Abstract
Breast cancer is a heterogeneous disease consisting of different biological subtypes, with differences in terms of incidence, response to diverse treatments, risk of disease progression, and sites of metastases. In the last years, several molecular targets have emerged and new drugs, targeting PI3K/Akt/mTOR and cyclinD/CDK/pRb pathways and tumor microenvironment have been integrated into clinical practice. However, it is clear now that breast cancer is able to develop resistance to these drugs and the identification of the underlying molecular mechanisms is paramount to drive further drug development. Autophagy is a highly conserved homeostatic process that can be activated in response to antineoplastic agents as a cytoprotective mechanism. Inhibition of autophagy could enhance tumor cell death by diverse anti-cancer therapies, representing an attractive approach to control mechanisms of drug resistance. In this manuscript, we present a review of autophagy focusing on its interplay with targeted drugs used for breast cancer treatment.
Keywords: breast cancer, autophagy, ATG, Chloroquine, Hydroxychloroquine, ACD
1. Introduction
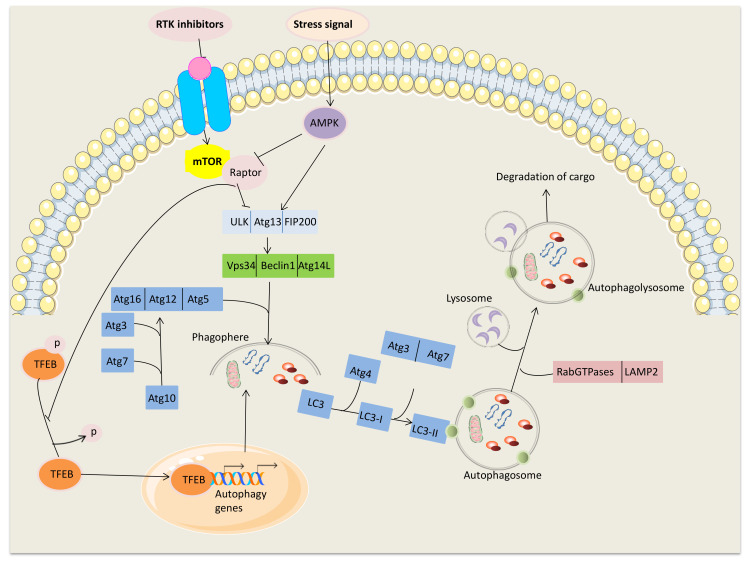
Autophagy literally means “self-eating”. It is a complex multi-faceted process in which a cell destroys old or defective cellular components, which can later be recycled by the cell to meet its metabolic needs [1]. In mammalian cells, autophagy can be classified into three main categories: macroautophagy, microautophagy, and chaperone-mediated autophagy. In particular, the lysosomal degradation pathway macroautophagy (henceforth, autophagy), represents an adaptive response to different forms of stress, such as nutrient deprivation, growth factor depletion, infection, hypoxia or cellular damages induced by chemical agents [2]. In this context, autophagy could exert cytoprotection, by selectively eliminate potential cytotoxic materials such as misfolding proteins or damaged mitochondria, in a process defined “selective autophagy”, or supplying nutrients and energy during fasting and other forms of stress [3]. In addition to the cytoprotective function, autophagy has been proposed as a mechanism of cell death, given that features of autophagy have been observed in dying cells: this has been referred to autophagic cell death (ACD) [4]. Accordingly, autophagic activity modulates many pathologies, including neurodegeneration, cancer, and infectious diseases [5,6]. During autophagy, cellular proteins and organelles undergo a catabolic process in which they are engulfed by autophagosomes, digested in lysosomes, and recycled to sustain cellular metabolism. In mammals, autophagy is regulated by ~16–20 high conserved autophagy-related genes (ATG), that control the process of autophagosome formation and its fusion with lysosomes. This process involves four steps, so-called initiation, nucleation, maturation, and degradation, in which different ATG proteins act at different levels [7]. The initiation and nucleation proteins promote the formation of the autophagic vesicle membrane, with the recruitment of ATGs proteins to a specific subcellular location termed the phagophore assembly site (PAS). Gradual elongation of the curved isolation membrane results in expansion of the phagophore into a double-membraned vesicle, termed the autophagosome, thereby trapping the engulfed cytosolic material as autophagic cargo. Then the autophagosome fuses with the lysosomal membrane to form an autolysosome, followed by the degradation of the autophagic body together with its cargo [8]. The main ATGs complexes, such as the Unc-51 like autophagy activating kinase (ULK1) complex, the Beclin–ATG4-1-VPS34 class III phosphoinositide 3-kinase (PI3K) complex, the ATG5-ATG12-ATG16 complex, and ATG8/LC3 complex, involved in this process, have been extensively reported in other review [9,10,11]. An overview of the autophagy process [12], its induction [13,14] and regulation [15] have been reported in Figure 1.
Figure 1.
The Unc-51 like autophagy activating kinase (ULK1) serine threonine kinase complex, involving ULK1, FIP200, and ATG13, acts in the initiation process, phosphorylating multiple downstream factors. Then, the complex involving Beclin1, VPS34, a class III phosphatidylinositol-3-kinase (PI3K) and ATG14 acts in autophagosome nucleation step. During the maturation step, the ATG7 and ATG10 conjugate ATG5 to ATG12, and ATG7 and ATG3 conjugate microtubule-associated protein 1 light chain 3 (LC3) (ATG8) to the lipid phosphatidylethanolamine (PE). The ATG5-ATG12 conjugate forms a complex with ATG16, and the ATG5-ATG12-ATG16 complex gets anchored onto phosphoinositol 3-phosphate generated by VPS34 on emerging autophagosomal membranes through WD repeat domain, phosphoinositide-interacting 2b (WIPI-2b). The complex is efficient for the LC3 conjugation system. LC3 is first conjugated with PE. ATG4 plays a role in lipoxidating LC3 to LC3-I and exposes the C-terminal glycine of LC3 for the subsequent conjugation of PE. PE is conjugated to the C-terminal glycine of LC3-I, and this conjugation needs to be catalyzed by the E1-like enzyme ATG7 and the E2-like enzyme ATG3. The autophagosomal membrane is conjugated with LC3-PE. During fusion with the lysosome, the LC3-II bound to the outer membrane is cleaved and recycled by ATG4, while LC3-PE associated with the inner membrane is degraded by lysosomal proteases along with the cargo of the autophagosome, thus recycling amino acids, fatty acids, and nucleotides.
2. Main Mechanisms Inducing Autophagy
A major regulator of autophagy is the mammalian target of rapamycin (mTOR) pathway, which consists of 2 distinct signaling complexes known as mTORC1 and mTORC2, but only mTORC1 directly regulates autophagy. In high-nutrient conditions, ATG13 and ULK1 are both directly bound and phosphorylated by mTORC1 and remain inactive in this phosphorylated form. Upon amino-acids deprivation, mTOR repression shifts cellular metabolism toward autophagy and recycling of cytosolic constituents. mTORC1 sites on ULK1 are dephosphorylated and ULK1 dissociates from mTORC1. Concomitantly, ULK1 undergoes autophosphorylation, followed by phosphorylation of ATG13 and FIP200 [3,7,11].
A decrease in intracellular energy results in activation of homeostasis regulatory kinases 5′ AMP- activated protein kinase (AMPK), a serine/threonine-protein kinase acting as a sensor of low cellular energy levels as in glucose starvation. The activation of AMPK can induce the autophagic process through two different mechanisms: (1) inhibition of the mTOR protein kinase complex, or (2) by bypassing mTOR by directly phosphorylation of ULK1, VPS34 and Beclin1 [16,17].
In the last years, the research group of Ballabio et al. identified a new key regulator of autophagy in TFEB (transcription factor EB), which is a master transcription factor controlling the expression of target genes involved in autophagy. Also, TFEB is negatively regulated by mTORC1 and released upon starvation, when TFEB translocates from the cytoplasm (where it normally resides) to the nucleus where it is active and regulates the expression of its target genes involved in lysosomal biogenesis [14,18,19,20] (Figure 1).
For a long time, autophagy was considered a non-selective pathway activated in response to cellular stresses such as starvation or nutrient deprivation in order to provide essential amino acids and nutrients to sustain cellular growth. However, in the last years, extensive evidence has reported the key role of autophagy in selective degradation of dysfunctional organelles, protein aggregates, and intracellular pathogens [7,21]. Selective processes including removal of peroxisomes (pexophagy) [22] endoplasmic reticulum (ERphagy) [23], ribosomes (ribophagy) [24], lipid droplets (lipophagy) [25], invading microbes (xenophagy) [26], protein aggregates (aggrephagy) [27,28] and damaged mitochondria (mitophagy) [29] have been reported. As well, selective cargo receptors, such as p62/SQSTM 1, BNIP3L (BCL2-interacting protein 3-like) NBR1 (Next to BRCA1), CALCOCO2 (calcium-binding and coiled-coil domain-containing protein 2), and OPTN (optineurin), needed for selective degradation process, have been widely studied. Indeed, mutations in the molecules involved in selective autophagy have been associated with susceptibility to a variety of human diseases [7].
3. Role of Autophagy in Cancer Diseases
Autophagy has dual roles in cancer, acting both as tumor suppressor by preventing the accumulation of damaged proteins and organelles and as a mechanism of cell survival that can promote the growth of established tumors [30,31]. Selective autophagy, by inhibiting accumulation of oncogenic p62/SQSTM 1 protein aggregates and damaged organelles like mitochondria, exerts a key role in cellular quality control, reducing the production of reactive oxygen species (ROS), preventing chronic tissue damage and inflammation, and promoting genome stability [32,33]. In this regard, the knockdown of p62/SQSTM 1 in autophagy-defective cells prevented ROS and the DNAdamage response [34]. Autophagy defects, e.g., heterozygous knockdown Beclin1 and ATG7 in mice, are associated with susceptibility to metabolic stress, genomic damage, and tumorigenesis [35]. In contrast, excessive stimulation of autophagy due to Beclin1 overexpression can inhibit tumor development [36].
Other evidence indicates that the predominant role of autophagy in cancer cells is to confer stress tolerance, which serves to maintain tumor cell survival [37,38]. Cancer cells seem to activate autophagy to sustain high proliferation level that require high ATP demand. Cytotoxic and metabolic stresses, such as hypoxia and nutrient deprivation, can activate autophagy to recycle cellular components to sustain survival of cancer cells [37,39,40]. Elevated levels of autophagy have been observed in many tumors. In particular, Beclin1 was upregulated in colorectal cancer, gastric cancer, liver cancer, breast cancer, and cervical cancer [41,42,43,44], suggesting that the enhancement of autophagy can promote tumorigenesis. Inhibition of autophagy in tumor cells has been shown to increase the efficacy of anti-cancer drugs, supporting its key role in cancer [45,46]. Other findings reported that mutations in H-RAS or K-RAS in cancer cells can trigger a high level of autophagy [47,48], and in these cells, suppression of essential autophagy proteins inhibit cell growth, indicating that autophagy is an essential survival pathway in these tumors [49]. A relevant role of autophagy in cancer metastasis development has also been reported. This association was found in breast cancer metastasis [50,51], melanoma metastases [52], hepatocellular carcinoma [53], and glioblastoma [54]. In addition, autophagy seems to play a role in the survival of distant colonies in response to various environmental stress including hypoxia, nutrient deprivation, and detachment from the extracellular matrix (ECM) [55,56,57,58]. Beyond the role in tumorigenesis, supported by extensive evidence, autophagy may also play a role in cancer cell death and tumor suppression. The induction of ACD has been proposed as a mechanism of cell death, because accumulation of autophagosomes and autolysosomes have been observed in the cytoplasm of dying cells, without activation of apoptosis [59]. ACD could occur in conditions of prolonged stress, and consequently high activation of autophagy, which could cause an excessive protein and organelle turnover that overwhelm the capacity of the cell [60]. A form of ACD, called ‘‘autosis’’, has been reported during starvation in dying cells that showed not apoptosis or necrosis, but morphological characteristics of autophagy; this cell death could be prevented by inhibition of autophagy. [61,62]. In this context, an interplay between autophagy and apoptosis is supposed by the finding that in cancer models, when autophagy is inhibited, apoptosis is promoted [63]. Else, in preclinical models with defective apoptosis, other forms of cell death, such ACD, could occur [64,65,66,67,68,69,70,71,72].
Targeting Autophagy as a Cancer Treatment
Because autophagy plays a role in tumor suppression, the modulation of autophagy may be an important strategy for cancer treatment. Several agents are being testing in preclinical and clinical trials, either to inhibit the cytoprotective role of autophagy or to induce ACD in apoptosis-resistant cells [73]. However, only some autophagy inhibitors have, so far, shown some efficacy in advanced cancer. Common autophagy- inhibiting molecules could be categorized into four groups according to their mode of action: (1) Repressors of autophagosome formation, class III PI3K inhibitors, e.g., 3-methyladenine (3-MA), Wortmannin, LY294002, SAR405 and Viridiol that block the formation of autophagosome. (2) Repressors of lysosomal acidification: Lysosomotropic agents, such as Cloroquine (CQ), Hydroxychloroquine (HCQ), Lys0569, and Monensin prevent acidification of lysosomes and thus inhibit degradation of the cargo in the autophagosomes; (3) Inhibitors of autophagosome-lysosome fusion: Vacuolar-ATPase inhibitors, Bafilomycin and Concanamycin variants that interfere with the fusion of autophagosomes with lysosomes; (4) Silencers of transcription of autophagy-related genes: it has been demonstrated, indeed, that by means of siRNA- or miRNA-mediated silencing strategies, knockdown of autophagy-related genes can be achieved with subsequent inhibition of autophagic activity [74,75,76,77,78,79].
In this regard, CQ and HCQ have been extensive studied as anti-cancer therapy, both in combination with conventional anti-cancer treatments or with new target-therapies, since they are able to sensitize tumor cells to a variety of drugs [80]. It has been fully described that CQ and HCQ exert anti-cancer effects due to their anti-autophagy activities. CQ and HCQ inhibit the autophagic flux at a late stage, increasing lysosomal pH, which inhibits lysosomal degradative enzymes. Moreover, other preclinical anti-cancer activities, like activation of TLR9/nuclear factor kappa B (NF-κB) signaling and p53 pathways, or inhibition of CXCL12/CXCR4 signaling pathway, have been described [80,81,82].
Adding CQ to chemotherapy increases sensitivity of various cancers, including lung cancer, ovarian cancer, glioma cancer, gastric cancer, bladder cancer, and endometrial cancer cells [83,84,85,86,87]. Else, autophagy seems to limit the therapeutic effects of PI3K/Akt/mTOR signaling inhibitors; therefore, autophagy inhibition can theoretically overcome tumor resistance to these agents: combination of mTORC1 inhibitors plus CQ, show a synergistic effect on cell death in different cancers such as renal cancer [88] colon cancer [89] hepatocellular carcinoma [90] and glioma [91]. In aggressive prostate cancers, the efficacy of the AKT inhibitor AZD5363 is limited, while the blocking of autophagy using 3-MA, CQ, and Bafilomycin enhance cell death [92]. Moreover, CQ has been shown to synergize with tyrosine kinase inhibitors such as Imatinib, Sorafenib, and Sunitinib by inducing apoptosis, in different preclinical models [93,94,95]. Beyond the speculative use of these drugs in preclinical studies, their use in clinical trials remains controversial. CQ is often cytotoxic at high doses and can promote cell cycle arrest [96] or DNA damage that induces cancerogenesis [97]. According to http://clinicaltrials.gov, there are several ongoing phase I/II trials evaluating the combination of HCQ/CQ alone or with chemotherapeutic and targeted agents in different cancers. The availability of clinical results is limited now, as most trials are still recruiting or ongoing, and those that have been completed focused primarily on safety and tolerability of CQ and HCQ in cancer.
4. Targeting Autophagy in Breast Cancer
Breast cancer is the most diagnosed cancer in women [98], characterized by different outcome depending on histopathological characteristics and molecular profile [99].
Based on gene expression profile, breast cancer can be classified into at least three subtypes: luminal tumors, which are positive for estrogen (ER+) and/or progesterone receptors (PR+); HER2-enriched, which overexpress the ERBB2 oncogene; and triple-negative tumors (TNBC), which lack of hormone receptors and HER2 amplification. Each subtype has different characteristics, in terms of incidence, response to treatment, risk of disease progression, and sites of metastases [100]. Hormone receptor-positive (HR+) breast cancer subtypes (luminal A and B) represent approximately 60–75% of all breast cancers and respond well to endocrine therapy (ET), in both adjuvant and metastatic setting. However, the majority of these patients show de-novo resistance (primary) or develop acquired resistance (secondary) to ET, which requires the administration of sequential endocrine-based therapy, both as monotherapy and in combination with targeted therapy, before switching to chemotherapy-based regimens [101].
The HER2 subtype is associated with an aggressive behavior. However, this sub-group of patients could benefit from several anti-HER2 targeted therapies, including Trastuzumab, Pertuzumab, Trastuzumab emtansine (T-DM1), Lapatinib, Neratinib, etc. [102].
Finally, TNBC are more likely to exhibit an aggressive phenotype that become metastatic and resistant to various chemotherapeutics [103]. Atezolizumab, an immunocheckpoint inhibitor targeting the protein programmed cell death-ligand 1 (PDL1), was recently approved in combination with nab-paclitaxel, in the treatment of patients with unresectable locally advanced or metastatic PDL1-positive TNBC [104], however, chemotherapy is still the standard of care in many early or advanced TNBC tumors.
In the last years, preclinical and clinical research focused on targeting different pathways involved in tumor growth, such as PI3K/Akt/mTOR, cyclinD/CDK/pRb pathways and tumor microenvironment [103,105,106]. In this regard, inhibition of autophagy has proven to improve the drug response and reduce the mechanism of drug resistance [11,32,41].
4.1. ER+, PgR+, HER2-Subtype
ET with tamoxifen, a selective estrogen receptor modulator, or aromatase inhibitors (AIs) is recommended for HR+ breast cancer patients in pre and post-menopause [107]. Several mechanisms of resistance to ET have been identified [108]. In this regard, tamoxifen therapy has been reported to induce autophagy-mediated resistance in ER+ cellular model, MCF-7 and T-47D cells [109,110], while, in in vivo models, autophagy inhibitors restored antiestrogen sensitivity in resistant tumors [111].
At the same time, Exemestane (Exe), a powerful steroidal AI, promoted a cytoprotective autophagy in acquired resistant breast cancer cell models. In these models, the inhibition of autophagy and/or of PI3K pathway reverted Exe-resistance through apoptosis promotion, disruption of cell cycle, and inhibition of cell survival pathways [112]. Alterations of PI3K/AKT/mTOR pathway, such as gain-of-function mutations of the Phosphatidylinositol-4, 5-bisphosphate 3-kinase, catalytic subunit, alpha (PIK3CA) and loss/low expression of Phosphatase and Tensin Homolog (PTEN), could result in over-activation of this pathway, responsible of aberrant proliferation and cell survival [113]. In the last years, several PI3K/AKT/mTOR inhibitors were developed. It is well known that Everolimus, an mTOR inhibitor approved to be used in combination with Exemestane in a metastatic setting, could induce autophagy [112,114,115,116,117]. The increase in autophagy, induced by PI3K/AKT/mTOR inhibitors, is not surprising due to their inhibition of mTOR signaling, control of autophagy initiation and ULK1/2 phosphorylation. Recently, Alpelisib, an oral α-specific PI3K inhibitor, was approved, in metastatic setting [118], while other PI3K/AKT/mTOR inhibitors, such as Taselisib (PI3K inhibitor), Ipatasertib, and Capivasertib (AKT inhibitors) are in clinical investigation [103]. Recently, Zhai et al. reported that Ipatasertib was able to induce ACD in hepatocellular carcinoma models [119], while, Zorea et al. showed that an ovarian cancer cell line treated with Taselisib activates autophagy to avoid cell death [120].
Recently, CDK4/6 inhibitors in combination with ET have been approved both for AI-sensitive and AI-resistant patients [121]. Disregulation in cyclinD/CDK/Rb pathway is frequent in many types of human cancers, including breast cancer, particularly HR-positive. CDK4/6 kinases phosphorylate retinoblastoma protein (Rb) at serine 807/811, leading to E2F release, thus triggering G1--to--S transition. Currently, there are three approved oral highly selective CDK4/6 inhibitors, namely, Palbociclib (PD0332991), Ribociclib (LEE011), and Abemaciclib (LY2835219) [105,122]. These drugs have been widely studied in preclinical models. There is evidence that autophagy can occur after CDK4/6 inhibitors exposure in different cancer models with different response [30,123]: in hepatocellular carcinoma models, Palbociclib can induce apoptosis and ACD by activating AMPK [124]. In gastric cancer models, Palbociclib induced autophagy that occurred as an adaptive mechanism of cell survival in response to senescence, as the simultaneous blockade of CDK4/6 and autophagy exacerbated the senescence phenotype [125]. Else, in breast cancer models, cells activate autophagy in response to Palbociclib, and blockade of autophagy significantly improved the efficacy of CDK4/6 inhibition in in vitro and in vivo breast cancers models with an intact G1/S transition [126]. Other findings suggested that the CDK4 inhibitors could induce autophagy only in some cellular models of solid cancer, since autophagy induction was observed in selective cell lines, where the combination of CDK4 inhibitors and autophagy inhibitors induced apoptosis [127]. Also, Abemaciclib was reported to induce autophagy in multiple myeloma and renal cell carcinoma models [128,129]. However, it was recently reported that, in different cancer models, Abemaciclib induced an atypical cell death, independent from autophagy, accompanied by lysosomal dysfunction, block of autophagic flux, and accumulation of autophagosomes [130]. Recently, a relevant finding suggested that CDK4/6 kinases regulate lysosome biogenesis during cell cycle, through phosphorylation of TFEB/TFE3 in the nucleus, thereby promoting their shuttling to the cytoplasm. Otherwise, inhibition of CDK4/6 kinases increases lysosomal numbers by activating the lysosome and autophagy transcription factors TFEB and TFE3 [131].
4.2. HER2-Positive Subtype
HER2 is a transmembrane receptor tyrosine kinase of the epidermal growth factor receptor (EGFR) family. Its overexpression is associated with the hyperactivation of pathways, such as MAPK, JAK/STAT, RAS/MEK/ERK and PI3K/AKT/mTOR pathways, involved in cellular proliferation and differentiation of different cancers [132]. Anti-HER2 drugs could be divided into two classes: (1) HER2-directed antibodies, such as Trastuzumab, Pertuzumab and TDM1 and (2) small-molecule inhibitors targeting the kinase activity of the receptor, such as Lapatinib, Neratinib and Tucatinib [133]. Generally, HER2+ tumors have been shown to exhibit low levels of autophagy. A significant association between Beclin1 loss or decreased Beclin1 mRNA expression and HER2 amplification was found in breast cancer tumors [134,135]. In vitro studies confirmed that human and mouse HER2+ breast cancer cells presented low Beclin1 mRNA and a low autophagy-genes expression [136]. Finally, other evidence showed that the eHER2 receptor directly represses autophagy by interaction with Beclin1 [137]. At the same time, the signaling cascade activated by HER2 also activates mTORC1 signaling, a negative regulator of autophagy. Therefore HER2-target therapy has been reported to induce autophagy and its inhibition could be used to reduce the drug resistance [138]. In different preclinical models, mechanisms of resistance to Trastuzumab were associated to increased autophagy pathway [139]; while in Lapatinib resistant models, the inhibition of autophagy led to reversion of resistance [140,141]. Furthermore, Trastuzumab-resistant tumors present a higher level of AMPK expression [142].
4.3. TNBC Subtype
As mentioned above, TNBC tumors are characterized by a more aggressive behavior and early relapse [103]. Higher levels of basal autophagy were found in the metastatic cell lines when compared to the non-metastatic, suggesting that autophagy could promote invasiveness and possibly increase tolerance to the cellular stress occurring during the metastatic process [143]. TNBC tumors are more hypoxic than non-TNBC and it has been suggested that they are less sensitive to hypoxic conditions because of perpetually higher levels of autophagy [144]. Indeed, several reports have indicated that TNBC tumors exhibit a higher level of autophagy than other breast cancer subtypes. Expression of the autophagy-related microtubule-associated proteins, including Beclin1, LC3A and LC3B are higher in TNBC cells compared to the other breast cancer subtypes, with the lowest expression in the stroma of TNBC [145]. High expression of LC3B has also been associated with tumor progression and poor outcome in TNBC [51], demonstrating its role as a potential prognostic marker in TNBC. High expression of Autophagy marker ATG9 was found in TNBC breast cancer tissue, while its inhibition led to the inhibition of pro-cancer phenotypes [146]. Furthermore, knockdown of autophagy-related genes (LC3 and Beclin1) inhibited autophagy and significantly suppressed cell proliferation, colony formation, migration/invasion and induced apoptosis in MDA-MB-231 and BT-549 TNBC cells [147], while silencing of ATG5, ATG7 and Beclin1 reduced the proliferation of different TNBC cell lines (basal and claudin-low) [148]. Else, In 4T1 and 67NR mouse mammary tumor cells, CQ increased sensitivity to the mTOR inhibitor rapamycin and to the PI3K inhibitor LY294002, even in the absence of Beclin1 and ATG12, suggesting that CQ could also exert tumor growth suppression beyond autophagy [149]. Recently, it was reported an association between the hippo signaling pathway yes-associated protein (YAP) and autophagy in TNBC. In particular, autophagy induction promoted the YAP nuclear translocation and activation in TNBC models [150]. Overall, these data strongly suggest that autophagy is essential to the survival of TNBC cells and that blocking autophagy would lead to cell death [144,151].
Recently, the immunocheckpoint inhibitor Atezolizumab, a monoclonal antibody against PDL1, was approved for treatment in metastatic PDL1-positive TNBC [104]. Interestingly, emerging evidence points to the prominent role of autophagy in tumor immunoescape by multiple overlapping mechanisms [152]. Some evidence has shown an inter-play between autophagy and PDL1 expression: a study by Clark et al. showed that tumor-intrinsic PDL1 signals regulate cell proliferation and autophagy in ovarian cancer and melanoma. Tumor cells with high levels of PDL1 expression were more sensitive to autophagy inhibitors than cells with lower PDL1 [153]; in gastric cancer, autophagy regulates PDL1 expression through the p62/SQSTM1-NF-κB pathway, while pharmacological inhibition of autophagy increased the levels of PDL1 in gastric cancer cells and in xenografts models, influencing the therapeutic efficacy of PDL1 inhibitors [154]. These data suggest that autophagy could represent an attractive future therapeutic target to develop innovative and effective cancer immunotherapeutic approaches.
Extensive evidence highlighted the contribution of cancer stem cells (CSCs) in tumorigenic potential, high risk of metastasis, and drug resistance of TNBC [155]. CSCs represent a small population of cancer cells with staminal phenotype; they expose CD44+/CD24- and high ALDH expression [156,157,158]. This signature is associated with high capacity of self-renewal, of proliferation, and mammalian spheroids forming [159,160,161]. These CD44+/CD24- cells show an EMT phenotype, with a great tumorigenic potential and invasion and metastasis ability [156,162,163], that is responsible of capacity of CSCs to survive in hard metabolic conditions [164,165,166]. A relevant feature of CSCs is their ability to obtain energy from different sources and, therefore, to survive in hostile microenvironments, or in unfavorable circumstances encountered during tumor progression and in metastatic sites. Hypoxia-inducible factor-1 (HIF-1) is responsible of response to hypoxia in cancer cells [167,168], and it has been reported that the expression of EMT and stemness activators such as WNT, Hedgehog, and NOTCH pathways, or stemness markers such as FOXA2, cMET, CD133, NANOG, SOX2, SOX17, and PDX1 could also occur through the activation of HIF-1 [169,170]. Indeed, the exposure of TNBC cells to hypoxia increased the percentage of CSCs in a HIF-1–dependent manner [171,172]. Another study reported that in paclitaxel- or gemcitabine-treated TNBCs, the induction of HIF was required for enrichment of CSCs both in vitro and in vivo [169], while, in patients with breast cancer, HIF-1 overexpression was associated with increased mortality [173], poor prognosis [174], and drugs resistance [175]. It is well known that hypoxia induces autophagy, mediated by HIF-1α [176]. In this regard, autophagy has been shown to impact on CSCs generation, differentiation, plasticity, migration/invasion, and drug resistance [177]. Autophagy has been related to a variety of CSCs in several cancers, including breast, and its inhibition affected the expression of staminal markers and the cell self-renewal capacity [178,179]. The expression of Beclin1 and ATG4 was found upregulated in mammospheres and needed for their maintenance and expansion [179]. In two breast cancer stem-like cell lines, autophagy was found act through EGFR/Stat3 and Tgfβ/Smad signaling, to induce stem phenotype [180]. In CSCs originated by TNBC, autophagy inhibition decreased the secretion of IL-6, through the STAT3/JAK2 pathway, impairing their maintenance and stem phenotype [148]. While, inhibition of autophagy by CQ impaired cell migration and invasion, increased expression of the epithelial marker CD24, and decreased vimentin in CSCs [181].
4.4. Clinical Trials
Several clinical trials are investigating the use of CQ or HCQ in different cancer types. Results from early phase clinical trials with CQ or HCQ have been, so far, controversial [182,183,184,185]. A recent meta-analysis of clinical trials evaluating treatment with HCQ alone or in combination with chemotherapy or radiation in different cancers, showed that the addition of autophagy-inhibitor-based therapy has a better treatment response compared to chemotherapy or radiation alone. In particular, autophagy inhibition exerted the best survival benefit in patients with glioblastoma [186].
On website clinicaltrials.gov, about 11 clinical trials are reported to investigate the use of CQ or HCQ with different dosage, alone or in addition to other therapies, in breast cancer (Table 1). Recently, Arnaout et al. (NCT02333890) published the results from a randomized, double-blind clinical trial evaluating the effects of treatment with single-agent CQ 500 mg daily for 2- to 6-weeks prior to breast surgery. Results showed that in the preoperative setting, the treatment was not associated with significant effects on breast cancer cell proliferation, while it was associated with toxicity that may affect its broader use in oncology [187]. The disappointing results achieved in clinical trials could be in part ascribed to the choose of the wrong target. Else, the clinical usefulness of CQ may have been limited because high doses are required to compensate for its nonselective distribution in vivo, with high intersubject variability in the steady state blood concentrations of CQ and accumulation of its metabolites, desethylchloroquine, and bisdesethylchloroquine, responsible for related adverse effects [188,189].
Table 1.
Clinical trials in breast cancer with Chloroquine or Hydroxychloroquine by clinicaltrial.gov website.
| Breast Cancer Stage/Subtype/Setting | Intervention | Study Design | Dosage | Clinicaltrial.gov |
|---|---|---|---|---|
| Invasive Breast Cancer/Neoadjuvant | Drug: Chloroquine Drug: Placebo |
Phase 2 Randomized, Double-blind, placebo-controlled |
chloroquine 500 mg daily as an oral capsule during the wait time to surgery. | * NCT02333890 |
| Advanced or Metastatic Breast Cancer | Chloroquine in Combination with taxane or taxane-like Chemo Agents | Phase II Non-Randomized, open label |
chloroquine 250 mg daily as an oral capsule | NCT01446016 |
| Carcinoma, Intraductal, Noninfiltrating DCIS Ductal Carcinoma In Situ Neoadjuvant |
Chloroquine | Phase 1 Phase 2 Non-Randomized open label |
chloroquine standard dose (500mg/week) or chloroquine low dose (250mg/week) for 1 month prior to surgical removal of the tumor. | NCT01023477 |
| Metastatic Breast Cancer | Hydroxychloroquine + Ixabepilone | Phase 1 Phase 2 Non-Randomized, open label |
Dose escalation from 200 mg po qd to 200 mg po bid. | # NCT00765765 |
| Estrogen Receptor-Positive Metastatic Breast Cancer | Hydroxychloroquine in combination with the current hormonal therapy | Phase Ib/II Study Non-Randomized, open label |
Not available | NCT02414776 |
| Invasive breast cancer | Arm A: Abemaciclib Arm B: Abemaciclib + Hydroxychloroquine |
Phase II Pilot Trial, Randomized, open lable | Hydroxychloroquine 600 mg BID | NCT04523857 |
| Recurrent Breast Cancer | Phase II Arm A: Hydroxychloroquine for 24 weeks Arm B: Hydroxychloroquine for 24 weeks + Gedatolisib and × 2 weeks administered weekly Arm C: Hydroxychloroquine for 24 weeks + Gedatolisib and × 6 weeks administered weekly Arm D: Hydroxychloroquine for 24 weeks + Gedatolisib and × 12 weeks administered weekly Phase Ib: Hydroxychloroquine for 24 weeks + Gedatolisib and × 6 weeks administered weekly |
Phase 1 Phase 2 Randomized open label |
Hydroxychloroquine 600 mg BID | NCT03400254 |
| Breast Cancer Stage IIB | Arm A: Hydroxychloroquine Arm B: Everolimus Arm C: Hydroxychloroquine + Everolimus Arm D: Observation |
Phase 2 Pilot Randomized | Not available | NCT03032406 |
| HR+/Her 2- Advanced Breast Cancer | Arm A: Abemaciclib + Hydroxychloroquine 200 mg b.i.d. Arm B. Abemaciclib + Hydroxychloroquine 400 mg b.i.d. Arm C: Abemaciclib + Hydroxychloroquine 600 mg b.i.d. Arm D: Abemaciclib + Hydroxychloroquine + endocrine therapy. |
Phase 1 Non-Randomized Sequential Assignment Open Label |
Hydroxychloroquine 200 mg Hydroxychloroquine 400 mg Hydroxychloroquine 600 mg |
NCT04316169 |
| Participants with advanced, metastatic (stage IV) breast cancer (phase I) Participants with early stage (stage I-III) breast cancer (phase II) |
Hydroxychloroquine, Palbociclib, and Letrozole | Phase 1 Phase 2 Open label |
Hydroxychloroquine 400 mg | NCT03774472 |
| Breast Cancer Neoadijuvant | Hydrochloroquine | Phase II Pilot Open Label |
Hydrochloroquine 800 mg per os once, and then 400 mg per day |
NCT01292408 |
* This study achieved negative results [187]. # This study achieved a Response Rate of 30%.
5. Conclusions
Autophagy plays a complex role in cancer, and its inhibition could, in theory, represent an effective therapeutic strategy. In the last years, new targeted drugs, such as CDK4/6 inhibitors, PI3K/AKT inhibitors, new anti-HER2 drugs and immunocheckpoint inhibitors have emerged and have been integrated in the clinical practice. We have reviewed how these therapies could modulate autophagy. We have also highlighted how this modulation may play a key role in the mechanisms of resistance to these drugs. In this perspective, inhibition of autophagy may help reversing or overcoming drug resistance. However, clinical applications with autophagy inhibitors, have been so far disappointing, due either to the non-efficient in vivo autophagy inhibition or to the toxicity of the tested drugs.
Based on preclinical data, however, targeting autophagy remains a promising approach in breast cancer, and the development of new agents with a higher therapeutic index should be regarded as a worthwhile achievement in pharmacological research.
Abbreviations
| 3-MA | 3-methyladenine |
| AI | Aromatase Inhibitor |
| ALDH | Aldehyde Dehydrogenase |
| AMPK | 5’ AMP-activated protein kinase |
| ATG | Autophagy related genes |
| ACD | Autophagic cell death |
| BC | Breast Cancer |
| CQ | Chloroquine |
| CSCs | Cancer Stem Cells |
| DNA | Deoxyribonucleic Acid |
| ECM | Extracellular matrix |
| EGFR | Epidermal Growth Factor Receptor |
| EMT | Epithelial Mesenchymal Transition |
| ER | Estrogen Receptor |
| EXE | Exemestane |
| HCQ | Hidroxychloroquine |
| HER2 | Human Epidermal Growth Factor Receptor 2 |
| HIF-1 | Hipoxia-inducible factor 1 |
| YAP | Yes-associated protein |
| LC3 | Light chain 3 |
| MAPK | Mitogen-activated protein kinase |
| MSL | Mesenchymal Stem-Like |
| mTOR | Mammalian Target Of Rapamycin |
| mTORC1 | mTOR complex 1 |
| NFKB | Nuclear factor kappa-light-chain-enhancer of activated B cells |
| PAS | Phagophore assembly site |
| PDL1 | Programmed Cell Death Ligand-1 |
| PDX | Patient-Derived-Xenograft |
| PE | Phosphatidylethanolamine |
| PI3K | Phosphatidylinositol 3-kinase |
| PIK3CA | Phosphatidylinositol-4, 5-bisphosphate 3-kinase, catalytic subunit, alpha |
| PIP3 | Phosphatidylinositol (3,4,5)-trisphosphate |
| PTEN | Phosphatase and Tensin Homolog |
| PgR | Progesteron Receptor |
| RB | Retinoblastoma protein |
| ROS | Reactive Oxygen Species |
| T-DM1 | Trastuzumab emtansine |
| TFEB | Transcription factor EB |
| TNBC | Triple-Negative Breast Cancer |
| ULK1 | Unc-51 like autophagy activating kinase |
Author Contributions
S.C., A.L. and M.P. conceived, designed the present study and wrote the manuscript. A.B. and M.D.L. reviewed, edited and supervised. R.C., V.D.L., F.D.R., conducted the bibliography research and supported the clinical perspectives. G.F., M.C. and G.d.G. conducted the bibliography research and supported the preclinical perspectives. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Galluzzi L., Baehrecke E.H., Ballabio A., Boya P., Pedro J.M.B.-S., Cecconi F., Choi A.M., Chu C.T., Codogno P., Colombo M.I., et al. Molecular Definitions of Autophagy and Related Processes. EMBO J. 2017;36:1811–1836. doi: 10.15252/embj.201796697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kroemer G., Mariño G., Levine B. Autophagy and the Integrated Stress Response. Mol. Cell. 2010;40:280–293. doi: 10.1016/j.molcel.2010.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dikic I., Elazar Z. Mechanism and Medical Implications of Mammalian Autophagy. Nat. Rev. Mol. Cell Biol. 2018;19:349–364. doi: 10.1038/s41580-018-0003-4. [DOI] [PubMed] [Google Scholar]
- 4.Jung S., Jeong H., Yu S.-W. Autophagy as a Decisive Process for Cell Death. Exp. Mol. Med. 2020;52:921–930. doi: 10.1038/s12276-020-0455-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Khandia R., Dadar M., Munjal A., Dhama K., Karthik K., Tiwari R., Yatoo M.I., Iqbal H.M.N., Singh K.P., Joshi S.K., et al. A Comprehensive Review of Autophagy and Its Various Roles in Infectious, Non-Infectious, and Lifestyle Diseases: Current Knowledge and Prospects for Disease Prevention, Novel Drug Design, and Therapy. Cells. 2019;8:674. doi: 10.3390/cells8070674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bonam S.R., Wang F., Muller S. Autophagy: A New Concept in Autoimmunity Regulation and a Novel Therapeutic Option. J. Autoimmun. 2018;94:16–32. doi: 10.1016/j.jaut.2018.08.009. [DOI] [PubMed] [Google Scholar]
- 7.Levine B., Kroemer G. Biological Functions of Autophagy Genes: A Disease Perspective. Cell. 2019;176:11–42. doi: 10.1016/j.cell.2018.09.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang Z., Klionsky D.J. An Overview of the Molecular Mechanism of Autophagy. Curr. Top. Microbiol. Immunol. 2009;335:1–32. doi: 10.1007/978-3-642-00302-8_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mizushima N., Komatsu M. Autophagy: Renovation of Cells and Tissues. Cell. 2011;147:728–741. doi: 10.1016/j.cell.2011.10.026. [DOI] [PubMed] [Google Scholar]
- 10.Mercer T.J., Gubas A., Tooze S.A. A Molecular Perspective of Mammalian Autophagosome Biogenesis. J. Biol. Chem. 2018;293:5386–5395. doi: 10.1074/jbc.R117.810366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cecconi F., Levine B. The Role of Autophagy in Mammalian Development: Cell Makeover Rather than Cell Death. Dev. Cell. 2008;15:344–357. doi: 10.1016/j.devcel.2008.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bonam S.R., Bayry J., Tschan M.P., Muller S. Progress and Challenges in The Use of MAP1LC3 as a Legitimate Marker for Measuring Dynamic Autophagy In Vivo. Cells. 2020;9:1321. doi: 10.3390/cells9051321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Corona Velazquez A.F., Jackson W.T. So Many Roads: The Multifaceted Regulation of Autophagy Induction. Mol. Cell. Biol. 2018;38:e00303-18. doi: 10.1128/MCB.00303-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Settembre C., Zoncu R., Medina D.L., Vetrini F., Erdin S., Erdin S., Huynh T., Ferron M., Karsenty G., Vellard M.C., et al. A Lysosome-to-Nucleus Signalling Mechanism Senses and Regulates the Lysosome via MTOR and TFEB. EMBO J. 2012;31:1095–1108. doi: 10.1038/emboj.2012.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ao X., Zou L., Wu Y. Regulation of Autophagy by the Rab GTPase Network. Cell Death Differ. 2014;21:348–358. doi: 10.1038/cdd.2013.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Alers S., Löffler A.S., Wesselborg S., Stork B. Role of AMPK-MTOR-Ulk1/2 in the Regulation of Autophagy: Cross Talk, Shortcuts, and Feedbacks. Mol. Cell. Biol. 2012;32:2–11. doi: 10.1128/MCB.06159-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li Y., Chen Y. AMPK and Autophagy. Adv. Exp. Med. Biol. 2019;1206:85–108. doi: 10.1007/978-981-15-0602-4_4. [DOI] [PubMed] [Google Scholar]
- 18.Settembre C., Di Malta C., Polito V.A., Garcia Arencibia M., Vetrini F., Erdin S., Erdin S.U., Huynh T., Medina D., Colella P., et al. TFEB Links Autophagy to Lysosomal Biogenesis. Science. 2011;332:1429–1433. doi: 10.1126/science.1204592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Settembre C., De Cegli R., Mansueto G., Saha P.K., Vetrini F., Visvikis O., Huynh T., Carissimo A., Palmer D., Klisch T.J., et al. TFEB Controls Cellular Lipid Metabolism through a Starvation-Induced Autoregulatory Loop. Nat. Cell Biol. 2013;15:647–658. doi: 10.1038/ncb2718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sardiello M., Palmieri M., di Ronza A., Medina D.L., Valenza M., Gennarino V.A., Di Malta C., Donaudy F., Embrione V., Polishchuk R.S., et al. A Gene Network Regulating Lysosomal Biogenesis and Function. Science. 2009;325:473–477. doi: 10.1126/science.1174447. [DOI] [PubMed] [Google Scholar]
- 21.Stolz A., Ernst A., Dikic I. Cargo Recognition and Trafficking in Selective Autophagy. Nat. Cell Biol. 2014;16:495–501. doi: 10.1038/ncb2979. [DOI] [PubMed] [Google Scholar]
- 22.Germain K., Kim P.K. Pexophagy: A Model for Selective Autophagy. Int. J. Mol. Sci. 2020;21 doi: 10.3390/ijms21020578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Smith M., Wilkinson S. ER Homeostasis and Autophagy. Essays Biochem. 2017;61:625–635. doi: 10.1042/EBC20170092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Denton D., Kumar S. Ribophagy: New Receptor Discovered. Cell Res. 2018;28:699–700. doi: 10.1038/s41422-018-0054-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kounakis K., Chaniotakis M., Markaki M., Tavernarakis N. Emerging Roles of Lipophagy in Health and Disease. Front. Cell Dev. Biol. 2019;7 doi: 10.3389/fcell.2019.00185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Levine B. Eating Oneself and Uninvited Guests: Autophagy-Related Pathways in Cellular Defense. Cell. 2005;120:159–162. doi: 10.1016/j.cell.2005.01.005. [DOI] [PubMed] [Google Scholar]
- 27.Ding W.-X., Ni H.-M., Gao W., Yoshimori T., Stolz D.B., Ron D., Yin X.-M. Linking of Autophagy to Ubiquitin-Proteasome System Is Important for the Regulation of Endoplasmic Reticulum Stress and Cell Viability. Am. J. Pathol. 2007;171:513–524. doi: 10.2353/ajpath.2007.070188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Matsumoto G., Wada K., Okuno M., Kurosawa M., Nukina N. Serine 403 Phosphorylation of P62/SQSTM1 Regulates Selective Autophagic Clearance of Ubiquitinated Proteins. Mol. Cell. 2011;44:279–289. doi: 10.1016/j.molcel.2011.07.039. [DOI] [PubMed] [Google Scholar]
- 29.Bernardini J.P., Lazarou M., Dewson G. Parkin and Mitophagy in Cancer. Oncogene. 2017;36:1315–1327. doi: 10.1038/onc.2016.302. [DOI] [PubMed] [Google Scholar]
- 30.Yang Z.J., Chee C.E., Huang S., Sinicrope F.A. The Role of Autophagy in Cancer: Therapeutic Implications. Mol. Cancer Ther. 2011;10:1533. doi: 10.1158/1535-7163.MCT-11-0047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen P., Cescon M., Bonaldo P. Autophagy-Mediated Regulation of Macrophages and Its Applications for Cancer. Autophagy. 2014;10:192–200. doi: 10.4161/auto.26927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Barnard R.A., Regan D.P., Hansen R.J., Maycotte P., Thorburn A., Gustafson D.L. Autophagy Inhibition Delays Early but Not Late-Stage Metastatic Disease. J. Pharmacol. Exp. Ther. 2016;358:282–293. doi: 10.1124/jpet.116.233908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang K., Klionsky D.J. Mitochondria Removal by Autophagy. Autophagy. 2011;7:297–300. doi: 10.4161/auto.7.3.14502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mathew R., Karp C.M., Beaudoin B., Vuong N., Chen G., Chen H.-Y., Bray K., Reddy A., Bhanot G., Gelinas C., et al. Autophagy Suppresses Tumorigenesis through Elimination of P62. Cell. 2009;137:1062–1075. doi: 10.1016/j.cell.2009.03.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Anding A.L., Baehrecke E.H. Cleaning House: Selective Autophagy of Organelles. Dev. Cell. 2017;41:10–22. doi: 10.1016/j.devcel.2017.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liang X.H., Jackson S., Seaman M., Brown K., Kempkes B., Hibshoosh H., Levine B. Induction of Autophagy and Inhibition of Tumorigenesis by Beclin 1. Nature. 1999;402:672–676. doi: 10.1038/45257. [DOI] [PubMed] [Google Scholar]
- 37.Mathew R., Karantza-Wadsworth V., White E. Role of Autophagy in Cancer. Nat. Rev. Cancer. 2007;7:961–967. doi: 10.1038/nrc2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Das C.K., Mandal M., Kögel D. Pro-Survival Autophagy and Cancer Cell Resistance to Therapy. Cancer Metastasis Rev. 2018;37:749–766. doi: 10.1007/s10555-018-9727-z. [DOI] [PubMed] [Google Scholar]
- 39.Thomas M., Davis T., Loos B., Sishi B., Huisamen B., Strijdom H., Engelbrecht A.-M. Autophagy Is Essential for the Maintenance of Amino Acids and ATP Levels during Acute Amino Acid Starvation in MDAMB231 Cells. Cell Biochem. Funct. 2018;36:65–79. doi: 10.1002/cbf.3318. [DOI] [PubMed] [Google Scholar]
- 40.Martin S., Dudek-Peric A.M., Garg A.D., Roose H., Demirsoy S., Van Eygen S., Mertens F., Vangheluwe P., Vankelecom H., Agostinis P. An Autophagy-Driven Pathway of ATP Secretion Supports the Aggressive Phenotype of BRAFV600E Inhibitor-Resistant Metastatic Melanoma Cells. Autophagy. 2017;13:1512–1527. doi: 10.1080/15548627.2017.1332550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li X., He S., Ma B. Autophagy and Autophagy-Related Proteins in Cancer. Mol. Cancer. 2020;19:12. doi: 10.1186/s12943-020-1138-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ahn C.H., Jeong E.G., Lee J.W., Kim M.S., Kim S.H., Kim S.S., Yoo N.J., Lee S.H. Expression of Beclin-1, an Autophagy-Related Protein, in Gastric and Colorectal Cancers. APMIS Acta Pathol. Microbiol. Immunol. Scand. 2007;115:1344–1349. doi: 10.1111/j.1600-0463.2007.00858.x. [DOI] [PubMed] [Google Scholar]
- 43.Tang H., Da L., Mao Y., Li Y., Li D., Xu Z., Li F., Wang Y., Tiollais P., Li T., et al. Hepatitis B Virus X Protein Sensitizes Cells to Starvation-Induced Autophagy via up-Regulation of Beclin 1 Expression. Hepatol. Baltim. MD. 2009;49:60–71. doi: 10.1002/hep.22581. [DOI] [PubMed] [Google Scholar]
- 44.Sun Y., Liu J.-H., Jin L., Lin S.-M., Yang Y., Sui Y.-X., Shi H. Over-Expression of the Beclin1 Gene Upregulates Chemosensitivity to Anti-Cancer Drugs by Enhancing Therapy-Induced Apoptosis in Cervix Squamous Carcinoma CaSki Cells. Cancer Lett. 2010;294:204–210. doi: 10.1016/j.canlet.2010.02.001. [DOI] [PubMed] [Google Scholar]
- 45.Qu X., Yu J., Bhagat G., Furuya N., Hibshoosh H., Troxel A., Rosen J., Eskelinen E.-L., Mizushima N., Ohsumi Y., et al. Promotion of Tumorigenesis by Heterozygous Disruption of the Beclin 1 Autophagy Gene. J. Clin. Invest. 2003;112:1809–1820. doi: 10.1172/JCI20039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yang Y., Hu L., Zheng H., Mao C., Hu W., Xiong K., Wang F., Liu C. Application and Interpretation of Current Autophagy Inhibitors and Activators. Acta Pharmacol. Sin. 2013;34:625–635. doi: 10.1038/aps.2013.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Guo J.Y., Chen H.-Y., Mathew R., Fan J., Strohecker A.M., Karsli-Uzunbas G., Kamphorst J.J., Chen G., Lemons J.M.S., Karantza V., et al. Activated Ras Requires Autophagy to Maintain Oxidative Metabolism and Tumorigenesis. Genes Dev. 2011;25:460–470. doi: 10.1101/gad.2016311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang Y., Xiong H., Liu D., Hill C., Ertay A., Li J., Zou Y., Miller P., White E., Downward J., et al. Autophagy Inhibition Specifically Promotes Epithelial-Mesenchymal Transition and Invasion in RAS-Mutated Cancer Cells. Autophagy. 2019;15:886–899. doi: 10.1080/15548627.2019.1569912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Seton-Rogers S. Eliminating Protective Autophagy in KRAS-Mutant Cancers. Nat. Rev. Cancer. 2019;19:247. doi: 10.1038/s41568-019-0137-5. [DOI] [PubMed] [Google Scholar]
- 50.Lazova R., Camp R.L., Klump V., Siddiqui S.F., Amaravadi R.K., Pawelek J.M. Punctate LC3B Expression Is a Common Feature of Solid Tumors and Associated with Proliferation, Metastasis, and Poor Outcome. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2012;18:370–379. doi: 10.1158/1078-0432.CCR-11-1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhao H., Yang M., Zhao J., Wang J., Zhang Y., Zhang Q. High Expression of LC3B Is Associated with Progression and Poor Outcome in Triple-Negative Breast Cancer. Med. Oncol. Northwood Lond. Engl. 2013;30:475. doi: 10.1007/s12032-013-0475-1. [DOI] [PubMed] [Google Scholar]
- 52.Lazova R., Klump V., Pawelek J. Autophagy in Cutaneous Malignant Melanoma. J. Cutan. Pathol. 2010;37:256–268. doi: 10.1111/j.1600-0560.2009.01359.x. [DOI] [PubMed] [Google Scholar]
- 53.Peng Y.-F., Shi Y.-H., Ding Z.-B., Ke A.-W., Gu C.-Y., Hui B., Zhou J., Qiu S.-J., Dai Z., Fan J. Autophagy Inhibition Suppresses Pulmonary Metastasis of HCC in Mice via Impairing Anoikis Resistance and Colonization of HCC Cells. Autophagy. 2013;9:2056–2068. doi: 10.4161/auto.26398. [DOI] [PubMed] [Google Scholar]
- 54.Galavotti S., Bartesaghi S., Faccenda D., Shaked-Rabi M., Sanzone S., McEvoy A., Dinsdale D., Condorelli F., Brandner S., Campanella M., et al. The Autophagy-Associated Factors DRAM1 and P62 Regulate Cell Migration and Invasion in Glioblastoma Stem Cells. Oncogene. 2013;32:699–712. doi: 10.1038/onc.2012.111. [DOI] [PubMed] [Google Scholar]
- 55.Fung C., Lock R., Gao S., Salas E., Debnath J. Induction of Autophagy during Extracellular Matrix Detachment Promotes Cell Survival. Mol. Biol. Cell. 2008;19:797–806. doi: 10.1091/mbc.e07-10-1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chaffer C.L., Weinberg R.A. A Perspective on Cancer Cell Metastasis. Science. 2011;331:1559–1564. doi: 10.1126/science.1203543. [DOI] [PubMed] [Google Scholar]
- 57.Valastyan S., Weinberg R.A. Tumor Metastasis: Molecular Insights and Evolving Paradigms. Cell. 2011;147:275–292. doi: 10.1016/j.cell.2011.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ravanan P., Srikumar I.F., Talwar P. Autophagy: The spotlight for cellular stress responses. Life Sci. 2017;188:53–67. doi: 10.1016/j.lfs.2017.08.029. [DOI] [PubMed] [Google Scholar]
- 59.Kroemer G., Levine B. Autophagic Cell Death: The Story of a Misnomer. Nat. Rev. Mol. Cell Biol. 2008;9:1004–1010. doi: 10.1038/nrm2529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mariño G., Niso-Santano M., Baehrecke E.H., Kroemer G. Self-Consumption: The Interplay of Autophagy and Apoptosis. Nat. Rev. Mol. Cell Biol. 2014;15:81–94. doi: 10.1038/nrm3735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Liu Y., Shoji-Kawata S., Sumpter R.M., Wei Y., Ginet V., Zhang L., Posner B., Tran K.A., Green D.R., Xavier R.J., et al. Autosis Is a Na+,K+-ATPase-Regulated Form of Cell Death Triggered by Autophagy-Inducing Peptides, Starvation, and Hypoxia-Ischemia. Proc. Natl. Acad. Sci. USA. 2013;110:20364–20371. doi: 10.1073/pnas.1319661110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Xie C., Ginet V., Sun Y., Koike M., Zhou K., Li T., Li H., Li Q., Wang X., Uchiyama Y., et al. Neuroprotection by Selective Neuronal Deletion of Atg7 in Neonatal Brain Injury. Autophagy. 2016;12:410–423. doi: 10.1080/15548627.2015.1132134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gump J.M., Thorburn A. Autophagy and apoptosis: What is the connection? Trends Cell Biol. 2011;21:387–392. doi: 10.1016/j.tcb.2011.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Napoletano F., Baron O., Vandenabeele P., Mollereau B., Fanto M. Intersections between Regulated Cell Death and Autophagy. Trends Cell Biol. 2019;29:323–338. doi: 10.1016/j.tcb.2018.12.007. [DOI] [PubMed] [Google Scholar]
- 65.Bialik S., Dasari S.K., Kimchi A. Autophagy-dependent cell death - where, how and why a cell eats itself to death. J. Cell Sci. 2018;131 doi: 10.1242/jcs.215152. [DOI] [PubMed] [Google Scholar]
- 66.Luciani M.-F., Giusti C., Harms B., Oshima Y., Kikuchi H., Kubohara Y., Golstein P. Atg1 Allows Second-Signaled Autophagic Cell Death in Dictyostelium. Autophagy. 2011;7:501–508. doi: 10.4161/auto.7.5.14957. [DOI] [PubMed] [Google Scholar]
- 67.Zhao Y., Yang J., Liao W., Liu X., Zhang H., Wang S., Wang D., Feng J., Yu L., Zhu W.-G. Cytosolic FoxO1 Is Essential for the Induction of Autophagy and Tumour Suppressor Activity. Nat. Cell Biol. 2010;12:665–675. doi: 10.1038/ncb2069. [DOI] [PubMed] [Google Scholar]
- 68.Law B.Y.K., Michelangeli F., Qu Y.Q., Xu S.-W., Han Y., Mok S.W.F., de Seabra Rodrigues Dias I.R., Javed M.-H., Chan W.-K., Xue W.-W., et al. Neferine Induces Autophagy-Dependent Cell Death in Apoptosis-Resistant Cancers via Ryanodine Receptor and Ca2+-Dependent Mechanism. Sci. Rep. 2019;9:20034. doi: 10.1038/s41598-019-56675-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yoshikawa N., Liu W., Nakamura K., Yoshida K., Ikeda Y., Tanaka H., Mizuno M., Toyokuni S., Hori M., Kikkawa F., et al. Plasma-Activated Medium Promotes Autophagic Cell Death along with Alteration of the MTOR Pathway. Sci. Rep. 2020;10:1614. doi: 10.1038/s41598-020-58667-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Shimizu S., Kanaseki T., Mizushima N., Mizuta T., Arakawa-Kobayashi S., Thompson C.B., Tsujimoto Y. Role of Bcl-2 Family Proteins in a Non-Apoptotic Programmed Cell Death Dependent on Autophagy Genes. Nat. Cell Biol. 2004;6:1221–1228. doi: 10.1038/ncb1192. [DOI] [PubMed] [Google Scholar]
- 71.Law B.Y.K., Chan W.K., Xu S.W., Wang J.R., Bai L.P., Liu L., Wong V.K.W. Natural Small-Molecule Enhancers of Autophagy Induce Autophagic Cell Death in Apoptosis-Defective Cells. Sci. Rep. 2014;4:5510. doi: 10.1038/srep05510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bauckman K.A., Haller E., Flores I., Nanjundan M. Iron Modulates Cell Survival in a Ras- and MAPK-Dependent Manner in Ovarian Cells. Cell Death Dis. 2013;4:e592. doi: 10.1038/cddis.2013.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kocaturk N.M., Akkoc Y., Kig C., Bayraktar O., Gozuacik D., Kutlu O. Autophagy as a Molecular Target for Cancer Treatment. Eur. J. Pharm. Sci. Off. J. Eur. Fed. Pharm. Sci. 2019;134:116–137. doi: 10.1016/j.ejps.2019.04.011. [DOI] [PubMed] [Google Scholar]
- 74.Bao J., Shi Y., Tao M., Liu N., Zhuang S., Yuan W. Pharmacological Inhibition of Autophagy by 3-MA Attenuates Hyperuricemic Nephropathy. Clin. Sci. Lond. Engl. 2018;132:2299–2322. doi: 10.1042/CS20180563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Del Bel M., Abela A.R., Ng J.D., Guerrero C.A. Enantioselective Chemical Syntheses of the Furanosteroids (−)-Viridin and (−)-Viridiol. J. Am. Chem. Soc. 2017;139:6819–6822. doi: 10.1021/jacs.7b02829. [DOI] [PubMed] [Google Scholar]
- 76.Yang W., Jiang C., Xia W., Ju H., Jin S., Liu S., Zhang L., Ren G., Ma H., Ruan M., et al. Blocking Autophagy Flux Promotes Interferon-Alpha-Mediated Apoptosis in Head and Neck Squamous Cell Carcinoma. Cancer Lett. 2019;451:34–47. doi: 10.1016/j.canlet.2019.02.052. [DOI] [PubMed] [Google Scholar]
- 77.Florey O., Gammoh N., Kim S.E., Jiang X., Overholtzer M. V-ATPase and Osmotic Imbalances Activate Endolysosomal LC3 Lipidation. Autophagy. 2015;11:88–99. doi: 10.4161/15548627.2014.984277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Mauvezin C., Neufeld T.P. Bafilomycin A1 Disrupts Autophagic Flux by Inhibiting Both V-ATPase-Dependent Acidification and Ca-P60A/SERCA-Dependent Autophagosome-Lysosome Fusion. Autophagy. 2015;11:1437–1438. doi: 10.1080/15548627.2015.1066957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Mauthe M., Langereis M., Jung J., Zhou X., Jones A., Omta W., Tooze S.A., Stork B., Paludan S.R., Ahola T., et al. An SiRNA Screen for ATG Protein Depletion Reveals the Extent of the Unconventional Functions of the Autophagy Proteome in Virus Replication. J. Cell Biol. 2016;214:619–635. doi: 10.1083/jcb.201602046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Verbaanderd C., Maes H., Schaaf M.B., Sukhatme V.P., Pantziarka P., Sukhatme V., Agostinis P., Bouche G. Repurposing Drugs in Oncology (ReDO)—Chloroquine and Hydroxychloroquine as Anti-cancer Agents. [(accessed on 25 September 2020)]; doi: 10.3332/ecancer.2017.781. Available online: http://ecancer.org/en/journal/article/781-repurposing-drugs-in-oncology-redo-chloroquine-and-hydroxychloroquine-as-anti-cancer-agents. [DOI] [PMC free article] [PubMed]
- 81.Zhang Y., Wang Q., Ma A., Li Y., Li R., Wang Y. Functional Expression of TLR9 in Esophageal Cancer. Oncol. Rep. 2014;31:2298–2304. doi: 10.3892/or.2014.3095. [DOI] [PubMed] [Google Scholar]
- 82.Kim J., Yip M.L.R., Shen X., Li H., Hsin L.-Y.C., Labarge S., Heinrich E.L., Lee W., Lu J., Vaidehi N. Identification of Anti-Malarial Compounds as Novel Antagonists to Chemokine Receptor CXCR4 in Pancreatic Cancer Cells. PLoS ONE. 2012;7:e31004. doi: 10.1371/journal.pone.0031004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zhan Z., Li Q., Wu P., Ye Y., Tseng H.-Y., Zhang L., Zhang X.D. Autophagy-Mediated HMGB1 Release Antagonizes Apoptosis of Gastric Cancer Cells Induced by Vincristine via Transcriptional Regulation of Mcl-1. Autophagy. 2012;8:109–121. doi: 10.4161/auto.8.1.18319. [DOI] [PubMed] [Google Scholar]
- 84.Wang J., Wu G.S. Role of Autophagy in Cisplatin Resistance in Ovarian Cancer Cells. J. Biol. Chem. 2014;289:17163–17173. doi: 10.1074/jbc.M114.558288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Bao L., Jaramillo M.C., Zhang Z., Zheng Y., Yao M., Zhang D.D., Yi X. Induction of Autophagy Contributes to Cisplatin Resistance in Human Ovarian Cancer Cells. Mol. Med. Rep. 2015;11:91–98. doi: 10.3892/mmr.2014.2671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Fukuda T., Oda K., Wada-Hiraike O., Sone K., Inaba K., Ikeda Y., Miyasaka A., Kashiyama T., Tanikawa M., Arimoto T., et al. The Anti-Malarial Chloroquine Suppresses Proliferation and Overcomes Cisplatin Resistance of Endometrial Cancer Cells via Autophagy Inhibition. Gynecol. Oncol. 2015;137:538–545. doi: 10.1016/j.ygyno.2015.03.053. [DOI] [PubMed] [Google Scholar]
- 87.Wu T., Wang M.-C., Jing L., Liu Z.-Y., Guo H., Liu Y., Bai Y.-Y., Cheng Y.-Z., Nan K.-J., Liang X. Autophagy Facilitates Lung Adenocarcinoma Resistance to Cisplatin Treatment by Activation of AMPK/MTOR Signaling Pathway. Drug Des. Devel. Ther. 2015;9:6421–6431. doi: 10.2147/DDDT.S95606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Grimaldi A., Santini D., Zappavigna S., Lombardi A., Misso G., Boccellino M., Desiderio V., Vitiello P.P., Di Lorenzo G., Zoccoli A., et al. Antagonistic Effects of Chloroquine on Autophagy Occurrence Potentiate the Anticancer Effects of Everolimus on Renal Cancer Cells. Cancer Biol. Ther. 2015;16:567–579. doi: 10.1080/15384047.2015.1018494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wang L., Zhu Y.-R., Wang S., Zhao S. Autophagy Inhibition Sensitizes WYE-354-Induced Anti-Colon Cancer Activity in Vitro and in Vivo. Tumour Biol. J. Int. Soc. Oncodev. Biol. Med. 2016;37:11743–11752. doi: 10.1007/s13277-016-5018-x. [DOI] [PubMed] [Google Scholar]
- 90.Elmansi A.M., El-Karef A.A., El-Shishtawy M.M., Eissa L.A. Hepatoprotective Effect of Curcumin on Hepatocellular Carcinoma Through Autophagic and Apoptic Pathways. Ann. Hepatol. 2017;16:607–618. doi: 10.5604/01.3001.0010.0307. [DOI] [PubMed] [Google Scholar]
- 91.Misirkic M., Janjetovic K., Vucicevic L., Tovilovic G., Ristic B., Vilimanovich U., Harhaji-Trajkovic L., Sumarac-Dumanovic M., Micic D., Bumbasirevic V., et al. Inhibition of AMPK-Dependent Autophagy Enhances in Vitro Antiglioma Effect of Simvastatin. Pharmacol. Res. 2012;65:111–119. doi: 10.1016/j.phrs.2011.08.003. [DOI] [PubMed] [Google Scholar]
- 92.Lamoureux F., Thomas C., Crafter C., Kumano M., Zhang F., Davies B.R., Gleave M.E., Zoubeidi A. Blocked Autophagy Using Lysosomotropic Agents Sensitizes Resistant Prostate Tumor Cells to the Novel Akt Inhibitor AZD5363. Clin. Cancer Res. 2013;19:833. doi: 10.1158/1078-0432.CCR-12-3114. [DOI] [PubMed] [Google Scholar]
- 93.PRIME PubMed|Antitumor Activity of Chloroquine in Combination with Cisplatin in Human Gastric Cancer Xenografts. [(accessed on 28 September 2020)]; Available online: https://www.unboundmedicine.com/medline/citation/25987058/Antitumor_activity_of_chloroquine_in_combination_with_Cisplatin_in_human_gastric_cancer_xenografts_.
- 94.Abdel-Aziz A.K., Shouman S., El-Demerdash E., Elgendy M., Abdel-Naim A.B. Chloroquine Synergizes Sunitinib Cytotoxicity via Modulating Autophagic, Apoptotic and Angiogenic Machineries. Chem. Biol. Interact. 2014;217:28–40. doi: 10.1016/j.cbi.2014.04.007. [DOI] [PubMed] [Google Scholar]
- 95.Honma Y., Harada M. Sorafenib Enhances Proteasome Inhibitor-Mediated Cytotoxicity via Inhibition of Unfolded Protein Response and Keratin Phosphorylation. Exp. Cell Res. 2013;319:2166–2178. doi: 10.1016/j.yexcr.2013.05.023. [DOI] [PubMed] [Google Scholar]
- 96.Chang M.-C., Lin L.-D., Wu M.-T., Chan C.-P., Chang H.-H., Lee M.-S., Sun T.-Y., Jeng P.-Y., Yeung S.-Y., Lin H.-J., et al. Effects of Camphorquinone on Cytotoxicity, Cell Cycle Regulation and Prostaglandin E2 Production of Dental Pulp Cells: Role of ROS, ATM/Chk2, MEK/ERK and Hemeoxygenase-1. PLoS ONE. 2015;10:e0143663. doi: 10.1371/journal.pone.0143663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Michael R.O., Williams G.M. Choloroquine Inhibition of Repair of DNA Damage Induced in Mammalian Cells by Methyl Methanesulfonate. Mutat. Res. Mol. Mech. Mutagen. 1974;25:391–396. doi: 10.1016/0027-5107(74)90068-2. [DOI] [PubMed] [Google Scholar]
- 98.Siegel R.L., Miller K.D., Jemal A. Cancer Statistics, 2020, CA. Cancer J. Clin. 2020;70:7–30. doi: 10.3322/caac.21590. [DOI] [PubMed] [Google Scholar]
- 99.Redig A.J., McAllister S.S. Breast Cancer as a Systemic Disease: A View of Metastasis. J. Intern. Med. 2013;274:113–126. doi: 10.1111/joim.12084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Prat A., Perou C.M. Deconstructing the Molecular Portraits of Breast Cancer. Mol. Oncol. 2011;5:5–23. doi: 10.1016/j.molonc.2010.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Cardoso F., Costa A., Senkus E., Aapro M., André F., Barrios C.H., Bergh J., Bhattacharyya G., Biganzoli L., Cardoso M.J., et al. 3rd ESO-ESMO International Consensus Guidelines for Advanced Breast Cancer (ABC 3) Ann. Oncol. Off. J. Eur. Soc. Med. Oncol. 2017;28:16–33. doi: 10.1093/annonc/mdw544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Wang J., Xu B. Targeted Therapeutic Options and Future Perspectives for HER2-Positive Breast Cancer. Signal Transduct. Target. Ther. 2019;4:34. doi: 10.1038/s41392-019-0069-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Cocco S., Piezzo M., Calabrese A., Cianniello D., Caputo R., Lauro V.D., Fusco G., di Gioia G., Licenziato M., de Laurentiis M. Biomarkers in Triple-Negative Breast Cancer: State-of-the-Art and Future Perspectives. Int. J. Mol. Sci. 2020;21 doi: 10.3390/ijms21134579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Schmid P., Adams S., Rugo H.S., Schneeweiss A., Barrios C.H., Iwata H., Diéras V., Hegg R., Im S.-A., Shaw Wright G., et al. IMpassion130 Trial Investigators. Atezolizumab and Nab-Paclitaxel in Advanced Triple-Negative Breast Cancer. N. Engl. J. Med. 2018;379:2108–2121. doi: 10.1056/NEJMoa1809615. [DOI] [PubMed] [Google Scholar]
- 105.Piezzo M., Chiodini P., Riemma M., Cocco S., Caputo R., Cianniello D., Di Gioia G., Di Lauro V., Rella F.D., Fusco G., et al. Progression-Free Survival and Overall Survival of CDK 4/6 Inhibitors Plus Endocrine Therapy in Metastatic Breast Cancer: A Systematic Review and Meta-Analysis. Int. J. Mol. Sci. 2020;21 doi: 10.3390/ijms21176400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Heimes A.-S., Schmidt M. Atezolizumab for the Treatment of Triple-Negative Breast Cancer. Expert Opin. Investig. Drugs. 2019;28:1–5. doi: 10.1080/13543784.2019.1552255. [DOI] [PubMed] [Google Scholar]
- 107.Pistelli M., Mora A.D., Ballatore Z., Berardi R. Aromatase Inhibitors in Premenopausal Women with Breast Cancer: The State of the Art and Future Prospects. Curr. Oncol. Tor. Ont. 2018;25:e168–e175. doi: 10.3747/co.25.3735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Arpino G., De Angelis C., Giuliano M., Giordano A., Falato C., De Laurentiis M., De Placido S. Molecular Mechanism and Clinical Implications of Endocrine Therapy Resistance in Breast Cancer. Oncology. 2009;77:23–37. doi: 10.1159/000258493. [DOI] [PubMed] [Google Scholar]
- 109.Duan L., Motchoulski N., Danzer B., Davidovich I., Shariat-Madar Z., Levenson V.V. Prolylcarboxypeptidase Regulates Proliferation, Autophagy, and Resistance to 4-Hydroxytamoxifen-Induced Cytotoxicity in Estrogen Receptor-Positive Breast Cancer Cells. J. Biol. Chem. 2011;286:2864–2876. doi: 10.1074/jbc.M110.143271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Samaddar J.S., Gaddy V.T., Duplantier J., Thandavan S.P., Shah M., Smith M.J., Browning D., Rawson J., Smith S.B., Barrett J.T., et al. A Role for Macroautophagy in Protection against 4-Hydroxytamoxifen–Induced Cell Death and the Development of Antiestrogen Resistance. Mol. Cancer Ther. 2008;7:2977. doi: 10.1158/1535-7163.MCT-08-0447. [DOI] [PubMed] [Google Scholar]
- 111.Cook K.L., Wärri A., Soto-Pantoja D.R., Clarke P.A., Cruz M.I., Zwart A., Clarke R. Hydroxychloroquine Inhibits Autophagy to Potentiate Antiestrogen Responsiveness in ER+ Breast Cancer. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2014;20:3222–3232. doi: 10.1158/1078-0432.CCR-13-3227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Amaral C., Augusto T.V., Tavares-da-Silva E., Roleira F.M.F., Correia-da-Silva G., Teixeira N. Hormone-Dependent Breast Cancer: Targeting Autophagy and PI3K Overcomes Exemestane-Acquired Resistance. J. Steroid Biochem. Mol. Biol. 2018;183:51–61. doi: 10.1016/j.jsbmb.2018.05.006. [DOI] [PubMed] [Google Scholar]
- 113.Fruman D.A., Rommel C. PI3K and Cancer: Lessons, Challenges and Opportunities. Nat. Rev. Drug Discov. 2014;13:140–156. doi: 10.1038/nrd4204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Loehberg C.R., Strissel P.L., Dittrich R., Strick R., Dittmer J., Dittmer A., Fabry B., Kalender W.A., Koch T., Wachter D.L., et al. Akt and P53 Are Potential Mediators of Reduced Mammary Tumor Growth by Cloroquine and the MTOR Inhibitor RAD001. Biochem. Pharmacol. 2012;83:480–488. doi: 10.1016/j.bcp.2011.11.022. [DOI] [PubMed] [Google Scholar]
- 115.Wang Y., Liu J., Qiu Y., Jin M., Chen X., Fan G., Wang R., Kong D. ZSTK474, a Specific Class I Phosphatidylinositol 3-Kinase Inhibitor, Induces G1 Arrest and Autophagy in Human Breast Cancer MCF-7 Cells. Oncotarget. 2016;7:19897–19909. doi: 10.18632/oncotarget.7658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Cai J., Xia J., Zou J., Wang Q., Ma Q., Sun R., Liao H., Xu L., Wang D., Guo X. The PI3K/MTOR Dual Inhibitor NVP-BEZ235 Stimulates Mutant P53 Degradation to Exert Anti-Tumor Effects on Triple-Negative Breast Cancer Cells. FEBS Open Bio. 2020;10:535–545. doi: 10.1002/2211-5463.12806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Ji Y., Di W., Yang Q., Lu Z., Cai W., Wu J. Inhibition of Autophagy Increases Proliferation Inhibition and Apoptosis Induced by the PI3K/MTOR Inhibitor NVP-BEZ235 in Breast Cancer Cells. Clin. Lab. 2015;61:1043–1051. doi: 10.7754/clin.lab.2015.150144. [DOI] [PubMed] [Google Scholar]
- 118.André F., Ciruelos E., Rubovszky G., Campone M., Loibl S., Rugo H.S., Iwata H., Conte P., Mayer I.A., Kaufman B., et al. SOLAR-1 Study Group. Alpelisib for PIK3CA-Mutated, Hormone Receptor-Positive Advanced Breast Cancer. N. Engl. J. Med. 2019;380:1929–1940. doi: 10.1056/NEJMoa1813904. [DOI] [PubMed] [Google Scholar]
- 119.Zhai B., Hu F., Jiang X., Xu J., Zhao D., Liu B., Pan S., Dong X., Tan G., Wei Z., et al. Inhibition of Akt Reverses the Acquired Resistance to Sorafenib by Switching Protective Autophagy to Autophagic Cell Death in Hepatocellular Carcinoma. Mol. Cancer Ther. 2014;13:1589–1598. doi: 10.1158/1535-7163.MCT-13-1043. [DOI] [PubMed] [Google Scholar]
- 120.IGF1R Upregulation Confers Resistance to Isoform-specific Inhibitors of PI3K in PIK3CA-driven Ovarian Cancer—Abstract—Europe PMC. [(accessed on 28 September 2020)]; doi: 10.1038/s41419-018-1025-8. Available online: https://europepmc.org/article/pmc/pmc6148236. [DOI] [PMC free article] [PubMed]
- 121.Im S.-A., Lu Y.-S., Bardia A., Harbeck N., Colleoni M., Franke F., Chow L., Sohn J., Lee K.-S., Campos-Gomez S., et al. Overall Survival with Ribociclib plus Endocrine Therapy in Breast Cancer. N. Engl. J. Med. 2019;381:307–316. doi: 10.1056/NEJMoa1903765. [DOI] [PubMed] [Google Scholar]
- 122.Piezzo M., Cocco S., Caputo R., Cianniello D., Gioia G.D., Lauro V.D., Fusco G., Martinelli C., Nuzzo F., Pensabene M., et al. Targeting Cell Cycle in Breast Cancer: CDK4/6 Inhibitors. Int. J. Mol. Sci. 2020;21 doi: 10.3390/ijms21186479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Wang R., Zhang Q., Peng X., Zhou C., Zhong Y., Chen X., Qiu Y., Jin M., Gong M., Kong D. Stellettin B Induces G1 Arrest, Apoptosis and Autophagy in Human Non-Small Cell Lung Cancer A549 Cells via Blocking PI3K/Akt/MTOR Pathway. Sci. Rep. 2016;6:27071. doi: 10.1038/srep27071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Hsieh F.-S., Chen Y.-L., Hung M.-H., Chu P.-Y., Tsai M.-H., Chen L.-J., Hsiao Y.-J., Shih C.-T., Chang M.-J., Chao T.-I., et al. Palbociclib Induces Activation of AMPK and Inhibits Hepatocellular Carcinoma in a CDK4/6-Independent Manner. Mol. Oncol. 2017;11:1035–1049. doi: 10.1002/1878-0261.12072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Valenzuela C.A., Vargas L., Martinez V., Bravo S., Brown N.E. Palbociclib-Induced Autophagy and Senescence in Gastric Cancer Cells. Exp. Cell Res. 2017;360:390–396. doi: 10.1016/j.yexcr.2017.09.031. [DOI] [PubMed] [Google Scholar]
- 126.Vijayaraghavan S., Karakas C., Doostan I., Chen X., Bui T., Yi M., Raghavendra A.S., Zhao Y., Bashour S.I., Ibrahim N.K., et al. CDK4/6 and Autophagy Inhibitors Synergistically Induce Senescence in Rb Positive Cytoplasmic Cyclin E Negative Cancers. Nat. Commun. 2017;8:15916. doi: 10.1038/ncomms15916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Okada Y., Kato S., Sakamoto Y., Oishi T., Ishioka C. Synthetic Lethal Interaction of CDK Inhibition and Autophagy Inhibition in Human Solid Cancer Cell Lines. Oncol. Rep. 2017;38:31–42. doi: 10.3892/or.2017.5684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Iriyama N., Hino H., Moriya S., Hiramoto M., Hatta Y., Takei M., Miyazawa K. The Cyclin-Dependent Kinase 4/6 Inhibitor, Abemaciclib, Exerts Dose-Dependent Cytostatic and Cytocidal Effects and Induces Autophagy in Multiple Myeloma Cells. Leuk. Lymphoma. 2018;59:1439–1450. doi: 10.1080/10428194.2017.1376741. [DOI] [PubMed] [Google Scholar]
- 129.Small J., Washburn E., Millington K., Zhu J., Holder S.L. The Addition of Abemaciclib to Sunitinib Induces Regression of Renal Cell Carcinoma Xenograft Tumors. Oncotarget. 2017;8:95116–95134. doi: 10.18632/oncotarget.19618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Hino H., Iriyama N., Kokuba H., Kazama H., Moriya S., Takano N., Hiramoto M., Aizawa S., Miyazawa K. Abemaciclib Induces Atypical Cell Death in Cancer Cells Characterized by Formation of Cytoplasmic Vacuoles Derived from Lysosomes. Cancer Sci. 2020;111:2132–2145. doi: 10.1111/cas.14419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Yin Q., Jian Y., Xu M., Huang X., Wang N., Liu Z., Li Q., Li J., Zhou H., Xu L., et al. CDK4/6 Regulate Lysosome Biogenesis through TFEB/TFE3. J. Cell Biol. 2020;219:e201911036. doi: 10.1083/jcb.201911036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Hsu J.L., Hung M.-C. The Role of HER2, EGFR, and Other Receptor Tyrosine Kinases in Breast Cancer. Cancer Metastasis Rev. 2016;35:575–588. doi: 10.1007/s10555-016-9649-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Meric-Bernstam F., Johnson A.M., Dumbrava E.E.I., Raghav K., Balaji K., Bhatt M., Murthy R.K., Rodon J., Piha-Paul S.A. Advances in HER2-Targeted Therapy: Novel Agents and Opportunities Beyond Breast and Gastric Cancer. Clin. Cancer Res. 2019;25(7):2033–2041. doi: 10.1158/1078-0432.CCR-18-2275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Negri T., Tarantino E., Orsenigo M., Reid J.F., Gariboldi M., Zambetti M., Pierotti M.A., Pilotti S. Chromosome Band 17q21 in Breast Cancer: Significant Association between Beclin 1 Loss and HER2/NEU Amplification. Genes. Chromosomes Cancer. 2010;49:901–909. doi: 10.1002/gcc.20798. [DOI] [PubMed] [Google Scholar]
- 135.Tang H., Sebti S., Titone R., Zhou Y., Isidoro C., Ross T.S., Hibshoosh H., Xiao G., Packer M., Xie Y., et al. Decreased BECN1 MRNA Expression in Human Breast Cancer Is Associated with Estrogen Receptor-Negative Subtypes and Poor Prognosis. EBioMedicine. 2015;2:255–263. doi: 10.1016/j.ebiom.2015.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Lozy F., Cai-McRae X., Teplova I., Price S., Reddy A., Bhanot G., Ganesan S., Vazquez A., Karantza V. ERBB2 Overexpression Suppresses Stress-Induced Autophagy and Renders ERBB2-Induced Mammary Tumorigenesis Independent of Monoallelic Becn1 Loss. Autophagy. 2014;10:662–676. doi: 10.4161/auto.27867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Vega-Rubín-de-Celis S., Zou Z., Fernández Á.F., Ci B., Kim M., Xiao G., Xie Y., Levine B. Increased Autophagy Blocks HER2-Mediated Breast Tumorigenesis. Proc. Natl. Acad. Sci. USA. 2018;115:4176–4181. doi: 10.1073/pnas.1717800115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Janser F.A., Adams O., Bütler V., Schläfli A.M., Dislich B., Seiler C.A., Kröll D., Langer R., Tschan M.P. Her2-Targeted Therapy Induces Autophagy in Esophageal Adenocarcinoma Cells. Int. J. Mol. Sci. 2018;19:3069. doi: 10.3390/ijms19103069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Autophagy-related Gene 12 (ATG12) is a Novel Determinant of Primary Resistance to HER2-targeted Therapies: Utility of Transcriptome Analysis of the Autophagy Interactome to Guide Breast Cancer Treatment—Abstract—Europe PMC. [(accessed on 28 September 2020)]; doi: 10.18632/oncotarget.742. Available online: https://europepmc.org/article/pmc/3681498. [DOI] [PMC free article] [PubMed]
- 140.Chen S., Zhu X., Qiao H., Ye M., Lai X., Yu S., Ding L., Wen A., Zhang J. Protective Autophagy Promotes the Resistance of HER2-Positive Breast Cancer Cells to Lapatinib. Tumour Biol. J. Int. Soc. Oncodev. Biol. Med. 2016;37:2321–2331. doi: 10.1007/s13277-015-3800-9. [DOI] [PubMed] [Google Scholar]
- 141.Chen S., Li X., Feng J., Chang Y., Wang Z., Wen A. Autophagy Facilitates the Lapatinib Resistance of HER2 Positive Breast Cancer Cells. Med. Hypotheses. 2011;77:206–208. doi: 10.1016/j.mehy.2011.04.013. [DOI] [PubMed] [Google Scholar]
- 142.Hardie D.G., Ross F.A., Hawley S.A. AMPK: A Nutrient and Energy Sensor That Maintains Energy Homeostasis. Nat. Rev. Mol. Cell Biol. 2012;13:251–262. doi: 10.1038/nrm3311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Cotzomi-Ortega I., Rosas-Cruz A., Ramírez-Ramírez D., Reyes-Leyva J., Rodriguez-Sosa M., Aguilar-Alonso P., Maycotte P. Autophagy Inhibition Induces the Secretion of Macrophage Migration Inhibitory Factor (MIF) with Autocrine and Paracrine Effects on the Promotion of Malignancy in Breast Cancer. Biology. 2020;9:20. doi: 10.3390/biology9010020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.O’Reilly E.A., Gubbins L., Sharma S., Tully R., Guang M.H.Z., Weiner-Gorzel K., McCaffrey J., Harrison M., Furlong F., Kell M., et al. The Fate of Chemoresistance in Triple Negative Breast Cancer (TNBC) BBA Clin. 2015;3:257–275. doi: 10.1016/j.bbacli.2015.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Overgaard J., Yilmaz M., Guldberg P., Hansen L.L., Alsner J. TP53 Mutation Is an Independent Prognostic Marker for Poor Outcome in Both Node-Negative and Node-Positive Breast Cancer. Acta Oncol. Stockh. Swed. 2000;39:327–333. doi: 10.1080/028418600750013096. [DOI] [PubMed] [Google Scholar]
- 146.ATG9A Is Overexpressed in Triple Negative Breast Cancer and Its In Vitro Extinction Leads to the Inhibition of Pro-Cancer Phenotypes—Abstract—Europe PMC. [(accessed on 28 September 2020)]; doi: 10.3390/cells7120248. Available online: https://europepmc.org/article/med/30563263. [DOI] [PMC free article] [PubMed]
- 147.Hamurcu Z., Delibaşı N., Geçene S., Şener E.F., Dönmez-Altuntaş H., Özkul Y., Canatan H., Ozpolat B. Targeting LC3 and Beclin-1 Autophagy Genes Suppresses Proliferation, Survival, Migration and Invasion by Inhibition of Cyclin-D1 and UPAR/Integrin Β1/ Src Signaling in Triple Negative Breast Cancer Cells. J. Cancer Res. Clin. Oncol. 2018;144:415–430. doi: 10.1007/s00432-017-2557-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Maycotte P., Gearheart C.M., Barnard R., Aryal S., Mulcahy Levy J.M., Fosmire S.P., Hansen R.J., Morgan M.J., Porter C.C., Gustafson D.L., et al. STAT3-Mediated Autophagy Dependence Identifies Subtypes of Breast Cancer Where Autophagy Inhibition Can Be Efficacious. Cancer Res. 2014;74:2579. doi: 10.1158/0008-5472.CAN-13-3470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Liang D.H., Choi D.S., Ensor J.E., Kaipparettu B.A., Bass B.L., Chang J.C. The Autophagy Inhibitor Chloroquine Targets Cancer Stem Cells in Triple Negative Breast Cancer by Inducing Mitochondrial Damage and Impairing DNA Break Repair. Cancer Lett. 2016;376:249–258. doi: 10.1016/j.canlet.2016.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Chen W., Bai Y., Patel C., Geng F. Autophagy Promotes Triple Negative Breast Cancer Metastasis via YAP Nuclear Localization. Biochem. Biophys. Res. Commun. 2019;520:263–268. doi: 10.1016/j.bbrc.2019.09.133. [DOI] [PubMed] [Google Scholar]
- 151.Maycotte P., Thorburn A. Targeting Autophagy in Breast Cancer. World J. Clin. Oncol. 2014;5:224–240. doi: 10.5306/wjco.v5.i3.224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Viry E., Noman M.Z., Arakelian T., Lequeux A., Chouaib S., Berchem G., Moussay E., Paggetti J., Janji B. Hijacker of the Antitumor Immune Response: Autophagy Is Showing Its Worst Facet. Front. Oncol. 2016;6:246. doi: 10.3389/fonc.2016.00246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Clark C.A., Gupta H.B., Sareddy G., Pandeswara S., Lao S., Yuan B., Drerup J.M., Padron A., Conejo-Garcia J., Murthy K., et al. Tumor-Intrinsic PD-L1 Signals Regulate Cell Growth, Pathogenesis, and Autophagy in Ovarian Cancer and Melanoma. Cancer Res. 2016;76:6964–6974. doi: 10.1158/0008-5472.CAN-16-0258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Wang X., Wu W.K.K., Gao J., Li Z., Dong B., Lin X., Li Y., Li Y., Gong J., Qi C., et al. Autophagy Inhibition Enhances PD-L1 Expression in Gastric Cancer. J. Exp. Clin. Cancer Res. CR. 2019;38:140. doi: 10.1186/s13046-019-1148-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Park S.-Y., Choi J.-H., Nam J.-S. Targeting Cancer Stem Cells in Triple-Negative Breast Cancer. Cancers. 2019;11 doi: 10.3390/cancers11070965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Al-Hajj M., Wicha M.S., Benito-Hernandez A., Morrison S.J., Clarke M.F. Prospective Identification of Tumorigenic Breast Cancer Cells. Proc. Natl. Acad. Sci. USA. 2003;100:3983–3988. doi: 10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Ginestier C., Hur M.H., Charafe-Jauffret E., Monville F., Dutcher J., Brown M., Jacquemier J., Viens P., Kleer C.G., Liu S., et al. ALDH1 Is a Marker of Normal and Malignant Human Mammary Stem Cells and a Predictor of Poor Clinical Outcome. Cell Stem Cell. 2007;1:555–567. doi: 10.1016/j.stem.2007.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Li W., Ma H., Zhang J., Zhu L., Wang C., Yang Y. Unraveling the Roles of CD44/CD24 and ALDH1 as Cancer Stem Cell Markers in Tumorigenesis and Metastasis. Sci. Rep. 2017;7:13856. doi: 10.1038/s41598-017-14364-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Van Phuc P., Nhan P.L., Nhung T.H., Tam N.T., Hoang N.M., Tue V.G., Thuy D.T., Ngoc P.K. Downregulation of CD44 Reduces Doxorubicin Resistance of CD44CD24 Breast Cancer Cells. OncoTargets Ther. 2011;4:71–78. doi: 10.2147/OTT.S21431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Bartucci M., Dattilo R., Moriconi C., Pagliuca A., Mottolese M., Federici G., Benedetto A.D., Todaro M., Stassi G., Sperati F., et al. TAZ Is Required for Metastatic Activity and Chemoresistance of breast cancer stem cells. Oncogene. 2015;34:681–690. doi: 10.1038/onc.2014.5. [DOI] [PubMed] [Google Scholar]
- 161.Palomeras S., Ruiz-Martínez S., Puig T. Targeting Breast Cancer Stem Cells to Overcome Treatment Resistance. Molecules. 2018;23:2193. doi: 10.3390/molecules23092193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Liu S., Cong Y., Wang D., Sun Y., Deng L., Liu Y., Martin-Trevino R., Shang L., McDermott S.P., Landis M.D., et al. Breast Cancer Stem Cells Transition between Epithelial and Mesenchymal States Reflective of Their Normal Counterparts. Stem Cell Rep. 2014;2:78–91. doi: 10.1016/j.stemcr.2013.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Liu H., Patel M.R., Prescher J.A., Patsialou A., Qian D., Lin J., Wen S., Chang Y.F., Bachmann M.H., Shimono Y., et al. Cancer Stem Cells from Human Breast Tumors Are Involved in Spontaneous Metastases in Orthotopic Mouse Models. Proc. Natl. Acad. Sci. USA. 2010;107:18115–18120. doi: 10.1073/pnas.1006732107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Ohi Y., Umekita Y., Yoshioka T., Souda M., Rai Y., Sagara Y., Sagara Y., Sagara Y., Tanimoto A. Aldehyde Dehydrogenase 1 Expression Predicts Poor Prognosis in Triple-Negative Breast Cancer. Histopathology. 2011;59:776–780. doi: 10.1111/j.1365-2559.2011.03884.x. [DOI] [PubMed] [Google Scholar]
- 165.Wu Q., Wang J., Liu Y., Gong X. Epithelial Cell Adhesion Molecule and Epithelial-Mesenchymal Transition Are Associated with Vasculogenic Mimicry, Poor Prognosis, and Metastasis of Triple Negative Breast Cancer. Int. J. Clin. Exp. Pathol. 2019;12:1678–1689. [PMC free article] [PubMed] [Google Scholar]
- 166.Mani S.A., Guo W., Liao M.J., Eaton E.N., Ayyanan A., Zhou A.Y., Brooks M., Reinhard F., Zhang C.C., Shipitsin M., et al. The Epithelial-Mesenchymal Transition Generates Cells with Properties of Stem Cells. Cell. 2008;133:704–715. doi: 10.1016/j.cell.2008.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Schito L., Semenza G.L. Hypoxia-Inducible Factors: Master Regulators of Cancer Progression. Trends Cancer. 2016;2:758–770. doi: 10.1016/j.trecan.2016.10.016. [DOI] [PubMed] [Google Scholar]
- 168.Michiels C. Physiological and Pathological Responses to Hypoxia. Am. J. Pathol. 2004;164:1875–1882. doi: 10.1016/S0002-9440(10)63747-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Samanta D., Gilkes D.M., Chaturvedi P., Xiang L., Semenza G.L. Hypoxia-Inducible Factors Are Required for Chemotherapy Resistance of Breast Cancer Stem Cells. Proc. Natl. Acad. Sci. USA. 2014;111:E5429–E5438. doi: 10.1073/pnas.1421438111. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 170.Liu L., Salnikov A.V., Bauer N., Aleksandrowicz E., Labsch S., Nwaeburu C., Mattern J., Gladkich J., Schemmer P., Werner J., et al. Triptolide Reverses Hypoxia-Induced Epithelial-Mesenchymal Transition and Stem-like Features in Pancreatic Cancer by NF-KappaB Downregulation. Int. J. Cancer. 2014;134:2489–2503. doi: 10.1002/ijc.28583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Conley S.J., Gheordunescu E., Kakarala P., Newman B., Korkaya H., Heath A.N., Clouthier S.G., Wicha M.S. Antiangiogenic Agents Increase Breast Cancer Stem Cells via the Generation of Tumor Hypoxia. Proc. Natl. Acad. Sci. USA. 2012;109:2784–2789. doi: 10.1073/pnas.1018866109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Schwab L.P., Peacock D.L., Majumdar D., Ingels J.F., Jensen L.C., Smith K.D., Cushing R.C., Seagroves T.N. Hypoxia-Inducible Factor 1alpha Promotes Primary Tumor Growth and Tumor-Initiating Cell Activity in Breast Cancer. Breast Cancer Res. 2012;14:R6. doi: 10.1186/bcr3087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Semenza G.L. Oxygen Sensing, Hypoxia-Inducible Factors, and Disease Pathophysiology. Annu. Rev. Pathol. 2014;9:47–71. doi: 10.1146/annurev-pathol-012513-104720. [DOI] [PubMed] [Google Scholar]
- 174.Buffa F.M., Harris A.L., West C.M., Miller C.J. Large Meta-Analysis of Multiple Cancers Reveals a Common, Compact and Highly Prognostic Hypoxia Metagene. Br. J. Cancer. 2010;102:428–435. doi: 10.1038/sj.bjc.6605450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Cao Y., Eble J.M., Moon E., Yuan H., Weitzel D.H., Landon C.D., Yu-Chih Nien C., Hanna G., Rich J.N., Provenzale J.M., et al. Tumor Cells Upregulate Normoxic HIF-1α in Response to Doxorubicin. Cancer Res. 2013;73:6230. doi: 10.1158/0008-5472.CAN-12-1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Daskalaki I., Gkikas I., Tavernarakis N. Hypoxia and Selective Autophagy in Cancer Development and Therapy. Front. Cell Dev. Biol. 2018;6:104. doi: 10.3389/fcell.2018.00104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Nazio F., Bordi M., Cianfanelli V., Locatelli F., Cecconi F. Autophagy and Cancer Stem Cells: Molecular Mechanisms and Therapeutic Applications. Cell Death Differ. 2019;26:690–702. doi: 10.1038/s41418-019-0292-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Chaterjee M., van Golen K.L. Breast Cancer Stem Cells Survive Periods of Farnesyl-Transferase Inhibitor-Induced Dormancy by Undergoing Autophagy. Bone Marrow Res. 2011;2011:362938. doi: 10.1155/2011/362938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Gong C., Song E., Codogno P., Mehrpour M. The Roles of BECN1 and Autophagy in Cancer Are Context Dependent. Autophagy. 2012;8:1853–1855. doi: 10.4161/auto.21996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Sk Y., J W., S C., Jl G. Autophagy Differentially Regulates Distinct Breast Cancer Stem-like Cells in Murine Models via EGFR/Stat3 and Tgfβ/Smad Signaling. [(accessed on 28 September 2020)]; doi: 10.1158/0008-5472.CAN-15-2946. Available online: https://pubmed.ncbi.nlm.nih.gov/27197172/ [DOI]
- 181.Golden E.B., Cho H.-Y., Jahanian A., Hofman F.M., Louie S.G., Schönthal A.H., Chen T.C. Chloroquine Enhances Temozolomide Cytotoxicity in Malignant Gliomas by Blocking Autophagy. Neurosurg. Focus. 2014;37:E12. doi: 10.3171/2014.9.FOCUS14504. [DOI] [PubMed] [Google Scholar]
- 182.Xie L., Xia L., Klaiber U., Sachsenmaier M., Hinz U., Bergmann F., Strobel O., Büchler M.W., Neoptolemos J.P., Fortunato F., et al. Effects of Neoadjuvant FOLFIRINOX and Gemcitabine-Based Chemotherapy on Cancer Cell Survival and Death in Patients with Pancreatic Ductal Adenocarcinoma. Oncotarget. 2019;10:7276–7287. doi: 10.18632/oncotarget.27399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Karasic T.B., O’Hara M.H., Loaiza-Bonilla A., Reiss K.A., Teitelbaum U.R., Borazanci E., De Jesus-Acosta A., Redlinger C., Burrell J.A., Laheru D.A., et al. Effect of Gemcitabine and Nab-Paclitaxel With or Without Hydroxychloroquine on Patients With Advanced Pancreatic Cancer: A Phase 2 Randomized Clinical Trial. JAMA Oncol. 2019;5:993–998. doi: 10.1001/jamaoncol.2019.0684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Malhotra J., Jabbour S., Orlick M., Riedlinger G., Guo Y., White E., Aisner J. Phase Ib/II Study of Hydroxychloroquine in Combination with Chemotherapy in Patients with Metastatic Non-Small Cell Lung Cancer (NSCLC) Cancer Treat. Res. Commun. 2019;21:100158. doi: 10.1016/j.ctarc.2019.100158. [DOI] [PubMed] [Google Scholar]
- 185.A Phase I Trial of MK-2206 and Hydroxychloroquine in Patients with Advanced Solid Tumors—Abstract—Europe PMC. [(accessed on 28 September 2020)]; Available online: https://europepmc.org/article/med/31463691.
- 186.Xu R., Ji Z., Xu C., Zhu J. The Clinical Value of Using Chloroquine or Hydroxychloroquine as Autophagy Inhibitors in the Treatment of Cancers: A Systematic Review and Meta-Analysis. Medicine. 2018;97:e12912. doi: 10.1097/MD.0000000000012912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Arnaout A., Robertson S.J., Pond G.R., Lee H., Jeong A., Ianni L., Kroeger L., Hilton J., Coupland S., Gottlieb C., et al. A Randomized, Double-Blind, Window of Opportunity Trial Evaluating the Effects of Chloroquine in Breast Cancer Patients. Breast Cancer Res. Treat. 2019;178:327–335. doi: 10.1007/s10549-019-05381-y. [DOI] [PubMed] [Google Scholar]
- 188.Augustijns P., Geusens P., Verbeke N. Chloroquine Levels in Blood during Chronic Treatment of Patients with Rheumatoid Arthritis. Eur. J. Clin. Pharmacol. 1992;42:429–433. doi: 10.1007/bf00280130. [DOI] [PubMed] [Google Scholar]
- 189.Schrezenmeier E., Dörner T. Mechanisms of Action of Hydroxychloroquine and Chloroquine: Implications for Rheumatology. Nat. Rev. Rheumatol. 2020;16:155–166. doi: 10.1038/s41584-020-0372-x. [DOI] [PubMed] [Google Scholar]