Summary
Deep learning, a set of approaches using artificial neural networks, has generated rapid recent advancements in machine learning. Deep learning does, however, have the potential to reduce the reproducibility of scientific results. Model outputs are critically dependent on the data and processing approach used to initially generate the model, but this provenance information is usually lost during model training. To avoid a future reproducibility crisis, we need to improve our deep-learning model management. The FAIR principles for data stewardship and software/workflow implementation give excellent high-level guidance on ensuring effective reuse of data and software. We suggest some specific guidelines for the generation and use of deep-learning models in science and explain how these relate to the FAIR principles. We then present dtoolAI, a Python package that we have developed to implement these guidelines. The package implements automatic capture of provenance information during model training and simplifies model distribution.
Keywords: data, data management, AI, artificial intelligence, deep learning, machine learning, reproducibility, provenance, FAIR data
Highlights
-
•
We provide guidelines for improving the reproducibility of deep-learning models
-
•
Our Python package, dtoolAI, is a proof-of-concept implementation of these guidelines
-
•
dtoolAI supports automatic provenance annotation for DL models
The Bigger Picture
Science has made use of machine learning, a way of teaching computers to understand patterns in data, for a long time. Deep learning, based on the way that real brains process data, has brought enormous improvements in the speed and accuracy of image and language processing over the last few years. However, the “black box” nature of deep-learning models makes scientific analyses that make use of them difficult to reproduce.
In this work, we show how we might be able to improve long-term reproducibility for data analyses that rely on deep-learning models. We do this by giving guidance on how specific aspects of the FAIR principles for data management can be applied to training and using these models. We also present dtoolAI, a software tool and code library we have developed. We hope that in the future, adoption of our guidelines or similar principles will improve our collective trust in results that arise from deep learning.
Deep learning has brought impressive advances in our ability to extract information from data. However, models produced by these techniques are often difficult to reproduce or interpret. We provide guidelines for improving the reproducibility of deep-learning models, together with the Python package dtoolAI, a proof-of-concept implementation of these guidelines.
Introduction
Machine learning (ML) is a discipline involving algorithms, models, and analysis techniques that carry out tasks by making use of patterns in data, with minimal explicit rules. Deep learning (DL) approaches are a subset of ML, which is itself a subdiscipline of more general artificial intelligence.1 DL techniques make use of artificial neural networks, simulated systems that mirror aspects of the way that real neurons work. These are responsible for many of the recent advances in ML as a whole, particularly in domains such as image recognition2 and natural language processing.3 These advances have resulted in great excitement about the possibilities of DL approaches within scientific workflows. Within our own discipline (biology), DL has been applied to a wide range of problems such as cell image segmentation,4 genomic variant calling,5 and transcription factor binding site prediction,6 among others.
Reproducibility is a key pillar of scientific integrity. Results that support hypotheses must be replicable by others, within reasonable parameters.7 This reproducibility has come under close scrutiny recently, with initial attention directed toward the reproducibility of studies in psychology8 before widening to science as a whole.9
The complexity of modern data analysis pipelines complicates reproducibility. Data analysis often involves the application of many different computational tools. The output of these pipelines (and hence the results that support or contradict scientific hypotheses) are often critically dependent on the precise functioning of these tools, which can make reproduction of their results difficult without detailed description of all parts of the pipeline.
While DL holds great potential for faster and more powerful analyses, it also presents a set of new challenges that combine these two problems of analysis pipeline complexity and reproducibility. In this paper, we explain the problems that DL can create. We also discuss how the FAIR principles for software and data have provided solutions to avoid or mitigate these problems in other domains. We then explain our guidelines for implementing specific FAIR principles in the domain of DL and present the software tool we have developed to implement these guidelines.
Reproducibility: Concepts and Terminology
Scientific research has always been dependent on our ability to repeat experiments and reproduce their results. As the use of computational approaches has developed, the reproducible research movement for software and data10 has grown to become an important part of modern research.11
Reproducibility can carry different meanings in the context of science, particularly where computational approaches are concerned. Three similar terms, reproducibility, repeatability, and replicability, are used by different groups in different contexts, often with different sets of meanings. This problem is discussed in depth by the National Academies of Science, Engineering, and Medicine11 and Barba.12 Here we will clarify our understanding of these terms and explain what we intend by them throughout the paper.
We will use the terms as follows, corresponding to classification B1 in Barba's system:
Reproducibility is the ability to regenerate results using the original researchers' data, software, and parameters. Replicability is the ability to arrive at the same result using new data. Repeatability is the ability to rerun a published analysis pipeline and arrive at the same results (see, e.g., Krishnamurthi and Vitek13).
With this classification, we are most concerned with reproducibility and repeatability. We care about the ability to repeat the generation of a DL model in order to reproduce its results. Since exact weight values (which define the trained model) usually depend on random initialization, we expect only to reproduce a model's results within some given tolerance.
Provenance
The provenance of a computational object, such as a trained DL model, is the history of the processes used to produce it, together with their input data.14 Provenance is a key pillar of reproducibility, since providing the information necessary to allow analysis to be rerun and artifacts to be regenerated requires recording the processes of creating those artifacts and analyses.
We can distinguish between prospective provenance, whereby we capture the specification for how we will generate data, and retrospective provenance, which captures past data derivation.15 When we look at the problems inherent in DL model training, we will be primarily concerned about retrospective provenance. In particular, we will argue that the provenance of a DL model must include the data used to train that model as well as the training parameters and hyperparameters.
How Deep Learning Works
A DL model has two parts, a model architecture and a set of model weights.
The model architecture describes the components of the model and how they will take the inputs to the model and transform them to produce outputs. DL models often consist of many layers of artificial neurons. A single model might have tens of thousands of neurons in total, and the architecture describes how these are connected to each other.
Each of these connections can have a different strength, and collectively a set of connection strengths is called model weights. The model architecture and model weights together constitute a usable model.
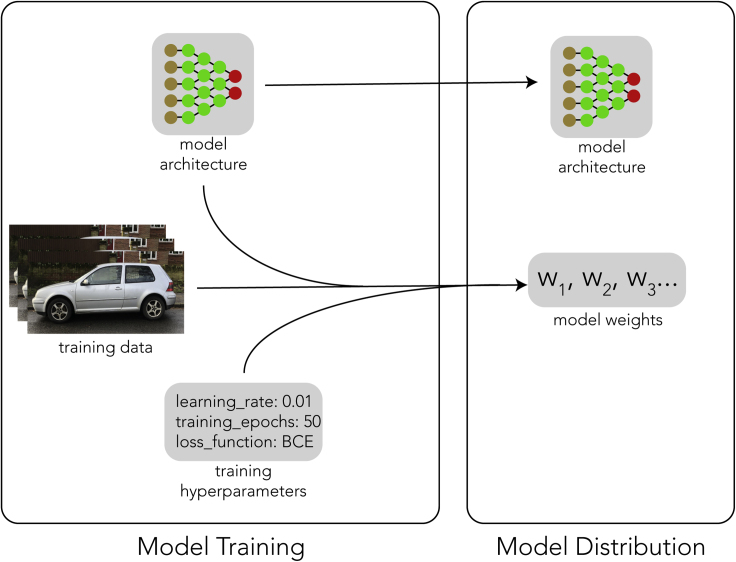
The process of training a model involves repeatedly supplying the model with data and some instructions as to how the model's response to that data should be used to update the model weights. The end result of this process is a specific set of model weights (Figure 1).
Figure 1.
The DL Model Training Process
Model training uses a model architecture, weights, and hyperparameters in order to produce model weights. The model architecture and model weights are distributed together as a usable model. Training data and hyperparameters are not, generally, extractable from the model.
ML and Reproducibility: Data
The first challenge that ML poses to reproducibility involves the training data and the training process. Since model weights depend on training data, and the operation of the model depends on those weights, we cannot reproduce the model without the training data. While the model weights arise from these data, the training process is (in general) not reversible and we cannot extract the data from the weights.
Introductory examples and tutorials in ML often use well-understood and "ready-packaged" datasets such as ImageNet,16 CIFAR,17 or MNIST.18 However, when more specialized models are trained, the input data are usually highly specific, hand-curated datasets. This problem extends beyond reproducibility. The power of ML models lies in their ability to generalize beyond their training data. This generalization is very dependent on the range of those data. Without knowing the data on which a model was trained, it can be difficult to understand what the limitations of the model will be.
ML and Reproducibility: Training
While model weights depend on training data, they also depend on the parameters of that training process. These parameters are often referred to as hyperparameters to distinguish them from the model weights themselves. Some of these hyperparameters and other factors that we need to know in order to reproduce models are:
-
•
The loss function used during training. This function determines how the model's performance against data with known results is scored.
-
•
The type of optimizer used during training. The optimizer determines how the model weights are updated in response to the loss of function.
-
•
The learning rate applied during the training. This determines how fast model weights are updated in response to the optimizer output.
-
•
How input data are preprocessed. Often "data augmentation" is applied to training data in image processing networks, for example. This involves applying randomly selected transforms such as rotation, cropping, or zooming to images to prevent the model from learning very specific features of input data.
Each of these hyperparameters may have its own parameters—for example, some optimizers have many different parameters, or learning rates might change over the training life cycle.
Model Distribution
Training a model is usually a much more expensive operation (in terms of computation cost) than using it. For this reason, models are usually trained on much more specialized computer hardware than that where they are applied. This distribution process needs to transfer both the model architecture and model weights. Models are often updated (i.e., retrained) with new data and these updated model weights also need to be transferred.
Current Solutions
Because DL model training and application involves both data and software, existing work on the reproducibility of both is an important step toward developing better DL.
Data Management
Effective data management in science is an important subdiscipline in its own right, in which substantial progress has been made. The FAIR principles (Findability, Accessibility, Interoperability, Reusability) have crystallized a set of high-level guidelines for how data can be stored in a way that best encourages reproducibility and reuse.19
While these high-level principles are a critical overarching guide, they do not provide specific guidance on detailed domain-specific implementation, which is needed in the case of DL. We will discuss how to apply the FAIR principles to DL specifically when we turn to potential improvements to DL reproducibility.
Software and Workflow
DL models are trained from data, and the model weights produced by their training are also data. However, they require software for instantiation, and are both trained and used as part of wider workflows.
Recent developments have looked at how to incorporate FAIR principles in computational workflows. The challenges of doing this are described in Lamprecht et al.,20 and specific suggestions as to how to make FAIR computational workflows in Goble et al.21 Steps toward this process include ensuring that the metadata used and generated during workflows are recorded with analysis results and artifacts created. Ivie and Thain22 provide a comprehensive overview of the challenges of both the theoretical and practical challenges of creating reproducible workflows. These authors also describe a range of tools, components, and concepts that can be used to solve these problems, at least in part.
Many computational experiments and analyses are carried out by scripts rather than full workflow management systems. Scripts offer a quick and flexible way to get analyses up and running.23 However, the lack of a systematic way to manage versions and metadata within scripts can make determining the provenance of results and artifacts more difficult, a problem analyzed in detail by Pimentel et al.24
ML Model Management
There are a number of solutions aimed at managing the process of experimenting with model training in ML. For example, cometML25 and similar systems provide online tools for recording training experiments. These tools are primarily aimed at keeping track of model evaluation scores and hyperparameters for different models. This is important for finding the best model architecture and hyperparameters for a given problem, but is a different set of concerns from reproducibility and data provenance.
ML Schema26 proposes an ontology for representing ML models and environments. Such a schema is an important step toward developing reproducible DL models if applied within suitable ML model training and application systems.
Model Distribution
Model distribution is often managed by providing model architecture as source code in a hosting platform such as GitHub, and model weights via cloud storage such as Amazon S3. This provides easy access to the weights but does not provide a mechanism to associate them with the input data that produced them. This is a critical missing piece of the provenance information of that trained model.
Summary
-
•
We cannot reproduce a DL model, or even properly understand its limitations, without access to the data on which it was trained.
-
•
We also need the details of that training process, particularly hyperparameters, to reproduce the model.
-
•
At the moment, model distribution does not usually include these key metadata.
-
•
Currently, most DL models in use are not reproducible, at least by their end users, nor do these models include the provenance information we need to properly understand their strengths and limitations.
-
•
FAIR data and software principles offer high-level solutions to these problems. They have been applied successfully in many domains of computational science, and providing specific implementations for DL would be beneficial.
Results
Guidelines and Practice
The FAIR principles for data management, and recent developments in adapting and applying these principles to software, scripts, and workflows, provide an excellent framework for developing domain-specific solutions for better reproducibility. In this section, we discuss first the high-level guidelines that we have found useful for applying specific FAIR principles to solve or mitigate the problems we have described above. Second, we present the tool that we have developed to implement these guidelines.
Guidelines for Reproducible Deep Learning
Annotate Model Training Data with Metadata
Good data management practices necessitate that data have appropriate metadata to allow them to be understood.27 Since ML models are derived from their input data, we need to ensure that these input data have suitable metadata before models based on them are trained. These metadata should follow a schema appropriate to the setting within which they will be used. Standards such as Bioschemas28 provide a good general scheme for biological datasets (with specific adaptations to particular biological domains).
Give Those Data Persistent URIs
Reproducibility and model data provenance require us to be able to consistently refer to data on which models were trained. Achieving this consistency requires us to be able to refer uniquely to models' input data, which requires unique identifiers.
When models, or their downstream results, are published, the data used to train them should be made publicly available with a persistent identifier. Where possible, ML specific repositories, such as OpenML,29 should be used for this to increase discoverability of data. These repositories will require specific schemas which should be kept in mind during model development.
Capture Training Parameters at Model Training Time
While the data used to train a model are the primary determinant of what that model will do, model and training hyperparameters are also a key input into the model without which the model cannot be reproduced. Hence, it is critical to record those hyperparameters at training time. This should be done using schemas that will be consistent across training and application of multiple models, for example that proposed in ML-Schema,26 which includes defining and annotating hyperparameters.
Store These Training Parameters and Data Inputs Together with the Model
Model hyperparameters are usually defined in the program code used to train the model. While effective tools to manage program source code exist, it is very easy for the information to be either lost or separated from the model. For example, if several updates to the training code are made, each of which results in a different set of parameters and therefore a different model, associating each model with its version of program code is difficult.
Ensuring that we store these parameters together with the model avoids this danger. This means that we require a storage format (or mechanism) for ML models that also incorporates metadata about how they were trained.
Summary
Together, we can summarize these guidelines as:
-
1
Provide appropriate metadata (with domain-appropriate schema) and persistent URIs for model training data.
-
2
Add this information, together with training hyperparameters, as metadata to the generated model.
Relationship with the FAIR Principles
These guidelines implement specific aspects of the FAIR principles, giving particular focus to those elements that we consider most critical for the specific problems of DL model training and distribution. Here, we clarify this relationship using the definitions of specific subprinciples in Box 2 of Wilkinson et al.19
The FAIR principles that are most important in training DL models in a way that support provenance annotation and reproducibility are:
-
•
F1. (Meta)data are assigned a globally unique and persistent identifier.
-
•
F2. Data are described with rich metadata.
-
•
A1. (Meta)data are retrievable by their identifier using a standardized communications protocol.
-
•
R1. Metadata are richly described with a plurality of accurate and relevant attributes.
We require F1 to ensure that DL model training data are persistently identifiable, together with F2 to ensure that model consumers can understand the model's provenance. A1 and R1 allow those metadata to be used by model training software.
When creating a trained model, we also rely on:
-
•
I3. (Meta)data include qualified references to other (meta)data.
-
•
R1.2. (Meta)data are associated with detailed provenance.
I3 ensures that the trained model references its training data, and R1.2, in the context of DL model training, requires encoding training hyperparameters and the details of any data preprocessing applied.
Implementation in dtoolAI
To demonstrate application of these guidelines and to improve reproducibility of DL models in both our own work and within our institution, we have developed dtoolAI. To do this, we made use of the existing dtool library30 to take advantage of its features.
dtool is a software application programming interface (API) and set of tools to make managing heterogeneous data easier without requiring expensive centralized infrastructure. It provides the ability to annotate data with metadata that is both human readable and programmatically accessible. It also allows attaching unique URIs (Universal Resource Identifiers) to datasets. These URIs can refer to cloud-hosted data (for example, Amazon S3 or Azure storage) allowing datasets to be both uniquely identifiable and widely accessible.
We developed dtoolAI on top of dtool to take advantage of dtool's capabilities for managing metadata programmatically, as well as providing URIs. The key feature of dtoolAI is that it makes it easier to automatically capture data inputs and model hyperparameters at model training time and to distribute those metadata with the model.
Code Architecture
In this section, we explain how dtoolAI is arranged and packaged. dtoolAI is a package for the popular Python programming language. Python is widely used in the scientific community. It is an interpreted language and generally slower than compiled languages for direct execution of numerical code. However, most DL model work is carried out with frameworks such as Tensorflow or Pytorch in which actual computation is carried out in highly optimized code written in a compiled language but accessed via a binding language such as Python. dtoolAI uses the Pytorch framework.31 This provides the advantages of working with a user-friendly language suitable for rapid development without compromising on execution speed. Another advantage of Python is its simple package management system, pip. We can use this to install dtoolAI with the command line instruction:
pip install dtoolai
dtoolAI then provides a set of useful tools and functions that can be employed by inclusion in Python scripts and programs. The most important are:
-
1
A base class that allows encapsulation of data in a form that both dtool and Pytorch understand, together with developed subclasses showing how to use this for image and tensor data.
-
2
Functions to train a DL model while capturing metadata about the model inputs and training parameters.
-
3
Classes/functions to use the models trained in this way by applying them either to fully constructed datasets or to individual data, such as images.
dtoolAI, dtool, and Pytorch
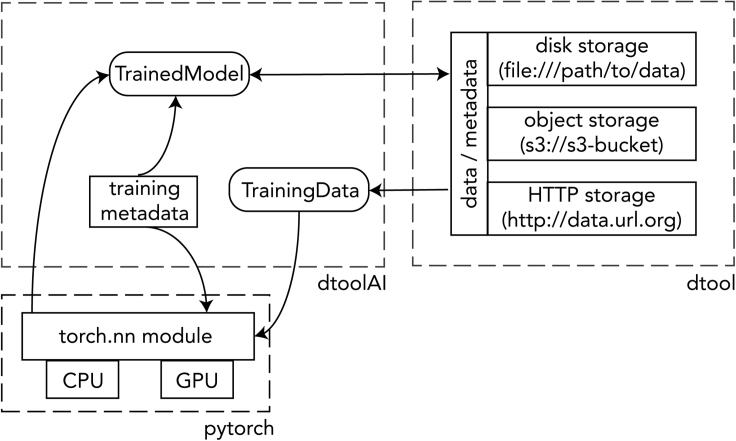
Functionally, dtoolAI acts as a bridge between dtool and Pytorch. Here we explain the relationship between the three libraries, with Figure 2 for illustration.
Figure 2.
Relationship between dtoolAI, dtool, and pytorch
End users of the library interact directly with the dtoolAI code. dtoolAI provides the classes and functions necessary to load model training data, train DL models, and store the resulting models together with key metadata providing the provenance of those models.
dtoolAI provides the direct interface for user code. It has responsibility for managing the process of transforming training data and parameters into a trained DL model while capturing training metadata and ensuring that references to input data are maintained.
dtool provides two key functions:
-
1
Storage abstraction, in particular the ability for users of dtoolAI to interact with both DL model training data and trained model weights in persistent, world-accessible storage (object storage, or data served via HTTP(S) requests).
-
2
A programmatic interface for storage and retrieval of both data and metadata associated with a DL model, allowing dtoolAI to programmatically use annotations of input data to guide model training, as well as encode that training process in the trained model objects. This programmatic access allows setting and validation of particular schemas for both for model training data and model metadata.
Pytorch provides the core neural network calculation functions for training and application of DL models. dtoolAI provides it with either suitably formatted input data and parameters to train models, or trained model weights and unlabeled data for classification. It can run on either CPU or GPU, allowing for accelerated model training and application.
ML Model Workflows
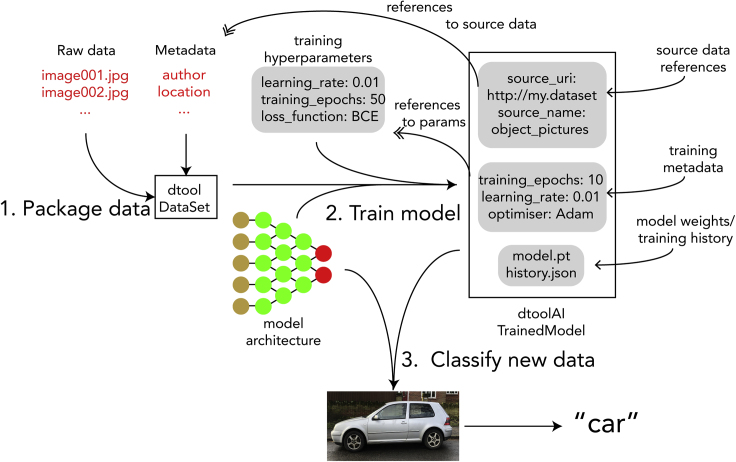
The workflow of using dtoolAI to train, distribute, and use ML models is:
-
1
Use dtoolAI's helper functions, or the dtool library itself to create a suitable training dataset annotated with relevant metadata. Through its templating system, dtool allows specification of metadata schemas, to be entered either manually or through its API.
-
2
Use the provided dtoolAI library functions to train the model, during which process the training data identifiers and chosen training hyperparameters are automatically captured and recorded. The resulting model is another dtool dataset.
-
3
Use the model, by employing the dtoolAI API functions. The model can be distributed as a self-contained dataset with model weights, training hyperparameters, and references to the original training data.
This workflow is illustrated in Figure 3.
Figure 3.
Creating Reproducible Models with dtoolAI
(1) We first create a dataset that combines both training data and metadata. (2) We then train a model architecture with our training dataset and hyperparameters. The resulting model dataset captures these parameters as well as references to the training data. (3) We can then use the resulting model for predictions or distribute it.
When training, it is possible to use more than one dataset to train the model as long as those datasets share the same set of metadata. For example, multiple datasets containing images of flowers could be used to train an image categorization network if each of those datasets provided the same metadata labeling the images.
Discussion
ML approaches have been an integral part of many scientific workflows and analysis pipelines since they were first developed.32 DL's impressive achievements in image recognition, natural language processing, and reinforcement learning have already started to translate to scientific advances.33 DL models are, however, difficult to interpret. Models operate on input data and produce results, but determining how they produce these results can be very difficult. Understandable ML has become a research field in its own right.34
A further problem is possible bias. ML models attempt to generalize the input data they are given. Therefore, if there are biases in the training data for a model, those biases will be reflected in the model's performance on real data. Similarly, if the real data to which the model is applied are too different from its training data, the model may not be able to generalize enough to give good results. These challenges are particularly problematic for scientific applications of ML. We need the results of scientific experiments to be reproducible. We also often apply existing techniques to new sets of data or problem domains and so need to understand the limitations of those techniques.
For all of these reasons, we need our ML models to carry retrospective provenance information with them, particularly unique identifiers for their training data, together with the parameters for preprocessing those data and training the model.
The FAIR principles—Findability, Accessibility, Interoperability, and Reusability—are designed to enhance the reuse of data. While originally developed for data management, recent work has shown how these principles can be adapted to software and workflows.20,21 We have provided DL-specific guidance as to how the FAIR principles should be applied to improve reproducibility without compromising the speed of experimentation.
Ensuring that model training data are annotated with appropriate metadata and available at a persistent URI ensures that models trained from those data can reference that URI as part of their provenance information. During that training process, we also need to capture training hyperparameters and any preprocessing applied to the data during training. Together, these aspects of provenance provide sufficient information to reproduce the model within tolerances.
The final step to reproducibility is establishing a self-contained distribution format for ML models that combines both model weights and provenance metadata. Ideally we want both to be able to read these data directly as humans (and cross-reference the training data, so we can better understand the model) and also programmatically, such that model retraining or more detailed analysis of a model's inputs can be performed automatically.
We showed that dtool datasets work well for this purpose: they are a lightweight wrapper around the raw data (in this case model weights) that provide both human- and machine-readable metadata. dtoolAI ties dtool enhanced input data together with dtool output datasets by providing library code and routines for processing these datasets, capturing training parameters and encoding them in the output.
Current Limitations and Further Work
It is important to recognize the limitations of our current solution. We have made use of the FAIR principles to improve the reproducibility of workflows based on DL models through automated model provenance annotation. However, we are far from a full FAIR implementation, our focus being immediate reproducibility improvements to DL model training and distribution workflows within research.
Our specific implementation provides only limited interoperability. The trained models produced by dtoolAI can be used only as part of software systems or workflows that either include the dtoolAI library or access the model weights directly through Pytorch. Interoperability of trained DL models is generally limited by the implementation frameworks for those models (i.e., Pytorch models work in Pytorch, Tensorflow models work in Tensorflow). Improving this by better standards for specifying model architectures and training parameters, rather than trained models, would be a possible direction for improvement (essentially focusing on prospective rather than retrospective provenance).
dtoolAI also provides no direct findability. Trained model artifacts produced by the library can be either written directly to widely accessible storage systems or uploaded by dtool, providing a persistent URI for the model. However, this provides no direct discoverability mechanism; the URI must be shared or linked as part of a workflow description. This supports reproducibility by provenance recording, our immediate goal, but not general model sharing. A natural step to improve this would be enforcement of schemas specific to a particular model repository together with code for uploading models to that repository.
Wider Community Use
Adoption of common practices and standards for metadata and workflows is a community process. Our tool grew out of our development of internal guidelines within our own group and wider institute for managing DL models. Our colleagues who have tested dtoolAI have found considerable benefits for their workflows, particularly in distributing models to others while keeping provenance information for those models.
As a next step, we would like to expand use throughout the wider research software engineering community. As the group within which models must be interoperable grows, the importance of adherence to shared schemas within that group also grows. As community schemas such as Bioschemas or ML-Schema become more widely adopted, the flexible nature of dtoolAI allows end users to decide how rigorously to enforce these schemas, supporting this growth.
Model Fine-Tuning
A common approach to applying DL networks to problems that are close to the original domain on which the network trained is "fine-tuning." This takes a pretrained model (i.e., an existing set of weights) and modifies them by the application of new training data. Sometimes parts of the model are "frozen," i.e., a subset of weights is not allowed to change.
This works because the structure of DL models results in the early processing of parts of the model learning general features, while the later parts in turn associate these features with categories. We can reuse the general feature parts of the model while learning new categories.
Providing support for tracking both the original training data and later application of subsequent retraining data would allow better understanding of the training history of these models.
Recording Training Environment
Although we propose recording of training data and parameters, these are, in general, not enough to precisely reproduce model weights, i.e., to produce identical weight values from identical input. This is because model training usually relies on random elements. Initial model weights are usually chosen randomly: the order in which training data are presented to the model is often randomized, and so forth. Model training is also dependent on the specifics of the computing hardware on which training happens. Usually we are more concerned with repeatability than strict reproducibility (see Krishnamurthi and Vitek13 for discussion of the distinction); however, there are some circumstances whereby we may wish to be able to specify more rigorously for reproduction.
It is usually impractical to replicate the whole hardware environment in which a model was trained. However, we could at least record the details of this environment for future reference. The challenges of doing so are discussed in detail by Ivie and Thain.22
Conclusion
Progress in science relies on reproducibility. Without the ability to verify and repeat results, we cannot establish scientific consensus. ML, particularly recent advances in the DL family of techniques, has brought enormous advances in speed and accuracy to a range of data-processing problems. These techniques show great promise for application to scientific analyses. However, without careful attention to how retrospective provenance information is captured during model training and distributed together with models, they also pose a substantial risk of reducing the reproducibility of those analyses and introducing bias.
Managing DL data and models is both a software and data problem. The FAIR data principles provide core guidance on management of computational data. Recent developments in adaptation of those principles to workflows and individual software components are pointing the way toward computationally based science that embraces reproducibility and repeatability.
To apply these advances to the domain of DL requires specifying how these principles should be implemented for DL model training and distribution workflows. We have suggested specific guidelines, linked to the FAIR principles, for how to do this. These guidelines involve annotation of model training data with appropriate metadata, ensuring that those training data have persistent identifiers recording training hyperparameters and storing and distributing models in a form that retains all of this information.
We have also developed a set of tools that allow practical application of these guidelines. dtoolAI, a Python library, makes use of dtool for the formation of training data into datasets with metadata (including the specification of suitable schemas) and persistent identifiers. It then enables easy recording of training hyperparameters and input dataset identifiers in a form that automatically stores those data together with model weights, giving a packaged artifact that includes key provenance information. The artifact can then be easily distributed and its metadata can be programmatically accessed.
dtoolAI has improved the reproducibility of the ML workflows we have built on top of it, and we hope it will do the same for others.
Experimental Procedures
Resource Availability
Lead Contact
Matthew Hartley, Matthew.Hartley@jic.ac.uk is the lead contact for this work.
Materials Availability
This work generated no non-code materials.
Data and Code Availability
All code associated with the work is available at https://github.com/jic-csb/dtoolai.
dtoolAI Workflows
In the Results section, we explained the internal architecture and design of dtoolAI. Here we will look at some examples of how we can use it to generate reproducible ML models. Examples in this paper consist of short snippets to highlight features; full code examples are provided in the dtoolAI repository on GitHub (https://github.com/jic-csb/dtoolai) in the form of scripts and Jupyter notebooks, as well as library code documentation and examples of use at https://dtoolai.readthedocs.io.
Training a Simple Model
In this example, we will look at a common neural network tutorial example, recognition of handwritten digits from the MNIST dataset. In this case, we have already marked up the MNIST dataset with the metadata we need to be able to train from it directly.
Firstly, we will look at how we load the data from a persistent identifier.
train_dataset_uri = "http://bit.ly/2NVFGQd"
train_ds = TensorDataSet(train_dataset_uri)
This code uses the data URI ("http://bit.ly/2NVFGQd") to load the data. After defining a model, loss function, and optimizer, we can then train a network from this dataset:
model = GenNet(∗∗params.init_params)
loss_fn = torch.nn.NLLLoss()
optimiser = torch.optim.SGD(model.parameters(), lr=params.learning_rate)
with DerivedDataSet(base_uri, "mnist_model", train_ds) as output_ds:
train_model_with_metadata_capture(
model,
tds_train,
optimiser,
loss_fn,
params,
output_ds
)
This constructs the output network as a dataset. When we run this code, the output will be a trained model dataset, with name mnist_model and stored at the base URI base_uri. Metadata about the training data and the parameters used for training (contained in the params object) are recorded in this dataset and can be viewed either through the dtool API or using a helper script, dtoolai-provenance, provided:
$ dtoolai-provenance example/mnistcnn/
Network architecture name: dtoolai.simpleScalingCNN
Model training parameters: {'batch_size': 128,
'init_params': {'input_channels': 1, 'input_dim': 28},
'input_channels': 1,
'input_dim': 28,
'learning_rate': 0.01,
'loss_func': 'NLLLoss',
'n_epochs': 1,
'optimiser_name': 'SGD'}
Source dataset URI: http://bit.ly/2uqXxrk
Source dataset name: mnist.train
Source dataset readme:
---
dataset_name: MNIST handwritten digits
project: dtoolAI demonstration datasets
authors:
- Yann LeCun
- Corinna Cortes
- Christopher J.C. Burges
usetype: train
We can see how the name, unique identifier (UUID), and persistent resource identifier (URI) are part of the model metadata, as well as the training parameters. We also see how we can train a model from the input data without having to explicitly download it. This helps the practical process of reproducibility.
Using the Model
Our model can now be used as part of an analysis script. We can use dtoolAI's model helper class, TrainedTorchModel to load the model from a URI and apply it:
model = TrainedTorchModel("http://bit.ly/2tbPzSB")
This will automatically download the model weights and load the model into memory. We can then apply the model with:
my_image = Image.open("handwritten_8.png")
result = model.convert_and_predict(my_image)
print(f"Classified image as {result}")
We can also access the model's history:
print(model.get_readme_content())
in which we would see the same data that were accessible to us after we trained the model.
Filesystem URIs and File Paths
In general, when we create model training datasets and trained models, we want to store these in permanent HTTP accessible object storage with persistent URIs. However, since this requires setting up Amazon S3 or Microsoft Azure storage credentials, for simplicity we can work with filesystem URIs in some of these examples.
For convenience's sake, we allow file URIs to be expressed as filesystem paths, such that file:///path/to/data can be addressed simply as /path/to/data/ and dtool will internally convert this into a full URI. When working with real data and models, we can either write directly to HTTP addressable object storage or upload our local filesystem data after creation using dtool, both of which result in persistent URIs.
Training a New Model with New Data
Now we will look at how we can train a model on novel data. To do this, we will firstly see how to mark up that data as a dataset. The dtoolAI package provides a helper script create-image-dataset-from-dirtree to create an input dataset from a directory of images. For example, if we have the following images:
$ tree image_dirtree/
image_dirtree/
|-- car
| |-- image0001.jpg
| `-- image0002.jpg
|-- chair
| |-- image0001.jpg
| `-- image0002.jpg
`-- mug
|-- image0001.jpg
`-- image0002.jpg
We can then run:
$ create-image-dataset-from-dirtree image_dirtree base_uri objects
Created image dataset at base_uri/objects
This will create a training dataset at the base URI "base_uri" with the name "objects." For testing and development purposes, we can use file URIs, for creating distributable persistent models, we would use HTTP accessible object storage. Now we use a very similar training script to the one we saw for training on the MNIST data:
train_ds = ImageDataSet("base_uri/objects")
model = GenNet(∗∗params.init_params)
loss_fn = torch.nn.NLLLoss()
optimiser = torch.optim.SGD(model.parameters(), lr=params.learning_rate)
with DerivedDataSet(base_uri, "objects_model", train_ds) as output_ds:
train_model_with_metadata_capture(
model,
tds_train,
optimiser,
loss_fn,
params,
output_ds
)
This will train a classifier and save the trained model, references to our input data, and training metadata.
Acknowledgments
We would like to thank the Patterns editorial team and the reviewers of our manuscript for their constructive feedback that resulted in a much-improved paper.
Author Contributions
Conceptualization, M.H.; Data Curation, T.S.G.O. and M.H.; Investigation, M.H.; Methodology, M.H; Resources, T.S.G.O. and M.H.; Software, T.S.G.O. and M.H.; Writing – Original Draft, M.H; Writing – Review & Editing, T.S.G.O. and M.H.
Declaration of Interests
The authors declare no competing interests.
Published: July 23, 2020
References
- 1.LeCun Y., Bengio Y., Hinton G. Deep learning. Nature. 2015;521:436–444. doi: 10.1038/nature14539. [DOI] [PubMed] [Google Scholar]
- 2.Krizhevsky A., Sutskever I., Hinton G.E. ImageNet classification with deep convolutional neural networks. Commun. ACM. 2017;60:84–90. [Google Scholar]
- 3.Collobert R., Weston J., Bottou L., Karlen M., Kavukcuoglu K., Kuksa P. Natural language processing (almost) from scratch. J. Machine Learn. Res. 2011;12:2493–2537. [Google Scholar]
- 4.Ronneberger O., Fischer P., Brox T. U-Net: convolutional networks for biomedical image segmentation. arXiv. 2015 1505.04597 [cs] [Google Scholar]
- 5.Poplin R., Chang P.-C., Alexander D., Schwartz S., Colthurst T., Ku A., Newburger D., Dijamco J., Nguyen N., Afshar P.T. A universal SNP and small-indel variant caller using deep neural networks. Nat. Biotechnol. 2018;36:983–987. doi: 10.1038/nbt.4235. [DOI] [PubMed] [Google Scholar]
- 6.Angermueller C., Pärnamaa T., Parts L., Stegle O. Deep learning for computational biology. Mol. Syst. Biol. 2016;12:878. doi: 10.15252/msb.20156651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sandve G.K., Nekrutenko A., Taylor J., Hovig E. Ten simple rules for reproducible computational research. PLoS Comput. Biol. 2013;9:e1003285. doi: 10.1371/journal.pcbi.1003285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Aarts A.A., Anderson J.E., Anderson C.J., Attridge P., Attwood A., Axt J., Babel M., Bahnik S., Baranski E., Barnett-Cowan M. Estimating the reproducibility of psychological science. Science. 2015;349:943–950. doi: 10.1126/science.aac4716. [DOI] [PubMed] [Google Scholar]
- 9.Fanelli D., Costas R., Ioannidis J.P.A. Meta-assessment of bias in science. Proc. Natl. Acad. Sci. U S A. 2017;114:3714–3719. doi: 10.1073/pnas.1618569114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Claerbout J.F., Karrenbach M. SEG Technical Program Expanded Abstracts 1992 SEG Technical Program Expanded Abstracts. Society of Exploration Geophysicists; 1992. Electronic documents give reproducible research a new meaning; pp. 601–604. [Google Scholar]
- 11.Committee on Reproducibility and Replicability in Science. Board on Behavioral, Cognitive, and Sensory Sciences. Committee on National Statistics. Division of Behavioral and Social Sciences and Education. Nuclear and Radiation Studies Board. Division on Earth and Life Studies. Board on Mathematical Sciences and Analytics. Committee on Applied and Theoretical Statistics. Division on Engineering and Physical Sciences. Board on Research Data and Information . National Academies Press; 2019. Reproducibility and Replicability in Science. [Google Scholar]
- 12.Barba L.A. Terminologies for reproducible research. arXiv. 2018 1802.03311 [cs] [Google Scholar]
- 13.Krishnamurthi S., Vitek J. The real software crisis: repeatability as a core value. Commun. ACM. 2015;58:34–36. [Google Scholar]
- 14.Moreau L., Groth P., Miles S., Vazquez-Salceda J., Ibbotson J., Jiang S., Munroe S., Rana O., Schreiber A., Tan V. The provenance of electronic data. Commun. ACM. 2008;51:52–58. [Google Scholar]
- 15.Lim C., Lu S., Chebotko A., Fotouhi F. 2010 IEEE International Conference on Services Computing. 2010. Prospective and retrospective provenance collection in scientific workflow environments; pp. 449–456. [Google Scholar]
- 16.Deng J., Dong W., Socher R., Li L., Kai L., Li F.F. 2009 IEEE Conference on Computer Vision and Pattern Recognition. IEEE; 2009. ImageNet: a large-scale hierarchical image database; pp. 248–255. [Google Scholar]
- 17.Krizhevsky A. University of Toronto; 2009. Learning Multiple Layers of Features from Tiny Images. Technical Report TR-2009.https://www.cs.toronto.edu/∼kriz/learning-features-2009-TR.pdf [Google Scholar]
- 18.Lecun Y., Bottou L., Bengio Y., Haffner P. Gradient-based learning applied to document recognition. Proc. IEEE. 1998;86:2278–2324. [Google Scholar]
- 19.Wilkinson M.D., Dumontier M., Aalbersberg I.J., Appleton G., Axton M., Baak A., Blomberg N., Boiten J.-W., da Silva Santos L.B., Bourne P.E. The FAIR Guiding Principles for scientific data management and stewardship. Sci. Data. 2016;3:160018. doi: 10.1038/sdata.2016.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lamprecht A.-L., Garcia L., Kuzak M., Martinez C., Arcila R., Martin Del Pico E., Dominguez Del Angel V., van de Sandt S., Ison J., Martinez P.A. Towards FAIR principles for research software. Data Sci. 2020;3:37–59. [Google Scholar]
- 21.Goble C., Cohen-Boulakia S., Soiland-Reyes S., Garijo D., Gil Y., Crusoe M.R., Peters K., Schober D. FAIR computational workflows. Data Intelligence. 2019;2:108–121. [Google Scholar]
- 22.Ivie P., Thain D. Reproducibility in scientific computing. ACM Comput. Surv. 2018;51 doi: 10.1145/3186266. [DOI] [Google Scholar]
- 23.Leipzig J. A review of bioinformatic pipeline frameworks. Brief. Bioinform. 2017;18:530–536. doi: 10.1093/bib/bbw020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pimentel J.F., Freire J., Murta L., Braganholo V. A survey on collecting, managing, and analyzing provenance from scripts. ACM Comput. Surv. 2019;52 doi: 10.1145/3311955. [DOI] [Google Scholar]
- 25.Comet.ml Comet.ml: supercharging machine learning. 2019. https://medium.com/comet-ml
- 26.Publio G.C., Esteves D., Ławrynowicz A., Panov P., Soldatova L., Soru T., Vanschoren J., Zafar H. ML-schema: exposing the semantics of machine learning with schemas and ontologies. arXiv. 2018 1807.05351 [cs, stat] [Google Scholar]
- 27.Goodman A., Pepe A., Blocker A.W., Borgman C.L., Cranmer K., Crosas M., Stefano R.D., Gil Y., Groth P., Hedstrom M. Ten simple rules for the care and feeding of scientific data. PLoS Comput. Biol. 2014;10:e1003542. doi: 10.1371/journal.pcbi.1003542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gray A.J.G., Goble C., Jimenez . 16th International Semantic Web Conference. 2017. Bioschemas: from potato salad to protein annotation.https://iswc2017.ai.wu.ac.at/wp-content/uploads/papers/PostersDemos/paper579.pdf [Google Scholar]
- 29.Vanschoren J., van Rijn J.N., Bischl B., Torgo L. OpenML: networked science in machine learning. ACM SIGKDD Explor. Newsl. 2014;15:49–60. [Google Scholar]
- 30.Olsson T.S.G., Hartley M. Lightweight data management with dtool. PeerJ. 2019;7:e6562. doi: 10.7717/peerj.6562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Paszke A., Gross S., Massa F., Lerer A., Bradbury J., Chanan G., Killeen T., Lin Z., Gimelshein N., Antiga L. PyTorch: an imperative style, high-performance deep learning library. In: Wallach H., Larochelle H., Beygelzimer A., Alché-Buc F.D., Fox E., Garnett R., editors. Advances in Neural Information Processing Systems 32. Curran Associates; 2019. pp. 8024–8035. [Google Scholar]
- 32.Rosenblatt F. The perceptron: a probabilistic model for information storage and organization in the brain. Psychol. Rev. 1958;65:386–408. doi: 10.1037/h0042519. [DOI] [PubMed] [Google Scholar]
- 33.Hutson M. Bringing machine learning to the masses. Science. 2019;365:416–417. doi: 10.1126/science.365.6452.416. [DOI] [PubMed] [Google Scholar]
- 34.Murdoch W.J., Singh C., Kumbier K., Abbasi-Asl R., Yu B. Interpretable machine learning: definitions, methods, and applications. arXiv. 2019 doi: 10.1073/pnas.1900654116. 1901.04592 [cs, stat] [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All code associated with the work is available at https://github.com/jic-csb/dtoolai.