In the yeast Hansenula polymorpha, Krikken et al. find that Inp1 tethers peroxisomes to the plasma membrane for organelle retention. The C terminus of Inp1 associates with peroxisomes, while the N terminus, which contains a PH-like domain, is important for plasma membrane binding.
Abstract
Retention of peroxisomes in yeast mother cells requires Inp1, which is recruited to the organelle by the peroxisomal membrane protein Pex3. Here we show that Hansenula polymorpha Inp1 associates peroxisomes to the plasma membrane. Peroxisome–plasma membrane contact sites disappear upon deletion of INP1 but increase upon INP1 overexpression. Analysis of truncated Inp1 variants showed that the C terminus is important for association to the peroxisome, while a stretch of conserved positive charges and a central pleckstrin homology-like domain are important for plasma membrane binding. In cells of a PEX3 deletion, strain Inp1-GFP localizes to the plasma membrane, concentrated in patches near the bud neck and in the cortex of nascent buds. Upon disruption of the actin cytoskeleton by treatment of the cells with latrunculin A, Inp1-GFP became cytosolic, indicating that Inp1 localization is dependent on the presence of an intact actin cytoskeleton.
Introduction
In budding yeast cells, organelles are precisely partitioned over mother cells and buds. This involves transport of a subset of organelles to developing buds concomitant with retention of others in the mother cells. Yeast organelle inheritance machineries consist of several components. For active transport, the cytoskeleton and motor proteins are required. Mother cells have organelle retention systems in conjunction with capturing devices in buds that prevent return of organelles to mother cells.
For yeast peroxisomes, detailed information on various aspects of organelle inheritance is available (Knoblach and Rachubinski, 2016). Peroxisomes are cell organelles that occur in almost all eukaryotic cells and are involved in a large variety of functions. In yeast they play crucial roles in the metabolism of certain carbon and nitrogen sources such as oleic acid (in Saccharomyces cerevisiae) and methanol (in Hansenula polymorpha and Pichia pastoris).
In S. cerevisiae, it has been shown that peroxisome transport involves the actin cytoskeleton and the class V myosin motor protein Myo2 (Hoepfner et al., 2001). Association of Myo2 to peroxisomes occurs via INheritance of Peroxisomes (Inp) 2, a cell cycle–regulated peroxisomal membrane protein (PMP; Fagarasanu et al., 2006). The cytosolic peroxin Pex19 stabilizes this Inp2–Myo2 interaction in S. cerevisiae (Otzen et al., 2012). S. cerevisiae Inp1 tethers peroxisomes to the cortex of mother cells, but also is important for capturing organelles in buds (Fagarasanu et al., 2005). Inp1 is a soluble protein that is recruited to peroxisomes by the PMP Pex3 (Munck et al., 2009; Knoblach et al., 2013; Knoblach and Rachubinski, 2019). It mainly associates to mature peroxisomes that are retained in mother cells. Inp2 associates to the newer organelles that are transported to the bud (Kumar et al., 2018).
S. cerevisiae Inp1 was proposed to connect peroxisomes to the cortical ER by simultaneously binding ER- and peroxisome-localized Pex3 (Knoblach et al., 2013). This model is based on the view that Pex3 traffics to peroxisomes via the ER (Kim and Hettema, 2015; Mayerhofer, 2016), resulting in a transient localization of a portion of the cellular Pex3 at the ER (Knoblach et al., 2013). The N and C termini of S. cerevisiae Inp1 can bind Pex3 independently. Therefore, Inp1 can associate peroxisomes to the ER by forming a hinge between ER- and peroxisome-localized Pex3 (Knoblach et al., 2013).
Pex3 is a highly conserved peroxin. The C-terminal domain of Pex3 is exposed to the cytosol. This domain binds several components that play important roles in peroxisome biology (Jansen and van der Klei, 2019). In addition to Inp1, it binds Pex19 to facilitate PMP sorting and Atg proteins that are required for peroxisome degradation by autophagy (Farré et al., 2008; Motley et al., 2012). The C-terminal domain of Pex3 also can bind directly to membrane lipids (Pinto et al., 2009; Chen et al., 2014).
We recently reported on a novel function of H. polymorpha Pex3, namely in formation of peroxisome-vacuole contacts (Wu et al., 2019). The vacuole-bound components of this contact site are not yet known. At peroxisome–vacuole contacts, Pex3-GFP accumulates in large patches. In that study, we noticed that in budding cells, there is often a second Pex3-GFP spot of enhanced fluorescence on peroxisomes (at ∼30% of the organelles). This patch typically localized to the cell cortex (Wu et al., 2019).
We previously demonstrated that, like S. cerevisiae Inp1, H. polymorpha Inp1 is important in peroxisome retention (Krikken et al., 2009). Here, we show that the cortical Pex3-GFP patches in H. polymorpha represent peroxisome–plasma membrane (PM) contact sites that also harbor Inp1. In the absence of Inp1, the contacts are lost, while upon overproduction, they expand. Based on the analysis of various Inp1 mutants, we conclude that an internal conserved domain in H. polymorpha Inp1 together with an N-terminal region rich in positively charged residues are important for association to the PM, whereas the C-terminal half of Inp1 is required for peroxisome binding. The predicted structure of the conserved internal domain is compatible with a divergent pleckstrin homology (PH)-like domain. Probably this domain binds either proline-rich peptides or an isolated helix. Based on latrunculin A experiments, we conclude that Inp1 localization is dependent on an intact actin cytoskeleton.
Results and discussion
Inp1 colocalizes with Pex3 at peroxisome–PM contacts
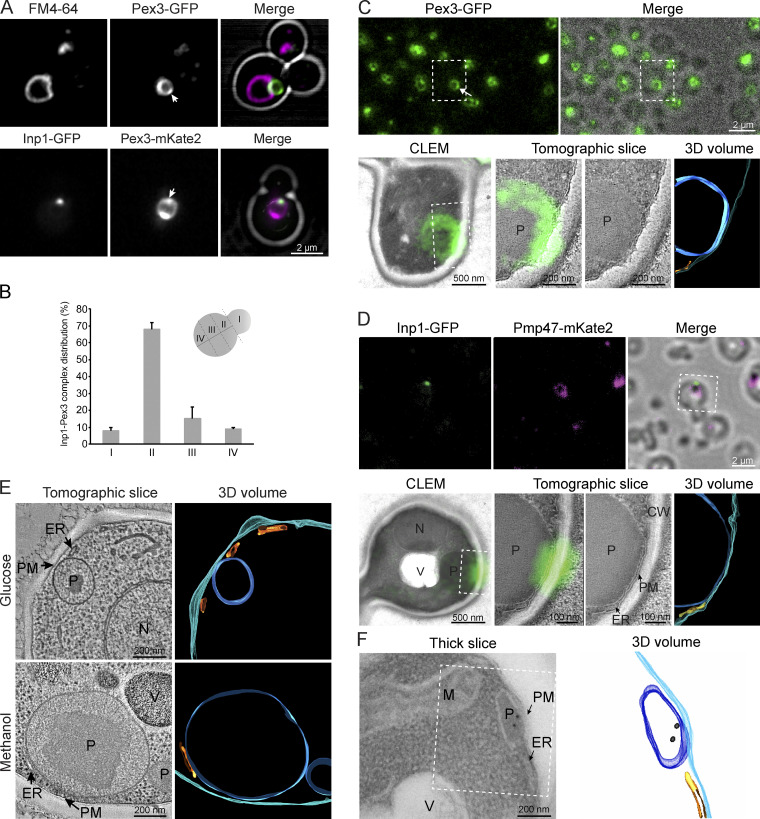
To analyze whether the cortical Pex3 patches represent contact sites involved in Inp1-dependent peroxisome retention, we performed colocalization studies using a strain producing Inp1-GFP and Pex3-mKate2, both under control of their endogenous promoters. As shown in Fig. 1 A, both proteins colocalized in the cortical Pex3 patches. Quantification revealed that 74% ± 2% of the Inp1-GFP spots colocalized with a Pex3-mKate2 patch. The Inp1-GFP patches typically are present close to the bud neck (Fig. 1 B). Correlative light and EM (CLEM) showed that the cortical patch of Pex3-GFP (Haan et al., 2002; Fig. 1 C) or Inp1-GFP (Fig. 1 D) localizes to a region where the peroxisomal membrane is in close vicinity of the PM. The localization of Inp1-GFP at these contacts was confirmed by immuno-labeling (Fig. 1 F). Electron tomography of glucose- or methanol-grown WT (Sudbery et al., 1988) cells showed that associations of peroxisomes with the PM occurred at both peroxisome-repressing and -inducing growth conditions (Fig. 1 E). Membrane contacts with the cortical ER were also observed, but Pex3-GFP or Inp1-GFP were not enriched at these sites.
Figure 1.
Inp1 colocalizes with the peripheral Pex3 patch at peroxisome-PM contacts. (A) Single focal plane confocal laser scanning microscopy Airyscan images of cells producing the indicated proteins under control of their endogenous promoters. Cells were grown for 8 h on methanol medium. Vacuoles were stained with FM4-64. The cell contours are indicated in white. The arrows indicate peripheral Inp1 patches. (B) Quantitative analysis of the position of Inp1-Pex3 spots in budding cells. The budding cell is segmented in four regions as indicated in the scheme. The positions of 2 × 50 spots were determined from two independent experiments. Error bars indicate SD. (C) CLEM analysis of single focal plane Pex3-GFP localization. The upper panel shows an FM image and merged phase contrast/FM image of a 150 nm cryo-section from cells producing Pex3-GFP and grown for 8 h on methanol. The arrowhead indicates the cortical Pex3-GFP patch. The lower panel shows the CLEM image of the Pex3-GFP patch at the PM–peroxisome contact. The dashed square indicates the region shown in the tomographic slice. P, peroxisome. (D) Single focal plane CLEM analysis as described in C of cells producing Inp1-GFP under control of the PTEF1 together with the peroxisomal membrane marker Pmp47-mKate2. CW, cell wall; N, nucleus; V, vacuole. (E) Tomographic reconstruction of peroxisomes in glucose- or methanol-grown WT cells. (F) Immuno-EM using anti-HA antibodies in WT cells producing Inp1-2HA under control of the PADH1. The dashed region indicates the region used for tomography. 3D view of the tomogram. Colors in tomograms indicate the following: cyan, PM; blue, peroxisome; orange, ER.
Inp1 is required for the formation of peroxisome–PM contacts
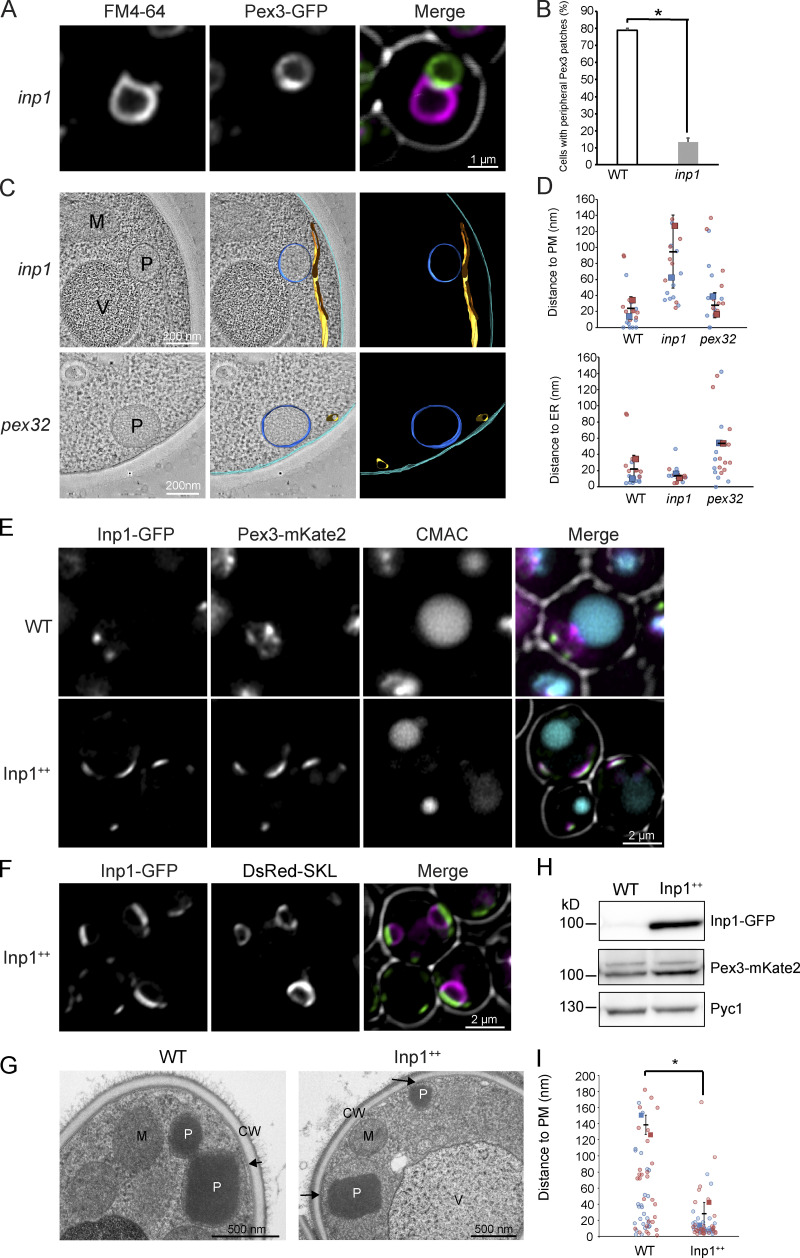
Fluorescence microscopy (FM) of an INP1 deletion strain (inp1) revealed that the percentage of cells containing a cortical Pex3-GFP patch had significantly decreased relative to WT controls (Fig. 2, A and B). Moreover, EM showed that in inp1 cells, tight connections between peroxisomes and the PM were lost, while peroxisome associations with the ER and vacuoles were unaffected (Fig. 2 C). Quantification of the distance between the peroxisomal and PM showed that the distance between the peroxisomal membrane and PM, but not between peroxisomal and ER membranes, increased in inp1 cells (Fig. 3 D). In line with our earlier observations, peroxisome–ER contacts require Pex32 (Wu et al., 2020). The distance between peroxisomes and ER increased in the absence of Pex32, whereas associations with the PM were unaffected in these cells (Fig. 2, C and D).
Figure 2.
Deletion of INP1, but not PEX32, affects peroxisome-PM contact formation, whereas INP1 overexpression increases these contacts. (A) Single focal plane confocal laser scanning microscopy Airyscan images of inp1 cells grown for 8 h on methanol and producing Pex3-GFP under control of the endogenous promoter. Vacuoles are marked with FM4-64. (B) Quantification of peripheral Pex3 patches in WT and inp1 cells. 2 × 45 cells were quantified from two independent experiments. Two-tailed Student’s t test was performed. *, P < 0.05. Error bar represents SD. (C) Tomographic reconstruction of glucose-grown inp1 and pex32 cells. Blue, peroxisomal membrane; orange, ER; cyan, PM. (D) SuperPlots showing the distance between the peroxisomal membrane and the PM or the ER in the indicated strains. 2 × 10 cell sections were quantified from two independent experiments. The duplicate experiments are color-coded. The circles represent individual data points. The squares are the averages from each experiment. The error bars indicate the SD. (E and F) Single focal plane confocal laser scanning microscopy Airyscan images from cells grown for 16 h on a mixture of glycerol and methanol and producing Inp1-GFP controlled by the endogenous promoter (WT) or PADH1 (Inp1++) together with Pex3-mKate2 or DsRed-SKL (peroxisomal matrix marker). The vacuole lumen is marked with CMAC. Note that upon growth in these conditions, the cells contain multiple peroxisomes; therefore, in the WT control cells, more Pex3-mKate2 signal and Inp1-GFP spots are present. Because in glycerol/methanol-grown cells peroxisomes harbor an alcohol oxidase crystalloid, the red fluorescence of DsRed-SKL is only present at the periphery of the organelle. Cells were grown on a mixture of glycerol and methanol because Inp1 overproduction affects peroxisome function. (G) Electron micrographs of cryofixed WT or Inp1++ cells, grown for 16 h on methanol/glycerol-containing medium. The arrows indicate peroxisome–PM contact sites. CW, cell wall; P, peroxisome; V, vacuole; M, mitochondrion. (H) Western blot analysis of Inp1-GFP levels in the indicated strains grown for 16 h on methanol/glycerol. Blots were decorated with α-GFP, α-Pex3, or α-Pyc1 antibodies. Pyc1 serves as a loading control. (I) Quantification of the distance between peroxisome membrane and PM in the indicated strains grown in medium containing methanol/glycerol. Error bars indicate SD. 2 × 34 cell sections from two independent cultures were counted. The data are plotted in a SuperPlot. The duplicate experiments are color-coded. The circles represent the single peroxisomes. Two-tailed Student’s t test was performed. *, P < 0.05.
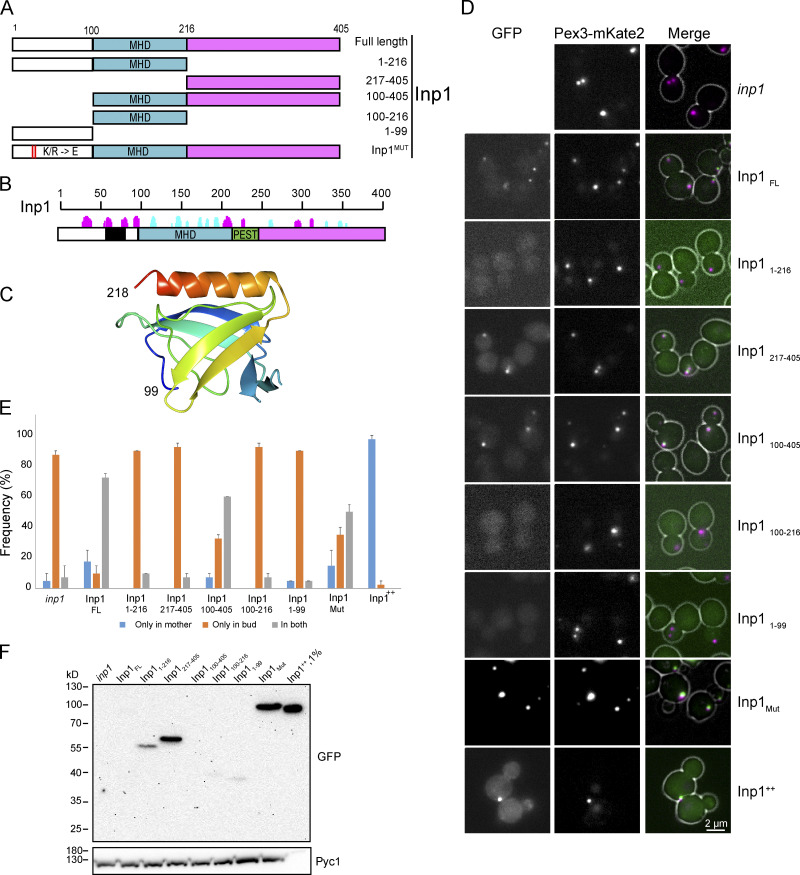
Figure 3.
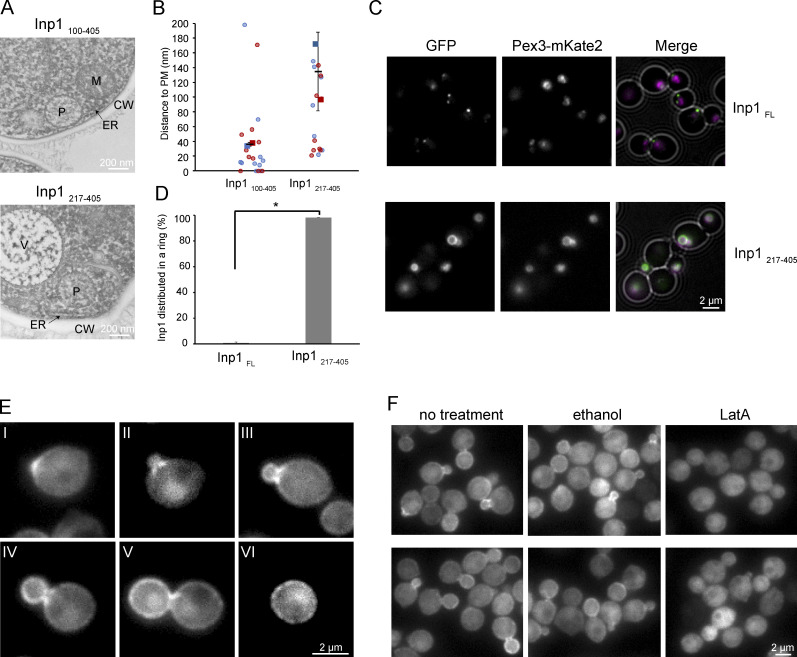
Analysis of the three domains of Inp1. (A) Schematic representation of truncated and mutant forms of Inp1. White, N terminus with conserved positive charges; blue, MHD/PH-like domain; purple, C-terminal domain. In InpMut 13 lysines (K) and arginines (R) were substituted by glutamic acids (E). (B) Predicted secondary structures of H. polymorpha Inp1 obtained with Foundation (Bordin et al., 2018) and sequence features. The black horizontal line represents the protein sequence. The predicted β-strands and α-helices are depicted by bars above the line in cyan and magenta, respectively, with the height of the bars representing the confidence of the prediction. The black box represents the stretch of positively charged residues. The green box represents the predicted conserved PEST sequence. White, blue, and purple blocks are as in A. (C) Predicted structure of H. polymorpha Inp1 residues 99–218 (MHD). The alignment of this region with human Ran-binding protein 2 (1XKE_A) was used to seed Modeller to predict the structure. Residues 121–134 between strands 1 and 2 are omitted as they do not align to any known structure, have no conserved residues, and are predicted to form an unstructured loop. (D) Single focal plane FM images of glucose-grown cells producing Pex3-mKate2 and the indicated Inp1 variants C-terminally tagged with GFP and produced under control of the INP1 promoter. The GFP fluorescence images were processed differently in order to visualize the fluorescence optimally. (E) Quantification of peroxisome inheritance in the indicated strains. 2 × 20 cells were quantified from two independent experiments. Error bars indicate SD. (F) Western blot analysis of cells producing the indicated Inp1 truncations. Blots were decorated with α-GFP or α-Pyc1 antibodies. Pyc1 was used as a loading control.
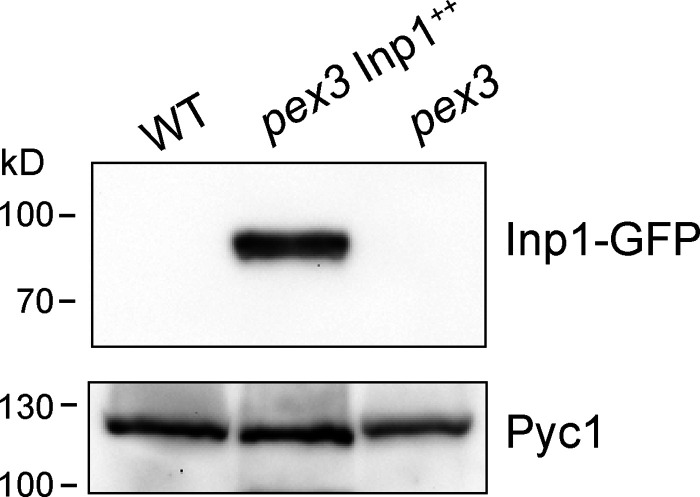
Overproduction of Inp1-GFP (Inp1++) resulted in enlarged patches of Inp1-GFP and Pex3-mKate2 (Fig. 2, E and F), accompanied by a complete loss in Pex3-GFP patches at peroxisome vacuole contacts (Fig. 2 E). Western blot analysis confirmed that Inp1 was overproduced, together with a relatively minor increase in Pex3 protein levels (Fig. 2 H). These data indicate that the bulk of the Pex3 protein was recruited to peroxisome–PM contacts upon massive Inp1 overproduction.
In methanol-grown Inp++ cells, peroxisomes were positioned at the cell cortex, in contrast to WT controls, where peroxisomes are also present in the cell center (Fig. 2, E–G). Quantification of the distance between peroxisomal membrane and PM confirmed the enhanced PM association of the organelles (Fig. 2 I).
Taken together, our data show that deletion of INP1 results in a loss in peroxisome–PM contacts, whereas overexpression leads to expansion of these contacts.
A stretch of positive charges in the N terminus, together with an internal domain, is responsible for Inp1 association to the PM
H. polymorpha Inp1 can be divided in three major domains, the N terminus (amino acids 1–99), the middle homology domain (MHD), which is relatively conserved in yeast (residues 100–216; Fig. S1; Saraya et al., 2010), and the C terminus (residues 217–405), which contains a predicted peptide sequence that is rich in proline (P), glutamic acid (E), serine (S), and threonine (T; PEST). The extreme N terminus harbors a stretch of conserved positive charges (Fig. 3, A and B).
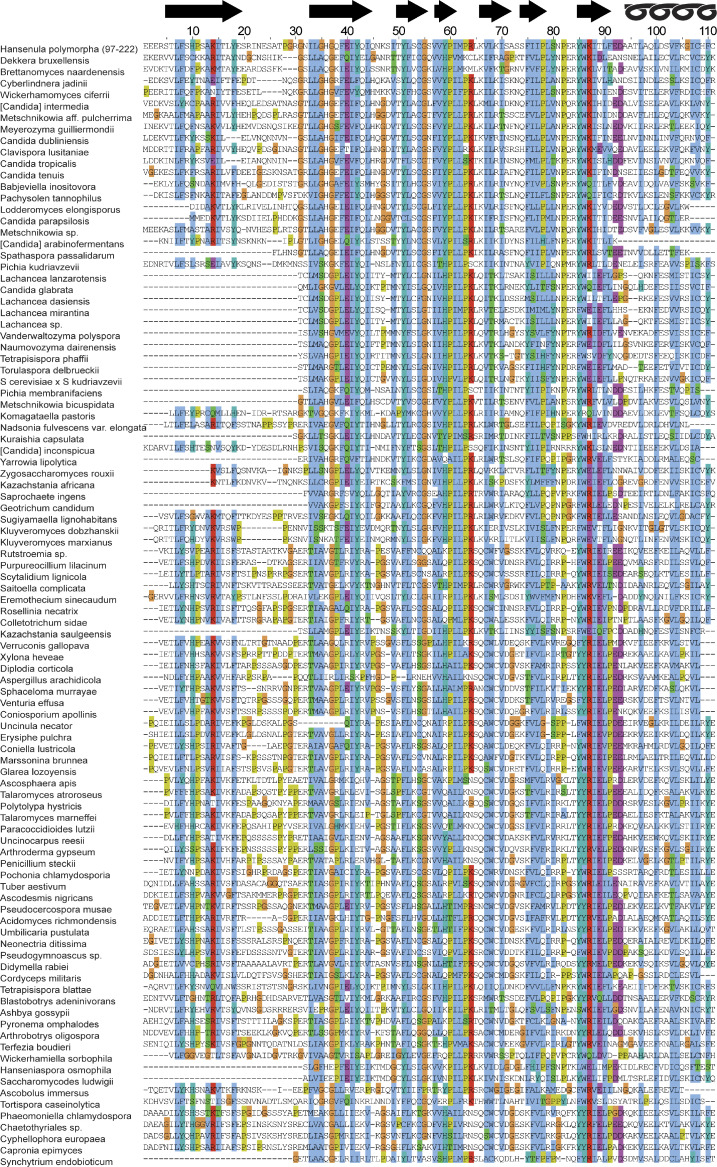
Figure S1.
The multiple sequence alignment obtained by HHblits with standard settings (three iterations, searching into a UNiREF database at 30% nonredundancy) contained Inp1 from H. polymorpha (residues 97–222) and 97 other sequences. These were aligned to remove all gaps in the seed sequence, and colored according to the CLUSTAL scheme, with predicted strands 1 to 7 and α helix indicated at the top. Note the lack of conserved positively charged residues from any loop.
The MHD contains seven β-sheets and one α-helix (Fig. 3 B and Fig. S1). Remote comparative modeling of the MHD predicted that it folds as a divergent Ran binding domain from the PH-like domain superfamily (Fig. 3 C; Fidler et al., 2016; Zimmermann et al., 2018). PH-like domains are usually involved in protein–protein interactions or binding to phospholipids (Scheffzek and Welti, 2012).
To analyze the role of the different domains in Inp1, GFP-tagged, truncated variants were constructed and tested for their function and localization upon growth of cells on glucose. All truncations were produced under control of the INP1 promoter and introduced in an inp1 deletion strain.
In budding inp1 control cells, peroxisomes occur mostly in the buds. Introduction of GFP-tagged full-length (FL) Inp1 (Inp1FL) resulted in the complementation of the peroxisome retention defect of inp1 cells. In Inp1-GFP overproducing cells (Inp1++), the organelles were almost exclusively present in the mother cells, as expected (Fig. 3, D and E).
The N- (1–216) or C-terminal (217–405) halves of Inp1 as well as constructs consisting of only the N terminus (1–99) or the MHD (100–216) were unable to complement the inp1 phenotype (Fig. 3 E). Upon production of an N-terminally truncated variant (100–405) that harbors the PH-like domain but lacks the N-terminal region with the conserved positively charged residues, the peroxisome retention defect of inp1 cells was partially restored. To further analyze the function of the positively charged residues, we constructed an Inp1 variant in which 13 of these residues were mutated (Mut) into negatively charged ones (designated Inp1Mut). As shown in Fig. 3 D, also Inp1Mut partially suppressed the inp1 phenotype. These data indicate that all three major domains are important for Inp1 function.
Western blot analysis using anti-GFP antibodies showed that the protein level of Inp1FL was below the limit of detection (Fig. 3 F), indicating that low protein levels are sufficient for peroxisome retention. Upon overproduction of FL Inp1 (Inp1++), a protein band of the expected size was detected. Like the FL protein, constructs 100–405, 100–216, and 1–99 were below the limit of detection, while constructs 1–216, 217–405, and Inp1Mut were detected. The enhanced levels of these three constructs relative to Inp1FL should be taken into account by the data interpretation. While the levels of constructs 1–216 and 217–405 are enhanced, these constructs do not suppress the inp1 phenotype, indicating that both proteins are indeed not functional. Because Inp1 overproduction results in enhanced peroxisome retention, the partial complementation by Inp1Mut, which is present at relatively high levels, indicates that the conserved positive charges contribute to Inp1 function.
Analysis of the localization of the mutant variants indicated that Inp1FL, Inp1++, Inp1Mut, and the truncations 217–405 and 100–405 colocalize with the peroxisome marker Pex3-mKate2 (Fig. 3 D). This indicates that region 217–405 is sufficient for association of Inp1 to peroxisomes. We were unable to properly assess the localization of the other constructs because of the very low GFP fluorescence levels (Fig. 3 D).
To test whether the MHD is important to associate Inp1 to the PM, we analyzed the strains producing truncations 100–405 or 217–405 by EM. Quantification of the distance between the peroxisomal membrane and the PM indicated that the absence of the MHD in 217–405 led to a loss of the close associations between both membranes (Fig. 4, A and B). Therefore, the MHD is important for PM association of peroxisomes.
Figure 4.
The N-terminal half of Inp1 is important for cortical association. (A) Electron micrographs of cryofixed glucose-grown cells of the indicated strains. P, peroxisome; V, vacuole; M, mitochondrion; CW, cell wall. (B) Quantification of the distance between the peroxisomal membrane and the PM in 2 × 10 peroxisomes from two independent experiments. The data are plotted in a SuperPlot, and the duplicate experiments are color coded. The circles represent the single data points. Error bars represent SD. (C) Single focal plane FM images of cells producing GFP tagged FL Inp1 or the indicated Inp1 truncation. Cells were grown for 8 h on methanol. (D) Quantification of the percentage of peroxisomes in which Inp1-GFP is distributed equally over the surface of the peroxisomes. 2 × 150 peroxisomes from two independent experiments were quantified. Error bars indicate the SD. Two-tailed Student’s t test was performed. *, P < 0.05. (E) Single focal plane FM images of glucose-grown pex3 cells producing Inp1-GFP under the control of the PTEF1. Different stages of budding are shown. At the onset of budding, Inp1-GFP accumulates at the site of bud emergence (I). Next, fluorescence accumulates in patches close to the bud neck (II–IV). At a later stage, the highest fluorescence signal was observed at the cortex of the buds (IV and V), while after separation of the bud, patches of enhanced fluorescence were detected over the entire cell cortex (VI). (F) Single focal plane FM images of the same cells as in E treated for 15 min with ethanol or LatA, respectively. Untreated cells were used as a control.
As shown in Fig. 1 A, Inp1FL colocalizes in a peripheral spot with a patch of Pex3-mKate2 in methanol-grown cells. In methanol-grown cells producing truncation 217–405 this spot disappeared. Instead, this Inp1 variant was evenly distributed over the peroxisomal membrane, resulting in a ring-like fluorescence pattern. Moreover, the peripheral Pex3-mKate2 patch was lost in these cells (Fig. 4, C and D). These data suggest that region 217–405 of Inp1 associates with Pex3 at peroxisomes.
Summarizing, our data indicate that the C terminus of Inp1 associates with peroxisomes, while the N terminus (including the positive charges and the MHD) contribute to the association of Inp1 to the PM.
Inp1 associates to the PM in the absence of Pex3
S. cerevisiae Inp1 is recruited to peroxisomes by Pex3 (Munck et al., 2009; Knoblach et al., 2013). To analyze whether H. polymorpha Inp1 associates to the PM in the absence of Pex3, we analyzed the localization of Inp1-GFP in a pex3 deletion strain. Produced from the endogenous promoter Inp1-GFP, levels were below the limit of detection. Upon overproduction (using PTEF1), Inp1-GFP fluorescence and protein were readily detected (Fig. 4 E and Fig. S2). FM revealed enhanced fluorescence at the cell cortex in conjunction with cytosolic fluorescence. Fluorescence was infrequently also detected in large, faint spots in the cell center, which probably represent vacuoles or nuclei. The pattern of fluorescence was dependent on the stage of bud formation. At the onset of budding, Inp1-GFP accumulated at the site of bud emergence (Fig. 4 E, I). In cells with early buds, fluorescence accumulated in patches close to the bud neck (Fig. 4 E, II–IV). At a later stage, the highest fluorescence signal was observed at the cortex of the buds (Fig. 4 E, IV and V), while after separation of the bud, patches of enhanced fluorescence were detected over the entire cell cortex (Fig. 4 E, VI). Accumulation of Inp1 near the bud neck is in agreement with the positioning of peroxisomes in budding WT cells (compare Fig. 1 B). Inp1 protein at the cortex of developing buds is probably important for capturing peroxisomes in the newly forming bud, as reported for S. cerevisiae (Fagarasanu et al., 2005).
Figure S2.
Western blot analysis of Inp1-GFP levels in the indicated strains using α-GFP or α-Pyc1 antibodies. Pyc1 was used as a loading control.
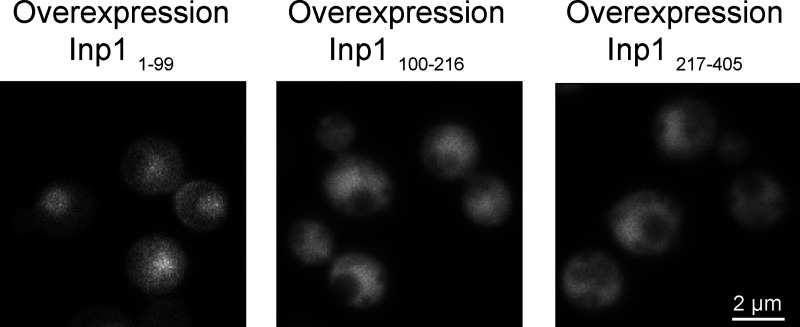
Overproduction of the extreme N terminus (1–99) of Inp1 in pex3 inp1 cells resulted in a predominantly cytosolic localization, but this truncation also slightly accumulated at the cell cortex. Possibly, this region of the protein directly associates to lipids in the PM via the conserved stretch of positive charges. An overproduced construct consisting of only the PH-like domain (MHD) was only detected in the cytosol of pex3 inp1 cells (Fig. S3). However, because of the strong fluorescence in the cytosol, we cannot exclude that a portion of the protein is associated to the PM.
Figure S3.
Overexpression of Inp1 truncations in glucose grown pex3 inp1cells. Single focal plane FM images of indicated Inp1 truncations in pex3 inp1 cells. Expression was controlled by the PTEF1 promoter.
To test whether the localization and formation of Inp1-GFP patches in pex3 cells depend on an intact actin cytoskeleton, we treated the cells with latrunculin A. As shown in Fig. 4 F, Inp1-GFP became cytosolic, and cortical Inp1-GFP patches close to the bud neck were lost. This was not observed in the ethanol-treated controls or in untreated cells, indicating that disassembly of the actin cytoskeleton caused the dissociation of Inp1-GFP from the PM. Quantification of the percentage of budding cells that contain Inp1-GFP patches revealed that treatment with ethanol (in which the latrunculin A stock solution was prepared) did not affect the patches (71% ± 3% in the untreated sample, 69% ± 3% in the ethanol-treated sample). However, upon treatment with latrunculin A, the percentage dropped to 8% ± 2%.
Together with the lack of any conserved positive amino acid in the loops of the PH-like domain (Fig. S1), this suggests that the MHD most likely does not bind a PM anionic lipid.
Inp1-dependent peroxisome–PM contacts are crucial for peroxisome retention
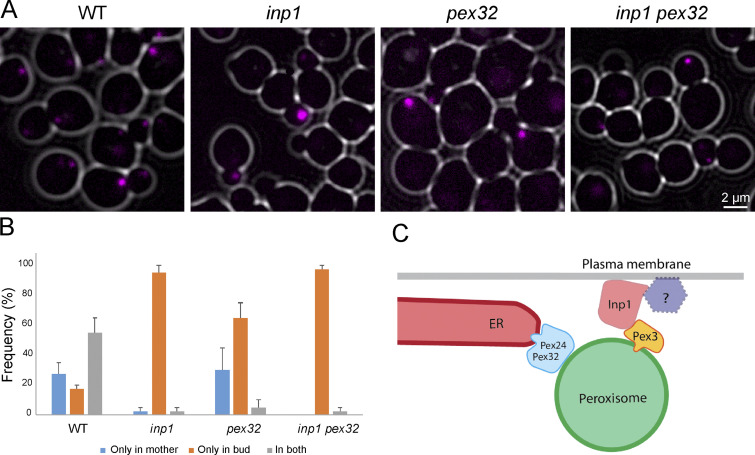
Studies in S. cerevisiae indicated that peroxisome–ER contacts are important for Inp1-dependent peroxisome inheritance (Knoblach et al., 2013). We recently showed that in H. polymorpha ER proteins of the Pex23 family are important for peroxisome–ER contact sites (Wu et al., 2020). To analyze the contribution of ER and PM contact sites, we compared peroxisome inheritance in inp1, pex32, and inp1 pex32 strains. As shown in Fig. 5, inp1 and inp1 pex32 show similar defects in peroxisome retention, indicating that in the absence of Inp1, the ER contact is not sufficient to retain peroxisomes in mother cells. In pex32 cells, a peroxisome partitioning defect was observed in that enhanced numbers of budding cells with either peroxisomes only in the bud or only in the mother occur. These findings indicate that in H. polymorpha, Inp1-dependent peroxisome–PM contacts play a key role in peroxisome retention.
Figure 5.
Inp1 plays a key role in peroxisome retention. (A) Single focal plane FM images of WT, inp1, pex32, and inp1 pex32 cells producing Pex3-mKate2, grown for 4 h in glucose-containing medium. (B) Quantitative analysis of peroxisome inheritance in the indicated strains. Peroxisomes from 2 × 20 cells were counted from two independent experiments. Error bars represent SD. (C) Hypothetical model showing Inp1-dependent peroxisome–PM contacts and peroxisome–ER contacts that require the Pex23 family proteins Pex32 and Pex24. Inp1 is recruited to the peroxisome by the PMP Pex3 and associates to the PM and a yet unknown PM protein (indicated with the question mark).
Concluding remarks
This study for the first time describes peroxisome–PM contact sites in yeast. We show that H. polymorpha Inp1 is important for the formation of these contacts, rather than for peroxisome–ER contacts. In the absence of Inp1, peroxisomes still associate to the ER. The peroxisome–ER contact is not sufficiently strong to fully retain peroxisomes in the mother cell of INP1 deletion mutants. We show that Inp1 localization is dependent on an intact cytoskeleton. Moreover, we report for the first time the detection of a divergent PH-like domain in Inp1.
Materials and methods
Strains and growth conditions
All H. polymorpha strains used in this study are listed in Table S1. Yeast cells were cultivated at 37°C in mineral medium (van Dijken et al., 1976) supplemented with 0.5% glucose or 0.5% methanol or a mixture of 0.5% methanol and 0.05% glycerol. Leucine was added to a final concentration of 60 µg/ml, if necessary. Selection of positive transformants was performed on YPD plates (1% Yeast extract, 1% Peptone, and 1% Dextrose) containing 100 µg/ml zeocin (Invitrogen), nourseothricin (Werner Bioagents), or 300 µg/ml hygromycin (Invitrogen).
Molecular and biochemical techniques
Plasmids and primers are listed in Table S2 and Table S3, respectively. Total cell extracts were prepared for Western blot analysis as described previously (Baerends et al., 2000). Equal amounts of proteins were loaded per lane. Blots were probed with mouse monoclonal antiserum against GFP (sc-9996; Santa Cruz Biotechnology, Inc.), pyruvate carboxylase-1 (Pyc1, Ozimek et al., 2003) or Pex3 (Baerends et al., 1996) and secondary goat anti-rabbit or goat anti-mouse antibodies conjugated to horseradish peroxidase (Thermo Fisher Scientific).
Construction of H. polymorpha strains
Plasmid pHIPN Pex3-mKate2 was constructed as follows: a fragment encoding the C-terminal PEX3 gene was obtained from pHIPZ Pex3-GFP (pSEM61, Wu et al., 2019) upon digestion with HindIII and BglII, then inserted between the HindIII and BglII sites of plasmid pHIPN Inp1-mKate2, resulting in plasmid pHIPN Pex3-mKate2. Subsequently, StuI-linearized pHIPN Pex3-mKate2 was integrated into the genome of strains including WT yku80 (Saraya et al., 2012), WT::PINP1 Inp1-GFP, inp1, pex32, or inp1 pex32. In parallel, pSEM61 was digested with EcoRI and transformed into the inp1 mutant.
To obtain plasmid pHIPN Inp1-mKate2, a PCR fragment was amplified with primers Nat_F and Nat_R using pHIPN4 (Cepińska et al., 2011) as a template. The PCR fragment was digested with Bpu10I and NotI, and ligated in Bpu10I and NotI digested plasmid pHIPZ Inp1-mKate2, resulting in plasmid pHIPN Inp1-mKate2. Plasmid pHIPZ Inp1-mKate2 was obtained as follows: the fragment encoding the C terminus of the INP1 gene was digested with BglII and HindIII from plasmid pHIPZ Inp1-GFP (pAMK6, Krikken et al., 2009), then inserted between the BglII and HindIII sites of pHIPZ Pex14-mKate2 (Chen et al., 2018), producing pHIPZ Inp1-mKate2.
To construct plasmid pHIPZ7 Inp1-GFP, PCR was performed with primers HLW045 and HLW046 using genomic DNA of strain WT:: PINP1 Inp1-GFP:: PTEF1 DsRed-SKL (Krikken et al., 2009) as a template. The produced PCR fragment encoding the FL of INP1 gene fused with GFP was digested with HindIII and SalI, then inserted between HindIII and SalI sites in plasmid pHIPZ7 (Baerends et al., 1997), resulting in pHIPZ7 Inp1-GFP. The constructed plasmid was linearized with MunI, and integrated into yku80 or pex3 mutant. Subsequently, the SpeI-linearized pHIPX Pmp47-mKate2 was transformed into WT:: PTEF1 Inp1-GFP. For the construction of pex3:: pHIPZ Inp1-GFP, pAMK6 was linearized with Bpu1102I and integrated into pex3 cells.
To obtain plasmid pHIPX Pmp47-mKate2, pHIPX Pmp47-mGFP was digested with BglII and MluI, and ligated with the fragment between BglII and MluI in pHIPZ Pmp47-mKate2 (pAMK142). A plasmid encoding PMP47 with monomeric GFP (mGFP) was constructed as follows: first, a PCR fragment containing Candida albicans LEU2 was amplified with primers Leucine-F and Leucine-R using pENTR221-LEU2Ca (Nagotu et al., 2008) as a template. The obtained PCR fragment was digested with XhoI and NotI, and inserted between the XhoI and NotI sites of pHIPZ Pmp47-mGFP (pMCE7), resulting in plasmid pHIPX PMP47-mGFP. Plasmid pAMK142 was constructed as follows: a PCR fragment containing PMP47 was amplified with primers PMP47_fw and PMP47_rev using WT genomic DNA as a template. The obtained PCR fragment was digested with BamHI and HindIII, and inserted between the BglII and HindIII sites of pHIPZ Pex14-mKate2 (Chen et al., 2018), resulting in plasmid pAMK142.
Plasmid pHIPZ24 Inp1-2HA is made by performing a PCR with primer Inp1-10 and Inp1-11 on yku80 chromosomal DNA. The PCR fragment and plasmid pHIPZ24 were digested with HindIII and SalI and ligated to obtain pHIPZ24 Inp1-2HA. Subsequently, plasmid pHIPZ24 Inp1-2HA and plasmid pAMK106 were digested with HindIII and NotI and ligated to obtain pHIPN18 Inp1-2HA. This plasmid was linearized with EcoRI and integrated in yku80.
To obtain plasmid pHIPZ24, PCR was performed using primer Pinp1 fw and Pinp1 rev on yku80 genomic DNA. The obtained PCR fragment and plasmid pHIPZ4 (Salomons et al., 2000) were digested with HindIII and NotI and ligated to obtain plasmid pHIPZ24.
For the construction of plasmid pHIPZ18 Inp1-GFP, a HindIII/NotI ADH1 promoter fragment was cut from pHIPN18 eGFP-SKL (pAMK106) and inserted between HindIII and NotI of pHIPZ7 Inp1-GFP, resulting in plasmid pHIPZ18 Inp1-GFP. EcoRI-linearized pHIPZ18 Inp1-GFP was integrated into genome strain yku80. In this strain, StuI-linearized pHIPN Pex3-mKate2 or DraI-linearized pHIPX7 DsRed-SKL (pAMK15, Krikken et al., 2009) was integrated. To construct plasmid pHIPZ18 eGFP-SKL (pAMK94), PCR was performed on H. polymorpha NCYC495 genomic DNA using primers Adh1-F and Adh1-R. The PCR product was digested with HindIII and NotI and the resulting fragment inserted between the HindIII and NotI sites of pHIPZ4 GFP-SKL (Leão-Helder et al., 2003), resulting in plasmid pHIPZ18 eGFP-SKL. Subsequently, pAMK94 was digested with NotI and XbaI and inserted in pHIPN4 (Cepińska et al., 2011), which was digested with the same enzymes, resulting in pHIPN18 eGFP-SKL (pAMK106).
Plasmid pHIPZ24 Inp1-GFP was obtained by digestion of plasmid pHIPZ24 with HindIII and NotI to obtain the INP1 promoter. This part was ligated in plasmid pHIPZ7 Inp1-GFP, also digested with HindIII and NotI to remove the TEF1 promoter.
Plasmid pHIPZ24 Inp11-216GFP was made by generating a PCR fragment using primer con1 fw and con2 rev on chromosomal yku80 DNA. This PCR fragment and plasmid pHIPZ24 Inp1100-405GFP were digested with HindIII and BglII and ligated.
Plasmids pHIPZ24 Inp11-99 GFP, pHIPZ24 Inp1100-216GFP, pHIPZ24 Inp1217-405GFP, and pHIPZ24 Inp1100-405GFP were created by changing the promoter region from plasmid pHIPZ7 Inp11-99GFP, pHIPZ7 Inp1100-216GFP, pHIPZ7 Inp1217-405GFP, and pHIPZ7 Inp1100-405GFP with the INP1 promoter from plasmid pHIPZ24 by restriction with HindIII and NotI.
To construct plasmids containing truncated Inp1 variants fused with GFP, pHIPZ7 Inp11-99GFP, pHIPZ7 Inp1100-216GFP, pHIPZ7 Inp1217-405GFP, and pHIPZ7 Inp1100-405GFP, PCR was performed with the corresponding primers: con1 fw and con1 rev, con2 fw and con2 rev, con3 fw and con3 rev, and con2 fw and con3 rev, respectively. WT genomic DNA was used as a template. All the obtained PCR products were digested with HindIII and BglII and inserted in pHIPZ7 Inp1-GFP, which was digested with the same enzymes, respectively.
To obtain a fragment containing 13 mutations (residues 56, 57, 58, 60, 64, 65, 68, 69, 70, 86, 87, 89, and 90 K/R to E), a synthetic DNA fragment was made (Genscript) and inserted in plasmid pUC57. This plasmid and plasmid pHIPZ24 Inp1GFP were digested with NruI and HindIII and ligated to obtain pHIPZ24 Inp1MutGFP.
All pHIPZ24 plasmids were linearized with MunI and integrated in the inp1 strain containing Pex3-mKate2. To construct the inp1 pex3 mutant, a PCR fragment containing the INP1 deletion cassette was amplified with primers Inp1 del forward and Inp1 del reverse using plasmid pAMK18 as a template. This INP1 deletion fragment was transformed in pex3 cells. Correct deletion of the INP1 gene was confirmed by PCR and Southern blotting. Plasmids pHIPZ7 Inp11-99GFP, pHIPZ7 Inp1100-216GFP, and pHIPZ7 Inp1217-405GFP were linearized with MunI and integrated in pex3 inp1.
Latrunculin A treatment
A 24 mM stock solution of Latrunculin A (LatA) in 100% ethanol was prepared (Enzo Life Sciences). LatA was added to a suspension of glucose-grown cells to a final concentration of 200 µM. In control cultures, the same amount of 100% ethanol was added. Cells were incubated for 15 min before imaging. The percentage of budding cells containing Inp1-GFP was quantified for 2 × 50 cells from two independent experiments. The error represents SD.
Structure prediction
Inp1 was submitted to foundation (Bordin et al., 2018) for secondary structure prediction, and to HHpred (Söding et al., 2005) with default parameters for structure prediction. Comparative models were built with the Modeller software (Šali and Blundell, 1993) called within HHsuite (Zimmermann et al., 2018). A full atom 3D model was built with the 1xkeA template with default parameters. The loop between Inp1 strands 1 and 2 was omitted due to the lack of template.
FM
A Zeiss Axioscope A1 fluorescence microscope (Carl Zeiss), Micro-Manager 1.4 software, and a digital camera (CoolSNAP HQ2) were used for capturing images of living cells. A 100 × 1.30 NA objective (Carl Zeiss) was applied for acquiring widefield fluorescence images. The GFP signal was visualized with a 470/40 nm bandpass excitation filter, a 495 nm dichromatic mirror, and a 525/50 nm band pass emission filter. The DsRed fluorescence and the signal of FM4-64, a vacuolar staining dye (Invitrogen), were visualized with a 546⁄12-nm bandpass excitation filter, a 560-nm dichromatic mirror, and a 575–640-nm bandpass emission filter. The mKate2 fluorescence were visualized with a 587/25-nm bandpass excitation filter, a 605-nm dichromatic mirror, and a 647/70-nm bandpass emission filter. Cells were incubated in 2 µM FM4-64 for 8 h at 37°C. ImageJ and Adobe Illustrator were applied for image analysis.
For Airyscan imaging, cells were fixed in 1% formaldehyde for 10 min on ice. Airyscan images were captured with a confocal microscope (LSM800; Carl Zeiss) equipped with a 32-channel gallium arsenide phosphide photomultiplier tube (GaAsP-PMT), Zen 2009 software (Carl Zeiss), and a 63 × 1.40 NA objective (Carl Zeiss). 7-amino-4-chloromethylcoumarin (CMAC) was visualized by excitation with a 405 nm laser. The GFP signal was visualized by excitation with a 488 nm laser, and DsRed and mKate2 were visualized with a 561 nm laser. For staining the vacuolar lumen, cells were incubated with 100 µM CMAC for 1 h at 37°C (Thermo Fisher Scientific) before fixation.
Quantification of Pex3-GFP patches was performed as described previously (Wu et al., 2019). For analyzing the distribution of patches in which Inp1 and Pex3 colocalize, budding cells were divided in four regions. The bud was determined as region I. Considering the bud neck as a baseline, in mother cells, the diameter that is vertical to the baseline was divided into three identical lengths, resulting in three regions including regions II (close to the bud neck), III (in the middle of mother cells), and IV (most distant to the bud neck; Fig. 1 B).
Co-localization of Inp1-GFP and Pex3-mKate2 patches was performed on 2 × 100 peroxisomes from two independent experiments.
Quantification of peroxisome inheritance
Assuming that the cells are spherical, the volume of the mother and bud was determined, and only dividing cells having Pex3-mKate2 signal and a bud smaller than 25% were used for quantification.
EM
Cells were cryo-fixed and freeze-substituted as described before (Wu et al., 2019). Epon-embedded cells were sectioned and collected on formvar-coated and carbon-evaporated copper grids. A CM12 (Philips) transmission electron microscope was used to inspect the grids. ImageJ was used for measuring the distance between membranes.
CLEM was performed for localization analysis as described previously (Knoops et al., 2015). 150-nm-thick cryo-sections were imaged with a widefield fluorescence microscope as described above. The corresponding fluorescence signals were visualized using the same filter sets as mentioned before. The grid was post-stained and embedded in a mixture containing 0.5% uranyl acetate and 0.5% methylcellulose. A CM12 transmission electron microscope under 100 kV was applied for the generation of double-tilt tomography series including a tilt range of 40° to −40° with 2.5° increments. To make CLEM images, FM and EM images were aligned using the eC-CLEM plugin (Paul-Gilloteaux et al., 2017) in Icy (http://icy.bioimageanalysis.org). The IMOD software package was used for reconstructing the tomograms.
Immuno-EM was performed as described previously (Thomas et al., 2018). Labeling of HA was performed using monoclonal antibodies (Sigma-Aldrich, H9658) followed by goat-anti-mouse antibodies conjugated to 6 nm gold (Aurion).
Online supplemental material
Fig. S1 shows the multiple sequence alignment of the PH domain of Inp1 obtained by HHblits. Fig. S2 shows Western blot analysis of Inp1-GFP protein levels in WT, pex3, and pex3 cells overproducing Inp1. Fig. S3 Shows the localization of overproduced Inp1 1–99, 100–216, and 217–405 in an inp1 pex3 strain. Table S1 contains all H. polymorpha strains used in this study. Table S2 contains all plasmids used in this study. Table S3 contains all primers used in this study.
Supplementary Material
contains all H. polymorpha strains used in this study.
contains all plasmids used in this study.
contains all primers used in this study.
Acknowledgments
This work was supported by a grant from the China Scholarship Council to H. Wu.
The authors declare no competing financial interests.
Author contributions: A.M. Krikken, R. de Boer, H. Wu, and I.J. van der Klei conceived the project. H. Wu, A.M. Krikken, R. de Boer, T.P. Levine, and D.P. Devos performed the experiments, analyzed the data, and prepared the figures. I.J. van der Klei and H. Wu wrote the original draft. All authors contributed to reviewing and editing the manuscript.
References
- Baerends R.J.S., Rasmussen S.W., Hilbrands R.E., van der Heide M., Faber K.N., Reuvekamp P.T.W., Kiel J.A.K.W., Cregg J.M., van der Klei I.J., and Veenhuis M.. 1996. The Hansenula polymorpha PER9 gene encodes a peroxisomal membrane protein essential for peroxisome assembly and integrity. J. Biol. Chem. 271:8887–8894. 10.1074/jbc.271.15.8887 [DOI] [PubMed] [Google Scholar]
- Baerends R.J.S., Salomons F.A., Kiel J.A.K.W., van der Klei I.J., and Veenhuis M.. 1997. Deviant Pex3p levels affect normal peroxisome formation in Hansenula polymorpha: a sharp increase of the protein level induces the proliferation of numerous, small protein-import competent peroxisomes. Yeast. 13:1449–1463. [DOI] [PubMed] [Google Scholar]
- Baerends R.J.S., Faber K.N., Kram A.M., Kiel J.A.K.W., van der Klei I.J., and Veenhuis M.. 2000. A stretch of positively charged amino acids at the N terminus of Hansenula polymorpha Pex3p is involved in incorporation of the protein into the peroxisomal membrane. J. Biol. Chem. 275:9986–9995. 10.1074/jbc.275.14.9986 [DOI] [PubMed] [Google Scholar]
- Bordin N., González-Sánchez J.C., and Devos D.P.. 2018. PVCbase: an integrated web resource for the PVC bacterial proteomes. Database (Oxford). 2018 10.1093/database/bay042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cepińska M.N., Veenhuis M., van der Klei I.J., and Nagotu S.. 2011. Peroxisome fission is associated with reorganization of specific membrane proteins. Traffic. 12:925–937. 10.1111/j.1600-0854.2011.01198.x [DOI] [PubMed] [Google Scholar]
- Chen Y., Pieuchot L., Loh R.A., Yang J., Kari T.M.A., Wong J.Y., and Jedd G.. 2014. Hydrophobic handoff for direct delivery of peroxisome tail-anchored proteins. Nat. Commun. 5:5790 10.1038/ncomms6790 [DOI] [PubMed] [Google Scholar]
- Chen X., Devarajan S., Danda N., and Williams C.. 2018. Insights into the Role of the Peroxisomal Ubiquitination Machinery in Pex13p Degradation in the Yeast Hansenula polymorpha. J. Mol. Biol. 430:1545–1558. 10.1016/j.jmb.2018.03.033 [DOI] [PubMed] [Google Scholar]
- Fagarasanu M., Fagarasanu A., Tam Y.Y.C., Aitchison J.D., and Rachubinski R.A.. 2005. Inp1p is a peroxisomal membrane protein required for peroxisome inheritance in Saccharomyces cerevisiae. J. Cell Biol. 169:765–775. 10.1083/jcb.200503083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagarasanu A., Fagarasanu M., Eitzen G.A., Aitchison J.D., and Rachubinski R.A.. 2006. The peroxisomal membrane protein Inp2p is the peroxisome-specific receptor for the myosin V motor Myo2p of Saccharomyces cerevisiae. Dev. Cell. 10:587–600. 10.1016/j.devcel.2006.04.012 [DOI] [PubMed] [Google Scholar]
- Farré J.-C., Manjithaya R., Mathewson R.D., and Subramani S.. 2008. PpAtg30 tags peroxisomes for turnover by selective autophagy. Dev. Cell. 14:365–376. 10.1016/j.devcel.2007.12.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fidler D.R., Murphy S.E., Courtis K., Antonoudiou P., El-Tohamy R., Ient J., and Levine T.P.. 2016. Using HHsearch to tackle proteins of unknown function: A pilot study with PH domains. Traffic. 17:1214–1226. 10.1111/tra.12432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haan G.J., Faber K.N., Baerends R.J.S., Koek A., Krikken A., Kiel J.A.K.W., van der Klei I.J., and Veenhuis M.. 2002. Hansenula polymorpha Pex3p is a peripheral component of the peroxisomal membrane. J. Biol. Chem. 277:26609–26617. 10.1074/jbc.M108569200 [DOI] [PubMed] [Google Scholar]
- Hoepfner D., van den Berg M., Philippsen P., Tabak H.F., and Hettema E.H.. 2001. A role for Vps1p, actin, and the Myo2p motor in peroxisome abundance and inheritance in Saccharomyces cerevisiae. J. Cell Biol. 155:979–990. 10.1083/jcb.200107028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen R.L.M., and van der Klei I.J.. 2019. The peroxisome biogenesis factors Pex3 and Pex19: multitasking proteins with disputed functions. FEBS Lett. 593:457–474. 10.1002/1873-3468.13340 [DOI] [PubMed] [Google Scholar]
- Kim P.K., and Hettema E.H.. 2015. Multiple pathways for protein transport to peroxisomes. J. Mol. Biol. 427(6 Pt A):1176–1190. 10.1016/j.jmb.2015.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoblach B., and Rachubinski R.A.. 2016. How peroxisomes partition between cells. A story of yeast, mammals and filamentous fungi. Curr. Opin. Cell Biol. 41:73–80. 10.1016/j.ceb.2016.04.004 [DOI] [PubMed] [Google Scholar]
- Knoblach B., and Rachubinski R.A.. 2019. Determinants of the assembly, integrity and maintenance of the endoplasmic reticulum-peroxisome tether. Traffic. 20:213–225. 10.1111/tra.12635 [DOI] [PubMed] [Google Scholar]
- Knoblach B., Sun X., Coquelle N., Fagarasanu A., Poirier R.L., and Rachubinski R.A.. 2013. An ER-peroxisome tether exerts peroxisome population control in yeast. EMBO J. 32:2439–2453. 10.1038/emboj.2013.170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoops K., de Boer R., Kram A., and van der Klei I.J.. 2015. Yeast pex1 cells contain peroxisomal ghosts that import matrix proteins upon reintroduction of Pex1. J. Cell Biol. 211:955–962. 10.1083/jcb.201506059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krikken A.M., Veenhuis M., and van der Klei I.J.. 2009. Hansenula polymorpha pex11 cells are affected in peroxisome retention. FEBS J. 276:1429–1439. 10.1111/j.1742-4658.2009.06883.x [DOI] [PubMed] [Google Scholar]
- Kumar S., de Boer R., and van der Klei I.J.. 2018. Yeast cells contain a heterogeneous population of peroxisomes that segregate asymmetrically during cell division. J. Cell Sci. 131 jcs207522 10.1242/jcs.207522 [DOI] [PubMed] [Google Scholar]
- Leão-Helder A.N., Krikken A.M., van der Klei I.J., Kiel J.A.K.W., and Veenhuis M.. 2003. Transcriptional down-regulation of peroxisome numbers affects selective peroxisome degradation in Hansenula polymorpha. J. Biol. Chem. 278:40749–40756. 10.1074/jbc.M304029200 [DOI] [PubMed] [Google Scholar]
- Mayerhofer P.U. 2016. Targeting and insertion of peroxisomal membrane proteins: ER trafficking versus direct delivery to peroxisomes. Biochim. Biophys. Acta. 1863:870–880. 10.1016/j.bbamcr.2015.09.021 [DOI] [PubMed] [Google Scholar]
- Motley A.M., Nuttall J.M., and Hettema E.H.. 2012. Pex3-anchored Atg36 tags peroxisomes for degradation in Saccharomyces cerevisiae. EMBO J. 31:2852–2868. 10.1038/emboj.2012.151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munck J.M., Motley A.M., Nuttall J.M., and Hettema E.H.. 2009. A dual function for Pex3p in peroxisome formation and inheritance. J. Cell Biol. 187:463–471. 10.1083/jcb.200906161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagotu S., Saraya R., Otzen M., Veenhuis M., and van der Klei I.J.. 2008. Peroxisome proliferation in Hansenula polymorpha requires Dnm1p which mediates fission but not de novo formation. Biochim. Biophys. Acta. 1783:760–769. 10.1016/j.bbamcr.2007.10.018 [DOI] [PubMed] [Google Scholar]
- Otzen M., Rucktäschel R., Thoms S., Emmrich K., Krikken A.M., Erdmann R., and van der Klei I.J.. 2012. Pex19p contributes to peroxisome inheritance in the association of peroxisomes to Myo2p. Traffic. 13:947–959. 10.1111/j.1600-0854.2012.01364.x [DOI] [PubMed] [Google Scholar]
- Ozimek P., van Dijk R., Latchev K., Gancedo C., Wang D.Y., van der Klei I.J., and Veenhuis M.. 2003. Pyruvate carboxylase is an essential protein in the assembly of yeast peroxisomal oligomeric alcohol oxidase. Mol. Biol. Cell. 14:786–797. 10.1091/mbc.e02-07-0417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul-Gilloteaux P., Heiligenstein X., Belle M., Domart M.C., Larijani B., Collinson L., Raposo G., and Salamero J.. 2017. eC-CLEM: flexible multidimensional registration software for correlative microscopies. Nat. Methods. 14:102–103. 10.1038/nmeth.4170 [DOI] [PubMed] [Google Scholar]
- Pinto M.P., Grou C.P., Fransen M., Sá-Miranda C., and Azevedo J.E.. 2009. The cytosolic domain of PEX3, a protein involved in the biogenesis of peroxisomes, binds membrane lipids. Biochim. Biophys. Acta. 1793:1669–1675. 10.1016/j.bbamcr.2009.08.007 [DOI] [PubMed] [Google Scholar]
- Šali A., and Blundell T.L.. 1993. Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 234:779–815. 10.1006/jmbi.1993.1626 [DOI] [PubMed] [Google Scholar]
- Salomons F.A., Kiel J.A., Faber K.N., Veenhuis M., and van der Klei I.J.. 2000. Overproduction of Pex5p stimulates import of alcohol oxidase and dihydroxyacetone synthase in a Hansenula polymorpha Pex14 null mutant. J. Biol. Chem. 275:12603–12611. 10.1074/jbc.275.17.12603 [DOI] [PubMed] [Google Scholar]
- Saraya R., Cepińska M.N., Kiel J.A.K.W., Veenhuis M., and van der Klei I.J.. 2010. A conserved function for Inp2 in peroxisome inheritance. Biochim. Biophys. Acta. 1803:617–622. 10.1016/j.bbamcr.2010.02.001 [DOI] [PubMed] [Google Scholar]
- Saraya R., Krikken A.M., Kiel J.A.K.W., Baerends R.J.S., Veenhuis M., and van der Klei I.J.. 2012. Novel genetic tools for Hansenula polymorpha. FEMS Yeast Res. 12:271–278. 10.1111/j.1567-1364.2011.00772.x [DOI] [PubMed] [Google Scholar]
- Scheffzek K., and Welti S.. 2012. Pleckstrin homology (PH) like domains - versatile modules in protein-protein interaction platforms. FEBS Lett. 586:2662–2673. 10.1016/j.febslet.2012.06.006 [DOI] [PubMed] [Google Scholar]
- Söding J., Biegert A., and Lupas A.N.. 2005. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 33(Web Server):W244–W248. 10.1093/nar/gki408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudbery P.E., Gleeson M.A., Veale R.A., Ledeboer A.M., and Zoetmulder M.C.M.. 1988. Hansenula polymorpha as a novel yeast system for the expression of heterologous genes. Biochem. Soc. Trans. 16:1081–1083. 10.1042/bst0161081a [DOI] [PubMed] [Google Scholar]
- Thomas A.S., Krikken A.M., de Boer R., and Williams C.. 2018. Hansenula polymorpha Aat2p is targeted to peroxisomes via a novel Pex20p-dependent pathway. FEBS Lett. 592:2466–2475. 10.1002/1873-3468.13168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dijken J.P., Otto R., and Harder W.. 1976. Growth of Hansenula polymorpha in a methanol-limited chemostat. Physiological responses due to the involvement of methanol oxidase as a key enzyme in methanol metabolism. Arch. Microbiol. 111:137–144. 10.1007/BF00446560 [DOI] [PubMed] [Google Scholar]
- Wu H., de Boer R., Krikken A.M., Akşit A., Yuan W., and van der Klei I.J.. 2019. Peroxisome development in yeast is associated with the formation of Pex3-dependent peroxisome-vacuole contact sites. Biochim. Biophys. Acta Mol. Cell Res. 1866:349–359. 10.1016/j.bbamcr.2018.08.021 [DOI] [PubMed] [Google Scholar]
- Wu F, de Boer R., Krikken A.M., Aksit A., Bordin N., Devos D.P. and van der Klei I.J.. 2020. Pex24 and Pex32 are required to tether peroxisomes to the ER for organelle biogenesis, positioning and segregation. J. Cell Sci. jcs.246983. doi: 10.1242/jcs.246983 [DOI] [PubMed]
- Zimmermann L., Stephens A., Nam S.-Z., Rau D., Kübler J., Lozajic M., Gabler F., Söding J., Lupas A.N., and Alva V.. 2018. A Completely Reimplemented MPI Bioinformatics Toolkit with a New HHpred Server at its Core. J. Mol. Biol. 430:2237–2243. 10.1016/j.jmb.2017.12.007 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
contains all H. polymorpha strains used in this study.
contains all plasmids used in this study.
contains all primers used in this study.