Summary
To bridge the gap between in vivo and in vitro molecular mechanisms, we dissected the transcriptional control of the endogenous histone gene cluster (His-C) by single live cell imaging. A combination of quantitative immunofluorescence, RNA FISH and FRAP measurements revealed atypical promoter recognition complexes and differential transcription kinetics directing histone H1 versus core histone gene expression. While H1 is transcribed throughout S-phase, core histones are only transcribed in a short pulse during early S-phase. Surprisingly, no TFIIB or TFIID was detectable or functionally required at the initiation complexes of these promoters. Instead, a highly stable preloaded TBP/TFIIA “pioneer” complex primes the rapid initiation of His-C transcription during early S-phase. These results provide mechanistic insights for the role of gene specific core promoter factors and implications for cell-cycle regulated gene expression.
Introduction
Transcription initiation constitutes a first critical step in the regulation of gene expression. This highly choreographed process requires the coordinated interplay of three types of protein factors: trans-activators, co-activators and core promoter recognition factors that assemble into the Pre-initiation complex (PIC) (Hampsey, 1998). Most of the proteins involved in this process have been identified through decades of in vitro biochemical studies and genetic analysis. The general transcription factors (GTFs), TFIIA, TFIIB, TFIID, TFIIE, TFIIF, and TFIIH associate with RNA polymerase II (Pol II) at the promoter to form a PIC that directs accurate transcription initiation. Sequence specific trans-activators bound to cis-regulatory elements of the DNA template can influence PIC formation via two major conduits holo-TFIID and/or the Mediator complex (Albright and Tjian, 2000; Myers and Kornberg, 2000). Many of these core promoter factors are themselves multi-subunit complexes resulting in a PIC greater than 2 Mega-daltons. Because of their gene and promoter specificity trans-activators have generally been thought to act as the primary gatekeepers of transcriptional specificity and gene regulation. However, recent studies suggest that differential usage of core promoter components and multiple levels of selectivity within the PIC can dramatically expand the repertoire of regulatory mechanisms that drive cell type-specific expression (Zhou et al., 2013). Here we will address the question of whether changes in the core apparatus can also contribute to gene specific regulation within a given cell-type.
For decades, detailed reductionist biochemistry provided many insights regarding general mechanisms of transcriptional control (Orphanides et al., 1996; Roeder, 1996). However, in vitro biochemistry primarily informs us what is possible or even likely but not always what actually occurs in living cells. On the other hand, powerful genetic approaches inform us about in vivo pathways but rarely provide molecular mechanisms of complex regulatory processes particularly in higher eukaryotic systems. The availability of high resolution and quantitative methods of imaging fixed and living cells affords us new opportunities to probe in vivo mechanisms of transcriptional regulation and gain new insights into metazoan gene expression (Darzacq et al., 2009). In this report, we set out to probe transcriptional control of an endogenous gene in live cells as a step toward a greater understanding of in vivo mechanisms modulating gene regulation. We chose the histone gene cluster (His-C) in drosophila cells for our studies because it offered a number of advantages including strong signal to noise for imaging at the single cell level and a highly efficient RNAi mediated knockdown response.
The five His genes (H1, H2A, H2B, H3, H4) are clustered into a highly repeated unit in the Drosophila genome and are transcribed during S-phase to produce new nucleosome octamers in preparation to be wrapped by newly synthetized DNA to form chromatin (Marzluff and Duronio, 2002). At the same time, linker His1 is produced to instigate the formation of higher order and/or more condensed chromatin (Thoma et al., 1979). The S-phase restricted expression of the histone genes is achieved by cell cycle regulated activation of transcription coupled with dynamic control of mRNA stability and splicing through a unique mechanism involving His-C specific processing factors (White et al., 2007). All five genes are clustered within a 5kb fragment punctuated with short promoter sequences that are shared between genes encoding pairs of heterodimer forming proteins (i.e. H2A-H2B, H3-H4) with the exception that H1 has its own promoter (Kedes, 1979). This 5kb fragment is repeated as many as 100 times in closely spaced units at the same location on chromosome 2, near a centromere. It remains unclear to this day how exactly the transcription of His-C is regulated and if or how the genes within the cluster are coordinately regulated. Previous studies found that Histone H1 transcription depends on an alternative PIC component named TRF2 that replaces the prototypic TATA Binding Protein (TBP) (Isogai et al., 2007). Why the H1 gene should utilize a distinct promoter recognition factor remained unclear. The silencing of histone genes at the end of S-phase has recently been suggested to involve various mechanisms: the building of heterochromatin over the repeats (Ito et al., 2012); phosphorylation of H2B at Tyr37 upstream of the locus (Mahajan et al., 2012); and/or the binding of metabolic factors to single stranded regions of His-C DNA (Kozhevnikova et al., 2012). Indeed, the Drosophila histone genes have been the subjects of numerous studies over the years, but to our knowledge, despite repeated efforts, including ours, none have successfully developed an in vitro transcription activation assay for the His genes. Consequently, there has been little progress in fully dissecting either factor requirements or the mechanisms governing His-C transcription.
Here, we have opted to explore the regulatory mechanisms governing His-C locus transcription by single cell molecular imaging methodologies. To study transcriptional control of the histone genes at single cell resolution we first employed quantitative FISH to measure RNA production and localization in Drosophila S2 cells. Because His-C transcription only occurs during S-Phase, we developed strategies to analyze transcription at specific stages of the replicative cycle in S2-cells. For these studies we purposely avoided drug induced S-phase synchronization that can be prone to experimental artifacts such as perturbation of DNA replication. We instead, designed a single cell pulse chase double labeling assay that allowed us to visualize His gene expression with good time resolution during S-phase in unsynchronized cells. Our studies revealed evidence for distinct timing and patterns of transcription for different genes within the His-C cluster. Our analysis also unexpectedly uncovered unusual kinetics, stability and composition of general transcription factors at the PIC of His-C by single live cell imaging.
Results
Imaging transcription at the drosophila histone locus
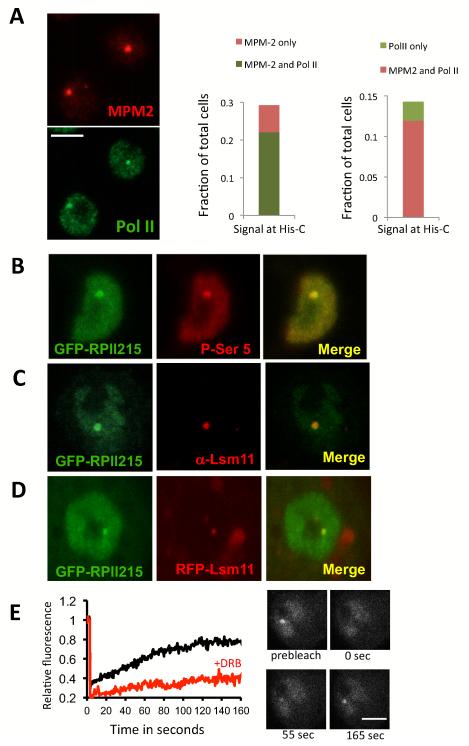
In order to probe transcriptional control of an endogenous gene in live cells we chose the highly reiterated (100X) histone gene cluster in drosophila. Although Drosophila S2 cells offer a number of advantages, this system also poses some challenges since the histone locus is only transcribed during S-phase of the cell cycle and drosophila S2 cells are difficult to synchronize and the processes of synchronization perturbs the timing of transcription (data not shown). We therefore opted to study transcription regulation of the histone genes at the single cell level in non-synchronized cells. We devised a transcription factor and gene locus co-localization strategy based on quantitating the radial profile of specific fluorescently labeled proteins and transcripts associated with the histone gene cluster (His-C) (Fig S1, and supplemental methods). First, we sought to establish a means to reliably identify those cells in the asynchronous population wherein RNA polymerase II (pol II) was actively engaged in transcribing the His genes. We made use of the well accepted marker Mxc for identifying actively transcribing S-phase engaged His genes by using the anti-phospho Mxc antibody MPM-2. It had been well established that Mxc becomes phosphorylated at the histone locus exclusively during S-phase when transcription of His-C also occurs (White et al., 2007; 2011). Using MPM-2 staining as a reference marker, we found that 74% of the cells staining for MPM-2 were also positive for Pol II P-ser5 accumulation at the histone locus (Figure 1A Middle panel) confirming S-phase co-localization of transcription at the His-C locus in the asynchronous population. Cells stained with a Pol II P-Ser5 specific antibody display a consistent and characteristic pattern at the histone locus that is intense and easy to score as a “dot” of ~0.5 μm radius (Figure 1A, left panel), usually seen as a single dot because of somatic homologous chromosome pairing in Drosophila cells, when both alleles are brought together. A similar Pol II staining pattern at His-C was also observed using a Pol II P-Ser2 antibody as well as other antibodies directed against Pol II (Figure S2). Using these Pol II patterns as a reference, we found that they show a strong co-localization with MPM-2 staining (Figure 1A Right panel). These findings indicate that Pol lI indeed only becomes associated with the His-C gene cluster during S-phase.
Figure 1.
Visualizing RNA polymerase II transcription at the histone gene locus. (A) Left panel, Co-staining of S2 cells with MPM2 antibody (red) and Pol II P-Ser5 antibody in green. Middle panel, fraction of cells showing MPM-2 staining (light Red) and MPM-2/Pol II co-localization (light green). Right panel, fraction of cells showing Pol II His-C staining (light green) and MPM-2/Pol II co-localization (light red) (N=336). Bar = 5μm. (B) A stable cell line expressing GFP-RPII215 (Pol II-green) stained with P-Ser5 antibody (red); and showing co-localization. (C) A stable cell line expressing GFP-RPII215 is stained with Lsm11 (red) antibody, showing co-localization. (D) A stable cell line expressing both GFP-RPII215 (green) and RFP-Lsm11 (red); and showing co-localization. (E) FRAP experiment tracking GFP-RPII215 at the histone locus. Left panel: Black line shows recovery of signal from untreated cells. The red line shows recovery of signal from cells treated with DRB. Average of 10-15 FRAP experiments, standard error <15%. Right panel: Example of of GFP-RPII215 FRAP at the histone locus. Bar = 5μm.
In an effort to further establish the equivalency of Pol II staining and active transcription at the His-C loci and to ascertain that the presence of Pol II does not merely represent a stalled, poised or otherwise inactive but bound enzyme complex, we next tracked the behavior and movement of Pol II at the His-C loci by live cell imaging and fluorescence recovery after photo-bleaching (FRAP). First, we generated S2 cell lines with a GFP fused to the N-Terminus of the largest subunit of RNA Pol II (RPII215). These clonal lines were isolated by using a RPII215αr carrying a naturally occurring α-amanitin resistant mutation for selecting clones expressing the tagged enzyme (supplemental methods). We next confirmed that α-amanitin resistant clones grow normally and that the GFP-RPII215αr fully replaced the endogenous subunit rendering the cells completely dependent on a functional α-amanitin resistant Pol II (not shown). GFP-RPII215αr expression and localization in these cells is indistinguishable from those of endogenous Pol II as determined by staining with the P-Ser5 antibody (Figure 1B). As expected, GFP-RPII215αr accumulates at the histone genes as shown by counterstaining with Lsm11, a mRNA splicing factor specific to Histone mRNAs (Figure 1C). To follow the localization of GFPRPII215αr on the histone genes we generated another cell line that co-expresses Lsm11 fused to a red fluorescent protein (Figure 1D). Fluorescence recovery after photo-bleaching (FRAP) was subsequently measured by bleaching the GFP-RPII215αr signal enriched at the His-C transcription sites (Figure 1E). The recovery curves revealed two populations of RNA Pol II, a mobile population representing ~80% of the total, and a smaller (~20%) immobile population. The addition of DRB, a potent inhibitor of transcription elongation, efficiently blocks the recovery during FRAP, strongly suggesting that at least 80% of the RNA Pol II molecules at the His-C locus are engaged in active transcription elongation (Figure 1E), suggesting that initiation is not limiting at His-C. The remaining immobile fraction (10-20%) could represent RNA Pol II binding to sites near or at the His-C promoter but not actively transcribing. Alternatively the small immobile population could represent newly reinitiating transcription complexes trapped by the high density of promoters and PICs stacked up at this tandemly repeated loci. We favor this explanation for the “stalled” RNA pol II population because DRB treatment for longer than 5 minutes effectively eliminated the entire Pol II signal at His-C (not shown). This suggests that most, if not all of the polymerases at these sites become rapidly engaged in active transcription elongation and therefore are sensitive to DRB. These results taken in aggregate provide a firm basis for concluding that RNA pol II association is in fact a good indicator of active transcription at the drosophila His-C locus which is also entirely consistent with the MPM-2 staining.
Measuring the Transcription Kinetics of His Genes in the Locus
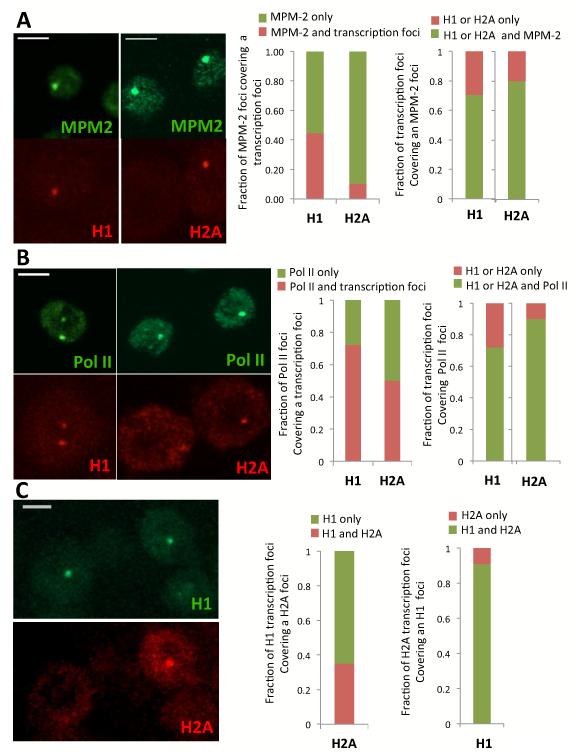
To directly assess mRNA production from the histone locus we designed sets of tiling probes targeting the 5 individual histone genes using FISH and fixed S2 cells. We were able to detect the histone gene transcripts in both the cytoplasm and nucleus of asynchronous S2 cells for all five genes (Figure 2 and Figure S3). Using MPM-2 and/or Pol II staining as markers for the His-C locus, we could easily detect co-localization of Histone H1 and H2A transcript signals in the nucleus (Figure 2A left panel) confirming that we can visualize nascent RNA at these transcription sites. When using H1 or H2A as a reference, the majority (80%) of transcription foci colocalized with MPM-2 or Pol II (Figure 2 A,B Right panel), confirming that His-C transcription is restricted to S-phase. Surprisingly, however, only half of the cells with MPM-2/Pol II signals were found to be associated with H1 transcripts and even less (10-40%) with H2A transcript signals (Figure 2 A,B Middle panel). It was possible that some H1 and H2A signals were simply too weak to be detected by our methods. However, the signal detected at the His-C locus was far stronger than that of the signal detected in the cytoplasm, where RNA are mostly in dispersed single copies. Thus we believe that our threshold of detection is close to single molecule resolution and therefore the absence of signal at His-C is noteworthy. To further analyze these findings we set up a double FISH experiment for H1 and H2A using a different fluorophore for H1 (but with the same specific activity). When H1 is used as a reference, H2A transcription signal is found in only 30% of the nuclei containing H1 transcripts (Figure 2C Middle panel). But when H2A is used as a reference, up to 90% of the nuclei showing H2A transcription foci also contain H1 transcription signal (Figure 2C Right panel). These findings suggest that there are indeed more cells transcribing H1 than H2A, and that the period of H1 transcription encompasses H2A transcription. The same trend appears to hold true for the other core histone genes, since double FISH experiment with H2B, H3 and H4 referenced against H2A all showed a consistent and systematic co-localization which was distinct from the H1 pattern of transcription frequency (Figure S3 and not shown). These initial studies establish that we can detect and track active transcription at the Histone Locus in drosophila S2 cells by following RNA Polymerase II or nascent transcripts. Surprisingly, however, despite all 5 genes being arrayed in a cluster the core histone genes appear to display a distinct transcription timing from that of Histone H1. At this stage it is not possible to tell if the more frequent transcription of H1 is due to out of S-phase transcription or due to a truly distinct pattern of expression for H1 during S-phase relative to the core histone genes.
Figure 2.
Visualizing nascent transcription of the histone genes by FISH in single S2 cells. (A) Co-staining of MPM2 antibody (green) with H1 or H2A mRNA FISH (red), bar=5μm. Middle panel shows fraction of MPM-2 signal co-localized with H1 or H2A. Right panel shows fraction of H1 and H2A foci colocalized with MPM-2 staining (N=60-116) (B) Co-staining of Pol II P-Ser5 (green) with H1 or H2A mRNA FISH (red), bar=5μm. Middle panel shows fraction of Pol II His-C signal co-localized with H1 or H2A. Right panel shows fraction of H1 and H2A foci colocalized with Pol II His-C staining (N=40-93) (C) Co-staining through FISH of H1 (green) and H2A (red). Middle panel shows fraction of co-localized signals of H1 with H2A, bar=5μm. Right panel shows fraction of co-localized signals of H2A with H1.
Since we are sampling a highly reiterated array of genes, we expect that the stochastic expression of each individual gene within the array should give rise to a continuous expression level over the active phase much as we have seen for histone H1. This is apparently not the case for the core histone genes, suggesting perhaps that there is an additional level of regulation being imposed on the core histone genes at this locus. To address this intriguing possibility, we needed to design an experiment to capture and identify different stages of the S-phase in asynchronous cell cultures and correlate these to transcriptional activity at the different histone genes.
Differential Transcription regulation of the histone genes during S-phase
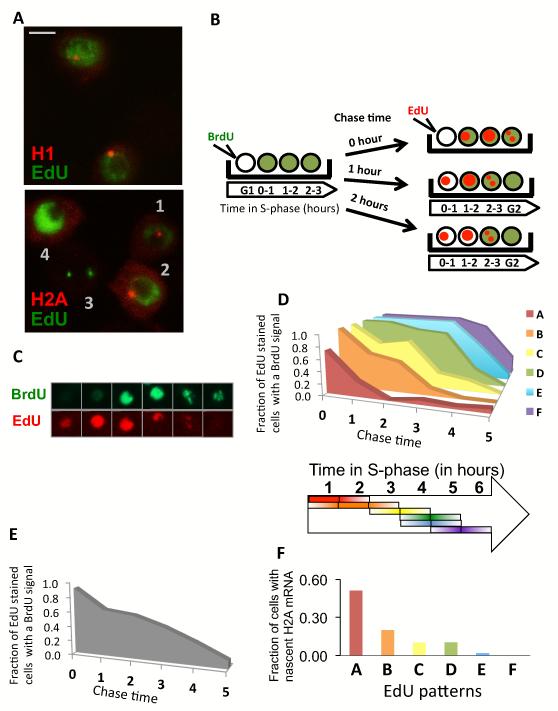
To compare H1 to H2A transcription during S-phase, we devised a scheme using two different nucleotide analogs, each tagged with a different fluorescent dye to carry out a pulse chase experiment. First we used EdU incorporation to detect cells carrying out S-phase specific transcription. As expected, most S-phase cells that incorporated EdU show H1 nascent RNA signals (Figure 3A), indicating that H1 transcription is active essentially throughout the duration of S-phase. By contrast, only a restricted sub-population of S-phase cells shows active H2A transcription consistent with the previous observation of differential transcription patterns between H1 and the core histones. Cells positive for H2A transcription foci also displayed distinct EdU incorporation patterns and/or intensities compared to cells without foci (see Figure 3A cells 1, 2 vs to 3, 4). This result suggests that the H2A pattern of expression is perhaps restricted to a limited period during the course of S-phase. To test this notion, we designed a two-step pulse chase labeling procedure that would allow a more refined parsing of S-phase based on nucleotide incorporation staining patterns, and that could be coupled to visualizing nascent transcript production, without perturbing the cell cycle as imposed by standard drug induced synchronization methods. As schematized in figure 3B, we used increasing chase times between the incorporation of two different color-coded nucleotide analogs EdU-red and BrdU-green. Thus, within a given group of unsynchronized cells, the first nucleotide analog (BrdU-green) that was added should be incorporated during replication. These cells were then washed to remove the unincorporated 1st nucleotide analog and allowed to grow for a “chase period” of one hour or longer. The second nucleotide analog (EdU-red) was then added so that it would be incorporated by the cells in S-phase rendering them red. Because of the predetermined chase times (i.e. 1hr, 2hr, 3hr, etc.), we can extrapolate for the one hour chase time that the cells stained only in red were fixed in the first hour of S-phase. Likewise, if we use a 2 hours chase period, the cells stained only in red represent those labeled within the first 2 hours of S-phase. Thus, by incorporating incrementally longer chase times we can determine roughly which period within S-phase each cell was fixed simply by observing the pattern of red and green stained cells. Using this procedure we were able to temporally order the different patterns of EdU and BrDU stained cells in the asynchronous population (Figure 3B,C). As expected, with increasing chase times, the proportion of Edu stained cells overlapping with BrdU positive cells decreases (Figure 3E). There is no remaining overlap when chase times exceed the length of S-phase which in our culture condition is between 5 and 6 hours. By analyzing the variegated patterns of red only (EdU) stained cells, we distinguished 6 EdU staining patterns representing periods of the entire S-phase from early S to late S (Figure 3D). We then quantitated the number of H2A nascent RNA transcripts found in any given sub-population of cells caught in a specific period of the cell cycle (Figure 3F). Combining EdU with H2A mRNA FISH, this procedure allowed us to unambiguously determine that drosophila S2 cells are most likely to transcribe high levels of H2A during the first two hours of early S-phase and very little during the last 2 hours of late S phase (Figure 3F). By contrast H1 transcription occurs throughout the S-phase with equal frequency within all 5 stages. These findings strongly suggest that indeed H1 transcription is regulated differentially from the core histone genes and extends our previous finding that H1 uses TRF2 at its core promoter while H2A, H2B, H3 and H4 use TBP (Isogai et al., 2007).
Figure 3.
Differential Expression of H1 and H2A during S-phase. (A) Co-staining of H1 or H2A mRNA through FISH (Red) with EdU incorporation (green), marking S-phase. (B) Schematic of the double pulse chase experiment for S-phase timing. (C) Co-staining of BrdU (green) with EdU (red) when using a 90 minutes chase between BrdU and EdU incubations. (D) Upper panel, Association of EdU staining patterns with BrdU staining as a function of chase time. Each colored graph (A to F) represents one of 6 distinct EdU staining patterns. Lower panel, Repartition of EdU staining patterns across S-phase. Each colored bar represents one independently detectable EdU staining pattern (N=80-166). Lower Panel, The length of the bar on the arrow represents approximately when an EdU pattern is occurring the most during S-phase. (E) Fraction of cells that are stained by Edu and BrdU both in function of chase time. (F) Fraction of cells with nascent H2A mRNA for cells within each different EdU staining pattern (N=68-103). Each colored bar corresponds to a EdU staining pattern with the same color code from panel (D).
Non Prototypic Core Promoter Complex Assembled at the Histone Locus
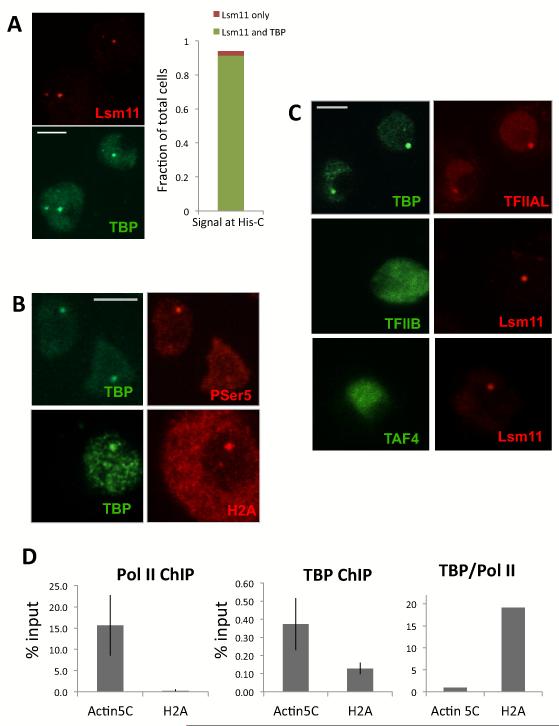
In order to determine the composition of the Pre-Initiation Complex (PIC) and General Transcription Factors (GTFs) at the His-C locus we first used Drosophila antibodies specific against the TATA Binding Protein (TBP) to detect binding to the histone gene promoters. These experiments confirmed a strong TBP signal associated with the His-C locus (Figure 4A). Surprisingly, however, the association of TBP with the histone locus was not dependent on the phase of the cell cycle since its co-localization with Lsm11 is coincident 90% of the cases, and Lsm11 itself is present in most cells (90%) of an asynchronous population. Thus unlike RNA Pol II, TBP does not appear to be recruited to the His-C locus only during S-phase. As expected, TBP is also present at the His-C locus when it is transcribed as it co-localizes with the P-ser5 Pol II and H2A transcript signals in the nucleus (Figure 4B). We then tested various antibodies targeting Pol II, GTFs and the TAFs (TFIIE, TFIIF, TAF4, not shown). Remarkably, TFIIA was the only other PIC member we tested that could be detected at the histone locus. As expected TFIIA co-localizes precisely with TBP, strongly indicating its association with His-C locus also does not depend on the cell cycle (Figure 4C). We were particularly surprised to find that no detectable TFIIB or TAF4 could be found associated with the His-C locus (Figure 4C and Figure S5) despite both antibodies signal being specific (Figure S4).
Figure 4.
TBP and TFIIA accumulation at the histone locus in interphase. (A) Left panel, co-staining of Lsm11 antibody (red) with TBP antibody (green). Right panel, distribution of Lsm11 and TBP signal at His-C among the total number of cells, using Lsm11 signal as reference. (B) Co-staining of TBP (right) with Pol II P-Ser5 or with H2A mRNA FISH (left). (C) Upper panel, co-staining of TBP (green) with TFIIAL (red). middle panel, co-staining of TFIIB (green) with Lsm11 (red). Lower panel, co-staining of TAF4 (green) with Lsm11 (red). (D) Left and middle panel, chromatin immunoprecipitation of Pol II and TBP on Actin5C and H2A promoter, error bar is Standard Deviation of three independent ChIP. Right panel, ratio of TBP ChIP to Pol II ChIP on Actin5c and H2A promoter, normalized to 1 for Actin5C.
To confirm the existence of a preloaded TBP at the histone gene promoter, we used an independent Chromatin Immunoprecipitation method. To this end, we obtained specific and potent polyclonal antibodies directed against RNA Pol II and dTBP and carried out ChIP-PCR experiments at the His-C locus as well as some control genes. As expected, both TBP and Pol II were easily detected occupying the promoter fragments of the conventional very active gene Actin5C (Figure 4D). Whereas little Pol II could be found at the H2A promoter, H2A being mostly inactive in asynchronous cells, a TBP signal could be detected in the same order of magnitude as on Actin5C. The ratio of TBP over Pol II is 20:1 at the H2A promoter compared to the “house keeping” Actin5C promoter (Figure 4D) confirming a preloaded TBP during interphase.
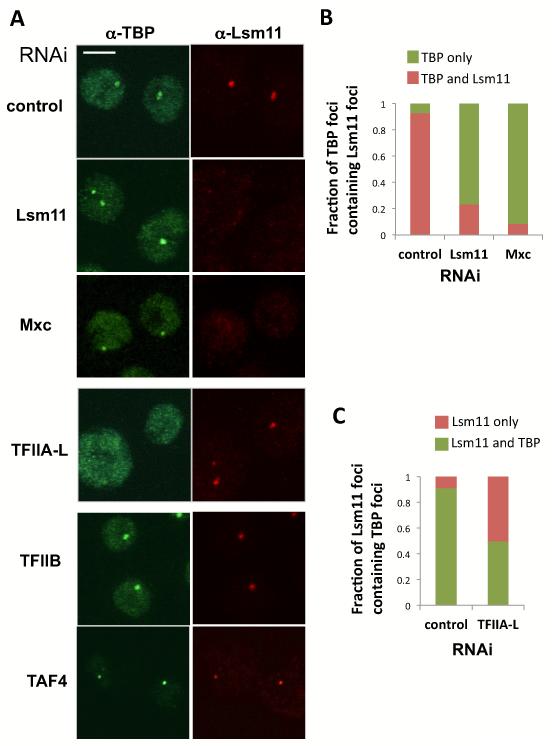
We next assessed which factors are required for TBP to co-localize at the His-C locus (Figure 5). The histone locus is associated with a self-organized structure composed of multiple proteins called the Histone Locus Body (HLB). As a first step in probing the polypeptide requirements for a functional transcriptional complex associated with the HLB, we knocked down two well established components of the HLB, Lsm11 and Mxc (White et al., 2007; 2011). Depleting S2 cells of these 2 HLB markers had no detectable effect on TBP accumulation. Indeed, we could easily detect the same intense TBP signal in Lsm11 and Mxc knock down cells as in control RNAi treated cells with a strong TBP/His-C signal found at the periphery of the nucleolus where His-C is usually located (Figure 5A Row 1 to 3). Consistent with this finding, we detected very few cells with an Lsm11 signal associated with TBP in the knocked down cells (Figure 5A and B). Our results also indicate that loss of Mxc indeed disrupts the HLB, but not the binding of TBP to the His genes. Next we monitored the presence of TBP at the His-C while knocking down factors known to interact with TBP but that should not affect HLB assembly. Our first surprise was that the TAF subunits or holo-TFIID also did not appear to be required for TBP-promoter association since complete depletion of the cornerstone subunit TAF4 whose loss has been shown to disrupt the entire core TFIID complex (Wright et al., 2006) had little or no effect on the localization or intensity of TBP at the His-C locus. As expected TAF4 knock down did noticeably reduce overall TBP levels in the nucleus where it forms the functional TFIID complex but without reducing TBP levels at His-C (Figure 5A). In stark contrast, knock-down depletion of TFIIA-L severely reduced the proportion of cells with TBP at the His-C locus (Figure 5 A,C) with half of the cells showing a total absence of TBP accumulation at His-C while the rest of the cells had a noticeably weakened TBP signal (not shown). As expected, control knockdown cells showed 90% co-localization of TBP with Lsm11 (Figure 5A,C) whereas KD cells showed very little co-localization. Remarkably, depletion of TFIIB had no detectable effect on TBP localization at the His-C locus (Figure 5) consistent with its absence at these loci. These data suggest a strong functional role for a TBP-TFIIA specific complex at His-C while the other TAF subunits of TFIID and TFIIB appear to be largely dispensable.
Figure 5.
TBP depends on TFIIA but not HLB factors, TFIIB or TFIID for binding to the histone locus. (A) Representative co-staining of TBP (green) with Lsm11 (red) in S2 cells after dsRNA treatment. From first to last row, cells after dsRNA treatment against control, Lsm11, Mxc, TFIIA-L, TFIIB, TAF4. (B) Fraction of TBP dots at His-C showing co-localization with a Lsm11 signal in cells depleted of Lsm11 or Mxc. Average of 2-3 RNAi experiments with standard deviations (N=96-169). (C) Fraction of Lsm11 dots at His-C showing co-localization with a TBP signal in cells depleted of TFIIA-L. Average of 2-3 RNAi experiments with standard deviations.
Functional requirement for His-C transcription
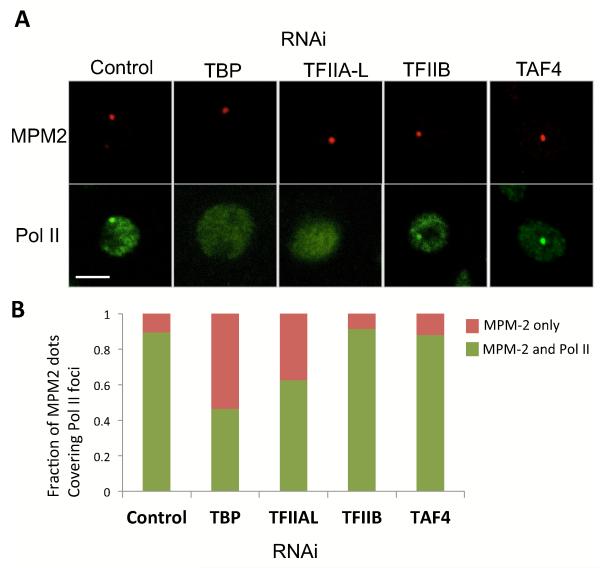
We next sought to directly test the potential functional requirements of various PIC components in driving histone gene transcription. However, as expected, knock down of many of the PIC subunits indirectly but significantly blocked cell cycle progression circumventing our ability to distinguish histone transcription effects that occur exclusively during S-phase from a more general indirect effect on the cell cycle (not shown). In order to bypass this complication we designed experiments to assess His-C transcription activity at the single cell level. Our approach involved fixing cells at various time points after RNAi treatment and then staining them for both MPM-2 and P-Ser5-Pol II to score for the sub-population of actively dividing cells that remained. MPM-2 staining at His-C depends on the phosphorylation of Mxc by the Cdk2-CycE kinase complex that occurs only at the onset of S-phase. Therefore, cells with positive MPM2 staining are likely to have entered S-phase as illustrated in Figure 1. We again used the MPM-2 staining as a reference to find the Pol II “transcription” signal at the His-C locus. In this manner, we could score just those cells that have not yet been blocked in the cell cycle but may have already been depleted of specific transcription factors by RNAi treatment. As shown in Figure 6, the knock down of TBP and TFIIA-L significantly inhibited Pol II accumulation at the histone genes as detected by our single cell assay. TBP knock down reduces the total proportion of MPM-2 positive cells from 30% to 10% (not shown) suggesting that TBP depletion likely indirectly blocks the cell cycle. Remarkably, among the cells that manage to enter S-phase, as marked by MPM-2 staining, more than half show no detectable Pol II accumulation. This single cell analysis thus provides evidence that loss of TBP blocks His-C transcription of cells that are still able to enter S-phase, confirming a major direct role for TBP in the transcription of the His genes. A similar functional requirement can also be assigned to TFIIA-L as we find that 40% of the cells depleted of TFIIA enter S-phase without transcribing the His genes. By contrast TAF4 knock down had little or no effect on His-C transcription while TFIIB knock down showed minimal effects with approximately 90% of Pol II accumulation still active at the histone locus in the sub-population of cells that remain attached to the cover slip after RNAi treatment. These findings are consistent with the notion that the PIC at the histone locus likely involves an atypical Pol II machinery wherein TFIIA and TBP play a critical functional role while TFIIB and the TAFs appear largely dispensable. To gain further insight into the functional role of the TBP-TFIIA complex at His-C we next sought to image the movement of TBP within the Histone locus in live cells.
Figure 6.
TBP and TFIIA are required for Pol II transcription of the His genes. (A) Unsynchronized S2 cells were stained for Pol II P-Ser5 (green) and MPM2 (red) after treatment with dsRNA for knock down of TBP, TFIIA-L, TFIIB, and TAF4. Upper panel shows representative pictures. (B) Fraction of MPM-2 dot that co-localized with Pol II signal dots after depletion of different PIC components. Average of 2-3 RNAi experiments (N=72-103, 26 for TFIIB).
Imaging TBP and TRF2 Binding Kinetics at the Histone Locus in Live Cells
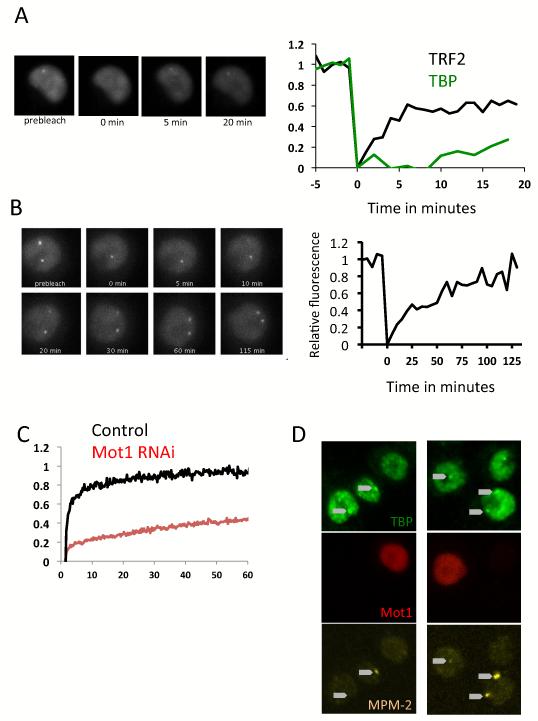
We generated cell lines carrying TBP or TRF2 fused to GFP at the N-Terminus. The GFP-TBP is found in the nucleus but not the nucleolus and accumulates as a bright dot at His-C in most cells of an asynchronous population (Figure S6A). GFP-TRF2 could also be localized at His-C (Figure S6B), but the enrichment is significantly lower than that of TBP, in keeping with TRF2 being present only at the H1 promoter (Isogai et. al., 2007). We next tracked the mobility of TBP and TRF2 at the His-C by FRAP. During initial attempts where we tracked them for only 20 minutes (Figure 7A left panel), we could detect 60% recovery for TRF2 in the first 5 minutes, whereas TBP showed less than 15% recovery. This was in contrast to the behavior of photo-bleached GFP-TBP at random locations in the nucleoplasm other than the His-C (Figure 7C, black line) where we observed a 95% recovery in 60 seconds. As has been reported previously, FRAP in the nucleoplasm gives information about various types of TBP populations - freely diffusing, interacting with non-specific DNA and more stably interacting with specific promoter DNA sequences (TATA Boxes). Our results suggest that the behavior of TBP globally in the nucleoplasm is quite distinct from its behavior at the His-C locus and that it also behaves quite differently from TRF2 at the His-C locus. To maximize the likelihood that the behavior of our GFP-TBP would largely reflect the behavior of endogenous TBP, we expressed it at levels comparable to that of endogenous TBP (Figure S6C) In addition, we confirmed that like endogenous TBP, the GFP-TBP depends on TFIIA for its accumulation at His-C as shown by RNAi of TFIIAL (Figure S6A). To further assess the functionality of GFP-TBP we tested the viability of cells whose only source of TBP is GFP-TBP. We generated a cell line expressing a GFP-TBP version wherein the gene encoding the C-Terminal half of the melanogaster molecule has been replaced by the coding region of Drosophila Grimshawi. This construct encodes the same amino acid sequence but uses a different DNA coding sequence that is no longer targeted by the DsRNA we employed to knock down endogenous TBP. After a few days of treatment, cells that only expressed endogenous TBP died, whereas clones transfected with the chimeric TBP grew exponentially (Figure S6D), demonstrating full functionality of the GFP-TBP fusion.
Figure 7.
FRAP analysis of TBP and TRF2 binding kinetics in the nucleoplasm and at the histone gene locus. (A) FRAP recovery over 20 minutes of GFP-TRF2 (left and right panel) or GFP-TBP at the histone locus, average of 10 cells. (B) Representative FRAP recovery of GFP-TBP at the histone locus. GFP-TBP at His-C fully recovers in 120 minutes. (C) FRAP recovery of GFP-TBP in random locations of the nucleoplasm +/− RNAi of Mot1. Average of 20 FRAP, standard errors are <10%. GFP-TBP in the nucleoplasm fully recovers in one minute. (D) Overexpression of Drosophila Mot1 in S2 cells. Mot1 appears in red TBP is stained in green, and MPM-2 in Yellow. Arrow point to the TBP foci location. Images are maximum projections of Z-stacks.
We then repeated our FRAP experiment with GFP-TBP at His-C, but now continued to measure recovery for hours rather than minutes (Figure 7B), anticipating that perhaps TBP, especially in the presence of TFIIA, could be very long lived at the His-C array. With these longer time scale experiments we found that S2 cells creep along the surface in a macrophage like manner. These movements are of limited scale and speed and seem to revolve around the initial position of the cell. We did not find any procedure that would eliminate this cell movement but instead we implemented tracking procedures to follow both the loci and the cell from frame to frame. This approach allowed us to track and image GFP-TBP at His-C revealing that a full recovery after photo-bleaching is attained only after 1 to 2 hours (Figure 7B). Despite the inherent limitations of our tracking method, we could approximate the residence time of TBP at the His-C and it is clearly an order of magnitude longer than TBP in the rest of the nucleoplasm and also much longer than TRF2 at the histone locus. These finding suggest an unusually stable TBP-TFIIA complex formed at the histone gene promoters consistent with the well established finding that TFIIA significantly stabilizes the TBP-DNA interaction in vitro (Weideman et al., 1997).
Previous studies have found that TBP binding to promoter DNA is also influenced by Mot1/BTAF1/HEL89b, an ATPase that helps catalyze the removal of TBP from DNA. We therefore wondered whether part of this unusually long lived binding of TBP/IIA to His-C could be due in part to a limited or restricted effect of at these promoters. To test this notion, we performed both loss of function and gain of function experiments with Mot1. RNAi mediated depletion of Mot1 (Figure 7C) substantially slows down the FRAP recovery of a majority of the TBP population in these cells. It indicates that in the absence of Mot1 it is possible to establish a more stable binding of TBP to DNA throughout the nucleoplasm. Next, to test the idea that perhaps limited access of Mot1 to His-C is what allows our observed stable binding of TBP, we transiently over-expressed a RFP fused Drosophila Mot1/HEL89b in S2 cells (Figure 7D). Many of the cells over-expressing Mot1 showed an enlarged nucleus that could be an indication of difficulties in traversing through mitosis, as well as some reduction of the TBP staining. Importantly, we found that most Mot1 over-expressing cells also did not show the typical accumulation of TBP on the His-C loci (Figure 7D left column). In those very few cases were we could find a TBP foci in Mot1 over-expressing cells, it was generally associated with MPM-2 staining raising the possibility that during S-phase, other factors may also contribute stable binding of TBP at His-C in addition to TFIIA. These experiments taken in aggregate indicate that a TBP containing PIC is preloaded at core histone promoters and that TBP binding is stabilized by and dependent on TFIIA while subject to destabilization by the over-expression of Mot1. By contrast, the PIC formed at the His1 promoter within the His-C locus is comprised of TRF2 instead of TBP and its binding is considerably less stable reflecting a distinct transcriptional regulatory system. In any case, the TBP/TFIIA complex appears to behave in an unusual manner at the His-C locus that significantly alters our view of how distinct PICs can contribute to gene specific regulatory mechanisms.
Discussion
To gain a more complete understanding of the physiologically relevant mechanisms regulating eukaryotic gene expression we set out to probe the transcriptional control of an endogenous gene cluster by single live cell imaging. Using a combination of immunofluorescence, RNA FISH, GFP-tagged factors and FRAP we characterized the composition of initiation complexes and unusual transcription kinetics of the histone genes in Drosophila S2 cells. We found that whereas the 4 core histones genes are co-transcribed, the timing and pattern of H1 expression is quite distinct despite the tight promoter arrangement of all 5 His genes (Figure S7). The core histone genes are expressed through short but intense burst in the very beginning of S-phase whereas Histone H1 is expressed throughout S-phase. We speculate that this separation in the timing, distinct factor requirements and pattern of H1 expression is in keeping with the differential functions assigned to these gene products. The core histone genes encode proteins destined to form a tight nucleosomal octamer whereas histone H1 operates alone by binding to inter-nucleosomal spacer regions to mediate higher order condensed chromatin (Thoma et al., 1979; Lu et al., 2009).
The differential pattern of H1 transcription versus core histones could reflect a mechanism to achieve greater flexibility and access to chromatin during DNA replication to allow S-phase checkpoint control. For example, a common event that can instigate S-phase checkpoint arrest is DNA-damage requiring repair. The continuous transcription of H1 throughout S-phase coupled with its much shorter mRNA half-life makes the system more responsive to checkpoint arrest. One can imagine that a tight but highly responsive control of H1 could be a simple mechanism to avoid a build up of excess H1 protein pools thereby limiting the formation of higher order less accessible chromatin for repair enzymes to reach damaged regions of DNA. On the other hand, building an abundant and stable pool of H2A, H2B, H3 and H4 mRNA early in S-phase could be a way to maintain core nucleosome mediated chromatin integrity during S-phase arrest. We speculate that perhaps such a constant flux of nucleosome production might be a mechanism to prevent the loss of valuable epigenetic information encoded in pools of modified nucleosomes that must be transmitted to newly formed chromatin.
The transcription of both H1 and core histone genes becomes activated at early stages of S-phase suggesting that they are both likely responsive to S-phase signaling initiated by the CyclinE-Cdk2 complex (White et al., 2007). However, it is unclear how core histone gene transcription is shut down early in S-phase while the arrest of H1 transcription only occurs at the end of S-phase. It has also been proposed that the DNA binding factor HERS silences histone gene transcription by formation of heterochromatin at the His-C locus towards the end of S-phase (Ito et al., 2012). This mechanism, if involved, could not account for the early S-phase silencing of the core histone genes, but could represent a more long term silencing of the locus outside of S-phase.
Another intriguing feature of core histone gene expression revealed by our studies is the unusual pattern of transcription dynamics during S-phase. A number of studies have concluded that transcription is largely a stochastic process (Larson et al., 2009). By contrast, our studies of the His-C cluster points to a more concerted and deterministic type of mechanism that may be at play. Various studies measured the in vivo binding rates of transcription factors (McNally et al., 2000; Yao et al., 2006; Elf et al., 2007; Karpova et al., 2008). Other studies of RNA Pol II transcription in living cells have found that initiation can in some case be rather inefficient relative to elongation (Dundr et al., 2002; Darzacq et al., 2007). However, to our knowledge, the binding of lynch pin core promoter factors such as TBP to specific endogenous genes in living cells had not been measured. Our studies of the endogenous histone genes in drosophila cells revealed highly efficient initiation by RNA Pol II and very long dwell times (hours) for TBP. Indeed, we found that TBP is involved in an unusually stable TBP-TFIIA complex that enhances the TBP dwell time at the His locus.
We imagine that such a stable marking of the His-C locus that occurs well before the onset of transcription during S-phase can act to rapidly deploy the transcription machinery by serving as a landing pad for the other PIC components including RNA pol II (Figure S7). Such a preloaded highly stable pioneer TBP-TFIIA complex may provide an efficient mechanism to rapidly direct high levels of His gene transcription activation needed during the short window of early S-phase expression. A stable bookmarked TBP/TFIIA complex could also serve as an insulator protecting the His genes promoter from heterochromatin formation thereby facilitating transcription initiation at the start of S-phase. Such a mechanism could likewise be useful at certain stages of development when spikes of gene activation must occur in narrow time intervals that are followed by efficient shut down. Curiously, there has been one other example of a TBP/TFIIA complex that can be purified from P19 embryonal carcinoma cells but not from more differentiated cells (Mitsiou and Stunnenberg, 2000). Why this dimer complex is so abundant in these cells is not clear, but it may point to a more general role of TBP-TFIIA substituting for TFIID as a physiologically relevant alternative transcription initiation complex.
The rapid and coordinated mechanism of on/off switches modulating transcription initiation we report here is quite distinct from the popular models invoking paused polymerases and reliance on post initiation events to regulate transcription output described in a number of recent studies. It seems likely that diverse mechanisms may have evolved to deal with timing and dynamics of transcription dependent on cell-type and developmental context.
Finally, we were most surprised by the use of alternative PIC subunits even within the His-C locus. First, we confirmed the dependence of H1 transcription initiation on the TBP related factor TRF2, whereas H2A, H2B, H3 and H4 depend on the prototypic TATA Binding Protein (TBP) for their expression. This differential use of core promoter recognition factors could play a key role in the transcriptional control of the His genes, particularly in combination with other gene specific promoter binding factors responsible for directing S-phase triggered His gene transcription. Perhaps even more surprising was the observation that no TFIIB or TAFs could be detected binding to the His gene promoters. We cannot entirely rule out that TFIIB has such a fast on/off rate that we are unable to measure its presence at this locus. However, an equally plausible explanation would be that the PIC at these promoters is substantially different from canonical initiation complexes and neither TFIIB nor TAFs are required to form an active PIC at these promoters. It is also possible that there is another, as yet unidentified functional substitute for TFIIB operating at these promoters.
At this stage we cannot rationalize why the His genes would have evolved to require such a seemingly distinct PIC instead of merely adapting more classical mechanisms such as gene specific DNA binding activators to orchestrate the differential expression of H1 versus core histones during S-phase. In any case, the pre-eminence of a highly stable pre-loaded TBP/TFIIA complex and potential changes in PIC subunit composition suggests that whatever form of PIC does assemble at the His-C locus they represent a significant departure in composition and likely functional specificity from the prototypic house keeping or canonical RNA Poll II promoter recognition apparatus. Perhaps the notion that a universal invariant “basal or general” core promoter machinery is all that is required to mediate transcriptional regulation within a single cell-type in eukaryotic organisms is a concept that has outlived its usefulness.
In summary, single live cell gene expression analysis unmasked a set of unexpected mechanisms regulating transcription of an endogenous gene cluster. Our single cell imaging of core promoter factors in unsynchronized populations of living cells also revealed a surprisingly efficient temporal control of transcription executed by a unique set of stably pre-bound promoter recognition complexes that significantly expands our understanding of the diverse molecular mechanisms that have evolved to accommodate gene specific transcription controlling physiologically critical processes in animal cells.
Experimental procedures
Cell culture
S2 cells were used for all experiments. They were maintained as exponentially growing cultures in M3BPYE unless indicated otherwise (Baum and Cherbas, 2008).
Preparation of stable cell lines expressing transgenes
Expression vectors for transgenes were cotransfected with selectable markers. pCoPuro or pCoBlast (Invitrogen) were used as cotransfectable selectable markers. Stable cell lines were selected with puromycine or blasticidine (concentrations). Transfection was performed with Effectene (Qiagen).
Immunofluorescence
Cells were cultured on uncoated coverslips and fixed with 4% PFA in 1X PBS. Fixed cells were incubated with various concentrations of primary antibody for 2 hours at 25 deg C in PBS 1X Triton 0.1%. Washed in PBS and incubated with secondary antibodies for 1 hour at 25 deg C. Washed in PBS and mounted in Vectashield (Vector Labs). Primary antibodies: Pol II P-Ser5, rabbit polyclonal NB100-1806 from Novus.MPM-2, mouse monoclonal, Fisher Millipore. Lsm11, (Liu et al., 2006). DmTBP, 3C3, mouse monoclonal Tjian lab. DmTFIIB, rabbit polyclonal Tjian lab. DmTFIIAL, rabbit polyclonal Tjian lab (Yokomori et al., 1993). Secondary antibodies were Alexa488, 546, 647 from Molecular Probes.
Gene cloning
cDNA were amplified through PCR (PFX polymerase, Invitrogen), and cloned with the Gateway cloning system (Invitrogen) producing Entry clones which were fully sequenced. Fusion with Fluorescent Protein (FP) under the control of various promoters was obtained through recombination of Entry clones with destination vectors from the Drosophila Gateway Vector Collection (DGVC,).
Gene, cDNA (DGRC), destination vector, Description:
TBP, LD44083, pCGW, N-terminal fusion of GFP to TBP under the control of a Copia promoter
Lsm11, SD11312, pCRW, N-terminal fusion of mRFP1 to TBP under the control of a Copia promoter
TFIIB, RE29729,pCGW, N-terminal fusion of GFP to TBP under the control of a Copia promoter
RpII215-R741H, RE17843,pAGW, N-terminal fusion of GFP to Pol II biggest subunit rendered resistant to alpha-amanitin under the control of a Actin5C promoter
To obtain RpII215-R741H, RpII215, the biggest subunit of Drosophila melanogaster RNA Pol II, was cloned in a Gateway Entry clone. The insert comes from DGRC cDNA RE17843. It contained 4 missense mutations that were reversed and 2 silent mutations (C3192T, A4764G). The 2221CGT codon was changed for CAC, imitating the R741H mutation of the pPC4 vector. It renders the subunit resistant to alpha-amanitin.
pCGW and pCRW are Gateway destination vectors obtained from modification of vectors from the DGVC by introduction of a Copia promoter cloned from pCoPuro. Cloning of hybrid TBP. The 3′ conserved region of Drosophila melanogaster (starting at basepair 571/residue 191) was replaced by the sequence of Drosophila Grimshawi (EST 56956424 from DGRC).
Supplementary Material
Highlights for B. Guglielmi et al. Molecular Cell
Atypical Transcription Complex at His-C.
Probing Transcription Coupled to Cell Cycle.
Acknowledgments
The authors wish to thank Y. Isogai, M. Marr and K. Wright for cells vectors and various materials. J. Gall for anitbodies. T. Lionnet D. Larson and R. Singer and other members of the Janelia Farm Imaging Transcription Consortium for protocols, software and advices. H. White at JFRC Tissue Culture Facility. S. Ruzin, D. Schichnes at CNR Biological Imaging Facility (UC Berkeley) and H. Aaron at Molecular Imaging Center (UC Berkeley) for microscopy material and technical help. D. Rio and X. Darzacq for advices and critical reading of the manuscript. All members of our laboratory for technical help, comments and advice.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Albright SR, Tjian R. TAFs revisited: more data reveal new twists and confirm old ideas. Gene. 2000;242:1–13. doi: 10.1016/s0378-1119(99)00495-3. [DOI] [PubMed] [Google Scholar]
- Baum B, Cherbas L. Drosophila cell lines as model systems and as an experimental tool. Methods Mol Biol. 2008;420:391–424. doi: 10.1007/978-1-59745-583-1_25. [DOI] [PubMed] [Google Scholar]
- Darzacq X, Shav-Tal Y, De Turris V, Brody Y, Shenoy SM, Phair RD, Singer RH. In vivo dynamics of RNA polymerase II transcription. Nat Struct Mol Biol. 2007;14:796–806. doi: 10.1038/nsmb1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darzacq X, Yao J, Larson DR, Causse SZ, Bosanac L, De Turris V, Ruda VM, Lionnet T, Zenklusen D, Guglielmi B, et al. Imaging transcription in living cells. Annual Review of Biophysics. 2009;38:173–196. doi: 10.1146/annurev.biophys.050708.133728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dundr M, Hoffmann-Rohrer U, Hu Q, Grummt I. A Kinetic Framework for a Mammalian RNA Polymerase in Vivo. Science. 2002 doi: 10.1126/science.1076164. [DOI] [PubMed] [Google Scholar]
- Elf J, Li G-W, Xie XS. Probing transcription factor dynamics at the single-molecule level in a living cell. Science. 2007;316:1191–1194. doi: 10.1126/science.1141967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampsey M. Molecular genetics of the RNA polymerase II general transcriptional machinery. Microbiology and Molecular Biology Reviews. 1998;62:465–503. doi: 10.1128/mmbr.62.2.465-503.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isogai Y, Keles S, Prestel M, Hochheimer A, Tjian R. Transcription of histone gene cluster by differential core-promoter factors. Genes Dev. 2007;21:2936–2949. doi: 10.1101/gad.1608807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito S, Fujiyama-Nakamura S, Kimura S, Lim J, Kamoshida Y, Shiozaki-Sato Y, Sawatsubashi S, Suzuki E, Tanabe M, Ueda T, et al. Epigenetic Silencing of Core Histone Genes by HERS in Drosophila. Mol Cell. 2012;45:494–504. doi: 10.1016/j.molcel.2011.12.029. [DOI] [PubMed] [Google Scholar]
- Karpova TS, Kim MJ, Spriet C, Nalley K, Stasevich TJ, Kherrouche Z, Heliot L, Mcnally JG. Concurrent Fast and Slow Cycling of a Transcriptional Activator at an Endogenous Promoter. Science. 2008;319:466–469. doi: 10.1126/science.1150559. [DOI] [PubMed] [Google Scholar]
- Kedes LH. Histone genes and histone messengers. Annu Rev Biochem. 1979;48:837–870. doi: 10.1146/annurev.bi.48.070179.004201. [DOI] [PubMed] [Google Scholar]
- Kozhevnikova EN, van der Knaap JA, Pindyurin AV, Ozgur Z, van Ijcken WFJ, Moshkin YM, Verrijzer CP. Metabolic enzyme IMPDH is also a transcription factor regulated by cellular state. Mol Cell. 2012;47:133–139. doi: 10.1016/j.molcel.2012.04.030. [DOI] [PubMed] [Google Scholar]
- Larson DR, Singer RH, Zenklusen D. A single molecule view of gene expression. Trends in Cell Biology. 2009;19:630–637. doi: 10.1016/j.tcb.2009.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J-L, Buszczak M, Gall JG. Nuclear bodies in the Drosophila germinal vesicle. Chromosome Res. 2006;14:465–475. doi: 10.1007/s10577-006-1062-5. [DOI] [PubMed] [Google Scholar]
- Lu X, Wontakal SN, Emelyanov AV, Morcillo P, Konev AY, Fyodorov DV, Skoultchi AI. Linker histone H1 is essential for Drosophila development, the establishment of pericentric heterochromatin, and a normal polytene chromosome structure. Genes Dev. 2009;23 doi: 10.1101/gad.1749309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahajan K, Bin Fang, Koomen JM, Mahajan NP. H2B Tyr37 phosphorylation suppresses expression of replication-dependent core histone genes. Nat Struct Mol Biol. 2012:1–10. doi: 10.1038/nsmb.2356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marzluff WF, Duronio RJ. Histone mRNA expression: multiple levels of cell cycle regulation and important developmental consequences. Curr Opin Cell Biol. 2002;14:692–699. doi: 10.1016/s0955-0674(02)00387-3. [DOI] [PubMed] [Google Scholar]
- McNally JG, Müller WG, Walker D, Wolford R, Hager GL. The Glucocorticoid Receptor: Rapid Exchange with Regulatory Sites in Living Cells. Science Signaling. 2000;287:1262. doi: 10.1126/science.287.5456.1262. [DOI] [PubMed] [Google Scholar]
- Mitsiou DJ, Stunnenberg HG. TAC, a TBP-sans-TAFs complex containing the unprocessed TFIIAalphabeta precursor and the TFIIAgamma subunit. Mol Cell. 2000;6:527–537. doi: 10.1016/s1097-2765(00)00052-6. [DOI] [PubMed] [Google Scholar]
- Myers LC, Kornberg RD. Mediator of transcriptional regulation. Annu Rev Biochem. 2000;69:729–749. doi: 10.1146/annurev.biochem.69.1.729. [DOI] [PubMed] [Google Scholar]
- Orphanides G, Lagrange T, Reinberg D. The general transcription factors of RNA polymerase II. Genes Dev. 1996;10:2657–2683. doi: 10.1101/gad.10.21.2657. [DOI] [PubMed] [Google Scholar]
- Roeder RG. Nuclear RNA polymerases: role of general initiation factors and cofactors in eukaryotic transcription. Meth Enzymol. 1996;273:165–171. doi: 10.1016/s0076-6879(96)73016-1. [DOI] [PubMed] [Google Scholar]
- Thoma F, Koller T, Klug A. Involvement of histone H1 in the organization of the nucleosome and of the salt-dependent superstructures of chromatin. J Cell Biol. 1979 doi: 10.1083/jcb.83.2.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weideman CA, Netter RC, Benjamin LR, McAllister JJ, Schmiedekamp LA, Coleman RA, Pugh BF. Dynamic interplay of TFIIA, TBP and TATA DNA. Journal of Molecular Biology. 1997;271:61–75. doi: 10.1006/jmbi.1997.1152. [DOI] [PubMed] [Google Scholar]
- White AE, Burch BD, Yang X-C, Gasdaska PY, Dominski Z, Marzluff WF, Duronio RJ. Drosophila histone locus bodies form by hierarchical recruitment of components. J Cell Biol. 2011;193:677–694. doi: 10.1083/jcb.201012077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White AE, Leslie ME, Calvi BR, Marzluff WF, Duronio RJ. Developmental and cell cycle regulation of the Drosophila histone locus body. Mol Biol Cell. 2007;18:2491–2502. doi: 10.1091/mbc.E06-11-1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright KJ, Marr MT, Tjian R. TAF4 nucleates a core subcomplex of TFIID and mediates activated transcription from a TATA-less promoter. Proc Natl Acad Sci USA. 2006;103 doi: 10.1073/pnas.0605499103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao J, Munson KM, Webb WW, Lis JT. Dynamics of heat shock factor association with native gene loci in living cells. Nature. 2006;442:1050–1053. doi: 10.1038/nature05025. [DOI] [PubMed] [Google Scholar]
- Yokomori K, Admon A, Goodrich JA, Chen JL, Tjian R. Drosophila TFIIA-L is processed into two subunits that are associated with the TBP/TAF complex. Genes Dev. 1993;7:2235–2245. doi: 10.1101/gad.7.11.2235. [DOI] [PubMed] [Google Scholar]
- Zhou H, Kaplan T, Li Y, Grubisic I, Zhang Z, Wang PJ, Eisen MB, Tjian R. Dual functions of TAF7L in adipocyte differentiation. Elife. 2013;2:e00170. doi: 10.7554/eLife.00170. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.