Summary
Fibrosis, or the accumulation of extracellular matrix, is a common feature of many chronic diseases. To interrogate core molecular pathways underlying fibrosis, we cross-examine human primary cells from various tissues treated with TGF-β, as well as kidney and liver fibrosis models. Transcriptome analyses reveal that genes involved in fatty acid oxidation are significantly perturbed. Furthermore, mitochondrial dysfunction and acylcarnitine accumulation are found in fibrotic tissues. Substantial downregulation of the PGC1α gene is evident in both in vitro and in vivo fibrosis models, suggesting a common node of metabolic signature for tissue fibrosis. In order to identify suppressors of fibrosis, we carry out a compound library phenotypic screen and identify AMPK and PPAR as highly enriched targets. We further show that pharmacological treatment of MK-8722 (AMPK activator) and MK-4074 (ACC inhibitor) reduce fibrosis in vivo. Altogether, our work demonstrate that metabolic defect is integral to TGF-β signaling and fibrosis.
Keywords: fibrosis, metabolism, fatty acid oxidation, TGF-β, PGC1α, MK-8722, MK-4074, AMPK, PPAR
Graphical Abstract

Highlights
Profiling of multiple fibrosis models reveals a common fatty acid oxidation defect
Downregulation of PGC1α expression in both in vitro and in vivo fibrosis models
Compound library screen identifies AMPK and PPAR as enriched targets for fibrosis
MK-8722 and MK-4074 treatment reduces liver fibrosis in a NASH model
Fibrosis, also known as scarring of the tissue, is a common feature of many chronic diseases. Zhang et al. combine transcriptome profiling, metabolomics, metabolic flux analyses, and high-throughput compound library phenotypic screening to show that metabolic defect is a common feature of tissue fibrosis.
Introduction
Despite recent advances in drug development, cardiovascular and metabolic diseases remain the leading cause of death and disability, posing a huge public health burden globally.1 The biggest challenge today is to develop novel therapies that modify disease progression and improve patient outcome, while retaining a superior safety profile for chronic treatment. Many of the drugs currently used in the clinic reduce cardiometabolic risk factors, including hypertension, hyperglycemia, and hyperlipidemia.2, 3, 4, 5, 6 However, risk factor management alone is insufficient to prevent end-stage organ damage, as many patients still progress to end-stage renal disease (ESRD), nonalcoholic steatohepatitis (NASH) or heart failure, even while on intensive blood pressure control or euglycemic treatment.7, 8, 9, 10 A deeper understanding of the complex biology underlying these disease associations would greatly help the development of newer transformative therapies.
Notably, most of the progressive forms of cardiometabolic diseases are characterized by the presence of extensive fibrosis.11,12 Fibrosis is an evolutionarily conserved mechanism developed by an organism to survive chronic injury.13 Excessive fibrosis, however, leads to disruption of organ function and is a common feature of many chronic diseases. In the heart, fibrosis contributes to systolic and diastolic dysfunction and correlates with the incidence of arrhythmia and cardiac death events.14 In the kidney, tubulointerstitial fibrosis is a hallmark of ESRD and is strongly associated with renal function decline in patients with chronic kidney disease (CKD).15, 16, 17 A recent study demonstrated that genetic ablation of a subset of fibroblasts in mice leads to a reduction of cardiac fibrosis and an improvement of cardiac function,18 providing exciting preclinical evidence that an anti-fibrotic therapy could be used as a disease-modifying agent for end-stage organ damage control. There are many promising therapies currently being developed for fibrosis.19, 20, 21 Several agents have demonstrated proof-of-concept fibrosis reduction in humans, including Obeticholic acid, Cenicriviroc, NGM282, and Liraglutide.22, 23, 24, 25, 26 Those findings have greatly stimulated interest in developing more anti-fibrotic drugs.
Many studies have investigated the molecular mechanisms underlying fibrosis and a wide range of targets have been tested for drug discovery and clinical development.11,13,16,28 Regardless of disease etiology, fibrosis in the heart, lung, kidney, liver, and skin share many similar features, such as increases in myofibroblast activation, collagen production, and inflammation, suggesting the existence of common core pathways or mechanisms among all fibrotic tissues. We sought to interrogate these core pathways and capitalized on the use of multiple orthogonal molecular and pharmacological approaches, including genome-wide transcriptome analysis, metabolomics, metabolic flux analyses, and high-throughput phenotypic screening. The results we obtained through profiling several in vitro and in vivo fibrosis models have provided strong experimental support that common metabolic defects are integral to fibrosis development in multiple tissue types.
Results
Genome-wide Transcriptome Profiling Reveals a Close Link between Metabolic Defects and TGF-β Response
Among many factors that regulate fibrosis, transforming growth factor β (TGF-β) plays an essential role in both myofibroblast activation and extracellular matrix remodeling;29 it has often been used to induce fibrotic response in cultured cells. To obtain insights into the pathways and mechanisms modulating TGF-β, we carried out genome-wide transcriptome analysis in a variety of human primary cells treated with TGF-β, including cardiac fibroblasts, lung fibroblasts (NHLFs, normal human lung fibroblasts), hepatic stellate cells (HSCs), and renal proximal tubular epithelial cells (RPTECs) (Figure 1A; GEO: GSE152250). RT-PCR analysis of fibrotic marker genes, such as α-smooth muscle actin (α-SMA), PAI-1, and Collagen 1A1, in these cells revealed a robust response to TGF-β, which was almost completely reversed by the presence of SB-525334, a TGF-β type I receptor (ALK5) inhibitor (Figure 1B; Figure S1A). At 6, 24, and 48 h after the TGF-β treatment, human primary cells were collected for RNA sequencing (RNA-seq) analysis. Among the four cell types, many genes were significantly changed upon TGF-β treatment across the fold change cutoff values, with a small set of genes identified as common genes in all cells. At the 24 h time point, there were a total of 1,603 genes identified in all four cell types that met the ±1.2-fold change and false discovery rate (FDR) <0.1 selection criteria (Table S1A–C). Their relative expression levels compared to vehicle treatment are shown in a heatmap (Figure 1C). Many collagen isoforms and inflammation-related gene were drastically changed upon TGF-β treatment (Figures S1B and S1C). Interestingly, Gene Ontology (GO) term enrichment analysis of the 1,603 genes revealed that dysregulated fatty acid metabolism was a highly conserved feature in all cell types (Figure 1D; Table S1D). To delineate this further, schematic diagram of fatty acid metabolic pathways as well as enzymes involved in each reaction were illustrated in Figure 1E. The gene expression ratios (+/−TGF-β) of each enzyme were indicated by colored squares in a Log2 scale. Many fatty acid β-oxidation related genes were substantially decreased upon TGF-β treatment in various cell types, including isoforms of the rate-limiting enzyme Carnitine Palmitoyltransferse 1 (CPT1) (Figure 1E; Table S1E). Transcription factors regulating fatty acid oxidation, such as peroxisome proliferator-activated receptors (PPARα/γ), peroxisome proliferator-activated receptor gamma coactivator 1α (Ppargc1a or PGC1α), and PGC1β were also suppressed by TGF-β (Figure 1E, inset). In particular, downregulation of PGC1α by TGF-β was so profound in lung fibroblasts, which was partially reversed by the addition of SB-525334 (Figure S1D), suggesting a direct TGF-β-mediated effect. Our results found that metabolic defect could be a common feature of TGF-β-induced fibrotic response.
Figure 1.
Transcriptome Analysis Identifies a Common Metabolic Signature in Human Primary Cells Treated with TGF-β
(A) Schematic overview of RNA-seq analyses of four human primary cell types with TGF-β treatment. 3 independent replicates were used for each time point collection.
(B) RT-PCR analysis of αSMA gene expression in human primary cells upon TGF-β treatment (5 ng/mL for 24 h), which was almost completely reversed by the presence of ALK5 inhibitor, SB-525334 (10 μM). n = 3, Mean ± SD.
(C) Common effects of TGF-β across four cell types. Shown are the 1,603 genes that met the ±1.2-fold change and FDR_BH p < 0.1 threshold at 24 h in all four cell types. The color gradient represents fold change compared to vehicle-treated cells (−3.0 to 3.0-fold).
(D) GO term enrichment analysis of the 1,603 genes using the PANTHER enrichment test (http://pantherdb.org).
(E) Gene expression ratios of fatty acid oxidation enzymes in human primary cells and fibrotic tissues. From left: cardiac fibroblasts, hepatic stellate cells (HSCs), NHLF, RPTEC, UUO kidney, CCl4 liver, and BDL liver. Heatmap was generated by using Morpheus software (https://software.broadinstitute.org/morpheus/). The color gradient represents fold change compared treatment (TGF-β 24 h, surgery, or CCl4 treatment) to control (−8.5 to 8.5-fold). Gray, below detection limit. ∗Peroxisomal enzymes.
Metabolic Defects in Multiple Tissue Fibrosis Models
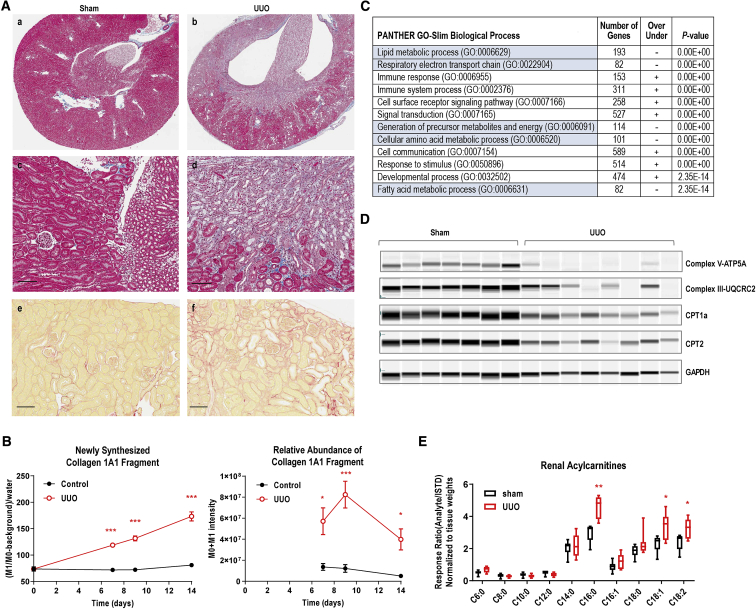
To explore this molecular signature in vivo, we examined a number of fibrotic animal models. Unilateral ureteral obstruction (UUO) is a well-established model of renal fibrosis.30 Complete ureter ligation resulted in tubular dilation and atrophy, inflammatory cell infiltration, and tubulointerstitial fibrosis in 7 days (Figure 2A, a–d). Automated imaging analysis showed a massive increase of Sirius red staining in UUO kidneys (Figure 2A, e and f), indicative of collagen accumulation. We previously reported a method to measure newly synthesized collagen in mouse liver by using stable isotope labeling and targeted peptide quantitation by liquid chromatography-mass spectrometry (LC-MS).31 While comparable stable isotope labeling was achieved in Sham and UUO mice, there was a dramatic increase in newly synthesized Collagen 1A1 fragment (GAAGPpGATGFpGAAGR) in UUO kidneys at day 7 following the surgery (Figure 2B, left panel), as well as its relative abundance (Figure 2B, right panel). The synthesis of collagen remained unchanged in sham-operated kidneys but was accelerated in UUO kidneys, consistent with histological observations of collagen accumulation. Our biochemical analysis also found substantial activation of TGF-β signaling in UUO kidneys, as measured by Smad3 nuclear translocation and downstream gene expression (Figures S2A and S2B).
Figure 2.
Downregulation of Fatty Acid Oxidation in UUO Kidneys
(A) Histological analyses of normal (sham) and UUO kidneys at day 9. (a–d) trichrome staining revealed renal pelvis and calyces blunting, cortical and medullary atrophy, tubular dilation, tubular epithelial cell apoptosis/necrosis, inflammatory cell infiltration, and diffuse interstitial fibrosis in UUO kidney. (e and f) Sirius red staining. Representative images taken from n = 6∼9 animal/group. Scale bar, 100 μm.
(B) Increased newly synthesized collagen in UUO kidneys as measured by 2H-water labeling and proteomic analyses. Collagen 1A1 fragment GAAGPPGATGFPGAAGR was shown. Left panel, newly synthesized; right panel, relative abundance. Mean ± SEM (time point: day 0, n = 3/group; days 7 and 9, and 14, n = 4 for Sham, and n = 7 for UUO). ∗p < 0.05, ∗∗∗p < 0.005, Student t test.
(C) GO term enrichment analysis of the 5,031 genes shown in Figure S2C, using the PANTHER enrichment test. Shown here are the top 12 biological process terms with metabolic-related terms highlighted in blue. n = 5 per treatment group or time point for RNA-seq analysis.
(D) The protein expression of key fatty acid oxidation regulators by Sally Sue simple western.
(D) Acylcarnitine profile of Sham (n = 6) versus UUO (n = 9) kidneys at day 10 following the surgery. Min to Max, ∗p < 0.05, ∗∗p < 0.01, Student t test.
To obtain insights into mechanisms underlying fibrosis, we carried out whole-genome transcriptome analysis in UUO kidneys at days 3 and 9 after the surgery. Among a total of 46,603 genes sequenced, 5,031 genes were dysregulated at least 1.5-fold (up or down compared to sham animals) with FDR <0.1 at both time points (Figure S2C; Table S1F–H; GEO: GSE152250). From the heatmap analysis, changes at the transcriptional level were comparable at both day 3 and day 9, while fibrosis remained progressive at day 9, suggesting that the injury response occurs immediately after the insult and persists over a prolonged period of time. To dissect out the pathways that were most significantly dysregulated in UUO kidneys, we next performed GO term enrichment analysis of the 5,031 genes. The top GO biological process terms are shown in Figure 2C, while the full list is shown in Table S1I. GO terms significantly enriched in the signature included lipid metabolism, mitochondrial respiratory electron transport chain, and fatty acid metabolism genes, resonating with the metabolic signature observed in human primary cells treated with TGF-β (Figure 1D). Moreover, gene expression ratios of fatty acid oxidation-related enzymes were found markedly decreased in UUO kidneys compared to sham-operated animals (Figure 1E; Table S1J). Substantial reduction of PGC1α gene was also observed in UUO kidneys (Figure 1E, inset; Figure S2E), suggesting a common metabolic signature underlying fibrosis from mouse to human.
To further probe metabolic defects in this model, we next examined the expression of key lipid metabolism players at the protein level. As shown in Figure 2D, mitochondrial electron transport chain proteins, ATP synthase V (ATP5A) of complex V, as well as complex III component UQCRC2, was almost completely abrogated in UUO kidneys, suggesting that mitochondrial oxidative phosphorylation machinery was not operational. CPT1a and CPT2 were also reduced in UUO kidneys, which are consistent with gene expression results and confirmed the existence of defective fatty acid oxidation. Furthermore, we carried out LC-MS analysis of acylcarnitines in total kidney extract (Figure 2E). While short- to medium-chain acylcarnitines (C6:0, C8:0, C10:0, C12:0, and C14:0) were comparable in sham and UUO kidneys, the more abundant long-chain acylcarnitines (C16:0, C16:1, C18:0, C18:1, and C18:2) were substantially upregulated in UUO kidneys. The acylcarnitine accumulation was consistent with defects in CPT1/2 and mitochondrial oxidative phosphorylation, confirming the existence of fatty acid oxidation block in UUO kidneys. PGC1α is an important regulator of mitochondrial biogenesis and oxidative phosphorylation.32, 33, 34 Inhibition of PGC1α likely contributes to the mitochondrial dysfunction observed in UUO kidneys.
We next examined molecular signatures of other fibrosis models. Two commonly used liver fibrosis models are carbon tetrachloride (CCl4)-induced hepatocyte injury and bile duct ligation (BDL)-induced portal inflammation and fibrosis.35 Chronic administration of CCl4 in mice led to marked hepatocellular necrosis, fatty degeneration, inflammatory cell infiltration, and fibrous tissue proliferation over the course of 9 weeks (Figure 3A). Collagen deposition was profoundly increased at 7 weeks as indicated by the quantification of trichrome staining (Figure 3B). To further characterize the temporal regulation of fibrosis in vivo, we used stable isotope-labeling technology and monitored collagen synthesis over the duration of the study. A substantial increase in Collagen 1A1 synthesis was detected as early as 1 week after the administration of CCl4 (Figure 3C). The production of other collagen isoforms were also elevated (data not shown). In addition to the CCl4 model, we carried out BDL surgery on rats to induce liver fibrosis. 5 weeks after the surgery, animals developed significant bile duct proliferation, hypertrophy of sinusoidal lining cells, and extensive peribiliary-interstitial collagen deposition (Figures 3E and 3F). Collagen 1A1 synthesis was augmented from 3 weeks after the surgery (Figure 3G). Additionally, we performed whole-genome transcriptome analysis on fibrotic livers from these two models (GEO: GSE152250). GO term enrichment analysis revealed an enrichment of fatty acid metabolic-related genes (Figures 3D and 3H; Figures S3A and S3C). Fatty acid oxidation genes and PGC1α were significantly downregulated in fibrotic livers, regardless of the cause of injury (Figure 1E; Table S1K–O and P–T).
Figure 3.
Defective Metabolic Signature in Liver Fibrosis Models
(A) Masson’s trichrome staining of mouse liver found significant collagen deposition around pericentral areas at week 7 following CCl4 treatment. (a and b) Lower magnification; (c and d), higher magnification. Scale bar, 100 μm.
(B) Quantitative analysis of trichrome positive area from whole-liver scan. Mean ± SEM, n = 5, ∗∗∗p < 0.005, Student t test.
(C) Increased newly synthesized collagen in CCl4-treated liver as measured by 2H-water labeling and proteomic technology. Collagen 1A1 fragment DGLNGLpGPIGppGPR was shown. Mean ± SEM, n = 4∼5, ∗p < 0.05, ∗∗∗p < 0.005, Student t test.
(D) RNA-seq analyses of livers from vehicle or CCl4-treated mice at week 5. GO term enrichment analysis revealed metabolic-related terms. n = 5 per treatment group.
(E) Masson’s trichrome staining of rat liver found extensive peribiliary and interstitial collagen deposition at week 5. (a and b) lower magnification; (c and d) higher magnification. Scale bar, 100 μm.
(F) Quantitative analysis of trichrome positive area from whole-liver scan. Mean ± SEM, n = 5, Student t test, ∗∗∗p < 0.005.
(G) Increased newly synthesized collagen in bile duct-ligated liver as measured by 2H-water labeling and proteomic analyses. Collagen 1A1 fragment DGLNGLpGPIGppGPR was shown. Mean ± SEM, n = 5∼6, ∗∗∗p < 0.005, Student t test.
(H) RNA-seq analyses of livers from sham-operated or bile duct-ligated rats at week 4. GO term enrichment analysis revealed metabolic-related terms. n = 5 per treatment group.
We also carried out gene expression analysis of fibrotic lungs in a bleomycin-induced lung injury mouse model.36 At 21 days after bleomycin instillation, mouse lungs developed multifocal inflammatory cell infiltration and fibrosis. Modified Ashcroft lung fibrosis score was dramatically increased, as well as αSMA-positive myofibroblasts (Figures S3E and S3F). Key lipid metabolism genes PGC1α and PPARα were also significantly reduced in fibrotic lungs (Figure S3G). Together, cross-examination of in vitro and in vivo fibrosis models over multiple tissue and cell types revealed a similar metabolic defect, suggesting that this is likely a common pathway involved in the development of fibrosis.
Fibrosis Phenotypic Screen Identifies Metabolic Modulators
As part of a major effort to identify new suppressors for fibrosis, we carried out high-throughput phenotypic screens against an annotated compound library as previously described.37 Normal human lung fibroblasts were treated with TGF-β and subsequently incubated with 3H-proline to monitor collagen production (Figure 4A). The potency of suppressors was determined by 3-point dose titration. After the primary screen, the anti-fibrotic effect of selected compounds was verified by an αSMA imaging assay. Among the top-ranked compounds that were found to inhibit fibrosis, PPAR and AMPK (Adenosine Monophosphate-activated Protein Kinase) were identified as highly enriched targets. The potency of a PPARγ agonist MRL-24,38,39 a PPARδ agonist GW501506,40 and a pan-AMPK activator MK-872241,42 on TGF-β-induced collagen production is shown in Figure 4B. Furthermore, we examined the expression of fibrosis marker genes in lung fibroblasts following MK-8722 treatment. As shown in Figure 4C, MK-8722 inhibited αSMA, PAI-1, and Collagen 1A1 (COL1A1) with IC50 of 1.87, 70.7, and 12.6 μM, respectively. In RPTECs, we measured type I pro-collagen level in cell-culture supernatant. After stimulating with TGF-β, cells have increased procollagen level and MK-8722 treatment was able to suppress the production of secreted procollagen similar to that of the ALK5 inhibitor SB-525334 (Figure 4D). To determine whether modulating fatty acid oxidation will reduce fibrotic response, we also treated cells with an ACC (Acetyl-CoA Carboxylase) inhibitor. ACC is an important regulator of fatty acid oxidation by converting acetyl-CoA to malonyl-CoA, the latter of which is an allosteric inhibitor of CPT1.43 MK-4074 is a potent liver-specific inhibitor of two ACC isoforms, ACC1 and ACC2.43 In RPTECs, we found that MK-4074 strongly inhibited TGF-β-induced procollagen production, confirming the importance of metabolic regulators.
Figure 4.
Reverse Fibrosis by Metabolic Modulators
(A) Fibroblast phenotypic screen identified targets highly enriched in metabolic regulation. For αSMA imaging, representative stitched images of four views per well are shown. αSMA immunofluorescence staining, green; Hoechst 33342 nuclear staining, blue.
(B) Inhibition of 3H-proline incorporation in normal human lung fibroblasts by MRL-24 (PPARγ agonist), GW501506 (PPARδ agonist), and MK-8722 (pan-AMPK activator) in a 3-point dose titration study. 3 independent replicates were obtained.
(C) The effect of AMPK activator (MK-8722) on fibrotic marker gene expression in lung fibroblasts. 3 independent replicates were obtained.
(D) The effect of MK-8722 and MK-4074 (ACC inhibitor) on pro-collagen I production in RPTECs. 3 independent replicates were obtained.
(E) MK-8722, MK-4074, and Telmisartan reduced liver fibrosis in STAM mouse NASH model. Top panel, liver triglyceride (TG) content; bottom panel, fibrosis as determined by Sirius red staining positive area in the liver. Mean ± SEM, n = 6∼8, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.005, one-way ANOVA followed by Tukey’s test.
Suppression of Fibrosis In Vivo by Metabolic Modulators
Our transcriptome profiling analyses in human primary cells and rodent models revealed a common metabolic signature underlying fibrosis. This agrees well with our phenotypic screen findings. To further investigate metabolic regulators in vivo, we next examined the anti-fibrotic efficacy of MK-8722, MK-4074, and Telmisartan. Telmisartan is a PPARγ and angiotensin II receptor dual-agent developed for hypertension (ONTARGET44). In a mouse NASH model, we carried out a side-by-side comparison of these three compounds. After a single low dose of Streptozocin injection and 32% high-fat diet treatment, animals develop progressive NASH and fibrosis in a relatively short period of time (6∼20 weeks).45 The compounds were given at 9–12 weeks of age when fibrosis became evident (Figure S4A). While food intake, body weight, plasma glucose, insulin, and triglyceride levels were rather comparable among treatment groups (Figures S4B–S4D), liver triglyceride (TG) content was significantly reduced after 3 weeks of MK-8722 (↓45%), MK-4074 (↓ 43%), and Telmisartan (↓76%) treatment (Figure 4E, top panel). Furthermore, histological analysis revealed that fibrosis was significantly decreased following MK-8722 (↓34%) and Telmisartan (↓41%) treatment (Figure 4E, bottom panel). MK-4074 treatment also lowered fibrosis by 19%, albeit at a statistically insignificant level. The NAS scores (nonalcoholic fatty liver disease/NAFLD activity scores) on steatosis, inflammation, and ballooning degeneration were also evaluated. Telmisartan was able to substantially reduce hepatocyte ballooning injury (Figure S4E). Our results found that the three metabolic regulators reduce fibrosis in vivo. Among which, Telmisartan appears to be more effective at suppressing hepatic steatosis and inflammation. Additionally, our results also found that Telmisartan was able to inhibit fibrosis in UUO, CCl4, and BDL models (Figures S2D, S3B, and S3D). Together, our results support the use of metabolic regulators for the treatment of fibrosis.
Discussion
In this study, we endeavored to identify pathways that share a commonality in the development of fibrosis in multiple tissues, hypothesizing that therapeutic agents targeting these key driver(s) of fibrosis would have a higher probability of success in modifying the disease in human. Our results from whole-genome transcriptome analysis of four different human primary cell types (lung, heart, liver, and kidney) demonstrated that fatty acid oxidation genes were significantly downregulated in all cells upon TGF-β treatment, suggesting that metabolic defect is a common feature of fibrosis. We further developed and profiled three independent animal models of kidney and liver fibrosis. By cross-referencing sequencing profiles, we identified defective fatty acid oxidation as one of the top dysregulated pathways, which was further supported by the observation that metabolic regulators reduced fibrosis in phenotypic screening assays and in vivo fibrosis models. Collectively, these observations suggest that a common metabolic defect presents across multiple fibrotic indications.
This finding is of clinical relevance, as similar metabolic-fibrosis signatures have been observed in patients with cardiometabolic diseases. A recent elegant study from the Sustzak group performed transcriptome analyses on human kidney samples from patients with diabetic or hypertensive CKD, underpinning the important role of dysfunctional fatty acid oxidation in renal fibrosis in human.46 Furthermore, comparison of gene expression patterns between mouse models and liver tissues from NASH/NAFLD patients identified pathways associated with lipid metabolism.47 Cross-sectional analysis of a community-dwelling twin cohort reveals highly significant shared genetic effects between hepatic steatosis and fibrosis,48 providing strong human genetic evidence that links metabolic dysfunction to fibrosis.
Fibrosis is an evolutionarily conserved mechanism developed by an organism to survive injury. The interplay between metabolic signature and fibrotic response remains mechanistically intriguing. As a master regulator of fibrosis, TGF-β has been shown modulating metabolic homeostasis in metabolic active tissues. Blockade of TGF-β/Smad3 signaling protects animals from obesity and diabetes and Smad3−/− adipocytes demonstrate a marked increase in mitochondrial biogenesis and PGC1α upregulation.49 In human primary fibroblasts, we have observed a profound suppression of PGC1α gene expression upon TGF-β treatment, which can be reversed by the administration of ALK5 inhibitor (Figure S1D). Downregulation of PGC1α was also evident in kidney, liver, and lung fibrosis models and treatment with the ALK5 inhibitor or Telmisartan relieved the suppression (Figures S2E and S3G), suggesting that TGF-β affects fatty acid oxidation and mitochondrial function through PGC1α in response to fibrotic signals. It will be of interest to determine whether Smad3 binds to the promoter of PGC1α and directly controls its transcription.
From therapeutic standpoints, we are interested in targeting common fibrotic pathway(s) for multiple indications. A number of TGF-β-focused therapies have been tested in the clinic, including LY2382770 (TGF-β1 antibody) for diabetic nephropathy and Fresolimumab (pan-TGF-β antibody) for systemic sclerosis and focal segmental glomerulosclerosis.50, 51, 52 However, no substantial clinical benefit has been observed to date. Our results suggested that metabolic defect is a common feature of TGF-β-induced fibrotic response. A successful anti-fibrotic therapy may potentially require targeting both metabolic defects and TGF-β signaling.
Our results highlighted an important role for PGC1α in fibrosis. While it has been challenging to develop PGC1α modulator for therapeutics, our compound library phenotypic screen identified metabolic regulators, such as AMPK and PPARs, as highly enriched targets to suppress fibrosis. Impaired AMPK activity has been found in multiple tissue fibrosis.53, 54, 55, 56, 57 PF-06409577, an AMPK β1-biased activator, has been recently shown to reduce hepatic lipid accumulation and the expression of liver fibrosis genes in preclinical animals.58 MK-8722 was developed previously as a pan-AMPK activator for diabetes.41,42 In this study, we provided preclinical evidence that MK-8722 reduced hepatic lipid accumulation and fibrosis, confirming that modulating lipid metabolism is indeed beneficial for fibrosis. Historically, it has been challenging to develop metabolic regulators with good long-term safety profiles. Chronic systemic administration of MK-8722 improves glucose homeostasis but induces physiological cardiac hypertrophy in rodents and non-human primates.42 The ACC inhibitor MK-4074 increases plasma triglycerides level by 200% in human.43 Developing new generation of metabolic regulators with desired selectivity profiles, optimized durations of action, or local delivery could potentially constitute approaches to achieve a better safety margin.
Our fibrosis profiling dataset also provides insights into other pathways underlying fibrosis. Immune response represents another highly enriched GO term through pathway analysis. While many inflammatory genes remained undetectable in human primary cells or fibrotic tissues, the expression of a small subset of innate and adaptive immune genes was consistently upregulated in all tissue/cell types profiled (Figure S1C). Among which, Tnfrsf12a or Fn14, a receptor for the TNF superfamily ligand TWEAK, has been shown involving in cell proliferation, differentiation, and inflammation.59 Therapeutic antibodies against both ligand and receptor have been developed for oncology and autoimmune indications.60, 61, 62, 63 Deficiency of Fn14 or TWEAK in mice protects animals against kidney and liver fibrosis,64,65 supporting the notion of targeting TWEAK-Fn14 pathway for fibrosis and a broader applicability of our dataset.
Altogether, our work has revealed a common mode of metabolic signature implicated in multiple fibrotic tissues. Targeting this common pathway could represent a promising therapeutic approach for fibrosis and cardiometabolic complications. Data mining of multiple fibrosis models will likely lead to the identification of new targets for therapeutic intervention.
Limitations of Study
In this study, we focused on common metabolic defects that are integral to fibrosis development in multiple tissue types. However, the contribution of other pathways, such as innate immunity, reactive oxidative species, and apoptosis, was not extensively examined during our studies. For future work, the relationship between TGF-β and lipid metabolism will be further interrogated to obtain a more thorough review of metabolic changes underlying fibrosis induction. The process of drug discovery and development from original idea to the launch of a finished product is long and costly. The lack of translation of preclinical findings is one of the main reasons underlying clinical failures. Obtaining an in-depth molecular profiling of fibrotic tissues from patients will increase the likelihood to succeed for newer anti-fibrotic therapies.
STAR★Methods
Key Resource Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit anti-ATP5A monoclonal antibody (EPR13030B) | Abcam | Cat#Ab176569; RRID:AB_2801536 |
| Mouse anti-UQCRC2 monoclonal antibody (13G12AF12BB11) | Abcam | Cat#Ab14745; RRID:AB_2213640 |
| Mouse anti-CPT1a monoclonal antibody (8F6AE9) | Abcam | Cat#Ab128568; RRID:AB_11141632 |
| Rabbit anti-CPT2 monoclonal antibody (EPR13626) | Abcam | Cat#Ab181114; RRID:AB_2687503 |
| Rabbit anti-GAPDH monoclonal antibody (14C10) | Cell Signaling Transduction | Cat#2118; RRID:AB_10693448 |
| Rabbit anti-Smad2/3 monoclonal antibody (D7G7) | Cell Signaling Transduction | Cat#8685; RRID:AB_10889933 |
| Rabbit anti-αSMA monoclonal antibody (EPR5368) IHC | Abcam | Cat#Ab124964; RRID:AB_11129103 |
| Mouse anti-αSMA monoclonal antibody (1A4) IF | Sigma | Cat#A5228; RRID:AB_262054 |
| Alexa Fluor 488-conjugated goat anti-mouse secondary antibody | Thermo Fisher Scientific | Cat#A-11001; RRID:AB_2534069 |
| Bacterial and Virus Strains | ||
| Biological Samples | ||
| Mouse kidneys from UUO model | This study | N/A |
| Mouse livers from CCl4 model | This study | N/A |
| Rat livers from bile duct ligation model | This study | N/A |
| Mouse lungs from Bleomycin model | This study | N/A |
| Mouse livers from STAM™ NASH model | This study | N/A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| recombinant human TGFβ1 | BioLegend | Cat#580702 |
| Olive Oil | Sigma | Cat#O1514 |
| Carbon tetrachloride | Sigma | Cat#1601168 |
| Normal saline | Baxter | Cat#BAX2F7124 |
| Bleomycin lyophilized powder injection SDV 15IU 10ml/Vl | Henry Schein | Cat#8700000 |
| Methylcellulose | Sigma | Cat#M0555 |
| Telmisartan | Alfa Aesar | Cat#J61441 |
| MK-8722 | Merck & Co., Inc., Kenilworth, NJ, USA | N/A |
| MK-4074 | Merck & Co., Inc., Kenilworth, NJ, USA | N/A |
| DMSO | Sigma | Cat#D2650 |
| SB-525334 | Sigma | Cat#S8822 |
| MRL-24 | Focus Biomolecules | Cat#10-1365 |
| GW501506 | Sigma | Cat#SML1491 |
| Tween-80 | Sigma | Cat#P4870 |
| cOmplete Protease inhibitor, mini-pellet, EDTA free | Roche | Cat#4693159001 |
| phosphatase inhibitor cocktails | Sigma | Cat#P5726 |
| Bradford reagent | Bio-Rad | Cat#5000006 |
| Deuterium oxide | Thermo Fisher Scientific | Cat#AC166300010 |
| NP-40/IGEPAL® CA-630 | Sigma | Cat#I8896 |
| Triton X-100 | Sigma | Cat#T9284 |
| Ammonium bicarbonate (AMBIC) | Sigma | Cat#A6141 |
| DTT (DL-Dithiothrietol) | Sigma | Cat#D9779 |
| Iodoacetamide | Sigma | Cat#I6125 |
| ProteaseMax Surfactant | Promega | Cat3V2071 |
| Trypsin gold, mass spectrometry grade | Promega | Cat#V5280 |
| stable isotope-labeled acylcarnitine standards | Cambridge Isotope Labs | Cat#NSK-B-1 |
| Formic acid | Sigma | Cat#33015 |
| Acetonitrile | Sigma | Cat#34851 |
| Methanol | Sigma | Cat#646377 |
| 3H-proline | Perkin Elmer | Cat#NET483001MC |
| Ascorbic acid | Acros Organics | Cat#352681000 |
| Opti-MEM I | Thermo Fisher Scientific | Cat#31985088 |
| Bovine Serum Albumin (BSA) | Sigma | Cat#A9418 |
| Penicillin-Streptomycin-Glutamine | Thermo Fisher Scientific | Cat#10378016 |
| MEM Non-essential amino acids | Thermo Fisher Scientific | Cat#11140076 |
| Hoechst 33342 | Thermo Fisher Scientific | Cat#62249 |
| Critical Commercial Assays | ||
| Picosirius Red staining kit | American Mastertech | Cat#KTPSRPT |
| RNeasy Mini QIAcube Kit | QIAGEN | Cat#74116 |
| SuperScript VILO cDNA Synthesis kit | Thermo Fisher Scientific | Cat#11754-050 |
| TaqMan Universal PCR Master mix | Thermo Fisher Scientific | Cat#4364338 |
| NE-PER nuclear and cytoplasmic extraction reagents | Thermo Scientific | Cat#78833 |
| Truseq stranded total RNA library preparation kit with Ribo-Zero human/mouse/rat | Illumina | Cat#RS-122-2201 |
| Human Procollagen type I HRTF assay | Cisbio | Cat#63ADK014PEG |
| Ultra-sensitive mouse Insulin ELISA kit | Morinaga Institute Biological Science, Inc., Japan | Cat#M1104 |
| Triglyceride E-test | Wako Pure Chemical Industries, Ltd., Japan | Cat#290-63701 |
| Deposited Data | ||
| Raw and analyzed RNA-seq data | This paper | GEO: GSE152250https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE152250 |
| Experimental Models: Cell Lines | ||
| Normal human lung fibroblasts | Lonza | Cat#CC-2512, Lot # 0000343490 |
| Human primary cardiac fibroblasts | Sciencell | Cat# 6300, Lot #5433 |
| Human primary renal proximal tubule epithelial cells | Lonza | Cat#CC-2553, Lot#0000362300 |
| Primary human hepatic stellate cells | Sciencell | Cat#5300, Lot#10279 |
| Experimental Models: Organisms/Strains | ||
| C57BL/6J male mice | Jackson Laboratory | Cat#JAX 000664; RRID:IMSR_JAX:000664 |
| Sprague Dawley male rats | Taconic | Cat#NTac:SD; RRID:RGD_1566440 |
| STAM™ NASH mice | Stelic/SMC Laboratories | https://www.smccro-lab.com/service/service_disease_area/stam.html |
| Oligonucleotides | ||
| Taqman probes | Life Technologies | See Table S3 |
| Recombinant DNA | ||
| Software and Algorithms | ||
| Aperio Spectrum and ImageScope (v12.2.1.5005) | Leica Biosystems | https://www.leicabiosystems.com/ |
| Compass for Simple Western (version 4.0.0) | Protein Simple | https://www.proteinsimple.com/compass/downloads/ |
| Omicsoft Array Studio (version 9.0.8.92) | QIAGEN | https://www.qiagen.com/us/ |
| PANTHER GO term enrichment test (version 15.0) | Gene Ontology Phylogenetic Annotation Project | http://pantherdb.org |
| Morpheus matrix visualization and analysis software | Broad Institute | https://software.broadinstitute.org/morpheus/ |
| CHEMGENIE | Merck & Co., Inc., Kenilworth, NJ, USA | Kutchukian et al., 201837 |
| Pipeline Pilot (version 9.2) | BIOVIA | https://www.3dsbiovia.com/products/collaborative-science/biovia-pipeline-pilot/ |
| GraphPad Prism (version 7) | GraphPad | https://www.graphpad.com/scientific-software/prism/ |
| TargetLynx (v4.1) | Waters corp., MA | https://www.waters.com/waters/en_US/TargetLynx-/nav.htm?cid=513791&locale=en_US |
Resource Availability
Lead Contact
Further information and requests for resources and reagents can be directed to the Lead Contact, Ji Zhang (ji.zhang1@merck.com)
Materials Availability
This study did not generate new unique reagents.
Data and Code Availability
The RNA-seq datasets generated during this study are available at NCBI GEO repository, The accession number reported in this paper is GEO:GSE152250 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE152250).
Experimental Model and Subject Details
Cultured Cells
Normal human lung fibroblasts (Lonza, #CC-2512, Lot # 0000343490, passage 3) were cultured in fibroblast basal medium (Lonza, #CC-3131) with growth supplements (Lonza, #CC-4126). Human primary cardiac fibroblasts (Sciencell, #6300, Lot #5433, passage 4) were cultured in FM2 media (Sciencell, #2331), supplemented with FBS (Sciencell, #0025), FGS-2 (Sciencell, #2382), and Penicillin-Streptomycin (Life Technologies, #15070-063). Human primary renal proximal tubule epithelial cells (Lonza, #CC-2553, Lot#0000362300, passage 3) were cultured in renal epithelial cell growth basal medium (Lonza, #CC-3191) supplemented with REGM SingleQuots (Lonza, #CC-4127). Primary human hepatic stellate cells (Sciencell, #5300, Lot#10279, passage 4) were cultured in Stellate Cell Medium (Sciencell, #5301) supplemented with FBS (Sciencell, #0010), stellate cell growth supplement (Sciencell, #5352), and Penicillin-Streptomycin solution (Sciencell, #0503). All cells were Incubated at 37°C in the presence of 5% CO2. For TGFβ treatment, cells were plated and serum-starved for overnight. The next day, 5ng/ml of recombinant human TGFβ1 (BioLegend, #580702) was added to the culture medium and cells were incubated at 37°C for the indicated time. For inhibitor treatment, cells were treated with recombinant human TGFβ1 in the presence of vehicle DMSO (Sigma, #D2650) or compounds at various concentrations.
Animals
Adult male C57BL/6 mice (Jackson Laboratory, Bar Harbor, ME) and adult male Sprague Dawley/SD rats (Taconic, Rensselaer, NY) were housed in a temperature- and humidity-controlled facility with a 12 hour light: 12-hour dark cycle. Animals had ad libitum access to food (LabDiet, Purina Rodent Chow 5053, St. Louis, MO) and water. All procedures utilizing experimental animals were conducted in accordance with the Guide for the Care and Use of Laboratory Animals, and experimental procedures were reviewed and approved by the Institutional Animal Care and Use Committee at Merck & Co., Inc., Kenilworth, NJ, USA.
UUO kidney fibrosis model
Mice at 12∼13 week of age were randomized to 3∼4 groups: sham (n = 6), UUO (n = 9), and UUO with therapeutic agents (n = 9). UUO was induced by complete ligation in the upper two-thirds of the left ureter. In sham animals, the kidney and ureter were exposed, but the ureter was not tied. For compound treatment, animals were given control or medicated diet (Telmisartan 10mpk, formulated at Research Diet Inc., New Brunswick, NJ) on the day before the surgery. The compound in-feed concentration was determined by body weight and daily food intake.
Bleomycin lung fibrosis model
Mice at 12∼13 weeks of age were randomized to 5 groups: saline instillation, bleomycin instillation with vehicle (0.5% methylcellulose) treatment, and bleomycin instillation with SB-525334 treatment. All mice were anaesthetized with isoflurane. Bleomycin was dosed by intra-tracheal instillation in a volume of 50 μl, at a dose of 2U/kg bw. After instillation, mice were kept in a heads-up position for 2-5 minutes before putting into cages. Vehicle or SB-525334 was administered via oral gavage bid from day 5 to day 21.
CCl4 liver fibrosis model
Mice at 12∼13 weeks of age were treated twice each week for 9 weeks with either olive oil or CCl4 (Sigma, #1601168, 1:4 dilution in olive oil) through intraperitoneal injection (2ml/kg BW). At the indicated time, the liver was collected for histology or stable isotope analysis. Two weeks into CCl4 treatment, animals were dosed with either vehicle (0.5% methylcellulose) or Telmisartan (10mpk) via po qd for 4 weeks. The severity of inflammation, ballooning degeneration and fibrosis were evaluated semiquantitatively using a predetermined score system (Table S2).
Bile duct ligation liver fibrosis model
For SD rats at 6∼8 weeks of age, the common bile duct was ligated twice at one cm distal to the duodenum and transected between the ligatures. For animals with shame operation, the surgical procedure is the same except the bile duct is not ligated or transected. At the indicated time, rat livers were collected for further analyses. Two days post bile duct ligation surgery, animals were dosed with either vehicle (0.5% methylcellulose) or Telmisartan (10mpk) via po qd for 2 weeks. The severity of inflammation, ballooning degeneration and fibrosis were evaluated semiquantitatively using a predetermined score system (Table S2).
STAM™ NASH model
STAM™ mouse model (Stelic, Japan) develops hyperglycemia, hypoinsulinemia, hepatic steatosis, NASH, and liver fibrosis, with an onset of baseline NAFLD activity score at 6 weeks of age45. To probe the anti-fibrotic effect of therapeutic agent, animals were given vehicle (0.25% methylcellulose, 5% Tween-80, 0.02% SDS), MK-8722 10mpk, MK-4074 50mpk, or Telmisartan 10mpk, at 9 weeks of age. The compound treatment was via oral gavage daily for 3 weeks. The body weight and food consumption were monitored daily. Individual weight of liver, heart, and kidney were measured at the end of study; organ to body weight was compared. Plasma was collected for biochemical analyses, and liver was subjected for triglyceride content measurement and histological evaluation (HE stain for NAFLD activity score, and Sirius red stain for fibrosis)45.
Method Details
Histopathology Analysis
Upon completion of the study, animals were euthanized and tissues were collected for histology assessment. Kidney or liver tissues were fixed in 10% formalin and then paraffin embedded. Tissue sections were stained with hematoxylin & eosin (H&E) and Masson’s trichrome, subsequently evaluated under light microscope. Lung tissues were perfused with 10% formalin, fixed for 24 hours, and then paraffin embedded. The severity of histopathologic changes and fibrosis in renal tubules, interstitium, and glomeruli were graded on a 1 to 5 scale corresponding to minimal, mild, moderate, marked, and severe as described previously66. The severity of liver inflammation, ballooning degeneration and fibrosis were evaluated semiquantitatively using a predetermined score system (Table S2). The severity of histopathologic changes and fibrosis in the lung were graded as described previously by pathologists67,68.
Following deparaffinization and rehydration, each tissue section was processed to identify αSMA (Sigma, #F3777) deposition by immunohistochemistry. Picosirius Red staining kit was purchased from American Mastertech (#KTPSRPT) and was carried out according to manufacturer’s protocol. The Aperio ScanScope XT Slide Scanner (Aperio Technologies) system was used to capture whole slide digital images with a 20 × objective. Digital images were managed using Aperio Spectrum. The positive stains were identified and quantified using a macro created from a color deconvolution algorithm (Aperio Technologies).
Whole blood and plasma biochemistry
Non-fasting blood glucose in whole blood was measured weekly by LIFE CHECK glucose meter (EIDIA Co. Ltd., Japan) during the treatment period (6 to 9 weeks of age for NASH stage, and 9 to 12 weeks of age for fibrosis stage). For plasma biochemistry, blood was collected in polypropylene tubes with anticoagulant (Novo-Heparin, Mochida Pharmaceutical Co. Ltd., Japan) and centrifuged at 1,000xg for 15 minutes at 4°C. The supernatant was collected and stored at −80°C until use. Plasma ALT, AST, triglyceride and total cholesterol levels were measured by FUJI DRI-CHEM 7000 automatic clinical chemistry analyzer (Fujifilm Corporation, Japan). Plasma insulin levels were quantified by the ultra-sensitive mouse Insulin ELISA kit (Morinaga Institute Biological Science, Inc., Japan, #M1104).
Liver TG measurement
Liver total lipid-extracts were obtained by Folch’s method69. Liver samples were homogenized in chloroform-methanol (2:1, v/v) and incubated overnight at room temperature. After washing with chloroform-methanol-water (8:4:3, v/v/v), the extracts were evaporated to dryness, and dissolved in isopropanol. Liver triglyceride content was measured by Triglyceride E-test (Wako Pure Chemical Industries, Ltd., Japan, #290-63701).
Sally Sue simple western analysis
Cells or tissue samples were lyzed with assay buffer (50 mM Tris, pH7.4, 150 mM NaCl, 1 mM EDTA, 10% glycerol, 0.5% NP-40) with the addition of protease inhibitor (Roche cOmplete mini-pellet, EDTA free, #4693159001) and phosphatase inhibitor cocktails (Sigma, #P5726). The total protein concentration was determined by Bradford reagent (Bio-Rad, #5000006). 1 mg/ml of cell/tissue lysates were subsequently subjected for Sally Sue Simple Western analysis (Protein Simple)70. The antibodies used for detecting GAPDH and Smad3 antibodies were from Cell Signaling Transduction, and anti-CPT1a, CPT2, ATP5A, and UQCRC2 antibodies were from Abcam.
Gene Expression Taqman Analysis
Total RNA was extracted from tissue or cultured cells following the instruction of RNeasy Mini QIAcube Kit (QIAGEN, #74116). 2 μg RNA was then reverse-transcribed using SuperScript VILO cDNA Synthesis kit (Thermo Fisher Scientific, #11754-050). 2 μL diluted cDNA was added to quantitative real-time PCR mix with TaqMan Universal PCR Master mix (Thermo Fisher Scientific, #4364338) and Taqman probes (Life Technologies). PCR amplification (one cycle at 50°C for 2 minutes, one cycle at 95°C for 10 minutes and 40 cycles at 95°C for 15 s and 60°C for 1 minute) was done in an Applied Biosystems 7900HT Fast Real-Time PCR System (Applied Biosystem). Ct value of each sample was normalized to GAPDH level and quantified based on 2e-delta (deltaCT) method (comparing with no treatment or sham group).
Subcellular fractionation
Frozen kidney tissues were grinded into powder in liquid nitrogen and homogenized in lysis buffer: (50 mM Tris-Cl, pH7.5, 150 mM NaCl, 10% glycerol, 0.4% Triton X-100, protease and phosphatase inhibitors). The samples were subsequently centrifuged at 1,000 rpm to pellet down crude nuclei. NE-PER nuclear extraction buffer (Thermo Scientific, #78833) was used to solubilize nuclear fraction. The supernatant was spinned down again and transfer the supernatant to new tubes as cytoplasmic fraction. 0.5-1 mg/ml lysates were subjected to Sally Sue simple western analysis.
RNA Isolation and Transcriptome Profiling
RNA sequencing analysis was performed as previously described71. Briefly sequencing was performed using the Truseq stranded total RNA RiboZero library preparation kit (Illumina, #RS-122-2201) according to the manufacturer’s instructions. The resulting cDNA libraries were sequenced on an Illumina HiSeqTM 4000 using a 50 base paired-end run. Alignment and differential gene expression analysis was performed in Omicsoft Array Studio version 9.0.8.92. Briefly, cleaned reads were aligned to the mouse and human B38 genome references, for the UUO kidney and primary cells respectively, and the rat B6.0 genome reference for the BDL model, using the Omicsoft Aligner with a maximum of 2 allowed mismatches. Gene level counts, and FPKM, were determined by the OSA algorithm as implemented in Omicsoft Array Studio and using Ensembl.R82 gene models. At least 90% of reads across all samples mapped to the respective genome (corresponding to 55-80 million reads). Differential gene expression analysis was performed by the DESeq2 algorithm as implemented in Omicsoft Array Studio with the samples from the sham operated, or vehicle treated, animals serving as reference, yielding fold change and corrected p values (False Discovery Rate, Benjamini–Hochberg; FDR_BH). GO term enrichment analysis was performed using the PANTHER enrichment test (http://pantherdb.org). Heatmaps and boxplots were created using Omicsoft Array Studio.
2H-water labeling and proteomic analysis
To maintain a stable enrichment of body water for the 14-day duration of UUO time course, mice were given a bolus of 2H-labeled water (Thermo Fisher Scientific, #AC166300010) through intraperitoneal injection (10 mL/kg), and then allowed free access to food and 10% 2H-labeled drinking water. The collagen extract was prepared as described previously31. Kidney and/or liver tissue samples were homogenized in ∼20X volume of 0.1 M NaOH (in ddH2O). The homogenates were filtered through a 100 μM pore size strainer and subsequently spinned at 4,000 rpm. The insoluble pellets contained collagen fibers and were washed with 25 mM ammonium bicarbonate (AMBIC), pH8.0 once. The pellets were resuspended with 25 mM AMBIC, followed by sonication, denaturation in the presence of 10mM DTT, 20mM iodoacetamide incubation, solubilization with 0.025% ProteaseMax Surfactant (Promega, #V2071) and trypsin (Promega, #V5280) digestion at 30°C for 24 hours. The collagen peptides were purified by reverse-phase columns and subjected to LC-MS analysis31.
Acylcarnitine LC-MS Profiling
The acylcarnitine analysis was performed on an Acquity UPLC system coupled to a Xevo-G2-XS mass spectrometer (Waters Corp., MA, USA). Briefly, 50 mg of kidney tissue was weighed into 1 mL homogenization tubes. 800 μL (weight normalized) of working internal standard solution containing stable labeled acylcarnitines (Cambridge Isotope Labs, #NSK-B-1) in 0.3% formic acid in 3:1 Acetonitrile: Methanol was added to this tube. Tissue was homogenized twice at 5,500 rpm for 15 s using a Precellys tissue homogenizer. Homogenized tissue extracts were centrifuged at 14,000 rpm and at 5°C for 10 minutes and 600 μL of supernatant were transferred to an LC/MS vial. 2 μL were injected onto LC/MS system. Mass spectrometry data was processed by using TargetLynx v4.1 (Waters Corp., MA) and analyzed by GraphPad Prism 7.
3H-proline incorporation assay
Normal human lung fibroblasts were plated and serum-starved overnight. The cells were treated with 5 ng/ml of recombinant human TGFβ1 in the presence of 2 μCi/ml of 3H-proline (Perkin Elmer, #NET483001MC), and 50 μg/ml freshly-made ascorbic acid (Acros Organics, #352681000). After 24 hours of incubation, cells were washed with PBS twice, lyzed with lysis buffer (0. 5M NaOH, 0.3% SDS), and incubated at 37°C for 2 hours. After the addition of neutralization buffer (1 M HCl), the cell lysates were mixed with scintillation cocktail and counted for radioactivity by Perkin Elmer Tri-Carb 2900 scintillation counter.
αSMA imaging
Normal human lung fibroblasts were plated in 384-well poly-D-lysine coated microplates (Corning, #4587) in 20 μL volumes of assay medium (Opti-MEM I, Thermo Fisher Scientific, #31985088; 2% BSA, Sigma, #A9418; 1% Penicillin-Streptomycin-Glutamine, Thermo Fisher Scientific, #10378016; and 1% Non-essential amino acids, Thermo Fisher Scientific, #11140076). Following overnight incubation at 37°C in 5% CO2, 5 ng/mL of recombinant human TGFβ1 and compounds were added to a final DMSO concentration of 0.4%. After 48 hours of incubation at 37°C in 5% CO2, cells were fixed and permeabilized. After removal of fixative/permeabilization solution, cells were blocked for 1 hour and incubated with primary antibody αSMA mAb (Sigma, #A5228) at 4°C overnight. An Alexa Fluor 488-conjugated goat anti-mouse secondary antibody (Thermo Fisher Scientific, #A-11001) was used with 5 μM Hoechst 33342 (Thermo Fisher Scientific, #62249) to stain nuclei. Plates were scanned using the laser scanning plate cytometer, acumen eX3 (TTP Labtech). Object data was collected using the 405 laser, bandwidth 420-500 nm for detection of nuclei, and the 488 laser, bandwidth 500-530 nm for detection of αSMA antibody stain. Object data was reduced to produce 2 readouts: cell counts (Hoechst objects) and total intensity of Alexa Fluor488 αSMA antibody stain. Compound IC50 values were determined by plotting total intensity of Alexa Fluor 488 (normalized by cell counts) versus the log of the compound concentration, using a 4-parameter logistic fit based on the Levenberg-Marquardt algorithm.
Target Enrichment for phenotypic screen
All calculations were carried out in Pipeline Pilot (version 9.2). Compounds were first categorized as hits or non-hits. The hit calling from the primary screen (single concentration) was used for categorization. Once categorized, the descriptors for training were then added to each compound. In this case, we used unique target annotations as the descriptors. In order to do this, we added all target annotations from the CHEMGENIE database to each compound37 and kept all dose response values with a potency of < 1 uM and a qualifier of “=” or “<.” We also selected all non-numeric target associations, for example, from the PDB. We then kept all unique target identifiers for each compound (Entrez Gene ID if available, if not then the UniProt ID). We employed the “Learn Good from Bad” component in PLP, building a Naive Bayesian model using “hit” and “non-hit” as our categories, and the target identifiers as the user defined descriptors. We then extracted the feature weights for each target identifier and used these weights to prioritize enriched targets.
Human Procollagen type I HRTF assay
RPTEC cells were plated in 384-well plate. The next day, cells were treated with or without the compound in the presence of 5 ng/ml recombinant human TGFβ1. After 48 hours of incubation, culture media (supernatant) were collected and subjected to the human procollagen type I HRTF kit (Cisbio, #63ADK014PEG) for detection following manufacturer’s protocol.
Quantification and Statistical Analysis
All data are presented as mean ± standard error of the mean (SE). One-way ANOVA post hoc Tukey or Student’s t test was used for data comparisons as indicated in the figure legends. All analyses were done using GraphPad Prism 7 software. A p value of < 0.05 was considered to be statistically significant.
Acknowledgments
The authors thank Jennifer Cho for help with the RNA-seq, Kenny Wong, Hong-Ping Guan, David Kelley, and Ajay Chawla for their valuable feedback throughout the work.
Author Contributions
S.P., K.J., P.C., T-Q.C., T.E.A., J.Z., S.F.P., J.I., and A.M.P. conceived the experiments; J.Z., S.H., Y.Z., Y.K., H.Z., V.S., Y.H., P.C., A.S., T.E.A., X-L.S., K.S., E.Z., L.Y., Y.T., Y.C., E.S., and K.D. performed experiments; E.S.M., P.S.K., and J.C. performed data curation and visualization; E.C-J. coordinated external studies; J.Z., E.S.M., P.S.K.., P.C., D.G.M., S.F.P., and S.P. wrote the manuscript; L-J.M., M.H., S.T., and D.G.M. provided expertise and feedback.
Declaration of Interests
The authors are current or former employees of Merck Sharp & Dohme Corp., a subsidiary of Merck & Co., Inc., Kenilworth, NJ, USA, and/or shareholders of Merck & Co., Inc., Kenilworth, NJ, USA. P.S.K. is a current employee and/or shareholder of Jnana Therapeutics. P.C. is a current employee and/or shareholder of PRA Health Sciences. Y.Z. is a current employee and/or shareholder of HistoBridge, LLC. Y.H. and T.E.A. are current employees of FDA. T-Q.C. is a current employee and/or shareholder of Calico Life Sciences. J.C. is a current employee and/or shareholder of AstraZeneca. L-J.M. is a current employee and/or shareholder of Johnson and Johnson. M.H. is a current employee and/or shareholder of Maze Therapeutics. K.K.J. is a current employee and/or shareholder of Shrὄdinger, Inc. I.K.S and S.P. are current employees and/or shareholders of Kallyope, Inc.
Published: July 21, 2020
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.xcrm.2020.100056.
Contributor Information
Ji Zhang, Email: ji.zhang1@merck.com.
Eric S. Muise, Email: eric_muise@merck.com.
Shirly Pinto, Email: shirly@kallyope.com.
Supplemental Information
References
- 1.WHO . World Health Organization; 2014. Global Status Report on Noncommunicable Diseases 2014. [Google Scholar]
- 2.Krumholz H.M. Treatment of Cholesterol in 2017. JAMA. 2017;318:417–418. doi: 10.1001/jama.2017.6753. [DOI] [PubMed] [Google Scholar]
- 3.Coffman T.M. The inextricable role of the kidney in hypertension. J. Clin. Invest. 2014;124:2341–2347. doi: 10.1172/JCI72274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jellinger P.S., Handelsman Y., Rosenblit P.D., Bloomgarden Z.T., Fonseca V.A., Garber A.J., Grunberger G., Guerin C.K., Bell D.S.H., Mechanick J.I. American Association of Clinical Endocrinologists and American College of Endocrinology Guidelines for Management of Dyslipidemia And Prevention of Cardiovascular Disease - Executive SUMMARY Complete Appendix to Guidelines available at. Endocr. Pract. 2017;23:479–497. doi: 10.4158/EP171764.GL. http://journals.aace.com [DOI] [PubMed] [Google Scholar]
- 5.American Diabetes Association Pharmacologic Approaches to Glycemic Treatment: Standards of Medical Care in Diabetes-2018. Diabetes Care. 2018;41(Suppl 1):S73–S85. doi: 10.2337/dc18-S008. [DOI] [PubMed] [Google Scholar]
- 6.Whelton P.K., Carey R.M. The 2017 Clinical Practice Guideline for High Blood Pressure. JAMA. 2017;318:2073–2074. doi: 10.1001/jama.2017.18209. [DOI] [PubMed] [Google Scholar]
- 7.Appel L.J., Wright J.T., Jr., Greene T., Agodoa L.Y., Astor B.C., Bakris G.L., Cleveland W.H., Charleston J., Contreras G., Faulkner M.L., AASK Collaborative Research Group Intensive blood-pressure control in hypertensive chronic kidney disease. N. Engl. J. Med. 2010;363:918–929. doi: 10.1056/NEJMoa0910975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wright J.T., Jr., Williamson J.D., Whelton P.K., Snyder J.K., Sink K.M., Rocco M.V., Reboussin D.M., Rahman M., Oparil S., Lewis C.E., SPRINT Research Group A Randomized Trial of Intensive versus Standard Blood-Pressure Control. N. Engl. J. Med. 2015;373:2103–2116. doi: 10.1056/NEJMoa1511939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nathan D.M., Genuth S., Lachin J., Cleary P., Crofford O., Davis M., Rand L., Siebert C., Diabetes Control and Complications Trial Research Group The effect of intensive treatment of diabetes on the development and progression of long-term complications in insulin-dependent diabetes mellitus. N. Engl. J. Med. 1993;329:977–986. doi: 10.1056/NEJM199309303291401. [DOI] [PubMed] [Google Scholar]
- 10.Gregg E.W., Li Y., Wang J., Burrows N.R., Ali M.K., Rolka D., Williams D.E., Geiss L. Changes in diabetes-related complications in the United States, 1990-2010. N. Engl. J. Med. 2014;370:1514–1523. doi: 10.1056/NEJMoa1310799. [DOI] [PubMed] [Google Scholar]
- 11.Friedman S.L., Sheppard D., Duffield J.S., Violette S. Therapy for fibrotic diseases: nearing the starting line. Sci. Transl. Med. 2013;5:167sr1. doi: 10.1126/scitranslmed.3004700. [DOI] [PubMed] [Google Scholar]
- 12.Friedman S.L., Sheppard D., Duffield J.S., Violette S. Therapy for fibrotic diseases: nearing the starting line. Sci. Transl. Med. 2013;5:167sr1. doi: 10.1126/scitranslmed.3004700. [DOI] [PubMed] [Google Scholar]
- 13.Wynn T.A., Ramalingam T.R. Mechanisms of fibrosis: therapeutic translation for fibrotic disease. Nat. Med. 2012;18:1028–1040. doi: 10.1038/nm.2807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rockey D.C., Bell P.D., Hill J.A. Fibrosis--a common pathway to organ injury and failure. N. Engl. J. Med. 2015;372:1138–1149. doi: 10.1056/NEJMra1300575. [DOI] [PubMed] [Google Scholar]
- 15.Breyer M.D., Susztak K. Developing Treatments for Chronic Kidney Disease in the 21st Century. Semin. Nephrol. 2016;36:436–447. doi: 10.1016/j.semnephrol.2016.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Duffield J.S. Cellular and molecular mechanisms in kidney fibrosis. J. Clin. Invest. 2014;124:2299–2306. doi: 10.1172/JCI72267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lim B.J., Yang J.W., Zou J., Zhong J., Matsusaka T., Pastan I., Zhang M.Z., Harris R.C., Yang H.C., Fogo A.B. Tubulointerstitial fibrosis can sensitize the kidney to subsequent glomerular injury. Kidney Int. 2017;92:1395–1403. doi: 10.1016/j.kint.2017.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kramann R., Schneider R.K., DiRocco D.P., Machado F., Fleig S., Bondzie P.A., Henderson J.M., Ebert B.L., Humphreys B.D. Perivascular Gli1+ progenitors are key contributors to injury-induced organ fibrosis. Cell Stem Cell. 2015;16:51–66. doi: 10.1016/j.stem.2014.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wattacheril J., Issa D., Sanyal A. Nonalcoholic Steatohepatitis (NASH) and Hepatic Fibrosis: Emerging Therapies. Annu. Rev. Pharmacol. Toxicol. 2018;58:649–662. doi: 10.1146/annurev-pharmtox-010617-052545. [DOI] [PubMed] [Google Scholar]
- 20.Koyama Y., Xu J., Liu X., Brenner D.A. New Developments on the Treatment of Liver Fibrosis. Dig. Dis. 2016;34:589–596. doi: 10.1159/000445269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang P., Koyama Y., Liu X., Xu J., Ma H.Y., Liang S., Kim I.H., Brenner D.A., Kisseleva T. Promising Therapy Candidates for Liver Fibrosis. Front. Physiol. 2016;7:47. doi: 10.3389/fphys.2016.00047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Neuschwander-Tetri B.A., Loomba R., Sanyal A.J., Lavine J.E., Van Natta M.L., Abdelmalek M.F., Chalasani N., Dasarathy S., Diehl A.M., Hameed B., NASH Clinical Research Network Farnesoid X nuclear receptor ligand obeticholic acid for non-cirrhotic, non-alcoholic steatohepatitis (FLINT): a multicentre, randomised, placebo-controlled trial. Lancet. 2015;385:956–965. doi: 10.1016/S0140-6736(14)61933-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Friedman S.L., Ratziu V., Harrison S.A., Abdelmalek M.F., Aithal G.P., Caballeria J., Francque S., Farrell G., Kowdley K.V., Craxi A. A randomized, placebo-controlled trial of cenicriviroc for treatment of nonalcoholic steatohepatitis with fibrosis. Hepatology. 2018;67:1754–1767. doi: 10.1002/hep.29477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ratziu V., Harrison S.A., Francque S., Bedossa P., Lehert P., Serfaty L., Romero-Gomez M., Bouirseier J., Abdelmalek M., Caldwell Elafibranor, an Agonist of the Peroxisome Proliferator-Activated Receptor-alpha and -delta, Induces Resolution of Nonalcoholic Steatohepatitis Without Fibrosis Worsening. Gastroenterology. 2016;150:1147–1159. doi: 10.1053/j.gastro.2016.01.038. [DOI] [PubMed] [Google Scholar]
- 25.Harrison S.A., Rinella M.E., Abdelmalek M.F., Trotter J.F., Paredes A.H., Arnold H.L., Kugelmas M., Bashir M.R., Jaros M.J., Ling L. NGM282 for treatment of non-alcoholic steatohepatitis: a multicentre, randomised, double-blind, placebo-controlled, phase 2 trial. Lancet. 2018;391:1174–1185. doi: 10.1016/S0140-6736(18)30474-4. [DOI] [PubMed] [Google Scholar]
- 26.Armstrong M.J., Gaunt P., Aithal G.P., Barton D., Hull D., Parker R., Hazlehurst J.M., Guo K., Abouda G., Aldersley M.A., LEAN trial team Liraglutide safety and efficacy in patients with non-alcoholic steatohepatitis (LEAN): a multicentre, double-blind, randomised, placebo-controlled phase 2 study. Lancet. 2016;387:679–690. doi: 10.1016/S0140-6736(15)00803-X. [DOI] [PubMed] [Google Scholar]
- 28.Nanthakumar C.B., Hatley R.J., Lemma S., Gauldie J., Marshall R.P., Macdonald S.J. Dissecting fibrosis: therapeutic insights from the small-molecule toolbox. Nat. Rev. Drug Discov. 2015;14:693–720. doi: 10.1038/nrd4592. [DOI] [PubMed] [Google Scholar]
- 29.Akhurst R.J., Hata A. Targeting the TGFβ signalling pathway in disease. Nat. Rev. Drug Discov. 2012;11:790–811. doi: 10.1038/nrd3810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Klahr S. New insights into the consequences and mechanisms of renal impairment in obstructive nephropathy. Am. J. Kidney Dis. 1991;18:689–699. doi: 10.1016/s0272-6386(12)80611-1. [DOI] [PubMed] [Google Scholar]
- 31.Zhou H., Wang S.P., Herath K., Kasumov T., Sadygov R.G., Previs S.F., Kelley D.E. Tracer-based estimates of protein flux in cases of incomplete product renewal: evidence and implications of heterogeneity in collagen turnover. Am. J. Physiol. Endocrinol. Metab. 2015;309:E115–E121. doi: 10.1152/ajpendo.00435.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lin J., Handschin C., Spiegelman B.M. Metabolic control through the PGC-1 family of transcription coactivators. Cell Metab. 2005;1:361–370. doi: 10.1016/j.cmet.2005.05.004. [DOI] [PubMed] [Google Scholar]
- 33.Jornayvaz F.R., Shulman G.I. Regulation of mitochondrial biogenesis. Essays Biochem. 2010;47:69–84. doi: 10.1042/bse0470069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Luo C., Widlund H.R., Puigserver P. PGC-1 Coactivators: Shepherding the Mitochondrial Biogenesis of Tumors. Trends Cancer. 2016;2:619–631. doi: 10.1016/j.trecan.2016.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Iredale J.P. Models of liver fibrosis: exploring the dynamic nature of inflammation and repair in a solid organ. J. Clin. Invest. 2007;117:539–548. doi: 10.1172/JCI30542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Moore B.B., Lawson W.E., Oury T.D., Sisson T.H., Raghavendran K., Hogaboam C.M. Animal models of fibrotic lung disease. Am. J. Respir. Cell Mol. Biol. 2013;49:167–179. doi: 10.1165/rcmb.2013-0094TR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kutchukian P.S., Chang C., Fox S.J., Cook E., Barnard R., Tellers D., Wang H., Pertusi D., Glick M., Sheridan R.P. CHEMGENIE: integration of chemogenomics data for applications in chemical biology. Drug Discov. Today. 2018;23:151–160. doi: 10.1016/j.drudis.2017.09.004. [DOI] [PubMed] [Google Scholar]
- 38.Acton J.J., 3rd, Black R.M., Jones A.B., Moller D.E., Colwell L., Doebber T.W., Macnaul K.L., Berger J., Wood H.B. Benzoyl 2-methyl indoles as selective PPARgamma modulators. Bioorg. Med. Chem. Lett. 2005;15:357–362. doi: 10.1016/j.bmcl.2004.10.068. [DOI] [PubMed] [Google Scholar]
- 39.Acton J.J., 3rd Discovery of (2R)-2-(3-{3-[(4-Methoxyphenyl)carbonyl]-2-methyl-6-(trifluoromethoxy)-1H-indol-1 -yl}phenoxy)butanoic acid (MK-0533): a novel selective peroxisome proliferator-activated receptor gamma modulator for the treatment of type 2 diabetes mellitus with a reduced potential to increase plasma and extracellular fluid volume. J. Med. Chem. 2009;52:3846–3854. doi: 10.1021/jm900097m. [DOI] [PubMed] [Google Scholar]
- 40.Oliver W.R., Jr., Shenk J.L., Snaith M.R., Russell C.S., Plunket K.D., Bodkin N.L., Lewis M.C., Winegar D.A., Sznaidman M.L., Lambert M.H. A selective peroxisome proliferator-activated receptor delta agonist promotes reverse cholesterol transport. Proc. Natl. Acad. Sci. USA. 2001;98:5306–5311. doi: 10.1073/pnas.091021198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Feng D., Biftu T., Romero F.A., Kekec A., Dropinski J., Kassick A., Xu S., Kurtz M.M., Gollapudi A., Shao Q. Discovery of MK-8722: A Systemic, Direct Pan-Activator of AMP-Activated Protein Kinase. ACS Med. Chem. Lett. 2017;9:39–44. doi: 10.1021/acsmedchemlett.7b00417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Myers R.W., Guan H.P., Ehrhart J., Petrov A., Prahalada S., Tozzo E., Yang X., Kurtz M.M., Trujillo M., Gonzalez Trotter D. Systemic pan-AMPK activator MK-8722 improves glucose homeostasis but induces cardiac hypertrophy. Science. 2017;357:507–511. doi: 10.1126/science.aah5582. [DOI] [PubMed] [Google Scholar]
- 43.Kim C.W., Addy C., Kusunoki J., Anderson N.N., Deja S., Fu X., Burgess S.C., Li C., Ruddy M., Chakravarthy M. Acetyl CoA Carboxylase Inhibition Reduces Hepatic Steatosis but Elevates Plasma Triglycerides in Mice and Humans: A Bedside to Bench Investigation. Cell Metab. 2017;26:394–406. doi: 10.1016/j.cmet.2017.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yusuf S., Teo K.K., Pogue J., Dyal L., Copland I., Schumacher H., Dagenais G., Sleight P., Anderson C., ONTARGET Investigators Telmisartan, ramipril, or both in patients at high risk for vascular events. N. Engl. J. Med. 2008;358:1547–1559. doi: 10.1056/NEJMoa0801317. [DOI] [PubMed] [Google Scholar]
- 45.Saito T., Muramatsu M., Ishii Y., Saigo Y., Konuma T., Toriniwa Y., Miyajima K., Ohta T. Pathophysiological analysis of the progression of hepatic lesions in STAM mice. Physiol. Res. 2017;66:791–799. doi: 10.33549/physiolres.933592. [DOI] [PubMed] [Google Scholar]
- 46.Kang H.M., Ahn S.H., Choi P., Ko Y.A., Han S.H., Chinga F., Park A.S., Tao J., Sharma K., Pullman J. Defective fatty acid oxidation in renal tubular epithelial cells has a key role in kidney fibrosis development. Nat. Med. 2015;21:37–46. doi: 10.1038/nm.3762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Teufel A., Itzel T., Erhart W., Brosch M., Wang X.Y., Kim Y.O., von Schönfels W., Herrmann A., Brückner S., Stickel, F Comparison of Gene Expression Patterns Between Mouse Models of Nonalcoholic Fatty Liver Disease and Liver Tissues From Patients. Gastroenterology. 2016;151:513–525. doi: 10.1053/j.gastro.2016.05.051. [DOI] [PubMed] [Google Scholar]
- 48.Cui J., Chen C.H., Lo M.T., Schork N., Bettencourt R., Gonzalez M.P., Bhatt A., Hooker J., Shaffer K., Nelson K.E., For The Genetics Of Nafld In Twins Consortium Shared genetic effects between hepatic steatosis and fibrosis: A prospective twin study. Hepatology. 2016;64:1547–1558. doi: 10.1002/hep.28674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yadav H., Quijano C., Kamaraju A.K., Gavrilova O., Malek R., Chen W., Zerfas P., Zhigang D., Wright E.C., Stuelten C. Protection from obesity and diabetes by blockade of TGF-β/Smad3 signaling. Cell Metab. 2011;14:67–79. doi: 10.1016/j.cmet.2011.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rice L.M., Padilla C.M., McLaughlin S.R., Mathes A., Ziemek J., Goummih S., Nakerakanti S., York M., Farina G., Whitfield M.L. Fresolimumab treatment decreases biomarkers and improves clinical symptoms in systemic sclerosis patients. J. Clin. Invest. 2015;125:2795–2807. doi: 10.1172/JCI77958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Trachtman H., Fervenza F.C., Gipson D.S., Heering P., Jayne D.R., Peters H., Rota S., Remuzzi G., Rump L.C., Sellin L.K. A phase 1, single-dose study of fresolimumab, an anti-TGF-β antibody, in treatment-resistant primary focal segmental glomerulosclerosis. Kidney Int. 2011;79:1236–1243. doi: 10.1038/ki.2011.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Voelker J., Berg P.H., Sheetz M., Duffin K., Shen T., Moser B., Greene T., Blumenthal S.S., Rychlik I., Yagil Y. Anti-TGF-β1 Antibody Therapy in Patients with Diabetic Nephropathy. J. Am. Soc. Nephrol. 2017;28:953–962. doi: 10.1681/ASN.2015111230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Romero-Vasquez F., Shah D., Stafstrom W., Summer R.S. Impaired AMP-Activated Protein Kinase (AMPK) Activity And Increased Lipogenesis Are Early Events In The Development Of Bleomycin-Induced Pulmonary Fibrosis. Am. J. Respir. Crit. Care Med. 2013;187:A5659. [Google Scholar]
- 54.Rangarajan S., Bone N.B., Zmijewska A.A., Jiang S., Park D.W., Bernard K., Locy M.L., Ravi S., Deshane J., Mannon R.B. Metformin reverses established lung fibrosis in a bleomycin model. Nat. Med. 2018;24:1121–1127. doi: 10.1038/s41591-018-0087-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Declèves A.E., Zolkipli Z., Satriano J., Wang L., Nakayama T., Rogac M., Le T.P., Nortier J.L., Farquhar M.G., Naviaux R.K., Sharma K. Regulation of lipid accumulation by AMP-activated kinase [corrected] in high fat diet-induced kidney injury. Kidney Int. 2014;85:611–623. doi: 10.1038/ki.2013.462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kikuchi H., Kawai K., Nakashiro Y., Yonezawa T., Kawaji K., Kodama E.N., Oshima Y. Construction of a Meroterpenoid-Like Compounds Library Based on Diversity-Enhanced Extracts. Chemistry. 2019;25:1106–1112. doi: 10.1002/chem.201805417. [DOI] [PubMed] [Google Scholar]
- 57.Garcia D., Hellberg K., Chaix A., Wallace M., Herzig S., Badur M.G., Lin T., Shokhirev M.N., Pinto A.F.M., Ross D.S. Genetic Liver-Specific AMPK Activation Protects against Diet-Induced Obesity and NAFLD. Cell Rep. 2019;26:192–208. doi: 10.1016/j.celrep.2018.12.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Esquejo R.M., Salatto C.T., Delmore J., Albuquerque B., Reyes A., Shi Y., Moccia R., Cokorinos E., Peloquin M., Monetti M. Activation of Liver AMPK with PF-06409577 Corrects NAFLD and Lowers Cholesterol in Rodent and Primate Preclinical Models. EBioMedicine. 2018;31:122–132. doi: 10.1016/j.ebiom.2018.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Winkles J.A. The TWEAK-Fn14 cytokine-receptor axis: discovery, biology and therapeutic targeting. Nat. Rev. Drug Discov. 2008;7:411–425. doi: 10.1038/nrd2488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ye S., Fox M.I., Belmar N.A., Sho M., Chao D.T., Choi D., Fang Y., Zhao V., Keller S.F., Starling G.C., Culp P.A. Enavatuzumab, a Humanized Anti-TWEAK Receptor Monoclonal Antibody, Exerts Antitumor Activity through Attracting and Activating Innate Immune Effector Cells. J. Immunol. Res. 2017;2017:5737159. doi: 10.1155/2017/5737159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lam E.T., Eckhardt S.G., Messersmith W., Jimeno A., O’Bryant C.L., Ramanathan R.K., Weiss G.J., Chadha M., Oey A., Ding H.T. Phase I Study of Enavatuzumab, a First-in-Class Humanized Monoclonal Antibody Targeting the TWEAK Receptor, in Patients with Advanced Solid Tumors. Mol. Cancer Ther. 2018;17:215–221. doi: 10.1158/1535-7163.MCT-17-0330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yin X., Luistro L., Zhong H., Smith M., Nevins T., Schostack K., Hilton H., Lin T.A., Truitt T., Biondi D. RG7212 anti-TWEAK mAb inhibits tumor growth through inhibition of tumor cell proliferation and survival signaling and by enhancing the host antitumor immune response. Clin. Cancer Res. 2013;19:5686–5698. doi: 10.1158/1078-0432.CCR-13-0405. [DOI] [PubMed] [Google Scholar]
- 63.Yadava R.S., Foff E.P., Yu Q., Gladman J.T., Kim Y.K., Bhatt K.S., Thornton C.A., Zheng T.S., Mahadevan M.S. TWEAK/Fn14, a pathway and novel therapeutic target in myotonic dystrophy. Hum. Mol. Genet. 2015;24:2035–2048. doi: 10.1093/hmg/ddu617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gomez I.G., Roach A.M., Nakagawa N., Amatucci A., Johnson B.G., Dunn K., Kelly M.C., Karaca G., Zheng T.S., Szak S. TWEAK-Fn14 Signaling Activates Myofibroblasts to Drive Progression of Fibrotic Kidney Disease. J. Am. Soc. Nephrol. 2016;27:3639–3652. doi: 10.1681/ASN.2015111227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wilhelm A., Shepherd E.L., Amatucci A., Munir M., Reynolds G., Humphreys E., Resheq Y., Adams D.H., Hübscher S., Burkly L.C. Interaction of TWEAK with Fn14 leads to the progression of fibrotic liver disease by directly modulating hepatic stellate cell proliferation. J. Pathol. 2016;239:109–121. doi: 10.1002/path.4707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhou X., Zhang J., Haimbach R., Zhu W., Mayer-Ezell R., Garcia-Calvo M., Smith E., Price O., Kan Y., Zycband E. An integrin antagonist (MK-0429) decreases proteinuria and renal fibrosis in the ZSF1 rat diabetic nephropathy model. Pharmacol. Res. Perspect. 2017;5 doi: 10.1002/prp2.354. Published online September 11, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ashcroft T., Simpson J.M., Timbrell V. Simple method of estimating severity of pulmonary fibrosis on a numerical scale. J. Clin. Pathol. 1988;41:467–470. doi: 10.1136/jcp.41.4.467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hubner R.H., Gitter W., El Mokhtari N.E., Mathiak M., Both M., Bolte H., Freitag-Wolf S., Bewig B. Standardized quantification of pulmonary fibrosis in histological samples. Biotechniques. 2008;44:507–511. doi: 10.2144/000112729. 514–507. [DOI] [PubMed] [Google Scholar]
- 69.Folch J., Lees M., Sloane Stanley G.H. A simple method for the isolation and purification of total lipides from animal tissues. J. Biol. Chem. 1957;226:497–509. [PubMed] [Google Scholar]
- 70.Harris V.M. Protein detection by Simple Western™ analysis. Methods Mol. Biol. 2015;1312:465–468. doi: 10.1007/978-1-4939-2694-7_47. [DOI] [PubMed] [Google Scholar]
- 71.Schlessinger K., Li L., Tan Y., Liu F., Souza S.C., Tozzo E., Liu K., Thompson J.R., Wang L., Muise E.S. Gene expression in WAT from healthy humans and monkeys correlates with FGF21-induced browning of WAT in mice. Obesity. 2015;23:1818–1829. doi: 10.1002/oby.21153. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The RNA-seq datasets generated during this study are available at NCBI GEO repository, The accession number reported in this paper is GEO:GSE152250 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE152250).