Summary
Image analysis is key to extracting quantitative information from scientific microscopy images, but the methods involved are now often so refined that they can no longer be unambiguously described by written protocols. We introduce BIAFLOWS, an open-source web tool enabling to reproducibly deploy and benchmark bioimage analysis workflows coming from any software ecosystem. A curated instance of BIAFLOWS populated with 34 image analysis workflows and 15 microscopy image datasets recapitulating common bioimage analysis problems is available online. The workflows can be launched and assessed remotely by comparing their performance visually and according to standard benchmark metrics. We illustrated these features by comparing seven nuclei segmentation workflows, including deep-learning methods. BIAFLOWS enables to benchmark and share bioimage analysis workflows, hence safeguarding research results and promoting high-quality standards in image analysis. The platform is thoroughly documented and ready to gather annotated microscopy datasets and workflows contributed by the bioimaging community.
Keywords: image analysis, software, benchmarking, deployment, reproducibility, web application, community, bioimaging, deep learning
Highlights
-
•
Image analysis is inescapable in extracting quantitative data from scientific images
-
•
It can be difficult to deploy and apply state-of-the-art image analysis methods
-
•
Comparing heterogeneous image analysis methods is tedious and error prone
-
•
We introduce a platform to deploy and fairly compare image analysis workflows
The Bigger Picture
Image analysis is currently one of the major hurdles in the bioimaging chain, especially for large datasets. BIAFLOWS seeds the ground for virtual access to image analysis workflows running in high-performance computing environments. Providing a broader access to state-of-the-art image analysis is expected to have a strong impact on research in biology, and in other fields where image analysis is a critical step in extracting scientific results from images. BIAFLOWS could also be adopted as a federated platform to publish microscopy images together with the workflows that were used to extract scientific data from these images. This is a milestone of open science that will help to accelerate scientific progress by fostering collaborative practices.
While image analysis is becoming inescapable in the extraction of quantitative information from scientific images, it is currently challenging for life scientists to find, test, and compare state-of-the-art image analysis methods compatible with their own microscopy images. It is also difficult and time consuming for algorithm developers to validate and reproducibly share their methods. BIAFLOWS is a web platform addressing these needs. It can be used as a local solution or through an immediately accessible and curated online instance.
Introduction
As life scientists collect microscopy datasets of increasing size and complexity,1 computational methods to extract quantitative information from these images have become inescapable. In turn, modern image analysis methods are becoming so complex (often involving a combination of image-processing steps and deep-learning methods) that they require expert configuration to run. Unfortunately, the software implementations of these methods are commonly shared as poorly reusable and scarcely documented source code and seldom as user-friendly packages for mainstream bioimage analysis (BIA) platforms.2, 3, 4 Even worse, test images are not consistently provided with the software, and it can hence be difficult to identify the baseline for valid results or the critical adjustable parameters to optimize the analysis. Altogether, this does not only impair the reusability of the methods and impede reproducing published results5,6 but also makes it difficult to adapt these methods to process similar images. To improve this situation, scientific datasets are now increasingly made available through public web-based applications7, 8, 9 and open-data initiatives,10 but existing platforms do not systematically offer advanced features such as the ability to view and process multidimensional images online or to let users assess the quality of the analysis against a ground-truth reference (also known as benchmarking). Benchmarking is at the core of biomedical image analysis challenges and it a practice known to sustain the continuous improvement of image analysis methods and promote their wider diffusion.11 Unfortunately, challenges are rather isolated competitions and they suffer from known limitations12: each event focuses on a single image analysis problem, and it relies on ad hoc data formats and scripts to compute benchmark metrics. Both challenge organizers and participants are therefore duplicating efforts from challenge to challenge, whereas participants' workflows are rarely available in a sustainable and reproducible fashion. Additionally, the vast majority of challenge datasets come from medical imaging, not from biology: for instance, as of January 2020, only 15 out of 198 datasets indexed in Grand Challenge13 were collected from fluorescence microscopy, one of the most common imaging modalities for research in biology. As a consequence, efficient BIA methods are nowadays available but their reproducible deployment and benchmarking are still stumbling blocks for open science. In practice, end users are faced with a plethora of BIA ecosystems and workflows to choose from, and they have a hard time reproducing results, validating their own analysis, or ensuring that a given method is the most appropriate for the problem they face. Likewise, developers cannot systematically validate the performance of their BIA workflows on public datasets or compare their results to previous work without investing time-consuming and error-prone reimplementation efforts. Finally, it is challenging to make BIA workflows available to the whole scientific community in a configuration-free and reproducible manner.
Results
Conception of Software Architecture for Reproducible Deployment and Benchmarking
Within the Network of European Bioimage Analysts (NEUBIAS COST [www.cost.eu] Action CA15124), an important body of work focuses on channeling the efforts of bioimaging stakeholders (including biologists, bioimage analysts, and software developers) to ensure a better characterization of existing bioimage analysis workflows and to bring these tools to a larger number of scientists. Together, we have envisioned and implemented BIAFLOWS (Figure 1), a community-driven, open-source web platform to reproducibly deploy and benchmark bioimage analysis workflows on annotated multidimensional microscopy data. Whereas some emerging bioinformatics web platforms14,15 simply rely on “Dockerized” (https://www.docker.com/resources/what-container) environments and interactive Python notebooks to access and process scientific data from public repositories, BIAFLOWS offers a versatile and extensible integrated framework to (1) import annotated image datasets and organize them into BIA problems, (2) encapsulate BIA workflows regardless of their target software, (3) batch process the images, (4) remotely visualize the images together with the results, and (5) automatically assess the performance of the workflows from widely accepted benchmark metrics.
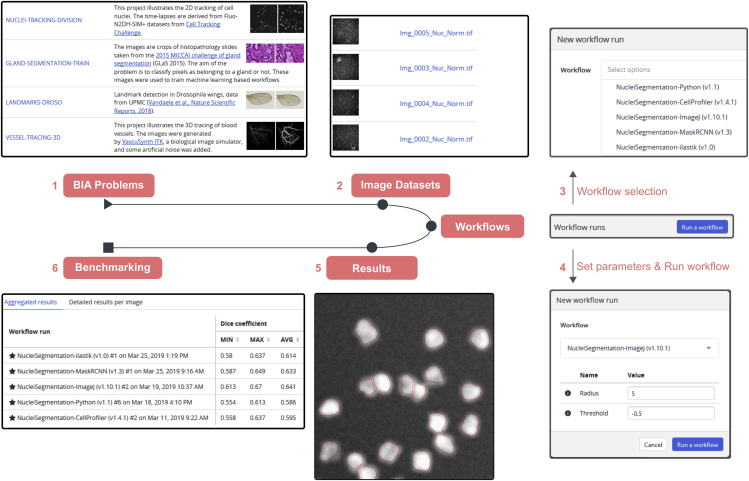
Figure 1.
BIAFLOWS Web Interface
(1) Users select a BIA problem (Table S1) and (2) browse the images illustrating this problem, for instance to compare them with their own images, then (3) select a workflow (Table S1) and associated parameters (4) to process the images. The results can then be overlaid on the original images from the online image viewer (5), and (6) benchmark metrics can be browsed, sorted, and filtered both as overall statistics or per image.
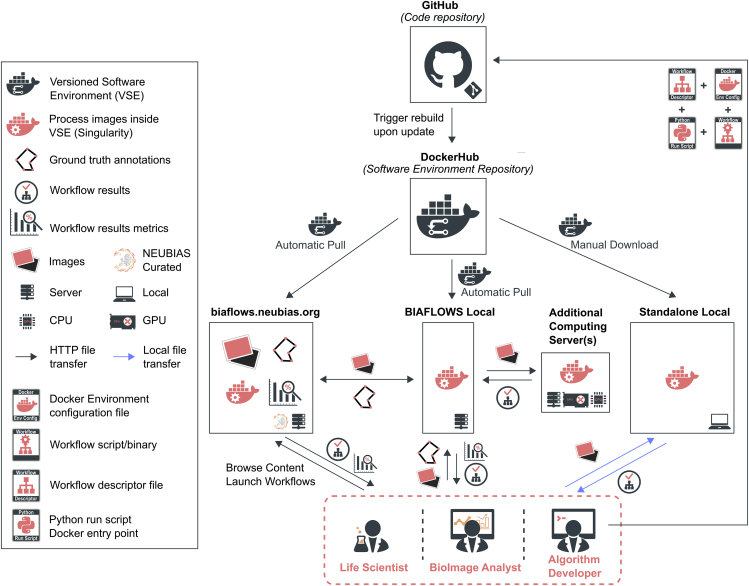
BIAFLOWS content can be interactively explored and triggered (Box 1) from a streamlined web interface (Figure 1). For a given problem, a set of standard benchmark metrics (Supplemental Experimental Procedures section 6) are reported for every workflow run, with accompanying technical and interpretation information available from the interface. One main metric is also highlighted as the most significant metric to globally rank the performance of the workflows. To complement benchmark results, workflow outputs can also be visualized simultaneously from multiple annotation layers or synchronized image viewers (Figure 2). BIAFLOWS is open-source and thoroughly documented (https://biaflows-doc.neubias.org/), and extends Cytomine,16 a web platform originally developed for the collaborative annotation of high-resolution bright-field bioimages. BIAFLOWS required extensive software development and content integration to enable the benchmarking of BIA workflows; accordingly, the web user interface has been completely redesigned to streamline this process (Figure 1). First, a module to upload multidimensional (C, Z, T) microscopy datasets and a fully fledged remote image viewer were implemented. Next, the architecture was refactored to enable the reproducible remote execution of BIA workflows encapsulated with their original software environment in Docker images (workflow images). To abstract out the operations performed by a workflow, we adopted a rich application description schema17 describing its interface (input, output, parameters) and default parameter values (Supplemental Experimental Procedures section 3). The system was also engineered to monitor trusted user spaces hosting a collection of workflow images and to automatically pull new or updated workflows (Figure 3, DockerHub). In turn, workflow images are built and versioned in the cloud whenever a new release is triggered from their associated source code repositories (Figure 3, GitHub). To ensure reproducibility, we enforced that all versions of the workflow images are permanently stored and accessible from the system. Importantly, the workflows can be run on any computational resource, including high-performance computing and multiple server architectures. This is achieved by seamlessly converting the workflow images to a compatible format (Singularity18), and dispatching them to the target computational resources over the network by SLURM19 (Figure 3, additional computing servers). To enable interoperability between all components, some standard object annotation formats were specified for important classes of BIA problems (Supplemental Experimental Procedures section 4). We also developed a software library to compute benchmark metrics associated with these problem classes by adapting and integrating the code from existing biomedical challenges13 and scientific publications.20 With this new design, benchmark metrics are automatically computed after every workflow run. BIAFLOWS can also be deployed on a local server to manage private images and workflows and to process images locally (Figure 3, BIAFLOWS local; Supplemental Experimental Procedures section 2). To simplify the coexistence of these different deployment scenarios, we developed migration tools (Supplementary Experimental Procedures section 5) to transfer content between existing BIAFLOWS instances (including the online instance described hereafter). Importantly, all content from any instance can be accessed programmatically through a RESTful interface, which ensures complete data accessibility and interoperability. Finally, for full flexibility, workflows can be downloaded manually from DockerHub to process local images independently of BIAFLOWS (Figure 3, standalone local; Supplemental Experimental Procedures section 5).
Box 1. How to Get Started with BIAFLOWS.
-
•
Watch BIAFLOWS video tutorial (https://biaflows.neubias.org).
-
•
Visit BIAFLOWS documentation portal (https://biaflows-doc.neubias.org).
-
•
Access BIAFLOWS online instance (https://biaflows.neubias.org) in read-only mode.This public instance is curated by NEUBIAS (http://neubias.org) and backed by bioimage analysts and software developers across the world. You can also access BIAFLOWS sandbox server (https://biaflows-sandbox.neubias.org/) without access restriction.
-
•
Install your own BIAFLOWS instance on a desktop computer or a server to manage images locally or process them with existing BIAFLOWS workflows. Follow “Installing and populating BIAFLOWS locally” from the documentation portal.
-
•
Download a workflow to process your own images locally. Follow “Executing a BIAFLOWS workflow without BIAFLOWS server” from the documentation portal.
-
•
Share your thoughts and get help on our forum (https://forum.image.sc/tags/biaflows), or write directly to our developer team at biaflows@neubias.org.
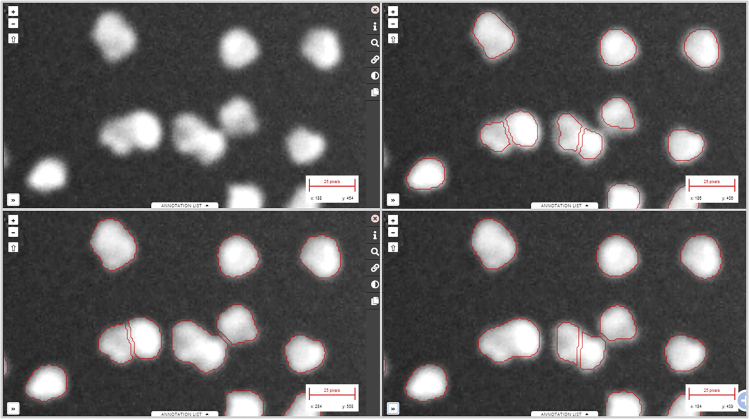
Figure 2.
Synchronizing Image Viewers Displaying Different Workflow Results
Region from one of the sample images available in NUCLEI-SEGMENTATION problem (accessible from the BIAFLOWS online instance). Original image (upper left), same image overlaid with results from: custom ImageJ macro (upper right), custom CellProfiler pipeline (lower left), and custom Python script (lower right).
Figure 3.
BIAFLOWS Architecture and Possible Deployment Scenarios
Workflows are hosted in a trusted source code repository (GitHub). Workflow (Docker) images encapsulate workflows together with their execution environments to ensure reproducibility. Workflow images are automatically built by a cloud service (DockerHub) whenever a new workflow is released or an existing workflow is updated from its trusted GitHub repository. Different BIAFLOWS instances monitor DockerHub and pull new or updated workflow images, which can also be downloaded to process local images without BIAFLOWS (Standalone Local).
BIAFLOWS Online Curated Instance for Public Benchmarking
An online instance of BIAFLOWS is maintained by NEUBIAS and available at https://biaflows.neubias.org/ (Figure 3). This server is ready to host community contributions and is already populated with a substantial collection of annotated image datasets illustrating common BIA problems and several associated workflows to process these images (Table S1). Concretely, we integrated BIA workflows spanning nine important BIA problem classes illustrated by 15 image datasets imported from existing challenges (DIADEM,21 Cell Tracking Challenge,22 Particle Tracking Challenge,23 Kaggle Data Science Bowl 201824), created from synthetic data generators25 (CytoPacq,26 TREES toolbox,27 Vascusynth,28 SIMCEP29), or contributed by NEUBIAS members.30 The following problem classes are currently represented: object detection/counting, object segmentation, and pixel classification (Figure 4); particle tracking, object tracking, filament network tracing, filament tree tracing, and landmark detection (Figure 5). To demonstrate the versatility of the platform we integrated 34 workflows, each targeting a specific software or programming language: ImageJ/FIJI macros and scripts,31 Icy protocols,32 CellProfiler pipelines,33 Vaa3D plugins,34 ilastik pipelines,35 Octave scripts,36 Jupyter notebooks,15 and Python scripts leveraging Scikit-learn37 for supervised learning algorithms, and Keras38 or PyTorch39 for deep learning. This list, although already extensive, is not limited, as BIAFLOWS core architecture enables one to seamlessly add other software as long as they fulfill minimal requirements (Supplemental Experimental Procedures section 3). To demonstrate the potential of the platform to perform open benchmarking, a case study has been performed with (and is available from) BIAFLOWS to compare workflows identifying nuclei in microscopy images. The content from the BIAFLOWS online instance (https://biaflows.neubias.org) can be viewed in read-only mode from the guest account, while the workflows can be launched from the sandbox server (https://biaflows-sandbox.neubias.org/). An extensive user guide and video tutorial are available online from the same URLs. To enhance their visibility, all workflows hosted in the system are also referenced from NEUBIAS Bioimage Informatics Search Index (http://biii.eu/). BIAFLOWS online instance is fully extensible and, with minimal effort, interested developers can package their own workflows (Supplemental Experimental Procedures section 3) and make them available for benchmarking (Box 2). Similarly, following our guidelines (Supplemental Experimental Procedures section 2), scientists can make their images and ground-truth annotations available online through the online instance or through a local instance they manage (Box 2). Finally, all online content can be seamlessly migrated to a local BIAFLOWS instance (Supplemental Experimental Procedures section 5) for further development or to process local images.
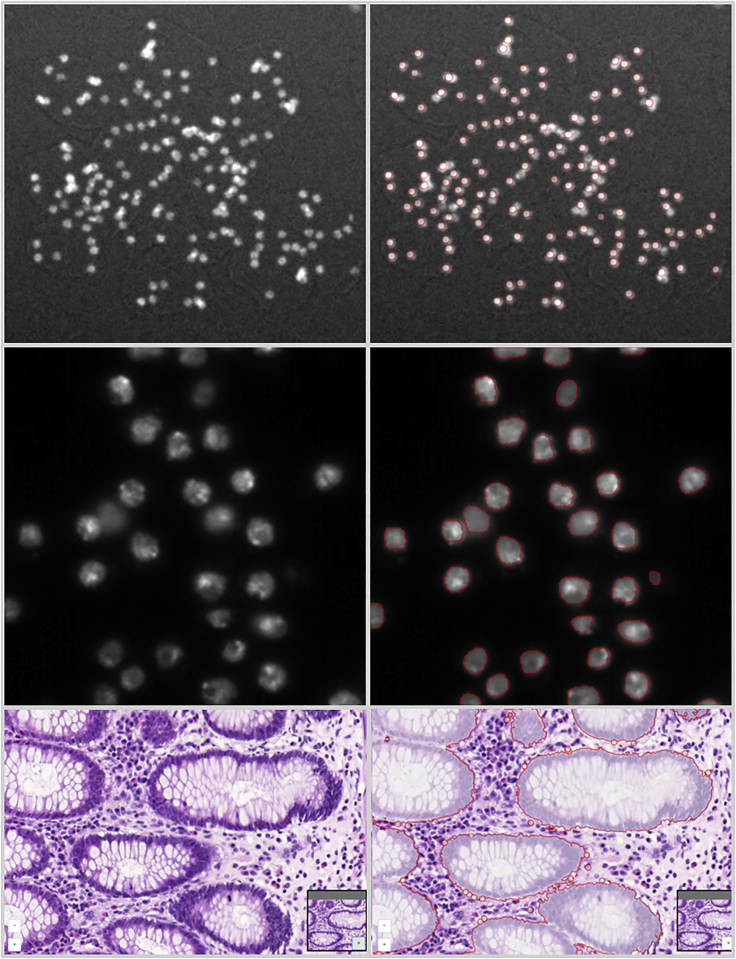
Figure 4.
Sample Images from the BIAFLOWS Online Instance Illustrating Several BIA Problem Classes, and Results from Associated Workflows
Original image (left) and workflow results (right), from top to bottom: (1) spot detection in synthetic images (SIMCEP29); (2) nuclei segmentation in images from Kaggle Data Science Bowl 2018;24 (3) pixel classification in images from 2015 MICCAI gland segmentation challenge.40
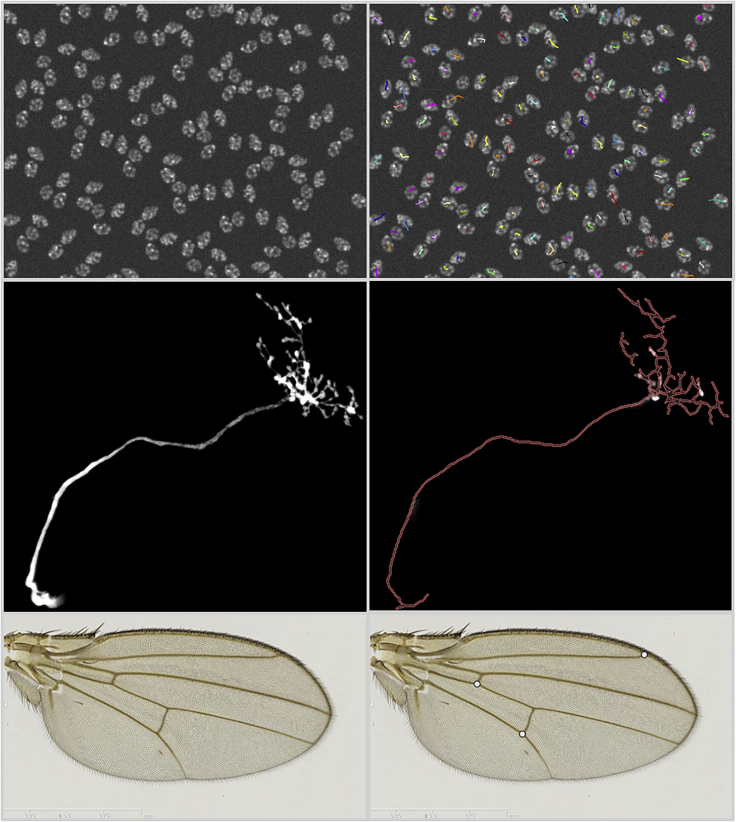
Figure 5.
Sample Images from the BIAFLOWS Online Instance Illustrating Several BIA Problem Classes, and Results from Associated Workflows
Original image (left) and workflow results (right), from top to bottom: (1) particle tracking in synthetic time-lapse displaying non-dividing nuclei (CytoPACQ26), single frame + dragon-tail tracks; (2) neuron tree tracing in 3D image stacks from DIADEM challenge,21 average intensity projection (left), traced skeleton z projection (dilated, red); (3) landmark detection in Drosophila wing images.30
Box 2. How to Contribute to BIAFLOWS.
-
•
Scientists can contribute published annotated microscopy images to BIAFLOWS online instance. See “Problem classes, ground truth annotations and reported metrics” from the documentation portal for information on the expected images and ground-truth annotations formats, and contact us through the dedicated thread on https://forum.image.sc/tags/biaflows.
-
•
To showcase a workflow in the BIAFLOWS online instance, developers can encapsulate their source code, test it on a local BIAFLOWS instance or BIAFLOWS sandbox server (https://biaflows-sandbox.neubias.org/), and open an issue in this GitHub repository: https://github.com/Neubias-WG5/SubmitToBiaflows. Follow “Creating a BIA workflow and adding it to a BIAFLOWS instance” from the documentation portal.
-
•
Feature requests or bug reports can be posted to BIAFLOWS GitHub (https://github.com/neubias-wg5).
-
•
Users can contribute to the documentation by submitting a pull request to https://github.com/Neubias-WG5/neubias-wg5.github.io.
-
•
Any user can share data and results, e.g., accompanying scientific publications, via “Access BIAFLOWS from a Jupyter notebook” from the documentation portal or by directly linking the content of a BIAFLOWS instance.
To further increase the content currently available in BIAFLOWS online instance, calls for contribution will be shortly launched to gather more annotated microscopy images and encourage developers to package their own workflows. The support of new problem classes is also planned, for example, to benchmark the detection of blinking events in the context of super-resolution localization microscopy or the detection of landmark points for image registration. There is no limitation in using BIAFLOWS in other fields where image analysis is a critical step in extracting scientific results from images, for instance material or plant science and biomedical imaging.
Case Study: Comparing the Performance of Nuclei Segmentation by Classical Image Processing, Classical Machine Learning, and Deep-Learning Methods
To illustrate how to use BIAFLOWS for the open benchmarking of BIA workflows, we integrated seven nuclei segmentation workflows (Supplemental Experimental Procedures section 1). All content (images, ground-truth annotations, workflows, benchmark results) is readily accessible from the BIAFLOWS online instance. The workflows were benchmarked on two different image datasets: a synthetic dataset of ten images generated29 for the purpose of this study, and a subset of 65 images from an existing nuclei segmentation challenge (Kaggle Data Science Bowl 201824). The study was articulated in three parts: (1) evaluating the performance of three BIA workflows implementing classical methods to identify nuclei (synthetic dataset); (2) evaluating the performance of three ubiquitous deep-learning workflows on the same dataset; and (3) evaluating the performance of these deep-learning workflows (and a classical machine-learning workflow) on Kaggle Data Science Bowl 2018 (KDSB2018) subset. As a baseline, the classical workflows were manually tuned to obtain the best performance on the synthetic dataset while the machine-learning workflows were trained on generic nuclei image datasets with no further tuning for the synthetic dataset. Despite this, the deep-learning methods proved to be almost as accurate, or in some cases more accurate, than the best classical method (Tables S2 and S3). It was also evidenced that a set of benchmark metrics is generally to be favored over a single metric, since some widely used metrics only capture a single aspect of a complex problem. For instance, object segmentation does not only aim at accurately discriminating foreground from background pixels (assessed by DICE-like metrics) but overall at identifying independent objects (for instance to further measure their geometrical properties). Also, the visual inspection of workflow results proved useful in understanding the underlying errors evidenced by poor benchmark metrics results (Figure S1). All these features are readily available in BIAFLOWS, which swiftly enables to link workflow source code, benchmark metrics results, and visual results. The same methodology can be easily translated to other experiments.
Discussion
BIAFLOWS addresses a number of critical requirements to foster open image analysis for life sciences: (1) sharing and visualizing annotated microscopy images illustrating commonly faced BIA problems; (2) sharing reproducible BIA workflows; (3) exposing workflow parameters and associated default values; (4) computing relevant benchmark metrics to compare workflows performance; and (5) providing a standard way to store, visualize, and share BIA workflows results. As such, BIAFLOWS is a central asset for biologists and bioimage analysts to leverage state-of-the-art bioimaging methods and efficiently reuse them in a different context. It is also a tool of choice for algorithm developers and challenge organizers to benchmark bioimage analysis workflows. Challenge participants traditionally reported workflow predictions on websites such as Kaggle and grand-challenge.org. The latter is currently developing a Docker-based mechanism (https://grand-challengeorg.readthedocs.io/en/latest/evaluation.html#) to package workflows (mostly coming from medical imaging), but these platforms do not offer a complete integrated web environment to host image datasets, automatically import workflows from open-source repositories, automate benchmark metric computation, and remotely visualize all results in a streamlined web interface such as BIAFLOWS. We believe BIAFLOWS could be made interoperable with the grand-challenge.org Docker-based mechanism to package workflows, and used by challenge organizers as a fully integrated platform to automate benchmarking and share challenge results in a more reproducible way. Finally, BIAFLOWS provides a solution to authors willing to share online supporting data, methods, and results associated with their published scientific results.
With respect to sustainability and scalability, BIAFLOWS is backed by a team of senior bioimage analysts and software developers. The software is compatible with high-performance computing environments and is based on Cytomine architecture,16 which has already proved itself capable of serving large datasets to many users simultaneously.41 We invested a large amount of effort in documenting BIAFLOWS, and the online instance is ready to receive hundreds of new image datasets and workflows as community contributions (Box 2). To increase the content of BIAFLOWS online instance, we will briefly launch calls for contributions targeting existing BIAFLOWS problem classes. We propose that BIAFLOWS becomes a hub for BIA methods developers, bioimage analysts, and life scientists to share annotated datasets, reproducible BIA workflows, and associated results from benchmark and research studies. In future work, we will work toward interoperability with existing European image storage and workflow management infrastructures such as BioImage Archive,42 https://www.eosc-life.eu/, and Galaxy,15 and further improve the scalability and sustainability of the platform.
Experimental Procedures
Resource Availability
Lead Contact
Further information and requests for resources should be directed to the Lead Contact, Sébastien Tosi (sebastien.tosi@irbbarcelona.org).
Materials Availability
No materials were used in this study.
Data and Code Availability
BIAFLOWS is an open-source project and its source code can be freely downloaded at https://github.com/Neubias-WG5.
All images and annotations described and used in this article can be downloaded from the BIAFLOWS online instance at https://biaflows.neubias.org/.
A sandbox server from which all workflows available in BIAFLOWS online instance can be launched remotely, and new workflows/datasets appended for testing are available at https://biaflows-sandbox.neubias.org/.
The documentation to install, use, and extend the software is available at https://neubias-wg5.github.io/.
Acknowledgments
This project is funded by COST CA15124 (NEUBIAS). BIAFLOWS is developed by NEUBIAS (http://neubias.org) Workgroup 5, and it would not have been possible without the great support from the NEUBIAS vibrant community of bioimage analysts and the dedication of Julien Colombelli and Kota Miura in organizing this network. Local organizers of the Taggathons who have fostered the development of BIAFLOWS are also greatly acknowledged: Chong Zhang, Gabriel G. Martins, Julia Fernandez-Rodriguez, Peter Horvath, Bertrand Vernay, Aymeric Fouquier d’Hérouël, Andreas Girod, Paula Sampaio, Florian Levet, and Fabrice Cordelières. We also thank the software developers who helped us integrating external code, among others Jean-Yves Tinevez (Image Analysis Hub of Pasteur Institute) and Anatole Chessel (École Polytechnique). We thank the Cytomine SCRL FS for developing additional open-source modules and Martin Jones (Francis Crick Institute) and Pierre Geurts (University of Liège) for proofreading the manuscript and for their useful comments. R.V. was supported by ADRIC Pôle Mecatech Wallonia grant and U.R. by ADRIC Pôle Mecatech and DeepSport Wallonia grants. R. Marée was supported by an IDEES grant with the help of the Wallonia and the European Regional Development Fund. L.P. was supported by the Academy of Finland (grant 310552). M.M. was supported by the Czech Ministry of Education, Youth and Sports (project LTC17016). S.G.S. acknowledges the financial support of UEFISCDI grant PN-III-P1-1.1-TE-2016-2147 (CORIMAG). V.B. and P.P.-G. acknowledge the France-BioImaging infrastructure supported by the French National Research Agency (ANR-10-INBS-04).
Author Contributions
S.T. and R. Marée conceptualized BIAFLOWS, supervised its implementation, contributed to all technical tasks, and wrote the manuscript. U.R. worked on the core implementation and web user interface of BIAFLOWS with contributions from G.M. and R.H. R. Mormont implemented several modules to interface bioimage analysis workflows and the content of the system. S.T., M.M., and D.Ü. implemented the module to compute benchmark metrics. S.T., V.B., R. Mormont, L.P., B.P., M.M., R.V., and L.A.S. integrated their own workflows or adapted existing workflows, and tested the system. S.T., D.Ü., O.G., and G.B. organized and collected content (image datasets, simulation tools). S.G.S., N.S., and P.P.-G. provided extensive feedback on BIAFLOWS and contributed to the manuscript. All authors took part in reviewing the manuscript.
Declaration of Interests
R Marée and R.H. are co-founders and members of the board of directors of the non-profit cooperative company Cytomine SCRL FS.
Published: June 3, 2020
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.patter.2020.100040.
Supplemental Information
References
- 1.Ouyang W., Zimmer C. The imaging tsunami: computational opportunities and challenges. Curr. Opin. Syst. Biol. 2017;4:105–113. [Google Scholar]
- 2.Eliceiri K.W., Berthold M.R., Goldberg I.G., Ibáñez L., Manjunath B.S., Martone M.E., Murphy R.F., Peng H., Plant A.L., Roysam B. Biological imaging software tools. Nat. Methods. 2012;9:697–710. doi: 10.1038/nmeth.2084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carpenter A.E., Kamentsky L., Eliceiri K.W. A call for bioimaging software usability. Nat. Methods. 2012;9:666–670. doi: 10.1038/nmeth.2073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schneider C.A., Rasband W.S., Eliceiri K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Munafò M.R., Nosek B.A., Bishop D.V.M., Button K.S., Chambers C.D., Percie du Sert N., Simonsohn U., Wagenmakers E.-J., Ware J.J., Ioannidis J.P.A. A manifesto for reproducible science. Nat. Hum. Behav. 2017;1:0021. doi: 10.1038/s41562-016-0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hutson M. Artificial intelligence faces reproducibility crisis. Science. 2018;359:725–726. doi: 10.1126/science.359.6377.725. [DOI] [PubMed] [Google Scholar]
- 7.Ellenberg J., Swedlow J.R., Barlow M., Cook C.E., Sarkans U., Patwardhan A., Brazma A., Birney E. A call for public archives for biological image data. Nat. Methods. 2018;15:849–854. doi: 10.1038/s41592-018-0195-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Allan C., Burel J.M., Moore J., Blackburn C., Linkert M., Loynton S., Macdonald D., Moore W.J., Neves C., Patterson A. OMERO: flexible, model-driven data management for experimental biology. Nat. Methods. 2012;9:245–253. doi: 10.1038/nmeth.1896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kvilekval K., Fedorov D., Obara B., Singh A., Manjunath B.S. Bisque: a platform for bioimage analysis and management. Bioinformatics. 2010;26:544–552. doi: 10.1093/bioinformatics/btp699. [DOI] [PubMed] [Google Scholar]
- 10.Williams E., Moore J., Li S.W., Rustici G., Tarkowska A., Chessel A., Leo S., Antal B., Ferguson R.K., Sarkans U. Image Data Resource: a bioimage data integration and publication platform. Nat. Methods. 2017;14:775–781. doi: 10.1038/nmeth.4326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vandewalle P. Code sharing is associated with research impact in image processing. Comput. Sci. Eng. 2012;14:42–47. [Google Scholar]
- 12.Maier-Hein L., Eisenmann M., Reinke A., Onogur S., Stankovic M., Scholz P., Arbel T., Bogunovic H., Bradley A.P., Carass A. Why rankings of biomedical image analysis competitions should be interpreted with care. Nat. Commun. 2018;9:5217. doi: 10.1038/s41467-018-07619-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Meijering E., Carpenter A., Peng H., Hamprecht F.A., Olivo-Marin J.C. Imagining the future of bioimage analysis. Nat. Biotechnol. 2016;34:1250–1255. doi: 10.1038/nbt.3722. [DOI] [PubMed] [Google Scholar]
- 14.Perkel J.M. A toolkit for data transparency takes shape. Nature. 2018;560:513–515. doi: 10.1038/d41586-018-05990-5. [DOI] [PubMed] [Google Scholar]
- 15.Grüning B.A., Rasche E., Rebolledo-Jaramillo B., Eberhard C., Houwaart T., Chilton J., Coraor N., Backofen R., Taylor J., Nekrutenkoet A. Jupyter and Galaxy: easing entry barriers into complex data analyses for biomedical researchers. PLoS Comput. Biol. 2017;13:e1005425. doi: 10.1371/journal.pcbi.1005425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marée R., Rollus L., Stévens B., Hoyoux R., Louppe G., Vandaele R., Begon J.M., Kainz P., Geurts P., Wehenkel L. Collaborative analysis of multi-gigapixel imaging data with Cytomine. Bioinformatics. 2016;32:1395–1401. doi: 10.1093/bioinformatics/btw013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Glatard T., Kiar G., Aumentado-Armstrong T., Beck N., Bellec P., Bernard R., Bonnet A., Brown S.T., Camarasu-Pop S., Cervenansky F. Boutiques: a flexible framework to integrate command-line applications in computing platforms. GigaScience. 2018;7:giy016. doi: 10.1093/gigascience/giy016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kurtzer G.M., Sochat V., Bauer M.W. Singularity: scientific containers for mobility of compute. PLoS One. 2017;12:e0177459. doi: 10.1371/journal.pone.0177459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yoo A., Jette M., Grondona M. SLURM: simple Linux utility for resource management, job scheduling strategies for parallel processing. Lect. Notes Comput. Sci. 2003;2862:44–60. [Google Scholar]
- 20.Kozubek M. Challenges and benchmarks in bioimage analysis. Adv. Anat. Embryol. Cell Biol. 2016;219:231–262. doi: 10.1007/978-3-319-28549-8_9. [DOI] [PubMed] [Google Scholar]
- 21.Brown K.M., Barrionuevo G., Canty A.J., De Paola V., Hirsch J.A., Jefferis G.S., Lu J., Snippe M., Sugihara I., Ascoli G.A. The DIADEM data sets: representative light microscopy images of neuronal morphology to advance automation of digital reconstructions. Neuroinformatics. 2011;9:143–157. doi: 10.1007/s12021-010-9095-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ulman V., Maška M., Magnusson K.E.G., Ronneberger O., Haubold C., Harder N., Matula P., Matula P., Svoboda D., Radojevic M. An objective comparison of cell-tracking algorithms. Nat. Methods. 2017;14:1141–1152. doi: 10.1038/nmeth.4473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chenouard N., Smal I., de Chaumont F., Maška M., Sbalzarini I.F., Gong Y., Cardinale J., Carthel C., Coraluppi S., Winter M. Objective comparison of particle tracking methods. Nat. Methods. 2014;11:281–289. doi: 10.1038/nmeth.2808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Caicedo J.C., Goodman A., Karhohs K.W., Cimini B.A., Ackerman J., Haghighi M., Heng C., Becker T., Doan M., McQuin C. Nucleus segmentation across imaging experiments: the 2018 data science bowl. Nat. Methods. 2019;16:1247–1253. doi: 10.1038/s41592-019-0612-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Svoboda D., Kozubek M., Stejskal S. Generation of digital phantoms of cell nuclei and simulation of image formation in 3D image cytometry. Cytometry A. 2009;75:494–509. doi: 10.1002/cyto.a.20714. [DOI] [PubMed] [Google Scholar]
- 26.Wiesner D., Svoboda D., Maška M., Kozubek M. CytoPacq: a web-interface for simulating multi-dimensional cell imaging. Bioinformatics. 2019;35:4531–4533. doi: 10.1093/bioinformatics/btz417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cuntz H., Forstner F., Borst A., Häusser M. One rule to grow them all: a general theory of neuronal branching and its practical application. PLoS Comput. Biol. 2010;6:e1000877. doi: 10.1371/journal.pcbi.1000877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jassi P., Hamarneh G. VascuSynth: vascular tree synthesis software. Insight J. 2011 http://hdl.handle.net/10380/3260 [Google Scholar]
- 29.Lehmussola A., Ruusuvuori P., Selinummi J., Huttunen H., Yli-Harja O. Computational framework for simulating fluorescence microscope images with cell populations. IEEE Trans. Med. Imaging. 2007;26:1010–1016. doi: 10.1109/TMI.2007.896925. [DOI] [PubMed] [Google Scholar]
- 30.Vandaele R., Aceto J., Muller M., Péronnet F., Debat V., Wang C.W., Huang C.T., Jodogne S., Martinive P., Geurts P. Landmark detection in 2D bioimages for geometric morphometrics: a multi-resolution tree-based approach. Sci. Rep. 2018;8:538. doi: 10.1038/s41598-017-18993-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schneider C.A., Rasband W.S., Eliceiri K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.de Chaumont F., Dallongeville S., Chenouard N., Hervé N., Pop S., Provoost T., Meas-Yedid V., Pankajakshan P., Lecomte T., Le Montagner Y. Icy: an open bioimage informatics platform for extended reproducible research. Nat. Methods. 2012;9:690–696. doi: 10.1038/nmeth.2075. [DOI] [PubMed] [Google Scholar]
- 33.McQuin C., Goodman A., Chernyshev V., Kamentsky L., Cimini B.A., Karhohs K.W., Doan M., Ding L., Rafelski S.M., Thirstrup D. CellProfiler 3.0: next-generation image processing for biology. PLoS Biol. 2018;16:e2005970. doi: 10.1371/journal.pbio.2005970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Peng H., Ruan Z., Long F., Simpson J.H., Myers E.W. V3D enables real-time 3D visualization and quantitative analysis of large-scale biological image data sets. Nat. Biotechnol. 2010;28:348–353. doi: 10.1038/nbt.1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Berg S., Kutra D., Kroeger T., Straehle C.N., Kausler B.X., Haubold C., Schiegg M., Ales J., Beier T., Rudy M. ilastik: interactive machine learning for (bio)image analysis. Nat. Methods. 2019;16:1226–1232. doi: 10.1038/s41592-019-0582-9. [DOI] [PubMed] [Google Scholar]
- 36.Eaton J.W., Bateman D., Hauberg S., Wehbring R. Free Software Foundation; 2016. GNU Octave Version 4.2.0 Manual: A High-Level Interactive Language for Numerical Computations.https://octave.org/doc/octave-4.2.0.pdf [Google Scholar]
- 37.Pedregosa F., Varoquaux G., Gramfort A., Michel V., Thirion B., Grisel O., Blondel M., Prettenhofer P., Weiss R., Dubourg V. Scikit-learn: machine learning in Python. J. Mach. Learn. Res. 2011;12:2825–2830. [Google Scholar]
- 38.Chollet F. Manning; 2017. Deep Learning with Python. [Google Scholar]
- 39.Paszke, A., Gross, S., Chintala, S., Chanan, G., Yang, E., DeVito, Z., Lin, Z., Desmaison, A., Antiga, L., Lerer, A. (2017). Automatic Differentiation in PyTorch. NIPS Autodiff Workshop, 2017.
- 40.Sirinukunwattana K., Pluim J.P.W., Chen H., Qi X., Heng P.A., Guo Y.B., Wang L.Y., Matuszewski B.J., Bruni E., Sanchez U. Gland segmentation in colon histology images: the glas challenge contest. Med. Image Anal. 2017;35:489–502. doi: 10.1016/j.media.2016.08.008. [DOI] [PubMed] [Google Scholar]
- 41.Multon S., Pesesse L., Weatherspoon A., Florquin S., Van de Poel J.F., Martin P., Vincke G., Hoyoux R., Marée R., Verpoorten D. A Massive Open Online Course (MOOC) on practical histology: a goal, a tool, a large public! Return on a first experience. Ann. Pathol. 2018;38:76–84. doi: 10.1016/j.annpat.2018.02.002. [DOI] [PubMed] [Google Scholar]
- 42.Ellenberg J., Swedlow J.R., Barlow M., Cook C.E., Sarkans U., Patwardhan A., Brazma A., Birney E. A call for public archives for biological image data. Nat. Methods. 2018;15:849–854. doi: 10.1038/s41592-018-0195-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
BIAFLOWS is an open-source project and its source code can be freely downloaded at https://github.com/Neubias-WG5.
All images and annotations described and used in this article can be downloaded from the BIAFLOWS online instance at https://biaflows.neubias.org/.
A sandbox server from which all workflows available in BIAFLOWS online instance can be launched remotely, and new workflows/datasets appended for testing are available at https://biaflows-sandbox.neubias.org/.
The documentation to install, use, and extend the software is available at https://neubias-wg5.github.io/.