GuA1-containing AMPA receptors play a key role in synaptic plasticity and cognition. Casas et al. demonstrate that the sensing of nutrients by CPT1C regulates the phosphatase activity of SAC1, and consequently PI(4)P levels, at the contact sites between the endoplasmic reticulum and the trans-Golgi network to control the trafficking of GluA1 receptors to the cell surface of neurons.
Abstract
Carnitine palmitoyltransferase 1C (CPT1C) is a sensor of malonyl-CoA and is located in the ER of neurons. AMPA receptors (AMPARs) mediate fast excitatory neurotransmission in the brain and play a key role in synaptic plasticity. In the present study, we demonstrate across different metabolic stress conditions that modulate malonyl-CoA levels in cortical neurons that CPT1C regulates the trafficking of the major AMPAR subunit, GluA1, through the phosphatidyl-inositol-4-phosphate (PI(4)P) phosphatase SAC1. In normal conditions, CPT1C down-regulates SAC1 catalytic activity, allowing efficient GluA1 trafficking to the plasma membrane. However, under low malonyl-CoA levels, such as during glucose depletion, CPT1C-dependent inhibition of SAC1 is released, facilitating SAC1’s translocation to ER-TGN contact sites to decrease TGN PI(4)P pools and trigger GluA1 retention at the TGN. Results reveal that GluA1 trafficking is regulated by CPT1C sensing of malonyl-CoA and provide the first report of a SAC1 inhibitor. Moreover, they shed light on how nutrients can affect synaptic function and cognition.
Introduction
α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid receptors (AMPARs) are ionotropic glutamate receptors that mediate the majority of excitatory synaptic transmissions in the mammalian central nervous system. Functional AMPARs are tetramers, with each subunit having four transmembrane domains, two extracellular regions that bind glutamate, and a cytosolic C-terminal regulatory domain that determines interactions with other binding proteins. GluA1 is the major AMPAR subunit mediating fast excitatory neurotransmission in the brain and plays a critical role in synaptic strength and plasticity, with key features encoding many brain functions such as learning and memory (reviewed by Jacobi and von Engelhardt, 2018). For this reason, GluA1 surface expression at the plasma membrane (PM) is tightly regulated, since its down-regulation depresses neuronal activity and overexpression leads to excitotoxicity (Henley and Wilkinson, 2016).
External environmental signals, including situations of oxygen and glucose deprivation, caloric restriction or high-fat diets, and exposure to orexigenic and anorexigenic hormones (Moult et al., 2010; Ribeiro et al., 2014; Dennis et al., 2011; Fernandes et al., 2014; Spinelli et al., 2017; Ouyang et al., 2017), have the capacity to influence GluA1 subunit abundance at the PM. At the molecular level, they do so by tuning intracellular trafficking patterns. To date, three trafficking pathways have been documented as regulating GluA abundance at the PM: (1) the canonical ER–TGN secretory pathway, which requires passage through an ER-Golgi intermediate compartment; (2) a recycling pathway, in which endocytosed GluA1 subunits are recycled back to the PM through recycling endosomes; and (3) the Golgi-independent route, in which GluA1-containing vesicles exiting the ER are routed to recycling endosomes without transiting through the Golgi apparatus (Hangen et al., 2018; Bowen et al., 2017). Despite this information, there have been few molecular targets identified within these trafficking pathways that influence GluA1 expression at the PM. Uncovering such mechanisms is essential for understanding how our brain responds and behaves under energy deficiencies, such as in metabolic disorders or anorexic subjects.
One intracellular protein that has recently been found be part of AMPAR macrocomplexes is the phosphoinositide phosphatase Suppressor of Actin 1 (SAC1; Schwenk et al., 2012; Chen et al., 2014). SAC1 is a ubiquitously expressed, ER-localized, phosphatidyl-inositol-4-phosphate (PI(4)P) phosphatase that tightly regulates PI(4)P levels at different cellular compartments, including the TGN, by dephosphorylating PI(4)P into phosphatidylinositol (PI). Since PI(4)P at the TGN is necessary for the recruitment of coat molecules and transport carriers that control cargo segregation toward endosomal destinations or directly to the PM, it follows that SAC1 plays a crucial regulatory role in vesicular trafficking. Indeed, under stressful conditions, such as glucose or serum deprivation, SAC1 accumulates at Golgi membranes to deplete PI(4)P pools in order to halt secretory transport toward the PM (Faulhammer et al., 2007; Blagoveshchenskaya et al., 2008). Specifically related to AMPAR trafficking, it has been described that upon N-methyl-D-aspartate (NMDA)–induced stress, which causes excitotoxicity, SAC1 is retained within the TGN regions, causing Golgi PI(4)P dephosphorylation and a down-regulation of GluA1 trafficking to the PM in neurons (Yang et al., 2013). It remains controversial whether SAC1 modulates the TGN pool of PI(4)P in a cis- or in a trans-conformation, with evidence that SAC1 can act in trans by directly dephosphorylating PI(4)P in the TGN across ER-TGN junctions (Venditti et al., 2019) or in cis combined with the actions of Oxysterol-binding protein (OSBP; Mesmin et al., 2013). Despite numerous reports exploring the spatial localization of SAC1 across subcellular compartments, little is known about possible mechanisms that may influence its cellular localization or even its phosphatase activity.
A recently described interactor of SAC1 is the carnitine palmitoyltransferase 1C (CPT1C) protein, which is also part of AMPAR macrocomplexes (Brechet et al., 2017). CPT1C belongs to a family of enzymes classically involved in regulating the transport of long-chain fatty acids into mitochondria for their β-oxidation. However, the CPT1C isoform has no catalytic activity, is found only in the ER (not in mitochondria), and is exclusively expressed in mammalian neurons, cancer cells, and stem cells (Price et al., 2002; Sierra et al., 2008; Carrasco et al., 2012; Roa-Mansergas et al., 2018; Zaugg et al., 2011). In neurons, CPT1C together with ferric-chelate reductase 1-like drives multimerization of GluAs into functional tetrameters at the ER (Schwenk et al., 2019). Moreover, it is necessary for proper trafficking of GluA1-containing AMPARs to the PM (Fadó et al., 2015; Gratacòs-Batlle et al., 2018, 2015). In the brain, CPT1C has been found to be critical for correct regulation of food intake and peripheral lipid metabolism in the hypothalamus (Gao et al., 2011, 2009; Ramírez et al., 2013; Pozo et al., 2017; Rodríguez-Rodríguez et al., 2019). In addition, its rich expression in the cerebral motor cortex and cerebellum makes it relevant in motor coordination and activity (Carrasco et al., 2013), underscored by human CPT1C mutations being associated with hereditary spastic paraplegia (Rinaldi et al., 2015; Hong et al., 2019). Finally, due to its abundance in the hippocampus and in the cerebral cortex, CPT1C also plays a pivotal role in cognition (Carrasco et al., 2012). Here, CPT1C regulates spatial learning via modifications in spine morphology and synaptic plasticity.
CPT1C contains two transmembrane domains connected by a short loop that allows the N-terminus and C-terminus to face the cytoplasm. The C-terminal region of CPT1C is around 30 residues longer than that of the other 1A and 1B isoforms, suggesting that it may possess an additional or differing cellular role than its family counterparts. Additionally, the N-terminal regulatory domain of CPT1C is known to undergo a malonyl-CoA–dependent switch between the Nα and the Nβ conformations (López-Viñas et al., 2007; Rao et al., 2011; Samanta et al., 2014). Malonyl-CoA is an intermediate in the novo synthesis of long-chain fatty acids and is synthetized by acetyl-CoA carboxylase (ACC), a downstream target of the master energy sensor AMP-activated protein kinase (AMPK). Crucially, malonyl-CoA levels undergo brain-specific fluctuations depending on nutritional state, becoming reduced during fasting and increased following feeding (Tokutake et al., 2010). Given the capacity of CPT1C to weakly bind malonyl-CoA, several authors have proposed that CPT1C is a malonyl-CoA sensor (Price et al., 2002; Wolfgang et al., 2006; Casals et al., 2016). Despite this, nothing is known about nutritional-dependent regulation of CPT1C function or downstream molecular interactions that enable it to control GluA1 trafficking.
Here, we demonstrate that CPT1C regulates surface GluA1 abundance in cortical neurons in response to metabolic challenges, such as glucose depletion, through the sensing of malonyl-CoA levels. Moreover, we uncovered that CPT1C controls SAC1 translocation to ER-TGN contact sites, as well as SAC1 PI(4)P phosphatase activity in a malonyl-CoA–dependent manner. In basal conditions, CPT1C decreases SAC1 activity, which maintains normal PI(4)P levels at the TGN and GluA1 trafficking to the PM. However, under energy stressful conditions, CPT1C releases SAC1 inhibition and promotes SAC1 translocation to ER-TGN junctions, where it decreases TGN PI(4)P pools to halt GluA1 trafficking. Collectively, we describe a novel regulator of SAC1 phosphatase activity in neurons and unravel a nutritionally dependent mechanism by which CPT1C regulates GluA1 trafficking.
Results
CPT1C regulates GluA1 trafficking through malonyl-CoA sensing
Since CPT1C has been proposed to be a malonyl-CoA sensor that regulates the function of other proteins (Price et al., 2002; Wolfgang et al., 2006; Casals et al., 2016) and is well known to interact with the AMPAR GluA1 (Fadó et al., 2015; Gratacòs-Batlle et al., 2015), we decided to study if CPT1C regulates GluA1 trafficking under different nutrient and hormonal challenges known to modify intracellular malonyl-CoA levels in cortical cultured neurons.
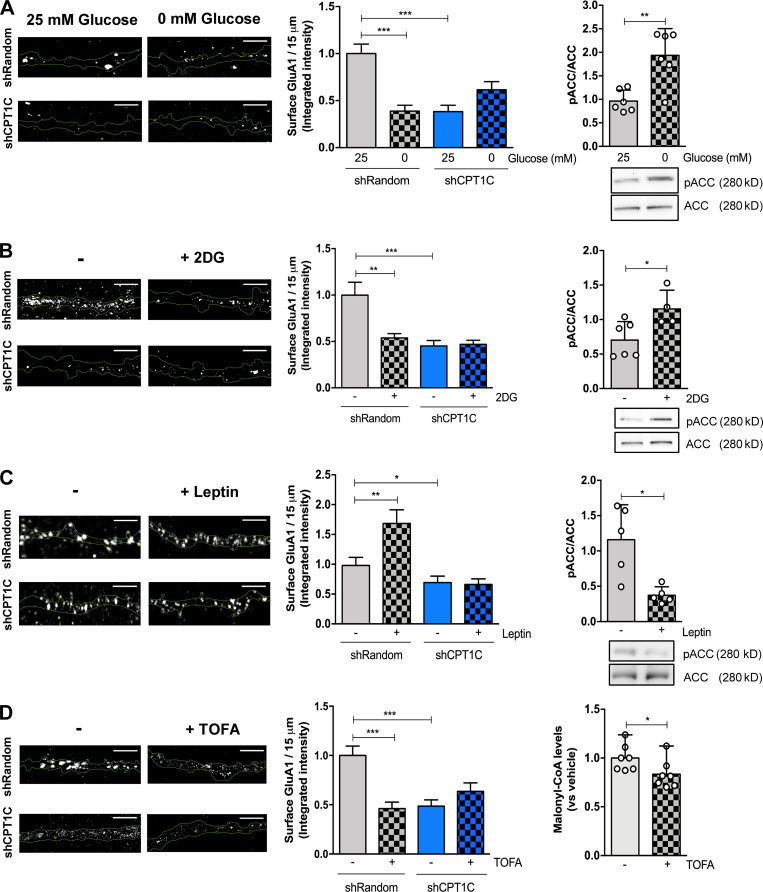
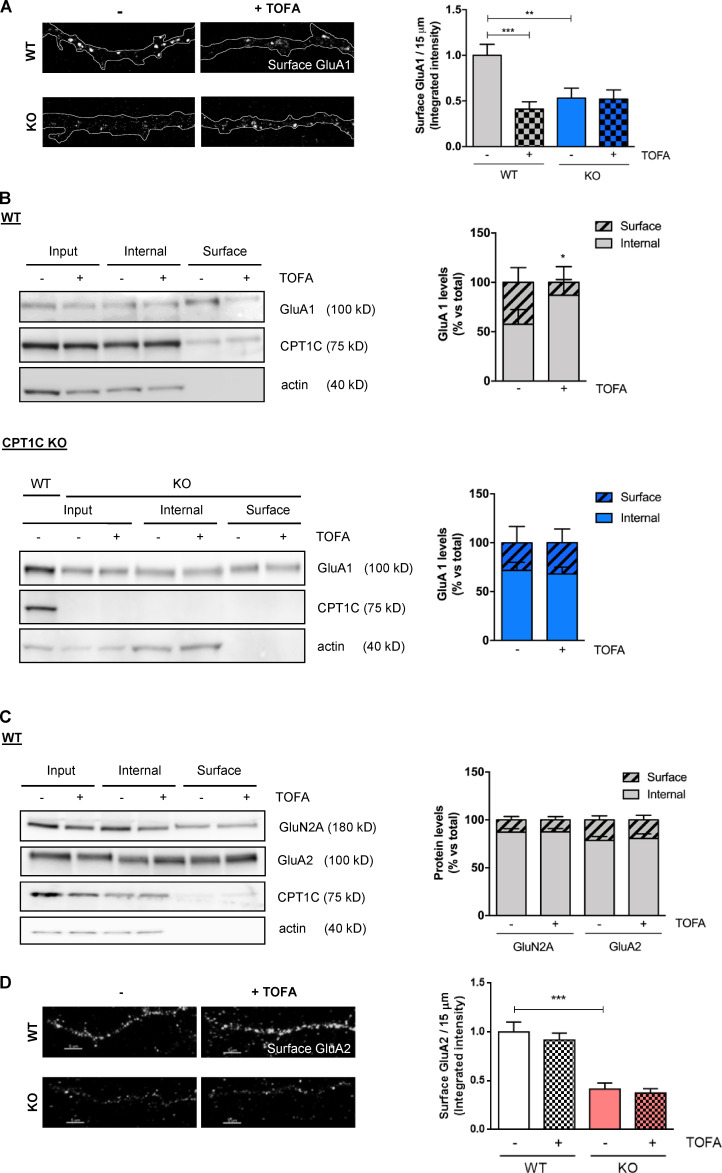
We first analyzed the effect of glucose deprivation. Glucose deprivation for 2 h remarkably decreased the amount of surface GluA1 in cultured cortical neurons. Interestingly, CPT1C-silenced neurons showed lower basal GluA1 levels at the PM, as previously published (Fadó et al., 2015), but they did not further decrease with glucose depletion (Fig. 1 A; Fig. S1 A; and Fig. 2, A and B). Similar findings were obtained with 15 mM 2-deoxy-D-glucose (2DG) treatment, a glucose analogue that cannot be metabolized and competitively inhibits glycolysis (Fig. 1 B and Fig. S1 B). Glucose deprivation or 2DG treatments are known to reduce intracellular malonyl-CoA levels through phosphorylation of ACC (Wolfgang et al., 2007). Consistent with previous results, ACC, the enzyme that synthesizes malonyl-CoA, showed elevated levels of phosphorylation under glucose depletion or 2DG treatment (Fig. 1, A and B, right graphs), which is indicative of its inhibition. Importantly, in both cases, these treatments resulted in no change in neuronal survival (Fig. S2, C and D). Next, we analyzed the effect of leptin addition, an anorexigenic hormone that is known to increase malonyl-CoA levels (Gao et al., 2007; Wolfgang et al., 2007). Consistent with malonyl-CoA levels playing a role in GluA1 surface expression, 50 nM leptin for 30 min promoted an increase in GluA1 surface levels (Fig. 1 C, left; and Fig. S1 C), as previously reported (Moult et al., 2010), and a decrease in ACC phosphorylation (Fig. 1 C, right). This effect was abolished in neurons where CPT1C was knocked down (Fig. 1 C). These results are consistent with the hypothesis that malonyl-CoA regulates GluA1 trafficking and, specifically, that malonyl-CoA down-regulation leads to a CPT1C-dependent decrease in GluA1 at the PM.
Figure 1.
CPT1C is required for metabolic regulation of GluA1 surface levels. (A–D) Surface GluA1 quantification. CPT1C-silenced (shCPT1C; see Fig. S2 A) or control (short hairpin-loop RNA with a random sequence; shRandom) cortical neurons at 14–15 DIV were submitted to glucose deprivation for 2 h (A), 2DG treatment (15 mM, 2 h; B), leptin treatment (50 nM; 30 min; C), or TOFA treatment (20 µg/ml, 2 h; D), respectively. Then, nonpermeabilized cells were fixed and processed by immunocytochemistry using an anti-GluA1 antibody against the N-terminal domain (extracellular). Lentivirus plasmids (pLVTHM-Random-IRES-GFP and pLVTHM-CPT1C-IRES-GFP) used in these experiments produced soluble GFP; therefore, it was feasible to draw masks for the dendrites using Imaris software. Representative images are shown. In each condition, we analyzed 40 neurons from two independent experiments performed by biological duplicates. Results in left graphs are given as the mean ± SEM (two-way ANOVA followed by Bonferroni’s comparison test; **, P < 0.05; ***, P < 0.001). (A) shRandom 25 mM glucose (1.00 ± 0.08, n = 50), shRandom 0 mM glucose (0.55 ± 0.08, n = 45); shCPT1C 25 mM glucose (0.58 ± 0.08, n = 40), and shCPT1C 0 mM glucose (0.82 ± 0.08, n = 44). (B) shRandom + vehicle (1.00 ± 0.07, n = 53), shRandom + 2DG (0.73 ± 0.056, n = 50), shCPT1C + vehicle (0.36 ± 0.05, n = 44), and shCPT1C + 2DG (0.78 ± 0.05, n = 51). (C) shRandom + vehicle (1.00 ± 0.14, n = 51), shRandom + leptin (1.65 ± 0.23, n = 52), shCPT1C + vehicle (0.69 ± 0.11, n = 52), and shCPT1C + leptin (0.66 ± 0.09, n = 52). (D) shRandom + vehicle (1.00 ± 0.10, n = 57), shRandom + TOFA (0.46 ± 0.07, n = 52), shCPT1C + vehicle (0.49 ± 0.06, n = 50), and shCPT1C + TOFA (0.64 ± 0.09, n = 55). Treatment efficiency was determined by phosphorylated ACC (pACC) using Western blotting method in cortical neurons under glucose deprivation (n = 6; 0.96 ± 0.23 for 25 mM glucose and 1.94 ± 0.57 for 0 mM glucose), 2DG treatment (n = 3–6; 0.97 ± 0.28 for vehicle and 2.09 ± 0.21 for 2DG), or leptin treatment (n = 5; 1.16 ± 0.50 for vehicle and 0.37 ± 0.12 for leptin). Results in right graphs are the mean ± SD (Mann-Whitney U test; **, P < 0.01 and *, P < 0.05). TOFA efficiency was evaluated in cortical neurons by malonyl-CoA recycling assay. Values are the mean ± SD of four independent experiments, three of them performed by biological duplicates (n = 7 samples per condition; two-sided Student’s t test; P = 0.049; 1.00 ± 0.05 for vehicle and 0.83 ± 0.06 for TOFA). Scale bars = 5 µm.
Figure S1.
Whole-cell images of cortical neurons showing GluA1 surface levels under different treatments. (A–D) CPT1C-silenced or control cortical neurons were submitted to glucose deprivation for 2 h (A), 2DG treatment (15 mM, 2 h; B), leptin treatment (50 nM, 30 min; C), or TOFA treatment (20 µg/ml, 2 h; D). A short hairpin-loop RNA with a random sequence (shRandom) was used as a control. Surface GluA1 was detected by immunocytochemistry in nonpermeabilized cells. Lentiviral infection was confirmed by the detection of the GFP tag. Complete images used in Fig. 1 are shown. The selected ROI in each image is identified with a rectangular box. Scale bars = 7 µm for A, B, and D and 10 µm for C.
Figure 2.
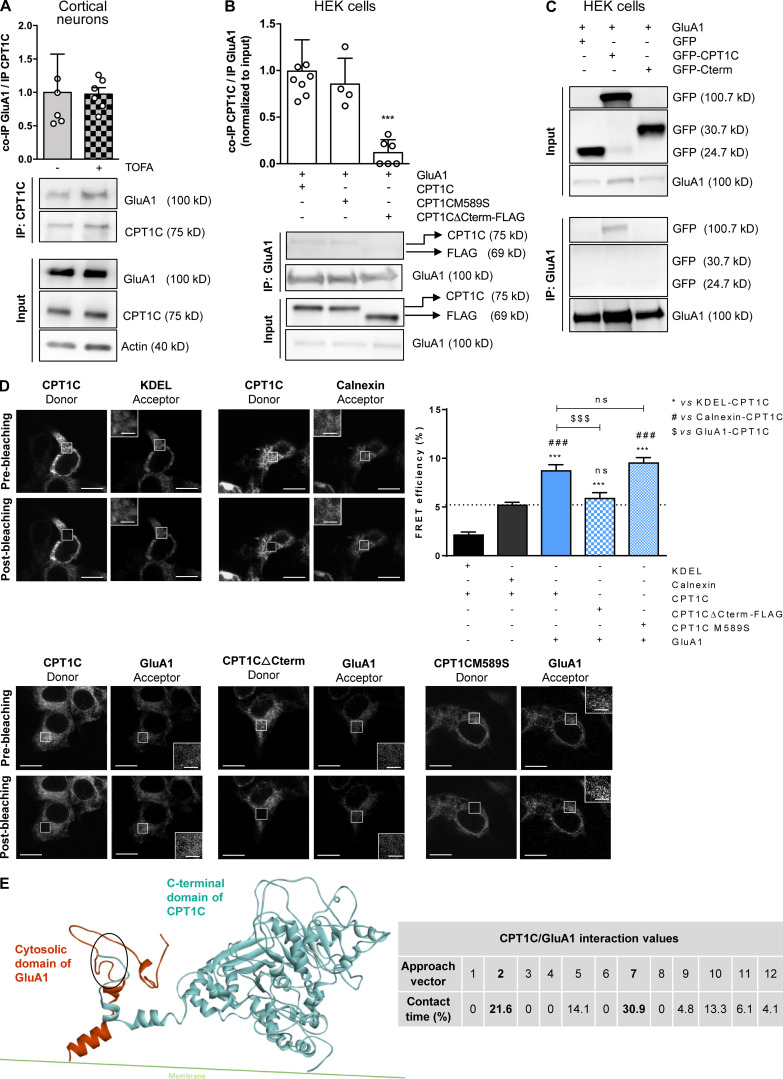
CPT1C–GluA1 interaction is dependent on CPT1C C-terminal domain but not on malonyl-CoA levels. (A) Quantification of coimmunoprecipitated GluA1 after CPT1C immunoprecipitation in TOFA-treated cortical neurons. Results are the mean ± SD from three independent experiments performed by biological duplicates (n = 5 or 6 samples per condition; Mann-Whitney U test; P = 0.6991; 1.00 ± 0.57 for vehicle and 0.97 ± 0.24 for TOFA). (B) Coimmunoprecipitated CPT1C quantification versus immunoprecipitated GluA1 in HEK cells cotransfected with GluA1 and WT CPT1C, CPT1CM589S (a mutated form insensitive to malonyl-CoA), or CPT1CΔCterm-FLAG (a truncated form in which the last 39 aa have been substituted by the tag FLAG). Whole lysates (input) were used to corroborate a proper transfection. Western blots using antibodies against CPT1C, FLAG tag, and GluA1 were performed. Values are the mean ± SD from three independent experiments performed by biological replicates (one-way ANOVA followed by Bonferroni’s comparison test; ***, P < 0.001). GluA1 + WT CPT1C (1.00 ± 0.34, n = 8), GluA1 + CPT1CM589S (0.85 ± 0.28, n = 4), and GluA1 + CPT1CΔCterm-FLAG (0.12 ± 0.14, n = 6). (C) Coimmunoprecipitated GFP, CPT1C-GFP, or GFP-Cterm (GFP fused with the last 36 aa of CPT1C) in GluA1 immunoprecipitated samples from transfected HEK cells. Representative Western blots from four independent experiments are shown. (D) Percentage of FRET effciency between CFP-GluA1 and different YFP-CPT1C forms in transfected HEK cells (blue bars). KDEL (a short sequence with ER localization)/CPT1C and calnexin (an ER resident protein)/CPT1C pairs were used as negative controls for CPT1C interaction (black bars). GluA1/WT CPT1C pair was used as a positive control (solid blue). The dashed line represents the higher percentage of FRET of the negative control pairs. Results are the mean ± SEM from three independent experiments performed by biological duplicates (one-way ANOVA followed by Bonferroni’s comparison test; ***, P < 0.001 versus KDEL/CPT1C pair; ###, P < 0.001 versus calnexin/CPT1C pair; $$$, P < 0.001 versus GluA1/CPT1C pair). KDEL + CPT1C (2.13 ± 0.31, n = 36), calnexin + CPT1C (5.18 ± 0.30, n = 36), GluA1 + CPT1C (8.70 ± 0.65, n = 30), GluA1 + CPT1CΔCterm-FLAG (5.87 ± 0.59, n = 30), and GluA1 + CPT1CM589S (9.51 ± 0.56, n = 26). Scale bars = 10 µm; scale bars of inset magnifications = 2 µm. ns, not significant. (E) C-terminal CPT1C region encompassing the catalytic globular domain and the cytosolic GluA1 domain was used for protein–protein interaction simulations. The percentage of contact time for each of the 12 approaching vectors analyzed is shown in the table. The two approaching vectors with the highest contact time are shown in bold. A ribbon representation of the GluA1 (orange) and CPT1C (blue) interaction frame from approach vector 7 is shown. The main contact region is highlighted by the oval and corresponds to residues 843–851 for GluA1 and 786–792 for CPT1C (localized in the C-terminal domain, which ends in residue 803).
Figure S2.
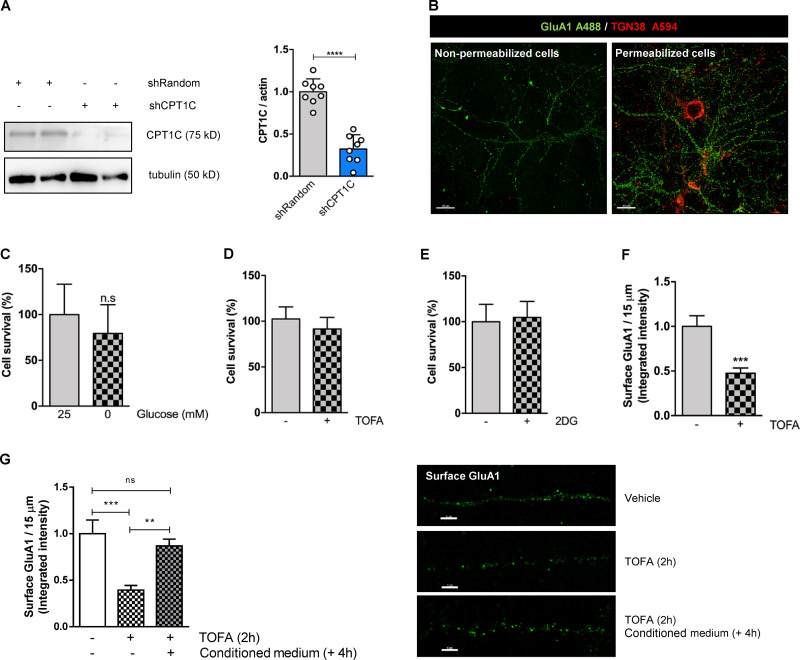
Glucose deprivation, 2DG, and TOFA treatments have no effect on cortical neuron survival. (A) CPT1C silencing in cortical neurons. Cells were infected at 7 DIV with a shCPT1C-carrying lentivirus or a short hairpin-loop RNA with a random sequence (shRandom) was used as a control. At 14–15 DIV, CPT1C down-regulation was confirmed by Western blot. A representative image is shown. Graph shows the mean ± SD from three independent experiments performed by biological replicates (n = 8 per condition; two-sided Student’s t test; ****, P < 0.0001; 1.00 ± 0.15 for shRandom and 0.32 ± 0.17 for shCPT1C). (B) GluA1 and TGN38 detection in nonpermeabilized and permeabilized neurons. To make sure that chemical fixation with 4% of paraformaldehyde does not induce permeabilization itself, GluA1 and TGN38 were immunodetected by immunocytochemistry in cultured cortical neurons permeabilized or nonpermeabilized with 0.1% (vol/vol) of Triton X-100. As expected, only surface GluA1 was stained in nonpermeabilized neurons, since TGN38 (a Golgi-resident protein) and total GluA1 (surface and internal) were detected in permeabilized cells. Scale bars = 20 µm. (C–E) Cell survival after 2 h of glucose starvation (C), 2DG treatment (15 mM; D), or TOFA treatment (20 µg/ml; E) in cortical neurons was determined by MTT assay. Values are the mean ± SD from two independent experiments performed by biological replicates (two-sided Student’s t test; P > 0.05). (C) 25 mM glucose (100.00 ± 33.24, n = 17) and 0 mM glucose (75.25 ± 17.33, n = 15). n.s, not significant. (D) Vehicle (100.00 ± 13.22, n = 12) and TOFA (91.55 ± 12.63, n = 11). (E) Vehicle (100.00 ± 18.99, n = 10) and 2DG (104.7 ± 17.45, n = 7). (F) TOFA treatment (microgram per milliliter, 2 h) down-regulates GluA1 surface levels in hippocampal neurons. All data represent mean ± SEM of two independent experiments (n = 20 cells per condition; two-sided Student’s t test; ***, P = 0.0004; 1.00 ± 0.12 for vehicle and 0.47 ± 0.06 for TOFA). (G) Reversibility of TOFA-induced effects on GluA1 surface levels. Cultured cortical neurons were treated at 14 DIV with TOFA (microgram per milliliter) or vehicle (DMSO; 1:500). 2 h later, TOFA was removed, replacing the medium with conditioned media (Cond. medium) from sibling plates, and cells were incubated for an additional 4 h. The rest of the wells were fixed at 2 h after treatment. In all conditions, surface GluA1 levels were detected by immunocytochemistry. Data represent mean ± SEM of one experiment performed by biological duplicates (one-way ANOVA; **, P < 0.01 and ***, P < 0.001). Vehicle (1.00 ± 0.15, n = 33), TOFA (0.39 ± 0.05, n = 37), and reversion (0.87 ± 0.07, n = 34). Scale bars = 5 µm. ns, not significant.
To test this hypothesis, we decided to directly down-regulate malonyl-CoA levels through treatment with an ACC inhibitor, 5-tetradecyloxy-2-furoic acid (TOFA). TOFA is an acetyl-CoA analogue that cannot be further metabolized to malonyl-CoA. In consequence, TOFA does not modify ACC phosphorylation levels. We verified that 20 µg/ml TOFA treatment for 2 h did reduce malonyl-CoA levels in cortical neurons (Fig. 1 D, right) and that it did not produce neuronal death (Fig. S2 E). Fig. 1 D and Fig. S1 D show that neurons treated with TOFA had significantly reduced levels of GluA1 at the PM compared with control cells. Importantly, this response was not detected in CPT1C silenced neurons. The regulation of GluA1 trafficking by TOFA treatment was reversible and was also observed in cultured hippocampal neurons (Fig. S2, F and G). To increase rigor and further underscore these results, immunocytochemistry and surface biotinylation studies were also conducted in cortical neurons derived from CPT1C knockout (KO) mice embryos (CPT1C−/−), with comparable results observed (Fig. S3, A and B). Interestingly, this regulation was specific to the GluA1 subunit, as no changes in surface levels of AMPAR subunit GluA2 or NMDA receptor subunit 2A (GluN2A) were observed after TOFA treatment (Fig. S3, C and D). Collectively, these results strongly link CPT1C and malonyl-CoA levels as part of a rheostat for controlling GluA1 subunit surface expression.
Figure S3.
TOFA treatment decreases the levels of GluA1 but not GluA2 and GluN2A at the surface of cortical neurons in a CPT1C-dependent manner. (A) Surface GluA1 quantification by immunochemistry in nonpermeabilized WT and CPT1C KO cortical neurons treated with TOFA. Values are the mean ± SEM of two independent experiments (two-way ANOVA followed by Bonferroni’s comparison test; **, P < 0.01 and ***, P < 0.001). WT + vehicle (1.00 ± 0.12, n = 67), WT + TOFA (0.41 ± 0.08, n = 45), KO + vehicle (0.53 ± 0.11, n = 36), and KO + TOFA (0.52 ± 0.10, n = 38). (B) Surface GluA1 levels measured by the biotinylation assay in WT and CPT1C KO cortical neurons. Results are given as the mean ± SD from three independent experiments performed by biological replicates (n = 4 samples per condition; two-way ANOVA followed by Bonferroni’s comparison test; *, P < 0.05 for surface levels). WT + vehicle (57.54 ± 7.47 for surface and 42.46 ± 7.47 for internal, n = 4), WT + TOFA (13.05 ± 7.96 for surface and 86.95 ± 7.96 for internal, n = 4), KO + vehicle (28.31 ± 16.55 for surface and 71.69 ± 16.55 for internal, n = 4), and KO + TOFA (31.95 ± 14.05 for surface and 68.07 ± 14.05 for internal, n = 4). (C) Surface GluN2A and GluA2 quantification by biotinylation in cortical neurons. Results are mean ± SD from two independent experiments performed by biological replicates (two-way ANOVA followed by Bonferroni’s comparison test; P > 0.05). GluN2A vehicle (12.78 ± 7.17 for surface and 87.22 ± 7.17 for internal, n = 4), GluN2A TOFA (12.55 ± 6.87 for surface and 87.45 ± 6.87 for internal, n = 4), GluA2 vehicle (22.59 ± 10.24 for surface and 77.40 ± 10.24 for internal, n = 5), and GluA2 TOFA (19.22 ± 12.43 for surface and 80.78 ± 12.43 for internal, n = 5). (D) Surface GluA2 quantification by immunocytochemistry in WT and CPT1C KO cortical neurons. Results are mean ± SEM of one experiment performed by biological duplicates (two-way ANOVA followed by Bonferroni’s comparison test; ***, P < 0.001). WT + vehicle (1.00 ± 0.10, n = 37), WT + TOFA (0.92 ± 0.07, n = 34), KO + vehicle (0.41 ± 0.06, n = 32), and KO + TOFA (0.37 ± 0.04, n = 24). Scale bars = 5 µm.
C-terminal domain of CPT1C is necessary for interaction with GluA1
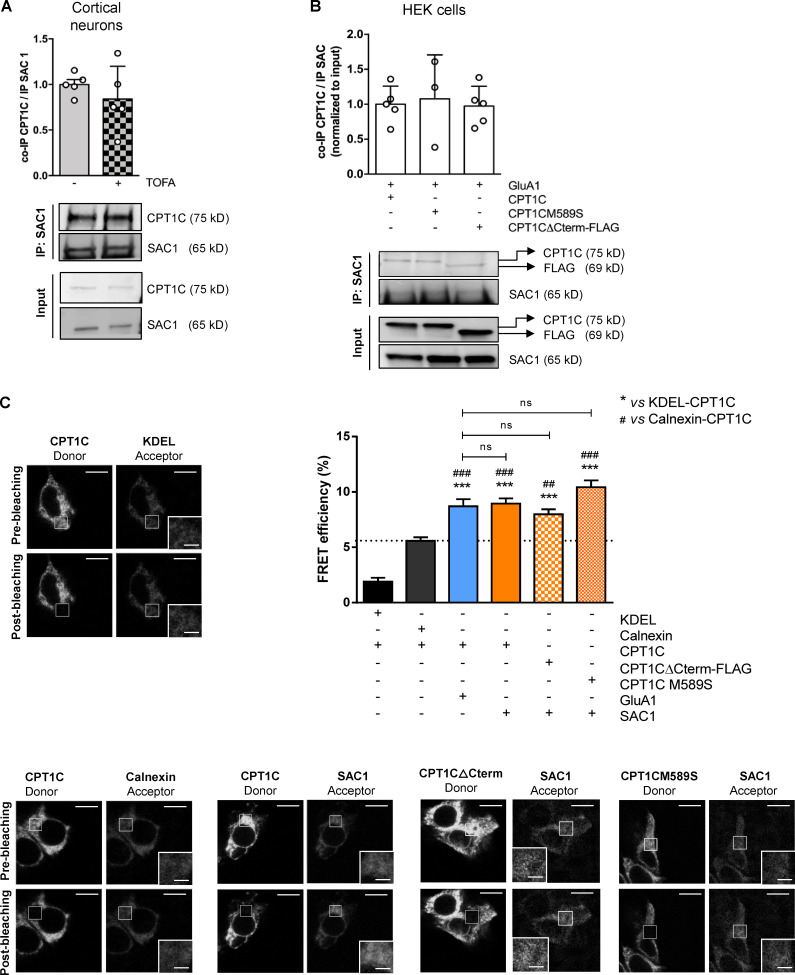
Given that CPT1C strongly interacts with GluA1 (Schwenk et al., 2012; Fadó et al., 2015) and, as outlined above, malonyl-CoA is a regulator of GluA1 trafficking in a CPT1C-dependent manner, we next analyzed whether malonyl-CoA could regulate CPT1C–GluA1 interaction. Moreover, since the last 36 aa of the protein are one of the main differences between CPT1C and the mitochondrial isoforms CPT1A and CPT1B, which lack that region (Price et al., 2002; Sierra et al., 2008), we considered the C-terminus as a possible region facilitating CPT1C-GluA1 interactions.
To address this, a mutant CPT1C insensitive to malonyl-CoA (CPT1CM589S; Morillas et al., 2004; Rodríguez-Rodríguez et al., 2019) and a truncated CPT1C in which the last 39 aa were replaced by the FLAG tag (CPT1CΔCterm-FLAG) were transfected into HEK293 cells. Coimmunoprecipitation assays showed that malonyl-CoA does not seem to facilitate CPT1C-GluA1 interactions, since no differences were observed after TOFA treatment (Fig. 2 A) or with the CPT1CM589S mutant compared with the WT CPT1C (Fig. 2 B). However, CPT1C lacking the C-terminus (CPT1CΔCterm) hardly coimmunoprecipitated with GluA1 (Fig. 2 B), suggesting that CPT1C was interacting with this AMPA subunit through its C-terminal domain. When only the last 36 aa of CPT1C were fused directly to GFP (GFP-Cterm), the coimmunoprecipitation was not observed (Fig. 2 C), indicating that the last 36 aa alone are necessary but not enough for the interaction. Similar interaction results were observed using fluorescence resonance energy transfer (FRET) assays in HEK cells: a positive interaction between GluA1 and CPT1C or CPT1CM589S, but not with CPT1CΔCterm-FLAG, was observed (Fig. 2 D).
To further identify a putative interaction region between the two proteins, in silico molecular dynamic simulations were conducted taking the C-terminal cytosolic domain of GluA1 and the C-terminal plus the catalytic globular cytosolic domain of CPT1C under consideration. 12 different starting positions, each of them differing in the initial GluA1 fragment positioning relative to the CPT1C fragment and referred to as “approaching vectors,” were employed in order to most efficiently map all the potential contact spaces (Knapp et al., 2018). Fig. 2 E shows that five of the approaching vectors presented a contact time percentage >5%, which according to previous literature can be considered a statistically significant amount of interaction, enough to determine that contact exists (see Convertino et al., 2009; Domínguez et al., 2015). A ribbon representation for the GluA1 and CPT1C interaction frame from the approaching vector 7 (the one with the highest contact time) is shown and provides further evidence supporting a physical interaction between GluA1 and CPT1C.
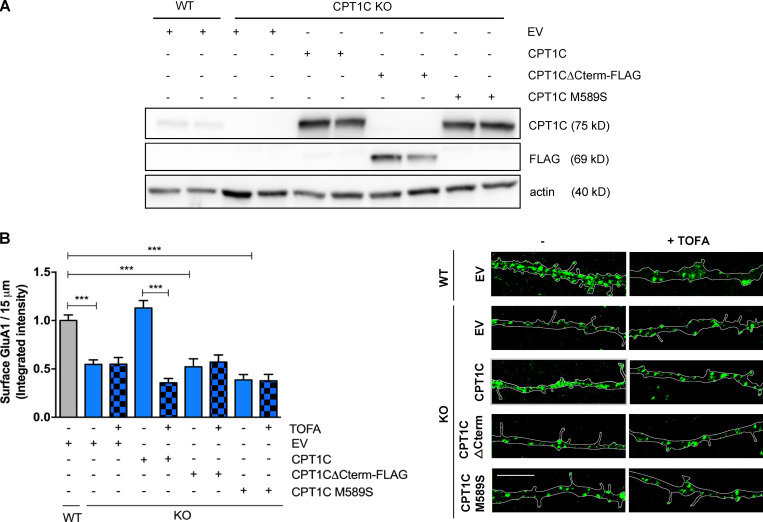
Next, we tested if (1) the CPT1C C-terminal domain was required for GluA1 transport to the PM and (2), even though malonyl-CoA binding is not a determinant for CPT1C-GluA1 interactions, if its sensing is necessary to properly regulate GluA1 trafficking. To this end, CPT1C−/− neurons were infected at 7 d in vitro (DIV) with lentiviral vectors carrying the truncated CPT1CΔCterm-FLAG, the CPT1CM589S mutant, or the WT CPT1C. The expression of these constructs was confirmed a week later (Fig. 3 A). Transfected neurons were then treated with TOFA or vehicle, and GluA1 surface levels were analyzed by immunostaining. As expected, KO neurons showed a robust reduction of GluA1 levels at the PM compared with WT, which was rescued by full-length CPT1C (Fig. 3 B). However, neither the C-terminal–truncated nor the malonyl-CoA–insensitive mutated forms were able to restore GluA1 at the PM. Likewise, TOFA-mediated down-regulation of GluA1 surface levels was only effective in full-length CPT1C-expressing neurons. These data support the hypothesis that the C-terminal domain of CPT1C and malonyl-CoA binding are necessary for the interaction and trafficking of GluA1.
Figure 3.
CPT1C C-terminal and binding to malonyl-CoA are needed to regulate GluA1 trafficking. (A and B) Surface GluA1 level recovery by CPT1C reexpression in KO neurons. CPT1C KO cortical neurons were transduced at 7 DIV with lentiviral vectors carrying CPT1C, CPT1CM589S, CPT1CΔCterm-FLAG, or an EV. WT neurons transduced with the EV were used as the control. At 14-15 DIV, cells were treated with TOFA (20 µg/ml, 2 h) or the vehicle (DMSO; 1:500). Surface GluA1 levels were detected and quantified as in Fig. 1. Proper overexpression of different CPT1C forms was corroborated by Western blot in A. Data in B represent the mean ± SEM from three independent experiments performed by biological duplicates (one-way ANOVA followed by Bonferroni’s comparison test; ***, P < 0.001). WT + EV + vehicle (1.00 ± 0.02, n = 17), KO + EV + vehicle (0.55 ± 0.05, n = 80), KO + EV + TOFA (0.54 ± 0.069, n = 63), KO + CPT1C + vehicle (1.13 ± 0.07, n = 85), KO + CPT1C + TOFA (0.35 ± 0.04, n = 68), KO + CPT1CΔCterm-FLAG + vehicle (0.52 ± 0.08, n = 54), KO + CPT1CΔCterm-FLAG + TOFA (0.57 ± 0.07, n = 56), KO + CPT1CM589S + vehicle (0.38 ± 0.05, n = 52), and KO + CPT1CM589S + TOFA (0.38 ± 0.07, n = 33). Scale bars = 5 µm.
CPT1C silencing or decreased malonyl-CoA production trigger GluA1 retention at the TGN by decreasing PI(4)P
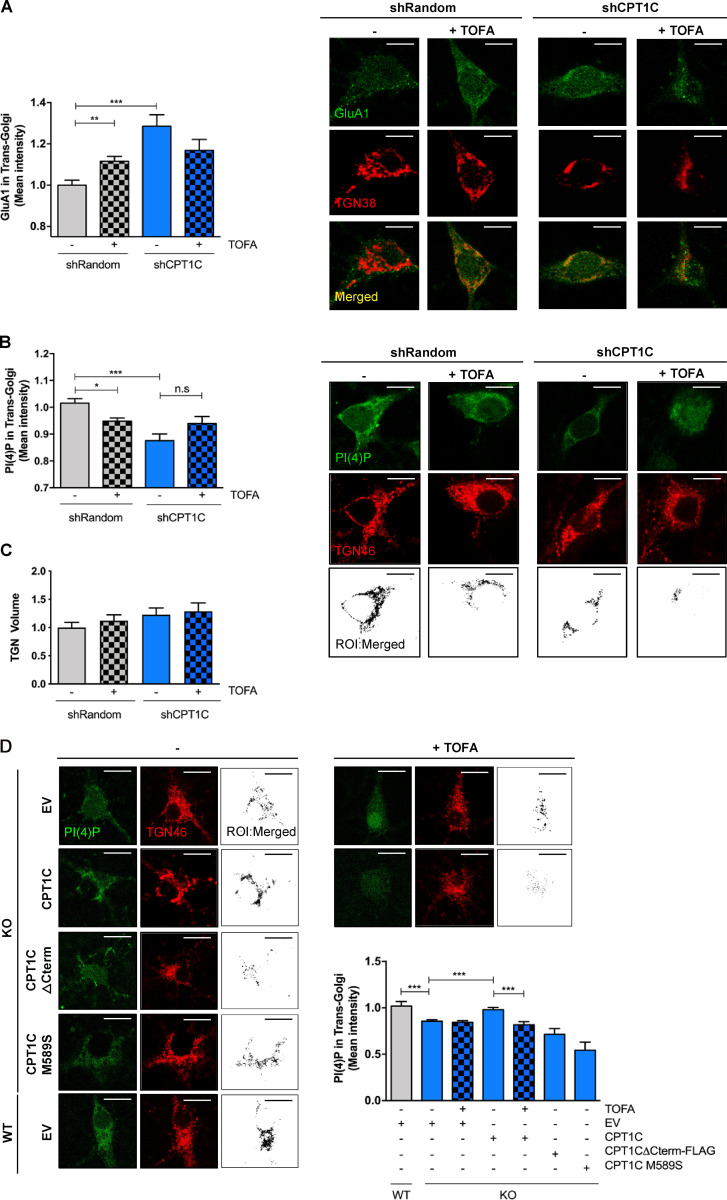
Next, we wanted to determine the precise molecular mechanism through which CPT1C regulates GluA1 trafficking. To begin, we focused on the TGN, a central trafficking hub for GluA1 receptors on their way to the PM (reviewed by Jacobi and von Engelhardt, 2018; Yang et al., 2013). Treating cortical neurons with TOFA to decrease malonyl-CoA production resulted in the significant accumulation of GluA1 in the TGN (Fig. 4 A). Additionally, knockdown of CPT1C resulted in a similar GluA1 retention pattern at the TGN. These data point to the TGN as a potential location where CPT1C and malonyl-CoA exert influence over GluA1 trafficking.
Figure 4.
CPT1C regulates GluA1 and PI(4)P levels at the TGN in a malonyl-CoA–dependent manner. (A) GluA1 retention at the TGN. CPT1C was silenced in WT cortical neurons by the transduction of shCPT1C-carrying lentivirus. A short hairpin-loop RNA with a random sequence (shRandom) was used as a control. At 14–15 DIV, cells were treated with TOFA (20 µg/ml, 2 h), fixed, and permeabilized in order to analyze the levels of GluA1 in the TGN by coimmunocytochemistry. TGN38 protein was used to localize and define the TGN compartment. The mean intensity of GluA1 in the TGN compartment per each neuron soma was analyzed. Graph shows the mean ± SEM of 20 cells from two independent experiments performed by biological duplicates (two-way ANOVA followed by Bonferroni’s comparison test; **, P < 0.01 and ***, P < 0.001). shRandom + vehicle (1.00 ± 0.02, n = 19), shRandom + TOFA (1.12 ± 0.03, n = 27), shCPT1C + vehicle (1.29 ± 0.06, n = 23), and shCPT1C + TOFA (1.17 ± 0.06, n = 21). Scale bars = 7 µm. (B) Quantification of PI(4)P levels in the TGN compartment by immunocytochemistry in control or CPT1C-silenced neurons after TOFA treatment (20 µg/ml, 2 h). PI(4)P was detected by immunocytochemistry using a specific antibody purchased from Echelon (Z-P004). TGN46 protein was used to localize and define the TGN compartment. Below, black-and-white images show the ROI generated by TGN-PI(4)P colocalizing pixels. The mean intensity of PI(4)P in the TGN compartment per each neuron soma was analyzed. Data represent the mean ± SEM from four independent experiments performed by biological duplicates (two-way ANOVA followed by Bonferroni’s comparison test; *, P < 0.05 and ***, P < 0.001). shRandom + vehicle (1.00 ± 0.02, n = 42), shRandom + TOFA (0.95 ± 0.01, n = 48), shCPT1C + vehicle (0.88 ± 0.03, n = 39), and shCPT1C + TOFA (0.94 ± 0.04, n = 37). Scale bars = 7 µm. n.s, not significant. (C) Quantification of the TGN volume per each neuron soma in the same experiments and conditions as in B. Values are the mean ± SEM from two independent experiments performed by biological replicates (two-way ANOVA; P > 0.05). shRandom + vehicle (1.00 ± 0.09, n = 22), shRandom + TOFA (1.12 ± 0.10, n = 23), shCPT1C + vehicle (1.23 ± 0.11, n = 19), and shCPT1C + TOFA (1.29 ± 0.13, n = 18). (D) Quantification of PI(4)P levels in the TGN compartment in CPT1C KO cortical neurons overexpressing or not CPT1C full or different mutated CPT1C forms and submitted to TOFA treatment (20 µg/ml, 2 h). Analysis was performed as in Fig. 4 B. On the right, black-and-white images show the ROI generated by TGN-PI(4)P colocalizing voxels. Results are shown as the mean ± SEM from two independent experiments performed by biological duplicates (one-way ANOVA followed by Bonferroni’s comparison test; ***, P < 0.001). WT + EV + vehicle (1.00 ± 0.05, n = 7), KO + EV + vehicle (0.86 ± 0.01, n = 43), KO + EV + TOFA (0.84 ± 0.02, n = 40), KO + CPT1C + vehicle (0.98 ± 0.02, n = 36), KO + CPT1C + TOFA (0.81 ± 0.03, n = 46), KO + CPT1CΔCterm-FLAG + vehicle (0.71 ± 0.06, n = 26), and KO + CPT1CM589S + vehicle (0.54 ± 0.08, n = 25). Scale bars = 7 µm.
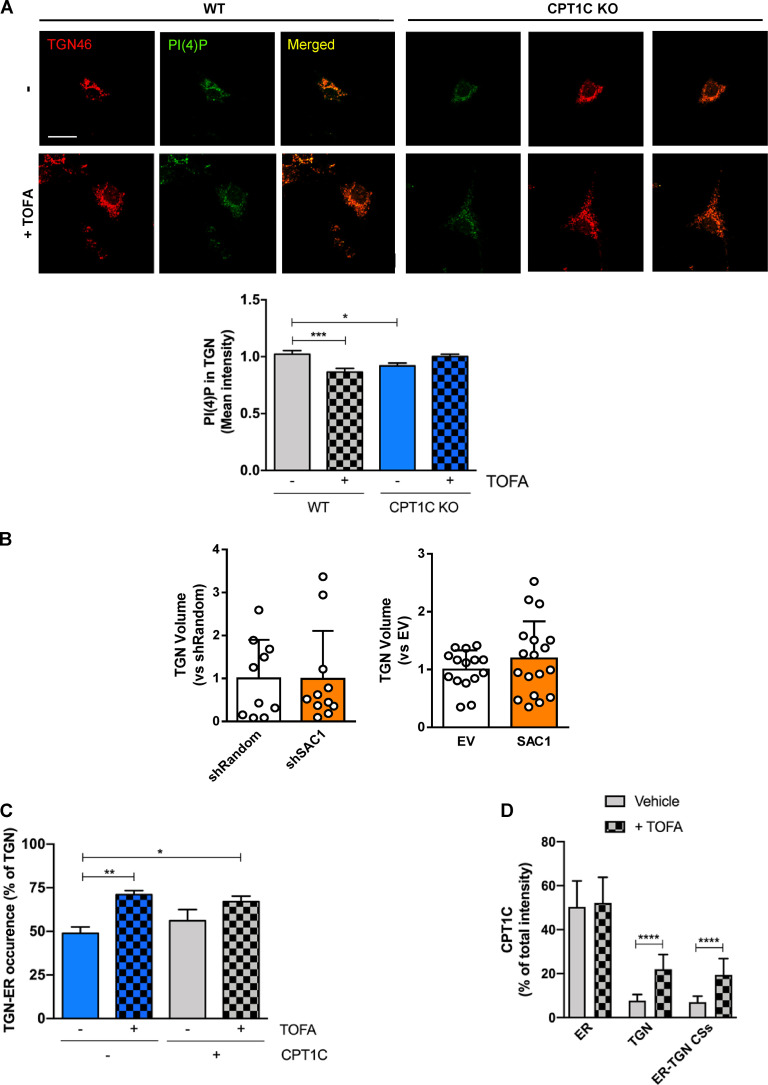
Previously, it was demonstrated that TGN PI(4)P controls GluA1 vesicular release from the TGN (Yang et al., 2013); therefore, we asked if CPT1C or malonyl-CoA fluctuations can alter TGN PI(4)P levels. To test this hypothesis, WT cortical neurons were treated with TOFA or the short hairpin-loop RNA of CPT1C (shCPT1C), and TGN PI(4)P levels were quantified using immunocytochemistry. As shown in Fig. 4 B, both knocking down CPT1C or treatment with TOFA resulted in a significant decrease of PI(4)P levels at the TGN. TOFA-dependent decreases in TGN pools of PI(4)P were detected only in WT neurons, with no further decrease observed in CPT1C-silenced neurons. Changes in PI(4)P occurred independently of alterations in the size of the TGN (Fig. 4 C). Underscoring these data, similar results were obtained when comparing WT and CPT1C−/− neurons and using a specific PI(4)P biosensor (mCherry-P4M; Fig. S4 A). To further corroborate the involvement of CPT1C in controlling Golgi PI(4)P pools in a malonyl-CoA–dependent manner, we expressed either the insensitive malonyl-CoA CPT1CM589S, the truncated CPT1CΔCterm-FLAG, or the WT form of CPT1C in KO neurons. As expected, the full-length CPT1C form was able to rescue PI(4)P levels in the TGN compartment (Fig. 4 D), while neither the malonyl-CoA–insensitive nor the truncated form could restore Golgi PI(4)P pools in CPT1C−/− neurons.
Figure S4.
Effects of TOFA treatment or SAC1 modulation on PI(4)P levels, TGN volume, and TGN-ER occurrence. (A) Quantification of PI(4)P levels using the specific plasmid mCherry-P4M at the TGN compartment (TGN46) in WT or CPT1C KO neurons after TOFA treatment (20 µg/ml, 2 h). Data represent the mean ± SEM from two independent experiments performed by biological duplicates (two-way ANOVA followed by Bonferroni’s comparison test; *, P < 0.05 and ***, P < 0.001). WT + vehicle (1.00 ± 0.03, n = 26), WT + TOFA (0.87 ± 0.03, n = 26), KO + vehicle (0.92 ± 0.02, n = 20), and KO + TOFA (1.00 ± 0.02, n = 28). Scale bar = 7 µm. (B) SAC1 modulation (overexpressing, OE, or knocking down, KD) does not change TGN volume. The same images used in Fig. 6, B and E were taken to analyze TGN volume in KD or OE SAC1 neurons. Values are the mean ± SD from two independent experiments (Mann-Whitney U test and P = 0.8444 for KD experiment; two-sided Student’s t test and P = 0.3062 for OE experiment). For KD experiment, short hairpin-loop RNA with a random sequence (shRandom; 1.00 ± 0.90, n = 10) and shSAC1 (0.99 ± 1.11, n = 11). For OE experiment, EV (1.00 ± 0.33, n = 15) and SAC1 (1.20 ± 0.64, n = 18). (C) TGN-ER occurrence increases upon TOFA treatment. The same images used in Fig. 7, B–D were taken to analyze the percentage of TGN that was forming contacts with the ER. Results are the ratio between the TGN-ER contact area and the TGN area per cell, given as a percentage. Data represent the mean ± SD of two independent experiments performed by biological duplicates (two-way ANOVA followed by Bonferroni’s comparison test; *, P < 0.05 and **, P < 0.01). EV + vehicle (0.49 ± 0.13, n = 13), EV + TOFA (0.71 ± 0.08, n = 13), CPT1C + vehicle (0.56 ± 0.23, n = 13), and CPT1C + TOFA (0.67 ± 0.11, n = 13). (D) CPT1C intensity at ER-TGN contact sites. The images used in Fig. 7 E were also used for analyzing CPT1C intensity inside the ROIs of the ER, TGN, and ER-TGN contacts (CSs) relative to the total CPT1C intensity in the whole cell (expressed in percentage). The graph represents the mean ± SD from two independent experiments performed by biological duplicates (two-way ANOVA followed by Bonferroni’s comparison test; ****, P < 0.001). ER vehicle (50.37 ± 11.83, n = 13), ER TOFA (52.22 ± 11.69, n = 15), TGN vehicle (7.74 ± 2.73, n = 13), TGN TOFA (21.99 ± 7.02, n = 15), ER-TGN vehicle (7.04 ± 2.72, n = 13), and ER-TGN TOFA (19.43 ± 7.66, n = 15).
Taken together, our data indicate that CPT1C sensing of malonyl-CoA regulates GluA1 retention at the TGN through the regulation of PI(4)P levels. Moreover, results suggest that the C-terminal domain of CPT1C is necessary for this regulation.
Characterization of CPT1C–SAC1 interaction
As mentioned above, CPT1C regulates PI(4)P levels in the TGN compartment. This finding can be explained by the involvement of the TGN lipid phosphatase SAC1, which is a strong regulator of PI(4)P (reviewed by Del Bel and Brill, 2018) and has been reported to interact with CPT1C (Brechet et al., 2017). Thus, we explored a potential role for malonyl-CoA sensing and the CPT1C C-terminus in tuning CPT1C-SAC1 interactions.
Coimmunoprecipitation studies were performed in cortical neurons with endogenous proteins and in HEK cells overexpressing SAC1 and WT CPT1C, CPT1CM589S, or CPT1CΔCterm-FLAG. Results presented in Fig. 5 A confirm the previously published interaction between SAC1 and CPT1C. Treatment with TOFA (to reduce malonyl-CoA; Fig. 5 A) or CPT1CM589S expression (Fig. 5 B) both failed to alter interactions between CPT1C and SAC1. Moreover, the last 36 aa of CPT1C were not required for an efficient interaction between CPT1C and SAC1 (Fig. 5 B). Similar results were obtained using FRET assays (Fig. 5 C). These results indicate that the interaction of CPT1C with SAC1 is not modulated by malonyl-CoA and that the C-terminal domain of CPT1C does not appear to play a critical role.
Figure 5.
CPT1C–SAC1 interaction is dependent on neither malonyl-CoA levels nor CPT1C C-terminal domain. (A) Quantification of coimmunoprecipitated CPT1C after SAC1 immunoprecipitation in TOFA-treated cortical neurons. Results are the mean ± SD from two independent experiments performed by biological replicates (n = 5 samples per condition; Mann-Whitney U test; P = 0.4206; 1.00 ± 0.12 for vehicle and 0.84 ± 0.36 for TOFA). (B) Quantification of coimmunoprecipitated CPT1C by SAC1 immunoprecipitation in cotransfected HEK cells. Western blot using antibodies against CPT1C, FLAG tag, and SAC1 was performed. Values are the mean ± SD from three independent experiments (one-way ANOVA followed by Bonferroni’s comparison test; P > 0.05; SAC1 + WT CPT1C 1.00 ± 0.26, n = 5; SAC1 + CPT1CM589S 1.08 ± 0.63, n = 3; and SAC1 + CPT1CΔCterm-FLAG 0.97 ± 0.28, n = 5). (C) Percentage of FRET efficiency between SAC1 and different CPT1C mutated forms in transiently transfected HEK293 cells (orange bars). KDEL/CPT1C and calnexin/CPT1C pairs were used as negative controls for CPT1C interaction (black bars). The GluA1/WT CPT1C pair was used as a positive control (blue bar). The dashed line indicates the highest percentage of FRET corresponding to a negative interaction. Results are the mean ± SEM from three independent experiments performed by biological replicates (n = 30 cells per condition; one-way ANOVA followed by Bonferroni’s comparison test; ***, P < 0.001 versus KDEL/CPT1C pair; ##, P < 0.01, and ###, P < 0.001 versus calnexin/CPT1C pair). KDEL + CPT1C (1.91 ± 0.33, n = 27), calnexin + CPT1C (5.58 ± 0.32, n = 31), GluA1 + CPT1C (8.70 ± 0.65, n = 30), SAC1 + CPT1C (8.94 ± 0.48, n = 31), SAC1 + CPT1CΔCterm-FLAG (7.98 ± 0.45, n = 36), and SAC1 + CPT1CM589S (10.42 ± 0.62, n = 31). Scale bars = 10 µm; scale bars of inset magnifications = 2 µm. ns, not significant.
SAC1 is a key modulator of GluA1 trafficking downstream of the malonyl-CoA/CPT1C axis
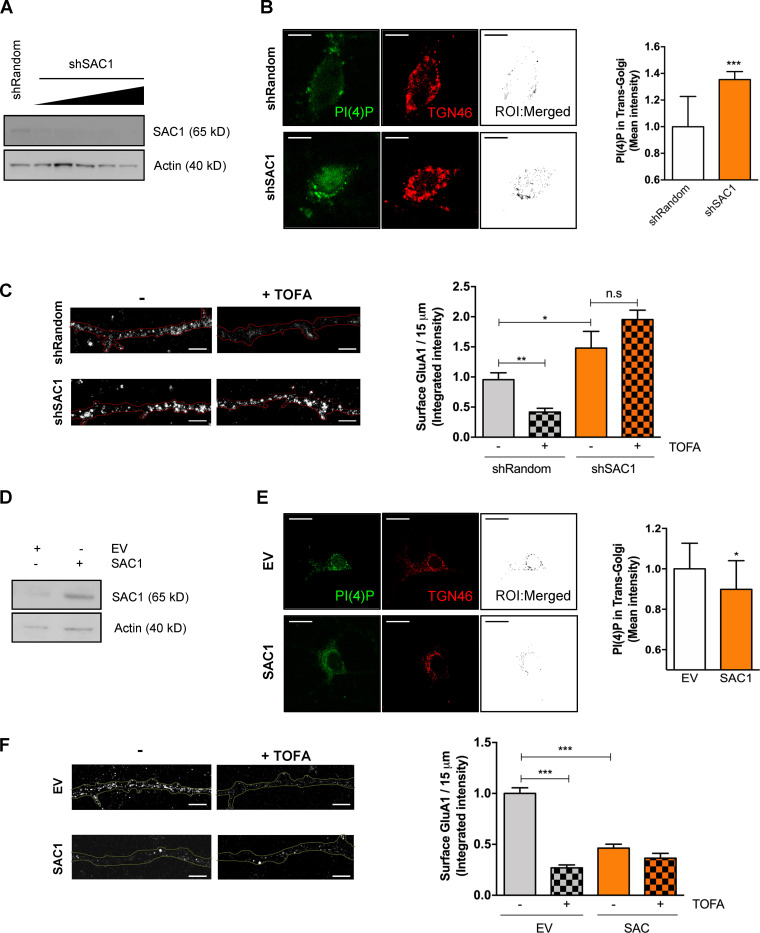
Next, we wanted to confirm whether SAC1 was actually regulating surface GluA1 levels downstream of the malonyl-CoA/CPT1C axis. To this end, surface GluA1 levels were quantified by immunostaining in SAC1-silenced and SAC1-overexpressing cortical neurons submitted to TOFA treatment to block the synthesis of malonyl-CoA.
In agreement with Yang et al. (2013), SAC1-silencing enhanced Golgi PI(4)P pools, as well as GluA1 surface levels (Fig. 6, A–C). Interestingly, the silencing of SAC1 abrogated the effects of TOFA on surface GluA1 expression. Conversely, SAC1 overexpression reduced both TGN PI(4)P levels and GluA1 levels at the PM compared with control neurons. Moreover, TOFA treatment did not have a significant effect on SAC1-overexpressing cells (Fig. 6, D–F). Neither SAC1 silencing nor SAC1 overexpression changed TGN volume in neurons (Fig. S4 B). These data indicate that SAC1 is downstream of the malonyl-CoA/CPT1C axis, which is crucial for the maintenance of GluA1 levels at the PM in neurons.
Figure 6.
SAC1 is downstream of malonyl-CoA/CPT1C axis in cultured cortical neurons. (A and B) Increasing volumes of shSAC1 lentiviruses were used to knock down SAC1 expression in cultured cortical neurons at 8 DIV. A short hairpin-loop RNA with a random sequence (shRandom) was used as a control. shSAC1 silencing efficiency was checked at 15 DIV by quantifying the decrease of SAC1 protein levels by Western blot (A) and by the increase in TGN PI(4)P levels measured by immunocytochemistry (B; scale bars = 7 µm). TGN46 was used as a marker of TGN. Values are the mean ± SD from two independent experiments (n = 8–12 cells per condition; two-sided Student’s t test; ***, P = 0.0009; 1.00 ± 0.23 for shRandom and 1.35 ± 0.18 for shSAC1). (C) SAC1 silencing increased GluA1 surface levels and abolished the effect of TOFA treatment. Surface GluA1 after TOFA treatment (20 µg/ml, 2 h) was detected in nonpermeabilized neurons by immunocytochemistry. Results are the mean ± SEM from two independent experiments performed by biological duplicates (two-way ANOVA followed by Bonferroni’s comparison test; *, P < 0.05 and **, P < 0.01). shRandom + vehicle (1.00 ± 0.09, n = 35), shRandom + TOFA (0.44 ± 0.06, n = 32), shSAC1 + vehicle (0.90 ± 0.13, n = 34), and shSAC1 + TOFA (1.48 ± 0.09, n = 20). Scale bars = 5 µm. n.s, not significant. (D) SAC1 overexpression in cortical neurons. A lentiviral vector carrying SAC1 was used to infect the cells at 7 DIV. At 14–15 DIV, SAC1 overexpression was corroborated by Western blot. (E) PI(4)P levels in the TGN compartment decreased by SAC1 overexpression. TGN46 was used as a marker of TGN. Values are the mean ± SD from two independent experiments performed by biological replicates (n = 16–22 cells per condition; two-sided Student’s t test; *, P = 0.0269; 1.00 ± 0.13 for EV and 0.89 ± 0.14 for SAC1). (F) SAC1 overexpression decreased surface levels of GluA1 and abolished the effect of TOFA treatment in cortical neurons. Neurons, transduced as explained above, were treated with TOFA at 14–15 DIV. Results are the mean ± SEM from two independent experiments performed by biological duplicates (two-way ANOVA followed by Bonferroni’s comparison test; ***, P < 0.001). EV + vehicle (1.00 ± 0.05, n = 36), shRandom + TOFA (0.27 ± 0.03, n = 36), SAC1 + vehicle (0.46 ± 0.04, n = 39), and shSAC1 + TOFA (0.36 ± 0.05, n = 36). Scale bars = 5 µm.
CPT1C regulates SAC1 translocation to ER-TGN contacts
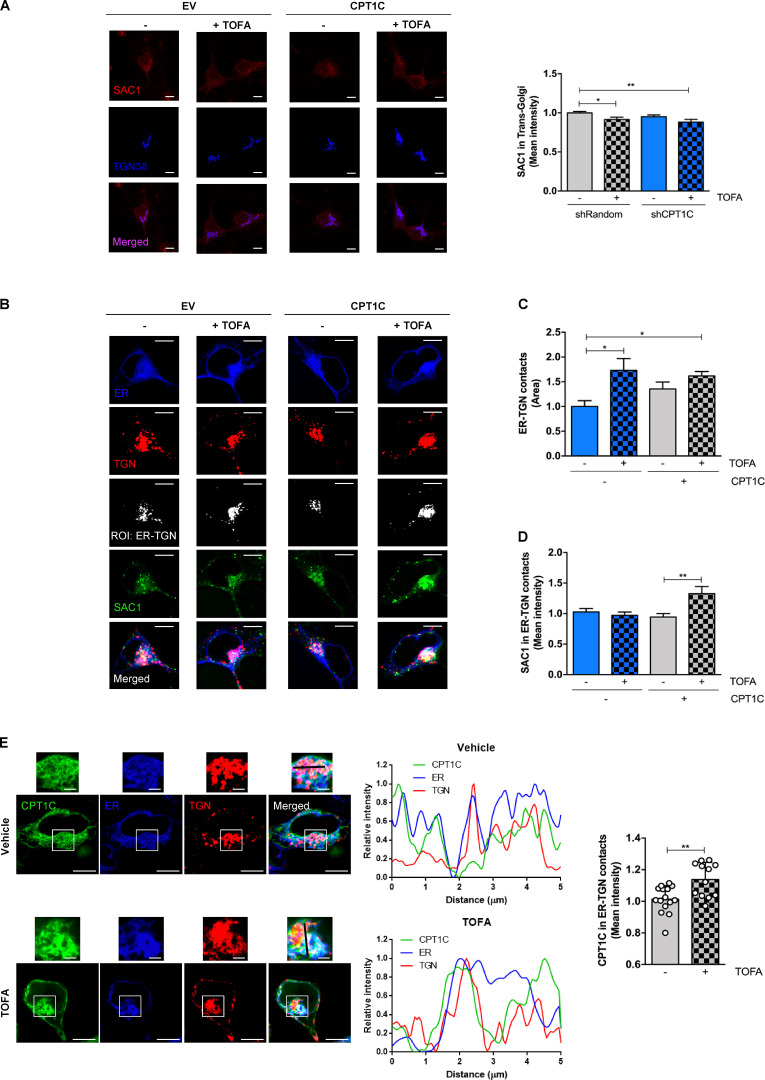
Previous studies described SAC1 translocation to the TGN under stressful conditions, which correlated with a decrease in Golgi PI(4)P levels and impaired vesicular transport to the PM (Blagoveshchenskaya et al., 2008; Yang et al., 2013). For this reason, we hypothesized that CPT1C could promote SAC1 transport to the TGN in low malonyl-CoA conditions and, consequently, decrease both Golgi PI(4)P pools and GluA1 trafficking to the PM. However, our data do not support such model, since SAC1 did not increase either at the TGN after TOFA treatment or in CPT1C knockdown neurons (Fig. 7 A).
Figure 7.
CPT1C regulates SAC1 recruitment to TGN-ER contacts. (A) CPT1C knockdown or TOFA treatment does not increase SAC1 in the TGN. Quantification of SAC1 levels at the TGN in shCPT1C-transduced cortical neurons by immunocytochemistry. A short hairpin-loop RNA with a random sequence (shRandom) was used as a control. All data represent the mean ± SEM from two independent experiments performed by biological duplicates (two-way ANOVA followed by Bonferroni’s comparison test; *, P < 0.05; **, P < 0.01). shRandom + vehicle (1.00 ± 0.02, n = 40), shRandom + TOFA (0.91 ± 0.03, n = 38), shCPT1C + vehicle (0.95 ± 0.02, n = 39), and shCPT1C + TOFA (0.88 ± 0.04, n = 29). Scale bars = 7 µm. (B–D) SAC1 localization at ER-TGN contact sites. HEK-SAC1-GFP cells were transfected with CPT1C or an EV together with expressing plasmids for ER (iRFP-Sec61β) and TGN (mTurquoise-TGN38-FRB) markers. 24 h later, cells were treated with TOFA (20 µg/ml, 2 h) or vehicle and live imaged. Representative images are shown in B. Black-and-white images show the ROI generated by TGN-PI(4)P colocalizing pixels. ER-TGN contacts were quantified as the total area of positive colocalization between TGN and ER signals (C). Endogenous SAC1-GFP mean intensity at these junctions was quantified (D). Data represent the mean ± SEM from two independent experiments performed by biological duplicates (two-way ANOVA followed by Bonferroni’s comparison test; *, P < 0.05 and **, P < 0.01). Scale bars = 7 µm. (C) EV + vehicle (1.00 ± 0.12, n = 37), EV + TOFA (1.73 ± 0.24, n = 24), CPT1C + vehicle (1.44 ± 0.15, n = 37), and CPT1C + TOFA (1.62 ± 0.09, n = 33). (D) EV + vehicle (1.00 ± 0.06, n = 37), EV + TOFA (0.97 ± 0.05, n = 24), CPT1C + vehicle (0.94 ± 0.06, n = 37), and CPT1C + TOFA (1.33 ± 0.12, n = 33). (E) CPT1C mean intensity at ER-TGN contact sites in HEK293 cells overexpressing CPT1C-SYFP2. Contact sites were determined as in B. Representative images of each condition are shown. The graphs on the left represent the linear fluorescence profiles of the black line drawn on enlarged images: CPT1C in green, ER (Sec61β) in blue, and TGN (TGN38) in red. The right graph shows the mean intensity of CPT1C in the ER-TGN ROIs, and data represent the mean ± SD from two independent experiments performed by biological duplicates (n = 15–12 cells per condition; Mann-Whitney U test; **, P = 0.0037; 1.00 ± 0.09 for vehicle and 1.14 ± 0.11 for TOFA). Scale bars = 7 µm; scale bars of inset magnifications = 1.75 µm.
More recently, other authors have demonstrated that SAC1 can act in trans on the TGN PI(4)P pool, across ER-TGN junctions (Venditti et al., 2019). To address whether CPT1C can modulate SAC1 recruitment to these contact sites under low malonyl-CoA levels, we studied SAC1 in ER-TGN junctions using CRISPR-Cas–edited HEK cells that have endogenous SAC1 tagged with GFP (Zewe et al., 2018). Our results clearly demonstrate that TOFA treatment favors ER-TGN junction creation in a CPT1C-independent manner (Fig. 7, B and C; and Fig. S4 C), with endogenous SAC1 only being enriched at these contact sites under low malonyl-CoA levels and CPT1C overexpression (Fig. 7 D). Considering that the CPT1C–SAC1 interaction is not affected by malonyl-CoA (Fig. 6), we decided to analyze whether CPT1C also accumulated at ER-TGN contact sites under low malonyl-CoA levels. In line with this hypothesis, CPT1C levels at ER-TGN junctions increased after TOFA treatment (Fig. 7 E). The percentage of CPT1C found in the ER-TGN junctions (<10%) was similar to that found in the TGN (Fig. S4 D), suggesting that CPT1C does not translocate to the TGN but accumulates at the ER side of the ER-TGN junctions. These findings suggest that low malonyl-CoA levels increase ER-TGN contact sites favoring CPT1C/SAC1 recruitment to these junctions.
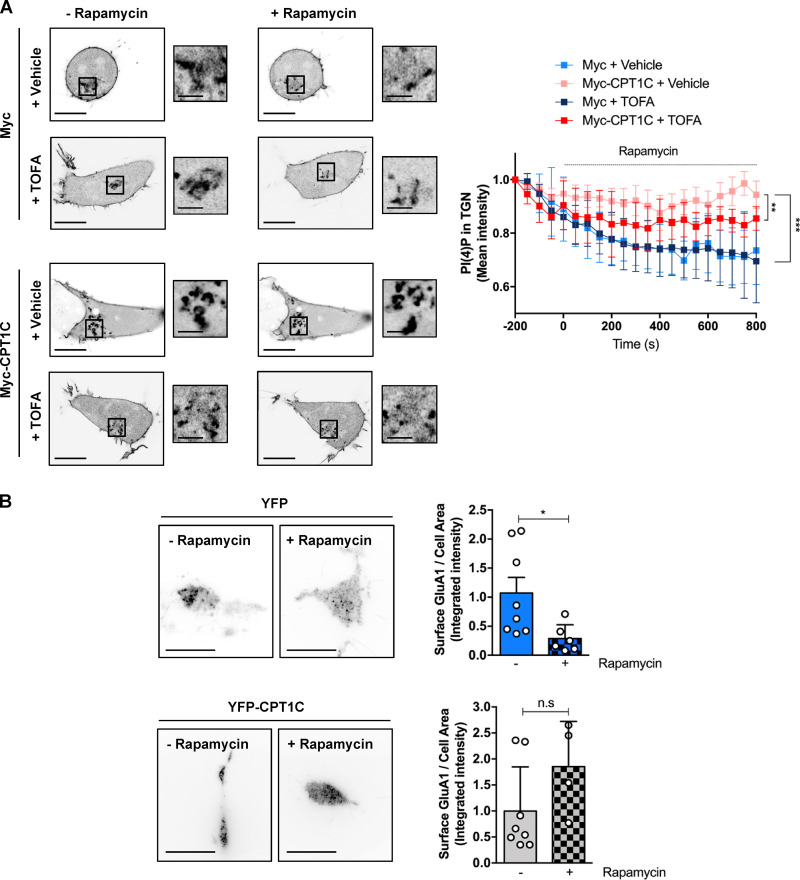
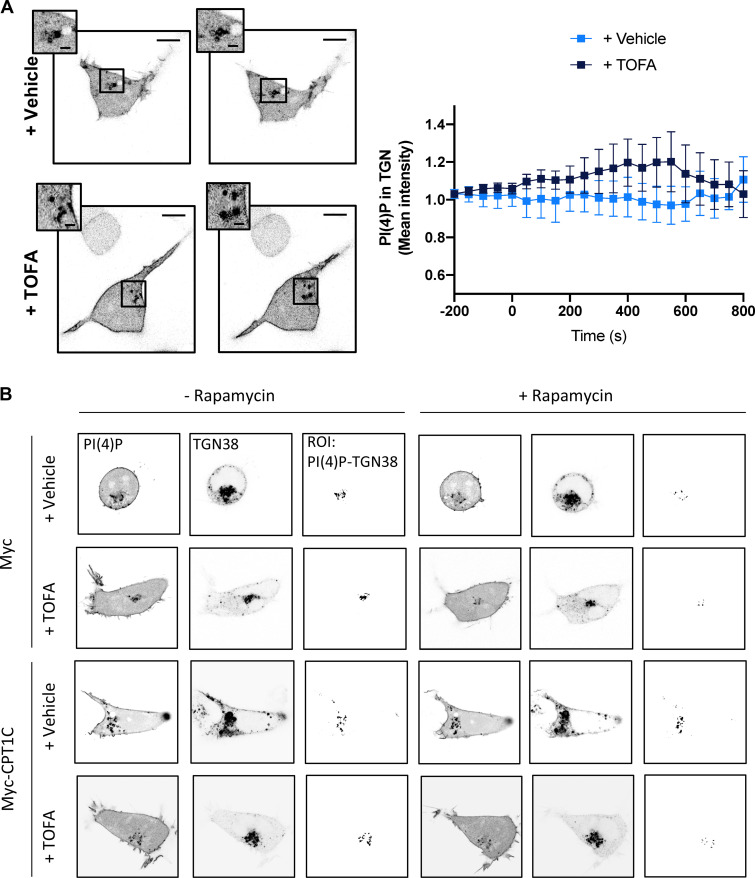
To verify that GluA1 trafficking is regulated by ER-TGN junctions, we next decided to artificially enhance ER-TGN contacts using the rapamycin-induced dimerization system (Dickson et al., 2016, 2014). Increasing ER-TGN junctions resulted in the expected decrease of TGN PI(4)P levels following the addition of rapamycin (Fig. 8 A; and Fig. S5, A and B) and a corresponding decrease in the levels of GluA1 at the PM (Fig. 8 B), indicating that GluA1 trafficking can be influenced by ER-TGN–dependent regulation of TGN PI(4)P levels.
Figure 8.
Artificially formed ER-TGN contacts decrease surface GluA1 levels. (A) PI(4)P at the TGN after artificial ER-TGN contact creation. HEK cells overexpressing or not Myc-CPT1C were transfected with the mCherry-pHR-TcRb-FKBP and CFP-TGN38-FRB plasmids and treated with rapamycin (5 µM, 30 min) in order to induce ER-TGN contacts. PI(4)P was labeled with the lipid probe YFP-P4M. A Myc EV was used as a control. Cells were treated with TOFA 24 h after transfection. Cells were live imaged with confocal microscopy. Values are the mean ± SD from four independent experiments (n = 6 cells per condition; two-way ANOVA followed by Bonferroni’s comparison test; **, P < 0.01 and ***, P < 0.001). (B) Surface GluA1 levels after artificial ER-TGN contact creation. Live HEK cells were transfected and treated as in A. Surface GluA1 was imaged by TIRF microscopy. Data represent the mean ± SD from two independent experiments (n = 4–8 cells per condition; two-sided Student’s t test; *, P < 0.05 in YFP control cells and P = 0.0503 in YFP-CPT1C overexpressing cells). EV + vehicle (1.00 ± 0.76, n = 8), EV + rapamycin (0.29 ± 0.10, n = 6), CPT1C + vehicle (1.00 ± 0.85, n = 8), and CPT1C + rapamycin (1.85 ± 0.87, n = 4). Scale bars = 7 µm; scale bars of inset magnifications = 1.75 µm. n.s, not significant.
Surprisingly, Myc-CPT1C expression totally abolished rapamycin-dependent PI(4)P decay and surface GluA1 decrease. This result suggests that CPT1C could be an inhibitor of SAC1 activity. Interestingly, in this context TOFA treatment did not exert any effect in control cells, but it partially abolished the effects of CPT1C on PI(4)P levels. These results suggest that CPT1C may be regulating not only SAC1 subcellular distribution but also its PI(4)P phosphatase activity in response to malonyl-CoA changes to control GluA1 trafficking patterns to the PM.
CPT1C regulates SAC1 PI(4)P phosphatase activity in a malonyl-CoA–dependent manner
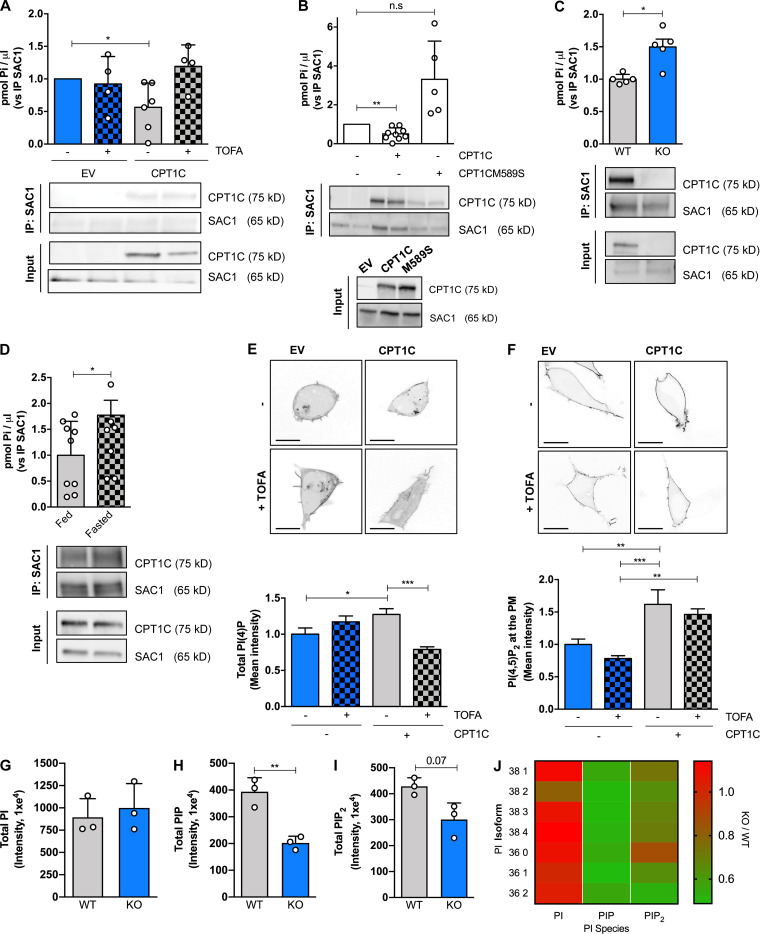
To further investigate a role for CPT1C in regulating SAC1 phosphatase activity, we took several independent approaches. First, we directly measured the specific PI(4)P phosphatase activity of SAC1 (Mani et al., 2007) in Hela cells stably expressing CPT1C or CPT1CM589S under different malonyl-CoA conditions. As shown in Fig. 9 A, CPT1C expression decreased SAC1 activity, a reduction that was antagonized by TOFA treatment and was absent following the expression of the malonyl-CoA–insensitive CPT1C (CPT1CM589S; Fig. 9 B). In parallel experiments performed in mice brains, we determined that SAC1 phosphatase activity is significantly increased in CPT1C−/− brains compared with WTs (Fig. 9 C). Moreover, brains from 16 h–fasted animals, a protocol that strongly decreases malonyl-CoA levels in the cortex, hippocampus, hypothalamus, and cerebellum (Tokutake et al., 2010), had increased SAC1 phosphatase activity (Fig. 9 D).
Figure 9.
CPT1C negatively regulates SAC1 PI(4)P phosphatase activity. (A) SAC1 phosphatase activity regulation by CPT1C and TOFA. Hela cells permanently expressing CPT1C or the EV were treated by TOFA (20 µg/ml, 2 h) or vehicle. After TOFA treatment, SAC1 was immunoprecipitated from cellular lysates, and PI(4)P phosphatase activity was measured as the Pi released from PI(4)P substrate using a green malachite assay. All data were normalized by the amount of immunoprecipitated SAC1 in each sample detected by Western blot. Values are the mean ± SD of three independent experiments performed by technical replicates (one-sample t test; *, P < 0.05). EV + vehicle (1.00 ± 0, n = 6), EV + TOFA (0.56 ± 0.37, n = 6), CPT1C + vehicle (0.92 ± 0.42, n = 4), and CPT1C + TOFA (1.19 ± 0.33, n = 4). (B) SAC1 phosphatase activity regulation by CPT1C-sensing of malonyl-CoA. HeLa cells permanently expressing CPT1C, malonyl-CoA–insensitive CPT1C form (M589S), or EV were analyzed for PI(4)P phosphatase activity as in A. Results are the mean ± SD of three independent experiments performed by technical replicates (n = 4–8 samples per condition; one sample t test; **, P < 0.01). EV (1.00 ± 0.00, n = 9), CPT1C (0.50 ± 0.31, n = 9), and CPT1CM589S (3.31 ± 0.1.96, n = 5). n.s, not significant. (C) SAC1 phosphatase activity in brains from WT and CPT1C KO adult mice. Total brain lysates were processed for SAC1 immunoprecipitation. PI(4)P phosphatase activity was measured in immunoprecipitated pellets as in A. Values are the mean ± SD (n = 5 animals per genotype; two-sided Student’s t test; *, P = 0.0353; 1.00 ± 0.08 for WT and 1.50 ± 0.27 for KO). (D) SAC1 phosphatase activity in brains from fed and fasted mice. WT mice were subjected to 24 h of fasting. PI(4)P phosphatase activity was measured as in C. Values are the mean ± SD (n = 9 or 10 animals per condition; one-sided Student’s t test; *, P = 0.0256; 1.00 ± 0.66 for fed and 1.77 ± 0.91 for fasted). (E) Total PI(4)P levels were quantified with a lipid probe (mCherry-P4M) in live HEK cells overexpressing or not CPT1C after TOFA or vehicle addition. Graph shows the mean ± SEM from three independent experiments performed by biological triplicates (two-way ANOVA followed by Bonferroni’s comparison test; *, P < 0.05 and ***, P < 0.001). EV + vehicle (1.00 ± 0.08, n = 16), EV + TOFA (1.17 ± 0.08, n = 18), CPT1C + vehicle (1.27 ± 0.08, n = 22), and CPT1C + TOFA (0.79 ± 0.04, n = 20). Scale bars = 10 µm. (F) Total PI(4,5)P2 levels were quantified with a lipid probe (YFP-PHPLCδ1) in live HEK cells overexpressing or not CPT1C after TOFA or vehicle addition. Graph shows the mean ± SEM from three independent experiments performed by biological triplicates two-way ANOVA followed by Bonferroni’s comparison test; **, P < 0.01 and ***, P < 0.001). EV + vehicle (1.00 ± 0.08, n = 18), EV + TOFA (0.78 ± 0.04, n = 18), CPT1C + vehicle (1.62 ± 0.23, n = 18), and CPT1C + TOFA (1.46 ± 0.08, n = 18). Scale bars = 10 µm. (G) Summary histogram of UPLC-MS/MS PI measurements from WT and CPT1C−/− brains (n = 3 brains per condition; 894.51 ± 208.68 for WT and 999.26 ± 273.00 for KO). (H) Same experiments as in G only measuring PIP. n = 3 brains per condition; two-sided Student’s t test; **, P < 0.01; 393.40 ± 52.77 for WT and 202.40 ± 24.76 for KO. (I) Same experiment as in G only measuring PIP2. n = 3 brains per condition; two-sided Student’s t test; P = 0.07; 428.60 ± 32.77 for WT and 300.80 ± 63.45 for KO. Graphs in G, H, and I show the mean ± SD from one experiment performed by biological replicates. (J) Heat map detailing the relative changes in each PI isoform from CPT1C−/− brains relative to WT brains.
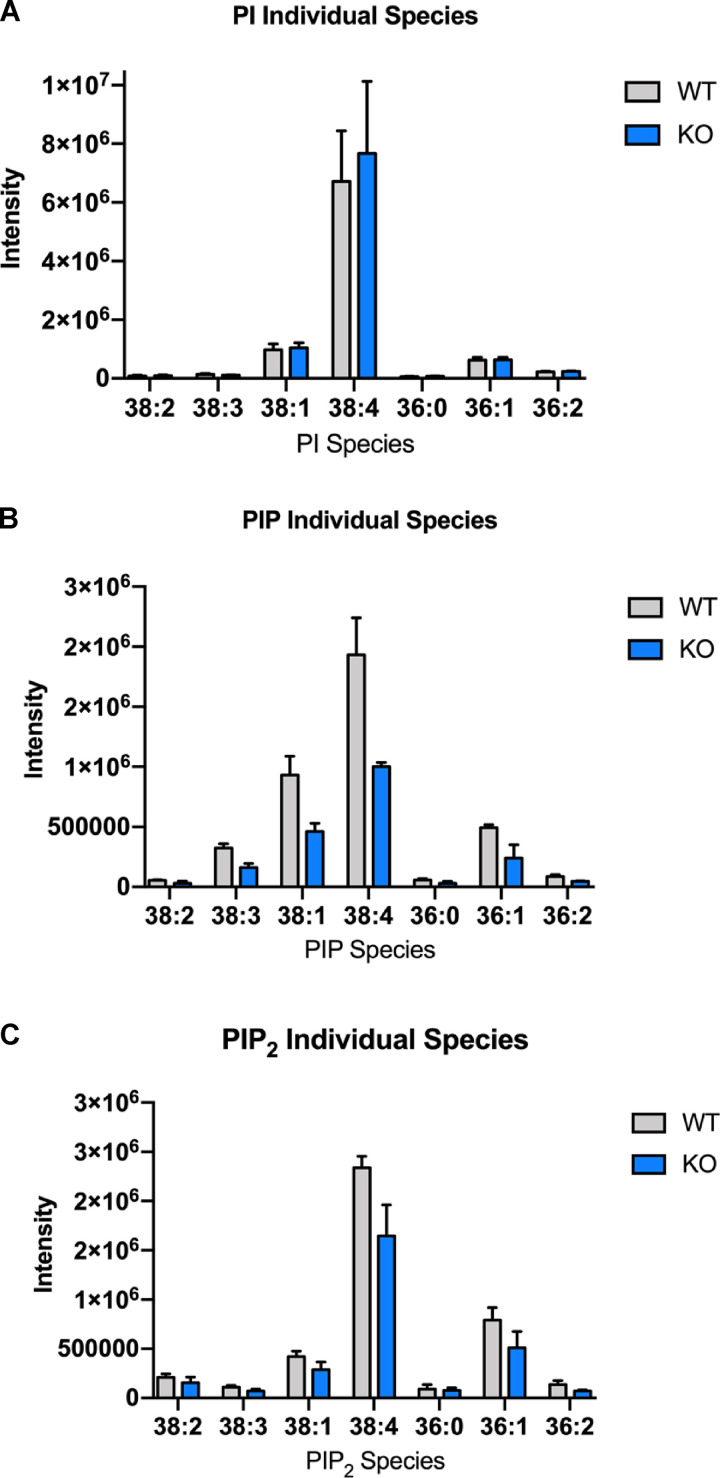
As a second, indirect approach to measure SAC1 activity, we measured the intensity and distribution of a cellular PI(4)P biosensor (mCherry-P4M) in live HEK293 cells overexpressing CPT1C, with and without the addition of TOFA. Quantitative analysis from these cells revealed that CPT1C overexpression increased total cellular PI(4)P levels, consistent with CPT1C inhibiting SAC1 activity, with this increase in PI(4)P inhibited following TOFA treatment (Fig. 9 E). The increase in cellular PI(4)P was paralleled by a significant increase in PM PI(4,5)P2 (Fig. 9 F) and suggests that TGN PI(4)P is a source for the maintenance of PM PI(4,5)P2 (Dickson et al., 2014). To confirm that the CPT1C-dependent control of SAC1 can regulate PI(4)P and PI(4,5)P2 levels in the brain, ultra performance liquid chromatography–tandem mass spectrometer (UPLC MS/MS) was performed on lipids extracted from WT and CPT1C−/− brains. Consistent with biosensor data, mass-spec analysis of phosphoinositides revealed that CPT1C−/− brains have significantly reduced levels of phosphatidylinositol monophosphate (PIP) and phosphatidylinositol bisphosphate (PIP2), relative to WT animals (Fig. 9, H–J; and Fig. S6).
Figure S6.
CPT1C deficiency decreases the brain levels of different PIP and PIP2 species. (A–C) Mass spectrometry analysis for PI, PIP, and PIP2 species. Histogram of the most abundant PI (A), PIP (B), and PIP2 (C) species in WT (gray bars) and CPT1C−/−(blue bars) brains (mean ± SEM; n = 3).
Collectively, these data support the hypothesis that CPT1C is a metabolic regulator of SAC1 PI(4)P phosphatase activity in a malonyl-CoA–dependent manner.
Discussion
AMPARs mediate excitatory neurotransmission in the brain and play a key role in most brain functions, including cognition. Recent studies have demonstrated the involvement of CPT1C in GluA1 trafficking, the main component of AMPARs (Brechet et al., 2017; Fadó et al., 2015; Gratacòs-Batlle et al., 2015, 2018). However, little is known about the precise molecular mechanism and whether this function is linked to CPT1C’s capacity to bind and sense malonyl-CoA fluctuations in the brain across different nutritional conditions. In the present work, we address both questions and demonstrate that CPT1C regulates GluA1 trafficking from the TGN toward the PM by modulating SAC1 phosphatase activity in a malonyl-CoA–dependent manner (see the proposed model at Fig. 10).
Figure 10.
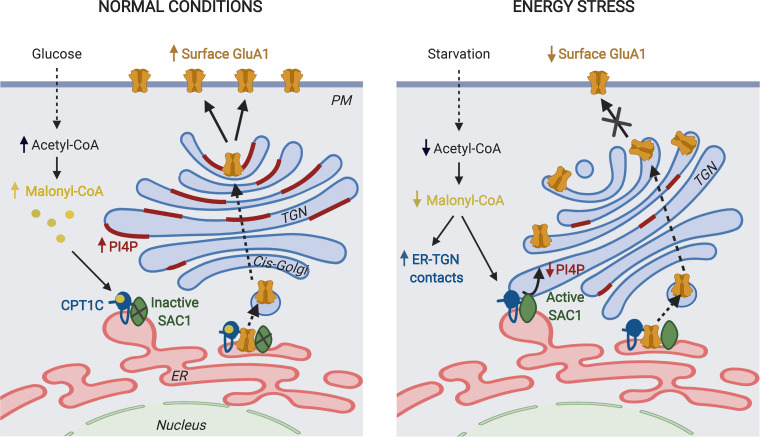
A working model for CPT1C regulation of GluA1 trafficking. CPT1C and SAC1 form part of the AMPAR complex at the ER of neurons. We do not discard the existence of CPT1C/SAC1 pairs in the ER that do not form part of AMPAR complexes. In normal nutrient conditions, malonyl-CoA–bound CPT1C down-regulates SAC1 activity, allowing the accumulation of PI(4)P at the TGN. This favors the trafficking of GluA1-containing AMPARs from the TGN to the PM through the secretory pathway. In energy stress conditions that decrease malonyl-CoA levels, such as glucose deficiency, ER-TGN contacts are increased and CPT1C favors the transport of SAC1 to these junctions. At the same time, malonyl-CoA–free CPT1C releases its inhibition on SAC1 activity. This results in dephosphorylation of PI(4)P at the TGN by SAC1 in trans, which halts GluA1-containing AMPAR trafficking. The figure was drawn using BioRender web-based software.
We demonstrate that glucose depletion, glycolysis inhibition, or leptin addition—metabolic challenges known to modify intracellular malonyl-CoA levels (Gao et al., 2007; Wolfgang et al., 2007)—modulate GluA1 surface expression in cortical neurons. Moreover, direct down-regulation of malonyl-CoA levels through the inhibition of ACC recapitulates these effects, confirming the important role of malonyl-CoA in GluA1 trafficking. Interestingly, CPT1C-deficient neurons did not respond to any of the treatments above, and the malonyl-CoA–insensitive CPT1C was unable to restore GluA1 at the PM in CPT1C KO neurons. These findings clearly indicate that surface GluA1 is finely regulated by CPT1C-sensing of malonyl-CoA. Importantly, malonyl-CoA is an intermediary shared by the metabolism of lipids and glucose, which makes it a precise indicator of nutritional conditions. In fact, malonyl-CoA levels in the brain highly decrease by fasting, while they increase after refeeding (Tokutake et al., 2010). Therefore, we can say that nutrients impact GluA1 trafficking through CPT1C-sensing of malonyl-CoA. Since the energy sensor AMPK phosphorylates and inhibits the activity of ACC, the enzyme that catalyzes the conversion of acetyl-CoA to malonyl-CoA, it suggests that the CPT1C/SAC1 axis is downstream of the AMPK pathway. To date, very few articles have reported the mechanism by which nutrients regulate GluA1 trafficking. A recent paper demonstrates that a long-time high-fat diet (during 6 wk in mice or for 24 h in cultured neurons) hinders GluA1 trafficking to the PM by a mechanism involving GluA1 palmitoylation (Spinelli et al., 2017). Here, we demonstrate that short-term fluctuations of malonyl-CoA, such as glucose depletion for 2 h, regulates GluA1 trafficking by a mechanism involving CPT1C regulation of SAC1.
Although the brain only accounts for 2% of body mass, it consumes up to 25% of total glucose metabolized by the body. Moreover, neurons use 85% of this energy mainly to maintain glutamatergic transmission (Harris et al., 2012). Therefore, neurons need to sense their energy status in a very precise way to turn on or turn off synaptic activity and preserve brain function. Interestingly, one of the most important cell energy sensors and regulators during synaptic activation is AMPK (Marinangeli et al., 2018). Pathological conditions that undergo with deficits in energy supply to the brain have been associated with impaired cognitive performance. Examples of this are severe undernutrition in anorexic patients (reviewed by Beilharz et al., 2015), exhaustive exercise (Rosa et al., 2007; Zimmer et al., 2017), brain metabolic hypoactivity in Alzheimer’s disease, and sleep deprivation (Dworak et al., 2010; Nikonova et al., 2010). Recent integrative analyses drawing on multiple sources of data imply that AMPAR trafficking is one of the crucial determinants underlying the negative effects of insufficient sleep (Diessler et al., 2018). However, it is well known that aerobic exercise and caloric-restricted diets promote synaptic incorporation of GluA1 and improve cognition (Thacker et al., 2019; Ouyang et al., 2017). This contradiction can be explained by the existence of several mechanisms for organ and location-specific control of energy balance and supply to the brain, including mechanisms independent of malonyl-CoA. In any case, our results clearly demonstrate that GluA1 trafficking in primary cultured cortical neurons is sensitive to cell-autonomous intracellular malonyl-CoA fluctuations.
The second crucial result of our work is that CPT1C regulates GluA1 trafficking through SAC1-dependent modulation of TGN PI(4)P levels. To our knowledge, this is the first report of a negative regulator of SAC1 enzymatic activity (for an exhaustive review on SAC1, see Del Bel and Brill, 2018). SAC1 PI(4)P phosphatase activity assays in cell lines and brain tissues demonstrate that SAC1 activity is regulated by CPT1C in an energy-dependent manner. When malonyl-CoA levels are decreased (by TOFA treatment in cultured neurons or by fasting in whole animals), the CPT1C-mediated inhibition of SAC1 is released and phosphatase activity restored. Moreover, SAC1 activity regulation correlates with PI(4)P levels in cell lines and in the total brain. This is of crucial importance, since SAC1 has been involved in the regulation of other PI(4)P pools and functions in the cell, such as the regulation of cholesterol transport at the ER-TGN contacts (Mesmin et al., 2013), phosphatidylserine transport at the ER-PM contacts (Chung et al., 2015; Von Filseck et al., 2015), the retromer function at the ER-endosome contacts (Dong et al., 2016), or ceramide de novo synthesis at the ER (Breslow et al., 2010). Therefore, it is reasonable to think that CPT1C sensing of malonyl-CoA could also regulate some other functions of SAC1. In fact, this could account for the reduced levels of ceramide found in brain tissues of CPT1C KO mice (Carrasco et al., 2013).
One controversial issue that remains is how SAC1 dephosphorylates PI(4)P at the TGN. Some authors claim that SAC1 dephosphorylates PI(4)P in trans at membrane contact sites, while others suggest it acts in cis after its translocation to the TGN. Additionally, these two models of SAC1 activity do not have to be mutually exclusive, with a clear compromise between the two models being that SAC1 can act in both configurations. Our stressful conditions in cortical neurons were not able to induce a shuttling of SAC1 from the ER to the TGN, contrary to some previous studies performed in nonexpressing CPT1C cells (Faulhammer et al., 2007; Blagoveshchenskaya et al., 2008; Yang et al., 2013). Our data align with recent literature reporting the ability of SAC1 to metabolize the TGN PI(4)P pool across ER-TGN junctions (Venditti et al., 2019). Underscoring this statement, artificial induction of ER-TGN contacts using the rapamycin platform induced TGN PI(4)P depletion and a consequent reduction of GluA1 at the PM, which was abolished by CPT1C only under high malonyl-CoA levels. Notably, we found that inhibition of malonyl-CoA synthesis by TOFA increased ER-TGN contacts and triggered CPT1C translocation to ER-TGN junctions together with SAC1, suggesting that both proteins translocate together. These findings indicate that GluA1 trafficking in neurons is regulated by SAC1 activity at ER-TGN junctions under the control of CPT1C.
The fact that GluA1 abundance at the PM decreased by 50% after TOFA treatment or in CPT1C-silenced neurons while at the TGN it only increased around 10% suggests that CPT1C not only regulates the canonical trafficking pathway but also may be involved in other pathways, such as the Golgi-bypass route. Additionally, it has been published that PI(4)P contributes to the PI(4,5)P2 pool at the PM (Dickson et al., 2014), which is known to directly bind and stabilize surface GluA1 and therefore avoid its internalization (Seebohm et al., 2014). The fact that CPT1C favors an increase in PI(4,5)P2 at the PM in a malonyl-CoA–dependent manner reveals that CPT1C probably modulates GluA1 surface expression by also regulating its PI(4,5)P2-dependent stabilization at the PM.
Surprisingly, neither GluA2 nor GluN2A surface levels in cortical neurons were altered after 2 h of TOFA treatment, suggesting that CPT1C-mediated regulation of GluA1 trafficking is specific to this AMPAR subunit. Therefore, we think that CPT1C is regulating specifically GluA1 homomeric AMPARs, which have higher calcium permeability and larger channel conductance than heteromeric receptors containing calcium-impermeable GluA2 subunits. In the case of GluA2, it is established that the exit from the ER toward the PM is slower than GluA1 (Henley and Wilkinson, 2013); for GluN2A, not much is known about the timing of its turnover. Perhaps longer times of TOFA treatment are required to observe changes in their transport to the PM, but TOFA treatment can become toxic over prolonged treatment periods, impairing our ability to test this idea. It has been suggested that coat molecules and post-Golgi carriers may be sensitive to different PI(4)P levels and pools at the TGN, leading to a selective vesicular releasing (Venditti et al., 2019). Moreover, different motor proteins are involved in the vesicular transport of GluA1, GluA2, and NMDA receptor (Kneussel et al., 2014), which could also favor individual/selective traffcking. Clearly, additional experiments are warranted to further unravel the mechanism of this selective trafficking.
The last interesting issue is that both CPT1C and SAC1 form part of the AMPAR macrocomplexes at the ER in neurons. Recently, Schwenk et al. (2019) showed that CPT1C, specifically bound to ferric-chelate reductase 1-like, drives the assembly of GluAs into functional tetramers at the ER. Moreover, a role for CPT1C has been suggested in the palmitoylation state of GluA1 (Gratacòs-Batlle et al., 2018). Here, we complete the picture of CPT1C function and describe a new role of CPT1C linked to SAC1 at the ER-TGN contacts. We show that the CPT1C–SAC1 complex regulates the trafficking of GluA1-containing AMPAR from the TGN to the PM in response to nutritional changes and unravel the molecular mechanism involved. Interestingly, SAC1 is not found in AMPAR macrocomplexes of CPT1C KO mice (Brechet et al., 2017), indicating that the role of SAC1 is necessarily linked to CPT1C. The interacting region between CPT1C and SAC1 is currently unknown. Using experimental and in silico approaches, we have identified the C-terminal region of CPT1C as being essential for GluA1 interaction and its transport to the PM. Other authors have demonstrated (using nuclear magnetic resonance spectroscopy) that the N-terminal domain of CPT1C (a peptide of 50 residues) can switch between two different conformations depending on malonyl-CoA (Rao et al., 2011; Samanta et al., 2014). Collectively, results support a model in which CPT1C binds GluA1 through its C-terminal domain, while its N-terminal domains acts as a sensor of malonyl-CoA. CPT1C will change its conformation in response to nutritional scarcity or energy supply deficits, which will regulate SAC1 activity and, in consequence, block the exit of GluA1 from the TGN.
In summary, our study reveals that CPT1C sensing of malonyl-CoA regulates GluA1-containing AMPAR trafficking in neurons through the regulation of SAC1 activity. Moreover, it discloses the first inhibitor of SAC1 PI(4)P phosphatase activity and supports the model of trans-activity of SAC1 at the ER-TGN contact sites. Considering that CPT1C has been involved in synaptogenesis, glutamatergic transmission, and learning processes, the present work provides new molecular evidence regarding how CPT1C senses nutritional conditions to control synaptic function and cognition.
Materials and methods
Animals
CPT1C KO mice were developed as described previously (Carrasco et al., 2012). After eight backcrosses with C57BL/6J mice, littermate homozygous CPT1C KO and WT mice were crossed separately to obtain a line for each genotype that was used in the experiments. All animal procedures met the guidelines in Spanish legislation (Boletín oficial del estado 32/2007) and were approved by the local ethics committee.
Cell culture and treatments
Primary cortical mouse neurons were prepared from E16 WT or CPT1C KO embryos, cultured, and maintained as described previously (Fadó et al., 2015) in Neurobasal (21103049; Gibco) supplemented with B27 (17504044; Gibco), glutaMAX (35050061; Gibco), and antibiotics. Cell lines were grown according to American Type Culture Collection. HeLa and HEK cells were maintained in DMEM (D5671; Sigma-Aldrich) supplemented with 10% FBS (F7524; Sigma-Aldrich), 2 mM glutamine (X0551-100; Biowest), 100 U/ml penicillin, and 100 mg/ml streptomycin (P0781; Sigma-Aldrich) at 37°C with 5% CO2. Cell lines were authenticated by genotyping and regularly tested for mycoplasma contamination. For glucose starvation, cell medium was replaced by DMEM without glucose (A14430-01; Gibco) supplemented with (control cells) or without (glucose-depleted cells) 25 mM glucose for 2 h. To inhibit malonyl-CoA synthesis, 20 µg/ml TOFA (ab141578; Abcam) was added for 2 h. 15 mM 2DG (D8375; Sigma-Aldrich) was added for 2 h to inhibit glycolysis. For the leptin treatment, leptin (450–31; Bionova PrepoTech) was added to a final concentration of 50 nM for 30 min. For rapamycin-dependent recruitment, 5 µM rapamycin (R8781; Sigma-Aldrich) was used at room temperature for 15–30 min.
Plasmids and transfection
Human CPT1C-SYFP2 and mTurquoise-CPT1C were gifts from Dr. Serra’s group (School of Pharmacy and Food Sciences, Universitat de Barcelon, Barcelona, Spain). The mTurquoise-TGN38-FRB plasmid, the PI(4)P probes mCherry-P4M and YFP-P4M, and the PI(4,5)P2 probe YFP-PHPLCδ1 were gifts from E. Dickson’s group (School of Medicine, University of California, Davis, CA). mTurquoise-calnexin (#55539), mTurquoise-ER (#55550), mCherry-ER-FKBP (#72904), and iRFP-Sec61β (near-infrared fluorescent protein-Sec61β; #108125) were purchased from Addgene. pcDNA3-Myc-CPT1C (Sierra et al., 2008) and pWPI-CPT1C-IRES-GFP (Fadó et al., 2015) plasmids were engineered in our laboratory as published. CPT1CM589S-mTurquoise2 and pWPI-CPT1CM589S-IRES-GFP (Rodríguez-Rodríguez et al., 2019) encoded a malonyl-CoA–insensitive CPT1C form (generated by site-directed mutagenesis at methionine 589). Plasmids containing the C-terminal truncated form of CPT1C (mTurquoise2-CPT1CΔCterm-FLAG and pWPI-CPT1CΔCterm-FLAG-IRES-GFP) were constructed by replacing the last 39 aa (772–810) of the CPT1C sequence for the FLAG epitope (DYKDDDDK). SAC1 and GluA1 cDNAs were obtained by RT-PCR from mice brain tissue, using TRIzol (15596018; Life Technologies) for RNA isolation, Mu-MLV (#3022-1; Lucigen) as a reverse transcription, and the Q5 DNA polymerase (M0491L; New England Biolabs). SAC1 and GluA1 were cloned in pmTurquoise2 (mTurquoise2-SAC1 and mTurquoise2-GluA1). SAC1 was also cloned in the lentiviral pWPI-IRES-GFP plasmid (pWPI-SAC1-IRES-GFP). For CPT1C silencing, the pVTHM vector encoding a silencing sequence was used (Fadó et al., 2015). For SAC1 silencing, the sequence 5′-gtagcaaatcacatggatg-3′ (Yang et al., 2013) was cloned in the vector psi-LVRU6MP (Tebu-Bio) to generate the plasmid short hairpin-loop RNA of SAC1 (shSAC1); and as a control (shRandom), the random sequence 5′-gcttcgcgccgtagtctta-3′ was used (CSHCTR001-LVRU6MP; Tebu-Bio). All plasmids were verified by sequencing. FuGENE6 (E2692; Promega) was used for transient transfection in HeLa cells, the calcium phosphate method in HEK293T cells, and Lipofectamine 2000 reagent (11668019; Invitrogen) in both HEK-SAC1GFP cells and cortical neurons at 13–14 DIV. All transfections with cell lines were conducted at 80% of confluence.
Lentiviral infection
For protein overexpression in cultures of primary cortical neurons, pWPI-SAC1-IRES-GFP, pWPI-CPT1C-IRES-GFP, pWPI-CPT1CΔCterm-FLAG-IRES-GFP, pWPI-CPT1CM589S-IRES-GFP, and pWPI-IRES-GFP (empty vector; EV) were used to generate lentiviral particles, as previously described (Fadó et al., 2015). To silence CPT1C and SAC1, the pVTHM and LVRU6MP plasmids described above were employed. Neurons were transduced with low virus titles (MOI = 2) at 8 DIV.
Generation of stable cell lines
HeLa stably expressing mouse CPT1C or CPT1CM589S was generated by lentiviral transduction of pWPI-CPT1C-IRES-GFP (CPT1C cells) and pWPI-CPT1CM589S-IRES-GFP (CPT1CM589S cells), respectively. Cells were transduced with low virus titles (MOI = 5), and stable cells showing green fluorescence were isolated by cell-sorter (FACS Aria Fusion; Becton Dickinson). GFP was excited at 488 nm and green fluorescence collected at 530/30 nm. pWPI-IRES-GFP EV was used to obtain control cells (EV cells). HEK-SAC1GFP cells were genetically modified using CRISPR-Cas (CRISPR-associated proteins) technology as previously described (Zewe et al., 2018).
Viability assay
To test cellular viability, the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay was performed in cortical neurons at 14–15 DIV. After treatments, MTT solution was added at a final concentration of 0.2 mg/ml, and cells were incubated for 1 h at 37°C. The mitochondrial activity of live cells was measured by spectrophotometry at an absorbance wavelength of 570 nm. Cellular viability was given as a percentage versus control cells.
Analysis of malonyl-CoA levels
Intracellular malonyl-CoA levels were quantified in cortical neurons at 15 DIV. After treatment, neurons were washed with PBS, scraped, and collected. After centrifugation at 500 ×g for 3 min, the cell pellets were frozen in liquid nitrogen. Malonyl-CoA levels in the collected cells were determined using the acyl-CoA cycling method (Tokutake et al., 2010) and normalized by cellular wet weight (milligram wet weight). Briefly, intracellular malonyl-CoA was extracted by adding 0.6 M sulfuric acid per each 100 mg wet cells. After being kept at 4°C overnight and centrifuged, 1 M Tris (0.05 volume of supernatant) was added to the supernatant, and the pH of the solution was adjusted to around 6.5 with NaOH on ice. Malonyl-CoA levels in the extracts were measured by the enzymatic cycling assay using malonate decarboxylase after elimination of acetyl-CoA by citrate synthase. The reaction mixture for the citrate synthase treatment was composed of 50 mM Tris-HCl (pH 7.2), 2 mM oxaloacetate, 5 mg recombinant citrate synthase from Bacillus subtilis, and the tissue extracts in 1 ml. The reaction was performed at 25°C for 20 min, and an aliquot of the mixture was transferred to the reaction mixture for the acyl-CoA cycling method. The mixture contained 50 mM Tris-HCl (pH 7.2), 1 mM 2-mercaptoethanol, 10 mM MgSO4, 50 mM malonate, 10 mM ATP, 1 U of malonate decarboxylase, and the solution from citrate synthase treatment in 400 µl. The mixture was incubated at 30°C for 20 min, and 1 U of recombinant acetate kinase from Escherichia coli was added. After 20 min of incubation, 0.2 ml of 2.5 M neutralized hydroxylamine was added, and the incubation was continued for an additional 20 min at 30°C. The reaction was terminated by adding 0.6 ml of 10 mM ferric chloride dissolved in 25 mM trichloroacetic acid–1 M HCl. The A540 of the acetohydroxamate formed was measured.
Immunostaining
Immunostaining of proteins and lipids was used in fixed cells with 4% (wt/vol) paraformaldehyde. For surface proteins, nonpermeabilized cells were directly blocked with goat serum and then incubated with the appropriate primary antibody. For internal proteins and lipids, cells were permeabilized with 0.1% (vol/vol) Triton X-100 before blocking and incubation with the primary antibody. Control experiments were performed to make sure that chemical fixation did not induce permeabilization (see Fig. S1 B). For surface GluA1 analysis, isolated primary dendrites close to the soma of the neuron were selected. Rabbit anti-GluA1 (1:30; PC246) was from Calbiochem. Mouse anti-PI(4)P (1:100; Z-P004) was from Echelon. Mouse anti-TGN38 (1:100; NB300-575) was from Novus Biologicals. Rabbit anti-TGN46 (1:100; ab16059) and chicken anti-GFP (1:1,000; ab13970) were from Abcam. Rabbit anti-SAC1 (1:100; 13033–1-AP) was from Proteintech. Incubation with the appropriate Alexa Fluor–conjugated secondary antibodies was also performed. Anti-mouse Alexa633 (1:100; A21052), anti-rabbit Alexa488 (1:300; A11008), anti-chicken Alexa488 (1:1,000; A11039), and anti-mouse Alexa568 (1:150; A11011) were from Invitrogen. Coverslips were mounted with Fluoromount Aqueous Mounting Medium (F4680; Sigma-Aldrich).
Immunoblotting
Western blot was performed as previously described (Fadó et al., 2015). Rabbit anti-CPT1C was developed in our laboratory (1:1,000; Sierra et al., 2008). Rabbit anti-ACC (1:1,000; 3676) and rabbit anti-phosphorylated ACC (Ser79; 1:2,000; 3661) were from Cell Signaling. Rabbit anti-SAC1 (1:500; 13033–1-AP) was from Proteintech. Mouse anti–β-actin (1:1,000; ab6276) and chicken anti-GFP (1:5,000; ab13970) were from Abcam. Rabbit anti-GluA1 (1:1,000; ab1504) and rabbit anti-GluA2 (1:1,000; ab397) were from Millipore. Rabbit anti-GluN2A (1:1,000; G9038), mouse anti–β-tubulin (1:2,000; T5201), and goat anti-FLAG (1:500; F3165) were from Sigma-Aldrich. HRP-conjugated secondary antibodies anti-mouse or anti-rabbit (1:10,000) were from DAKO. Blots were developed using Luminata Forte Western HRP substrate (Millipore). Semiquantitative analysis was performed using densitometry with Fiji software.
Cell-surface biotinylation assay
Surface protein levels were measured in cortinal neurons at 14-15 DIV by the biotin covalent binding method and processed as described previously (Miñano-Molina et al., 2011). Briefly, live cells were transferred to ice-cold PBS-Ca2+-Mg2+ buffer (pH 7.4, 1 mM CaCl2, and 0.1 mM MgCl2), followed by incubation with 0.5 mg/ml biotin (b10185; Life Technologies). Free biotin was quenched by three washes in cold PBS-Ca2+-Mg2+ + 0.1 M glycine. Cells were next scraped in cold 1% Triton X-100 homogenization buffer (NaCl, 50 mM; EDTA, 10 mM; EGTA, 10 mM; Na3VO4, 1 mM; NaF, 50 mM; NaPPi, 25 mM; glycerophosphate, 1 mM; PMSF, 1 mM; and Hepes, 50 mM, pH 7.5) supplemented with phosphatase and protease inhibitor cocktails. Homogenates were centrifuged at 10,000 ×g for 20 min to pellet insoluble fraction. 75 µl of the supernatants was mixed and heated with 25 µl of 2× SDS buffer (input). The remaining supernatants were pulled down with 40 µl of 50% avidin-agarose beads (53150; Pierce) overnight at 4°C. Beads were pelleted, washed three times with homogenization buffer, and heated in 100 µl of 2 x SDS buffer (surface fraction). 75 µl of the supernatants was mixed and heated with 25 µl of 2× SDS buffer (internal fraction). Equal volumes of the input, internal, and biotinylated fractions were processed by immunoblotting.
Immunoprecipitation assay
Cells were washed in ice-cold PBS and immediately scraped in cold lysis buffer (500 µl/2 × 100-mm plate; 1% Triton buffer for CPT1C immunoprecipitations, Tris 50 mM, NaCl 150 mM, and Triton X-100 1% at pH 8; or CL-47 buffer for Sac1 immunoprecipitations). Homogenates from cultures were solubilized 30 min at 4°C in the orbital and centrifuged at 10,000 ×g for 10 min to pellet insoluble fraction. 75 µl of the supernatant was mixed and heated with 25 µl of 4× SDS sample buffer (input). 1 mg of total protein from the remaining supernatants was incubated 1 h in the orbital with 10 µl of slurry protein-G–Sepharose beads in order to retain protein that bind beads unspecifically. Meanwhile, 2 µg of anti-CPT1C, 1 µg of anti-GluA1, or 2.5 µg of anti-SAC1 antibodies was incubated with 10 µl of protein G–Sepharose beads (Ab-beads). Both samples were centrifuged at 4,000 g for 2 min. Clean supernatants were immunoprecipitated with the beads attached to the antibody (Ab-beads) overnight at 4°C, since protein G–Sepharose beads (preclear fraction) were mixed with 100 µl of 2× SDS buffer and 1 µl of DTT at 1 M and heated. The beads were pelleted, and 75 µl of the supernatant (unbound fraction) was mixed and heated with 25 µl of 4× SDS sample buffer. The beads were then rinsed three times with lysis buffer and heated in 100 µl of 2× SDS sample buffer and 1 µl of DTT at 1 M (immunoprecipitation fraction; IP). 30 µl of the preclear fraction and IP and 10–20 µl of the input and the unbound fractions were detected by immunoblotting.
Protein–protein interaction simulations
For the CPT1C-GluA1 interactions, the GluA1 cytosolic region (residues from 827 to 903) was modeled with the available GluA2 crystal structure (Protein Data Bank accession no. 3KG2), and the CPT1C C-terminal plus its globular regions (residues from 157 to 803) was modeled with the COT crystal structure (Protein Data Bank accession no. 1XL7). The whole process was performed on a simulated aqueous solution (tip4p-D cubic waterbox; optimization by Best et al., 2012) with physiological concentration of NaCl and enough ions to neutralize the charge of the system. The mathematical function Charmm36 was employed. It was calculated as interaction time percentage (when low energies are required for protein approach). Next, a minimization function with 10,000 steps was used in order to get rid of bad contacts or unsuitable torsion angles from both interacting molecules that may have been inevitably introduced when setting up the whole system. A final two-step equilibration process was also employed. We submitted the CPT1C-GluA1 system to an aggregated 300 ns of production runs in which 12 different starting points were employed, each of them differing in the initial GluA1 fragment positioning relative to CPT1C fragment. The 12 approaching vectors were split into two groups of six each, where each of the group presented one of two “faces” of the GluA1 cytosolic fragment (as defined by an arbitrary plane intersecting lengthwise with the protein fragment) to the CPT1C fragment, and each approaching vector on the group was displaced 60 degrees in a clockwise manner from the previous. The utility of multiple replicas is discussed in Knapp et al. (2018). The molecular dynamics package used for the minimization, equilibration, and production processes was GROMACS (Van Der Spoel et al., 2005).
FRET analysis
All FRET imaging experiments were performed on a Leica TCS SP2 microscope. A 63×/1.40 NA objective with immersion oil and argon lasers with 458 and 514 lines (for donor, mTurquoise2, and acceptor, YFP, respectively) was used for all image acquisitions. The pinhole size was set to 2 (airy units), and the region of interest (ROI) to 5 × 5 (mm). 458-nm laser power was set up to 23%–60% and 514-nm laser up to 5%–10% so that no pixel saturation occurred. The number of bleaches was between three and four, all at 40%. For the calculation of FRET efficiency from acceptor to donor, we used the following formula operated at pixel basis:
where Ifret refers to FRET intensity, Idrefers to donor intensity in the absence of acceptor, Ia is the acceptor intensity in the presence of donor, Idcrossem is the fraction of donor emission cross-emitted into the acceptor channel, and Icrossexis the fraction of acceptor intensity cross-excited by donor excitation.
Confocal microscopy
Confocal imaging was performed in fixed cortical neurons with the confocal laser-scanning microscope ZEISS LSM 700 or Leica DMi8 using a ×63 1.4 NA oil objective. Approximately 18 stacks with 0.5-µm distance were taken per each imaged cell using identical settings, and all quantifications were done on a reconstructed 3D image with IMARIS 9.2 software (Bitplane). For quantification, sets of cells were cultured and stained simultaneously and imaged using identical settings. For surface GluA1 quantification, primary dendrites were taken, being considered as the prolongations first branching from soma. The axon was identified and discarded according to morphological criteria (thickness and position relative to the nucleus). A 15 µm × 5 µm ROI was randomly selected, and integrated intensity was measured inside it using the Spot tool. For the measurements of SAC1, GluA1, and PI(4)P intensities at the TGN compartment, the mean intensity of each staining in the 3D TGN-englobing ROI close to the soma was quantified using the Surface tool of Imaris software. The same threshold values were used in all the samples.
Live-cell imaging
Live-cell imaging was used to detect lipids or proteins in nonfixed transfected HEK-SAC1GFP cells (kind gift of Dr. Gerry Hammond, University of Pittsburgh, Pittsburgh, PA). 1 d after transfection, cells were transferred from culture medium to a recording chamber containing modified Krebs–Ringer solution (160 mM NaCl, 2.5 mM KCl, 2 mM CaCl2, 1 mM MgCl2, and 8 mM glucose, pH 7.4). Cells were kept in this solution up to ∼1 h and 30 min while imaging.
Total internal reflection fluorescence (TIRF) microscopy
TIRF imaging was performed in GluA1-transfected live HEK-SAC1GFP cells. Footprints were acquired in a Nikon TiE microscope equipped with a TIRF 60×/1.25 oil differential interference contrast H objective and Photometrics QuantEM electron-multiplying charge-coupled device camera (Nikon). The incident angle settled in all experiments was 110 degrees. All images were quantified with the ImageJ/Fiji (National Institutes of Health) software and surface GluA1 mean intensity was measured per cell.
Super-resolution imaging
Super-resolution studies were conducted using HEK-SAC1GFP cells transfected with TGN and ER organelle markers (mTurquoise-TGN38-FRB and iRFP-Sec61β, respectively) and a CPT1C overexpressing plasmid (CPT1C-SYFP2). Images were acquired in a ZEISS LSM 880 with Airyscan using 63× oil-immersion objective. Quantification of contact sites, endogenous SAC1, and overexpressed CPT1C at these junctions was performed cell per cell in one-stack images by using ImageJ/Fiji software. The amount of ER-TGN contacts was measured as a positive colocalization between the ER and the TGN signals and was always normalized to ER area. To measure SAC1 or CPT1C in the ER-TGN contacts, we drew two 2D ROIs corresponding to the ER and the TGN signals and then applied the colocalization plugin of ImageJ/Fiji software in order to determine the ER-TGN contact area per cell and calculate the mean intensity of SAC1 or CPT1C in that colocalized ROI. For the analysis of linear fluorescence profiles, a line 1 µm wide and 5 µm long was drawn inside the cell, and the sum intensity histograms of CPT1C, TGN marker, and ER marker from 2D images was obtained using ImageJ/Fiji software.
Rapamycin-dependent ER-TGN contact site creation
To explore ER-TGN contact sites, the rapamycin system was used to produce a tight association between the FKBP and FRB domains (Bohdanowicz and Fairn, 2011). Based on this, transfection of HEK-SAC1GFP cells with mCherry-pHR-TcRb-FKBP, CFP-TGN38-FRB, and YFP-P4M was performed followed by rapamycin addition (5 µM at room temperature; R8781; Sigma-Aldrich). This synthetic system allows ER recruitment to the TGN and a consequent decrease in Golgi PI(4)P (Dickson et al., 2014). Under constant perfusion, cells were tracked for ∼15 min with images taken every ∼5 s. Positive ER-TGN junction creation was only considered when the ER signal localized at the TGN increased with respect to its signal outside the TGN. This increase was measured with ImageJ/Fiji software as described above in one-stack images. Mean PI(4)P intensity was also quantified in the TGN compartment in each cell by creating a TGN-englobing mask and then applying it on the PI(4)P image. PI(4)P inside the TGN was always normalized to the cytoplasm signal in order to discard any difference due to the transfection efficiency.
PI(4)P-dependent phosphatase activity assay of SAC1
The activity assay of SAC1 was performed in stable transfected HeLa cells and in mice brain tissues. In cell lines, SAC1 was overexpressed by FuGENE6 transfection 48 h before the assay. Mice brain tissues were obtained from 4-mo-old male mice. HeLa cells or brain tissue was homogenized with chilled CL-47 lysis buffer supplemented with a protein inhibitor cocktail. 2 mg or 10 mg of protein (for HeLa cells and brain tissue, respectively) was immunoprecipitated with anti–SAC1 conjugated beads as described above. The immunoprecipitated protein-containing pellet was processed as described previously (Mani et al., 2007). Released free phosphate, an indicator of SAC1 activity on PI(4)P, was measured at 650 nm and was always normalized to immunoprecipitated SAC1 detected by Western blot.
UPLC MS/MS
Lipids were extracted with butanol and chloroform from age- and sex-matched WT and CPT1C−/− brains. Samples were neutralized by methylation with trimethylsilyldiazomethane, infused with sodium formate, and analyzed using a Waters XEVO TQ-S MS/MS in multiple reaction monitoring mode using electrospray and positive ion mode (Vivas et al., 2019). Elution profiles plotting the intensity (arbitrary units) as a function of elution time were analyzed to determine relative changes between samples. Total integrated areas under peaks from samples and standards were quantified using MassLynx software (Research Resource Identifier: SCR_014271; Waters). Samples were normalized to total brain protein levels and nonphysiological lipid standards: PI(4)P, 37:4, and PI(4,5)P2, 37:4 (Avanti Polar Lipids).
Statistical analysis
As indicated in each figure legend, the results were obtained from two to four independent experiments performed (at least) by biological duplicates and are given as the mean ± SEM or SD. When individual cells were analyzed, we decided to evaluate around 30 per condition for FRET assays and between 20 and 40 cells for immunostaining studies (at least) seeded in two independent coverslips per experiments. Exceptionally, the outliers were excluded if the values were above or below the mean ± two SDs. Statistical analysis was performed using PRISM (GraphPad Software). Data normality was determined in all the experiments according to the Shapiro-Wilk test and D’Agostino and Pearson test. Significance between groups was detected using Student’s t test (one or two sided, as indicated in each figure legend) or a Mann-Whitney U test (indicated in each figure legend) according to normality and a 95% confidence interval. For comparisons among three or four groups, ANOVA was performed (one-way or two-way; indicated in each figure legend), followed by the Bonferroni’s multiple comparison post-test.
Online supplemental material
Fig. S1 presents whole cell images of cortical neurons showing GluA1 surface levels under different treatments. Fig. S2 shows that glucose deprivation, 2DG, and TOFA treatments have no effect on cortical neuron survival. Fig. S3 shows TOFA treatment decreases GluA1 but not GluA2 and GluN2A at the surface of cortical neurons in a CPT1C-dependent manner. Fig. S4 shows the effects of TOFA treatment or SAC1 modulation on PI(4)P levels, TGN volume, and TGN-ER occurrence. Fig. S5 shows rapamycin-induced ER-TGN contacts. Fig. S6 shows that CPT1C deficiency decreases the brain levels of different PIP and PIP2 species.
Figure S5.
Rapamycin-induced ER-TGN contacts. (A) No artificial contacts were formed without the addition of rapamycin. HEK cells were transfected and treated as in Fig. 8 A. PI(4)P levels in the TGN were live imaged with confocal microscopy. TOFA addition in these conditions does not have any effect. Data represent the mean ± SD from one experiment performed by biological replicates (n = 5 or 6 cells per condition). Scale bars = 7 µm; scale bars of inset magnifications = 1.75 µm. (B) Effect of CPT1C expression and TOFA treatment on PI(4)P levels at the TGN after induction of artificial ER-TGN contacts. HEK cells were transfected with mCherry-pHR-TcRb-FKBP and CFP-TGN38-FRB plasmids to induce artificial ER-TGN by rapamycin (5 µM, 30 min). PI(4)P was labeled with the lipid probe YFP-P4M. Cells were treated for 1 h with TOFA or vehicle 24 h after transfection. Cells were live imaged with confocal microscopy. PI(4)P, TGN, and the ROI of PI(4)P inside the TGN are shown. Individual cells are the same as the ones analyzed in Fig. 8 A.
Acknowledgments
This work was partially supported by Ministerio de Economia y Competitividad from Spain (grant SAF2017-82813-C3-3R to N. Casals, grant SAF2017-89271-R to J. Rodríguez-Álvarez, cofunded by the European Regional Development Fund), Centro de Investigación Biomédica en Red Fisiopatología de la Obesidad y la Nutrición (grant CB06/03/0001 to N. Casals), Centro de Investigación Biomédica en Red sobre Enfermedades Neurodegenerativas (grant CB06/05/0042 to J. Rodríguez-Álvarez), Generalitat de Catalunya (2014SGR465 to N. Casals, SGR2014-0984 to J. Rodríguez-Álvarez), Fundació La Marató de TV3 (grant 87/C/2016 to N. Casals, grant 2014-3610 to J. Rodríguez-Álvarez), University of California funds (to E.J. Dickson), and National Institutes of Health grant R01 GM127513 (to E.J. Dickson).
The authors declare no competing financial interests.
Author contributions: M. Casas conducted conceptualization, formal analysis, investigation, methodology, validation, visualization, and review and editing. R. Fadó conducted conceptualization, formal analysis, investigation, validation, visualization, supervision, original draft, and review and editing. J.L. Domínguez and A. Roig conducted investigation and methodology. M. Kaku and S. Chohnan conducted methodology, investigation, resources and review, and editing. M. Solé and M. Unzeta conducted review and editing. A.J. Miñano-Molina, J. Rodríguez-Álvarez, and E.J. Dickson conducted conceptualization, methodology, resources, supervision, and review and editing. N. Casals conceived the study and conducted conceptualization, formal analysis, funding acquisition, project administration, resources, supervision, and review and editing.
References
- Beilharz J.E., Maniam J., and Morris M.J.. 2015. Diet-Induced Cognitive Deficits: The Role of Fat and Sugar, Potential Mechanisms and Nutritional Interventions. Nutrients. 7:6719–6738. 10.3390/nu7085307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Best R.B., Zhu X., Shim J., Lopes P.E.M., Mittal J., Feig M., and Mackerell A.D. Jr.. 2012. Optimization of the additive CHARMM all-atom protein force field targeting improved sampling of the backbone φ, ψ and side-chain χ(1) and χ(2) dihedral angles. J. Chem. Theory Comput. 8:3257–3273. 10.1021/ct300400x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blagoveshchenskaya A., Cheong F.Y., Rohde H.M., Glover G., Knödler A., Nicolson T., Boehmelt G., and Mayinger P.. 2008. Integration of Golgi trafficking and growth factor signaling by the lipid phosphatase SAC1. J. Cell Biol. 180:803–812. 10.1083/jcb.200708109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohdanowicz M., and Fairn G.D.. 2011. Rapamycin-based inducible translocation systems for studying phagocytosis. Methods Mol. Biol. 748:183–193. 10.1007/978-1-61779-139-0_13 [DOI] [PubMed] [Google Scholar]
- Bowen A.B., Bourke A.M., Hiester B.G., Hanus C., and Kennedy M.J.. 2017. Golgi-independent secretory trafficking through recycling endosomes in neuronal dendrites and spines. eLife. 6 e27362 10.7554/eLife.27362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brechet A., Buchert R., Schwenk J., Boudkkazi S., Zolles G., Siquier-Pernet K., Schaber I., Bildl W., Saadi A., Bole-Feysot C., et al. . 2017. AMPA-receptor specific biogenesis complexes control synaptic transmission and intellectual ability. Nat. Commun. 8:15910 10.1038/ncomms15910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breslow D.K., Collins S.R., Bodenmiller B., Aebersold R., Simons K., Shevchenko A., Ejsing C.S., and Weissman J.S.. 2010. Orm family proteins mediate sphingolipid homeostasis. Nature. 463:1048–1053. 10.1038/nature08787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrasco P., Sahún I., McDonald J., Ramírez S., Jacas J., Gratacós E., Sierra A.Y., Serra D., Herrero L., Acker-Palmer A., et al. . 2012. Ceramide levels regulated by carnitine palmitoyltransferase 1C control dendritic spine maturation and cognition. J. Biol. Chem. 287:21224–21232. 10.1074/jbc.M111.337493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrasco P., Jacas J., Sahún I., Muley H., Ramírez S., Puisac B., Mezquita P., Pié J., Dierssen M., and Casals N.. 2013. Carnitine palmitoyltransferase 1C deficiency causes motor impairment and hypoactivity. Behav. Brain Res. 256:291–297. 10.1016/j.bbr.2013.08.004 [DOI] [PubMed] [Google Scholar]
- Casals N., Zammit V., Herrero L., Fadó R., Rodríguez-Rodríguez R., and Serra D.. 2016. Carnitine palmitoyltransferase 1C: From cognition to cancer. Prog. Lipid Res. 61:134–148. 10.1016/j.plipres.2015.11.004 [DOI] [PubMed] [Google Scholar]
- Chen N., Pandya N.J., Koopmans F., Castelo-Székelv V., van der Schors R.C., Smit A.B., and Li K.W.. 2014. Interaction proteomics reveals brain region-specific AMPA receptor complexes. J. Proteome Res. 13:5695–5706. 10.1021/pr500697b [DOI] [PubMed] [Google Scholar]
- Chung J., Torta F., Masai K., Lucast L., Czapla H., Tanner L.B., Narayanaswamy P., Wenk M.R., Nakatsu F., and De Camilli P.. 2015. PI4P/phosphatidylserine countertransport at ORP5- and ORP8-mediated ER–plasma membrane contacts. Science. 349:428–432. 10.1126/science.aab1370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Convertino M., Pellarin R., Catto M., Carotti A., and Caflisch A.. 2009. 9,10-Anthraquinone hinders beta-aggregation: how does a small molecule interfere with Abeta-peptide amyloid fibrillation? Protein Sci. 18:792–800. 10.1002/pro.87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Bel L.M., and Brill J.A.. 2018. Sac1, a lipid phosphatase at the interface of vesicular and nonvesicular transport. Traffic. 19:301–318. 10.1111/tra.12554 [DOI] [PubMed] [Google Scholar]
- Dennis S.H., Jaafari N., Cimarosti H., Hanley J.G., Henley J.M., and Mellor J.R.. 2011. Oxygen/glucose deprivation induces a reduction in synaptic AMPA receptors on hippocampal CA3 neurons mediated by mGluR1 and adenosine A3 receptors. J. Neurosci. 31:11941–11952. 10.1523/JNEUROSCI.1183-11.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickson E.J., Jensen J.B., and Hille B.. 2014. Golgi and plasma membrane pools of PI(4)P contribute to plasma membrane PI(4,5)P2 and maintenance of KCNQ2/3 ion channel current. Proc. Natl. Acad. Sci. USA. 111:E2281–E2290. 10.1073/pnas.1407133111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickson E.J., Jensen J.B., Vivas O., Kruse M., Traynor-Kaplan A.E., and Hille B.. 2016. Dynamic formation of ER-PM junctions presents a lipid phosphatase to regulate phosphoinositides. J. Cell Biol. 213:33–48. 10.1083/jcb.201508106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diessler S., Jan M., Emmenegger Y., Guex N., Middleton B., Skene D.J., Ibberson M., Burdet F., Götz L., Pagni M., et al. . 2018. A systems genetics resource and analysis of sleep regulation in the mouse. PLoS Biol. 16 e2005750 10.1371/journal.pbio.2005750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domínguez J.L., Fernández-Nieto F., Castro M., Catto M., Paleo M.R., Porto S., Sardina F.J., Brea J.M., Carotti A., Villaverde M.C., et al. . 2015. Computer-aided structure-based design of multitarget leads for Alzheimer’s disease. J. Chem. Inf. Model. 55:135–148. 10.1021/ci500555g [DOI] [PubMed] [Google Scholar]
- Dong R., Saheki Y., Swarup S., Lucast L., Harper J.W., and De Camilli P.. 2016. Endosome-ER Contacts Control Actin Nucleation and Retromer Function through VAP-Dependent Regulation of PI4P. Cell. 166:408–423. 10.1016/j.cell.2016.06.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dworak M., McCarley R.W., Kim T., Kalinchuk A.V., and Basheer R.. 2010. Sleep and brain energy levels: ATP changes during sleep. J. Neurosci. 30:9007–9016. 10.1523/JNEUROSCI.1423-10.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fadó R., Soto D., Miñano-Molina A.J., Pozo M., Carrasco P., Yefimenko N., Rodríguez-Álvarez J., and Casals N.. 2015. Novel Regulation of the Synthesis of α-Amino-3-hydroxy-5-methyl-4-isoxazolepropionic Acid (AMPA) Receptor Subunit GluA1 by Carnitine Palmitoyltransferase 1C (CPT1C) in the Hippocampus. J. Biol. Chem. 290:25548–25560. 10.1074/jbc.M115.681064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faulhammer F., Kanjilal-Kolar S., Knödler A., Lo J., Lee Y., Konrad G., and Mayinger P.. 2007. Growth control of Golgi phosphoinositides by reciprocal localization of sac1 lipid phosphatase and pik1 4-kinase. Traffic. 8:1554–1567. 10.1111/j.1600-0854.2007.00632.x [DOI] [PubMed] [Google Scholar]
- Fernandes J., Vieira M., Carreto L., Santos M.A.S., Duarte C.B., Carvalho A.L., and Santos A.E.. 2014. In vitro ischemia triggers a transcriptional response to down-regulate synaptic proteins in hippocampal neurons. PLoS One. 9 e99958 10.1371/journal.pone.0099958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao L., Chiou W., Tang H., Cheng X., Camp H.S., and Burns D.J.. 2007. Simultaneous quantification of malonyl-CoA and several other short-chain acyl-CoAs in animal tissues by ion-pairing reversed-phase HPLC/MS. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 853:303–313. 10.1016/j.jchromb.2007.03.029 [DOI] [PubMed] [Google Scholar]
- Gao X.F., Chen W., Kong X.P., Xu A.M., Wang Z.G., Sweeney G., and Wu D.. 2009. Enhanced susceptibility of Cpt1c knockout mice to glucose intolerance induced by a high-fat diet involves elevated hepatic gluconeogenesis and decreased skeletal muscle glucose uptake. Diabetologia. 52:912–920. 10.1007/s00125-009-1284-0 [DOI] [PubMed] [Google Scholar]
- Gao S., Zhu G., Gao X., Wu D., Carrasco P., Casals N., Hegardt F.G., Moran T.H., and Lopaschuk G.D.. 2011. Important roles of brain-specific carnitine palmitoyltransferase and ceramide metabolism in leptin hypothalamic control of feeding. Proc. Natl. Acad. Sci. USA. 108:9691–9696. 10.1073/pnas.1103267108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gratacòs-Batlle E., Yefimenko N., Cascos-García H., and Soto D.. 2015. AMPAR interacting protein CPT1C enhances surface expression of GluA1-containing receptors. Front. Cell. Neurosci. 8:469. https://doi.org/ 10.3389/fncel.2014.00469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gratacòs-Batlle E., Olivella M., Sánchez-Fernández N., Yefimenko N., Miguez-Cabello F., Fadó R., Casals N., Gasull X., Ambrosio S., and Soto D.. 2018. Mechanisms of CPT1C-Dependent AMPAR Trafficking Enhancement. Front. Mol. Neurosci. 11:275 10.3389/fnmol.2018.00275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hangen E., Cordelières F.P., Petersen J.D., Choquet D., and Coussen F.. 2018. Neuronal Activity and Intracellular Calcium Levels Regulate Intracellular Transport of Newly Synthesized AMPAR. Cell Rep. 24:1001–1012.e3. 10.1016/j.celrep.2018.06.095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris J.J., Jolivet R., and Attwell D.. 2012. Synaptic energy use and supply. Neuron. 75:762–777. 10.1016/j.neuron.2012.08.019 [DOI] [PubMed] [Google Scholar]
- Henley J.M., and Wilkinson K.A.. 2013. AMPA receptor trafficking and the mechanisms underlying synaptic plasticity and cognitive aging. Dialogues Clin. Neurosci. 15:11–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henley J.M., and Wilkinson K.A.. 2016. Synaptic AMPA receptor composition in development, plasticity and disease. Nat. Rev. Neurosci. 17:337–350. 10.1038/nrn.2016.37 [DOI] [PubMed] [Google Scholar]
- Hong D., Cong L., Zhong S., Liu L., Xu Y., and Zhang J.. 2019. A novel CPT1C variant causes pure hereditary spastic paraplegia with benign clinical course. Ann. Clin. Transl. Neurol. 6:610–614. 10.1002/acn3.717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobi E., and von Engelhardt J.. 2018. AMPA receptor complex constituents: Control of receptor assembly, membrane trafficking and subcellular localization. Mol. Cell. Neurosci. 91:67–75. 10.1016/j.mcn.2018.05.008 [DOI] [PubMed] [Google Scholar]
- Knapp B., Ospina L., and Deane C.M.. 2018. Avoiding False Positive Conclusions in Molecular Simulation: The Importance of Replicas. J. Chem. Theory Comput. 14:6127–6138. 10.1021/acs.jctc.8b00391 [DOI] [PubMed] [Google Scholar]
- Kneussel M., Triller A., and Choquet D.. 2014. SnapShot: receptor dynamics at plastic synapses. Cell. 157:1738–1738.e1. 10.1016/j.cell.2014.06.002 [DOI] [PubMed] [Google Scholar]
- López-Viñas E., Bentebibel A., Gurunathan C., Morillas M., de Arriaga D., Serra D., Asins G., Hegardt F.G., and Gómez-Puertas P.. 2007. Definition by functional and structural analysis of two malonyl-CoA sites in carnitine palmitoyltransferase 1A. J. Biol. Chem. 282:18212–18224. 10.1074/jbc.M700885200 [DOI] [PubMed] [Google Scholar]
- Mani M., Lee S.Y., Lucast L., Cremona O., Di Paolo G., De Camilli P., and Ryan T.A.. 2007. The dual phosphatase activity of synaptojanin1 is required for both efficient synaptic vesicle endocytosis and reavailability at nerve terminals. Neuron. 56:1004–1018. 10.1016/j.neuron.2007.10.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marinangeli C., Didier S., Ahmed T., Caillerez R., Domise M., Laloux C., Bégard S., Carrier S., Colin M., Marchetti P., et al. . 2018. AMP-Activated Protein Kinase Is Essential for the Maintenance of Energy Levels during Synaptic Activation. iScience. 9:1–13. 10.1016/j.isci.2018.10.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesmin B., Bigay J., Moser von Filseck J., Lacas-Gervais S., Drin G., and Antonny B.. 2013. A four-step cycle driven by PI(4)P hydrolysis directs sterol/PI(4)P exchange by the ER-Golgi tether OSBP. Cell. 155:830–843. 10.1016/j.cell.2013.09.056 [DOI] [PubMed] [Google Scholar]
- Miñano-Molina A.J., España J., Martín E., Barneda-Zahonero B., Fadó R., Solé M., Trullás R., Saura C.A., and Rodríguez-Alvarez J.. 2011. Soluble oligomers of amyloid-β peptide disrupt membrane trafficking of α-amino-3-hydroxy-5-methylisoxazole-4-propionic acid receptor contributing to early synapse dysfunction. J. Biol. Chem. 286:27311–27321. 10.1074/jbc.M111.227504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morillas M., López-Viñas E., Valencia A., Serra D., Gómez-Puertas P., Hegardt F.G., and Asins G.. 2004. Structural model of carnitine palmitoyltransferase I based on the carnitine acetyltransferase crystal. Biochem. J. 379:777–784. 10.1042/bj20031373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moult P.R., Cross A., Santos S.D., Carvalho A.-L., Lindsay Y., Connolly C.N., Irving A.J., Leslie N.R., and Harvey J.. 2010. Leptin regulates AMPA receptor trafficking via PTEN inhibition. J. Neurosci. 30:4088–4101. 10.1523/JNEUROSCI.3614-09.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikonova E.V., Naidoo N., Zhang L., Romer M., Cater J.R., Scharf M.T., Galante R.J., and Pack A.I.. 2010. Changes in components of energy regulation in mouse cortex with increases in wakefulness. Sleep. 33:889–900. 10.1093/sleep/33.7.889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouyang J., Carcea I., Schiavo J.K., Jones K.T., Rabinowitsch A., Kolaric R., Cabeza de Vaca S., Froemke R.C., and Carr K.D.. 2017. Food restriction induces synaptic incorporation of calcium-permeable AMPA receptors in nucleus accumbens. Eur. J. Neurosci. 45:826–836. 10.1111/ejn.13528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pozo M., Rodríguez-Rodríguez R., Ramírez S., Seoane-Collazo P., López M., Serra D., Herrero L., and Casals N.. 2017. Hypothalamic Regulation of Liver and Muscle Nutrient Partitioning by Brain-Specific Carnitine Palmitoyltransferase 1C in Male Mice. Endocrinology. 158:2226–2238. 10.1210/en.2017-00151 [DOI] [PubMed] [Google Scholar]
- Price N., van der Leij F., Jackson V., Corstorphine C., Thomson R., Sorensen A., and Zammit V.. 2002. A novel brain-expressed protein related to carnitine palmitoyltransferase I. Genomics. 80:433–442. 10.1006/geno.2002.6845 [DOI] [PubMed] [Google Scholar]
- Ramírez S., Martins L., Jacas J., Carrasco P., Pozo M., Clotet J., Serra D., Hegardt F.G., Diéguez C., López M., et al. . 2013. Hypothalamic ceramide levels regulated by CPT1C mediate the orexigenic effect of ghrelin. Diabetes. 62:2329–2337. 10.2337/db12-1451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao J.N., Warren G.Z.L., Estolt-Povedano S., Zammit V.A., and Ulmer T.S.. 2011. An environment-dependent structural switch underlies the regulation of carnitine palmitoyltransferase 1A. J. Biol. Chem. 286:42545–42554. 10.1074/jbc.M111.306951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro L.F., Catarino T., Santos S.D., Benoist M., van Leeuwen J.F., Esteban J.A., and Carvalho A.L.. 2014. Ghrelin triggers the synaptic incorporation of AMPA receptors in the hippocampus. Proc. Natl. Acad. Sci. USA. 111:E149–E158. 10.1073/pnas.1313798111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinaldi C., Schmidt T., Situ A.J., Johnson J.O., Lee P.R., Chen K.L., Bott L.C., Fadó R., Harmison G.H., Parodi S., et al. . 2015. Mutation in CPT1C Associated With Pure Autosomal Dominant Spastic Paraplegia. JAMA Neurol. 72:561–570. 10.1001/jamaneurol.2014.4769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roa-Mansergas X., Fadó R., Atari M., Mir J.F., Muley H., Serra D., and Casals N.. 2018. CPT1C promotes human mesenchymal stem cells survival under glucose deprivation through the modulation of autophagy. Sci. Rep. 8:6997 10.1038/s41598-018-25485-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez-Rodríguez R., Miralpeix C., Fosch A., Pozo M., Calderón-Domínguez M., Perpinyà X., Vellvehí M., López M., Herrero L., Serra D., et al. . 2019. CPT1C in the ventromedial nucleus of the hypothalamus is necessary for brown fat thermogenesis activation in obesity. Mol. Metab. 19:75–85. 10.1016/j.molmet.2018.10.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosa E.F., Takahashi S., Aboulafia J., Nouailhetas V.L.A., and Oliveira M.G.M.. 2007. Oxidative stress induced by intense and exhaustive exercise impairs murine cognitive function. J. Neurophysiol. 98:1820–1826. 10.1152/jn.01158.2006 [DOI] [PubMed] [Google Scholar]
- Samanta S., Situ A.J., and Ulmer T.S.. 2014. Structural characterization of the regulatory domain of brain carnitine palmitoyltransferase 1. Biopolymers. 101:398–405. 10.1002/bip.22396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwenk J., Harmel N., Brechet A., Zolles G., Berkefeld H., Müller C.S., Bildl W., Baehrens D., Hüber B., Kulik A., et al. . 2012. High-resolution proteomics unravel architecture and molecular diversity of native AMPA receptor complexes. Neuron. 74:621–633. 10.1016/j.neuron.2012.03.034 [DOI] [PubMed] [Google Scholar]
- Schwenk J., Boudkkazi S., Kocylowski M.K., Brechet A., Zolles G., Bus T., Costa K., Kollewe A., Jordan J., Bank J., et al. . 2019. An ER Assembly Line of AMPA-Receptors Controls Excitatory Neurotransmission and Its Plasticity. Neuron. 104:680–692.e9. 10.1016/j.neuron.2019.08.033 [DOI] [PubMed] [Google Scholar]
- Seebohm G., Wrobel E., Pusch M., Dicks M., Terhag J., Matschke V., Rothenberg I., Ursu O.N., Hertel F., Pott L., et al. . 2014. Structural basis of PI(4,5)P2-dependent regulation of GluA1 by phosphatidylinositol-5-phosphate 4-kinase, type II, alpha (PIP5K2A). Pflugers Arch. 466:1885–1897. 10.1007/s00424-013-1424-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sierra A.Y., Gratacós E., Carrasco P., Clotet J., Ureña J., Serra D., Asins G., Hegardt F.G., and Casals N.. 2008. CPT1c is localized in endoplasmic reticulum of neurons and has carnitine palmitoyltransferase activity. J. Biol. Chem. 283:6878–6885. 10.1074/jbc.M707965200 [DOI] [PubMed] [Google Scholar]
- Spinelli M., Fusco S., Mainardi M., Scala F., Natale F., Lapenta R., Mattera A., Rinaudo M., Li Puma D.D., Ripoli C., et al. . 2017. Brain insulin resistance impairs hippocampal synaptic plasticity and memory by increasing GluA1 palmitoylation through FoxO3a. Nat. Commun. 8:2009 10.1038/s41467-017-02221-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thacker J.S., Xu Y., Tang C., Tupling A.R., Staines W.R., and Mielke J.G.. 2019. A Single Session of Aerobic Exercise Mediates Plasticity-Related Phosphorylation in both the Rat Motor Cortex and Hippocampus. Neuroscience. 412:160–174. 10.1016/j.neuroscience.2019.05.051 [DOI] [PubMed] [Google Scholar]
- Tokutake Y., Onizawa N., Katoh H., Toyoda A., and Chohnan S.. 2010. Coenzyme A and its thioester pools in fasted and fed rat tissues. Biochem. Biophys. Res. Commun. 402:158–162. 10.1016/j.bbrc.2010.10.009 [DOI] [PubMed] [Google Scholar]
- Van Der Spoel D., Lindahl E., Hess B., Groenhof G., Mark A.E., and Berendsen H.J.C.. 2005. GROMACS: fast, flexible, and free. J. Comput. Chem. 26:1701–1718. 10.1002/jcc.20291 [DOI] [PubMed] [Google Scholar]
- Venditti R., Masone M.C., Rega L.R., Di Tullio G., Santoro M., Polishchuk E., Serrano I.C., Olkkonen V.M., Harada A., Medina D.L., et al. . 2019. The activity of Sac1 across ER-TGN contact sites requires the four-phosphate-adaptor-protein-1. J. Cell Biol. 218:783–797. 10.1083/jcb.201812021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vivas O., Tiscione S.A., Dixon R.E., Ory D.S., and Dickson E.J.. 2019. Niemann-Pick Type C Disease Reveals a Link between Lysosomal Cholesterol and PtdIns(4,5)P2 That Regulates Neuronal Excitability. Cell Rep. 27:2636–2648.E4. 10.1016/j.celrep.2019.04.099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Von Filseck J.M., Čopič A., Delfosse V., Vanni S., Jackson C.L., Bourguet W., and Drin G.. 2015. Phosphatidylserine transport by ORP/Osh proteins is driven by phosphatidylinositol 4-phosphate. Science. 349:432–436. 10.1126/science.aab1346 [DOI] [PubMed] [Google Scholar]
- Wolfgang M.J., Kurama T., Dai Y., Suwa A., Asaumi M., Matsumoto S., Cha S.H., Shimokawa T., and Lane M.D.. 2006. The brain-specific carnitine palmitoyltransferase-1c regulates energy homeostasis. Proc. Natl. Acad. Sci. USA. 103:7282–7287. 10.1073/pnas.0602205103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfgang M.J., Cha S.H., Sidhaye A., Chohnan S., Cline G., Shulman G.I., and Lane M.D.. 2007. Regulation of hypothalamic malonyl-CoA by central glucose and leptin. Proc. Natl. Acad. Sci. USA. 104:19285–19290. 10.1073/pnas.0709778104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang G., Zhou X., Zhu J., Liu R., Zhang S., Coquinco A., Chen Y., Wen Y., Kojic L., Jia W., et al. . 2013. JNK3 couples the neuronal stress response to inhibition of secretory trafficking. Sci. Signal. 6:ra57 10.1126/scisignal.2003727 [DOI] [PubMed] [Google Scholar]
- Zaugg K., Yao Y., Reilly P.T., Kannan K., Kiarash R., Mason J., Huang P., Sawyer S.K., Fuerth B., Faubert B., et al. . 2011. Carnitine palmitoyltransferase 1C promotes cell survival and tumor growth under conditions of metabolic stress. Genes Dev. 25:1041–1051. 10.1101/gad.1987211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zewe J.P., Wills R.C., Sangappa S., Goulden B.D., and Hammond G.R.. 2018. SAC1 degrades its lipid substrate PtdIns4P in the endoplasmic reticulum to maintain a steep chemical gradient with donor membranes. eLife. 7 e35588 10.7554/eLife.35588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmer P., Binnebößel S., Bloch W., Hübner S.T., Schenk A., Predel H.G., Wright P., Stritt C., and Oberste M.. 2017. Exhaustive exercise alters thinking times in a tower of london task in a time-dependent manner. Front. Physiol. 7:694 10.3389/fphys.2016.00694 [DOI] [PMC free article] [PubMed] [Google Scholar]