During the collective migration of epithelial cells, interior cells produce “cryptic” lamellipodia to advance. Ozawa et al. show that formation of cryptic lamellipodia is regulated by WAVE and Arp2/3 complexes that accumulate at the adherens junctions. Without this system, cells lose control of their migration.
Abstract
Collective migration of epithelial cells plays crucial roles in various biological processes such as cancer invasion. In migrating epithelial sheets, leader cells form lamellipodia to advance, and follower cells also form similar motile apparatus at cell–cell boundaries, which are called cryptic lamellipodia (c-lamellipodia). Using adenocarcinoma-derived epithelial cells, we investigated how c-lamellipodia form and found that they sporadically grew from around E-cadherin–based adherens junctions (AJs). WAVE and Arp2/3 complexes were localized along the AJs, and silencing them not only interfered with c-lamellipodia formation but also prevented follower cells from trailing the leaders. Disruption of AJs by removing αE-catenin resulted in uncontrolled c-lamellipodia growth, and this was brought about by myosin II activation and the resultant contraction of AJ-associated actomyosin cables. Additional observations indicated that c-lamellipodia tended to grow at mechanically weak sites of the junction. We conclude that AJs not only tie cells together but also support c-lamellipodia formation by recruiting actin regulators, enabling epithelial cells to undergo ordered collective migration.
Introduction
Animal cells migrate as a collective in many morphogenetic processes, as well as in pathological events such as cancer invasion (Cheung and Ewald, 2014; De Pascalis and Etienne-Manneville, 2017; Friedl and Gilmour, 2009). It is therefore important to understand why cells move together rather than as single cells, and how the movement of individual cells is controlled and coordinated to allow their collective migration. Various types of cells require cadherin-mediated cell–cell adhesion for their orderly migration not only in vivo (Cai et al., 2014; Gritsenko et al., 2020; Niewiadomska et al., 1999), but also in vitro (Camand et al., 2012; Desai et al., 2009; Dupin et al., 2009; Grimsley-Myers et al., 2020; Ladoux and Mège, 2017; Mayor and Etienne-Manneville, 2016). This suggests that cadherins regulate cell behavior that is necessary for collective migration. However, the precise mechanisms of how epithelial cells require cadherins for their collective migration are not yet known.
Cells of “simple epithelia” are connected to each other via a junctional complex, which consists of a tight junction (TJ), an adherens junction (AJ, formally zonula adherens), and a desmosome, at the apical-most end of cell–cell contacts (Farquhar and Palade, 1963). Because of the observation that the TJ and AJ are closely adjoined to one another, this set of junctions is often called the apical junctional complex (AJC; Anderson et al., 2004; Vogelmann and Nelson, 2005). The AJC associates with a bundle of actin cables, called the circumferential actin belt or cable, which encircles individual cells at their apical ends, resulting in a honeycomb-like pattern of distribution. Below the AJC, nonspecialized junctions, for convenience termed the lateral cell–cell contacts (LCs), extend to the basal end of the cell, which actually occupies most areas of the cell junction. E-cadherin is a main adhesion receptor at the AJ of epithelial cells, which also functions at LCs. It binds β-catenin or plakoglobin and in turn αE-catenin, forming the cadherin–catenin complex. αE-catenin interacts with F-actin directly, or indirectly via binding to vinculin. In the absence of αE-catenin, E-cadherin is unable to maintain the AJC, indicating that the interaction of the cadherin–catenin complex with F-actin is crucial for epithelial-specific junction organization (Mege and Ishiyama, 2017; Takeichi, 2014).
The lamellipodium is a major structure in cell motility. At its front edge, actin polymerization and its network formation are initiated under the control of numerous regulators, including Rac1 and its effectors (Ridley, 2015), and these processes result in generation of a force for the cellular margin to advance. When cells migrate as a collective, leader cells, which occupy the front edge of a cell sheet, generate lamellipodia to move forward, and are trailed by follower cells (Haeger et al., 2015; Omelchenko et al., 2003). The followers also organize protrusions or lamellipodium-like structures, called cryptic lamellipodia (c-lamellipodia), most likely to chase the leaders (Farooqui and Fenteany, 2005). Similar structures related to cell movement are also detectable when epithelial cells move without any leader cells (Barlan et al., 2017; Krndija et al., 2019; Squarr et al., 2016).
Cadherin-mediated cell–cell contacts are known to be a mechanism that interferes with cell motility, particularly in the process of contact inhibition of cell locomotion (Roycroft and Mayor, 2016; Theveneau et al., 2010). This reported role of cadherins seemingly contradicts the observation that c-lamellipodia still form at cell–cell boundaries. In the present study we investigated how epithelial cells manage their motility at cell–cell contact zones, using adenocarcinoma-derived cell lines. Our observations indicate that AJs not only function to tie cells together and prevent random movement, but they also serve to regulate c-lamellipodium formation by recruiting the WAVE regulatory complex (WRC) and its effectors. Thus, we revealed an unexpected function of AJs: to support the migration of epithelial cells as a sheet.
Results
Epithelial cells require cell junctions to migrate
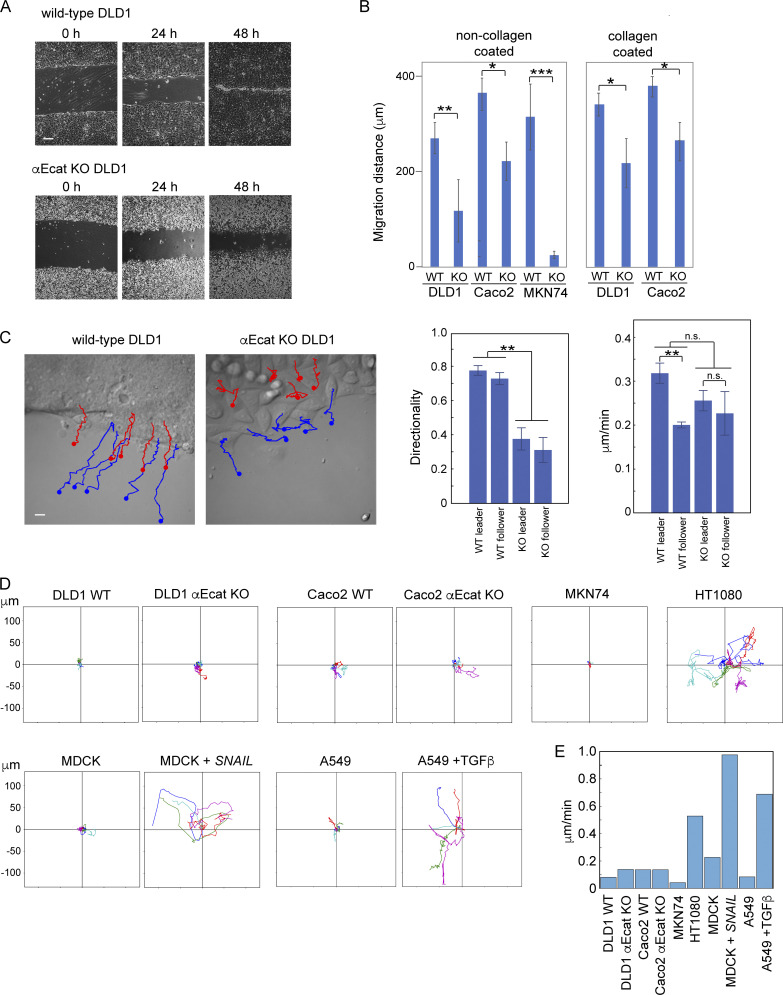
To reexamine the role of AJs in epithelial cell migration, we disrupted them by removing the αΕ-catenin gene (CTNNA1) in three adenocarcinoma lines: DLD1, Caco2 (both derived from colon carcinoma), and MKN74 (derived from gastric carcinoma) using the CRISPR/Cas9 method (Fig. S1 A). We isolated CTNNA1-deleted clones for each line and subjected them to a classic wound healing assay using collagen-coated or noncoated substrates. The results showed that αΕ-catenin removal caused an overall delay in wound healing for any cell line (Fig. 1, A and B), although their initial speed of migration varied from experiment to experiment, probably because of multiple factors that affect wound healing movement (De Pascalis and Etienne-Manneville, 2017). Time-lapse tracking of wild-type DLD1 cells located at the front row of their sheets (leader cells) and those located within a few rows behind the leaders (follower cells) showed that all these cells exhibited directed movement, whereas CTNNA1-knockout DLD1 (αEcat KO DLD1) cells moved in an nonstraight fashion at every position of the cell sheet (Fig. 1 C) and Video 1), as noted before (Seddiki et al., 2018). On the other hand, the average migration speed of these cells did not significantly differ between the wild-type and αEcat KO cell sheets (Fig. 1 C), although leaders apparently moved faster than followers in the wild-type sheets; this difference was recorded as a result of faster spreading of the leaders than followers during the early phases of wound healing. These results suggest that removal of αΕ-catenin caused reduced directionality in cell movement, explaining why αEcat KO cells were delayed in wound healing. We additionally confirmed that αΕ-catenin removal did not significantly alter the proliferation of these cells. For example, in the case of DLD1 and Caco2 cells, the percentages of mitotic cells in culture were 3.42 ± 1.81 and 4.07 ± 1.64 for wild-type (n = 12) and αEcat KO DLD1 (n = 10, P = 0.2), respectively; and 3.43 ± 0.33 and 3.96 ± 0.17 for wild-type (n = 6) and CTNNA1-deleted (αEcat KO) Caco2 (n = 6, P = 0.2), respectively. These results suggest that the epithelial cells used here require the cadherin-based AJs, whose formation is dependent on αΕ-catenin (Watabe et al., 1994; Watabe-Uchida et al., 1998), for their efficient migration, as shown for other cell types (Mayor and Etienne-Manneville, 2016).
Figure S1.
Western blotting analysis of CRISPR/Cas9-mediated deletion of proteins. (A) αE-catenin. (B) E-cadherin. (C) Myosin IIA (myo IIA) and IIB (myo IIB).
Figure 1.
Junction-dependent migration of epithelial cells. (A) Wound healing assays of wild-type and CTNNA1-deleted (αEcat KO) DLD1 cells. (B) Migration distance of the marginal cells at 24 h after scratching of the culture. KO, αEcat KO cells. Values were obtained at 25 points in a culture photographed with a 4× objective. Three cultures were analyzed. *, P < 0.05; **, P < 0.01; ***, P < 0.001 (t test, one-sided). (C) Trajectories of five independent leader (blue) or follower (red) cells during a 6.5-h time span are drawn over a montage image of Video 1. Follower cells were chosen from the zone consisting of two to three cell rows right behind the leader cell zone. Graphs show migration speed of the individual cells (right) and directionality in their migration (left). Here, directionality is the ratio of the net path length to the total path length in 200 min. The migration speed and directionality were obtained for individual cells as temporal averages. **, P < 0.01 (t test, one-sided). (D) Trajectories of singly isolated cells recorded for 20-h MDCK + SNAIL, MDCK cells stably transfected with SNAIL. A549 + TGFβ, cells cultured in 5 ng/ml of TGF-β. Values in the vertical and horizontal axes are identical. (E) Migration speed of isolated cells. Videos used for D were analyzed to obtain migration speed of each cell by temporal averaging of its instantaneous speed, and then by averaging of the values across the ensemble. Error bars represent SEM. Scale bars, 200 µm for A; 10 µm for C.
Video 1.
Time-lapse images of wild-type (left) and αEcat KO (right) DLD1 cells at the edge of a wound. The images were acquired at 10-min intervals, displayed at 12 fps.
To further study the role of cell–cell adhesion in cell migration, we observed the behavior of singly isolated DLD1 or Caco2 in time-lapse videos. Contrary to the observations using other epithelial types such as keratinocytes (Euteneuer and Schliwa, 1984), isolated DLD1 or Caco2 cells did not show any extensive migration, only movement around a fixed position (Fig. 1 D and Video 2). To test whether this is a general property of simple epithelium-derived cells, we examined other cell lines (MKN74, A549 [lung adenocarcinoma], and MDCK cells) and confirmed that they also do not migrate when isolated (Fig. 1 D). As mesenchymal cells are known to self-migrate, we examined the effect of epithelial-mesenchymal transition on the migration of these cells, using MDCK cells transfected with SNAIL cDNA (Ozawa and Kobayashi, 2015) or A549 cells treated with TGF-β (Thiery, 2003). The resultant mesenchyme-like cells became highly migratory, as observed with the fibroblastic line HT1080 (Fig. 1 C), exhibiting an increase of their migration speed (Fig. 1 E). Thus, these epithelial cells were unable to migrate alone unless transformed into the mesenchymal type. For further experiments, we chose either αEcat KO DLD1 or αEcat KO Caco2 cells, taking advantage of the unique characteristics of each line.
Video 2.
Time-lapse images of an isolated wild-type DLD1 cell. The images were acquired at 5-min intervals, displayed at 7 fps.
Junctional defects in αE-catenin-deleted cells
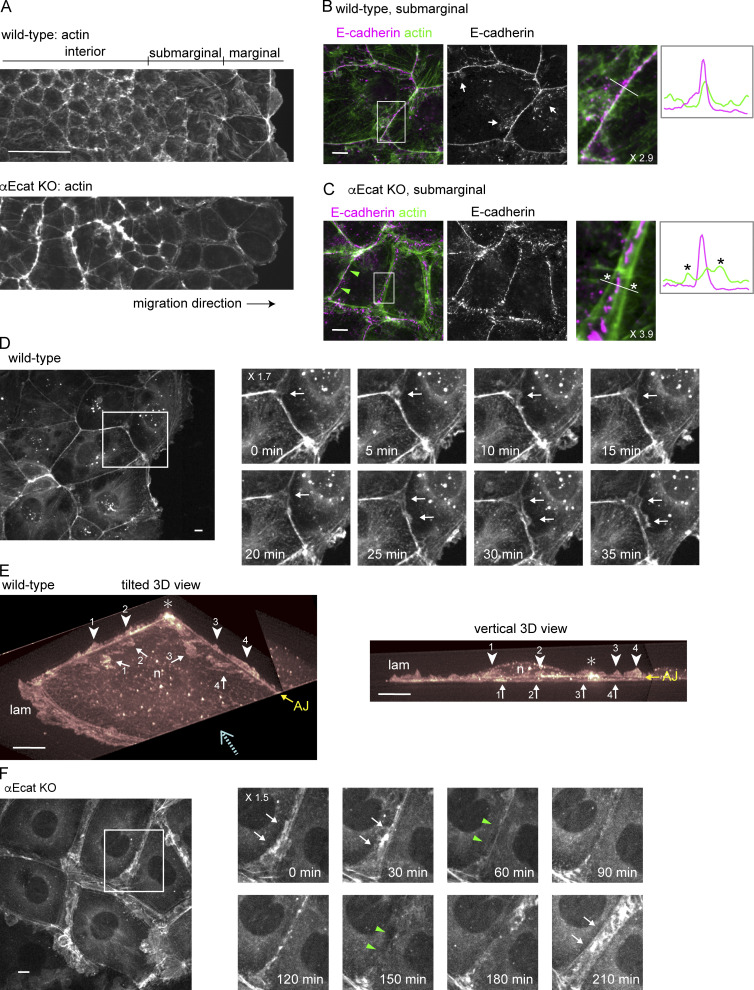
We next examined how cell junctions were affected by αΕ-catenin removal. As previously reported (Watabe-Uchida et al., 1998), DLD1 cells organized a typical AJC network, whereas αEcat KO DLD1 cells did not, showing dotted distribution of E-cadherin and TJ proteins at their cell–cell boundaries (Fig. S2 A). These αEcat KO DLD1 cells tended to round up, preventing us from observing their peripheral structures closely. For detailed microscopic analysis, therefore, we mainly used Caco2 cells, which show flatter morphology. In a migrating sheet of wild-type Caco2 cells, cells located at the front and subfront regions, defined as marginal and submarginal cells, respectively, spread more extensively than those located in deeper (interior) regions (Fig. 2 A). In any region of the sheet, their AJs were characterized by linear distribution of E-cadherin and associated actin cables along the apical cell–cell boundaries (Fig. 2 B). E-cadherin also distributed to LCs, which were generally slanted toward either side of the junction, exhibiting a strand-like or dotted pattern (Fig. 2 B, arrows), as reported previously (Kametani and Takeichi, 2007; Nishimura et al., 2016; Otani et al., 2006).
Figure S2.
Junctional proteins in DLD1 and Caco2 cells. (A) Coimmunostaining for E-cadherin and ZO-1 in wild-type or αE-cat KO DLD1 cells. (B–E) Coimmunostaining for the indicated proteins in wild-type or αEcat KO Caco2 cells. Arrowheads indicate closed junctions in αEcat KO cells. p120, p120-catenin; β-cat, β-catenin. Scale bars, 10 µm.
Figure 2.
Actin assembly and protrusion formation at junctions. (A) Low-magnification view of a wild-type (A) or αEcat KO (B) Caco2 cell sheet that is engaging in wound-healing movement. Stained for actin. (B and C) Costaining for E-cadherin and actin in wild-type (B) and αEcat KO (C) Caco2 cells. Fluorescence signals in the boxed regions, which are enlarged at the right, were scanned along the white bar. Asterisks indicate split actin cables. White arrows indicate examples of E-cadherin–positive LCs. Green arrowheads point to closed junction. (D) Montage of Video 3. Time-lapse images of the boxed region are shown. Arrows indicate emergence of protrusions. (E) LLSM images of a marginal wild-type Caco2 cell expressing LifeAct-RFP are displayed in 3D and viewed from two distinct angles. These were picked out from the images of Video 4, which had been edited to observe from desired angles. Arrows indicate protrusions that crawl under the cell shown here, and arrowheads point at another form of protrusions that grow upward. Numbers indicate identical structures at the left and right images. AJ shows the position of this junction, which is 1.5–2.0 µm high above the bottom of the cell. Dotted arrow shows an approximate direction for the vertical observation at the right panel. lam, lamellipodium; n, the position of nucleus; asterisk, the position where two bicellular junctions merge. The images were cropped three-dimensionally for better visualization of vertical protrusions. (F) Oscillation of the junctional space in αEcat KO cells. Montage of Video 9. Time-lapse images of the boxed region are shown. Arrows and green arrowheads indicate the open and closed stages of the junction, respectively. At 60, 120, and 180 min, the junction is only slightly open. Magnification is shown in the partly enlarged images. Scale bars, 100 µm for A; 10 µm for B–F.
αEcat KO Caco2 cells retained intercellular contacts but exhibited various defects in the junctions. Actin cables running along AJs were split into parallel lines at cell–cell contact sites, resulting in formation of a gap between them (Fig. 2 C). The gaps are often filled with irregular F-actin networks, and such actin reorganization occurred throughout the cell sheet (Fig. 2 A). E-cadherin was detected within this gap in a fragmentary pattern, but it did not show any consistent colocalization with actin filaments (Fig. 2 C), agreeing with the model that cadherins are normally linked to F-actin via αΕ-catenin (Takeichi, 2014). E-cadherin partners, β- and p120-catenin, showed distributions nearly identical to that of E-cadherin, as expected (Fig. S2 B). Desmosomal proteins were also concentrated within these gaps (Fig. S2 C). These observations suggest that, in the absence of αE-catenin, E-cadherin still helps bind the plasma membranes together, along with desmosomes, despite its failure to associate with F-actin. On the other hand, TJ proteins were localized along the split actin cables (Fig. S2, D and E).
Importantly, the actin-cable splitting did not always occur in the αEcat KO Caco2 cell sheets: that is, a given single cell often had nonsplit junctions, too, at its borders with other neighbors (Fig. 2 C, green arrowheads). In these junctions, not only E-cadherin but also desmosomal and TJ proteins accumulated together (Fig. S2, C and D). For convenience, we hereafter refer to the junctions where actin cables split and did not split as “open” and “closed” junctions, respectively.
c-Lamellipodium formation from AJ zones
To investigate how the cadherin-mediated junctions or AJs control epithelial sheet migration, we observed actin dynamics using wild-type or αEcat KO Caco2 cells that were stably transfected with LifeAct-RFP, an F-actin binding peptide (Riedl et al., 2008), since actin plays a central role in cell migration. Live imaging of a wild-type cell sheet, which was undergoing wound healing, showed that all migrating cells were firmly connected together at the level of AJs, as assessed by the stable appearance of AJ-associated actin belts (Video 3). Closer observation, however, indicated that fan-shaped protrusions sporadically emerged from the AJ-associated actin cables, which likely correspond to c-lamellipodia (Farooqui and Fenteany, 2005). Such protrusions occurred at the junctions of marginal and submarginal cells but were less clearly detectable in interior cells that were taller and migrating more slowly than the front cells, as noted before (Farooqui and Fenteany, 2005). Frequently, protrusions initially arose from a multicellular junction, then extended toward the bicellular sites of the junction (Fig. 2 D). For more detailed analysis of protrusion formation, we used lattice light-sheet microscopy (LLSM). Three-dimensional imaging by LLSM unexpectedly revealed that there are two forms of protrusion (Video 4 and Fig. 2 E). One was a flat projection (with a thickness <1 μm) that invaded underneath the adjoining cell, which structurally corresponds to c-lamellipodium. The other form of protrusion arose upward from the AJ zone, whose orientation was similar to that of ruffling membranes that occur at the front edges (or lamellipodia) of migrating cells. Whether these upward protrusions grew from both sides of the junction or only from a single side was not clear. These observations suggested that AJs function not only in tying cells together, but also as a site to remodel actin filaments required for c-lamellipodium formation and other dynamic protrusions.
Video 3.
Time-lapse images of a sheet of wild-type Caco2 cells, expressing LifeAct-RFP, at the wound edge. The images were acquired at 5-min intervals, displayed at 12 fps.
Video 4.
3D time-lapse images of wild-type Caco2 cells, expressing LifeAct-RFP, at a marginal zone of a cell sheet, collected with an LLSM. The video is replayed while being tilted. Time scale, h:m:s.
We additionally observed how the actin cytoskeleton changes during AJ formation, and how the AJs enable cells to migrate. To this end, we took videos of singly isolated cells labeled with LifeAct-RFP and their descendants. Isolated wild-type Caco2 cells tended to display a disc-like shape, generating lamellipodia all along the cell periphery (Video 5). When a cell divided into a pair, the descendants promptly organized AJ-associated actin cables between them. Such pairs of cells often showed rotating movement but never displayed migration. After further divisions, they came to form a multicellular colony. A marginal cell in the colony, which was surrounded by two to three neighbors, sporadically began migration using its lamellipodia, which formed at the free edges (Video 6). Thus, cells begin to remodel the actin cytoskeleton at the two-cell stage through AJ formation, but more cells are required to organize a polarized sheet to conduct directed migration.
Video 5.
Time-lapse images of an isolated wild-type Caco2 cell, expressing LifeAct-RFP. The cell undergoes mitosis during imaging. The images were acquired at 5-min intervals, displayed at 12 fps.
Video 6.
Time-lapse images of a small colony of wild-type Caco2 cells, expressing LifeAct-RFP. The images were acquired at 5-min intervals, displayed at 12 fps.
In the case of αEcat KO Caco2 sheets, moving cells exhibited lamellipodia-like protrusions everywhere at their periphery (Video 7). AJ-like actin cables were detectable, but not stable. The behavior of isolated αEcat KO Caco2 cells was indistinguishable from that of isolated wild-type cells. However, when they formed a pair, vigorous membrane ruffling continued even at the cell–cell contact sites (Video 8), and this feature persisted after further division of the cells (Video 9). Videos also revealed that, in colonies of αEcat KO Caco2, the open and closed junctions dynamically converted from one to the other, and the junctional closure resulted in a temporary suppression of membrane ruffling (Fig. 2 F), which implies that a certain form of cell–cell contacts is sufficient for suppressing membrane protrusion when αE-catenin is absent. Thus, in the absence of αE-catenin, it seems that cells are unable to control c-lamellipodium formation but exhibit an altered form of contact-dependent regulation of protrusion formation.
Video 7.
Time-lapse images of a sheet of αEcat KO Caco2 cells, expressing LifeAct-RFP, at the wound edge. The images were acquired at 5-min intervals, displayed at 12 fps.
Video 8.
Time-lapse images of an isolated αEcat KO Caco2 cell, expressing LifeAct-RFP. The cell undergoes mitosis during imaging. The images were acquired at 5-min intervals, displayed at 12 fps.
Video 9.
Time-lapse images of a small colony of αEcat KO Caco2 cells, expressing LifeAct-RFP. The images were acquired at 5-min intervals, displayed at 12 fps.
WAVE complex is required for junctional membrane protrusion
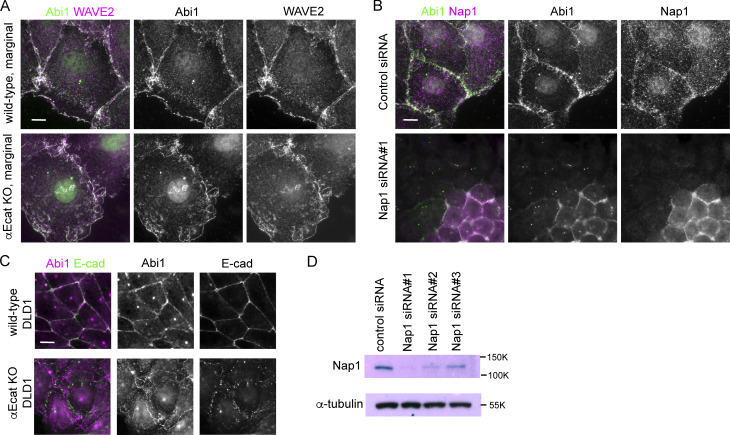
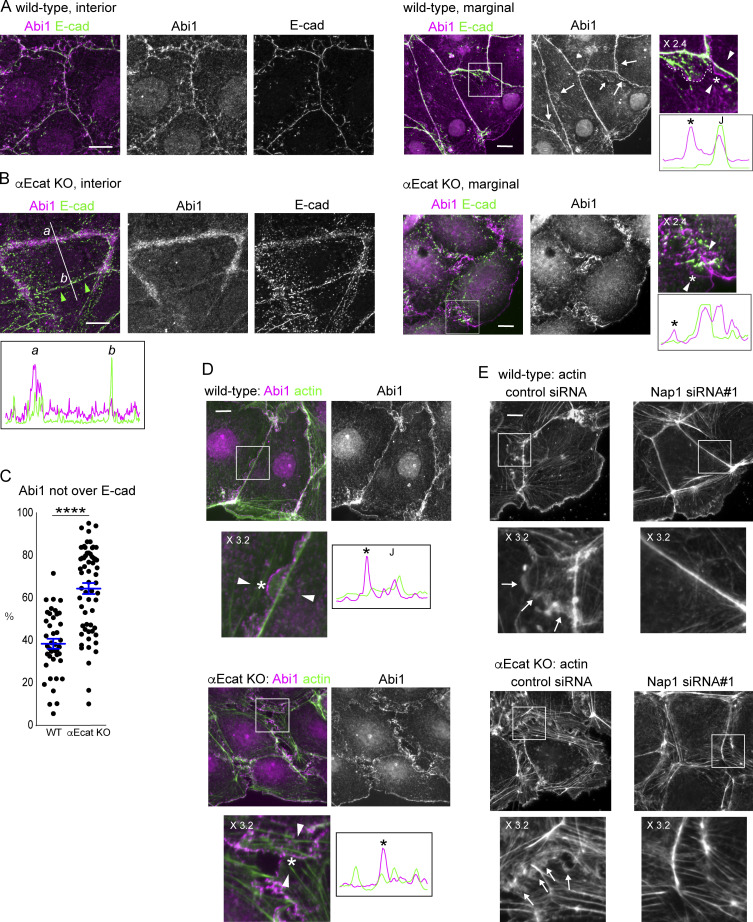
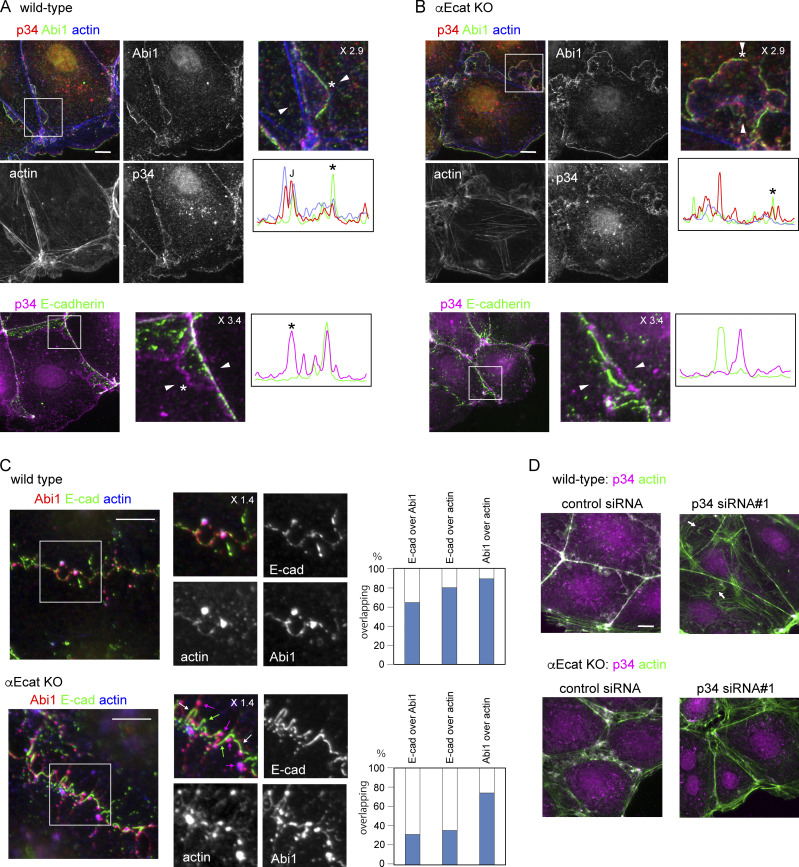
We began to explore how c-lamellipodia form. Because the WRC is known as a major regulator of actin assembly in lamellipodia (Takenawa and Suetsugu, 2007), we examined their potential contribution to c-lamellipodium formation using Caco2 cells. To determine subcellular distribution of WRC components, we initially observed three of them, Abi1, WAVE2, and Nap1, finding that they were essentially identical in distribution (Fig. S3, A and B). In the ensuing experiments, we observed a representative one of the three, unless otherwise noted. Consistent with previous reports (Han et al., 2014; Nishimura et al., 2016; Verma et al., 2012; Yamazaki et al., 2007), Abi1 was localized to AJs, overlapping with E-cadherin, in interior cells of wild-type cell sheets (Fig. 3 A, left). In marginal and submarginal cells whose junctions showed protrusions, however, Abi1 was detected not only along the junctions, but also at the edges of the protrusions (Fig. 3 A, right). The frequency of the Abi1-positive protrusions considerably varied from junction to junction, as expected from their dynamic nature observed in videos. E-cadherin did not localize at the edges of such protrusions, although it was detected inside the larger protrusions in particular, an example of which is shown in Fig. 3 A, rightmost panel.
Figure S3.
Detection of WRC components in Caco2 and DLD1 cells. (A) Coimmunostaining for Abi1 and WAVE2 in wild-type or αEcat KO Caco2 cells. (B) Coimmunostaining for Abi1 and Nap1 in wild-type Caco2 cells that were treated with siRNA for Nap1. Junctional Abi1 was removed as a result of Nap1 depletion, whereas centrosomal staining with anti-Abi1 antibody was not, which suggests that this staining is due to nonspecific binding of the antibody to centrosomes. Consistently, antibodies for WAVE2 or Nap2 did not detect centrosomes. (C) Coimmunostaining for Abi1 and E-cadherin in wild-type and αEcat KO DLD1 cells. (D) Western blots for Nap1 in Nap1 siRNA-treated cells. Scale bars, 10 µm.
Figure 3.
WAVE complex at junctions. (A and B) Coimmunostaining for Abi1 and E-cadherin in wild-type (A) and αEcat KO (B) Caco2 cells. IF signals in the boxed regions, which are enlarged at the right, were scanned along the line (not depicted) drawn between the white arrowheads. Asterisk in the image indicates a position roughly corresponding to the peak labeled with the same symbol on the scan, throughout the figures. J, the position of cell junction. In B, IF signals were also scanned along the line marked a and b. Larger arrows point to Abi-positive protrusions; small arrow, a protrusion having E-cadherin, which is outlined with a broken line at the rightmost image. Green arrowheads point to a closed junction. (C) IF signals for Abi1 that overlap or do not overlap those for E-cadherin were measured using interior cells, and the ratio of the nonoverlapping to the total signals was plotted. For measurement, a few points in each of the bicellular junctions were randomly selected, using four wild-type and three αE-cat KO cells. ****, P < 0.0001 (t test, one-sided). (D) Coimmunostaining for Abi1 and actin in wild-type or αEcat KO Caco2 cells. IF signals were scanned as explained in A. Marginal and submarginal cells are shown. J, the position of cell junction. (E) Effect of siRNA-mediated Nap1 depletion on actin assembly. The boxed areas are enlarged, being placed under each panel. Arrows indicate protrusions. Marginal and submarginal cells are shown. Magnification is shown in the partly enlarged images. Scale bars, 10 µm.
In αEcat KO Caco2 cell sheets, the open junctions in interior cells were filled with Abi1-bearing amorphous structures (Fig. 3 B, left), which were detected as fan-shaped membranes in marginal and submarginal cells (Fig. 3 B, right). Quantitative measurement indicated that Abi1-positive membranes, which do not overlap with E-cadherin, were increased at those junctions (Fig. 3 C). In the closed junctions, on the other hand, the relative level of Abi1 was reduced (Fig. 3 B, left scan), in accordance with the observation that membrane ruffling was suppressed at these sites. DLD1 cells also showed a similar overlapping of Abi1 with E-cadherin at wild-type junctions and a separation of Abi1-postive membranes from E-cadherin in the absence of αE-catenin (Fig. S3 C). To summarize, Abi1 localizes at E-cadherin–positive AJs, but it becomes distributed also to protrusions when they form, and the latter fraction of Abi is increased at the open junction of αEcat KO cells.
Double-staining for Abi1 and actin confirmed that they overlap with one another at both protrusions and junctions in wild-type cells (Fig. 3 D, upper). Live imaging of Nap1-GFP introduced into LifeAct-RFP transfectants showed that Nap1/actin double-positive membranes dynamically protruded from AJ regions (Video 10), suggesting that WRC redistributes from AJ to the protrusion during its formation. In αEcat KO Caco2 cells, Abi1-positive membranes lost any defined relation with particular actin cables (Fig. 3 D, lower), preventing us from identifying the origin of their formation.
Video 10.
Time-lapse images of Nap1-GFP (green) and LifeAct-RFP (magenta) at a junction of wild-type Caco2 cells. The images were acquired at 5-min intervals, displayed at 6 fps.
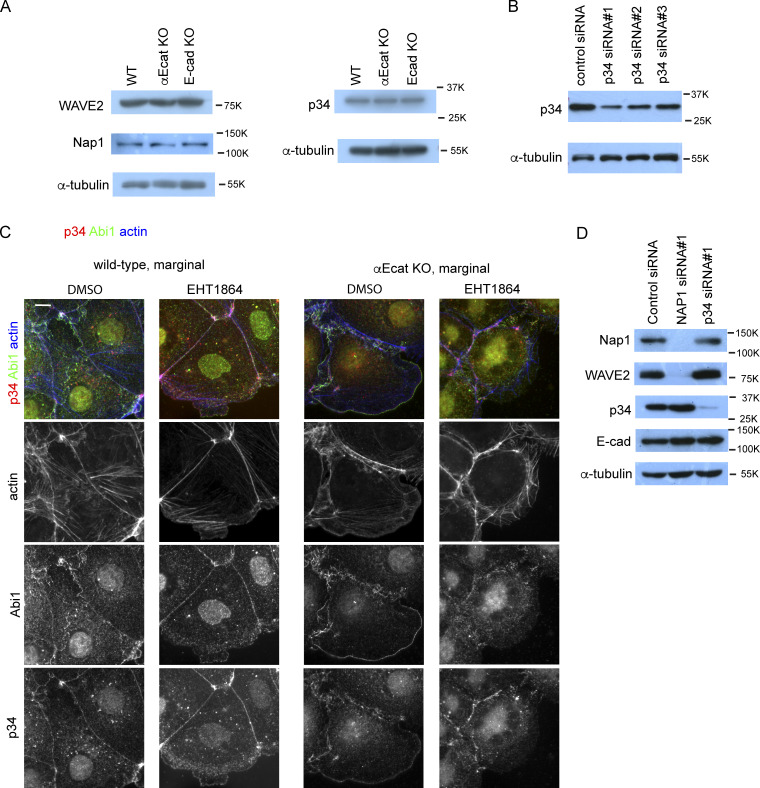
To confirm the role of WRC in c-lamellipodium formation, we depleted Nap1 in Caco2 cells using siRNAs (Fig. S3 D). Nap1 depletion resulted in disappearance of Abi1 from junctions (Fig. S3 B), suggesting that WRC was disorganized there, and this treatment eliminated actin-positive protrusions that are associated with AJs (Fig. 3 E, top). The open junctions in αEcat KO Caco2 cells also lost fan-shaped protrusions, becoming filled with only fibrous actins (Fig. 3 E, bottom). Thus, WRC is important for c-lamellipodium formation, supporting previous observations (Palamidessi et al., 2019).
Arp2/3 complex cooperates with WRC for protrusion formation
A major function of WRC is to activate the Arp2/3 complex, which mediates actin nucleation (Krause and Gautreau, 2014; Rotty et al., 2013), prompting us to test whether Arp2/3 is also involved in c-lamellipodium formation. Immunostaining for p34/ARPC2, a subunit of this complex, showed that it localized along AJs (Fig. 4 A), consistent with previous observations (Kovacs et al., 2002; Verma et al., 2004, 2012). This p34/ARPC2 overlapped with Abi1, E-cadherin, and associated actin cables, although it did not distribute to LCs, unlike WRC. p34/ARPC2 was also detectable in actin-positive protrusions, distributing more diffusely than Abi1. In αEcat KO Caco2 cells, similar overlapping of p34/ARPC2 and Abi1 was seen along protrusions in the open junctions (Fig. 4 B). At basal regions of these protrusions, p34/ARPC2 and Abi1 irregularly aggregated together with actin clusters, instead of localizing at AJs. Notably, E-cadherin lost its overlapping with p34/ARPC2 in these cells (Fig. 4 B, bottom). Thus, the codistribution of these molecules at AJs was disorganized in the absence of αE-catenin. The expression levels of WRC components and p34/ARPC2 were not affected by αE-catenin loss (Fig. S4 A).
Figure 4.
Arp2/3 complex at junctions and its interaction with WRC and F-actin. (A and B) Coimmunostaining for p34/ARPC2 (p34), Abi1 and actin (top), and p34/ARPC2 and E-cadherin (bottom) in wild-type (A) or αEcat KO (B) Caco2 cells. J, the position of junction. Marginal or marginal plus submarginal cells are shown throughout this figure. (C) Coimmunostaining for Abi1, E-cadherin, and actin in wild-type or αE-cat KO Caco2 cells, which were incubated with 10 µM latrunculin A for 60 min. The boxed regions are enlarged. The overlapping ratios of these molecules were quantified. Magenta arrows, Abi1–actin coclusters; white arrows, Abi1–E-cadherin–actin coclusters; green arrows, E-cadherin fragments that do not overlap with other molecules. (D) Effect of p34/ARPC2 siRNA treatment on actin assembly. Arrows point to actin-positive structures that are not closely associated with the junctions. Magnification is shown in the partly enlarged images. Scale bars, 10 µm.
Figure S4.
Expression of WRC and Arp2/3 complex, effect of Rac1 inhibitor, and effect of NAP1 or p34/ARPC2 depletion. (A) Western blots for WAVE2, Nap1, or p34/ARPC2 (p34) in wild-type, αEcat KO, and E-cad KO Caco2 cells. The Nap1 blot was obtained separately from the WAVE2 and α-tubulin blots. (B) Western blots for p34/ARPC2 in p34/ARPC2 siRNA-treated cells. (C) Effect of the Rac1 inhibitor EHT1864 on actin, Abi1, and p34/ARPC2 distribution in wild-type or αE-cat KO Caco2 cells. Cells were incubated with 100 µM EHT1864 for 5 h 30 min. (D) Effect of Nap1 or p34/ARPC2 depletion on the level of other proteins. Scale bars, 10 µm.
Previous studies suggested that WRC, Arp2/3, and E-cadherin physically interact with each other via cortactin (Han et al., 2014; Kovacs et al., 2002). To gain further insights into how these molecules associate with AJs, we examined the effect of actin disturbance on their distribution, because all of them directly or indirectly interact with F-actin. Treatment of wild-type Caco2 cells with an actin polymerization inhibitor, latrunculin A, resulted in clustering of actin, leaving strand-like actin linkages between the clusters (Fig. 4 C). Abi1 well coclustered with these reorganized actin molecules, and E-cadherin also showed some level of overlapping with actin or Abi1, even after the severe disturbance of actin assembly. In αEcat KO cells, however, although Abi1 still overlapped with actin clusters, E-cadherin became less coaggregative with Abi1 or actin. p34/ARPC2 behaved in a way similar to Abi1 in these latrunculin A–treated cells. These observations suggest that, in normal AJs, actin filaments serve as a core structure for E-cadherin, WRC, and Arp2/3 to assemble together, and in the absence of αE-catenin, E-cadherin leaves from this assemblage, owing to the loss of its actin-binding partner.
To confirm the role of Arp2/3 in c-lamellipodium formation, we treated cells with siRNA for p34/ARPC2 (Fig. S4 B). This treatment caused regression of actin-positive protrusions in both wild-type and αEcat KO cells, although some irregular actin-positive structures, which were separated from AJs, remained in wild-type cells (Fig. 4 D). We also examined the effect of an Arp2/3 inhibitor, CK666, using LifeAct-RFP–expressing Caco2 cells. Videos showed that this inhibitor suppressed dynamic protrusion of fan-shaped membranes, although it permitted extension of some flat structures (Video 11, right). Because WRC-Arp2/3 signaling is regulated by Rac1 GTPase (Chen et al., 2017), we also examined the effect of a Rac inhibitor, EHT 1864 (Shutes et al., 2007). After treatment with this inhibitor, actin-positive protrusions disappeared, as reported before (Malinverno et al., 2017), and both Abi1 and p34/ARPC2 came to accumulate only along AJ-associated actin filaments in wild-type cells (Fig. S4 C). In the case of αEcat KO cells treated with the inhibitor, actin-bearing protrusions were reduced, and Abi1 and p34/ARPC2 came to aggregate on irregular actin clusters. These observations confirmed that the Rac1-dependent WRC-Arp2/3 system is important for c-lamellipodium formation, and also suggested that AJs can hold nonactivated WRC and Arp2/3 components.
Video 11.
Time-lapse images of wild-type Caco2 cells not treated (left) or treated with 200 µM CK666 (right). Imaging started 14.5 h after addition of the reagent. The images were acquired at 15-min intervals, displayed at 6 fps.
WRC-Arp2/3 system is required for collective migration of epithelial cells
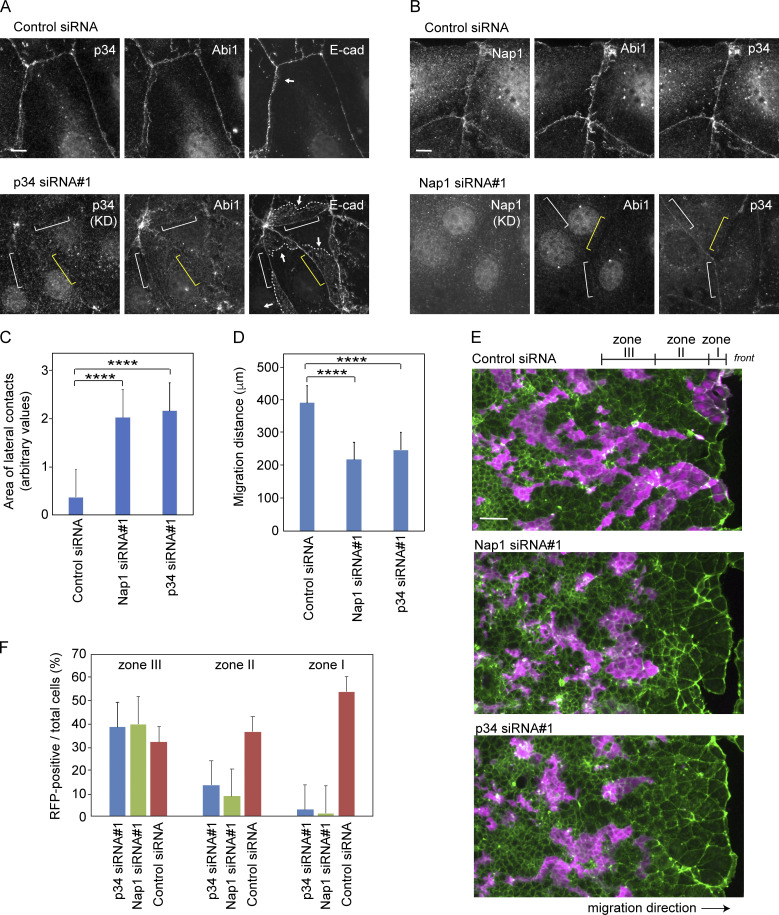
We then tested whether c-lamellipodium formation is really required for the collective migration of Caco2 cells. Before this test, we collected data regarding whether Nap1 or p34/ARPC2 depletion affected cell junctions. Depletion of each molecule did not affect the expression of the other molecule, nor E-cadherin, except that Nap1 depletion also removed its partner, WAVE2 (Fig. S4 D), as was found for Abi1. Immunocytological analysis, however, showed that depletion of Nap1 or p34/ARPC2 caused reduction of the other at the junctions, suggesting that their junctional recruitment is interdependent (Fig. 5, A and B). Notably, the E-cadherin–positive areas that extend below the AJ dramatically increased in these cells (Fig. 5, A and C), which suggests that static LCs form in place of c-lamellipodia when the WRC-Arp2/3 system is inactive.
Figure 5.
Effects of p34/ARPC2 or Nap1 depletion on collective cell migration. (A and B) p34/ARPC2 (A) or Nap1 (B) was depleted using siRNAs, followed by coimmunostaining for the indicated molecules. Yellow brackets indicate a region of cell junction in which the siRNA-targeted molecule is undetectable in immunostaining. White brackets indicate a region in which p34/ARPC2 or the Nap1 partner Abi is still faintly visible. Arrows point to LCs that are identified by E-cadherin distribution, and broken lines trace the basal margins of these LCs. Marginal and submarginal cells are shown. (C) Areas of E-cadherin–positive LCs in marginal and submarginal cells. The entire area of LC expanding below from each bilateral AJ was measured and compared. 31 junctions in control cells and 23 junctions in Nap1 or p34/ARPC2 siRNA-treated cells were analyzed. ****, P < 0.0001 (t test, one sided). (D) Migration distance of the marginal cells during 24 h in wound healing cultures. Values were obtained using four and six independent areas for control and Nap1- or p34/ARPC2 siRNA-treated cells, respectively. ****, P < 0.0001 (t test, one sided). (E and F) Sheets of wild-type Caco2 cells that were mixed in a 1-to-1 ratio with those expressing LifeAct-RFP, in which Nap1 or p34 p34/ARPC2 has been depleted with siRNAs. Magenta, LifeAct-RFP; green, actin. Photographed 48 h after wounding. In F, the ratio of siRNA-treated (LifeAct-RFP–labeled) cells to total cells in the three tandem zones of a culture was measured. Zones I, II, and III roughly correspond to a row of marginal cells, rows of submarginal cells, and an anterior group of interior cells, which span 58, 188, and 188 µm in width, respectively. We analyzed 11, 13, and 5 images for control and Nap1- and p34 p34/ARPC2 siRNA-treated cultures, respectively. Error bars represent SEM. Scale bars, 10 µm in A and B; 100 µm in E.
Next, by wound healing methods, we examined the migration of Caco2 cells in which Nap1 or p34/ARPC2 was depleted and found that their migration was delayed (Fig. 5 D). In this experiment, however, depletion of the actin regulators must have also affected the functions of genuine lamellipodia formed by the leader cells. To examine specifically the role of Nap1 or p34/ARPC2 in the migration of follower cells, we prepared LifeAct-RFP–transfected Caco2 cells in which these molecules were depleted, and mixed them with nonlabeled wild-type cells in a 1-to-1 ratio, subjecting the cell mixture to wound healing assays. The results showed that Nap1- or p34/ARPC2-depleted cells were left behind during the movement of cell sheets (Fig. 5, E and F), confirming that the WRC-Arp2/3 system is required for the migration of follower cells. In these experiments, cell proliferation rates did not differ between control and Nap1- and p34/ARPC2 siRNA-treated cells, in which the percentage of mitotic cells in culture were 7.69 ± 0.70 (n = 6), 8.54 ± 0.25 (n = 6, P = 0.3 versus control), and 7.07 ± 0.36 (n = 6, P = 0.6 versus control), respectively.
AJ disruption induces myosin II activation
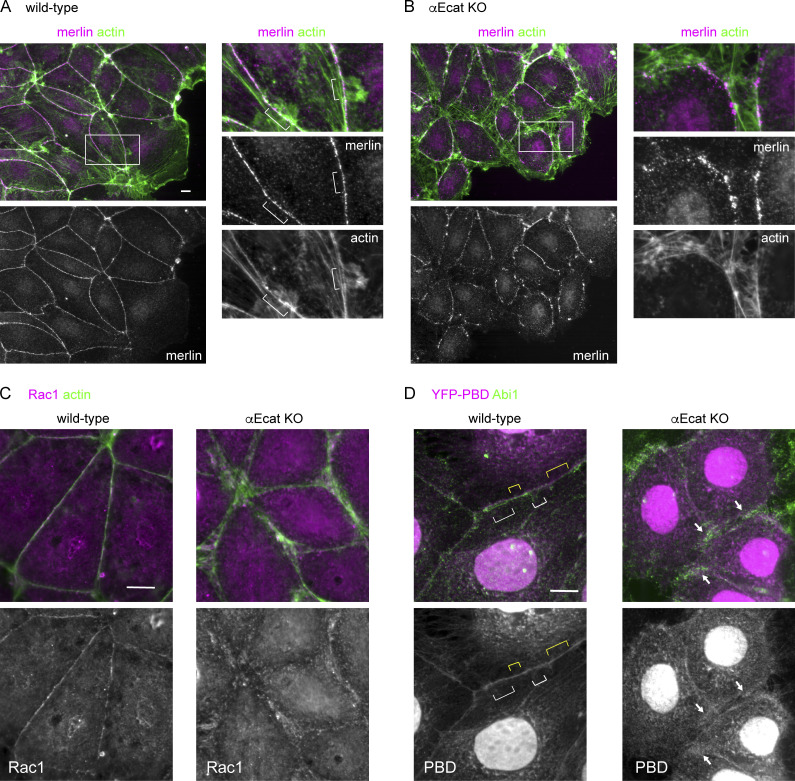
Next, we investigated whether there is any mechanism to “control” c-lamellipodium formation, as it occurred in a nonpersistent way in wild-type cells, compared with the constant production in αEcat KO cells. Previous studies indicated that merlin, which generally localizes at cell–cell junctions, relocalizes to the cytoplasm responding to intercellular pulling force, and this triggers Rac1-dependent c-lamellipodium formation (Das et al., 2015), prompting us to test the potential involvement of merlin in the present system. Immunostaining showed that merlin localized along the AJ-associated actin cables in wild-type Caco2 cells, and it was also detected along split actin cables in αEcat KO cells (Fig. S5, A and B). However, we did not observe any cytoplasmic relocalization of merlin in these cells that were undergoing wound healing movement. Furthermore, the density of merlin on the junctional actin cables did not particularly change at the sites where actin-positive protrusions appeared. Thus, we could not find any possible functional relation between merlin distribution and c-lamellipodium formation in our cell model. We also examined the distribution of Rac1 by immunostaining, as well as its active form, by transiently introducing a YFP–PBD (PAK-binding domain) construct that binds Rac-GTP (Hoppe and Swanson, 2004) into Caco2 cells. In wild-type cells, both Rac1 and YFP–PBD were detected along the AJs, irrespective of whether or not protrusions occurred there, whereas in αEcat KO cells, Rac1 and YFP–PBD came to overlap with actin-positive or Abi1-positive structures at the open junctions (Fig. S5, C and D). These observations indicate that Rac1 accumulates at the AJs in wild-type cells, but it redistributes to protrusions in the absence of αE-catenin, just as observed for its effectors. Importantly, we did not detect any noticeable difference in the overall intensity of YFP–PBD bound to these subcellular structures between wild-type and αEcat KO cells, nor between protrusion-positive and -negative regions at AJs, implying that altered Rac1 activity may not be a key to explain the differences in c-lamellipodium behavior between the different adhesive sites.
Figure S5.
Distribution of merlin, Rac1, and YFP-PBD. (A and B) Coimmunostaining for merlin and actin in a layer of wild-type (A) or E-cad KO (B) Caco2 cells that is engaging in wound healing. The squared portion is enlarged at the right in each of the figure sets. Brackets indicate junctional sites where protrusions emerge. (C) Coimmunostaining for Rac1 and actin. Rac1 immunostaining signals disappeared when cells were pretreated with Rac1 siRNA, verifying the significance of the data. (D) Covisualization of Abi1 and YFP-PBD transiently introduced into the cells. White and yellow brackets indicate examples of the junctional sites where Abi1-positive protrusions are detected and undetected, respectively. Arrows point to representative sites where Abi1 and YFP-PBD overlap. Nuclear stains in YFP-PBD–transfected cells are not identified. Scale bars, 10 µm.
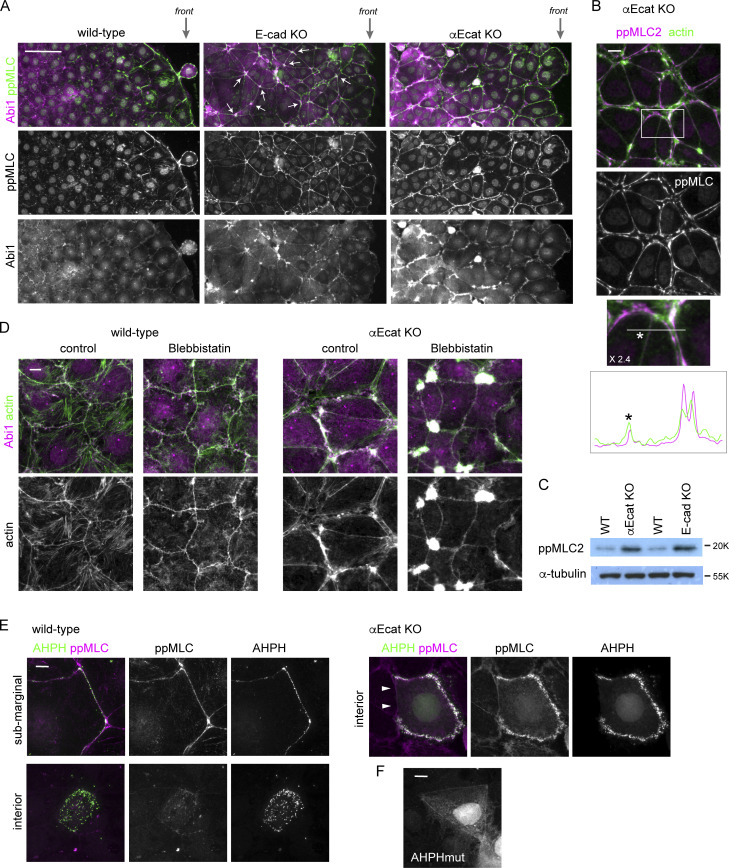
As mentioned already, a hot spot to generate membrane protrusion was multicellular junctions where multiple bicellular junctions met (Fig. 2 D), and the multicellular junctions are thought to be prone to disruption, as they receive tensile force from the bicellular junctions (Higashi and Miller, 2017; Stephenson et al., 2019). Such force is generated by contraction of junction-associated actomyosin, and therefore we examined whether there is any relation between myosin II activation and c-lamellipodium formation. Myosin II is activated by Rho kinase/ROCK via phosphorylation of myosin regulatory light chain 2 (MLC2; Vicente-Manzanares et al., 2009). Immunostaining for Thr18/Ser19-phosphorylated MLC2 (ppMLC2) showed that, in wild-type Caco2 cells, ppMLC2 was detected along some junctions of marginal and submarginal cells, but not in those of interior cells (Fig. 6 A, leftmost), as noted before for other types of epithelial cells (Ng et al., 2012), suggesting that cells in the marginal and submarginal zones receive higher tension than interior cells, as myosin II activation is known to be a tension-sensitive process (Fernandez-Gonzalez et al., 2009). However, ppMLC2 localization did not perfectly correlate with the abundance of Abi1-posive membranes at AJs.
Figure 6.
Junction disruption induces myosin II activation. (A) Low-magnification view of the indicated Caco2 cell sheets, which are engaging in wound healing, coimmunostained for ppMLC2 and Abi1. ppMLC2 and Abi are cocondensed in ∼40% of multicellular junctions in E-cad KO cells, examples of which are indicated by arrows, whereas the junctions in wild-type cells do not show such features. front, the front regions of the migrating cell sheet. (B) Coimmunostaining for ppMLC2 and actin in αE-cat KO Caco2 cells. The boxed area was enlarged to 2.4× at the bottom, and ppMLC2 and actin were scanned along the line indicated. (C) Western blots for ppMLC2 in the indicated Caco2 lines. (D) Wild-type or αEcat KO Caco2 cells were incubated with 10 µM blebbistatin for 6 h and double-immunostained for Abi1 and actin. Cells located around the inner to submarginal zones were photographed. (E) Distribution of the Rho-GTP localization sensor GFP-AHPH. Wild-type or αEcat KO Caco2 cells were transiently transfected with GFP-AHPH and then double-immunostained for GFP and ppMLC2. Arrowheads indicate a closed junction in an αE-cat KO Caco2 cell. (F) A wild-type cell transiently transfected with a function-deficient mutant of AHPH (AHPHmut) tagged with GFP. Scale bars, 100 µm for A; 10 µm for B and D–F.
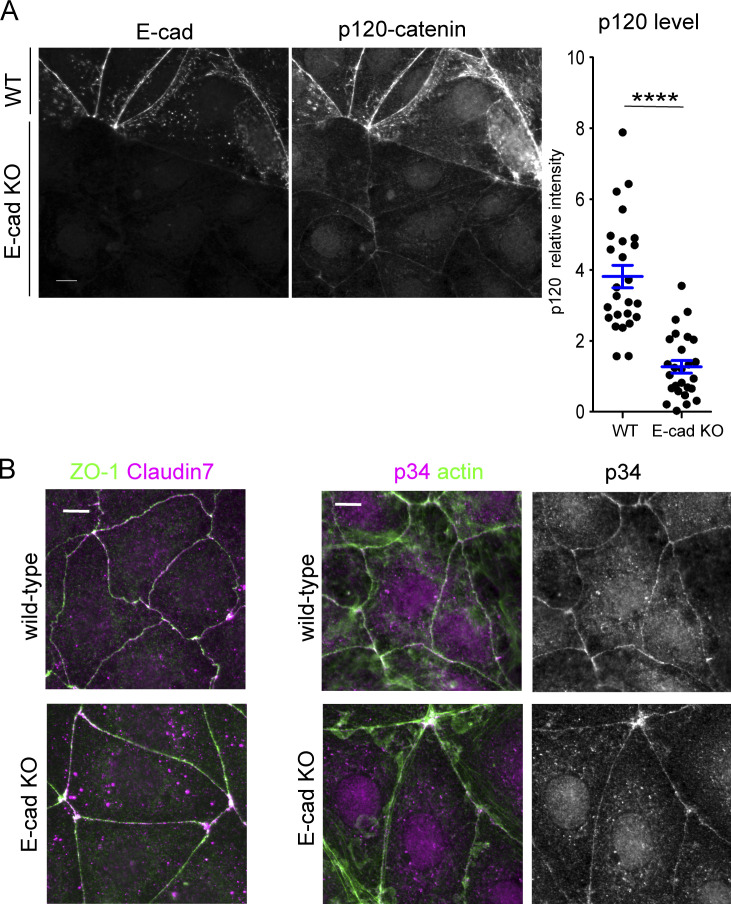
To further test the potential involvement of myosin II in c-lamellipodium regulation, we introduced E-cad KO Caco2 cells to the experiments (Fig. S1 B). These cells still maintain epithelial sheets, in which p120-catenin, a cadherin partner, normally localized along cell junctions, although its level was greatly reduced (Fig. S6 A), suggesting that they sustain AJs, likely because of the presence of some classic cadherins other than E-cadherin. Consistently, TJ network and p34/ARPC2 distribution also looked normal (Fig. S6 B). We used these cells as a model that has normal-looking junctions but reduced adhesive molecules. Immunostaining showed that ppMLC2-positive cells extended to deeper regions of their sheets than in wild-type cell sheets (Fig. 6 A, middle), indicating that the junctions with fewer cadherins are more susceptible to myosin II activation. Then, we examined the distribution of Abi1 in these cells and found that it was condensed at multicellular junctions, along with condensed ppMLC2, whereas Abi level at the bilateral junctions was comparable to that in wild-type cells. These findings support the idea that WRC-dependent protrusion occurs most easily at mechanically weak points of the junction. Furthermore, in αEcat KO Caco2 cells, ppMLC2 was detected throughout the cell layer along the split actin cables in the open junctions, although the closed junctions exhibited lower levels of ppMLC2 (Fig. 6, A and B). Consistent with these observations, total ppMLC2 level was increased in both E-cad KO and αEcat KO cells (Fig. 6 C). Thus, myosin II activation occurred in correlation with reduction of cadherins or junction disruption.
Figure S6.
Effect of E-cadherin deletion on other proteins. (A) Effect of E-cadherin deletion on p120-catenin level. Wild-type and E-cad KO Caco2 cells were mixed and coimmunostained for E-cadherin (E-cad) and p120-catenin, which allows direct comparison of the level of these proteins between the two cell populations. Graph, relative immunostaining intensity of p120-catenin. A few points in each of the bicellular junctions were randomly selected for measurement, using six wild-type and seven E-cad KO cells. ****, P < 0.0001 (t test, one-sided). (B) Coimmunostaining for ZO-1 and claudin 7 or 34/ARPC2 and actin in wild-type and E-cad KO Caco2 cells. Scale bars, 10 µm.
To confirm the impact of myosin II activation on junction or protrusion formation, we treated cells with blebbistatin, an inhibitor of myosin II (Straight et al., 2003). Although this treatment did not much affect wild-type junctions in terms of their integrity and Abi1 distribution, it induced closure of junctions in αEcat KO Caco2 cells, except at multicellular junctions where amorphous actin and Abi remained in clusters (Fig. 6 D). At the blebbistatin-induced closed junctions, Abi1 level and protrusion frequency became comparable to those in the closed junctions of untreated αEcat KO cells (Fig. 6 D). These results suggest that AJ disruption enhances myosin II activation; that the resultant contraction of actomyosin cables induces cell separation, leading to uncontrolled production of c-lamellipodia; and that these processes are prevented by blebbistatin treatment. This inhibitor-mediated myosin II inactivation, however, was insufficient for closing multicellular junctions, which probably requires the intact cadherin/αE-catenin–dependent adhesion system. On the other hand, it remains unclear whether this myosin II–dependent mechanism controls c-lamellipodium formation in wild-type cells.
Because myosin II is activated by RhoA and its effectors, we observed active RhoA localization. We transiently transfected Caco2 cells with a biosensor for active RhoA containing the RhoA-binding domain of anillin, GFP-AHPH (Anillin homology and Pleckstrin homology domains; Tse et al., 2012) and found that, in wild-type cells, this probe was detected along some junctions of marginal or submarginal cells, but not interior cells (Fig. 6 E, left). In the case of αEcat KO Caco2 cells, GFP-AHPH was detected in the open junctions even at interior portions of a cell sheet, whereas it never came to the closed junctions present in the same cell (Fig. 6 E, right). Importantly, these distributions of GFP-AHPH correlated with the ppMLC2 level at the junctions. The specificity of the GFP-AHPH probe used in these experiments was confirmed using its mutated, nonfunctional version, which mostly diffuses in the cytoplasm (Fig. 6 F). These results suggest that myosin II activation induced by junction disruption was probably mediated by RhoA activation at cell–cell contact areas.
AJ disruption interferes with epithelial cell migration through myosin IIA activation
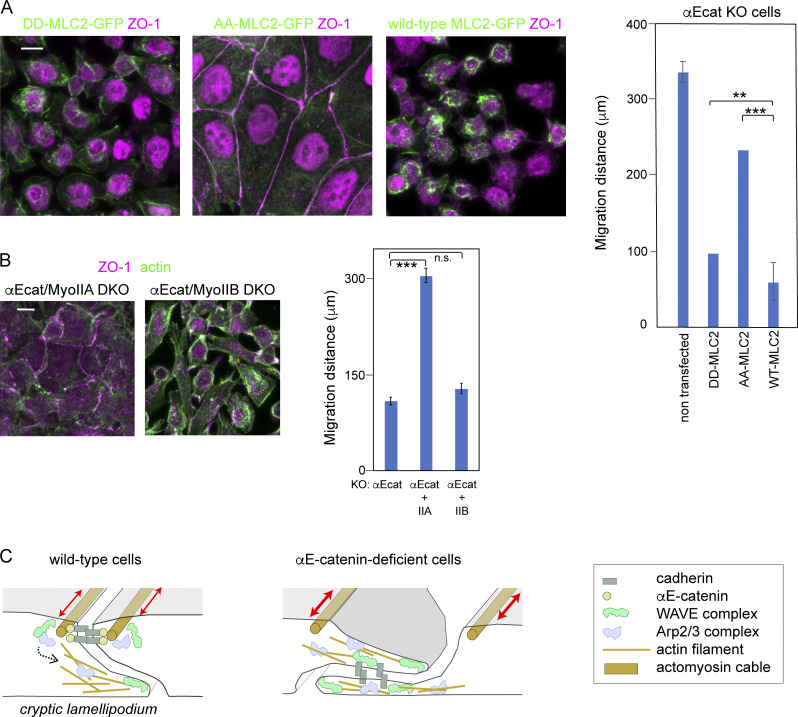
We finally tested whether myosin II–dependent AJ disruption was involved in the impaired migration of epithelial cells, using the DLD1 line, as migration of this line is more sensitive to AJ loss than that of Caco2 (Fig. 1 B). Initially we checked whether myosin II activation is responsible for AJ disruption also in this cell line. We prepared GFP-tagged MLC2 mutants in which Thr18 and Ser19 were replaced with Ala (AA-MLC2) and Asp (DD-MLC2) to generate constitutively inactive and active constructs, respectively. When AA-MLC2 was introduced into αEcat KO DLD1 cells, it restored a normal-looking AJC, whereas DD-MLC2 expression had no such effect (Fig. 7 A). Introduction of these constructs into wild-type cells, however, showed no particular effects on junction organization (Fig. S7 A), suggesting that the observed myosin II–dependent changes of junctions is detectable only when the cadherin–catenin adhesion system is impaired. We next explored which myosin IIA and IIB heavy chains was important as the partner for ppMLC2 in the regulation of AJs. When MHY9 (NMMHC-IIA) was deleted in αEcat KO DLD1 cells, AJC-like reorganization was partly recovered, whereas removal of MHY10 (NMMHC-IIB) had no clear effect (Fig. 7 B), indicating that myosin IIA–based actomyosin contraction is more important in inducing junction disruption. Then, we tested whether these changes in myosin II affected cell migration using wound-healing assays, and found that AA-MLC2 expression in αEcat KO DLD1 cells promoted their wound healing more vigorously than DD-MLC2 expression (Fig. 7 A), although their migration was still retarded compared with that of wild-type DLD1 cells. Likewise, deletion of myosin IIA, but not myosin IIB, promoted migration of αEcat KO DLD1 cells (Fig. 7 B). As expected from the morphological observations, wild-type cells transfected with the MLC2 mutants did not show any changes in their migration ability (Fig. S7 B). These findings suggest that the molecular events that interfere with the collective migration of epithelial cells whose AJs have been disrupted include myosin IIA activation.
Figure 7.
Tests for the role of myosin II activation in migration of αEcat KO DLD1 cells and summary of the results. (A) Stable lines of αEcat KO DLD1 cells transfected with the indicated GFP-tagged MLC2 mutants were immunostained for GFP and ZO-1. Staining of nuclei is due to nonspecific binding of antibodies. (B) Myosin IIA or IIB was deleted in αE-cat KO DLD1 cells, as shown in Fig. S1 C. Cells were immunostained for ZO-1 and actin. Graphs, migration distance of cells at the wound edges 24 h after scratching of the culture, in both A and B. Data were analyzed as explained in the legend of Fig. 1 B. **, P < 0.01; ***, P < 0.001 (t test, one-sided). (C) Illustrated summary of the results. In wild-type cells, WRC and Arp2/3 components are anchored to AJ and are involved in c-lamellipodia formation when activated by undefined mechanisms (dotted arrow). In the open junction of αEcat KO cells, WRC and Arp2/3 components appear to be constitutively active, without having any specific anchoring structures. Contraction of AJ-associated actomyosin cables (two-way arrows) causes AJ disruption in αEcat KO cells. Error bars represent SEM. Scale bars, 10 µm.
Figure S7.
Effects of expression of MLC2 mutants in wild-type DLD1 cells. (A) Stable lines of wild-type DLD1 cells transfected with the indicated GFO-tagged MLC2 mutants were immunostained for GFP and ZO-1. (B) Migration distance of these cells at the wound edges 24 h after scratching of the culture. Data were analyzed as explained in the legend of Fig. 1 B. Error bars represent SEM. Scale bars, 10 µm.
Discussion
Mechanisms of cell migration have been studied extensively using various cell types (De Pascalis and Etienne-Manneville, 2017). The behavior of “isolated” epithelial cells, however, has not been observed so closely, except for restricted cell types. Our results showed that, when singly isolated, adenocarcinoma-derived epithelial cells as well as MDCK cells were unable to migrate by themselves, at least in the studied conditions, as reported before (Desai et al., 2009). In these isolated cells and also in AJ-disrupted cells, lamellipodia (or c-lamellipodia) randomly emerged from multiple sites of the cell periphery, explaining why they are unable to move toward a particular direction. By contrast, in wild-type cell sheets, the leader cells were polarized to protrude lamellipodia only at the free edges, and within the sheets, AJs restrained individual cells from free movement, resulting in well-ordered migration of the cells.
For the entire epithelial sheet to move, however, not only the leader cells but also follower cells must actively move. Actually, in a moving cell sheet, individual cells are known to form protrusions at their basal sides (Barlan et al., 2017; Farooqui and Fenteany, 2005; Krndija et al., 2019; Squarr et al., 2016). Our present study has now provided evidence for the importance of such protrusions or c-lamellipodia for adenocarcinoma cells to move as a collective. c-Lamellipodia were generated as dynamic protrusions of the plasma membranes located around AJs, and this process depended on WRC and Arp2/3, whose components were localized along the AJs (Fig. 7 C, left). Given the well-known function of these molecules at lamellipodia, we can infer that junctional WRC–Arp2/3 serves for actin nucleation so as to induce c-lamellipodia formation. c-Lamellipodia were morphologically similar to slanted LCs, as larger c-lamellipodia actually contained E-cadherin. Of note, silencing of WRC or Arp2/3 resulted in an increase of E-cadherin–positive LCs, along with suppression of dynamic protrusions. These suggest that the lateral plasma membranes normally organize into static LCs, but when cells move as a collective, the membranes acquire motile functions to become “protrusions,” responding to activation of the WRC–Arp2/3 system. It remains to be determined whether the AJ-dependent mechanism uncovered here using “thin” epithelial cells also works for protrusion formation in cuboidal or columnar epithelial cells, in which AJs are located quite distant from the basal sides. LLSM analysis revealed that protrusions also grow upward at AJs, as seen during the retraction of ruffling membranes at the leading edge of migrating cells. This suggests that similar molecular events occur at both AJs and free edges. The biological function of upward protrusions, however, remains to be investigated in the future. Previous studies indicated that WRC and Arp2/3 are necessary for the integrity of zonula adherens (Verma et al., 2012), suggesting the possibility that this actin-regulating system might differently take part in junction dynamics in moving and stationary cells.
Because c-lamellipodia formation is an intermittent process, there could be a mechanism to control WRC–Arp2/3 activity during cell migration. In the case of αEcat KO cells, c-lamellipodia freely grew at open junctions, whereas they disappeared at closed junctions. In the latter junctions, Abi1 level was reduced, likely because AJs as a center for WRC recruitment were absent, explaining why protrusion growth halted there. In normal cells, however, WRC was always detectable along AJs; nevertheless, extension and retraction of protrusions occurred sporadically. This suggests that the activity of WRC and Arp2/3 held by AJs is controlled by some intermittent signals. Observations of Rac1, an upstream regulator of them, did not indicate that its activity change is involved in this regulation, as its active form appears to localize through the AJs. We suspect that mechanical signals might play a role in this hypothetical process, as our results indicated that mechanical weakening or disruption of AJs enhances protrusion formation: that is, in E-cadherin–deleted cells and even in wild-type cells, Abi-positive membranes tended to grow from multicellular junctions, where cell–cell contacts are thought to be prone to disruption (Higashi and Miller, 2017). Our results also suggested that WRC and Arp2/3 bound to AJs are not necessarily always active, as they stayed in AJs even when Rac1 was inhibited. Based on these observations, we propose that mechanical changes in AJs may trigger activation of the WRC–Arp2/3 system. Such changes could be brought about by tensile force that is exerted to AJs. Actually, marginal and submarginal cells appeared to receive such force, as myosin II activation, which is known to be a tension-sensitive process (Fernandez-Gonzalez et al., 2009), was observed in their junctions. In the open junctions of αEcat KO cells, on the other hand, WRC and Arp2/3 were apparently not anchored to any special structures except for their target, F-actin, and this might cause a constitutive activation of these actin regulators and in turn uncontrolled protrusion growth (Fig. 7 C, right), just as seen in the lamellipodia of free cell edges.
We demonstrated that myosin II activation played a crucial role in disrupting AJs under the αE-catenin–free condition. Inhibition of myosin II with inhibitors or MLC2 mutations led to partial restoration of cell–cell contacts in αEcat KO cells. Furthermore, active RhoA was detected only along the disrupted junctions, where myosin II was also activated. These observations suggest the presence of a signaling pathway in which αE-catenin loss triggers activation of RhoA, and this in turn leads to MLC2 activation. The resultant contraction of AJ-associated actomyosin cables, apparently, was thus the primary cause of AJ disruption. How RhoA–MLC2 signals are activated by AJ loss remains to be elucidated. On the other hand, in the normal cadherin-bearing junctions, it appears that myosin II activation per se is not directly involved in WRC-Arp2/3 regulation, as the localization of ppMLC2 did not correlate with Abi1 level in wild-type cells. It is of note that junctional myosin II activation, which depends on cadherin reduction, was also observed in vivo (Hashimoto and Munro, 2019), suggesting that such a mechanism might be used as a physiological tool for tissue reorganization.
Previous studies on the contact inhibition of cell locomotion suggested that the motility of nonepithelial cells, such as neural crest and glioma cells, is inhibited through cadherin-mediated cell–cell adhesion (Hayashi et al., 2014; Theveneau et al., 2010). However, the contact-dependent control of cell motility appears to be more complicated in epithelial cells, as they keep producing c-lamellipodia at the cell–cell contact sites. Our results suggest that the epithelial AJ has a dual role: it works for stable linking of cells on the one hand, but on the other hand it supports generation of motile apparatus. Importantly, whereas WRC and Arp2/3 components accumulate at AJs in epithelial cells, these actin regulators are excluded from the cell–cell contact sites of other cells such as glioma, unless special mechanisms operate (Hayashi et al., 2014). This explains at least in part why different cell types differently respond to cell–cell contacts in motility regulation.
Cell migration plays a crucial role in cancer invasion and metastasis. Mechanisms for how tumor cells relocate themselves is a topic of controversy. E-cadherin or associated proteins have been found to be mutated in various types of carcinomas (Fanjul-Fernández et al., 2013; Morrogh et al., 2012; Wang et al., 2014), and E-cadherin loss promotes metastasis (Cai et al., 2014). Dysfunction of E-cadherin indeed causes tumor dissemination in certain cancers (Derksen et al., 2006; Nanki et al., 2018). On the other hand, cancer invasion often proceeds in the way of collective cell migration rather than free movement of dispersed cells (Friedl et al., 2012; Pandya et al., 2017), implying that invading tumor cells do not always lose cell–cell adhesion molecules. Recent studies showed that ductal carcinoma metastasis is driven by E-cadherin–positive cells but not –negative cells in a mouse model (Padmanaban et al., 2019). Our in vitro observations reported here would provide a clue to our deeper understanding of how E-cadherin or AJs are really involved in the invasive behavior of carcinoma cells.
Materials and methods
Cell cultures and wound healing assay
DLD1 (a gift from Shintaro Suzuki, Kwansei Gakuin University, Hyogo, Japan), Caco2 (ATCC), MKN74 (JCRB Cell Bank), HT1080 (a gift from Kiyotoshi Sekiguchi, Osaka University, Osaka, Japan), MDCK (a gift from Yasushi Daikuhara, Kagoshima University, Kagoshima, Japan), and A549 (a gift from Varisa Pongrakhananon, Chulalongkorn University, Bangkok, Thailand) were cultured in DMEM/Ham’s F12 medium (Wako) supplemented with 10% FCS (CCB Cell Culture Bioscience; 171012, lot 10L015) at 37°C, 5% CO2. For wound healing assays, cells were plated in a six-well culture plate (IWAKI collagen type-I microplate 6well; 4810-010) and cultured for 24 h. A plastic pipette tip was used to draw a wound area across the center of the plate. Culture medium was then replaced with fresh medium. At 0, 12, 24, and 48 h of culture, cells were photographed. The distances of cell migration from the original wound edge to the leading edge of the migrating cells were measured. For wound healing assays using siRNA-treated cells, cells were treated with siRNA for 6 h, followed by replacement with fresh medium. After 48 h, cells were dispersed by trypsinization, plated, and cultured overnight, before wound healing assays. When a mixed culture of siRNA-treated and nontreated cells was used, they were mixed in a 1-to-1 ratio before plating.
To detect mitotic cells in culture, cells were incubated for 24 h, and their monolayers were wounded. After another 24 h, cells were fixed and immunostained for phosphorylated Histone H3 and DAPI. For detecting mitotic cells in siRNA-treated cultures, cells were precultured overnight and treated with siRNAs for 6 h. After another 42-h incubation with fresh medium, cells were trypsinized, replated, and cultured overnight before measurement of mitotic cells.
Isolation of cells lines using CRISPR/Cas9 plasmid-mediated gene KO
For CRISPR/Cas9-mediated KO of genes, we used the pCGsapI vector developed by Takayuki Sakurai (Shinshu University, Nagano, Japan; Ozawa, 2018). The vector contains the hCas9 gene under the control of the CAG promoter and a unique cloning site, SapI, for insertion of the guide RNA under the control of the U6 promoter. All synthetic oligonucleotides corresponding to the guide RNA and complementary chain, therefore, contain the adaptor sequence for SapI. The following oligonucleotides were used to construct guide RNAs (lowercase letters represent the adaptor sequences): αΕ-catenin, 5′-accgGAAATGACTGCTGTCCATGCg-3′ and 5′-aaacGCATGGACAGCAGTCATTTCc-3′; 5′-accgTCTGGCAGTTGAGAGACTGTg-3′ and 5′-aaacACAGTCTCTCAACTGCCAGAc-3′; 5′-accgGAAGCGAGGCAACATGGTTCg-3′ and 5′-aaacGAACCATGTTGCCTCGCTTCc-3′; and 5′-accgGTCAGCCAAAATCAGCAACCg-3′ and 5′-aaacGGTTGCTGATTTTGGCTGACc-3′; for E-cadherin, 5′-accgCCCTTGGAGCCGCAGCCTCTg-3′ and 5′-aaacAGAGGCTGCGGCTCCAAGGGc-3′; 5′-accgGAGCCGGAGCCCTGCCACCCg-3′ and 5′-aaacGGGTGGCAGGGCTCCGGCTCc-3′; for Myosin IIA, 5′-accgCCCGCCCAAGTTCTCCAAGGg-3′ and 5′-aaacCCTTGGAGAACTTGGGAGGGc-3′; and for Myosin IIB, 5′-accgCCCACCTAAGTTTTCCAAGGg-3′ and 5′-aaacCCTTGGAAAACTTAGGTGGGc-3′. The vectors were introduced into cells together with the pCAG/bsr-7 vector, which confers blasticidin resistance (Ozawa, 2018). After selection with blasticidin (8 µg/ml), multiple colonies were isolated and tested for expression of the gene products by immunofluorescence (IF) staining and Western blotting. The isolated multiple clones were then examined to detect the abnormalities common to all the clones, and one of them was chosen as a representative for further analysis. We also verified whether the properties of cells that had changed after gene KO were due to the removal of this gene, by reintroducing the corresponding cDNA into the KO lines. All gene KO cell lines used in the present experiments restored the original properties after this treatment.
Transfection of cells with siRNA oligos and expression vectors
Protein depletion was achieved using Stealth siRNAs (Invitrogen). The following oligos were used: NCKAP1 HSS116652 (#1), NCKAP1 HSS116650 (#2), and NCKAP1 HSS173885 (#3) for Nap1, and ARPC2 HSS173400 (#1), ARPC2 HSS115366 (#2), and ARPC2 HSS115367 (#3) for p34-Arc/ARPC2. We also used negative control siRNAs (Thermo Fisher Scientific; 12935-300; Invitrogen; 46-2000). All siRNAs were transfected using Lipofectamine RNAiMAX Reagent (Invitrogen; 13778-150), according to the manufacturer’s protocol. The efficiency of protein depletion was verified by Western blot and IF staining. Data obtained using one of the multiple siRNAs are shown as representatives, after confirming that others showed similar effects.
To construct LifeAct-RFP, we replaced the region between Sal1 and HindIII sites of the pCAH-LifeAct-GFP plasmid, which was published previously (Nishimura et al., 2016; Riedl et al., 2008), with the same region of pCANw-RFP plasmid. GFP-AHPH-DM (Tse et al., 2012; Addgene plasmid 71368), as well as its control mutant version GFP-AHPHA740D/E758K-DM, were a gift from A. Yap (University of Queensland, St. Lucia, Australia). Nap1-GFP was described previously (Hayashi et al., 2014). Wild-type and mutated MLC2, AA- and DD-MLC2, tagged with EGFP at the C terminus, were a generous gift of Shigenobu Yonemura (Tokushima University, Tokushima, Japan; Watanabe et al., 2007). YFP-PBD (Hoppe and Swanson, 2004) was obtained from Addgene (plasmid 11407). Plasmids were introduced into cells using Lipofectamine LTX Plus Reagent (Invitrogen; 15338-100), according to the manufacturer’s protocol.
Antibodies and other reagents
We used the following primary antibodies: mouse anti–β-catenin (BD Transduction; 610153, 1:100 for IF); rabbit anti–β-catenin (Cell Signaling Cat#8480S, 1:100 for IF); rabbit anti–α-catenin (Sigma Cat#C2081, 1:100 for IF); mouse anti–α-catenin (ENZO Life Sciences; ALX-804-101-C100, 1:100 for IF); mouse anti–P-cadherin (Takara; M127, 1:100 for IF); mouse anti-Rac1 (Abcam; ab33186, 1:100 for IF); mouse anti-claudin 7 (Invitrogen; 37-4800, 1:100 for IF); rabbit anti-ARPC2 (Abcam; ab133315, 1:300 for Western blot); rabbit anti-p34-Arc/ARPC2 (EMD Millipore; 07-227, 1:100 for IF); mouse anti-p120-catenin (BD Transduction; 610134, 1:100 for IF); mouse anti–ABI-1 (MBL; D147-3, 1:100 for IF); rabbit anti-phospho-Myosin Light Chain 2 (Thr18/Ser19; Cell Signaling; 3674S, 1:100 for IF); rabbit anti-Myosin IIA (Sigma-Aldrich; M8064, 1:100 for IF); rabbit anti-Myosin IIB (Sigma-Aldrich; M7939, 1:100 for IF); mouse anti-Rac1 (23A8; Abcam; 33186, 1:100 for IF); rabbit anti-GFP (MBL; 598, 1:500–1,000 for IF); rat anti–E-cadherin (ECCD2, ascites fluid, 1:100 for IF); mouse anti–E-cadherin (HECD1, ascites fluid, 1:100 for IF); mouse anti–α-tubulin (Sigma-Aldrich; T9026, 1:500 for WB); rabbit anti-phospho-Histone H3 (Ser10; Sigma-Aldrich; 06-570, 1:100 for IF); and rabbit anti-merlin antibody (Cell Signaling, D1D8; 6995, 1:100 for IF). Secondary antibodies used were Alexa Fluor 488 donkey anti-mouse IgG (Life Technologies; A21202); Alexa Fluor 594 goat anti-mouse IgG (Life Technologies; A11032); Alexa Fluor 647 goat anti-mouse IgG (Invitrogen Molecular Probes; A21236); Alexa Fluor 488 goat anti-rabbit IgG (Invitrogen; A11034); Alexa Fluor 594 goat anti-rabbit IgG (Life Technologies; A11037); Alexa Fluor 647 goat anti-rabbit IgG (Invitrogen Molecular Probes; A21245); Alexa Fluor 488 chicken anti-rat IgG (Invitrogen Molecular Probes; A21470); and Alexa Fluor 594 donkey anti-rat IgG (Invitrogen Molecular Probes; A21209).
For actin staining, we used phalloidin-488 (Invitrogen; A12379, 1:500), phalloidin-594 (Invitrogen; A12381, 1:1,000), and phalloidin-647 (Invitrogen; A22287, 1:500). The following inhibitors were used: blebbistatin (Sigma-Aldrich; B0560); EHT1864 (Abcam; AB229172); CK666 (MERK; 182515); and latrunculin A (Sigma-Aldrich; L5163).
Western blotting
Cell lysates were boiled in an SDS sample buffer (0.0625 M Tris-HCl, pH 6.8, 2.3% SDS, 10% glycerol, and 5% 2-mercaptethanol), separated by SDS-PAGE, and transferred to Immobilon-P or nitrocellulose membranes (Millipore). Membranes were blocked with 5% skim milk in TBS for 1 h and subsequently exposed to primary antibodies for 2 h or overnight, and then to secondary antibodies for 1 h. The proteins were detected by use of the ECL Plus system (GE Healthcare). Reproductivity of the results was confirmed by repeated experiments, and a representative blot is shown.
IF staining and microscopy
Cells were fixed with 2% PFA in PBS (pH 7.4) for 10–20 min at room temperature, permeabilized with 0.25 or 0.5% Triton X-100 in PBS for 10 min, blocked for 30 min with 3% BSA in PBS, and incubated with primary antibodies (2 h), followed by incubation with secondary antibodies and/or phalloidin (1 h) in a blocking buffer. Three 10-min washes with PBS were performed after first and secondary antibody incubations. After washing with distilled water, samples were mounted in FluorSave Reagent (Calbiochem; 345789-20ML). All steps were performed at room temperature. Samples were analyzed with a Zeiss Axiopan 2 or Axio Imajor.Z2 through Plan-Apochromat 63×/1.4 Oil differential interference contrast (DIC) or Plan-Neofluar 40×/1.3 Oil DIC objectives, or a laser scanning confocal microscope (LSM780) on an inverted Axio Observer.Z1 through Plan-A Apochromat 63× 1.4 Oil DIC objectives. Generally, we used conventional optical microscopes for photographing the marginal and submarginal zones of a colony, as they are thin, and confocal microscopes for photographing interior cells, which are thicker than marginal cells. Photographic images were processed with ImageJ/Fiji software (National Institutes of Health).
Time-lapse videos
For analysis of wound healing and cell migration by live imaging, we used a LCV100 (Olympus) equipped with a UAPO 40×/340× objective lens (Olympus), an LED light source, a DP30 camera (Olympus), and DIC optical components and interference filters, except Video 2, for which we used an inverted fluorescence microscope (IX-81, Olympus) equipped with a spinning disk confocal imaging unit (CSU-X1, Yokogawa), a 40×/1.35 oil-immersion objective (UApo/340, Olympus), and electron-multiplying charge coupled device (EMCCD; iXon+, Andor Technology). To observe actin dynamics, we isolated stable lines of wild-type and αEcat KO Caco-2 cells expressing LifeAct-RFP. A wild-type line of these transfectants was additionally transfected with Nap1-GFP in a transient way. These cells were observed using an inverted fluorescence microscope (IX-81, Olympus) equipped with a spinning disk confocal imaging unit (CSU-X1, Yokogawa), a 40×/1.35 oil-immersion objective (UApo/340, Olympus), and a 561-nm laser (Sapphire LP, Coherent) for RFP excitation or a 488-nm laser (Sapphire LP, Coherent) for GFP excitation. We took fluorescence images with multiple z-stacks by EMCCD (iXon+, Andor Technology) at the specified time intervals and then made maximum-intensity Z projections.
LLSM
The LLSM was home-built in the Kiyosue laboratory at RIKEN Center for Biosystems Dynamics Research following the design of the Betzig laboratory (Chen et al., 2014) under a research license agreement from Howard Hughes Medical Institute. Electric wiring was performed at RIKEN Advanced Manufacturing Support Team. Metal parts were processed by Maeda Precision Manufacturing Ltd. and Zera Development Co. To create a lattice light sheet, a dithered square lattice was used through a spatial light modulator (Fourth Dimension Displays) in combination with an annular mask with 0.55 outer and 0.44 inner numerical apertures (Photo-Sciences) and a custom NA 0.65 excitation objective (Special Optics). Images were acquired through a CFI Apo LWD 25XW 1.1-NA detection objective (Nikon) and a scientific sCMOS camera, Orca Flash 4.0 v3 (Hamamatsu Photonics). Caco2 cells expressing LifeAct-RFP were seeded on a collagen-coated coverslip 5 d before imaging. During imaging, cells were maintained in L-15 medium (Thermo Fisher Scientific) supplemented with 10% serum at 25°C. For live imaging of LifeAct-RFP, a 560-nm laser (MPB Communications) and a longpass emission filter BLP02-561R-25 (Semrock) were used. Image stacks were collected with a 200-nm step size between planes with 20-ms per plane exposure times and 24.099-s intervals. After acquisition, images were deskewed and deconvolved using LLSpy (Lambert and Shao, 2019). After deskew processing, the voxel pitch was 0.104 × 0.104 × 0.103 µm. Images were represented in 3D using Imaris software (Bitplane).
Image analysis and quantification
Tracking of individual cells was performed with an ImageJ/Fiji plugin, Manual Tracking. Speed and directionality were calculated by a custom code in Matlab (MathWorks). Intensity of IF antibody stains was measured using MetaMorph Image Analysis Software, and the overlapping of the stains derived from different antigens was analyzed by a MetaMorph application, Measure Colocalization. The areas in an image were measured using ImageJ/Fiji. For statistical analysis of data, we used a one-sided t test. Data distribution was assumed to be normal, but this was not formally tested.
Online supplemental material
Fig. S1 shows immunoblot analysis of CRISPR/Cas9-mediated deletion of proteins. Fig. S2 shows analysis of various junctional proteins. Fig. S3 shows the distribution of WRC components and the effects of their siRNAs. Fig. S4 shows the expression of WRC and Arp2/3 components and the effect of their depletion or Rac1 inhibitor treatment. Fig. S5 shows the distribution of merlin, Rac1, and a Rac-GTP sensor. Fig. S6 shows the effect of E-cadherin deletion on other proteins. Fig. S7 shows the effect of MLC2 mutant expression on cell junctions and migration. Video 1 shows time-lapse images of wild-type and αEcat KO DLD1 cells at the edge of a wound. Video 2 shows time-lapse images of an isolated wild-type DLD1 cell. Video 3 shows time-lapse images of a sheet of wild-type Caco2 cells at the wound edge. Video 4 presents a 3D time-lapse of wild-type Caco2 cells at a marginal zone of a cell sheet, collected with an LLSM. Video 5 shows time-lapse images of an isolated wild-type Caco2 cell expressing LifeAct-RFP. Video 6 presents time-lapse images of a small colony of wild-type Caco2 cells expressing LifeAct-RFP. Video 7 presents time-lapse images of a sheet of αEcat KO Caco2 cells, expressing LifeAct-RFP, at the wound edge. Video 8 presents time-lapse images of an isolated αEcat KO Caco2 cell expressing LifeAct-RFP. Video 9 shows time-lapse images of a small colony of αEcat KO Caco2 cells expressing LifeAct-RFP. Video 10 shows time-lapse images of Nap1-GFP and LifeAct-RFP at a junction of wild-type Caco2 cells. Video 11 presents time-lapse images of wild-type Caco2 cells not treated or treated with 200 µM CK666.
Supplementary Material
Acknowledgments
We thank S. Ito for advice on time-lapse imaging; T. Kimura for help in statistical analysis; M. Uemura and S. Hirano (Kansai Medical University) for help in microscopy; S. Ohno (Yokohama City University, Yokohama, Japan), K. Yamashita (Tohoku University, Sendai, Japan), and K. Tamura (RIKEN Center for Biosystems Dynamics Research, Kobe, Japan) for anti-merlin antibody; and the RIKEN Kobe light microscopy facility for imaging experiments. We thank E. Betzig and W. Legant (Howard Hughes Medical Institute, Janelia Research Campus) for their generous support in providing technical information and operational know-how. We also thank M. Yamaguchi (Carl Zeiss Microscopy Co.) for support using Imaris software.
This work was supported by intramural funds of RIKEN Center for Biosystems Dynamics Research to M. Takeichi; Grant-in-Aid for Japan Society for the Promotion of Science Fellows (grant no. 18J01239) to T. Yamamoto; the Japan Society for the Promotion of Science–NEXT program (no. LS128), Takeda Science Foundation, the Uehara Memorial Foundation, a Grant-in-Aid for Challenging Exploratory Research (Japan Society for the Promotion of Science KAKENHI no. 20K20379), and Japan Science and Technology Agency Core Research for Evolutional Science and Technology (no. JPMJCR1863) to Y. Mimori-Kiyosue. S. Upadhyayula acknowledges the Philomathia Foundation and Chan Zuckerberg Initiative Imaging Scientist Program (no. 2019-198142) for support.
The authors declare no competing financial interests.
Author contributions: M. Ozawa and M. Takeichi conceived and designed the study. M. Ozawa and S. Hiver performed biological experiments and data analysis of wound healing experiments. T. Yamamoto performed fluorescence microscopy imaging presented in Videos 2, 3, and 5–11. T. Yamamoto and T. Shibata performed data analysis in the experiments presented in Fig. 1. Y. Mimori-Kiyosue performed LLSM and image processing. S. Upadhyayula provided critical advice on operation of LLSM and edited LLSM images. M. Takeichi analyzed the data and wrote the manuscript.
References
- Anderson J.M., Van Itallie C.M., and Fanning A.S.. 2004. Setting up a selective barrier at the apical junction complex. Curr. Opin. Cell Biol. 16:140–145. 10.1016/j.ceb.2004.01.005 [DOI] [PubMed] [Google Scholar]
- Barlan K., Cetera M., and Horne-Badovinac S.. 2017. Fat2 and Lar Define a Basally Localized Planar Signaling System Controlling Collective Cell Migration. Dev. Cell. 40:467–477.e5. 10.1016/j.devcel.2017.02.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai D., Chen S.C., Prasad M., He L., Wang X., Choesmel-Cadamuro V., Sawyer J.K., Danuser G., and Montell D.J.. 2014. Mechanical feedback through E-cadherin promotes direction sensing during collective cell migration. Cell. 157:1146–1159. 10.1016/j.cell.2014.03.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camand E., Peglion F., Osmani N., Sanson M., and Etienne-Manneville S.. 2012. N-cadherin expression level modulates integrin-mediated polarity and strongly impacts on the speed and directionality of glial cell migration. J. Cell Sci. 125:844–857. 10.1242/jcs.087668 [DOI] [PubMed] [Google Scholar]
- Chen B., Chou H.T., Brautigam C.A., Xing W., Yang S., Henry L., Doolittle L.K., Walz T., and Rosen M.K.. 2017. Rac1 GTPase activates the WAVE regulatory complex through two distinct binding sites. eLife. 6 e29795 10.7554/eLife.29795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B.C., Legant W.R., Wang K., Shao L., Milkie D.E., Davidson M.W., Janetopoulos C., Wu X.S., Hammer J.A. III, Liu Z., et al. 2014. Lattice light-sheet microscopy: imaging molecules to embryos at high spatiotemporal resolution. Science. 346 1257998 10.1126/science.1257998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung K.J., and Ewald A.J.. 2014. Illuminating breast cancer invasion: diverse roles for cell-cell interactions. Curr. Opin. Cell Biol. 30:99–111. 10.1016/j.ceb.2014.07.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das T., Safferling K., Rausch S., Grabe N., Boehm H., and Spatz J.P.. 2015. A molecular mechanotransduction pathway regulates collective migration of epithelial cells. Nat. Cell Biol. 17:276–287. 10.1038/ncb3115 [DOI] [PubMed] [Google Scholar]
- De Pascalis C., and Etienne-Manneville S.. 2017. Single and collective cell migration: the mechanics of adhesions. Mol. Biol. Cell. 28:1833–1846. 10.1091/mbc.e17-03-0134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derksen P.W., Liu X., Saridin F., van der Gulden H., Zevenhoven J., Evers B., van Beijnum J.R., Griffioen A.W., Vink J., Krimpenfort P., et al. 2006. Somatic inactivation of E-cadherin and p53 in mice leads to metastatic lobular mammary carcinoma through induction of anoikis resistance and angiogenesis. Cancer Cell. 10:437–449. 10.1016/j.ccr.2006.09.013 [DOI] [PubMed] [Google Scholar]
- Desai R.A., Gao L., Raghavan S., Liu W.F., and Chen C.S.. 2009. Cell polarity triggered by cell-cell adhesion via E-cadherin. J. Cell Sci. 122:905–911. 10.1242/jcs.028183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupin I., Camand E., and Etienne-Manneville S.. 2009. Classical cadherins control nucleus and centrosome position and cell polarity. J. Cell Biol. 185:779–786. 10.1083/jcb.200812034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Euteneuer U., and Schliwa M.. 1984. Persistent, directional motility of cells and cytoplasmic fragments in the absence of microtubules. Nature. 310:58–61. 10.1038/310058a0 [DOI] [PubMed] [Google Scholar]
- Fanjul-Fernández M., Quesada V., Cabanillas R., Cadiñanos J., Fontanil T., Obaya A., Ramsay A.J., Llorente J.L., Astudillo A., Cal S., et al. 2013. Cell-cell adhesion genes CTNNA2 and CTNNA3 are tumour suppressors frequently mutated in laryngeal carcinomas. Nat. Commun. 4:2531 10.1038/ncomms3531 [DOI] [PubMed] [Google Scholar]
- Farooqui R., and Fenteany G.. 2005. Multiple rows of cells behind an epithelial wound edge extend cryptic lamellipodia to collectively drive cell-sheet movement. J. Cell Sci. 118:51–63. 10.1242/jcs.01577 [DOI] [PubMed] [Google Scholar]
- Farquhar M.G., and Palade G.E.. 1963. Junctional complexes in various epithelia. J. Cell Biol. 17:375–412. 10.1083/jcb.17.2.375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Gonzalez R., Simoes S.M., Röper J.C., Eaton S., and Zallen J.A.. 2009. Myosin II dynamics are regulated by tension in intercalating cells. Dev. Cell. 17:736–743. 10.1016/j.devcel.2009.09.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedl P., and Gilmour D.. 2009. Collective cell migration in morphogenesis, regeneration and cancer. Nat. Rev. Mol. Cell Biol. 10:445–457. 10.1038/nrm2720 [DOI] [PubMed] [Google Scholar]
- Friedl P., Locker J., Sahai E., and Segall J.E.. 2012. Classifying collective cancer cell invasion. Nat. Cell Biol. 14:777–783. 10.1038/ncb2548 [DOI] [PubMed] [Google Scholar]
- Grimsley-Myers C.M., Isaacson R.H., Cadwell C.M., Campos J., Hernandes M.S., Myers K.R., Seo T., Giang W., Griendling K.K., and Kowalczyk A.P.. 2020. VE-cadherin endocytosis controls vascular integrity and patterning during development. J. Cell Biol. 219 e201909081 10.1083/jcb.201909081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gritsenko P.G., Atlasy N., Dieteren C.E.J., Navis A.C., Venhuizen J.H., Veelken C., Schubert D., Acker-Palmer A., Westerman B.A., Wurdinger T., et al. 2020. p120-catenin-dependent collective brain infiltration by glioma cell networks. Nat. Cell Biol. 22:97–107. 10.1038/s41556-019-0443-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haeger A., Wolf K., Zegers M.M., and Friedl P.. 2015. Collective cell migration: guidance principles and hierarchies. Trends Cell Biol. 25:556–566. 10.1016/j.tcb.2015.06.003 [DOI] [PubMed] [Google Scholar]
- Han S.P., Gambin Y., Gomez G.A., Verma S., Giles N., Michael M., Wu S.K., Guo Z., Johnston W., Sierecki E., et al. 2014. Cortactin scaffolds Arp2/3 and WAVE2 at the epithelial zonula adherens. J. Biol. Chem. 289:7764–7775. 10.1074/jbc.M113.544478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto H., and Munro E.. 2019. Differential Expression of a Classic Cadherin Directs Tissue-Level Contractile Asymmetry during Neural Tube Closure. Dev. Cell. 51:158–172.e4. 10.1016/j.devcel.2019.10.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi S., Inoue Y., Kiyonari H., Abe T., Misaki K., Moriguchi H., Tanaka Y., and Takeichi M.. 2014. Protocadherin-17 mediates collective axon extension by recruiting actin regulator complexes to interaxonal contacts. Dev. Cell. 30:673–687. 10.1016/j.devcel.2014.07.015 [DOI] [PubMed] [Google Scholar]
- Higashi T., and Miller A.L.. 2017. Tricellular junctions: how to build junctions at the TRICkiest points of epithelial cells. Mol. Biol. Cell. 28:2023–2034. 10.1091/mbc.e16-10-0697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoppe A.D., and Swanson J.A.. 2004. Cdc42, Rac1, and Rac2 display distinct patterns of activation during phagocytosis. Mol. Biol. Cell. 15:3509–3519. 10.1091/mbc.e03-11-0847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kametani Y., and Takeichi M.. 2007. Basal-to-apical cadherin flow at cell junctions. Nat. Cell Biol. 9:92–98. 10.1038/ncb1520 [DOI] [PubMed] [Google Scholar]
- Kovacs E.M., Goodwin M., Ali R.G., Paterson A.D., and Yap A.S.. 2002. Cadherin-directed actin assembly: E-cadherin physically associates with the Arp2/3 complex to direct actin assembly in nascent adhesive contacts. Curr. Biol. 12:379–382. 10.1016/S0960-9822(02)00661-9 [DOI] [PubMed] [Google Scholar]
- Krause M., and Gautreau A.. 2014. Steering cell migration: lamellipodium dynamics and the regulation of directional persistence. Nat. Rev. Mol. Cell Biol. 15:577–590. 10.1038/nrm3861 [DOI] [PubMed] [Google Scholar]
- Krndija D., El Marjou F., Guirao B., Richon S., Leroy O., Bellaiche Y., Hannezo E., and Matic Vignjevic D.. 2019. Active cell migration is critical for steady-state epithelial turnover in the gut. Science. 365:705–710. 10.1126/science.aau3429 [DOI] [PubMed] [Google Scholar]
- Ladoux B., and Mège R.M.. 2017. Mechanobiology of collective cell behaviours. Nat. Rev. Mol. Cell Biol. 18:743–757. 10.1038/nrm.2017.98 [DOI] [PubMed] [Google Scholar]
- Lambert T., and Shao L.. 2019. tlambert03/LLSpy: v0.4.8. Zenodo. 10.5281/zenodo.3554482 [DOI]
- Malinverno C., Corallino S., Giavazzi F., Bergert M., Li Q., Leoni M., Disanza A., Frittoli E., Oldani A., Martini E., et al. 2017. Endocytic reawakening of motility in jammed epithelia. Nat. Mater. 16:587–596. 10.1038/nmat4848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayor R., and Etienne-Manneville S.. 2016. The front and rear of collective cell migration. Nat. Rev. Mol. Cell Biol. 17:97–109. 10.1038/nrm.2015.14 [DOI] [PubMed] [Google Scholar]
- Mège R.M., and Ishiyama N.. 2017. Integration of Cadherin Adhesion and Cytoskeleton at Adherens Junctions. Cold Spring Harb. Perspect. Biol. 9 a028738 10.1101/cshperspect.a028738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrogh M., Andrade V.P., Giri D., Sakr R.A., Paik W., Qin L.X., Arroyo C.D., Brogi E., Morrow M., and King T.A.. 2012. Cadherin-catenin complex dissociation in lobular neoplasia of the breast. Breast Cancer Res. Treat. 132:641–652. 10.1007/s10549-011-1860-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nanki K., Toshimitsu K., Takano A., Fujii M., Shimokawa M., Ohta Y., Matano M., Seino T., Nishikori S., Ishikawa K., et al. 2018. Divergent Routes toward Wnt and R-spondin Niche Independency during Human Gastric Carcinogenesis. Cell. 174:856–869.e17. 10.1016/j.cell.2018.07.027 [DOI] [PubMed] [Google Scholar]
- Ng M.R., Besser A., Danuser G., and Brugge J.S.. 2012. Substrate stiffness regulates cadherin-dependent collective migration through myosin-II contractility. J. Cell Biol. 199:545–563. 10.1083/jcb.201207148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niewiadomska P., Godt D., and Tepass U.. 1999. DE-Cadherin is required for intercellular motility during Drosophila oogenesis. J. Cell Biol. 144:533–547. 10.1083/jcb.144.3.533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura T., Ito S., Saito H., Hiver S., Shigetomi K., Ikenouchi J., and Takeichi M.. 2016. DAAM1 stabilizes epithelial junctions by restraining WAVE complex-dependent lateral membrane motility. J. Cell Biol. 215:559–573. 10.1083/jcb.201603107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omelchenko T., Vasiliev J.M., Gelfand I.M., Feder H.H., and Bonder E.M.. 2003. Rho-dependent formation of epithelial “leader” cells during wound healing. Proc. Natl. Acad. Sci. USA. 100:10788–10793. 10.1073/pnas.1834401100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otani T., Ichii T., Aono S., and Takeichi M.. 2006. Cdc42 GEF Tuba regulates the junctional configuration of simple epithelial cells. J. Cell Biol. 175:135–146. 10.1083/jcb.200605012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozawa M. 2018. Nonmuscle myosin IIA is involved in recruitment of apical junction components through activation of α-catenin. Biol. Open. 7 bio031369 10.1242/bio.031369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozawa M., and Kobayashi W.. 2015. Reversibility of the Snail-induced epithelial-mesenchymal transition revealed by the Cre-loxP system. Biochem. Biophys. Res. Commun. 458:608–613. 10.1016/j.bbrc.2015.02.012 [DOI] [PubMed] [Google Scholar]
- Padmanaban V., Krol I., Suhail Y., Szczerba B.M., Aceto N., Bader J.S., and Ewald A.J.. 2019. E-cadherin is required for metastasis in multiple models of breast cancer. Nature. 573:439–444. 10.1038/s41586-019-1526-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palamidessi A., Malinverno C., Frittoli E., Corallino S., Barbieri E., Sigismund S., Beznoussenko G.V., Martini E., Garre M., Ferrara I., et al. 2019. Unjamming overcomes kinetic and proliferation arrest in terminally differentiated cells and promotes collective motility of carcinoma. Nat. Mater. 18:1252–1263. 10.1038/s41563-019-0425-1 [DOI] [PubMed] [Google Scholar]
- Pandya P., Orgaz J.L., and Sanz-Moreno V.. 2017. Modes of invasion during tumour dissemination. Mol. Oncol. 11:5–27. 10.1002/1878-0261.12019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridley A.J. 2015. Rho GTPase signalling in cell migration. Curr. Opin. Cell Biol. 36:103–112. 10.1016/j.ceb.2015.08.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riedl J., Crevenna A.H., Kessenbrock K., Yu J.H., Neukirchen D., Bista M., Bradke F., Jenne D., Holak T.A., Werb Z., et al. 2008. Lifeact: a versatile marker to visualize F-actin. Nat. Methods. 5:605–607. 10.1038/nmeth.1220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rotty J.D., Wu C., and Bear J.E.. 2013. New insights into the regulation and cellular functions of the ARP2/3 complex. Nat. Rev. Mol. Cell Biol. 14:7–12. 10.1038/nrm3492 [DOI] [PubMed] [Google Scholar]
- Roycroft A., and Mayor R.. 2016. Molecular basis of contact inhibition of locomotion. Cell. Mol. Life Sci. 73:1119–1130. 10.1007/s00018-015-2090-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seddiki R., Narayana G.H.N.S., Strale P.O., Balcioglu H.E., Peyret G., Yao M., Le A.P., Teck Lim C., Yan J., Ladoux B., et al. 2018. Force-dependent binding of vinculin to α-catenin regulates cell-cell contact stability and collective cell behavior. Mol. Biol. Cell. 29:380–388. 10.1091/mbc.E17-04-0231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shutes A., Onesto C., Picard V., Leblond B., Schweighoffer F., and Der C.J.. 2007. Specificity and mechanism of action of EHT 1864, a novel small molecule inhibitor of Rac family small GTPases. J. Biol. Chem. 282:35666–35678. 10.1074/jbc.M703571200 [DOI] [PubMed] [Google Scholar]
- Squarr A.J., Brinkmann K., Chen B., Steinbacher T., Ebnet K., Rosen M.K., and Bogdan S.. 2016. Fat2 acts through the WAVE regulatory complex to drive collective cell migration during tissue rotation. J. Cell Biol. 212:591–603. 10.1083/jcb.201508081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephenson R.E., Higashi T., Erofeev I.S., Arnold T.R., Leda M., Goryachev A.B., and Miller A.L.. 2019. Rho Flares Repair Local Tight Junction Leaks. Dev. Cell. 48:445–459.e5. 10.1016/j.devcel.2019.01.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straight A.F., Cheung A., Limouze J., Chen I., Westwood N.J., Sellers J.R., and Mitchison T.J.. 2003. Dissecting temporal and spatial control of cytokinesis with a myosin II Inhibitor. Science. 299:1743–1747. 10.1126/science.1081412 [DOI] [PubMed] [Google Scholar]
- Takeichi M. 2014. Dynamic contacts: rearranging adherens junctions to drive epithelial remodelling. Nat. Rev. Mol. Cell Biol. 15:397–410. 10.1038/nrm3802 [DOI] [PubMed] [Google Scholar]
- Takenawa T., and Suetsugu S.. 2007. The WASP-WAVE protein network: connecting the membrane to the cytoskeleton. Nat. Rev. Mol. Cell Biol. 8:37–48. 10.1038/nrm2069 [DOI] [PubMed] [Google Scholar]
- Theveneau E., Marchant L., Kuriyama S., Gull M., Moepps B., Parsons M., and Mayor R.. 2010. Collective chemotaxis requires contact-dependent cell polarity. Dev. Cell. 19:39–53. 10.1016/j.devcel.2010.06.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiery J.P. 2003. Epithelial-mesenchymal transitions in development and pathologies. Curr. Opin. Cell Biol. 15:740–746. 10.1016/j.ceb.2003.10.006 [DOI] [PubMed] [Google Scholar]
- Tse Y.C., Werner M., Longhini K.M., Labbe J.C., Goldstein B., and Glotzer M.. 2012. RhoA activation during polarization and cytokinesis of the early Caenorhabditis elegans embryo is differentially dependent on NOP-1 and CYK-4. Mol. Biol. Cell. 23:4020–4031. 10.1091/mbc.e12-04-0268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma S., Han S.P., Michael M., Gomez G.A., Yang Z., Teasdale R.D., Ratheesh A., Kovacs E.M., Ali R.G., and Yap A.S.. 2012. A WAVE2-Arp2/3 actin nucleator apparatus supports junctional tension at the epithelial zonula adherens. Mol. Biol. Cell. 23:4601–4610. 10.1091/mbc.e12-08-0574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma S., Shewan A.M., Scott J.A., Helwani F.M., den Elzen N.R., Miki H., Takenawa T., and Yap A.S.. 2004. Arp2/3 activity is necessary for efficient formation of E-cadherin adhesive contacts. J. Biol. Chem. 279:34062–34070. 10.1074/jbc.M404814200 [DOI] [PubMed] [Google Scholar]
- Vicente-Manzanares M., Ma X., Adelstein R.S., and Horwitz A.R.. 2009. Non-muscle myosin II takes centre stage in cell adhesion and migration. Nat. Rev. Mol. Cell Biol. 10:778–790. 10.1038/nrm2786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogelmann R., and Nelson W.J.. 2005. Fractionation of the epithelial apical junctional complex: reassessment of protein distributions in different substructures. Mol. Biol. Cell. 16:701–716. 10.1091/mbc.e04-09-0827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K., Yuen S.T., Xu J., Lee S.P., Yan H.H., Shi S.T., Siu H.C., Deng S., Chu K.M., Law S., et al. 2014. Whole-genome sequencing and comprehensive molecular profiling identify new driver mutations in gastric cancer. Nat. Genet. 46:573–582. 10.1038/ng.2983 [DOI] [PubMed] [Google Scholar]
- Watabe M., Nagafuchi A., Tsukita S., and Takeichi M.. 1994. Induction of polarized cell-cell association and retardation of growth by activation of the E-cadherin-catenin adhesion system in a dispersed carcinoma line. J. Cell Biol. 127:247–256. 10.1083/jcb.127.1.247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watabe-Uchida M., Uchida N., Imamura Y., Nagafuchi A., Fujimoto K., Uemura T., Vermeulen S., van Roy F., Adamson E.D., and Takeichi M.. 1998. alpha-Catenin-vinculin interaction functions to organize the apical junctional complex in epithelial cells. J. Cell Biol. 142:847–857. 10.1083/jcb.142.3.847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe T., Hosoya H., and Yonemura S.. 2007. Regulation of myosin II dynamics by phosphorylation and dephosphorylation of its light chain in epithelial cells. Mol. Biol. Cell. 18:605–616. 10.1091/mbc.e06-07-0590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki D., Oikawa T., and Takenawa T.. 2007. Rac-WAVE-mediated actin reorganization is required for organization and maintenance of cell-cell adhesion. J. Cell Sci. 120:86–100. 10.1242/jcs.03311 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.