Summary
Hepatitis delta virus (HDV) requires hepatitis B virus (HBV) to complete its infection cycle and causes severe hepatitis, with limited therapeutic options. To determine the prospect of T cell therapy in HBV/HDV co-infection, we study the impact of HDV on viral antigen processing and presentation. Using in vitro models of HBV/HDV co-infection, we demonstrate that HDV boosts HBV epitope presentation, both in HBV/HDV co-infected and neighboring mono-HBV-infected cells through the upregulation of the antigen processing pathway mediated by IFN-β/λ. Liver biopsies of HBV/HDV patients confirm this upregulation. We then validate in vitro and in a HBV/HDV preclinical mouse model that HDV infection increases the anti-HBV efficacy of T cells with engineered T cell receptors. Thus, by unveiling the effect of HDV on HBV antigen presentation, we provide a framework to better understand HBV/HDV immune pathology, and advocate the utilization of engineered HBV-specific T cells as a potential treatment for HBV/HDV co-infection.
Keywords: viral hepatitis, chronic liver disease, immunotherapy, CAR-T cell therapy, human liver chimeric mice, RNA viruses, viroid
Graphical Abstract
Highlights
HDV infection affects viral antigen processing and presentation
HDV boosts HBV epitope presentation on HBV/HDV and mono-HBV-infected hepatocytes
Anti-HBV efficacy of T cells engineered with T cell receptors is enhanced by HDV
Tham et al. demonstrate that HDV activates hepatocyte antigen processing and presentation capacity, increasing their activation of virus-specific T cells. HDV, a defective RNA virus that relies on HBV envelope proteins to infect hepatocytes, exacerbates HBV-related liver pathology. This immunological adjuvant-like feature can increase the anti-HBV efficacy of immunological therapy.
Introduction
The major etiological agent of chronic viral hepatitis worldwide is hepatitis B virus (HBV), a hepatotropic non-directly cytopathic DNA virus that establishes chronic infection in ∼240 million people. The pathological consequences of chronic HBV infection are aggravated by co- or super-infection with the hepatitis delta virus (HDV), a defective RNA virus that relies on the envelope proteins of HBV to infect human hepatocytes.1 HDV is the only known viroid infecting humans, and according to the World Health Organization (WHO), ∼20 million individuals are HBV/HDV co-infected worldwide, while new epidemiological estimations have suggested that the number of HDV+ individuals may be even substantially higher (70 million people worldwide).2 Patients with persistent HBV/HDV co-infection develop liver cirrhosis at a younger age and are more likely to progress toward hepatocellular carcinoma.3 Conventional antiviral therapies used in HBV mono-infection (nucleoside analogs and interferon [IFN]-α) have low efficacy in HBV/HDV co-infection.4,5 Novel treatments targeting different steps of HDV infection and replications are being tested, alone or in combination with IFN-α, with some encouraging results.6, 7, 8 However, there is still ample space and urgent need to develop new therapeutic approaches for the majority of HBV/HDV-co-infected patients.
We and others have advocated the use of strategies able to boost virus-specific adaptive immunity for the treatment of chronic HBV (CHB) infection.9,10 HBV-specific CD8 T cells are associated with the control of HBV replication in animal models11 and in patients.12 Furthermore, the demonstration that transplantation of bone marrow containing HBV-specific T and B cells into patients with CHB can lead to an HBV functional cure13,14 highlights the therapeutic potential of HBV-specific immunity reconstitution. The defect of HBV-specific B and T cell responses present in CHB patients15, 16, 17, 18, 19 renders the restoration of HBV immunity, through vaccine therapy or checkpoint inhibitors, unlikely to succeed. However, reconstitution of HBV-specific immunity through the adoptive transfer of autologous T cells engineered with chimeric antigen receptors (CARs) or T cell receptors (TCRs) specific for HBV,20,21 may bypass such defects and could lead to specific targeting of HBV-infected hepatocytes.22 Recent studies in chimeric mice with HBV-infected human livers have shown the efficacy of these specific strategies.21,23, 24, 25 We reasoned that such approaches possess therapeutic potential also in HBV/HDV co-infection. The demonstration that HDV can persist intracellularly in replicating human hepatocytes despite blocking re-infection with Myrcludex B,26 suggests that immune-mediated destruction of a substantial fraction of HDV-infected cells is required to substantially lower intrahepatic HDV infection. Targeting HDV/HBV-co-infected or HDV-mono-infected cells with engineered HDV-specific T cells is hindered, however, by the high mutation rate of HDV. Two recent studies have shown that HDV generates a high frequency of mutations leading to the selection of HDV variants able to escape HDV-specific CD8 T cell recognition in HBV/HDV-co-infected patients.27,28 Thus, targeting directly the pool of HBV/HDV-co-infected cells by transferring engineered T cells specific to the more conserved HBV-derived CD8 T cell epitopes may develop into a promising alternative strategy. However, this approach can only be successful if HDV does not interfere with the processing and presentation of HBV epitopes.
To answer this question, we established two in vitro HBV/HDV co-infection models, based on HepG2 cells transduced with human NTCP (HepG2-hNTCP) cells29 and with normal primary human hepatocytes (PHHs). We quantified the expression of the genes associated with antigen presentation in HBV-mono-infected cells. Subsequently, we tested whether HDV co-infection modulates the processing and presentation of two distinct HBV CD8 T cell epitopes (one immunoproteasome-dependent [human leukocyte antigen {HLA}-A0201/HBs183-91] and one immunoproteasome-independent [HLA-A0201/HBc18-27]30), using two readouts: (1) direct quantification of epitope complexes with TCR-like antibodies and (2) testing the ability of HBV/HDV-co-infected cells to activate HBV-specific CD8 T cells. Finally, we used the human liver chimeric mouse model to test directly in vivo whether HBV/HDV co-infection alters the antiviral efficiency of adoptive T cell therapy.
Results
Establishing In Vitro HBV/HDV Co-infection in Primary Human Hepatocytes and in HepG2-NTCP Cell Lines
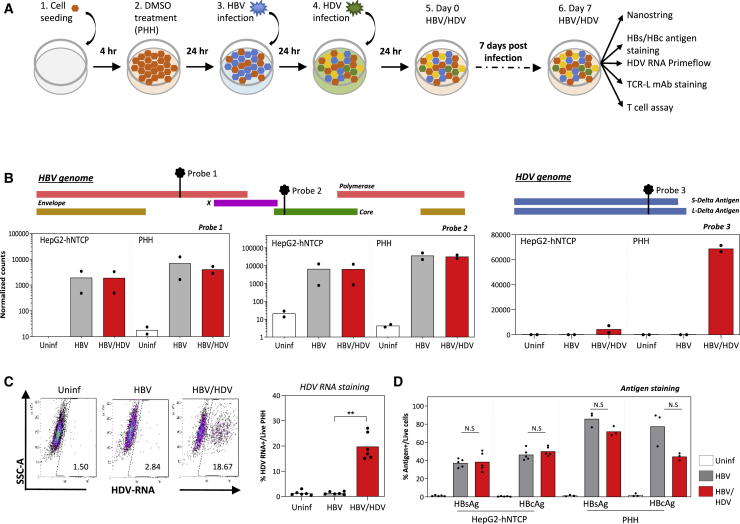
We used two in vitro models of HBV/HDV co-infection established with PHHs or HepG2-hNTCP cells29 (Figure 1A). Briefly, 24 h after HBV infection (MOI 3,000 genome equivalents [GE]/cell), HDV was added at an MOI of 500 GE/cell. Seven days post-co-infection, HBV and HDV infections were tested by measuring HBV and HDV mRNA levels using NanoString technology. Customized probe sets targeting 2 specific regions in the HBV genome (genotype D) and 1 region in the HDV genome (genotype 1) were used (Figure 1B).
Figure 1.
Establishment of an In Vitro HBV/HDV Infection System in HepG2-hNTCP Cells and PHHs
(A) Schematic of the experimental procedure. HepG2-hNTCP cells or PHHs were seeded and treated with 2% DMSO for 4 h. Cells were then inoculated with HBV at a MOI of 3,000 genome equivalents (GE) per cell for 24 h and subsequently with HDV at a MOI of 500 GE/cell for another 24 h. Infection status of the cells was analyzed 7 days post-infection.
(B) HBV and HDV mRNA expression in infected target cells (HepG2-hNTCP and PHH) analyzed using customized NanoString probes. The relative positions of each NanoString probe targeting the HBV and HDV genome are annotated as probes 1 to 3. Bar graphs show the average normalized counts of probes 1 and 2 expressed on a log10 scale and probe 3 expressed on a linear scale (n = 2 for each cell type).
(C) Expression of HDV RNA was quantified by the PrimeFlow RNA assay. A representative dot plot is shown (left), and bars on the right show the average frequency of HDV RNA+ cells in infected PHH (n = 6; p = 0.0073).
(D) Quantification of HBsAg and HBcAg expression in infected HepG2-hNTCP cells (n = 5) and PHHs (n = 3) by flow cytometry. Bars indicate the average frequency of HBsAg+ and HBcAg+ cells in the respective infection, and each dot represents a single experiment.
∗p = 0.01–0.05 and ∗∗p = 0.001–0.01. Non-significant p values are indicated as N.S.
See also Figure S1.
HBV replication was confirmed in both HBV-mono- and HBV/HDV-co-infected HepG2-hNTCP cells and PHHs, as seen from the high levels of HBV RNA expression (Figure 1B, left and center), while HDV infection was detected only in HBV/HDV-co-infected HepG2-hNTCP cells and PHHs (Figure 1B, right column). Although HDV RNA levels differed dramatically between PHHs and HepG2-hNTCP cells (4,425 mRNA counts in HepG2-hNTCP versus 68,863 mRNA counts in PHHs), HBV RNAs were only slightly higher in PHHs, showing that HBV infection was similar in both cell types. To quantify HDV infection at a single-cell level and determine the frequency of infected PHH-producing HDV, PrimeFlow RNA assay, a flow cytometry-based method for detecting HDV RNA, was applied. HDV RNA was detected in ∼20% of HBV/HDV-co-infected PHHs (Figure 1C), while no co-infected cells were visualized with this technology in HepG2-NTCP cells (Figure S1). Furthermore, we analyzed the expression of HBV antigens in HBV-mono- and HBV/HDV-co-infected cultured HepG2-hNTCP cells and PHHs by staining with antibodies specific for HBV surface antigen (HBsAg) and core antigen (HBcAg). Flow cytometry analysis showed that HepG2-hNTCP cells either HBV mono- or HBV/HDV co-infected were on average 35% HBsAg+ and 48% HBcAg+. HBV-mono-infected PHH cultures were 90% HBsAg+ and 80% HBcAg+, which was reduced to 75% HBsAg+ and 45% HBcAg+ in HBV/HDV-co-infected PHHs, indicating that HDV infection lowers HBV antigen expression, which was more evident for HBcAg (Figure 1D).
HDV Activates the Antigen Processing and Presentation Machinery of HBV-Infected Hepatocytes
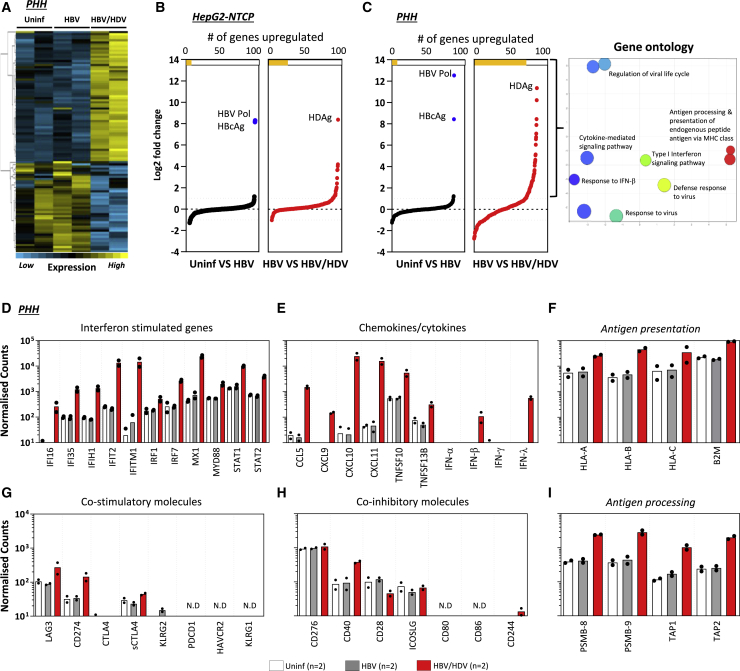
The ability of HDV to trigger the expression of innate immune genes has been shown in both in vitro31,32 and in vivo models.33, 34, 35 To further characterize the transcriptional profile of immune signaling in HBV/HDV infection, we comprehensively evaluated the impact of HDV infection on HBV-infected cells using NanoString technology. The expression level of 579 human immune-related genes was analyzed in HBV-mono-infected and HBV/HDV-infected PHHs and HepG2-hNTCP cells. Globally, uninfected and HBV-mono-infected cells showed a very similar gene expression profile, while HDV infection modified profoundly the expression of immune genes. Figure 2A shows the heatmap of immune genes expressed in cultured PHHs, while Figures 2B and 2C display a pairwise comparison of gene expression between the different infection statuses of HepG2-hNTCP cells and PHHs, respectively. HBV RNAs were detected exclusively in HBV-infected HepG2-hNTCP cells and PHHs compared to uninfected cells, and the RNA of HDV antigen (HDAg) was detected only in HDV-infected cells. Consistent with the data in Figure 1B, the level of mRNA coding for HDAg was lower in HepG2-NTCPs than in PHHs (4,425 mRNA counts in HepG2-hNTCP versus 68,863 mRNA counts in PHHs).
Figure 2.
Increased Expression of Immune Genes upon HDV Infection
Uninfected, HBV mono-infected-, and HBV/HDV-infected PHH and HepG2-hNTCP cells were lysed 7 days post-infection, and mRNA expression of 579 immune-related genes was analyzed using NanoString.
(A) Heatmap showing immune gene expression in uninfected, HBV mono-infected-, and HBV/HDV-infected PHH cultures.
(B and C) A side-by-side comparison of immune genes differentially expressed in HBV mono-infected- and HBV/HDV-infected HepG2-hNTCP cells (B) and in PHHs (C). The number of genes that are differentially expressed (≥2-fold) between either uninfected and HBV mono-infected- or HBV mono-infected- and HBV/HDV-infected cells are depicted with yellow bars at the top of each panel. Immune-related genes that are upregulated in HBV/HDV-infected PHHs are clustered by their involvement in different biological processes, using Gene Ontology (C, right panel).
(D–I) Normalized counts of mRNA expression of genes related to (D) interferon; (E) chemokines or cytokines; (F) antigen presentation; (G) co-stimulatory; (H) co-inhibitory and (I) antigen processing in uninfected, HBV mono-infected-, and HBV/HDV-infected PHHs. Normalized counts are expressed on a log10 scale, and bars represent the mean normalized count. Each dot represents a single experiment (n = 2). Probes with no detected counts are labeled as not detected (N.D.).
See also Figure S2.
HBV infection alone did not induce any significant changes in the immune gene expression in both cell types. In contrast, 30 and 78 immune-related genes were upregulated (≥2-fold) in HBV/HDV-co-infected HepG2-hNTCPs (Figure 2B) and PHHs (Figure 2C), respectively. The immune-related gene expression changes were more pronounced in PHH compared to HepG2-hNTCP cells (the range of fold change in HBV/HDV-co-infected HepG2-hNTCP cells was between 2- and 4-fold compared to 2- and 10-fold in co-infected PHHs), reflecting the different infection rates and/or the lower integrity of immune response genes in hepatoma cells.
A Gene Ontology analysis classifying all of the upregulated immune-related genes in HBV/HDV-infected PHHs revealed that the biological processes related to antiviral immunity, cytokine production, and antigen processing and presentation pathways were activated upon HDV infection (Figure 2C, right panel). A comparative analysis of the expression of individual genes in PHHs is depicted in Figures 2D–2I. HDV activated a plethora of IFN-stimulated genes such as Mx1, IRF7, STAT1, and STAT2, as previously described31,33,34 (Figure 2D). The previously reported ability of HDV to induce the production of IFN-β and IFN-λ but not IFN-α34 was confirmed at the mRNA level (Figure 2E). We detected in HBV/HDV-co-infected PHHs a robust increase in CXCL-10 and CXCL-11 (∼104 normalized counts) mRNA, chemokines involved in the recruitment of inflammatory cells, and activated T cells34,36 (Figure 2E). The expression of genes coding for co-inhibitory and co-stimulatory molecules was also analyzed. Most co-inhibitory molecules were not detected, with only a minimal increased value (<500) of mRNA levels of LAG3 and CD274 (programmed death-ligand 1 [PD-L1]) in HBV/HDV-co-infected compared to HBV-mono-infected PHH cultures (Figure 2G). Across the co-stimulatory molecules analyzed, only CD40 mRNA expression was increased in HBV/HDV-co-infected hepatocytes (Figure 2H).
Notably, all of the genes analyzed related to antigen presentation and processing were clearly upregulated upon HDV infection in PHH (Figures 2F and 2I). Beta 2 microglobulin (B2M) and the proteasome subunits PSMB-8 and PSMB-9 reflected the largest mRNA count differences between HBV/HDV-co-infected and HBV-mono-infected PHHs. Differential upregulation in these 3 genes ranged between 4- and 7-fold. A similar pattern of gene expression changes was also confirmed in HepG2-hNTCP HBV/HDV-co-infected cells, although at a lower level (Figure S2).
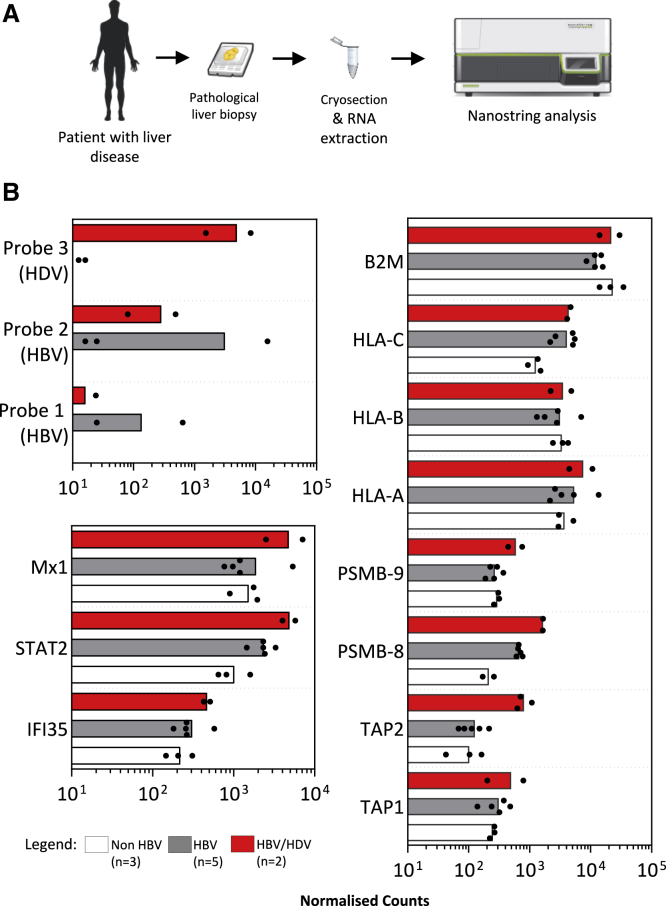
Increased Expression of Antigen Processing and Presentation Genes in HBV/HDV-Co-infected Human Liver Biopsies
To confirm our in vitro findings, we analyzed the liver biopsies of patients with hepatic diseases presenting different etiologies (HBV, hepatitis C virus [HCV], HDV chronic infection, and non-viral related alcoholic and non-alcoholic steatohepatitis [NASH] pathologies). Liver biopsy samples from 10 patients with chronic hepatitis/cirrhosis (3 without viral hepatitis, 5 HBV, and 2 HBV/HDV) that were previously embedded in an optimal cutting temperature (OCT) compound, were cryosectioned and tissues were extracted for RNA analysis using the NanoString human immunology panel supplemented with HBV and HDV RNA-specific probes (Figure 3A). The presence of HBV RNA in the HBV-infected livers and of HDV RNA in the two HDV/HBV-co-infected livers confirmed the ability of our method to assess the mRNA expression profile in cryopreserved liver specimens. Despite the fact that all of these hepatic tissues are derived from the liver biopsies of patients with ongoing inflammatory events (chronic active hepatitis or cirrhosis), the two HBV/HDV-infected livers displayed a pattern of gene expression showing a superior activation of type I IFN genes in comparison to non-HDV-infected livers. The level of HLA class I molecules did not differ between HDV- and non-HDV-infected livers. In contrast, the expression of genes involved in antigen processing (TAP1, TAP2, PSMB-8, and PSMB-9) were upregulated in HBV/HDV livers in comparison to mRNA levels detected in the liver biopsies of different etiologies (Figure 3B), even though the limited number of samples did not allow us to perform any statistical analysis.
Figure 3.
Expression Profile of Antigen Processing and Presentation Genes in Liver Biopsies
(A) Schematic experimental procedure of liver biopsy samples obtained from non-HBV-related (n = 3), HBV mono-infected- (n = 5), or HBV/HDV-co-infected patients (n = 2) stored under optimal cutting temperature (OCT) compound. OCT sections were subsequently cryosectioned, and RNA was extracted for analysis by NanoString.
(B) Bar graphs show the mean normalized counts of the indicated NanoString probes, including customized probes 1, 2 (HBV RNA), and 3 (HDV RNA), as annotated in Figure 1B, IFN-stimulated genes (bottom left panel), or genes involved in antigen processing and presentation (right panel). Normalized counts are expressed on a log10 scale, and each dot represents a single liver biopsy.
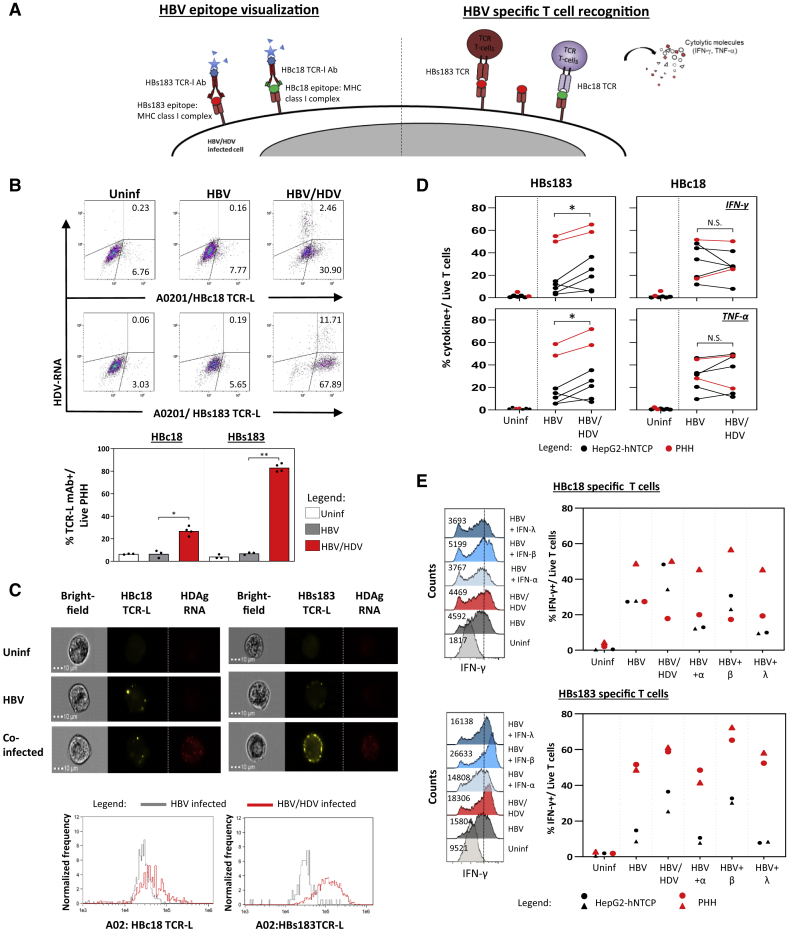
HBV/HDV Infection Increases HBV-Epitope Presentation
Next, we investigated whether the increased expression of genes associated with antigen processing and presentation had functional consequences. We quantified HBV-epitope/HLA class I complexes on HBV-mono-infected and HBV/HDV-infected PHHs by using antibodies specific for two HLA-A0201/HBV epitope complexes, HLA-A0201/HBc18-27 (HBc18 TCR-L monoclonal antibody [mAb]) and HLA-A0201/HBs183-91 (HBs183 TCR-L mAb) that were previously used in the liver biopsies of HLA-A0201+ CHB patients37 and in HBV-mono-infected hepatocytes in vitro.30,37 Notably, we recently demonstrated that the processing and presentation of these two different HBV epitopes is differentially regulated.30 While the HBc18 epitope was constitutively presented in HBV-infected hepatocytes, HBs183 epitope processing and presentation appear to require the induction of the immunoproteasome within the infected cells, since IFN-γ treatment of HBV mono-infected hepatocytes was required for its visualization.30 Here, we stained HBV-mono-infected and HBV/HDV-co-infected HLA-A0201 hepatocytes (Figures 4B and 4C) with the two TCR-L antibodies and HDV PrimeFlow RNA probes. Single-cell analysis performed with flow cytometry allowed us to decipher whether the HBV epitopes presentation was incremented preferentially by HDV replication within the HBV/HDV-infected cells or by the effect of cytokines released by HDV-infected cells in the cell culture media. Presentation of both HBV epitopes was clearly boosted by HDV infection; however, it was detected not only on HBV/HDV-infected hepatocytes but also on hepatocytes that were negative for HDV RNA (Figure 4B). A more robust effect was observed for the presentation of the HBs183 epitope (p = 0.001), even though HBc18 epitope presentation was also increased (p = 0.03). Direct visualization of HDV-infected cells confirmed the boosting of HLA-A0201/HBs183 complexes on HBV/HDV-infected cells appearing as punctuated structures lining the infected cells (Figure 4C). Thus, HDV infection boosts the presentation of HBV epitopes on HBV/HDV-co-infected hepatocytes and neighboring HBV-mono-infected cells.
Figure 4.
Quantification of HBV Epitope Presentation and HBV-Specific CD8 T Cell Activation
(A) Schematic of HBV epitope visualization using TCR-like antibodies (left) and of HBV-specific CD8 T cell activation (right).
(B) Representative dot plots of infected PHHs stained with antibodies specific for HLA-A0201/HBc18-27 (HBc18 TCR-L) and HLA-A0201/HBs183-91 (HBs183 TCR-L) complexes and with HDV PrimeFlow RNA probes. Bar graph below shows the frequency of indicated TCR-L Ab+ cells out of total PHHs. Each dot represents a single experiment (n = 3 for mock and HBV infected, n = 4 for HBV/HDV-infected PHHs).
(C) PHHs were stained with TCR-like mAb and HDV PrimeFlow RNA probes and then analyzed using ImageStream. Representative bright-field (left), yellow fluorescent (indicated TCR-L antibody, center), and red fluorescent (anti-HDAg RNA probes, right) images of respective PHHs (uninfected, HBV mono-infected, and HBV/HDV co-infected) are shown. The histograms below show the average geometric mean fluorescence intensities (MFIs) of the TCR-like mAb staining for the HBV- and HBV/HDV-infected PHHs.
(D) HBs183- and HBc18-specific CD8 T cells were tested against the indicated target cells (HepG2-NTCP and PHH) uninfected, infected with HBV, or infected with HBV/HDV. Frequency of IFN-γ+ CD8+, and TNF-α+ CD8+ T cells out of the total live T cells were determined by flow cytometry analysis after 5 h of incubation at an effector:target (E:T) ratio of 1:2.
(E) HBV-mono-infected PHHs were treated with 100 IU/mL IFN-α, 5 ng/mL IFN-β, or 100 ng/mL IFN-λ for 48 h and co-cultured for 5 h with HBc18- or HBs183-specific T cell clones and then stained for intracellular cytokines. Representative histogram plots of IFN-γ expression in CD8+ T cells co-cultured with uninfected, HBV-mono-infected, HBV/HDVco-infected, and IFN-treated HBV-mono-infected cells are shown.
The average frequency of IFN-γ+ CD8+T cells out of the total live T cells are shown in the bar graphs. Each dot represents a single experiment (n = 2 per cell type). ∗p = 0.01–0.05 and ∗∗p = 0.001–0.01. Non-significant p values are indicated as N.S.
See also Figure S3.
HDV Infection of HBV-Infected Hepatocytes Increases the Activation of HBV-Specific CD8 T Cells
To test directly the effect of HDV infection on HBV-specific CD8 T cell function, we co-cultured previously characterized38 HLA-A0201 restricted HBc18 and HBs183-specific CD8 T cells with the HBV-mono- and HBV/HDV-co-infected HLA-A0201+ target cells. After a 5 h incubation, T cell function was measured by intracellular cytokine staining (ICS) of T cells using anti-IFN-γ- and anti-TNF-α-specific antibodies (Figure 4A, right panel). The frequency of IFN-γ and TNF-α producing HBs183-specific CD8 T cells was significantly higher (p = 0.03) upon co-culture with HBV/HDV-infected PHHs or HepG2-hNTCP cells compared to HBV-mono-infected cells (Figure 4D, left panel). HBc18-specific CD8 T cell activation by HBV/HDV-infected cells was instead not significantly augmented in all of the experiments in comparison with HBV-mono-infected cells (Figure 4D, right panel). HDV infection of HBV-infected PHHs and HepG2-hNTCP cells does not suppress the activation of HBV-specific CD8 T cells, and in addition, could enhance the ability of specific CD8 T cells to recognize the immunoproteasome-dependent HBs183 epitope.
IFN-β Increases HBV Epitope Presentation
HDV infection triggers the production of IFN-β and IFN-λ in hepatocytes.31, 32, 33, 34, 35 The direct quantification of HBV epitopes on cells infected with HBV and HDV (Figures 4B and 4C) showed an increased HBV epitope presentation not only on co-infected hepatocytes but also on mono-infected cells. This suggests that antiviral cytokines triggered by HDV infection can boost the presentation of some HBV epitopes in a bystander fashion on HBV-mono-infected cells. To assess this possibility, we measured directly the ability of IFN-β and IFN-λ to boost HBV-specific CD8 T cell recognition of HBV-infected HLA-A0201+ hepatocytes and HepG2-hNTCP cells (Figure 4E). A titration assay using recombinant human IFN-α, IFN-β, and IFN-λ was performed on HepG2-hNTCP cells to gauge their ability to increase HLA class I expression (Figure S3). The doses of cytokines able to maximally increase HLA class I expression on HepG2-hNTCP cells (100 IU/mL IFN-α, 5 ng/mL IFN-β, and 100 ng/mL IFN-λ) were thereafter selected for use in CD8 T cell-recognition experiments. After the respective cytokine incubation, HBV-mono-infected PHHs and HepG2-hNTCP cells were co-cultured with HBc18- and HBs183-specific T cells. Their ability to produce IFN-γ or TNF-α was analyzed by ICS. Incubation of HBV-mono-infected PHHs and HepG2-hNTCP cells with all 3 types of IFNs had minimal effect on HBc18-specific CD8 T cell activation (Figure 4E, upper panel). In contrast, IFN-β- but not IFN-α- or IFN-λ-treated cells increased the activation of HBs183-specific CD8 T cells (Figure 4E, lower panel). These data confirmed previous evidence showing that HBs183 epitope processing is dependent on the activation of immunoproteasomes mediated by type I and II IFNs30 and show that IFN-β can boost the antigen presentation of selected HBV epitopes.
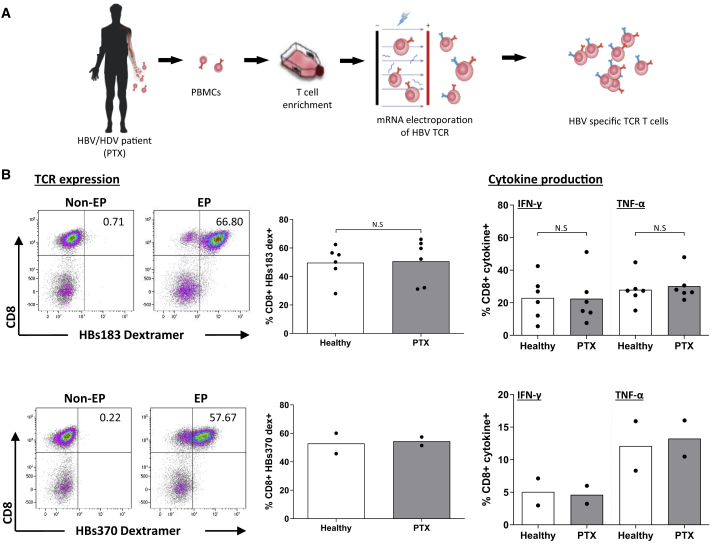
T Cells of Patients Chronically Co-infected with HBV/HDV Can Be Efficiently Used to Engineer HBV-Specific TCR-Redirected T Cells
To test whether the reconstitution of HBV-specific T cell responses through the adoptive transfer of HBV-specific TCR-redirected T cells could have therapeutic potential, we tested the ability of T cells obtained from 6 chronic HBV/HDV-co-infected patients to expand in vitro, express the HBs183 TCR, and produce cytokines after antigen-specific recognition (Figure 5A). These were compared with the T cells of 6 healthy individuals. Our results show that T cells of healthy controls and of chronically HBV/HDV-co-infected patients expressed the introduced TCRs at the same frequency and displayed similar levels of activation and cytokine production after antigen-specific recognition. The ability of T cells from HBV/HDV patients to be engineered into efficient HBV-specific TCR-redirected T cells was confirmed also with a different TCR specific for the HBs370-379 (HBs370) epitope (Figure 5B).
Figure 5.
Function and TCR Expression of HBV-Specific TCR-Redirected T Cells of Healthy and HBV/HDV Chronically Infected Subjects
(A) Schematic illustrating HBV-specific TCR T cells produced in vitro from HBV/HDV patient (PTX) PBMCs.
(B) Dot plots show the representative expression of indicated TCR on T cells of HBV/HDV-co-infected patients measured with the respective dextramers (left panel). The frequency of dextramer+ CD8+ T cells are indicated in the center panel. Graph bars depict the mean frequency of IFN-γ+ and TNF-α+ CD8+ cells out of the total CD8+ T cells obtained by ICS. HBV-TCR T cells were incubated with HLA-A02+ HepG2 cells pulsed or unpulsed with the corresponding peptide (HBV peptide 183-91 and HBV peptide 370-79).
Each dot represents a single individual (6 donors from each group; healthy control and HBV/HDV patients tested for HBs183 TCR-T cells and 2 donors from each group tested for HBs 370-TCR). Non-significant p values are indicated as N.S.
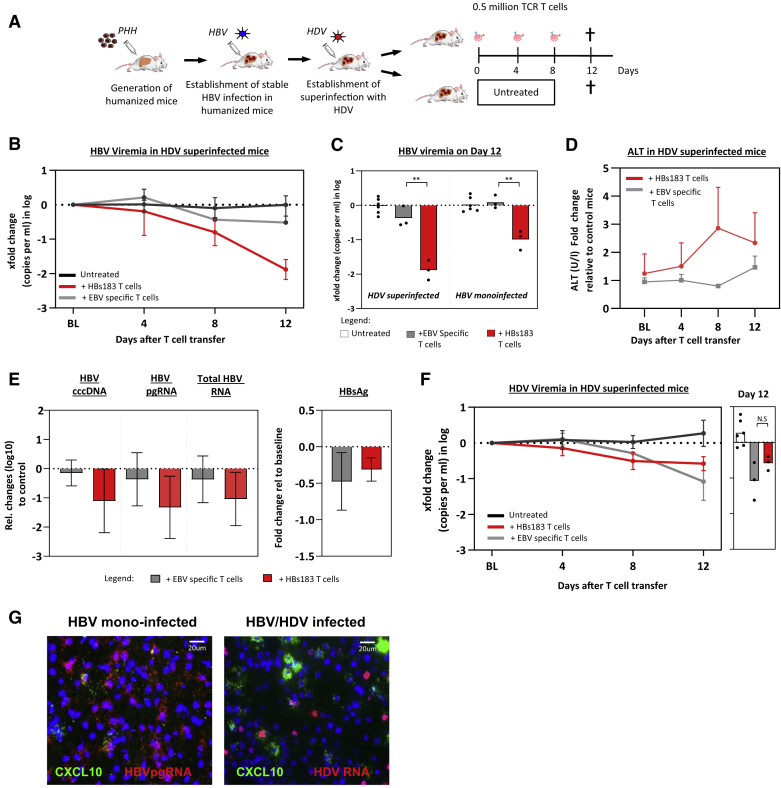
Antiviral Efficacy of TCR-Redirected T Cells In Vivo
We then tested in an animal model of HBV infection whether HDV modulates the antiviral efficacy of adoptive T cell therapy. We generated humanized chimeric mice reconstituted with HLA-A0201 hepatocytes and infected them with HBV and HDV viruses.39 The humanized mice were infected with HDV after having established stable HBV viremia titers (8–10 weeks post-HBV inoculation) (Figure 6A). T cell therapy was performed using TCR-redirected T cells engineered from T cells of a healthy individual, expanded in vitro with anti-CD3 and interleukin-2 (IL-2), and electroporated with mRNA coding for the HBs183-specific TCR or with the EBV-LMP2A 426-34 (EBV)-specific TCR. Both HBs183-TCR and EBV-TCR recognize their cognate epitope (HBs183-91 and LMP2A426-34) in association with HLA-A0201 molecules. A detailed description of the production of mRNA TCR-redirected T cells is described in the STAR Methods and was previously published.21,40 Note that due to ethical and practical restrictions, human hepatocytes and lymphocytes used to reconstitute the mice were not derived from the same donor but were matched based on presenting an HLA-A02 haplotype. HBV/HDV-infected mice received 3 doses of 0.5 million HBV-specific TCR T cells (n = 3) or EBV-specific TCR T cells (n = 3) at the indicated intervals (0, 4, and 8 days), while mice that were infected in parallel with similar viral titers were left untreated and served as controls (Figure 6B). A progressive reduction in HBV DNA was observed after the infusion of HBs183-specific TCR T cells with a mean reduction in HBV viremia of 1.9log at day 12 (Figure 6B). Notably, this was a 10-fold greater reduction in HBV viremia than what was observed in HBV-mono-infected mice (Figure 6C).21 Adoptive transfer of HBs183-specific TCR T cells in HBV/HDV-infected mice also caused a substantial reduction in intrahepatic covalently closed circular DNA (cccDNA) (mean 1.1log), HBV pregenomic RNA (pgRNA) (mean 1.3log), and total HBV RNA levels (mean 1.0log) in only 12 days of treatment (Figure 6E). We also observed a moderate decrease in circulating HBsAg levels in 3/3 mice receiving 3 infusions of HBs183-specific TCR T cells (0.3log decrease; Figure 6E). The antiviral effect of HBs183-specific TCR T cells in HBV/HDV-co-infected mice was linked with a mild elevation in ALT (up to 3-fold) in comparison to ALT levels determined over time in untreated matched control mice (Figure 6D).
Figure 6.
HDV/HBV Infection Increases the Antiviral Efficacy of HBs183-Specific TCR-Redirected T Cells In Vivo
(A) Schematic representation of the experimental design: human liver chimeric mice were stably infected with HBV and 10 weeks after with HDV. After 8 weeks of double infection, mice were treated with the indicated number of TCR T cells or left untreated.
(B) Median HBV viremia changes relative to baseline levels in HBV/HDV-co-infected mice at days 4, 8, and 12 after the first injection of HBs183-specific TCR T cells (n = 3), EBV-specific TCR T cells (n = 3), or untreated animals (n = 6).
(C) Bars represent the fold changes of HBV DNA viremia in the HBV-mono-infected or HBV/HDV-infected mice observed at day 12 (4 days after the last TCR-T cell infusion).
(D) Changes in ALT levels in HBV/HDV-co-infected mice after treatment with indicated TCR T cells in relation to day 0 levels (BL, baseline) (n = 3 mice per group).
(E) Fold changes of cccDNA, HBV pgRNA, total RNA, and HBsAg in the HBV/HDV-infected mice (n = 3 per group) observed 4 days after the last infusion of the indicated TCR-T cells.
(F) Median HDV viremia changes relative to baseline levels in HBV/HDV-co-infected mice after 3 injections of the indicated TCR T cells (n = 6 for untreated, n = 3 for both EBV- and HBs183-specific T cells).
(G) RNA in situ hybridization in liver tissues of humanized mice chronically infected with HBV (left) or HBV/HDV (right). HBV pgRNA (left) and antigenomic HDV RNA (right) are depicted in red and hCXCL10 is shown in green. Nuclei are stained with DAPI (blue).
Adoptive transfer of EBV-specific TCR-T cells also exerted a degree of antiviral effect against HBV. While the reduction of cccDNA (0.15log) was not triggered by the adoptive transfer of EBV-TCR T cells (Figure 6E), we observed a moderate decrease in HBV viremia (mean 0.5log), intrahepatic HBV RNA (0.4log), and HBsAg in 2/3 HBV/HDV-co-infected mice (mean 0.48log reduction; Figures 6B and 6E). HDV viremia was also lowered by TCR T cell transfer, but without a substantial difference between the adoptive transfer of HBV-TCR T cells and EBV-TCR T cells (mean 0.6 and 1.0log, respectively; Figure 6F). This antiviral effect of adoptively transferred T cells independent of TCR specificity was unexpected and likely due to an alloreactive T cell activation that is detected only in HBV/HDV-co-infected mice. The adoptive transfer of T cells engineered with a non-HBV-specific TCR did not cause any alteration in HBV virological parameters in comparison to non-treated mice in HBV-mono-infected mice (Figure 6C).21
The differential effect of activated T cells engineered with a non-HBV-specific TCR confirmed the ability of HDV to modify the liver environment and to increase, through the upregulation of inflammatory chemokines, the intrahepatic recruitment and activation of specific and also alloreactive T cells. This was confirmed using RNA in situ hybridization technology showing a more pronounced enhancement of human chemokines such as CXCL10 (Figure 6G) in human hepatocytes of HBV/HDV-infected mice in comparison to HBV-mono-infected mice. These in vivo data corroborate the findings observed in our NanoString analysis, which showed a robust upregulation of CXCL10 transcripts in HBV/HDV-infected hepatocytes (Figure 2E). These data show that HDV co-infection in vivo increases the function of adoptively transferred engineered T cells.
Discussion
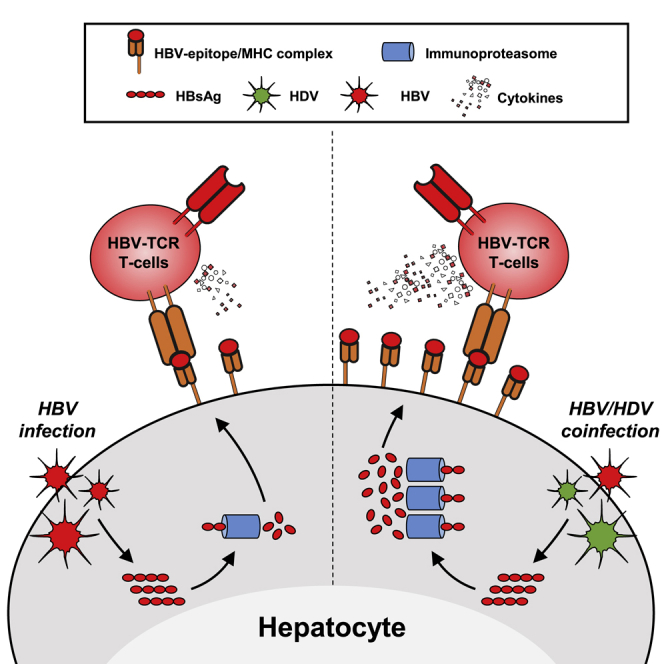
Viruses that establish chronic infection usually develop strategies to dampen host immunity. HDV appears to use a different strategy. In addition to its known ability to activate IFN responses,31, 32, 33, 34, 35 we demonstrated here that HDV increases the efficiency of the antigen processing and presentation machinery of the infected and neighboring hepatocytes. By collecting data in in vitro HDV infection systems, liver biopsies, and mice with humanized livers, we showed that HDV infection not only enhances the gene expression of HLA class I molecules, B2M, immunoproteasome, and co-stimulatory molecules genes but it also increases the presentation of viral epitopes, and as a direct consequence, the efficiency of T cell recognition of infected hepatocytes.
The enhanced HLA class I presentation of viral epitopes could explain the high incidence of escape mutations found in HDV epitopes,28 but it can have a different effect in HBV. The compact nature of the HBV DNA genome poses a limit on the generation of mutated HBV viruses, and this makes HBV unlikely to generate virus progeny able to escape HBV-specific CD8 T cells. This can therefore increase the chances to control HBV in HBV/HDV-co-infected individuals. Epidemiological evidence supports this hypothesis: a longitudinal study of HBV-mono- and HBV/HDV-infected patients demonstrated that within 4 years, 10% of the dual-infected patients became HBsAg−, a much higher rate than in HBV-mono-infected patients, in whom only 3% lost serum HBsAg.41 Furthermore, HBV clearance in HBV/HDV-co-infected patients was associated with an increased number of HDV antigen-expressing hepatocytes,42 a historical observation that supports the concept that HDV can act as an adjuvant for the immunological control of HBV. Nevertheless, this positive outcome occurs only in a minority of HBV/HDV-co-infected patients,43 since most adult CHB patients have severe functional impairment of HBV-specific adaptive immunity.9
Consequently, in the presence of a low frequency of HBV-specific CD8 T cells, the increased activation of intrahepatic T cells would not result in more chances of HBV clearance, but only in increased liver inflammation. This scenario explains the higher intrahepatic inflammatory levels and more severe liver pathology frequently present in patients with HDV/HBV co-infection in comparison to those harboring only HBV.3 We think, however, that the ability of HDV to boost hepatic antigen presentation could, apart from the described pathogenic effects, be used to increase the therapeutic efficacy of T cell-based therapies in HBV/HDV-co-infected patients. T cell therapy with lymphocytes modified with CAR or with HLA class I-restricted TCR is starting to be considered as a therapeutic option not only for cancer but also for chronic viral diseases.44 We have recently shown that the adoptive transfer of HBV-specific TCR-redirected T cells in a patient with HBsAg+ HCC relapses drastically reduced the HBsAg values.45 The data provided in this work, showing that HDV infection of HBV-infected humanized chimeric mice increased the anti-HBV activity of adoptively transferred HBV-TCR redirected T cells, encourage the use of HBV-TCR T cells in HBV/HDV-co-infected patients. This possibility is further supported by the demonstration that the T cells of HBV/HDV chronically co-infected patients are amenable to TCR-T cell engineering and can be expanded in vitro to reach a functionality that is identical to that of healthy subjects.
Although the overall decrease in HBV and HDV viral loads observed in our experiments was objectively limited, we used a low number of human T cells (only 0.5 million HBV-TCR T cells per mouse) that only transiently expressed the virus-specific TCRs. This strategy was selected to limit inflammatory responses in the liver.21 Nevertheless, increasing the number of infused lymphocytes or using treatment strategies that combine the use of TCR T cells with a blocker of re-infection, such as neutralizing antibodies or entry inhibitors (i.e., Myrcludex B), could increase the antiviral efficacy of such therapies.25
A caveat of using a T cell therapy-targeting HBV to address the problem of HDV co-infection is the fact that HDV can persist in non-HBV-infected hepatocytes46 and endure cell division.26 Therefore, HDV-mono-infected cells will not be targeted by HBV-specific TCR T cells, unless HBV envelope proteins are produced from HBV DNA integrations into the host genome. However, the pathological impact of the presence of clusters of HDV-only-infected cells in the natural history of HBV/HDV chronic infection is still unknown.
In summary, by demonstrating that HDV acts as an immunogenic adjuvant in HBV-infected hepatocytes, we unveil a previously unknown feature of HDV that can explain its ability to increase liver inflammation in HBV/HDV-co-infected patients. In addition, we show that its ability to boost HBV epitope presentation increases the efficacy of T cell-based therapy in a in vivo mouse model of HBV/HDV infection. We believe that our findings broaden the therapeutic strategies that can be applied to HBV/HDV-co-infected patients, who are still the only patients with hepatotropic infections without any US Food and Drug Administration (FDA)-approved therapy.5
Limitations of Study
This work describes experiments done in vitro and in humanized chimeric mouse models that demonstrate the ability of HDV to increase the antigen presentation of HBV epitopes and HBV-specific T cell activation. Whether HDV could enhance the efficacy of HBV-specific TCR T cells in the treatment of chronic HBV/HDV infection needs to be directly tested in patients. The humanized chimeric mouse model has limitations in the measurement of the efficacy of TCR T cell therapy: there is a partial HLA match between human hepatocytes and human T cells, a situation that is different from the proposed therapy in which patients will be treated with autologous lymphocytes.
STAR★Methods
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Pacific Orange live/dead stain | Invitrogen | L34959 |
| Anti-human HBcAg | Abcam | #7841 |
| Anti-human HBsAg | Raybiotech | #MD050186 |
| Goat-anti-rabbit CF555 | Sigma Aldrich | SAB46000068 |
| Goat-anti-mouse APC | Life Technologies | AB_2536211 |
| Goat-anti-mouse PE | Life Technologies | AB_2539848 |
| HBs183-191 TCR-like antibody | Sastry et al., 2011 | N/A |
| HBc18-27 TCR-like antibody | Sastry et al., 2011 | N/A |
| Anti-human CD107a FITC | BD Biosciences | 560949 |
| Anti-human CD8 V500 | BD Horizon | 560774 |
| Anti-human IFN-γ PeCy7 | eBiosciences | AB_469682 |
| Anti-human TNF-α PE | BD Bioscience | 559321 |
| HBs183-191 PE pentamer | ProImmune | WB3290-PE |
| HBs370 PE pentamer | ProImmune | WB4073-PE |
| EBV PE pentamer | ProImmune | WB2144-PE |
| Biological Samples | ||
| Human pathological liver tissue | Asian American Liver Centre, Gleneagles Hospital | https://www.aamg.co/liver/ |
| Human HBV/HDV patient blood | Fonsazione IRCCS Ca’Granda Ospedale Maggiore Policlinico | https://www.policlinico.mi.it/ |
| Human healthy blood | Health Sciences Authority | https://www.hsa.gov.sg/ |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Recombinant Human IL-2 | R&D systems | 202-1L-050 |
| Anti-CD3 | eBioscience | AB_468854 |
| Ficoll-Paque PLUS | GE Healthcare | 17-1440-02 |
| BD Cytofix/Cytoperm Fixation | BD Bioscience | 51-2090 KZ |
| Recombinant Human IFN-α2a | PBL | 11100-1 |
| Recombinant Human IFN-β | Peprotech | 300-02BC |
| Recombinant Human IL-29/IFN-λ1 | R&D systems | 1598-IL-025 |
| Critical Commercial Assays | ||
| Primeflow assay with customized probeset | Invitrogen | EB 16488 |
| Nanostring human immunology version 2 and customized probes assay | Nanostring technologies | XT-CSO-HIM2-12 |
| FASER PE amplification kit | Miltenyi | 130-091-764 |
| One Shot Top10 E. coli kit | Invitrogen | C404003 |
| QIAamp minELute virus spin kit | QIAGEN | 57704w |
| Rneasy Mini kit | QIAGEN | 74004 |
| mMESSAGE mMACHINE T7 Ultra kit | Ambion | AM1345 |
| Nucelofactor kit | Lonza | V4XP-3024 |
| Human serum albumin ELISA quantification kit | Bethyl Laboratories | E88-129 |
| RNAScope Fluorescent Multiplex kit | Advanced Cell Diagnostics | 320850 |
| HBV Artus kit | QIAGEN | 4506265 |
| Quantitative HBsAg kit | Abbott Architect Platform | 6C36 |
| Experimental Models: Cell Lines | ||
| Primary human hepatocytes (Fresh) | Invitrocue | 5M-000S |
| Primary human hepatocytes (Cryopreserved) | Lonza | HUM4012 |
| HepG2-hNTCP cells | Laboratory of Stephan Urban | N/A |
| HepG2 cells | ATCC | HB-8065 |
| Huh7-hNTCP cells | Laboratory of Camile Sureau | N/A |
| HepAD38 cells | Laboratory of Christoph Seeger | N/A |
| HBV HBc18-27 and HBs183-191 T cell clones | 38 | N/A |
| Experimental Models: Organisms/Strains | ||
| uPA/SCID/ILγR2 (USG) mice | 21 | N/A |
| Oligonucleotides | ||
| HDV forward and reverse primers | 49 | N/A |
| RNA in situ primers | 34 | N/A |
| Recombinant DNA | ||
| pSVLD-3 plasmid | Laboratory of Camile Sureau | Addgene: #29335 |
| pT7HB2.7 plasmid | N/A | |
| pVAC-HBs183 and -HBs370 | 50,51 | N/A |
| Software and Algorithms | ||
| FACSDiva | BD Bioscience | https://www.bdbiosciences.com/en-us/instruments/research-instruments/research-software/flow-cytometry-acquisition/facsdiva-software |
| GraphPad Prism 8 | GraphPad | https://www.graphpad.com/scientific-software/prism/ |
| IDEAS software | Amnis | https://www.luminexcorp.com/imagestreamx-mk-ii/ |
| Kaluza software | Beckman Coulter | https://www.beckman.com/flow-cytometry/software/kaluza |
| nSolver version 4.0 | Nanostring techonologies | https://www.nanostring.com/products/analysis-software/nsolver |
Resource Availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Antonio Bertoletti (antonio@duke-nus.edu.sg).
Materials availability
This study did not generate new, unique reagents but other specific reagents used in this study are available from the Lead Contact with a complete Materials Transfer Agreement.
Data and code availability
This study did not generate any unique datasets or code.
Experimental Model and Subject Details
Cell lines
HepG2-hNTCP cells (HepG2 cells transduced with human NTCP (a gift from Dr Stephan Urban, Heidelberg) were cultured in DMEM medium supplemented with 10% heat-inactivated fetal bovine serum (FBS), 100 IU/ml penicillin, 100 μg/ml streptomycin, Glutamax (Invitrogen). HepG2-hNTCP cells were selected using 5mg/ml puromycin (BD Biosciences). Huh7 cells overexpressing hNTCP used for HDV viral production were cultured in William’s medium E supplemented with 10% FBS and selected using 250ug/ml gentamicin (Sigma-Aldrich). HepAD38 cells used for HBV viral production were cultured in DMEM with 10% tetracycline-free FBS, 100U/ml penicillin/streptomycin. 2mM L-glutamine and 0.4mg/ml doxycycline. Two HLA-A0201 CD8 T cells specific for HBc18-27 and HBs183-191 were generated from HLA-A0201+ patients with acute hepatitis infection as described previously47. CD8+ T cell lines were cultured for 10 to 15 days in AIM-V+2% human AB serum supplemented with 20 IU/ml recombinant human IL-2 and 10ng/ml IL-7 and IL-15 before functional test against different targets.
Human subjects and primary cell cultures
Peripheral blood mononuclear cells (PBMCs) from chronic HBV/HDV infected patients (average age: 55.5 ± 14.8 years old, 33% females and 67% males) and were collected under informed consent from Fondazione IRCCS Ca’Granda Ospedale Maggiore Policlinico at the University of Milan. PBMCs from healthy donors were reviewed and approved for use by the institutional review board of the Health Sciences Authority. PBMCs were isolated by gradient centrifugation using Ficoll-plaque and frozen in FBS (Life Technologies) supplemented with 10% DMSO (Sigma Aldrich) until analysis.
Pathological human liver biopsy samples from patients with different advanced cirrhosis and/or HCC etiologies were kept frozen in OCT (VWR chemicals) before RNA extraction. Tissues were obtained from living donor liver transplantations (Asian American Liver Centre, Gleneagles Hospital, Singapore). All clinical samples were collected with patient consent under the Declaration of Helsinki and local research ethics committee approval. Tissues were sectioned (5mm), and OCT was removed as much as possible before lysis in buffer RLT following manufacturer’s instructions for RNA extraction (QIAGEN). 100ng of total RNA was used for Nanostring analysis.
Primary human hepatocyte (PHH) culture. Fresh HLA-A0201 PHH were obtained from humanized FRG mouse (Invitrocue). PHH were cultured in Hepacur media containing 2% DMSO, according to the commercial vendor’s instructions.
Animals
Human liver chimeric uPA/SCID/ILγR2 (USG) mice were generated by transplanting 1 million thawed human hepatocytes from one donor with haplotype HLA-A0201(Lonza) into 3 to 4 week-old mice anesthetized with isofluorane. Mice were housed under specific pathogen-free conditions in accordance with the European Communities Council Directive (86/EEC) and protocols approved by the Ethical Committee of the city and state of Hamburg. Levels of human liver chimerism were determined 8-10 weeks after transplantation by measuring human serum albumin (HSA) levels in mouse serum using the human serum albumin ELISA quantification kit according to manufacturer’s recommendations.
Method Details
HBV Virus production
HBV (genotype D) was derived from HepAD38 cells, which harbor a tetracycline (tet)-inducible more-than-genomic-length genomic integrate48. Cell culture supernatants without antibiotic selection were collected for virus precipitation beginning on day 20 after the removal of tetracycline. Virus supernatants were concentrated as described using a commercial polyethylene glycol kit (Abcam). HBV viral titers were quantified using HBV DNA. HBV DNA was extracted from viral supernatants using the QIAamp minElute virus spin kit according to the manufacturer’s instructions. HBV titers were then measured using the QIAGEN HBV Artus PCR kit.
HDV virus production
HDV was produced in HuH7-hNTCP cells by transfection of two plasmids pSVLD3 (HDV genotype 1) and pT7HB2.7 (HBV genotype D) to produce infectious HDV particles. Virus supernatants were collected from day 13 post-transfection and clarified by centrifugation. HDV viral titers were measured using HDV RNA extracted first using the QIAamp viral RNA mini kit then quantified by reverse transcription and quantitative real-time PCR. Briefly, 5 ul RNA was denatured at 95°C for 10 min, immediately cooled down at −20°C and reverse transcribed at 50°C for 5 min with HDV specific primers and probes. After inactivation of the reverse transcriptase at 95°C for 20 s, amplification was performed under the following conditions: 40 cycles at 95°C for 3 s and 60°C for 30 s HDV viremia was measured using the Superscript III One-step RT-PCR system (Invitrogen), published HDV specific primers and probes on a Lightcycler 480 (Roche)49. Known amounts of HDV containing plasmids were used as standards for quantification.
HBV/HDV viral infection
PHH were treated with 2% DMSO 24 hours prior to HBV infection. After DMSO treatment, approximately multiplicities of genome equivalents (GE) of 3000/well HBV in medium containing 2% DMSO and 4% PEG for 24 hours. After which the infection inoculum was removed, cells were washed three times with PBS. For HDV infection, HBV infected cells were then inoculated with approximate multiplicities of genome equivalent of 500/well, 2% DMSO and 4% PEG. After 24 hours, the HDV inoculum was removed and washed as mentioned above. Infected hepatocytes were then cultured for 7 days, with media change every other day. HepG2-hNTCP cells were infected in the same manner, without DMSO pre-treatment.
Nanostring
Custom probesets specific to two regions across the HBV genotype D genome and one region in the HDV antigen were designed by Nanostring. 20,000 cells were harvested from the respective infected PHH and lysed using RLT buffer and β-mercaptoethanol for each analysis. Gene expression was quantified together with the nCounter Human Immunology panel V2 and customized probes according to the manufacturer’s instructions. Raw data were then analyzed using the nSolver version 4.0.
IFNs treatment of PHH and HepG2-hNTCP cells
For interferon treatment conditions, mono-HBV infected PHH were treated as follows: IFN-α (100 IU/ml), IFN-β (5ng/ml) and IFN-λ (100ng/ml) for 48 hours. Cells were then washed 3 times with 1x PBS before co-culture experiments.
Intracellular HBV antigen staining
Infected PHH and HepG2-hNTCP cells were trypsinized, quenched with media followed by live/dead Pacific Orange (Invitrogen) staining. Cells were fixed and permeabilized using BD Cytofix/Cytoperm, then stained with primary antibody of rabbit anti-HBcAg or mouse anti-HBsAg for 30 minutes on ice, followed by secondary antibodies goat anti-rabbit-CF555 and goat anti-mouse-APC for 30 minutes on ice. All acquisitions were made using BD LSR-II flow cytometer and data analyzed in Kaluza (Beckman Coulter) software unless otherwise stated.
HBV epitope/HLA-A0201 complex coupled with PrimeFlow RNA staining
HLA-A0201 TCR like (TCR-L) monoclonal antibody specific to HBs183 or HBc18 were used to quantify the cell surface expression of HBV peptide/HLA-A0201 specific complexes37. PHH were first stained using live/dead Pacific Orange followed by HBV TCR-L antibodies at 1:100 for 1 hour on ice then secondary staining with goat anti-mouse PE at 1:100 dilution. After this staining step, FASER PE amplification was carried out; three rounds of amplification for HBs183 and four rounds of amplification for HBc18. Cells were then prepared according to the manufacturer’s instructions for the PrimeFlow RNA assay. Briefly, cells were fixed and permeabilized, followed by target probe hybridization. The signal of the target probe was amplified using type I probes to be acquired. Data acquisition was performed using the Amnis Imagestream and BD LSR-II flow cytometer. Data were analyzed in the IDEAS and Kaluza software, respectively.
Intracellular cytokine staining
HBV specific CD8+ T cells were incubated with infected PHH at 1:2 effector to target ratio in the presence of 10mg/ml Brefeldin-A and CD107a-FITC for 5 hours. Following which T cells were stained for live/dead Pacific Orange, surface stained for CD8-V500 then fixed and permeabilized using BD Cytofix/Cytoperm. Intracellular cytokine staining using IFN-γ-PE-Cy7 and TNF-α-PE was then carried out for 30 minutes on ice. HepG2 cell positive controls were peptide-pulsed with 1 μg/ml of the respective peptide for 1 hour at 37°C prior to the co-culture with T cells.
Production of TCR mRNA
Virus-specific TCRs were cloned in pVAX1 and propagated from Escherichia coli using the One Shot Top10 E. coli kit purified using the QIAGEN Endo Free plasmid maxi kit (QIAGEN). TCR Plasmids were linearized using the Xbal restriction enzyme (ThermoFisher). TCR mRNA was then produced in vitro using the mMESSAGE mMACHINE T7 Ultra kit as previously described50.
Preparation of engineered T cells via mRNA electroporation
HBs183-191 and HBc18-27 TCR mRNA as well as EBV-specific TCR mRNA were produced using in vitro transcription. A detailed report describing the selection of HBV-specific T cells and the cloning of their corresponding TCRs is described51. Activated T cells for mRNA TCR electroporation were derived from PBMCs stimulated with 600 IU/ml IL-2 and 50 ng/ml anti-CD3 in AIM-V supplemented with 2% human AB serum for 8 days, and rIL-2 was increased to 1000 IU/ml 1 day before electroporation. For mRNA electroporation 1 × 107 activated T cells were suspended in 100μl Cell Line Nucleofector Solution V and TCR mRNA was added at 20μg. The mixture was placed in a certified cuvette and electroporated using the transduction program “X-001” present in the nucleofector device 2B. After electroporation, cells were resuspended in AIM-V supplemented with 10% human AB serum and 100 IU/ml IL-2 at 37°C and 5% CO2 until analysis. T cells expressing the introduced TCR were quantified by flow cytometry using the specific HLA class I pentamers (HLA-A0201/HBs183-PE pentamer, HLA-A0201/HBs370-PE pentamer or HLA-A0201/EBV LMP2a426-34-PE pentamer).
In vivo HBV infection and T cell administration
Generation of stably HBV-infected human liver chimeric mice was performed as previously reported21. Briefly, animals received a single intraperitoneal injection of HBV-infectious serum (1 × 107 HBV DNA copies/mouse; genotype D). HBV-infected mice displaying high levels of viremia (≥1 × 109 HBV DNA copies/ml) were used for the adoptive transfer of engineered human T cells. To establish HBV/HDV infection, animals received one single intraperitoneal injection of serum obtained from mice stably infected with HBV (1x107 HBV DNA genome equivalents, genotype D) and 10 weeks later they were superinfected with HDV (1x108 genome equivalents/mouse, genotype 3, cell culture-derived).
Human T cells isolated from PBMCs (HLA-A0201) were expanded in vitro and electroporated as described in the previous section. After analysis of TCR expression on T cells, 0.5 × 106 activated HBs183 specific T cells, corresponding to an approximately 1:40 E:T ratio, were injected in the HBV/HDV co-infected mouse three times at 4 days intervals. Blood was withdrawn at indicated times. Liver specimens removed at sacrifice (12 days after the first T cell injection) were snap-frozen in 2-methyl butane for histological and molecular analyses.
Analysis of biochemical and virological parameters
ALT was measured using the Roche Cobas c111 System (Roche). Due to the limited amount of serum that can be obtained from the mouse model during the treatment phase, 5μl of serum was used for the longitudinal measurement. DNA was extracted from liver specimens using the Master Pure DNA purification kit (Epicenter) and RNA was isolated using the RNeasy RNA purification kit. Intrahepatic virological parameters (HBV DNA and HDV RNA) were quantified using specific primers and probes as previously described39,49. Briefly, viral nucleic acids were extracted from mouse serum samples using the QiAmp MinElute Virus spin kit and liver tissue by using the RNeasy Micro and Mini RNA purification kit (QIAGEN), respectively. HDV viremia was determined via reverse transcription and qRT-PCR using the ABI Fast 1-Step Virus Master (Applied Biosystems) on an ABI Viia7 (Applied Biosystems)49,52. The plasmid pBluescript II SK(þ), containing one copy of the HDV genome, was used as a standard for HDV cDNA quantification39. cccDNA levels were determined in the whole cell liver DNA extracted by Hirt, after digestion with plasmid-safe ATP-dependent DNase (PSD), T5 exonuclease or a combination of Exonuclease I and II as previously described53.
RNA in situ hybridization
RNA in situ hybridization was performed on paraformaldehyde-fixed, cryo-preserved liver sections using the RNAScope Fluorescent Multiplex Kit according to the manufacturer’s instructions. Briefly, liver sections were dehydrated and pretreated with Pretreat 4 (Pretreatment Kit, ACD) for 30 min. Samples were incubated with pregenomic HBV RNA (GT-D, assay number 442741), antigenomic HDV RNA (assay number 433471) and hCXCL10 RNA (assay number 311851-C2) specific target probes for 2 h at 40°C (HybEZ oven, ACD). DAPI staining was performed to visualize nuclei. Stained sections were analyzed by fluorescence microscopy using a 60 × /1.40 NA oil objective. Merged z stack images were prepared using the same settings for all groups.
Quantification and Statistical Analysis
Analysis of the Amnis Imagestream and BD LSR-II flow cytometer data were analyzed in IDEAS and Kaluza software respectively. Statistical significance were determined by paired, two-tailed, parametric Student’s t test for Figures 1, 2, 3, 4, and 5. Figure 6 was analyzed using non-paired, two tailed, parametric Student’s t test. P values of < 0.05 were considered statistically significant. Data was plotted using Graphpad Prism 8 software as mean values. Asterisks indicate ∗ p = 0.01-0.05 and ∗∗ p = 0.001-0.01.
Acknowledgments
This work was supported by a Singapore Translational Research Investigator Award of the Singapore Ministry of Health’s National Medical Research Council MOH-000019 (MOH-StaR17Nov-0001), to A.B., and the German Research Foundation (DFG; SFB 841 A5, A8) and the German Center for Infection Research (DZIF-BMBF; TTU-hepatitis 05.815 and 05.816), to M.D. and M.L. We would like to thank all of the patients from the Asian American Liver Centre, Gleneagles Hospital, Singapore, for the liver biopsy samples, and the Fondazione IRCCS Ca’ Granda Ospedale Maggiore Policlinico at the University of Milan; healthy donors at the Health Sciences Authority; Dr Silvia Giovanelli and Dr Elena Trombetta for peripheral blood mononuclear cell (PBMC) isolation and preservation management of HDV samples; and Roswitha Reusch and Corinna Eggers for their technical support with the in vivo experiments.
Author Contributions
A.B. and M.D. designed the study. A.B., M.D., and M.L. provided the funding. C.Y.L.T. and A.T.T. designed and performed the in vitro experiments and analyzed the data. J.K., T.V., and Y.L. designed and performed the in vivo studies. M.L. and K.G. designed and performed the HDV analysis and analyzed the data. A.C. performed the in vitro experiments and provided technical support. A.L., M.B., and P.L. selected and provided the clinical samples and contributed to the analysis of in vitro experiments. C.S. provided technical information for HDV production and in vitro HDV infection. A.B., C.T., and M.D. wrote the manuscript.
Declaration of Interests
A.B. is a co-founder of LION TCR, a biotech company developing T cell receptors for the treatment of virus-related cancers and chronic viral diseases. P.L. advises and is on the speakers’ bureaus for Janssen, MYR Pharmaceuticals, GlaxoSmithKline, Gilead Sciences, AbbVie, Roche, Eiger, Alnylam, Bristol-Myers Squibb, and Merck Sharp & Dohme. A.L. is on the speakers’ bureaus of MYR Pharmaceuticals and Gilead. The other authors declare no competing interests.
Published: July 21, 2020
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.xcrm.2020.100060.
Supplemental Information
References
- 1.Sureau C., Negro F. The hepatitis delta virus: replication and pathogenesis. J. Hepatol. 2016;64(1, Suppl):S102–S116. doi: 10.1016/j.jhep.2016.02.013. [DOI] [PubMed] [Google Scholar]
- 2.Chen H.-Y., Shen D.-T., Ji D.-Z., Han P.-C., Zhang W.-M., Ma J.-F., Chen W.-S., Goyal H., Pan S., Xu H.-G. Prevalence and burden of hepatitis D virus infection in the global population: a systematic review and meta-analysis. Gut. 2019;68:512–521. doi: 10.1136/gutjnl-2018-316601. [DOI] [PubMed] [Google Scholar]
- 3.Wedemeyer H. Re-emerging interest in hepatitis delta: new insights into the dynamic interplay between HBV and HDV. J. Hepatol. 2010;52:627–629. doi: 10.1016/j.jhep.2010.02.001. [DOI] [PubMed] [Google Scholar]
- 4.Rizzetto M. Targeting Hepatitis D. Semin. Liver Dis. 2018;38:66–72. doi: 10.1055/s-0037-1621711. [DOI] [PubMed] [Google Scholar]
- 5.Koh C., Heller T., Glenn J.S. Pathogenesis of and New Therapies for Hepatitis D. Gastroenterology. 2019;156:461–476.e1. doi: 10.1053/j.gastro.2018.09.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bazinet M., Pântea V., Cebotarescu V., Cojuhari L., Jimbei P., Albrecht J., Schmid P., Le Gal F., Gordien E., Krawczyk A. Safety and efficacy of REP 2139 and pegylated interferon alfa-2a for treatment-naive patients with chronic hepatitis B virus and hepatitis D virus co-infection (REP 301 and REP 301-LTF): a non-randomised, open-label, phase 2 trial. Lancet Gastroenterol. Hepatol. 2017;2:877–889. doi: 10.1016/S2468-1253(17)30288-1. [DOI] [PubMed] [Google Scholar]
- 7.Yurdaydin C., Keskin O., Kalkan Ç., Karakaya F., Çalişkan A., Karatayli E., Karatayli S., Bozdayi A.M., Koh C., Heller T. Optimizing lonafarnib treatment for the management of chronic delta hepatitis: the LOWR HDV-1 study. Hepatology. 2018;67:1224–1236. doi: 10.1002/hep.29658. [DOI] [PubMed] [Google Scholar]
- 8.Wedemeyer H., Schöneweis K., Bogomolov P.O., Voronkova N., Chulanov V., Stepanova T., Bremer B., Allweiss L., Dandri M., Burhenne J. GS-13-Final results of a multicenter, open-label phase 2 clinical trial (MYR203) to assess safety and efficacy of myrcludex B in cwith PEG-interferon Alpha 2a in patients with chronic HBV/HDV co-infection. J. Hepatol. 2019;70:e81. [Google Scholar]
- 9.Fanning G.C., Zoulim F., Hou J., Bertoletti A. Therapeutic strategies for hepatitis B virus infection: towards a cure. Nat. Rev. Drug Discov. 2019;18:827–844. doi: 10.1038/s41573-019-0037-0. [DOI] [PubMed] [Google Scholar]
- 10.Gehring A.J., Protzer U. Targeting Innate and Adaptive Immune Responses to Cure Chronic HBV Infection. Gastroenterology. 2019;156:325–337. doi: 10.1053/j.gastro.2018.10.032. [DOI] [PubMed] [Google Scholar]
- 11.Thimme R., Wieland S., Steiger C., Ghrayeb J., Reimann K.A., Purcell R.H., Chisari F.V. CD8(+) T cells mediate viral clearance and disease pathogenesis during acute hepatitis B virus infection. J. Virol. 2003;77:68–76. doi: 10.1128/JVI.77.1.68-76.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Maini M.K., Boni C., Lee C.K., Larrubia J.R., Reignat S., Ogg G.S., King A.S., Herberg J., Gilson R., Alisa A. The role of virus-specific CD8(+) cells in liver damage and viral control during persistent hepatitis B virus infection. J. Exp. Med. 2000;191:1269–1280. doi: 10.1084/jem.191.8.1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lau G.K., Lok A.S., Liang R.H., Lai C.L., Chiu E.K., Lau Y.L., Lam S.K. Clearance of hepatitis B surface antigen after bone marrow transplantation: role of adoptive immunity transfer. Hepatology. 1997;25:1497–1501. doi: 10.1002/hep.510250631. [DOI] [PubMed] [Google Scholar]
- 14.Ilan Y., Nagler A., Adler R., Naparstek E., Or R., Slavin S., Brautbar C., Shouval D. Adoptive transfer of immunity to hepatitis B virus after T cell-depleted allogeneic bone marrow transplantation. Hepatology. 1993;18:246–252. [PubMed] [Google Scholar]
- 15.Salimzadeh L., Le Bert N., Dutertre C.-A., Gill U.S., Newell E.W., Frey C., Hung M., Novikov N., Fletcher S., Kennedy P.T., Bertoletti A. PD-1 blockade partially recovers dysfunctional virus-specific B cells in chronic hepatitis B infection. J. Clin. Invest. 2018;128:4573–4587. doi: 10.1172/JCI121957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Burton A.R., Pallett L.J., McCoy L.E., Suveizdyte K., Amin O.E., Swadling L., Alberts E., Davidson B.R., Kennedy P.T.F., Gill U.S. Circulating and intrahepatic antiviral B cells are defective in hepatitis B. J. Clin. Invest. 2018;128:4588–4603. doi: 10.1172/JCI121960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Boni C., Fisicaro P., Valdatta C., Amadei B., Di Vincenzo P., Giuberti T., Laccabue D., Zerbini A., Cavalli A., Missale G. Characterization of hepatitis B virus (HBV)-specific T-cell dysfunction in chronic HBV infection. J. Virol. 2007;81:4215–4225. doi: 10.1128/JVI.02844-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kurktschiev P.D., Raziorrouh B., Schraut W., Backmund M., Wächtler M., Wendtner C.M., Bengsch B., Thimme R., Denk G., Zachoval R. Dysfunctional CD8+ T cells in hepatitis B and C are characterized by a lack of antigen-specific T-bet induction. J. Exp. Med. 2014;211:2047–2059. doi: 10.1084/jem.20131333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schuch A., Salimi Alizei E., Heim K., Wieland D., Kiraithe M.M., Kemming J., Llewellyn-Lacey S., Sogukpinar Ö., Ni Y., Urban S. Phenotypic and functional differences of HBV core-specific versus HBV polymerase-specific CD8+ T cells in chronically HBV-infected patients with low viral load. Gut. 2019;68:905–915. doi: 10.1136/gutjnl-2018-316641. [DOI] [PubMed] [Google Scholar]
- 20.Bohne F., Chmielewski M., Ebert G., Wiegmann K., Kürschner T., Schulze A., Urban S., Krönke M., Abken H., Protzer U. T cells redirected against hepatitis B virus surface proteins eliminate infected hepatocytes. Gastroenterology. 2008;134:239–247. doi: 10.1053/j.gastro.2007.11.002. [DOI] [PubMed] [Google Scholar]
- 21.Kah J., Koh S., Volz T., Ceccarello E., Allweiss L., Lütgehetmann M., Bertoletti A., Dandri M. Lymphocytes transiently expressing virus-specific T cell receptors reduce hepatitis B virus infection. J. Clin. Invest. 2017;127:3177–3188. doi: 10.1172/JCI93024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bertoletti A., Le Bert N. Immunotherapy for Chronic Hepatitis B Virus Infection. Gut Liver. 2018;12:497–507. doi: 10.5009/gnl17233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Koh S., Kah J., Tham C.Y.L., Yang N., Ceccarello E., Chia A., Chen M., Khakpoor A., Pavesi A., Tan A.T. Nonlytic Lymphocytes Engineered to Express Virus-Specific T-Cell Receptors Limit HBV Infection by Activating APOBEC3. Gastroenterology. 2018;155:180–193.e6. doi: 10.1053/j.gastro.2018.03.027. [DOI] [PubMed] [Google Scholar]
- 24.Kruse R.L., Shum T., Tashiro H., Barzi M., Yi Z., Whitten-Bauer C., Legras X., Bissig-Choisat B., Garaigorta U., Gottschalk S., Bissig K.-D. HBsAg-redirected T cells exhibit antiviral activity in HBV-infected human liver chimeric mice. Cytotherapy. 2018;20:697–705. doi: 10.1016/j.jcyt.2018.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wisskirchen K., Kah J., Malo A., Asen T., Volz T., Allweiss L., Wettengel J.M., Lütgehetmann M., Urban S., Bauer T. T cell receptor grafting allows virological control of Hepatitis B virus infection. J. Clin. Invest. 2019;129:2932–2945. doi: 10.1172/JCI120228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Giersch K., Bhadra O.D., Volz T., Allweiss L., Riecken K., Fehse B., Lohse A.W., Petersen J., Sureau C., Urban S. Hepatitis delta virus persists during liver regeneration and is amplified through cell division both in vitro and in vivo. Gut. 2019;68:150–157. doi: 10.1136/gutjnl-2017-314713. [DOI] [PubMed] [Google Scholar]
- 27.Karimzadeh H., Kiraithe M.M., Oberhardt V., Salimi Alizei E., Bockmann J., Schulze Zur Wiesch J., Budeus B., Hoffmann D., Wedemeyer H., Cornberg M. Mutations in Hepatitis D Virus Allow It to Escape Detection by CD8+ T Cells and Evolve at the Population Level. Gastroenterology. 2019;156:1820–1833. doi: 10.1053/j.gastro.2019.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kefalakes H., Koh C., Sidney J., Amanakis G., Sette A., Heller T., Rehermann B. Hepatitis D Virus-Specific CD8+ T Cells Have a Memory-Like Phenotype Associated With Viral Immune Escape in Patients With Chronic Hepatitis D Virus Infection. Gastroenterology. 2019;156:1805–1819.e9. doi: 10.1053/j.gastro.2019.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hoh A., Heeg M., Ni Y., Schuch A., Binder B., Hennecke N., Blum H.E., Nassal M., Protzer U., Hofmann M. Hepatitis B Virus-Infected HepG2hNTCP Cells Serve as a Novel Immunological Tool To Analyze the Antiviral Efficacy of CD8+ T Cells In Vitro. J. Virol. 2015;89:7433–7438. doi: 10.1128/JVI.00605-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Khakpoor A., Ni Y., Chen A., Ho Z.Z., Oei V., Yang N., Giri R., Chow J.X., Tan A.T., Kennedy P.T. Spatiotemporal Differences in Presentation of CD8 T Cell Epitopes during Hepatitis B Virus Infection. J. Virol. 2019;93:e01457-18. doi: 10.1128/JVI.01457-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang Z., Filzmayer C., Ni Y., Sültmann H., Mutz P., Hiet M.-S., Vondran F.W.R., Bartenschlager R., Urban S. Hepatitis D virus replication is sensed by MDA5 and induces IFN-β/λ responses in hepatocytes. J. Hepatol. 2018;69:25–35. doi: 10.1016/j.jhep.2018.02.021. [DOI] [PubMed] [Google Scholar]
- 32.Alfaiate D., Lucifora J., Abeywickrama-Samarakoon N., Michelet M., Testoni B., Cortay J.-C., Sureau C., Zoulim F., Dény P., Durantel D. HDV RNA replication is associated with HBV repression and interferon-stimulated genes induction in super-infected hepatocytes. Antiviral Res. 2016;136:19–31. doi: 10.1016/j.antiviral.2016.10.006. [DOI] [PubMed] [Google Scholar]
- 33.He W., Ren B., Mao F., Jing Z., Li Y., Liu Y., Peng B., Yan H., Qi Y., Sun Y. Hepatitis D Virus Infection of Mice Expressing Human Sodium Taurocholate Co-transporting Polypeptide. PLoS Pathog. 2015;11:e1004840. doi: 10.1371/journal.ppat.1004840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Giersch K., Allweiss L., Volz T., Helbig M., Bierwolf J., Lohse A.W., Pollok J.M., Petersen J., Dandri M., Lütgehetmann M. Hepatitis Delta co-infection in humanized mice leads to pronounced induction of innate immune responses in comparison to HBV mono-infection. J. Hepatol. 2015;63:346–353. doi: 10.1016/j.jhep.2015.03.011. [DOI] [PubMed] [Google Scholar]
- 35.Suárez-Amarán L., Usai C., Di Scala M., Godoy C., Ni Y., Hommel M., Palomo L., Segura V., Olagüe C., Vales A. A new HDV mouse model identifies mitochondrial antiviral signaling protein (MAVS) as a key player in IFN-β induction. J. Hepatol. 2017;67:669–679. doi: 10.1016/j.jhep.2017.05.010. [DOI] [PubMed] [Google Scholar]
- 36.Cole K.E., Strick C.A., Paradis T.J., Ogborne K.T., Loetscher M., Gladue R.P., Lin W., Boyd J.G., Moser B., Wood D.E. Interferon-inducible T cell alpha chemoattractant (I-TAC): a novel non-ELR CXC chemokine with potent activity on activated T cells through selective high affinity binding to CXCR3. J. Exp. Med. 1998;187:2009–2021. doi: 10.1084/jem.187.12.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sastry K.S.R., Too C.T., Kaur K., Gehring A.J., Low L., Javiad A., Pollicino T., Li L., Kennedy P.T.F., Lopatin U. Targeting hepatitis B virus-infected cells with a T-cell receptor-like antibody. J. Virol. 2011;85:1935–1942. doi: 10.1128/JVI.01990-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tan A.T., Loggi E., Boni C., Chia A., Gehring A.J., Sastry K.S.R., Goh V., Fisicaro P., Andreone P., Brander C. Host ethnicity and virus genotype shape the hepatitis B virus-specific T-cell repertoire. J. Virol. 2008;82:10986–10997. doi: 10.1128/JVI.01124-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lütgehetmann M., Mancke L.V., Volz T., Helbig M., Allweiss L., Bornscheuer T., Pollok J.M., Lohse A.W., Petersen J., Urban S., Dandri M. Humanized chimeric uPA mouse model for the study of hepatitis B and D virus interactions and preclinical drug evaluation. Hepatology. 2012;55:685–694. doi: 10.1002/hep.24758. [DOI] [PubMed] [Google Scholar]
- 40.Koh S., Shimasaki N., Bertoletti A. Redirecting T Cell Specificity Using T Cell Receptor Messenger RNA Electroporation. Methods Mol. Biol. 2016;1428:285–296. doi: 10.1007/978-1-4939-3625-0_19. [DOI] [PubMed] [Google Scholar]
- 41.Niro G.A., Gravinese E., Martini E., Garrubba M., Facciorusso D., Conoscitore P., Di Giorgio G., Rizzetto M., Andriulli A. Clearance of hepatitis B surface antigen in chronic carriers of hepatitis delta antibodies. Liver. 2001;21:254–259. doi: 10.1034/j.1600-0676.2001.021004254.x. [DOI] [PubMed] [Google Scholar]
- 42.Negro F., Baldi M., Bonino F., Rocca G., Demartini A., Passarino G., Maran E., Lavarini C., Rizzetto M., Verme G. Chronic HDV (hepatitis delta virus) hepatitis. Intrahepatic expression of delta antigen, histologic activity and outcome of liver disease. J. Hepatol. 1988;6:8–14. doi: 10.1016/s0168-8278(88)80457-4. [DOI] [PubMed] [Google Scholar]
- 43.Romeo R., Del Ninno E., Rumi M., Russo A., Sangiovanni A., de Franchis R., Ronchi G., Colombo M. A 28-year study of the course of hepatitis Δ infection: a risk factor for cirrhosis and hepatocellular carcinoma. Gastroenterology. 2009;136:1629–1638. doi: 10.1053/j.gastro.2009.01.052. [DOI] [PubMed] [Google Scholar]
- 44.Bertoletti A., Tan A.T. Challenges of CAR- and TCR-T cell-based therapy for chronic infections. J. Exp. Med. 2020;217:e20191663. doi: 10.1084/jem.20191663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Qasim W., Brunetto M., Gehring A.J., Xue S.-A., Schurich A., Khakpoor A., Zhan H., Ciccorossi P., Gilmour K., Cavallone D. Immunotherapy of HCC metastases with autologous T cell receptor redirected T cells, targeting HBsAg in a liver transplant patient. J. Hepatol. 2015;62:486–491. doi: 10.1016/j.jhep.2014.10.001. [DOI] [PubMed] [Google Scholar]
- 46.Giersch K., Helbig M., Volz T., Allweiss L., Mancke L.V., Lohse A.W., Polywka S., Pollok J.M., Petersen J., Taylor J. Persistent hepatitis D virus mono-infection in humanized mice is efficiently converted by hepatitis B virus to a productive co-infection. J. Hepatol. 2014;60:538–544. doi: 10.1016/j.jhep.2013.11.010. [DOI] [PubMed] [Google Scholar]
- 47.Gehring A.J., Sun D., Kennedy P.T.F., Nolte-’t Hoen E., Lim S.G., Wasser S., Selden C., Maini M.K., Davis D.M., Nassal M., Bertoletti A. The level of viral antigen presented by hepatocytes influences CD8 T-cell function. J. Virol. 2007;81:2940–2949. doi: 10.1128/JVI.02415-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ladner S.K., Otto M.J., Barker C.S., Zaifert K., Wang G.H., Guo J.T., Seeger C., King R.W. Inducible expression of human hepatitis B virus (HBV) in stably transfected hepatoblastoma cells: a novel system for screening potential inhibitors of HBV replication. Antimicrob. Agents Chemother. 1997;41:1715–1720. doi: 10.1128/aac.41.8.1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ferns R.B., Nastouli E., Garson J.A. Quantitation of hepatitis delta virus using a single-step internally controlled real-time RT-qPCR and a full-length genomic RNA calibration standard. J. Virol. Methods. 2012;179:189–194. doi: 10.1016/j.jviromet.2011.11.001. [DOI] [PubMed] [Google Scholar]
- 50.Koh S., Shimasaki N., Suwanarusk R., Ho Z.Z., Chia A., Banu N., Howland S.W., Ong A.S.M., Gehring A.J., Stauss H. A practical approach to immunotherapy of hepatocellular carcinoma using T cells redirected against hepatitis B virus. Mol. Ther. Nucleic Acids. 2013;2:e114. doi: 10.1038/mtna.2013.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Banu N., Chia A., Ho Z.Z., Garcia A.T., Paravasivam K., Grotenbreg G.M., Bertoletti A., Gehring A.J. Building and optimizing a virus-specific T cell receptor library for targeted immunotherapy in viral infections. Sci. Rep. 2014;4:4166. doi: 10.1038/srep04166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Homs M., Giersch K., Blasi M., Lütgehetmann M., Buti M., Esteban R., Dandri M., Rodríguez-Frias F. Relevance of a full-length genomic RNA standard and a thermal-shock step for optimal hepatitis delta virus quantification. J. Clin. Microbiol. 2014;52:3334–3338. doi: 10.1128/JCM.00940-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Allweiss L., Volz T., Giersch K., Kah J., Raffa G., Petersen J., Lohse A.W., Beninati C., Pollicino T., Urban S. Proliferation of primary human hepatocytes and prevention of hepatitis B virus reinfection efficiently deplete nuclear cccDNA in vivo. Gut. 2018;67:542–552. doi: 10.1136/gutjnl-2016-312162. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
This study did not generate any unique datasets or code.