Summary
Autoimmune destruction of pancreatic β cells underlies type 1 diabetes (T1D). To understand T cell-mediated immune effects on human pancreatic β cells, we combine β cell-specific expression of a model antigen, CD19, and anti-CD19 chimeric antigen receptor T (CAR-T) cells. Coculturing CD19-expressing β-like cells and CD19 CAR-T cells results in T cell-mediated β-like cell death with release of activated T cell cytokines. Transcriptome analysis of β-like cells and human islets treated with conditioned medium of the immune reaction identifies upregulation of immune reaction genes and the pyroptosis mediator GSDMD as well as its activator CASP4. Caspase-4-mediated cleaved GSDMD is detected in β-like cells under inflammation and endoplasmic reticulum (ER) stress conditions. Among immune-regulatory genes, PDL1 is one of the most upregulated, and PDL1 overexpression partially protects human β-like cells transplanted into mice. This experimental platform identifies potential mechanisms of β cell destruction and may allow testing of therapeutic strategies.
Keywords: human pluripotent stem cells, regenerative medicine, pancreatic beta cells, autoimmune diabetes, CAR-T cells, pyroptosis, inflammation, chimeric mice
Graphical Abstract
Highlights
CD19-expressing β-like cells differentiated from human ES cells are functional
Tractable in vitro and in vivo killing of CD19-expressing β-like cells by CAR-T cells
Upregulation of pyroptosis factors GSDMD and CAPS4 during β-like cell inflammation
PDL1-overexpressing in β-like cells partially protects against reactive T cells
Ma et al. combined human ESC genome engineering and CAR-T cells to construct synthetic immune responses resembling autoimmune diabetes. This system demonstrates involvement of the pyroptosis factors GSDMD and caspase-4 in β-like cell destruction during inflammation and could potentially be used to test β cell protection strategies, including PDL1 overexpression.
Introduction
Type 1 diabetes (T1D) is caused by destruction of pancreatic β cells by self-reactive immune cells.1, 2, 3 The majority of risk loci for T1D map to genes and enhancer elements active in T cells, suggesting that T cells play important roles in the disease.4 In addition to genetic predisposition, environmental factors could contribute to T1D progression by engaging immune cells.5 Although β-like cells differentiated from T1D patient-derived induced pluripotent stem cells (iPSCs) did not show an apparent phenotype compared with control β-like cells,6,7 they could contribute to immune responses by mechanisms such as expressing antigenic fusion peptides.8 Therefore, a better understanding of how pancreatic β cells are affected by autoimmune T cells is important for understanding the disease.
T1D can progress over a long period without detectable symptoms, resulting in extensive β cells loss at the time of diagnosis, making it difficult to study and modulate the interactions between human β cells and T cells during early T1D.1, 2, 3 Multiple β cell proteins, including insulin, glutamic acid decarboxylase, and SLC30A8, have been identified as β cell autoantigens.9, 10, 11, 12 The variety of β cell autoantigens and the complex interactions between different immune components make human T1D progression challenging to investigate. Therefore, robust and tractable systems are needed to study β cell responses caused by autoimmunity.
Human pluripotent stem cells (PSCs), with their virtually unlimited replicative capability and remarkable developmental potential, can differentiate into diverse cell types, facilitating basic and translational research.13,14 Additionally, PSCs provide opportunities to model human diseases either in vitro or in humanized mice.15 Human PSCs have been differentiated into β-like cells with gene expression and functional similarities to primary β cells.6,16, 17, 18, 19, 20, 21, 22 Human PSC based models could contribute to understanding the general mechanisms related to human T1D and for developing potential therapeutic approaches.
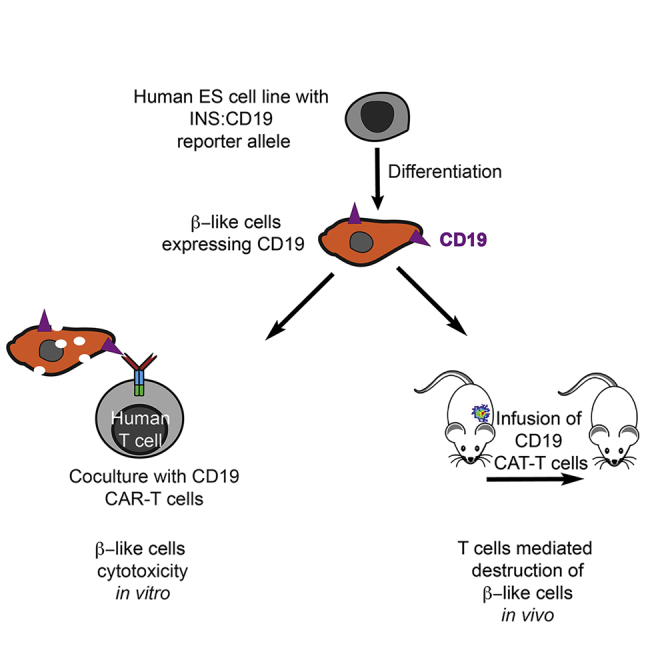
As an initial step to investigate mechanisms regulating the human β cell response during T cell-mediated β cell destruction, we developed a tractable experimental system by combining β cell-specific expression of CD19 as a model antigen and anti-CD19 chimeric antigen receptor (CAR) T cells.23 Upon activation by the extracellular antigen-binding domain, intracellular components of the intracellular domains of CARs provide activating and costimulatory signals through fusion of CD28 and CD3 ζ chain signaling domains, which are activated separately in T cells.24 The similar signaling components of CAR-T cells and T cells prompted us to use CAR-T cells and a model antigen as a proxy to infer T cell-mediated effects on β cells under highly defined conditions. Bypassing the mechanisms leading to β cell-reactive T cell activation enabled us to develop a disease-simulating model. We focused on studying interactions between human T cells and β-like cells in vitro and in humanized mice. This tractable system recapitulated the prediabetic transcriptional program in T cell-mediated β cell response, such as rapid upregulation of inflammation genes, antigen presentation components, and immune-regulatory genes, including PDL1 (CD274), CD47, and the CD277-encoding BTN3A1, BTN3A2, and BTN3A3. Overexpression of one of the most upregulated immune checkpoint genes, PDL1, partially protected β-like cells in mice that received transplants. Furthermore, we found that the pyroptosis components GSDMD25,26 and its enzymatic activator CASP4 were upregulated in β-like cells and primary islets when treated with activated T cell-conditioned medium, implicating pyroptosis as a possible process in disease progression and a possible target for treatment.
Results
Differentiation of INS:tdT and INS:CD19 Reporter PSCs into Functional β-like Cells
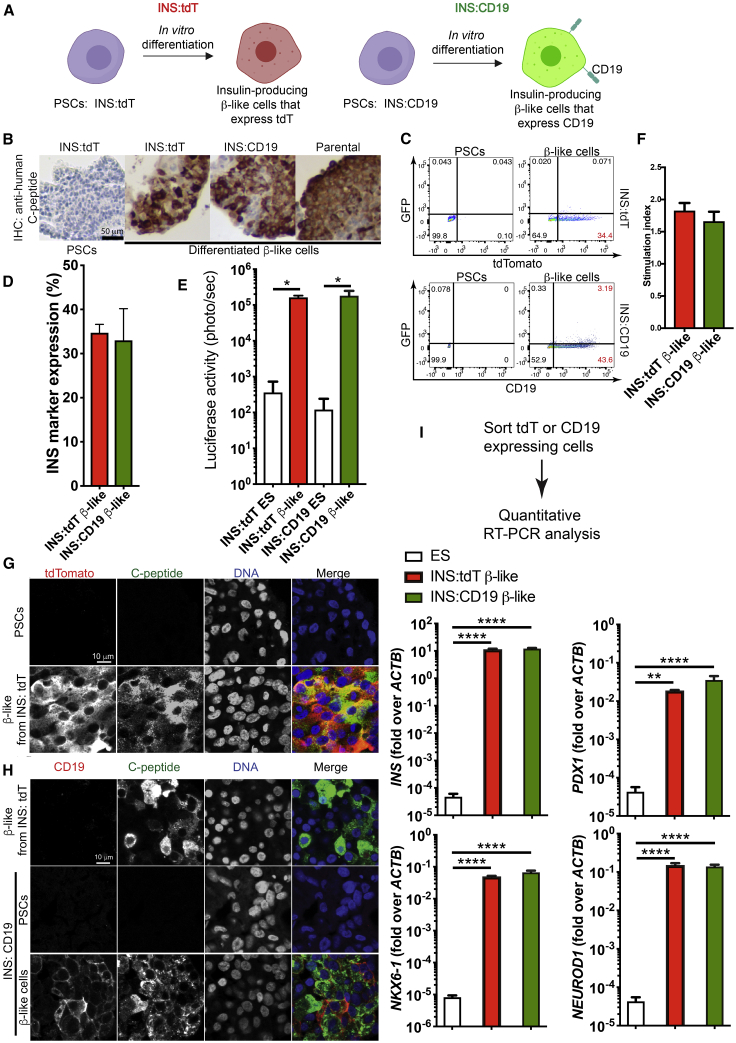
To trace insulin-producing β cells in vitro and in vivo, we generated a multicistronic vector with a constitutive CAGGS (CMV enhancer, chicken β-actin promoter, rabbit β-globin intron) promoter-driven CRIPSR/Cpf127 and U6 promoter-driven single guide RNA (sgRNA) targeting the human insulin (INS) locus (sgINS). H1 human PSCs28 were electroporated with the Cpf1/sgINS construct and one of two targeting constructs (Figure S1A). The first targeting construct introduced a T2A-luciferase-T2A-tdTomato (tdT) cassette into the 3′ end of the INS coding sequence (INS:tdT) to generate a control cell line (Figure S1A, control [CTL]) that would express tdTomato controlled by the endogenous INS regulatory element (Figure 1A), and the second construct utilized a T2A-luciferase-T2A-CD19-T2A-GFP cassette (INS:CD19) (Figure S1A, experiment [EXP]) to generate cells with cell surface expression of CD19 (Figure 1A). We identified edited clones by genotyping PCR (Figure S1A, bottom right panel) and Southern blotting (Figures S1B and S1C). The CTL and experimental cells are referred to as INS:tdT and INS:CD19, respectively. Both cell lines formed teratomas consisting of all three germ layers (Figure S1D), indicating that gene editing did not interfere with pluripotency of the PSCs.
Figure 1.
Differentiation of Human PSC INS Reporter Cells
(A) A diagram showing control (left panel) and experimental (right panel) INS reporter cells.
(B) Immunohistochemistry (IHC) staining with an anti-human-C-peptide antibody in β-like cells differentiated from INS:tdT, INS:CD19, and parental PSCs (right three panels) and undifferentiated PSCs (left panel). Scale bar, 50 μm.
(C) Representative flow cytometry results of PSCs (left panels) and β-like cells (right panels) differentiated from INS:tdT (top panel) and INS:CD19 (bottom panel), showing the GFP signal on the vertical axis and the tdTomato fluorescence signal (top panel) or anti-CD19 signal on the horizontal axis (bottom panel).
(D) Quantifications of the proportion of INS reporter-expressing cells at the end of differentiation (n = 3).
(E) Quantification of the luciferase signal in undifferentiated ESCs and β-like cells differentiated from the INS reporter cells (n = 4).
(F) In vitro glucose-stimulated insulin secretion (GSIS) assays of PSC-differentiated β-like cells. The stimulation index shows the ratio of the amounts of secreted human insulin released when β-like cells were treated with 20 mM glucose compared with 2.5 mM glucose (n = 4).
(G) Confocal micrographs with an anti-tdTomato antibody (red) and an anti-C-peptide antibody (green) of β-like cells differentiated from INS:tdT PSCs. Scale bar, 10 μm.
(H) Confocal micrographs with an anti-CD19 antibody (red) and an anti-C-peptide antibody (green) of INS:tdT β-like cells (top panel), INS:CD19 PSCs (center panel), and differentiated β-like cells (bottom panel). Scale bar, 10 μm.
(I) Quantitative RT-PCR analysis of the β-cell marker genes on RNA samples extracted from sorted INS marker-expressing cells differentiated from INS:tdT and INS:CD19 PSCs (n = 4).
∗p < 0.05, ∗∗p < 0.01, ∗∗∗∗p < 0.0001, non-paired t tests.
See also Figures S1–S3.
Based on previous differentiation protocols,6,18,19,21,22 we developed an in vitro β-like cell differentiation system composed of 2D culture (stages 1–4) followed by 3D suspension culture (stages 5–7) (Figure S2A). Flow cytometry analyses demonstrated expression of stage-specific marker proteins (Figures S2B–S2E). As expected, quantitative RT-PCR (Figure S2F) and RNA sequencing (RNA-seq) (Figures S2G and S2H) showed gene expression similar to that of β cells. Moreover, vesicles with characteristics of insulin-containing secretory granules were detected in β-like cells (Figure S2I).
INS:tdT PSCs showed a tdTomato fluorescence signal at day 2 of stage 5 , a stage of pancreatic endocrine precursor differentiation characterized by upregulation of NGN3.19,29 At the end of differentiation, β-like cells differentiated from the INS reporter cells showed C-peptide expression similar to parental cells (Figure 1B). In addition, flow cytometry revealed that the tdTomato fluorescence signal increased as INS:tdT PSCs differentiated (Figure 1C, top panel). Similarly, INS:CD19 cells gained cell surface expression of CD19 as cells differentiated into β-like cells (Figure 1C, bottom panel). The proportion of INS reporter-expressing cells were similar in differentiation to INS:tdT or INS:CD19 PSCs (Figure 1D). Furthermore, we detected a significant increase in luciferase signals in differentiated cells from INS:tdT and INS:CD19 PSCs (Figure 1E). Glucose-stimulated insulin secretion in in-vitro-differentiated β-like cells showed no significant difference between tdTomato- and CD19-expressing β-like cells (Figure 1F).
To examine the correlation between INS marker expression and expression of INS, we performed immunofluorescence staining and confocal microscopy. We detected expression of C-peptide only when the INS:tdT PSCs were differentiated into tdTomato-positive β-like cells (Figure 1G). Similarly, CD19 and C-peptide were expressed in β-like cells differentiated from INS:CD19 cells (Figure 1H). Importantly, CD19 expression did not significantly affect expression of the β cell marker genes INS, PDX1, NKX6-1, and NEUROD1 (Figure 1I). Finally, immunofluorescence staining of in-vitro-differentiated cells and cells transplanted into immunocompromised mice did not show obvious differences between β-like cells differentiated from INS:tdT or INS:CD19 PSCs (Figure S3A), and C-peptide-positive β-like cells did not express the proliferation marker Ki-67 (Figure S3B).
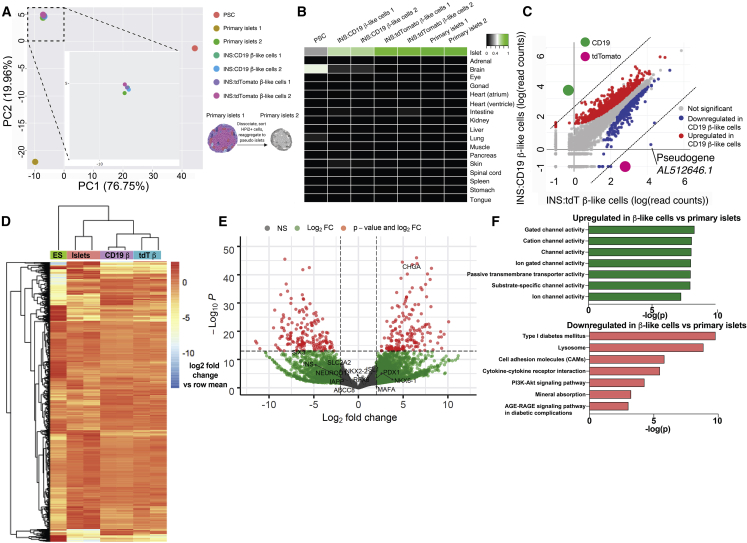
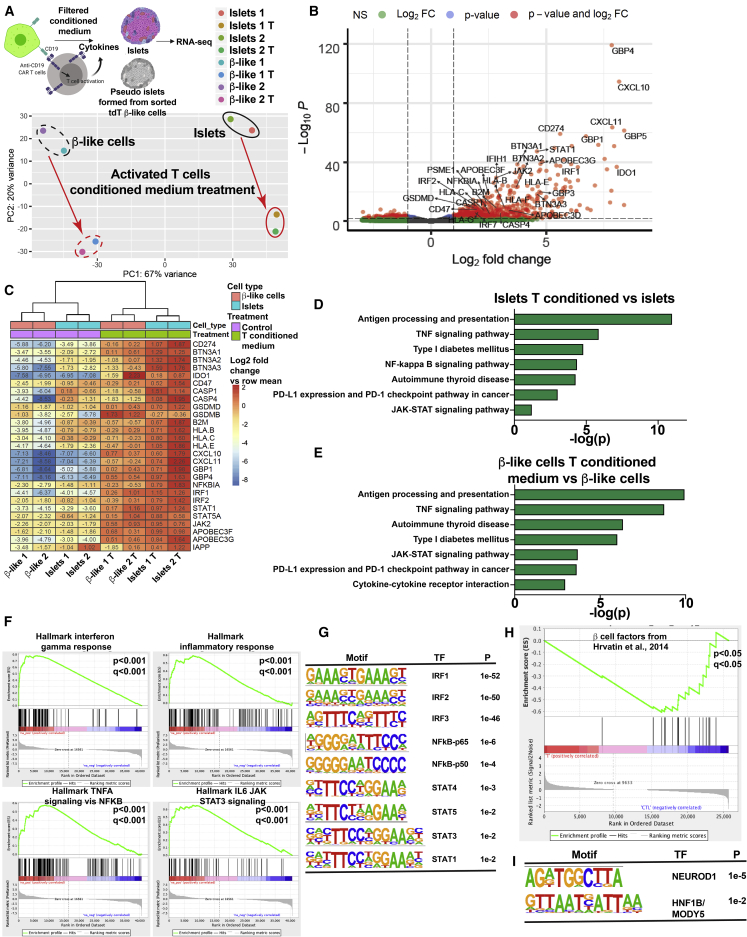
To further characterize the β-like INS reporter cells, we extracted RNA from human primary islets and pseudoislets formed by reaggregating sorted HPi2-positive endocrine cells30 for RNA-seq. Principal-component analysis showed that INS:tdT and INS:CD19 β-like cells overlapped with the pseudoislets formed from primary pancreatic endocrine cells and away from PSCs (Figure 2A). Sorted INS reporter cell samples showed the highest similarities to islets rather than other tissue types, based on the Keygenes analysis (Figure 2B), a test for cell type identity and specificity.31 The most differentially expressed genes in INS:CD19 and INS:tdT β-like cells were CD19 and tdTomato (Figure 2C), indicating that expression of CD19 did not interfere with expression of β cell-specific genes. This was confirmed by hierarchical clustering of the approximately 1,500 most differentially expressed genes between embryonic stem cells (ESCs), primary islets, and β-like INS reporter cells, showing gene expression similarity between primary islets and β-like INS reporter cells (Figure 2D).
Figure 2.
Gene Expression of β-like Cells Differentiated from INS:tdT and INS:CD19 PSCs
(A) Principal-component analysis (PCA) of undifferentiated human pluripotent stem cells (PSCs), CD19-expressing β-like cells (INS:CD19 β-like cells 1 and 2), tdTomato-expressing β-like cells (INS:tdTomato β-like cells 1 and 2), and human primary islets. Primary islet 1 RNA was extracted from primary islets, and primary islet 2 RNA was extracted from pseudoislets reaggregated from sorted endocrine cells. The boxed area was magnified for better sample separation.
(B) Keygene plots of RNA-seq samples, showing islet identity of tdTomato- and CD19-expressing β-like cells and primary islets but not for PSCs.
(C) Scatterplot of log10 (read counts) values for transcripts present in tdTomato-expressing β-like cells (x axis) and CD19-expressing β-like cells (y axis). The most differentially expressed genes, CD19 and tdTomato, are labeled in green and magenta, respectively.
(D) Heatmap and hierarchical clustering of the approximately 1,500 most differentially expressed genes between ESCs, primary islets, and β-like INS reporter cells.
(E) Volcano plot showing genes differentially expressed in β-like cells (right) and in primary islets (left). Genes with a log2 fold change of more than 2 and p < 10−14 are shown in red.
(F) Selected KEGG pathways enriched in genes upregulated in β-like cells compared with primary islets (green) and in genes downregulated in β-like cells compared with primary islets (red).
See also Figures S2 and S3 and Tables S1 and S2.
Gene expression analysis demonstrated that approximately 1,500 genes were differentially expressed (log2 fold change > 2 and p < 10−14; Figure S2G; Table S1) between undifferentiated PSCs and differentiated β-like cells. KEGG (Kyoto Encyclopedia of Genes and Genomes) analysis showed that the most upregulated genes in the β-like cells were maturity onset of diabetes of the young (MODY) genes, INS secretion, Ca2+ signaling pathway, circadian entrainment, protein processing in the endoplasmic reticulum, and type 2 diabetes (Figure S2H). In contrast, the downregulated genes were in DNA replication, cell cycle, and Hippo signaling and PSC pathways (Figure S2H). These results are consistent with differentiation from PSCs to pancreatic β-like cells. Compared with primary islets, approximately 290 genes were differentially expressed in β-like cells (log2 fold change > 2 and p < 10−14; Figure 2E; Table S2), which were enriched in diabetes pathogenesis signature genes. Many β cell marker genes were expressed at similar levels (Figure 2E). Other genes, such as the age-dependent β cell maturation factor SIX3,32 were expressed at lower levels in β-like cells compared with primary islets (Figure 2E). KEGG pathways of differentially expressed genes include ion channels activities, T1D, and lysosomes (Figure 2F). These results suggest that, even though there were gene expression differences between PSC-differentiated β-like cells and primary islets, the β-like cells generated here showed transcriptional similarities to primary islets.
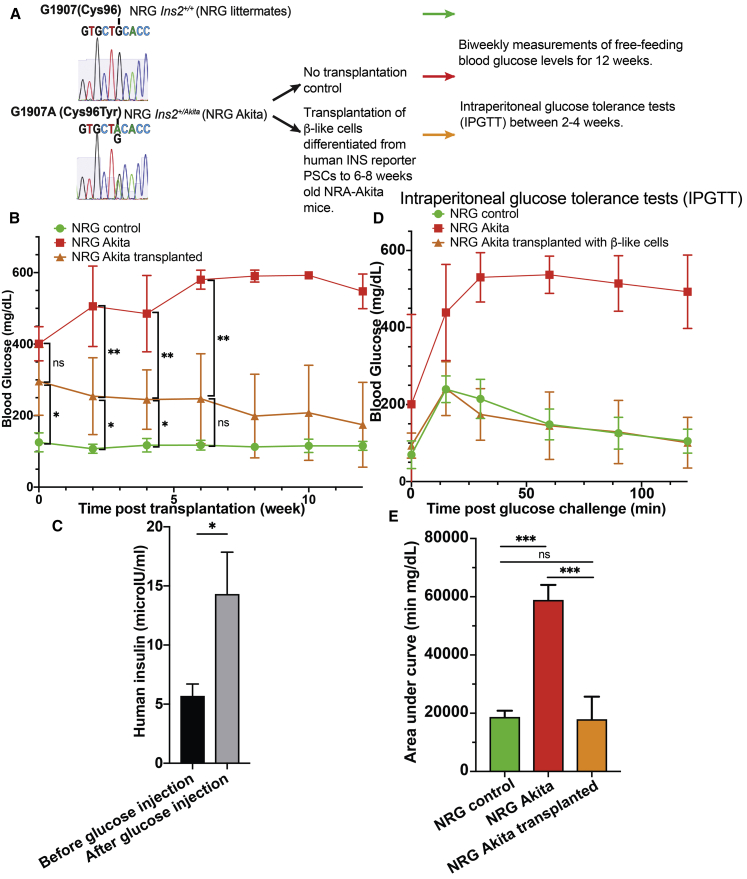
To characterize β-like cells in vivo, we transplanted the cells under the kidney capsule of immunocompromised diabetic non-obese diabetic (NOD).Cg-Rag1tm1Mom Ins2Akita Il2rgtm1Wjl/SzJ (NRG-Akita) mice,33 and their ability to rescue the elevated glucose levels was assessed (Figure 3A). After a 2-week surgery recovery period, NRG-Akita mice with kidney capsular transplantation of 5 million β-like INS reporter cells showed reduced blood glucose levels under ad libitum feeding conditions compared with CTL NRG-Akita mice (Figure 3B). ELISA measurement of human insulin in the mouse blood circulation showed increased human insulin secretion after glucose injection compared with mice before glucose injection (Figure 3C). Although the blood glucose levels were still higher than in nondiabetic NRG CTL mice 2 weeks after transplantation, the ad libitum feeding glucose levels reduced further over time and became similar to NRG CTL mice 6 weeks after transplantation (Figure 3B). Likewise, although between 2–4 weeks after transplantation when the ad libitum feeding glucose levels in NRG-Akita mice that received the transplant were still statistically significantly higher than in NRG CTL mice, intraperitoneal glucose tolerance tests (IPGTT) showed that transplanted mice were able to efficiently clear introduced glucose (Figures 3D and 3E). We also noted that the fasting glucose levels of some NRG-Akita mice 2–4 weeks after transplantation appeared to be very similar to the fasting glucose levels of NRG CTL mice (Figure 3D). These results are consistent with the in vitro glucose-stimulated insulin secretion (GSIS) results (Figure 1F) and gene expression analysis (Figure 2). Thus, the in vitro and in vivo data suggest that the β-like INS reporter cells are functional.
Figure 3.
Restoration of Glucose Homeostasis in Diabetic NRG-Akita Mice upon Transplantation of INS Reporter β-like Cells
(A) A schematic representation of the experimental design.
(B) Biweekly glucose measurements of ad libitum-fed NRG mice (green, n = 3), NRG-Akita mice transplanted with 5 million hPSC-differentiated β-like cells (orange, n = 8 from weeks 0–4, n = 7 from weeks 5–12), or CTL NRG-Akita mice (red, n = 3) before transplantation (week 0) to 12 weeks after transplantation. Data are plotted as mean ± SD.
(C) Measurement of human insulin in mice transplanted with β-like cells 2–4 weeks after transplantation in in vivo GSIS experiments (n = 8).
(D) Intraperitoneal glucose tolerance test (IPGTT) results of NRG mice (green, n = 3), NRG-Akita mice transplanted with hPSC-differentiated β-like cells (orange, n = 6), or CTL NRG-Akita mice (red, n = 3) between 2–4 weeks after transplantation.
(E) The area-under-the-curve analysis of (D).
In (C): ∗p < 0.05, paired t test. Other: ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, non-paired t tests.
In Vitro Interaction between CD19-Expressing Human β-like Cells and CD19 CAR-T Cells
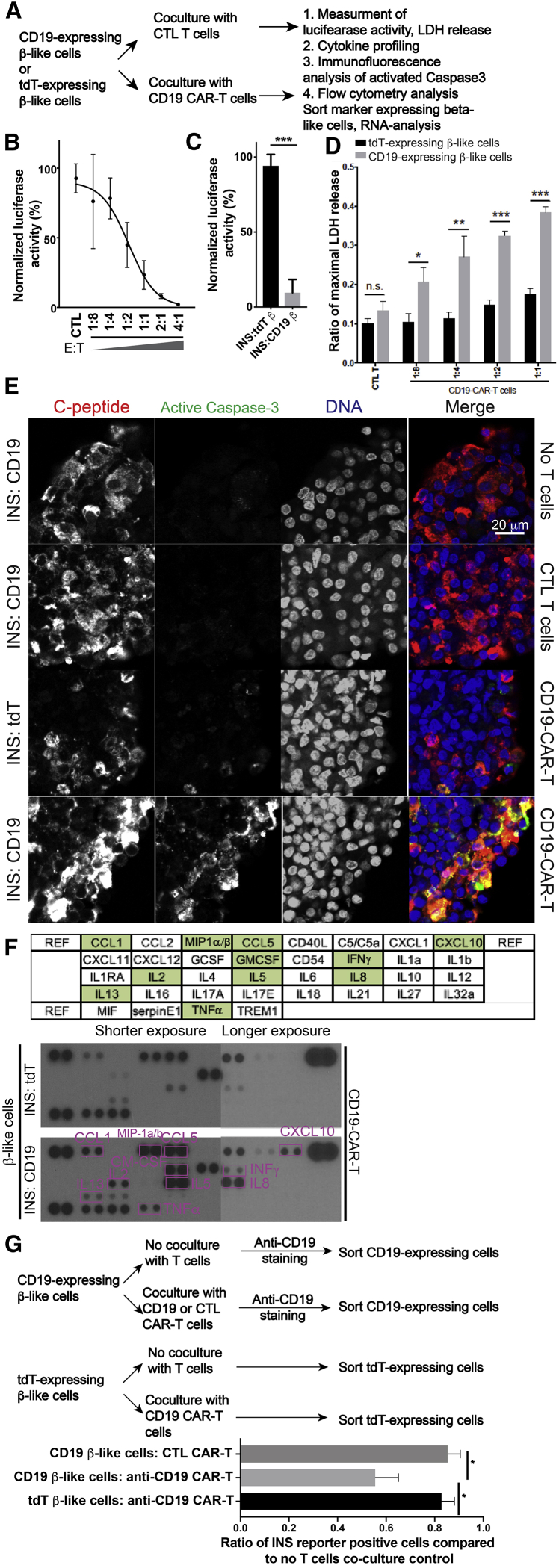
To establish a model system for studying the interaction between β-like cells and cytotoxic T cells, we generated CD-19 CAR-T cells and CTL cells carrying a truncated CAR with a disrupted CD19-binding ectodomain from primary human T cells (Figure S4A and S4B). Although CD19 CAR-T cells showed CD19-mediated cytotoxicity, CTL CAR-T cells did not show significant lysis of CD19-expressing cells (Figures S4C and S4D), demonstrating CD19-specific cytotoxic effects conferred by CD19 CAR-T cells.
To test whether the cytotoxic effect of CD19 CAR-T could be directed against β-like INS cells, we cocultured CD19-expressing β-like cells with CD19 CAR-T or CTL CAR-T cells, followed by luciferase imaging, a lactate dehydrogenase (LDH) release-based cytotoxicity assay, cytokine screening, and immunofluorescence and flow cytometry analyses (Figure 4A). Coculture of CD19 CAR-T cells, but not CD19-binding domain-truncated CTL CAR-T cells, in vitro reduced the luciferase signal from CD19-expressing β-like cells (Figure 4B). Furthermore, coculture of CD19 CAR-T cells and CD19-expressing β-like cells resulted in significantly reduced luciferase activity compared with coculture of CD19 CAR-T cells and tdTomato-expressing β-like cells (Figure 4C). To verify cytotoxicity in in vitro coculture experiments with a different assay, we quantified the enzymatic activity of released LDH, a large protein complex present in most cells whose leakage indicates cell membrane disruption and cell lysis.34 Although coculture of CTL-CAR-T cells with CD19-expressing β-like cells yielded LDH levels similar to coculture with tdTomato-expressing β-like cells, coculture of CD19-CAR-T cells with CD19-expressing β-like cells released significantly higher levels of LDH than coculture with tdTomato-expressing β-like cells (Figure 4D). Similar to luciferase signal reduction, LDH release increased with a higher effector:target (E:T) ratio. Coculture experiments of CD19-expressing β-like cells with CD4 CAR-T cells and CD8 CAR-T cells showed that CD8 CAR-T cell-mediated cytotoxicity was more pronounced than that of CD4 T cells (Figures S4E–S4G). These results demonstrate that in vitro coculture between CD19-CAR-T cells and CD19-expressing β-like cells provides an antigen-specific immune response.
Figure 4.
In Vitro Interaction between CD19-CAR-T Cells and CD19-Expressing β-like Cells
(A) Experimental design to study interaction between CD19-CAR-T cells and CD19-expressing β-like cells in vitro.
(B) CD19-expressing β-like cells were cocultured with CAR-positive T cells at different effector:target (E:T) ratios or with CTL CAR-T cells (CTL-T). Remaining luciferase activities were normalized to signals from samples without T cell treatment (n = 3).
(C) Quantification of coculture experiments with CD19-CAR-T cells and β-like cells (n = 5).
(D) LDH release assay after coculture of β-like cells differentiated from INS:tdT PSCs (black bars) or from INS:CD19 cells (gray bars) with CTL-CAR-T cells or CD19 CAR-T cells (n = 3).
(E) Representative confocal microscopy analyzing active caspase-3 (green) in β-like cells after coculture with T cells. Scale bar, 20 μm.
(F) Cytokine array-based (layout on the top panel) screening of secreted cytokines during coculture of CD19-CAR-T cells and β-like cells differentiated from INS:tdT PSCs (center) or from INS:CD19 PSCs (bottom).
(G) Top panel: experiment design. Bottom panel: quantification of INS reporter-expressing cells after coculture experiments. ∗p < 0.05, non-paired t tests (n = 4).
∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, non-paired t tests.
See also Figure S4.
T cell-mediated β cell death is considered to be mostly mediated by apoptosis.35 To test β-like cell apoptosis in the in vitro coculture system, we collected cells after coculture and performed immunofluorescence analysis with an antibody recognizing cleaved and activated caspase-3.36 Under CTL conditions, we observed background levels of cleaved caspase-3, and similar levels of apoptosis were observed with β-like cells incubated with CTL CAR-T cells (Figure 4E). However, apoptosis was increased more when CD19-expressing β-like cells and CD19-CAR-T cells were cocultured than when tdTomato-expressing β-like cells and CD19-CAR-T cells were cocultured (Figure 4E). These results suggest apoptosis as a mechanism through which β-like cells died in the coculture system.
To identify secreted cytokines, we collected conditioned medium from coculture of CD19-CAR-T cells and CD19-expressing β-like cells and of CD19-CAR-T cells and tdTomato-expressing β-like cells. The cytokine assay with conditioned medium showed release of T cell cytokines, including interferon γ (INFγ), tumor necrosis factor alpha (TNF-α), and interleukin-2 (IL-2) (Figure 4F). In addition, CD19-expressing β-like cells, when cocultured with CD19-CAR-T cells, secreted more CCL1, CCL5, CXCL10, IL-8, IL-5, GM-CSF, IL-13, and MIP1α/β than when tdTomator-expressing β-like cells were cocultured with CD19-CAR-T cells (Figure 4F).
Finally, we tested the effects of coculturing T cells with β-like cells on INS reporter expression by flow cytometry. We incubated β-like cells in base medium as CTL and with CTL CAR-T or anti-CD19 CAR-T cells, followed by cell sorting for CD19 or tdTomato expression (Figure 4G, top panel). Coculturing CD19-expressing β-like cells with CD19-CAR-T cells reduced the ratio of CD19-expressing cells significantly more than tdTomato-expressing β-like cells cocultured with anti-CD19 CAR-T cells or CD19-expressing β-like cells cocultured with CTL CAR-T cells (Figure 4G, bottom panel). Thus, different methods characterizing the effect of T cells on β-like cells under coculture conditions demonstrated a robust in vitro culture system to examine human T cell and human β-like cell interactions in an antigen-specific manner.
Inflammatory Response of β-like Cells to T Cell Cytokines
In T1D, direct contact between β cells and responsive CD8 T cells has been shown to result in β cell destruction by perforin and granzyme-containing cytotoxic granules.2 In addition, β cells may be affected by exposure to cytokines released from T cells during the prediabetic insulitis phase.2,3,9 To understand the transcriptional response in PSC differentiated β-like cells upon exposure to T cell cytokines, we added conditioned medium from T cells cocultured with target cells or CTL medium to pseudoislets formed by aggregating sorted INS:tdTomato-positive β-like cells (Figure 5A). We also treated primary human islets or pseudoislets formed from sorted pancreatic endocrine cells with activated T cell-conditioned medium or CTL medium and performed RNA-seq experiments after 1 day of treatment (Figure 5A). Differential gene expression analysis showed predominant upregulation of approximately 370 genes (log2 fold change > 1 and p < 10−4; Table S3), whereas about 20 genes (log2 fold change < −1 and p < 10−4; Table S3) showed downregulation (Figure 5B). Pathway analysis with GSEA showed a cohort of β cell marker genes37 that were coordinately downregulated (Figure 5H). Motif analysis of the promoter regions of approximately 260 downregulated genes (log2 fold change < −1) showed enrichment of motifs for NEUROD1 and HNF1B (Figure 5I).
Figure 5.
RNA-Seq Analysis of Transcriptional Changes of β-like Cells and Primary Islets upon Treatment with Medium Conditioned by Reactive T Cells
(A) Top panel: experimental design. Bottom panel: PCA plot showing gene expression changes of PSC-differentiated β-like cells and primary islets upon treatment with conditioned medium of CD19-expressing β-like cells and CD19-CAR-T cells. Representative results of two experiments are shown.
(B) Volcano plot of genes up- and downregulated in primary islets and PSC-differentiated β-like cells treated with CTL medium versus treated with reactive T cell-conditioned medium. Genes with a log2 fold change of more than 1 and p < 10−3 are shown in red.
(C) Heatmap and hierarchical clustering of representative genes, showing differential expression upon treatment with medium conditioned by reactive T cells.
(D and E) Selected KEGG pathways enriched in genes upregulated in human islets (D) or human PSC (hPSC)-differentiated β-like cells (E) treated with reactive T cell-conditioned medium compared with CTL medium.
(F) GSEA of upregulation of TNF-α, INFγ, and inflammation genes upon exposure to reactive T cell-conditioned medium.
(G) Transcription factor binding motifs enriched in the promoter regions of genes upregulated upon treatment with reactive T cell-conditioned medium.
(H) GSEA of downregulation of β cell marker genes upon exposure to reactive T cell-conditioned medium.
(I) Transcription factor binding motifs enriched in the promoter regions of genes downregulated upon treatment with reactive T cell-conditioned medium.
See also Table S3.
Consistent with the similar pattern in the PCA analysis upon T cell cytokine treatment (Figure 5A), genes differentially expressed in PSC-differentiated β-like cell islets and primary islets upon T cell cytokine treatment showed a high degree of correlation (Figure 5C), indicating that β-like cells responded to T cell cytokines similarly as primary islets. Analysis of differentially expressed genes suggested upregulation of inflammation response genes upon treatment with conditioned medium from activated T cells, with multiple factors in the antigen presentation process among the most upregulated. These included major histocompatibility complex (MHC) class I molecules such as HLA-B, HLA-C, HLA-E, and β2-microglobulin (Figure 5C), which were overexpressed in T1D islets compared with CTLs.38 In addition, multiple inflammation response genes (including the cytokine genes CXCL10 and CXCL11, the multiple guanylate binding proteins GBP1 and GBP4, and the IFN-regulatory transcription factors IRF1 and IRF2) were upregulated (Figure 5C). KEGG analysis placed the significantly upregulated genes in T1D pathogenesis, antigen processing and presentation, the JAK-STAT pathway, the immune checkpoint pathway, and the TNF signaling and nuclear factor κB (NF-κB) pathways (Figures 5D and 5E). Upregulated pathways in the gene set enrichment analysis (GSEA) included INFγ response and TNF-α signaling (Figure 5F). These results are consistent with the in vitro interaction and cytokine release results (Figures 4F and 4G). As expected, motif analysis of the promoter regions of upregulated genes showed enrichment of motifs of NF-κb, IRF, and STAT transcription factors (Figure 5G). Significantly upregulated genes also included PDL1, GSDMD, and CASP4 (Figure 5C) and will be discussed below.
In Vivo Modeling of Human T Cell-Mediated β Cell Destruction
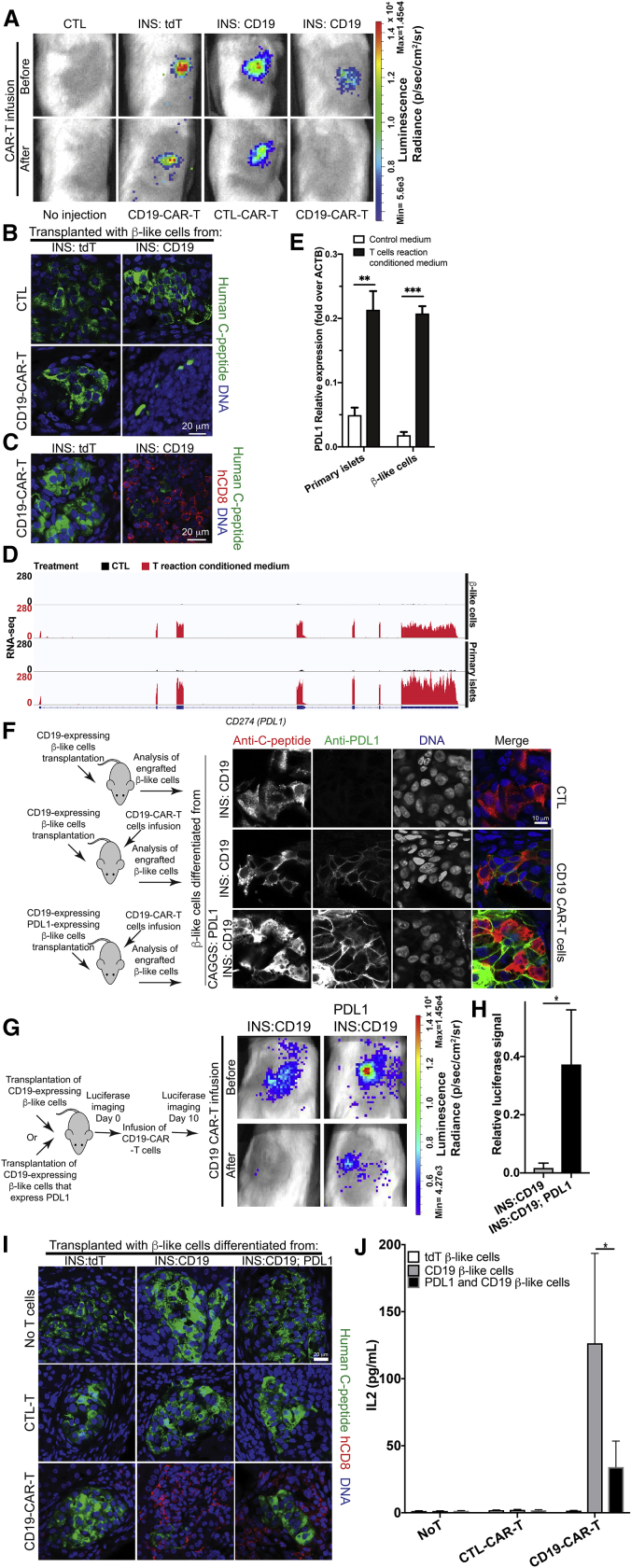
We generated an experimental platform to examine the interactions between human β-like cells and T cells in immunocompromised NSG mice. CD19- and tdTomato-expressing β-like cells were transplanted into mice with similar luciferase signals (Figure 6A). To test the interaction between CD19-CAR-T cells and CD19-expressing β-like cells, we infused human CAR-T cells into the tail vein, which resulted in a reduction of the luminescence signal in mice injected with CD19-expressing β-like engrafted cells but not in mice injected with tdTomato-expressing β-like CTL cells (Figure 6A). In contrast, infusion of CTL-CAR-T cells into mice transplanted with CD19-expressing β-like cells did not result in a luminescence reduction (Figure 6A). These data suggest that this experimental platform allows studying antigen-specific interactions of human β-like cells with human CAR-T cells in mice.
Figure 6.
Interaction between CD19-CAR-T Cells and CD19-Expressing β-like Cells in Transplanted Mice
(A) Top panel: in vivo detection of β-like INS reporter cells engrafted under the kidney capsule of immunocompromised NSG mice. Bottom panel: luciferase signal changes in mice that received a transplant upon infusion of CD19-CAR-T cells or CTL-CAR-T cells, as labeled.
(B) Representative confocal microscopy of transplanted β-like cells. Scale bar, 20 μm.
(C) Infiltration of human CD8-T cells (red) in the kidney transplanted with INS:tdT β-like CTL cells (left panel) or with INS:CD19 β-like cells (right panel). Scale bar, 20 μm.
(D) Representative RNA-seq results of the CD274 (PDL1) locus in β-like cells (top two panels) and primary islets (bottom two panels) when treated with CTL medium (black tracks) or reactive T cell-conditioned medium (red tracks).
(E) qRT-PCR analysis of PDL1 expression in primary human islets (left panels) and β-like cells (right panels) when treated with CTL medium (open bars) or reactive T cell-conditioned medium (filled bars) (n = 4).
(F) Left panel: experimental design. Right panels: representative immunofluorescence micrographs of CD19-expressing β-like cells engrafted in mice without CD19 CAR-T cell infusion (top panels) and upon CD19-CAR-T cell infusion (center panels). Bottom panels: mice engrafted with PDL1- and CD19-expressing β-like cells. Scale bar, 10 μm.
(G) Left panel: experimental design. Right panels: in vivo luciferase imaging before T cell infusion (day 0) or 10 days after T cells infusion (day 10). The results are quantified in (H) (n = 4).
(I) Representative confocal micrographs of C-peptide-expressing β-like cells (green) and infiltrating human CD8+ T cells (red). Scale bar, 20 μm.
(J) Coculture of PDL1- and CD19- expressing β-like cells with CD19-CAR-T cells showed reduced IL-2 secretion compared with CD19-expressing β-cells that do not overexpress PDL1 (n = 4).
In (H): ∗p < 0.05, Wilcoxon-Mann-Whitney test. Other: ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, non-paired t test.
See also Figures S5 and S6.
To assess whether the interactions between transplanted β-like cells and infused CAR-T cells would mimic aspects of T1D pathology, we performed immunofluorescence staining on mice transplanted with β-like cells. The morphology of INS-producing β-like cells clusters was compromised in mice transplanted with CD19-expressing β-like cells followed by CD19-CAR-T cells infusion but not in mice transplanted with tdTomato-expressing β-like cells and infused with CD19-CAR-T cells (Figure 6B). To examine infiltration of donor human T cells, we performed immunostaining with an anti-human-CD8 antibody. CD8 T cells were enriched in regions with remaining human C-peptide-expressing β-like cells in mice transplanted with CD19-expressing β-like cells and CD19-CAR-T cells (Figure 6C, right panel). In contrast, no enrichment of human CD8 T cells was observed in β-like cell clusters in mice transplanted with tdTomato-expressing β-like cells and CD19-CAR-T cells (Figure 6C, left panel). Thus, this experimental platform provides an animal model to characterize interactions of human T cells and β-like cells.
PDL1 Partially Protects β-like Cells from Cytotoxic T Cells
The results summarized in Figures 5B and 5C suggest that exposure of β-like cells and primary islets to activated T cell-conditioned medium stimulated expression of multiple immunomodulatory genes, including IDO1, BTN3A1, BTN3A2, BTN3A3, and CD47, consistent with β cells actively recruiting immune-protective mechanisms that may modify the course of T1D progression. Among the activated immunomodulatory genes, the immune checkpoint gene PDL1/CD274 was one of the most upregulated (Figures 5B and 6D). Human primary islets and PSC-differentiated β-like cells treated with activated T cell-conditioned medium showed upregulation of PDL1, as measured by qRT-PCR experiments (Figure 6E). PDL1 upregulation was confirmed in engrafted CD19-expressing β-like cells after infusion of CD19-CAR-T cells (Figure 6F), which is similar to PDL1 upregulation in β-cells observed in early-onset T1D patients.39 To test whether PDL1 expression in β-like cells would protect against destruction by CD19-CAR-T cells, we inserted a PDL1 expression cassette into the AAVS1 locus (Figure S5A). Correct insertion was verified by Southern blotting (Figure S5B), and transgene expression was confirmed by PDL1 cell surface staining (Figure S5C), and the presence of PDL1 mRNA (Figure S5D). PDL1-expression did not interfere with pluripotency, as shown by formation of teratomas (Figure S5E). When PDL1-overexpressing INS:CD19 cells were differentiated, PDL1 protein was detected in β-like cells (Figure S5F) and immunofluorescence analysis confirmed proper expression of β cell marker proteins upon in vitro differentiation or after transplantation into immunocompromised mice (Figures S5G and S5H).
INS:CD19 or PDL1-overexpressing INS:CD19 β-like cells were transplanted into immunocompromised mice, followed by CAR-T cell infusion. CD19 CAR-T cells efficiently eliminated luciferase signals from transplanted INS:CD19 β-like cells, whereas PDL1 expression conferred partial protection (Figures 6G and 6H). Histological analysis of engrafted cells showed improved morphology of β-like cell clusters and less infiltrating CD8 T cells for PDL1 CD19 β-like cells compared with the CD19 counterpart (Figure 6I). We also performed in vitro coculture of T cells and β-like cells differentiated from INS:tdT, INS:CD19, or PDL1-overexpressing INS:CD19 PSCs. Quantification of the secreted cytokines showed that PDL1 significantly reduced IL2 secretion during the in vitro coculture experiment (Figure 6J), whereas other tested cytokines also showed some reduction but not at significant levels (Figure S6). In summary, our results demonstrate that overexpression of PDL1 conferred partial protection against reactive T cells.
Induction of Caspase-4-Mediated Cleavage of GSDMD by Activated T Cell Cytokines
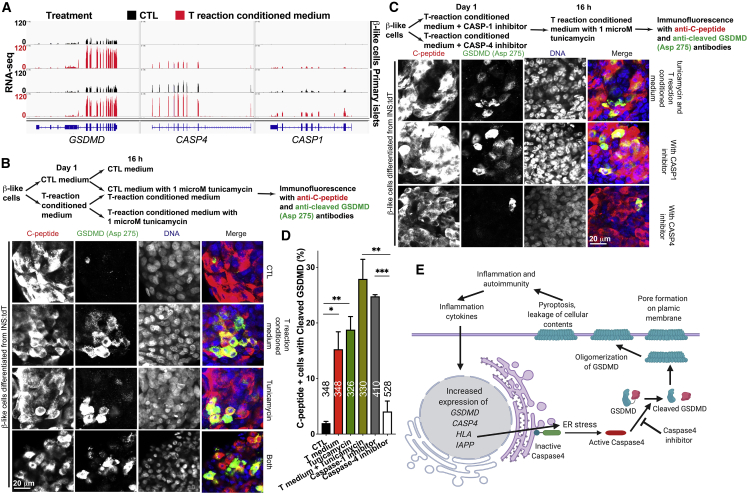
In addition to PDL1, expression of the Gasdermin family genes GSDMB, GSDMD, and GSDME was highly induced, with GSDMD showing the highest expression level (Figures 5B and 5C). Upregulation of GSDMD, a component regulating inflammatory pyroptotic cell death in macrophages,25,26 was detected upon treatment with activated T cell-conditioned medium of β-like cells and primary islets (Figure 7A, left panel). GSDMD is proteolytically cleaved by caspase-1 or caspase-4/5 in the canonical and noncanonical inflammasome activation pathways, respectively, and the cleaved amino-terminal fragment of GSDMD oligomerizes and forms pores on the plasma membrane, resulting in pyroptosis and inflammatory pathology.25,26,40 In addition to GSDMD, increased expression of CASP4 and CASP1 was detected upon reactive T cell-conditioned medium treatment in β-like cells and in human islets, with CASP4 showing higher expression levels than CASP1 (Figures 5C and 7A). To test whether cleaved GSDMD was expressed in β-like cells, we performed immunofluorescence with an antibody specific for GSDMD when cleaved at Asp275 and detected cleaved and activated GSDMD in some C-peptide-expressing β-like cells (Figures 7B–7D).
Figure 7.
Caspase-4-Mediated Cleavage of GSDMD in β-like Cells
(A) Representative RNA-seq results of GSDMD, CASP4, and CASP1 loci in β-like cells (top two panels) and primary islets (bottom two panels) when treated with CTL medium (black tracks) or with reactive T cell-conditioned medium (red tracks).
(B) Top panel: experimental design. Bottom panel: representative immunofluorescence staining with anti-C-peptide antibody (red) and anti-cleaved GSDMD (Asp275) antibody (green). Scale bar, 20 μm.
(C) Top panel: experimental design. Bottom panel: representative immunofluorescence staining with anti-C-peptide antibody (red) and anti-cleaved GSDMD (Asp275) antibody (green). Scale bar, 20 μm.
(D) Quantification of the percentage of cleaved GSDMD (Asp275)-positive cells in C-peptide-expressing cells from (B) and (C). The number of C-peptide-expressing cells from three experiments are labeled (n = 3).
(E) A diagram showing caspase-4-mediated, ER stress-regulated cleavage of GSDMD during β cell inflammation. Inflammation leads to upregulation of GSDMD and its activator CASP4. Furthermore, concomitant upregulation of MHC class I and IAPP contributes to ER stress, activatinges caspase-4, which cleaves and activates GSDMD. The amino-terminal fragment of GSDMD could form membrane-destructing oligomers, which results in autoimmunity, propelling pyroptosis.
∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, unpaired t test.
See also Figure S7.
We hypothesized that inflammation-mediated endoplasmic reticulum (ER) stress leads to caspase-4 activation,41 resulting in GSDMD cleavage. To test this hypothesis, we treated β-like cells with activated T cell-conditioned medium for 24 h, followed by treatment with the ER stress compound tunicamycin. Compared with the CTL, tunicamycin treatment resulted in a significant increase in cleaved GSDMD in β-like cells (Figures 7B and 7D). Furthermore, treatment with activated T cell-conditioned medium and tunicamycin resulted in an increased proportion of C-peptide-expressing cells with cleaved GSDMD (Figures 7B and 7D), demonstrating that inflammation and ER stress regulate GSDMD cleavage in β-like cells. Also, immunofluorescence staining revealed increased levels of cleaved GSDMD in pancreatic islets of NRG-Akita mice compared with NRG littermate CTLs (Figure S7). To test the contribution of caspase-4 and caspase-1, we treated the cells with the caspase-1 inhibitor VX-765 or the caspase-4 inhibitor Z-LEVD-fmk and observed that the caspase-4 inhibitor more effectively reduced the proportion of β-like cells positive for activated GSDMD than the caspase-1 inhibitor (Figures 7C and 7D), suggesting that ER stress activation of caspase-4 rather than CAPS1 activation mediates activation of GSDMD. Our results suggest the presence of caspase-4 mediated cleavage of GSDMD in β-like cells under conditions of inflammation and ER stress (Figure 7E).
Discussion
Here we established an experimental platform to study the interaction of activated human T cells and human pancreatic β-like cells. We generated human PSCs carrying a β-cell-specific CD19-expression cassette. CD19-expressing β-like cells displayed β cell signature genes and, upon transplanted into diabetic NRG Akita mice, rescued the elevated glucose levels. When cocultured with human CD19 CAR-T cells, CD19-expressing β-like cells elicited a strong T cell response, resulting in secretion of immune-stimulatory cytokines. Also, GSDMD, a component regulating inflammatory pyroptotic cell death,25,26 was upregulated. Similarly, human CD19 CAR-T cells, when infused into humanized mice transplanted with β-like cells, infiltrated the transplants and caused their destruction. In addition to inflammatory cytokines, PDL1 was induced in β-like cells exposed to activated T cell-conditioned medium, and PDL1 overexpression in transplanted CD19-positive β-like cells partially protected against CD19 CAR-T cells.
Gene expression analysis of β-like cells and primary islets upon treatment with activated T cell-conditioned medium revealed upregulation of genes involved in the immune response. Upregulated genes included the immunomodulatory molecules PDL1 and CD47. This is consistent with a recent study where human primary islets or a β cell line were treated with INFγ and IL1β.42 Thus, the model exhibited features of human islet inflammation and identified a caspase-4-mediated inflammation- and ER stress-promoted cleavage and activation of pyroptosis mediator, GSDMD, as a potential additional immune response mechanism of β-like cells. CASP4 transcription was also upregulated, corroborating previous results.41 Our data suggest a model of caspase-4-mediated cleavage and activation of GSDMD via inflammation→ ER stress → caspase-4 activation → GSDMD activation (Figure 7E).
The role of pyroptosis in non-immune cells, such as terminally differentiated β cells, in diabetes remains to be defined. Upregulation of cleaved GSDMD in β cells suggests that pyroptosis might be a mechanism mediating β cell death during the initial phase of T1D. We postulate that, under inflammation conditions, β cells may undergo inflammatory pyroptosis, leading to leakage of cellular components including insulin granules, which could be presented to immune cells, which further propel the inflammation process of autoimmunity against β cells. The function of β cells to produce insulin and islet amyloid polypeptide (IAPP), exacerbated by ER stress, which can be caused by obligatory ER-mediated antigen presentation, could lead to activation of caspase-4 and activation of GSDMD, enhancing inflammatory cell death and accelerating autoimmunity.
Substantial progress in cancer treatment is based on immune checkpoint blockade therapies using antibodies against CTLA4- or PD1/PDL1.43,44 However, PDL1 expression is not restricted to cancer cells, and mice with defects in PD1/PDL1 pathways develop autoimmune diseases.45,46 Anti-PDL1 therapy in patients has been reported to result in autoimmune diabetes,47 and genetic variants in PDL1 have been linked to diabetes.48 A study of patient islets showed upregulation of PDL1 in T1D patients but not in non-T1D individuals.39 These results are consistent with PDL1 being upregulated in β-like cells when treated with activated T cell-conditioned medium or cocultured with cognitive T cells (Figure 6). Because PDL1 overexpression did not interfere with human β-like cell differentiation and function, PDL1 induction in β-cells may have therapeutic effects by reducing β-cell-targeted autoimmunity.
Studying pancreatic islets from patients at the onset of T1D would be valuable to provide insights into the initiating events of the chronic disorder if not for the dangerous consequences associated with biopsy procedures.49 By combining genome editing in human PSCs and CAR-T cells, this study provides a tractable platform to study human β-like cell/T cell interactions in vitro and in vivo. Complementing other systems to investigate interactions of β cells and immune cells (Table S4), the experimental platform established here is an initial step to understand T1D mechanisms and may provide insights important for potential therapies.
Limitations of Study
Several model systems have been established for studying β cell/T cell interactions (Table S4). Although using CAR-T cells allows a robust immune response in a timely manner that is conducive for experimentation, whether β cell responses are similar in our model and in patients remains to be carefully evaluated. We detected increased levels of cleaved GSDMD in islets of NRG-Akita mice carrying an ER stress-causing mutation in the Ins2 gene (Figure S7), but we did not test whether cleaved GSDMD could be detected in islets of T1D patients. Additionally, future studies are needed to examine whether pyroptosis contributes to T1D progression in patients and whether inhibiting caspase-4 could be a safe and effective therapeutic strategy for patients.
STAR★Methods
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Alexa Fluor 647 conjugated HPi2 antibody | Novus Biologicals | NBP1-18946AF647; RRID:AB_2868415 |
| DyLight 488 conjugated HPx1 antibody | Novus Biologicals | NBP1-18951G; RRID:AB_1723850 |
| PE conjugated anti-CD19 antibody | Affymetrix | 12-0199-42; RRID:AB_1834376 |
| Anti-human C-peptide antibody | DBHB | GN-ID4; RRID:AB_2255626 |
| Anti-NKX6-1 antibody | DBHB | F55A12; RRID:AB_532379 |
| Anti-PDX1 antibody | R & D Systems | AF2419; RRID:AB_355257 |
| Anti-glucagon antibody | Cell signaling | D16G10; RRID:AB_10859908 |
| Anti-RFP antibody | Rockland | 600-401-379; RRID:AB_2209751 |
| Anti-CD19 antibody | Abcam | 134114; RRID:AB_2801636 |
| Anti-PDL1 antibody | Abcam | 205921; RRID:AB_2687878 |
| PE conjugated anti-human-PDL1 antibody | BD biosciences | BD561787; RRID:AB_2868414 |
| Anti-human-CD8 antibody | Thermo Fisher Scientific | RM-9116-S0; RRID:AB_149958 |
| FITC conjugated anti-human CD8 antibody (clone HIT8A) | Thermo Fisher Scientific | BDB555634; RRID:AB_2868416 |
| PE conjugated anti-human CD4 antibody (clone RPA-T4) | Thermo Fisher Scientific | BDB555347; RRID:AB_2868417 |
| Anti-LNGFR antibody | BD biosciences | 557194; RRID:AB_2152663 |
| Anti-insulin antibody | Thermo Fisher | PA1-26938; RRID:AB_794668 |
| Anti-cleaved GSDMD antibody | Cell signaling | 36425; RRID:AB_2799099 |
| Alexa Fluor 647 conjugated goat guinea pig IgG secondary antibody | Life Technologies | A21450; RRID:AB_141882 |
| Alexa Fluor 647 conjugated goat anti-rat IgG secondary antibody | Life Technologies | A21247; RRID:AB_141778 |
| Alexa Fluor 594 conjugated donkey anti-rat IgG secondary antibody | Life Technologies | A21209; RRID:AB_2535795 |
| Alexa Fluor 594 conjugated donkey anti-mouse IgG secondary antibody | Life Technologies | A21203; RRID:AB_141633 |
| Alexa Fluor 647 conjugated donkey anti-mouse IgG secondary antibody | Life Technologies | A31571; RRID:AB_162542 |
| Alexa Fluor 488 conjugated donkey anti-goat IgG secondary antibody | Life Technologies | A11055; RRID:AB_2534102 |
| Alexa Fluor 594 conjugated donkey anti-rabbit IgG secondary antibody | Life Technologies | A21207; RRID:AB_141637 |
| Alexa Fluor 488 conjugated donkey anti-rabbit IgG secondary antibody | Life Technologies | A21206; RRID:AB_2535792 |
| DPAI | Life Technologies | D3571 |
| Bacterial and Virus Strains | ||
| Stable 3 Chemically Competent E. coli | Invitrogen | C737303 |
| Anti-CD19-28z-2A-LNGFR retrovirus | Eyquem et al.50 | N/A |
| 28z-2A-LNGFR retrovirus | This study | N/A |
| Biological Samples | ||
| Human primary islets | Prodo labs | HP-19226-01 |
| Human buffy coat | Research blood components | 002 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| DEME-F12 medium | Life Technologies | 11330-057 |
| KSR | Lift Technologies | 10828-028 |
| FGF | Lift Technologies | PHG0261 |
| 2-Mercaptoethanol | Lift Technologies | 21985-023 |
| 100x L-glutamine | Lift Technologies | 25030-081 |
| 100x glutamax | Lift Technologies | 35050-061 |
| 100X MEM-NEAA | Lift Technologies | 11140-050 |
| 100X penicillin/streptomycin | Lift Technologies | 15140-122 |
| 100X Insulin-transferrin-selenium-ethanolamine | Lift Technologies | 51500-056 |
| Collagenase IV | Lift Technologies | 17104019 |
| mTeSR1 medium | STEMCELL Technologies | mTeSR1 |
| Accutase | STEMCELL Technologies | 07920 |
| MCDB131 medium | Life Technologies | 10372019 |
| Y-27632 | Thermo Fisher Scientific | 688000-100MG |
| Puromycin | Sigma Aldrich | P7255-100MG |
| CHIR99021 | Cayman Chemical | 252917-06-9 |
| Matrigel | Thermo Fisher Scientific | CB40234 |
| Versene | Life Technologies | 15040066 |
| Activin A | R & D Systems | 338-AC-050 |
| KGF | PeproTech | 100-19-250UG |
| Vitamin C | Sigma Aldrich | A4544-25G |
| Retinoic acid | Sigma Aldrich | R2625-100MG |
| SANT-1 | Sigma Aldrich | S4572-5MG |
| LDN193189 | Sigma Aldrich | SML0559-5MG |
| Heparin | Sigma Aldrich | H3149 |
| TPB | Tocris | 5343 |
| EGF | PeproTech | AF-100-15 |
| T3 | EMD | 64245 |
| ALK5 inhibitor II | Axxora | ALX-270-445-M005 |
| Gamma secretase inhibitor XX | VWR | 82602-300 |
| Trace Elements A | Corning | 25-021-CI |
| Trace Elements B | Corning | 25-022-CI |
| Fatty acid free BSA | Thermo Fisher Scientific | 50412870 |
| D-Luciferin potassium salt | Perkin Elmer | 122799 |
| RPMI 1640 | GIBCO | 31800-089 |
| Human serum A/B | Gemini Bio | 100512 |
| Phytohaemagglutinin | Remel | R30852801 |
| IL2 | Miltenyi Biotec | 130-097-745 |
| Ficoll-Paque Plus | GE healthcare | 17-1440-02 |
| Retronectin | Takara Bio | T100A |
| Tunicamycin | Sigma Aldrich | T7765 |
| VX-765 | Thermo Fisher Scientific | 508389 |
| Z-LEVD-FMK | VWR | 10187-344 |
| Critical Commercial Assays | ||
| CytoTox 96 Non-Radioactive Cytotoxicity Assay | Promega | G1780 |
| Pan-T cells purification kit | Miltenyi Biotec | 130-096-535 |
| Luminex assay (custom kit) | R&D Systems | tHB2l7Hu |
| Human cytokine array | R&D Systems | ARY005B |
| Accu-check Aviva Plus glucometer test kit | Roche | GTIN: 00365702438101 |
| Ultrasensitive Insulin ELISA kit | Mercordia | 10-1132-01 |
| RNeasy Plus Micro Kit | QIAGEN | 74034 |
| SuperScript III First-Strand Synthesis SuperMix | Life Technologies | 18080400 |
| Fast SYBR Green Master Mix | Life Technologies | 4385618 |
| SMART-Seq v4 Ultra Low Input RNA Kit | Takara Bio | R400752 |
| Nextera XT Library Preparation kit | Illumina | FC-131-1024 |
| Prime-It II Random Primer Labeling Kit | Agilent | 300385 |
| AmpliTaq Gold 360 Master Mix | Life Technologies | 4398881 |
| Deposited Data | ||
| RNA-seq data | This paper | GSE155713 |
| Experimental Models: Cell Lines | ||
| H1 human pluripotent stem cells (Thomson et al.28) | WiCell Research Institute | NIHhESC-10-0043 |
| H1-INS:tdT | This study | N/A |
| H1-INS:CD19 | This study | N/A |
| H1-INS:CD19; AAVS1:PDL1 | This study | N/A |
| H29 cells (Stephan et al.51) | A gift provided by Prof. Sadelain (MSKCC). | N/A |
| PG13 cells (Miller et al.52) | A gift provided by Prof. Sadelain (MSKCC). | N/A |
| HEK293T cells | ATCC | CRL-11268 |
| Experimental Models: Organisms/Strains | ||
| NOD.Cg-PrkdcscidIl2rgtm1Wjl/SzJ (NSG) mice | Jackson Laboratory | 005557 |
| NOD.Cg-Rag1tm1Mom Ins2Akita Il2rgtm1Wjl/SzJ (NRG-Akita) mice | Jackson Laboratory | 014568 |
| NOD.Cg-Rag1tm1Mom Il2rgtm1Wjl/SzJ (NRG) mice | Jackson Laboratory | 007799 |
| Recombinant DNA | ||
| pY010 (Zetsche et al.27) | Addgene | Addgene 69982 |
| pX458 (Ran et al.53) | Addgene | Addgene 48138 |
| INS-Tdtm (Liu et al.54) | A gift provided by Prof. Deng (Peking University). | N/A |
| INS-2A-luciferase-2A-tdT | This study | Addgene 159348 |
| INS-2A-luciferase-2A-CD19-GFP | This study | Addgene 159349 |
| CAGGS-AsCpf1-2A-GFP-U6-sgRNA-cloning vector | This study | Addgene 159281 |
| CAGGS-AsCpf1-U6-INS-sgRNA | This study | Addgene 159283 |
| AAVS1-PDL1 | This study | Addgene 159280 |
| AAVS1-tdTomato (Ma et al.55) | Addgene | Addgene 159275 |
| SFG-1928z-P2A-LNGFR (Zhao et al.56) | A gift provided by Prof. Sadelain (MSKCC). | N/A |
| SFG-28z-P2A-LNGFR | This study | N/A |
| CAGGS: Cre-2A-tdTomato | This study | N/A |
| Software and Algorithms | ||
| Prism (Version 8) | Graphpad Prism | https://www.graphpad.com/scientific-software/prism/ |
| Living Image (Version 4.5.4) | Perkin Elmer | https://www.perkinelmer.com/lab-products-and-services/resources/in-vivo-imaging-software-downloads.html |
| Zeiss ZEN software | Carl Zeiss | https://www.zeiss.com/microscopy/us/products/microscope-software/zen.html#downloads |
| STAR (Version 2.7.1a; Dobin et al.57) | Github | https://github.com/alexdobin/STAR |
| featureCounts (Version 1.6.2; Liao et al.58) | SourceForge | http://subread.sourceforge.net/ |
| DEBrowser (Kucukural et al.59) | Bioconductor | https://www.bioconductor.org/packages/release/bioc/html/debrowser.html |
| DEseq2 (Love et al.60) | Bioconductor | https://bioconductor.org/packages/release/bioc/html/DESeq2.html |
| Homer (Version 4.11; Heinz et al.61) | http://homer.ucsd.edu/homer/ | N/A |
| R (Version 3.5.2) | R project | https://www.r-project.org/ |
| Adobe Illustrator (CC) | Adobe | https://www.adobe.com/products/illustrator.html |
| GSEA (Version 4.0.1; Subramanian et al.62) | https://www.gsea-msigdb.org/gsea/index.jsp | N/A |
| FlowJo (Version 10.5.3) | FlowJo | https://www.flowjo.com/solutions/flowjo/ |
| Other | ||
| AggreWell 400 plates | Stem Cell Technology | 34415 |
| Ultra-Low Attachment Multiple Well Plates (flat bottom, 6-well) | Corning | 3471 |
| Ultra-Low Attachment Multiple Well Plates (round bottom, 96-well) | Sigma Aldrich | CLS7007 |
Resource Availability
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Rudolf Jaenisch (jaenisch@wi.mit.edu).
Materials Availability
Plasmids generated in this study have been deposited to Addgene (INS-2A-luciferase-2A-tdT: 159348, INS-2A-luciferase-2A-CD19-GFP: 159349, CAGGS-AsCpf1-2A-GFP-U6-sgRNA-cloning vector: 159281, CAGGS-AsCpf1-U6-INS-sgRNA: 159283, AAVS1-PDL1: 159280). Cell lines generated in this study are available from the Lead Contact with a completed materials transfer agreement.
Data and Code Availability
The accession number for the RNA-seq data reported in this paper is GEO: GSE155713.
Experimental Model and Subject Details
Primary human islets
Primary human islets from a healthy donor were obtained from Prodo Laboratories (HP-19226-01). After recovery by culturing in PIM complete medium (Prodo Laboratories) in 6-well ultra-low attachment plates (Corning 3471) overnight, islet samples were washed with PBS-, incubated with 1:1 PBS diluted Accutase in 37°C for 2 min, and at room temperature for 5 min, with gentle agitation. Then 6 mL PBS with 1% BSA was added, and islets clusters were gentle dissociated with a 1 mL pipette. After centrifugation, cells were resuspended in PBS with 1% BSA, and stained with 1:100 diluted A647-conjugated HPi2 antibody (Novus Biologicals NBP1-18946AF647) and DyLight 488 conjugated HPx1 Antibody (Novus Biologicals NBP1-18951G), for 30 min at 4°C. After washing, HPi2 positive and HPx1 negative endocrine cells were sorted with a FACSAria (BD Biosciences) cell sorter, and about 2000 cells were plated in 0.1 mL PIM medium supplemented with 10 μM Rho kinase (ROCK) inhibitor Y-27632 to each well of 96-well round bottom ultra-low attachment plates (Sigma Aldrich, CLS7007) to form aggregated pseudoislets.
Human PBMC isolation, T cell enrichment, and preparation of CAR-T cells
PBMCs were isolated from buffy coat samples (Research blood components, item number 002) from healthy volunteers with Ficoll-paque plus (GE Healthcare) centrifugation. T cells were enriched from PBMC with Pan-T cells purification kit (Miltenyi 130-096-535) and Miltenyi LS MACS columns. T cells were stimulated with R-10 medium (RPMI medium supplemented with 10% human serum A/B (Gemini Bio # 100512), 1x penicillin/streptomycin, 1x glutamax, 1x MEM-NEAA) supplemented with 50 unit/mL IL2 (Miltenyi 130-097-745), and PHA (5 μg/mL) for 2 days, before spin transduction with 0.45 μm filter filtered viral supernatant from PG13 viral production cells51 (a gift generously provided by Prof. Sadelain, MSKCC) in Retronectin (Takara Bio T100A) coated non-tissue culture treated plates for two days. Then cells were passaged for 3-4 days before analysis of CAR-expression and infusion to animals. For examining CAR expressing, mouse anti-LNGFR antibody (BD 557194) was used to incubate with T cells for 10 min at 4°C (1:200) dilution. After washing, Alexa Fluor-647 conjugated anti-mouse IgG secondary antibody (1:400, 10 min incubation at 4°C) was used to detect primary antibody. After washing, the samples were analyzed with a BD LSRFortessa analyzer, and results were analyzed with FlowJo software (Version 10.5.3).
Human PSC culture
For gene targeting, H1 human pluripotent stem cells28 were maintained in DMEM-F12 (Life Technologies cat# 11330-057) supplemented with 20% KSR (Life Technologies cat# 10828-028), 4 ng/mL FGF (Life Technologies cat# PHG0261), 0.1 mM 2-mercaptoethanol (Life Technologies cat #21985-023), 1x L-glutamine (Life Technologies cat# 25030-081), 1x MEM-NEAA (Life Technologies cat# 11140-050), 1x penicillin/streptomycin (Life Technologies cat# 15140-122). Cells were passaged with 1 mg/mL collagenase IV (Life Technologies cat# 17104019) every 4-6 days on mitomycin C inactivated MEF feeders cultured in DMEM medium supplemented with 10% FBS, 1x MEM-NEAA, 1x penicillin/streptomycin, and 1x L-glutamine. For differentiation to β-like cells, H1 cells and targeted H1 cells were adapted for feeder-free conditions by culturing in Matrigel (Corning) coated tissue culture plates in mTeSR (Stem Cell Technology). For coating plates, Matrigel (Corning) was 1:100 diluted in cold DMEM-F12 medium, and 2 mL of diluted Matrigel solution was added to one well of a 6-well plate well, and coat for overnight at room temperature before pre-warming in a 37°C incubator for 1 hour before using. Cells were cultured in mTeSR medium in 6-well plates and passaged very 3-4 days with a 1:4-6 passage ratio.
Mice
Immunodeficient NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ (NSG) mice were obtained from the Jackson Laboratory and housed in autoclaved cages with autoclaved food and water in environmentally controlled rooms at the Whitehead Institute animal facility. NOD.Cg-Rag1tm1Mom Ins2Akita Il2rgtm1Wjl/SzJ (NRG-Akita) mice were purchased from Jackson Laboratory, and housed under conditions similar to NSG mice except high absorbance bedding was used. NRG-Akita mice were maintained by crossing male NRG-Akita with female NOD.Cg-Rag1tm1Mom Il2rgtm1Wjl/SzJ (NRG) mice. For genotyping NRT-Akita mice, DNA were extracted from mouse tail tissues with proteinase K digestion and isopropanol precipitation. A total of 0.5-1 μg of DNA was used in PCR reactions with the primers (5′- TGCTGATGCCCTGGCCTGCT-3′, and 5′-TGGTCCCACATATGCACATG) with 57°C annealing and 68°C extension for 30 s, and amplify for 35 cycles with AmpliTaq Gold DNA Polymerase (Life Technology). PCR products were purified and sequenced with 5′- TGCTGATGCCCTGGCCTGCT-3′. Animals showing the following sequence were identified as NRG-Akita mice: GGGAGCAGATGCTGGTG(C/T)AGCACTGATCTACAATG. Both male and female mice (6-8 weeks) were used for β-like cell clusters transplantation experiments. Both male and female mice (8-16 weeks) were used for teratoma assays. All mice experiments were in accordance with the protocols approved by the Animal Research Regulation Committee at the Whitehead Institute and guidelines from the Department of Comparative Medicine at Massachusetts Institute of Technology.
Method Details
Molecular cloning
To generate a multicistronic plasmid with CAGGS:As-Cpf1-2A-GFP and U6:sgRNA cassettes,
PCR products using pcDNA3.1-hAsCpf1 (pY010 Addgene 6998227) as the template and the oligoes (5′-TAACCGGTCCACCATGGCCCCAAAGAAGAAGCGGAAG-3′ and 5′-GCCTTAATTAATCAGGCATAGTCGGGGACATCATATGGGTATG-3′) as primers were cloned between the AgeI and PacI sites of the AAVS1-tdTomato targeting construct,55 generating the AAVS1-AsCpf1 targeting construct (#66). The annealed oligos (5′-CACCGTAATTTCTACTCTTGTAGATGTCTTCGAGAAGACTTTTTTTT-3′ and
5′-AAACAAAAAAAAGTCTTCTCGAAGACATCTACAAGAGTAGAAATTAC-3′) were cloned into pX458 (Addgene 4813853) between the BbsI sites. Then the resulted plasmid was used as a temple to generate PCR products with the following primers (5′-GACGAGATCCTCGCCGTCGGCATGCCTAGAGCCATTTGTCTGCAGAATTG-3′ and
5′-ACGTGCCTGATTATGCATACCCATATGATGTCCCCGACTATGCCGGCAGTGGAGAGGGCA-3′). The purified PCR products were ligated to the 6.6 kb SphI and NsiI fragment from #66 by Gibson assembly,63 generating CAGGS-AsCpf1-2A-GFP-U6-sgRNA-cloning vector containing the CAGGS-AsCpf1-2A-GFP and U6:sgRNA cassettes. For Cpf1 sgRNAs targeting the INS loci (CAGGS-AsCpf1-U6-INS-sgRNA), annealed products from the following oligos, were cloned between the BbsI sites of CAGGS-AsCpf1-2A-GFP-U6-sgRNA-cloning vector based on previous cloning methods64:
INS: 5′-GTAGATTTCCATCTCTCTCGGTGCAGGAG-3′ and 5′- AAAACTCCTGCACCGAGAGAGATGGAAAT-3′.
Generation of INS targeting constructs:
The puromycin resistance gene was cloned into the AAVS1-tdTomator plasmid55 under CAGGS promoter control between the AgeI and PacI site using the following primers: 5′-TAACCGGTCCACCATGACCGAGTACAAGCCCAC-3′, and 5′-GCCTTAATTAATCAGGCACCGGGCTTGCGG-3′ to generate an intermediate vector CAGGS:puro. Then the PCR product from CAGGS:puro as a template and the primers (5′-GGAGGGGCGCGGCGGCCCCCGGAGC-3′, and 5′-CGCTATGTCCTGATAGCGTCAGGCACCGGGCTTGCGGG-3′) were ligated to the ~17.8 kb RsrII-SgrAI fragment from INS-Tdtm targeting construct54 by Gibson assembly, giving rise to INS-tdTomato-puro (#82). The firefly luciferase-T2A-CD19-T2A-GFP cassette was cloned between the AgeI and PacI sites of AAVS1-tdTomato targeting construct,55 generating the AAVS1-Luciferase-T2A-CD19-t2A-GFP targeting construct (#95). The PCR product was generated with #95 as template with the following primers: 5′- TACTTCGAAATGTCCGTTCGGTTGGCA-3′, and 5′-AAGTTAACAACAACAATTGTTAATTAATTACTTGTACAGCTCGTCC-3′. The purified PCR products, the KpnI and MfeI fragment from #82, and the geneblock (synthesized by IDT:
CTGGAGAACTACTGCAACGGTACCTACCCATACGATGTTCCAGATTACGCTGGCAGTGGAGAGGGCAGAGGAAGTCTGCT
AACATGCGGTGACGTCGAGGAGAATCCTGGCCCAATGGAAGACGCCAAAAACATAAAGAAAGGCCCGGCGCCATTCTATCCGCTGGAAGATGGAACCGCTGGAGAGCAACTGCATAAGGCTATGAAGAGATACGCCCTGGTTCCTGGAACAATTGCTTTTACAGATGCACATATCGAGGTGGACATCACTTACGCTGAGTACTTCGAAATGTCCGTTCGGTTGGCA) were ligated by Gibson assembly, giving rise to INS-2A-luciferase-2A-CD19-GFP.
A luciferase-tdTomato-t2A cassette was generated by cloning the PCR product (with AAVS-tdTomato55 construct as template and the following primers 5′-ACGATGCATATGGTGAGCAAGGGCGAGGAGGTCATCAAAG-3′, and 5′-ACGCGGCCGCTTACTTGTACAGCTCGTCCATGCCGTACAGG-3′) to #95 between NsiI and NotI site, giving rise to an intermediate plasmid #98. Then the PCR product was generated with #98 as template with the following primers: 5′- TACTTCGAAATGTCCGTTCGGTTGGCA-3′, and 5′-AAGTTAACAACAACAATTGTTAATTAATTACTTGTACAGCTCGTCC-3′. The purified PCR products, the KpnI and MfeI fragment from #82, and the geneblock (synthesized by IDT:
CTGGAGAACTACTGCAACGGTACCTACCCATACGATGTTCCAGATTACGCTGGCAGTGGAGAGGGCAGAGGAAGTCTGCT
AACATGCGGTGACGTCGAGGAGAATCCTGGCCCAATGGAAGACGCCAAAAACATAAAGAAAGGCCCGGCGCCATTCTATCCGCTGGAAGATGGAACCGCTGGAGAGCAACTGCATAAGGCTATGAAGAGATACGCCCTGGTTCCTGGAACAATTGCTTTTACAGATGCACATATCGAGGTGGACATCACTTACGCTGAGTACTTCGAAATGTCCGTTCGGTTGGCA) were ligated by Gibson assembly, giving rise to INS-2A-luciferase-2A-tdT.
To generate a control CAR construct SFG-28z-P2A-LNGFR that lacks the CD19-binding domain, the ~7.9 kb AfeI-MfeI fragment from SFG-1928z-P2A-LNGFR (a gift provided by Prof. Sadelain from MSKCC) was purified and ligated to the annealed products of primers 5′-GCTTCTCCTGCATGCACGGGCGGCCGC-3′ and 5′-AATTGCGGCCGCCCGTGCATGCAGGAGAAGC-3′.
PCR products generated from the human PDL1 coding sequence as template with the primers 5′- TAACCGGTCCAACCATGAGGATATTTGCTGTCTTTATATTC-3′ and 5′- GCCTTAATTAATTACGTCTCCTCCAAATGTGTATCA
−3′ were cloned between AgeI and PacI sites of AAVS1-tdTomato targeting construct,55 giving rise to AAVS1-PDL1.
T7EI assay
Indel formation was tested in HEK293T cells by the T7EI assay. HEK293T cells were cultured in DMEM high glucose medium (GIBCO) supplemented with 10% FBS, 1X L-glutamine, 1X MEM-NEAA, and 1X penicillin/streptomycin (Life Technologies) in 37°C incubators under 5% CO2. Cells cultured in 12-well plates to 60%–80% confluence were transfected with 0.5 μg CAGGS-AsCpf1-U6-INS-sgRNA plasmids using FuGENE 6 transfection reagent (Promega) based on the manufacturer’s guideline. Two to three days post transfection, cells were dissociated with trypsin, and GFP positive cells were sorted on a FACSAria (BD Biosciences), followed by DNA purification with DNeasy Blood & Tissue Kit (QIAGEN). For T7EI assay, PCR reactions were performed with 0.5 μg genomic DNA as template, and PCR with AmpliTaq Gold DNA Polymerase (Life technology) with the following primers:
INS: 5′-TGGGGCAGGTGGAGCTGGGCGG-3′, and 5′-ACCACCCCTGGCCCCTCAGAGACC-3′.
A total of 200 ng PCR products were annealed in NEB buffer 2 and processed with 5 units of T7EI (NEB) for 1 hour at 37°C before resolved on 10% TBE PAGE gels (BioRad) run at 100 V for about 2 hours. The PAGE gels were stained with ethidium bromide, and images were taken with an AlphaImager gel documentation system (Alpha Innotech). Mutation efficiency were measured as previously reported.64
CRISPR-mediated genome editing in human PSCs
Targeting of the H1 pluripotent cell line was performed using modified protocols based on established methods.65 One day before electroporation, cells were cultured in medium supplemented with 10 μM Rho kinase (ROCK) inhibitor Y-27632 (Thermo Fisher Scientific). About 10 million PSCs were dissociated into single cells with Accutase (StemCell Technologies, Inc.), filtered with 40 μm cell strainers (Corning), washed with medium containing Y-27632, spun down, and resuspended in 800 μl of PBS with 100 μg CAGGS-AsCpf1-U6-INS-sgRNA, and 40 μg donor construct (INS-2A-luciferase-2A-tdT or INS-2A-luciferase-2A-CD19-GFP). The mixture was loaded into a 0.4-cm cuvette (BioRad), incubated on ice for 5 minutes before electroporation with a Gene Pulser Xcell System (Bio-Rad) with 1 pulse of 250 V, 500 μF. Then the cuvette was incubated on ice for 5 minutes before plating cells on 2 6-well plates of DR4 MEF.66 Puromycin containing medium (0.5 μg/mL) was added 3-4 days after electroporation. Individual colonies were manually passaged onto MEF coated 12-well plate wells, expanded and genotyped with PCR.
About 0.3-1 μg genomic DNA was used for each genotyping reaction with Kapa HiFi 2X ready mix. For identifying clones with potential knockin at the INS locus, the primers 5′-GGCCTTTGGTGCAGTGACCAGAGTGTCAGG-3′, and 5′-GATTCTCCTCGACGTCACCGCATGTTAGC-3′ were used with the PCR condition: 95°C 3 min; 40 cycles of 98°C 20 s, 58°C 30 s, 72°C 5.5 min; 72°C for 10 min. Amplicons of 5 kb were purified with gel purification kit (Omega Biotech) and sequenced with the primer: 5′-GGCCTTTGGTGCAGTGACCAGAGTGTCAGG-3′. Clones that yielded amplicons with correct sequences were subjected to Southern blotting analysis. In clones with correctly targeted INS locus, the untargeted INS allele was amplified with primers 5′-TGGGGCAGGTGGAGCTGGGCGG-3′, and 5′-ACCACCCCTGGCCCCTCAGAGACC-3′ and sequenced to verify that the allele without knockin was not altered.
Generation of PD1L1 expressing cells
To remove the floxed puromycin-resistance cassette from heterozygous INS knockin cells, the cells were electroporated with a CAGGS: Cre-2A-tdTomato plasmid with the electroporation condition described above. Then tdTomato-expressing cells were sorted 1-2 days post electroporation and clonally expanded. Clones that had lost puromycin-resistance were used for PDL1 knockin at the AAVS1 locus with a similar procedure as described above. Expression of PDL1 was visualized with 1:5 diluted PE-conjugated anti-CD274 (BD561787) antibody according to manufacturer’s guideline.
Southern blot
For INS targeted cells, 15 μg of genomic DNA was digested with SpeI and HindIII-HF followed by agarose gel electrophoresis and transfer to Hybond-XL membrane (GE Healthcare Life Sciences RPN203S). Then the membrane was hybridized with 5′ and 3′ DNA probes randomly primed with 32P- dCTP (50 μCi). For AAVS1 locus targeted cells, Southern blots were carried out by digesting 15 μg of genomic DNA with SphI, followed by hybridization with random primed 5′ and 3′ DNA probes with 32P- dCTP (50 μCi).
Probes for Southern blot
AAVS1 internal probe was generated by PCR using AAVS1-tdTomato plasmid as template and the following primers: 5′-GAATTCGCCCTTTGCTTTCTCTGAC-3′, and 5′- TGAGCTCTCGGACCCCTGGAAGAT-3′. AAVS1 external probe was generated by PCR with H1 genomic DNA and the following primer: 5′-ACAGGTACCATGTGGGGTTC-3′, and 5′- CCCTTGCCTCACCTGGCGAT-3′. A fragment from firefly luciferase coding sequence was amplified with the following primers: forward (5′-ATGGAAGACGCCAAAAACATAAAGAAAGGCCC-3′, and reverse: 5′-CACGGCGATCTTTCCGCCCTT-3′), and was used as 5′ internal probe for the INS locus. H1 ES cells genomic DNA was used to generate INS 3′ external probe by PCR with the following primers: 5′-GACTCCCCACTTCCTGCCCATCT-3′, and 5′-TCTTCTCCCAGCCCCGTCCTCAC-3′.
Teratoma assay
Teratoma assays of targeted PSCs cells were performed as before55 with slight modifications. Briefly, PSCs harvested from two near confluent six-well plates were dissociated with collagenase, washed, and resuspended in DMEM without serum. The suspension (about 100–200 μL) was injected subcutaneously or intramuscularly into NSG mice between 8-16 weeks of age using 1-mL syringes with 23-gauge needles. Formation of teratomas was monitored twice-weekly for about 2 months. After palpable tumors reached between 7 and 10 mm in diameter, teratoma tissues were dissected and fixed in buffer-neutralized formalin solution. Paraffin embedding and histological analysis (H&E staining) were performed with standard protocols67.
Differentiation of PSCs to β-like cells
PSCs were treated with mTeSR medium supplemented with 10 μM Y-27632 for overnight, then the cells were dissociated into single cells with Accutase (Stem Cell Technology). Cells were dislodged by tapping the sides of the plates. Then Y-27632-supplemented mTeSR was added to dissociate cells to single cells. After cell counting with Countess (Invitrogen), about 1.8-2 million live cells were plated to each well of a 6-well Matrigel-coated plates in mTeSR medium supplemented with 10 μM Y-27632. After overnight culture, Y-27632-containing mTeSR medium was replaced by mTeSR medium. Differentiation was initiated when cells reach around 90%–95% confluency (usually between 1-2 days after plating cells). The cells were washed with PBS-, and then daily medium changes were the following (Figure S2A).
S1 (3 days) base medium: MCDB131 with 0.5% FAF-BSA, 1.5 g/L NaHCO3, 10 mM glucose, 1x glutamax, 1x penicillin/streptomycin. Freshly supplemented factors: 100 ng/mL Activin A, and 3 μM CHIR99021 (day 1); 100 ng/mL Activin A, and 0.3 μM CHIR99021 (day 2); 100 ng/mL Activin A (day 3).
S2 (2 days) base medium: MCDB131 with 0.5% FAF-BSA, 1.5 g/L NaHCO3, 10 mM glucose, 1x penicillin/streptomycin, 1x glutamax, and 0.25 mM Vc. Freshly supplemented factors: 50 ng/mL KGF.
S3 (2 days) base medium: MCDB131 with 2% FAF-BSA, 1.5 g/L NaHCO3, 10 mM glucose, 1x penicillin/streptomycin, 1x glutamax, 0.25 mM Vc, and 0.5x insulin-transferrin-selenium-ethanolamine. Freshly supplemented factors: 50 ng/mL KGF, 0.25 μM SANT-1, 1 μM retinoic acid, 100 nM LDN193189, and 200 nM TPB.
S4 (4 days) base medium: MCDB131 with 2% FAF-BSA, 1.5 g/L NaHCO3, 10 mM glucose, 1x penicillin/streptomycin, 1x glutamax, 0.25 mM Vc, and 0.5x insulin-transferrin-selenium-ethanolamine. Freshly supplemented factors: 50 ng/mL KGF, 50 ng/mL EGF, 0.25 μM SANT-1, 0.1 μM retinoic acid, 200 nM LDN193189, and 100 nM TPB. For the last medium change, 10 μM Y-27632 was added to the S4 medium.
S5 (3 day) started with transition from 2D culture to 3D culture. Cells were washed with PBS-, dissociated with Accutase (Stem Cell Technology), washed with H1 S5 base medium (see below), and about 6 million live cells were plated in 3 mL S5 medium (see below) supplemented with 10 μM Y-27632 in each well of 6-well AggreWell 400 plates (Stemcell Technology, 34415). After overnight culture, the clusters were moved to 6-well ultralow adherent culture plates placed on an orbital shaker set at 95-100 rpm for 2 more days in S5 medium, and the rest of the differentiation were carried out in ultralow attachment plates placed on an orbital shaker with the same setting.
S5 Base medium: MCDB131 with 2% FAF-BSA, 1.5 g/L NaHCO3, 20 mM glucose, 1x penicillin/streptomycin, 1x glutamax, 0.25 mM Vc, 0.5x insulin-transferrin-selenium-ethanolamine, 10 μg/mL heparin, and 10 μM ZnSO4. Freshly supplemented factors: 10 μM ALK5 inhibitor II, 0.25 μM SANT-1, 0.05 μM retinoic acid, 100 nM LDN193189, and 1 μM T3.
S6 (7 days) base medium: MCDB131 with 2% FAF-BSA, 1.5 g/L NaHCO3, 20 mM glucose, 1x penicillin/streptomycin, 1x glutamax, 0.25 mM Vc, 0.5x 100X insulin-transferrin-selenium-ethanolamine, 10 μg/mL heparin, 10 μM ZnSO4. Freshly supplemented factors: 10 μM ALK5 inhibitor II, 100 nM gamma secretase inhibitor XX, 100 nM LDN193189, and 1 μM T3.
S7 (7-14 days) medium: MCDB131 with 2% FAF-BSA, 1.5 g/L NaHCO3, 8 mM glucose, 1x penicillin/streptomycin, 1x glutamax, 1x MEM-NEAA, 10 μg/mL heparin, 10 μM ZnSO4, 1x trace element A, and 1x trace element B.
In vitro glucose induced insulin secretion (GSIS)
Differentiated stage 7 clusters were washed with KRB medium (128 mM NaCl, 5 mM KCl, 2.7 mM CaCl2, 1.2 mM MgSO4, 1 mM NaHPO4, 1.2 mM KH2PO4, 5 mM NaHCO3, 10 mM HEPES (pH 7.2), 0.1% FAF-BSA) twice, with KRB medium with 2.5 mM glucose once. Then the cell clusters were incubated in KRB with 2.5 mM glucose for 30 minutes on an orbital shaker set at 95-100 rpm in a cell culture incubator. The conditioned KRB buffer was collected, and clusters were incubated in KRB 20 mM glucose for 30 minutes followed by buffer collection. For some experiments, clusters were washed twice with KRB with 2.5 mM glucose, followed by another round of incubation and buffer collection. Lastly, the clusters were incubated in KRB buffer with 2.5 mM glucose and 30 mM KCl for 30 minutes followed by buffer collection. All collected KRB buffers were centrifuged at 3000 g for 3 min, and the clear supernatants were diluted 1:10 with KRB buffer, and human INS concentrations were determined with Ultrasensitive Human Insulin ELISA kits (Mercodia) based on the manufacturer’s protocol. Standard curves were fitted with a linear model.
Transmission electron microscopy (TEM)
Differentiated Stage 7 β-like cells were fixed in 2.5% glutaraldehyde, 3% paraformaldehyde with 5% sucrose in 0.1M sodium cacodylate buffer (pH 7.4), pelleted, and post fixed in 1% OsO4 in veronal-acetate buffer. The cells were stained overnight with 0.5% uranyl acetate in veronal-acetate buffer (pH 6.0), then dehydrated and embedded in Embed-812 resin. Sections were cut on a Reichert Ultracut E microtome with a Diatome diamond knife at a thickness setting of 50 nm, stained with uranyl acetate, and lead citrate. The sections were examined using a FEI Tecnai spirit at 80KV and photographed with an AMT CCD camera.
Co-culture setup between β-like cells and T cells, cytokine profiling, and quantification
CD19-CAR-T cells were washed with RPMI medium supplemented with 5% human serum, 1% glutamax, 1% MEM-NEAA, 1x penicillin/streptomycin (R-5 medium), then resuspended in R-5 medium at a concentration of 1 million/mL. Stage 7 β-like cells differentiated from INS: tdT, INS:CD19, or INS:CD19; PDL1 PSCs were washed with PBS-. Then R-5 medium with different numbers of CD19-CAR-T cells were added to β-like cells based on designated E/T ratios. After overnight coculture, medium samples were collected and centrifuged at 1500 g for 5 min, and the supernatant were analyzed with human cytokine array (R&D ARY005B) based on the manufacturer’s instructions. Cytokines showing difference between CD19-CAR-T cells cocultured with INS:tdt β-like cells and INS:CD19 β-like cells were selected to for quantification with a Luminex assay (custom assay from R&D to analyze IL2, TNFalpha, INFgamma, GM-CSF, CCL3, CCL4, and CCL5) with a Bio-Rad Bioplex3D Luminex equipment with the cytokine standards and suggested settings provided by the manufacturer.
In vitro luciferase imaging
Coculture of β-like cells differentiated from INS:tdT or INS:CD19 cells and T cells were setup in 96-well black-walled plates. Before imaging with an IVIS Spectrum system (PerkinElmer), 30 μL 15mg/mL D-luciferin were added and mixed 1 min before imaging acquisition with an exposure of 1 min and mid-bin setting. Imaging quantification were performed with Living Image (Version 4.5.4). The same sized regions of interest (ROI) of a background well were used to subtract background signal from the measurements of samples.
LDH (lactate dehydrogenase) release assay
About 80,000 β-like cells (stage 7) were planted in 100 μL R-5 medium. Then different numbers of T cells were added β-like cells in 20 μL R-5 medium. Conditioned medium samples were harvested after 8 hours, centrifuged at 250 g for 4 min, and 50 μL cleared supernatant medium were transferred to a black-walled 96-well plate, followed by CytoTox 96 Non-Radioactive Cytotoxicity Assay based on the manufacturer’s instructions (Promega G1780). OD490 were measured with a Multiskan Microplate Photometer (Thermo Fisher Scientific). R-5 medium, T cells control sample values were used to calculated LDH release. β-like cells not treated with T cells were lysed with lysis buffer, and used to calculating percentage of maximal lysis.
RNA-seq of human islets samples
RNA samples from islets, reaggregated pseudoislets, or samples treated with cell-depleted T reaction conditioned medium for 1 day were prepared with RNeasy plus micro kit (QIAGEN), and sequencing library was prepared with SMART-Seq v4 Ultra Low Input RNA Kit (Clontech) and sequenced with an Illumina HiSeq 2500 sequencer.
Fluorescent activated cell sorting of INS reporter expressing cells, aggregation into pseudo-islets, RNA purification, quantitative RT-PCR, and RNA-seq
Stage 7 β-like cells differentiated from INS reporter cells were washed twice with PBS-, then dissociated with 1:1 PBS diluted Accutase in 37°C with gentle agitation until clusters start to disintegrate. After centrifugation collection and resuspension, tdTomato-expressing β-like cells differentiated from INS:tdt PSCs were sorted on a FACSAria (BD Biosciences) cell sorter. CD19-expressing β-like cells were stained with an anti-CD19 antibody (PE conjugated Affymetrix # 12-0199-42) at 4°C for 30 min, and sorted with a FACSAria (BD Biosciences) cell sorter. Approximately 1.2 million sorted cells were plated into 1 well of a 24-well plate in S7 medium supplemented with 10 μM Y-27632. After overnight clustering formation, the clusters were transferred to 3 mL of S7 medium in an Ultra-low Attachment 6-well plate (Corning 3471) placed on an orbital shaker set for 95-100 rpm. S7 medium were changed every other day. Clusters were treated with control RPMI1640 medium with 10% human serum AB, or 0.45 μm filtered conditioned medium for one day, and collected for RNA extraction with RNeasy plus micro kit (QIAGEN). Reverse transcription was performed using SuperScript III first-strand synthesis Supermix (Invitrogen 18080400). Quantitative PCR experiments were performed with SYBR Green Master Mix reagent (Life Technologies 4385618) with a QuantStudio 6 Flex Real-Time PCR System (Applied Biosystems). Relative quantifications were determined with the above primer pairs using delta delta threshold cycle methods with the following primers:
| Gene | Forward primer | Reverse primer |
|---|---|---|
| PD1L1 | GGTGGTGCCGACTACAAGCGA | CCTTGGGGTAGCCCTCAGCCT |
| ACTB | CATGTACGTTGCTATCCAGGC | CTCCTTAATGTCACGCACGAT |
| PDX1 | CCTTTCCCATGGATGAAGTCTAC | TTCAACATGACAGCCAGCTCC |
| NKX6-1 | CTGGCCTGTACCCCTCATCA | CTTCCCGTCTTTGTCCAACAA |
| NEUROD1 | GCGTCTTAGAATAGCAAGGCA | GCGTCTTAGAATAGCAAGGCA |
| INS | AAGAGGCCATCAAGCAGATCA | CAGGAGGCGCATCCACA |
| NKX2-2 | GGCCTTCAGTACTCCCTGCA | GGGACTTGGAGCTTGAGTCCT |
| MAFB | TCAAGTTCGACGTGAAGAAGG | GTTCATCTGCTGGTAGTTGCT |
| OCT4 | GTGTTCAGCCAAAAGACCATCT | GGCCTGCATGAGGGTTTCT |
| SOX17 | GTGGACCGCACGGAATTTG | GGAGATTCACACCGGAGTCA |
| FOXA2 | GGAGCAGCTACTATGCAGAGC | CGTGTTCATGCCGTTCATCC |
| PDCD1 | GGCCAGGATGGTTCTTAGACT | GAAGCTGCAGGTGAAGGTG |
Sequencing library was prepared with SMART-Seq v4 Ultra Low Input RNA Kit (Clontech) and sequenced with an Illumina HiSeq 2500 sequencer.
For co-culture of CD19-CAR-T cells and β-like cells, CD19-CAR-T cells were washed twice with RPMI medium, then resuspended in RPMI medium at a concentration of 1 million/mL. Stage 7 β-like cells differentiated from INS: CD19 were washed twice with PBS-. RPMI medium was added to β-like cells as a control, and CD19 CAR-T cells were added to experimental well. After 6 hours, cells were washed with PBS twice before dissociation into single cells with Accutase. Then cells were stained with a PE-conjugated anti-CD19 antibody and PE positive cells were quantified with a FACSAria (BD Biosciences) cell sorter.
Immunofluorescence staining
Immunostaining were performed with a modified protocol based on previous method.67 Briefly, mouse tissues were fixed in 10% buffer neutralized formalin solution overnight at 4°C before tissue processing and paraffin embedding. Four-micron thick slices were cut and attached to glass slides for antigen retrieval followed by immunohistochemistry (IHC) staining or immunofluorescence.
For differentiated β-like cell clusters, cells were fixed in 4% PFA for 2 hours at room temperature, followed by twice wash with PBS. Then cells were incubated in PBS for 20 min, then PBS with 0.5% Tx100 for 20 min. Afterward, the samples were blocked in PBS with 0.1% Tween (PBST) with 5% donkey serum for 1 hour, followed by overnight incubation with primary antibody (1:150 rat-anti-c-peptide DBHB GN-ID4; 1:2000 rabbit anti-RFP antibody, Rockland 600-401-379; 1:40 mouse-anti-NKX6.1 antibody DBHB F55A12; 1: 200 goat-anti-PDX1 antibody, R&D af2419; 1:200 rabbit Anti-CD19 antibody, ab134114; 1:500, rabbit Anti-PDL1 antibody Ab205921; 1:200 rabbit anti-glucagon antibody, Cell Signaling D16G10; 1:50, rabbit anti-human CD8 antibody, Thermo Fisher Scientific, RM-9116-S0; 1:200 rabbit anti-cleaved GSDMD antibody, Cell signaling 36425) overnight at 4°C. After washing four times with PBST, secondary antibodies were added. Then pellets were washed and mounted in Fluoromount-G mounting medium (Electron Microscopy Sciences 1798425) with coverslip gently covered on top of samples. Confocal images were obtained with a Zeiss LSM 710 confocal microscope.
Transplantation and glucose measurements in mice
Stage 7 β-like cell clusters were transplanted under the kidney capsules of NSG mice (6-8 weeks) with a 1 mL syringe and a 23-gauge butterfly needle (Terumo Corporation SV-23BLK).68 Approximately 5 million cells in about 50 μL were injected. Transplanted mice were IVIS imaged after rested for about 2-3 weeks for engraftment of cells expressing human insulin. Mice were intraperitoneally injected with D-luciferin (PerkinElmer 122799, 0.195 mg per gram weight). Five to eight min after IP injection, mice were anesthetized with Isoflurane, and the flanks were shaved with a hair clipper. The IVIS imager (PerkinElmer) was used with mid-bin, and exposure for 5 min. Imaging analyses were performed with Living Image (Version 4.5.4).
After genotyping, 6-8 weeks old NRG-Akita mice were transplanted with about 5 million cells as described above. Transplanted mice were allowed to recover for 2 weeks after transplantation surgery, then tail blood glucose levels under ad libitum feeding conditions were measured every two weeks with an Accu-Chek glucometer (Roche) for about 3 months. Non-transplanted NRG-Akita and NRG littermates were used as controls.
For intraperitoneal glucose tolerance test (IPGTT), mice were starved in water-supply only cages for 16 hours, then glucose levels from tail blood were measured with an Accu-Chek glucometer (Roche) before intraperitoneal injection of glucose (2g glucose/kg body weight), 15 min, 30 min, 60 min, 90 min and 120 min after IP glucose injection.
Tail vein infusion of T cells
Freshly transduced CD19-28z CAR-T cells, and control CAR-T cells were analyzed with expression of LNGFR. Then about 5 million cells were washed with RPMI medium and resuspended in RPMI medium. Mice were anesthetized with Avertin and tail vein was dilated with warming pad before T cells were injected with 1 mL syringes with 30 g needles (injection volume about 200 μL).
Measurement of human insulin levels in mouse blood
Human insulin levels in mouse blood were measured based on a previously reported method.55 Mouse blood samples were collected into heparin-coated blood collection tubes (Sarstedt Microvette; CB 300 LH). Samples were centrifuged at 3000 g for 3 min, and plasma samples were collected into PCR tubes. Human insulin level measurements were performed with an Ultrasensitive Human Insulin ELISA kit (Mercodia) based on the manufacturer’s protocol. Standard curves were fitted with a linear model.
Chemical treatments of β-like clusters
β-like cells clusters were treated with 1 μM Tunicamycin (Sigma Aldrich T7765) for 16-20 hours, 1 μM VX-765 (Thermo Fisher Scientific 508389) for 2 days, or 2 μM Z-LEVD-FMK (BioVision 1108-100) for 2 days, followed by fixation in 4% PFA, and immunofluorescence staining as described above.
Quantification and Statistical Analysis
For statistical analyses of RNA-seq data, reads were aligned to the human (GRCh38) genome with tdTomato and Luciferase sequences, using STAR V2.7.1a (indexed using Ensembl 93 transcript models and ‘–sjdbOverhang 100’). Gene counts were obtained using featureCounts58 (with ‘-s 0’) and the same Ensembl 93 transcript models (with tdTomato and Luciferase). Then raw counts tables were imported to DEBrowser59 for KEGG analysis after low count filter of (Max < 1). Volcano plots and heatmaps were generated in R (Version 3.5.2). Gene set enrichment analysis62 were performed with GSEA 4.0.1 with gene symbols and corresponding DESeq2 normalized counts table as input files. Motif analyses were performed with Homer61 (Version 4.11) with promoter sequences from 264 downregulated genes and 332 upregulated genes. Other statistical analyses were performed using GraphPad Prism software (Version 8). Statistical significance was reached when p ≤ 0.05.
Acknowledgments
We thank Drs. M. Sadelain, H. Deng, and F. Zhang for sharing research reagents; Drs. L. Griffith and D. Lauffenburger for sharing research equipment; E. de Zwaan, E. Ness-Cohn, D. Fu, and R. Flannery for assistance; Dr. C. Garrett-Engele for critical reading of the manuscript; members of the R.J. laboratory for discussions; Drs. G. Gu, A. Rezania, T. Kieffer, R. Goldsby, H. Ploegh, H. Yang, and S. Elledge for advice and discussions; Drs. D. Melton and J. Kenty and other members of the D.A.M. laboratory for discussions; G. Gunset of the M.S. laboratory for discussions and shipping materials; Dr. G. Bell (Bioinformatics and Research Computing Group of the Whitehead Institute) for assisting with the bioinformatics analysis; W. Salmon and N. Watson (W.M. Keck Biological Imaging Facility of the Whitehead Institute) for assistance with fluorescence imaging and transmission electron microscopy, respectively; P. Wisniewski and H. Aharonov (Whitehead Institute) for assisting with cell sorting; members of the Genome Technology Core at the Whitehead Institute for assisting with RNA-seq; and P. Macfarlane of the Whitehead Institute IT for assistance with bioinformatics software. This study was supported by a generous gift from Liliana and Hillel Bachrach, a collaborative research agreement from Novo Nordisk, and NIH grant 1R01-NS088538 (to R.J.).
Author Contributions
H.M. and R.J. designed the experiments, interpreted the results, and wrote the manuscript. H.M. performed research and analyzed results. J.F.J. had major intellectual input into the project.
Declaration of Interests
R.J. is a cofounder of Fate, Fulcrum, and Omega Therapeutics and an advisor to Dewpoint and Camp4 Therapeutics. J.F.J. is an employee of Novo Nordisk.
Published: September 22, 2020
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.xcrm.2020.100097.
Supporting Citations
The following references appear in the Supplemental Information: Tan et al.69 and Furuyama et al.70
Supplemental Information
References
- 1.Bluestone J.A., Herold K., Eisenbarth G. Genetics, pathogenesis and clinical interventions in type 1 diabetes. Nature. 2010;464:1293–1300. doi: 10.1038/nature08933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.van Belle T.L., Coppieters K.T., von Herrath M.G. Type 1 diabetes: etiology, immunology, and therapeutic strategies. Physiol. Rev. 2011;91:79–118. doi: 10.1152/physrev.00003.2010. [DOI] [PubMed] [Google Scholar]
- 3.Mathis D., Vence L., Benoist C. beta-Cell death during progression to diabetes. Nature. 2001;414:792–798. doi: 10.1038/414792a. [DOI] [PubMed] [Google Scholar]
- 4.Onengut-Gumuscu S., Chen W.M., Burren O., Cooper N.J., Quinlan A.R., Mychaleckyj J.C., Farber E., Bonnie J.K., Szpak M., Schofield E., Type 1 Diabetes Genetics Consortium Fine mapping of type 1 diabetes susceptibility loci and evidence for colocalization of causal variants with lymphoid gene enhancers. Nat. Genet. 2015;47:381–386. doi: 10.1038/ng.3245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wen L., Ley R.E., Volchkov P.Y., Stranges P.B., Avanesyan L., Stonebraker A.C., Hu C., Wong F.S., Szot G.L., Bluestone J.A. Innate immunity and intestinal microbiota in the development of Type 1 diabetes. Nature. 2008;455:1109–1113. doi: 10.1038/nature07336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Millman J.R., Xie C., Van Dervort A., Gürtler M., Pagliuca F.W., Melton D.A. Generation of stem cell-derived β-cells from patients with type 1 diabetes. Nat. Commun. 2016;7:11463. doi: 10.1038/ncomms11463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sui L., Danzl N., Campbell S.R., Viola R., Williams D., Xing Y., Wang Y., Phillips N., Poffenberger G., Johannesson B. β-Cell Replacement in Mice Using Human Type 1 Diabetes Nuclear Transfer Embryonic Stem Cells. Diabetes. 2018;67:26–35. doi: 10.2337/db17-0120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Delong T., Wiles T.A., Baker R.L., Bradley B., Barbour G., Reisdorph R., Armstrong M., Powell R.L., Reisdorph N., Kumar N. Pathogenic CD4 T cells in type 1 diabetes recognize epitopes formed by peptide fusion. Science. 2016;351:711–714. doi: 10.1126/science.aad2791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Burrack A.L., Martinov T., Fife B.T. T Cell-Mediated Beta Cell Destruction: Autoimmunity and Alloimmunity in the Context of Type 1 Diabetes. Front. Endocrinol. (Lausanne) 2017;8:343. doi: 10.3389/fendo.2017.00343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Roep B.O., Peakman M. Antigen targets of type 1 diabetes autoimmunity. Cold Spring Harb. Perspect. Med. 2012;2:a007781. doi: 10.1101/cshperspect.a007781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Purcell A.W., Sechi S., DiLorenzo T.P. The Evolving Landscape of Autoantigen Discovery and Characterization in Type 1 Diabetes. Diabetes. 2019;68:879–886. doi: 10.2337/dbi18-0066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tisch R., Yang X.D., Singer S.M., Liblau R.S., Fugger L., McDevitt H.O. Immune response to glutamic acid decarboxylase correlates with insulitis in non-obese diabetic mice. Nature. 1993;366:72–75. doi: 10.1038/366072a0. [DOI] [PubMed] [Google Scholar]
- 13.Saha K., Jaenisch R. Technical challenges in using human induced pluripotent stem cells to model disease. Cell Stem Cell. 2009;5:584–595. doi: 10.1016/j.stem.2009.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Soldner F., Jaenisch R. Stem Cells, Genome Editing, and the Path to Translational Medicine. Cell. 2018;175:615–632. doi: 10.1016/j.cell.2018.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kieffer T.J. Closing in on Mass Production of Mature Human Beta Cells. Cell Stem Cell. 2016;18:699–702. doi: 10.1016/j.stem.2016.05.014. [DOI] [PubMed] [Google Scholar]
- 16.D’Amour K.A., Bang A.G., Eliazer S., Kelly O.G., Agulnick A.D., Smart N.G., Moorman M.A., Kroon E., Carpenter M.K., Baetge E.E. Production of pancreatic hormone-expressing endocrine cells from human embryonic stem cells. Nat. Biotechnol. 2006;24:1392–1401. doi: 10.1038/nbt1259. [DOI] [PubMed] [Google Scholar]
- 17.Kroon E., Martinson L.A., Kadoya K., Bang A.G., Kelly O.G., Eliazer S., Young H., Richardson M., Smart N.G., Cunningham J. Pancreatic endoderm derived from human embryonic stem cells generates glucose-responsive insulin-secreting cells in vivo. Nat. Biotechnol. 2008;26:443–452. doi: 10.1038/nbt1393. [DOI] [PubMed] [Google Scholar]
- 18.Pagliuca F.W., Millman J.R., Gürtler M., Segel M., Van Dervort A., Ryu J.H., Peterson Q.P., Greiner D., Melton D.A. Generation of functional human pancreatic β cells in vitro. Cell. 2014;159:428–439. doi: 10.1016/j.cell.2014.09.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rezania A., Bruin J.E., Arora P., Rubin A., Batushansky I., Asadi A., O’Dwyer S., Quiskamp N., Mojibian M., Albrecht T. Reversal of diabetes with insulin-producing cells derived in vitro from human pluripotent stem cells. Nat. Biotechnol. 2014;32:1121–1133. doi: 10.1038/nbt.3033. [DOI] [PubMed] [Google Scholar]
- 20.Veres A., Faust A.L., Bushnell H.L., Engquist E.N., Kenty J.H., Harb G., Poh Y.C., Sintov E., Gürtler M., Pagliuca F.W. Charting cellular identity during human in vitro β-cell differentiation. Nature. 2019;569:368–373. doi: 10.1038/s41586-019-1168-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Russ H.A., Parent A.V., Ringler J.J., Hennings T.G., Nair G.G., Shveygert M., Guo T., Puri S., Haataja L., Cirulli V. Controlled induction of human pancreatic progenitors produces functional beta-like cells in vitro. EMBO J. 2015;34:1759–1772. doi: 10.15252/embj.201591058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Velazco-Cruz L., Song J., Maxwell K.G., Goedegebuure M.M., Augsornworawat P., Hogrebe N.J., Millman J.R. Acquisition of Dynamic Function in Human Stem Cell-Derived β Cells. Stem Cell Reports. 2019;12:351–365. doi: 10.1016/j.stemcr.2018.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.June C.H., Sadelain M. Chimeric Antigen Receptor Therapy. N. Engl. J. Med. 2018;379:64–73. doi: 10.1056/NEJMra1706169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sadelain M., Brentjens R., Rivière I. The basic principles of chimeric antigen receptor design. Cancer Discov. 2013;3:388–398. doi: 10.1158/2159-8290.CD-12-0548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shi J., Zhao Y., Wang K., Shi X., Wang Y., Huang H., Zhuang Y., Cai T., Wang F., Shao F. Cleavage of GSDMD by inflammatory caspases determines pyroptotic cell death. Nature. 2015;526:660–665. doi: 10.1038/nature15514. [DOI] [PubMed] [Google Scholar]
- 26.Kayagaki N., Stowe I.B., Lee B.L., O’Rourke K., Anderson K., Warming S., Cuellar T., Haley B., Roose-Girma M., Phung Q.T. Caspase-11 cleaves gasdermin D for non-canonical inflammasome signalling. Nature. 2015;526:666–671. doi: 10.1038/nature15541. [DOI] [PubMed] [Google Scholar]
- 27.Zetsche B., Gootenberg J.S., Abudayyeh O.O., Slaymaker I.M., Makarova K.S., Essletzbichler P., Volz S.E., Joung J., van der Oost J., Regev A. Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell. 2015;163:759–771. doi: 10.1016/j.cell.2015.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thomson J.A., Itskovitz-Eldor J., Shapiro S.S., Waknitz M.A., Swiergiel J.J., Marshall V.S., Jones J.M. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282:1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- 29.Gu G., Dubauskaite J., Melton D.A. Direct evidence for the pancreatic lineage: NGN3+ cells are islet progenitors and are distinct from duct progenitors. Development. 2002;129:2447–2457. doi: 10.1242/dev.129.10.2447. [DOI] [PubMed] [Google Scholar]
- 30.Dorrell C., Schug J., Canaday P.S., Russ H.A., Tarlow B.D., Grompe M.T., Horton T., Hebrok M., Streeter P.R., Kaestner K.H., Grompe M. Human islets contain four distinct subtypes of β cells. Nat. Commun. 2016;7:11756. doi: 10.1038/ncomms11756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Roost M.S., van Iperen L., Ariyurek Y., Buermans H.P., Arindrarto W., Devalla H.D., Passier R., Mummery C.L., Carlotti F., de Koning E.J. KeyGenes, a Tool to Probe Tissue Differentiation Using a Human Fetal Transcriptional Atlas. Stem Cell Reports. 2015;4:1112–1124. doi: 10.1016/j.stemcr.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Arda H.E., Li L., Tsai J., Torre E.A., Rosli Y., Peiris H., Spitale R.C., Dai C., Gu X., Qu K. Age-Dependent Pancreatic Gene Regulation Reveals Mechanisms Governing Human β Cell Function. Cell Metab. 2016;23:909–920. doi: 10.1016/j.cmet.2016.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brehm M.A., Bortell R., Diiorio P., Leif J., Laning J., Cuthbert A., Yang C., Herlihy M., Burzenski L., Gott B. Human immune system development and rejection of human islet allografts in spontaneously diabetic NOD-Rag1null IL2rgammanull Ins2Akita mice. Diabetes. 2010;59:2265–2270. doi: 10.2337/db10-0323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Burd J.F., Usategui-Gomez M. Immunochemical studies on lactate dehydrogenase. Biochim. Biophys. Acta. 1973;310:238–247. doi: 10.1016/0005-2795(73)90026-3. [DOI] [PubMed] [Google Scholar]
- 35.Cnop M., Welsh N., Jonas J.C., Jörns A., Lenzen S., Eizirik D.L. Mechanisms of pancreatic beta-cell death in type 1 and type 2 diabetes: many differences, few similarities. Diabetes. 2005;54(Suppl 2):S97–S107. doi: 10.2337/diabetes.54.suppl_2.s97. [DOI] [PubMed] [Google Scholar]
- 36.Nicholson D.W., Ali A., Thornberry N.A., Vaillancourt J.P., Ding C.K., Gallant M., Gareau Y., Griffin P.R., Labelle M., Lazebnik Y.A. Identification and inhibition of the ICE/CED-3 protease necessary for mammalian apoptosis. Nature. 1995;376:37–43. doi: 10.1038/376037a0. [DOI] [PubMed] [Google Scholar]
- 37.Hrvatin S., O’Donnell C.W., Deng F., Millman J.R., Pagliuca F.W., DiIorio P., Rezania A., Gifford D.K., Melton D.A. Differentiated human stem cells resemble fetal, not adult, β cells. Proc. Natl. Acad. Sci. USA. 2014;111:3038–3043. doi: 10.1073/pnas.1400709111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Richardson S.J., Rodriguez-Calvo T., Gerling I.C., Mathews C.E., Kaddis J.S., Russell M.A., Zeissler M., Leete P., Krogvold L., Dahl-Jørgensen K. Islet cell hyperexpression of HLA class I antigens: a defining feature in type 1 diabetes. Diabetologia. 2016;59:2448–2458. doi: 10.1007/s00125-016-4067-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Colli M.L., Hill J.L.E., Marroquí L., Chaffey J., Dos Santos R.S., Leete P., Coomans de Brachène A., Paula F.M.M., Op de Beeck A., Castela A. PDL1 is expressed in the islets of people with type 1 diabetes and is up-regulated by interferons-α and-γ via IRF1 induction. EBioMedicine. 2018;36:367–375. doi: 10.1016/j.ebiom.2018.09.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ruan J., Xia S., Liu X., Lieberman J., Wu H. Cryo-EM structure of the gasdermin A3 membrane pore. Nature. 2018;557:62–67. doi: 10.1038/s41586-018-0058-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hitomi J., Katayama T., Eguchi Y., Kudo T., Taniguchi M., Koyama Y., Manabe T., Yamagishi S., Bando Y., Imaizumi K. Involvement of caspase-4 in endoplasmic reticulum stress-induced apoptosis and Abeta-induced cell death. J. Cell Biol. 2004;165:347–356. doi: 10.1083/jcb.200310015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ramos-Rodríguez M., Raurell-Vila H., Colli M.L., Alvelos M.I., Subirana-Granés M., Juan-Mateu J., Norris R., Turatsinze J.V., Nakayasu E.S., Webb-Robertson B.M. The impact of proinflammatory cytokines on the β-cell regulatory landscape provides insights into the genetics of type 1 diabetes. Nat. Genet. 2019;51:1588–1595. doi: 10.1038/s41588-019-0524-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wei S.C., Duffy C.R., Allison J.P. Fundamental Mechanisms of Immune Checkpoint Blockade Therapy. Cancer Discov. 2018;8:1069–1086. doi: 10.1158/2159-8290.CD-18-0367. [DOI] [PubMed] [Google Scholar]
- 44.Chen L., Han X. Anti-PD-1/PD-L1 therapy of human cancer: past, present, and future. J. Clin. Invest. 2015;125:3384–3391. doi: 10.1172/JCI80011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nishimura H., Okazaki T., Tanaka Y., Nakatani K., Hara M., Matsumori A., Sasayama S., Mizoguchi A., Hiai H., Minato N., Honjo T. Autoimmune dilated cardiomyopathy in PD-1 receptor-deficient mice. Science. 2001;291:319–322. doi: 10.1126/science.291.5502.319. [DOI] [PubMed] [Google Scholar]
- 46.Nishimura H., Nose M., Hiai H., Minato N., Honjo T. Development of lupus-like autoimmune diseases by disruption of the PD-1 gene encoding an ITIM motif-carrying immunoreceptor. Immunity. 1999;11:141–151. doi: 10.1016/s1074-7613(00)80089-8. [DOI] [PubMed] [Google Scholar]
- 47.Way J., Drakaki A., Drexler A., Freeby M. Anti-PD-L1 therapy and the onset of diabetes mellitus with positive pancreatic autoantibodies. BMJ Case Rep. 2017;2017 doi: 10.1136/bcr-2017-220415. bcr2017220415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Qian C., Guo H., Chen X., Shi A., Li S., Wang X., Pan J., Fang C. Association of PD-1 and PD-L1 Genetic Polymorphyisms with Type 1 Diabetes Susceptibility. J. Diabetes Res. 2018;2018:1614683. doi: 10.1155/2018/1614683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Atkinson M.A. Pancreatic biopsies in type 1 diabetes: revisiting the myth of Pandora’s box. Diabetologia. 2014;57:656–659. doi: 10.1007/s00125-013-3159-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Eyquem J., Mansilla-Soto J., Giavridis T., van der Stegen S.J., Hamieh M., Cunanan K.M., Odak A., Gönen M., Sadelain M. Targeting a CAR to the TRAC locus with CRISPR/Cas9 enhances tumour rejection. Nature. 2017;543:113–117. doi: 10.1038/nature21405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stephan M.T., Ponomarev V., Brentjens R.J., Chang A.H., Dobrenkov K.V., Heller G., Sadelain M. T cell-encoded CD80 and 4-1BBL induce auto- and transcostimulation, resulting in potent tumor rejection. Nat. Med. 2007;13:1440–1449. doi: 10.1038/nm1676. [DOI] [PubMed] [Google Scholar]
- 52.Miller A.D., Garcia J.V., von Suhr N., Lynch C.M., Wilson C., Eiden M.V. Construction and properties of retrovirus packaging cells based on gibbon ape leukemia virus. J. Virol. 1991;65:2220–2224. doi: 10.1128/jvi.65.5.2220-2224.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ran F.A., Hsu P.D., Wright J., Agarwala V., Scott D.A., Zhang F. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 2013;8:2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liu H., Yang H., Zhu D., Sui X., Li J., Liang Z., Xu L., Chen Z., Yao A., Zhang L. Systematically labeling developmental stage-specific genes for the study of pancreatic β-cell differentiation from human embryonic stem cells. Cell Res. 2014;24:1181–1200. doi: 10.1038/cr.2014.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ma H., Wert K.J., Shvartsman D., Melton D.A., Jaenisch R. Establishment of human pluripotent stem cell-derived pancreatic β-like cells in the mouse pancreas. Proc. Natl. Acad. Sci. USA. 2018;115:3924–3929. doi: 10.1073/pnas.1702059115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhao Z., Condomines M., van der Stegen S.J.C., Perna F., Kloss C.C., Gunset G., Plotkin J., Sadelain M. Structural Design of Engineered Costimulation Determines Tumor Rejection Kinetics and Persistence of CAR T Cells. Cancer Cell. 2015;28:415–428. doi: 10.1016/j.ccell.2015.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Liao Y., Smyth G.K., Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30:923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 59.Kucukural A., Yukselen O., Ozata D.M., Moore M.J., Garber M. DEBrowser: interactive differential expression analysis and visualization tool for count data. BMC Genomics. 2019;20:6. doi: 10.1186/s12864-018-5362-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Heinz S., Benner C., Spann N., Bertolino E., Lin Y.C., Laslo P., Cheng J.X., Murre C., Singh H., Glass C.K. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S., Mesirov J.P. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gibson D.G., Young L., Chuang R.Y., Venter J.C., Hutchison C.A., 3rd, Smith H.O. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods. 2009;6:343–345. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- 64.Yang Y.P., Ma H., Starchenko A., Huh W.J., Li W., Hickman F.E., Zhang Q., Franklin J.L., Mortlock D.P., Fuhrmann S. A Chimeric Egfr Protein Reporter Mouse Reveals Egfr Localization and Trafficking In Vivo. Cell Rep. 2017;19:1257–1267. doi: 10.1016/j.celrep.2017.04.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hockemeyer D., Soldner F., Beard C., Gao Q., Mitalipova M., DeKelver R.C., Katibah G.E., Amora R., Boydston E.A., Zeitler B. Efficient targeting of expressed and silent genes in human ESCs and iPSCs using zinc-finger nucleases. Nat. Biotechnol. 2009;27:851–857. doi: 10.1038/nbt.1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tucker K.L., Wang Y., Dausman J., Jaenisch R. A transgenic mouse strain expressing four drug-selectable marker genes. Nucleic Acids Res. 1997;25:3745–3746. doi: 10.1093/nar/25.18.3745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Li C., Ma H., Wang Y., Cao Z., Graves-Deal R., Powell A.E., Starchenko A., Ayers G.D., Washington M.K., Kamath V. Excess PLAC8 promotes an unconventional ERK2-dependent EMT in colon cancer. J. Clin. Invest. 2014;124:2172–2187. doi: 10.1172/JCI71103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Brissova M., Fowler M., Wiebe P., Shostak A., Shiota M., Radhika A., Lin P.C., Gannon M., Powers A.C. Intraislet endothelial cells contribute to revascularization of transplanted pancreatic islets. Diabetes. 2004;53:1318–1325. doi: 10.2337/diabetes.53.5.1318. [DOI] [PubMed] [Google Scholar]
- 69.Tan S., Li Y., Xia J., Jin C.H., Hu Z., Duinkerken G., Li Y., Khosravi Maharlooei M., Chavez E., Nauman G. Type 1 diabetes induction in humanized mice. Proc. Natl. Acad. Sci. USA. 2017;114:10954–10959. doi: 10.1073/pnas.1710415114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Furuyama K., Chera S., van Gurp L., Oropeza D., Ghila L., Damond N., Vethe H., Paulo J.A., Joosten A.M., Berney T. Diabetes relief in mice by glucose-sensing insulin-secreting human α-cells. Nature. 2019;567:43–48. doi: 10.1038/s41586-019-0942-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The accession number for the RNA-seq data reported in this paper is GEO: GSE155713.