Abstract
Sporothrix schenckii is a dimorphic fungus existing as mould in the environment and as yeast in the host. The morphological shift between mycelial/yeast phases is crucial for its virulence, but the transcriptional networks implicated in dimorphic transition are still not fully understood. Here, we report the global transcriptomic differences occurring between mould and yeast phases of S. schenckii, including changes in gene expression profiles associated with these distinct cellular phenotypes. Moreover, we also propose a new genome annotation, which reveals a more complex transcriptional architecture than previously assumed. Using RNA-seq, we identified a total of 17 307 genes, of which 11 217 were classified as protein-encoding genes, whereas 6090 were designated as non-coding RNAs (ncRNAs). Approximately ~71 % of all annotated genes were found to overlap and the different-strand overlapping type was the most common. Gene expression analysis revealed that 8795 genes were differentially regulated among yeast and mould forms. Differential gene expression was also observed for antisense ncRNAs overlapping neighbouring protein-encoding genes. The release of transcriptome-wide data and the establishment of the Sporothrix Genome DataBase (http://sporothrixgenomedatabase.unime.it) represent an important milestone for Sporothrix research, because they provide a strong basis for future studies on the molecular pathways involved in numerous biological processes.
Keywords: Sporothrix schenckii, sporotrichosis, gene expression, RNA-seq, overlapping genes, long non-coding RNAs
Data Summary
The data supporting the conclusions of this article have been deposited in the National Center for Biotechnology Information (NCBI) Gene Expression Omnibus (GEO) database and are accessible through GEO series accession number GSE145856 (www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE145856). The Illumina raw reads have also been submitted into the NCBI Sequence Read Archive (SRA) database under the following study accession number SRP194160 (www.ncbi.nlm.nih.gov/sra/?term=SRP194160) associated with BioProject ID PRJNA539953.
All supplementary material (Supplementary File S1 and Tables S1–S6) has been deposited in Figshare and is available within the project ‘Sporothrix schenckii RNA-Seq’ at https://doi.org/10.6084/m9.figshare.12894761.v1.
All genomic data, including gene models and expression data, can also be viewed, inspected and/or downloaded from the Sporothrix Genome DataBase (http://sporothrixgenomedatabase.unime.it).
Impact Statement.
The genetic knowledge of Sporothrix schenckii is, at present, very poor, and even less is known about the transcriptional organization and regulation of the genes that control the myriad of biological processes, including dimorphism and the associated saprophytic/pathogenic change, that make this fungus a successful pathogen of humans and animals. Our results expand current knowledge on the content, organization, structure and expression of S. schenckii genes by providing, for what is believed to be the first time, fundamental information on the transcriptional activity of its non-coding genome.
Introduction
Sporothrix is a fungal genus comprising over 50 species [1, 2] that are largely known as environmental saprophytes, which can be isolated from soil, plants and even animals [3]. A few species are also able to infect humans and other mammals, causing a sub-acute or chronic disease called sporotrichosis [2, 4].
In a recent taxonomic revision of the genus Sporothrix [1], the disease-causing species have been grouped in the so-called ‘pathogenic clade’, which contains the four human pathogens (Sporothrix schenckii, Sporothrix brasiliensis, Sporothrix globosa and Sporothrix luriei) responsible for almost all cases of sporotrichosis occurring worldwide [2, 5]. One of the most striking phenotypic traits of these species is their ability to switch reversibly between two different morphologies in response to temperature shifts [6]. In the environment, or at 25 °C, they grow as moulds, forming conidia, whereas at 37 °C, or when acquired by humans, they are capable of converting into pathogenic yeasts that effectively overcome the host immune defences and cause disease [7]. Surprisingly, despite their very close phylogenetic relationships, the members of the pathogenic clade showed remarkable differences in many aspects of their basic biology, including epidemiology, antifungal resistance, virulence and pathogenicity [8–12]. Nevertheless, the study of pathogenic Sporothrix species is still mainly limited to their clinical aspects, laboratory diagnosis and therapy [6], and little progress has been made in understanding the genetics and gene regulatory networks that control many biological processes, including dimorphism and the associated saprophytic/pathogenic change.
In this study, we report, for what is believed to be the first time, a comprehensive transcriptomic analysis of S. schenckii in relation to two different cellular morphologies (yeast-form and hyphal-form) obtained by growing the fungus at 37 and 25 °C, respectively. Moreover, we also present the Sporothrix Genome DataBase (SGDB; http://sporothrixgenomedatabase.unime.it), a web-based genomic platform for integrating RNA-seq expression data and exploring gene models in the S. schenckii genome.
Methods
Fungal strain and culture conditions
In this study, we examined the reference strain S. schenckii ATCC MYA-4821 (formerly referred to as strain 1099–18), as its genome was already sequenced and in silico annotated in 2014 (GenBank assembly accession number GCA_000961545.1) [13]. To induce the formation of the yeast-like and mould phases, as well as the expression of genes associated with these two different morphologies, six test tubes, each containing 25 ml YPD broth (1 % yeast extract, 2 % peptone and 2 % dextrose), were inoculated with 50 µl of a standardized suspension of S. schenckii conidia from a 4-day-old YPD agar culture grown at 28 °C. Conidia were harvested by washing the agar plate with PBS 1x, pH 7.4, and removed from the mycelium with a sterile glass rod. Then, the conidial suspension was filtered through sterile glass wool to remove mycelial fragments and standardized to McFarland’s number 1 (~107 cells ml−1) using a Den-1B benchtop densitometer (bioSan). Inoculated YPD broth tubes were incubated in triplicate at 37 and 25°C, and monitored daily up to 6 days to confirm the expected morphology of the organism (yeast-like cells at 37 °C and hyphae at 25 °C).
RNA extraction, Illumina TruSeq stranded library preparation and sequencing
Total RNA extraction was performed by using a RiboPure yeast extraction kit (Thermo Fisher Scientific), following the manufacturer’s instructions. Purified RNA was quantified spectrophotometrically at 260 and 280 nm, and the RNA integrity was evaluated with an Agilent 2100 Bioanalyzer instrument using an RNA 6000 Nano kit (Agilent Technologies). High-quality total RNA (OD260/280 ≥2.0; RNA Integrity Number, RIN value ≥8.0) was sent to IGATech (https://igatechnology.com) for library preparation using TruSeq stranded mRNA chemistry (Illumina) and paired-end (2×150 bp) sequencing on the Illumina NextSeq 500 platform. A total of three biological replicates were sequenced for each culture condition tested.
Transcriptome assembly and genome annotation
For each sequenced library, Illumina raw reads were pre-processed with the program Trimmomatic (v.0.39) [14] to remove adapters and sequences with low Phred-scores (minimum Phred-quality score ≥25; minimum length 35 bp), and then used for downstream bioinformatic analyses including genome annotation and differential gene expression analysis. The software star (v.2.7.1a) [15] was used to map cleaned reads on the previously published and annotated S. schenckii reference genome (GenBank accession no. GCA_000961545.1). Mapping quality was evaluated using Qualimap 2 software (v.2.2.1) [16].
Although over 99 % of the reads mapped uniquely to the reference genome, a significant fraction of these (>30 %) were located in unannotated genomic regions. Therefore, before gene expression analysis, we decided to re-annotate the existing S. schenckii ATCC MYA-4821 genome using our RNA-seq data. Genome annotation was performed using the software maker (v.3.00.0) [17] and a combination of other resources, including snap (v.2.39) [18] and augustus (v. 3.3.1) [19] ab initio gene predictors. In addition, a genome-guided transcriptome was generated with Trinity software (v.2.5.1) [20] using our RNA-seq data (minimum coverage 10 reads).
To obtain the first draft of the annotation, in the first round of the maker pipeline we provided the original S. schenckii genome annotation (GCF_000961545.1_S_schenckii_v1_genomic.gff), the whole transcriptome created by Trinity and two whole data sets of Ophiostomatales [National Center for Biotechnology Information (NCBI) txid5151] expressed sequence tags (ESTs), and protein sequences downloaded from the NCBI Protein and Nucleotide databases (www.ncbi.nlm.nih.gov). However, before using these reference data sets in the annotation pipeline, the sequence redundancy was removed by cd-hit software v.4.8.1 [21] using a sequence similarity threshold of 90 %. Subsequently, to improve the accuracy of the S. schenckii gene models, we performed four iterations of training and prediction in order to generate hidden Markov models (HMMs) for training ab initio gene-finding software (first round with snap, then augustus) included in the maker pipeline.
Since many transcripts were not automatically annotated by maker, we manually extracted all transcript evidence reported by maker as ‘match’. GenomeTools (v.1.5.10) [22] was first used to properly format the annotation, and then to extract transcripts and corresponding proteins. Transcripts were aligned against the UniProt database (www.uniprot.org; release 2019_03) using the blast algorithm (v.2.8.1) [23]. Transcripts without any match in the UniProt database were further investigated by using InterProScan (v. 5.35–74.0) [24], Coding Potential Calculator 2 (cpc2) [25] and Infernal (v.1.1.2) [26] programs in order to evaluate the presence of protein domains, non-coding and potential coding sequences. However, transcripts without matched known protein-encoding functions were predicted as novel, potential, long non-coding RNA (lncRNA) genes based on the following criteria: (i) the assembled transcripts showed definite strand information; (ii) their length was ≥200bp; and (iii) their coding potential was predicted by the cpc2 program using specific sequence intrinsic features: ORF, length, ORF integrity and isoelectric point [25]. Only transcripts labelled as ‘noncoding’ in the cpc2 output were kept.
RepeatMasker (v.4.0.9) (http://www.repeatmasker.org) and Tandem Repeat Finder (v.4.09) (https://tandem.bu.edu/trf/trf.html) software were used to predict repetitive and transposable elements. The tRNAs were detected using tRNAscan-SE software (v.1.3.1) [27]. All the proteins were functionally annotated with pannzer2 software [28].
Identification of cis-sense–antisense (cis-SAS) gene pairs, same-strand overlapping genes and overlapping gene groups
Stranded RNA-seq data obtained in this study provided potentially valuable information to map sense and antisense transcripts, and thereby accurately predict genomic regions containing genes arranged on opposite DNA strands (Fig. 1). We defined overlapping sense–antisense regions as those genomic traits in which one sense transcript overlaps (at least 1 nucleotide) with an antisense transcript (Fig. 2). However, it should be pointed out that a single sense transcript could overlap with two or more different antisense transcripts by generating multiple overlapping areas (Fig. 2a). Nevertheless, each overlap site within these composite gene regions was counted once. According to this criterion, we searched the S. schenckii genome for overlapping regions by using the BEDtools intersect utility (v2.28.0) [29] and custom bash scripts. Subsequently, we counted the number of cis-SAS genes occurring in these regions and classified them into different groups (paired, triple, quadruple, quintuple and so on), based on the number of sense–antisense transcripts involved in each single uninterrupted genomic region (Fig. 2a). However, in addition to the type of overlapping transcripts described above, generically known as different-strand overlapping type, there is also another type of overlapping genes called same-strand overlapping type, in which the genes involved are transcribed from the same DNA strand [30] (Fig. 2b). Therefore, we also searched for same-strand overlapping genes by using BEDtools [29], and the boundaries of newly annotated genes defined by the start and end gene positions reported on assembled genome scaffolds. Genes were considered to overlap with each other if they shared at least 1 nucleotide.
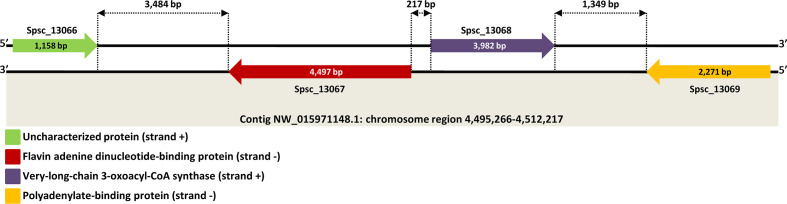
Fig. 1.
A representative genomic region showing four different protein-encoding genes transcribed from opposite DNA strands. The image was built based on the information provided by the strand-specific RNA-seq data obtained in this study.
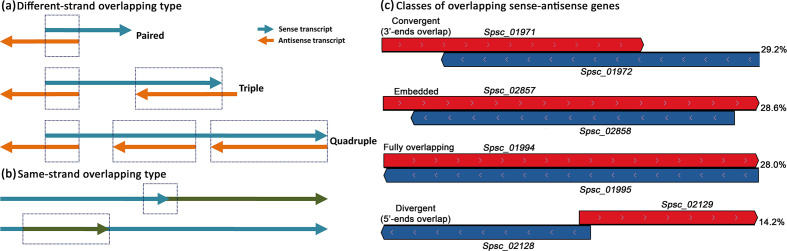
Fig. 2.
Different types of overlapping genes. (a) Some examples of different-strand overlapping genes in which two or more genes are transcribed from opposite DNA strands. Gene groups (paired, triple, quadruple) were established based on the number of sense–antisense transcripts involved in each single uninterrupted genomic region. (b) Same-strand overlapping genes in which two adjacent genes overlap partially (or entirely) with each other by sharing a common genomic region. (c) Schematic representation of paired cis-SAS genes according to the orientation of the overlapping transcripts and their transcriptional direction. The percentage of each configuration found in the S. schenckii genome is shown on the right.
Differential gene expression and gene ontology (GO) enrichment analyses
Using our new comprehensive genome annotation, we carried out a global gene expression analysis for identifying genes specifically expressed during yeast-like growth at 37 °C and/or in mould-producing cultures at 25 °C. The high-quality reads of each sample were mapped onto the newly annotated reference genome with the star program (v.2.7.1a) [15] and the quality of mapping was assessed using Qualimap 2 software (v.2.2.1) [16]. The number of RNA-seq reads or read-pairs overlapping the genomic features was counted by featureCounts software integrated into the Subreads package (v.1.6.4) [31]. For each RNA-seq library, fragments per kilobase of transcript per million mapped reads (FPKM) values were calculated and used to estimate the number of transcripts detected. According to another study [32], a FPKM cut-off >1 in at least one of the six libraries was used to establish whether a transcript was expressed by S. schenckii cells from which RNA was extracted. The transcripts whose FPKM value was below the threshold value of 1 were considered not detected by the RNA-seq method employed.
The normalization of raw counts was performed using the trimmed means of M-values (TMM) method in the HTSFilter package [33]. Differentially expressed genes (DE genes) were identified using the edgeR package [34]. The genes whose expression levels showed an absolute log2 fold-change (log2FC) value ≥1 and a false discovery rate (FDR) ≤0.05 were considered to be significantly differentially expressed. GO enrichment analysis was performed on the DE genes using in-house scripts following the methods outlined in agriGO (v.2.0) [35].
Quantitative real-time PCR (qRT-PCR)
RNA-seq data were validated by qRT-PCR using a panel of randomly selected coding- and non-coding transcripts (Table S1) representing DE genes and non-differentially expressed genes for the two conditions tested. PCR primers were designed and validated using Vector NTI software (version 10.3.0; Invitrogen). For the quantification of transcripts encoded by overlapping loci, primers were designed in non-overlapping regions of the gene to avoid the amplification and quantitation of the corresponding complementary RNA sequence.
Total RNA (2 µg) was digested with DNase I (Sigma-Aldrich), following manufacturer’s instructions, and retrotranscribed by a RevertAid first strand cDNA synthesis kit (Thermo Fisher Scientific) using oligo(dT) at 42 °C for 1 h, followed by a reverse transcriptase denaturation step at 70 °C for 10 min. qRT-PCR was performed using the StepOnePlus real-time PCR system (Applied Biosystem) with PowerUp SYBR Green master mix (Thermo Fisher Scientific) and the primer sequences listed in Table S1.
Relative gene expression quantification was calculated by the 2−ΔΔ C tmethod [36] using the β-tubulin gene as an endogenous housekeeping gene. Spearman’s correlation coefficient (rs) was used to estimate the degree of correlation between gene expression levels measured by qRT-PCR and RNA-seq methods.
SGDB
In order to make our data easily accessible and available to the scientific community, we developed the SGDB (http://sporothrixgenomedatabase.unime.it), an online resource that offers an integrated toolset for accessing, analysing and exploring genomics data. Currently, SGDB provides gene expression data as well as protein and genomic sequences for S. schenckii only, but it could be integrated in the future with additional genomic/transcriptomic data from other Sporothrix species.
The web server was built using Node JS (version 8.9.4) with the Express framework (version 4.16) in the back-end. Data are stored in files read by the back-end and supplied to the front-end, which consists of mainly html/css and JavaScript, in the corresponding format (e.g. tables or images). A JavaScript-based genome browser (JBrowse v.1.12.3) [37] was also integrated to allow users to quickly view large-scale RNA-seq data and gene annotations along the reference genome in an interactive manner. The SGDB also incorporates the blast search tool allowing investigators to query any sequence against the genome, transcriptome or proteome of S. schenckii using either blastn or blastp algorithms.
Results
RNA sequencing, transcriptome statistics and genome annotation
Illumina whole-transcriptome shotgun sequencing produced a total of 98 949 246 reads of which 98 837 150 (~99.9 %) were high-quality reads (clean reads) used for further genomics analysis (Table 1). The total size of the sequenced reads was 245.92 Gbp. The overall transcriptome statistics of the six processed biological samples are shown in Table 1.
Table 1.
Statistics of the RNA-seq data generated in this study
|
Sequence characteristic |
Mould-form |
Yeast-form |
Total |
||||
|---|---|---|---|---|---|---|---|
|
SSC_1 |
SSC_2 |
SSC_3 |
SSC_4 |
SSC_5 |
SSC_6 |
||
|
Raw reads |
15 840 139 |
15 759 028 |
13 714 380 |
19 217 684 |
18 303 397 |
16 114 618 |
98 949 246 |
|
Cleaned reads |
15 821 431 |
15 743 831 |
13 692 566 |
19 196 851 |
18 283 034 |
16 099 437 |
98 837 150 |
|
Read 1 (unpaired) |
8668 |
6371 |
9843 |
8277 |
7517 |
6166 |
46 842 |
|
Read 2 (unpaired) |
3620 |
3382 |
4401 |
4124 |
4722 |
2917 |
23 166 |
|
Total mapped* |
15 766 109 |
15 688 507 |
13 619 058 |
19 156 060 |
18 246 967 |
15 831 742 |
98 308 443 |
|
Uniquely assigned† |
12 095 953 |
12 325 194 |
10 882 445 |
16 019 278 |
14 055 739 |
13 021 760 |
78 400 369 |
|
Multiply mapped‡ |
482 067 |
483 333 |
432 399 |
802 480 |
715 040 |
692 013 |
3 607 332 |
|
No features mapped§ |
3 188 089 |
2 879 980 |
2 304 214 |
2 334 302 |
3 476 188 |
2 117 969 |
16 300 742 |
|
G+C content (mol%) |
50.70 |
51.35 |
52.34 |
54.68 |
54.46 |
56.19 |
53.28|| |
*Total number of reads aligned on the S. schenckii reference genome (v2 annotation).
†Total number of reads mapped to unique features in the newly annotated S. schenckii genome (v2 annotation).
‡Total number of reads mapped to multiple locations in the S. schenckii genome (v2 annotation).
§Total number of reads mapped to unannotated (v2 annotation) genomic regions.
||Mean value.
The first level of difference between the biological replicates of the two conditions examined was that the data set of the RNA-seq reads produced by the mycelia showed a lower mean G+C content (~51.5 mol%) than that associated with the yeast-form (mean G+C content 55.1 mol%) (Student-t P value=0.003634) (Table 1). The raw RNA-seq reads have been deposited in the NCBI Sequence Read Archive (SRA) database and are available under BioProject ID PRJNA539953.
Using our RNA-seq data, we were able to refine the initial set of S. schenckii ATCC MYA-4821 gene models (referred to hereafter as the v1 annotation) that was originally generated using only computational methods and, therefore, consisted solely of ab initio gene predictions (10 293 protein-encoding genes and 139 tRNAs) [13] (Table 2). Our genome-wide reannotation (v2 annotation) provides an extended and improved annotation of the S. schenckii genome by unravelling unreported aspects of its transcriptional architecture (Table 2). Overall, 17 307 genes were identified by integrating our RNA-seq data into the genome annotation pipeline. Most of them (64.8 %; 11 217) showed a hit on at least one of the four databases inspected and were classified as protein-encoding genes, whereas 5929 (34.2 %) lacked evidence for a canonical or functional ORF and were predicted as potential lncRNAs. Moreover, a total of 24 rRNAs, 134 tRNAs and 3 snRNAs (small nuclear RNAs) were also identified (Table 2). The updated v2 annotation of the S. schenckii ATCC MYA-4821 genome is available as supplementary material (Supplementary File S1) in the form of a GFF file or can be downloaded from the SGDB (http://sporothrixgenomedatabase.unime.it).
Table 2.
Comparison of genome-wide statistics for the v1 and v2 annotations of the S. schenckii ATCC MYA-4821 genome
|
Sequence characteristic |
Annotation version |
|
|---|---|---|
|
v1 |
v2 |
|
|
Total genes |
10 432 |
17 307 |
|
Protein-encoding genes |
10 293 |
11 217 |
|
Gene length |
||
|
Mean (bp) |
~1612 |
~2101 |
|
Total (bp) |
16 590 731 |
21 625 318 |
|
Gene length/genome (%) |
51.23 |
66.78 |
|
Total exons |
20 756 |
30 392 |
|
Exon length |
||
|
Mean (bp) |
~757 |
~1132 |
|
Total (bp) |
7 796 060 |
11 657 420 |
|
Exon length/genome (%) |
24.07 |
36.00 |
|
lncRNAs |
na |
5929 |
|
rRNA loci |
na |
24 |
|
tRNA loci |
139 |
134 |
|
snRNA loci |
na |
3 |
na, Not available.
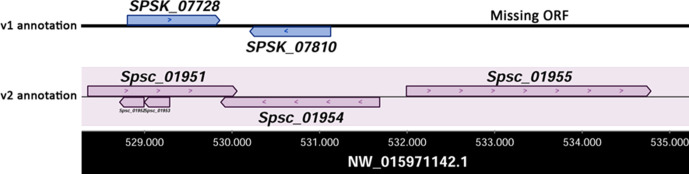
According to our new genome annotation, we confirmed 98.9 % (10 180/10 293) of the previously reported protein-encoding genes [13] and identified 132 novel protein-encoding sequences within genomic regions that were not annotated in the previous published v1 version (GenBank accession no. GCA_000961545.1) (Table S1). An example is presented for the ORF Spsc_01955, reported in Fig. 3, encoding a heat-shock protein, Hsp98. Table 2 shows the comparison of genome-wide statistics for the v1 and v2 annotations of the S. schenckii ATCC MYA-4821 genome.
Fig. 3.
Schematic comparison of v1 and v2 genome annotations illustrating the discovery of additional gene models and the improvement of gene model boundaries.
Strand-specific RNA-seq reveals a large number of antisense RNAs (asRNAs) overlapping protein-encoding and non-coding genes
In this study, we performed strand-specific RNA sequencing to retain the strand origin of the transcripts; thus, allowing us to predict sense RNAs and asRNAs (Fig. 1), identifying exactly the overlapping regions of transcription and estimating accurately the expression levels of sense and antisense genes. Using this approach, we identified a number of genomic regions, generically known as ‘transcriptionally active regions’ (TARs) or ‘transcribed fragments’ (transfrags) [38], that contain a pair (or more) of distinct genes mapping to opposite DNA strands (Table S1). Technically, two genes that reside on opposite genomic strands within the same locus and share sequence overlap (at least 1 nucleotide) can be defined as a cis-SAS gene pair [39] that can be structurally classified into four different types (divergent, convergent, embedded and fully overlapping) based on the overlapping ends of the sense–antisense transcripts and on their reciprocal direction of transcription [40] (Fig. 2c). In our study, the extent of the overlapping region between sense transcripts and complementary asRNAs was highly variable, ranging from 1 to 22 569 nucleotides (Table S1). Of the 7820 overlapping regions detected, 2066 held only one pair of complementary transcripts (called paired sense–antisense overlapping genes) (Table 3, Fig. 2a) produced from opposite DNA strands (Table S1). Most of them (603/2066; ~29.2 %) showed a convergent configuration (3′-ends overlap), followed by embedded (591/2066;~28.6 %), fully overlapping (578/2066;~28.0 %) and divergent conformations (5′-ends overlap) (294/2066;~14.2 %) (Fig. 2c). The remaining genomic regions were assorted in different and more complex configurations containing multiple sense–antisense transcripts or overlapping gene clusters (Table 3). Some examples are depicted in Fig. 4. However, for a more in-depth structural exploration of S. schenckii gene models, it is possible to inspect the new genome annotation (v2) at http://sporothrixgenomedatabase.unime.it.
Table 3.
List of gene groups found among different- and same-strand overlapping genes detected in this study
|
Gene group (no. of genes within each group) |
No. of each gene group found in S. schenckii |
Total no. of genes detected within each gene group |
Frequency (%)* |
|---|---|---|---|
|
Gene groups obtained from different-strand overlapping genes | |||
|
Paired (2) |
2066 |
4132 |
35.2 |
|
Triple (3) |
885 |
2655 |
22.6 |
|
Quadruple (4) |
433 |
1732 |
14.8 |
|
Quintuple (5) |
236 |
1180 |
10.1 |
|
Sextuple (6) |
114 |
684 |
5.8 |
|
Septuple (7) |
72 |
504 |
4.3 |
|
Octuple (8) |
43 |
344 |
2.9 |
|
Nonuple (9) |
25 |
225 |
1.9 |
|
Decuple (10) |
9 |
90 |
0.8 |
|
Undecuple (11) |
3 |
33 |
0.3 |
|
Duodecuple (12) |
7 |
84 |
0.7 |
|
Tredecuple (13) |
4 |
52 |
0.4 |
|
Quattuordecuple (14) |
1 |
14 |
0.1 |
|
Gene groups obtained from same-strand overlapping genes | |||
|
Paired (2) |
781 |
1562 |
73.6 |
|
Triple (3) |
127 |
381 |
18.0 |
|
Quadruple (4) |
35 |
140 |
6.6 |
|
Quintuple (5) |
5 |
20 |
0.9 |
|
Sextuple (6) |
3 |
18 |
0.8 |
*Percentage of genes of each group for total different-strand overlapping genes (11 729) and for total same-strand overlapping genes (2121).
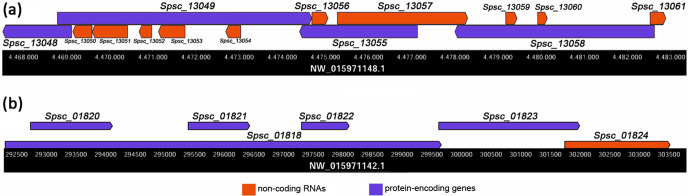
Fig. 4.
Snapshots of the newly annotated genomic regions of S. schenckii. (a) Uninterrupted chromosome region containing multiple sense–antisense transcripts assorted in a complex configuration. The image shows the largest gene group detected containing 14 different-strand overlapping genes. (b) Uninterrupted chromosome region containing one of the three largest gene groups involving six same-strand overlapping genes.
Examining the various cis-SAS gene pairs occurring in the S. schenckii genome, we found that they contain both protein-encoding genes and non-coding RNA (ncRNA) genes, albeit the most common model includes one coding strand and one non-coding strand in each pair (Table S1). However, the bi-directional transcription of a generic cis-SAS gene pair can lead to both coding and/or non-coding asRNAs that overlap, partially or completely, with other coding or non-coding sequences; thus, highlighting the extremely sophisticated organization of the S. schenckii genome. In fact, up to ~39 % (3034/7820) of sense–antisense overlapping regions detected produced protein-encoding asRNAs (transfrags with significant alignment to proteins deposited in the UniProt database) with a partial (or complete) overlap with the corresponding sense protein-encoding sequence (Table S1). Interestingly, a total of 243 of these overlapping regions showed a fully overlapping configuration (Fig. 2c), indicating that they are entirely transcribed in both directions and that two different mRNAs are produced (Table S1). As a consequence, we found that ~4 % (486/11 217) of the S. schenckii protein-encoding genes are bi-directionally transcribed with opposite polarity. The remaining overlapping regions (4786) encoded a number of transcripts that were predicted to be potential antisense ncRNAs (ancRNAs) by the cpc2 program. Over 87 % of these regions (4171/4786) transcribed ncRNAs (3544 non-redundant non-coding transcripts) (Table S1) that overlap neighbouring protein-encoding genes, whereas ~13 % (615/4786) produced 1211 unique (non-redundant) ncRNAs that overlap each other (Table S1).
Overlapping gene groups in the S. schenckii genome
To investigate the occurrence of overlapping gene types in S. schenckii, we used 11 217 protein-encoding and 5929 lncRNA genes annotated in this study by using strand-specific RNA-seq data (Table 1). Most of the overlapping genes detected (11 729) belong to the different-strand overlapping type (Fig. 2a, Table 3) as their transcripts map within 7820 overlapping regions that can be transcribed in both sense and antisense directions. By estimating the gene density in each of these uninterrupted overlapping regions, we found different clusters of overlapping cis-SAS genes ranging from 2 to 14 genes (Table 3). The paired sense–antisense overlapping genes constituted the most common type (4132/11 729; 35.2 %), followed by the triple (2655/11 729; 22.6 %), quadruple (1732/11 729; 14.8 %), quintuple (1180/11 729; 10.1 %), sextuple (684/11 729; 5.8 %) and septuple overlapping genes (504/11 729; 4.3 %), and other minor groups containing from 8 to 14 gene clusters that together make up ~7 % of all overlapping sense–antisense gene groups detected (Table 3). The chromosome region reported in Fig. 4(a) contains the unique largest overlapping gene group (14 genes involved) found in the S. schenckii genome. Based on these data, we estimated that over 68 % (11 729/17 146) of the S. schenckii genes are found to overlap and many of them are tightly compacted into genomic regions in which transcription may occur from either strand of the DNA molecule. However, we also identified a total of 1171 genomic regions containing 2121 same-strand overlapping genes (Tables 3 and S1), although it should be noted that our approach for classifying the different gene overlaps introduces the possibility of redundancy, as some transcripts may be represented more than once in the different categories of overlap (the same gene may share both different- and same-strand overlaps). Consequently, we estimated that approximately ~71 % (12 111 non-redundant genes) of the 17 146 S. schenckii protein-encoding/lncRNA genes were found to overlap in some way.
Similarly to different-strand, for same-strand overlapping genes, the paired configuration was the most common type (1562/2121; 73.6 %), although the same-strand overlapping genes were grouped into only five groups (Table 3). The triple and quadruple overlapping gene groups accounted for 18 and 6.6%, respectively, whereas the quintuple and sextuple overlapping gene groups were the least abundant among all the same-strand overlapping genes detected (<1 % each; Table 3). The chromosome region displayed in Fig. 4(b) shows the organization of one of the three gene clusters found involving six same-strand overlapping genes.
Differential gene expression analysis
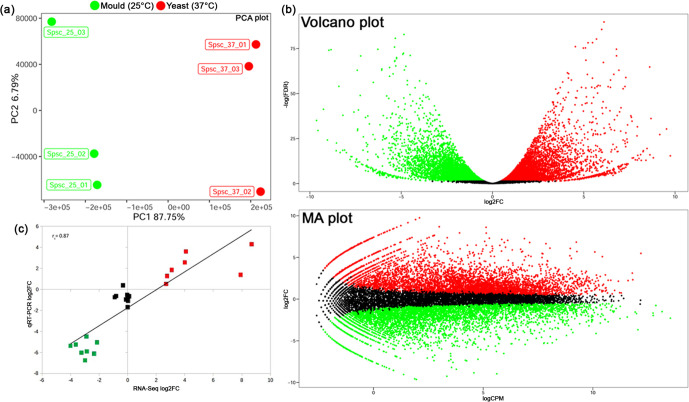
The main goal of this study was the characterization of the transcriptomic differences between yeast and mycelial forms of S. schenckii. The principal component analysis clearly separated the yeast and mycelial samples, along the first axis, into two well-defined groups (Fig. 5a), indicating that the observed variation (87.75 %), measured in terms of gene expression, was due to dimorphism. There was only a small variation (6.79 %) between biological replicates within each morphotype (Fig. 5a).
Fig. 5.
Sample variability, validation of RNA-seq data by qRT-PCR and differential gene expression between yeast and mould samples of S. schenckii examined in this study. (a) Principal component analysis (PCA) plot showing transcriptional profiles of S. schenckii cells grown at 25 °C (SSC_1–3 green) and 37 °C (SSC_4–6 red). The two groups are visually well-separated by the PC1 dimension and there is only a minor variation among the biological replicates as evidenced by the tenfold smaller scale for dimension PC2 compared to PC1. (b) Volcano plot and MA plot displaying differential gene expression between yeast and mould forms of S. schenckii. Significantly up-regulated and down-regulated genes in yeast cells compared with mould cells are indicated by red and green dots, respectively. Black dots indicate genes with no significant differences in their expression levels between the two tested conditions. (c) Scatter plot showing the correlation between RNA-seq and qRT-PCR expression data for 24 selected genes. The log2FC value is displayed for both RNA-seq data, as obtained from edgeR software, and qRT-PCR results, obtained from log2(2−ΔΔCt). The Spearman’s correlation coefficient rs is displayed. Squares denote up-regulated (red), down-regulated (green) and not regulated (black) genes.
Using our new genome annotation, global gene expression analysis revealed a catalogue of 12 104 and 12 911 genes expressed (FPKM >1) in the yeast-phase and mould-form, respectively. Of these, 10 443 genes were found to be expressed in both morphotypes (Table S2), while 1661 and 2468 were yeast-specific and mould-specific genes, respectively (Table S2). However, in total, 8795 genes were found to be differentially regulated among yeast and mould forms of S. schenckii when an absolute log2FC value ≥1 and an FDR threshold of 0.05 were used (Fig. 5b, Table S3). Collectively, these genes represent ~51 % (8795/17 307) of all annotated S. schenckii genes and more than half of them (4494 genes) were found to be up-regulated in the yeast-form (Fig. 5b, Table S3).
qRT-PCR results confirmed the RNA-seq expression patterns observed for the selected differentially up-/down-regulated and unregulated genes. Comparison of edgeR estimated expression data with those determined by qRT-PCR showed a significant positive correlation (rs=0.87) between the two measures of gene expression levels (Fig. 5c).
RNA-seq analysis also revealed a large amount of variation in ancRNA expression. In fact, among the 4171 ancRNA overlapping protein-encoding loci, 2236 unique ancRNAs were found to be statistically differentially expressed (absolute log2FC ≥1; FDR ≤0.05) between the two morphological types examined (Table S4). Interestingly, 1117 protein-encoding genes that were not differentially expressed (FDR>0.05) among yeast and mould forms showed their antisense transcript partners to be differentially regulated in the two conditions (Table S4).
GO term enrichment analysis of DE genes
GO enrichment analysis was performed to identify and classify the potential molecular function exerted by DE genes, including the different biological processes in which they are involved and the cellular locations where these occur (Table S5). Based on GO analysis of 8795 DE genes, we found that 5624 (~64 %) were categorized in a total of 2629 functional groups or classes (GO terms) (Table S5). However, when data were filtered using an FDR cut-off <0.05, we detected a total of 1318 GO terms in the entire data set (Table S5). More specifically, 1030 genes were classified into 542 functional groups within the main domain of biological process, 1455 genes into 525 functional groups within molecular function, and 835 genes were categorized into 251 classes within the cellular component domain (Table S5).
Next, we performed a GO enrichment analysis for genes that were specifically up-regulated (4494 genes) and down-regulated (4301 genes) between the two morphological stages of S. schenckii (yeast versus mould) (Table S3). The results are reported in Table S6. After FDR filtering (cut-off <0.05), up-regulated genes were categorized in a total of 664 functional groups, of which 270 belonged to biological process, 270 to molecular function, and 124 to the cellular component domain.
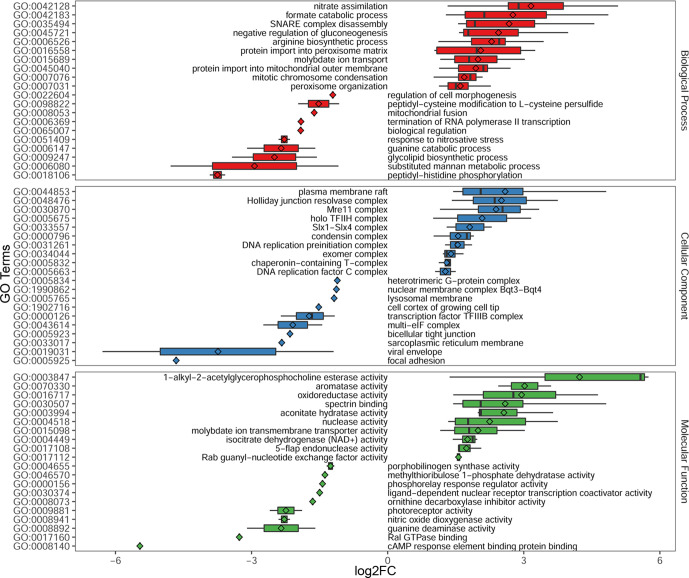
Regarding down-regulated genes, a total of 321 GO terms were found at the FDR <0.05 threshold. Of these, 136 were within the domain of biological processes, 132 within molecular function and 53 in the cellular component category. Fig. 6 displays the top 10 GO terms assigned to up- and down-regulated genes in the three main ontologies.
Fig. 6.
Box plot representation of the top 10 enriched GO terms of DE genes for each ontology category (biological process, cellular component and molecular function). The rhombuses inside each box indicate the mean of the log2FC values of genes included in the respective GO term.
Discussion
In this study, we generated high-quality RNA-seq data that were used for obtaining a complete view of the genes expressed by the dimorphic fungus S. schenckii under hyphae- and yeast-inducing conditions. This comprehensive transcriptome dataset can be queried and explored at http://sporothrixgenomedatabase.unime.it, where RNA-seq data, including the new genome annotation and gene expression profiles, can be also freely downloaded for further investigation. A point of special interest in this study is the detailed genomic description of thousands (5929) of lncRNA loci that make up over 34 % of the total genes occurring in the S. schenckii genome. A similar number has been reported recently in other fungal genomes [41, 42], but their abundance may vary widely among different species with different genome sizes [43]. However, the real occurrence of lncRNAs in fungi is, currently, likely underestimated, because the detection of these regulatory elements largely depends on the type of experimental condition examined and on the sequenced transcripts which, as in our study, are often poly(A) selected; thus, precluding the possibility of capturing the non-polyadenylated lncRNAs [44]. Anyhow, by uncovering the expansive landscape of lncRNAs associated with the yeast and/or mould forms of S. schenckii, we provide the scientific community with a powerful starting point to begin investigating their biological relevance.
Undoubtedly, an interesting aspect of lncRNA biogenesis in S. schenckii is that most lncRNAs (~77 %) are transcribed from overlapping loci (cis-SAS gene pairs) and only a small fraction originates from intergenic or other genomic regions (~23 %). These results are in contrast with the very few findings dealing with lncRNAs in other fungi [43, 45, 46], in which intergenic regions appear to be the main sources of these RNA molecules. However, our data are in line with the results of two recent studies in Schizosaccharomyces pombe [41] and Nosema ceranae [47], where most of the discovered lncRNAs were transcribed as natural antisense transcripts (NATs). NATs are RNA molecules that originate from opposite DNA strands of the same genomic locus (cis-NAT) or unlinked loci (trans-NAT), and can be generically defined as coding RNAs or ncRNAs that are complementary to, and overlap with, either protein-encoding or non-coding transcripts [48]. Many cis-NATs encode lncRNAs that have been reported as important regulators of gene expression in a wide range of ascomycete and basidiomycete fungi [40]. In S. schenckii, we found a total of 11 729 cis-SAS genes which represent, to our knowledge, the largest number of overlapping sense–antisense genes reported in fungi to date. However, through comprehensive transcriptomic studies, thousands of NATs have been detected in other fungi also, including Magnaporthe oryzae (4215), Schizosaccharomyces pombe (4384) and Schizophyllum commune (5635), but their functional roles remain to be investigated [40]. Interestingly, in our study, we observed different clusters of overlapping cis-SAS genes ranging from 2 (paired overlapping genes) to 14 genes. Similar multigene-transcript overlapping regions have also recently been described in the human genome, where about 72 % of the overlapping protein-encoding genes showed a paired configuration, followed by more complex overlapping groups until reaching 22 overlapping genes in a protocadherin gene cluster on chromosome 5 [49]. Consequently, in terms of gene overlap, our data suggest that the physical chromosomal arrangement of the S. schenckii genes follows, to some extent, the pattern observed in other eukaryotic genomes where the different-strand overlapping type seems to be the most commonly adopted gene model [30]. In this regard, it is worth noting that this contrasts with what has been observed in bacteria, where the genes that overlap on the same-strand are by far the most abundant [50]. This different gene disposition could probably be linked to the presence of a high number of antisense lncRNAs occurring in eukaryotes compared to prokaryotic genomes in which this class of RNA molecules is rare [51]. Furthermore, the existence of overlapping genes in S. schenckii and other eukaryotic genomes should be carefully considered in future gene manipulation and knockout studies, because possible polar effects could appear by manipulation of one overlapping locus, especially those in which both DNA strands encode different proteins.
Another important aspect of our study concerns the genome-wide analysis of gene expression, which emphasizes the extraordinary complexity of the molecular pathways that control either genesis and maintenance of yeast and hyphae cell morphologies, which could be governed by the expression of different lncRNA molecules and by a number of still uncharacterized protein-encoding genes (Table S3). Interestingly, GO enrichment analysis using DE genes revealed an overall up-regulation of genes involved in nitrate assimilation and arginine biosynthesis (Fig. 6). In fungi, nitric oxide and l-arginine have been implicated in several cellular processes, including the regulation of morphogenesis and reproduction [52, 53]. In particular, in Coniothyrium minitans and M. oryzae, l-arginine plays an important role in the asexual developmental process and is essential for conidiation [52, 54]. In fact, in Coniothyrium minitans, the disruption of the carbamoyl-phosphate synthase (CSP1)-encoding gene (a key gene in the biosynthesis of l-arginine) yielded a conidiation deficiency mutant that could only grow hyphae in culture [52]. Disruption of the gene ortholog in M. oryzae (CPA2 gene) also impairs conidiogenesis and its transcript levels were significantly lower in mycelial growth compared to the conidial stage [54]. Similarly, we found that the expression of the S. schenckii carbamoyl-phosphate synthase gene (ORF Spsc_00280) was down-regulated in the mould-form compared to the yeast-form, suggesting a possible involvement of the amino acid biosynthetic pathways in hyphae-to-yeast transition, an assumption supported by a recent work exploring the crucial role of metabolic pathways in Candida albicans morphogenesis [55].
In S. schenckii, few studies have been conducted to identify key genes involved in triggering and maintaining the dimorphic changes. One gene (DRK1), encoding a hybrid histidine kinase (HHK), has been described as a master regulator of the hyphae-to-yeast transition and virulence in several dimorphic fungi, including S. schenckii [56, 57]. The expression of this gene was reported to be up-regulated in the yeast-phase compared to the mould-phase [56, 57] but, in our study, we found it (ORF Spsc_00605) to be not differentially expressed between mould and yeast forms of S. schenckii (Table S3). This result was also confirmed by using a qRT-PCR assay (Table S1). In our opinion, this discrepancy in DRK1 expression levels could be explained by the different experimental conditions employed for obtaining the yeast and mould phases by various studies. In fact, in our study, both morphologies were obtained by growing the fungus in YPD medium up to 6 days, while in previous studies [56, 57], the mould and yeast forms were obtained by using different incubation times (≤4 days) and culture media: Sabouraud liquid medium for mould growth and brain heart infusion broth for the yeast development. However, based on our data, it is possible that in S. schenckii, after induction of the yeast-phase, the DRK1 gene expression level decreases and other factors contribute to maintaining the yeast form. Interestingly, in this study, we also found an additional HHK encoded by the ORF Spsc_09813. This new HHK was found to be differentially expressed between the two examined S. schenckii morphologies, it was up-regulated in the yeast-form. Bioinformatics analysis of the Spsc_09813 transcript revealed that it encodes a protein carrying all structural domains specific for fungal HHKs [58]: (i) a N-terminus Per-Arnt-Sim (PAS_9) sensing domain; followed by (ii) a central region composed of a histidine kinase (dimerization/phosphoacceptor) domain (HisKA) and a cognate histidine kinase-like ATPase catalytic domain (HATPase_c); and (iii) a conserved C-terminus region referred to as receiver domain [58]. However, the molecular mechanisms leading to dimorphic switching in fungi are very complex and depend on the expression of many genes. In fact, this phenomenon can be induced by different biophysical stimuli that operate through specific or cross-talk molecular signalling pathways [59], which, in turn, activate specific downstream proteins whose transcription can also be finely regulated by ncRNAs. This latter assumption has been recently proven by a study in Cryptococcus in which a specific lncRNA, encoded by the RZE1 gene, controls the yeast-to-hypha transition in this fungus [60]. Moreover, many other lncRNAs seem to have important roles in fungal development and stress response [61, 62]; therefore, the functional characterization of the thousands of lncRNAs differentially expressed in S. schenckii (Table S3) could give new insight on the capacity of these non-coding elements in regulating morphogenesis in this species also. The transcriptomic data reported in this study will not only serve as a valuable reference for investigation of genes involved in the dimorphic transition and other biological processes, but also expand our knowledge beyond what has been learnt through recent genomic studies of different Sporothrix species [6, 13, 63, 64].
Funding information
This work was supported by the University of Messina (CT_ROMEO_GOM – grant agreement no. 36036 to O.R.); the Italian Ministry of Education, University and Research (MIUR) (FFABRA2017 to G.C.); the National Nature Foundation of China [81371746 and 81974300 to H.H.]; and Sun Yat-sen University, Guangzhou, China [82000-31143405 to H.H.].
Author contributions
Conceptualization: O. R., D. G., M. R. F. Methodology: D. G., L. G., C. L. P., C. B. Investigation: M. R. F., L. G., C. L. P., C. B. Software: D. G., L. G., R. A. C., A. P.-G., O. R. Formal analysis: D. G., M. R. F., E. D. A., H. H. Supervision: O. R., H. M. M.-M., S.d.H., H.H. Validation: M.R.F., C.L.P., C.B., L.G. Resources: O.R., H. H., E.D.A., G.C. Data curation: D. G., L. G., R. A. C., A. P.-G. Writing
– original draft: O. R., G. C. Writing – review and editing: O. R., R. A. C., H. M. M.-M., S. d. H., H. H. Funding: O. R., G. C., H. H.
Conflicts of interest
The authors declare that there are no conflicts of interest.
Footnotes
Abbreviations: ancRNA, antisense non-coding RNA; asRNA, antisense RNA; cis-SAS, cis-sense–antisense; DE gene, differentially expressed gene; FDR, false discovery rate; FPKM, fragments per kilobase of transcript per million mapped reads; GO, gene ontology; lncRNA, long non-coding RNA; log2FC, log2fold-change; NAT, natural antisense transcript; NCBI, National Center for Biotechnology Information; ncRNA, non-coding RNA; qRT-PCR, quantitative real-time PCR; SGDB, Sporothrix Genome DataBase.
All supporting data, code and protocols have been provided within the article or through supplementary data files. Six supplementary tables and one supplementary file are available online at Figshare (https://doi.org/10.6084/m9.figshare.12894761.v1).
The raw Illumina reads have been deposited in the NCBI SRA database under BioProject PRJNA539953. The GEO accession number is GSE145856.
References
- 1.de Beer ZW, Duong TA, Wingfield MJ. The divorce of Sporothrix and Ophiostoma: solution to a problematic relationship. Stud Mycol. 2016;83:165–191. doi: 10.1016/j.simyco.2016.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rodrigues AM, Della Terra PP, Gremião ID, Pereira SA, Orofino-Costa R, et al. The threat of emerging and re-emerging pathogenic Sporothrix species. Mycopathologia. 2020 doi: 10.1007/s11046-020-00425-0. [DOI] [PubMed] [Google Scholar]
- 3.Ramírez-Soto MC, Aguilar-Ancori EG, Tirado-Sánchez A, Bonifaz A. Ecological determinants of sporotrichosis etiological agents. J Fungi. 2018;4:95. doi: 10.3390/jof4030095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arenas R, Sánchez-Cardenas CD, Ramirez-Hobak L, Ruíz Arriaga LF, Vega Memije ME. Sporotrichosis: from KOH to molecular biology. J Fungi. 2018;4:62. doi: 10.3390/jof4020062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chakrabarti A, Bonifaz A, Gutierrez-Galhardo MC, Mochizuki T, Li S. Global epidemiology of sporotrichosis. Med Mycol. 2015;53:3–14. doi: 10.1093/mmy/myu062. [DOI] [PubMed] [Google Scholar]
- 6.Mora-Montes HM, Dantas Ada S, Trujillo-Esquivel E, de Souza Baptista AR, Lopes-Bezerra LM. Current progress in the biology of members of the Sporothrix schenckii complex following the genomic era. FEMS Yeast Res. 2015;15:fov065. doi: 10.1093/femsyr/fov065. [DOI] [PubMed] [Google Scholar]
- 7.Téllez MD, Batista-Duharte A, Portuondo D, Quinello C, Bonne-Hernández R, et al. Sporothrix schenckii complex biology: environment and fungal pathogenicity. Microbiology. 2014;160:2352–2365. doi: 10.1099/mic.0.081794-0. [DOI] [PubMed] [Google Scholar]
- 8.Marimon R, Serena C, Gené J, Cano J, Guarro J. In vitro antifungal susceptibilities of five species of Sporothrix . Antimicrob Agents Chemother. 2008;52:732–734. doi: 10.1128/AAC.01012-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Arrillaga-Moncrieff I, Capilla J, Mayayo E, Marimon R, Mariné M, et al. Different virulence levels of the species of Sporothrix in a murine model. Clin Microbiol Infect. 2009;15:651–655. doi: 10.1111/j.1469-0691.2009.02824.x. [DOI] [PubMed] [Google Scholar]
- 10.Castro RA, Kubitschek-Barreira PH, Teixeira PAC, Sanches GF, Teixeira MM, et al. Differences in cell morphometry, cell wall topography and gp70 expression correlate with the virulence of Sporothrix brasiliensis clinical isolates. PLoS One. 2013;8:e75656. doi: 10.1371/journal.pone.0075656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang Y, Hagen F, Stielow B, Rodrigues AM, Samerpitak K, et al. Phylogeography and evolutionary patterns in Sporothrix spanning more than 14 000 human and animal case reports. Persoonia. 2015;35:1–20. doi: 10.3767/003158515X687416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Batista-Duharte A, Téllez-Martínez D, Roberto de Andrade C, Portuondo DL, Jellmayer JA, et al. Sporothrix brasiliensis induces a more severe disease associated with sustained Th17 and regulatory T cells responses than Sporothrix schenckii sensu stricto in mice. Fungal Biol. 2018;122:1163–1170. doi: 10.1016/j.funbio.2018.08.004. [DOI] [PubMed] [Google Scholar]
- 13.Teixeira MM, de Almeida LGP, Kubitschek-Barreira P, Alves FL, Kioshima ES, et al. Comparative genomics of the major fungal agents of human and animal sporotrichosis: Sporothrix schenckii and Sporothrix brasiliensis . BMC Genomics. 2014;15:943. doi: 10.1186/1471-2164-15-943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Okonechnikov K, Conesa A, García-Alcalde F. Qualimap 2: advanced multi-sample quality control for high-throughput sequencing data. Bioinformatics. 2016;32:292–294. doi: 10.1093/bioinformatics/btv566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Campbell MS, Holt C, Moore B, Yandell M. Genome annotation and curation using MAKER and MAKER-P. Curr Protoc Bioinformatics. 2014;48:4.11.1–4.11.39. doi: 10.1002/0471250953.bi0411s48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Korf I. Gene finding in novel genomes. BMC Bioinformatics. 2004;5:59. doi: 10.1186/1471-2105-5-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stanke M, Morgenstern B. AUGUSTUS: a web server for gene prediction in eukaryotes that allows user-defined constraints. Nucleic Acids Res. 2005;33:W465–W467. doi: 10.1093/nar/gki458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol. 2011;29:644–652. doi: 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fu L, Niu B, Zhu Z, Wu S, Li W. CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics. 2012;28:3150–3152. doi: 10.1093/bioinformatics/bts565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gremme G, Steinbiss S, Kurtz S. GenomeTools: a comprehensive software library for efficient processing of structured genome annotations. IEEE/ACM Trans Comput Biol Bioinform. 2013;10:645–656. doi: 10.1109/TCBB.2013.68. [DOI] [PubMed] [Google Scholar]
- 23.Boratyn GM, Camacho C, Cooper PS, Coulouris G, Fong A, et al. BLAST: a more efficient report with usability improvements. Nucleic Acids Res. 2013;41:W29–W33. doi: 10.1093/nar/gkt282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jones P, Binns D, Chang HY, Fraser M, Li W, et al. InterProScan 5: genome-scale protein function classification. Bioinformatics. 2014;30:1236–1240. doi: 10.1093/bioinformatics/btu031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kang YJ, Yang DC, Kong L, Hou M, Meng YQ, et al. CPC2: a fast and accurate coding potential calculator based on sequence intrinsic features. Nucleic Acids Res. 2017;45:W12–W16. doi: 10.1093/nar/gkx428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nawrocki EP, Eddy SR. Infernal 1.1: 100-fold faster RNA homology searches. Bioinformatics. 2013;29:2933–2935. doi: 10.1093/bioinformatics/btt509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chan PP, Lowe TM. tRNAscan-SE: searching for tRNA genes in genomic sequences. Methods Mol Biol. 2019;1962:1–14. doi: 10.1007/978-1-4939-9173-0_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Törönen P, Medlar A, Holm L. PANNZER2: a rapid functional annotation web server. Nucleic Acids Res. 2018;46:W84–W88. doi: 10.1093/nar/gky350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sanna CR, Li WH, Zhang L. Overlapping genes in the human and mouse genomes. BMC Genomics. 2008;9:169. doi: 10.1186/1471-2164-9-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30:923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 32.Donaldson ME, Ostrowski LA, Goulet KM, Saville BJ. Transcriptome analysis of smut fungi reveals widespread intergenic transcription and conserved antisense transcript expression. BMC Genomics. 2017;18:340. doi: 10.1186/s12864-017-3720-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rau A, Gallopin M, Celeux G, Jaffrézic F. Data-based filtering for replicated high-throughput transcriptome sequencing experiments. Bioinformatics. 2013;29:2146–2152. doi: 10.1093/bioinformatics/btt350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tian T, Liu Y, Yan H, You Q, Yi X, et al. agriGO v2.0: a GO analysis toolkit for the agricultural community, 2017 update. Nucleic Acids Res. 2017;45:W122–W129. doi: 10.1093/nar/gkx382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCt method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 37.Buels R, Yao E, Diesh CM, Hayes RD, Munoz-Torres M, et al. JBrowse: a dynamic web platform for genome visualization and analysis. Genome Biol. 2016;17:66. doi: 10.1186/s13059-016-0924-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kahl G. The Dictionary of Genomics, Transcriptomics and Proteomics. 5th edn. Weinheim: Wiley-VCH; 2015. [Google Scholar]
- 39.Wood EJ, Chin-Inmanu K, Jia H, Lipovich L. Sense-antisense gene pairs: sequence, transcription, and structure are not conserved between human and mouse. Front Genet. 2013;4:183. doi: 10.3389/fgene.2013.00183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Donaldson ME, Saville BJ. Natural antisense transcripts in fungi. Mol Microbiol. 2012;85:405–417. doi: 10.1111/j.1365-2958.2012.08125.x. [DOI] [PubMed] [Google Scholar]
- 41.Atkinson SR, Marguerat S, Bitton DA, Rodríguez-López M, Rallis C, et al. Long noncoding RNA repertoire and targeting by nuclear exosome, cytoplasmic exonuclease, and RNAi in fission yeast. RNA. 2018;24:1195–1213. doi: 10.1261/rna.065524.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Han G, Cheng C, Zheng Y, Wang X, Xu Y, et al. Identification of long non-coding RNAs and the regulatory network responsive to arbuscular mycorrhizal fungi colonization in maize roots. Int J Mol Sci. 2019;20:4491. doi: 10.3390/ijms20184491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Borgognone A, Sanseverino W, Aiese Cigliano R, Castanera R. Distribution, characteristics, and regulatory potential of long noncoding RNAs in brown-rot fungi. Int J Genomics. 2019;2019:9702342. doi: 10.1155/2019/9702342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fernandes JCR, Acuña SM, Aoki JI, Floeter-Winter LM, Muxel SM. Long non-coding RNAs in the regulation of gene expression: physiology and disease. Noncoding RNA. 2019;5:17. doi: 10.3390/ncrna5010017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Linde J, Duggan S, Weber M, Horn F, Sieber P, et al. Defining the transcriptomic landscape of Candida glabrata by RNA-Seq. Nucleic Acids Res. 2015;43:1392–1406. doi: 10.1093/nar/gku1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cemel IA, Ha N, Schermann G, Yonekawa S, Brunner M. The coding and noncoding transcriptome of Neurospora crassa . BMC Genomics. 2017;18:978. doi: 10.1186/s12864-017-4360-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Guo R, Chen DF, Xiong CL, Hou CS, Zheng YZ, et al. First identification of long non-coding RNAs in fungal parasite Nosema ceranae . Apidologie. 2018;49:660–670. doi: 10.1007/s13592-018-0593-z. [DOI] [Google Scholar]
- 48.Latgé G, Poulet C, Bours V, Josse C, Jerusalem G. Natural antisense transcripts: molecular mechanisms and implications in breast cancers. Int J Mol Sci. 2018;19:123. doi: 10.3390/ijms19010123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chen C-H, Pan C-Y, Lin W. Overlapping protein-coding genes in human genome and their coincidental expression in tissues. Sci Rep. 2019;9:13377. doi: 10.1038/s41598-019-49802-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sabath N, Graur D, Landan G. Same-strand overlapping genes in bacteria: compositional determinants of phase bias. Biol Direct. 2008;3:36. doi: 10.1186/1745-6150-3-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Harris KA, Breaker RR. Large noncoding RNAs in bacteria. Microbiol Spectr. 2018;6:RWR-0005-2017. doi: 10.1128/microbiolspec.RWR-0005-2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gong X, Fu Y, Jiang D, Li G, Yi X, et al. L-Arginine is essential for conidiation in the filamentous fungus Coniothyrium minitans . Fungal Genet Biol. 2007;44:1368–1379. doi: 10.1016/j.fgb.2007.07.007. [DOI] [PubMed] [Google Scholar]
- 53.Cánovas D, Marcos JF, Marcos AT, Strauss J. Nitric oxide in fungi: is there NO light at the end of the tunnel? Curr Genet. 2016;62:513–518. doi: 10.1007/s00294-016-0574-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liu X, Cai Y, Zhang X, Zhang H, Zheng X, et al. Carbamoyl phosphate synthetase subunit MoCpa2 affects development and pathogenicity by modulating arginine biosynthesis in Magnaporthe oryzae . Front Microbiol. 2016;7:2023. doi: 10.3389/fmicb.2016.02023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Han T-L, Cannon RD, Gallo SM, Villas-Bôas SG. A metabolomic study of the effect of Candida albicans glutamate dehydrogenase deletion on growth and morphogenesis. NPJ Biofilms Microbiomes. 2019;5:13. doi: 10.1038/s41522-019-0086-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hou B, Zhang Z, Zheng F, Liu X. Molecular cloning, characterization and differential expression of DRK1 in Sporothrix schenckii . Int J Mol Med. 2013;31:99–104. doi: 10.3892/ijmm.2012.1193. [DOI] [PubMed] [Google Scholar]
- 57.Zhang Z, Hou B, Wu YZ, Wang Y, Liu X, et al. Two‑component histidine kinase DRK1 is required for pathogenesis in Sporothrix schenckii . Mol Med Rep. 2018;17:721–728. doi: 10.3892/mmr.2017.8005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Defosse TA, Sharma A, Mondal AK, Dugé de Bernonville T, Latgé J-P, et al. Hybrid histidine kinases in pathogenic fungi. Mol Microbiol. 2015;95:914–924. doi: 10.1111/mmi.12911. [DOI] [PubMed] [Google Scholar]
- 59.Pathan EK, Ghormade V, Panmei R, Deshpande MV. Biochemical and molecular aspects of dimorphism in fungi. In: Satyanarayana T, Deshmukh S, Deshpande M, editors. Advancing Frontiers in Mycology and Mycotechnology. Singapore: Springer; 2019. pp. 69–94. [Google Scholar]
- 60.Chacko N, Zhao Y, Yang E, Wang L, Cai JJ, et al. The lncRNA RZE1 controls cryptococcal morphological transition. PLoS Genet. 2015;11:e1005692. doi: 10.1371/journal.pgen.1005692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Till P, Mach RL, Mach-Aigner AR. A current view on long noncoding RNAs in yeast and filamentous fungi. Appl Microbiol Biotechnol. 2018;102:7319–7331. doi: 10.1007/s00253-018-9187-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang Z, Jiang Y, Wu H, Xie X, Huang B. Genome-wide identification and functional prediction of long non-coding RNAs involved in the heat stress response in Metarhizium robertsii . Front Microbiol. 2019;10:2336. doi: 10.3389/fmicb.2019.02336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.D'Alessandro E, Giosa D, Huang L, Zhang J, Gao W, et al. Draft genome sequence of the dimorphic fungus Sporothrix pallida, a nonpathogenic species belonging to Sporothrix, a genus containing agents of human and feline sporotrichosis. Genome Announc. 2016;4:e00184-16. doi: 10.1128/genomeA.00184-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Huang L, Gao W, Giosa D, Criseo G, Zhang J, et al. Whole-genome sequencing and in silico analysis of two strains of Sporothrix globosa . Genome Biol Evol. 2016;8:3292–3296. doi: 10.1093/gbe/evw230. [DOI] [PMC free article] [PubMed] [Google Scholar]