Abstract
The quantification of the total microbial content in metagenomic samples is critical for investigating the interplay between the microbiome and its host, as well as for assessing the accuracy and precision of the relative microbial composition which can be strongly biased in low microbial biomass samples. In the present study, we demonstrate that digital droplet PCR (ddPCR) can provide accurate quantification of the total copy number of the 16S rRNA gene, the gene usually exploited for assessing total bacterial abundance in metagenomic DNA samples. Notably, using DNA templates with different integrity levels, as measured by the DNA integrity number (DIN), we demonstrated that 16S rRNA copy number quantification is strongly affected by DNA quality and determined a precise correlation between quantification underestimation and DNA degradation levels. Therefore, we propose an input DNA mass correction, according to the observed DIN value, which could prevent inaccurate quantification of 16S copy number in degraded metagenomic DNAs. Our results highlight that a preliminary evaluation of the metagenomic DNA integrity should be considered before performing metagenomic analyses of different samples, both for the assessment of the reliability of observed differential abundances in different conditions and to obtain significant functional insights.
Keywords: microbiome content, DNA integrity, metabarcoding, digital PCR
Data Summary
All supporting data and protocols have been provided within the article or through supplementary data files. Six supplementary figures and six supplementary tables are available with the online version of this article. The sequencing data generated in this study are available from the National Center for Biotechnology Information (NCBI) database under the accession number PRJNA622512 (www.ncbi.nlm.nih.gov/bioproject/?term=PRJNA622512).
Impact Statement.
The identification and functional characterization of microbial communities present in a variety of environments are generally carried out through sequencing-based approaches, which provide relative quantifications of taxon occurrence or metabolic pathways, although they may not allow reliable biological interpretation of the functional interplay of microbial species among themselves or with their eventual specific host. This limitation may be overcome through quantification of the microbial content, which could help to provide a more accurate overview of the microbial dynamics in the investigated environments. Here we demonstrate that accurate quantification of the total 16S rDNA copy number in metagenomic DNAs can be performed by digital droplet PCR (ddPCR) and that it is strongly affected by DNA quality, as measured by the DNA integrity number (DIN). Therefore, our study suggests that preliminary accurate quantitative evaluation of the total microbial content by ddPCR is highly recommended before performing metagenomic analyses of different samples; both for the assessment of the reliability of observed differential abundances in different conditions and to obtain significant functional insights.
Introduction
The recent impressive blooming of metagenomics through the concurrent development of high-throughput sequencing platforms has opened up unprecedented avenues for studying the microbiome in a variety of physio-pathological contexts. The identification and functional characterization of microbial communities are generally performed using genome shotgun or amplicon-based sequencing approaches. Independently of the adopted approach, metagenomic projects generally provide relative quantifications of taxon or metabolic pathway abundance [1], limiting reliable biological interpretation of the functional interplay between present microbial species or with their eventual host organism [2]. Indeed, as relative abundance data are mutually dependent, they can lead to misinterpretations and false discoveries and may generate artefactual statistical inferences [3]. Therefore, microbial relative abundance could be reliably interpreted if related to the total bacterial content. In this context, the absolute quantification of the microbial species can assist in the reliable assessment of the functional impact of the microbiome as well as in evaluating pathogenicity [4]. For example, the absolute concentration of a pathogen is a specific marker of disease severity and could suggest the most adequate therapeutic strategy [5]. Total bacterial abundance is generally based on the quantification of the total number of copies of the 16S rRNA gene – a conserved prokaryotic gene with hypervariable sequences that differ between species – present in the metagenomic sample [6, 7]. Several aspects need to be carefully considered in microbial quantification studies in order to avoid the misinterpretation of experimental results [8, 9]. First, there is the sensitivity and specificity of the quantitative approach used. Quantitative PCR (qPCR) of a target gene has been widely used for the absolute quantification of bacterial content in recent years [10, 11]. While accurate, qPCR is limited by the need for a standard curve or a reference gene and many technical replicates, and has the problem that the detectable copy number is affected by the presence of inhibitors that are commonly present in metagenomic samples [12, 13]. Recently, these limitations have been overcome through the development of droplet digital PCR (ddPCR), which allows absolute DNA quantification without a standard curve and functions over a wide dynamic range with high sensitivity in low copy number detection [14–19]. Indeed, several examples of the application of ddPCR have recently been reported in the literature for the absolute quantification of microbial species [16, 20, 21].
Then, for PCR-based methods, accurate selection of the primers targeting the 16S rRNA gene is required in order to broadly and appropriately sample the prokaryotic diversity characterizing the community under investigation, and to reduce biases and preserve the accuracy of abundance estimates [22–24].
Furthermore, from complex matrices, such as those used for most microbial studies, for example soil, stool or swabs, the accuracy of bacterial quantification is affected by the efficiency of the DNA extraction method, which can lead to underestimation of genome abundances if DNA loss occurs [25–27].
Finally, the overall quality of the metagenomic DNA, such as its integrity as measured by the DNA integrity number (DIN) and the absence of contaminants, is another crucial – but still largely unattended – aspect to be considered in total bacterial abundance studies, as low quality can lead to significantly inaccurate bacterial quantification, especially when comparisons between different communities are performed [28].
In the present study, we applied ddPCR to DNAs from single bacterial strains, bacterial mocks and metagenomic samples, demonstrating that it allows accurate absolute quantification of 16S rRNA gene copy numbers. Moreover, using DNA templates with different integrity levels, we demonstrated that 16S copy number quantification is strongly affected by DNA quality, a relevant issue that has not been addressed in previous studies [16, 20, 21, 29, 30]. Remarkably, we determined a precise correlation between quantification underestimation and DNA degradation levels, measured as DIN values, and demonstrated that a correction of the DNA mass, based on the underestimation value, allows one to estimate the 16S copy number accurately even in the most degraded metagenomic DNAs.
Our results provide a novel insight into the effect of metagenomic DNA quality on the quantification of microbial abundances and highlight that a preliminary evaluation of the metagenomic DNA integrity is required to assess the reliability of observed differential abundances in different conditions and obtain significant functional insights.
Methods
DNA samples
A plasmid (PGEM3.1) containing a single copy of a portion of the 16S rRNA gene (600 bp long), including the V5–V6 hypervariable regions, was synthesized by a gene synthesis service (GenScript Biotech, NJ, USA). Four different genomic DNA from bacterial strains ( Bacteriovorax stolpii , Deinococcus radiodurans , Bacteroides vulgatus , Lactobacillus plantarum ) were purchased from Leibniz Institute DSMZ – German Collection of Microorganisms and Cell Cultures, (Braunschweig, Germany). ZymoBIOMICS Microbial Community DNA Standard, a mixture of genomic DNA isolated from pure cultures of eight bacterial and two fungal strains, was purchased from Zymo Research (Irvine, CA, USA) (Table 1). A laboratory mock was prepared by mixing together 2.1 pg of B. stolpii , 1 pg of D. radioduran s, 2.2 pg of B. vulgatus and 1.2 pg of L. plantarum . A mixture of ZymoBIOMICS mock and human genomic DNA (ZymoBIOMICS/humDNA mixture) was prepared by combining 16 pg of ZymoBIOMICS DNA Standard with 200 ng of human genomic DNA.
Table 1.
Bacterial strains and the microbial community used in 16S absolute quantification by ddPCR. For each species, the genome size, the 16S rRNA gene copy number per genome and 16S rRNA gene copies ng−1 of DNA are reported
|
Bacterial species |
Genome size (Mb) |
16S rRNA copies/genome |
16S rRNA copies ng−1 |
|---|---|---|---|
|
Bacteriovorax stolpii (DSM12778) |
3.81 |
2 |
477 893 |
|
Deinococcus radiodurans (DSM20539) |
3.28 |
3 |
834 550 |
|
Lactobacillus plantarum (DSM2601) |
3.26 |
5 |
1 398 888 |
|
Bacteroides vulgatus (DSM1447) |
5.16 |
7 |
1 239 104 |
|
ZymoBIOMICS mock |
|||
|
6.79 |
4 |
538 000 |
|
|
4.87 |
7 |
1 309 000 |
|
|
4.76 |
7 |
1 344 000 |
|
|
1.90 |
5 |
2 395 000 |
|
|
2.84 |
4 |
1 284 000 |
|
|
2.73 |
6 |
2 004 000 |
|
|
2.99 |
6 |
1 830 000 |
|
|
4.04 |
10 |
2 260 000 |
|
|
Saccharomyces cerevisiae |
12.1 |
/ |
/ |
|
Cryptococcus neoformans |
18.9 |
/ |
/ |
Stool samples from 13 volunteers from the laboratory group were collected and stored at −80 °C until use. Metagenomic DNA was extracted using the Fast DNA Spin Kit for Soil (MP Biomedicals, Santa Ana, CA, USA), according to the manufacturer’s instructions. A 40 s bead-beating step at speed 6 was applied using the FastPrep Instrument (BIO 101, Carlsbad, Canada). DNA was eluted in 100 µl and stored at −80 °C.
Assessment of DNA integrity and concentration
DNA integrity was evaluated using the Agilent TapeStation 2200 System (Agilent, Santa Clara, CA, USA) and the Genomic DNA ScreenTape assay (Agilent, Santa Clara, CA, USA). The DIN was determined for each sample using Agilent 2200 TapeStation software (controller version A.01.05). DNA concentration was assessed by fluorimetry using the Quant-iT PicoGreen dsDNA Assay Kit (Invitrogen, Carlsbad, CA, USA) on a NanoDrop 3300 Fluorospectrometer (Thermo Fisher Scientific, Waltham, MA, USA).
Generation of DNA samples with decreasing DIN
DNA samples with increasing fragmentation rates were obtained using the Covaris M220 focused-ultrasonicator (Thermo Fisher Scientific, Waltham, MA, USA), setting variable time (seconds) and duty factor (DF) in combination with fixed peak incident power (PIP) of 50 W and 200 cycles per burst.
For the ZymoBIOMICS/humDNA mixture, starting from the untreated sample (DIN9, DIN value of 9.1±0.1), six samples (each with five independent replicates) with increasing levels of degradation (7.5±0.3, 6.2±0.4, 4.5±0.3, 3.5±0.3, 2.2±0.3, 1.5±0.3) were prepared and named DIN7, DIN6, DIN4, DIN3, DIN2 and DIN1, respectively. The time and DF settings were: DIN7 : 5 s and 1 % DF; DIN6 : 5 s and 2 % DF; DIN4 : 3–5 s and 10 % DF; DIN3 : 20 s and 10 % DF; DIN2 : 20 s and 12 % DF; DIN1 : 35 s and 20 % DF (Fig. S1, available in the online version of this article).
Metagenomic DNAs, with an average DIN of 6.5±0.6 (DIN6), were diluted at a concentration of 4 ng µl−1 and concentration was confirmed by fluorimetric assay. Approximately two hundred nanograms of DNA were transferred in a Covaris microTube-50 (Thermo Fisher Scientific, Waltham, MA, USA) and sonicated to prepare three progressive degradation points, using the time and DF settings as reported: DIN 4 : 5–10 s and 3 % DF; DIN 2 : 10–20 s and 12 % DF; DIN 1 : 15–30 s and 20 % DF. The final samples had the following average DIN values: 4.5±1 (DIN4), 2.05±0.35 (DIN2) and 1.35±0.35 (DIN1) (Fig. S2).
ddPCR experiments
ddPCR (Bio-Rad, Hercules, CA, USA) was performed to determine the total number of 16S copies by using universal primers targeting the V5–V6 regions of 16S rDNA [31], as they have been used successfully in other DNA metabarcoding analyses (primer sequences: forward, B-V5 : 5′-ATTAGATACCCYGGTAGTCC-3′; reverse, A-V6, 5′-ACGAGCTGACGACARCCATG-3′) [32, 33]. For the quantification of the gene copy numbers for Akkermansia muciniphila 16S rRNA, specific primers targeting the V7–V8 regions were used (primer sequence: forward, AM1 : 5′-CAGCACGTGAAGGTGGGGAC-3′; reverse, AM2 : 5′-CCTTGCGGTTGGCTTCAGAT-3′) [34].
Each DNA sample was diluted to a concentration of 1 ng µl−1 and the concentration was confirmed by fluorimetric assay. Starting from this concentration, serial dilutions were made for each DNA. The final dilution factor used in the ddPCR reaction was chosen on the basis of preliminary tests in order to balance positive events versus negative events and optimize quantification reliability.
A reaction volume of 22 µl was prepared by combining the diluted DNA with 11 µl of 2× Evagreen Supermix (Bio-Rad, Hercules, CA, USA), 0.39 µl of 10 µM forward and reverse universal 16S primers or 0.44 µl of 10 µM forward and reverse specific 16S A. muciniphila primers, and water. Emulsion was produced in the QX200 Droplet Generator (Bio-Rad, Hercules, CA, USA) according to the manufacturer’s instructions. The thermal cycling conditions were as follows: for total 16S copies quantification: 1 cycle at 95 °C for 5 min, 40 cycles at 95 °C for 30 s and 58 °C for 1 min, 1 cycle at 4 °C for 5 min, 1 cycle at 90 °C for 5 min, final hold at 4 °C; for A. muciniphila 16S copy quantification: 1 cycle at 95 °C for 5 min, 40 cycles at 95 °C for 30 s and 60 °C for 1 min, 1 cycle at 4 °C for 5 min, 1 cycle at 90 °C for 5 min, final hold at 4 °C. Each DNA sample was analysed at least in duplicate, preparing independent dilutions. For each experiment, a negative control (no template control) was used. Absolute quantification was performed using QuantaSoft version 7.4.1 software (Bio-Rad, Hercules, CA, USA,) and the negative/positive thresholds were set manually, excluding samples with a number of droplets <10 000. Output results were expressed in 16S copies µl−1.
Estimation of 16S rRNA gene copy number
The expected copy number ng−1 DNA of 16S rDNA for the one-copy 16S plasmid, the commercial bacterial species and the ZymoBIOMICS mock was calculated on the basis of the known genome sizes and the 16S rRNA copy number/genome, as reported in Table 1.
The ddPCR measured copy number ng−1 DNA was calculated considering the DNA dilution factor used in ddPCR reaction, the fluorimetric DNA concentration verified after dilution to 1 ng µl−1 and the final ddPCR reaction volume (22 µl) as reported in the formula below:
A polynomial regression model to predict the percentage underestimation of 16S rDNA copies as a function of the measured DIN was inferred based on the experimental data obtained by using the ZymoBIOMICS/hum DNA mixture (Fig. S3). The model training was carried out in R by using the lm and poly functions belonging to the stat package and plotted with ggplot2. Finally, by using the regression estimation we corrected the input DNA mass in ddPCR experiments on metagenomic DNAs.
Metabarcoding sequencing and data analysis
The protocol used for amplicon library preparation was described previously [32, 33]. The V5–V6 region was amplified using the same primers pair as were used in the ddPCR experiments in five metagenomic DNAs from stool samples (F9–F13), at their high-quality (DIN6), low-quality (DIN1) and mass corrected low-quality (DIN1 input corrected, DIN1-IC) levels. Each DNA was analysed in triplicate and the prepared libraries were sequenced on Illumina MiSeq platform (Illumina, San Diego, CA, USA) to generate 2×250 bp paired end (PE) reads. 30% of the PhiX genome library was loaded in the run to compensate for low base diversity in the amplicon libraries.
Overall, 5.6 M PE reads were generated, with an average of 125 000 PE reads per sample (sd 18 000, min 58 783, max 156 957). Raw sequencing data are available in SRA (PRJNA622512). The raw sequencing data were denoised using DADA2, a tool that applies a statistical approach to discriminate between the biological diversity and the noise introduced by both PCR and sequencing, in order to remove the latter [35]. Moreover, it removed reads derived from the PhiX genome and PCR chimeras. About 72 % of the raw reads (sd 10 %, min 49.35 %, max 86.30 %) passed the denoising step and were used to infer 1298 amplicon sequences variants (ASVs). The BioMaS pipeline was applied to annotate the inferred ASVs taxonomically using release 11.5 of the RDP database and the National Center for Biotechnology Information (NCBI) taxonomy for the 16S rRNA reference collection and taxonomy, respectively [36–38]. In particular, comparison of the ASV sequences with the reference collection was performed by means of Bowtie 2 and the resulting alignments were filtered according to query coverage (≥70 %) and identity percentage (≥90 %) [39]. The classification was performed using TANGO: for ASVs obtaining matches with an identity percentage equal to or higher than 97 %, the taxonomic classification at species level was assigned; otherwise, they were classified to higher taxonomic ranks [40, 41].
Statistics
Statistical analyses of ddPCR data were performed using GraphPad Prism 5.0 software (GraphPad Software, San Diego, CA, USA). Student’s t-test was used for statistical comparisons and data were presented as mean±standard deviation (sd). P-values <0.05 were considered to be indicative of statistically significant differences.
The qualitative comparison of taxa observed in next-generation sequencing (NGS) data was performed by using an ad hoc-developed Python script. In particular, to infer the number of common and uncommon taxa among the DIN6, DIN1 and DIN-IC DNA samples for each subject (F9-F13), all the taxa observed in only one replicate or with an average relative abundance lower than 0.5 % were filtered out. The retained taxa were collected in three sets corresponding to the tested DIN values. The qualitative comparison between sets was summarized at the six main taxonomic ranks level (i.e. phylum, class, order, family, genus and species).
Moreover, a quantitative comparison was performed by measuring for each sample the Pearson correlation between the relative frequencies of observed taxa in DIN6, DIN1 and DIN-IC samples.
Finally, by using the Wilcoxon rank test, the A. muciniphila relative abundances observed by using ddPCR and NGS data were compared.
Results
ddPCR allows accurate quantification of 16s rRNA gene copy number
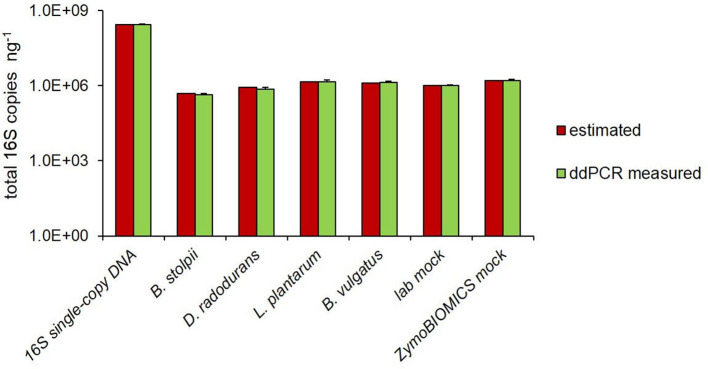
The number of copies of the 16S rRNA gene (16S rDNA) were quantified by ddPCR technology, using the universal primers pair targeting the V5–V6 hypervariable regions [31], previously used for DNA metabarcoding investigations [32, 33]. First, we evaluated the specificity of the selected primers pair, performing the analysis on DNA samples with a known number of 16S rDNA copies. In particular, we used a plasmidic DNA, containing one copy of a synthetic 16S rRNA sequence, four individual bacterial species and a mock community (ZymoBIOMICS) with eight bacterial strains (Table 1). As reported in Fig. 1, perfect concordance was observed between the ddPCR estimated and the expected number of 16S copies in all samples, including the 16S single-copy DNA, the four bacterial DNAs analysed both individually and mixed together (lab mock) and the ZymoBIOMICS mock. The expected number of copies of 16S rDNA was calculated for 1 ng of DNA, taking into account both genome size and genomic 16S copy number. Hence, these results demonstrate the reliability of the V5–V6 primers pair for ddPCR-based absolute quantification of 16S rDNA copies.
Fig. 1.
Estimated vs ddPCR-measured total 16S copy number. Comparison between estimated and ddPCR-measured total 16S copies in the plasmid containing a single copy of the V5–V6 region, in the four bacterial DNAs, analysed individually and mixed together (labmock), and in the ZymoBIOMICS mock. 16S copy number is reported per ng of DNA. Each sample was analysed at least in triplicate and results are expressed as mean±sd.
DNA integrity significantly affects 16S rRNA copy number quantification
The extraction of high-quality DNA from complex matrices is often a challenging task, and this, in turn, may affect its accurate quantification [25]. Apart the extraction efficiency, another critical issue is the integrity of the extracted DNA [42], which is usually measured by the DIN using the Agilent 2200 TapeStation system [43].
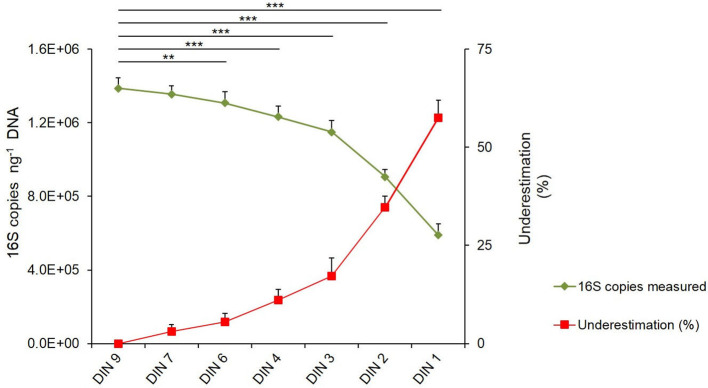
In order to evaluate how DNA integrity affects 16S rDNA copy number quantification in metagenomic DNAs by ddPCR, we first prepared a DNA sample resembling a metagenomic DNA of human origin, consisting of the ZymoBIOMICS DNA mock and human DNA (ZymoBIOMICS/humDNA mock; see the Methods section). This sample was subjected to gradients of DNA shearing by sonication to obtain DNA with increasing levels of degradation, evaluated by measuring the DIN value. We obtained samples with DIN values of 9.1±0.1, 7.5±0.3, 6.2±0.4, 4.5±0.3, 3.5±0.3, 2.2±0.3 and 1.5±0.3 that we named DIN9, DIN7, DIN6, DIN4, DIN3, DIN2 and DIN1, respectively (Fig. S1). Copy number quantification of 16S rDNA was performed for each of these ZymoBIOMICS/humDNA mocks. As shown in Fig. 2, the higher the DNA degradation rate, the greater the underestimation of the measured 16S copy number., The results were statistically significant for all samples, with the exception of the DIN7 sample, whose integrity level was not significantly different from that of the high-quality DIN9 sample (Fig. 2). The rate of 16S underestimation with respect to the DIN9 sample increased progressively up to 57 % in the DIN1 sample (Fig. 2, Table S1). Thus, these results confirm that DNA integrity affects target quantification accuracy to a remarkable degree and suggest that a preliminary evaluation of DNA integrity, by estimating the DIN value, can provide a reliable assessment of the quantification underestimation, which can then be properly corrected.
Fig. 2.
16S rRNA gene copy number quantification is influenced by the quality of the DNA mock. ZymoBIOMICS/humDNA mock at original DIN9 value and its six degradation levels corresponding to DIN7, DIN6, DIN4, DIN3, DIN2 and DIN1 were analysed for 16S copy quantification by ddPCR. The green line represents the total 16S copies ng−1 DNA, with this decreasing with the increase of the degradation level. The red line represents the 16S copy underestimation percentage, with this increasing with the DNA degradation level. 16S copies are reported as mean of at least three replicates for each sample±sd. 16S copy underestimation was calculated as a percentage compared to the highest quality (DIN 9) sample. **, P<0.01; ***, P<0,001.
Assessment of the underestimation rate for 16s rRNA gene copy number at variable integrity levels of metagenomic DNAs
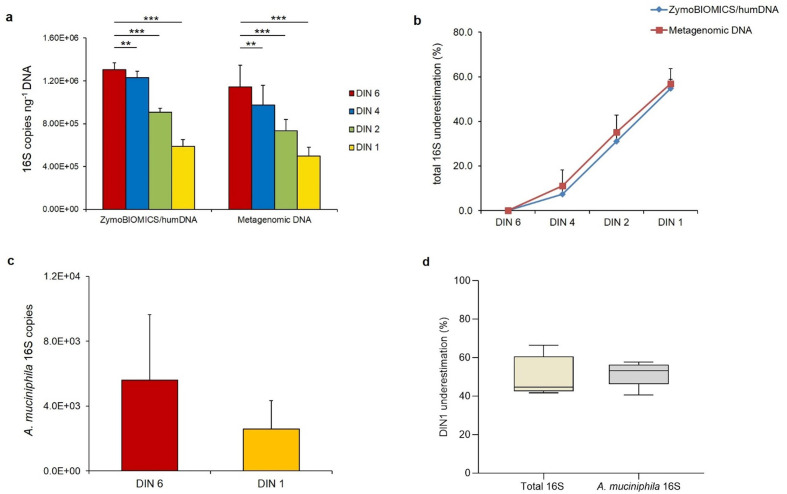
To evaluate the possibility of quantifying the underestimation rate of ddPCR determination as a function of the degradation state of metagenomic DNA samples, we analysed 13 metagenomic DNAs extracted from stool samples, with an average DIN of 6.5±06 (DIN6). From each of these DNA samples, three additional levels of degradation, corresponding to DIN values of 4.5±1 (DIN4), 2.05±0.35 (DIN2) and 1.35±0.35 (DIN1), were generated by sonication (Fig. S2). For each DNA sample, the number of 16S copies was quantified by ddPCR. We observed a progressive decrease in the 16S copy number correlated with degradation levels (Table S2). Interestingly, we observed a similar extent of copy number decrease for both the ZymoBIOMICS/humDNA mock and the faecal metagenomic samples (Fig. 3a, b). In order to investigate the species specificity of this pattern, we further investigated the 16S copy number in DIN6 and DIN1 faecal DNAs for a specific bacterium, A. muciniphila , one of the most promising probiotics for gut microbiota-related diseases [44]. As shown in Fig. 3c, in the DIN1 sample, we observed a reduction of the A. muciniphila 16S copy number with respect to the DIN6 sample and, interestingly, this reduction (~55 %) was comparable to that of the total 16S copy number in the same sample (Fig. 3d).
Fig. 3.
Total and species-specific 16S rRNA gene copy number quantification is affected by metagenomic DNA quality. (a) 16S copy quantification in 13 metagenomic DNAs at their original DIN6 value and at three degradation levels, corresponding to DIN4, DIN2 and DIN1 values, compared to the ZymoBIOMICS/humDNA mock at the same integrity levels. (b) 16S copies underestimation percentage for the ZymoBIOMICS/humDNA mock and for the 13 metagenomic DNAs at the same integrity levels. (c) A. muciniphila 16S copies measured in five metagenomic DNAs (f9-f13) at their original DIN6 value and at the DIN1 degradation level. (d) A. muciniphila 16S copy underestimation percentage (56%) is in accordance with that measured for total 16S copies in the same metagenomic DNAs (f9–f13) at the DIN1 degradation level. 16S copies are reported as the mean of at least two replicates for each sample±sd. 16S copy underestimation was calculated as a percentage compared to the highest quality (DIN 6) samples (considered as 100%). *, P<0.05, **,P<0.01, ***, P<0,001 vs DIN6.
Taken together, these data robustly confirm that 16S copy number estimates are remarkably biased by the DNA degradation level as measured by the DIN and suggest a simple correction based on visual or mathematical interpolation (Fig. S3) of the standard curve in Fig. 2.
DNA input correction in ddPCR for an accurate quantification of 16S copy number in degraded metagenomic DNA samples
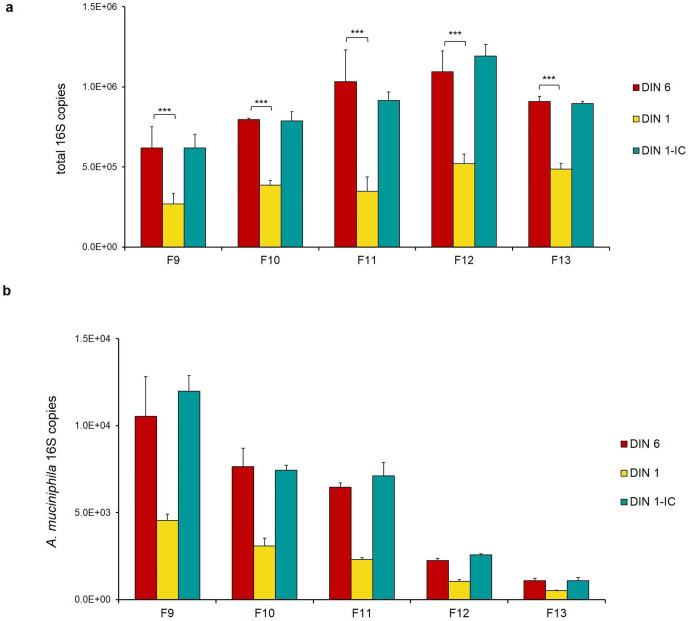
To account for the levels of DNA integrity in the quantification of the 16S copy number, for five DIN1 faecal DNAs (samples F9–F13), we adopted a mass correction of the DNA template in the ddPCR reaction, based on the 55 % underestimation percentage calculated (DIN1 input corrected, DIN1-IC). As shown in Fig. 4a, with this correction, we obtained a number of total 16S copies roughly corresponding to that measured in the original DIN6 DNA samples. Remarkably, the same DNA mass correction also allowed us to recover A. muciniphila 16S copies in these degraded DNAs (Fig. 4b, Table S3).
Fig. 4.
Input DNA mass correction allows accurate quantification of the 16S copy number in degraded metagenomic DNAs. For five DIN1 faecal DNAs (samples F9–F13), a mass correction of the DNA template in the ddPCR reaction, based on the 55 % underestimation percentage (DIN1 input corrected, DIN1-IC), allowed us to obtain the total number of 16S copies (a) and the number of A. muciniphila 16S copies (b) corresponding to those measured in the original DIN6 DNA samples. *, P<0.05, **, P<0.01, ***, P<0,001 vs DIN1.
Overall, these results demonstrate that the input DNA mass correction, calculated on the ZymoBIOMICS/humDNA mock standard curve (Figs 2 and S3), can mitigate inaccurate quantification of 16S copy number in degraded metagenomic DNAs.
Relative bacterial abundance is not affected by the integrity level in metagenomic DNA samples
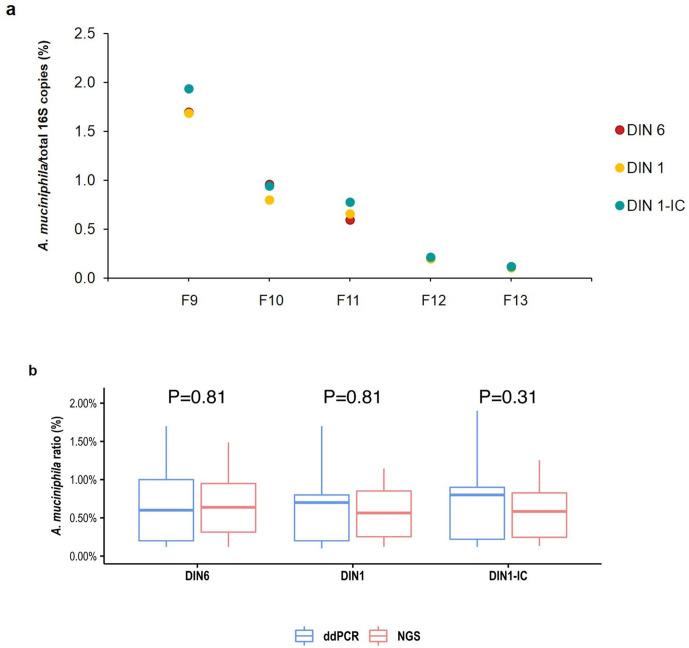
In order to investigate whether relative bacterial abundance may also be affected by the degradation of metagenomic DNA, we calculated the relative abundance of A. muciniphila 16S copies measured by ddPCR in a subset of five faecal DNAs (samples F9–F13), at DIN6 and DIN1 integrity levels and at DIN1 with mass correction (DIN1-IC). As shown in Fig. 5a and in Table S3, for all samples, the relative abundance of A. muciniphila 16S copies was not affected by the level of DNA integrity.
Fig. 5.
A. muciniphila relative abundance is not affected by the integrity level of metagenomic DNAs. (a) A. muciniphila 16S copies/total 16S copies measured by ddPCR in DIN6, DIN1 and DIN1 with mass correction (DIN1-IC) for faecal DNAs (samples F9–F13). (b) A. muciniphila relative abundance derived by ddPCR compared to DNA metabarcoding data. A. muciniphila 16S copies measured by ddPCR and in NGS data analysis in DIN6, DIN1 and DIN1 with mass correction (DIN1-IC) for faecal DNAs (samples F9–F13) were box-plotted together. p-values of pairwise comparison are indicated.
Next, we sequenced the V5–V6 amplicon in the same subset of metagenomic DNA samples at DIN6 and DIN1 integrity levels and at DIN1 with mass correction. We evaluated whether DNA integrity may influence the number of observed taxa in metabarcoding analysis. After the removal of marginally abundant taxa (average relative abundance among replicates ≤0.5 %) as well as those observed in only one out of three replicates, we enumerated common and uncommon taxa at different DNA integrity levels and collected the results for the F9–F13 samples at the six main taxonomic ranks level (i.e. phylum, class, order, family, genus and species). As expected, the higher the taxonomic rank, the smaller the differences in the observed taxa. In particular, a core set of shared taxa among DIN6, DIN1 and DIN1-IC samples was observed. The percentage of shared taxa was 100 % at phylum level and decreased up to 74 % at the order rank (Tables S4 and S5). The observation that the DNA integrity level does not significantly affect the relative abundance of detected taxa was confirmed by the high Pearson correlation obtained by comparing taxon relative abundance at phylum, class, order, family, genus and species level, for DIN6, DIN1 and DIN1-IC samples (Table S6, Figs S4–S6).
Finally, in order to assess the species-specificity of DNA metabarcoding data, we compared A. muciniphila relative abundances measured by both ddPCR and NGS analysis, by using the pairwise Wilcoxon rank test. Interestingly, we found a perfect concordance between ddPCR and DNA metabarcoding data (Fig. 5b), suggesting that the relative proportions of A. muciniphila are preserved in NGS data analysis, regardless of the initial DNA quality.
Discussion
The large majority of microbiome investigations adopt the DNA metabarcoding approach, which provides relative estimates of the composition of microbial taxa, which are intrinsically mutually dependent. This introduces a systematic bias as the increase of one taxon in a specific condition inevitably leads to the decrease of all others. This bias is particularly relevant in the case of low microbial contents with a consequent high risk of false discoveries or artefactual results [45]. Indeed, measuring the absolute microbial content in a metagenomic sample is critical for reliable investigation of the interplay between the microbiome and its host, which is affected by quantitative parameters (e.g. metabolite concentration) [46], as well as for assessing the functional impact of microbial dynamics in a variety of environments and conditions [4].
In the present study, we present data showing that ddPCR provides accurate quantification of 16S rDNA copy number in metagenomic DNAs. ddPCR is characterized by precise quantification, higher reproducibility compared to qPCR and higher sensitivity in low-copy-number detection [14, 20, 47]. Furthermore, ddPCR mitigates the effects of the presence of PCR inhibitors that may affect PCR sensitivity in 16S metabarcoding sequencing analysis [27, 48].
For the quantification of 16S copy number in metagenomic DNAs by PCR, the choice of the primers pair is crucial, as it should be able to cover the entire microbial representation of a large variety of complex matrices [22–24]. For our study, we selected the universal primers pair targeting the V5–V6 hypervariable regions of the 16S rDNA [31], successfully used in DNA metabarcoding investigations [32, 33, 49–53]. We demonstrated the reliability of these primers pair as, in different DNA samples with a known number of 16S rDNA copies, a perfect concordance was observed between the expected 16S copy number and that estimated by ddPCR, both in DNAs from single bacterial strains and from bacterial communities.
Furthermore, we showed that 16S copy number quantification is strongly affected by DNA quality, as measured by its DIN. By analysing a DNA mock, with a known 16S rDNA copy number, at different DIN values, we calculated a percentage of underestimation of 16S rDNA copy number correlated to the degradation state. Interestingly, in faecal metagenomic DNAs, we observed a similar degree of decrease both in total and species-specific 16S copy number, at the same level of DNA integrity. In degraded DNAs, we demonstrated that reliable quantification of 16S copy number can be obtained through a correction of the input DNA mass to use as template, based on the underestimation percentage calculated on the mock standard curve. Our results robustly confirmed that 16S copy number estimates are consistently biased by the DNA degradation level and indicate the necessity for a preliminary evaluation of metagenomic DNA integrity in quantitative analyses. For degraded DNAs, we recommend a correction of the DNA mass to use as template, calculated on the mock standard curve or on a mathematical interpolation.
On the other hand, our results demonstrated that relative bacterial quantification is not affected by the integrity level in metagenomic DNAs, as we showed that the relative abundance of A. muciniphila 16S rDNAs measured by ddPCR did not vary in different faecal metagenomic DNAs, at DIN6 and DIN1 integrity levels. These data were confirmed by NGS analysis of the same samples, which showed perfect concordance between ddPCR results and DNA metabarcoding data, suggesting that the relative proportions of specific taxa are also preserved in NGS data analysis, regardless of the initial DNA quality. Moreover, consistent with the proposal that DNA quality does not affect relative bacterial quantification, we found a high Pearson correlation by comparing taxon relative abundance, at phylum, class, order, family, genus and species level, for DIN6, DIN1 and DIN1-IC samples.
While metabarcoding analyses allow estimation of the relative abundances of the different taxa, regardless of the quality of the input DNA, they do not provide any indication regarding the overall microbial content, which can be a very relevant factor. Indeed, even relative estimates of different taxa are unreliable when the overall microbial content is too low [54]. On the other hand, significant variations of the microbial content in different samples (e.g. 2–3-fold or more) may have remarkable effects on functional interpretations. Indeed, it is well known that an altered gut permeability of the microbiome is correlated with several diseases [55], and in this respect a quantitative assessment of systemic microbial leaks is mandatory for functional studies.
Our study highlights that preliminary quantitative evaluation of the total microbial content by ddPCR is strongly indicated before performing metagenomic analyses of samples, for both assessment of the reliability of observed differential abundances in different conditions and to obtain significant functional insights. The method we propose to accurately quantify the total microbial content also makes suitable adjustments for DNA quality. Indeed, although ddPCR has already been applied for absolute quantification of bacteria [16, 20, 21] and viruses [29, 30], a suitable correction of input mass for DNA quality, which may vary substantially in different samples and conditions, has never been addressed.
Supplementary Data
Funding information
This work was supported by the Italian Ministero dell’Istruzione, Università e Ricerca (MIUR): PRIN 2017 to G. P. and A. M. D.
Acknowledgements
We thank Drs Anita Annese and Marianna Intranuovo for collaborating on some experiments, Professor David S. Horner for his expert advice and Mrs Annarita Armenise for technical assistance. We thank ELIXIR-IIB for providing computational facilities.
Author contributions
C.M., A.O.G.P. and A.M.D. conceived and designed the study; C.M., A.O. and E.P. performed experiments; B.F. performed metabarcoding analysis; A.M.D. and G.P. provided supervision. C.M., A.O., A.M.D. and G.P. wrote the manuscript. All authors reviewed and edited the manuscript.
Conflicts of interest
The authors declare that there are no conflicts of interest
Footnotes
Abbreviations: ddPCR, droplet digital PCR; DF, duty factor; DIN, DNA integrity number; DIN1-IC, DIN1 input corrected; humDNA, human DNA; PE, paired end.
All supporting data, code and protocols have been provided within the article or through supplementary data files. Six supplementary tables and six supplementary figures are available with the online version of this article.
The sequencing data generated in this study are available in the NCBI database under the BioProject accession number: PRJNA622512.
References
- 1.Noecker C, McNally CP, Eng A, Borenstein E. High-resolution characterization of the human microbiome. Translational Research. 2017;179:7–23. doi: 10.1016/j.trsl.2016.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Props R, Kerckhof F-M, Rubbens P, De Vrieze J, Hernandez Sanabria E, et al. Absolute quantification of microbial taxon abundances. ISME J. 2017;11:584–587. doi: 10.1038/ismej.2016.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jian C, Luukkonen P, Yki-Järvinen H, Salonen A, Korpela K. Quantitative PCR provides a simple and accessible method for quantitative microbiota profiling. PLoS One. 2020;15:e0227285. doi: 10.1371/journal.pone.0227285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Abasıyanık MF, Wolfe K, Van Phan H, Lin J, Laxman B, et al. Ultrasensitive digital quantification of cytokines and bacteria predicts septic shock outcomes. Nat Commun. 2020;11:2607. doi: 10.1038/s41467-020-16124-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ricchi M, Bertasio C, Boniotti MB, Vicari N, Russo S, et al. Comparison among the quantification of bacterial pathogens by qPCR, dPCR, and cultural methods. Front Microbiol. 2017;8:1174. doi: 10.3389/fmicb.2017.01174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang Y, Qian P-Y. Conservative fragments in bacterial 16S rRNA genes and primer design for 16S ribosomal DNA amplicons in metagenomic studies. PLoS One. 2009;4:e7401. doi: 10.1371/journal.pone.0007401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tkacz A, Hortala M, Poole PS. Absolute quantitation of microbiota abundance in environmental samples. Microbiome. 2018;6:110. doi: 10.1186/s40168-018-0491-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fiedorová K, Radvanský M, Němcová E, Grombiříková H, Bosák J, et al. The impact of DNA extraction methods on stool bacterial and fungal microbiota community recovery. Front Microbiol. 2019;10:821. doi: 10.3389/fmicb.2019.00821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brooks JP, Edwards DJ, Harwich MD, Rivera MC, Fettweis JM, et al. The truth about metagenomics: quantifying and counteracting bias in 16S rRNA studies. BMC Microbiol. 2015;15:66. doi: 10.1186/s12866-015-0351-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sinha R, Abnet CC, White O, Knight R, Huttenhower C. The microbiome quality control project: baseline study design and future directions. Genome Biol. 2015;16:276. doi: 10.1186/s13059-015-0841-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brukner I, Longtin Y, Oughton M, Forgetta V, Dascal A. Assay for estimating total bacterial load: relative qPCR normalisation of bacterial load with associated clinical implications. Diagn Microbiol Infect Dis. 2015;83:1–6. doi: 10.1016/j.diagmicrobio.2015.04.005. [DOI] [PubMed] [Google Scholar]
- 12.Cavé L, Brothier E, Abrouk D, Bouda PS, Hien E, et al. Efficiency and sensitivity of the digital droplet PCR for the quantification of antibiotic resistance genes in soils and organic residues. Appl Microbiol Biotechnol. 2016;100:10597–10608. doi: 10.1007/s00253-016-7950-5. [DOI] [PubMed] [Google Scholar]
- 13.Wang M, Yang J, Gai Z, Huo S, Zhu J, et al. Comparison between digital PCR and real-time PCR in detection of Salmonella typhimurium in milk. Int J Food Microbiol. 2018;266:251–256. doi: 10.1016/j.ijfoodmicro.2017.12.011. [DOI] [PubMed] [Google Scholar]
- 14.Hindson CM, Chevillet JR, Briggs HA, Gallichotte EN, Ruf IK, et al. Absolute quantification by droplet digital PCR versus analog real-time PCR. Nat Methods. 2013;10:1003–1005. doi: 10.1038/nmeth.2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Elmahalawy ST, Halvarsson P, Skarin M, Höglund J. Droplet digital polymerase chain reaction (ddPCR) as a novel method for absolute quantification of major gastrointestinal nematodes in sheep. Vet Parasitol. 2018;261:1–8. doi: 10.1016/j.vetpar.2018.07.008. [DOI] [PubMed] [Google Scholar]
- 16.Gobert G, Cotillard A, Fourmestraux C, Pruvost L, Miguet J, et al. Droplet digital PCR improves absolute quantification of viable lactic acid bacteria in faecal samples. J Microbiol Methods. 2018;148:64–73. doi: 10.1016/j.mimet.2018.03.004. [DOI] [PubMed] [Google Scholar]
- 17.Kanagal-Shamanna R. Digital PCR: principles and applications. Methods Mol Biol. 2016;1392:43–50. doi: 10.1007/978-1-4939-3360-0_5. [DOI] [PubMed] [Google Scholar]
- 18.Pinheiro LB, Coleman VA, Hindson CM, Herrmann J, Hindson BJ, et al. Evaluation of a droplet digital polymerase chain reaction format for DNA copy number quantification. Anal Chem. 2012;84:1003–1011. doi: 10.1021/ac202578x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sze MA, Abbasi M, Hogg JC, Sin DD. A comparison between droplet digital and quantitative PCR in the analysis of bacterial 16S load in lung tissue samples from control and COPD gold 2. PLoS One. 2014;9:e110351. doi: 10.1371/journal.pone.0110351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ziegler I, Lindström S, Källgren M, Strålin K, Mölling P. 16S rDNA droplet digital PCR for monitoring bacterial DNAemia in bloodstream infections. PLoS One. 2019;14:e0224656. doi: 10.1371/journal.pone.0224656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dreo T, Pirc M, Ramšak Živa, Pavšič J, Milavec M, et al. Optimising droplet digital PCR analysis approaches for detection and quantification of bacteria: a case study of fire blight and potato brown rot. Anal Bioanal Chem. 2014;406:6513–6528. doi: 10.1007/s00216-014-8084-1. [DOI] [PubMed] [Google Scholar]
- 22.Klindworth A, Pruesse E, Schweer T, Peplies J, Quast C, et al. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res. 2013;41:e1. doi: 10.1093/nar/gks808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sambo F, Finotello F, Lavezzo E, Baruzzo G, Masi G, et al. Optimizing PCR primers targeting the bacterial 16S ribosomal RNA gene. BMC Bioinformatics. 2018;19:343. doi: 10.1186/s12859-018-2360-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ghyselinck J, Pfeiffer S, Heylen K, Sessitsch A, De Vos P. The effect of primer choice and short read sequences on the outcome of 16S rRNA gene based diversity studies. PLoS One. 2013;8:e71360. doi: 10.1371/journal.pone.0071360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bag S, Saha B, Mehta O, Anbumani D, Kumar N, et al. An improved method for high quality Metagenomics DNA extraction from human and environmental samples. Sci Rep. 2016;6:26775. doi: 10.1038/srep26775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kumar J, Kumar M, Gupta S, Ahmed V, Bhambi M, et al. An improved methodology to overcome key issues in human fecal metagenomic DNA extraction. Genomics Proteomics Bioinformatics. 2016;14:371–378. doi: 10.1016/j.gpb.2016.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Videnska P, Smerkova K, Zwinsova B, Popovici V, Micenkova L, et al. Stool sampling and DNA isolation kits affect DNA quality and bacterial composition following 16S rRNA gene sequencing using MiSeq Illumina platform. Sci Rep. 2019;9:13837. doi: 10.1038/s41598-019-49520-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Krehenwinkel H, Fong M, Kennedy S, Huang EG, Noriyuki S, et al. The effect of DNA degradation bias in passive sampling devices on metabarcoding studies of arthropod communities and their associated microbiota. PLoS One. 2018;13:e0189188. doi: 10.1371/journal.pone.0189188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jahne MA, Brinkman NE, Keely SP, Zimmerman BD, Wheaton EA, et al. Droplet digital PCR quantification of norovirus and adenovirus in decentralized wastewater and graywater collections: Implications for onsite reuse. Water Res. 2020;169:115213. doi: 10.1016/j.watres.2019.115213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Martinez-Hernandez F, Garcia-Heredia I, Lluesma Gomez M, Maestre-Carballa L, Martínez Martínez J, et al. Droplet digital PCR for estimating absolute abundances of widespread Pelagibacter viruses. Front Microbiol. 2019;10:1226. doi: 10.3389/fmicb.2019.01226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stecher B, Chaffron S, Käppeli R, Hapfelmeier S, Freedrich S, et al. Like will to like: abundances of closely related species can predict susceptibility to intestinal colonization by pathogenic and commensal bacteria. PLoS Pathog. 2010;6:e1000711. doi: 10.1371/journal.ppat.1000711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Manzari C, Fosso B, Marzano M, Annese A, Caprioli R, et al. The influence of invasive jellyfish blooms on the aquatic microbiome in a coastal lagoon (Varano, Se Italy) detected by an Illumina-based deep sequencing strategy. Biol Invasions. 2015;17:923–940. doi: 10.1007/s10530-014-0810-2. [DOI] [Google Scholar]
- 33.Leoni C, Ceci O, Manzari C, Fosso B, Volpicella M, et al. Human endometrial microbiota at term of normal pregnancies. Genes. 2019;10:971. doi: 10.3390/genes10120971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Collado MC, Derrien M, Isolauri E, de Vos WM, Salminen S. Intestinal integrity and Akkermansia muciniphila, a mucin-degrading member of the intestinal microbiota present in infants, adults, and the elderly. Appl Environ Microbiol. 2007;73:7767–7770. doi: 10.1128/AEM.01477-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJA, et al. DADA2: high-resolution sample inference from Illumina amplicon data. Nat Methods. 2016;13:581–583. doi: 10.1038/nmeth.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cole JR, Wang Q, Cardenas E, Fish J, Chai B, et al. The ribosomal database project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res. 2009;37:D141–D145. doi: 10.1093/nar/gkn879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cole JR, Wang Q, Fish JA, Chai B, McGarrell DM, et al. Ribosomal database project: data and tools for high throughput rRNA analysis. Nucleic Acids Res. 2014;42:D633–D642. doi: 10.1093/nar/gkt1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fosso B, Santamaria M, Marzano M, Alonso-Alemany D, Valiente G, et al. BioMaS: a modular pipeline for bioinformatic analysis of metagenomic amplicons. BMC Bioinformatics. 2015;16:203. doi: 10.1186/s12859-015-0595-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Alonso-Alemany D, Barré A, Beretta S, Bonizzoni P, Nikolski M, et al. Further steps in TANGO: improved taxonomic assignment in metagenomics. Bioinformatics. 2014;30:17–23. doi: 10.1093/bioinformatics/btt256. [DOI] [PubMed] [Google Scholar]
- 41.Fosso B, Pesole G, Rosselló F, Valiente G. Unbiased taxonomic annotation of metagenomic samples. J Comput Biol. 2018;25:348–360. doi: 10.1089/cmb.2017.0144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fidler G, Tolnai E, Stagel A, Remenyik J, Stundl L, et al. Tendentious effects of automated and manual metagenomic DNA purification protocols on broiler gut microbiome taxonomic profiling. Sci Rep. 2020;10:3419. doi: 10.1038/s41598-020-60304-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Padmanaban A. Dna integrity number (DIN) for the assessment of genomic DNA samples in real-time quantitative PCR (qPCR) experiments. 2015.
- 44.Zhang T, Li Q, Cheng L, Buch H, Zhang F. Akkermansia muciniphila is a promising probiotic. Microb Biotechnol. 2019;12:1109–1125. doi: 10.1111/1751-7915.13410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Karstens L, Asquith M, Davin S, Fair D, Gregory WT, et al. Controlling for contaminants in Low-Biomass 16S rRNA gene sequencing experiments. mSystems. 4 doi: 10.1128/mSystems.00290-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vandeputte D, Kathagen G, D’hoe K, Vieira-Silva S, Valles-Colomer M, et al. Quantitative microbiome profiling links gut community variation to microbial load. Nature. 2017;551:507–511. doi: 10.1038/nature24460. [DOI] [PubMed] [Google Scholar]
- 47.Wouters Y, Dalloyaux D, Christenhusz A, Roelofs HMJ, Wertheim HF, et al. Droplet digital polymerase chain reaction for rapid broad‐spectrum detection of bloodstream infections. Microb Biotechnol. 2020;13:657–668. doi: 10.1111/1751-7915.13491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Verhaegen B, De Reu K, De Zutter L, Verstraete K, Heyndrickx M, et al. Comparison of droplet digital PCR and qPCR for the quantification of Shiga toxin-producing Escherichia coli in bovine feces. Toxins. 2016;8:157. doi: 10.3390/toxins8050157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rigoni R, Fontana E, Guglielmetti S, Fosso B, D'Erchia AM, et al. Intestinal microbiota sustains inflammation and autoimmunity induced by hypomorphic RAG defects. J Exp Med. 2016;213:355–375. doi: 10.1084/jem.20151116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Perruzza L, Gargari G, Proietti M, Fosso B, D’Erchia AM, et al. T follicular helper cells promote a beneficial gut ecosystem for host metabolic homeostasis by sensing Microbiota-Derived extracellular ATP. Cell Rep. 2017;18:2566–2575. doi: 10.1016/j.celrep.2017.02.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Perruzza L, Strati F, Gargari G, D’Erchia AM, Fosso B, et al. Enrichment of intestinal Lactobacillus by enhanced secretory IgA coating alters glucose homeostasis in P2rx7−/− mice. Sci Rep. 2019;9:9315. doi: 10.1038/s41598-019-45724-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fransen F, Zagato E, Mazzini E, Fosso B, Manzari C, et al. Balb/C and C57BL/6 mice differ in polyreactive IgA abundance, which impacts the generation of antigen-specific IgA and microbiota diversity. Immunity. 2015;43:527–540. doi: 10.1016/j.immuni.2015.08.011. [DOI] [PubMed] [Google Scholar]
- 53.Pinto-Ribeiro I, Ferreira RM, Pereira-Marques J, Pinto V, Macedo G, et al. Evaluation of the use of formalin-fixed and paraffin-embedded archive gastric tissues for microbiota characterization using next-generation sequencing. Int J Mol Sci. 2020;21:1096. doi: 10.3390/ijms21031096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Thorsen J, Brejnrod A, Mortensen M, Rasmussen MA, Stokholm J, et al. Large-scale benchmarking reveals false discoveries and count transformation sensitivity in 16S rRNA gene amplicon data analysis methods used in microbiome studies. Microbiome. 2016;4:62. doi: 10.1186/s40168-016-0208-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Spadoni I, Zagato E, Bertocchi A, Paolinelli R, Hot E, et al. A gut-vascular barrier controls the systemic dissemination of bacteria. Science. 2015;350:830–834. doi: 10.1126/science.aad0135. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.