Abstract
The family Erythrobacteraceae , belonging to the order Sphingomonadales , class Alphaproteobacteria , is globally distributed in various environments. Currently, this family consist of seven genera: Altererythrobacter , Croceibacterium , Croceicoccus , Erythrobacter , Erythromicrobium , Porphyrobacter and Qipengyuania . As more species are identified, the taxonomic status of the family Erythrobacteraceae should be revised at the genomic level because of its polyphyletic nature evident from 16S rRNA gene sequence analysis. Phylogenomic reconstruction based on 288 single-copy orthologous clusters led to the identification of three separate clades. Pairwise comparisons of average nucleotide identity, average amino acid identity (AAI), percentage of conserved protein and evolutionary distance indicated that AAI and evolutionary distance had the highest correlation. Thresholds for genera boundaries were proposed as 70 % and 0.4 for AAI and evolutionary distance, respectively. Based on the phylo-genomic and genomic similarity analysis, the three clades were classified into 16 genera, including 11 novel ones, for which the names Alteraurantiacibacter, Altericroceibacterium, Alteriqipengyuania, Alteripontixanthobacter, Aurantiacibacter, Paraurantiacibacter, Parerythrobacter, Parapontixanthobacter, Pelagerythrobacter, Tsuneonella and Pontixanthobacter are proposed. We reclassified all species of Erythromicrobium and Porphyrobacter as species of Erythrobacter . This study is the first genomic-based study of the family Erythrobacteraceae , and will contribute to further insights into the evolution of this family.
Keywords: Erythrobacteraceae, phylogenomic reconstruction, AAI, evolutionary distance
Introduction
The family Erythrobacteraceae , belonging to the order Sphingomonadales , class Alphaproteobacteria [1], is distributed globally, inhabiting various environments including subterrestrial, lake, intertidal areas, mangrove, coastal and deep-sea sediments [2–10], soil [11–13], desert sands [14, 15], a stadium seat [16], seawater [17–19], estuary water [20–22], fresh water [23, 24], hot springs [25–27], air [28] as well as plants and animals [29–36] (Table S1, available in the online version of this article). The members of the family Erythrobacteraceae are Gram-stain-negative, rod or pleomorphic coccoid-shaped, pink-, red-, orange- or yellow-pigmented, and aerobic chemoorganotrophs [1]. The majority require NaCl for growth [1]. Ubiquinone-10 (Q-10) is the major respiratory quinone [1, 2, 30].
The family was established by Lee et al. who included the genera Erythrobacter (Erb. litoralis and Erb. longus), Erythromicrobium (Erm. ramosum) and Porphyrobacter ( Por. neustonensis and Por. tepidarius ) based on 16S rRNA gene phylogeny in 2005 [37]. Four other genera including Altererythrobacter (Aeb.), Croceibacterium (Crb.), Croceicoccus (Ccc.) and Qipengyuania (Qpy.) were later proposed by Kwon et al. [38], Liu et al. [30], Xu et al. [4] and Feng et al. [2], respectively, based on 16S rRNA gene phylogeny [2, 4, 30, 38]. At the time of writing (September 2019), the genera Altererythrobacter , Croceibacterium , Croceicoccus , Erythrobacter , Erythromicrobium , Porphyrobacter and Qipengyuania consist of 41, two, four, 23, one, eight and one species, respectively [9, 10, 17, 19, 22, 30, 35, 39–43]. With the increase in the number of species proposed, the taxonomic status of the family Erythrobacteraceae should be revised in view of the polyphyletic nature of the group based on 16S rRNA gene sequence comparison [16, 30, 44]. The family Erythrobacteraceae includes aerobic anoxygenic phototrophic bacteria (AAPB), which can harvest light energy and play a significant role in the carbon cycling of the oceans globally [45–47]. Members of the family also show bioremediation and industrial potential, such as degradation of benzo[a]pyrene [48] and oil [49], and production of erythrazoles [50] and erythrolic acids [51]. A comprehensively taxonomic investigation of the family Erythrobacteraceae could not only lead to an improved classification of its members, but also broaden our understanding of their ecology and potential biotechnological applications.
Development of genome sequencing technologies has made bacterial genomic data more and more accessible, resulting in a revolution in bacterial taxonomy [52–54]. Phylogenomic reconstruction can provide a higher-resolution phylogeny than that based on 16S rRNA gene or several housekeeping genes [55–58]. In addition, genomic similarity calculations including average nucleotide identity (ANI), average amino acid identity (AAI) and percentage of conserved protein (POCP) provide numerical thresholds for delineation of each taxon [59–61]. Therefore, a genome-wide investigation of the taxonomy of the family Erythrobacteraceae was performed to revise the taxonomic status of this family.
Methods
Collection of Erythrobacteraceae type strains
In addition to the 47 Erythrobacteraceae type strains for which genome sequences were available, 27 type strains were obtained from culture collections including the China General Microbiological Culture Collection (CGMCC), the Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH (DSMZ), the Japan Collection of Microorganisms (JCM), the Korean Collection for Type Cultures (KCTC), the Korea Environmental Microorganism Bank (KEMB), the Collection of the Laboratorium voor Microbiologie en Microbiele Genetica (LMG) and the Marine Culture Collection of China (MCCC) or received as gifts from other scholars (Table 1 and Acknowledgements). These type strains were cultivated under appropriate conditions proposed previously [3, 12, 14, 20, 28, 31–33, 36, 44, 62–77] for subsequent genomic sequencing.
Table 1.
Genomic information for Erythrobacteraceae strains included in this study
DOE-JGI, U.S. Department of Energy, Joint Genome Institute; KRIBB, Korea Research Institute of Bioscience and Biotechnology; SDU, Shandong University.
|
Strain |
NCBI GenBank Accession |
Genome size (Mbp) |
Gene count |
Contig count |
G+C content (%) |
Reference |
|---|---|---|---|---|---|---|
|
Aeb. aerius 100921-2T |
2.75 |
2793 |
2 |
66.3 |
This study |
|
|
Aeb. aerophilus Ery1T |
3.65 |
3638 |
19 |
65.4 |
[17] |
|
|
Aeb. aestiaquae KCTC 42006T |
2.87 |
2825 |
2 |
57.2 |
This study |
|
|
Aeb. aestuarii JCM 16339T |
2.24 |
2497 |
5 |
62.6 |
This study |
|
|
Aeb. amylolyticus NS1T |
2.79 |
2791 |
1 |
67.0 |
[10] |
|
|
Aeb. aquaemixtae KCTC 52763T |
2.98 |
2933 |
4 |
58.5 |
This study |
|
|
Aeb. aquimixticola SSKS-13T |
3.43 |
3349 |
5 |
63.9 |
[40] |
|
|
Aeb. atlanticus 26DY36T |
3.51 |
3425 |
2 |
61.9 |
[120] |
|
|
Aeb. aurantiacus MCCC 1A09962T |
2.90 |
2907 |
7 |
61.2 |
This study |
|
|
Aeb. buctensis M0322T |
3.77 |
3734 |
22 |
66.0 |
This study |
|
|
Aeb. confluentis KCTC 52259T |
2.93 |
2892 |
5 |
59.1 |
This study |
|
|
Aeb. dongtanensis KCTC 22672T |
3.01 |
2976 |
1 |
65.8 |
[121] |
|
|
Aeb. endophyticus LMG 29518T |
3.47 |
3314 |
13 |
58.6 |
This study |
|
|
Aeb. epoxidivorans CGMCC 1.7731T |
2.79 |
2819 |
1 |
61.5 |
[48] |
|
|
Aeb. flavus MS1-4T |
3.28 |
3154 |
29 |
60.5 |
[7] |
|
|
Aeb. gangjinensis JCM 17802T |
2.89 |
2889 |
1 |
55.5 |
This study |
|
|
Aeb. halimionae LMG 29519T |
2.81 |
2778 |
2 |
63.6 |
This study |
|
|
Aeb. indicus DSM 18604T |
3.11 |
3011 |
20 |
55.8 |
This study |
|
|
Aeb. insulae BPTF-M16T |
3.32 |
3997 |
1055 |
52.8 |
[41] |
|
|
Aeb. ishigakiensis NBRC 107699T |
2.68 |
2670 |
1 |
56.9 |
[122] |
|
|
Aeb. luteolus SW-109T |
2.89 |
2841 |
3 |
59.3 |
This study |
|
|
Aeb. lutipelagi GH1-16T |
3.10 |
3114 |
2 |
60.6 |
[42] |
|
|
Aeb. mangrovi C9-11T |
2.70 |
2650 |
1 |
63.5 |
[6] |
|
|
Aeb. marensis KCTC 22370T |
2.88 |
2784 |
1 |
64.7 |
KRIBB |
|
|
Aeb. marinus H32T |
3.00 |
2898 |
16 |
68.2 |
This study |
|
|
Aeb. maritimus HME9302T |
2.68 |
2737 |
2 |
60.8 |
[43] |
|
|
Aeb. namhicola JCM 16345T |
2.59 |
2590 |
1 |
65.0 |
This study |
|
|
Aeb. oceanensis MCCC 1A09965T |
2.87 |
2892 |
14 |
63.9 |
This study |
|
|
Aeb. rigui KCTC 42620T |
2.86 |
2903 |
30 |
66.7 |
[112] |
|
|
Aeb. salegens MCCC 1K01500T |
3.63 |
3630 |
69 |
64.6 |
This study |
|
|
Aeb. sediminis KCTC 42453T |
3.16 |
3102 |
6 |
61.5 |
This study |
|
|
Aeb. soli MCCC 1K02066T |
3.08 |
2998 |
15 |
67.0 |
This study |
|
|
Aeb. troitsensis JCM 17037T |
2.90 |
2848 |
9 |
64.7 |
[123] |
|
|
Aeb. xiamenensis CGMCC 1.12494T |
3.09 |
3064 |
5 |
61.8 |
DOE-JGI |
|
|
Aeb. xinjiangensis CCTCC AB 207166T |
3.11 |
3153 |
59 |
64.2 |
[17] |
|
|
Aeb. xixiisoli S36T |
3.88 |
3768 |
9 |
63.3 |
This study |
|
|
Crb. ferulae SX2RGS8T |
3.61 |
3434 |
36 |
66.5 |
[30] |
|
|
Crb. mercuriale CoronadoT |
3.48 |
3205 |
10 |
67.3 |
[124] |
|
|
Ccc. marinus E4A9T |
4.11 |
3956 |
3 |
64.5 |
[125] |
|
|
Ccc. mobilis Ery22T |
4.21 |
4061 |
32 |
62.5 |
[5] |
|
|
Ccc. naphthovorans PQ-2T |
3.86 |
4007 |
3 |
62.6 |
[110] |
|
|
Ccc. pelagius Ery9T |
3.31 |
3264 |
40 |
62.8 |
[5] |
|
|
Erb. aquimaris JCM 12189T |
2.66 |
2680 |
3 |
61.8 |
This study |
|
|
Erb. aquimixticola JSSK-14T |
2.55 |
2633 |
2 |
63.0 |
[35] |
|
|
Erb. arachoides RC4-10-4T |
2.94 |
2929 |
1 |
65.4 |
This study |
|
|
Erb. atlanticus s21-N3T |
3.23 |
3296 |
2 |
58.3 |
[109] |
|
|
Erb. citreus CGMCC 1.8703T |
3.03 |
3045 |
24 |
64.2 |
This study |
|
|
Erb. gaetbuli DSM 16225T |
2.78 |
2752 |
4 |
64.1 |
This study |
|
|
Erb. gangjinensis CGMCC 1.15024T |
2.72 |
2695 |
2 |
62.7 |
[126] |
|
|
Erb. jejuensis JCM 16677T |
4.15 |
4124 |
1 |
60.2 |
This study |
|
|
Erb. litoralis DSM 8509T |
3.25 |
3164 |
1 |
65.2 |
[127] |
|
|
Erb. longus DSM 6997T |
3.60 |
3430 |
14 |
57.4 |
[127] |
|
|
Erb. luteus KA37T |
2.89 |
2876 |
22 |
67.2 |
[118] |
|
|
Erb. lutimaris S-5T |
3.31 |
3219 |
12 |
65.5 |
SDU |
|
|
Erb. marinus KCTC 23554T |
2.84 |
2818 |
5 |
59.1 |
This study |
|
|
Erb. marisflavi KEM-5T |
2.67 |
2656 |
18 |
61.7 |
[22] |
|
|
Erb. nanhaisediminis CGMCC 1.7715T |
2.90 |
2870 |
12 |
62.0 |
DOE-JGI |
|
|
Erb. odishensis KCTC 23981T |
3.19 |
3137 |
25 |
63.7 |
[9] |
|
|
Erb. pelagi JCM 17468T |
3.03 |
2936 |
9 |
64.2 |
This study |
|
|
Erb. seohaensis SW-135T |
2.94 |
2919 |
1 |
61.7 |
[78] |
|
|
Erb. spongiae HN-E23T |
2.86 |
2867 |
2 |
65.5 |
[35] |
|
|
Erb. vulgaris DSM 17792T |
3.23 |
3212 |
19 |
60.6 |
This study |
|
|
Erb. xanthus CCTCC AB 2015396T |
4.38 |
4320 |
151 |
64.5 |
[9] |
|
|
Erb. zhengii V18T |
3.80 |
3812 |
29 |
62.7 |
[9] |
|
|
Erm. ramosum JCM 10282T |
3.24 |
3175 |
10 |
64.3 |
This study |
|
|
Por. algicida KEMB 9005-328T |
3.22 |
3255 |
21 |
60.7 |
This study |
|
|
Por. colymbi JCM 18338T |
4.31 |
4092 |
53 |
66.5 |
[85] |
|
|
Por. cryptus DSM 12079T |
2.95 |
2902 |
36 |
67.9 |
DOE-JGI |
|
|
Por. dokdonensis DSM 17193T |
3.00 |
2885 |
13 |
64.8 |
[85] |
|
|
Por. donghaensis DSM 16220T |
3.37 |
3199 |
11 |
66.2 |
[85] |
|
|
Por. neustonensis DSM 9434T |
3.09 |
2955 |
1 |
65.3 |
[128] |
|
|
Por. sanguineus JCM 20691T |
3.02 |
2931 |
34 |
63.6 |
[85] |
|
|
Por. tepidarius DSM 10594T |
3.22 |
3151 |
32 |
65.9 |
[85] |
|
|
Qpy. sediminis CGMCC 1.12928T |
2.42 |
2400 |
1 |
66.87 |
[129] |
Sequencing and assembly of genomic sequences
Genomic sequencing and assembly were performed as described previously [78]. Cells were harvested by centrifuge at 12,000 g for 30 s. Genomic DNA was extracted by using AxyPre Bacterial Genomic DNA Miniprep Kit (Corning Life Sciences) according to its manual. Genomes were sequenced on the HiSeq 2000 system (Illumina) by Solexa paired-end sequencing technology with a paired-end library with insert length of 500 bp by the Novogene Corporation (Beijing, PR China). Draft genomes were assembled by using SPAdes version 3.11.1 [79] based on clean reads generated from raw reads by quality trimming. The collection of assembled and obtained genomes covered 92 % (74/80) of the Erythrobacteraceae type strains, comprising 88 % (36/41), 100 % (2/2), 100 % (4/4), 96 % (22/23), 100 % (1/1), 100 % (8/8) and 100 % (1/1) of the genera Altererythrobacter , Croceibacterium , Croceicoccus , Erythrobacter , Erythromicrobium , Porphyrobacter and Qipengyuania .
Genomic annotation and comparative genomic analysis
Genomes for annotation and comparative analysis were selected following assessment of genomic completeness (>95 %) and contamination (<5 %) using CheckM software version 1.0.7 [80] with the command ‘checkm lineage_wf -x fasta bins/ checkm/’. rRNA and tRNA genes were searched by the command RNAmmer 1.2 package [81] and the tRNAscan-SE web server (http://lowelab.ucsc.edu/tRNAscan-SE/) [82], respectively. Annotated 16S rRNA genes were used to compare sequence identities on the EzBioCloud web server (www.ezbiocloud.net/identify) [83] to confirm that a genome represented its corresponding type strain. Coding sequences (CDSs) were predicted and annotated by using Rapid Annotation using Subsystem Technology (RAST) web server version 2.0 (http://rast.nmpdr.org/rast.cgi) [84]. The DNA G+C contents were also calculated on the RAST web server version 2.0.
Comparative genomic analysis was performed as previously described [85, 86]. Orthologous clusters (OCs) were identified by comparing whole protein sequences translated from CDSs pairwise with the execution of Proteinortho version 5.16b [87] with command ‘-e 1e-5 -cov=50 -identity=50’, which is accordance with the threshold values for a group of OCs sharing identities more than 50 % and coverage longer than half of their sequence lengths. Subsequently, single-copy OCs were filtered by an in-house Perl script.
16s rRNA gene phylogenetic and phylogenomic reconstructions
In accordance with previous polyphasic taxonomic studies of the members in the family Erythrobacteraceae [19, 22, 35, 40, 88], Rhodospirillum rubrum ATCC 11170T was chosen as an outgroup, with its 16S rRNA gene sequence and genomic sequences obtained from the NCBI GenBank database under the accession numbers D30778 and CP000230–CP000231, respectively. 16S rRNA gene phylogeny was reconstructed as described by Xu et al. [89]. Gene sequences of 80 Erythrobacteraceae type strains and an outgroup were aligned with clustal_w [90] built in in the mega7 software [91]. Then, aligned sequences were processed into maximum-likelihood phylogenetic analysis [92], using mega7 software with the substitution model and the bootstrap value set as Kimura's two-parameter model [93] and 1000 replicates, respectively.
Protein and gene sequences of filtered single-copy OCs were both performed in the phylogenomic analyses. Protein sequences were aligned by using mafft version 7 [94] with the parameter ‘-auto’, while gene sequences were aligned by mapping nucleotides on amino acids based on aligned protein sequences through PAL2NAL program version 14 [95]. Aligned sequences were refined to select the most reliable positions through trimAL version 1.4.1 [96] with the parameter ‘-automated1’ and concatenated through our in-house perl script. Concatenation and partition methods were both applied in this study. The best substitution models for were proposed by IQ-Tree 1.6.1 software [97] with the command ‘-m MFP’. Subsequently, LG+F+R9 and GTR+F+R8 were estimated as the best substitution models for concatenations of amino acid and nucleotide sequences, respectively, and the best substitution models for partition methods are listed in Table S2. Maximum-likelihood phylogenomic trees were reconstructed by using IQ-Tree 1.6.1 software [97] with the bootstrap value set to 100 replicates.
Genomic similarity analysis
ANI, AAI and POCP values were used to calculate genomic similarities. ANI values were calculated by the orthologous average nucleotide identity tool (OrthoANI version 0.93.1) [98] implemented with the blast algorithm [99]. AAI values were obtained using the Kostas lab AAI calculator web server (http://enve-omics.ce.gatech.edu/aai/) [100]. POCP values were obtained according to the formula ‘POCP=(C1 +C2)/(T1 +T2)×100 %’ where C1 and C2 indicated the conserved number of predicted proteins in the two pairwise compared genomes, respectively, as well as T1 and T2 stands for the total number of predicted proteins in the two pairwise compared genomes, respectively [59], following comparative genomic analysis by using Proteinortho version 5.16b with the command ‘-e=1e-5 -cov=50 -identity=40’. In addition, the t-tests of AAI, ANI and POCP values of inter- and intra-group were calculated by using the function ‘t.text’ within R version 3.4.2 [101].
Discussion
Characteristics of Erythrobacteraceae genomes
All obtained genomes were of high quality with genomic completeness of 97.6–99.9 % (average 99.3 %; median 99.4 %) and contamination of 0–4.9 % (average 0.7 %; median 0.4 %), as shown in Table S2. Sequence identity analysis of annotated 16S rRNA genes from the genomes sequenced in this study indicated that each represented its type strain with high identities of 99.2–100.0 % (Table S3). Several strains, including Aeb. confluentis KCTC 52259T, Aeb. indicus DSM 18604T, Aeb. luteolus SW-109T, Erb. citreus CGMCC 1.8703T and Erb. jejuensis JCM 16677T, had multi-copy 16S rRNA genes, whose sequences were identical.
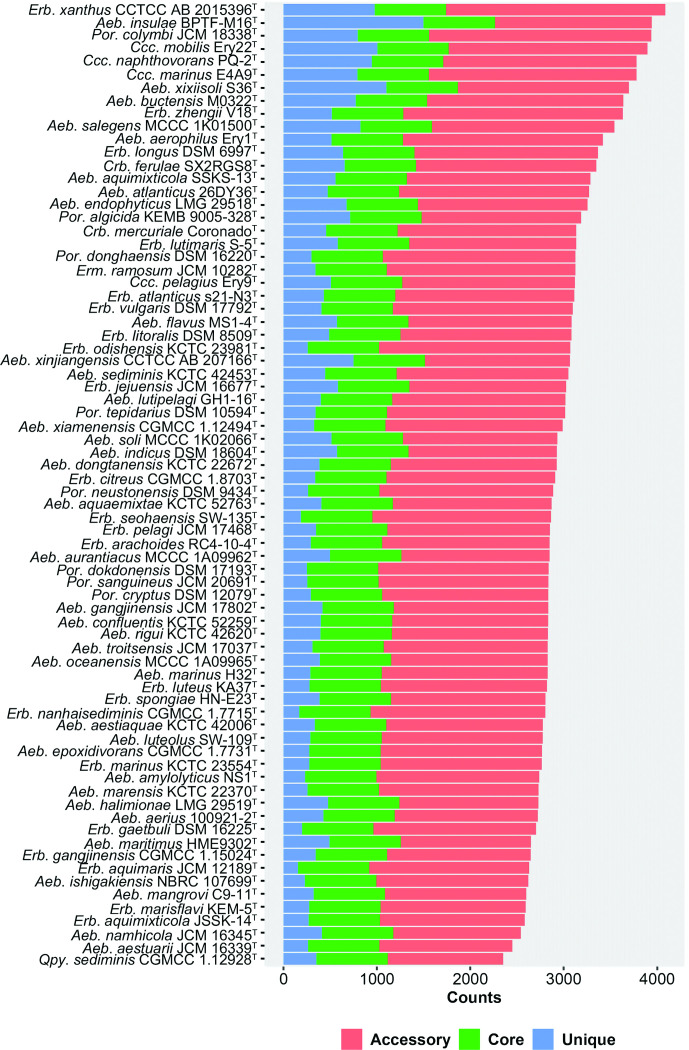
Genomic sizes, gene counts and G+C contents were 2.24–4.38 Mbp (average 3.16 Mbp; median 3.06 Mbp), 2400–4320 (average 3117; median 2987) and 52.8–68.2 % (average 63.0 %; median 63.6 %), respectively (Table 1). Comparative genomic analysis revealed that the pan-genome of the family Erythrobacteraceae harboured 49, 006 OCs, among which 763 OCs were shared by all type strains, which also had 1,233–2,375 accessory and 157–1,500 unique OCs (Fig. 1). The percentages of accessory, core and unique OCs in each type strain varied greatly with values of 18.7–32.5, 42.6–66.7 and 6.0–38.1 %, respectively, which showed a rich genetic diversity in this family. A total of 288 single-copy OCs (Table S4) were included in our phylogenomic analyses.
Fig. 1.
Accessory, core and unique OCs distributed in each type strain belonging to the family Erythrobacteraceae .
16s rRNA gene phylogeny
As stated before, several genera within the family Erythrobacteraceae did not form an independent clade in the 16S rRNA gene phylogenetic tree (Fig. S1): (1) the genus Erythrobacter , being the type genus of the family, could be divided into four clades, one of which was grouped with the genera Erythromicrobium and Porphyrobacter ; (2) the genus Altererythrobacter showed five clades which also included the genera Croceicoccus and Qipengyuania ; (3) the genera Erythromicrobium and Qipengyuania , each consisting of a single species, were clustered in clades mostly containing of Porphyrobacter and Altererythrobacter , respectively. The genera Croceibacterium and Croceicoccus formed two independent clades, and they did not belong to monophyletic clades which could be separated from other genera. Thus, 16S rRNA gene sequences did not confirm monophyletic relationships within the genera of the family [2, 4, 30, 37, 38]. Only 19 nodes accounting for 26.0 % exhibited bootstrap values higher than 70 %, indicating that this phylogenetic tree was not reliable enough to correctly reveal the taxonomic status of the genera of the family.
Phylogenomic and genomic similarity analyses proposing three clades
Four phylogenomic trees, based on 288 single-copy OC, amino acid or nucleotide sequences, with annotations and substitution models are shown in Table S4 and had similar topological structures with 59 nodes accounting for 80.8 %, and were identical in all four calculated phylogenetic relationship parameters (Fig. 2 and Figs. S2–4). In all four phylogenomic trees, the bootstrap value of most nodes (66/73–68/73) exceeded 70 %, indicating those phylogenies were robust. Compared with 16S rRNA gene phylogeny, those similar and robust phylogenomic trees could provide a reliable taxonomic status for the family Erythrobacteraceae .
Fig. 2.
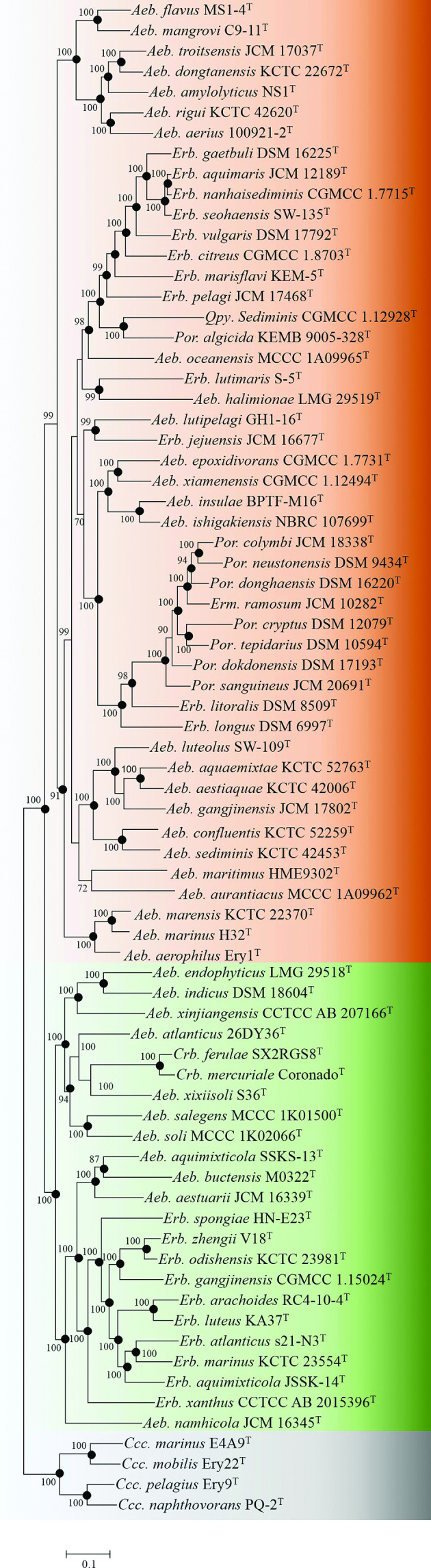
A maximum-likelihood tree based on the partition of 288 single-copy OC protein sequences showing the phylogenetic relationship of type strains belonging to the family Erythrobacteraceae . Bootstrap values are based on 100 replicates. Bar., 0.1 substitutions per nucleotide position. The backgrounds coloured brown, green and grey indicate Clades I, II and III, respectively. Rhodospirillum rubrum ATCC 11170T was used as an outgroup (not shown).
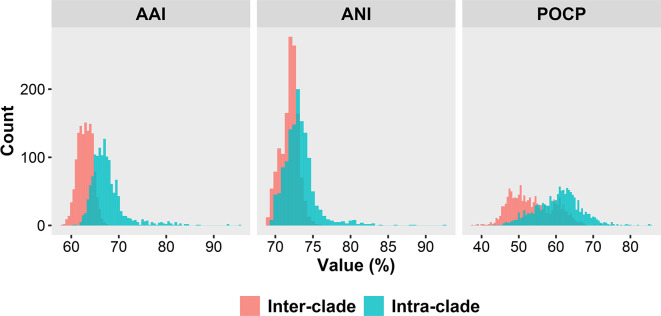
Based on these phylogenomic trees, the family Erythrobacteraceae can be divided into three separate clades, Clades I, II and III, consisting of 47, 23 and four species, respectively (Fig. 2). Genomic similarity analyses by AAI, ANI and POCP calculations also supported that the three clades were significantly separated with p value<2.2×10−16 (Fig. 3). Clades I and II contained most species. Clade III only contained four Croceicoccus species, indicating that the taxonomic status of this genus should not be changed.
Fig. 3.
Histograms of AAI, ANI and POCP values regarding inter- and intra-clade. Red and blue indicate inter-clade and intra-clade, respectively.
AAI value and evolutionary distance classifying genera
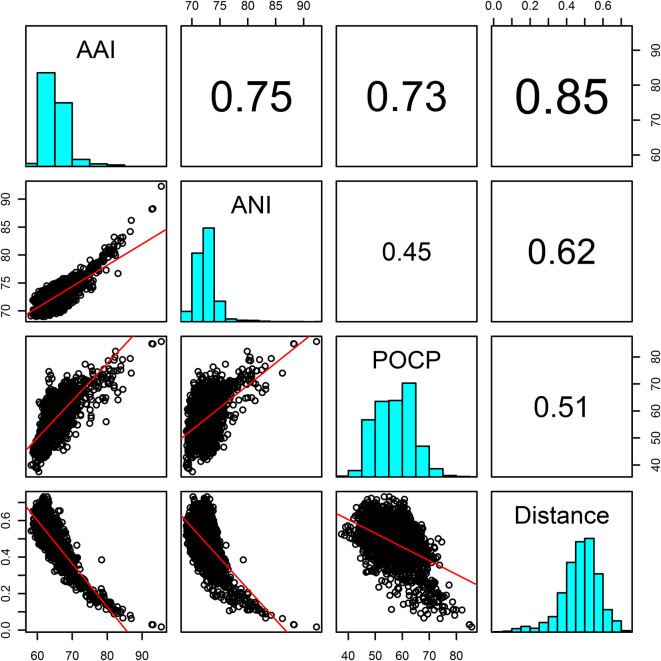
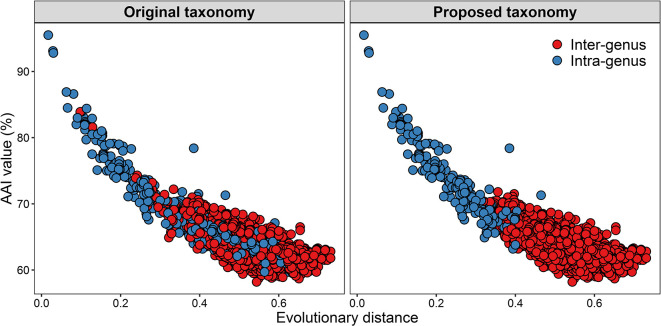
A robust core-genome phylogeny of the family was obtained. Although there is no generally recognized genus boundary, recent studies suggested AAI (60–80 %) and POCP (50 %) could be thresholds for distinguishing genera [59, 61]. Evolutionary distance is also a relatively conserved criterion for inferring evolutionary relationships [102]. Pairwise comparisons of ANI, AAI, POCP and evolutionary distance indicated that the pair of AAI and evolutionary distance had a much higher correlation coefficient (rcc=0.85) than other pairs (Fig. 4). Type strains shared pairwise >50 % of POCP values, which is similar to the result of a phylogenomic study of the Roseobacter group [58], suggesting that POCP values could not be applied for delineating genera within the family Erythrobacteraceae . AAI was more suitable to distinguish each taxon in the family than ANI and POCP (Figs. S5–S7). Thus, AAI and evolutionary distance were selected to classify genera of Erythrobacteraceae .
Fig. 4.
Pairwise correlations of ANI, AAI, POCP and evolutionary distance calculated from the genomes of Erythrobacteraceae type strains.
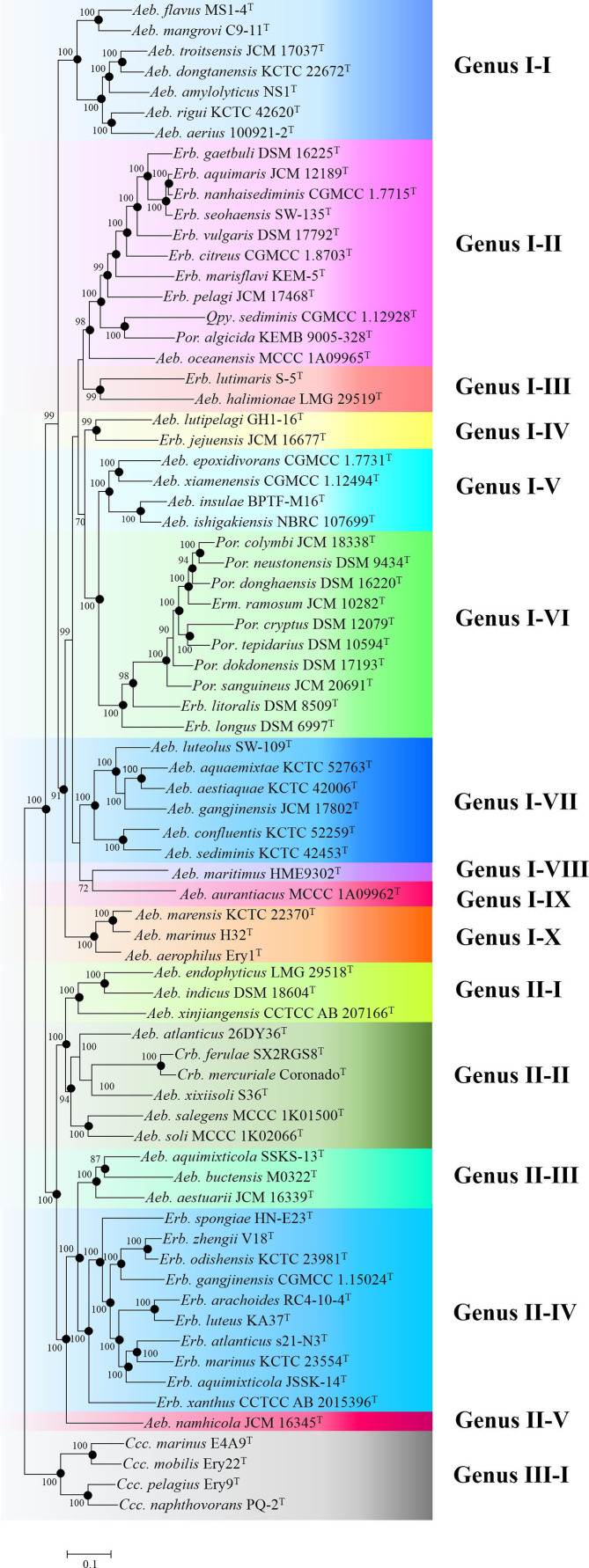
Since phylogenetic tree topology is a major criterion for classifying genera, we propose that one genus should be clustered into one group only. Based on this criterion, Clade I is then composed of 10 genera (Fig. 5), including Genus I-I (Aeb. flavus MS1-4T, Aeb. mangrovi C9-11T, Aeb. troitsensis JCM 17037T, Aeb. dongtanensis KCTC 22672T, Aeb. amylolyticus NS1T, Aeb. rigui KCTC 42620T and Aeb. aerius 10092102T), Genus I-II (Erb. gaetbuli DSM 16225T, Erb. aquimaris JCM 12189T, Erb. nanhaisediminis CGMCC 1.7715T, Erb. seohaensis SW-135T, Erb. vulgaris DSM 17792T, Erb. citreus CGMCC 1.8703T, Erb. marisflavi KEM-5T, Erb. pelagi JCM 17468T, Qpy. sediminis M1T, Por. algicida KEMB 9005-328T and Aeb. oceanensis MCCC 1A09965T), Genus I-III (Erb. lutimaris S-5T and Aeb. halimionae LMG 29519T), Genus I-IV (Aeb. lutipelagi GH1-6T and Erb. jejuensis JCM 16677T), Genus I-VI (Aeb. epoxidivorans CGMCC 1.7731T, Aeb. xiamensis CGMCC 1.12494T, Aeb. insulae BPTF-M16T and Aeb. ishigakiensis NBRC 107699T), Genus I-VII ( Por. colymbi JCM 18338T, Por. neustonensis DSM 9434T, Por. donghaensis DSM 16220T, Erm. ramosum JCM 10282T, Por. cryptus DSM 12079T, Por. tepidarius DSM 10594T, Por. dokdonensis DSM 17193T, Por. sanguineus JCM 20691T, Erb. litoralis DSM 8509T and Erb. longus DSM 6997T), Genus I-VIII (Aeb. luteolus SW-109T, Aeb. aquaemixtae KCTC 52763T, Aeb. aestiaquae KCTC 42006T, Aeb. gangjinensis JCM 17802T, Aeb. confluentis KCTC 52259T and Aeb. sediminis KCTC 42453T), Genus I-IX (Aeb. maritimus HME 9302T) and Genus I-X (Aeb. aurantiacus MCCC 1A09962T). The type strains of each of these genera exhibited pairwise evolutionary distance <0.4 %, except for Aeb. oceanensis MCCC 1A09965T and Qpy. sediminis M1T. These two type strains also showed a pairwise AAI value of 67.3 %, while the pairwise AAI value for the majority of this clade (96.8 %, 1047/1081) were higher than 70%. Clade III consisted of one genus, whose species had AAI values of 68.1–77.5 % and evolutionary distances of 0.13–0.27. Based on the analysis of these two clades, the genus boundary for the family is here proposed as AAI values of 70% and an evolutionary distance of 0.4 (Fig. 6).
Fig. 5.
Proposed taxonomic status for the family Erythrobacteraceae . Maximum-likelihood tree based on the partition of 288 single-copy OC protein sequences. Bootstrap values are based on 100 replicates. Bar, 0.1 substitutions per nucleotide position.
Fig. 6.
Comparison of original and proposed taxonomy for the family Erythrobacteraceae based on AAI values and evolutionary distances. Red and blue indicate inter-genus and intra-genus, respectively.
Based on these criteria, Clade II with all nodes with bootstrap values of >85 % could be divided into five genera (Fig. 5), consisting of Genus II-I (Aeb. endophyticus LMG 29518T, Aeb. indicus DSM 18604T and Aeb. xinjiangensis CCTCC AB 207166T), Genus II-II (Aeb. atlanticus 26DY36T, Crb. ferulae SX2RGS8T, Crb. mercuriale CoronadoT, Aeb. xixiisoli S36T, Aeb. salegens MCCC 1K01500T, Aeb. soli MCCC 1K02066T), Genus II-III (Aeb. aquimixticola SSKS-13T, Aeb. buctensis M0322T and Aeb. aestuarii JCM 16339T), Genus II-IV (Erb. spongiae HN-E23T, Erb. zhengii V18T, Erb. odishensis KCTC 23981T, Erb. gangjiensis CGMCC 1.15204T, Erb. arachoides RC4-10-4T, Erb. luteus KA37T, Erb. atlanticus s21-N3T, Erb. marinus KCTC 23554T, Erb. aquimixticola JSSK-14T and Erb. xanthus CCTCC AB 2015396T) and Genus II-V (Aeb. namhicola 16345T).
We therefore propose that phylogenomic topology supplemented with AAI values and evolutionary distance values could replace the phylogeny based on 16S rRNA gene sequences in the taxonomy of the family Erythrobacteraceae .
Genotype and phenotype support the proposal of new genera
Comparison of genomic contents within the family Erythrobacteraceae revealed that 12 443 OCs could be indicators for distinguishing newly proposed genera. While considerable metabolic diversity is found within the family, metabolic pathways involving carbon, nitrogen, phosphorus and sulfur could not be applied to refine the taxonomic status of this family. Therefore, the pathways of aerobic anoxygenic photosynthesis and flagella biosynthesis, which contain multiple genes and reactions [47, 103], were selected to investigate their value as indicators for their taxonomic status.
Aerobic anoxygenic photosynthesis is encoded by a series of genes that were found in all Genus I-VI species, Aeb. ishigakiensis NBRC 107699T (Genus I-V), Erb. marinus KCTC 23554T (Genus II-III) and Erb. odishensis KCTC 239891T (Genus II-III), as shown in Table 2. Phenotypic characteristics revealed that Genus I-VI consisted of AAPB [23, 24, 26, 27, 29, 31, 104–106], while other genera did not include AAPB. Moreover, phylogenetic analysis indicated that genes for aerobic anoxygenic photosynthesis of Aeb. ishigakiensis NBRC 107699T, Erb. marinus KCTC 23554T and Erb. odishensis KCTC 239891T were paralogs of such genes in Genus I-VI (Fig. S8).
Table 2.
Comparisons of genotype and phenotype regarding aerobic anoxygenic photosynthesis and flagella biosynthesis in the family Erythrobacteraceae
Genera: 1, Genus I-I; 2, Genus I-II; 3, Genus I-III; 4, Genus I-IV; 5, Genus I-V; 6, Genus I-VI; 7, Genus I-VII; 8, Genus I-VIII; 9, Genus I-IX; 10, Genus I-X; 11, Genus II-I; 12, Genus II-II; 13, Genus II-III; 14, Genus II-IV; 15, Genus II-V; 16, Genus III. +, Gene detected; −, gene not detected.
|
Pathway |
1 |
2 |
3 |
4 |
5 |
6 |
7 |
8 |
9 |
10 |
11 |
12 |
13 |
14 |
15 |
16 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
Aerobic anoxygenic photosynthesis |
||||||||||||||||
|
bchD |
− |
− |
+ |
− |
+ |
+ |
+ |
− |
− |
− |
− |
− |
− |
+ |
− |
− |
|
bchE |
− |
− |
+ |
− |
+ |
+ |
+ |
− |
− |
− |
− |
− |
− |
+ |
− |
− |
|
bchG |
− |
− |
+ |
− |
+ |
+ |
+ |
− |
− |
− |
− |
− |
− |
+ |
− |
− |
|
bchL |
− |
− |
+ |
− |
+ |
+ |
+ |
− |
− |
− |
− |
− |
− |
+ |
− |
− |
|
bchM |
− |
− |
+ |
− |
+ |
+ |
+ |
− |
− |
− |
− |
− |
− |
+ |
− |
− |
|
bchN |
− |
− |
+ |
− |
+ |
+ |
+ |
− |
− |
− |
− |
− |
− |
+ |
− |
− |
|
bchP |
− |
− |
+ |
− |
+ |
+ |
+ |
− |
− |
− |
− |
− |
− |
+ |
− |
− |
|
bchX |
− |
− |
+ |
− |
+ |
+ |
+ |
− |
− |
− |
− |
− |
− |
+ |
− |
− |
|
bchY |
− |
− |
+ |
− |
+ |
+ |
+ |
− |
− |
− |
− |
− |
− |
+ |
− |
− |
|
bchZ |
− |
− |
+ |
− |
+ |
+ |
+ |
− |
− |
− |
− |
− |
− |
+ |
− |
− |
|
hemD |
+ |
+ |
− |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
|
hemF |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
− |
− |
− |
+ |
+ |
+ |
+ |
− |
+ |
|
pufL |
− |
− |
− |
− |
+ |
+ |
− |
− |
− |
− |
− |
− |
− |
+ |
− |
− |
|
Phenotype |
− |
− |
− |
− |
− |
+ |
− |
− |
− |
− |
− |
− |
− |
− |
− |
− |
|
Flagella biosynthesis |
||||||||||||||||
|
flgA |
− |
+ |
− |
− |
− |
− |
− |
− |
− |
− |
− |
− |
− |
− |
− |
− |
|
flgB |
− |
+ |
− |
− |
− |
+ |
− |
− |
− |
+ |
− |
+ |
− |
+ |
− |
+ |
|
flgC |
− |
+ |
− |
− |
− |
+ |
− |
− |
− |
+ |
− |
+ |
− |
+ |
− |
+ |
|
flgD |
− |
+ |
− |
− |
− |
+ |
− |
− |
− |
+ |
− |
+ |
− |
+ |
− |
+ |
|
flgE |
− |
+ |
− |
− |
− |
+ |
− |
− |
− |
+ |
− |
+ |
− |
+ |
− |
+ |
|
flgF |
− |
+ |
− |
− |
− |
+ |
− |
− |
− |
+ |
− |
+ |
− |
+ |
− |
+ |
|
flgG |
− |
+ |
− |
− |
− |
+ |
− |
− |
− |
+ |
− |
+ |
− |
+ |
− |
+ |
|
flgH |
− |
+ |
− |
− |
− |
+ |
− |
− |
− |
+ |
− |
+ |
− |
+ |
− |
+ |
|
flgI |
− |
+ |
− |
− |
− |
+ |
− |
− |
− |
+ |
− |
+ |
− |
+ |
− |
+ |
|
flgJ |
− |
+ |
− |
− |
− |
+ |
− |
− |
− |
+ |
− |
+ |
− |
+ |
− |
+ |
|
flgK |
− |
+ |
− |
− |
− |
− |
− |
− |
− |
+ |
− |
+ |
− |
+ |
− |
+ |
|
flgL |
− |
+ |
− |
− |
− |
− |
− |
− |
− |
+ |
− |
+ |
− |
+ |
− |
+ |
|
flhA |
− |
+ |
− |
− |
− |
+ |
− |
− |
− |
+ |
− |
+ |
− |
+ |
− |
+ |
|
flhB |
− |
+ |
− |
− |
− |
+ |
− |
− |
− |
+ |
− |
+ |
− |
+ |
− |
+ |
|
fliC |
− |
+ |
− |
− |
− |
+ |
− |
− |
− |
+ |
− |
+ |
− |
+ |
− |
+ |
|
fliD |
− |
+ |
− |
− |
− |
+ |
− |
− |
− |
+ |
− |
+ |
− |
+ |
− |
+ |
|
fliE |
− |
+ |
− |
− |
− |
+ |
− |
− |
− |
+ |
− |
+ |
− |
+ |
− |
+ |
|
fliF |
− |
+ |
− |
− |
− |
+ |
− |
− |
− |
+ |
− |
+ |
− |
+ |
− |
+ |
|
fliG |
− |
+ |
− |
− |
− |
+ |
− |
− |
− |
+ |
− |
+ |
− |
+ |
− |
+ |
|
fliL |
− |
+ |
− |
− |
− |
+ |
− |
− |
− |
+ |
− |
+ |
− |
+ |
− |
+ |
|
fliM |
− |
+ |
− |
− |
− |
− |
− |
− |
− |
− |
− |
− |
− |
− |
− |
− |
|
fliN |
− |
+ |
− |
− |
− |
+ |
− |
− |
− |
+ |
− |
+ |
− |
+ |
− |
+ |
|
fliP |
− |
+ |
− |
− |
− |
+ |
− |
− |
− |
+ |
− |
+ |
− |
+ |
− |
+ |
|
fliQ |
− |
+ |
− |
− |
− |
+ |
− |
− |
− |
+ |
− |
+ |
− |
+ |
− |
+ |
|
fliR |
− |
+ |
− |
− |
− |
+ |
− |
− |
− |
+ |
− |
+ |
− |
+ |
− |
+ |
|
Phenotype |
− |
+ |
− |
− |
− |
+ |
− |
− |
− |
+ |
− |
+ |
− |
+ |
− |
+ |
Flagella can be used for locomotion and sensing, which improves the survival of prokaryotes [107, 108]. Comparison of gene contents showed that several strains in Genus I-II ( Por. algicida KEMB 9005-328T), Genus I-VI (Erb. litoralis DSM 8509T, Erb. longus DSM 6997T, Erm. ramosum JCM 10282T, Por. colymbi JCM 18338T, Por. cryptus DSM 12079T, Por. neustonensis DSM 9434T and Por. sanguineus JCM 20691T), Genus I-X (Aeb. marinus H32T), Genus II-II (Aeb. atlanticus 26DY36T and Aeb. soli MCCC 1K02066T), Genus II-IV (Erb. atlanticus s21-N3T) and Genus III (Ccc. mobilis Ery22T and Ccc. naphthovorans PQ-2T) had genes related to flagella biosynthesis. Microscopic observations showed flagella in those strains [5, 8, 14, 23, 24, 26, 29, 31, 44, 104, 109, 110], except for Aeb. marinus H32T.
Based on the phylogenomic and genomic similarity analyses, we propose that the family Erythrobacteraceae could be reclassfied into 16 genera including 11 novel genera, for which the names Alteraurantiacibacter, Altericroceibacterium, Alteriqipengyuania, Alteripontixanthobacter, Aurantiacibacter, Paraurantiacibacter, Parapontixanthobacter, Parerythrobacter, Pelagerythrobacter, Pontixanthobacter and Tsuneonella are proposed. Because the species of Erythromicrobium and Porphyrobacter were merged into the genus Erythrobacter , those two genera are no longer necessary in taxonomic discussions, but their names remain validly published and can still be used.
Description of Tsuneonella gen. nov.
Tsuneonella (Tsu.ne.o.nel'la. N.L. fem. n. Tsuneonella, named in honour of Tsuneo Shiba who established genus Erythrobacter ).
Cells are Gram-stain-negative, ovoid to rod, non-spore-forming and non-motile. Aerobic or facultatively aerobic. Contains carotenoid pigments but not bacteriochlorophyll a. The predominant ubiquinone is Q-10. The major fatty acid (>10 %) is summed feature 8 (C18 : 1 ω7c and/or C18 : 1 ω6c). The major polar lipids are diphosphatidylglycerol and phosphatidylethanolamine. The genus represents a distinct branch in the family Erythrobacteraceae of the class Alphaproteobacteria based on the core-genomic phylogeny. The DNA G+C content is 60.5–67.0 % (by genome). The type species is Tsuneonella dongtanensis.
Description of Tsuneonella aeria comb. nov.
Tsuneonella aeria (a.e'ri.a. L. fem. adj. aeria pertaining to the air, aerial).
Basonym: Altererythrobacter aerius Xue et al. 2016.
The description is the same as for Aeb. aerius [28]. Core-genomic phylogenetic analysis strongly supported the placement of this species in the genus Tsuneonella. The type strain, 100921-2T (=CFCC 14287T=KCTC 42844T), was isolated from air at the foot of Xiangshan Mountain, Beijing, PR China. The DNA G+C content of the type strain is 66.3 % (by genome).
Description of Tsuneonella amylolytica comb. nov.
Tsuneonella amylolytica (a.my.lo.ly'ti.ca. Gr. neut. n. amylon starch; Gr. fem. adj. lytikê able to loosen, able to dissolve; N.L. fem. adj. amylolytica starch dissolving).
Basonym: Altererythrobacter amylolyticus Qu et al. 2019.
The description is the same as for Aeb. amylolyticus [10]. Core-genomic phylogenetic analysis strongly supported the placement of this species in the genus Tsuneonella. The type strain, NS1T (=CGMCC 1.13679T=NBRC 113553T), was isolated from sediment of Taihu Lake in Jiangsu Province, PR China. The DNA G+C content of type strain 67.0 % (by genome).
Description of Tsuneonella dongtanensis comb. nov.
Tsuneonella dongtanensis (dong.tan.en'sis. N.L. fem. adj. dongtanensis pertaining to Dongtan, a wetland region in Chongming Island, Shanghai, PR China).
Basonym: Altererythrobacter dongtanensis Fan et al. 2011.
The description is the same as for Aeb. dongtanensis [111]. Core-genomic phylogenetic analysis strongly supported the placement of this species in the genus Tsuneonella. The type strain, JM27T (=KCTC 22672T=CCTCC AB 209199T), was isolated from a tidal flat (Dongtan Wetland, Chongming Island, Shanghai, PR China). The DNA G+C content of the type strain is 65.8 % (by genome).
Description of Tsuneonella flava comb. nov.
Tsuneonella flava (fla'va. L. fem. adj. flava yellow, the colour of colonies and pigments of the bacterium).
Basonym: Altererythrobacter flavus Ma et al. 2018.
The description is the same as for Aeb. flavus [44]. Core-genomic phylogenetic analysis strongly supported the placement of this species in the genus Tsuneonella. The type strain, MS1-4T (=MCCC 1K02683T=NBRC 112977T), was isolated from mangrove sediment of the Jiulong River Estuary, Fujian Province, PR China. The DNA G+C content of the type strain is 60.5 % (by genome).
Description of Tsuneonella mangrovi comb. nov.
Tsuneonella mangrovi (man.gro'vi. N.L. gen. n. mangrovi of or belonging to a mangrove wetland).
Basonym: Altererythrobacter mangrovi Liao et al. 2017.
The description is the same as for Aeb. mangrovi [6]. Core-genomic phylogenetic analysis strongly supported the placement of this species in the genus Tsuneonella. The type strain, C9-11T (=MCCC 1K03311T=JCM 32056T), was isolated from a mangrove sediment sample collected from Yunxiao Mangrove National Nature Reverse in Zhangzhou, Fujian Province, PR China. The DNA G+C content of the type strain is 63.5 % (by genome).
Description of Tsuneonella rigui comb. nov.
Tsuneonella rigui (ri’gu.i. L. gen. n. rigui of a well-watered place).
Basonym: Altererythrobacter rigui Kang et al. 2016.
The description is the same as for Aeb. rigui [112]. Core-genomic phylogenetic analysis strongly supported the placement of this species in the genus Tsuneonella. The type strain, WW3T (=KCTC 42620T=JCM 30975T), was isolated from freshwater of Woopo wetland, Republic of Korea. The DNA G+C content of the type strain is 66.7 % (by genome).
Description of Tsuneonella troitsensis comb. nov.
Tsuneonella troitsensis (troi.tsen'sis. N.L. fem. adj. troitsensis referring to Troitsa Bay, from where the organism was isolated).
Basonym: Altererythrobacter troitsensis Nedashkovskaya et al. 2013.
The description is the same as for Aeb. troitsensis [34]. Core-genomic phylogenetic analysis strongly supported the placement of this species in the genus Tsuneonella. The type strain, KMM 6042T (=KCTC 12303T=JCM 17037T), was isolated from the sea urchin Strongylocentrotus intermedius. The DNA G+C content of the type strain is 64.7 % (by genome).
Emended description of the genus Qipengyuania Feng et al. 2015
The description is as given by Feng et al. [2] with the following amendment. Cells are aerobic or facultatively aerobic. Contains carotenoid pigments but not bacteriochlorophyll a. Positive or negative for oxidase. The major fatty acid (>10%) is summed feature 8 (C18 : 1 ω7c and/or C18 : 1 ω6c). The major polar lipids are phosphatidylcholine, phosphatidylethanolamine and phosphatidylglycerol. The genus represents a distinct branch in the family Erythrobacteraceae of the class Alphaproteobacteria based on the core-genomic phylogeny. The DNA G+C content is 60.6–66.7 % (by genome). The type species for the genus is Qipengyuania sediminis .
Description of Qipengyuania algicida comb. nov.
Qipengyuania algicida (al.gi.ci’da. L. fem. n. alga alga; L. suff. –cida from L. v. caedere to kill; N.L. fem. n. algicida a killer of algae).
Basonym: Porphyrobacter algicida Kristyanto et al. 2017.
The description is the same as for Por. algicida [44]. Core-genomic phylogenetic analysis strongly supported the placement of this species in the genus Qipengyuania . The type strain, Yeonmyeong 2-22T (=KEMB 9005–328T=JCM 31499T), was isolated from surface seawater collected from Geoje Island in the South Sea, Republic of Korea. The DNA G+C content of the type strain is 60.7 % (by genome).
Description of Qipengyunia aquimaris comb. nov.
Qipengyuania aquimaris (a.qui.ma'ris. L. fem. n. aqua water; L. neut. n. mare the sea; N.L. gen. n. aquimaris of the water of the sea).
Basonym: Erythrobacter aquimaris Yoon et al. 2004.
The description is the same as for Erb. aquimaris [73]. Core-genomic phylogenetic analysis strongly supported the placement of this species in the genus Qipengyuania . The type strain, SW-110T (=KCCM 41818T=JCM 12189T), was isolated from sea water of a tidal flat of the Yellow Sea in the Republic of Korea. The DNA G+C content of the type strain is 61.8 % (by genome).
Description of Qipengyuania citrea comb. nov.
Qipengyuania citrea (ci'tre.a. L. fem. adj. citrea, describing the lemon-yellow pigmentation).
Basonym: Erythrobacter citreus Denner et al. 2002.
The description is the same as for Erb. citreus [75]. Core-genomic phylogenetic analysis strongly supported the placement of this species in the genus Qipengyuania . The type strain, RE35/F1T (=CIP 107092T=DSM 14432T=JCM 21816T), was isolated from the western Mediterranean Sea (Bay of Calvi, Corsica, France). The DNA G+C content of the type strain is 64.2 % (by genome).
Description of Qipengyuania gaetbuli comb. nov.
Qipengyuania gaetbuli (gaet.bu'li. N.L. gen. n. gaetbuli of gaetbul, the Korean name for a tidal flat).
Basonym: Erythrobacter gaetbuli Yoon et al. 2005.
The description is the same as for Erb. gaetbuli [3]. Core-genomic phylogenetic analysis strongly supported the placement of this species in the genus Qipengyuania . The type strain, SW-161T (=KCTC 12227T=DSM 16225T), was isolated from a tidal flat of the Yellow Sea in the Republic of Korea. The DNA G+C content of the type strain is 64.1 % (by genome).
Description of Qipengyuania marisflavi comb. nov.
Qipengyuania marisflavi (ma.ris.fla'vi. L. neut. n. mare the sea; L. masc. adj. flavus yellow; N.L. gen. n. marisflavi of the Yellow Sea).
Basonym: Erythrobacter marisflavi Park et al. 2019.
The description is the same as for Erb. marisflavi [22]. Core-genomic phylogenetic analysis strongly supported the placement of this species in the genus Qipengyuania . The type strain, KEM-5T (=KACC 19865T=KCTC 62896T=NBRC 113546T), was isolated from water collected from an estuary environment where the ocean and a river meet at Seocheon, Republic of Korea. The DNA G+C content of the type strain is 61.7 % (by genome).
Description of Qipengyuania nanhaisediminis comb. nov.
Qipengyuania nanhaisediminis (nan.hai.se.di'mi.nis. Chin. n. nanhai meaning 'the South China Sea'; L. gen. n. sediminis of a sediment; N.L. gen. n. nanhaisediminis of a sediment from the South China Sea).
Basonym: Erythrobacter nanhaisediminis Xu et al. 2010.
The description is the same as for Erb. nanhaisediminis [113]. Core-genomic phylogenetic analysis strongly supported the placement of this species in the genus Qipengyuania . The type strain, T30T (=CGMCC 1.7715T=JCM 16125T), was isolated from the South China Sea. The DNA G+C content of the type strain is 62.0 % (by genome).
Description of Qipengyuania oceanensis comb. nov.
Qipengyuania oceanensis (o.ce.a.nen'sis. L. fem. adj. oceanensis, belonging to the ocean).
Basonym: Altererythrobacter oceanensis Yang et al. 2014.
The description is the same as for Aeb. oceanensis [70]. Core-genomic phylogenetic analysis strongly supported the placement of this species in the genus Qipengyuania . The type strain, Y2T (=CGMCC 1.12752T=LMG 28109T), was isolated from a deep-sea sediment of the western Pacific Ocean. The DNA G+C content of the type strain is 63.9 % (by genome).
Description of Qipengyuania pelagi comb. nov.
Qipengyuania pelagi (pe'la.gi. L. gen. n. pelagi of/from the sea, reflecting isolation of the type strain from seawater of the Red Sea).
Basonym: Erythrobacter pelagi Wu et al. 2012.
The description is the same as for Erb. pelagi [77]. Core-genomic phylogenetic analysis strongly supported the placement of this species in the genus Qipengyuania . The type strain, UST081027-248T (=JCM 17468T=NRRL 59511T), was isolated from shallow seawater collected from the middle of the Red Sea. The DNA G+C content of the type strain is 64.2 % (by genome).
Emended description of Qipengyuania sediminis Feng et al. 2015
Qipengyuania sediminis (se.di'mi.nis. L. gen. n. sediminis of sediment)
The description is identical to that given for Qpy. sediminis [2], except for the DNA G+C content. The type strain, M1T (=CGMCC 1.12928T=JCM 30182T), was isolated from a borehole sediment sample collected from Qiangtang Basin in Qinghai-Tibetan Plateau, PR China. The DNA G+C content of the type strain is 66.7% (by genome).
Description of Qipengyuania seohaensis comb. nov.
Qipengyuania seohaensis (seo.ha.en'sis. N.L. fem. adj. seohaensis of Seohae, the Korean name of the Yellow Sea in Korea, from where the type strain was isolated).
Basonym: Erythrobacter seohaensis Yoon et al. 2005.
The description is the same as for Erb. seohaensis [3]. Core-genomic phylogenetic analysis strongly supported the placement of this species in the genus Qipengyuania . The type strain, SW-135T (=KCTC 12228T=DSM 16221T=JCM 21815T), was isolated from a tidal flat of the Yellow Sea in the Republic of Korea. The DNA G+C content of the type strain is 61.7 % (by genome).
Description of Qipengyuania vulgaris comb. nov.
Qipengyuania vulgaris (vul.ga'ris. L. fem. adj. vulgaris, ordinary, usual, common).
Basonym: Erythrobacter vulgaris Ivanova et al. 2006.
The description is the same as for Erb. vulgaris [36]. Core-genomic phylogenetic analysis strongly supported the placement of this species in the genus Qipengyuania . The type strain, 022-2-10T (=KMM 3465T=CIP 107841T=DSM 17792T), was isolated from the starfish Stellaster equestris collected from the East China Sea. The DNA G+C content of the type strain is 60.6 % (by genome).
Description of Alteriqipengyuania gen. nov.
Alteriqipengyuania (Al.te.ri.qi.peng.yu.an'i.a. L. adj. alter, another, other, different; N.L. fem. n. Qipengyuania , a genus name; N.L. fem. n. Alteriqipengyuania, another or different Qipengyuania ).
Cells are Gram-stain-negative, rod-shaped, non-spore-forming, non-motile and aerobic. Oxidase- and catalase-positive. Contains carotenoid pigments but not bacteriochlorophyll a. The predominant ubiquinone is Q-10. The major fatty acids (>10%) are C17 : 1 ω6c and summed feature 8 (C18 : 1 ω7c and/or C18 : 1 ω6c). The genus represents a distinct branch in the family Erythrobacteraceae of the class Alphaproteobacteria based on the core-genomic phylogeny. The DNA G+C content is 63.6–65.5 ol% (by genome). The type species is Alteriqipengyuania lutimaris.
Description of Alteriqipengyuania halimionae comb. nov.
Alteriqipengyuania halimionae (ha.li.mi.o'nae. N.L. gen. n. halimionae of the marsh plant Halimione portulacoides).
Basonym: Altererythrobacter halimionae Fidalgo et al. 2017.
The description is the same as for Aeb. halimionae [32]. Core-genomic phylogenetic analysis strongly supported the placement of this species in the genus Alteriqipengyuania. The type strain, CPA5T (=CECT 9130T=LMG 29519T), was isolated from the surface-sterilized aboveground tissues of the halophyte Halimione portulacoides. The DNA G+C content of the type strain is 65.5 % (by genome).
Description of Alteriqipengyuania lutimaris comb. nov.
Alteriqipengyuania lutimaris (lu.ti.ma'ris. L. neut. n. lutum mud; L. neut. n. mare the sea; N.L. gen. n. lutimaris of a marine mud).
Basonym: Erythrobacter lutimaris Jung et al. 2014.
The description is the same as for Erb. lutimaris [114]. Core-genomic phylogenetic analysis strongly supported the placement of this species in the genus Alteriqipengyuania. The type strain, S-5T (=KCTC 42109T=CECT 8624T), was isolated from a tidal flat sediment of Saemankum in the Republic ofKorea. The DNA G+C content of the type strain is 63.6 % (by genome).
Description of Parerythrobacter gen. nov.
Parerythrobacter (Par.e.ry.thro.bac'ter. Gr. prep. para, beside, alongside of, near, like; N.L. masc. n. Erythrobacter , a genus name; N.L. masc. n. Parerythrobacter, near or like Erythrobacter ).
Cells are Gram-stain-negative, rod-shaped, non-spore-forming, non-motile and strictly aerobic. Oxidase- and catalase-positive. Contains carotenoid pigments but not bacteriochlorophyll a. Requires NaCl for growth. The predominant ubiquinone is Q-10. The major fatty acids (>10%) are C17 : 1 ω6c and summed feature 8 (C18 : 1 ω7c and/or C18 : 1 ω6c). The major polar lipids are diphosphatidylglycerol, phosphatidylcholine, phosphatidylethanolamine, phosphatidylglycerol and sphingoglycolipid. The genus represents a distinct branch in the family Erythrobacteraceae of the class Alphaproteobacteria based on the core-genomic phylogeny. The DNA G+C content of the type strain is 60.2–60.6 % (by genome). The type species is Parerythrobacter jejueneis.
Description of Parerythrobacter jejuensis comb. nov.
Parerythrobacter jejuensis (je.ju.en'sis. N.L. masc. adj. jejuensis of or belonging to Jeju Island in the Republic of Korea, where the type strain was isolated).
Basonym: Erythrobacter jejuensis Yoon et al. 2013.
The description is the same as for Erb. jejuensis [76]. Core-genomic phylogenetic analysis strongly supported the placement of this species in the genus Parerythrobacter. The type strain, CNU001T (=KCTC 23090T=JCM 16677T), was isolated from seawater collected off Jeju Island, Republic of Korea. The DNA G+C content of the type strain is 60.2 % (by genome).
Description of Parerythrobacter lutipelagi comb. nov.
Parerythrobacter lutipelagi (lu.ti.pe.la'gi. L. neut. n. lutum, mud; L. neut. n. pelagus the sea; N.L. gen. n. lutipelagi of mud of the sea, where the type strain was isolated).
Basonym: Altererythrobacter lutipelagi Lee 2019.
The description is the same as for Aeb. lutipelagi [42]. Core-genomic phylogenetic analysis strongly supported the placement of this species in the genus Parerythrobacter. The type strain, GH1-16T (=KCTC 52845T=NBRC 113275T), was isolated from a tidal mudflat sample collected at the seashore of Gangwha Island, Republic of Korea. The DNA G+C content of the type strain is 60.6 % (by genome).
Emended description of the genus Altererythrobacter Kwon et al. 2007, emend. Xue et al. 2012, emend. Xue et al. 2016
The description is as given by Kwon et al. 2007 [38], Xue et al. [15] and Xue et al. 2016 [28] with the following amendment. Cells are aerobic and non-motile. Oxidase- and catalase-positive. Requires NaCl for growth. The major fatty acid (>10%) is C18 : 1 ω7c. The major polar lipids are phosphatidylethanolamine, phosphatidylglycerol and sphingoglycolipid. The genus represents a distinct branch in the family Erythrobacteraceae of the class Alphaproteobacteria based on the core-genomic phylogeny. The DNA G+C content is 52.0–61.8 % (by genome). The type species for the genus is Altererythrobacter epoxidivorans .
Emended description of Altererythrobacter epoxidivorans Kwon et al. 2007
Altererythrobacter epoxidivorans (e.po.xi.di.vo'rans. N.L. neut. n. epoxidum epoxide; L. pres. part. vorans devouring; N.L. part. adj. epoxidivorans epoxide-devouring).
The description is identical to that given for Aeb. epoxidivorans [38], except for the DNA G+C content. The type strain, JCS350T (=KCCM 42314T=JCM 13815T), was isolated from cold-seep sediments of Kagoshima Bay, Japan. The DNA G+C content of the type strain is 61.5 % (by genome).
Emended description of Altererythrobacter ishigakiensis Matsumoto et al. 2011
Altererythrobacter ishigakiensis (i.shi.ga.ki.en'sis. N.L. masc. adj. ishigakiensis of or belonging to Ishigaki island, Okinawa, Japan, where the type strain was isolated).
The description is identical to that given for Aeb. ishigakiensis [115], except for the DNA G+C content. The type strain, JPCCMB0017T (=NITE-AP48T=ATCC BAA-2084T= NBRC 107699T), was isolated from the coastal area of Okinawa, Japan. The DNA G+C content of the type strain is 56.9 % (by genome).
Emended description of Altererythrobacter xiamenensis Lei et al. 2014
Altererythrobacter xiamenensis (xia.men.en'sis. N.L. masc. adj. xiamenensis of Xiamen, a city in Fujian, PR China, where the type strain was first isolated).
The description is identical to that given for Aeb. xiamenensis [18], except for the DNA G+C content. The type strain, LY02T (=CGMCC 1.12494T=KCTC 32398T=NBRC 109638T), was isolated from red tide seawater in Xiamen, Fujian Province, PR China. The DNA G+C content of the type strain is 61.8 % (by genome).
Emended description of the genus Erythrobacter Shiba et al. 1982
The description is as given by Shiba et al. 1982 [29] with the following amendment. Cells are motile or non-motile. Positive or negative for oxidase. Requires NaCl for growth. The major fatty acids (>10%) are C18 : 1 ω7c and C17 : 1 ω6c. The major polar lipids include a sphingoglycolipid. The genus represents a distinct branch in the family Erythrobacteraceae of the class Alphaproteobacteria based on the core-genomic phylogeny. The DNA G+C content is 57.4–67.9 % (by genome). The type species for the genus is Erythrobacter longus .
Description of Erythrobacter colymbi comb. nov.
Erythrobacter colymbi (co.lym’bi. L. gen. n. colymbi, of a swimming pool, thus indicating the site of isolation of the type strain).
Basomym: Porphyrobacter colymbi Rainey et al. 2003.
The description is the same as for Por. colymbi [24]. Core-genomic phylogenetic analysis strongly supported the placement of this species in the genus Erythrobacter . The type strain, TPW-24T (=JCM 18338T= KCTC 32078T), was isolated from swimming pool water in Tokyo, Japan. The DNA G+C content of the type strain is 66.5 % (by genome).
Description of Erythrobacter cryptus comb. nov.
Erythrobacter cryptus (cryp'tus. N.L. masc. adj. cryptus from Gr. masc. adj. kryptos hidden, to indicate the cryptic relationship of this species to the closely related organisms).
Basomym: Porphyrobacter cryptus Rainey et al. 2003.
The description is the same as for Por. cryptus [26]. Core-genomic phylogenetic analysis strongly supported the placement of this species in the genus Erythrobacter . The type strain, ALC-2T (=DSM 12079T=ATCC BAA-386T), was isolated from the hot spring at Alcafache in Portugal. The DNA G+C content of the type strain is 67.9 % (by genome).
Description of Erythrobacter dokdonensis comb. nov.
Erythrobacter dokdonensis (dok.do.nen'sis. N.L. masc. adj. dokdonensis of Dokdo, from where the strain was isolated).
Basomym: Porphyrobacter dokdonensis Yoon et al. 2006.
The description is the same as for Por. dokdonensis [106]. Core-genomic phylogenetic analysis strongly supported the placement of this species in the genus Erythrobacter . The type strain, DSW-74T (=KCTC 12395T=DSM 17193T), was isolated from sea water off the island of Dokdo, Korea. The DNA G+C content of the type strain is 64.8 % (by genome).
Description of Erythrobacter donghaensis comb. nov.
Erythrobacter donghaensis (dong.ha.en'sis. N.L. masc. adj. donghaensis of Donghae, the Korean name for the East Sea in the Republic of Korea from which the strains were isolated).
Basomym: Porphyrobacter donghaensis Yoon et al. 2004.
The description is the same as for Por. donghaensis [105]. Core-genomic phylogenetic analysis strongly supported the placement of this species in the genus Erythrobacter . The type strain, SW-132T (=KCTC 12229T=DSM 16220T), was isolated from sea water from the East Sea in the Republic of Korea. The DNA G+C content of the type strain is 66.2 % (by genome).
Emended description of Erythrobacter litoralis Yurkov et al. 1994
Erythrobacter litoralis (li.to.ra'lis. L. masc. adj. litoralis, at the beach or coast, referring to the supralitoral habitat).
The description is identical to that given for Erb. litoralis [31], except for the DNA G+C content. The type strain, T4T (=ATCC 700002T=CIP 106926T=DSM 8509T=JCM 10281T=NBRC 102620T), was isolated from a marine cyanobacterial mat in a supralitoral zone. The DNA G+C content of the type strain is 65.2 % (by genome).
Emended description of Erythrobacter longus Shiba et al. 1982
Erythrobacter longus (lon'gus. L. masc. adj. longus, long).
The description is identical to that given for Erb. longus [29], except for the DNA G+C content. The type strain, Och01T (=ATCC 33941T=CIP 104268T=DSM 6997T= JCM 6170T=NBRC 14126T), was isolated from high-tidal seaweed Enteromorpha linza. The DNA G+C content of the type strain is 57.4 % (by genome).
Description of Erythrobacter neustonensis comb. nov.
Erythrobacter neustonensis [neu.sto.nen'sis. N.L. masc. adj. derived from Gr. n. neustos, swimming (floating), referring to occurrence of the bacterium as a member of the neuston (organisms floating at the air–water interface surface layer of a body of water)].
Basomym: Porphyrobacter neustonensis Fuerst et al. 1993.
The description is the same as for Por. neustonensis [23]. Core-genomic phylogenetic analysis strongly supported the placement of this species in the genus Erythrobacter . The type strain, ACM 2844T (=CIP 104070T=DSM 9434T), was isolated from air–water interface of freshwater subtropical pond in Brisbane, Australia. The DNA G+C content of the type strain is 65.3 % (by genome).
Description of Erythrobacter ramosus comb. nov.
Erythrobacter ramosus (ra.mo'sus. L. masc. adj. ramosus, ramifying, referring to the morphology of the cells).
Basomym: Erythromicrobium ramosum Yurkov et al. 1994.
The description is the same as for Erm. ramosum [31]. Core-genomic phylogenetic analysis strongly supported the placement of this species in the genus Erythrobacter . The type strain, E5T (=ATCC 700003T=CIP 106927T=DSM 8510T=JCM 10282T=NBRC 102621T), was isolated from a cyanobacterial mat from an alkaline spring. The DNA G+C content of the type strain is 64.3 % (by genome).
Description of Erythrobacter sanguineus comb. nov.
Erythrobacter sanguineus (san.gui'ne.us. L. masc. adj. sanguineus blood-coloured).
Basomym: Porphyrobacter sanguineus Hiraishi et al. 2002.
The description is the same as for Por. sanguineus [104]. Core-genomic phylogenetic analysis strongly supported the placement of this species in the genus Erythrobacter . The type strain, A91T (=ATCC 25659T=DSM 11302T=IAM 12620T=ICPB 4167T=NBRC 15763T=JCM 20691T), was isolated from sea water collected in Baltic Sea. The DNA G+C content of the type strain is 63.6 % (by genome).
Description of Erythrobacter tepidarius comb. nov.
Erythrobacter tepidarius (te.pi.da’ri.us. L. neut. n. tepidarium, a warm bath fed by natural thermal water; N.L. masc. adj. tepidarius, warm bathing).
Basomym: Porphyrobacter tepidarius Hanada et al. 1997.
The description is the same as for Por. tepidarius [27]. Core-genomic phylogenetic analysis strongly supported the placement of this species in the genus Erythrobacter . The type strain, OT3T (=DSM 10594T), was isolated from a cyanobacterial mat in brackish water of a hot spring in Shidzuoka Prefecture, Japan. The DNA G+C content of the type strain is 65.9 % (by genome).
Description of Pontixanthobacter gen. nov.
Pontixanthobacter (Pon.ti.xan.tho.bac'ter. L. masc. n. pontus, the sea; Gr. masc. adj. xanthos, yellow; N.L. masc. n. bacter, rod or staff; N.L. masc. n. Pontixanthobacter, a yellow bacterium from the sea).
Cells are Gram-stain-negative, ovoid to rod, non-spore-forming, non-motile and aerobic. Positive and negative for oxidase. Catalase-positive. Contains carotenoid pigments but not bacteriochlorophyll a. The predominant ubiquinone is Q-10. The major fatty acid (>10 %) is summed feature 8 (C18 : 1 ω7c and/or C18 : 1 ω6c). The major polar lipids are phosphatidylcholine, phosphatidylethanolamine, phosphatidylglycerol and sphingoglycolipid. The genus represents a distinct branch in the family Erythrobacteraceae of the class Alphaproteobacteria based on the core-genomic phylogeny. The DNA G+C content is 55.5–61.5 % (by genome). The type species is Pontixanthobacter luteolus.
Description of Pontixanthobacter aestiaquae comb. nov.
Pontixanthobacter aestiaquae (aes.ti.a'quae. L. masc. n. aestus the sea tide; L. fem. n. aqua water; N.L. gen. n. aestiaquae of the water of the sea tide).
Basonym: Altererythrobacter aestiaquae Jung et al. 2014.
The description is the same as for Aeb. aestiaquae [62]. Core-genomic phylogenetic analysis strongly supported the placement of this species in the genus Pontixanthobacter. The type strain, HDW-31T (=KCTC 42006T=CECT 8527T), was isolated from seawater of Hwang-do in the Republic of Korea. The DNA G+C content of the type strain is 57.2 % (by genome).
Description of Pontixanthobacter aquaemixtae comb. nov.
Pontixanthobacter aquaemixtae (a.quae.mi'xtae. L. fem. n. aqua water; L. fem. perf. part. mixta mixed; N.L. fem. gen. n. aquaemixtae of mixed waters).
Basonym: Altererythrobacter aquaemixtae Park et al. 2017.
The description is the same as for Aeb. aquaemixtae [64]. Core-genomic phylogenetic analysis strongly supported the placement of this species in the genus Pontixanthobacter. The type strain, JSSK-8T (=KCTC 52763T=NBRC 112764T), was isolated from the place where the ocean and a freshwater spring meet at Jeju Island, Republic of Korea. The DNA G+C content of the type strain is 58.5 % (by genome).
Description of Pontixanthobacter confluentis comb. nov.
Pontixanthobacter confluentis (con.flu.en'tis. L. gen. n. confluentis of a meeting place of waters).
Basonym: Altererythrobacter confluentis Park et al. 2016.
The description is the same as for Aeb. confluentis [20]. Core-genomic phylogenetic analysis strongly supported the placement of this species in the genus Pontixanthobacter. The type strain, KEM-4T (=KCTC 52259T=NBRC 112305T), was isolated from water collected from an estuary environment where the ocean and a river meet at Seocheon, Republic of Korea. The DNA G+C content of the type strain is 59.1 % (by genome).
Description of Pontixanthobacter gangjinensis comb. nov.
Pontixanthobacter gangjinensis (gang.jin.en'sis. N.L. masc. adj. gangjinensis pertaining to Gangjin bay where the type strain was isolated).
Basonym: Altererythrobacter gangjinensis Jeong et al. 2013.
The description is the same as for Aeb. gangjinensis [67]. Core-genomic phylogenetic analysis strongly supported the placement of this species in the genus Pontixanthobacter. The type strain, KJ7T (=KACC 16190T=JCM 17802T), was isolated from a tidal flat of the Gangjin bay in the Republic of Korea. The DNA G+C content of the type strain is 55.5 % (by genome).
Description of Pontixanthobacter luteolus comb. nov.
Pontixanthobacter luteolus (lu.te'o.lus. L. masc. adj. luteolus, yellowish).
Basonym: Altererythrobacter luteolus Yoon et al. 2005. emend. Kwon et al. 2007
The description is the same as for Aeb. luteolus [38, 68]. Core-genomic phylogenetic analysis strongly supported the placement of this species in the genus Pontixanthobacter. The type strain, SW-109T (=KCTC 12311T=JCM 12599T), was isolated from a tidal flat of the Yellow Sea in the Republic of Korea. The DNA G+C content of the type strain is 59.3 % (by genome).
Description of Pontixanthobacter sediminis comb. nov.
Pontixanthobacter sediminis (se.di'mi.nis. L. gen. n. sediminis of sediment).
Basonym: Altererythrobacter sediminis Kim et al. 2016.
The description is the same as for Aeb. sediminis [72]. Core-genomic phylogenetic analysis strongly supported the placement of this species in the genus Pontixanthobacter. The type strain, CAU 1172T (=KCTC 42453T=NBRC 110917T), was isolated from a sample of lagoon sediment from along the east coast of the Republic of Korea. The DNA G+C content of the type strain is 61.5 % (by genome).
Description of Alteripontixanthobacter gen. nov.
Alteripontixanthobacter (Al.te.ri.pon.ti.xan.tho.bac'ter. L. adj. alter, another, other, different; N.L. masc. n. Pontixanthobacter, a genus name; N.L. masc. n. Alteripontixanthobacter, another or different Pontixanthobacter).
Cells are Gram-stain-negative, rod, non-spore-forming, non-motile and aerobic. Oxidase- and catalase-positive. Contains carotenoid pigments but not bacteriochlorophyll a. Requires NaCl for growth. The predominant ubiquinone is Q-10. The major fatty acids (>10%) are summed feature 8 (C18 : 1 ω7c and/or C18 : 1 ω6c), summed feature 3 (C16 : 1 ω7c and/or C16 : 1 ω6c) and C16 : 0. The major polar lipids are diphosphatidylglycerol, phosphatidylcholine, phosphatidylethanolamine, phosphatidylglycerol and sphingoglycolipid. The genus represents a distinct branch in the family Erythrobacteraceae of the class Alphaproteobacteria based on the core-genomic phylogeny. The DNA G+C content is 60.8 % (by genome). The type species is Alteripontixanthobacter maritimus.
Description of Alteripontixanthobacter maritimus comb. nov.
Alteripontixanthobacter maritimus (ma.ri'ti.mus. L. masc. adj. maritimus of the marine environment).
Basonym: Altererythrobacter maritimus Kang et al. 2019.
The description is the same as for Aeb. maritimus [43]. Core-genomic phylogenetic analysis strongly supported the placement of this species in the genus Alteripontixanthobacter. The type strain, HME9302T (=KCTC 32463T=KACC 17617T=CECT 8417T), was isolated from seawater in the Republic of Korea. The DNA G+C content of the type strain is 60.8 % (by genome).
Description of Parapontixanthobacter gen. nov.
Parapontixanthobacter (Pa.ra.pon.ti.xan.tho.bac'ter. Gr. prep. para, beside, alongside of, near, like; N.L. masc. n. Pontixanthobacter, a genus name; N.L. masc. n. Parapontixanthobacter, near or like Pontixanthobacter).
Cells are Gram-stain-negative, coccoid, non-spore-forming, non-motile and strictly aerobic. Oxidase-negative and catalase-positive. Contains carotenoid pigments but not bacteriochlorophyll a. Requires NaCl for growth. Reduces nitrate to nitrite. The predominant ubiquinone is Q-10. The major fatty acids (>10 %) are summed feature 8 (C18 : 1 ω7c and/or C18 : 1 ω6c), summed feature 3 (C16 : 1 ω7c and/or C16 : 1 ω6c) and C16 : 0. The major polar lipids are diphosphatidylglycerol, phosphatidylethanolamine, phosphatidylglycerol and sphingoglycolipid. The genus represents a distinct branch in the family Erythrobacteraceae of the class Alphaproteobacteria based on the core-genomic phylogeny. The DNA G+C content is 61.2 % (by genome).The type species is Parapontixanthobacter aurantiacus.
Description of Parapontixanthobacter aurantiacus comb. nov.
Parapontixanthobacter aurantiacus (au.ran.ti'a.cus. N.L. masc. adj. aurantiacus, orange-coloured).
Basonym: Altererythrobacter aurantiacus Zhang et al. 2016.
The description is the same as for Aeb. aurantiacus [65]. Core-genomic phylogenetic analysis strongly supported the placement of this species in the genus Parapontixanthobacter. The type strain, O30T (=CGMCC 1.12762T=JCM 19853T=LMG 28110T=MCCC 1A09962T), was isolated from a deep-sea sediment of the west Pacific Ocean. The DNA G+C content of the type strain is 61.2 % (by genome).
Description of Pelagerythrobacter gen. nov.
Pelagerythrobacter (Pe.lag.e.ry.th.ro.bac'ter. L. neut. n. pelagus the sea; N.L. masc. n. Erythrobacter , a genus name; N.L. masc. n. Pelagerythrobacter, Erythrobacter from the sea).
Cells are Gram-stain-negative, rod-shaped, non-spore-forming and aerobic. Motile or non-motile. Positive and negative for oxidase. Catalase-positive. Contains carotenoid pigments but not bacteriochlorophyll a. Requires NaCl for growth. The predominant ubiquinone is Q-10. The major fatty acid (>10 %) is C18 : 1 ω7c. The major polar lipids are diphosphatidylglycerol, phosphatidylcholine, phosphatidylethanolamine, phosphatidylglycerol and sphingoglycolipid. The genus represents a distinct branch in the family Erythrobacteraceae of the class Alphaproteobacteria based on the core-genomic phylogeny. The DNA G+C content is 64.7–68.2 % (by genome).The type species is Pelagerythrobacter marinus.
Description of Pelagerythrobacter aerophilus comb. nov.
Pelagerythrobacter aerophilus (a.e.ro'phi.lus. Gr. masc. n. aer, air; N.L. adj. philus from Gr. masc. adj. philos friend, loving; N.L. masc. adj. aerophilus, air-loving).
Basonym: Altererythrobacter aerophilus Meng et al. 2019.
The description is the same as for Aeb. aerophilus [17]. Core-genomic phylogenetic analysis strongly supported the placement of this species in the genus Pelagerythrobacter. The type strain, Ery1T (=KCTC 62387T=CGMCC 1.16499T=MCCC 1A10037T), was isolated from deep-sea seawater of the Mariana Trench. The DNA G+C content of the type strain is 65.4 % (by genome).
Description of Pelagerythrobacter marinus comb. nov.
Pelagerythrobacter marinus (ma.ri'nus. L. masc. adj. marinus of the sea, marine).
Basonym: Altererythrobacter marinus Lai et al. 2009.
The description is the same as for Aeb. marinus [69]. Core-genomic phylogenetic analysis strongly supported the placement of this species in the genus Pelagerythrobacter. The type strain, H32T (=CCTCC AB 208229T=LMG 24629T=MCCC 1A01070T), was isolated from deep seawater of the Indian Ocean. The DNA G+C content of the type strain is 68.2 % (by genome).
Description of Pelagerythrobacter marensis comb. nov.
Pelagerythrobacter marensis (ma.ren'sis. N.L. masc. adj. marensis of Mara Island, Jeju, Republic of Korea, where the type strain was isolated).
Basonym: Altererythrobacter marensis Seo et al. 2010.
The description is the same as for Aeb. marensis [116]. Core-genomic phylogenetic analysis strongly supported the placement of this species in the genus Pelagerythrobacter. The type strain, MSW-14T (=KCTC 22370T=DSM 21428T), was isolated from seawater collected around Mara Island, Jeju, Republic of Korea. The DNA G+C content of the type strain is 64.7 % (by genome).
Description of Altericroceibacterium gen. nov.
Altericroceibacterium (Al.te.ri.cro.ce.i.bac.te'ri.um. L. masc. adj. alter another, other, different; N.L. neut. n. Croceibacterium , a genus name; N.L. neut. n. Altericroceibacterium, another or different Croceibacterium ).
Cells are Gram-stain-negative, rod-shaped, non-spore-forming, aerobic and non-motile. Positive and negative for oxidase. Catalase-positive. Contains carotenoid pigments but not bacteriochlorophyll a. Requires NaCl for growth. The predominant ubiquinone is Q-10. The major fatty acid (>10%) is summed feature 8 (C18 : 1 ω7c and/or C18 : 1 ω6c). The major polar lipid is phosphatidylethanolamine. The genus represents a distinct branch in the family Erythrobacteraceae of the class Alphaproteobacteria based on the core-genomic phylogeny. The DNA G+C content is 55.8–64.2 % (by genome). The type species is Altericroceibacterium indicum.
Description of Altericroceibacterium endophyticum comb. nov.
Altericroceibacterium endophyticum (en.do.phy'ti.cum. Gr. pref. endo within; Gr. n. phyton plant; L. neut. suff. -icum adjectival suffix used with the sense of belonging to; N.L. neut. adj. endophyticum within plant, endophytic).
Basonym: Altererythrobacter endophyticus Fidalgo et al. 2017.
The description is the same as for Aeb. endophyticus [32]. Core-genomic phylogenetic analysis strongly supported the placement of this species in the genus Altericroceibacterium. The type strain, BR75T (=CECT 9129T=LMG 29518T), was isolated from the surface-sterilized belowground tissues of the halophyte Halimione portulacoides. The DNA G+C content of the type strain is 58.6 % (by genome).
Description of Altericroceibacterium indicum comb. nov.
Altericroceibacterium indicum (in'di.cum. L. neut. adj. indicum pertaining to India, where the type strain was isolated).
Basonym: Altererythrobacter indicus Kumar et al. 2008.
The description is the same as for Aeb. indicus [33]. Core-genomic phylogenetic analysis strongly supported the placement of this species in the genus Altericroceibacterium. The type strain, MSSRF26T (=LMG 23789T=DSM 18604T), isolated from the rhizosphere of mangrove-associated wild rice (Porteresia coarctata Tateoka). The DNA G+C content of the type strain is 55.8 % (by genome).
Description of Altericroceibacterium xinjiangense comb. nov.
Altericroceibacterium xinjiangense (xin.jiang.en'se. N.L. neut. adj. xinjiangense of or pertaining to Xinjiang, an autonomous region in north-west China).
Basonym: Altererythrobacter xinjiangensis Xue et al. 2012.
The description is the same as for Aeb. xinjiangensis [15]. Core-genomic phylogenetic analysis strongly supported the placement of this species in the genus Altericroceibacterium. The type strain, S3-63T (=CCTCC AB 207166T=CIP 110125T), was isolated from sand from the desert of Xinjiang, PR China. The DNA G+C content of the type strain is 64.2 % (by genome).
Emended description of the genus Croceibacterium Liu et al. 2019
The description is as given by Liu et al. [30]with the following amendment. Cells are pleomorphic. Some species can motile by means of polar flagella. Positive or negative for oxidase. The major fatty acid (>10 %) is summed feature 8 (C18 : 1 ω7c and/or C18 : 1 ω6c). The major polar lipids are diphosphatidylglycerol, phosphatidylethanolamine and phosphatidylglycerol. The genus represents a distinct branch in the family Erythrobacteraceae of the class Alphaproteobacteria based on the core-genomic phylogeny. The DNA G+C content is 61.9–67.0 % (by genome). The type species for the genus is Croceibacterium ferulae .
Description of Croceibacterium atlanticum comb. nov.
Croceibacterium atlanticum (at.lan'ti.cum. L. neut. adj. atlanticum of or pertaining to the Atlantic Ocean, where the type strain was isolated).
Basonym: Altererythrobacter atlanticus Wu et al. 2014.
The description is the same as for Aeb. atlanticus [8]. Core-genomic phylogenetic analysis strongly supported the placement of this species in the genus Croceibacterium . The type strain, 26DY36T (=CGMCC 1.12411T=JCM 18865T), was isolated from a deep-sea sediment sample collected from the North Atlantic Rise. The DNA G+C content of the type strain is 61.9 % (by genome).
Description of Croceibacterium salegens comb. nov.
Croceibacterium salegens (sal.e'gens. L. masc. n. sal, salis salt; L. pres. part. egens needy; N.L. part. adj. salegens salt-needy).
Basonym: Altererythrobacter salegens Liang et al. 2017.
The description is the same as for Aeb. salegens [71]. Core-genomic phylogenetic analysis strongly supported the placement of this species in the genus Croceibacterium . The type strain, XY-R17T (=KCTC 52267T=MCCC 1K01500T), was isolated from the surface sediment of Mai Po Inner Deep Bay Ramsar Site in Hong Kong. The DNA G+C content of the type strain is 64.6% (by genome).
Description of Croceibacterium soli comb. nov.
Croceibacterium soli (so'li. L. gen. n. soli of soil).
Basonym: Altererythrobacter soli Zhao et al. 2017.
The description is the same as for Aeb. soli [14]. Core-genomic phylogenetic analysis strongly supported the placement of this species in the genus Croceibacterium . The type strain, MN-1T (=KCTC 52135T=MCCC 1K02066T), isolated from a desert sand sample collected from Tengger desert, north-western PR China. The DNA G+C content of the type strain is 67.0 % (by genome).
Description of Croceibacterium xixiisoli comb. nov.
Croceibacterium xixiisoli (xi.xi.i.so'li. L. gen. n. soli of soil. N.L. gen. n. xixiisoli from Xixi soil).
Basonym: Altererythrobacter xixiisoli Yuan et al. 2017.
The description is the same as for Aeb. xixiisoli [12]. Core-genomic phylogenetic analysis strongly supported the placement of this species in the genus Croceibacterium . The type strain, S36T (=CGMCC 1.12804T=NBRC 110413T), was isolated from soil of the Xixi wetland in Hangzhou, eastern PR China. The DNA G+C content of the type strain is 63.3 % (by genome).
Description of Alteraurantiacibacter gen. nov.
Alteraurantiacibacter (Al.ter.au.ran.ti.a.ci.bac'ter. L. masc. adj. alter another, other, different; N.L. masc. n. Aurantiacibacter, a genus name; N.L. masc. n. Alteraurantiacibacter, another or different Aurantiacibacter).
Cells are Gram-stain-negative, pleomorphic, non-spore-forming, aerobic and non-motile. Oxidase- and catalase-positive. Contains carotenoid pigments but not bacteriochlorophyll a. Requires NaCl for growth. The predominant ubiquinone is Q-10. The major fatty acid (>10%) is summed feature 8 (C18 : 1 ω7c and/or C18 : 1 ω6c). The major polar lipids are phosphatidylethanolamine and phosphatidylglycerol. The genus represents a distinct branch in the family Erythrobacteraceae of the class Alphaproteobacteria based on the core-genomic phylogeny. The DNA G+C content is 62.5–66.0 % (by genome). The type species is Alteraurantiacibacter aestuarii.
Description of Alteraurantiacibacter aestuarii comb. nov.
Alteraurantiacibacter aestuarii (aes.tu.a'ri.i. L. gen. n. aestuarii of a tidal flat, from where the type strain was isolated).
Basonym: Altererythrobacter aestuarii Park et al. 2011.
The description is the same as for Aeb. aestuarii [63]. Core-genomic phylogenetic analysis strongly supported the placement of this species in the genus Alteraurantiacibacter. The type strain, KYW147T (=KCTC 22735T=JCM 16339T), was isolated from a seawater sample collected from the South Sea, Republic of Korea. The DNA G+C content of the type strain is 62.5 % (by genome).
Description of Alteraurantiacibacter aquimixticola comb. nov.
Alteraurantiacibacter aquimixticola (a.qui.mix.ti’co.la. L. fem. n. aqua water; L. masc. perf. part. mixtus mixed; L. suff. -cola inhabitant; N.L. masc. n. aquimixticola an inhabitant of mixed waters).
Basonym: Altererythrobacter aquimixticola Park et al. 2019.
The description is the same as for Aeb. aquimixticola [40]. Core-genomic phylogenetic analysis strongly supported the placement of this species in the genus Alteraurantiacibacter. The type strain, SSKS-13T (=KACC 19863T=KCTC 62900T=NBRC 113545T), was isolated from sediment sampled at the junction between the ocean and a freshwater spring at Jeju island of the South Sea, Republic of Korea. The DNA G+C content of the type strain is 63.9 % (by genome).
Description of Alteraurantiacibacter buctensis comb. nov.
Alteraurantiacibacter buctensis (buc.ten'sis. N.L. masc. adj. buctensis referring to the acronym BUCT, Beijing University of Chemical Technology, where the strain was identified).
Basonym: Altererythrobacter buctensis Zhang et al. 2016.
The description is the same as for Aeb. buctensis [66]. Core-genomic phylogenetic analysis strongly supported the placement of this species in the genus Alteraurantiacibacter. The type strain, M0322T (=CGMCC 1.12871T=JCM 30112T), was isolated from the Mohe Basin, PR China. The DNA G+C content of the type strain is 66.0 % (by genome).
Description of Aurantiacibacter gen. nov
Aurantiacibacter (Au.ran.ti.a.ci.bac'ter. N.L. masc. adj. aurantiacus, orange-coloured; N.L. masc. n. bacter, rod or staff; N.L. masc. n. Aurantiacibacter, orange-coloured rod).
Cells are Gram-stain-negative, pleomorphic and non-spore-forming. Aerobic or facultative anaerobic. Positive or negative for oxidase. Catalase-positive. Contains carotenoid pigments but not bacteriochlorophyll a. The predominant ubiquinone is Q-10. The major fatty acid (>10 %) is summed feature 8 (C18 : 1 ω7c and/or C18 : 1 ω6c). The major polar lipid is phosphatidylethanolamine. The genus represents a distinct branch in the family Erythrobacteraceae of the class Alphaproteobacteria based on the core-genomic phylogeny. The DNA G+C content is 58.3–67.2 % (by genome). The type species is Aurantiacibacter gangjinensis.
Description of Aurantiacibacter aquimixticola comb. nov.
Aurantiacibacter aquimixticola (a.qui.mix.ti’co.la. L. fem. n. aqua water; L. masc. perf. part. mixtus mixed; L. suff. -cola from L. n. incola dweller, inhabitant; N.L. masc. n. aquimixticola an inhabitant of mixed waters).
Basonym: Erythrobacter aquimixticola Park et al. 2017.
The description is the same as for Erb. aquimixticola [88]. Core-genomic phylogenetic analysis strongly supported the placement of this species in the genus Aurantiacibacter. The type strain, JSSK-14T (=KCTC 52764T=NBRC 112765T), was isolated from water from the place where the ocean and a freshwater spring meet at Jeju island, Republic of Korea. The DNA G+C content of the type strain is 63.0 % (by genome).
Description of Aurantiacibacter arachoides comb. nov.
Aurantiacibacter arachoides (a.ra.cho'i.des. N.L. fem. n. arachis, peanut; L. suff. –oides, looking like; N.L. masc. adj. arachoides, looking like a peanut).
Basonym: Erythrobacter arachoides Xing et al. 2017.
The description is the same as for Erb. arachoides [74]. Core-genomic phylogenetic analysis strongly supported the placement of this species in the genus Aurantiacibacter. The type strain, RC4-10-4T (=CGMCC 1.15507T=JCM 31277T), was isolated from an ice core in the East Rongbuk Glacier, Tibetan Plateau. The DNA G+C content of the type strain is 65.4 % (by genome).
Description of Aurantiacibacter atlanticus comb. nov.
Aurantiacibacter atlanticus (at.lan'ti.cus. N.L. masc. adj. atlanticus referring to the Atlantic Ocean, where the type strain was isolated).
Basonym: Erythrobacter atlanticus Zhuang et al. 2015.
The description is the same as for Erb. atlanticus [109]. Core-genomic phylogenetic analysis strongly supported the placement of this species in the genus Aurantiacibacter. The type strain, s21-N3T (=MCCC 1A00519T=KCTC 42697T) was isolated from deep-sea sediment of the Atlantic Ocean. The DNA G+C content of the type strain is 58.3 % (by genome).
Description of Aurantiacibacter gangjinensis comb. nov.
Aurantiacibacter gangjinensis (gang.jin.en'sis. N.L. masc. adj. gangjinensis referring to Gangjin, the name of the bay in Korea from which the type strain was isolated).
Basonym: Erythrobacter gangjinensis Lee et al. 2010.
The description is the same as for Erb. gangjinensis [117]. Core-genomic phylogenetic analysis strongly supported the placement of this species in the genus Aurantiacibacter. The type strain, K7-2T (=KCTC 22330T=JCM 15420T), was isolated from seawater of Gangjin Bay, Republic of Korea. The DNA G+C content of the type strain is 62.7 % (by genome).
Description of Aurantiacibacter luteus comb. nov.
Aurantiacibacter luteus (lu'te.us. L. masc. adj. luteus orange-coloured, referring to the colour of the colony).
Basonym: Erythrobacter luteus Lei et al. 2015.
The description is the same as for Erb. luteus [118]. Core-genomic phylogenetic analysis strongly supported the placement of this species in the genus Aurantiacibacter. The type strain, KA37T (=MCCC 1F01227T=KCTC 42179T), was isolated from a mangrove sediment sample collected from Yunxiao mangrove National Nature Reserve, Fujian Province, PR China. The DNA G+C content of the type strain is 67.2 % (by genome).
Description of Aurantiacibacter marinus comb. nov.
Aurantiacibacter marinus (ma.ri'nus. L. masc. adj. marinus of the sea, marine).
Basonym: Erythrobacter marinus Jung et al. 2012.
The description is the same as for Erb. marinus [119]. Core-genomic phylogenetic analysis strongly supported the placement of this species in the genus Aurantiacibacter. The type strain, HWDM-33T (=KCTC 23554T=CCUG 60528T), was isolated from seawater of Hwang-do, an island of the Yellow Sea, Republic of Korea. The DNA G+C content of the type strain is 59.1 % (by genome).
Description of Aurantiacibacter odishensis comb. nov.
Aurantiacibacter odishensis (o.dish.en'sis. N.L. masc. adj. odishensis of or belonging to Odisha, a coastal state in India rich in bacterial diversity).
Basonym: Erythrobacter odishensis Subhash et al. 2013.
The description is the same as for Erb. odishensis [13]. Core-genomic phylogenetic analysis strongly supported the placement of this species in the genus Aurantiacibacter. The type strain, JA747T (=KCTC 23981T=NBRC 108930T), was isolated from a soil sample of a solar saltern at Humma, Odisha, India. The DNA G+C content of the type strain is 63.7 % (by genome).
Description of Aurantiacibacter spongiae comb. nov.
Aurantiacibacter spongiae (spon'gi.ae. L. gen. n. spongiae of a sponge, the source of the type strain).
Basonym: Erythrobacter spongiae Zhuang et al. 2019.
The description is the same as for Erb. spongiae [35]. Core-genomic phylogenetic analysis strongly supported the placement of this species in the genus Aurantiacibacter. The type strain, HN-E23T (=MCCC 1K03331T=LMG 30457T), was isolated from a sponge sample. The DNA G+C content of the type strain is 65.5 % (by genome).
Description of Aurantiacibacter xanthus comb. nov.
Aurantiacibacter xanthus (xan'thus. Gr. masc. adj. xanthos yellow; N.L. masc. adj. xanthus yellow).
Basonym: Erythrobacter xanthus Li et al. 2017.
The description is the same as for Erb. xanthus [19]. Core-genomic phylogenetic analysis strongly supported the placement of this species in the genus Aurantiacibacter. The type strain, SM1501T (=KCTC 42669T=CCTCC AB 2015396T), was isolated from surface seawater of the South China Sea. The DNA G+C content of the type strain is 64.5 % (by genome).
Description of Aurantiacibacter zhengii comb. nov.
Aurantiacibacter zhengii [zheng'i.i. N.L. gen. n. zhengii of TianLing Zheng (June 1955 to August 2017), a Chinese microbiologist, for his contributions to marine microorganism resources, biological control of harmful red tides, and our understanding of the interactions between bacteria and algae].
Basonym: Erythrobacter zhengii Fang et al. 2019.
The description is the same as for Erb. zhengii [9]. Core-genomic phylogenetic analysis strongly supported the placement of this species in the genus Aurantiacibacter. The type strain, V18T (=KCTC 62389T=MCCC 1K03475T), was isolated from deep-sea sediment of the Pacific Ocean. The DNA G+C content of the type strain is 62.7 % (by genome).
Description of Paraurantiacibacter gen. nov.
Paraurantiacibacter (Par.au.ran.ti.a.ci.bac'ter. Gr. prep. para, beside, alongside of, near, like; N.L. masc. n. Aurantiacibacter, a genus name; N.L. masc. n. Paraurantiacibacter, near or like Aurantiacibacter).
Cells are Gram-stain-negative, ovoid- to rod-shaped, non-spore-forming and aerobic. Oxidase- and catalase-positive. Requires NaCl for growth. Contains carotenoid pigments but not bacteriochlorophyll a. The predominant ubiquinone is Q-10. The major fatty acids (>10 %) are C18 : 1 ω7c and summed feature 3 (C16 : 1 ω7c and/or C16 : 1 ω6c). The major polar lipids are diphosphatidylglycerol, phosphatidylcholine, phosphatidylethanolamine, phosphatidylglycerol and sphingoglycolipid. The genus represents a distinct branch in the family Erythrobacteraceae of the class Alphaproteobacteria based on the core-genomic phylogeny. The DNA G+C content is 65.0 % (by genome). The type species is Paraurantiacibacter namhicola.
Description of Paraurantiacibacter namhicola comb. nov.
Paraurantiacibacter namhicola (nam.hi'co.la. N.L. n. namhae Namhae, the Korean name of the South Sea; L. suff. -cola from L. n. incola a dweller, inhabitant; N.L. masc. n. namhicola a dweller of the South Sea, referring to the isolation of the type strain).
Basonym: Altererythrobacter namhicola Park et al. 2011.
The description is the same as for Aeb. namhicola [63]. Core-genomic phylogenetic analysis strongly supported the placement of this species in the genus Paraurantiacibacter. The type strain, KYW48T (=KCTC 22736T=JCM 16345T), was isolated from a seawater sample collected from the South Sea, Republic of Korea. The DNA G+C content of the type strain is 65.0 % (by genome).
Emended description of the genus Croceicoccus Xu et al. 2009 emend. Huang et al. 2015
The description is as given by Xu et al. [4] and Huang et al. [110] with the following amendments. Cells are coccid or rods. The major fatty acid (>10 %) is C18 : 1 ω7c. The genus represents a distinct branch in the family Erythrobacteraceae of the class Alphaproteobacteria based on the core-genomic phylogeny. The DNA G+C content is 62.5–64.5 % (by genome). The type species for the genus is Croceicoccus marinus .
Emended description of Croceicoccus marinus Xu et al. 2009 emend. Huang et al. 2015
Croceicoccus marinus (ma.ri'nus. L. masc. adj. marinus of or belonging to the sea, marine)
The description is identical to that given for Ccc. marinus [4, 110], except for the DNA G+C content. The type strain, E4A9T (=CGMCC 1.6776T=JCM 14846T), was isolated from a deep-sea sediment sample collected from a polymetallic nodule region in the East Pacific Ocean. The DNA G+C content of the type strain is 64.5% (by genome).
Emended description of Croceicoccus naphthovorans Huang et al. 2015
Croceicoccus naphthovorans (naph.tho.vo'rans. Gr. fem. n. naphtha oil; L. pres. part. vorans devouring; N.L. part. adj. naphthovorans oil-degrading).
The description is identical to that given for Ccc. naphthovorans [110], except for the DNA G+C content. The type strain, PQ-2T (=CGMCC 1.12805T=NBRC 110381T) was isolated from marine biofilm collected from a boat shell at a harbour of Zhoushan island in Zhejiang Province, PR China. The DNA G+C content of the type strain is 62.6 % (by genome).
Supplementary Data
Funding information
This study was funded by the National Natural Science Foundation of China (No. 31770004 and No. 91851114), the Natural Science Foundation of Zhejiang Province (No. LR17D060001), the National Program for Support of Top-Notch Young Professionals.
Acknowledgements
Aeb. aerius 100921-2T, Aeb. buctensis M0322T, Aeb. luteolus SW-109T, Aeb. marinus H32T, Aeb. xixiisoli S36T and Erb. arachoides RC4-10-4T were generously donated by Professor Yong Li (Research Institute of Forest Ecology Environment and Protection, Chinese Academy of Forestry), Professor Jie Lv (Beijing University of Chemical Technology), Professor Jung-Hong Yoon (Korea Research Institute of Bioscience and Biotechnology), Professor Zongze Shao (Third Institute of Oceanography, Ministry of Natural Resources), Professor Zhiliang Yu (Zhejiang University of Technology) and Professor Yongqin Liu (Institute of Tibetan Plateau Research, Chinese Academy of Sciences), respectively.
Conflicts of interest
The authors declare that there are no conflicts of interest.
Footnotes
Abbreviations: AAI, average amino acid identity; AAPB, aerobic anoxygenic phototrophic bacteria; ANI, average nucleotide identity; CDS, coding sequence; OC, orthologous cluster; POCP, percentage of conserved protein; Q-10, ubiquinone-10.
The GenBank/EMBL/DDBJ accession numbers for the whole genome sequences of Erythrobacteraceae type strains are as follows: Aeb. aerius 100921-2T (WTZA00000000), Aeb. aestiaquae KCTC 42006T (WTYZ00000000), Aeb. aestuarii JCM 16339T (WTYY00000000), Aeb. aquaemixtae KCTC 52763T (WTYX00000000), Aeb. aurantiacus MCCC 1A09962T (WTYW00000000), Aeb. buctensis M0322T (WTYV00000000), Aeb. confluentis KCTC 52259T (WTYU00000000), Aeb. endophyticus LMG 29518T (WTYT00000000), Aeb. gangjinensis JCM 17802T (WTYS00000000), Aeb. halimionae LMG 29519T (WTYR00000000), Aeb. indicus DSM 18604T (WTYQ00000000), Aeb. luteolus SW-109T (WTYP00000000), Aeb. marinus H32T (WTYO00000000), Aeb. namhicola JCM 16345T (CP016545), Aeb. oceanensis MCCC 1A09965T (WTYN00000000), Aeb. salegens MCCC 1K01500T (WTYM00000000), Aeb. sediminis KCTC 42453T (WTYL00000000), Aeb. soli MCCC 1K02066T (WTYK00000000), Aeb. xixiisoli S36T (WTYJ00000000), Erb. aquimaris JCM 12189T (WTYI00000000), Erb. arachoides RC4-10-4T (WTYH00000000), Erb. citreus CGMCC 1.8703T (WTYG00000000), Erb. gaetbuli DSM 16225T (WTYF00000000), Erb. jejuensis JCM 16677T (WTYE00000000), Erb. marinus KCTC 23554T (LDCP01000000), Erb. pelagi JCM 17468T (WTYD00000000), Erb. vulgaris DSM 17792T (WTYC00000000), Erm. ramosum JCM 10282T (WTYB00000000) and Por. algicida KEMB 9005-328T (WTYA00000000).
Eight supplementary figures and four supplementary tables are available with the online version of this article.
References
- 1.Tonon LAC, Moreira APB, Thompson F, et al. The family Erythrobacteraceae. In: Rosenberg E, DeLong EF, Lory S, Stackebrandt E, Thompson F, et al., editors. The Prokaryotes: Alphaproteobacteria and Betaproteobacteria. Berlin, Heidelberg: Springer; 2014. pp. 213–235. [Google Scholar]
- 2.Feng X-M, Mo Y-X, Han L, Nogi Y, Zhu Y-H, et al. Qipengyuania sediminis gen. nov., sp. nov., a member of the family Erythrobacteraceae isolated from subterrestrial sediment. Int J Syst Evol Microbiol. 2015;65:3658–3665. doi: 10.1099/ijsem.0.000472. [DOI] [PubMed] [Google Scholar]
- 3.Yoon J-H, Oh T-K, Park Y-H. Erythrobacter seohaensis sp. nov. and Erythrobacter gaetbuli sp. nov., isolated from a tidal flat of the Yellow Sea in Korea. Int J Syst Evol Microbiol. 2005;55:71–75. doi: 10.1099/ijs.0.63233-0. [DOI] [PubMed] [Google Scholar]
- 4.Xu X-W, Wu Y-H, Wang C-S, Wang X-G, Oren A, et al. Croceicoccus marinus gen. nov., sp. nov., a yellow-pigmented bacterium from deep-sea sediment, and emended description of the family Erythrobacteraceae . Int J Syst Evol Microbiol. 2009;59:2247–2253. doi: 10.1099/ijs.0.004267-0. [DOI] [PubMed] [Google Scholar]
- 5.Wu Y-H, Li G-Y, Jian S-L, Cheng H, Huo Y-Y, et al. Croceicoccus pelagius sp. nov. and Croceicoccus mobilis sp. nov., isolated from marine environments. Int J Syst Evol Microbiol. 2016;66:4506–4511. doi: 10.1099/ijsem.0.001381. [DOI] [PubMed] [Google Scholar]
- 6.Liao H, Li Y, Zhang M, Lin X, Lai Q, et al. Altererythrobacter mangrovi sp. nov., isolated from mangrove sediment. Int J Syst Evol Microbiol. 2017;67:4851–4856. doi: 10.1099/ijsem.0.002393. [DOI] [PubMed] [Google Scholar]
- 7.Ma H, Ren H, Huang L, Luo Y. Altererythrobacter flavus sp. nov., isolated from mangrove sediment. Int J Syst Evol Microbiol. 2018;68:2265–2270. doi: 10.1099/ijsem.0.002822. [DOI] [PubMed] [Google Scholar]
- 8.Wu Y-H, Xu L, Meng F-X, Zhang D-S, Wang C-S, et al. Altererythrobacter atlanticus sp. nov., isolated from deep-sea sediment. Int J Syst Evol Microbiol. 2014;64:116–121. doi: 10.1099/ijs.0.052951-0. [DOI] [PubMed] [Google Scholar]
- 9.Fang C, Wu Y-H, Sun C, Wang H, Cheng H, et al. Erythrobacter zhengii sp. nov., a bacterium isolated from deep-sea sediment. Int J Syst Evol Microbiol. 2019;69:241–248. doi: 10.1099/ijsem.0.003136. [DOI] [PubMed] [Google Scholar]
- 10.Qu J-H, Ma W-W, Li H-F, Wang X-F, Lu B-B, et al. Altererythrobacter amylolyticus sp. nov., isolated from lake sediment. Int J Syst Evol Microbiol. 2019;69:1231–1236. doi: 10.1099/ijsem.0.003299. [DOI] [PubMed] [Google Scholar]
- 11.Dahal RH, Kim J. Altererythrobacter fulvus sp. nov., a novel alkalitolerant alphaproteobacterium isolated from forest soil. Int J Syst Evol Microbiol. 2018;68:1502–1508. doi: 10.1099/ijsem.0.002697. [DOI] [PubMed] [Google Scholar]
- 12.Yuan N, Zeng Y, Feng H, Yu Z, Huang Y. Altererythrobacter xixiisoli sp. nov., isolated from wetland soil. Int J Syst Evol Microbiol. 2017;67:3655–3659. doi: 10.1099/ijsem.0.002198. [DOI] [PubMed] [Google Scholar]
- 13.Subhash Y, Tushar L, Sasikala C, Ramana CV. Erythrobacter odishensis sp. nov. and Pontibacter odishensis sp. nov. isolated from dry soil of a solar saltern. Int J Syst Evol Microbiol. 2013;63:4524–4532. doi: 10.1099/ijs.0.052183-0. [DOI] [PubMed] [Google Scholar]
- 14.Zhao Q, Li H-R, Han Q-Q, He A-L, Nie C-Y, et al. Altererythrobacter soli sp. nov., isolated from desert sand. Int J Syst Evol Microbiol. 2017;67:454–459. doi: 10.1099/ijsem.0.001652. [DOI] [PubMed] [Google Scholar]
- 15.Xue X, Zhang K, Cai F, Dai J, Wang Y, et al. Altererythrobacter xinjiangensis sp. nov., isolated from desert sand, and emended description of the genus Altererythrobacter . Int J Syst Evol Microbiol. 2012;62:28–32. doi: 10.1099/ijs.0.025437-0. [DOI] [PubMed] [Google Scholar]
- 16.Coil DA, Flanagan JC, Stump A, Alexiev A, Lang JM, et al. Porphyrobacter mercurialis sp. nov., isolated from a stadium seat and emended description of the genus Porphyrobacter . PeerJ. 2015;3:e1400. doi: 10.7717/peerj.1400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Meng F-X, Li G, Fang C, Wu Y-H, Cheng H, et al. Altererythrobacter aerophilus sp. nov., isolated from deep-sea water of the north-west Pacific. Int J Syst Evol Microbiol. 2019;69:1689–1695. doi: 10.1099/ijsem.0.003377. [DOI] [PubMed] [Google Scholar]
- 18.Lei X, Li Y, Chen Z, Zheng W, Lai Q, et al. Altererythrobacter xiamenensis sp. nov., an algicidal bacterium isolated from red tide seawater. Int J Syst Evol Microbiol. 2014;64:631–637. doi: 10.1099/ijs.0.057257-0. [DOI] [PubMed] [Google Scholar]
- 19.Li D-D, Zhang Y-Q, Peng M, Wang N, Wang X-J, et al. Erythrobacter xanthus sp. nov., isolated from surface seawater of the South China Sea. Int J Syst Evol Microbiol. 2017;67:2459–2464. doi: 10.1099/ijsem.0.001991. [DOI] [PubMed] [Google Scholar]
- 20.Park S, Jung Y-T, Park J-M, Yoon J-H. Altererythrobacter confluentis sp. nov., isolated from water of an estuary environment. Int J Syst Evol Microbiol. 2016;66:4002–4008. doi: 10.1099/ijsem.0.001301. [DOI] [PubMed] [Google Scholar]
- 21.Jung Y-T, Park S, Lee J-S, Yoon J-H. Altererythrobacter aquiaggeris sp. nov., isolated from water of an estuary bank. Int J Syst Evol Microbiol. 2017;67:3410–3416. doi: 10.1099/ijsem.0.002128. [DOI] [PubMed] [Google Scholar]
- 22.Park S, Won SM, Yoon J-H. Erythrobacter marisflavi sp. nov., isolated from isolated from estuary water. Int J Syst Evol Microbiol. 2019;69:2696–2702. doi: 10.1099/ijsem.0.003510. [DOI] [PubMed] [Google Scholar]
- 23.Fuerst JA, Hawkins JA, Holmes A, Sly LI, Moore CJ, et al. Porphyrobacter neustonensis gen. nov., sp. nov., an aerobic bacteriochlorophyll-synthesizing budding bacterium from fresh water. Int J Syst Bacteriol. 1993;43:125–134. doi: 10.1099/00207713-43-1-125. [DOI] [PubMed] [Google Scholar]
- 24.Furuhata K, Edagawa A, Miyamoto H, Kawakami Y, Fukuyama M. Porphyrobacter colymbi sp. nov. isolated from swimming pool water in Tokyo, Japan. J Gen Appl Microbiol. 2013;59:245–250. doi: 10.2323/jgam.59.245. [DOI] [PubMed] [Google Scholar]
- 25.Yuan C-G, Chen X, Jiang Z, Chen W, Liu L, et al. Altererythrobacter lauratis sp. nov. and Altererythrobacter palmitatis sp. nov., isolated from a Tibetan hot spring. Antonie van Leeuwenhoek. 2017;110:1077–1086. doi: 10.1007/s10482-017-0882-y. [DOI] [PubMed] [Google Scholar]
- 26.Rainey FA, Silva J, Nobre MF, Silva MT, da Costa MS. Porphyrobacter cryptus sp. nov., a novel slightly thermophilic, aerobic, bacteriochlorophyll a-containing species. Int J Syst Evol Microbiol. 2003;53:35–41. doi: 10.1099/ijs.0.02308-0. [DOI] [PubMed] [Google Scholar]
- 27.Hanada S, Kawase Y, Hiraishi A, Takaichi S, Matsuura K, et al. Porphyrobacter tepidarius sp. nov., a moderately thermophilic aerobic photosynthetic bacterium isolated from a hot spring. Int J Syst Bacteriol. 1997;47:408–413. doi: 10.1099/00207713-47-2-408. [DOI] [PubMed] [Google Scholar]
- 28.Xue H, Piao C-G, Guo M-W, Wang L-F, Fang W, et al. Description of Altererythrobacter aerius sp. nov., isolated from air, and emended description of the genus Altererythrobacter . Int J Syst Evol Microbiol. 2016;66:4543–4548. doi: 10.1099/ijsem.0.001388. [DOI] [PubMed] [Google Scholar]
- 29.Shiba T, Simidu U. Erythrobacter longus gen. nov., sp. nov., an aerobic bacterium which contains bacteriochlorophyll a . Int J Syst Bacteriol. 1982;32:211–217. doi: 10.1099/00207713-32-2-211. [DOI] [Google Scholar]
- 30.Liu Y-H, Fang B-Z, Dong Z-Y, Li L, Mohamad OAA, et al. Croceibacterium gen. nov., with description of Croceibacterium ferulae sp. nov., an endophytic bacterium isolated from Ferula sinkiangensis K. M. Shen and reclassification of Porphyrobacter mercurialis as Croceibacterium mercuriale comb. nov. Int J Syst Evol Microbiol. 2019;69:2547–2554. doi: 10.1099/ijsem.0.003540. [DOI] [PubMed] [Google Scholar]
- 31.Yurkov V, Stackebrandt E, Holmes A, Fuerst JA, Hugenholtz P, et al. Phylogenetic positions of novel aerobic, bacteriochlorophyll a-containing bacteria and description of Roseococcus thiosulfatophilus gen. nov., sp. nov., Erythromicrobium ramosum gen. nov., sp. nov., and Erythrobacter litoralis sp. nov. Int J Syst Bacteriol. 1994;44:427–434. doi: 10.1099/00207713-44-3-427. [DOI] [PubMed] [Google Scholar]
- 32.Fidalgo C, Rocha J, Martins R, Proença DN, Morais PV, et al. Altererythrobacter halimionae sp. nov. and Altererythrobacter endophyticus sp. nov., two endophytes from the salt marsh plant Halimione portulacoides . Int J Syst Evol Microbiol. 2017;67:3057–3062. doi: 10.1099/ijsem.0.002079. [DOI] [PubMed] [Google Scholar]
- 33.Kumar NR, Nair S, Langer S, Busse H-J, Kämpfer P. Altererythrobacter indicus sp. nov., isolated from wild rice (Porteresia coarctata Tateoka) Int J Syst Evol Microbiol. 2008;58:839–844. doi: 10.1099/ijs.0.65523-0. [DOI] [PubMed] [Google Scholar]
- 34.Nedashkovskaya OI, Cho S-H, Joung Y, Joh K, Kim MN, et al. Altererythrobacter troitsensis sp. nov., isolated from the sea urchin Strongylocentrotus intermedius . Int J Syst Evol Microbiol. 2013;63:93–97. doi: 10.1099/ijs.0.038836-0. [DOI] [PubMed] [Google Scholar]
- 35.Zhuang L, Lin B, Xu L, Li G, Wu C-J, et al. Erythrobacter spongiae sp. nov., isolated from marine sponge. Int J Syst Evol Microbiol. 2019;69:1111–1116. doi: 10.1099/ijsem.0.003278. [DOI] [PubMed] [Google Scholar]
- 36.Ivanova EP, Bowman JP, Lysenko AM, Zhukova NV, Gorshkova NM, et al. Erythrobacter vulgaris sp. nov., a novel organism isolated from the marine invertebrates. Syst Appl Microbiol. 2005;28:123–130. doi: 10.1016/j.syapm.2004.11.001. [DOI] [PubMed] [Google Scholar]
- 37.Lee K-B, Liu C-T, Anzai Y, Kim H, Aono T, et al. The hierarchical system of the ‘Alphaproteobacteria’: description of Hyphomonadaceae fam. nov., Xanthobacteraceae fam. nov. and Erythrobacteraceae fam. nov. Int J Syst Evol Microbiol. 2005;55:1907–1919. doi: 10.1099/ijs.0.63663-0. [DOI] [PubMed] [Google Scholar]
- 38.Kwon KK, Woo J-H, Yang S-H, Kang J-H, Kang SG, et al. Altererythrobacter epoxidivorans gen. nov., sp. nov., an epoxide hydrolase-active, mesophilic marine bacterium isolated from cold-seep sediment, and reclassification of Erythrobacter luteolus Yoon et al. 2005 as Altererythrobacter luteolus comb. nov. Int J Syst Evol Microbiol. 2007;57:2207–2211. doi: 10.1099/ijs.0.64863-0. [DOI] [PubMed] [Google Scholar]
- 39.Parte AC. LPSN – list of prokaryotic names with standing in Nomenclature (bacterio.net), 20 years on. Int J Syst Evol Microbiol. 2018;68:1825–1829. doi: 10.1099/ijsem.0.002786. [DOI] [PubMed] [Google Scholar]
- 40.Park S, Park J-M, Yoon J-H. Altererythrobacter aquimixticola sp. nov., isolated from sediment sampled at the junction between the ocean and a freshwater spring. Int J Syst Evol Microbiol. 2019;69:2408–2414. doi: 10.1099/ijsem.0.003494. [DOI] [PubMed] [Google Scholar]
- 41.Park S, Park J-M, Oh T-K, Yoon J-H. Altererythrobacter insulae sp. nov., a lipolytic bacterium isolated from a tidal flat. Int J Syst Evol Microbiol. 2019;69:1009–1015. doi: 10.1099/ijsem.0.003260. [DOI] [PubMed] [Google Scholar]
- 42.Lee SD. Altererythrobacter lutipelagi sp. nov., isolated from a tidal mudflat, and emended description of the genus Altererythrobacter . Int J Syst Evol Microbiol. 2019;69:1980–1985. doi: 10.1099/ijsem.0.003414. [DOI] [PubMed] [Google Scholar]
- 43.Kang H, Kim H, Joh K. Altererythrobacter maritimus sp. nov., isolated from seawater. Int J Syst Evol Microbiol. 2019;69:1566–1572. doi: 10.1099/ijsem.0.003336. [DOI] [PubMed] [Google Scholar]
- 44.Kristyanto S, Lee SD, Kim J. Porphyrobacter algicida sp. nov., an algalytic bacterium isolated from seawater. Int J Syst Evol Microbiol. 2017;67:4526–4533. doi: 10.1099/ijsem.0.002324. [DOI] [PubMed] [Google Scholar]
- 45.Jiao N, Zhang Y, Zeng Y, Hong N, Liu R, et al. Distinct distribution pattern of abundance and diversity of aerobic anoxygenic phototrophic bacteria in the global ocean. Environ Microbiol. 2007;9:3091–3099. doi: 10.1111/j.1462-2920.2007.01419.x. [DOI] [PubMed] [Google Scholar]
- 46.Zheng Q, Lin W, Liu Y, Chen C, Jiao N. A comparison of 14 Erythrobacter genomes provides insights into the genomic divergence and scattered distribution of phototrophs. Front Microbiol. 2016;7:984. doi: 10.3389/fmicb.2016.00984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Béjà O, Suzuki MT, Heidelberg JF, Nelson WC, Preston CM, et al. Unsuspected diversity among marine aerobic anoxygenic phototrophs. Nature. 2002;415:630–633. doi: 10.1038/415630a. [DOI] [PubMed] [Google Scholar]
- 48.Li Z-Y, Wu Y-H, Huo Y-Y, Cheng H, Wang C-S, et al. Complete genome sequence of a benzo[a]pyrene-degrading bacterium Altererythrobacter epoxidivorans CGMCC 1.7731T . Mar Genomics. 2016;25:39–41. doi: 10.1016/j.margen.2015.11.009. [DOI] [PubMed] [Google Scholar]
- 49.Alonso-Gutiérrez J, Figueras A, Albaigés J, Jiménez Núria, Viñas M, et al. Bacterial communities from shoreline environments (Costa da Morte, northwestern Spain) affected by the Prestige oil spill. Appl Environ Microbiol. 2009;75:3407–3418. doi: 10.1128/AEM.01776-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hu Y, MacMillan JB, Erythrazoles A-B. Erythrazoles A-B, cytotoxic benzothiazoles from a marine-derived Erythrobacter sp. Org Lett. 2011;13:6580–6583. doi: 10.1021/ol202944g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hu Y, Legako AG, Espindola APDM, MacMillan JB. Erythrolic acids A–E, meroterpenoids from a marine-derived Erythrobacter sp. J Org Chem. 2012;77:3401–3407. doi: 10.1021/jo300197z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chun J, Oren A, Ventosa A, Christensen H, Arahal DR, et al. Proposed minimal standards for the use of genome data for the taxonomy of prokaryotes. Int J Syst Evol Microbiol. 2018;68:461–466. doi: 10.1099/ijsem.0.002516. [DOI] [PubMed] [Google Scholar]
- 53.Whitman WB. Genome sequences as the type material for taxonomic descriptions of prokaryotes. Syst Appl Microbiol. 2015;38:217–222. doi: 10.1016/j.syapm.2015.02.003. [DOI] [PubMed] [Google Scholar]
- 54.Konstantinidis KT, Tiedje JM. Prokaryotic taxonomy and phylogeny in the genomic era: advancements and challenges ahead. Curr Opin Microbiol. 2007;10:504–509. doi: 10.1016/j.mib.2007.08.006. [DOI] [PubMed] [Google Scholar]
- 55.Zhu Q, Mai U, Pfeiffer W, Janssen S, Asnicar F, et al. Phylogenomics of 10,575 genomes reveals evolutionary proximity between domains bacteria and archaea. Nat Commun. 2019;10:5477. doi: 10.1038/s41467-019-13443-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.de la Haba RR, López-Hermoso C, Sánchez-Porro C, Konstantinidis KT, Ventosa A. Comparative genomics and phylogenomic analysis of the genus Salinivibrio . Front Microbiol. 2019;10:2104. doi: 10.3389/fmicb.2019.02104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liu Y, Lai Q, Shao Z. Genome-based analysis reveals the taxonomy and diversity of the family Idiomarinaceae . Front Microbiol. 2018;9:2453. doi: 10.3389/fmicb.2018.02453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wirth JS, Whitman WB. Phylogenomic analyses of a clade within the roseobacter group suggest taxonomic reassignments of species of the genera Aestuariivita, Citreicella, Loktanella, Nautella, Pelagibaca, Ruegeria, Thalassobius, Thiobacimonas and Tropicibacter, and the proposal of six novel genera. Int J Syst Evol Microbiol. 2018;68:2393–2411. doi: 10.1099/ijsem.0.002833. [DOI] [PubMed] [Google Scholar]
- 59.Qin Q-L, Xie B-B, Zhang X-Y, Chen X-L, Zhou B-C, et al. A proposed genus boundary for the prokaryotes based on genomic insights. J Bacteriol. 2014;196:2210–2215. doi: 10.1128/JB.01688-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kim M, Oh H-S, Park S-C, Chun J. Towards a taxonomic coherence between average nucleotide identity and 16S rRNA gene sequence similarity for species demarcation of prokaryotes. Int J Syst Evol Microbiol. 2014;64:346–351. doi: 10.1099/ijs.0.059774-0. [DOI] [PubMed] [Google Scholar]
- 61.Luo C, Rodriguez-R LM, Konstantinidis KT. MyTaxa: an advanced taxonomic classifier for genomic and metagenomic sequences. Nucleic Acids Res. 2014;42:e73. doi: 10.1093/nar/gku169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jung Y-T, Park S, Lee J-S, Yoon J-H. Altererythrobacter aestiaquae sp. nov., isolated from seawater. Int J Syst Evol Microbiol. 2014;64:3943–3949. doi: 10.1099/ijs.0.066639-0. [DOI] [PubMed] [Google Scholar]
- 63.Park SC, Baik KS, Choe HN, Lim CH, Kim HJ, et al. Altererythrobacter namhicola sp. nov. and Altererythrobacter aestuarii sp. nov., isolated from seawater. Int J Syst Evol Microbiol. 2011;61:709–715. doi: 10.1099/ijs.0.021196-0. [DOI] [PubMed] [Google Scholar]
- 64.Park S, Jung Y-T, Choi SJ, Yoon J-H. Altererythrobacter aquaemixtae sp. nov., isolated from the junction between the ocean and a freshwater spring. Int J Syst Evol Microbiol. 2017;67:3446–3451. doi: 10.1099/ijsem.0.002136. [DOI] [PubMed] [Google Scholar]
- 65.Zhang G, Yang Y, Wang L. Altererythrobacter aurantiacus sp. nov., isolated from deep-sea sediment. Antonie van Leeuwenhoek. 2016;109:1245–1251. doi: 10.1007/s10482-016-0726-1. [DOI] [PubMed] [Google Scholar]
- 66.Zhang W, Yuan X, Feng Q, Zhang R, Zhao X, et al. Altererythrobacter buctense sp. nov., isolated from mudstone core. Antonie van Leeuwenhoek. 2016;109:793–799. doi: 10.1007/s10482-016-0679-4. [DOI] [PubMed] [Google Scholar]
- 67.Jeong SH, Jin HM, Lee HJ, Jeon CO. Altererythrobacter gangjinensis sp. nov., a marine bacterium isolated from a tidal flat. Int J Syst Evol Microbiol. 2013;63:971–976. doi: 10.1099/ijs.0.039024-0. [DOI] [PubMed] [Google Scholar]
- 68.Yoon J-H, Kang KH, Yeo S-H, Oh T-K. Erythrobacter luteolus sp. nov., isolated from a tidal flat of the Yellow Sea in Korea. Int J Syst Evol Microbiol. 2005;55:1167–1170. doi: 10.1099/ijs.0.63522-0. [DOI] [PubMed] [Google Scholar]
- 69.Lai Q, Yuan J, Shao Z. Altererythrobacter marinus sp. nov., isolated from deep seawater. Int J Syst Evol Microbiol. 2009;59:2973–2976. doi: 10.1099/ijs.0.008193-0. [DOI] [PubMed] [Google Scholar]
- 70.Yang Y, Zhang G, Sun Z, Cheung MK, Huang C. Altererythrobacter oceanensis sp. nov., isolated from the Western Pacific. Antonie van Leeuwenhoek. 2014;106:1191–1198. doi: 10.1007/s10482-014-0288-z. [DOI] [PubMed] [Google Scholar]
- 71.Liang X, Lin H, Wang K, Liao Y, Lai Q, et al. Altererythrobacter salegens sp. nov., a slightly halophilic bacterium isolated from surface sediment. Int J Syst Evol Microbiol. 2017;67:909–913. doi: 10.1099/ijsem.0.001708. [DOI] [PubMed] [Google Scholar]
- 72.Kim J-H, Yoon J-H, Kim W. Altererythrobacter sediminis sp. nov., isolated from lagoon sediments. Int J Syst Evol Microbiol. 2016;66:5424–5429. doi: 10.1099/ijsem.0.001535. [DOI] [PubMed] [Google Scholar]
- 73.Yoon J-H, Kang KH, Oh T-K, Park Y-H. Erythrobacter aquimaris sp. nov., isolated from sea water of a tidal flat of the Yellow Sea in Korea. Int J Syst Evol Microbiol. 2004;54:1981–1985. doi: 10.1099/ijs.0.63100-0. [DOI] [PubMed] [Google Scholar]
- 74.Xing T, Liu Y, Wang N, Xu B, Liu K, et al. Erythrobacter arachoides sp. nov., isolated from ice core. Int J Syst Evol Microbiol. 2017;67:4235–4239. doi: 10.1099/ijsem.0.002290. [DOI] [PubMed] [Google Scholar]
- 75.Denner EBM, Vybiral D, Koblízek M, Kämpfer P, Busse H-J, et al. Erythrobacter citreus sp. nov., a yellow-pigmented bacterium that lacks bacteriochlorophyll a, isolated from the western Mediterranean Sea . Int J Syst Evol Microbiol. 2002;52:1655–1661. doi: 10.1099/00207713-52-5-1655. [DOI] [PubMed] [Google Scholar]
- 76.Yoon B-J, Lee D-H, Oh D-C. Erythrobacter jejuensis sp. nov., isolated from seawater. Int J Syst Evol Microbiol. 2013;63:1421–1426. doi: 10.1099/ijs.0.038349-0. [DOI] [PubMed] [Google Scholar]
- 77.Wu H-X, Lai PY, Lee OO, Zhou X-J, Miao L, et al. Erythrobacter pelagi sp. nov., a member of the family Erythrobacteraceae isolated from the red sea. Int J Syst Evol Microbiol. 2012;62:1348–1353. doi: 10.1099/ijs.0.029561-0. [DOI] [PubMed] [Google Scholar]
- 78.Xu L, Wu Y-H, Cheng H, Sun C, Han B-N, et al. Complete genome sequence of Erythrobacter seohaensis SW-135T sheds light on the ecological role of the genus Erythrobacter for phosphorus cycle in the marine environment. Mar Genomics. 2018;40:21–24. doi: 10.1016/j.margen.2018.03.001. [DOI] [PubMed] [Google Scholar]
- 79.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012;19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Parks DH, Imelfort M, Skennerton CT, Hugenholtz P, Tyson GW. CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 2015;25:1043–1055. doi: 10.1101/gr.186072.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lagesen K, Hallin P, Rødland EA, Stærfeldt H-H, Rognes T, et al. RNAmmer: consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res. 2007;35:3100–3108. doi: 10.1093/nar/gkm160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lowe TM, Chan PP. tRNAscan-SE on-line: integrating search and context for analysis of transfer RNA genes. Nucleic Acids Res. 2016;44:W54–W57. doi: 10.1093/nar/gkw413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Yoon S-H, Ha S-M, Kwon S, Lim J, Kim Y, et al. Introducing EzBioCloud: a taxonomically United database of 16S rRNA gene sequences and whole-genome assemblies. Int J Syst Evol Microbiol. 2017;67:1613–1617. doi: 10.1099/ijsem.0.001755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Overbeek R, Olson R, Pusch GD, Olsen GJ, Davis JJ, et al. The seed and the rapid annotation of microbial genomes using subsystems technology (RAST) Nucleic Acids Res. 2014;42:D206–D214. doi: 10.1093/nar/gkt1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Xu L, Wu Y-H, Zhou P, Cheng H, Liu Q, et al. Investigation of the thermophilic mechanism in the genus Porphyrobacter by comparative genomic analysis. BMC Genomics. 2018;19:385. doi: 10.1186/s12864-018-4789-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Xu L, Ye K-X, Dai W-H, Sun C, Xu L-H, K-X Y, L-H X, et al. Comparative genomic insights into secondary metabolism biosynthetic gene cluster distributions of marine Streptomyces . Mar Drugs. 2019;17:498. doi: 10.3390/md17090498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lechner M, Findeiß S, Steiner L, Marz M, Stadler PF, et al. Proteinortho: Detection of (Co-)orthologs in large-scale analysis. BMC Bioinformatics. 2011;12:124. doi: 10.1186/1471-2105-12-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Park S, Jung Y-T, Choi SJ, Yoon J-H. Erythrobacter aquimixticola sp. nov., isolated from the junction between the ocean and a freshwater spring. Int J Syst Evol Microbiol. 2017;67:2964–2969. doi: 10.1099/ijsem.0.002055. [DOI] [PubMed] [Google Scholar]
- 89.Xu L, Wu Y-H, Jian S-L, Wang C-S, Wu M, et al. Pseudohongiella nitratireducens sp. nov., isolated from seawater, and emended description of the genus Pseudohongiella . Int J Syst Evol Microbiol. 2016;66:5155–5160. doi: 10.1099/ijsem.0.001489. [DOI] [PubMed] [Google Scholar]
- 90.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kumar S, Stecher G, Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Felsenstein J. Evolutionary trees from DNA sequences: a maximum likelihood approach. J Mol Evol. 1981;17:368–376. doi: 10.1007/BF01734359. [DOI] [PubMed] [Google Scholar]
- 93.Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 1980;16:111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- 94.Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Suyama M, Torrents D, Bork P. PAL2NAL: robust conversion of protein sequence alignments into the corresponding codon alignments. Nucleic Acids Res. 2006;34:W609–W612. doi: 10.1093/nar/gkl315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Capella-Gutiérrez S, Silla-Martínez JM, Gabaldón T. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics. 2009;25:1972–1973. doi: 10.1093/bioinformatics/btp348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Nguyen L-T, Schmidt HA, von Haeseler A, Minh BQ. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 2015;32:268–274. doi: 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Lee I, Ouk Kim Y, Park S-C, Chun J. OrthoANI: an improved algorithm and software for calculating average nucleotide identity. Int J Syst Evol Microbiol. 2016;66:1100–1103. doi: 10.1099/ijsem.0.000760. [DOI] [PubMed] [Google Scholar]
- 99.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 100.Rodriguez-R LM, Konstantinidis KT. Bypassing cultivation to identify bacterial species. Microbe. 2014;9:111–118. doi: 10.1128/microbe.9.111.1. [DOI] [Google Scholar]
- 101.Ihaka R, Gentleman R. R: a language for data analysis and graphics. J Comput Graph Stat. 1996;5:299–314. [Google Scholar]
- 102.Tamames J. Evolution of gene order conservation in prokaryotes. Genome Biol. 2001;2:research0020. doi: 10.1186/gb-2001-2-6-research0020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Patenge N, Berendes A, Engelhardt H, Schuster SC, Oesterhelt D. The fla gene cluster is involved in the biogenesis of flagella in Halobacterium salinarum . Mol Microbiol. 2001;41:653–663. doi: 10.1046/j.1365-2958.2001.02542.x. [DOI] [PubMed] [Google Scholar]
- 104.Hiraishi A, Yonemitsu Y, Matsushita M, Shin YK, Kuraishi H, et al. Characterization of Porphyrobacter sanguineus sp. nov., an aerobic bacteriochlorophyll-containing bacterium capable of degrading biphenyl and dibenzofuran. Arch Microbiol. 2002;178:45–52. doi: 10.1007/s00203-002-0423-5. [DOI] [PubMed] [Google Scholar]
- 105.Yoon J-H, Lee M-H, Oh T-K. Porphyrobacter donghaensis sp. nov., isolated from sea water of the East Sea in Korea. Int J Syst Evol Microbiol. 2004;54:2231–2235. doi: 10.1099/ijs.0.63226-0. [DOI] [PubMed] [Google Scholar]
- 106.Yoon J-H, Kang S-J, Lee M-H, Oh HW, Oh T-K. Porphyrobacter dokdonensis sp. nov., isolated from sea water. Int J Syst Evol Microbiol. 2006;56:1079–1083. doi: 10.1099/ijs.0.63840-0. [DOI] [PubMed] [Google Scholar]
- 107.Wang Q, Suzuki A, Mariconda S, Porwollik S, Harshey RM. Sensing wetness: a new role for the bacterial flagellum. EMBO J. 2005;24:2034–2042. doi: 10.1038/sj.emboj.7600668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Bardy SL, Ng SYM, Jarrell KF. Prokaryotic motility structures. Microbiology. 2003;149:295–304. doi: 10.1099/mic.0.25948-0. [DOI] [PubMed] [Google Scholar]
- 109.Zhuang L, Liu Y, Wang L, Wang W, Shao Z. Erythrobacter atlanticus sp. nov., a bacterium from ocean sediment able to degrade polycyclic aromatic hydrocarbons. Int J Syst Evol Microbiol. 2015;65:3714–3719. doi: 10.1099/ijsem.0.000481. [DOI] [PubMed] [Google Scholar]
- 110.Huang Y, Zeng Y, Feng H, Wu Y, Xu X. Croceicoccus naphthovorans sp. nov., a polycyclic aromatic hydrocarbons-degrading and acylhomoserine-lactone-producing bacterium isolated from marine biofilm, and emended description of the genus Croceicoccus . Int J Syst Evol Microbiol. 2015;65:1531–1536. doi: 10.1099/ijs.0.000132. [DOI] [PubMed] [Google Scholar]
- 111.Fan Z-Y, Xiao Y-P, Hui W, Tian G-R, Lee J-S, et al. Altererythrobacter dongtanensis sp. nov., isolated from a tidal flat. Int J Syst Evol Microbiol. 2011;61:2035–2039. doi: 10.1099/ijs.0.024380-0. [DOI] [PubMed] [Google Scholar]
- 112.Kang JW, Kim MS, Lee JH, Baik KS, Seong CN. Altererythrobacter rigui sp. nov., isolated from wetland freshwater. Int J Syst Evol Microbiol. 2016;66:2491–2496. doi: 10.1099/ijsem.0.001078. [DOI] [PubMed] [Google Scholar]
- 113.Xu M, Xin Y, Yu Y, Zhang J, Zhou Y, et al. Erythrobacter nanhaisediminis sp. nov., isolated from marine sediment of the South China Sea. Int J Syst Evol Microbiol. 2010;60:2215–2220. doi: 10.1099/ijs.0.014027-0. [DOI] [PubMed] [Google Scholar]
- 114.Jung Y-T, Park S, Lee J-S, Yoon J-H. Erythrobacter lutimaris sp. nov., isolated from a tidal flat sediment. Int J Syst Evol Microbiol. 2014;64:4184–4190. doi: 10.1099/ijs.0.067728-0. [DOI] [PubMed] [Google Scholar]
- 115.Matsumoto M, Iwama D, Arakaki A, Tanaka A, Tanaka T, et al. Altererythrobacter ishigakiensis sp. nov., an astaxanthin-producing bacterium isolated from a marine sediment. Int J Syst Evol Microbiol. 2011;61:2956–2961. doi: 10.1099/ijs.0.024729-0. [DOI] [PubMed] [Google Scholar]
- 116.Seo SH, Lee SD. Altererythrobacter marensis sp. nov., isolated from seawater. Int J Syst Evol Microbiol. 2010;60:307–311. doi: 10.1099/ijs.0.011031-0. [DOI] [PubMed] [Google Scholar]
- 117.Lee YS, Lee D-H, Kahng H-Y, Kim EM, Jung JS. Erythrobacter gangjinensis sp. nov., a marine bacterium isolated from seawater. Int J Syst Evol Microbiol. 2010;60:1413–1417. doi: 10.1099/ijs.0.015743-0. [DOI] [PubMed] [Google Scholar]
- 118.Lei X, Zhang H, Chen Y, Li Y, Chen Z, et al. Erythrobacter luteus sp. nov., isolated from mangrove sediment. Int J Syst Evol Microbiol. 2015;65:2472–2478. doi: 10.1099/ijs.0.000283. [DOI] [PubMed] [Google Scholar]
- 119.Jung Y-T, Park S, Oh T-K, Yoon J-H. Erythrobacter marinus sp. nov., isolated from seawater. Int J Syst Evol Microbiol. 2012;62:2050–2055. doi: 10.1099/ijs.0.034702-0. [DOI] [PubMed] [Google Scholar]
- 120.Wu Y-H, Cheng H, Zhou P, Huo Y-Y, Wang C-S, et al. Complete genome sequence of the heavy metal resistant bacterium Altererythrobacter atlanticus 26DY36T, isolated from deep-sea sediment of the North Atlantic Mid-ocean ridge. Mar Genomics. 2015;24:289–292. doi: 10.1016/j.margen.2015.10.004. [DOI] [PubMed] [Google Scholar]
- 121.Cheng H, Wu Y-H, Huo Y-Y, Zhou P, Liu Q, et al. Complete genome sequence of Altererythrobacter dongtanensis KCTC 22672T, isolated from a tidal flat. Mar Genomics. 2017;34:11–14. doi: 10.1016/j.margen.2017.02.003. [DOI] [Google Scholar]
- 122.Shi X-L, Wu Y-H, Cheng H, Zhang X-Q, Wang C-S, et al. Complete genome sequence of astaxanthin-producing bacterium Altererythrobacter ishigakiensis NBRC 107699. Mar Genomics. 2016;30:77–79. doi: 10.1016/j.margen.2016.08.001. [DOI] [PubMed] [Google Scholar]
- 123.Zhou P, Wu Y-H, Cheng H, Wang C-S, Xu X-W, Y-H W, X-W X. Draft genome sequence of Altererythrobacter troitsensis JCM 17037, isolated from the sea urchin Strongylocentrotus intermedius . Genome Announc. 2016;4:e01556–15. doi: 10.1128/genomeA.01556-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Coil DA, Eisen JA. Draft genome sequence of Porphyrobacter mercurialis (sp. nov.) strain Coronado. Genome Announc. 2015;3:e00856–15. doi: 10.1128/genomeA.00856-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Wu Y-H, Cheng H, Huo Y-Y, Xu L, Liu Q, et al. Complete genome sequence of esterase-producing bacterium Croceicoccus marinus E4A9T . Stand Genomic Sci. 2017;12:88. doi: 10.1186/s40793-017-0300-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Xu L, Wu Y-H, Zhou Y-G, Cheng H, Liu Q, et al. Complete genome sequence of Erythrobacter gangjinensis CGMCC 1.15024T with two chromosomes. Mar Genomics. 2017;34:15–18. doi: 10.1016/j.margen.2017.02.004. [DOI] [Google Scholar]
- 127.Wang Y, Zhang R, Zheng Q, Jiao N. Draft genome sequences of two marine phototrophic bacteria, Erythrobacter longus strain DSM 6997 and Erythrobacter litoralis strain DSM 8509. Genome Announc. 2014;2:e00677–14. doi: 10.1128/genomeA.00677-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Liu Q, Wu Y-H, Cheng H, Xu L, Wang C-S, et al. Complete genome sequence of bacteriochlorophyll-synthesizing bacterium Porphyrobacter neustonensis DSM 9434. Stand Genomic Sci. 2017;12:32. doi: 10.1186/s40793-017-0243-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Gao Y, Wu Y-H, Xu L, Cheng H, Wang C-S, et al. Complete genome sequence of Qipengyuania sediminis CGMCC 1.12928T, shed light on its role in matter-cycle and cold adaption mechanism of the genus Qipengyuania . Curr Microbiol. 2019;76:988–994. doi: 10.1007/s00284-019-01712-w. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.