Abstract
Motivation
An intuitive graphical interface that allows statistical analyses and visualizations of extensive data without any knowledge of dedicated statistical software or programming.
Implementation
EpiMetal is a single-page web application written in JavaScript, to be used via a modern desktop web browser.
General features
Standard epidemiological analyses and self-organizing maps for data-driven metabolic profiling are included. Multiple extensive datasets with an arbitrary number of continuous and category variables can be integrated with the software. Any snapshot of the analyses can be saved and shared with others via a www-link. We demonstrate the usage of EpiMetal using pilot data with over 500 quantitative molecular measures for each sample as well as in two large-scale epidemiological cohorts (N >10 000).
Availability
The software usage exemplar and the pilot data are open access online at [http://EpiMetal.computationalmedicine.fi]. MIT licensed source code is available at the Github repository at [https://github.com/amergin/epimetal].
Introduction
We are living in a multi-omics era of systems epidemiology.1,2 Quantitative high-throughput metabolomics3–6 and lipidomics7,8 have resulted in hundreds of molecular measures for up to tens of thousands of people in multiple cohorts and biobanks. Extensive and complex data create significant challenges for statistical analyses. It would therefore be beneficial, not only for omics beginners but for all epidemiologists, to have a simple visual tool for rapid exploratory analyses of these kinds of modern datasets without the immediate need of bioinformaticians fluent with currently available professional statistical analysis tools as, for example, the R software.9
To this end, we developed a web browser-based graphical software—EpiMetal—for standard statistical epidemiological analyses as well as for multivariate self-organizing maps (SOMs) for data-driven analyses, metabolic profiling and potentially for clinical subgrouping.10–15 EpiMetal is versatile and any dataset with an arbitrary number of continuous and categorical variables can be easily integrated with the software. Data from multiple cohorts can be imported for comparative analyses. The original datasets can be divided into subgroups via multiple ways, for example based on SOMs, histograms or scatterplots; the created subgroups can be saved and analysed separately or in comparison with any other dataset. Regression analyses with covariate adjustments are available with graphical visualization of the results. Publication quality visualizations can be made and exported. Any snapshot of the analyses pipeline can be saved and shared with others via a www-link. Though it might not be an optimal choice to use EpiMetal for final publishable results, an additional good usage might be to use it as a benchmarking tool for newly written scripts and functions in another software.
As a usage exemplar, we present explorative analyses in a pilot cohort of 190 samples16–18 for which serum nuclear magnetic resonance (NMR) metabolomics3–6 and mass spectrometry (MS) lipidomics8,19 data are available. The data include over 500 quantitative molecular measures for each individual from these complementary methodologies that are getting increasingly popular in epidemiological applications. This is apparently the first time these comprehensive data are combined in an epidemiological setting. The data are made public along with the software (https://github.com/amergin/epimetal/blob/master/python-api/api-docker/samples.tsv). The exemplar demonstrates how the graphical interface of EpiMetal can be used to visualize extensive data, select subgroups and ultimately gain epidemiological insights via a combination of various statistical analysis options. In the supplement we also illustrate comparative analyses for two large-scale epidemiological cohorts.
Implementation
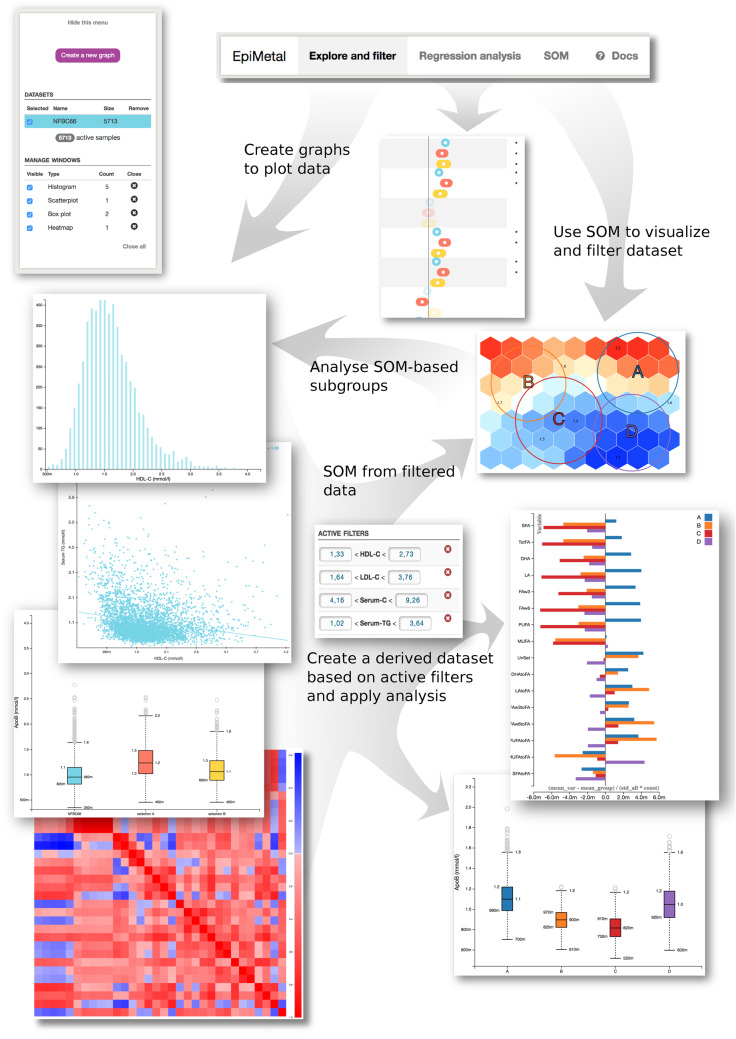
EpiMetal consists of three major components: (i) the database (MongoDB) is the long-term store for dataset samples, computational results and stored sessions; (ii) a single-page web application written in JavaScript (JS) that is accessed by users via a web browser; and (iii) a back-end software written in Python that serves as an intermediary between the web application and the database to retrieve data and record user sessions. The application uses third-party open-source libraries (versions and licensing information is available at Github). The software is encapsulated inside Docker containers to facilitate easy deployment across server platforms and to isolate the software from host system. Several Plotter instances can be run in parallel with differing configurations and data. The overall architecture of the software is presented in Supplementary Figure S1, available as Supplementary data at IJE online. Key data handling, visualization and statistical analyses features are summarized in Figure 1.
Figure 1.
Key data handling, visualization and statistical analyses features of the EpiMetal software illustrated using real epidemiological data (Northern Finland Birth Cohort 1966; N = 5713). A generalized flow of analysis begins by choosing a dataset(s) from user-uploaded options. This can be, for example, one population cohort but also a combination of many. Main analysis options are located in the top of the graphical interface and divided into three categories: ‘Explore and filter’, ‘Regression analysis’ and ‘SOM’. Under ‘Explore and filter’, the user can quickly generate basic plots to gain an overview of the data structure. Variables can be plotted and compared using histograms, scatterplots and boxplots. Heatmaps can also be created for an overall visualization of variable Spearman’s rank correlations. Active filters can also be applied to select subsets of the data. For example, one can choose to analyse only individuals with HDL-C <1.0 mmol/L in a given population cohort. The main category ‘Regression analysis’ allows the user to choose an outcome and exposure variables with an optional number of covariates and to generate a forest plot displaying the point estimate and 95% confidence intervals. Under ‘SOM’, the user can calculate a self-organizing map trained according to selected variables. The map can then be used to choose a subset of the entire dataset on the basis of this metabolic profiling. It should be noted that the analyses made in the ‘Explore and filter’ and ‘SOM’ sections are fully compatible with each other, enabling, for example, the SOM-based subgroups to be analysed via histograms and vice versa.
Installation
A step-by-step installation guide is provided in the EpiMetal software user guide. The source data are imported from a machine-readable format file (usually a tab-separated .tsv- or comma-separated values .csv-files) where each row corresponds to an individual sample. These samples have a unique identifier and usually belong to a single dataset. An import configuration file defines the column names and the column separator. A metadata file is needed for the source data to indicate variable name, free description, unit of measurement (e.g. mmol/L) and the group name for each variable. For variables that follow a common pattern, a regular expression pattern can be employed. Imported variable types are either numerical or categorical. Example files for the sample data are provided in the Github repository. There is no hard limit for the number of samples or variables that can be imported to the software, but the larger the dataset, the longer the download and processing times. The software has been tested with over 500 variables and some 30 000 samples, which present a realistic upper bound for current usage.
During the installation phase, a Docker container is set up that initializes the database from the source data file, along with the metadata descriptions. A second container is used for compiling the front-end application from the JS source files, forming a bundle. This container contains an http server to serve the bundle to user’s web browser. A third container runs the back end and its application programmatic interface (API). Using the Docker system, the EpiMetal software can be set up to serve an individual user locally, or to allow the software to be accessed by the public. An example configuration on how to limit the access to the software instance with a user name and a password is provided.
Architecture
The separation of concerns in EpiMetal is achieved by the common distinction between the presentation layer (front end) and the data access layer (back end). Modern desktop computers have considerable computing resources available, and thanks to the developments in web browser JS engines and web technologies, those resources can be fully appreciated in web applications. The philosophy of EpiMetal is to perform these calculations on the client side and store them to the database for later retrieval. This is achieved by asynchronously downloading chunks of the sample data and then performing the computations as requested by the user. Computationally heavy actions are processed in parallel using Web Workers, if supported by the browser.
AngularJS was chosen as the front-end web framework as it was popular at the time of starting the project, it had an active user base and several useful libraries, and its two-way data binding feature was appropriate considering the interactive nature of the application.
Back and front ends
The back end is a Python script developed with a Flask framework using Mongo Engine for object data mapping and served with Gunicorn HTTP server. The back end defines actions for retrieving the settings for the software, the metadata for variables, and samples for the requested variables. In addition, the back end is called to request previously stored SOM computations and SOM planes, and to store new ones.
The front end is a single-page application written in JS using AngularJS framework. Several open-source auxiliary JS libraries are employed, most notably DC.js for interactive charting throughout the application and Data-Driven Documents (D3.js) as a dependency for DC.js and for SOM planes and other chart types. Visual appearance and user interface (UI) stylings depend on Bootstrap framework and Angular-strap library. The UI allows the user to freely create, resize, move and close window-like objects containing figures. The front end fetches necessary data samples by querying the back end asynchronously as the user navigates on the page.
Figures produced with EpiMetal can be exported either in SVG or in PNG format. A particular state of the application can always be saved by creating a link to it and sharing the link with collaborators.
Usage exemplar: combined comprehensive metabolomics and lipidomics data
We present here explorative analyses in a unique pilot cohort of 190 blood samples16–18 for which serum NMR metabolomics3–6 and MS lipidomics8,19 data are available. All these molecular data, including basic clinical characteristics (age, sex, systolic and diastolic blood pressure, body mass index and height) have been made open access along with the EpiMetal software. The NMR metabolomics data comprise over 200 metabolic measures, including standard lipids, lipoprotein subclass and composition data, fatty acids, amino acids, ketones, glycolysis and gluconeogenesis-related substrates and an inflammatory marker, glycoprotein acetyls.3–6 The MS lipidomics data consist of over 350 individual lipid concentrations in 20 lipid classes including, for example, ceramides, sphingomyelins, phosphatidylcholines, phosphatidylinositols, cholesteryl esters and triacylglycerols.7,8,19 We used EpiMetal to conduct a multifaceted exploratory analysis of this pilot dataset. We sought to demonstrate some commonly known epidemiological and molecular features of these types of data.
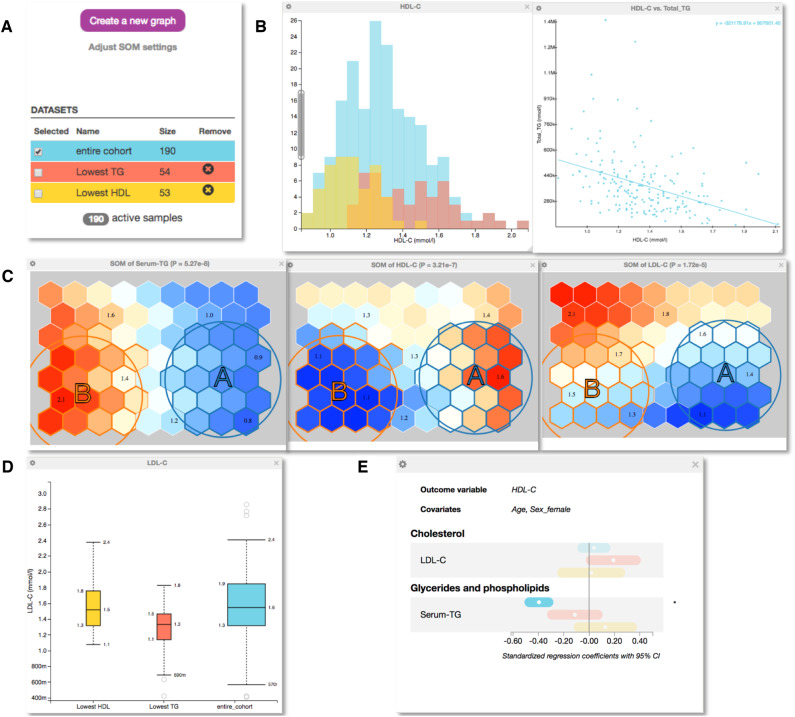
First, we plotted the distribution of high-density lipoprotein cholesterol (HDL-C) and the correlation of HDL-C with triglycerides (TG) in the entire dataset (Figure 2B). As expected, the histogram follows roughly a normal distribution. The scatterplot for HDL-C and TG association reveals the well-known negative population-level correlation.20
Figure 2.
Explorative analysis of a cohort of 190 samples with serum NMR metabolomics and mass spectrometry lipidomics measures available. A: The control panel of EpiMetal that contains clickable buttons for generating graphs and selecting, naming and generating subgroups. Colours indicate the entire cohort (cyan) and selected subgroups based on the self-organizing map (SOM) analysis. B: The histograms of HDL-C in the entire cohort and in the subgroups and the scatterplot of HDL-C vs triglycerides. C: The SOM component planes for serum triglycerides, HDL-C and LDL-C (note that the individuals in the entire cohort are identically distributed in each plane). Colours indicate high (red) and low (blue) concentration values of the variable in each plane. Individuals with similar metabolic profiles cluster close to each other in the SOM component planes. The user can specify and select different subgroups via the circular selection tools. D: A box plot for LDL-C in the entire cohort and in the two subgroups. E: Regression analyses with a forest plot showing standardized regression coefficients. Standardization means, that prior to analyses, all continuous, non-binary variables are normalized to zero mean and unit standard deviation. Point estimates are indicated by a dot surrounded by 95% confidence interval (CI). Plotting HDL-C as the outcome and triglycerides as an exposure illustrates the same negative association as already indicated via the scatterplot in B.
We then applied the SOM analysis to organize the samples in the dataset via their systemic metabolic profiles. Readers interested in the comparison of the SOM methodology with other subgrouping methods in epidemiology are referred to a recent Software Application Profile in the IJE.14 Additional details of the statistical issues in SOM analyses can be found in references.10–13 We based the SOM profiling on 26 metabolic measures including multiple amino acids, 14 lipoprotein subclasses, standard cholesterol measures, glycoprotein acetyls and glycolysis-related measures. It should be noted that users could freely modify the initial SOM training data according to their preferences and data characteristics. The SOM planes for low-density lipoprotein cholesterol (LDL-C), HDL-C and TG are shown in Figure 2C. These planes reveal, on average, that people with high circulating HDL-C (circle A in the SOM; subgroup marked lowest TG) are indeed those that have low TG and vice versa (area marked B in the SOM; subgroup marked lowest HDL), as expected by the previous scatter plot. The SOM analysis also reveals that circulating LDL-C concentrations are rather indifferent regarding HDL-C and TG in this pilot dataset; this is also emphasized in Figure 2D by the box plot for LDL-C. These associations can also be illustrated via formal regression analyses; we considered HDL-C as an outcome variable and LDL-C or TG as an exposure with age and sex as covariates. The results are given in Figure 2E for the entire cohort and the above-mentioned SOM-derived subgroups. The negative association between HDL-C and TG, depicted in Figure 2B, is well replicated in the formal regression analysis. These demonstrations indicate the internal consistency of various software functions and illustrate that the pilot cohort represents well-known features of lipoprotein metabolism with respect to lipoprotein lipid measures.
Exploration of associations between the NMR metabolomics and MS lipidomics data can be found in Supplementary Figure S2, available as Supplementary data at IJE online, which shows a heatmap of Spearman’s rank correlation coefficients between selected lipoprotein (NMR) and lipid variables (MS). Overall the correlations are very well in line with the known molecular characteristics of lipoprotein subclasses and their lipid compositions21 and demonstrate robust agreement between the NMR metabolomics and MS lipidomics platforms.
To additionally demonstrate the properties of EpiMetal, we performed an additional set of analyses using data from two large-scale population-based epidemiological cohorts including over 10 000 individuals (see the Supplement, available as Supplementary data at IJE online).
Conclusion
The new EpiMetal software is used via a modern web browser and it provides an intuitive easy-to-use graphical interphase for multiple statistical methods relevant in epidemiological analyses. It easily handles data for tens of thousands of people and for hundreds of measures—numbers that are a reality nowadays in many metabolomics applications. It provides instant data visualizations and allows convenient sharing of results and data via data captures accessible via an automatically created www-link. The datasets can be fully customized by the users. We illustrated the usage and opportunities of EpiMetal in real large-scale epidemiological datasets (Figure 1; and the Supplement, available as Supplementary data at IJE online). In addition, we provide an open access usage exemplar of EpiMetal for a pilot cohort in which over 500 quantitative molecular measures are available from each sample.
With increasing amounts of complex molecular data in epidemiology, sophisticated software is required for both convenient data handling and statistical analyses. Without statistical or programming expertise, the learning curve to conveniently use typical modern data analysis software, for example R,9 can be steep. From the epidemiology perspective, extensive molecular data may challenge traditional hypothesis-driven data analyses. These are common situations in which the EpiMetal software can help researchers. First, by enabling instant graphical exploration and analyses of a (new) dataset without the hurdles of programming-based data analyses; and second, by also allowing data-driven options to find unknown relations in the data without pre-existing hypotheses. As far as we are aware, the EpiMetal software is a first-of-a-kind versatile tool for both traditional and data-driven analyses of extensive large-scale epidemiological datasets.
Funding
M.A.K. is supported by a Senior Research Fellowship from the National Health and Medical Research Council (NHMRC) of Australia (APP1158958). He also works in a unit that is supported by the University of Bristol and UK Medical Research Council (MC_UU_12013/1). Q.W. is supported by the Novo Nordisk Foundation (NNF17OC0027034). B.A.K. is supported by a Senior Principal Research Fellowship from the NHMRC of Australia (APP1154331). The Sigrid Juselius Foundation and the Academy of Finland have also funded this work. The Northern Finland Birth Cohort 1966 and the Young Finns Study have been financially supported by multiple funding bodies (see the Supplement, available as Supplementary data at IJE online). The Baker Institute is supported in part by the Victorian Government’s Operational Infrastructure Support Program.
Conflict of interest: A.J.K. is an employee and shareholder of Nightingale Health Ltd, a company offering NMR-based metabolic profiling. Other authors declare no conflict of interest.
Supplementary Material
References
- 1. Sudlow C, Gallacher J, Allen N et al. UK Biobank: an open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLOS Med 2015;12:e1001779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ala-Korpela M, Davey Smith G. Metabolic profiling—multitude of technologies with great research potential, but (when) will translation emerge? Int J Epidemiol 2016;45:1311–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Soininen P, Kangas AJ, Würtz P, Suna T, Ala-Korpela M. Quantitative serum nuclear magnetic resonance metabolomics in cardiovascular epidemiology and genetics. Circ Cardiovasc Genet 2015;8:192–206. [DOI] [PubMed] [Google Scholar]
- 4. Würtz P, Kangas AJ, Soininen P, Lawlor DA, Davey Smith G, Ala-Korpela M. Quantitative serum nuclear magnetic resonance metabolomics in large-scale epidemiology: a primer on -omic technologies. Am J Epidemiol 2017;186:1084–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Würtz P, Wang Q, Soininen P et al. Metabolomic profiling of statin use and genetic inhibition of HMG-CoA reductase. J Am Coll Cardiol 2016;67:1200–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Sliz E, Kettunen J, Holmes MV et al. Metabolomic consequences of genetic inhibition of PCSK9 compared with statin treatment. Circulation 2018;138:2499–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Mundra PA, Shaw JE, Meikle PJ. Lipidomic analyses in epidemiology. Int J Epidemiol 2016;45:1329–38. [DOI] [PubMed] [Google Scholar]
- 8. Huynh K, Barlow CK, Jayawardana KS et al. High-throughput plasma lipidomics: detailed mapping of the associations with cardiometabolic risk factors. Cell Chem Biol 2019;26:71–84. [DOI] [PubMed] [Google Scholar]
- 9.R Development Core Team. R: A Language and Environment for Statistical Computing. Vienna: R Foundation for Statistical Computing, 2014. [Google Scholar]
- 10. Mäkinen V-P, Forsblom C, Thorn LM et al. Metabolic phenotypes, vascular complications, and premature deaths in a population of 4, 197 patients with type 1 diabetes. Diabetes 2008;57:2480–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kumpula LS, Mäkelä SM, Mäkinen V-P et al. Characterization of metabolic interrelationships and in silico phenotyping of lipoprotein particles using self-organizing maps. J Lipid Res 2010;51:431–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Mäkinen V-P, Tynkkynen T, Soininen P et al. Metabolic diversity of progressive kidney disease in 325 patients with type 1 diabetes (the FinnDiane Study). J Proteome Res 2012;11:1782–90. [DOI] [PubMed] [Google Scholar]
- 13. Lithovius R, Toppila I, Harjutsalo V et al. Data-driven metabolic subtypes predict future adverse events in individuals with type 1 diabetes. Diabetologia 2017;60:1234–43. [DOI] [PubMed] [Google Scholar]
- 14. Gao S, Mutter S, Casey A, Mäkinen V-P. Numero: a statistical framework to define multivariable subgroups in complex population-based datasets. Int J Epidemiol 2019;48:369–74. [DOI] [PubMed] [Google Scholar]
- 15. Mäkinen V-P, Kangas AJ, Soininen P, Würtz P, Groop P-H, Ala-Korpela M. Metabolic phenotyping of diabetic nephropathy. Clin Pharmacol Ther 2013;94:566–69. [DOI] [PubMed] [Google Scholar]
- 16. Khan AA, Mundra PA, Straznicky NE et al. Weight loss and exercise alter the high-density lipoprotein lipidome and improve high-density lipoprotein functionality in metabolic syndrome. Arteriosler Thromb Vasc Biol 2018;38:438–47. [DOI] [PubMed] [Google Scholar]
- 17. Straznicky NE, Lambert EA, Nestel PJ et al. Sympathetic neural adaptation to hypocaloric diet with or without exercise training in obese metabolic syndrome subjects. Diabetes 2010;59:71–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Straznicky NE, Grima MT, Sari CI et al. A randomized controlled trial of the effects of pioglitazone treatment on sympathetic nervous system activity and cardiovascular function in obese subjects with metabolic syndrome. J Clin Endocrinol Metab 2014;99:E1701–07. [DOI] [PubMed] [Google Scholar]
- 19. Weir JM, Wong G, Barlow CK et al. Plasma lipid profiling in a large population-based cohort. J Lipid Res 2013;54:2898–908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Schaefer EJ, Levy RI, Anderson DW, Danner RN, Brewer HB, Blackwelder WC. Plasma-triglycerides in regulation of HDL-cholesterol levels. Lancet 1978;2:391–93. [DOI] [PubMed] [Google Scholar]
- 21. Kumpula LS, Kumpula JM, Taskinen M-R, Jauhiainen M, Kaski K, Ala-Korpela M. Reconsideration of hydrophobic lipid distributions in lipoprotein particles. Chem Phys Lipids 2008;155:57–62. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.