Abstract
Mannoside phosphorylases are involved in the intracellular metabolization of mannooligosaccharides, and are also useful enzymes for the in vitro synthesis of oligosaccharides. They are found in glycoside hydrolase family GH130. Here we report on an analysis of 6308 GH130 sequences, including 4714 from the human, bovine, porcine and murine microbiomes. Using sequence similarity networks, we divided the diversity of sequences into 15 mostly isofunctional meta-nodes; of these, 9 contained no experimentally characterized member. By examining the multiple sequence alignments in each meta-node, we predicted the determinants of the phosphorolytic mechanism and linkage specificity. We thus hypothesized that eight uncharacterized meta-nodes would be phosphorylases. These sequences are characterized by the absence of signal peptides and of the catalytic base. Those sequences with the conserved E/K, E/R and Y/R pairs of residues involved in substrate binding would target β-1,2-, β-1,3- and β-1,4-linked mannosyl residues, respectively. These predictions were tested by characterizing members of three of the uncharacterized meta-nodes from gut bacteria. We discovered the first known β-1,4-mannosyl-glucuronic acid phosphorylase, which targets a motif of the Shigella lipopolysaccharide O-antigen. This work uncovers a reliable strategy for the discovery of novel mannoside-phosphorylases, reveals possible interactions between gut bacteria, and identifies a biotechnological tool for the synthesis of antigenic oligosaccharides.
Keywords: glycoside phosphorylases, GH130 CAZy family, sequence similarity networks, mannosides, gut microbiomes
Data Summary
The GH130 amino acid sequences were collected from the manually curated CAZy database and from four public gut microbial gene datasets from the human gut (GigaDB [1]), mouse gut (GigaDB [2]), pig gut (EBI, PRJEB11755 [3]) and bovine rumen (Giga DB [4]) automatically annotated with human supervision. The sequence accession numbers used in this study are listed in Table S1 (available in the online version of this article).
Impact Statement.
In recent years, several studies have revealed the ability of bacterial mannoside-phosphorylases classified in the glycoside hydrolase family 130 to degrade mannosides from plants, yeasts and mammals. In 2013, we proposed a classification of this family into subfamilies, which is still used today. Nevertheless, this previous study also revealed one heterogeneous sequence cluster, which was later shown to include both mannoside-phosphorylases and mannosidases, of various specificities. In this paper, we present a more accurate analysis of the GH130 family, using the greater sequence diversity available today, including that of metagenomes. We established a generic strategy based on the analysis of sequence similarity networks and conservation of active site residues to identify glycoside-phosphorylases from thousands of sequences and predict their specificity. We validated this predictive approach by biochemically characterizing members of yet uncharacterized sequence groups. This strategy allowed us to discover novel mannoside-phosphorylases from the human gut microbiome, which are able to degrade and synthesize glycosidic motifs found in pathogenic bacteria. This work opens up new perspectives for the synthesis of antigenic oligosaccharides and for the study of interactions between bacteria.
Introduction
Mannosides occur in many living organisms, from microbes to plants and animals, in the form of homo- or heteropolymers or glycoconjugates. They play a critical role in several cellular processes, such as cell wall structuring in plants [5], cell signalling, as key components of proteoglycans [6], lipopolysaccharides and capsular polysaccharides in bacteria, yeasts, mammals and plants [7], and also as a carbohydrate storage polymer in certain parasites [8]. Mannoside synthesis is mostly carried out by glycoside transferases (GTs) from mannose diphosphonucleotides, whereas mannoside degradation is carried out by glycoside hydrolases (GHs). These enzymes are very diverse in terms of structures, mechanisms and specificities, as indicated by their respective membership of 13 and 15 different GT and GH families, respectively, in the classification of carbohydrate-active enzymes (CAZy database, http://www.cazy.org/ [9]). In addition, mannoside synthesis and degradation can also be performed by mannoside phosphorylases (GPs). The latter enzymes, which are classified in the related CAZy families GH130 [10] and GT108 [11], breakdown β-mannosides in the presence of inorganic phosphate to generate α-mannose-1-phosphate (αMan1P) through an inverting phosphorolytic mechanism. They can also act as β-mannoside-synthases from αMan1P and glycosyl acceptors, in the so-called reverse phosphorolysis reaction. The newly created GT108 family only contains six characterized members, all acting on mannogen, a linear β-1,2-polymannoside. In contrast, the GH130 family is much more polyspecific, with 18 characterized members, of which 3 are β-1,2-mannosidases [12, 13] and 14 are glycoside phosphorylases acting on a large variety of substrates, such asβ-1,4-mannooligosaccharides [14–18], β-1,4-mannosyl-N-acetyl-glucosamine [19],β-1,4-mannosyl-N,N’-diacetylchitobiose [20],β-1,2-mannobiose [21, 22],β-1,2-oligomannans [21] andβ-1,3-mannobiose [23]. Their large diversity of specificities, the tolerance of some enzymes towards acceptors, and their ability to generate αMan1P through phosphorolysis of cheap substrates for the synthesis of various heteromannosides, makes GH130 glycoside phosphorylases attractive enzymes for the in vitro synthesis of β-mannosidic linkages, which are considered to be some of the most challenging glycosidic linkages in synthetic carbohydrate chemistry [24]. However, the functional and sequence diversity of GH130 enzymes, and their role in the functioning of ecosystems, are underinvestigated. Less than 1 % of the 2142 GH130 enzymes listed in the CAZy database (release 8 January 2020) have been biochemically characterized. In order to predict the specificity of enzymes in this family that are still uncharacterized, in 2013 we proposed a classification into two subfamilies, namely GH130_1, strictly acting on β-1,4-mannosyl-glucose, and GH130_2, targeting β-1,4-mannosyl-N,N′-diacetylchitobiose (the core oligosaccharide of N-glycans), or β-1,4-mannooligosaccharides [20]. This subfamily analysis also revealed one heterogeneous GH130_NC sequence cluster, which was later shown to include enzymes of various specificities and mechanisms, such as β-1,2-mannosidases [12, 13], β-1,2-mannobiose- and β-1,2-oligomannan-phosphorylases [21, 22], and a β-1,3-mannobiose-phosphorylase [23]. Even though these early subfamilies are still used [17], we wished to perform a new analysis based on the greater sequence diversity available today. Indeed the number of GH130 sequences listed in the CAZy database alone is now almost six times larger than in 2013. Because the CAZy database only lists sequences from daily releases from GenBank, we also wished to include a number of metagenomic GH130 sequences not present in CAZy in our novel analysis, as these may reveal a diversity not yet present in GenBank.
The Enzyme Function Initiative-Enzyme Similarity Tool (EFI-EST) was developed in 2015 (http://efi.igb.illinois.edu/efi-est/ [25]) to rationalize the search for sequences of interest in large datasets. This web tool automatically generates sequence similarity networks (SSNs) to visualize sequence relationships in protein families [26]. SSNs have recently been used to search for new functions in the glycyl radical enzyme superfamily, a class of prominent enzymes in the human gut microbiome [27], and to subdivide the multispecific GH16 family into 23 robust subfamilies [28].
In the present study, we used SSNs to analyse the diversity of GH130 sequences listed in the CAZy database, and in four metagenome datasets selected from mammalian guts, as ecosystems that are rich in mannosides of various origins and structures [29]. By segregating the GH130 sequences into iso-functional meta-nodes and analysing the conservation of the active site residues involved in catalysis and substrate accommodation, we uncovered a strategy to distinguish GPs from GHs and to predict their linkage specificity. We tested our predictions by biochemically characterizing members of the unexplored meta-nodes. This study allowed us to reveal a new function of heteromannoside degradation and synthesis.
Methods
GH130 sequence mining in mammal gut metagenomes
The GH130 sequences were identified from the metagenomic sequence datasets following a procedure previously described for other metagenomics analyses [30]. The procedure consisted of a blast [31] search of the protein sequences against the full-length GH130 sequences included in the CAZy database (http://www.cazy.org) in November 2018, using a cut-off E-value of 10−6. Sequences that aligned over their entire length with a GH130 sequence in the database with >50 % identity were assigned directly to the GH130 family. The remaining sequences were subjected to a sequence similarity search using the hidden Markov models (HMMER v3) built for each CAZy family, allowing identification of the CAZy family they belong to [9].
Sequence similarity networks
The web-based EFI-EST (http://efi.igb.illinois.edu/efi-est) [25] was used to construct SSNs from the GH130 sequences retrieved from the CAZy database in November 2018 (1592 sequences) and from the 4 metagenomes (4714 sequences), plus 2 sequences of recently characterized GPs (VCV21229.1 and VCV21228.1), with E-value thresholds between 10−30 and 10−90. The node networks were visualized using Cytoscape 3.2 (https://cytoscape.org/ [32]).
Analysis of sequence similarity
The CD-HIT program was used to analyse sequence redundancy within and between the datasets (http://weizhong-lab.ucsd.edu/cdhit-web-server/cgi-bin/index.cgi) [33]. For each GH130 sequence selected for biochemical characterization, the most similar sequence was identified from the CAZy database using blast, with the NIH blastp default search parameters (https://blast.ncbi.nlm.nih.gov). The conservation of active site residues was analysed using multiple sequence alignments performed using clustal Omega (https://www.ebi.ac.uk/Tools/msa/clustalo/). The alignments were corrected in the region containing residue K199 (reference sequence AAO78885.1), according to available structural information [22].
Phylogenetic analysis
First, a multiple structural alignment using mustang 3.2.2 [34] was performed with the 11 GH130 sequences associated to a PDB code (1VKD, 3QC2, 3R67, 3TAW, 3WAS [35], 4ONZ, 4UDG [36], 5A7V [12], 5AYC [16], 5AYE [16] and 5B0R [22]). Then, 10 sequences for each of the 15 meta-nodes (150 sequences in total) were chosen, including the 18 characterized sequences and 132 randomly chosen sequences. The multiple structural alignment was used to guide the multiple sequence alignment of the whole dataset (150 sequences) using MAFFT and the G-INS-i strategy (iterative refinement, using WSP and consistency scores, of pairwise Needleman–Wunsch global alignments) [37]. Finally, a phylogenetic tree was constructed by setting 1000 bootstrap replicates using the default neighbour-joining method [37, 38] and was visualized with iTOL [39].
Signal peptide detection
Signal peptides were predicted using the SignalP 4.1 server (http://www.cbs.dtu.dk/services/SignalP/) [40].
Analysis of sequence prevalence and abundance in the human gut metagenome
The prevalence and abundance of the GH130 genes in the human gut microbiome were calculated from the abundance table of 9.9 million genes from the human gut metagenomes of 1267 subjects (http://meta.genomics.cn/meta/dataTools [1]).
Chemicals
The reverse phosphorolysis reaction was tested using different acceptors and donors, as follows.
Acceptors: d-glucose (Glc) and d-fructose (Fru) (VWR Chemicals BDH); d-mannose (Man), d-galactose (Gal), N-acetyl-d-glucosamine (GlcNAc), N-acetyl-d-galactosamine (GalNAc), l-arabinose (Ara), l-rhamnose (Rha), d-xylose (Xyl), d-glucouronic acid (GlcA) (Sigma-Aldrich), d-glucosamine (GlcN), N-acetyl-d-mannosamine (ManNAc) and d-galacturonic acid (GalA) (Carbosynth).
Donors: α-d-glucose-1-phosphate (αGlc1P), α-d-mannose-1-phosphate (αMan1P), α-d-galactose-1-phosphate (αGal1P) and α-d-N-acetyl-glucosamine-1-phosphate (αGlcNAc1P) (Sigma Aldrich); β-d-glucose-1-phosphate (βGlc1P) (Carbosynth). β-1,4-mannobiose, β-1,4-mannotriose (Megazyme), β-1,3-mannobiose and β-1,2-mannobiose (Carbosynth) were used as standards. The sodium molybdate reagent and l-ascorbic acid were purchased from Sigma Aldrich.
The hydrolysis reaction was assessed using various pNP-substrates: 4-nitrophenyl α-l-arabinopyranoside; 4-nitrophenyl β-l-arabinopyranoside; 4-nitrophenyl α-d-mannopyranoside; 4-nitrophenyl β-d-mannopyranoside; 4-nitrophenyl α-d-glucopyranoside; 4-nitrophenyl β-d-glucopyranoside; 4-nitrophenyl α-d-galactopyranoside; 4-nitrophenyl β-d-galactopyranoside; 4-nitrophenyl β-d-galactofuranoside; 4-nitrophenyl α-l-fucopyranoside; 4-nitrophenyl β-l-fucopyranoside; 4-nitrophenyl β-d-fucopyranoside; 4-nitrophenyl-β-d-glucopyranosiduronic acid; 4-nitrophenyl β-d-xylopyranoside; and 4-nitrophenyl α-l-rhamnopyranoside (Megazyme).
Enzyme production
The genes encoding U3, U6 and U7 were synthesized by Biomatik Limited (Cambridge, ON, Canada), with optimization of codon usage for expression in Escherichia coli , and cloned in the pET-23a(+) vector, in fusion with N- and C-terminal (His)6 tags, and expressed in the E. coli BL21-DE3 star strain. The E. coli BL21-DE3 star cells transformed with the recombinant plasmids were cultured overnight at 37 °C in 400 ml of the auto-inducible medium ZYM5052 supplemented with 100 µg ml−1 ampicillin. The cells were harvested by centrifugation for 5 min at 5000 r.p.m., before being resuspended and concentrated in 20 mM Tris/HCl, pH 7.0, to obtain a final OD600 nm of 80. They were then lysed by sonication. The cell debris were centrifuged at 13 000 r.p.m. for 10 min and the cytoplasmic extracts were filtered using a 0.20 µm Minisart RC4 syringe filter.
The U1 and U4 enzymes were produced with an N-terminal (His)6 tag by Prozomix (Haltwhistle, Northumberland, UK), and provided in the form of cell-free extract powder.
Enzyme purification
The recombinant E. coli BL21-DE3 star cells were harvested, resuspended in the TALON buffer (20 mM Tris/HCl, pH 7.0, 300 mM NaCl) at OD600 nm 80 and lysed by sonication. The supernatant was collected after centrifugation at 13 000 r.p.m. for 10 min and the cytoplasmic extracts were filtered using a 0.20 µm Minisart RC4 syringe filter. For U1 and U4, the cell-free extract powder was solubilized in the TALON buffer at 10 mg ml−1.
The cytoplasmic extracts in the TALON buffer were subsequently applied to a TALON resin loaded with cobalt (GE Healthcare), washed with 20 mM Tris/HCl, pH 7.0, and 300 mM NaCl, and then eluted with 20 mM Tris/HCl, pH 7.0, 300 mM NaCl and 20 mM imidazole. The purified protein was eluted with 20 mM Tris/HCl, pH 7.0, 300 mM NaCl and 200 mM imidazole. Lastly, the purified protein was desalted using a PD-10 column (GE Healthcare) and recovered in 20 mM Tris/HCl, pH 7.0. The protein concentration was determined by spectrometry using a NanoDrop (Thermo Fisher Scientific, Waltham, MA, USA).
Acceptor and donor specificity screening
All enzymatic reactions were carried out at 37 °C. The molybdenum blue activity assay was used to assess the release of inorganic phosphate during reverse phosphorolysis [41–43]. The reaction mixture was composed of 45 µl of purified enzyme (final concentration 0.03 mg ml−1 for U1 and U3, and 0.0025 mg ml−1 for U7) in 255 µl of a solution containing 20 mM Tris/HCl (pH 7.0), 200 mM sodium molybdate, 10 mM sugar-1-phosphate and 10 mM monosaccharide (final concentrations). At sampling times, 50 µl of the reaction mixture was transferred to a 96-well plate containing 150 µl of a l-ascorbic acid solution 0.24 % (w/v) in 0.1 N HCl per well. After incubation at room temperature for 5 min, 150 µl of stop solution [2 % (v/v) acetic acid and 2 % (w/v) sodium citrate tribasic dihydrate] was added to stop the reaction. The absorbance was measured at 655 nm using a microplate reader (InfiniteM200pro, TECAN). One unit of activity corresponds to the amount of enzyme that catalyzes the production of 1 µmol of inorganic phosphate min−1 in the assay conditions.
Hydrolysis assays
The hydrolytic activity of the purified enzymes was quantified by monitoring the hydrolysis of pNP-glycosides (list above). Assays were performed for 1 h at 37 °C in 96-well microtitre plates. The reaction mixture was composed of 1 mM (final concentration) pNP-glycosides in 140 µl of 20 mM Tris/HCl buffer, pH 7.0, and 40 µl of 1 mg ml−1 purified enzyme. Para-nitrophenol (0 to 1 mM) was used as standard. The reaction was stopped by adding 180 µl of 200 mM Na2CO3. At the initial and final reaction times, the OD was measured at 405 nm with a microplate reader (InfiniteM200pro TECAN).
Mannoside synthesis
The synthesis of manno-oligosaccharides by reverse phosphorolysis was performed over 24 h at 37 °C in a total reaction volume of 100 µl, with 0.1 mg ml−1 purified protein, 10 mM αMan1P and 10 mM acceptor in 20 mM Tris/HCl buffer, pH 7.0. The variation of substrate and product amounts was assessed by high-performance anion exchange chromatography with pulsed amperometric detection (HPAEC-PAD).
HPAEC-PAD analysis
Carbohydrates and αMan1P were separated on a 4×250 mm DionexCarboPack PA100 column. A gradient of sodium acetate (from 0 to 150 mM in 15 min) and an isocratic step of 300 mM sodium acetate in 150 mM NaOH were applied at a flow rate of 1 ml min−1. Detection was performed by using a Dionex ED40 module with a gold working electrode and an Ag/AgCl pH reference.
NMR spectroscopy
All spectra were acquired using a Bruker Advance 500 MHz spectrometer with a 5 mm z-gradient TBI probe at 298 K, and an acquisition frequency of 500.13 MHz for 1H and 125.75 MHz for 13C NMR. The data were processed using TopSpin 3.0 software. The freeze-dried reaction media and standards were resuspended in deuterated water. Deuterium oxide was used as the solvent and the chemical shift scales were internally referenced to sodium 2,2,3,3-tetradeuterio-3-trimethylsilylpropanoate (d4-TSP) for 1H and to the non-deuterated acetone (singlet at 30.89 ppm) for 13C NMR. The various signals were assigned by comparison with signals obtained from d-mannose, d-glucose, αMan1P, β-1,4-mannobiose (Megazyme, Republic of Ireland), and β-1,3-mannobiose (Carbosynth, UK). The usual 2D techniques, such as TOCSY, HSQC and HMBC, were used.
Mass spectrometry
The reverse phosphorolysis reaction performed with U1 was analysed by mass spectrometry (MS) on a Synapt G2 Si HDMS (Waters Corp., Manchester, UK) equipped with a LockSpray Exact Mass Ionization Source (electrospray ion source, ESI). The instrument was operated in positive polarity in the sensitivity mode. The parameters used for electrospray were as follows: capillary voltage: 2.1 kV; sampling cone: 70; desolvation temperature: 120 °C; desolvation gas: 350 l h−1. The tandem MS (MS/MS) measurement was performed in the transfer cell of the triwave setting using an energy of 22. After drying, the sample was solubilized in H2O/MeOH and infused at a flow rate of 5 µl min−1. MS and MS/MS measurements were recorded for 30 s. Data were acquired using MassLynx 4.1 and processed using Mmass 5.5.0 [44]. The MS/MS spectrum was annotated according to the nomenclature of Domon and Costello [45].
Results
GH130 sequence mining in mammal gut microbiomes
In order to extend the GH130 diversity from the genomic to the metagenomic sequence space, 4 mammal gut metagenomes, totaling 33 962 473 genes, were searched for sequences containing at least 1 GH130 module. In total, 4714 GH130 sequences were retrieved from the human, mouse, pig and bovine gut metagenomes (1205, 467, 681 and 2361 sequences, respectively) (Table 1). The relative numbers of GH130 genes in these four datasets are of the same order of magnitude, varying from 6×10−3 to 26×10−3 % of the genes in each metagenome. The proportion of sequences that lack N- and/or C-terminal ends varies between 31–66 %. Logically, the larger is the N50 assembled contig length in the metagenomic dataset and the smaller is the percentage of fragmentary sequences. Most of the metagenomic sequences contain a single catalytic module. Only 19 combine a GH130 module with a GH2, GH43, GH92, GH127 or a GH142 module (Table S2). This very low number of multi-modular sequences is probably not due to gene fragmentation in the metagenomic datasets, since only 6 of the 1592 sequences listed in the CAZy database as of November 2018 are multi-modular (GH130–GT81 and GH130–GH130 modular combinations). None of the CAZy or metagenomic sequences contains a CBM domain. Of the 4714 metagenomic sequences, only 0.7 % (31 sequences) are fully identical to sequences already listed in the CAZy database. Among the metagenomic sequences, only 0.5 % (24 sequences) are identical. With a threshold of 90 % sequence identity, sequence redundancy between each metagenome and the CAZy database does not exceed 10.7 %. Between the metagenomes, it reaches 51.5 %, highlighting the similarities between mammal gut microbiomes regarding these mannoside-metabolizing enzymes (Table 1).
Table 1.
Diversity of metagenomic GH130 sequences in mammal gut metagenomes
|
Metagenome |
No. of genes |
N50 average contig length (kb) |
No. of GH130 sequences |
% of GH130 sequences |
No. of full-length GH130 sequences |
No. of multi-modular GH130 sequences |
No. of redundant GH130 sequences with those of the CAZy database |
|
|---|---|---|---|---|---|---|---|---|
|
100 % identity |
≥90 % identity |
|||||||
|
Human |
9 878 647 |
5.0 |
1205 |
12×10−3 |
676 (56 %) |
3 |
22 |
210 |
|
Mouse |
7 685 872 |
6.6 |
467 |
6×10−3 |
324 (69 %) |
0 |
7 |
53 |
|
Pig |
2 572 074 |
1.87 |
681 |
26×10−3 |
298 (44 %) |
0 |
1 |
62 |
|
Bovine |
13 825 880 |
0.91 |
2361 |
17×10−3 |
800 (34 %) |
16 |
1 |
180 |
|
Total |
33 962 473 |
– |
4714 |
– |
2098 |
19 |
31 |
505 |
|
Redundancy between metagenomes |
24 |
2426 |
||||||
Sequence diversity in the GH130 family
To analyse sequence diversity in the GH130 family, we constructed SSNs with the 1592 sequences from the CAZy database, plus 2 sequences corresponding to recently characterized GPs, and the 4714 metagenomic sequences. The low number of identical sequences does not affect clusterization, since they appear in the same nodes. We also checked that the presence of the four multi-modular GH130–GH130 sequences in our dataset (Table S2) did not create erroneous edges between otherwise distinct meta-nodes. Taken independently, the sequences of each of the modules do indeed gather in the same meta-node. Different E-value thresholds (10−30 and 10−90) were tested to construct the SSNs (Figs 1 and S1). For each SSN, we mapped the known functions of the 18 characterized GH130 sequences on the meta-nodes obtained. We defined isofunctional clusters as meta-nodes containing characterized members with the same osidic linkage and acceptor specificities. The optimal E-value threshold was defined as the highest threshold leading to the separation of the sequences into the maximum number of isofunctional meta-nodes. Optimal clusterization was obtained by setting an E-value threshold of 10−70, corresponding to 63 % sequence identity (Fig. 1).
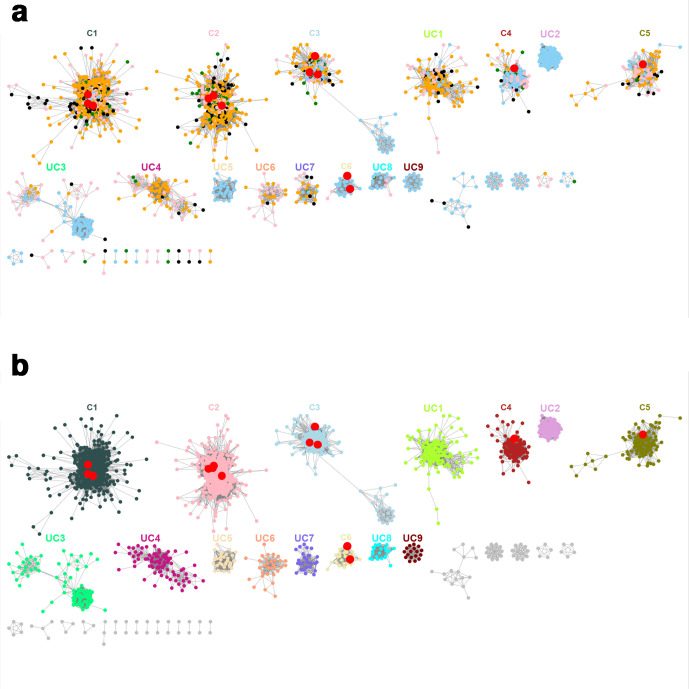
Fig. 1.
Protein SSN of the 6308 GH130 sequences from the CAZy database and from gut metagenomes, with an E-value threshold of 10−70. Each of the 3637 nodes contains sequences with more than 70 % identity. Nodes are connected by an edge when the pairwise sequence identity is over 63 %. In this figure, the 694 sequences appearing in 641 singletons have been omitted. Red nodes contain biochemically characterized members, belonging to meta-nodes C1 to C6. The meta-nodes containing at least 20 sequences and with no characterized members are labelled as UC1 to UC9. (a) Nodes coloured according to the origin of the sequences: sky blue, CAZy database; pink, human gut metagenome; green, mouse gut metagenome; black, pig gut metagenome; orange, bovine rumen metagenome. (b) Nodes coloured according to the meta-node they belong to.
In this optimal configuration, we obtained five isofunctional meta-nodes (meta-nodes C1 and C3 to C6, Fig. 2). The characterized GPs targeting β-1,2-mannosides appear in two different meta-nodes (C4 and C6). This result cannot be explained by differences of specificity towards β-1,2-mannosides of various chain lengths, since meta-node C6 contains both a β-1,2-mannobiose phosphorylase and a β-1,2-oligomannan phosphorylase. Conversely, the GPs acting on Man-β-1,4-GlcNAc and on β-1,4-mannosides (sequences belonging to the former GH130_2 subfamily) are grouped in the same meta-node (C2), apart from the those of the meta-node C1 targeting Man-β-1,4-Glc (former GH130_1 subfamily). It was not possible to obtain an SSN representation grouping GPs that act on β-1,2-mannosides into one single meta-node while splitting them out from those acting on β-1,3-mannosides (meta-node C5, Figs 2 and S1). In addition, even at the lowest E-value tested (10−90), the characterized sequences targeting Man-β-1,4-GlcNAc and β-1,4-mannosides are still grouped in the same meta-node. This may be due to the high promiscuity towards acceptors of these enzymes, as demonstrated for the RaMP2, BT1033 and UhgbMP enzymes [14, 19, 20].
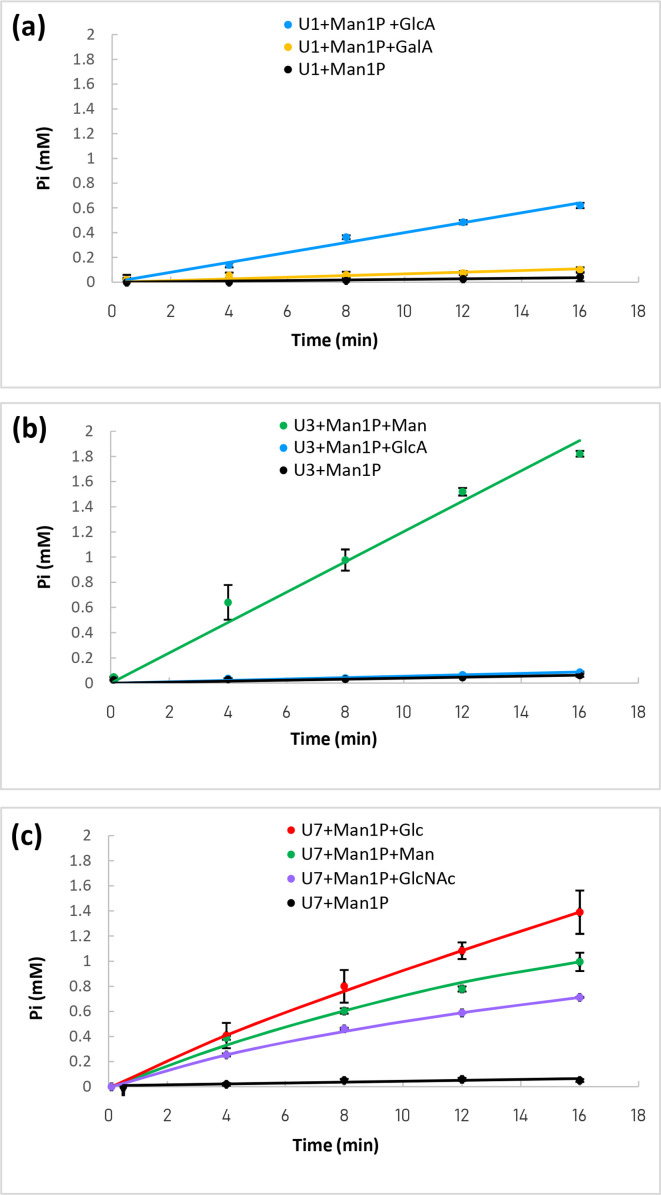
Fig. 2.
Determination of the acceptor specificity of the purified enzymes U1 (a), U3 (b) and U7 (c). The reactions were performed using 10 mM of αMan1P (glycosyl donor) and 10 mM of acceptor. The release of inorganic phosphate was followed using the molybdenum blue activity assay. Reactions were performed in triplicate. When no reverse phosphorolysis activity was detected (similar inorganic phosphate release rate to that in the presence of αMan1P as sole substrate), the data do not appear in this figure.
The optimal SSN contains 15 meta-nodes containing more than 20 sequences each; this is 1 of the criteria used to define subfamilies in the CAZy database [28]. Six meta-nodes contained characterized enzymes (meta-nodes C1 to C6). The other nine meta-nodes (UC1 to UC9) did not contain characterized sequences and were therefore investigated for their functions. In addition to these meta-nodes, the SSN also contains 20 meta-nodes with fewer than 20 sequences, and 641 singletons sharing less than 63 % identity with the sequences in meta-nodes. These small meta-nodes and singletons contain 798 sequences, which correspond to 12.7 % of the 6308 sequences used to construct the SSN. They correspond to small sequences of less than 292 amino acids in size, which is the length of the shortest full-length GH130 sequence indexed in the CAZy database (CEP78012.1). These short sequences appearing as singletons represent the majority (94%) of the incomplete metagenomic sequences. The longest sequences correspond to the few multi-modular proteins, which are distributed in different meta-nodes, in particular C3 and UC4 (Tables S1 and S2).
Prediction of mechanism
In order to support the functional assignment of meta-nodes containing characterized proteins, and to predict the function of the members of the uncharacterized SSN meta-nodes, we predicted the mechanism of osidic linkage breakdown (phosphorolysis or hydrolysis) for all the meta-nodes, based on three criteria. The first one was the presence or absence of a signal peptide in the sequences. Because GPs need intracellular phosphate to perform phosphorolysis, we assumed that they were all cytoplasmic enzymes. To test this hypothesis, we predicted the presence or absence of a signal peptide in all the sequences of characterized GPs from the GH3, GH13_18, GH65, GH94, GH112, GH130, GH149, GH161, GT4, GT35 and GT108 families, using the SignalP 4.1 server. No signal peptide was detected. In addition, none of the sequences from the GH130 SSN meta-nodes containing characterized GPs was predicted to have a signal peptide (Table 2). Conversely, the meta-node containing the characterized GHs (β-1,2-mannosidases) contained sequences with signal peptides (for example, two of the three characterized members). Based on this finding, we considered that the meta-nodes containing sequences with a predicted signal peptide do not include GPs.
Table 2.
Conservation of key residues in the sequences of each SSN meta-node. The proven activities are rendered in bold type. The presence of sequences with a signal peptide in each meta-node is also indicated. The reference sequences for mannoside-phosphorylases and mannosidases are ADD61463.1 [β-1,4-mannosyl-N,N’-diacetylchitobiose-phosphorylase (20)] and AAO78885.1 [β-1,2-mannosidase (12)], respectively. Both enzymes are functionally and structurally characterized. In meta-node C3, a signal peptide was detected for the characterized members ALJ48509.1 and ACT94389.1, but not for AAO78885.1
|
Meta-node |
Number of full-length sequences CAZy/metagenomes/total |
Sequences with signal peptide |
Catalytic residues |
Phosphate-binding residues |
+1 subsite |
Predicted mechanism and linkage specificity |
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
Proton donor |
Putative base |
|||||||||||
|
C1 |
221/836/1057 |
No |
D |
_ |
R |
H |
R |
Y |
R |
D |
GPs-β-1–4 |
|
|
C2 |
268/757/1025 |
No |
D D104 |
_ |
R R168 |
H H231 |
R R59 |
Y Y103 |
R R150 |
D D304 |
GPs-β-1–4 |
|
|
C3 |
113/152/265 |
Yes |
D D142 |
D/E E227 |
_ |
_ |
R R89 |
E E141 |
K K199 |
D D363 |
GHs-β-1–2 |
|
|
C4 |
290/76/366 |
295/366 |
No |
D |
_ |
R |
H |
R |
E |
K |
D |
GPs-β-1–2 |
|
70/366 |
R |
E |
R |
D |
GPs-β-1–3 |
|||||||
|
C5 |
42/70/112 |
No |
D |
_ |
R |
H |
R |
E |
R |
D |
GPs-β-1–3 |
|
|
C6 |
85/0/85 |
No |
D |
_ |
R |
H |
R |
E |
K |
D |
GPs-β-1–2 |
|
|
UC1 |
31/66/97 |
No |
D |
_ |
R |
H |
R |
Y |
R |
D |
GPs-β-1–4 |
|
|
UC2 |
199/0/199 |
No |
D |
_ |
R |
H |
R |
E |
K |
D |
GPs-β-1–2 |
|
|
UC3 |
135/18/153 |
124/153 |
No |
D |
_ |
R |
H |
R/D |
E |
K |
D |
GPs-β-1–2 |
|
7/153 |
R/D |
E |
R |
D |
GPs-β-1–3 |
|||||||
|
14/153 |
R/D |
Y |
R |
GPs-β-1–4 |
||||||||
|
UC4 |
0/53/53 |
Yes |
_ |
_ |
_ |
_ |
S |
M |
N |
_ |
GHs |
|
|
UC5 |
61/0/61 |
No |
D |
_ |
R |
H |
R |
E |
K |
D |
GPs-β-1–2 |
|
|
UC6 |
0/11/11 |
No |
D |
_ |
R |
H |
R |
I |
R |
D |
GPs |
|
|
UC7 |
9/10/19 |
No |
D |
_ |
R |
H |
R |
E |
R |
D |
GPs-β-1–3 |
|
|
UC8 |
26/7/33 |
No |
D |
_ |
R |
H |
R |
E |
K |
D |
GPs-β-1–2 |
|
|
UC9 |
29/0/29 |
No |
D |
_ |
R |
H |
R |
E |
K |
D |
GPs-β-1–2 |
|
The second criterion was the conservation of the catalytic machinery. As previously shown for GH130 enzymes [12, 20], inverting GPs indeed require a single catalytic amino acid to perform the catalytic event, with phosphate playing the role of a base. Conversely, inverting GHs require the presence of an additional carboxylic amino acid in their catalytic site acting as a base to catalyze the hydrolytic reaction. We thus examined the conservation of the catalytic proton donor aspartate (D), and searched for D or glutamate (E) residues that may act as a base by aligning the non-truncated sequences of each meta-node with those of the 18 phosphorylases and hydrolases characterized previously. The sequences ADD61463.1 and AAO78885.1 were chosen as references for amino acid numbering (Table 2). We assigned the GP activity to the meta-nodes where more than 95 % (to take into account the rare shifts in the automatic multiple sequence alignments) of sequences included the putative catalytic proton donor but lacking the base, and the GH activity to those where more than 95 % of sequences contained the two putative catalytic residues.
The final criterion was the conservation of the phosphate-binding residues in GP-encoding sequences. By examining the multiple alignments in each meta-node, we assigned the GP activity to the meta-nodes where more than 90 % of sequences included the arginine/histidine (R/H) pair described as being involved in Pi binding [36]. Conversely, all the sequences clustering with the characterized GHs lack the phosphate-binding residues. An overview of this analysis is given in Fig. S2, highlighting key regions in the multiple sequence alignment, such as proton donor, putative base,+1 subsite and Pi-binding site.
Based on the three criteria, we assigned all UC meta-nodes except UC4 to GPs. It is noted that meta-node UC3 has 15 sequences (<10 %), where the phosphate-binding residue H is replaced by tyrosine (Y). This variation in phosphate-binding residues has already been described in other GP-containing families, such as members of the family GH13_18 [46]. In the sequences belonging to the meta-node UC4, the catalytic and phosphate-binding residues are not conserved, but the presence of a signal peptide in some of these sequences suggests that they could be hydrolases.
Prediction of linkage specificity
In order to predict the linkage specificity of the enzymes assigned to uncharacterized meta-nodes, we studied the conservation of the residues involved in the recognition and accommodation of the acceptor at the +1 subsite (R59, Y103, R150, D304 residues, ADD61463.1 numbering, Table 2), based on the functional and structural knowledge of the previously characterized GH130 enzymes [12, 16, 22, 35, 36]. Firstly, the UC4 sequences do not contain any of these conserved residues. As they also lack the GH130 catalytic residues, we consider these sequences to be very distant from the other GH130 enzymes, which correlates with the fact that this meta-node was the first to separate from the other large meta-nodes in the SSN constructed with an E-value threshold of 10−30, corresponding to 32 % sequence identity (Fig. S1). This was confirmed by constructing a phylogenetic tree representing the distances between 10 sequences of each meta-node (Fig. S3).
In all the characterized meta-nodes and in UC1, UC3, UC7 and UC8, residue R59 is more than 98 % conserved, regardless of the linkage specificity of the enzymes. The Y103 residue is also conserved in more than 98 % of the sequences of the two meta-nodes containing characterized members targeting β-1,4-mannosides (C1 and C2), but is replaced by an E in more than 99 % of sequences of the three meta-nodes with members targeting β-1,2-mannosides (with a gap at this position for two sequences). An E is also observed in more than 80 % of the sequences of the C5 meta-node, otherwise it is a glutamine (Q). These isosteric Q and E residues thus may have the same function, although it is not yet proven. The residue at position Y103 thus seems to be involved in linkage specificity, although it is not its only determinant. Residue R150 is conserved in 99 % of the sequences of the meta-nodes targeting β-1,4 and β-1,3-mannosides. This residue is also strictly conserved in uncharacterized meta-nodes UC1, UC6 and UC7, and in 18 % of the UC3 sequences. It is replaced by a lysine (K) in 90 % of sequences of the meta-nodes targeting β-1,2-mannosides, with the remaining 10%, which belong to C4, having an R. A K is also present in meta-nodes UC2, UC5, UC8 and UC9, and in 82 % of UC3. It was previously proposed that this K residue confers β-1,2 linkage specificity to GH130 enzymes [12]. Lastly, D304 is conserved in more than 99 % of the sequences of all meta-nodes except UC4, and is thus not involved in linkage specificity.
Based on these results, we identified the pairs of residues corresponding to the residues Y103 and R150 in ADD61463.1 as the determinants of linkage specificity. The Y/R pair is conserved in all characterized GH130s that target β-1,4-mannosides, the E/K pair in all those that target β-1,2-mannosides, and the E/R pair in all those that target β-1,3-mannosides (+1 subsite region in Fig. S2). The conservation of this pair of residues was then used to predict the linkage specificity of the uncharacterized meta-nodes. Meta-node UC1 would thus target β-1,4-linkages, meta-node UC7 β-1,3 linkages and meta-nodes UC2, UC5, UC8 and UC9 the β-1,2 linkages. In meta-node UC3, again, 81 % of the sequences contain the E/K pair, and would thus target β-1,2 linkages, while those containing the E/R pair would target β-1,3 linkages and those containing the Y/R pair would target β-1,4 linkages. The prediction of linkage specificity for the UC6 sequences is not possible, since they have the conserved R150, but lack the Y103 or E, which is replaced by an isoleucine (I) in 10 of the 11 sequences and by a methionine (M) in the remaining 1. We thus propose that they may feature a new linkage specificity in the GH130 family.
Biochemical characterization of sequences from uncharacterized SSN meta-nodes
In order to experimentally validate our predictions regarding the mechanism and linkage specificity of the enzymes of the uncharacterized meta-nodes, and to discover potential new functions in the GH130 family, we selected one sequence to characterize in each meta-node containing at least 20 metagenomic sequences.
We thus selected five human gut metagenomic sequences (the most prevalent and abundant ones in the human gut microbiome) from meta-nodes UC1, UC3, UC4, UC6 and UC7, and named them U1, U3, U4, U6 and U7, respectively (Table S4). The remaining meta-nodes UC2, UC5, UC8 and UC9 contain no or only a few metagenomic sequences. The chosen sequences are very different from those of the 18 GH130 enzymes already characterized, with less than 40 % identity (Table S3). The target genes were synthesized and expressed in E. coli . Unfortunately, U6 was insoluble, and could not be characterized. The U1, U3, U4 and U7 enzymes were produced in soluble form and were further characterized.
Based on our mechanistic prediction, the members of meta-nodes UC1, UC3 and UC7 are probably phosphorylases, while the members of meta-node UC4 are expected to be hydrolases (although no signal peptide was predicted in the U4 sequence). In order to test these hypotheses, we screened for GP activity by means of an assay based on the measurement of released inorganic phosphate [42] in the direction of glycoside synthesis, using purified enzymes (Fig. 2). We first tested αMan1P as donor, and 11 different monosaccharides as acceptors, to detect phosphate release and identify the best acceptor for the validated GPs. Then, using the best acceptor, we assessed promiscuity towards glycosyl donors by comparing the phosphate release rates from αMan1P, αGlc1P, αGal1P, αGlcNAc1P and βGlc1P. The release of phosphate from αMan1P was proven for the U1, U3 and U7 enzymes (Fig. 2) in the presence of different acceptors. No other donor was identified for the synthesis reaction, indicating that these enzymes are strict mannoside-phosphorylases. In the assay conditions, U1’s best acceptor was GlcA (specific activity 1300±40 U g−1). A slight reverse phosphorolysis activity was also detected with GalA (specific activity 270±80 U g−1). U3’s best acceptor was Man (specific activity 3700±130 U g−1). It also reacted, very weakly, with GlcA (specific activity 130±20 U g−1). In contrast, U7 is a promiscuous enzyme, which is able to efficiently accommodate Glc (specific activity 46,480±4030 U g−1), but also, to a lesser extent, Man (specific activity 26 700±860 U g−1) and GlcNAc (specific activity 18 620±100 U g−1). Without acceptor, U1, U3 and U7 also displayed a slight activity ofαMan1P hydrolysis (specific activities 80±60, 100±50 and 440±80 U g−1, respectively), a side reaction already described for mannoside-phosphorylases [20]. No reverse phosphorolysis activity was detected with the U4 target, regardless of the acceptor tested. In case this enzyme might accommodate another sugar phosphate, we also tested it in the presence of various donors and Man, with this monosaccharide being the best acceptor for most characterized GH130 GPs. No reverse phosphorolysis activity was detected, indicating that U4 is probably not a GP, but rather, as hypothesized, a glycosidase. We further tested the ability of U4 to hydrolyzeβ-1,2/1,3 and 1,4-mannobiose and various pNP-glycosides, but no substrate consumption was observed by HPAEC-PAD analysis of the reaction media, or by the chromogenic assays performed with pNP-glycosides (data not shown).
To confirm the acceptor and donor specificity of U1, U3 and U7, and to determine the degree of polymerization of their products and their linkage specificity, the enzymes were purified and the glycoside synthesis reaction media were analysed by HPAEC-PAD, and 1H / 13C NMR or MS, as appropriate.
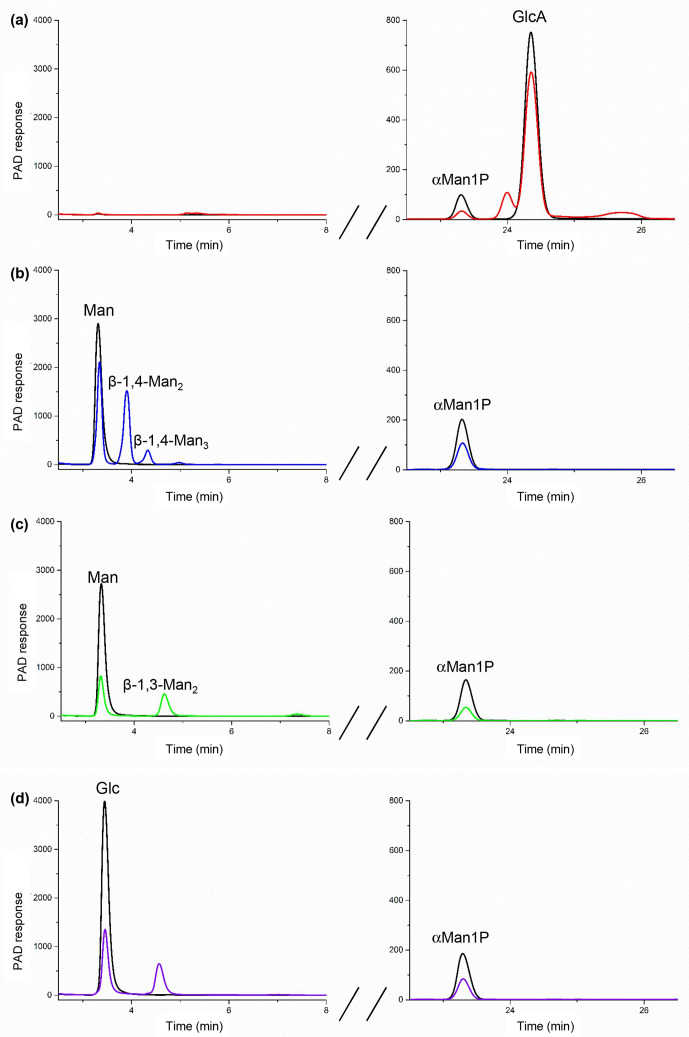
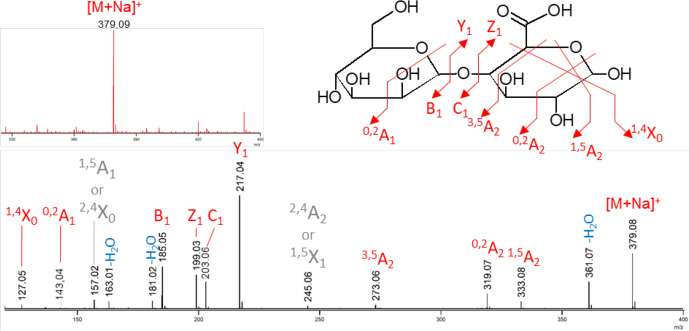
For U1 in the presence of αMan1P and GlcA, a 30 % decrease in substrates concentrations was observed on the HPAEC-PAD chromatograms after reaction, confirming that reverse-phosphorolysis occurred (Fig. 3a). MS analysis was then performed in order to elucidate the structure of the glycoside produced from αMan1P and GlcA. The ESI MS measurement in positive ionization mode highlighted a species at m/z 379.09 (Fig. 4, red spectrum). This ion corresponds to the [M+Na]+ of a disaccharide composed of a hexose and a uronic acid. The structural characterization of this disaccharide was performed by MS/MS. The intracyclic fragments 0,2A1 and 1,4X0 confirmed that the uronic acid is at the reducing end of the disaccharide and the 3,5A2 fragment proves that, as predicted, the linkage between the two subunits is of type 1,4 (Fig. 4). Since all the characterized GH130 enzymes share an inverting mechanism, we deduced from these results that the compound produced by U1 from αMan1P and GlcA is Man-β-1,4-GlcA.
Fig. 3.
HPAEC-PAD analysis of the reverse phosphorolysis reactional mixtures after incubation for 0 min (in black) and 24 h (in colour) with U1 (a), U3 (b) and U7 (c, d). The reactions were performed in the presence of 10 mM of αMan1P (glycosyl donor) and 10 mM of GlcA, Glc or Man (acceptors). Only the peaks at the retention times corresponding to commercial standards are labelled.
Fig. 4.
Mass spectrometry measurements of the U1 reverse phosphorolysis reactional mixture (substrates: 10 mM αMan1P and 10 mM glucuronic acid) in positive ionization mode. The upper spectrum in red corresponds to the ESI MS measurement. The lower spectrum in black corresponds to the ESI MS/MS of the precursor ion isolated as a [M+Na]+ at m/z 379.1. The detailed structure is shown with the specific fragments. Details of the annotation: red, unambiguous fragments; grey, ambiguous fragments (not reported on the corresponding structure); blue, water losses.
The HPAEC-PAD chromatogram of the U3 reaction in the presence of αMan1P and Man indicated that a disaccharide and a trisaccharide were produced from these substrates (Fig. 3b). Their retention times correspond to those of the β-1,4-mannobiose and β-1,4-mannotriose standards. The β-1,4 linkage specificity of U3 was confirmed by 1H NMR (Fig. S4). The signals of the anomeric protons (H1α, H1β, H1′) were assigned by comparison with signals obtained from β-1,4-mannobiose standard. This validates our prediction that U3 harbours the Y/R pair responsible for β-1,4 linkage specificity.
The U7 synthesis reactions were performed with αMan1P and Man or Glc acceptors. In both cases, a single product was observed on the HPAEC-PAD chromatograms (Fig. 3c, d), and its retention times did not correspond to that of β-1,4-mannobiose. By comparing the anomeric region in 1H NMR with standards, we proved that β-1,3-mannobiose was produced from Man as an acceptor. For the reaction performed in the presence of Glc as an acceptor, for which there is no available commercial standard, the 1H and 13C NMR (1D and 2D) analyses of the reaction medium proved that U7 produced the Man-β-1,3-Glc disaccharide (Fig. S4). This linkage specificity confirms our prediction for meta-node UC7′ sequences. The traces of free Man detected by 1H NMR in this reaction medium result from αMan1P hydrolysis.
Discussion
Use of SSNs to analyse the functional diversity of mono-modular CAZymes, and to predict GP functions
The efficiency of SSNs for rapid clustering of a large number of sequences has already been shown [27], and was recently applied to the creation of subfamilies within the large multi-functional CAZy family GH16 [28]. In the present paper, we describe the use of SSNs to: (i) analyse the functional diversity of CAZyme encoding metagenomic sequences, whose role in microbial ecosystems had never been investigated; (ii) identify new CAZyme functions.
Here, we have targeted a family that mainly contains putative glycoside phosphorylases in order to extend our knowledge of their structure–function relationships, to investigate their physiological functions, and because they are biotechnological tools of interest for glycoside synthesis. Mannoside-phosphorylases of the family GH130 are intracellular enzymes described as targeting, in cellulo, only di- or trisaccharides [36]. The breakdown of such simple carbohydrate structures does not require complementary catalytic modules or carbohydrate-binding modules, which are found to bind and depolymerize complex glycans in numerous cell surface-associated GHs. Thus, more than 99 % of the sequences proven or predicted in this study to encode mannoside-phosphorylases are monomodular. This specific feature means that GH130 sequences can be analysed directly with the EFI-EST web tool, without isolating the target catalytic domain, contrary to what has been done in respect of the often multi-modular GH16 sequences [28].
Despite these advantages, the SSN analysis presents two limitations. Firstly, as explained by Viborg et al. in 2019 [28], it cannot be used to establish evolutionary relationships between functional meta-nodes. The formation of stable subfamilies, whose boundaries will not evolve at the rate of the rapid increase of the number of genomic and metagenomic sequences, requires the use of complementary approaches, such as phylogenetic and hidden Markov model analyses. Secondly, as previously shown for the GH16 family and confirmed here with GH130s, SSNs do not sufficiently differentiate sequences with local structural differences, which can, however, have major implications in terms of enzyme specificity and mechanism. Regardless of the E-value threshold (10−30 to 10−90) used, it was impossible to cluster GH130 sequences according to linkage specificity (β-1,2, 1,3 or 1,4) or acceptor specificity (Figs 2 and S1). We thus had to devise another strategy to identify potential GPs and predict their specificity. This generic approach could be used regardless of CAZy family. The first stage is to eliminate hydrolases by excluding the meta-nodes that contain sequences with (i) signal peptides and (ii) conserved carboxylic acid residues that could act as bases in inverting hydrolases. This second criterion can only be used for inverting families (representing 7 of the 11 GP-containing families), because, like GHs, most of the retaining GPs involve a catalytic machinery that contains both an acid/base and a nucleophile. Another criterion that could be used for easy discrimination of hydrolases from phosphorylases could be to eliminate sequences containing CBMs, as it is indeed unlikely that intracellular enzymes acting on oligosaccharides possess CBMs. This hypothesis was not tested here, as none of the GH130 sequences contains a CBM. The second stage aims to predict linkage specificity. For the first time, thanks to the increasing number of crystallographic structures available in the GH130 family, we found that the linkage specificity of GH130 members, be they phosphorylases or hydrolases, is linked to the presence of a specific pair of amino acids located in the +1 subsite. These predictions were biochemically validated with sequences belonging to three meta-nodes, and allowed us to isolate, from thousands of GH130 sequences, GPs that catalyse a reaction never described before, namely the synthesis and degradation of Man-β-1,4-GlcA by the U1 enzyme. The reliability of these predictions remains to be validated for the members of the five meta-nodes that do not yet include any characterized GP member. Sequences of meta-node UC6 are of particular interest, since multiple sequence alignments show that the amino acids conferring specificity towards β-1,2/1,3/1,4 linkages are not conserved. These enzymes may target β-1,6-linked mannosyl residues, since heteromannoside structures containing this motif do exist in fungal polysaccharides, such as in Lentinus edodes [47] and in bacterial exopolysaccharides, in particular those of the enterobacterium Edwardsiella tarda [48] (Fig. S5).
GH130 enzymes ensure diverse ecological functions, targeting mammal, plant, mould, yeast and bacterial mannosides
The SSN analysis presented here allowed us to represent the diversity of GH130 enzymes in mammalian gut microbiomes. A large number of metagenomic sequences within a SSN meta-node indicates their dissemination in numerous bacteria, and thus, that the function might play a role in habitat colonization by these species. The largest meta-nodes by far are C1 and C2, with 36 and 31 % of the metagenomic sequences, respectively. These meta-nodes contain the enzymes that target plant cell wall mannans and N-glycans. This is not surprising, given the abundance of plant-derived fibres in the human, bovine, pig and mouse diet, and the fact that the gut microbes are in close contact with mammal cells harbouring N-glycans.
The meta-nodes containing the sequences shown or predicted to be involved in the degradation of β-1,2-mannosides, by either phosphorolysis or hydrolysis, are much more restricted, amounting to 8 % of the metagenomic sequences. In these meta-nodes, the only characterized member from a mammal gut bacterium has been shown to target Candida albicans mannan [12]. However, fungal mannosides are probably not the only targets of the enzymes from these eight meta-nodes, since several other yeast [49, 50], mould [47] and bacterial heteromannosides [51, 52] harbour β-1,2-linked mannosyl residues (Fig. S5). Furthermore, a fungal GH130 sequence is contained in meta-node UC9 (Fig. S6). These fungal enzymes may be involved in mannoside recycling, via the release of αMan1P, which in turn could be transformed by pyrophosphorylases into GDP-mannose, which would further be used as a GT substrate for heteromannan biosynthesis.
The UC1, UC7 and C5 meta-nodes are the final SSN meta-nodes for which we inferred a function from the analyses described here. Together, they only represent a few hundred of the sequences in mammal gut microbiomes. These meta-nodes also contain a few dozen genomic sequences from various bacterial phyla, including Firmicutes , Bacteroidetes and Actinobacteria , which are highly abundant in the gut microbiota (Fig. S6, Table S1). The UC1 meta-node, the U1 member of which was shown in this study to target the Man-β-1,4-GlcA disaccharide, only represents 4 % of our metagenomic dataset. The Man-β-1,4-GlcA motif is described in Nicotiana plumbaginifolia [53], but this tobacco plant species is not a dietary constituent for mammals. This motif also exists in bacterial lipopolysaccharides (Fig. S5), in particular the O-antigen polysaccharide of Shigella boydii [54], one of the four Shigella species considered to be major enteropathogens causing childhood diarrhoea worldwide [55]. The UC7 and the C5 meta-nodes, which target β-1,3-linked mannosides, together represent just 3 % of the metagenomic dataset. These two meta-nodes each contain promiscuous GPs (the U7 and zobellia_231 enzymes [23]), of which the substrates are the Man-β-1,3-Glc and Man-β-1,3-Man disaccharides. These motifs are found in some moulds [47, 56], and also in the lipopolysaccharides of certain pathogenic bacterial species, such as Pseudomonas aeruginosa (Fig. S5) [57]. Thanks to their ability to produce these particular oligosaccharides through reverse phosphorolysis, the U1 and U7 GPs characterized in this study are tools of interest for the synthesis of antigenic oligosaccharides.
In this study, we thus revisited the functional diversity of the GH130 CAZy family by integrating that from mammalian gut microbiomes. By combining a series of in silico and in vitro approaches, we predicted that the vast majority of the GH130 enzymes are glycoside-phosphorylases and not hydrolases. We identified that some of them are specific for oligosaccharidic motifs found in bacterial lipopolysaccharides, in particular those of certain pathogenic species. Co-culture and transcriptomic studies targeting the commensals that produce these enzymes will be needed to confirm the role of mannoside-phosphorylases in bacterial interactions.
Supplementary Data
Funding information
The authors received no specific grant from any funding agency.
Acknowledgements
The enzyme characterization was carried out using the equipment of the ICEO enzyme screening facility. ICEO is supported by grants from the Région Midi-Pyrénées, the European Regional Development Fund and France’s Institut National de la Recherche Agronomique (INRA). This research was funded by the European Union's Horizon 2020 framework programme (LEIT‐BIO‐2015‐685474, Metafluidics) and by the Agence Nationale de la Recherche (ANR-16-CE20-0006, Oligomet). A. L. is supported by INSA Toulouse and China Scholarship Council under the ‘CSC – UT/INSA Program’. We cordially thank Simon Charnock and Darren Cook (Prozomix) for their support with enzyme production.
Author contributions
Conceptualization, G.P.V.; Methodology, A.L., E.L., J.E., V.L.; Investigation, A.L., L.T., V.L., D.G., D.R., E.L., N.T., J.D., D.M., B.H.; Writing–Original Draft, A.L., E.L.; Writing – Review and Editing, G.P.V.; Funding Acquisition, G.P.V., A.L. ; Resources, G.P.V., D.R., B.H.; Supervision, G.P.V. and E.L.
Conflicts of interest
The authors declare that there are no conflicts of interest.
Footnotes
Abbreviations: Ara, L-arabinose; C, characterized meta-node; CAZyme, carbohydrate active enzyme; D, aspartic acid; E, glutamic acid; Fru, D-fructose; Gal, D-galactose; GalA, D-galacturonic acid; GalNAc, N-acetyl-D-galactosamine; αGal1P, α-D-galactose-1-phosphate; GH130, glycoside hydrolase family 130; GH, glycoside hydrolase; Glc, D-glucose; GlcA, D-glucouronic acid; GlcN, D-glucosamine; GlcNAc, N-acetyl-D-glucosamine; αGlcNAc1P, α-D-N-acetyl-glucosamine-1-phosphate; βGlc1P, β-D-glucose-1-phosphate; αGlc1P, α-D-glucose-1-phosphate; GP, glycoside phosphorylase; GT, glycosyltransferase; H, histidine; HCl, hydrochloric acid; HPAEC-PAD, high performance anion exchange chromatography with pulsed amperometric detection; K, lysine; Man, D-mannose; ManNAc, N-acetyl-D-mannosamine; αMan1P, α-D-mannose-1-phosphate; NaOH, sodium hydroxide; PDB, Protein Data Bank; Pi, inorganic phosphate; Q, glutamine; R, arginine; Rha, L-rhamnose; SSN, sequence similarity network; UC, uncharacterized meta-node; Xyl, D-xylose; Y, tyrosine.
All supporting data, code and protocols have been provided within the article or through supplementary data files. Six supplementary figures and four supplementary tables are available with the online version of this article.
References
- 1.Li J, Jia H, Cai X, Zhong H, Feng Q, et al. An integrated catalog of reference genes in the human gut microbiome. Nat Biotechnol. 2014;32:834–841. doi: 10.1038/nbt.2942. [DOI] [PubMed] [Google Scholar]
- 2.Xiao L, Feng Q, Liang S, Sonne SB, Xia Z, et al. A catalog of the mouse gut metagenome. Nat Biotechnol. 2015;33:1103–1108. doi: 10.1038/nbt.3353. [DOI] [PubMed] [Google Scholar]
- 3.Xiao L, Estellé J, Kiilerich P, Ramayo-Caldas Y, Xia Z, et al. A reference gene catalogue of the pig gut microbiome. Nat Microbiol. 2016;1:1–6. doi: 10.1038/nmicrobiol.2016.161. [DOI] [PubMed] [Google Scholar]
- 4.Li J, Zhong H, Ramayo-Caldas Y, Terrapon N, Lombard V, et al. A catalog of microbial genes from the bovine rumen unveils a specialized and diverse biomass-degrading environment. Gigascience. 2020;9 doi: 10.1093/gigascience/giaa057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brett CT, Waldron KW. Physiology and Biochemistry of Plant Cell Walls. Springer; 1996. p. 282. p. [Google Scholar]
- 6.Sharma V, Ichikawa M, Freeze HH. Mannose metabolism: more than meets the eye. Biochem Biophys Res Commun. 2014;453:220–228. doi: 10.1016/j.bbrc.2014.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zeleznick LD, Rosen SM, Saltmarsh-Andrew M, Osborn MJ, Horecker BL. Biosynthesis of bacterial lipopolysaccharide, IV. Enzymatic incorporation of mannose, rhamnose, and galactose in a mutant strain of Salmonella typhimurium. Proc Natl Acad Sci U S A. 1965;53:207–214. doi: 10.1073/pnas.53.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang K, Beverley SM. Mannogen-ing central carbon metabolism by Leishmania. Trends Parasitol. 2019;35:947–949. doi: 10.1016/j.pt.2019.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lombard V, Golaconda Ramulu H, Drula E, Coutinho PM, Henrissat B. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 2014;42:D490–D495. doi: 10.1093/nar/gkt1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Senoura T, Ito S, Taguchi H, Higa M, Hamada S, et al. New microbial mannan catabolic pathway that involves a novel mannosylglucose phosphorylase. Biochem Biophys Res Commun. 2011;408:701–706. doi: 10.1016/j.bbrc.2011.04.095. [DOI] [PubMed] [Google Scholar]
- 11.Sernee MF, Ralton JE, Nero TL, Sobala LF, Kloehn J, et al. A family of dual-activity glycosyltransferase-phosphorylases mediates mannogen turnover and virulence in Leishmania parasites. Cell Host Microbe. 2019;26:385–399. doi: 10.1016/j.chom.2019.08.009. [DOI] [PubMed] [Google Scholar]
- 12.Cuskin F, Baslé A, Ladevèze S, Day AM, Gilbert HJ, et al. The GH130 Family of Mannoside Phosphorylases Contains Glycoside Hydrolases That Target β-1,2-Mannosidic Linkages in Candida Mannan. J. Biol. Chem. 2015;290:25023–25033. doi: 10.1074/jbc.M115.681460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nihira T, Chiku K, Suzuki E, Nishimoto M, Fushinobu S, et al. An inverting β-1,2-mannosidase belonging to glycoside hydrolase family 130 from Dyadobacter fermentans . FEBS Lett. 2015;589:3604–3610. doi: 10.1016/j.febslet.2015.10.008. [DOI] [PubMed] [Google Scholar]
- 14.Kawahara R, Saburi W, Odaka R, Taguchi H, Ito S, et al. Metabolic mechanism of mannan in a ruminal bacterium, Ruminococcus albus, involving two mannoside phosphorylases and cellobiose 2-epimerase; discovery of a new carbohydrate phosphorylase, β-1,4-mannooligosaccharide phosphorylase. J Biol Chem. 2012;287:42389–42399. doi: 10.1074/jbc.M112.390336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chekan JR, Kwon IH, Agarwal V, Dodd D, Revindran V, et al. Structural and Biochemical Basis for Mannan Utilization by Caldanaerobius polysaccharolyticus Strain ATCC BAA-17. J Biol Chem. 2014;289:34965–34977. doi: 10.1074/jbc.M114.579904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ye Y, Saburi W, Odaka R, Kato K, Sakurai N, et al. Structural insights into the difference in substrate recognition of two mannoside phosphorylases from two GH130 subfamilies. FEBS Lett. 2016;590:828–837. doi: 10.1002/1873-3468.12105. [DOI] [PubMed] [Google Scholar]
- 17.La Rosa SL, Leth ML, Michalak L, Hansen ME, Pudlo NA, et al. The human gut firmicute Roseburia intestinalis is a primary degrader of dietary β-mannans. Nat Commun. 2019;10:1–14. doi: 10.1038/s41467-019-08812-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grimaud F, Pizzut-Serin S, Tarquis L, Ladevèze S, Morel S, et al. In Vitro Synthesis and Crystallization of β-1,4-Mannan. Biomacromolecules. 2019;20:846–853. doi: 10.1021/acs.biomac.8b01457. [DOI] [PubMed] [Google Scholar]
- 19.Nihira T, Suzuki E, Kitaoka M, Nishimoto M, Ohtsubo Ken'ichi, Ohtsubo K, et al. Discovery of β-1,4-d-Mannosyl- N -acetyl-d-glucosamine Phosphorylase Involved in the Metabolism of N -Glycans. J. Biol. Chem. 2013;288:27366–27374. doi: 10.1074/jbc.M113.469080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ladevèze S, Tarquis L, Cecchini DA, Bercovici J, André I, et al. Role of glycoside-phosphorylases in mannose foraging by human gut bacteria. J Biol Chem. 2013;483628:jbc.M113. doi: 10.1074/jbc.M113.483628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chiku K, Nihira T, Suzuki E, Nishimoto M, Kitaoka M, et al. Discovery of two β-1,2-mannoside phosphorylases showing different chain-length specificities from Thermoanaerobacter sp. X-514. PLoS One. 2014;9:e114882. doi: 10.1371/journal.pone.0114882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tsuda T, Nihira T, Chiku K, Suzuki E, Arakawa T, et al. Characterization and crystal structure determination of β-1,2-mannobiose phosphorylase from Listeria innocua . FEBS Lett. 2015;589:3816–3821. doi: 10.1016/j.febslet.2015.11.034. [DOI] [PubMed] [Google Scholar]
- 23.Awad FN, Laborda P, Wang M, AM L, Li Q, et al. Discovery and biochemical characterization of a mannose phosphorylase catalyzing the synthesis of novel β-1,3-mannosides. BBA-Gen Subjects. 1861;2017:3231–3237. doi: 10.1016/j.bbagen.2017.09.013. [DOI] [PubMed] [Google Scholar]
- 24.Tang S-L, Pohl NLB. Automated solution-phase synthesis of β-1,4-mannuronate and β-1,4-mannan. Org Lett. 2015;17:2642–2645. doi: 10.1021/acs.orglett.5b01013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gerlt JA, Bouvier JT, Davidson DB, Imker HJ, Sadkhin B, et al. Enzyme function Initiative-Enzyme similarity tool (EFI-EST): a web tool for generating protein sequence similarity networks. BBA-Proteins Proteom. 1854;2015:1019–1037. doi: 10.1016/j.bbapap.2015.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Atkinson HJ, Morris JH, Ferrin TE, Babbitt PC. Using sequence similarity networks for visualization of relationships across diverse protein superfamilies. PLoS One. 2009;4:e4345. doi: 10.1371/journal.pone.0004345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Levin BJ, Huang YY, Peck SC, Wei Y, Martínez-del Campo A, et al. A prominent glycyl radical enzyme in human gut microbiomes metabolizes trans -4-hydroxy-l-proline. Science. 2017;355:eaai8386. doi: 10.1126/science.aai8386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Viborg AH, Terrapon N, Lombard V, Michel G, Czjzek M, et al. A subfamily roadmap of the evolutionarily diverse glycoside hydrolase family 16 (GH16) J. Biol. Chem. 2019;294:15973–15986. doi: 10.1074/jbc.RA119.010619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ladevèze S, Laville E, Despres J, Mosoni P, Potocki-Véronèse G. Mannoside recognition and degradation by bacteria. Biol Rev. 2017;92:1969–1990. doi: 10.1111/brv.12316. [DOI] [PubMed] [Google Scholar]
- 30.Svartström O, Alneberg J, Terrapon N, Lombard V, de Bruijn I, et al. Ninety-nine de novo assembled genomes from the moose (Alces alces) rumen microbiome provide new insights into microbial plant biomass degradation. Isme J. 2017;11:2538–2551. doi: 10.1038/ismej.2017.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 32.Shannon P, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huang Y, Niu B, Gao Y, Fu L, Li W. CD-HIT suite: a web server for clustering and comparing biological sequences. Bioinformatics. 2010;26:680–682. doi: 10.1093/bioinformatics/btq003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Konagurthu AS, Whisstock JC, Stuckey PJ, Lesk AM. MUSTANG: a multiple structural alignment algorithm. Proteins. 2006;64:559–574. doi: 10.1002/prot.20921. [DOI] [PubMed] [Google Scholar]
- 35.Nakae S, Ito S, Higa M, Senoura T, Wasaki J, et al. Structure of novel enzyme in mannan biodegradation process 4-O-β-D-mannosyl-D-glucose phosphorylase MGP. J Mol Biol. 2013;425:4468–4478. doi: 10.1016/j.jmb.2013.08.002. [DOI] [PubMed] [Google Scholar]
- 36.Ladevèze S, Cioci G, Roblin P, Mourey L, Tranier S, et al. Structural bases for N-glycan processing by mannoside phosphorylase. Acta Crystallogr D. 2015;71:1335–1346. doi: 10.1107/S1399004715006604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 39.Letunic I, Bork P. Interactive tree of life (iTOL) V3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 2016;44:W242–W245. doi: 10.1093/nar/gkw290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nielsen H. Protein Function Prediction. Springer; 2017. Predicting secretory proteins with SignalP; pp. 59–73. [DOI] [PubMed] [Google Scholar]
- 41.Gawronski JD, Benson DR. Microtiter assay for glutamine synthetase biosynthetic activity using inorganic phosphate detection. Anal Biochem. 2004;327:114–118. doi: 10.1016/j.ab.2003.12.024. [DOI] [PubMed] [Google Scholar]
- 42.De Groeve MRM, Tran GH, Van Hoorebeke A, Stout J, Desmet T, et al. Development and application of a screening assay for glycoside phosphorylases. Anal Biochem. 2010;401:162–167. doi: 10.1016/j.ab.2010.02.028. [DOI] [PubMed] [Google Scholar]
- 43.Macdonald SS, Armstrong Z, Morgan-Lang C, Osowiecka M, Robinson K, et al. Development and application of a high-throughput functional metagenomic screen for glycoside phosphorylases. Cell Chem Biol. 2019;26:1001–1012. doi: 10.1016/j.chembiol.2019.03.017. [DOI] [PubMed] [Google Scholar]
- 44.Niedermeyer THJ, Strohalm M. mMass as a software tool for the annotation of cyclic peptide tandem mass spectra. PLoS One. 2012;7:e44913. doi: 10.1371/journal.pone.0044913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Domon B, Costello CE. A systematic Nomenclature for carbohydrate fragmentations in FAB-MS/MS spectra of glycoconjugates. Glycoconj J. 1988;5:397–409. doi: 10.1007/BF01049915. [DOI] [Google Scholar]
- 46.Wildberger P, Todea A, Nidetzky B. Probing enzyme–substrate interactions at the catalytic subsite of Leuconostoc mesenteroides sucrose phosphorylase with site-directed mutagenesis: the roles of Asp49 and Arg395. Biocatal Biotransfor. 2012;30:326–337. doi: 10.3109/10242422.2012.674720. [DOI] [Google Scholar]
- 47.Lee HH, Lee JS, Cho JY, Kim YE, Hong EK. Structural characteristics of immunostimulating polysaccharides from Lentinus edodes. J Microbiol Biotechnol. 2009;19:455–461. doi: 10.4014/jmb.0809.542. [DOI] [PubMed] [Google Scholar]
- 48.Guo S, Mao W, Han Y, Zhang X, Yang C, et al. Structural characteristics and antioxidant activities of the extracellular polysaccharides produced by marine bacterium Edwardsiella tarda. Bioresour Technol. 2010;101:4729–4732. doi: 10.1016/j.biortech.2010.01.125. [DOI] [PubMed] [Google Scholar]
- 49.Shibata N, Kobayashi H, Okawa Y, Suzuki S. Existence of novel beta-1,2 linkage-containing side chain in the mannan of Candida lusitaniae, antigenically related to Candida albicans serotype a. Eur J Biochem. 2003;270:2565–2575. doi: 10.1046/j.1432-1033.2003.03622.x. [DOI] [PubMed] [Google Scholar]
- 50.Mille C, Bobrowicz P, Trinel P-A, Li H, Maes E, et al. Identification of a New family of genes involved in β-1,2-mannosylation of glycans in Pichia pastoris and Candida albicans . J Biol Chem. 2008;283:9724–9736. doi: 10.1074/jbc.M708825200. [DOI] [PubMed] [Google Scholar]
- 51.Katzenellenbogen E, Kocharova NA, Toukach PV, Górska S, Korzeniowska-Kowal A, et al. Structure of an abequose-containing O-polysaccharide from Citrobacter freundii O22 strain PCM 1555. Carbohydr Res. 2009;344:1724–1728. doi: 10.1016/j.carres.2009.06.005. [DOI] [PubMed] [Google Scholar]
- 52.Liu B, Knirel YA, Feng L, Perepelov AV, Senchenkova Sof'ya N., et al. Structural diversity in Salmonella O antigens and its genetic basis. FEMS Microbiol Rev. 2014;38:56–89. doi: 10.1111/1574-6976.12034. [DOI] [PubMed] [Google Scholar]
- 53.Sims IM, Bacic A. Extracellular polysaccharides from suspension cultures of Nicotiana plumbaginifolia. Phytochemistry. 1995;38:1397–1405. doi: 10.1016/0031-9422(94)00832-E. [DOI] [Google Scholar]
- 54.Alberta MJ, Holme T, Lindberg B, Lindberg J, Mosihuzzaman M, et al. Structural studies of the Shigella boydii type 5 O-antigen polysaccharide. Carbohydr Res. 1994;265:121–127. doi: 10.1016/0008-6215(94)00223-1. [DOI] [PubMed] [Google Scholar]
- 55.Lima IFN, Havt A, Lima AAM. Update on molecular epidemiology of Shigella infection. Curr Opin Gastroen. 2015;31:30–37. doi: 10.1097/MOG.0000000000000136. [DOI] [PubMed] [Google Scholar]
- 56.Chen Y, Li X-H, Zhou L-Y, Li W, Liu L, et al. Structural elucidation of three antioxidative polysaccharides from Tricholoma lobayense. Carbohydr Polym. 2017;157:484–492. doi: 10.1016/j.carbpol.2016.10.011. [DOI] [PubMed] [Google Scholar]
- 57.Kocharova NA, Knirel YA, Shashkov AS, Kochetkov NK, Pier GB. Structure of an extracellular cross-reactive polysaccharide from Pseudomonas aeruginosa immunotype 4. J Biol Chem. 1988;263:11291–11295. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.