Abstract
Background
Mendelian randomization (MR) is widely used to unravel causal relationships in epidemiological studies. Whereas multiple MR methods have been developed to control for bias due to horizontal pleiotropy, their performance in the presence of other sources of bias, like non-random mating, has been mostly evaluated using simulated data. Empirical comparisons of MR estimators in such scenarios have yet to be conducted. Pleiotropy and non-random mating have been shown to account equally for the genetic correlation between height and educational attainment. Previous studies probing the causal nature of this association have produced conflicting results.
Methods
We estimated the causal effect of height on educational attainment in various MR models, including the MR-Egger and the MR-Direction of Causation (MR-DoC) models that correct for, or explicitly model, horizontal pleiotropy.
Results
We reproduced the weak but positive association between height and education in the Netherlands Twin Register sample (P= 3.9 × 10–6). All MR analyses suggested that height has a robust, albeit small, causal effect on education. We showed via simulations that potential assortment for height and education had no effect on the causal parameter in the MR-DoC model. With the pleiotropic effect freely estimated, MR-DoC yielded a null finding.
Conclusions
Non-random mating may have a bearing on the results of MR studies based on unrelated individuals. Family data enable tests of causal relationships to be conducted more rigorously, and are recommended to triangulate results of MR studies assessing pairs of traits leading to non-random mate selection.
Keywords: Mendelian randomization, non-random mating, pleiotropy, height, educational attainment
Key Messages
We compare empirically the performance of various Mendelian randomization (MR) models in the presence of non-random/assortative mating.
We show that non-random mating may generate data that are consistent with a causal relationship between height and educational attainment, and may have a bearing on the results of MR studies.
MR models based on family data allow tests of causal hypotheses to be conducted in the presence of non-random mating (and other confounders such as dynastic effects). Summary statistics-based MR methods might be confounded in such scenarios.
Introduction
Causal inference based on observational data is challenging because of genetic and environmental confounding. Mendelian randomization (MR) has tremendous potential in addressing causality in the presence of confounding, and currently enjoys considerable methodological and substantive interest (for example Hemani et al.1, Verbanck et al.2). MR employs genetic variants associated with a given exposure as randomization instruments to detect the causal effect of the exposure on disease/outcome in non-experimental settings.3,4 Specifically, MR exploits the fact that the genetic variants’ meiosis is random with respect to environmental and social confounders,5,6 and they are not modified by disease outcomes (i.e. are not subject to reverse causality). The MR design yields information about causality, under the assumptions that the (genetic) instrument: (i) is well-associated with the exposure; (ii) is independent of confounders; and (iii) affects the outcome exclusively via the exposure variable (i.e. there is no horizontal pleiotropy). Although assumptions (i) and (ii) are likely to be tenable,7,8 assumption (iii) is unlikely to hold on account of the ubiquity of pleiotropy.9–12 As a result, multiple MR methods have been developed to control for bias due to horizontal pleiotropy.8,13–17
Yet another source of bias that may have a bearing on the results of MR studies is assortative mating18 (i.e. ‘the tendency of like to marry like’). MR approaches assume that the genetic variants used to instrument the analysis are randomly distributed in the population. This assumption draws heavily on random mating,8 the absence of inbreeding and the absence of selection on the genetic instrument. There is extensive evidence for assortative mating, specifically for height and education (for example Courtiol et al.,19 Hugh-Jones et al.,20 Yengo et al.21) as well as cross-trait,22 but also within (and between) psychiatric disorders like substance use.23,24 Recently, Hartwig and colleagues18 showed by means of simulations that assortative mating may bias the causal estimate in two-sample methods that are robust to pleiotropy, like MR-Egger or inverse variance weighted regression. Empirical comparisons of MR estimators in such scenarios have yet to be conducted.
Here we assess the performance of several methods robust to horizontal pleiotropy in the presence of assortative mating. Pleiotropy and assortative mating have been shown to account equally for the genetic correlation between height and educational attainment.22 We therefore consider these traits for our illustrative analyses. Empirical evidence has shown that height predicts attained education at different ages throughout the lifetime.25–28 There are several explanations for the height-educational attainment association.
The first is that stature has a causal effect on education.22,29–35 The reverse hypothesis that educational attainment causally influences height is arguably too implausible to merit consideration (but see Supplementary material, available as Supplementary data at IJE online, for a sensitivity check). The third ‘underlying factor’ hypothesis is that the association is due to common factors influencing both traits. Twin studies22,32 and studies in unrelated individuals which used measured genetic variants to disentangle the sources of this correlation33 showed that the correlation between the two traits is entirely explained by shared genetic factors.22,32,33 This genetic correlation can arise from vertical pleiotropy (i.e. the genetic effects on height are manifest in educational attainment due to the direct causal effect of height on education). The correlation may also be due to horizontal pleiotropy, i.e. due to overlap in the set of genes affecting brain size, intelligence and height, given the significant bivariate correlations observed between these traits (note that both intelligence and height are genetically correlated with brain volume36–41; see also Figure S1, available as Supplementary data at IJE online).
Alternatively, cross-trait assortative mating,18,22 dynastic effects and population structure42 may also induce genetic correlation between these traits. We consider these competing explanations in several MR models including MR-Egger14 and MR-direction of causation (MR-DoC) models16,17 that correct for, or explicitly model, horizontal pleiotropy. We also use simulations to assess the MR-DoC model’s robustness in the presence of assortative mating and finally, we employ the latent causal variable (LCV) model43 to account for potential bias arising from participant overlap44 in the two-sample MR approaches.
Methods
Ethical approval
The NTR study was approved by the Central Ethics Committee on Research Involving Human Subjects of the VU University Medical Centre, Amsterdam, an Institutional Review Board certified by the US Office of Human Research Protections (IRB number IRB-2991 under Federal-wide Assurance-3703; IRB/institute codes, NTR 03–180). All subjects provided written informed consent.
Sample
We evaluated the association of height and educational attainment (EA) in a Dutch sample from the Netherlands Twin Register (NTR).45,46 For the phenotypic association analysis, we selected 5899 Dutch adults (of whom 3184 were twins) with information on height and EA, relevant covariates (i.e. year of birth and sex) and observed genotypic data. EA was measured using a seven-category variable (1 = primary school only; 2 = lower educational schooling; 3 = lower vocational schooling; 4 = intermediate vocational schooling; 5 = intermediate/higher secondary schooling; 6 = higher vocational schooling; and 7 = university). The availability of repeated measures allowed us to run a series of consistency checks (see Supplementary material, available as Supplementary data at IJE online). Given the choice of the scheme used to code EA (1 through 7), and the fact that the underlying assumption of equivalent distances between adjacent EA categories is unlikely to hold in the case of ordinal data,47 we converted the seven-category variable to sex- and birth-cohort specific ridit scores47,48 as described in van Dongen et al.49 The resulting scores reflect males’ and females’ hierarchy of attained education in accordance with the societal values and historical conditions that define a given birth cohort. In short, we calculated the sex- and cohort-specific ridit scores for each EA category j by summing the proportion of cases below that category and one half the proportion of cases in the category47,48 Supplementary Table S1, available as Supplementary data at IJE online, shows descriptive statistics for the sample used in the analyses. Genome-wide single nucleotide polymorphisms (SNP) data were collected in the NTR sample on various genotyping platforms.45 Missing data were cross-platform imputed with the Genome of the Netherlands as the reference set.50–52 MaCH-Admix53 was used for phasing and imputation. We refer to the Supplementary material, available as Supplementary data at IJE online, for details on quality control checks performed before and after imputation.
Statistical analyses
Phenotypic association
Education was regressed on height and on the following covariates: age, sex and birth cohort (see Supplementary material, available as Supplementary data at IJE online for details). To correct for the effect of family clustering, we employed a generalized estimation equation linear model54 with an exchangeable model for the background familial covariance matrix and robust standard errors.55–57
Causality testing: one- and two-sample MR
We tested the causal effect of height on education in one- and two-sample MR models. The one-sample MR was based on the two-stage least squares (TSLS) procedure (but standard MR can also be performed using the Wald estimator) and on the MR-DoC model.17 As an instrumental variable we used a polygenic score, i.e. a weighed linear combination of SNPs. The weights used in calculating the polygenic score were obtained from the largest to date GWAS of height58 (excluding the NTR participants, see Supplementary material, available as Supplementary data at IJE online). Causality was tested in a subset of the sample comprising 3184 twins born between 1926 and 1989. The covariates used in the causality analysis were the dummy coded birth cohorts, age, sex, and 10 principal components (to correct for potential Dutch population structure). To increase the ethnic homogeneity of the sample, we removed ethnic outliers (see59 and Supplementary material, available as Supplementary data at IJE online).
The two-sample MR was based on MR-Egger,14 the inverse variance weighted approach, mode-60 and median-based MR methods.13,15 We assessed the suitability of summary data for conducting MR-Egger using the I2GX statistics.61 We also employed heterogeneity tests and Rucker’s framework62 to further assess pleiotropy and for model selection. To account for potential sample overlap that may bias the MR results, we used the latent causal variable (LCV) model43 (see Supplementary material, available as Supplementary data at IJE online). The two-sample MR used summary statistics for the SNPs available for both traits, downloaded from Wood et al.58 and from Okbay et al.,41 respectively. We refer to [https://gitlab.com/camish/empiricalcomparisons] for the R-script used in the analyses and to the Supplementary material, available as Supplementary data at IJE online, for more details on the methods employed.
Simulation: checking the robustness of MR-DoC in the presence of assortative mating
To evaluate the effect of assortative mating in the MR-DoC model, we created genotypes based on 150 diallelic loci (all minor allele frequencies = 0.5) in 50 000 parent pairs. Of the 150 loci, 50 affected height, 50 affected education and 50 had pleiotropic effects (i.e. affecting directly both height and EA). All 150 loci were used to compute polygenic scores, which were subsequently sorted ascendingly and used to divide the samples of mothers and fathers into five quintiles. Mate selection was contingent upon genetic similarity: males and females with the same quantile score were allowed to form parent pairs. To control the degree of assortative mating, we used a mover-stayer transition probability matrix to allow the males and females to switch to any other quintiles; the probability of switching to any other quantile equalled 0.05. Based on the resulting matings, we created two offspring per parent pair. The offspring were subsequently randomly assigned to pairs and the assortative mating procedure was repeated. After 10 generations, the familial (co)variances reached the expected equilibrium values63 such that parent-parent genetic correlations equalled about 0.4 and the sibling correlations equaled about 0.7. Half of the resulting sibling pairs were used to create MZ twin pairs, by duplicating the first genotype. Phenotypic data were next created based on the genotypes and environmental variation. Polygenic scores computed using the 20 (horizontally) pleiotropic loci were used as the genetic instrument in the MR-DoC model.
Results
The observational regression analysis in the NTR sample showed that height is associated with educational attainment (Wald χ2(1) = 21.3; P = 3.9 × 10–6; R2 = ∼0.3%). TSLS indicated that height has a causal effect on educational attainment (bcausal = 0.004; standard error (SE) = 0.001; Wald χ2(1) =6.7; P = 0.0096). Table 1 includes the results obtained using two-sample MR methods. The I2GX statistic equalled 0.84—a value close to the recommended value of 1—providing no evidence of regression dilution, and supporting the use of MR-Egger. Consistent with the TSLS and with the other pleiotropy-robust methods results (see Table 1), MR-Egger implied a causal effect of height on EA. The MR-Egger intercept provided no evidence of detectable directional pleiotropy (resulting in the mean value of the pleiotropy distribution deviating from zero). In line with the MR-Egger intercept result, the funnel plot suggested the ‘no directional pleiotropy’ assumption is likely to be satisfied (Figure 1).
Table 1.
Two-sample MR results of the height-educational attainment analysis
| Analysis method | Estimate | SE | 95% CI-lower | 95% CI-upper | P-value |
|---|---|---|---|---|---|
| IVW fixed effects | 0.04 | 0.002 | 0.036 | 0.043 | 3 × 10−53 |
| IVW random effects | 0.04 | 0.0047 | 0.030 | 0.049 | 2 × 10−17 |
| MR-Egger regression fixed effects: slope | 0.03 | 0.006 | 0.018 | 0.041 | 9 × 10−6 |
| intercept | 0.0003 | 0.0001 | −0.00006 | 0.0006 | 0.10 |
| MR-Egger regression random effects: slope | 0.03 | 0.012 | 0.006 | 0.053 | 1.8 × 10−02 |
| intercept | 0.0003 | 0.0003 | −0.0004 | 0.001 | 0.39 |
| Simple median | 0.043 | 0.005 | 0.033 | 0.052 | 2 × 10−13 |
| Weighted median | 0.037 | 0.005 | 0.027 | 0.046 | 1 × 10−9 |
| Penalized weighted median | 0.033 | 0.005 | 0.023 | 0.042 | 2 × 10−8 |
| Simple mode | 0.058 | 0.027 | 0.005 | 0.110 | 2 × 10−2 |
| Weighted mode | 0.066 | 0.023 | 0.020 | 0.111 | 1.8 × 10−3 |
CI, Confidence intervals; SE, standard error; IVW, inverse variance weighted model; MR, Mendelian randomization.
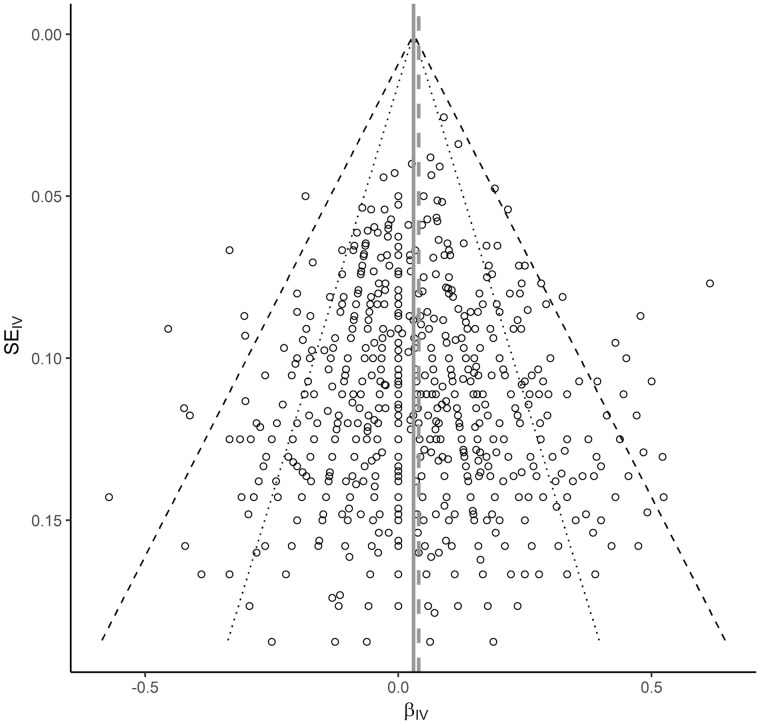
Figure 1.
Funnel plot for the height - educational attainment Mendelian Randomization analysis based on summary statistics. The dots represent the instrumental variable effects estimated based on each single nucleotide polymorphism (βIV), plotted against their precision (the standard error of the effect estimate, SEIV). The vertical solid gray line represents the MR-Egger intercept, the vertical gray dashed line represents the inverse variance weighted estimate. The plot resembles a symmetrical funnel suggesting there is no evidence for directional pleiotropy.
The Cochran’s Q test of heterogeneity64 detected heterogeneity (P <10–4). Here we assume this is due to balanced pleiotropy (i.e. the pleiotropy distribution has a zero mean); yet we note that heterogeneity may arise due to factors other than pleiotropy.62 The Rucker’s model selection framework (see Supplementary material, available as Supplementary data at IJE online, for details) showed there is no detectable directional pleiotropy, and indicated that an inverse variance weighted (IVW) random effects model adequately accommodated the heterogeneity present in the data (Qdiff(1) = 1.4; P = 0.2; where diff stands for the difference between the Cochran’s Q statistic—that tests for heterogeneity about the IVW model, and Rucker’s Q’ statistic—that tests for heterogeneity about the MR-Egger model). Consistent with these results, the MR-Egger intercept indicates that directional pleiotropy does not bias the estimate, and the Steiger test65 (which removes SNPs with potential reverse-causal effects) supports the conclusion that causation flows from height to EA. It can be seen from Figure 2 that all approaches based on summary statistics yielded consistent results.
Figure 2.

Scatterplot of the effect sizes on height and educational attainment of each of the 931 independent single nucleotide polymorphisms. Effect sizes are extracted from the genome-wide association study (GWAS) of height and from the GWAS of educational attainment.
The LCV model (see Supplementary material, available as Supplementary data at IJE online, for details), which is robust to sample overlap, found a genetic correlation of ρg = 0.15, and a genetic causality parameter GCP = 0.27 (SE = 0.08; z-score = 2.93; one-tailed P = 0.002), but did not reach the GCP value of at least 0.6 to claim robust positive finding.
MR-DoC: simulation and empirical results
Simulation results showed that non-random mate selection had no detectable effect on the causal effect estimate in the MR-DoC model when the pleiotropic effect is freely estimated (i.e. parameter b2 in Figure S2, available as Supplementary data at IJE online). The estimate of the causal effect (g1 in Figure S2, available as Supplementary data at IJE online) was close to its true value of zero (the statistical test of the hypothesis g1 = 0 was not rejected given alpha = 0.05). Interestingly, in the MR-DoC model with a non-pleiotropic polygenic score computed based on the 20 loci unique to height, non-random mating resulted in a pleiotropic effect (the statistical test of parameter b2 = 0 was rejected given alpha = 0.05). This suggests that assortative mating is accommodated by the b2 parameter.
The MR-DoC model17 explicitly estimates the amount of horizontal pleiotropy, but has to assume, for reasons of identification, that the unique environmental component influencing the exposure does not directly influence the outcome (other than through its effect on the exposure, i.e. re = 0, see Figure S2, available as Supplementary data at IJE online). With the parameter re constrained to 0, we tested the causal effect of height on EA while explicitly modelling horizontal pleiotropy (estimating the direct effect of the instrumental variable on the outcome). MR-DoC yielded a null finding: the causal effect was not different from zero at alpha = 0.05 (g1 = 0.0019; SE = 0.002; standardized g1 = 0.017; SE = 0.025; likelihood ratio test χ2(1) = 0.46; P = 0.49). When we assumed there is no horizontal pleiotropy (the parameter b2 in Figure S1, available as Supplementary data at IJE online, was fixed to zero), we detected a causal effect of height on EA (g1 = 0.004; SE = 0.001; standardized g1 = 0.039; SE = 0.015; χ2(1) = 6.9, P = 0.0085). The test of the pleiotropic effect failed to reject the null hypothesis of ‘no pleiotropy’ (b2 = 0.01; SE = 0.01; χ2(1) = 1.08; P = 0.29). Constraining b2 to zero and testing whether re = 0 suggested that the assumption is tenable (χ2(1) = 1.08; P = 0.29). However, the null findings concerning absence of non-shared environmental confounding and absence of directional pleiotropy may also reflect a lack of power (see Supplementary material, available as Supplementary data at IJE online). Importantly, constraining the parameter b2 to zero is not recommended in the light of ubiquity of pleiotropy across the human genome, and given that this parameter also accommodates potential non-random mating effects.
Discussion
We assessed the performance of several MR approaches in empirical data, in the presence of non-random mating. Recently, Hartwig and colleagues18 showed by means of simulations that non-random mating may bias the two-sample MR results based on summary statistics. Building on this work, the current study presented an empirical comparison of various one- and two-sample MR models when addressing the causal nature of the relationship between height and education (two traits extensively documented to lead to non-random mate selection). Previous studies addressing this relationship produced conflicting results. Tyrell et al.29 suggested that the observational association between height and educational attainment (indexed by a continuous variable representing time spent in full-time education) is causal in nature. Using MR-Egger regression and a TSLS model in the UK Biobank sample, they showed that taller individuals complete a larger number of years of formal education. Conversely, based on the IVW MR,15 Hagenaars et al.66 found no support for a causal effect flowing from height to education (indexed by a four-category variable representing degree level in the UK biobank). However, the authors noted that phenotype definition (i.e. using as the exposure a four-category variable instead of a continuous one as Tyrell et al. did), in conjunction with the use of weak genetic instruments may explain their null findings.66
Assuming all SNPs meet the three standard instrumental variable assumptions (see Introduction), the current one-sample TSLS results implied that height has a small positive causal effect on EA. This conclusion was reinforced by several alternative MR approaches robust to certain forms of pleiotropy, like IVW, median- and mode-based approaches and MR-Egger. Whereas the mode- and median-based approaches can each handle certain proportions of pleiotropic SNPs, MR-Egger explicitly tests for directional pleiotropy. The MR-Egger test showed there is no detectable directional pleiotropy. It is, however, important to note that the power of the MR-Egger test to detect pleiotropy may be low (see Table 1 in Bowden et al.14). We used heterogeneity tests as additional means to assess pleiotropy. Although pleiotropy might be present, we found that this is likely to be balanced, and may be adequately modelled by a random effects IVW model.
Sample overlap may affect the conclusions of summary statistics-based MR approaches, with large overlap biasing the causal effect estimate towards the observed association (because of weak instruments). The LCV model is robust to sample overlap and to other factors that may bias two-sample MR methods, like heritable confounding, differences in polygenicity or unequal sample sizes (see O’Connor and Price43 for details). This model provided weak evidence for the hypothesis that height affects education causally (genetic causality parameter GCP = 0.27; P = 0.002). Height and college completion are two of the traits studied by O'Connor and Price in their empirical applications in the UK Biobank.43 They found a genetic correlation between these traits of ρg = 0.17 and a GCP of 0.33. The positive GCP value lends some support to the hypothesis of height affecting education, but these values are rather too small to claim a robust positive finding. O’Connor and Price recommended absolute GCPs values larger than 0.6 as unlikely to be false-positives, and concluded that this low GCP value likely reflects shared developmental pathways rather than a direct causal effect. Further consideration of the GCP statistic supports this recommendation: a heritable latent confounder affecting height with a path coefficient of 0.55 and education with a path coefficient of 0.3 could yield a genetic correlation of ρg = 0.17 and a partial genetic causality parameter of GCP = 0.33. If the former coefficient were higher (say, at least 0.8), then one could reasonably argue that, although height is not literally a cause of education, it is nevertheless a very good proxy for some other variable that is a true cause.
The MR-DoC model is also useful when the confounding is predominantly genetic. The MR-DoC model has not detected a causal effect of height on EA (χ2(1) = 0.46; P = 0.49), unless we assumed directional pleiotropy was absent. The test of the pleiotropic effect could not reject the null hypothesis that the effect equals zero—a result consistent with the MR-Egger test result. The test of the parameter b2 suggests that the effect resulting from non-random mating is probably too weak to be picked up by the current sample. Notwithstanding this result, we argue against fixing this parameter to zero given the simulation results reported in Minică et al.16 (which showed that the estimate of the causal effect g1 is unbiased provided the pleiotropic effect b2 is freely estimated), and given the current simulation results (which showed assortative mating is accommodated by the b2 parameter). Estimating the b2 parameter might reduce power (if, indeed, pleiotropy is absent), but we rather err on the conservative side, given the ubiquity of pleiotropy across the human genome and given that non-random mating may presumably bias the causal effect if we do not model the directional pleiotropy path.
The usefulness of family data in addressing the non-random mating assumption was also demonstrated by Hartwig and colleagues.18 They showed, using data on trios, that height had no causal effect on education when correcting for possible effects of non-random mating (by using TSLS and adding the parental polygenic scores). However, as the authors acknowledge, it is worth noting that the small sample employed in this analysis may have resulted in imprecise estimates. The structural equation model developed by Warrington et al.,67 to estimate jointly maternal and fetal effects on birthweight, may also be extended to provide another means to address the potential bias induced by non-random mating.
Family data can additionally address bias in MR studies arising from dynastic/genetic nurture effects that may also confound the association between height and education.18,42,68 For instance, when available, trios data enable causality tests robust to dynastic effects;18,68 while providing a straightforward means to address dynastic effects and non-random mating; this approach requires trios data and needs to assume absence of horizontal pleiotropy. The within-family-based approach described by Brumpton et al.42 may also address dynastic effects. These within-family estimates of SNP outcome and SNP exposure effects obtained in separate sibling samples can be used, in a second step, with pleiotropy-robust summary data estimators; yet, as the authors note, these family-based estimates are likely to be underpowered to allow for both family structure and pleiotropy. It is important to note that the MR-DoC model in some scenarios has to make the strong assumption that there is no appreciable non-shared environmental confounding16 However, the model does account for environmental confounding shared by twins. In so doing, MR-DoC also controls for dynastic effects because twins share the environments created by parents. As twins share a womb and a household/neighbourhood, often until age 18, this is non-trivial. Dynastic effects are not controlled for in MR on unrelated individuals.69,70
To conclude, one should remain cautious when addressing the nature of the relationship between height and education as there are other confounders (beside horizontal pleiotropy) that may yield the observed association. This empirical example showed that non-random mating and, possibly, dynastic effects may generate data that are remarkably consistent with a causal model for the relationship between height and education. These factors—and others like fine-scale population structure42,71—may affect this relationship, and pleiotropy-robust methods are not necessarily robust to these forms of confounding. Structural equation models67 and MR models that exploit family data16,18,42 should enable tests of causal relationships to be conducted more rigorously, and are recommended to triangulate72 results of MR studies assessing pairs of complex traits leading to non-random mate selection.
Funding
This work was supported by the National Institute on Drug Abuse (grant number DA-018673).
Supplementary Material
Acknowledgements
We are grateful to our reviewers for their excellent comments, which substantially improved the paper. We are grateful to the Netherlands Twin Register participants whose data we analysed in this study. We thank the participants in the GIANT and the SSAG consortia for making the GWAS summary statistics for height and educational attainment publicly available.
Conflict of interest
None declared.
References
- 1. Hemani G, Zheng J, Elsworth B et al. The MR-Base platform supports systematic causal inference across the human phenome. Elife 2018;7:e34408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Verbanck M, Chen C-Y, Neale B, Do R. Detection of widespread horizontal pleiotropy in causal relationships inferred from Mendelian randomization between complex traits and diseases. Nat Genet 2018;50:693–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Davey Smith G, Hemani G. Mendelian randomization: genetic anchors for causal inference in epidemiological studies. Hum Mol Genet 2014;23:R89–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Evans DM, Davey Smith G. Mendelian randomization: new applications in the coming age of hypothesis-free causality. Annu Rev Genom Hum Genet 2015;16:327–50. [DOI] [PubMed] [Google Scholar]
- 5.Davey Smith G, Lawlor DA, Harbord R, Timpson N, Day I, Ebrahim S. Clustered environments and randomized genes: a fundamental distinction between conventional and genetic epidemiology. PLoS Med 2007;4:e352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Davey Smith G. Use of genetic markers and gene-diet interactions for interrogating population-level causal influences of diet on health. Genes Nutr 2011;6:27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. MacArthur J, Bowler E, Cerezo M et al. The new NHGRI-EBI Catalog of published genome-wide association studies (GWAS Catalog). Nucleic Acids Res 2017;45:D896–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Burgess S, Thompson SG. Mendelian Randomization: Methods for Using Genetic Variants in Causal Estimation. Boca Raton, FL: CRC Press, 2015. [Google Scholar]
- 9. Bulik-Sullivan B, Finucane HK, Anttila V et al. An atlas of genetic correlations across human diseases and traits. Nat Genet 2015;47:1236–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chesmore K, Bartlett J, Williams SM. The ubiquity of pleiotropy in human disease. Hum Genet 2018;137:1236–41. [DOI] [PubMed] [Google Scholar]
- 11. Visscher PM, Yang J. A plethora of pleiotropy across complex traits. Nat Genet 2016;48:707–08. [DOI] [PubMed] [Google Scholar]
- 12. Pickrell JK, Berisa T, Liu JZ, Ségurel L, Tung JY, Hinds DA. Detection and interpretation of shared genetic influences on 42 human traits. Nat Genet 2016;48:709–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Burgess S, Dudbridge F, Thompson SG. Re:“Multivariable Mendelian randomization: the use of pleiotropic genetic variants to estimate causal effects”. Am J Epidemiol 2015;181:290–91. [DOI] [PubMed] [Google Scholar]
- 14. Bowden J, Davey Smith G, Burgess S. Mendelian randomization with invalid instruments: effect estimation and bias detection through Egger regression. Int J Epidemiol 2015;44:512–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bowden J, Davey Smith G, Haycock PC, Burgess S. Consistent estimation in Mendelian randomization with some invalid instruments using a weighted median estimator. Genet Epidemiol 2016;40:304–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Minică CC, Dolan CV, Boomsma DI, de Geus E, Neale MC. Extending causality tests with genetic instruments: an integration of Mendelian randomization and the classical twin design. bioRxiv 2017:134585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Minică CC, Dolan CV, Boomsma DI, de Geus E, Neale MC. Extending causality tests with genetic instruments: an integration of Mendelian randomization with the classical twin design. Behav Genet 2018;48:337––49.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hartwig FP, Davies NM, Davey Smith G. Bias in Mendelian randomization due to assortative mating. Genet Epidemiol 2018;42:608–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Courtiol A, Raymond M, Godelle B, Ferdy JB. Mate choice and human stature: homogamy as a unified framework for understanding mating preferences. Evolution 2010;64:2189–203. [DOI] [PubMed] [Google Scholar]
- 20. Hugh-Jones D, Verweij KJ, Pourcain BS, Abdellaoui A. Assortative mating on educational attainment leads to genetic spousal resemblance for polygenic scores. Intelligence 2016;59:103–08. [Google Scholar]
- 21. Yengo L, Robinson MR, Keller MC et al. Imprint of assortative mating on the human genome. Nat Hum Behav 2018;2:948–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Keller MC, Garver-Apgar CE, Wright MJ et al. The genetic correlation between height and IQ: shared genes or assortative mating? PLoS Genet 2013;9:e1003451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Agrawal A, Heath AC, Grant JD et al. Assortative mating for cigarette smoking and for alcohol consumption in female Australian twins and their spouses. Behav Genet 2006;36:553–66. [DOI] [PubMed] [Google Scholar]
- 24. Merikangas KR, Risch N. Genomic priorities and public health. Science 2003;302:599–601. [DOI] [PubMed] [Google Scholar]
- 25. Magnusson PK, Rasmussen F, Gyllensten UB. Height at age 18 years is a strong predictor of attained education later in life: cohort study of over 950 000 Swedish men. Int J Epidemiol 2006;35:658–63. [DOI] [PubMed] [Google Scholar]
- 26. Huang Y, van Poppel F, Lumey LH. Differences in height by education among 371,105 Dutch military conscripts. Econ Hum Biol 2015;17(Suppl C):202–07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Gorry D. The influence of height on academic outcomes. Econ Educ Rev 2017;56(Suppl C):1–8. [Google Scholar]
- 28. Case A, Paxson C. Causes and consequences of early-life health. Demography 2010;47:S65–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Tyrrell J, Jones SE, Beaumont R et al. Height, body mass index, and socioeconomic status: Mendelian randomisation study in UK Biobank. BMJ 2016;352:i582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Carvalho L. Childhood circumstances and the intergenerational transmission of socioeconomic status. Demography 2012;49:913–38. [DOI] [PubMed] [Google Scholar]
- 31. Vogl TS. Education and health in developing economies. In: Encyclopedia of Health Economics. Amsterdam: Elsevier, 2012, pp. 246–9.
- 32. Silventoinen K, Posthuma D, Van Beijsterveldt T, Bartels M, Boomsma DI. Genetic contributions to the association between height and intelligence: evidence from Dutch twin data from childhood to middle age. Genes Brain Behav 2006;5:585–95. [DOI] [PubMed] [Google Scholar]
- 33. Marioni RE, Batty GD, Hayward C et al. Common genetic variants explain the majority of the correlation between height and intelligence: the Generation Scotland study. Behav Genet 2014;44:91–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Black RE, Allen LH, Bhutta ZA et al. Maternal and child undernutrition: global and regional exposures and health consequences. Lancet 2008;371:243–60. [DOI] [PubMed] [Google Scholar]
- 35. Perkins JM, Subramanian SV, Davey Smith G, Özaltin E. Adult height, nutrition, and population health. Nutr Rev 2016;74:149–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Posthuma D, De GE, Neale M et al. Multivariate genetic analysis of brain structure in an extended twin design. Behav Genet 2000;30:311–19. [DOI] [PubMed] [Google Scholar]
- 37. Posthuma D, De Geus EJ, Baaré WF, Pol HEH, Kahn RS, Boomsma DI. The association between brain volume and intelligence is of genetic origin. Nat Neurosci 2002;5:83–84. [DOI] [PubMed] [Google Scholar]
- 38. Rushton JP, Ankney CD. Whole brain size and general mental ability: a review. Int J Neurosci 2009;119:692–732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Deary IJ, Penke L, Johnson W. The neuroscience of human intelligence differences. Nat Rev Neurosci 2010;11:201–11. [DOI] [PubMed] [Google Scholar]
- 40. McDaniel MA. Big-brained people are smarter: a meta-analysis of the relationship between in vivo brain volume and intelligence. Intelligence 2005;33:337–46. [Google Scholar]
- 41. Okbay A, Beauchamp JP, Fontana MA et al. Genome-wide association study identifies 74 loci associated with educational attainment. Nature 2016;533:539–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Brumpton B, Sanderson E, Hartwig FP et al. Within-family studies for Mendelian randomization: avoiding dynastic, assortative mating, and population stratification biases. bioRxiv 2019:602516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. O’Connor LJ, Price AL. Distinguishing genetic correlation from causation across 52 diseases and complex traits. Nat Genet 2018;50:1728–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Burgess S, Davies NM, Thompson SG. Bias due to participant overlap in two‐sample Mendelian randomization. Genet Epidemiol 2016;40:597–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Willemsen G, De Geus EJ, Bartels M et al. The Netherlands Twin Register biobank: a resource for genetic epidemiological studies. Twin Res Hum Genet 2010;13:231–45. [DOI] [PubMed] [Google Scholar]
- 46. Willemsen G, Vink JM, Abdellaoui A et al. The Adult Netherlands Twin Register: twenty-five years of survey and biological data collection. Twin Res Hum Genet 2013;16:271–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Donaldson GW. Ridit scores for analysis and interpretation of ordinal pain data. Eur J Pain 1998;2:221–27. [DOI] [PubMed] [Google Scholar]
- 48. Bross ID. How to use ridit analysis. Biometrics 1958;14:18–38. [Google Scholar]
- 49. van Dongen J, Bonder MJ, Dekkers KF et al. DNA methylation signatures of educational attainment. NPJ Sci Learn 2018;3:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Boomsma DI, Wijmenga C, Slagboom EP et al. The Genome of the Netherlands: design, and project goals. Eur J Hum Genet 2014;22:221–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Fedko IO, Hottenga J-J, Medina-Gomez C et al. Estimation of genetic relationships between individuals across cohorts and platforms: application to childhood height. Behav Genet 2015;45:514–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Francioli LC, Menelaou A, Pulit SL et al. Whole-genome sequence variation, population structure and demographic history of the Dutch population. Nat Genet 2014;46:818–25. [DOI] [PubMed] [Google Scholar]
- 53. Liu EY, Li M, Wang W, Li Y. MaCH‐Admix: genotype imputation for admixed populations. Genet Epidemiol 2013;37:25–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Carey VJ, Lumley T, Ripley B. gee: Generalized Estimation Equation solver, R package version 4.13-18. 2012. http://CRAN.R-project.org/package=gee (2017, date last accessed).
- 55. Dobson A. An Introduction to Generalized Linear Models (Chatfield C, Zidek J, eds). London: Chapman & Hall/CRC, 2002. [Google Scholar]
- 56. Minică C, Boomsma D, Vink J, Dolan C. MZ twin pairs or MZ singletons in population family-based GWAS? More power in pairs. Mol Psychiatry 2014;19:1154–55. [DOI] [PubMed] [Google Scholar]
- 57. Minică CC, Dolan CV, Kampert MM, Boomsma DI, Vink JM. Sandwich corrected standard errors in family-based genome-wide association studies. Eur J Hum Genet 2015;23:388–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Wood AR, Esko T, Yang J et al. Defining the role of common variation in the genomic and biological architecture of adult human height. Nat Genet 2014;46:1173–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Abdellaoui A, Hottenga J-J, de Knijff P et al. Population structure, migration, and diversifying selection in the Netherlands. Eur J Hum Genet 2013;21:1277–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Hartwig FP, Davey Smith G, Bowden J. Robust inference in summary data Mendelian randomization via the zero modal pleiotropy assumption. Int J Epidemiol 2017;46:1285–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Bowden J, Del Greco MF, Minelli C, Davey Smith G, Sheehan NA, Thompson JR. Assessing the suitability of summary data for two-sample Mendelian randomization analyses using MR-Egger regression: the role of the I 2 statistic. Int J Epidemiol 2016;45:1961–974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Bowden J, Del Greco MF, Minelli C, Davey Smith G, Sheehan N, Thompson J. A framework for the investigation of pleiotropy in two‐sample summary data Mendelian randomization. Stat Med 2017;36:1783–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Frankham R. Introduction to Quantitative Genetics 4th edn (Falconer DS, Trudy FC, Longman M, eds). New York, NY: Elsevier Current Trends, 1996. [Google Scholar]
- 64. Greco M FD, Minelli C, Sheehan NA, Thompson JR. Detecting pleiotropy in Mendelian randomisation studies with summary data and a continuous outcome. Stat Med 2015;34:2926–40. [DOI] [PubMed] [Google Scholar]
- 65. Hemani G, Tilling K, Smith G. Orienting the causal relationship between imprecisely measured traits using GWAS summary data. PLoS Genet 2017;13:e1007081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Hagenaars SP, Gale CR, Deary IJ, Harris SE. Cognitive ability and physical health: a Mendelian randomization study. Sci Rep 2017;7: 2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Warrington NM, Freathy RM, Neale MC, Evans DM. Using structural equation modelling to jointly estimate maternal and fetal effects on birthweight in the UK Biobank. Int J Epidemiol 2018;47:1229–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Kong A, Thorleifsson G, Frigge ML et al. The nature of nurture: Effects of parental genotypes. Science 2018;359:424–28. [DOI] [PubMed] [Google Scholar]
- 69. Fletcher JM, Lehrer SF. Genetic lotteries within families. J Health Econ 2011;30:647–59. [DOI] [PubMed] [Google Scholar]
- 70. Pingault J-B, Cecil CA, Murray J, Munafò MR, Viding E. Causal inference in psychopathology: a systematic review of Mendelian randomisation studies aiming to identify environmental risk factors for psychopathology. Psychopathol Rev 2017;4:4–25. [Google Scholar]
- 71. Abdellaoui A, Hugh-Jones D, Yengo L et al. Genetic correlates of social stratification in Great Britain. Nat Hum Behav 2019;3:1332–42. [DOI] [PubMed] [Google Scholar]
- 72. Munafò MR, Smith G. Robust research needs many lines of evidence. Nature 2018;553:399–401. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.