-2 RNA found on particulate matter of Bergamo in Northern Italy: First evidence. Environ. Res. 2020;188:109754. doi: 10.1016/j.envres.2020.109754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Squizzato S., Masiol M., Agostini C., Visin F., Formenton G., Harrison R.M., et al. Factors, origin and sources affecting PM1 concentrations and composition at an urban background site. Atmos. Res. 2016;180:262–273. [Google Scholar]
- Suo T., Liu X., Feng J., Guo M., Hu M., Guo D., et al. ddPCR: a more accurate tool for SARS-CoV-2 detection in low viral load specimens. Emerg. Microbes Infect. 2020;9 doi: 10.1080/22221751.2020.1772678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Doremalen N., Bushmaker T., Morris D.H., Holbrook M.G., Gamble A., Williamson B.N., et al. Aerosol and surface stability of SARS-CoV-2 as compared with SARS-CoV-1. N. Engl. J. Med. 2020 doi: 10.1056/NEJMc2004973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan J., Grantham M., Pantelic J., Bueno de Mesquita P.J., Albert B., Liu F., et al. Infectious virus in exhaled breath of symptomatic seasonal influenza cases from a college community. Proc. Natl. Acad. Sci. 2018;115(5):1081–1086. doi: 10.1073/pnas.1716561115. [DOI] [PMC free article] [PubMed] [Google Scholar]