Abstract
Carbapenemases inactivate most β-lactam antibiotics, including carbapenems, and have frequently been reported among Enterobacteriaceae , Acinetobacter spp. and Pseudomonas spp. Traditionally, the horizontal gene transfer of carbapenemase-encoding genes (CEGs) has been linked to plasmids. However, given that integrative and conjugative elements (ICEs) are possibly the most abundant conjugative elements among prokaryotes, we conducted an in silico analysis to ascertain the likely role of ICEs in the spread of CEGs among all bacterial genomes (n=182 663). We detected 17 520 CEGs, of which 66 were located within putative ICEs among several bacterial species (including clinically relevant bacteria, such as Pseudomonas aeruginosa , Klebsiella pneumoniae and Escherichia coli ). Most CEGs detected within ICEs belong to the IMP, NDM and SPM metallo-beta-lactamase families, and the serine beta-lactamase KPC and GES families. Different mechanisms were likely responsible for acquisition of these genes. The majority of CEG-bearing ICEs belong to the MPFG, MPFT and MPFF classes and often encode resistance to other antibiotics (e.g. aminoglycosides and fluoroquinolones). This study provides a snapshot of the different CEGs associated with ICEs among available bacterial genomes and sheds light on the underappreciated contribution of ICEs to the spread of carbapenem resistance globally.
Keywords: antibiotic resistance, carbapenemases, clinically relevant bacteria, integrative and conjugative elements
Data Summary
All the bacterial genomes scanned in this study have been deposited previously in the National Center for Biotechnology Information (NCBI) genome database and are listed in the supplementary tables. The 66 extracted ICEs (in fasta format) and the outputs for the profile HMMs scanned on the 386 putative MGEs identified in this study have been deposited on figshare at https://figshare.com/projects/_Comprehensive_genome_data_analysis_establishes_a_triple_whammy_of_carbapenemases_ICEs_and_multiple_clinically-relevant_bacteria/78369.
Impact Statement.
Carbapenems are commonly used to treat severe infections in humans. Resistance is often mediated by carbapenemases. These enzymes degrade carbapenems and are frequently present in plasmids. Here, we demonstrate that common carbapenemase-encoding genes (CEGs) found in clinical isolates (e.g. bla KPC, bla GES, bla IMP, bla NDM, bla VIM) can also be located within integrative and conjugative elements (ICEs). CEG-bearing ICEs belong to three mating pair formation families. These mobile elements may be particularly important in bacteria where plasmids do not seem to play a significant role in the spread of antibiotic resistance genes, such as Pseudomonas spp. This study considerably expands our knowledge of the repertoire of CEGs-bearing ICEs among clinically relevant bacterial pathogens, such as Pseudomonas aeruginosa , Klebsiella pneumoniae and Escherichia coli .
Introduction
Due to the importance of carbapenems for the treatment of severe infections in humans, the World Health Organization (WHO) stated that these antibiotics should be reserved for infections caused by multidrug-resistant Gram-negative bacteria in humans [1]. Recently, the same agency presented a list of bacterial pathogens for which new antibiotics research and development are urgently required, and the top priority pathogens were the carbapenem-resistant strains of Acinetobacter baumannii , Pseudomonas aeruginosa and Enterobacteriaceae [2].
The evolution of carbapenem resistance in bacteria is often driven by the horizontal gene transfer (HGT) of carbapenemase-encoding genes (CEGs) [3, 4]. Carbapenemases are beta-lactamases that are able to hydrolyze carbapenems as well as most other beta-lactam antibiotics. These enzymes are members of serine beta-lactamases classes A and D, and the class B metallo-beta-lactamases [5]. The CEGs are often located on integrons or transposons that themselves target mobile genetic elements (MGEs) such as plasmids [3, 4], which makes the dissemination of these genes unpredictably complex within bacterial communities. Recently, it was demonstrated that another type of MGE, the integrative and conjugative elements (ICEs), are likely to play a significant role as vehicles for the dissemination of CEGs among P. aeruginosa [6]. Besides genes conferring antibiotic resistance, ICEs may harbour additional cargo genes that provide an adaptive advantage over other elements. Some of these examples include the presence of the siderophore yersiniabactin encoded within the ICEKp in hypervirulent clonal group CG23 from Klebsiella pneumoniae [7]; the Tn5252-related ICEs carrying bacteriocin clusters in Streptococcus suis [8]; the type I-C CRISPR-Cas systems identified within pKLC102-like ICEs in P. aeruginosa [9]; and the type III restriction–modification systems from SXT/R391-related ICEs in Shewanella spp. [10].
ICEs are self-transmissible MGEs that can integrate into and excise from the genome (like transposons and phages) and can exist as circular, sometimes replicable, extrachromosomal elements and be transferred by conjugation (like some plasmids) [11–14]. ICEclc from Pseudomonas knackmussii [15], SXT from Vibrio cholerae [16], pKLC102 from P. aeruginosa [17] and Tn4371 from Ralstonia oxalatica [18] are among the most well studied ICEs. ICEs appear to have a bipartite lifestyle that shifts between vertical and horizontal transmission [12, 19, 20]. HGT by conjugation requires three main components: a relaxase (MOB), a mating pair formation (MPF) system and a type IV coupling protein, with the last two forming a spanning-membrane multi-protein complex named the type IV secretion system (T4SS) [21]. To date, eight MPF classes have been proposed (B, C, F, FA, FATA, G, I and T), based on the phylogeny of VirB4, the only ubiquitous protein among the T4SS. The MPFT is widely distributed in both conjugative plasmids and ICEs, while MPFF is more prevalent in plasmids and MPFG on ICEs [11].
Given that ICEs have been identified in most bacterial clades and have been proposed to be more prevalent than conjugative plasmids [11], we conducted an in silico analysis to explore the distribution of CEG-bearing ICEs among all sequenced bacterial genomes available in the National Center for Biotechnology Information (NCBI). Our results demonstrate that CEG-bearing ICEs belong to three MPF families and are primarily located in several clinically relevant bacterial pathogens. Our analysis highlights the importance of investigating these elements thoroughly as important vehicles for the spread of antibiotic resistance (AR), particularly with respect to carbapenems.
Methods
Bacterial genome and carbapenemase search
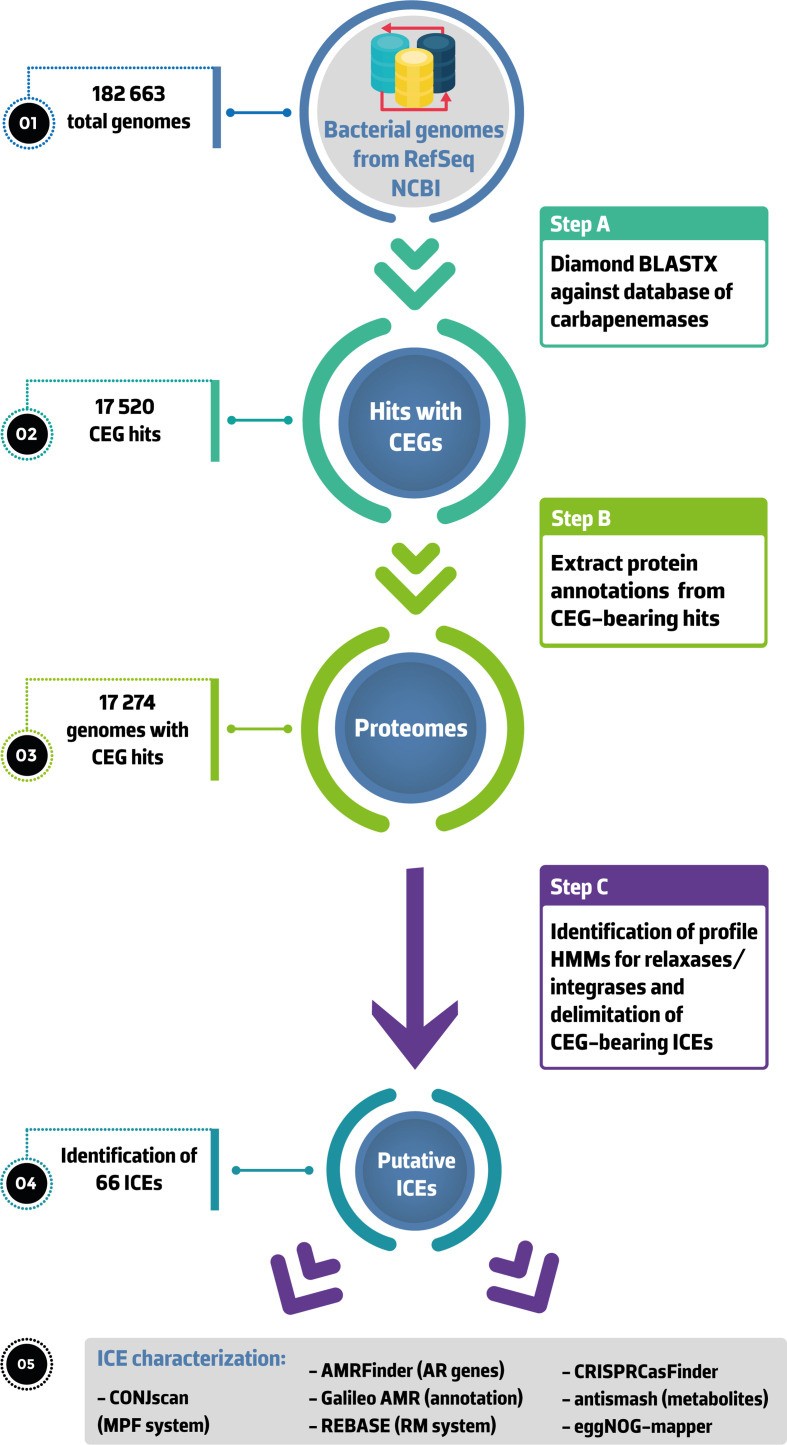
In Fig. 1, we present the workflow used in this study, from the acquisition of bacterial genomes to the identification and characterization of putative ICEs. We retrieved all bacterial genomes available in the NCBI Reference Sequence Database (RefSeq, accessed on 21 March 2020), including complete and draft genome sequences, using ncbi-genome-download v0.2.12 (https://github.com/kblin/ncbi-genome-download). We downloaded over 6000 curated AR protein sequences from the AMRfinder database (https://ftp.ncbi.nlm.nih.gov/pathogen/Antimicrobial_resistance/AMRFinderPlus/database/3.6/2020-01-22.1/) [22] and built an in-house database only including the proteins that code for a carbapenemase (n=1014, Table S1, available in the online version of this article). We then blasted the genomes against the extracted carbapenemases using diamond v0.9.29.130 (http://www.diamondsearch.org/index.php) [23], using minimum 100 % identity and subject cover and with the sensitive mode enabled.
Fig. 1.
Overview of the workflow followed in this study. All assemblies available in NCBI RefSeq were downloaded and blasted against an in-house database of carbapenemases using diamond blastx (step 1). NCBI annotated proteins from CGE-bearing genomes were then extracted (step 2) and used for the identification of relaxase and serine or tyrosine recombinase (step 3). Search of directed repeats and delimitation of putative ICEs was also performed. CONJscan was used to identify the MPF family of each element. We then looked for AR genes, restriction–modification systems, CRISPR arrays and their associated (Cas) proteins, as well as secondary metabolites within extracted ICEs. We also characterized the functional annotations of their proteomes and the MLST of the genomes carrying a CEG-bearing ICE. Abbreviations: AR, antibiotic resistance; CEG, carbapenemase-encoding gene; HMM, hidden Markov model; ICE, integrative and conjugative element; MPF, mating-pair formation; RM, restriction–modification.
Tracing ICEs among the bacterial genomes
The RefSeq protein files from the CEG-bearing genomes identified by diamond were extracted. We used the hmmsearch function of the HMMER3 software package v3.3 (http://hmmer.org/) [24] to search the proteomes against the standalone version of MOBfamDB, a curated hidden Markov models (HMM) relaxase database (https://castillo.dicom.unican.es/mobscan_about/) [25]. We also used this function to search the pfam v33.0 database for tyrosine or serine recombinase accessions numbers (Pfam IDs PF00589 and PF07508). The hmmsearch command was used with default parameters and an E-value threshold of 0.01. The CEG-bearing genomes with relaxase and integrase hits were further analysed. We used the Find Repeats tool from Geneious Prime 2020.0.4 (https://www.geneious.com) to inspect the hits for direct repeats. To delimit CEG-harbouring ICEs, we manually scanned candidate terminal regions with direct repetitions of the 3′ end from tRNA genes located next to the integrase-encoding gene. When no tRNA gene was identified next to this gene, we scanned the presence of direct repeats next to the integrase-encoding gene and next to candidate terminal regions. To assist in identifying putative terminal regions, we looked for blocks of DNA with variation in GC content. To predict the MPF families, the translated coding sequences of delimited ICEs were analysed on the standalone CONJscan module of MacSyFinder v1.0.5 (https://github.com/gem-pasteur/macsyfinder) [26, 27]. To identify the multi-locus sequence type of the genomes containing CEG-bearing ICEs, we used mlst v2.16.1 (https://github.com/tseemann/mlst), which scans the genomes against PubMLST typing schemes (https://pubmlst.org/) [28].
Characterization of the CEG-bearing ICEs
Screening of AR genes among ICEs was performed using amrfinder v3.6.10 (https://github.com/ncbi/amr) [22]. The genetic platforms involved in the acquisition of CEGs by ICEs were annotated using Galileo AMR (https://galileoamr.arcbio.com/mara/) (Arc Bio, Cambridge, MA, USA) [29]. We ran our extracted ICEs against REBASE (http://rebase.neb.com/rebase/rebase.html) to look for restriction–modification systems [30]. We used CRISPRCasFinder (https://crisprcas.i2bc.paris-saclay.fr/CrisprCasFinder/Index) to look for CRISPR (clustered regularly interspaced short palindromic repeats) arrays and their associated (Cas) proteins within ICE sequences [31]. Secondary metabolite biosynthetic gene clusters were traced using antismash v5.1.2 (https://antismash.secondarymetabolites.org/) [32]. We used eggNOG-mapper v2 (http://eggnog-mapper.embl.de/) for functional annotation based on orthology assignments of the ICE proteomes [33].
Results
Carbapenemase-encoding genes are mainly found in proteobacteria
We retrieved a total of 182 663 bacterial genomes from NCBI (16 798 complete genomes and 165 865 genomes assembled at the chromosome, scaffold or contig level). We identified a total of 17 520 CEGs, with 1422 CEGs on 1236 complete genomes (including 512 chromosomes and 724 plasmids) and 16 098 CEGs on 16 038 draft genomes (Table S2). We identified a total of 377 carbapenemase variants among the 17 520 hits. Our results show that CEGs are mostly located on Proteobacteria and dominated by clinically relevant pathogens such as A. baumannii , K. pneumoniae , Escherichia coli and P. aeruginosa (Table S2). These genomes encode a wide diversity of carbapenemases, including OXA-23 (15.6%, n=2 739/17 520), KPC-2 (13.2%), KPC-3 (8.9%) and NDM-1 (6.7%). As we are tracing MGEs integrated in the chromosome, the 724 plasmid hits in complete genomes were excluded from the analysis. Among the hits on the 16 038 draft genomes, we filtered out sequences with the word ‘plasmid’ present on the fasta header (n=131, Table S3). To maximize the chances of detecting entire ICEs, we filtered out sequences shorter than 40 kb (n=10 050). All the excluded ones are available for analysis in Table S4. The remaining sequences from draft genomes (n=5 857) were inspected for the presence of CEG-harbouring ICEs.
A large proportion of CEG-bearing ICEs belong to three families and target clinically relevant Gram-negative bacteria
We identified a total of 66 putative ICEs, including 42 newly characterized elements associated with 17 different CEGs (Table 1 and Fig. 2). We could predict the boundaries from 55 of these elements (Table S5). The terminal region of the remaining 11 putative ICEs could not be determined due to a fragmented contig or assembly gaps within the sequence. Nearly half of the putative ICEs (48.5%, n=32/66) were integrated at the 3′ end of a tRNAGly gene. Integration next to random genes was also observed (Fig. 3 and Table S5). The bacterial hosts housing these elements belong to 23 sequence types (STs) (Table S5).
Table 1.
Diversity and characterization of carbapenemase-encoding genes in integrative and conjugative element-associated genomes
|
MPF family |
CEG |
Integrase type |
Relaxase type |
Bacterial species |
MGEs flanking the CEGs |
ICE length (kb)* |
References |
|---|---|---|---|---|---|---|---|
|
MPFG (n=43) |
AFM-1 (n=1) |
INT_P4_C |
MOBH |
Flanked by IS91 family ISs |
130 |
This study |
|
|
DIM-1 (n=1) |
INT_P4_C |
MOBH |
Class I In flanked by IS6100 |
89 |
This study, [6] |
||
|
GES-5 (n=5) |
INT_P4_C |
MOBH |
Class I In within Tn3-like Tn |
93–116 |
This study, [6] |
||
|
GES-24 (n=1) |
INT_P4_C |
MOBH |
Class I In within Tn3-like Tn |
63 |
This study |
||
|
IMP-1 (n=4) |
INT_P4_C, INT_Rci_Hp1_C |
MOBH |
Class I In within Tn3-like Tn |
76–109 |
This study, [6] |
||
|
IMP-13 (n=9) |
INT_P4_C, INT_Rci_Hp1_C |
MOBH |
Class I In within Tn3-like Tn |
65–89 |
This study, [6] |
||
|
IMP-14 (n=2) |
INT_P4_C |
MOBH |
Class I In within Tn3-like Tn |
106–122 |
This study |
||
|
IMP-16 (n=1) |
INT_P4_C |
MOBH |
Class I In within Tn3-like Tn |
86 |
This study |
||
|
IMP-54 (n=1) |
INT_P4_C |
MOBH |
Class I In within Tn3-like Tn |
91 |
This study |
||
|
KPC-2 (n=1) |
INT_P4_C |
MOBH |
Tn4401 |
115 |
This study, [43] |
||
|
NDM-1 (n=12) |
INT_P4_C |
MOBH |
Pseudomonas aeruginosa, Pseudomonas asiatica, Morganella morganii |
Next to (or flanked by) IS91 family ISs |
97–167 |
This study, [44, 45] |
|
|
VIM-2 (n=1) |
INT_P4_C |
MOBH |
Class I In within Tn3-like Tn |
65 |
This study |
||
|
VIM-4 (n=4) |
INT_P4_C |
MOBH |
Pseudomonas aeruginosa, Klebsiella pneumoniae, Alcaligenes faecalis |
Class I In within Tn3-like Tn |
88–102 |
This study |
|
|
MPFT (n=16) |
AFM-1 (n=1) |
INT_P4_C |
MOBP |
Flanked by IS91 family ISs |
77 |
This study |
|
|
KPC-2 (n=1) |
INT_Rci_Hp1_C |
MOBP |
Pseudomonas sp. |
Next to ISKpn6 and IS26 |
62 |
This study, [6] |
|
|
NDM-1 (n=3) |
INT_P4_C, INT_Rci_Hp1_C |
MOBP |
Pseudomonas aeruginosa, Pseudomonas asiatica |
Flanked by IS91 family ISs |
73–74 |
This study, [35] |
|
|
SPM-1 (n=11) |
INT_Rci_Hp1_C |
MOBP |
Flanked by ISCR3-like elements |
44–58 |
This study, [36] |
||
|
MPFF (n=3) |
IMP-6 (n=1) |
DNA_BRE_C |
MOBF |
Class I In within Tn3-like Tn |
118 |
This study |
|
|
KPC-2 (n=2) |
DNA_BRE_C |
MOBF |
Tn4401 |
75 |
This study |
||
|
Incomplete MPFG (n=2) |
IMP-8 (n=1) |
DNA_BRE_C |
MOBH |
Class I In within Tn3-like Tn |
124 |
This study, [46] |
|
|
NDM-1 (n=1) |
INT_P4_C |
MOBH |
Flanked by ISCR3-like elements |
98 |
This study |
||
|
Incomplete MPFT (n=1) |
KPC-2 (n=1) |
INT_Rci_Hp1_C |
na |
Achromobacter sp. |
Next to ISKpn6 and IS26 |
43 |
This study |
|
Incomplete MPFF (n=1) |
NDM-5 (n=1) |
DNA_BRE_C |
na |
Flanked by ISCR1 and IS26 |
54 |
This study, [47] |
*Some sequences are not complete. For more details, please refer to Table S5.
CEG, carbapenemase-encoding gene; DNA_BRE_C, DNA breaking–rejoining enzymes, C-terminal catalytic domain; ICE, integrative and conjugative elements; In, integron; INT_P4_C, P4 integrase, C-terminal catalytic domain; INT_Rci_Hp1_C, shufflon-specific DNA recombinase Rci and bacteriophage Hp1_like integrase, C-terminal catalytic domain family; MPF, mating pair formation; na, no hit; ST, sequence type.
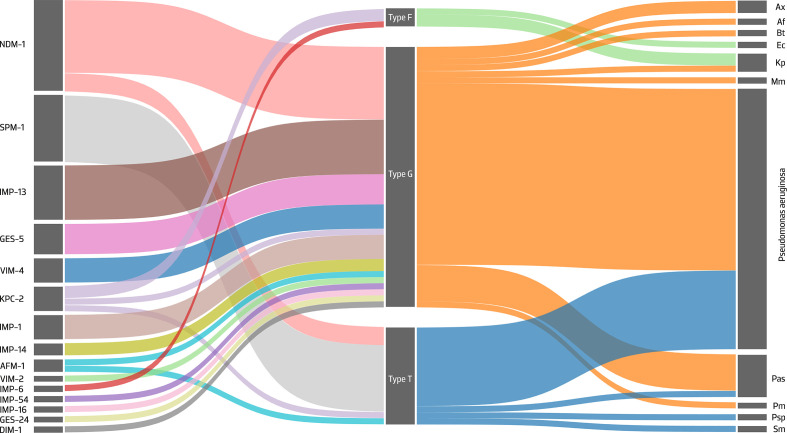
Fig. 2.
Sankey diagram showing the contribution of different MPF families to the spread of CEGs among several bacterial genomes. The left, centre and right axes represent the association between the identified carbapenemases, MPF type and the bacterial genus, respectively, while the width of each connection is proportional to the number of positive hits. Abbreviations: Ax, Achromobacter xylosoxidans; Af, Alcaligenes faecalis; Bt, Bordetella trematum ; Ec, Escherichia coli; Kp, Klebsiella pneumoniae; Mm, Morganella morganii ; Pas, Pseudomonas asiatica ; Pm, Pseudomonas monteilii ; Psp, Pseudomonas sp.; Sm, Stenotrophomonas maltophilia .
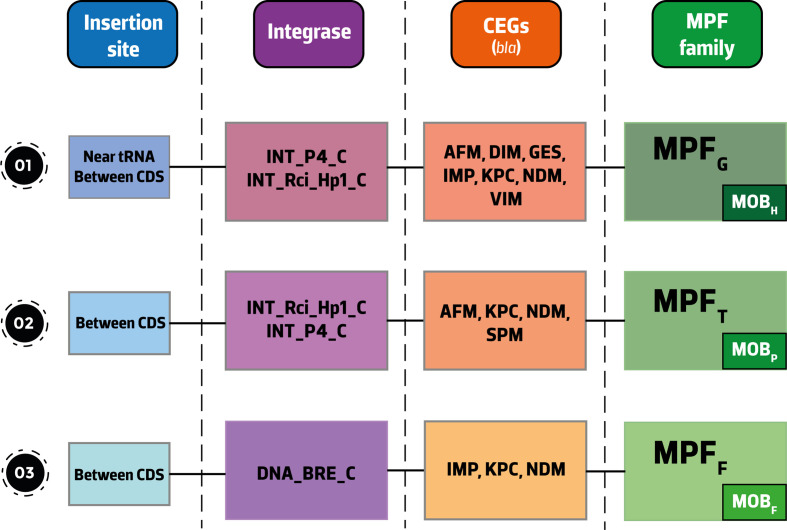
Fig. 3.
Schematic representation of the different insertion site/integrase/CEG/MPF/relaxase profiles identified in this study. The figure is not to scale and the relative position of the different modules (integration, conjugation, accessory CEGs) is for illustrative purposes to show the various relationships observed. Abbreviations: CDS, Coding Sequence; CEGs, carbapenemase-encoding genes; DNA_BRE_C, DNA breaking–rejoining enzymes, C-terminal catalytic domain; INT_P4_C, P4 integrase, C-terminal catalytic domain; INT_Rci_Hp1_C, shufflon-specific DNA recombinase Rci and bacteriophage Hp1_like integrase, C-terminal catalytic domain family; MPF, mating-pair formation; tRNA, transfer RNA gene.
Using the CONJscan module of MacSyFinder we identified the MPF family for 62 hits (incomplete MPF classes were predicted for the remaining 4 hits) and we noted that these hits belong to 3 families: MPFG (69%, n=43/62), MPFT (26%) and MPFF (5%) (Fig. 2, Tables 1, S5 and S6). In our results, the MPFG class was only associated with MOBH, the MPFT class with MOBP and the MPFF class with MOBF (Fig. 3). All ICEs identified here carried a tyrosine recombinase, with the majority of them (56%, n=37/66) belonging to the P4 integrase, C-terminal catalytic domain family (INT_P4_C). The shufflon-specific DNA recombinase Rci and bacteriophage Hp1_like integrase, C-terminal catalytic domain family (INT_Rci_Hp1_C) and the DNA breaking–rejoining enzymes, C-terminal catalytic domain family (DNA_BRE_C) integrases were also identified in our collection (36 and 8 %, respectively) (Tables 1 and S5). INT_P4_C and INT_Rci_Hp1_C integrases were associated with ICEs belonging to the MPFG and MPFT classes, while DNA_BRE_C was found on MPFF ICEs (Fig. 3). ICEs from the MPFG and MPFT classes were particularly promiscuous, being responsible for the spread of several CEGs of the metallo-beta-lactamase family such as bla NDM-1, bla SPM-1 and bla IMP variants among clinically relevant pathogens such as P. aeruginosa , K. pneumoniae and E. coli . ICEs of the MPFF class carrying blaIMP-6 or bla KPC-2 were restricted to E. coli and K. pneumoniae . The bla SPM-1 gene was exclusively identified in P. aeruginosa and in ICEs of the MPFT class (Table 1).
We analysed the types of integrase, relaxase and MPF classes present among four model ICEs: ICEclc, pKLC102, SXT and Tn4371. The MPFG-INT_P4_C ICEs identified here are related to ICEclc, since this ICE belongs to the same class and carries a MOBH relaxase and also an INT_P4_C integrase. The MPFT-INT_Rci_Hp1_C ICEs belong to the Tn4371 family, which also carries the MOBP relaxase. No conserved domain family could be attributed for SXT; however, the MPFF ICEs reported here should be related to this model ICE, which also uses a MOBH relaxase.
We also identified 386 hits encoding an integrase and a relaxase in the vicinity of CEGs (Table S7). For these hits, however, we could not predict if the CEG is located on a plasmid or an ICE, since the contig is fragmented and tracing the boundaries of the element is not possible, or the sequences have assembly gaps that make this prediction challenging. Some plasmids may also encode a tyrosine or serine recombinase, and some ICEs may encode replicases and partition systems that are typical of plasmids [34], which can hinder the accurate prediction of genetic platforms when the sequence has poor quality or is highly fragmented due to short-read sequencing approaches.
A variable repertoire of CEG-bearing integrons and transposons target ICEs
We identified 17 CEG variants among the 66 putative ICEs, dominated by bla NDM-1 and bla SPM-1 (Table 1 and Fig. 2). Insertion sequences (ISs; e.g. ISCR3-like elements) were frequently linked to the acquisition of bla SPM-1 and bla NDM-1 [35, 36], while bla IMP, bla VIM, and bla GES were found on class I integrons frequently integrated into Tn3 family transposons [6]. The bla KPC-2 gene was typically found within Tn4401-like transposons, which are capable of conferring a high frequency of transposition [37]. The recently identified AFM-1 metallo-beta-lactamase (GenBank accession number MK143105.1) was identified here in two ICEs inserted in Bordetella trematum and Stenotrophomonas maltophilia genomes (Table 1). We also found bla NDM-1 genes in ICEs integrated into the genomes of a recently proposed Pseudomonas asiatica species, which is spreading in hospital settings in Myanmar [38]. Besides CEGs, the ICEs identified in this study also harbour genes conferring resistance to other antibiotics, such as aminoglycosides, fluoroquinolones, macrolides and tetracyclines (Table S8), widening the spectrum of transmissible AR genes selectable by carbapenems due to linkage.
Acquisition of additional traits by ICEs, including competitive weapons such as bacteriocins and siderophores
Besides genes conferring AR, the CEG-bearing ICEs identified here harbour other cargo genes that may confer a selective advantage to the ICE host. We found DUF692 domains typical of bacteriocin producing genes among the six MPFG ICEs from P. asiatica strains and the P. aeruginosa N15-01092, 1334/14 and ST773 strains (Table S5). All these ICEs carry a bla NDM-1 gene and a similar copy of the bacteriocin-encoding gene. Curiously, the bacteriocin producing gene was not identified in the MPFT ICE from P. asiatica strain MY569. Additionally, we found the siderophore aerobactin operon within an ICE in E. coli strain E302 (Table S5). This operon is usually found in enterobacterial plasmids, and was also identified in a pathogenicity island in uropathogenic E. coli strain CFT073 [39]. We identified no CRISPR-Cas systems among the ICEs here identified. Nearly half of them (47.0%, n=31/66) carried complete or incomplete restriction–modification systems belonging to types II, III and IV (Table S9).
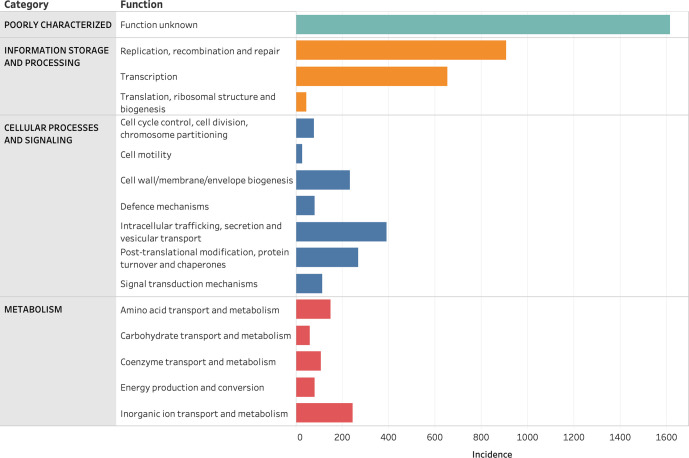
The majority of the proteins encoded within the 66 ICEs refer to replication, recombination, transcription and intracellular trafficking functions (Fig. 4). Several proteins, however, encoded for unknown functions (34.0%, n=1 615/4 754, Table S10), highlighting the lack of knowledge concerning the ICE proteome.
Fig. 4.
Grouped bar chart representing the incidence of each eggNOG function broken down by category (represented by different colours) in the CEG-bearing ICE sequences.
Discussion
We have set out to comprehensively identify the CEGs among all bacterial genomes deposited in the NCBI database and the CEG-bearing ICE sequences. Our study considerably expands our knowledge of the repertoire of CEGs-bearing ICEs. We uncovered 66 putative ICEs that may be involved in HGT of CEGs amongst bacterial genomes. To expand our predictions, we also used the CONJscan module of Macsyfinder to trace the MPF families likely to be involved in HGT. Our analysis on the co-occurrence of relaxases with MPF families (Tables 1 and S5) is in agreement with the combinations observed by Guglielmini and colleagues [11]; all ICEs belonging to the MPFG class carried a MOBH relaxase, and the MOBP and MOBF relaxases were linked to MPFT and MPFF, respectively. All of the MPFG class ICEs described here present a MPF class/relaxase/integrase profile that resembles that from ICEclc. Even though pKLC102 is also a representative of the MPFG class and carries a MOBH relaxase, it uses a DNA_BRE_C integrase instead of a INT_P4_C. The absence of CEG-carrying ICEs from the pKLC102 family has already been reported [6].
The scenario observed for the acquisition of the most important CEGs by ICEs (ISs for bla NDM-1 and bla SPM-1, class I integrons for bla IMP and bla VIM and Tn4401 for bla KPC-2) resembles that of plasmids [3] and provides additional support for the notion that the line separating these elements is blurred [13, 14]. We now show that besides plasmids, this promiscuous repertoire of integrons and transposons frequently targets ICEs of different MPF families. Even though CEGs might spread rapidly worldwide, local selection is likely required for them to reach fixation, as can be seen for the clonal expansion of P. aeruginosa ST277 harbouring bla SPM-1 [36]. Surprisingly, we noted that this gene was not detected beyond the same clonal lineage. Indeed, all hits were identified in P. aeruginosa ST277 strains from Brazil and within a MPFT ICE family, indicating that these ICEs are transferring vertically and/or horizontally within this STs. Understanding the limitations on HGT of this family of ICE may be translatable to other, more transferable, ICEs and could underpin a control strategy to prevent the spread of these elements in the future.
Although bla KPC and bla OXA were the most frequently identified CEGs within the analysed genomes (Table S2), we only found five ICEs carrying bla KPC-2 and two ICEs with bla OXA genes (Table S8). The bla KPC genes are mostly located in Tn4401 transposons that target K. pneumoniae plasmids, while the bla OXA genes are frequently associated with ISs that tend to target Acinetobacter spp. chromosomes and plasmids [3]. These genes may take advantage of the copy number and the higher genetic plasticity of plasmids [34]. This plasticity may increase the rate at which novel mutations appear and the high copy number may amplify the effect due to the increased gene dosage [40].
In addition to AR, ICEs can be involved in other adaptive traits such as carbon source utilization, symbiosis, restriction–modification, and siderophore and bacteriocin synthesis [14]. Since bacteria commonly inhabit highly competitive environments, the production of specific secondary metabolites (such as bacteriocins and siderophores) may confer a selective advantage to the host [41]. We speculate that the presence of these metabolites within the ICEs here characterized may promote their stability by preferentially selecting for cells harbouring the ICE. CRISPR-Cas systems are rarely found on MGEs, and the type I-C systems carried within pKLC102-related ICEs are one of the few examples [9]. Since none of the ICEs described here belong to the pKLC102 family, the absence of CRISPR-Cas systems within our dataset was expected.
One caveat of our studies is that these results do not yet expose the complete set of CEG-bearing ICEs present in all bacteria. There is an inherent bias in the number of times we detect a particular CEG in certain bacterial genomes, as some are over-represented in the database compared to others. It is possible that certain observations will flatten out as more genomes are analysed. Fewer than 10 % of the bacterial genomes currently present in the NCBI database are complete. This is a major drawback, since the putative ICEs present in draft genomes tend to be fragmented due to the presence of repetitive regions that are not resolved using short-read sequencing. Further, the relaxase and the T4SS encoded by ICEs resemble those of plasmids [11, 13, 42]. Plus, it is possible that ICEs and plasmids have swapped conjugation modules throughout their evolutionary history [11]. We believe that a more thorough exploration of this issue, especially regarding the precise delimitation of ICEs, will be an important further step toward an improved understanding of the contribution of these elements to bacterial adaptation and the evolution of AR.
While we have chosen to focus on CEG-bearing genomes, our computational approach can be applied to trace other relevant AR genes and other cargo genes that may confer a selective advantage to the ICE host. Leveraging knowledge linking the accurate prediction of ICE sequences to the carriage of AR genes will not only improve our understanding of HGT, but may also uncover potential approaches to tackle the spread of AR.
Supplementary Data
Funding information
This work was supported by the Applied Molecular Biosciences Unit – UCIBIO which is financed by national funds from FCT (UIDB/04378/2020).
Author contributions
J. B. conceptualized the project. J. B. ran the analyses. J. B. and J. M. analysed the data. J. B. wrote the manuscript. J. M., A. P. R. and L. P. edited and revised the manuscript. All authors read, commented on and approved the final manuscript.
Conflicts of interest
The authors declare that there are no conflicts of interest.
Footnotes
Abbreviations: AR, antibiotic resistance; CEG, carbapenemase-encoding gene; HGT, horizontal gene transfer; ICE, integrative and conjugative element; MGE, mobile genetic element; MPF, mating pair formation; NCBI, National Center for Biotechnology Information; T4SS, type IV secretion system; WHO, World Health Organization.
All supporting data has been provided within the article or through supplementary data files. Ten supplementary tables are available with the online version of this article.
The DNA sequences used in this study are available in the NCBI database, under the accession numbers provided in supplementary tables S1–S9.
References
- 1.EFSA Panel on Biological Hazards (BIOHAZ) Scientific opinion on carbapenem resistance in food animal ecosystems. EFSA J. 2013;11 [Google Scholar]
- 2.World Health Organization Global priority list of antibiotic-resistant bacteria to guide research, discovery, and development of new antibiotics. http://www.who.int/news-room/detail/27-02-2017-who-publishes-list-of-bacteria-for-which-new-antibiotics-are-urgently-needed 14 Aug 2020.
- 3.Partridge SR, Kwong SM, Firth N, Jensen SO. Mobile genetic elements associated with antimicrobial resistance. Clin Microbiol Rev. 2018;31:e00088–17. doi: 10.1128/CMR.00088-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Botelho J, Grosso F, Peixe L. Antibiotic resistance in Pseudomonas aeruginosa - Mechanisms, epidemiology and evolution. Drug Resist Updat. 2019;44:100640. doi: 10.1016/j.drup.2019.07.002. [DOI] [PubMed] [Google Scholar]
- 5.Queenan AM, Bush K. Carbapenemases: the versatile beta-lactamases. Clin Microbiol Rev. 2007;20:440–458. doi: 10.1128/CMR.00001-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Botelho J, Roberts AP, León-Sampedro R, Grosso F, Peixe L. Carbapenemases on the move: it’s good to be on ICEs. Mob DNA. 2018;9:37. doi: 10.1186/s13100-018-0141-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lam MMC, Wyres KL, Duchêne S, Wick RR, Judd LM, et al. Population genomics of hypervirulent Klebsiella pneumoniae clonal-group 23 reveals early emergence and rapid global dissemination. Nat Commun. 2018;9:2703. doi: 10.1038/s41467-018-05114-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Libante V, Nombre Y, Coluzzi C, Staub J, Guédon G, et al. Chromosomal conjugative and mobilizable elements in Streptococcus suis: major actors in the spreading of antimicrobial resistance and bacteriocin synthesis genes. Pathogens. 2019;9:22. doi: 10.3390/pathogens9010022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.van Belkum A, Soriaga LB, LaFave MC, Akella S, Veyrieras J-B, et al. Phylogenetic distribution of CRISPR-Cas systems in antibiotic-resistant Pseudomonas aeruginosa . mBio. 2015;6:e01796–15. doi: 10.1128/mBio.01796-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fang Y, Wang Y, Li Z, Liu Z, Li X, et al. Distribution and genetic characteristics of SXT/R391 integrative conjugative elements in Shewanella spp. from China. Front Microbiol. 2018;9:920. doi: 10.3389/fmicb.2018.00920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guglielmini J, Quintais L, Garcillán-Barcia MP, de la Cruz F, Rocha EPC. The repertoire of ICE in prokaryotes underscores the unity, diversity, and ubiquity of conjugation. PLoS Genet. 2011;7:e1002222. doi: 10.1371/journal.pgen.1002222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Carraro N, Burrus V. The dualistic nature of integrative and conjugative elements. Mob Genet Elements. 2015;5:98–102. doi: 10.1080/2159256X.2015.1102796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cury J, Touchon M, Rocha EPC. Integrative and conjugative elements and their hosts: composition, distribution and organization. Nucleic Acids Res. 2017;45:8943–8956. doi: 10.1093/nar/gkx607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Johnson CM, Grossman AD. Integrative and conjugative elements (ICEs): what they do and how they work. Annu Rev Genet. 2015;49:577–601. doi: 10.1146/annurev-genet-112414-055018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gaillard M, Vallaeys T, Vorhölter FJ, Minoia M, Werlen C, et al. The CLC element of Pseudomonas sp. strain B13, a genomic island with various catabolic properties. J Bacteriol. 2006;188:1999–2013. doi: 10.1128/JB.188.5.1999-2013.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Waldor MK, Tschäpe H, Mekalanos JJ. A new type of conjugative transposon encodes resistance to sulfamethoxazole, trimethoprim, and streptomycin in Vibrio cholerae O139. J Bacteriol. 1996;178:4157–4165. doi: 10.1128/JB.178.14.4157-4165.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Klockgether J, Reva O, Larbig K, Tümmler B. Sequence analysis of the mobile genome island pKLC102 of Pseudomonas aeruginosa C. J Bacteriol. 2004;186:518–534. doi: 10.1128/JB.186.2.518-534.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Springael D, Kreps S, Mergeay M. Identification of a catabolic transposon, Tn4371, carrying biphenyl and 4-chlorobiphenyl degradation genes in Alcaligenes eutrophus A5. J Bacteriol. 1993;175:1674–1681. doi: 10.1128/JB.175.6.1674-1681.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Botelho J, Schulenburg H. The role of integrative and conjugative elements in antibiotic resistance evolution. Trends Microbiol. 2020;S0966-842X(20)30137-2 doi: 10.1016/j.tim.2020.05.011. [DOI] [PubMed] [Google Scholar]
- 20.Delavat F, Miyazaki R, Carraro N, Pradervand N, van der Meer JR. The hidden life of integrative and conjugative elements. FEMS Microbiol Rev. 2017;41:512–537. doi: 10.1093/femsre/fux008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Smillie C, Garcillán-Barcia MP, Francia MV, Rocha EPC, de la Cruz F. Mobility of plasmids. Microbiol Mol Biol Rev. 2010;74:434–452. doi: 10.1128/MMBR.00020-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Feldgarden M, Brover V, Haft DH, Prasad AB, Slotta DJ, et al. Validating the AMRFinder tool and resistance gene database by using antimicrobial resistance genotype-phenotype correlations in a collection of isolates. Antimicrob Agents Chemother. 2019;63. doi: 10.1128/AAC.00483-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Buchfink B, Xie C, Huson DH. Fast and sensitive protein alignment using diamond. Nat Methods. 2015;12:59–60. doi: 10.1038/nmeth.3176. [DOI] [PubMed] [Google Scholar]
- 24.Eddy SR. Accelerated profile HMM searches. PLoS Comput Biol. 2011;7:e1002195. doi: 10.1371/journal.pcbi.1002195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Garcillán-Barcia MP, Redondo-Salvo S, Vielva L, de la Cruz F. MOBscan: automated annotation of MOB relaxases. Methods Mol Biol. 2020;2075:295–308.. doi: 10.1007/978-1-4939-9877-7_21. Humana Press Inc. [DOI] [PubMed] [Google Scholar]
- 26.Abby SS, Néron B, Ménager H, Touchon M, Rocha EPC. MacSyFinder: a program to mine genomes for molecular systems with an application to CRISPR-Cas systems. PLoS One. 2014;9:e110726. doi: 10.1371/journal.pone.0110726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Abby SS, Cury J, Guglielmini J, Néron B, Touchon M, et al. Identification of protein secretion systems in bacterial genomes. Sci Rep. 2016;6:23080. doi: 10.1038/srep23080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jolley KA, Bray JE, Maiden MCJ. Open-access bacterial population genomics: BIGSdb software, the PubMLST.org website and their applications. Wellcome Open Res. 2018;3:124. doi: 10.12688/wellcomeopenres.14826.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Partridge SR, Tsafnat G. Automated annotation of mobile antibiotic resistance in gram-negative bacteria: the multiple antibiotic resistance Annotator (MARA) and database. J Antimicrob Chemother. 2018;73:883–890. doi: 10.1093/jac/dkx513. [DOI] [PubMed] [Google Scholar]
- 30.Roberts RJ, Vincze T, Posfai J, Macelis D. REBASE–a database for DNA restriction and modification: enzymes, genes and genomes. Nucleic Acids Res. 2015;43:D298–D299. doi: 10.1093/nar/gku1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Couvin D, Bernheim A, Toffano-Nioche C, Touchon M, Michalik J, et al. CRISPRCasFinder, an update of CRISRFinder, includes a portable version, enhanced performance and integrates search for Cas proteins. Nucleic Acids Res. 2018;46:W246–W251. doi: 10.1093/nar/gky425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Blin K, Shaw S, Steinke K, Villebro R, Ziemert N, et al. antiSMASH 5.0: updates to the secondary metabolite genome mining pipeline. Nucleic Acids Res. 2019;47:W81–W87. doi: 10.1093/nar/gkz310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huerta-Cepas J, Szklarczyk D, Heller D, Hernández-Plaza A, Forslund SK, et al. eggNOG 5.0: a hierarchical, functionally and phylogenetically annotated orthology resource based on 5090 organisms and 2502 viruses. Nucleic Acids Res. 2019;47:D309–D314. doi: 10.1093/nar/gky1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cury J, Oliveira PH, de la Cruz F, Rocha EPC. Host range and genetic plasticity explain the coexistence of integrative and extrachromosomal mobile genetic elements. Mol Biol Evol. 2018;35:2230–2239. doi: 10.1093/molbev/msy123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ding Y, Teo JWP, Drautz-Moses DI, Schuster SC, Givskov M, et al. Acquisition of resistance to carbapenem and macrolide-mediated quorum sensing inhibition by Pseudomonas aeruginosa via ICETn4371 6385 . Commun Biol. 2018;1:57. doi: 10.1038/s42003-018-0064-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nascimento APB, Ortiz MF, Martins WMBS, Morais GL, Fehlberg LCC, et al. Intraclonal genome stability of the metallo-β-lactamase SPM-1-producing Pseudomonas aeruginosa ST277, an endemic clone disseminated in Brazilian hospitals. Front Microbiol. 2016;7:1946. doi: 10.3389/fmicb.2016.01946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cuzon G, Naas T, Nordmann P. Functional characterization of Tn4401, a Tn3-based transposon involved in bla KPC gene mobilization. Antimicrob Agents Chemother. 2011;55:5370–5373. doi: 10.1128/AAC.05202-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tohya M, Tada T, Watanabe S, Kuwahara-Arai K, Zin KN, et al. Emergence of carbapenem-resistant Pseudomonas asiatica producing NDM-1 and VIM-2 metallo-β-lactamases in Myanmar. Antimicrob Agents Chemother. 2019;63 doi: 10.1128/AAC.00475-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Banerjee R, Weisenhorn E, Schwartz KJ, Myers KS, Glasner JD, et al. Tailoring a global iron regulon to a uropathogen. mBio. 2020;11:e00351-20. doi: 10.1128/mBio.00351-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.San Millan A, Escudero JA, Gifford DR, Mazel D, MacLean RC. Multicopy plasmids potentiate the evolution of antibiotic resistance in bacteria. Nat Ecol Evol. 2017;1:0010. doi: 10.1038/s41559-016-0010. [DOI] [PubMed] [Google Scholar]
- 41.Inglis RF, Bayramoglu B, Gillor O, Ackermann M. The role of bacteriocins as selfish genetic elements. Biol Lett. 2013;9:20121173. doi: 10.1098/rsbl.2012.1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Guglielmini J, Néron B, Abby SS, Garcillán-Barcia MP, de la Cruz F, et al. Key components of the eight classes of type IV secretion systems involved in bacterial conjugation or protein secretion. Nucleic Acids Res. 2014;42:5715–5727. doi: 10.1093/nar/gku194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Abril D, Marquez-Ortiz RA, Castro-Cardozo B, Moncayo-Ortiz JI, Olarte Escobar NM, et al. Genome plasticity favours double chromosomal Tn4401b-bla KPC-2 transposon insertion in the Pseudomonas aeruginosa ST235 clone. BMC Microbiol. 2019;19:45. doi: 10.1186/s12866-019-1418-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Urbanowicz P, Izdebski R, Baraniak A, Żabicka D, Ziółkowski G, et al. Pseudomonas aeruginosa with NDM-1, DIM-1 and PME-1 β-lactamases, and RmtD3 16S rRNA methylase, encoded by new genomic islands. J Antimicrob Chemother. 2019;74:3117–3119. doi: 10.1093/jac/dkz262. [DOI] [PubMed] [Google Scholar]
- 45.Khan A, Shropshire WC, Hanson B, Dinh AQ, Wanger A, et al. Simultaneous Infection with Enterobacteriaceae and Pseudomonas aeruginosa Harboring Multiple Carbapenemases in a Returning Traveler Colonized with Candida auris . Antimicrob Agents Chemother. 2019;64 doi: 10.1128/AAC.01466-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhan Z, Hu L, Jiang X, Zeng L, Feng J, et al. Plasmid and chromosomal integration of four novel blaIMP-carrying transposons from Pseudomonas aeruginosa, Klebsiella pneumoniae and an Enterobacter sp. J Antimicrob Chemother. 2018;73:3005–3015. doi: 10.1093/jac/dky288. [DOI] [PubMed] [Google Scholar]
- 47.McCracken MG, Adam HJ, Blondeau JM, Walkty AJ, Karlowsky JA, et al. Characterization of carbapenem-resistant and XDR Pseudomonas aeruginosa in Canada: results of the CANWARD 2007-16 study. J Antimicrob Chemother. 2019;74:iv32–iv38. doi: 10.1093/jac/dkz285. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.