Wozniak et al. define a novel connection between cleavage of the Rab7 adaptor protein, RILP, and exosome secretion. They demonstrate that inflammasome-mediated RILP cleavage influences sequence-specific miRNA loading into exosomes via interactions with unique cargo vesicles and association with RNA binding proteins.
Abstract
Cells respond to inflammatory disease states by releasing exosomes containing highly specific protein and RNA cargos, but how inflammation alters cargo specificity and secretion of exosomes is unknown. We show that increases in exosome secretion induced by either viral infection or LPS/ATP exposure result from inflammasome activation and subsequent caspase-1–dependent cleavage of the trafficking adaptor protein RILP. This cleaved form of RILP promotes the movement of multivesicular bodies toward the cell periphery and induces selective exosomal miRNA cargo loading. We have identified a common short sequence motif present in miRNAs that are selectively loaded into exosomes after RILP cleavage. This motif binds the RNA binding protein FMR1 and directs miRNA loading into exosomes via interaction with components of the ESCRT (endosomal sorting complex required for transport) pathway. These results indicate that inflammasome-mediated RILP cleavage, and sequence-specific interactions between miRNAs and FMR1, play a significant role in exosome cargo loading and enhanced secretion during cellular inflammatory responses.
Introduction
There is considerable evidence that inflammatory diseases of many etiologies cause dramatic increases in exosome secretion. Furthermore, the exosomes that are produced after inflammatory stimuli do not simply reflect a random sample of cellular contents, but instead differ in both protein and miRNA content from those produced basally (Bala et al., 2011; McDaniel et al., 2014; Thomou et al., 2017). While several groups have correlated this exosome increase with inflammasome activation (Bala et al., 2012; Eguchi and Feldstein, 2018; He et al., 2016; Povero et al., 2014; Shen et al., 2017), the cellular mechanism by which inflammation results in increased exosome secretion remains to be determined.
Inflammasomes are multiprotein platforms that are activated after cellular infection or stresses that ultimately engage immune defenses (Guo et al., 2015; Kanneganti, 2010; Latz et al., 2013). Once activated, the caspase-1 component of the active inflammasome carries out any number of processes including cleavage of cellular proteins. These proteins/cargos can then be secreted from the cell through several cellular vesicular trafficking pathways including the multivesicular bodies (MVB), the site of exosome biogenesis.
Exosomes are formed by intraluminal budding of the MVB (Abrami et al., 2013; Alenquer and Amorim, 2015; Colombo et al., 2014), and the endosomal sorting complex required for transport (ESCRT) pathway is the best described mechanism for intraluminal vesicle (ILV) and MVB formation (Colombo et al., 2013; Raiborg and Stenmark, 2009; Sun et al., 2016; Woodman, 2016). The ESCRT machinery is a multiprotein complex that not only controls the invagination of ILVs into the MVB, but also recognizes, captures, and sorts ubiquitinated protein cargo into the ILV. ESCRT-0, -I, and -II complexes recognize and sequester ubiquitinated membrane proteins at the endosomal membrane, and ESCRT-III is responsible for inward budding and membrane scission. Not until the MVB fuses with the plasma membrane are the exosomes released in the extracellular milieu. The precise mechanism of the trafficking, and subsequent docking and fusion, of the exosome-containing MVB to the plasma membrane is unclear; however, several Rab GTPases within the endolysosomal trafficking pathway, particularly Rab27a and Rab27b, have been implicated in exosome loading and secretion (Fukuda, 2013; Jaé et al., 2015; Ostrowski et al., 2010). The recruitment of specific effector proteins by Rab GTPases ultimately drives cargo collection, organelle motility, and vesicle docking at target membranes. Therefore, Rab GTPases and their effectors are often used by both invading pathogens and the host as a means to promote the pathogen life cycle and/or modify immune responses.
We previously observed that hepatitis C virus (HCV) infection results in the cleavage of the Rab7 trafficking adaptor protein, RILP (rab interacting lysosomal protein; Wozniak et al., 2016). RILP is part of the complex that links the trafficking GTPase Rab7 to the dynein motor complex (Johansson et al., 2007; Jordens et al., 2001). Cleavage of RILP results in a truncated protein that binds Rab7 but cannot make the link to the dynein complex. This shifts vesicular trafficking and promotes kinesin-mediated movement toward the cell surface. RILP cleavage occurs as early as 3 d after infection, causes a redistribution of virion-containing intracellular vesicles, and increases the efficiency of virion secretion (Wozniak et al., 2016). We further showed that caspase-1 is the enzyme that cleaves RILP, thus linking inflammasome activation to efficient viral secretion (Adams et al., 2018). Formation of the cleaved form of RILP (cRILP) also induced a redistribution of other intracellular organelles, including the MVB (Adams et al., 2018).
Exosomes contain a variety of biomolecules including miRNAs. Cellular stresses, such as viral infections and inflammation, induce unique changes in the abundance of these exosomal molecules (Cypryk et al., 2018; Hirsova et al., 2016), indicating that cargo loading into exosomes cannot be a passive process and that specific mechanisms must exist to sort those miRNAs that are destined for export. Indeed, several pathways and molecules have been described to impact exosomal miRNA sorting, including lipids/ceramide (Cha et al., 2015; Kosaka et al., 2010), direct miRNA modification (Koppers-Lalic et al., 2014), and interaction with components of the MVB biogenesis pathway (Emerman and Blower, 2018; Hurley, 2015; Stuffers et al., 2009).
RNA binding proteins (RBPs) are key players in the regulation of the posttranscriptional processing and transport of RNA molecules, and their direct interaction with components of the MVB raises the possibility that RBPs may be relevant in exosomal miRNA sorting (Gibbings et al., 2009; Irion and St Johnston, 2007; Lee et al., 2009; Schnorrer et al., 2000). Therefore, RBPs present on MVBs could play a role in determining which RNAs are selected for inclusion into exosomes and secretion outside of the cell. However, the mechanisms by which RBP–RNA complexes are recognized by MVBs and how RNA species are sequestered in exosomes remain elusive.
Because cellular inflammation is often characterized by an increase in exosome secretion, and because caspase-1 activation is a consequence of inflammasome activation in many cells, we sought to determine if the inflammatory changes that result in enhanced exosome secretion result from caspase-1–mediated RILP cleavage. We found that the formation of cleaved RILP influences not only exosome secretion but also selective miRNA cargo loading by interacting with the machinery of exosome formation, repositioning the MVB, and changing the miRNA selectivity of exosome cargo loading through interaction with the RBP fragile X mental retardation 1 (FMR1).
Results
RILP cleavage links exosome secretion to inflammasome activation
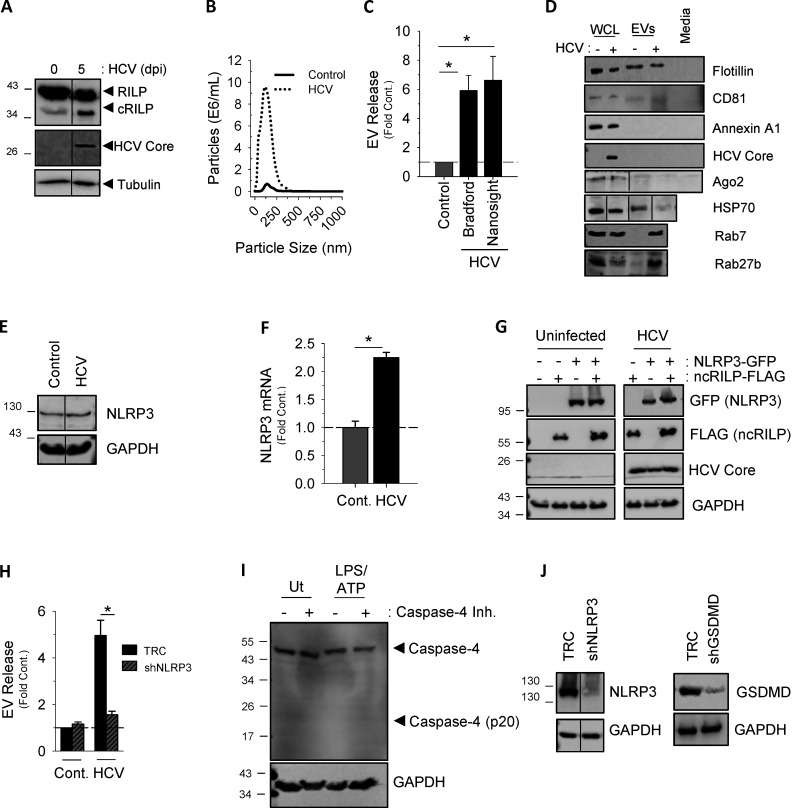
We first examined the effect of HCV infection on extracellular vesicle (EV) secretion. EVs were isolated from cell culture supernatants of both uninfected and Huh-7.5 cells that were infected with cell culture adapted HCV strain JFH-1 (JFH-1cc). HCV infection led to an approximately fivefold increase in EV secretion within 5 d after infection (Fig. 1 A), and this burst correlated with the appearance of the RILP cleavage product (Fig. S1 A). Analysis of purified EVs using Nanosight technology (Thery et al., 2006) showed that the HCV-induced EVs range in size from 40 to 150 nm (Fig. S1 B); similar results on EV quantification were obtained using Bradford protein assay of EV pellets (Fig. S1 C). Western blot analysis showed that they contain several exosomal markers (CD81, heat shock protein 70, Flotillin, Rab27, and Rab7) but do not contain the microvesicle marker annexin A1 or components of other organelles (Fig. S1 D). Because the vesicle preparations examined here possess the size and composition characteristics of exosomes (Hartjes et al., 2019; Théry et al., 2018), but not microvesicles or apoptotic bodies, we use the term exosome when describing these vesicles.
Figure 1.
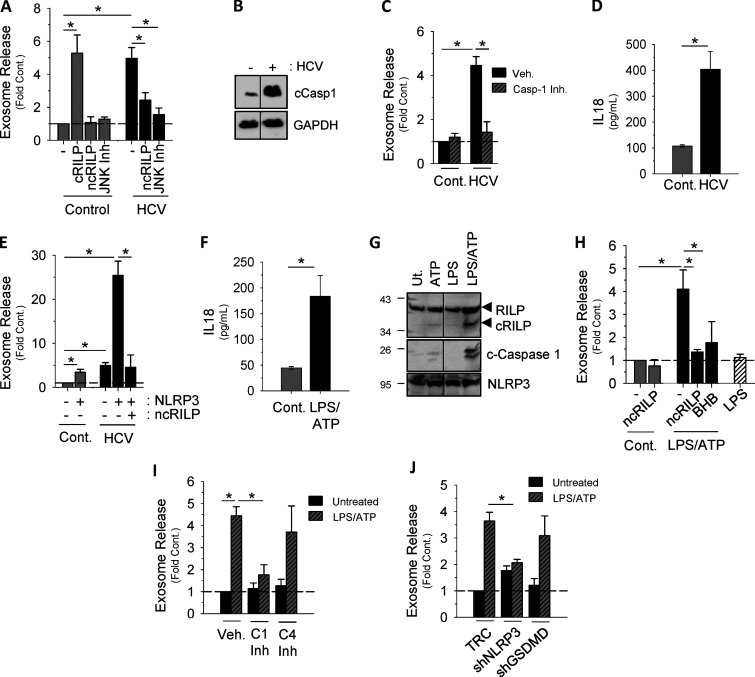
RILP cleavage links exosome secretion to inflammasome activation. (A) Exosome release is increased in HCV-infected cells. cRILP expression alone increases exosome output, while ncRILP blocks HCV-induced exosome secretion. When indicated, JNK inhibitor SP600125 (25 µM) was added 24 h before collection. (B) HCV infection activates the inflammasome as noted by the appearance of cleaved (active) caspase-1. (C) The caspase-1 inhibitor VI (Z-YVAD-FMK, 100 µM) blocks HCV-induced exosome secretion. Cont., control; Veh., vehicle. (D) IL-18 secretion in HCV-infected Huh7.5 cells. (E) NLRP3, in the context of HCV infection, increases exosome secretion nearly 25-fold, and this secretion is blocked by the expression of an ncRILP. (F) IL-18 secretion in LPS/ATP-treated THP-1 cells. (G) RILP is cleaved only after full inflammasome activation using both LPS and ATP. Ut, untreated. (H) Inhibition of either RILP function (ncRILP) or inflammasome activation (BHB) decreases LPS/ATP-induced exosome secretion. (I) The caspase-1 inhibitor VI (Z-YVAD-FMK, 100 µM) blocks LPS/ATP-induced exosome secretion. The caspase-4 inhibitor (Ac-LEVD-CHO, 100 µM) has no effect. (J) shRNA to NLRP3 inhibits LPS/ATP-induced exosome secretion, while shRNA to Gasdermin D has no effect. Data are shown as mean ± SD; *, P ≤ 0.05 for n = 3–8.
Figure S1.
Characterization of Inflammasome-induced exosome secretion. (A) EV release seen during HCV infection correlates with the appearance of a RILP cleavage product, as noted by Western blot. (B) Nanosight analysis of EVs isolated from virus-infected cells have a size range of 40–150 nm, consistent with that of exosomes. (C) EV protein content assessed by Bradford dye directly correlates with the total number of EVs as determined by NTA (Nanosight). Data are shown as mean ± SD; *, P ≤ 0.05 for n = 4–6. (D) EVs isolated from HCV-infected Huh-7.5 cells can be characterized by the exosomal markers CD81, flotillin, Rab7, and Rab27. They do not contain markers for microvesicles (annexin A1). WCL, whole-cell lysate. All markers were undetected in exosome-depleted medium. (E and F) HCV infection of Huh-7.5 cells increased expression of NLRP3, as noted by Western blot and pPCR. Cont., control. (G) Western blot analysis of Huh-7.5 cells expressing ncRILP-Flag with or without NLRP3-GFP showing equal protein expression. (H) shRNA to NLRP3 blocks HCV-induced exosome secretion. TRC, empty vector control. (I) LPS/ATP treatment of THP-1 cells does not up-regulate or activate caspase-4. The caspase-4 inhibitor Ac-LEVD-CHO was used at 100 µM. Ut, untreated. (J) Western blot analysis showing knockdown levels of NLRP3 and gasdermin D (GSDMD) in THP-1 cells.
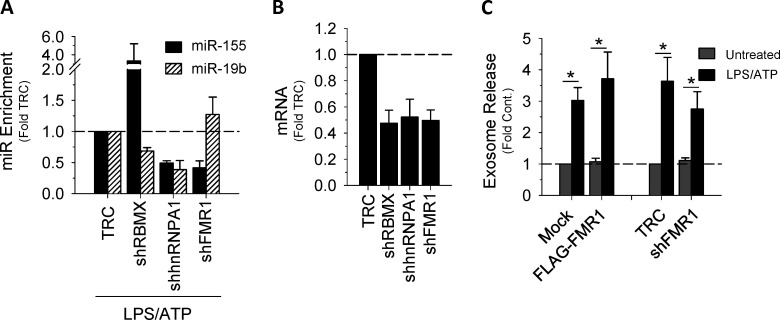
To determine whether RILP cleavage directly caused enhanced exosome production, we assessed the effect of expressing modified RILP constructs on exosome secretion. In the absence of viral infection, direct expression of cRILP caused a nearly sixfold increase in exosomal output, similar to that produced by virus (Fig. 1 A). To further assess the role of RILP cleavage in HCV-induced exosome secretion, a noncleavable form of RILP, ncRILP, was also expressed. As described previously (Adams et al., 2018; Wozniak et al., 2016), ncRILP simultaneously binds both Rab7 and dynein, thus preventing the trafficking changes induced by cRILP. Expression of ncRILP had no effect on basal exosome production, but it dramatically inhibited HCV-induced exosome release. In previous studies describing the requirements for RILP cleavage during virus infection, we showed that JNK was directly involved in the formation of a cleaved RILP product (Adams et al., 2018; Wozniak et al., 2016). In agreement, treatment with the JNK inhibitor SP600125 also blocked HCV-induced exosome secretion (Fig. 1 A). Altogether, the data confirm that the function of cRILP is both necessary and sufficient for HCV-induced exosome output.
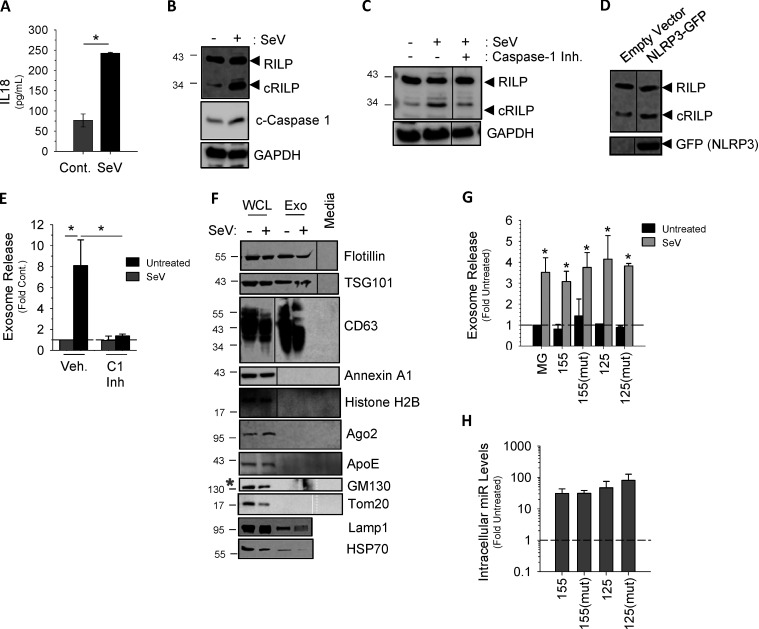
Previous work from our laboratory showed that caspase-1 is the enzyme responsible for cleaving RILP (Adams et al., 2018). Here we show that not only does viral infection increase caspase-1 cleavage (Fig. 1 B), but also caspase-1 inhibition prevented virus-induced exosome secretion (Fig. 1 C). Because caspase-1 activation is a consequence of inflammasome activation, we next examined the contribution of the NLRP3 (NOD-, LRR- and pyrin domain-containing protein 3) inflammasome to HCV-induced exosome secretion. HCV infection led to an increase in NLRP3 expression (Fig. S1, E and F) as well as an increase in IL-18 secretion (Fig. 1 D), both markers of inflammasome activation (He et al., 2016). We next assessed the effect of enhancing inflammasome activation via overexpression of NLRP3, a rate-limiting component of inflammasome activation (Guo et al., 2015; Kanneganti, 2010; Rathinam and Fitzgerald, 2016). In control, uninfected cells, overexpression of NLRP3 caused a modest increase in exosome secretion, not unlike that seen with HCV alone (Figs. 1 E and S1 G). In the context of HCV infection, however, NLRP3 overexpression increased exosome production nearly 25-fold, and this increase was completely blocked by coexpression of ncRILP. Conversely, knockdown of NLRP3 nearly completely inhibited HCV-induced exosome secretion (Fig. S1 H).
We next asked if activation of the inflammasome, independent from virus, would similarly cleave RILP and induce exosome secretion. We used THP-1 cells, a human monocytic cell line that express high levels of the NLRP3 inflammasome components, and differentiated them with PMA to induce a macrophage phenotype (Takashiba et al., 1999). LPS and ATP treatment led to inflammasome activation, as evidenced by the secretion of IL-18 (Fig. 1 F). Treatment with ATP or LPS alone increased NLRP3 protein levels, consistent with the primary signal of inflammasome activation, but RILP cleavage was seen only after full inflammasome activation using both LPS and ATP. This resulted in caspase-1 activation and RILP cleavage (Fig. 1 G). LPS/ATP treatment of THP-1 cells also enhanced exosome secretion fourfold (Fig. 1 H). This increase was abrogated by either the expression of the dominant-negative ncRILP or inhibition of the inflammasome using β-hydroxybutyrate (BHB). LPS alone had no effect on exosome secretion. Both shRNA to NLRP3 as well as inhibition of caspase-1 blocked LPS/ATP-induced exosome secretion, further demonstrating a role for the NLRP3 inflammasome and its downstream product, caspase-1 (Fig. 1, I and J; and Fig. S1 I). Inhibition of neither caspase-4 nor shRNA to gasdermin D had an effect on exosome secretion (Fig. 1, I and J; and Fig. S1, I and J). Taken together, these data show that caspase-1 activation, as a consequence of inflammasome activation, leads to RILP cleavage and exosome secretion.
RILP cleavage status regulates exosome cargo loading and specificity
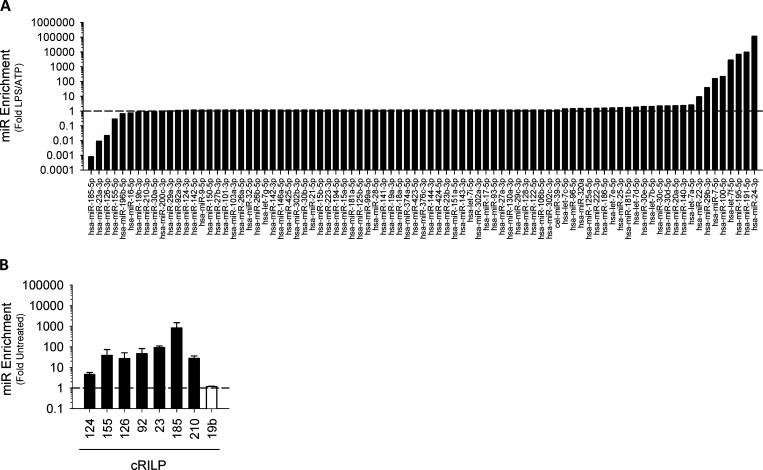
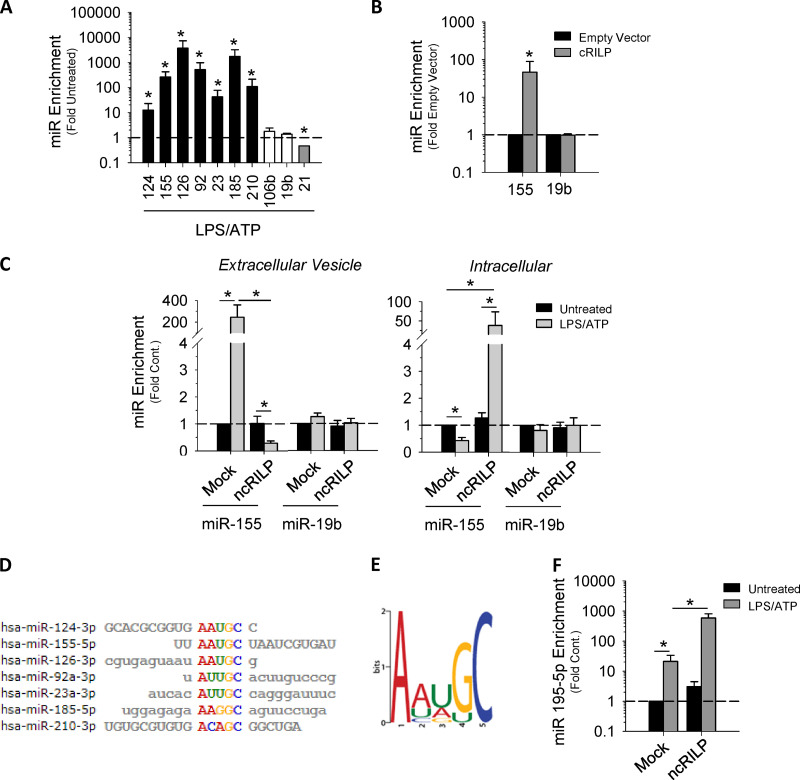
We next assessed the effect of inflammasome activation on exosome miRNA specificity. First, we performed an initial screen to determine which exosomal miRNAs are regulated by RILP cleavage. We treated mock-transfected THP-1 cells and ncRILP-expressing THP-1 cells with LPS/ATP and measured the exosomal enrichment of 84 different miRNAs by PCR array. The expression of ncRILP resulted in the differential regulation of exosomal miRNA content. While the majority of miRNAs tested were unaffected by ncRILP expression, ncRILP reduced the enrichment of some miRNAs and increased others (Fig. S2 A). Second, we further examined the miRNAs that were decreased by ncRILP and determined the effect of LPS/ATP on their secretion. Fig. 2 A shows that inflammasome activation by LPS/ATP led to dramatic changes in the exosomal enrichment of these miRNAs in the exosome. One group of miRNAs, including miR-155, which has previously been shown to be up-regulated in inflammation (Alexander et al., 2015; Bala et al., 2012; Eguchi et al., 2019), increased by several hundredfold. miRNAs that were not affected by ncRILP (Fig. S2 A) similarly were not affected by LPS/ATP, and one miRNA that was increased by ncRILP was decreased by LPS/ATP. Finally, we examined the role of cRILP in exosomal miRNA secretion. To do this, we expressed cRILP in the absence of LPS/ATP. This duplicated the changes seen by LPS/ATP, including the exosomal enrichment of miR-155 and no change in miR-19b (Figs. 2 B and S2 B). Thus, LPS/ATP-stimulated exosomal miRNAs appear to be under the control of RILP.
Figure S2.
The effect of blocking RILP cleavage on LPS/ATP-induced exosomal miRNA. (A) Mock-transfected THP-1 cells and ncRILP-expressing THP-1 cells were treated with LPS/ATP. Exosomes were isolated, and their miRNA content was assessed by PCR array. Data are shown as the miR enrichment in ncRILP-expressing exosomes over mock, LPS/ATP-treated. The majority of miRs were unchanged by ncRILP expression. However, several miRs were down-regulated, while others were up-regulated by ncRILP. (B) The expression of cRILP alone leads to the enrichment of several proinflammatory miRNAs within the exosome.
Figure 2.
RILP cleavage status regulates exosome cargo loading and specificity. (A) Inflammasome activation using LPS/ATP leads to the secretion of proinflammatory miRNAs within the exosome (black bars). Note that some miRs are unchanged by treatment with LPS/ATP (white bars), while at least one decreases (gray bars). *, P ≤ 0.05 compared with untreated for n = 4–8. (B) Exosomes derived from cRILP-expressing cells are enriched in miR-155, while miR19b remains unchanged. *, P ≤ 0.05 compared with empty vector for n = 4. (C) qPCR analysis of miRNAs in both exosome and cell-associated fractions showing that ncRILP blocks the secretion of a miR-155–containing exosome after inflammasome activation by LPS and ATP. miR19b is unaffected by the expression of ncRILP. (D) Sequence alignments of miRs that are up-regulated by cRILP. (E) A common motif is present in exosome miRNAs that are up-regulated by cRILP. (F) miR-195, an miRNA that does not contain the AAUGC motif, is enriched in exosomes isolated from LPS/ATP-treated cells. The expression of ncRILP further enriches this miR within the exosome. Data are shown as mean ± SD; *, P ≤ 0.05 for n = 4–8.
To further explore the requirement for RILP cleavage in the exosomal secretion of miR-155, we assessed the effect of the dominant-negative ncRILP on intracellular and secreted miRNA content. LPS/ATP treatment of control cells increased miR-155 in the exosome fraction by nearly 250-fold (Fig. 2 C). This effect was blocked by ncRILP expression. Increased exosomal content was miRNA specific, as neither LPS/ATP nor ncRILP expression changed the exosomal content of miR-19b. Intracellular miR-155 content was also affected by LPS/ATP and ncRILP. In the absence of ncRILP, LPS/ATP decreased intracellular miR-155, but after ncRILP expression, LPS/ATP increased it 50-fold. These data suggest that, as previously shown by others, LPS/ATP increased miR-155 synthesis (Bala et al., 2011; O’Connell et al., 2007), but the RILP-dependent secretion process was so efficient that overall intracellular content decreased. Taken together, the data suggest that different forms of RILP interact with mechanisms that control the active sorting of RNAs into exosomes.
To further examine this, we performed a multiple alignment analysis of the mature miRNA sequences of the cRILP-enhanced miRNAs (Fig. 2 D). The alignment detected a common motif of AAUGC (Fig. 2 E). To determine if this motif is involved in the RILP-mediated effects on exosomal miRNA specificity, we compared the previous results with the effect of ncRILP on relative abundance of an miRNA that is induced by inflammasome activation but does not contain the motif. LPS/ATP treatment of THP-1 cells caused the enrichment of miR-195 in the exosome. However, in contrast to the miRNAs that have the AAUGC motif, ncRILP does not block miR-195 secretion into exosomes, but rather increases it further (Fig. 2 F). Altogether, these data suggest that miRNA loading of vesicles that are destined for secretion during inflammatory conditions requires the specific recognition of miRNA sequence motifs.
Identification of a cRILP-associated RBP
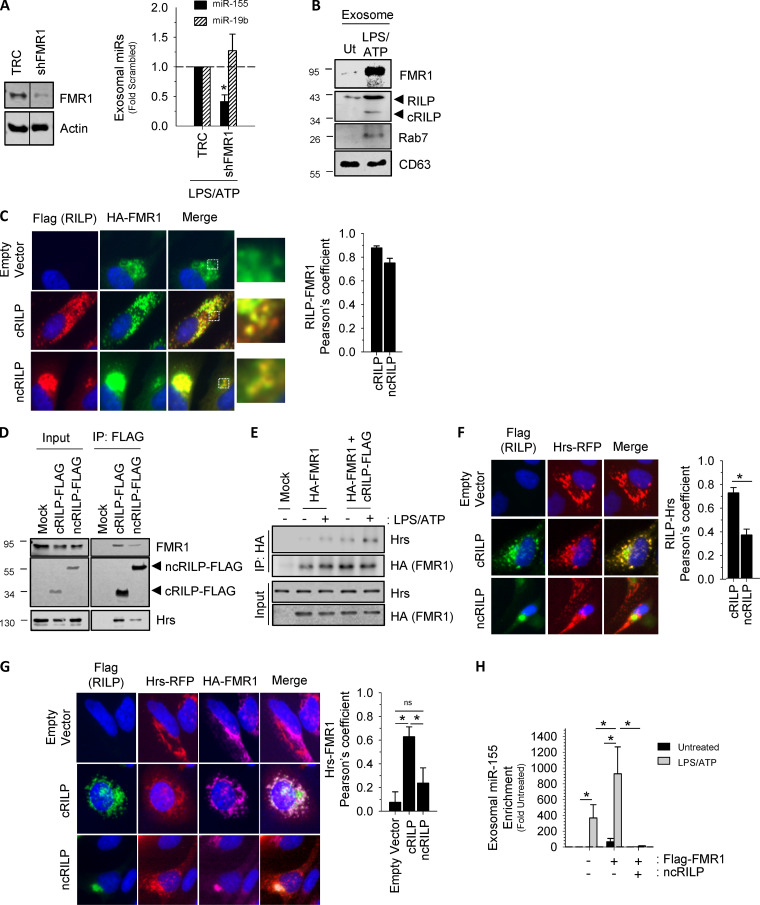
Exosome miRNA loading occurs in concert with ILV invagination at the MVB, where RBPs regulate RNA processing, localization, export, and stability (Janas et al., 2015; Lee et al., 2009). Therefore, we next examined potential RBPs predicted to interact with the short sequence motif shown in Fig. 2 E. To identify associating RBPs, the resultant motif, as well as the miRNAs that were up-regulated by cRILP, were uploaded into the ATtRACT database (a database of RBPs and associated motifs). To narrow the list, we excluded RBPs with the singular function of RNA splicing. Using the NCBI gene database, we further narrowed the list to those RBPs with identified roles in cellular trafficking. This resulted in three candidates. We then used shRNA to knock down the candidates and excluded RBPs that had no effect on exosomal miR-155 enrichment or caused a global reduction in exosomal miRNA when silenced. As shown in Fig. 3 A, shRNA knockdown of one of these proteins, FMR1, blocked LPS/ATP-induced exosome secretion of miR-155 but not miR-19b. The other tested RBPs did not show this effect (Fig. S3, A and B). FMR1 is known to play a role in transcriptional repression, but it has also been implicated in the trafficking of RNA in a kinesin-dependent manner (De Diego Otero et al., 2002; Schrier et al., 2004). FMR1 was also enriched in the exosomes induced by inflammasome activation (Fig. 3 B). Our data thus suggest a role for FMR1 in the selective loading of inflammatory miRNAs into exosomes.
Figure 3.
Identification of a cRILP-associated RBP. (A) Knockdown of candidate RBPs showing that shRNA to FMR1 blocks cRILP-associated miR-155 loading into exosomes after inflammasome activation by LPS and ATP, while not affecting miR-19b. TRC, empty vector control. (B) Western blot analysis of exosomes isolated from LPS/ATP-treated THP-1 cells shows that FMR1 is present in exosomes. These vesicles also contain cRILP and the RILP-associated GTPase, Rab7. Ut, untreated. (C) cRILP redistributes FMR1 throughout the cellular periphery. HeLa cells were transfected with HA-FMR with or without cRILP-Flag or ncRILP-Flag. HA-FMR1 (green) localizes loosely near the nucleus. Coexpression of cRILP (red) redistributes this localization toward the cell membrane, while ncRILP sequesters FMR1 in the mitotic center. Pearson’s coefficients for n = 10–15 cells. (D) cRILP is more tightly associated with FMR1 and the ESCRT-related protein Hrs than is ncRILP. cRILP-Flag or ncRILP-Flag was expressed in HeLa cells and immunoprecipitated (IP) using Flag-tagged magnetic beads. (E) FMR1 associates with Hrs, and this association is increased when cRILP is expressed. HA-FMR1 was expressed alone or with cRILP-Flag and immunoprecipitated using HA-tagged magnetic beads. (F) cRILP relocalizes Hrs. HeLa cells were transfected with Hrs-RFP with or without cRILP-Flag or ncRILP-Flag. Hrs localizes in larger structures throughout the cytoplasm. Coexpression of cRILP (green) redistributes this localization into smaller puncta, extending toward the cell membrane. ncRILP has no effect on the distribution of Hrs. Pearson’s coefficients for n = 12–15 cells. (G) cRILP induces the association between Hrs and FMR1. HeLa cells were transfected with Hrs-RFP and HA-FMR1 with or without cRILP-Flag or ncRILP-Flag. Coexpression of cRILP (green) enhances the interaction between Hrs and FMR1, while no association between the two is seen when ncRILP is expressed. Pearson’s coefficients for n = 10–15 cells. (H) miR-155 is slightly enriched in exosomes when FMR1 is overexpressed. This enrichment can be enhanced by treatment with LPS/ATP or blocked by expressing ncRILP. Data are shown as mean ± SD; ns, not significant; *, P ≤ 0.05 for n = 3–5.
Figure S3.
RNA binding proteins and exosome secretion. (A) The effect of shRNA to the RBPs RBMX, hnRNPA1, or FMR1 on exosomal miRs 155 and 19b. shRBMX had no effect on exosomal miR-155, while shhnRNPA1 caused a reduction in both miR-155 and miR-19b. (B) mRNA levels showing knockdown of RBPs. (C) The manipulation of FMR1 (either overexpression or knockdown) has no effect on exosome secretion.
We next looked at the effect of RILP cleavage on the intracellular localization of FMR1, which under basal conditions localizes loosely near the nucleus (Ascano et al., 2012; Fig. 3 C). cRILP localizes throughout the cytoplasm, and its presence redistributes FMR1 throughout the periphery where the two proteins colocalize. In contrast, ncRILP localizes in a tight distribution near the mitotic center, where it completely sequesters FMR1. Thus, FMR1 and RILP (either cleaved or uncleaved) share a cellular localization, and changes in the status of RILP cleavage can move FMR1 throughout the cell. The extent of colocalization was quantified by deriving the Pearson’s correlation coefficient for the two fluorophores. cRILP and FMR1 displayed a correlation coefficient of ∼0.8. For ncRILP and FMR1, the correlation coefficient was 0.75, both indicating colocalization.
To further define the interaction between FMR1 and cRILP, we performed immunoprecipitation of FLAG-tagged RILP constructs. While both cleaved and uncleaved RILP interact with FMR1, cRILP does so with more affinity (Fig. 3 D). Interestingly, cRILP also interacts with hepatocyte growth factor–regulated tyrosine kinase substrate (Hrs), a component of the ESCRT-0 complex. Exosomes originate at the MVB, and their formation requires components of the ESCRT pathway (Colombo et al., 2013). We therefore examined the association of FMR1 with ESCRT components and assessed whether RILP cleavage affects these interactions. Under basal conditions, the interaction between FMR1 and Hrs was just barely detectible by immunoprecipitation (Fig. 3 E). After inflammasome activation using LPS/ATP, a condition whereby RILP is cleaved, the interaction between FMR1 and Hrs increased. To test whether this association was cRILP dependent, HA-FMR1 was immunoprecipitated from THP-1 cells expressing both HA-FMR1 and cRILP-Flag. cRILP overexpression appeared to enhance this interaction, with the largest interaction observed when cRILP was expressed with HA-FMR1 in the presence of LPS/ATP. Immunofluorescence (IF) of an overexpressed Hrs shows distinct structures localized throughout the cytoplasm (Fig. 3 F). Similar to what is seen with FMR1, the presence of cRILP redistributes Hrs throughout the cell periphery. Yet the structures are reorganized into smaller, more punctate arrangements. However, unlike what is seen with FMR1, ncRILP has no effect on the localization or structure of Hrs. Taken together, the data suggest that cRILP brings FMR1 into association with Hrs, a component of the exosome loading and MVB biogenesis machinery. To assess this further, we examined the effect of RILP cleavage on the colocalization between Hrs and FMR1. At basal levels, minimal colocalization between FMR1 and Hrs is seen (Fig. 3 G). However, the expression of cRILP drives the association of these two proteins and gives a Pearson’s correlation coefficient of ∼0.6. In contrast, when ncRILP is expressed, there is no significant colocalization between Hrs and FMR1 (Pearson’s coefficient of ∼0.2).
Finally, we examined the effect of FMR1 manipulation on miR-155 loading into the exosome. Manipulation of FMR1, knockdown or overexpression, did not affect the number of exosomes secreted (Fig. S3 C). As shown previously, LPS/ATP induced miR-155 secretion in exosomes (Fig. 3 H). The overexpression of FMR1 alone resulted in EV enrichment for miR-155. However, after treatment with LPS/ATP, FMR1 overexpression had a much larger effect, leading to a nearly 1,000-fold enrichment of exosome miR-155. This enrichment could be blocked by simply expressing ncRILP, suggesting that FMR1 and cRILP cooperate to optimally load and secrete miR-155–containing exosomes.
cRILP, FMR1, and miRNA motifs
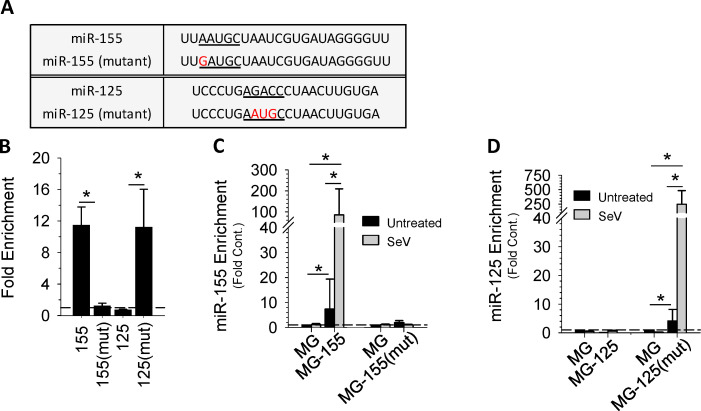
The data suggest that FMR1 is actively recruited by cRILP to the MVB to aid in the regulation of miRNA loading, possibly through small sequences present in miRNAs. To determine whether these motifs are important for RILP-mediated miRNA sorting, we performed RNA immunoprecipitation. We coexpressed HA-FMR1 with either a wild-type pre-miR-155 or a pre-miR-155(mutant) that was engineered to eliminate the AAUGC motif (Fig. 4 A). Quantitative PCR (qPCR) analysis of the FMR1-associated miRNAs shows that FMR1 binds miR-155, while the mutant miR-155 failed to bind (Fig. 4 B). To confirm these findings, we expressed a pre-miRNA corresponding to an miRNA whose secretion is not affected by RLIP, pre-miR-125, and also a modified version, pre-miR-125(mutant) containing the AAUGC motif. Wild-type miR-125 was unable to bind FMR1. However, addition of the short sequence motif to the miR-125(mutant) resulted in binding to FMR1 (Fig. 4 B).
Figure 4.
cRILP, FMR1, and miRNA motifs. (A) Schematic view of wild-type and mutant miRs showing the cRILP-associated motif (underlined) and mutations (in red). (B) RIP/chromatin immunoprecipitation of FMR1-associated miRNAs. HeLa cells were cotransfected with HA-Flag-FMR1 and the specified miRNA. FMR1 was immunoprecipitated, and qPCR analysis of the eluted miRNAs shows that FMR1 binds to the cRILP miR, miR-155. Mutation of the AAUGC motif abolishes this binding. Placing this motif into miR-125 enables its binding to FMR1. (C and D) Sequence specificity and exosomal miR loading. HeLa cells were transfected with the empty vector (MG) or the specified miRNA. After inflammasome activation by SeV, miR content in the exosomes was determined by qPCR. Wild-type miR-155 is enriched within the exosome in an inflammasome-mediated manner, while its mutant is not. miR-125 is not present in inflammasome-induced exosomes; however, the mutant miR-125 containing the AAUGC motif is selected for secretion within the exosome. Data are shown as mean ± SD; *, P ≤ 0.05 for n = 4–8.
To assess the sequence requirements for loading miRNAs into exosomes, we chose to use Sendai virus (SeV)-infected Hela cells owing to their ease of transfection. Similar to the effect of LPS/ATP on THP-1 cells, SeV infection resulted in inflammasome activation and caspase-1–induced RILP cleavage (Fig. S4, A–D; Adams et al., 2018; Wozniak et al., 2016). Fig. S4, E and F, show that SeV infection also causes a burst of exosome secretion. Both wild-type miR-155 and miR-155(mutant) were properly processed and expressed as mature miRNAs as well (Fig. S4, G and H); however, only the wild type was concentrated into secreted exosomes in response to viral infection (Fig. 4 C). The mutant miR-155, missing the AAUGC motif, was not enriched in exosomes even after inflammasome activation by virus. As expected, wild-type miR-125 was not loaded into SeV-induced exosome and was instead retained within the cell, while its mutant containing the AAUGC motif was strongly concentrated in the exosomes (Fig. 4 D). These data suggest that a short sequence motif present within miRNAs regulates its loading into the exosome via interactions with FMR1.
Figure S4.
Inflammasome-induced exosome secretion in HeLa cells. (A) Secretion of IL-18 in SeV-infected HeLa cells. Cont., control. (B) SeV infection activates the inflammasome as noted by the increase in cleaved caspase-1. (C) The caspase-1 inhibitor VI (Z-YVAD-FMK, 100 µM) inhibits (Inh.) RILP cleavage in SeV-infected HeLa cells. (D) Expression of the NLRP3 component of the inflammasome increases basal RILP cleavage. (E) SeV infection increases exosome secretion in HeLa cells by approximately eightfold. (F) EVs isolated from SeV-infected HeLa cells can be characterized by the exosome markers CD63, heat shock protein 70, Flotillin, and Tsg101. They do not contain Annexin A1 (microvesicles), histone H2B (apoptotic bodies), GM10 (Golgi), or Tom20 (mitochondria). All markers were undetected in exosome-depleted medium. WCL, whole-cell lysate. Exo, exosome. (G) Overexpression of empty vector (MG) or specific miRNAs does not alter exosome secretion. Data are shown as mean ± SD; *, P ≤ 0.05 compared with untreated for n = 4–6. (H) Intracellular expression of overexpressed miRNAs.
FMR1 and inflammatory liver disease
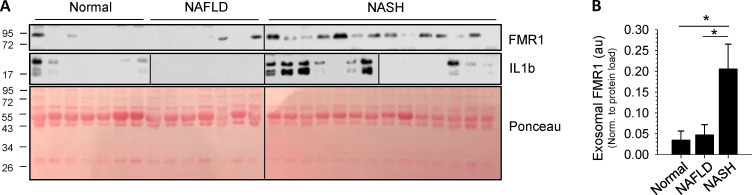
Our data directly link inflammasome activation to both increased RILP cleavage and the secretion of FMR1-enriched exosomes that contain a specific profile of miRNAs. Inflammasome activation is a common phenomenon in many diverse inflammatory diseases, and exosomes have been suggested to serve as ideal candidates for diagnostic biomarkers. To determine if inflammasome-mediated RILP cleavage and FMR1-associated exosome secretion occur in vivo in human disease, we isolated plasma exosomes from healthy individuals (normal), subjects with noninflammatory fatty liver disease (NAFLD), and patients with liver inflammation resulting from biopsy-proven nonalcoholic steatohepatitis (NASH). Exosomes were purified from human plasma by differential centrifugation. The resultant exosome pellet was resuspended in radioimmunoprecipitation assay (RIPA) lysis buffer and analyzed by Western blot. Fig. 5 shows that there was a dramatic increase in the levels of FMR1 in plasma exosome samples isolated from patients with NASH compared with both healthy controls and NAFLD subjects. Note that under noninflammatory conditions (normal, NAFLD), minimal exosomal FMR1 was seen. Furthermore, the plasma exosomes from NASH patients had increased levels of active IL1β, an indicator of inflammation. These data clearly show that increased levels of exosomal FMR1 are present in inflammatory liver disease.
Figure 5.
FMR1 is enriched in exosomes isolated from patients with inflammatory liver disease. (A) Exosomes were isolated from patient plasma by differential centrifugation, lysed in RIPA buffer, and analyzed by Western blot. Ponceau stain is shown as a loading control. (B) Densitometry analysis of the data in A showing that exosomes isolated from patients with NASH are enriched in FMR1 when compared with those from patients with NAFLD or healthy controls. au, arbitrary units. Data are shown as mean ± SD; *, P ≤ 0.05.
Discussion
Cells communicate with each other via the transfer of exosomal cargo, and the miRNA content can play a critical role in cell–cell communication, often determining the fate of the recipient cell (Alexander et al., 2015; Li et al., 2013). It is therefore critical for a cell to be able to control both the quantity of exosomes secreted and the precise cargo loaded into these exosomes. Our study establishes a direct mechanistic link between the cleavage of a trafficking adaptor protein and the enhanced exosome secretion seen during conditions of inflammation. We also identified an unsuspected role for RILP cleavage in determining exosome cargo specificity. We show that the selectivity of exosomal miRNA secretion is determined by a short sequence motif in the miRNA that binds to the RBP FMR1, which then associates with the ESCRT machinery. The cleavage of RILP enhances this interaction, resulting in the optimal enrichment of miRNA within the exosome.
Cells release EVs of various sizes and subcellular origins. Unlike microvesicles or apoptotic bodies, exosomes are small, ∼40–150 nm in size, and are formed by the intraluminal budding of the MVB. Not until the MVB fuses with the plasma membrane are the exosomes released in the extracellular milieu. To characterize the isolated EVs, we performed nanoparticle tracking analysis (NTA) to determine vesicle size, followed by Western blot analysis to identify exosome-associated proteins. The vesicle pellets contained several markers that are found in exosomes, including the tetraspanins CD63 and CD81 and the cytosolic proteins TSG101 and Flotillin. We further show the presence of MVB-associated ESCRT components as well as Rab trafficking proteins. The EV preparations were devoid of components such as nuclear proteins, indicative of apoptotic bodies, and annexin A1, a marker for microvesicles (Jeppesen et al., 2019). In accordance with current guidelines (Hartjes et al., 2019; Théry et al., 2018), we thus operationally defined these EVs as exosomes.
In this study, exosomes were isolated from donor cells cultured in medium containing FBS that was depleted of exosomes by centrifugation. Studies have suggested that this method may not fully deplete serum exosomes (Tosar et al., 2017). However, through a series of validation studies, we did not find serum-derived exosomes to negatively impact our findings should any remain after centrifugation. Even more, we have identical exosome-depleted serum present in all experiments. As such, we feel that the large increases in exosome number and the 500-fold increases in exosomal miRNA content in response to our treatments cannot be explained by a constant serum-derived background.
We establish the role of the NLRP3 inflammasome in exosome secretion and have systemically ruled out various alternative pathways leading to exosome secretion, including caspase-4 activation and gasdermin D. While we did not definitively determine which inflammasome was activated in HeLa cells, we do show that a SeV-induced process leading to caspase-1 activation and IL-18 secretion is responsible for induction of exosome secretion in this cell line. It is important to note that the common feature is caspase-1 activation, and all the exosome, trafficking, and miRNA effects flow from this.
The mechanism by which miRNA cargo are specifically loaded into the nascent exosome is not well understood. We find that the specificity of miRNA loading relies on FMR1, and our data suggest that FMR1 acts as a chaperone to package specific miRNAs into the ILVs. However, overexpression of FMR1 alone only slightly increased exosomal miR-155, suggesting that additional factors must exist to optimally load miRNA. FMR1 is not membrane bound and is typically associated with RNA granules within the cytoplasm (Feng et al., 1997; Zalfa et al., 2006). It therefore must interact with the MVB to deliver specific miRNAs to the ILV. Emerging and expanded roles for RBPs are becoming evident and now include their ability to govern trafficking to specific sites within the cell by directly or indirectly bridging the RNA to trafficking motor complexes (Blower, 2013; Schnorrer et al., 2000) or by complexing with components of the ESCRT pathway (Baietti et al., 2012; Gibbings et al., 2009). For example, the RBP swallow acts as an adaptor for the dynein complex, thereby enabling dynein to transport specific RNAs along microtubules (Schnorrer et al., 2000), and the heterogeneous nuclear ribonucleoprotein (hnRNP) Hrp36 links specific mRNAs to actin (Percipalle et al., 2001). A role for RBPs in dynein-based human immunodeficiency virus (HIV) virion trafficking has also been observed (Lehmann et al., 2009; Lévesque et al., 2006), giving credence to the idea that FMR1 may interact with components of cellular trafficking to aid in the secretion of specific cargo vesicles. In support of this hypothesis, RNA granules containing FMR1 have been shown to interact with kinesin heavy chain (Kanai et al., 2004), and FMR1 has been shown to play a role in the stimulus-induced localization of mRNAs in dendritic cells (Dictenberg et al., 2008). Our data show that FMR1 colocalizes with RILP (cRILP and ncRILP) throughout the cell (Fig. 3). It is therefore possible that the function of FMR1 to regulate miRNA loading into exosomes is dependent on its ability to interact with components of trafficking pathways such as cRILP.
Intracellular transport systems such as those involved in MVB biogenesis are highly regulated and conserved. Therefore, it is not surprising to see these components used and/or hijacked as mechanisms to initiate the loading and secretion of specific cargo. Viruses, including HCV and HIV, often recruit the ESCRT machinery to facilitate viral replication and budding (Cosset and Dreux, 2014; Lévesque et al., 2006; Medvedev et al., 2017; Nguyen et al., 2003). The MVB is also a site where several RNA biogenesis processes occur. GW182 and Argonaute 2 accumulate on the MVB, where they mediate the silencing of specific miRNAs. This silencing is dependent on their ability to interact with ESCRT proteins (Gibbings et al., 2009). While it is possible that FMR1 is ubiquitinated or modified in such a way as to directly associate with the components of the ESCRT machinery, our data suggest that the extent to which FMR1 interacts with the Hrs component of the ESCRT pathway is cRILP dependent, and that cRILP is bringing FMR1 to the ESCRTs and the MVB.
We show that inflammasome-induced exosomal secretion of specific miRNAs depends not only on the cleavage status of RILP but also on a short sequence motif that is present within the miRNAs. We detected the specific binding of FMR1 to miR-155 via this 5-nt sequence. Interestingly, this motif is also present in several FMR1-regulated mRNAs (Ascano et al., 2012). The ability of sequence motifs to govern RNA localization/enrichment to specific subcellular compartments has been reported. For example, a 20-nt stem-loop structure within nuclear-encoded mRNAs mediates their import into mitochondria (Wang et al., 2010), and a 6-nt sequence present within specific miRNAs can direct their enrichment within the nucleus (Hwang et al., 2007). We also demonstrated that FMR1, which is present in exosomes, regulates the loading of miR-155 in an inflammasome-dependent manner. More recently, the differential regulation of exosomal miRNA content has been observed in a models of alcoholic steatohepatitis (Eguchi et al., 2017, 2019). Examination of this data shows that the majority of the enriched exosomal miRNAs possess the AAUGC motif. Interestingly, this exosomal “barcode” was not seen in other models of injury that were void of inflammation, including noninflammatory alcoholic liver disease, bile duct ligation, and obesity, suggesting that exosomal cargo loading is highly injury specific. It is therefore conceivable that different stimuli use different RBPs with distinct binding preferences to secrete or retain miRNAs.
In conclusion, the phenomenon of increased EV secretion in pathological states is well documented, but the mechanism for how this occurs has remained unclear. We demonstrate a role of caspase-1 and the active inflammasome in intracellular trafficking and enhanced exosome secretion via the cleavage of the Rab7 effector protein RILP. Formation of cRILP not only increases total exosome output but also influences the miRNA selectivity of exosome cargo loading through interactions with the RBP FMR1, which directs the loading of certain miRNAs into exosomes through recognition of specific motifs and interactions with components of the ESCRT pathway. cRILP further enhances the interactions between FMR1 and ESCRTs, thereby promoting the optimal secretion of specialized exosomes. This work expands our understanding of the regulation of cellular trafficking and exosome biogenesis during conditions of inflammation, and thus provides important and novel insights into the potential application of exosomes as disease biomarkers and cell-free therapeutic agents.
Materials and methods
General materials and antibodies
General materials were purchased from Sigma-Aldrich or VWR. DMEM, FBS, Opti-MEM, and Lipofectamine 3000 were purchased from Thermo Fisher Scientific. Protease inhibitor cocktail (P8340; Sigma-Aldrich) was used at 1:100 dilution. Monoclonal antibody C7-50 to HCV core protein and monoclonal antibody JL-8 to GFP were purchased from Thermo Fisher Scientific. RILP (H85), ApoE (A1.4), CD63 (MX-49.129.5), CD81 (5A6), and GAPDH (FL-335) were purchased from Santa Cruz Biotechnology. Rab7 (D95F2), histone H2B (2722), β-Tubulin, Flotillin-1 (D2V7J), Argonaute 2 (C34C6), Hrs (D7T5N), NLRP3 (D4D8T), FMRP (4317S), GM130 (D6B1), Tom20 (D8T4N), and Caspase-4 (4450) were purchased from Cell Signaling Technology. FMR1 (F4055) and FLAG (F7425) were purchased from Sigma-Aldrich. Caspase-1 (EPR19675), IL-1β (EPR21086), TSG101 (EPR7130(B)), gasdermin D (EPR19829), and HA (HA.C5) were purchased from Abcam. Lamp1 (H4A3) was purchased from Developmental Studies Hybridoma Bank.
Plasmids
pTRE2-Bla-cRILP and pTRE2-Bla-ncRILP have been described (Wozniak et al., 2016). pEGFP-C2-NLRP3 was a gift from Christian Stehlik (Khare et al., 2012; Addgene plasmid 73955). MG (Addgene plasmid 26528), MG-155 (Addgene plasmid 26529), and pMG-miR-125b-1 (Addgene plasmid 58992) were gifts from David Baltimore (O’Connell et al., 2008; So et al., 2014). MG-155(mutant) and MG-125(mutant) were generated using the Q5 Site-Directed Mutagenesis Kit (NEB). All sequences were confirmed by DNA sequencing analysis. The pLKO-based plasmids to express shRNAs targeting RBMX (TRCN0000072673), hnRNPA1 (TRCN0000006586), FMR1 (TRCN0000059759), NLRP3 (TRCN0000432208), gasdermin D (TRCN0000179394), and TRC1 empty vector control (SHC001) were obtained from Sigma-Aldrich. pFRT-TODest-FLAGHAhFMRPiso1 (Addgene plasmid 48690) was a gift from Thomas Tuschl (Ascano et al., 2012). Flag-FMR1 and HA-FMR1 were generated by excising the gene from pFRT and inserting into pLVX-IRES-mCherry followed by site-directed mutagenesis (NEB) to remove the secondary tag. All sequences were confirmed by DNA sequencing analysis.
Cell culture and transfection
Huh-7.5 cells were obtained from Charles Rice (Rockefeller University, New York, NY) and cultured in DMEM containing 4.5 g/liter glucose, l-glutamine, and sodium pyruvate, 10% FBS, and 1% nonessential amino acids. THP-1 human monocyte cells were purchased from ATCC (TIB-202) and cultured in RPMI 1640 containing 10% FBS. HeLa cells were cultured in DMEM supplemented with 10% FBS and 1× MEM nonessential amino acids. All cells were maintained at 37°C/5% CO2. Transfections were performed using Lipofectamine 3000 according to manufacturer’s instructions. Drugs were used at the following concentrations: caspase 1 inhibitor VI (Z-YVAD-FMK), 100 µM; caspase-4 inhibitor (Ac-LEVD-CHO), 100 µM; JNK inhibitor (SP600125), 25 µM; and BHB (Thermo Fisher Scientific, AC121050050), 20 mM for 24 h before collection. These concentrations were not associated with toxicity.
HCV and SeV infection
JFH-1, a genotype 2a strain, was used for all experiments requiring HCV infection. JFH-1 plasmid was obtained from Takaji Wakita. Huh-7.5 cells were infected with supernatant containing JFH-1 virus at an MOI of 1.0 for 5 d as done previously (Wozniak et al., 2010, 2016). When indicated, SeV (Cantell strain, Charles River) was used at 400 hemagglutination units/ml for 24 h.
IF
Cells grown on coverslips were fixed in ice-cold methanol-acetone (1:1) at room temperature for 10 min and incubated in IF buffer (1% BSA and 2.5 mM EDTA in PBS) for 1 h at room temperature. After washing in PBS, the cells were incubated in primary antibody diluted in IF buffer for 2 h at room temperature. Primary antibodies used were rabbit anti-FLAG antibody (1 µg/ml, F7425; Sigma-Aldrich) and mouse anti-HA tag (HA.C5; 1:200, ab18181; Abcam). After washing with PBS, the coverslips were incubated with Alexa Fluor–conjugated goat secondary antibody (1:500) for 1 h in the dark at room temperature and mounted in ProLong Gold antifade reagent with DAPI (P36931; Thermo Fisher Scientific). Images were acquired by using an Eclipse Ti microscope (Nikon Americas) and analyzed using ImageJ2 (Rueden et al., 2017; Schindelin et al., 2012).
Lentivirus production and transduction
For lentivirus production, 293FT cells were plated in 10-cm2 dishes and transfected with 1.95 µg psPax2, 650 ng pMDG.2, and 2.6 µg of either the pLKO-based shRNAs or the pLVX-IRES-mCherry-based plasmids using 15.6 µl of X-tremeGENE HP transfection reagent (Roche). The next day, the medium was replaced with antibiotic-free complete medium (DMEM supplemented with 10% FBS, 1× MEM nonessential amino acids, 6 mM l-glutamine, and 1 mM sodium pyruvate). Supernatants were collected daily for the next 72 h. To transduce THP-1 cells, the cells were plated at a concentration of 0.5 × 106 cells per ml in 150-mm2 dishes. Lentivirus (10 ml) was incubated at room temperature with polybrene (8 µg/ml) for 15 min, then the virus was added to THP-1 cells and incubated overnight at 37°C/5% CO2. The next day, the cells were differentiated into macrophage-like cells using PMA (100 ng/ml) for 4 h, the cells were washed, and the medium was replaced. After a 48-h rest period, the cells were washed in PBS and the medium was replaced with EV-free complete medium. When indicated, the cells were primed with 1 µg/ml LPS (serotype O55:B5, Enzo) for 3 h and then stimulated with 5 mM ATP for an additional 60 min to activate the inflammasome.
RNA immunoprecipitation
HeLa cells were transfected with 10 µg of each pFRT-TODest-FLAGHAhFMRPiso1 (HA-FLAG-FMR1) and a pMG-based miRNA plasmid using Lipofectamine 3000 according to manufacturer’s instructions. After infection with SeV for 24 h, the cells were removed from the dish using trypsin, washed in PBS twice, and resuspended in 5 ml PBS. Formaldehyde was added to a final concentration of 1%, and the cells were left to rock at room temperature for 10 min. The formaldehyde was then quenched using 2 M glycine (270 mM final concentration) for 5 min, with rocking. After two washes in cold PBS, the cells were lysed in 1 ml of RIP lysis buffer (50 mM Hepes, pH 7.5, 0.4 M NaCl, 1 mM EDTA, 1 mM DTT, 0.5% Triton X-100, 10% glycerol, 1 U/µl SUPERase·In, and protease inhibitors), incubated on ice for 10 min, and subjected to one freeze/thaw cycle. HA-FLAG-FMR1 was immunoprecipitated from a clarified lysate (after 100 µl was reserved as input) using 50 µl FLAG-M2 magnetic beads, overnight at 4°C. The beads were washed five times using RIP lysis buffer, and the cross-link was reversed by resuspending the beads in 100 µl RIP reverse cross-link buffer (50 mM Hepes, pH 7.5, 0.1 M NaCl, 5 mM EDTA, 10 mM DTT, 0.5% Triton X-100, 10% glycerol, 1% SDS, 1 U/µl SUPERase·In, and protease inhibitors) and incubating at 70°C for 1 h. The cross-link was also reversed in the input sample. QIAzol was added (1 ml), and the miRNA was purified using the miRNeasy kit from Qiagen. Mature miRNAs were quantified with the miRCURY LNA RT kit and miRNA LNA primers.
NTA
To determine particle size and concentration in isolated EV samples, NTA was performed with a NanoSight LM10 (Malvern Instruments). EV preparations were first diluted in PBS to meet the optimal concentration of 105 to 108 particles/ml. At least 300 µl of diluted sample was needed for each analysis and was mixed by vortexing before injection into the chamber. Three individual videos were collected, and the resulting counts were averaged for each diluted sample. Triplicates of the same dilution were performed, and the overall average of these dilutions was used as the experimental result for each sample. A minimum of 200 particle tracks were completed for each video, and the data were analyzed using NTA 2.3 analytical software (Malvern Instruments). Data are presented as the average and SD of the triplicate.
EV isolation and analysis
Donor cells (0.5 × 106 cells per ml in 150-mm2 dishes) were cultured in 15 ml complete medium supplemented with 10% FBS (depleted of bovine EVs by overnight centrifugation at 100,000 g). EVs were isolated from cell culture supernatants via a series of differential ultracentrifugation steps as described in Thery et al. (2006). The final exosomal pellets were resuspended in PBS for further analysis. For total exosomal protein measurement, ≤5 µl of the resuspended EV pellet was assessed by standard Bradford dye assay. Before exosome collection, cell viability was assessed using trypan blue exclusion. To exclude the contribution of any remaining FBS-derived exosomes, an equal volume of growth medium was also subjected to differential centrifugation, and the resulting pellet was assessed by Western blot.
Human specimens
De-identified human plasma specimens were obtained from the Liver Center Tissue Bank at the University of Kansas Medical Center. All studies using human tissue samples were approved by the Human Subjects Committee of the University of Kansas Medical Center. To isolate EVs, 100 µl of plasma was diluted into 4 ml sterile PBS and centrifuged as stated above. The resultant EV pellet was resuspended in RIPA buffer (20 mM Tris, pH 7.4, 150 mM NaCl, 1 mM EDTA, 1 mM EGTA, 1% Triton X-100, 2.5 mM Na4P2O7, and protease inhibitors). EV content was assessed by Western blot.
Multiple alignment analysis
The multiple sequence alignment and sequence analysis was performed using MEME suite (Bailey et al., 2009) and a 0 or 1 occurrence per sequence model with a motif 4 to 8 nucleotides long. To identify associating RBPs, the resultant motif was then uploaded into the ATtRACT database (a database of RBPs and associated motifs).
miRNA isolation and qPCR
For miRNA analysis, the EV pellet was lysed in 1 ml QIAzol, and the miRNA was purified using the miRNeasy kit from Qiagen. cDNA was synthesized using the miScript II RT kit (Qiagen), making sure that equal amounts of miRNA input between samples were used. The levels of mature miRNAs were assessed using miScript primer assays. For the assessment of wild-type and mutant miRNAs, miRNAs were quantified with the miRCURY LNA RT kit and miRNA LNA primers for miR-155 (YP00204308), miR-155(mut) (YCP0043969), miR-125 (YP00205713), miR-125(mut) (YCP0044227), and miR-19b (YP00204450; Qiagen). PCR reactions were performed in triplicate in 10-µl volumes in 384-well plates. To assess extravesicular miRNA content via PCR array, the miScript miRNA PCR array human miFinder (Qiagen) was used.
Quantitation of exosomal miRNA enrichment
Exosomal content of specific miRNAs varied widely because of both the total amount of exosomes and the relative abundance of any particular miRNA under different conditions. Thus, when comparing two conditions, changes in exosomal miRNA are presented as “fold exosomal miR enrichment.” Because there is no accepted constant endogenous transcript control for exosomes, we accomplished this by normalizing miRNA content to the total RNA content of the exosomal preparation. Exosomal RNA was isolated from cell supernatants as described, and an equal RNA quantity (1 µg of total RNA) was used as input for cDNA synthesis. The relative miRNA enrichment after an experimental manipulation was then calculated as fold miRNA enrichment = miRNA quantityexperimental/miRNA quantitycontrol, where miRNA quantity was determined by qRT-PCR using a constant input of total RNA.
Western blotting and ELISA
Whole-cell lysates were prepared from cells lysed in RIPA buffer. Cell lysates were separated by SDS-PAGE and transferred to polyvinylidene difluoride membranes (Millipore). Membranes were blocked with 5% milk and 0.1% Tween-20 in TBS for 1 h at room temperature. After incubation with the appropriate primary antibodies overnight at 4°C, membranes were incubated with HRP-conjugated secondary antibodies and detected using the Pierce ECL Western blotting substrate kit (Thermo Fisher Scientific) and the Odyssey Fc, Dual-Mode Imaging system (Li-COR). The detection of IL-18 in medium was done using the Human IL-18 SimpleStep ELISA kit from Abcam (ab215539) according to manufacturer’s instructions.
Statistics
All experimental results represent observations from at least three biological replicates. Unless indicated otherwise, all data are presented as mean ± SD; the number of replicates is indicated in the legends. Student’s t test was used for statistical analyses. P ≤ 0.05 was considered significant.
Online supplemental material
Fig. S1 summarizes the link between RILP cleavage, exosome secretion, and inflammasome activation. Fig. S2 describes the effect of blocking RILP cleavage on LPS/ATP-induced exosomal miRNA. Fig. S3 examines potential RBPs involved in RILP-mediated miRNA loading. Fig. S4 investigates inflammasome activation, RILP cleavage, and exosome section in HeLa cells.
Acknowledgments
We thank Dan Dixon (University of Kansas) for comments on the manuscript. Liver specimens were obtained from the Liver Center Tissue Bank at the University of Kansas Medical Center.
The authors declare no competing financial interests.
Author contributions: A.L. Wozniak, A. Adams, and K.E. King performed experiments and acquired data. L.K. Christenson and W-T. Hung contributed key reagents for this study. W. Dunn helped to collect clinical samples. A.L. Wozniak and S.A. Weinman planned the experiments, analyzed the data, and wrote the paper. All authors approved the final manuscript.
References
- Abrami L., Brandi L., Moayeri M., Brown M.J., Krantz B.A., Leppla S.H., and van der Goot F.G.. 2013. Hijacking multivesicular bodies enables long-term and exosome-mediated long-distance action of anthrax toxin. Cell Rep. 5:986–996. 10.1016/j.celrep.2013.10.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams A., Weinman S.A., and Wozniak A.L.. 2018. Caspase-1 regulates cellular trafficking via cleavage of the Rab7 adaptor protein RILP. Biochem. Biophys. Res. Commun. 503:2619–2624. 10.1016/j.bbrc.2018.08.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alenquer M., and Amorim M.J.. 2015. Exosome Biogenesis, Regulation, and Function in Viral Infection. Viruses. 7:5066–5083. 10.3390/v7092862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander M., Hu R., Runtsch M.C., Kagele D.A., Mosbruger T.L., Tolmachova T., Seabra M.C., Round J.L., Ward D.M., and O’Connell R.M.. 2015. Exosome-delivered microRNAs modulate the inflammatory response to endotoxin. Nat. Commun. 6:7321 10.1038/ncomms8321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ascano M. Jr., Mukherjee N., Bandaru P., Miller J.B., Nusbaum J.D., Corcoran D.L., Langlois C., Munschauer M., Dewell S., Hafner M., et al. 2012. FMRP targets distinct mRNA sequence elements to regulate protein expression. Nature. 492:382–386. 10.1038/nature11737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baietti M.F., Zhang Z., Mortier E., Melchior A., Degeest G., Geeraerts A., Ivarsson Y., Depoortere F., Coomans C., Vermeiren E., et al. 2012. Syndecan-syntenin-ALIX regulates the biogenesis of exosomes. Nat. Cell Biol. 14:677–685. 10.1038/ncb2502 [DOI] [PubMed] [Google Scholar]
- Bailey T.L., Boden M., Buske F.A., Frith M., Grant C.E., Clementi L., Ren J., Li W.W., and Noble W.S.. 2009. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 37(Web Server):W202–W208. 10.1093/nar/gkp335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bala S., Marcos M., Kodys K., Csak T., Catalano D., Mandrekar P., and Szabo G.. 2011. Up-regulation of microRNA-155 in macrophages contributes to increased tumor necrosis factor alpha (TNFalpha) production via increased mRNA half-life in alcoholic liver disease. J. Biol. Chem. 286:1436–1444. 10.1074/jbc.M110.145870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bala S., Petrasek J., Mundkur S., Catalano D., Levin I., Ward J., Alao H., Kodys K., and Szabo G.. 2012. Circulating microRNAs in exosomes indicate hepatocyte injury and inflammation in alcoholic, drug-induced, and inflammatory liver diseases. Hepatology. 56:1946–1957. 10.1002/hep.25873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blower M.D. 2013. Molecular insights into intracellular RNA localization. Int. Rev. Cell Mol. Biol. 302:1–39. 10.1016/B978-0-12-407699-0.00001-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cha D.J., Franklin J.L., Dou Y., Liu Q., Higginbotham J.N., Demory Beckler M., Weaver A.M., Vickers K., Prasad N., Levy S., et al. 2015. KRAS-dependent sorting of miRNA to exosomes. eLife. 4 e07197 10.7554/eLife.07197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colombo M., Moita C., van Niel G., Kowal J., Vigneron J., Benaroch P., Manel N., Moita L.F., Théry C., and Raposo G.. 2013. Analysis of ESCRT functions in exosome biogenesis, composition and secretion highlights the heterogeneity of extracellular vesicles. J. Cell Sci. 126:5553–5565. 10.1242/jcs.128868 [DOI] [PubMed] [Google Scholar]
- Colombo M., Raposo G., and Théry C.. 2014. Biogenesis, secretion, and intercellular interactions of exosomes and other extracellular vesicles. Annu. Rev. Cell Dev. Biol. 30:255–289. 10.1146/annurev-cellbio-101512-122326 [DOI] [PubMed] [Google Scholar]
- Cosset F.L., and Dreux M.. 2014. HCV transmission by hepatic exosomes establishes a productive infection. J. Hepatol. 60:674–675. 10.1016/j.jhep.2013.10.015 [DOI] [PubMed] [Google Scholar]
- Cypryk W., Nyman T.A., and Matikainen S.. 2018. From Inflammasome to Exosome-Does Extracellular Vesicle Secretion Constitute an Inflammasome-Dependent Immune Response? Front. Immunol. 9:2188 10.3389/fimmu.2018.02188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Diego Otero Y., Severijnen L.A., van Cappellen G., Schrier M., Oostra B., and Willemsen R.. 2002. Transport of fragile X mental retardation protein via granules in neurites of PC12 cells. Mol. Cell. Biol. 22:8332–8341. 10.1128/MCB.22.23.8332-8341.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dictenberg J.B., Swanger S.A., Antar L.N., Singer R.H., and Bassell G.J.. 2008. A direct role for FMRP in activity-dependent dendritic mRNA transport links filopodial-spine morphogenesis to fragile X syndrome. Dev. Cell. 14:926–939. 10.1016/j.devcel.2008.04.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eguchi A., and Feldstein A.E.. 2018. Extracellular vesicles in non-alcoholic and alcoholic fatty liver diseases. Liver Res. 2:30–34. 10.1016/j.livres.2018.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eguchi A., Franz N., Kobayashi Y., Iwasa M., Wagner N., Hildebrand F., Takei Y., Marzi I., and Relja B.. 2019. Circulating Extracellular Vesicles and Their miR “Barcode” Differentiate Alcohol Drinkers With Liver Injury and Those Without Liver Injury in Severe Trauma Patients. Front. Med. (Lausanne). 6:30 10.3389/fmed.2019.00030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eguchi A., Lazaro R.G., Wang J., Kim J., Povero D., Willliams B., Ho S.B., Stärkel P., Schnabl B., Ohno-Machado L., et al. 2017. Extracellular vesicles released by hepatocytes from gastric infusion model of alcoholic liver disease contain a MicroRNA barcode that can be detected in blood. Hepatology. 65:475–490. 10.1002/hep.28838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emerman A.B., and Blower M.D.. 2018. The RNA-binding complex ESCRT-II in Xenopus laevis eggs recognizes purine-rich sequences through its subunit, Vps25. J. Biol. Chem. 293:12593–12605. 10.1074/jbc.RA118.003718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y., Absher D., Eberhart D.E., Brown V., Malter H.E., and Warren S.T.. 1997. FMRP associates with polyribosomes as an mRNP, and the I304N mutation of severe fragile X syndrome abolishes this association. Mol. Cell. 1:109–118. 10.1016/S1097-2765(00)80012-X [DOI] [PubMed] [Google Scholar]
- Fukuda M. 2013. Rab27 effectors, pleiotropic regulators in secretory pathways. Traffic. 14:949–963. 10.1111/tra.12083 [DOI] [PubMed] [Google Scholar]
- Gibbings D.J., Ciaudo C., Erhardt M., and Voinnet O.. 2009. Multivesicular bodies associate with components of miRNA effector complexes and modulate miRNA activity. Nat. Cell Biol. 11:1143–1149. 10.1038/ncb1929 [DOI] [PubMed] [Google Scholar]
- Guo H., Callaway J.B., and Ting J.P.. 2015. Inflammasomes: mechanism of action, role in disease, and therapeutics. Nat. Med. 21:677–687. 10.1038/nm.3893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartjes T.A., Mytnyk S., Jenster G.W., van Steijn V., and van Royen M.E.. 2019. Extracellular Vesicle Quantification and Characterization: Common Methods and Emerging Approaches. Bioengineering (Basel). 6:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y., Hara H., and Núñez G.. 2016. Mechanism and Regulation of NLRP3 Inflammasome Activation. Trends Biochem. Sci. 41:1012–1021. 10.1016/j.tibs.2016.09.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsova P., Ibrahim S.H., Krishnan A., Verma V.K., Bronk S.F., Werneburg N.W., Charlton M.R., Shah V.H., Malhi H., and Gores G.J.. 2016. Lipid-Induced Signaling Causes Release of Inflammatory Extracellular Vesicles From Hepatocytes. Gastroenterology. 150:956–967. 10.1053/j.gastro.2015.12.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurley J.H. 2015. ESCRTs are everywhere. EMBO J. 34:2398–2407. 10.15252/embj.201592484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang H.W., Wentzel E.A., and Mendell J.T.. 2007. A hexanucleotide element directs microRNA nuclear import. Science. 315:97–100. 10.1126/science.1136235 [DOI] [PubMed] [Google Scholar]
- Irion U., and St Johnston D.. 2007. bicoid RNA localization requires specific binding of an endosomal sorting complex. Nature. 445:554–558. 10.1038/nature05503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaé N., McEwan D.G., Manavski Y., Boon R.A., and Dimmeler S.. 2015. Rab7a and Rab27b control secretion of endothelial microRNA through extracellular vesicles. FEBS Lett. 589(20PartB, 20 Pt B):3182–3188. 10.1016/j.febslet.2015.08.040 [DOI] [PubMed] [Google Scholar]
- Janas T., Janas M.M., Sapoń K., and Janas T.. 2015. Mechanisms of RNA loading into exosomes. FEBS Lett. 589:1391–1398. 10.1016/j.febslet.2015.04.036 [DOI] [PubMed] [Google Scholar]
- Jeppesen D.K., Fenix A.M., Franklin J.L., Higginbotham J.N., Zhang Q., Zimmerman L.J., Liebler D.C., Ping J., Liu Q., Evans R., et al. 2019. Reassessment of Exosome Composition. Cell. 177:428–445.e18. 10.1016/j.cell.2019.02.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson M., Rocha N., Zwart W., Jordens I., Janssen L., Kuijl C., Olkkonen V.M., and Neefjes J.. 2007. Activation of endosomal dynein motors by stepwise assembly of Rab7-RILP-p150Glued, ORP1L, and the receptor betalll spectrin. J. Cell Biol. 176:459–471. 10.1083/jcb.200606077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordens I., Fernandez-Borja M., Marsman M., Dusseljee S., Janssen L., Calafat J., Janssen H., Wubbolts R., and Neefjes J.. 2001. The Rab7 effector protein RILP controls lysosomal transport by inducing the recruitment of dynein-dynactin motors. Curr. Biol. 11:1680–1685. 10.1016/S0960-9822(01)00531-0 [DOI] [PubMed] [Google Scholar]
- Kanai Y., Dohmae N., and Hirokawa N.. 2004. Kinesin transports RNA: isolation and characterization of an RNA-transporting granule. Neuron. 43:513–525. 10.1016/j.neuron.2004.07.022 [DOI] [PubMed] [Google Scholar]
- Kanneganti T.D. 2010. Central roles of NLRs and inflammasomes in viral infection. Nat. Rev. Immunol. 10:688–698. 10.1038/nri2851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khare S., Dorfleutner A., Bryan N.B., Yun C., Radian A.D., de Almeida L., Rojanasakul Y., and Stehlik C.. 2012. An NLRP7-containing inflammasome mediates recognition of microbial lipopeptides in human macrophages. Immunity. 36:464–476. 10.1016/j.immuni.2012.02.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koppers-Lalic D., Hackenberg M., Bijnsdorp I.V., van Eijndhoven M.A.J., Sadek P., Sie D., Zini N., Middeldorp J.M., Ylstra B., de Menezes R.X., et al. 2014. Nontemplated nucleotide additions distinguish the small RNA composition in cells from exosomes. Cell Rep. 8:1649–1658. 10.1016/j.celrep.2014.08.027 [DOI] [PubMed] [Google Scholar]
- Kosaka N., Iguchi H., Yoshioka Y., Takeshita F., Matsuki Y., and Ochiya T.. 2010. Secretory mechanisms and intercellular transfer of microRNAs in living cells. J. Biol. Chem. 285:17442–17452. 10.1074/jbc.M110.107821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latz E., Xiao T.S., and Stutz A.. 2013. Activation and regulation of the inflammasomes. Nat. Rev. Immunol. 13:397–411. 10.1038/nri3452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y.S., Pressman S., Andress A.P., Kim K., White J.L., Cassidy J.J., Li X., Lubell K., Lim D.H., Cho I.S., et al. 2009. Silencing by small RNAs is linked to endosomal trafficking. Nat. Cell Biol. 11:1150–1156. 10.1038/ncb1930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann M., Milev M.P., Abrahamyan L., Yao X.J., Pante N., and Mouland A.J.. 2009. Intracellular transport of human immunodeficiency virus type 1 genomic RNA and viral production are dependent on dynein motor function and late endosome positioning. J. Biol. Chem. 284:14572–14585. 10.1074/jbc.M808531200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lévesque K., Halvorsen M., Abrahamyan L., Chatel-Chaix L., Poupon V., Gordon H., DesGroseillers L., Gatignol A., and Mouland A.J.. 2006. Trafficking of HIV-1 RNA is mediated by heterogeneous nuclear ribonucleoprotein A2 expression and impacts on viral assembly. Traffic. 7:1177–1193. 10.1111/j.1600-0854.2006.00461.x [DOI] [PubMed] [Google Scholar]
- Li J., Liu K., Liu Y., Xu Y., Zhang F., Yang H., Liu J., Pan T., Chen J., Wu M., et al. 2013. Exosomes mediate the cell-to-cell transmission of IFN-α-induced antiviral activity. Nat. Immunol. 14:793–803. 10.1038/ni.2647 [DOI] [PubMed] [Google Scholar]
- McDaniel K., Herrera L., Zhou T., Francis H., Han Y., Levine P., Lin E., Glaser S., Alpini G., and Meng F.. 2014. The functional role of microRNAs in alcoholic liver injury. J. Cell. Mol. Med. 18:197–207. 10.1111/jcmm.12223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medvedev R., Hildt E., and Ploen D.. 2017. Look who’s talking-the crosstalk between oxidative stress and autophagy supports exosomal-dependent release of HCV particles. Cell Biol. Toxicol. 33:211–231. [DOI] [PubMed] [Google Scholar]
- Nguyen D.G., Booth A., Gould S.J., and Hildreth J.E.. 2003. Evidence that HIV budding in primary macrophages occurs through the exosome release pathway. J. Biol. Chem. 278:52347–52354. 10.1074/jbc.M309009200 [DOI] [PubMed] [Google Scholar]
- O’Connell R.M., Rao D.S., Chaudhuri A.A., Boldin M.P., Taganov K.D., Nicoll J., Paquette R.L., and Baltimore D.. 2008. Sustained expression of microRNA-155 in hematopoietic stem cells causes a myeloproliferative disorder. J. Exp. Med. 205:585–594. 10.1084/jem.20072108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connell R.M., Taganov K.D., Boldin M.P., Cheng G., and Baltimore D.. 2007. MicroRNA-155 is induced during the macrophage inflammatory response. Proc. Natl. Acad. Sci. USA. 104:1604–1609. 10.1073/pnas.0610731104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostrowski M., Carmo N.B., Krumeich S., Fanget I., Raposo G., Savina A., Moita C.F., Schauer K., Hume A.N., Freitas R.P., et al. 2010. Rab27a and Rab27b Control Different Steps of the Exosome Secretion Pathway In Nat. Cell Biol. Vol. 12 pp. 19–30. [DOI] [PubMed] [Google Scholar]
- Percipalle P., Zhao J., Pope B., Weeds A., Lindberg U., and Daneholt B.. 2001. Actin bound to the heterogeneous nuclear ribonucleoprotein hrp36 is associated with Balbiani ring mRNA from the gene to polysomes. J. Cell Biol. 153:229–236. 10.1083/jcb.153.1.229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Povero D., Eguchi A., Li H., Johnson C.D., Papouchado B.G., Wree A., Messer K., and Feldstein A.E.. 2014. Circulating extracellular vesicles with specific proteome and liver microRNAs are potential biomarkers for liver injury in experimental fatty liver disease. PLoS One. 9 e113651 10.1371/journal.pone.0113651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raiborg C., and Stenmark H.. 2009. The ESCRT machinery in endosomal sorting of ubiquitylated membrane proteins. Nature. 458:445–452. 10.1038/nature07961 [DOI] [PubMed] [Google Scholar]
- Rathinam V.A., and Fitzgerald K.A.. 2016. Inflammasome Complexes: Emerging Mechanisms and Effector Functions. Cell. 165:792–800. 10.1016/j.cell.2016.03.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rueden C.T., Schindelin J., Hiner M.C., DeZonia B.E., Walter A.E., Arena E.T., and Eliceiri K.W.. 2017. ImageJ2: ImageJ for the next generation of scientific image data. BMC Bioinformatics. 18:529 10.1186/s12859-017-1934-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., et al. 2012. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 9:676–682. 10.1038/nmeth.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnorrer F., Bohmann K., and Nüsslein-Volhard C.. 2000. The molecular motor dynein is involved in targeting swallow and bicoid RNA to the anterior pole of Drosophila oocytes. Nat. Cell Biol. 2:185–190. 10.1038/35008601 [DOI] [PubMed] [Google Scholar]
- Schrier M., Severijnen L.A., Reis S., Rife M., van’t Padje S., van Cappellen G., Oostra B.A., and Willemsen R.. 2004. Transport kinetics of FMRP containing the I304N mutation of severe fragile X syndrome in neurites of living rat PC12 cells. Exp. Neurol. 189:343–353. 10.1016/j.expneurol.2004.05.039 [DOI] [PubMed] [Google Scholar]
- Shen J., Huang C.K., Yu H., Shen B., Zhang Y., Liang Y., Li Z., Feng X., Zhao J., Duan L., et al. 2017. The role of exosomes in hepatitis, liver cirrhosis and hepatocellular carcinoma. J. Cell. Mol. Med. 21:986–992. 10.1111/jcmm.12950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- So A.Y., Sookram R., Chaudhuri A.A., Minisandram A., Cheng D., Xie C., Lim E.L., Flores Y.G., Jiang S., Kim J.T., et al. 2014. Dual mechanisms by which miR-125b represses IRF4 to induce myeloid and B-cell leukemias. Blood. 124:1502–1512. 10.1182/blood-2014-02-553842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuffers S., Brech A., and Stenmark H.. 2009. ESCRT proteins in physiology and disease. Exp. Cell Res. 315:1619–1626. 10.1016/j.yexcr.2008.10.013 [DOI] [PubMed] [Google Scholar]
- Sun S., Sun L., Zhou X., Wu C., Wang R., Lin S.H., and Kuang J.. 2016. Phosphorylation-Dependent Activation of the ESCRT Function of ALIX in Cytokinetic Abscission and Retroviral Budding. Dev. Cell. 37:581 10.1016/j.devcel.2016.06.002 [DOI] [PubMed] [Google Scholar]
- Takashiba S., Van Dyke T.E., Amar S., Murayama Y., Soskolne A.W., and Shapira L.. 1999. Differentiation of monocytes to macrophages primes cells for lipopolysaccharide stimulation via accumulation of cytoplasmic nuclear factor kappaB. Infect. Immun. 67:5573–5578. 10.1128/IAI.67.11.5573-5578.1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thery C., Amigorena S., Raposo G., and Clayton A.. 2006. Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Curr. Protoc. Cell Biol. Chapter 3:Unit 3.22. [DOI] [PubMed] [Google Scholar]
- Théry C., Witwer K.W., Aikawa E., Alcaraz M.J., Anderson J.D., Andriantsitohaina R., Antoniou A., Arab T., Archer F., Atkin-Smith G.K., et al. 2018. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): a position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J. Extracell. Vesicles. 7 1535750 10.1080/20013078.2018.1535750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomou T., Mori M.A., Dreyfuss J.M., Konishi M., Sakaguchi M., Wolfrum C., Rao T.N., Winnay J.N., Garcia-Martin R., Grinspoon S.K., et al. 2017. Adipose-derived circulating miRNAs regulate gene expression in other tissues. Nature. 542:450–455. 10.1038/nature21365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tosar J.P., Cayota A., Eitan E., Halushka M.K., and Witwer K.W.. 2017. Ribonucleic artefacts: are some extracellular RNA discoveries driven by cell culture medium components? J. Extracell. Vesicles. 6 1272832 10.1080/20013078.2016.1272832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G., Chen H.W., Oktay Y., Zhang J., Allen E.L., Smith G.M., Fan K.C., Hong J.S., French S.W., McCaffery J.M., et al. 2010. PNPASE regulates RNA import into mitochondria. Cell. 142:456–467. 10.1016/j.cell.2010.06.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodman P. 2016. ESCRT-III on endosomes: new functions, new activation pathway. Biochem. J. 473:e5–e8. 10.1042/BJ20151115 [DOI] [PubMed] [Google Scholar]
- Wozniak A.L., Griffin S., Rowlands D., Harris M., Yi M., Lemon S.M., and Weinman S.A.. 2010. Intracellular proton conductance of the hepatitis C virus p7 protein and its contribution to infectious virus production. PLoS Pathog. 6 e1001087 10.1371/journal.ppat.1001087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wozniak A.L., Long A., Jones-Jamtgaard K.N., and Weinman S.A.. 2016. Hepatitis C virus promotes virion secretion through cleavage of the Rab7 adaptor protein RILP. Proc. Natl. Acad. Sci. USA. 113:12484–12489. 10.1073/pnas.1607277113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zalfa F., Achsel T., and Bagni C.. 2006. mRNPs, polysomes or granules: FMRP in neuronal protein synthesis. Curr. Opin. Neurobiol. 16:265–269. 10.1016/j.conb.2006.05.010 [DOI] [PubMed] [Google Scholar]