Matikainen et al. preview work from Wozniak et al. that reveals that inflammasomes mediate sequence-specific miRNA loading into exosomes.
Abstract
Inflammasomes are multiprotein complexes of the innate immune system. Their activation leads to robust secretion of exosomes. In this issue, Wozniak et al. (2020. J. Cell Biol. https://doi.org/10.1083/jcb.201912074) reveal a connection between inflammasome-mediated cleavage of Rab-interacting lysosomal protein (RILP) and sequence-specific loading of miRNAs into exosomes.
Exosomes are nano-sized, membrane-bound extracellular vesicles originating from the endosomal pathway via the formation of multivesicular bodies (MVBs). They are released from the recipient cells into the extracellular space upon fusion of the MVBs with the plasma membrane. Once released by the cell, exosomes can be taken up by recipient cells delivering their cargo molecules such as proteins and multiple RNA species, including miRNAs, to the target cells (1). Exosomes mediate local and systemic intercellular communication, and the role of exosomes is emerging as an important new area of biomedical research. There is growing evidence that exosome secretion is increased in many inflammatory diseases, including cardiovascular and neurodegenerative diseases (1). In general, several inflammatory stimuli are known to induce exosome secretion, and exosomes that are secreted in response to inflammatory stimuli differ in their protein and miRNA content compared with exosomes produced under homeostasis (1).
The inflammasomes are protein complexes of the innate immune system that induce inflammation in response to microbial infection and endogenous danger signals. Inflammasomes are critical for both local and systemic inflammation and form the major signaling hub that regulates inflammation. The inflammasomes include both canonical and noncanonical inflammasomes (2). Canonical NLRP3 inflammasome activation is followed by the proteolytic processing and secretion of pro-inflammatory cytokines interleukin (IL)-1β and IL-18, which are important mediators of inflammatory response (3). It has been previously shown that several activators of the NLRP3 inflammasome trigger a robust vesicle-mediated protein secretion in human innate immune cells (4). This suggests that exosome secretion constitutes an essential part of the NLRP3 inflammasome-mediated immune response. The cellular mechanism by which inflammasomes trigger increased exosome secretion and regulate the content of exosomal cargo has remained uncharacterized. In this issue, Wozniak et al. explore how inflammasome activation regulates exosome production and modulates exosomal miRNA content (5). First, they show that exosome secretion is induced by inflammasome activation in response to hepatitis C virus (HCV) infection of hepatocellular carcinoma Huh-7.5 cells, and that this is dependent on the NLRP3 inflammasome. In a previous study, they had established that caspase-1 regulates cellular trafficking via cleavage of the Rab-interacting lysosomal protein (RILP) during HCV infection (6). Now they show that RILP cleavage is dependent on NLRP3 inflammasome activation and caspase-1. To understand the mechanism of inflammasome-induced exosomal secretion, the authors used PMA-differentiated macrophage-like THP-1 cells. Lipopolysaccharide (LPS) is known to enhance NLRP3 expression in macrophages, whereas ATP triggers NLRP3 inflammasome activation. LPS priming followed by ATP stimulation resulted in RILP cleavage in THP-1 cells, which was followed by an increase in exosome release. This increase was completely blocked by forced expression of a noncleavable form of RILP. After this, Wozniak and co-workers studied whether the NLRP3 inflammasome and RILP degradation affect miRNA content of the exosomes. The noncleavable form of RILP decreased the exosomal expression of some miRNAs, including miRNA-155. Remarkably, ATP stimulation of LPS-primed THP-1 cells led to a dramatic increase in the exosomal content of these same miRNAs. Collectively, the experiments demonstrate that ATP-stimulated exosomal miRNA enrichment is dependent on cleaved RILP (cRILP). This selective miRNA loading was dependent on the short sequence motif AAUGC present in miRNAs. In follow-up experiments, the authors made an intriguing observation that the cleaved form of RILP promoted the movement of MVBs toward the cell periphery, leading to selective exosomal miRNA cargo loading. The AAUGC motif was bound by fragile X mental retardation protein (FMRP) that contains a KH domain, an evolutionarily conserved sequence motif present in many RNA-binding proteins (7). FMRP directed miRNA loading into exosomes via interaction with components of the endosomal sorting complex required for transport (ESCRT) pathway. These results indicate that FMRP acts as a chaperone to package miRNA to intraluminal vesicles. Wozniak and colleagues further assessed the sequence requirements of miRNA’s exosomal loading during Sendai virus infection of HeLa cells, which is associated with caspase-1 activation. Similar to ATP stimulation in THP-1, Sendai virus infection of HeLa cells induced caspase-1–dependent RILP cleavage, which was followed by robust activation of exosome release (Fig. 1). Interestingly, whereas wild-type miRNA-155 was readily seen in exosomes, its mutant form lacking the AAUGC motif miRNA-155 was not loaded to exosomes. These results further confirm that the short AAUGC motif regulates loading of miRNAs to exosomes.
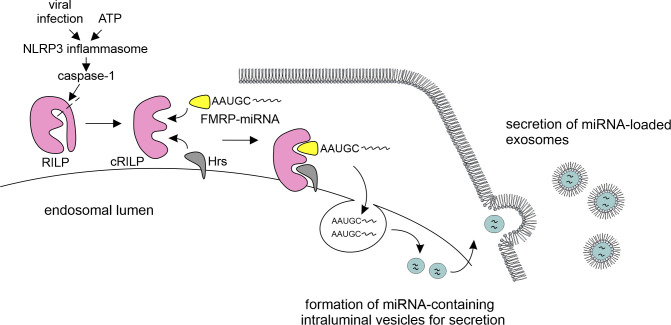
Figure 1.
NLRP3 inflammasome drives exosomal miRNA loading. Virus infection and ATP stimulation activates the NLRP3 inflammasome, leading to proteolytic processing of caspase-1. Activated caspase-1 in turn cleaves RILP. cRILP interacts with FMRP and with Hrs, a component of the ESCRT-0 complex. RNA-binding protein FMRP acts as a chaperone to package specific AAUGC motif–containing miRNAs into the intraluminal vesicles, which is followed by secretion of miRNA-loaded exosomes.
The present work by Wozniak et al. clarifies the mechanism by which miRNAs are loaded to exosomes in response to ATP-induced NLRP3 inflammasome activation (5). ATP-induced inflammasome activation has also been shown to induce robust exosome-mediated protein secretion in human macrophages (8). Based on the present study, it would be interesting to know how the caspase-1–mediated processing of RILP affects exosomal protein content, including quantitative and qualitative changes, after ATP stimulation. To address this question, high-throughput proteomics could be used. This approach could reveal the molecular machinery that is involved in loading proteins to exosome secretion following NLRP3 activation.
Inflammasome activation and exosome secretion are associated with many inflammatory diseases. Wozniak and colleagues purified exosomes from the plasma of healthy individuals as well as patients with noninflammatory fatty liver disease (NAFLD) and nonalcoholic steatohepatitis (NASH). They made an intriguing finding showing that the plasma exosomes of NASH patients had increased levels of FMRP and IL-1β compared with healthy individuals and NAFLD patients. It is tempting to speculate that patients having other inflammatory diseases may also have increased levels of inflammasome-induced exosomes in their circulation. It is well established that inflammation is a key pathogenic factor in cardiovascular and neurodegenerative diseases (9, 10), and secretion of exosomes is likely to be involved in the pathogenesis and development of these diseases. The findings described by Wozniak et al. now show that miRNAs are selectively targeted to exosomes by the action of inflammasomes, which constitutes a highly important and unprecedented discovery. The presented results open a new avenue of research to uncover mechanisms behind selective miRNA loading to exosomes.
Acknowledgments
This work was supported by the Academy of Finland, decision number 322638 (S. Matikainen), and by the National Science Centre, Poland grant 2018/28/C/NZ6/00069 (W. Cypryk).
The authors declare no competing financial interests.
References
- 1.Kalluri R., and LeBleu V.S.. 2020. Science. 10.1126/science.aau6977 [DOI] [Google Scholar]
- 2.Cypryk W., et al. 2018. Front. Immunol. 10.3389/fimmu.2018.02188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Broz P., and Dixit V.M.. 2016. Nat. Rev. Immunol. 10.1038/nri.2016.58 [DOI] [PubMed] [Google Scholar]
- 4.Matikainen S., et al. 2020. J. Immunol. 10.4049/jimmunol.2000373 [DOI] [Google Scholar]
- 5.Wozniak A.L., et al. 2020. J. Cell Biol. 10.1083/jcb.201912074 [DOI] [Google Scholar]
- 6.Adams A., et al. 2018. Biochem. Biophys. Res. Commun. 10.1016/j.bbrc.2018.08.013 [DOI] [Google Scholar]
- 7.Siomi H., et al. 1994. Cell. 10.1016/0092-8674(94)90232-1 [DOI] [Google Scholar]
- 8.Välimäki E., et al. 2016. J. Immunol. 10.4049/jimmunol.1501840 [DOI] [Google Scholar]
- 9.Ridker P.M., et al. 2017. N. Engl. J. Med. 10.1056/NEJMoa1707914 [DOI] [Google Scholar]
- 10.Chitnis T., and Weiner H.L.. 2017. J. Clin. Invest. 10.1172/JCI90609 [DOI] [PMC free article] [PubMed] [Google Scholar]