Summary
Heterogeneous and multidisciplinary data generated by research on sustainable global agriculture and agrifood systems requires quality data labeling or annotation in order to be interoperable. As recommended by the FAIR principles, data, labels, and metadata must use controlled vocabularies and ontologies that are popular in the knowledge domain and commonly used by the community. Despite the existence of robust ontologies in the Life Sciences, there is currently no comprehensive full set of ontologies recommended for data annotation across agricultural research disciplines. In this paper, we discuss the added value of the Ontologies Community of Practice (CoP) of the CGIAR Platform for Big Data in Agriculture for harnessing relevant expertise in ontology development and identifying innovative solutions that support quality data annotation. The Ontologies CoP stimulates knowledge sharing among stakeholders, such as researchers, data managers, domain experts, experts in ontology design, and platform development teams.
Keywords: ontologies, agriculture, agrifood systems, Big Data, FAIR data, data annotation, semantics for agriculture, Community of Practice, data labeling, knowledge representation
Graphical Abstract

Highlights
-
•
FAIR agricultural data must use ontologies that are popular in the knowledge domain
-
•
CGIAR Ontologies Community of Practice holds expertise for agricultural data annotation
-
•
The Community selects innovative solutions to assist the data annotation with ontologies
-
•
The Community develops multidisciplinary open-source ontologies for agricultural data
The Bigger Picture
Digital technology use in agriculture and agrifood systems research accelerates the production of multidisciplinary data, which spans genetics, environment, agroecology, biology, and socio-economics. Quality labeling of data secures its online findability, reusability, interoperability, and reliable interpretation, through controlled vocabularies organized into meaningful and computer-readable knowledge domains called ontologies. There is currently no full set of recommended ontologies for agricultural research, so data scientists, data managers, and database developers struggle to find validated terminology. The Ontologies Community of Practice of the CGIAR Platform for Big Data in Agriculture harnesses international expertise in knowledge representation and ontology development to produce missing ontologies, identifies best practices, and guides data labeling by teams managing multidisciplinary information platforms to release the FAIR data underpinning the evidence of research impact.
The deployment of digital technology in Agriculture and Food Science accelerates the production of large quantities of multidisciplinary data. The Ontologies Community of Practice (CoP) of the CGIAR Platform for Big Data in Agriculture harnesses the international ontology expertise that can guide teams managing multidisciplinary agricultural information platforms to increase the data interoperability and reusability. The CoP develops and promotes ontologies to support quality data labeling across domains, e.g., Agronomy Ontology, Crop Ontology, Environment Ontology, Plant Ontology, and Socio-Economic Ontology.
Introduction
The increasing application to agrifood research data of the FAIR (findable, accessible, interoperable, and reusable) principles1 has led to the research community's growing interest in using ontologies. FAIR principles indeed recommend that data must be described with commonly used, controlled vocabularies structured in thesauri and semantically rich ontologies. An ontology is a representation of a domain of knowledge where key concepts, as well as the relationships between those concepts, are defined.2 By providing standardized definitions for the terms used by scientists along with defined logical relationships among these terms, ontologies compile information about the content of a dataset that can be explicitly used by computers.3 Each concept has a Uniform Resource Identifier (URI) that uniquely identifies it as a web resource, accessible by anyone for data labeling, to efficiently support consistent use of ontology terms within and across disciplines and domains. Therefore, annotating data with quality and widely used ontologies increases the findability, interoperability, and reusability of data.
Despite the existence of robust ontologies in the Life Sciences, no agreed set of quality ontologies covering all agrifood research disciplines exists, because it is not easy to identify which ones are representative of community standards, what best practices exist for using ontologies, and how we can collectively fill domain gaps.4 Within this scenario, data managers often create their own customized controlled vocabularies, which fragment the global semantic framework and keep data in silos.
In 2013, the Interest Group on Agricultural Data (IGAD) (https://www.rd-alliance.org/groups/agriculture-data-interest-group-igad.html) was created within the Research Data Alliance to facilitate discussions on all aspects of agricultural information management. IGAD's Wheat Data Interoperability Working Group published guidelines recommending a set of standards and ontologies applicable to genetic, genomic, and phenotypic data (http://datastandards.wheatis.org) for wheat,5 while its Agrisemantics Working Group conducted a scoping study from which it produced list of global recommendations for the development, maintenance, and use of semantic resources in agriculture (https://rd-alliance.org/group/agrisemantics-wg/outcomes/39-hints-facilitate-use-semantics-data-agriculture-and-nutrition). IGAD does not directly engage in ontology development related to agriculture.
The CGIAR (https://www.cgiar.org/), the world's largest global agricultural innovation network dedicated to reducing poverty, enhancing food security, and improving natural resources, launched the Platform for Big Data in Agriculture (https://bigdata.cgiar.org/) in 2017. The aim is to increase the impact of agricultural research and development by turning FAIR data into a powerful tool for discovery, while integrating principles of responsible and ethical data use. Through the Platform on Big Data, CGIAR's primary objective is to annotate multidisciplinary research data with the appropriate ontologies for publishing on the GARDIAN platform (https://gardian.bigdata.cgiar.org/), CGIAR's metadata repository, and stimulate the ontology content gap filling rather than developing complete new ontologies.6 The Ontologies Community of Practice (CoP) was created to harness in-house and external expertise in the development of ontologies and support the five other CGIAR Platform CoPs (Agronomy Data, Crop Modeling, Geospatial Data, Livestock Data, and Socio-Economic Data) toward finding adequate ontologies for data description. The Ontologies CoP, hereafter referred to as “The CoP,” was also developed as a means to include data generated by the latest technologies (e.g., remote sensors) and expand beyond crops to encompass data on fisheries and aquaculture, livestock, socio-economics, water management, and agroecology (agroecology includes social, economic, and environmental aspects of the food production systems http://www.fao.org/agroecology/knowledge/definitions/en/). The Ontologies CoP's thematic working groups currently develop ontologies, such as the Crop Ontology (CO) (http://www.cropontology.org), the Agronomy Ontology (ArgO) (https://bigdata.cgiar.org/resources/agronomy-ontology/), and the Socio-Economic Ontology (SEOnt) (https://github.com/AgriculturalSemantics/SEOnt).
The CoP provides the ideal forum for co-learning and knowledge exchange on ontologies and for guiding consistent data annotation, as well as the deployment of quality ontologies in databases and repositories. The CoP stimulates exchanges between domain experts and experts in ontology design, knowledge modeling, ontology-driven applications, and semantic web technologies. While IGAD and the Ontologies CoP have members in common, only the Ontologies CoP aims to directly contribute to ontology development to ensure the quality of data mobilized by the CGIAR Platform for Big Data in Agriculture, its partners, as well as new players within the domains it covers. It includes researchers, modelers, information specialists, data managers, and ontology experts from the CGIAR research network, academia, and the private sector, thus creating a critical mass of expertise to tackle the major issues related to semantics for FAIR data in agrifood science.
Currently, the Ontologies CoP newsletter has 353 subscribers and a LinkedIn group “CGIAR Big Data-Ontologies CoP” (https://www.linkedin.com/groups/13707155/) with 144 active members: 35 from universities, 61 from public research institutes, and 48 from the private sector. We regularly organize webinars, which are recorded to build a public channel of online reference videos (https://www.youtube.com/c/OntologiesInAgriculture) and to which we have 118 subscribers. The CoP webpage (https://bigdata.cgiar.org/communities-of-practice/ontologies/) provides access to its objectives and yearly workplan developed with members' input.
In this paper we provide information on the ontology products that were developed by the CoP members, as well as the necessary perspectives to extend and cover all relevant domains for research on agriculture and food systems. We explain how the CoP supports and fosters the proper use of quality ontologies, the submission of missing terms by users, and collaboratively explore solutions to solving the complexity of data annotation. Finally, we stress the importance of partnering with industry in agriculture and food systems.
Results
The Ontologies CoP members play a direct role in ontology development and filling content gaps by compiling controlled vocabularies and requesting or mapping new terms to existing ontologies. Collaborative development of ontologies is a slow process but is a guarantee for quality and adoption. Currently, four thematic ontology working groups have been created for Agronomy, Fish and Fisheries, Plant phenotypes, and Socio-Economy. The CoP has begun to explore the use of new technologies in machine learning to create or improve ontologies and, in return, provides quality ontologies to support text mining. However, the use of artificial intelligence in the development of ontologies lags behind, largely due to the breadth and heterogeneous sets of expertise involved in quality assessment of the results.
Development of Ontologies for Agrifood Research Data
CGIAR currently has eight agrifood research programs (https://www.cgiar.org/research/research-portfolio/) focused on crop breeding, aimed at producing innovative technologies, such as improved crop varieties and advisory services to farmers. Producing FAIR data on plant genotypes and phenotypes, their environment, field management practices, and socio-economy is crucial to provide support information for the development and use of these technologies.
For several years, CGIAR and its partners have contributed to ontology development for plant phenotype studies and field management practices. The ontologies developed by the CoP provide validated concepts and formatted variables for direct integration in the design of field or lab books, thus supporting data aggregation into multidisciplinary platforms or use by analytical and modeling tools. The CoP provides wider communication and a formal framework for this work, stimulating new members' contributions, as in the case of PepsiCo Inc. and NIAB (a UK crop science organization) to the Oat Ontology development (https://www.cropontology.org/ontology/CO_350/Oat; https://bigdata.cgiar.org/blog-post/agricultural-ontologies-in-use-new-crops-and-traits-in-the-crop-ontology/) or interactions with other CoPs, such as the Data-driven Agronomy and the Socio-Economic Data (SED) CoPs.
Ontologies for Plant Traits and Agronomy Data
Crop breeding relies on collecting data on the desired traits for a new crop variety by testing it in multiple locations and diverse environments, linking phenotypes to genotypes, and drawing conclusions from meta-analyses. In addition, information produced by agronomic trials for field management practices applied by farmers is key to understanding how the significant differences in the practices underpin the performance of the variety. The quality and consistency of data collected during field trials are improved by the use of electronic field books and require the use of ontologies validated by end users.7,8
In 2008, CGIAR initiated the development of the CO (http://www.cropontology.org) in response to the need of breeding data management systems and field books to have access to valid lists of defined breeders' traits and variables. Currently, the CO comprises 4,235 traits and 6,151 variables for 31 plant species. By providing descriptions of agronomic, morphological, physiological, quality, and stress traits along with a standard for composing the variables, the CO enables digital capture and aggregation of crop trait data, as well as comparison across projects and locations.7 The CO was integrated into the Planteome's ontology project funded by the National Science Foundation, US (IOS:1340112 award; http://planteome.org) and was successfully adopted by the CGIAR Integrated Breeding Platform (https://www.integratedbreeding.net/) and by the Boyce Thompson Institute's Breedbase (https://breedbase.org/), both of which are comprehensive breeding management systems and analysis software, and by national databases, such as GnpIS (https://urgi.versailles.inra.fr/Tools/GnpIS)9 in France, or international projects, such as Emphasis (European Plant Phenotyping Infrastructures; https://emphasis.plant-phenotyping.eu/). Both the Minimum Information About a Plant Phenotype Experiment (https://www.miappe.org/) metadata schema (MIAPPE),10,11 and the Breeding Application Programming Interface (BrAPI) (https://brapi.org/),12 which enable the extraction of genotype and phenotype data across databases, are compliant with the CO format.
At the time CGIAR launched the CO, the Plant Trait Ontology (TO)13 did not include traits and definitions required for breeding data on the CGIAR mandate crops. To remediate this situation and create the necessary upper-level connection between the species-specific ontologies, CO trait terms were mapped to terms, thus enabling searches of annotated data across species, using a single trait term.14,15 As a result, Planteome Release 3.0 includes ten species-specific trait ontologies developed by the CO for the crops: cassava (Manihot esculenta), maize (Zea mays), pigeonpea (Cajanus cajan), rice (Oryza sativa), sweet potato (Ipomoea batatas), soybean (Glycine max), wheat (Triticum aestivum), lentil (Lens culinaris), sorghum (Sorghum bicolor), and yam (Dioscorea sp.). These mappings can be automatically created but still require manual curation, making them difficult to maintain considering that ontologies evolve over time.15 Planteome is developing a Plant Stress Ontology (https://github.com/Planteome/plant-stress-ontology) that will require support from the Ontologies CoP for content validation, particularly on the described pest and disease symptoms.
In 2014, CGIAR began developing the AgrO to support the new Agronomy Field Information System (AgroFIMS) (https://apps.cipotato.org/hidapagrofims/),8 which enables scientists to create their electronic field book. AgrO describes agronomic practices and techniques, and integrates variables used in agronomic experiments by agronomists of the Data-driven Agronomy CoP and by the International Consortium for Agricultural Systems Applications. Applying the principles of the Open Biological and Biomedical Ontology (OBO) Foundry,16 AgrO directly integrates terms and their original URIs taken from existing ontologies, such as the Environmental Ontology (ENVO) and the Chemical Ontology (ChEBI). For example, the definition of “tillage process” in AgrO uses the “soil” concept from ENVO in addition to AgrO's novel concept “tillage implement.” Missing terms or knowledge relevant to the agronomy domain were directly proposed to the ontologies. For instance, urea is a widely used fertilizer in agriculture, but the urea concept in ChEBI was not defined as having a fertilizer role. So, the missing link was requested by AgrO and added to ChEBI. More information about AgrO content can be found on the CGIAR Platform for Big Data in Agriculture Website (https://bigdata.cgiar.org/resources/agronomy-ontology/).
Socio-economic Data: Starting with Agricultural Household Surveys
CGIAR and its partners perform a large number of agricultural household surveys yielding important data and statistics on the socio-economic status, production and food systems, and environment of smallholders in the developing world. The SED CoP (https://bigdata.cgiar.org/communities-of-practice/socio-economic-data/) created the “100Q Working Group” that developed 100 core questions to be included in household surveys to collect consistent information on key socio-economic indicators. The set of questions consists of the following sections: household composition and characteristics, farm characteristics, land availability and use, livestock availability and use, income and assets, gender, food security and dietary diversity, and other aspects.17 The Ontologies and SED CoPs are working together to identify concepts from the survey questions and results, which will be used to form the new SEOnt. SEOnt will provide concepts and variables to the survey forms to annotate the data collected with the 100 questions, while taking into account the sensitive nature of the personal information. The first draft of SEOnt is available on GitHub (https://github.com/AgriculturalSemantics/SEOnt).
The use of ontologies in making data interoperable is also enhanced when metadata schemas are adopted, such as the metadata schema being developed by the SED CoP, which relies heavily on the work of the Ontologies CoP.
Expanding CoP Products to New Domains Relevant to Agriculture and Food Systems
CGIAR research also aims to improve the sustainability, productivity, and resilience of fish agrifood systems and collects fish-related datasets, which include fish health, diseases, breeding, genetics, and catch data, among others. Harmonizing fish data annotation with an ontology will enable easier data aggregation and analysis. One available ontology, FISHO (https://bioportal.bioontology.org/ontologies/FISHO),18 focuses on ichthyology, diversity, and adaptation. The Food and Agriculture Organization (FAO) of the United Nations initiated several fisheries ontologies, but the ontologies available remained drafts.19 Therefore, in May 2019, CGIAR and relevant partners formed the Fish Ontology Working Group to compile, update, and contribute fishery terms to existing ontologies. The working group plans to collaborate with the other animal science partners toward developing and adopting animal ontologies within CGIAR.
To enable the interoperability of data along the agricultural value chain, the Ontologies CoP members plan to foster a collaboration with the Food Ontology (https://foodon.org/) consortium, which aims at building a comprehensive global farm-to-fork ontology20 by contributing concepts on tropical and subtropical production systems and food products. A specific value chain ontology will be developed indicating the actors and their roles in the chain. The CoP could use the terminology compiled by CGIAR's Research Program on Policies, Institutions, and Markets for the Value Chains platform (http://tools4valuechains.org) as a source of concepts and invite social scientists and economists to contribute to this work.
Finally, CGIAR needs to demonstrate in a meaningful way the contribution of its research to the Sustainable Development Goals (SDGs). Integrating objectives, targets, and processes of the CGIAR Strategic Research Framework into the SDG Interface Ontology (SDGiO) (https://github.com/SDG-InterfaceOntology/sdgio), which is developed with the support of the United Nations Environment Program, will provide a new set of concepts to annotate data about agrifood innovations and their impact on stakeholders.
Identifying Criteria for the Adoption of Quality Ontologies by the Agrifood Research Community
In general, an increasing number of controlled vocabularies, structured taxonomies, and semantically rich ontologies are developed ex novo in an ad hoc manner to support research projects, often without drawing on concepts and definitions from existing ontologies. For example, the thematic repository AgroPortal (http://agroportal.lirmm.fr/),21 developed by the Laboratoire d'Informatique, de Robotique et de Microélectronique de Montpellier, currently compiles 121 ontologies and thesauri only for plants, agriculture, food, and biodiversity. This situation has led to a growing number of incompatible domain-specific ontologies impeding desirable data integration and interoperability. Consequently, scientists and data managers require guidance to unambiguously select the proper ontology terms in order to annotate data.
Taking a step closer toward identifying and agreeing upon the criteria that make an ontology a quality resource for data annotation, the Ontologies CoP organized a webinar with an Expert Panel (https://www.youtube.com/c/OntologiesInAgriculture) involving Christopher J. Mungall (Lawrence Berkeley National Laboratory) and Pier Luigi Buttigieg (Alfred Wegener Institute), who are both members of the OBO Foundry editorial board (http://www.obofoundry.org/docs/Membership.html), Pankaj Jaiswal, leader of the Planteome project (Oregon State University), and Alexandra Lafargue, Knowledge Manager (Syngenta). A list of 17 key criteria, inspired by the OBO Foundry principles (http://www.obofoundry.org/principles/fp-000-summary.html), was proposed by the Expert Panel (Table 1). CGIAR data managers and ontology curators were asked to rank the criteria to understand which were the most important to non-expert users and should therefore be documented as a priority to guide the selection for annotation.
Table 1.
Criteria Established by CoP Experts to Characterize the Quality of Ontologies for Data Annotation
| Criteria Classified by the Expert Panel | |
|---|---|
| 1 | Adhere to the OBO Foundry guidelines |
| 2 | Represent a unique non-overlapping knowledge domain (also known as orthogonality) |
| 3 | Willingness to express and integrate multiple, evidence-based classification systems in the chosen domain |
| 4 | Logically structured with a well-defined scope |
| 5 | May contain relationships and dependencies to other reference ontologies |
| 6 | Represent accurate science supported by evidence |
| 7 | Open source and Creative Commons CC-BY or CC-0 license (https://creativecommons.org/) |
| 8 | Must be widely used in annotation and data capture |
| 9 | Support both inter- and intra-specific needs with species agnostic (core) and specific (extensions) resources that work together |
| 10 | Sustainable funding sources |
| 11 | Human resources to manage (i.e., curators, editors, and developers) |
| 12 | Established ontology management system, including roles and responsibility |
| 13 | Must be designed to answer both the computing and community needs |
| 14 | Must explicitly identify the communities of reference |
| 15 | Centralized maintenance of the validated content, and distributed contribution and access |
| 16 | Ontology quality assurance by experts in the field of knowledge |
| 17 | Reducing reliance on internal processes and data stewardship networks |
The five top-ranking criteria selected were: (1) the domain-specific coverage of the ontology; (2) the ontology must be widely used in annotation and data capture (to reduce the cost of external data integration, which is routine work for data scientists and so was ranked higher by data managers and curators than by the Expert Panel); (3) indicators used for quality assurance should be available; (4) ontology maintenance is centralized, while contributions and access are distributed among users; and lastly (5) the existence of sustainable funding to support the ontology, funding being a real challenge and clearly of primary importance in securing the human resources necessary to manage the ontology. These five criteria are all part of the OBO Foundry principles of ontology design and format.
There are several ontologies applicable to agrifood science, which comply with many of the above quality criteria, available for modeling crops, livestock, and other animal species (Table 2). The most used ontologies for plants,22,23 aside from the Gene Ontology (GO), are the: Plant Ontology,24,25 TO, CO,7 Plant Experimental Conditions Ontology13—all included in the Planteome project (http://planteome.org/)—as well as the ENVO,26,27 AgrO,8 and NCBI Taxon Ontology28 (Table 2). The Sequence Ontology (SO)29 and the Unit Ontology (UO)30 are also widely used. Under the guidance of the Ontologies CoP, several of these ontologies have been adopted within CGIAR, thus progressively increasing the quality of the data annotation.
Table 2.
Widely Used Ontologies in Agricultural Science
| Ontology | Domain and URL |
|---|---|
| Agronomy Ontology8 | Agronomic practices, agronomic techniques, and agronomic variables used in agronomic experiments https://bigdata.cgiar.org/resources/agronomy-ontology/ http://obofoundry.org/ontology/agro |
| Crop Ontology7,13 | Species-specific phenotypic plant traits http://www.cropontology.org/ |
| Environment Ontology26,27 | Environmental features and habitats http://environmentontology.org/ http://obofoundry.org/ontology/envo |
| Evidence & Conclusion Ontology31 | Evidence of scientific events https://github.com/evidenceontology/evidenceontology/ |
| Gene Ontology32,33 | Molecular functions, biological processes, cellular components http://geneontology.org/ http://obofoundry.org/ontology/go |
| NCBI Taxon Ontology28 | Organismal taxonomy of National Center for Biotechnology Information https://github.com/obophenotype/ncbitaxon http://www.obofoundry.org/ontology/ncbitaxon.html |
| Plant Ontology13 | Plant anatomy, morphology, and growth and development http://browser.planteome.org/amigo http://obofoundry.org/ontology/po |
| Plant Experimental Conditions Ontology13 | Treatments and growth conditions used in plant science experiments http://browser.planteome.org/amigo http://obofoundry.org/ontology/peco |
| Plant Trait Ontology 13 | Phenotypic traits in plants http://browser.planteome.org/amigo http://obofoundry.org/ontology/to |
| Sequence Ontology29 | Features and attributes of biological sequence http://www.sequenceontology.org/ http://obofoundry.org/ontology/so |
| Units of Measurement Ontology30 | Units of measurement https://github.com/bio-ontology-research-group/unit-ontology http://www.obofoundry.org/ontology/uo.html |
Discussion
Improving User Experience in Selecting and Submitting Ontology Terms Used in Data Annotation
Because of the urgency to release data generated annually that support agricultural research questions and technological innovation, best practices for quality data annotation are not always systematically applied. The CoP plays a key role in providing guidance and interacting with teams developing solutions that can facilitate the annotation process. Developing or completing ontologies, as well as recommending annotation support tools, are tasks for the well-defined Ontologies CoP of the CGIAR Platform for Big Data in Agriculture.
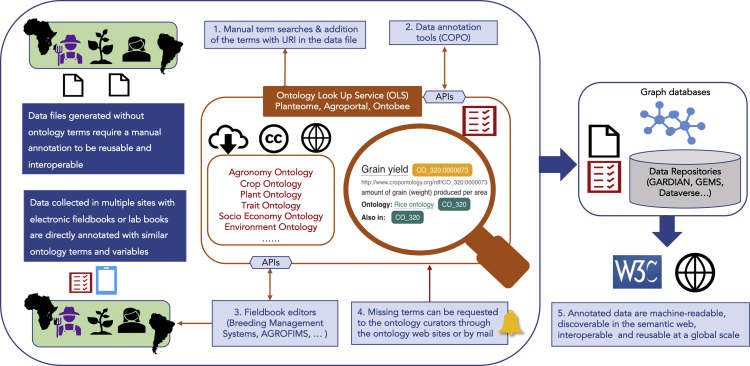
Figure 1 illustrates the current user's generic experience for selecting ontology terms for data annotation and submitting new concepts.
Figure 1.
Use of the CoP's Products and Tools for Data Annotation
Manual Ontology Term Searches
In general, when annotating datasets, scientists and data managers first need to manually check if relevant ontology terms exist (Figure 1, step 1). They also need to be familiar with the terms used in the original files because, for example, crop traits are included in a variety of nomenclatures, often decided by different groups of scientists without any coordination.
To illustrate step 1, we provide a specific example of data annotation for the evaluation and adoption by farmers of flood-tolerant rice varieties in Bangladesh.34 Submergence tolerance is a target trait for rice breeders because flooding is a major abiotic stress causing important yield losses in rice production areas in South and South-East Asia,35 and some parts of Africa.36 This annotation exercise was performed by a scientist using survey data collected by the International Rice Research Institute (IRRI) with the support of the Ontologies CoP experts (Havard Dataverse: https://dataverse.harvard.edu/dataset.xhtml?persistentId=doi:10.7910/DVN/26165). It was simplified for this paper by only selecting a sample of key concepts that could annotate data files at the level of their metadata and their variables (Table 3). We did not include all concepts or the finer annotation of the value, describing measurements or observation methods and scales or units.
Table 3.
Result of an Ontological Term Selection to Annotate Datasets about Submergence Tolerance of Rice Varieties for the Flood-Prone Lowlands in Nigeria
| Dataset Terms | Selected Ontology Terms | Definition | Source Ontologies | URI for Data Annotation | |
|---|---|---|---|---|---|
| Crop | Rice | Oryza sativa | (Rice), species, monocots | NCBI taxonomy | http://purl.obolibrary.org/obo/NCBITaxon_4530 |
| Variety Traits | |||||
| Genotype | Germplasm with the submergence tolerance “Sub1” gene | Response to flooding | Any process that results in a change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus indicating flooding, short-term immersion in water | Gene Ontology | http://purl.obolibrary.org/obo/GO_0009413 |
| Phenotype | Submergence tolerance | Rice submergence tolerance trait Submergence sensitivity |
The ability of plants to survive a period of submergence Measure of sensitivity of a plant if placed under submergence condition |
Crop Ontology (CO) Trait Ontology (TO) |
http://www.cropontology.org/rdf/CO_320:0000067a http://purl.obolibrary.org/obo/TO_0000286 |
| Field practices | Manual weeding | Hand picking weeding process | A mechanical weeding process in which unwanted organisms are removed by hands | Agronomy Ontology | http://purl.obolibrary.org/obo/AGRO_00002057 |
| Herbicide treatment | Chemical weeding process | A weeding process in which chemical is used to manage unwanted weeds | Agronomy Ontology | http://purl.obolibrary.org/obo/AGRO_00002053 | |
| Weeding application date | Term not found | ||||
| Farming system | Rain-fed rice production system | Rain-fed farming | Arable cultivation relying solely on rainfall | AGROVOC | http://aims.fao.org/aos/agrovoc/c_6436 |
| Abiotic stress | Flood-prone region exposure | Flood-prone region exposure Lowland region exposure |
A treatment in terms of a plant's exposure to the regional conditions found in the vicinity of the water bodies, such as sea, river, lake. Growth conditions may include aerobic to anaerobic soil, salinity or toxicity in tidal areas. Treatment may include standing or flash flooding Treatment involving the plant, or the populations grown in regions where the land level is slightly steep, noncontinuous flooding of variable depth and duration. Alternating conditions of aerobic to anaerobic soil |
Plant Experimental Conditions Ontology (PECO) |
http://purl.obolibrary.org/obo/PECO_0007396b http://purl.obolibrary.org/obo/PECO_0007391 |
| Geography | Bangladesh | Bangladesh | Gazetteer | http://purl.obolibrary.org/obo/GAZ_00000912 | |
| Agro-ecosystem | Lowland region | Lowland | None | AGROVOC | http://aims.fao.org/aos/agrovoc/c_4453 |
| Socio-economy | Farmers' income | Household income Agricultural income |
A demographic parameter indicating the amount of earnings made by a family Quantified household income using the sales information of agricultural products. This is gross income |
NCI thesaurus in Socio-economic ontology Socio-Economic Ontology |
http://purl.obolibrary.org/obo/NCIT_C70811 http://purl.obolibrary.org/obo/SURVO_00000200 |
| Fertilizer costs | Term not found | ||||
Annotation performed by Dr. Berta Miro, IRRI with the support of the CoP ontology experts.
CO term is mapped to a TO term so annotations using one or another are valid. CO will provide the format the variables measuring in the field the effect of the flood on the rice varieties.
PECO term is mapped to ENVO term “Floods (EO:0007172)” that has the definition: an unusual accumulation of water above the ground caused by high tide, heavy rain, melting snow, or rapid runoff from paved areas.
Users who are familiar with the domain-specific ontologies can perform a search directly on the relevant ontology website where they can visualize, browse, and download the ontology, and access direct term submission forms or templates, when available. If the user does not know any domain-specific ontology, consulting the quality ontology selection criteria recommended by the CoP on its web page is always good practice. Then a term search using ontology look-up services of the main registries (e.g., European Bioinformatics Institute [EBI] Ontology Lookup Service [OLS], Planteome, AgroPortal, Ontobee) will provide access to a large range of ontologies (Figure 1, step 1). These registries automatically synchronize their content using the Application Program Interfaces (APIs) of the ontologies' websites or of the open-source ontology project management tools.
In the example of flood-tolerant rice varieties, a search in the OLS returns the term response to flooding from GO that can annotate the presence of the Sub1 gene conferring the tolerance. The term identifier is GO:0009413 and is included in the URI. If the searched-for term is not found, looking for synonyms, such as submergence will help. For annotating the phenotypic evaluation results, the user can select submergence tolerance in the CO (CO_320:0000067) or TO (TO:0000286) as both ontologies are mapped. The CO will provide the rice-specific variables used to measure the effect of submergence.
The challenge lies in reading through the results of matching terms and checking for the most appropriate one. To see if the term fully corresponds to the search, users must check both the metadata of the term (e,g., definition, synonyms, context of use, note, evidence) and the ontology (e.g., domain, authority, curation, usage), and possibly linked terms. For example, a note in GO indicates that response to flooding (GO:0009413), which refers to short-term immersion should not be confused with response to deep water (GO:0030912), which refers to standing in water throughout an organism's life cycle.
Hybrid approaches involving both quality ontologies and largely used thesauri may offer a solution to data managers. Thesauri have a simpler semantic structure than ontologies, called a Knowledge Organization System, that use broader narrower relationships between concepts. The most popular thesauri in Agriculture are: AGROVOC (http://agrovoc.uniroma2.it/agrovoc/agrovoc/en/) maintained by FAO, the Center for Agriculture and Biosciences International Thesaurus (https://www.cabi.org/), and the US National Agricultural Library Thesaurus (https://agclass.nal.usda.gov/). For the rice data annotation example, the concept of lowland as a landform was only found in AGROVOC.
Once the term is identified, users can easily copy the URI and paste it in their file, ideally at the variable value level to increase the interoperability potential of the data.
Table 3 summarizes the results of a manual ontology term search using the EBI OLS. It shows only the key concepts that could be used to annotate the rice datasets relevant to the evaluation of flood-tolerant varieties. Datasets annotated with these ontological terms could then be retrieved through a query, such as: “Rice varieties that are flood-tolerant and can grow in Bangladesh in rain-fed lowlands subject to recurrent devastating flooding.” For example, annotated datasets on the rice strains with Sub1 gene disseminated in Bangladesh should systematically appear in a result list of such a search.
Although over time users will gain experience and confidence in the term selection and insertion of URIs in their files, such a manual process remains laborious and time consuming, often discouraging scientists and data managers from finding adequate terms. Consequently, they will limit their annotations to a strict minimum, such as a few keywords in metadata, which is insufficient for the interoperability of the data.
In an Ontologies CoP survey, members identified the development of an online hub of ontologies recommended for agriculture, food, and environment research domains as a necessary resource to improve their annotations. Indeed, repositories, such as GARDIAN, and data discovery platforms, such as GEMS (the platform of the AgroInformatics Consortium; https://agroinformatics.org), combine multidisciplinary data from biophysical studies to socio-economic surveys, which implies the use of several domain-specific ontologies to fully describe the data.23 Therefore, scientists and data managers need direct access to the set of quality ontologies recommended for the specific domain to upload their data and metadata in such repositories. The ideal solution does not yet exist, but ontology look-up services and ontology registries will be a part of it.
The OLS (https://www.ebi.ac.uk/ols/index), developed and hosted by the EBI, is the closest tool to this requirement as it provides a simple search function for finding specific concepts across 251 ontologies comprising over 6.1 million URIs representing concepts. The OLS API enables any database to access this wealth of ontologies. If users can restrict their term search to a single ontology, there is, however, no option for filtering the ones most used by agrifood domain experts.
AgroPortal provides a complementary solution focused on agronomy that, aside from quality ontologies, includes draft and specific community ontologies, therefore acting as an ontology project discovery tool. AgroPortal offers a set of ontology descriptive metadata and statistics on the ontology files downloaded and should add information on all the criteria listed in this paper that would guide users toward quality and popular ontologies for agricultural data.
To support such a work, the Ontologies CoP facilitates dialogue with the ontologies registries and promotes the use of ontology look-up services to users and to multidisciplinary data platforms, so that they can permanently access updated content from the ontologies.
Automation of Ontology Term Selection and Data Annotation
Ontology-driven data annotation tools enable the automation of the manual annotation process (Figure 1, step 2). The CoP members have identified and are testing COPO (collaborative open plant omics) (https://copo-project.org/), a promising tool currently being developed by the Earlham Institute, which provides metadata and ontology annotation capabilities, thus offering a platform for researchers to publish their research assets.37 COPO uses the EBI OLS to perform real-time look-up of ontology concepts when a user enters a term. The COPO tool goes further than simply adding keywords to metadata by supporting the tagging of column headings of data files where values of variables are stored, thus increasing the interoperability of the data. When further developed, COPO could fully describe the file's values drawing on terms from several ontologies.
A feature that the CoP members proposed was for COPO to preferentially indicate, at the top of the list, the ontologies and the terms that were most used in previous data annotations.
The CoP will continue conveying the members' needs to the developers of data annotation tools to ensure that they are fit for purpose and that developers understand users' priorities and requirements for ontology concept selection.
Many agricultural databases enabling the production of electronic field books for homogeneous quality data collection provide direct assistance with ontology term selection and data annotation through an ontology manager (Figure 1, step 3). Users simply need to select the ontology terms and variables directly in the database when designing their field books. Data will then be automatically labeled at the collection stage and uploaded back into the database along with their annotation. Any project database can automatically download and synchronize the versions of the ontologies through their APIs.
Submitting New Ontology Terms
If a term appears to be missing, users should contact the curation team of the domain-relevant ontology to confirm the gap (Figure 1, step 4). Sending questions to the CoP members via the CoP LinkedIn Group or website is good practice. For example, partners, such as GrainGenes (https://wheat.pw.usda.gov/GG3/), University of Cornell, US, and URGI (https://urgi.versailles.inra.fr/), and INRAe, France, holding specific projects' wheat traits and variables, developed their lists of traits and variables using the Trait Dictionary template of the CO and their integration into the CIMMYT wheat ontology is being performed under the supervision of the wheat ontology curator.
To maintain ontologies and consistent versioning, the CoP recommends using open-source tools for project management with version control systems that enable the management of released versions and can offer a publicly available tracker of issues posted by ontology curators and users. In general, an issue tracker enables subscribers of the open project management tool to directly insert their comments and suggestions, which will result in an email alert to all subscribers. For an ontologies project management tool, such as the Planteome GitHub (https://github.com/Planteome), any issue opened by a contributor will alert the ontology curators about new term submission or modification requests. Alternative options for submitting a term are the templates and forms proposed in the ontologies' websites. In this way, the new concepts are submitted to an established ontology and are correctly placed in the semantic graph by the ontology curator after its metadata is checked (synonyms, definition, context of use, reference) and is added with an URI.
In the rice data annotation example, the terms weeding application date and fertilizer cost were not found by the scientist. The gaps were confirmed by the respective curators of AgrO and SEOnt and the term weeding application date was then submitted by the scientist to the AgrO's GitHub issue tracker while fertilizer cost was submitted to SEOnt's tracker. The term weeding time was added into AgrO and will be included in the next ontology release.
In fact, annotation tools, such as COPO should include a feature enabling users to directly submit their missing terms to adequate ontologies' issue trackers, in a similar way that the Breedbases from BTI propose to use an online crop trait term submission form that directly creates an issue in the Planteome's open ontology management tool and alert the curators. This is an important feature that simplifies ontology term submission, requiring no specific technical knowledge on the use of an issue tracker.
Upload of Annotated Files into Repositories and Databases
Once the data file is described with appropriate metadata and ontologies, files can be uploaded into data repositories or a graph database (Figure 1, step 5). Data repositories archive datasets with their metadata and annotations for long-term storage and access. COPO allows the annotated data to be directly deposited in a range of repositories, including DSpace (https://duraspace.org/dspace/), CKAN (https://ckan.org/), and Dataverse (https://dataverse.org/), which are used by CGIAR.
If the URIs of the selected ontology terms are ideally present for each variable, the file can be uploaded into a database, such as a graph database. A graph database has no predefined structure constraining the data and is based on a graph that represents the semantic relationships between data, showing how each individual entity connects with or is related to the other (https://neo4j.com/developer/graph-database/), so semantic queries will use the ontological relationships to discover annotated data. To be efficient, the graph requires a quality and fine ontology annotation of the measured or observed variables.
Collaboration with the Agrifood Industry
For over 10 years, the agrifood industry has shown a strong interest in using ontologies and semantic web technologies to improve their data science activities (e.g., genomics data integration, data curation and annotation, responsible and ethical data management). The agrifood industry has progressed in the adoption of semantic tools and quality improvement of their data annotations faster than the public sector. Some success stories in industry and recurring challenges have been reported (https://f1000research.com/slides/5-348).38 The rise in digitalized farming has created several open challenges related to the application of ontologies and semantic web technologies. The Ontologies CoP provide an adequate space for discussing the most prominent concerns about best practices and data reusability in this sector. In particular, knowledge graphs are part of the new data science portfolio of advanced structures enabling data analysis in modern Research and Development. The industry sector largely uses the ontologies developed by the public sector and is progressively increasing its contribution to this collective effort.
Conclusion
The development of an Ontologies CoP for research on agrifood systems was necessary to harness the scattered ontology expertise and secure the quality, usability, and sustainability of a comprehensive set of semantic resources for agrifood science. CGIAR's Platform for Big Data in Agriculture realized the importance of ontologies to support FAIR data and knowledge sharing, investing financially in the creation of the Ontologies CoP.
The CoP members engage regularly across relevant networks to support the curation of data for biological, food and agronomic research, and socio-economics. They also play an advocacy role in sensitizing new donors, public institutions, and the agrifood industry to the importance of providing long-term financial support to this collaborative data curation effort, which contributes to breaking data silos and supporting the growing use of digital tools in agrifood systems. Long-term sustainable access to quality ontologies will increase the research community's confidence in using them and will improve the FAIR status of the data across research and development projects, in turn increasing their discoverability and value for re-use, and thus contributing to the return on investment for their collection and storage.
For any sector, including the agrifood industry, the development and maintenance of quality ontologies should go hand-in-hand with effective and responsible data governance, including data stewards, data owners, and a solid data policy. Information technology infrastructure (servers, connectivity, and underlying software) plays a crucial role in organizing the actual data structures in the form of ontologies, taxonomies, and controlled vocabularies. Therefore, sufficient resources should be allocated to developing those components when building a sustainable data management system.
The next set of priority ontologies to be developed for CGIAR's Platform for Big Data in Agriculture will be related to livestock, fisheries and aquaculture, water management, food systems, and value chains. To create the semantic framework that will support the evidence of CGIAR's and partners' contributions to the SDGs, the CoP will continue integrating concepts on agriculture and food systems into the SDGiO.
Based on emerging needs, the CoP will also create additional thematic working groups, for example to collaborate with the Geospatial Data CoP for the harmonization of data generated by remote sensors, such as drones. The CoP will stimulate collaboration on the development of knowledge graphs in agriculture that support graph databases, a domain in which the agrifood industry has made rapid progress.
Experimental Procedures
Resource Availability
Lead Contact
Elizabeth Arnaud, e.arnaud@cgiar.org, https://orcid.org/0000-0002-6020-5919.
Materials Availability
This study did not generate any physical material.
Data and Code Availability
All data held in the form of draft and final ontologies produced by the Ontologies CoP are accessible online on public repositories managing versioning—mainly in GitHub repositories. Final versions of the ontologies are published with a cc-by license.
| Ontology | URL |
|---|---|
| Agronomy Ontology |
https://bigdata.cgiar.org/resources/agronomy-ontology/ http://obofoundry.org/ontology/agro |
| Crop Ontology | http://www.cropontology.org/ |
| Environment Ontology |
http://environmentontology.org/ http://obofoundry.org/ontology/envo |
| Plant Ontology |
http://browser.planteome.org/amigo http://obofoundry.org/ontology/po |
| Plant Experimental Conditions Ontology |
http://browser.planteome.org/amigo http://obofoundry.org/ontology/peco |
| Plant Trait Ontology |
http://browser.planteome.org/amigo http://obofoundry.org/ontology/to |
| Plant Stress Ontology | https://github.com/Planteome/plant-stress-ontology |
| Planteome | https://github.com/Planteome |
| SEOnt | https://github.com/AgriculturalSemantics/SEOnt |
Velarde, Orlee. Dissemination of Submergence-Tolerant Varieties and Associated New Production Practices to Southeast Asia.(2014),Data set version 3, Harvard Dataverse, https://doi.org/10.7910/DVN/26165.
The code of the cited tools is publicly accessible:
| AgroFIMS | https://github.com/AGROFIMS |
| AgroPortal | https://github.com/agroportal |
| BrAPI | https://github.com/plantbreeding/API |
| Crop Ontology website | https://github.com/bioversity/Crop-Ontology |
| GARDIAN | https://github.com/SCiO-systems/CGIAR-BDP-GARDIAN |
| COPO | https://github.com/collaborative-open-plant-omics |
| MIAPPE | https://github.com/MIAPPE |
| Ontology Lookup Service | https://github.com/EBISPOT/OLS |
Acknowledgments
The Ontologies CoP and Socio-Economic Data CoP are financially supported by the CGIAR Platform for Big Data in Agriculture that is mainly supported by the CGIAR Trust Fund, (https://www.cgiar.org/funders/) and UKAID. The Crop Ontology is currently supported by the CGIAR Platform for Big Data in Agriculture and the CGIAR Research Programs on Roots, Tubers, and Bananas; Wheat, Maize, and Rice Programs; Grain Legumes and Dryland Cereals (CRP-GLDC); and by each CGIAR Center for its mandate crops. The rice example is based on data generated by the International Rice Research Institute (IRRI) for the RICE Research program. The Planteome Project, led by P.J. (Oregon State University), is funded by the National Science Foundation, USA (IOS:1340112 award). The coordinator of the Environment Ontology and SDG Interface Ontology is funded by the Frontiers in Arctic Marine Monitoring (FRAM) program of the Alfred Wegener Institute Helmholtz Centre for Polar and Marine Research, Helmholtz Centre for Polar and Marine Research (AWI). COPO was initially funded by a BBSRC Biological and Bioinformatics Resources (BBR) grant (BB/L024055/1, BB/L024101/1, and BB/L024071/1) and is now funded by the BBSRC Core Strategic Program grant awarded to the Earlham Institute (BBS/E/T/000PR9817). COPO is hosted within the CyVerse UK academic cloud, funded by BBSRC (BB/M018431/1 and BB/R000662/1). S.L. is funded by the Alan Turing Institute under the EPSRC grant EP/N510129/1. The Elixir and Emphasis contribution to the Crop Ontology and its adoption have been supported by the Infrastructure Biologie Santé “Phenome-FPPN′” supported by the French National Research Agency (ANR-11-INBS-0012), the TransPLANT project (EU 7th Framework Program, contract no. 283496), the H2020 ELIXIR-EXCELERATE project (funded by the European Commission within the Research Infrastructures program of Horizon 2020, grant agreement no. 676559), and the “Investments for the Future program” (PIA) (ANR-11-INBS-0012) as well as by INRAe. Developments of wheat, protein crops, rapeseed, and miscanthus ontologies have been supported by the Breedwheat (ANR-10-BTBR-03), BFF (11-BTBR-0006), Rapsodyn (11-BTBR-0004), and Peamust (11-BTBR-0002) PIA projects.
We acknowledge the contribution of Kate Dreher, data steward at CIMMYT for actively supporting discussions on semantics within the Data Management Working Group of the CGIAR Excellence in Breeding Platform. Aman Sidhu, consultant, for formatting and facilitating the CoP webinars. Olga Spellman, The Alliance Bioversity International-CIAT, for paper technical review and English editing.
Author Contributions
E. Arnaud, who oversees and leads the Ontologies Community of Practice (CoP) activity planning and execution, wrote the manuscript, . P.J. leads the Planteome project and secured funding for the work. C.A., E. Antezana, P.L.B., L.C., P.J., G.K., S.L., S.K., M.-A.L., J.M., and C.J.M., who lead investigation activities, develop ontologies and recommendations, contributed to the manuscript. B.M. provided the rice data example, performed the annotation, and contributed to the manuscript. A.A., G.B., R.D.D., J.P., V.H., J.M., N.M., C.P., A.R., R.S., and R.D. actively contribute to the development and curation of the ontologies for agriculture. S.A., E.B., B.C., I.D., E.D.Y., H.J., A.L., D.L., L.A.M., O.O., F.S., and K.S. are data managers and IT developers of ontology-supported tools and repositories. P.L.B. and C.J.M. provide expert advice to the CoP. M.D. and B.K. lead modules in the CGIAR Big Data Platform and actively and financially support the CoP. G.K. leads the Socio-Economic Data CoP. S.L. and L.A.M. are supportive project leaders. All authors are members of the Ontologies Community of Practice (CoP).
Declaration of Interests
The authors declare no competing interests.
Published: September 25, 2020
References
- 1.Wilkinson M.D., Dumontier M., Aalbersberg Ij.J., Appleton G., Axton M., Baak A., Blomberg N., Boiten J.-W., da Silva Santos L.B., Bourne P.E. The FAIR guiding principles for scientific data management and stewardship. Sci. Data. 2016;3:160018. doi: 10.1038/sdata.2016.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gruber T. Ontology. In: Liu L., Tamer Özsu M., editors. Encyclopedia of Database Systems. Springer-Verlag; 2009. [Google Scholar]
- 3.Walls R.L., Athreya B., Cooper L., Elser J., Gandolfo M.A., Jaiswal P., Mungall C.J., Preece J., Rensing S., Smith B. Ontologies as integrative tools for plant science. Am. J. Bot. 2012;99:1263–1275. doi: 10.3732/ajb.1200222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Leonelli S. Chicago University Press; 2016. Data-Centric Biology: A Philosophical Study. [Google Scholar]
- 5.Dzale Yeumo E., Alaux M., Arnaud E., Aubin S., Baumann U., Buche P., Cooper L., Ćwiek-Kupczyńska H., Davey R.P., Fulss Developing data interoperability using standards: a wheat community use case. F1000Res. 2017;6:1843. doi: 10.12688/f1000research.12234.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Leonelli S. Global data for local science: assessing the scale of data infrastructures in biological and biomedical research. BioSocieties. 2013;8:449–465. [Google Scholar]
- 7.Shrestha R., Matteis L., Skofic M., Portugal A., McLaren G., Hyman G., Arnaud E. Bridging the phenotypic and genetic data useful for integrated breeding through a data annotation using the Crop Ontology developed by the crop communities of practice. Front. Plant Physiol. 2012;3 doi: 10.3389/fphys.2012.00326. Article 326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Devare M., Aubert C., Laporte M.-A., Valette L., Arnaud E., Buttigieg P.L., (2016). Data-driven agricultural research for development: a need for data harmonization via semantics. International Conference on Biomedical Ontologies (ICBO), 2016.
- 9.Pommier C., Michotey C., Cornut G., Roumet P., Duchêne E., Flores R., Lebreton A., Alaux M., Durand S., Kimmel E. Applying FAIR principles to plant phenotypic data management in GnpIS. Plant Phenom. 2019;2019:15. doi: 10.34133/2019/1671403. 1671403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ćwiek-Kupczyńska H., Altmann T., Arend D., Arnaud E., Chen D., Cornut G., Fiorani F., Frohmberg W., Junker A., Klukas C. Measures for interoperability of phenotypic data: minimum information requirements and formatting. Plant Methods. 2016;12:44. doi: 10.1186/s13007-016-0144-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Papoutsoglou E.A., Faria D., Arend D., Arnaud E., Athanasiadis I.N., Chaves I., Coppens F., Cornut G., Costa B.V., Ćwiek-Kupczyńska Enabling reusability of plant phenomic datasets with MIAPPE 1.1. New Phytol. 2020 doi: 10.1111/nph.16544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Selby P., Abbeloos R., Backlund J.E., Basterrechea Salido M., Bauchet G., Benites-Alfaro O.E., Birkett C., Calaminos V.C., Carceller P., Cornut BrAPI consortium. BrAPI-an application programming interface for plant breeding applications. Bioinformatics. 2019;35:4147–4155. doi: 10.1093/bioinformatics/btz190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cooper L., Meier A., Laporte M.-A., Elser J.L., Mungall C.J., Sinn B.T., Cavaliere D., Carbon S., Dunn N.A., Smith B. The Planteome database: an integrated resource for reference ontologies, plant genomics and phenomics. Nucleic Acids Res. 2018;46:D1168–D1180. doi: 10.1093/nar/gkx1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Arnaud E., Cooper L., Shrestha R., Menda N., Nelson R.T., Matteis L., Skofic M., Bastow R., Jaiswal P., Mueller L. Proceedings of the International Conference on Knowledge Engineering and Ontology Development. SciTePress; 2012. Towards a reference Plant Trait Ontology for modeling knowledge of plant traits and phenotypes; pp. 220–225. [DOI] [Google Scholar]
- 15.Laporte M.-A., Valette L., Cooper L., Mungall C., Meier A., Jaiswal P., Arnaud E. Proceedings of the Joint International Conference on Biological Ontology and BioCreative. Oregon State University; 2016. Comparison of ontology mapping techniques to map plant trait ontologies. [Google Scholar]
- 16.Smith B., Ashburner M., Rosse C., Bard J., Bug W., Ceusters W., Goldberg L.J., Eilbeck K., Ireland A., Mungall C.J. The OBO Foundry: coordinated evolution of ontologies to support biomedical data integration. Nat. Biotechnol. 2007;25:1251–1255. doi: 10.1038/nbt1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Van Wijk M., Alvarez C., Anupama G., Arnaud E., Azzarri C., Burra D., Caracciolo F., Coomes D., Garbero A., Gotor E. Towards a core approach for cross-sectional farm household survey data collection: a tiered setup for quantifying key farm and livelihood indicators. Community of Practice on Socio-Economic Data Report COPSED-2019-001. CGIAR Platform for Big Data in Agriculture. 2019. https://cgspace.cgiar.org/handle/10568/105714
- 18.Ali N.M., Khan H.A., Then A.Y., Ving Ching C., Gaur M., Dhillon S.K. Fish Ontology framework for taxonomy-based fish recognition. PeerJ. 2017;5:e3811. doi: 10.7717/peerj.3811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Caracciolo C., Heguiabehere J., Gangemi A., Baldassarre C., Keizer J., Taconet M. Knowledge management at FAO: a case study on network of ontologies in fisheries. In: Suárez-Figueroa M., Gómez-Pérez A., Motta E., Gangemi A., editors. Ontology Engineering in a Networked World. Springer; 2012. pp. 383–405. [Google Scholar]
- 20.Dooley D.M., Griffiths E.J., Gosal G.S., Buttigieg P.L., Hoehndorf R., Lange M.C., Schriml L.M., Brinkman F.S.L., Hsiao W.W.L. FoodOn: a harmonized food ontology to increase global food traceability, quality control and data integration. NPJ Sci. Food. 2018;2:23. doi: 10.1038/s41538-018-0032-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jonquet C., Toulet A., Arnaud E., Aubin S., Dzalé Yeumo E., Emonet V., Graybeal J., Laporte M.-A., Musen M.A., Pesce P., Larmande P. AgroPortal: a vocabulary and ontology repository for agronomy. Comput. Electron. Agric. 2018;144:126–143. [Google Scholar]
- 22.Leonelli S., Davey R.P., Arnaud E., Parry G., Bastow R. Data management and best practice for plant science. Nat. Plants. 2017;3:17086. doi: 10.1038/nplants.2017.86. [DOI] [PubMed] [Google Scholar]
- 23.Harper L., Campbell J., Cannon E., Jung S., Poelchau M., Walls R., Andorf C., Arnaud E., Berardini T.Z., Birkett AgBioData consortium recommendations for sustainable genomics and genetics databases for agriculture. Database (Oxford) 2018;2018:bay088. doi: 10.1093/database/bay088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cooper L., Walls R.L., Elser J., Gandolfo M.A., Stevenson D.W., Smith B., Preece J., Athreya B., Mungall C.J., Rensing S. The Plant Ontology as a tool for comparative plant anatomy and genomic analyses. Plant Cell Physiol. 2013;54:e1. doi: 10.1093/pcp/pcs163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Walls R.L., Cooper L., Elser J., Gandolfo M.A., Mungall C.J., Smith B., Stevenson D.W., Jaiswal P. The plant ontology facilitates comparisons of plant development stages across species. Front. Plant Sci. 2019 doi: 10.3389/fpls.2019.00631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Buttigieg P.L., Morrison N., Smith B., Mungall C.J., Lewis S.E., Consortium Envo. The environment ontology: contextualising biological and biomedical entities. J. Biomed. Semant. 2013;4:43. doi: 10.1186/2041-1480-4-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Buttigieg P.L., Pafilis E., Lewis S.E., Schildhauer M.P., Walls R.L., Mungall C.J. The environment ontology in 2016: bridging domains with increased scope, semantic density, and interoperation. J. Biomed. Semant. 2016;7:57. doi: 10.1186/s13326-016-0097-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Federhen S. The NCBI taxonomy database. Nucleic Acids Res. 2012;40(D1):D136–D143. doi: 10.1093/nar/gkr1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mungall C.J., Batchelor C., Eilbeck K. Evolution of the sequence ontology terms and relationships. J. Biomed. Inform. 2011;44:87–93. doi: 10.1016/j.jbi.2010.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gkoutos G.V., Schofield P.N., Hoehndorf R. The Units Ontology: a tool for integrating units of measurement in science. Database. 2012;2012:bas033. doi: 10.1093/database/bas033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Giglio M., Tauber R., Nadendla S., Munro J., Olley D., Ball S., Mitraka E., Schriml L.M., Gaudet P., Hobbs E.T. ECO, the Evidence & Conclusion Ontology: community standard for evidence information. Nucleic Acids Res. 2019;47:D1186–D1194. doi: 10.1093/nar/gky1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.The Gene Ontology Consortium Expansion of the gene ontology knowledgebase and resources. Nucleic Acids Res. 2017;45:D331–D338. doi: 10.1093/nar/gkw1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.The Gene Ontology Consortium The gene ontology resource: 20 years and still going strong. Nucleic Acids Res. 2019;47:D330–D338. doi: 10.1093/nar/gky1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Singh U.S., Dar M.H., Singh S., Zaidi N.W., Bari M.A., Mackill D.J., Collard B.C.Y., Singh V.N., Singh J.P., Reddy J.N. Field performance, dissemination, impact and tracking of submergence tolerant (SUB1) rice varieties in South Asia. SABRAO J. Breed. Genet. 2015;45:112–131. [Google Scholar]
- 35.Mackill D.J., Ismail A.M., Singh U.S., Labios R.V., Paris T.R. Development and rapid adoption of submergence-tolerant (Sub1) rice varieties. Adv. Agron. 2012;115:303–356. [Google Scholar]
- 36.Africa Rice Center (AfricaRice). 2019. Africa rice center (Africa rice) annual report 2018: sustainable rice production in the face of climate emergency. Abidjan, Côte d’Ivoire: 28 pp.
- 37.Shaw F., Etuk A., Minotto A., Gonzalez-Beltran A., Johnson D., Rocca-Serra P., Laporte M.A., Arnaud E., Devare M., Kersey P.J. l. COPO: a metadata platform for brokering FAIR data in the life sciences [version 1; peer review: 1 approved] F1000Res. 2019;9:495. [Google Scholar]
- 38.Burkow, D., Hollunder, J., Heinrich, J., Abdallah, F., Rojas-Macias, M., Wiljes, C., Cimiano, P., Senger, P., (2018). A blueprint for semantically lifting field trial data: enabling exploration using knowledge graphs. In: proceedings of the 12th International Conference of Semantic Web Applications and Tools for Health Care and Life Science (SWAT4HCLS), 9-12 December, 2019, Edinburgh, Scotland.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data held in the form of draft and final ontologies produced by the Ontologies CoP are accessible online on public repositories managing versioning—mainly in GitHub repositories. Final versions of the ontologies are published with a cc-by license.
| Ontology | URL |
|---|---|
| Agronomy Ontology |
https://bigdata.cgiar.org/resources/agronomy-ontology/ http://obofoundry.org/ontology/agro |
| Crop Ontology | http://www.cropontology.org/ |
| Environment Ontology |
http://environmentontology.org/ http://obofoundry.org/ontology/envo |
| Plant Ontology |
http://browser.planteome.org/amigo http://obofoundry.org/ontology/po |
| Plant Experimental Conditions Ontology |
http://browser.planteome.org/amigo http://obofoundry.org/ontology/peco |
| Plant Trait Ontology |
http://browser.planteome.org/amigo http://obofoundry.org/ontology/to |
| Plant Stress Ontology | https://github.com/Planteome/plant-stress-ontology |
| Planteome | https://github.com/Planteome |
| SEOnt | https://github.com/AgriculturalSemantics/SEOnt |
Velarde, Orlee. Dissemination of Submergence-Tolerant Varieties and Associated New Production Practices to Southeast Asia.(2014),Data set version 3, Harvard Dataverse, https://doi.org/10.7910/DVN/26165.
The code of the cited tools is publicly accessible:
| AgroFIMS | https://github.com/AGROFIMS |
| AgroPortal | https://github.com/agroportal |
| BrAPI | https://github.com/plantbreeding/API |
| Crop Ontology website | https://github.com/bioversity/Crop-Ontology |
| GARDIAN | https://github.com/SCiO-systems/CGIAR-BDP-GARDIAN |
| COPO | https://github.com/collaborative-open-plant-omics |
| MIAPPE | https://github.com/MIAPPE |
| Ontology Lookup Service | https://github.com/EBISPOT/OLS |