Abstract
Matrix-assisted laser desorption/ionization mass spectrometry imaging (MALDI-MSI) is an established tool in drug development, which enables visualization of drugs and drug metabolites at spatial localizations in tissue sections from different organs. However, robust and accurate quantitation by MALDI-MSI still remains a challenge. We present a quantitative MALDI-MSI method using two instruments with different types of mass analyzers, i.e., time-of-flight (TOF) and Fourier transform ion cyclotron resonance (FTICR) MS, for mapping levels of the in vivo-administered drug citalopram, a selective serotonin reuptake inhibitor, in mouse brain tissue sections. Six different methods for applying calibration standards and an internal standard were evaluated. The optimized method was validated according to authorities’ guidelines and requirements, including selectivity, accuracy, precision, recovery, calibration curve, sensitivity, reproducibility, and stability parameters. We showed that applying a dilution series of calibration standards followed by a homogeneously applied, stable, isotopically labeled standard for normalization and a matrix on top of the tissue section yielded similar results to those from the reference method using liquid chromatography–tandem mass spectrometry (LC–MS/MS). The validation results were within specified limits and the brain concentrations for TOF MS (51.1 ± 4.4 pmol/mg) and FTICR MS (56.9 ± 6.0 pmol/mg) did not significantly differ from those of the cross-validated LC–MS/MS method (55.0 ± 4.9 pmol/mg). The effect of in vivo citalopram administration on the serotonin neurotransmitter system was studied in the hippocampus, a brain region that is the principal target of the serotonergic afferents along with the limbic system, and it was shown that serotonin was significantly increased (2-fold), but its metabolite 5-hydroxyindoleacetic acid was not. This study makes a substantial step toward establishing MALDI-MSI as a fully quantitative validated method.
Determining physiological distributions of drugs in tissues plays an important role in pharmaceutical research, toxicology, and medicine but is often difficult. Many imaging methods have limited capacity and are at best approximate. Hence, better methods for imaging drug distributions in tissues are needed to examine their targets and effects. Liquid chromatography–tandem mass spectrometry (LC–MS/MS) and ligand binding assays are widely applied for quantitative measurements of analytes in biological samples.1 However, during the sample preparation required for LC–MS/MS analyses, information about biomolecules and drug spatial distributions in tissues is inevitably lost. In contrast, quantitative whole-body autoradiography (QWBA) ligand-binding assays provide spatial information but have less molecular specificity than MS because both analytes and metabolites may contain the radioactive label. In addition, synthesis of the radioactive label is often costly and time-consuming.
Matrix-assisted laser desorption/ionization mass spectrometry imaging (MALDI-MSI)2 can overcome many of the obstacles of these methods while retaining chemical spatial information, high molecular specificity, and sensitivity. However, quantitation directly in a tissue section is challenging and not only depends on the limitations of the MSI instrument but also on the calibration and standardization protocol used. Ideally, the method should give a correct concentration value of the drug not only in the whole section of the imaged tissue but also in small, precise regions of interest (ROIs).
Different MSI strategies have been studied for the quantitative imaging of drugs directly in tissue sections.3 Three different methods have been exploited for applying calibration standards and internal standards: (i) the dilution series model,4 (ii) tissue extinction coefficient model,5 and (iii) mimetic tissue model.6 The dilution series model has been most commonly used because it is relatively straightforward and takes less time to prepare.3
Validation of bioanalytical methods has become an essential part of drug development and characterization. New analytical approaches must meet relevant authorities’ requirements to be widely accepted.7 Validated methods are crucial for robust pharmaceutical, nonclinical, and clinical pharmacology studies and for obtaining reliable data on drug safety and effectiveness. Such validation requires information on the methods’ selectivity, accuracy, precision, recovery, linearity, measurement range, and stability. In addition, results obtained with a new method should not significantly differ from those obtained by an established reference method, if one is available.8
Results from previously reported MALDI-MSI quantitative studies of drugs and their metabolites have been confirmed by comparative techniques such as LC–MS/MS4,5,9−12 and QWBA.5,13 However, none of these studies followed a full validation protocol but were performed using technical replicates of tissue dosed in vitro or reported limited validation parameters and procedures. Here, we report a comprehensive validation of a quantitative protocol for MALDI-MSI analysis of distributions of a drug in the brain using instruments with either time-of-flight (TOF) or Fourier transform ion cyclotron resonance (FTICR) mass analyzers. We used the dilution series model and evaluated different approaches for applying calibration standards and an internal standard. Citalopram, a lipophilic selective serotonin reuptake inhibitor that readily passes through the blood–brain barrier and widely distributes in the brain,14 was administered in vivo as a model substance to mice, and brain tissue was collected for analysis. The robustness of the analytical method was determined and evaluated according to general guidelines from the United States Food and Drug Administration (FDA), including cross-validation with LC–MS/MS.1 Furthermore, the effect of the administered drug on endogenous serotonin (5-HT) neurotransmitter system changes was investigated in the hippocampus region of the brain.
Experimental Section
Optimization of Calibration and Internal Standard Application
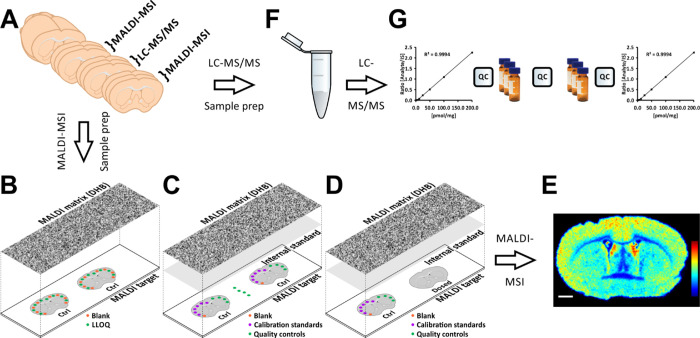
Before full validation of the MALDI-MSI method for the quantitative analysis of citalopram distributions and its application (using two types of MALDI-MSI instruments), we investigated the optimal method for applying calibration standards, an internal standard, and a matrix for MALDI-MSI. We also developed an LC–MS/MS method for the validation and quantitation of citalopram. These methodological developments are described in Note S1. The optimized protocol is illustrated in Figure 1.
Figure 1.
Validation, quantitation, and cross-validation of the MALDI-MSI methodology for quantitative analysis of distributions of the model substance citalopram. (A) Tissue sections were cut using a cryomicrotome at 14 μm thickness, and consecutive sections were collected for the different platforms. Sections collected for MALDI–MSI were thaw-mounted onto ITO-coated glass slides, while sections for LC–MS/MS were collected in microcentrifuge tubes. (B) Selectivity values were determined by measuring signal ratios between the lower limit of quantitation (LLOQ) and blank samples, deposited in 50 nL spots on control tissue and then coated with a uniform layer of MALDI matrix (2,5-dihydroxybenzoic acid, DHB) using an automated sprayer. (C) Accuracy, precision, and recovery values were obtained from measurements of 50 nL blank, calibration standard, and QC spots deposited on control tissue. QC samples were also spotted directly onto glass slides to measure recoveries of the analyte and internal standard (IS). For this, the MALDI target was coated with a uniform layer of IS followed by the MALDI matrix. (D) Arrangement of dosed and spotted control tissue for quantitation. Blank, calibration standard, and QC samples (50 nL) were deposited and subsequently coated with uniform layers of the IS and MALDI matrix. (E) Resulting ion intensity distributions of citalopram, normalized with respect to the IS, are presented using a rainbow scale. The scale bar is 1 mm. (F) Tissue cleanup protocol for LC–MS/MS. The tissue sections were homogenized, and the IS was added prior to liquid–liquid extraction and sample filtration. The sample was then evaporated and reconstituted before further analysis. (G) LC–MS/MS validation and quantitation were performed in a block design with every sample analyzed in triplicate, with QC samples between every block and standards for generating calibration curves at the start and end of the sequence.
Chemicals and Reagents
The following chemicals were used: LC–MS-grade methanol (MeOH), water, and acetonitrile (ACN) from Merck KGaA (Darmstadt, Germany); NaCl, acetic acid (HAc), formic acid (FA), and trifluoroacetic acid (TFA) from Sigma-Aldrich (Saint Louis, MO, USA); analysis-grade 2,5-dihydroxybenzoic acid (DHB) from Merck Schuchardt OHG (Hohenbrunn, Germany); and analysis-grade S-citalopram oxalate from Biotrend AG (Zürich, Switzerland). The internal standard for LC–MS/MS, (±)-citalopram-d4 hydrobromide (99% pure), was obtained from C/D/N Isotopes Inc. (Pointe-Claire, Quebec, Canada), whereas the internal standard for MALDI-MSI, (±)-citalopram-d6 oxalate (99% pure), was from ALSACHIM (Illkirch Graffenstaden, France). The reactive matrix 4-(anthracen-9-yl)-2-fluoro-1-methylpyridin-1-ium iodide (FMP-10) was synthesized and purified as previously reported.15
Preparation of Standard Solutions and the MALDI Matrix
A stock solution of 2 mM citalopram, used to prepare standards and quality control (QC) samples, was prepared by weighing citalopram on an analytical scale (Sartorius 1712, Sartorius AG, Göttingen, Germany) and dissolving it in 100% MeOH. Stock solutions of both internal standards, 2 mM citalopram-d4 and 2 mM citalopram-d6, were prepared in the same way. The stock solutions were stored at −20 °C until further use. To prepare standard solutions, a 50% b.w. (by weight) MeOH solution was prepared (relative density: 0.9156 g/mL at 20 °C)16 and added gravimetrically for high precision.
From the citalopram stock solution in 50% b.w. MeOH, we prepared six standards for MALDI-MSI quantitation and validation (at 2.5, 5, 7.5, 10, 15, and 20 μM), a 2 μM lower level of quantitation (LLOQ) solution, and three QC samples at 3.75, 8.75, and 17.5 μM. We also prepared a 5.0 μM internal standard solution for MALDI-MSI from the citalopram-d6 stock solution in 50% ACN, 0.2% TFA. A MALDI matrix was prepared from DHB (35 mg/mL) dissolved in 50% ACN, 0.2% TFA. The reactive matrix solution was prepared by dissolving 10 mg of FMP-10 in 5.5 mL of 70% ACN.
The following working stock solutions for LC–MS/MS were prepared from the citalopram and citalopram-d4 stock solutions, all with 100 μM citalopram-d4 in 100% MeOH: 1.6, 3.1, 12.5, 25, 50, 75, 100, 150, and 200 μM citalopram to generate a calibration curve; 0.8 μM LLOQ solution; and 2.3, 75, and 150 μM citalopram solutions for QC samples. We also prepared a 100 μM citalopram-d4 solution in 100% MeOH for extraction of brain tissues from animals administered citalopram in vivo.
Animals
Male mice (C57BL/6J, age 7–8 weeks, weight 23.8 ± 0.5 g, n = 9) were used in the studies. The animals were housed in air-conditioned rooms providing 12 h dark/12 h light cycles at 20 °C and 53% relative humidity with food ad libitum. The mice were administered S-citalopram oxalate (30 mg/kg, n = 6) or saline solution (n = 3) via intraperitoneal (i.p.) injection and euthanized by decapitation 30 min after injection. Following euthanasia, the brains were rapidly dissected out, frozen in dry ice-cooled isopentane, and stored at −80 °C until further use. Experiments were performed in accordance with European Communities Council Directives 86/609/EEC and 2010/63/EU on the ethical use of animals and were approved by the local ethical committee (Stockholm, approval no. N350/08 and N40/13).
Tissue Preparation
Coronal mouse brain sections (distance from bregma, 1.6 to −1.4 mm)17 were cut at a thickness of 14 μm in a cryomicrotome (CM1900, Leica Microsystems, Wetzlar, Germany) at a temperature of −20 °C according to a pre-defined cutting scheme. Three consecutive sections were collected in 1.5 mL microcentrifuge tubes for LC–MS/MS, the next three were thaw-mounted onto indium-tin-oxide (ITO) glass slides (Bruker Daltonics, Bremen, Germany) for MALDI-MSI18 (Figure 1A), and the procedure was repeated until we obtained the required number of samples. The weight of the tissue samples collected in the microcentrifuge tubes was measured on an analytical scale. The tissue sections (on ITO slides and in microcentrifuge tubes) were stored at −80 °C until further analysis. For validation of the MALDI-MSI method, ITO slides were prepared with six control tissue sections per slide, and for citalopram quantitation and neurotransmitter analysis by MALDI-MSI, slides were prepared with six differently dosed tissue sections and three control sections per slide. For the comparative study with LC–MS/MS, six microcentrifuge tubes were prepared for selectivity tests, four tubes for QC and LLOQ samples, nine tubes for calibration standards, and 3 × 6 tubes for dosed animal tissue extracts.
When preparing the tissue slides for either method validation or quantitation by MALDI-MSI, the slides were transferred on dry ice from −80 °C storage to a desiccator, where they were dried in vacuum for 30 min. Standard, QC, internal standard (citalopram-d6), and matrix solutions were freshly prepared. Then, the tissue slides were moved from the desiccator and mounted in a slide holder for application of calibration and QC standards by a CHIP-1000 chemical inkjet printer (Shimadzu, Tokyo, Japan) set to deposit 50 nL spots in 87 pL droplets. Measurements before validation and quantitation indicated that the true spot volume was 46.1 nL (Notes S1 and S2). Calibration standards were applied to the control tissues on the ITO slides from the lowest to highest citalopram concentration to avoid potentially significant carry-over effects of previous application of higher concentrations. Optical images of the slides were acquired at 3200 dpi using a flatbed scanner (Perfection V600 Photo, Seiko Epson, Nagano, Japan) prior to internal standard and MALDI matrix application. The internal standard was applied to all tissue sections except those used for the selectivity measurements, where internal standard must not be present,1 or those prepared with FMP-10 for neurotransmitter imaging. The internal standard was applied using an automatic spraying device (TM-Sprayer, HTX Technologies, LLC, Chapel Hill, NC, USA) in six passes at 80 °C, with 1100 mm/min nozzle speed, 2.0 mm track spacing, 70 mL/min flow rate, and an arbitrary flow rate of dry nitrogen at 6 psi pressure. DHB was applied as a MALDI matrix at 90 °C rather than 80 °C, but otherwise with identical settings. FMP-10 was applied in 20 passes at 80 °C with a flow rate of 80 μL/min, while all other parameters remained the same.
For the LC–MS/MS method validation and quantitation, the working stock solutions (for calibration standards, QC samples, and extraction of brain tissues from animals administered citalopram in vivo) were diluted 50-fold in 1% HAc solution and cooled on ice. The ice-cooled diluted working stock solutions were then added to the tissue samples at 25 μL/mg tissue. For samples containing dosed tissue, a diluted working stock solution of citalopram-d4 in 1% HAc was used. Ice-cooled ACN was then added at 50 μL/mg tissue followed by 10 mg NaCl/mg tissue to separate the sample into an aqueous and organic (ACN) phase. The tissue in each tube was homogenized using an ultrasonic probe (Sonics Vibra Cell, VC 130, Sonics & Materials Inc., Newtown, CT, USA). The two liquid phases were fully separated by centrifugation using a 5804R centrifuge (Eppendorf AG, Hamburg, Germany) at 14,000 relative centrifugation force (rcf) for 20 min at 4 °C. The organic (ACN) phase containing citalopram and citalopram-d4 was transferred to a second microcentrifuge tube. Residual analyte in the first tube was extracted by twice adding 200 μL portions of ACN, vortex-mixing using a Vortex-2 Genie (Scientific Industries Inc., Bohemia, NY, USA) for 1 min, centrifuging for 2 min in a Mini Centrifuge (Labnet International Inc., Woodbridge, NJ, USA), and transferring the resulting organic ACN phases to the second microcentrifuge tube. The pooled organic phase in the second microcentrifuge tube was evaporated to dryness in a Concentrator 5301 centrifugal evaporator (Eppendorf AG, Hamburg, Germany) at ambient temperature. The sample was reconstituted by adding 25 μL/mg tissue of 10% ACN and 0.25% HAc, vortex-mixing for 1 min, and then transferring the resulting mixture to a 10 kDa molecular-weight cut-off centrifugal filter (Micron Ultracel YM-10, Millipore Corp., Bedford, MA, USA) with a filter holder. After rinsing the second tube by adding a further 25 μL/mg tissue of 0.25% HAc, vortex-mixing, and transferring the rinsate to the centrifugal filter, the reconstituted sample was filtered by centrifugation at 14,000 rcf for 60–90 min at 4 °C. The filtrate was then stored in a freezer at −20 °C until further analysis. The sample preparation protocol for LC–MS/MS is described in Note S3.
MALDI Mass Spectrometry Imaging
MALDI-MSI analyses of the validation and quantitation tests as well as neurotransmitter analyses were performed in the positive ionization mode using a MALDI-TOF/TOF (ultrafleXtreme, Bruker Daltonics) or MALDI-FTICR (solariX XR 7 T-2ω, Bruker Daltonics) mass spectrometer. Both instruments were equipped with a Nd:YAG (Smartbeam-II, Bruker Daltonics) 2 kHz laser with laser spot size optimized for 70 μm spatial resolution analyses. The laser power was optimized at the start of each analysis and then held constant during each MALDI-MSI experiment. The methods were externally calibrated using red phosphorus and internally calibrated using the DHB molecular ion (m/z 155.0339) or FMP-10 cluster ion (m/z 555.2231) signals as lock masses. Tissue sections were analyzed in a random order to minimize possible bias due to factors such as matrix degradation or loss of sensitivity over time. The msIQuant 2.0.1.15 software package developed in-house19 was used for data processing, normalization, and quantitation. Regions of interest (ROIs) were manually defined in the analytical software using both optical images and MS imaging data.19 The concentrations of citalopram in defined ROIs were calculated, following normalization against the internal standard (citalopram-d6), from a standard calibration curve derived from measurements of known concentrations of citalopram applied on control tissue sections. For FTICR-MS analysis of derivatized neurotransmitters, spectra were normalized against the root mean square (RMS) of all data points. The brain density was set to 1.03 g/cm3.20
Liquid Chromatography Mass Spectrometry
In the liquid chromatography tandem MS (LC–MS/MS) validation and quantitation tests, a EASY-nLC pump (Thermo-Scientific, San Jose, CA, USA) set to the single column mode was used. The LC column was a 100 mm long silica column with an Integra Frit (New Objective, Woburn, MA, USA), 100 μm ID, packed in-house with 3 μm ACE C18-AR reverse-phase particles (Advanced Chromatography Technologies Ltd., Aberdeen, UK). After injecting 1 μL samples, analytes were eluted with a 20 min 2–100% ACN gradient and constant 0.1% FA (see Note S4 for detailed LC settings). They were then quantified by MS/MS using a linear ion trap mass spectrometer (LTQ, Thermo-Scientific, San Jose, California, USA) equipped with a nano-electrospray ionization interface and a Picotip fused silica emitter (Waters Corp., Milford, MA, USA). To maximize sensitivity, citalopram and citalopram-d4 were determined in the multiple reaction monitoring (MRM) mode in separate events using m/z 325.2 and 329.2 precursor ions, respectively. The three main product ions for citalopram (m/z 307.2, 280.1, and 262.1) and main analogue product ions for the internal standard citalopram-d4 (m/z 311.2, 284.1, and 266.1) were used for the quantitative measurements (see Note S4 for detailed MS/MS settings). Chemical structures of the precursor and product ions are illustrated in Figure S1.
Validation Procedure
The methodology applied for analyzing citalopram in mouse brain tissue sections was validated following the FDA guideline.1
Selectivity
Blank samples for MALDI-MSI validation were obtained by spotting blank solution on control tissues and covering the ITO slide with an even layer of matrix. To calculate selectivity values, LLOQ spots were also applied to the control tissues (Figure 1B). For LC–MS/MS blank samples, control samples that had not been spiked with analyte or internal standard were used.
Accuracy and Precision
Samples for accuracy and precision assessments were prepared by spiking control tissue with both the analyte and internal standard. For MALDI-MSI, citalopram was spotted at the LLOQ or three QC concentrations on control tissue before application of internal standard, whereas QC and LLOQ samples for LC–MS/MS were prepared by spiking control tissue with analyte and internal standard during sample preparation. Sets of six replicates were used to assess the methods’ accuracy and precision. Accuracy and precision are respectively defined as the deviation of the mean value from the true concentration and the coefficient of variation (CV), or RSD, of measured concentrations.1 The accuracy and precision thresholds for a fully validated method are relative standard deviations (RSDs) ≤ 15% at three QC concentrations and ≤ 20% at LLOQ.1
Recovery
For MALDI-MSI, recovery values were determined by comparing detector signals from QC spots applied directly onto the MALDI target to signals from QC spots on control tissue. For MALDI-TOF-MSI, the detector signal was normalized with respect to the total ion current (TIC), whereas for MALDI-FTICR-MSI, the detector signal was RMS-normalized.21 To determine LC–MS/MS recoveries, true concentrations of the analyte were prepared without any tissue homogenate. A hypothetical tissue mass of 5 mg was used to obtain adequate amounts of analyte and internal standard.
Stability
The stability of citalopram in the brain tissues under the −80 °C storage conditions was determined by quantitation of dosed animals on two separate occasions by MALDI-MSI. Tissue sections were cut and thaw-mounted on ITO-slides, as described in the sample preparation section, stored at −80 °C, and then analyzed 3 and 13 months later. The samples were considered stable if there was no significant difference between the amounts obtained on the two occasions.
Quantitation Procedure
In MALDI-MSI, the quantitation was performed on glass slides containing six dosed and three control tissues. On the control tissues, calibration curves, QCs, and LLOQ were applied. Subsequently, the internal standard and MALDI matrix were applied as previously described. For quantitation by LC–MS/MS, calibration standards were measured to generate standard curves before and after the analyses of homogenized tissue samples. The samples were analyzed in a randomized manner in blocks. Between the sample blocks, QCs were analyzed to monitor any drift in signal or quality of the quantitation procedure. For LC–MS/MS, each extract, standard, and QC sample were measured in duplicate, with a blank sample run between each case to avoid carry-over effects. Between every block, several blanks were run to clean the LC–MS/MS system.
Cross-validation
The LC–MS/MS method was treated as the reference bioanalytical method, whereas the investigated MALDI-MSI methods were considered comparator bioanalytical methods. To detect possible significant differences in quantitation results between the reference and comparator methods, unpaired two-tailed t tests with unequal variances were used.
Effect of Citalopram on the Hippocampal Serotonergic System
Tissue sections from citalopram administered animals and controls (distance from bregma, −5.6 mm) were coated with the derivatizing matrix FMP-10.15 MSI analysis was performed in the positive ionization mode using the FTICR-MS instrument. The acquired data were imported into msIQuant19 and relative intensity values of 5-HT, and its monoamine decarboxylated metabolite 5-hydroxyindoleacetic acid (5-HIAA) were extracted from the hippocampal area as RMS-normalized maximum intensity values. Student’s t test was performed to evaluate the statistical significance of the results.
Results
Optimization of Calibration and Internal Standard Application
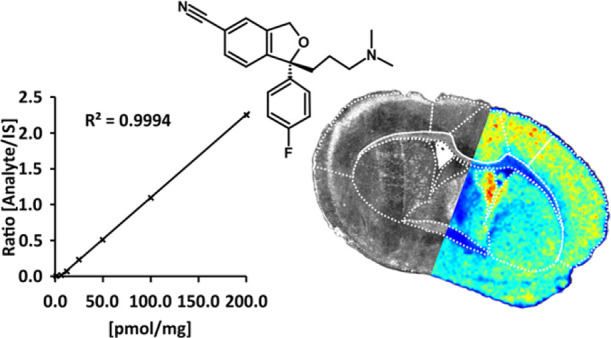
We compared six different methods to apply standard calibration spots and internal standard and compared the citalopram concentrations with results of LC–MS/MS analysis (see Note S1 and Figure S2–S4). Our approach showed that applying a dilution series of calibration standards, a homogeneously applied, stable, isotopically labeled standard for normalization, and subsequently a matrix on top of the tissue section showed similar results to the reference method using liquid chromatography–tandem mass spectrometry (LC–MS/MS).
Validation
Selectivity
When examining the matrix effects and selectivity of the MALDI-MSI methodology, no interfering peaks for either citalopram (m/z 325.171) or the deuterated internal standard citalopram-d6 (m/z 331.209) were found in signals from control (blank) tissue sections (Figure 1B, Figure S5). In addition, the MALDI-MSI methodology (with both instrument types) provided at least 10 times stronger responses for samples containing the analytes at the lower limit of quantitation (LLOQ) than for blank samples (Table 1), easily satisfying the general 5-fold selectivity criterion.1 Similarly, examination of the matrix effects and selectivity for the LC–MS/MS method showed no interfering peaks for citalopram or the internal standard (deuterated citalopram-d4) (m/z 329.20) (Table 1, Figure S6).
Table 1. Results of the MALDI-TOF-MSI, MALDI-FTICR-MSI Method Validation Using LC–MS/MS as the Reference Method and Citalopram as a Model Compounda.
| MALDI-TOF-MSI |
MALDI-FTICR-MSI |
||||||
|---|---|---|---|---|---|---|---|
| parameters | slide V1 | slide V2 | slide V3 | slide V4 | slide V5 | slide V6 | LC–MS/MS |
| Selectivity | |||||||
| ratio (LLOQ/blank sample) (≥5) | 11.1 | 33.0 | 5.6 | ||||
| Accuracy (n = 6) | |||||||
| LLOQ (≤ ±20%) [%] | –5.1 | 17.0 | 6.3 | –14.2 | 13.0 | –2.4 | –8.7 |
| LQC (≤ ±15%) [%] | 4.0 | –0.7 | –8.9 | –6.3 | 13.6 | 5.7 | –2.1 |
| MQC (≤ ±15%) [%] | 5.9 | –1.1 | 4.5 | 6.1 | –12.0 | 5.7 | 1.4 |
| HQC (≤ ±15%) [%] | 5.6 | –2.2 | 14.9 | 3.0 | 9.2 | 5.7 | 7.2 |
| Precision (n = 6) | |||||||
| LLOQ (≤20%) [%] | 14.8 | 10.1 | 9.9 | 13.1 | 16.4 | 3.8 | 10.8 |
| LQC (≤15%) [%] | 10.0 | 6.9 | 14.5 | 9.8 | 10.2 | 8.5 | 1.6 |
| MQC (≤15%) [%] | 10.6 | 11.0 | 7.3 | 12.6 | 10.8 | 9.6 | 2.3 |
| HQC (≤15%) [%] | 14.2 | 10.8 | 13.8 | 5.9 | 10.6 | 12.4 | 2.6 |
| Coefficient of determination | |||||||
| R2 | 0.990 | 0.977 | 0.984 | 0.996 | 0.989 | 0.998 | 0.999 |
| Recovery (analyte) | |||||||
| LQC [%] | 20 | 23 | 17 | 32 | 28 | 31 | 62 |
| MQC [%] | 35 | 32 | 26 | 34 | 39 | 38 | 82 |
| HQC [%] | 45 | 40 | 44 | 42 | 55 | 56 | 88 |
| Recovery (internal standard) | |||||||
| LQC [%] | 33 | 49 | 40 | 37 | 34 | 31 | 60 |
| MQC [%] | 33 | 47 | 43 | 36 | 45 | 34 | 87 |
| HQC [%] | 43 | 49 | 50 | 42 | 49 | 37 | 89 |
| MALDI-TOF-MSI | MALDI-FTICR-MSI | LC–MS/MS | |
|---|---|---|---|
| quantitation | (n = 3) | (n = 3) | (n = 6) |
| animal D1 [pmol/mg] | 54.1 ± 8.1, p = 0.739 | 60.7 ± 4.8, p = 0.218 | 55.9 ± 3.3 |
| animal D2 [pmol/mg] | 50.8 ± 6.8, p = 0.156 | 57.8 ± 5.2, p = 0.635 | 59.5 ± 0.8 |
| animal D3 [pmol/mg] | 53.2 ± 4.3, p = 0.177 | 59.2 ± 1.6, p = 0.409 | 58.3 ± 1.1 |
| animal D4 [pmol/mg] | 51.5 ± 7.3, p = 0.593 | 59.6 ± 5.4, p = 0.221 | 54.1 ± 0.7 |
| animal D5 [pmol/mg] | 52.9 ± 5.1, p = 0.372 | 56.1 ± 8.3, p = 0.971 | 56.3 ± 1.1 |
| animal D6 [pmol/mg] | 42.4 ± 4.6, p = 0.346 | 45.0 ± 2.3, p = 0.695 | 45.7 ± 1.1 |
| average concentration [pmol/mg] | 51.1 ± 4.4, p = 0.153 | 56.9 ± 6.0, p = 0.648 | 55.0 ± 4.9 |
Samples on three slides (whole brain) were analyzed in each MALDI-MSI mode, and extracts of corresponding samples were measured by LC–MS/MS. Selectivity, accuracy, precision, coefficient of determination (linearity), and recovery results are presented. The values in parentheses are thresholds for a valid method specified by the FDA.1 All the selectivity, accuracy, and precision results are within the limits. The coefficients of determination show good linearity, and both analyte and internal standard recoveries are reported, but no limits are defined. Results of citalopram quantitation in six dosed animals by MALDI-TOF-MSI, MALDI-FTICR-MSI, and LC–MS/MS are shown in terms of means ± SD. The p values were obtained by comparing the quantitation results obtained using the two MALDI-MSI modes with the LC–MS/MS results. LQC, MQC, and HQC refer to the quality control standards with the lowest, medium, and highest concentrations of citalopram, respectively.
Accuracy and Precision
For QC assessments of the methods’ precision, accuracy, and stability of the prepared samples, the analyte was applied at the LLOQ or three QC concentrations (low: LQC, medium: MQC, and high: HQC) to brain tissue sections from animals that had not received citalopram analyzed at approximately 1 week intervals. The results obtained using both MALDI-MSI instruments and LC–MS/MS met the acceptance criteria (Table 1).
Recovery
For our MALDI-MSI methodology, we defined recovery as the ratio of detector response to citalopram spotted on a tissue at three QC concentrations compared with the same amounts spotted directly on a MALDI glass slide (Figure 1C). Recovery of the internal standard was also measured to confirm consistency. The results showed that the two MALDI-MSI instruments provided similar recoveries, but only about half of the recoveries obtained by LC−MS/MS.
Stability
To assess the stability of the results, citalopram was quantified in coronal tissue sections from citalopram-administered mice twice at a 13 month interval. No significant difference was detected between the quantitative results at the two time points (Table S4).
Quantitation
For quantitation by MALDI-MSI, calibration curves with good linearity (Table 1) were obtained by applying six calibration standards and a zero blank directly onto control tissue sections prior to internal standard and matrix application. The calculated citalopram concentration in the tissue from six animals dosed did not differ significantly from each other according to the MALDI-TOF-MSI, MALDI-FTICR-MSI, and LC–MS/MS analyses (Table 1). The accuracy and precision were also assessed to gauge the robustness of the measurements. Both MALDI-FTICR-MSI and LC–MS/MS measurements generally met the relevant criteria,1 but there were minor deviations from the recommended accuracy and precision limits in the MALDI-TOF-MSI results (Table S1). Generally, samples can be analyzed by a single determination without duplicate or replicate analysis if the method has acceptable variability, as defined by validation data, for analyses under conditions where precision and accuracy are within acceptable limits. Despite the minor deviations in MALDI-TOF-MSI quantifications detected in the QC test, no significant effects on the quantitation results were observed. Moreover, no significant differences were detected between results obtained with the two MALDI-MSI methods and the LC–MS/MS method reference method in two comparative tests, at either the individual animal level or for obtained average concentrations (Table 1 and Tables S2 and S3).
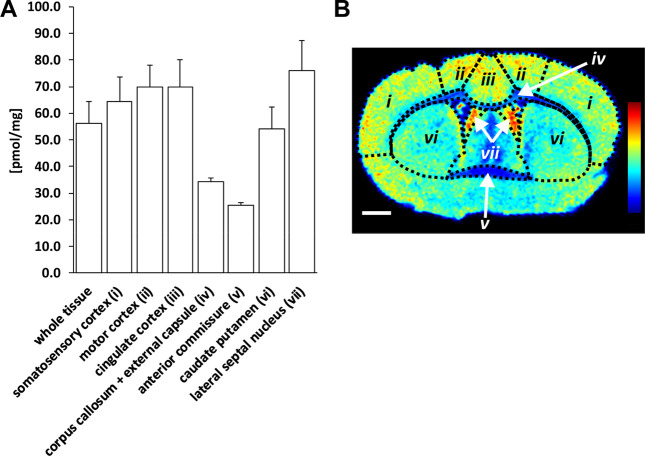
Citalopram was also quantified in selected brain regions (Figure 2). The grey matter brain regions, i.e., lateral septal nucleus, cingulate, motor and somatosensory cortex, and caudate-putamen, showed higher concentrations of citalopram (54–76 pmol/mg) compared to the white matter regions, i.e., corpus callosum, external capsule and anterior commissure (25–34 pmol/mg).
Figure 2.
Quantitation of citalopram in selected brain regions. (A) Bar chart showing average citalopram concentrations detected in coronal brain tissue sections (n = 3) of one dosed animal (designated D5). Concentrations in pmol/mg (mean ± SD) in whole tissue sections, (i) somatosensory cortex, (ii) motor cortex, (iii) cingulate cortex, (iv) corpus callosum and external capsule, (v) anterior commissure, (vi) caudate putamen, and (vii) lateral septal nucleus were 56.1 ± 8.3, 64.3 ± 9.3, 70.0 ± 8.0, 69.8 ± 10.3, 34.2 ± 1.6, 25.3 ± 1.0, 54.2 ± 8.3, and 76.2 ± 11.0, respectively. (B) Ion image of coronal brain tissue section showing the seven brain regions of interest, with heat map displaying citalopram distribution in ion intensities from lowest (blue) to highest (red), normalized with respect to the internal standard. Scale bar is 1 mm and coronal level is 0.20 mm (distance from bregma).
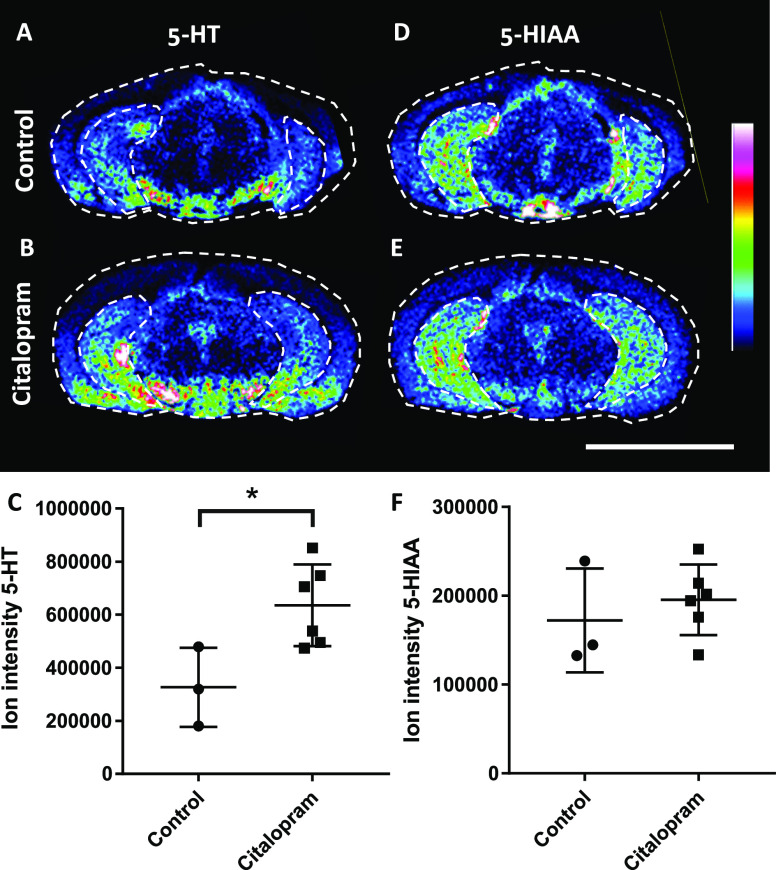
Serotonin Is Enhanced in the Hippocampus by Citalopram Administration
The effect of citalopram administration on the 5-HT system was investigated in the hippocampus, a principal target of the serotonergic afferents along with the limbic system.22,23The effect of citalopram in the hippocampus caused a 2-fold increase of 5-HT in the hippocampus (p < 0.05), whereas levels of 5-HIAA, the main metabolite of 5-HT remained unchanged (Figure 3). The citalopram concentration in the hippocampus was 63.0 ± 3.3 pmol/mg (±SD).
Figure 3.
Ion images of 5-HT and 5-HIAA in coronal brain tissue sections and quantitation in the hippocampus from citalopram administered animals and controls. Ion distribution images of 5-HT (m/z 444.21) in the (A) control and (B) citalopram-dosed tissue samples. The range of the color ion intensity scale is 0–50%. (C) Relative intensities of 5-HT in the hippocampus of controls (n = 3) and citalopram administered animals (n = 6). Ion distribution image of 5-HIAA (m/z 459.17) in (D) control tissue and (E) citalopram-dosed tissue. The range of the color ion intensity scale is 0–40%. (F) Relative intensities of 5-HIAA in the hippocampus of controls (n = 3) and citalopram-administered animals (n = 6). Data were normalized by the root-mean-square of all data points. Scale bar is 5 mm and coronal level is −5.6 mm (distance from bregma). *p < 0.05.
Discussion
It is generally accepted that a bioanalytical method must first be validated before it can be implemented for routine use in, e.g., pharmaceutical drug development, and authorities of various jurisdictions have published guidelines for such validation (for overview, see ref (24)). Generally, validated methods must include experiments on specificity, linearity, accuracy, precision, detection limit, quantitation limit, and robustness. Although performing thorough method validation can be a lengthy and tedious process, the quality of data generated with the validated method directly correlates to the standard of this process.25 In addition, a Good Laboratory Practices-validated bioanalytical method is needed to support all analytical studies when submitting new drug applications to the regulatory authorities.26
A conventional benchmark for quantitative MSI is comparison with LC–MS/MS of dissected tissue or serial tissue sections.4,6,11,27,28 However, no previous reports have performed a fully validated MSI study. Our study, following the FDA guideline,1 provides the first full validation of a MALDI-MSI method for quantifying an in vivo-administered drug, including cross-validation by LC–MS/MS, which is well established and widely used for quantitative analyses in drug development and characterization. Using citalopram as a model substance and the brain as a model tissue, the study provides a robust proof-of-concept for MALDI-MSI as a quantitative bioanalytical tool.
There are many aspects to consider when conducting a quantitative MSI study, such as how the molecule of interest is extracted, ionized, and detected from the complex matrix, e.g., tissue section (see ref (3) for review). A standard curve and internal standard must be applied in reasonably corresponding matrices for accurate quantitation. As mentioned above, three different experimental approaches have previously been proposed to apply the calibration standards.
In the dilution series approach, several dilutions of a standard solution are applied to tissue sections, using an on- or under-tissue strategy and a control section as a background matrix.4 In the tissue extinction coefficient approach, a control section is sprayed with a standard solution of the target analyte and the average intensity of the analyte signal from a selected tissue area is divided by the averaged intensity from the glass slide. A calibration curve is prepared by deposition of calibration solutions onto sample targets or glass slides next to the investigated tissue section, and the appropriate correction factor is applied for the regions of interest.5 In the mimetic tissue approach, tissue homogenates are spiked with standards to correct for variations in both ionization and extraction efficiency.6
The dilution series approach is relatively easy, offers several advantages, and has therefore been most commonly used.3 For example, its preparation and handling procedures are much less time-consuming, particularly when different types of tissues are being imaged. It is possible to apply calibration curves directly to specific regions (or subregions) of tissue organs with specific chemical and morphological characteristics (e.g., brain regions), preferably by using an automatic robotic spotter that can deliver droplets in the picoliter range. Using a homogeneously coated, stable, isotope-labeled internal standard, differences in tissue characteristics can be accounted for.19 There is no need to confirm that the compound of interest is properly merged or undergoes metabolism in a homogenate, as required in the mimetic tissue model.3 A direct comparison of the two methods has been reported has been reported and showed that there were no significant differences between them.11,29
Owing to the benefits described above, we used the dilution series model in the present study and investigated and optimized a protocol for applying calibration and internal standards. Numerous protocols for this have been described for the dilution series model. Commonly, the internal standard is added to the MALDI matrix solution and sprayed over the entire tissue section. The internal standard solution can also be deposited separately from the MALDI matrix, either beneath or on top of the tissue section, followed by matrix deposition.3,30 Of the six different methods tested in the present study, we found that applying calibration standard spots onto the tissue section prior to internal standard application was the procedure that correlated best to the LC–MS/MS analysis. The other methods tested were significantly different from the LC–MS/MS method and resulted in lower concentrations of citalopram. Our results are in agreement with Caprioli and coworkers, who also compared different internal standard deposition techniques and their influence on the quality (accuracy and precision) of quantitation.9 Notably, similar to our results, only application of internal standard on top of the tissue section prior to matrix application produced results that conformed to the LC–MS/MS analysis of the tissue extract.9
The brain is one of the most complex organs in the body, consisting of neurons of different types, and its use in the development and validation of robust bioanalytical methods can be challenging because of its heterogeneity. However, it may be crucial to map the distribution of drugs, e.g., citalopram, within it to check that they reach their targets and exert their desired effect. MALDI-MSI technology has major advantages in this respect because it can provide high-resolution lateral information of drugs, unlike most conventional quantitative analytical techniques. In addition, citalopram, an effective antidepressant drug, is a good model compound as it shows high levels of bioavailability and high rates of blood–brain barrier transport.31
Therefore, to confirm MALDI-MSI’s ability to quantify a drug in selected brain regions, citalopram concentrations were investigated. The results showed that the citalopram distribution correlated with serotonin transporter distribution32 and concentrations were the highest in the lateral septal nucleus and cortex regions, such as the cingulate and motor cortex. The lowest concentrations were found in the lipid-rich areas corpus callosum and anterior commissure.
In addition to measuring the concentration of citalopram in different brain regions, we studied its effect on the hippocampus of the brain, a principal target of the serotonergic afferents along with the limbic system.22 The hippocampus plays an important role in emotional and cognitive processing, and there is extensive evidence that depression and other stress-related conditions are associated with hippocampal dysfunction.33 It is densely innervated by serotonergic fibers, and most of the 5-HT receptor subtypes are expressed there.34 We imaged and measured serotonin and its metabolite 5-HIAA in the hippocampus and found that compared to untreated controls, serotonin levels increased, whereas 5-HIAA levels were unchanged (i.e., citalopram decreased the 5-HT turnover ratio). The effects of acute citalopram in the present study are in accordance with several previous studies demonstrating an increase in extracellular 5-HT following injections with various selective serotonin re-uptake inhibitors.35
In summary, we present fully validated methods for quantitative analyses of citalopram in mouse brain tissue using MALDI-MSI instruments with either a TOF or FTICR mass analyzer. The MALDI-MSI methods met the specified criteria, and no significant differences between results obtained with them and the LC–MS/MS reference method (used for cross-validation) were detected. Development of a robust and validated quantitative MSI method for regional determination of neuroactive drugs in the brain facilitates the study of their effect on neurotransmitter levels.
Acknowledgments
This work was supported by the Swedish Research Council (Medicine and Health, 2018–03320; Natural and Engineering Science 2018–05501), the Swedish Brain Foundation (FO2018-0292), the Swedish Foundation for Strategic Research (RIF14-0078), ARIADME (European FP7 ITN Community’s Seventh Framework Program, 607517), and Science for Life Laboratory (SciLifeLab).
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.analchem.0c03203.
Note S1: optimization of method for applying calibration standards, internal standard, and matrix for MALDI-MSI analysis of citalopram in mouse brain tissue sections; Note S2: determination of spotter volume by LC–MS/MS measurement; Note S3: LC–MS/MS sample preparation of standards, QC samples, and dosed tissue for citalopram quantitation; Note S4: LC and MS/MS settings; Figure S1: chemical structures of citalopram and internal standards (citalopram-d4 and citalopram-d6) used for method validation; Figure S2: investigation of optimal protocols for applying calibration and internal standards; Figure S3: citalopram concentrations determined by MALDI-MSI using the indicated application protocols and reference LC–MS/MS method; Figure S4: MALDI-MS images of citalopram distributions in brain sections following the indicated methods for applying the internal standard; Figure S5: investigation of citalopram and IS (citalopram-d6) MSI peak interference; Figure S6: ion chromatogram from LC–MS/MS analysis showing results of the selectivity test (blank sample vs citalopram at the lowest level of quantitation, LLOQ); Table S1: accuracy, precision, and coefficient of determination of citalopram measurements obtained by MALDI-MSI and LC–MS/MS analysis as part of quality control (QC) validation; Table S2: cross-validation of MALDI-TOF-MSI, MALDI-FTICR-MSI, and LC–MS/MS quantitation; Table S3: cross-validation of MALDI-TOF-MSI, MALDI-FTICR-MSI, and LC–MS/MS analyses of biological variance; Table S4: results of tests of citalopram’s long-term stability in brain tissue sections (PDF)
Data file S1: Method validation (ZIP)
Data file S2: Quantitation of citalopram (ZIP)
Data file S3: Calibration of chemical inkjet printer (XLSX)
Author Contributions
P.K. designed and performed experiments and analyzed data; T.V. performed experiments and statistical analysis; A.N. and R.S. designed and performed experiments; N.S. and M.P. performed animal experiments; F.B. performed experiments; H.W. performed experiments and analyzed data; P.S. provided tissue samples and supervised animal experiments; P.E.A. designed experiments, interpreted data, and supervised the project. P.K., T.V., A.N., R.S., and P.E.A. also wrote the manuscript; and all authors contributed to the editing of the final manuscript. P.E.A. is a principal investigator for the grants that funded this research.
The authors declare no competing financial interest.
Supplementary Material
References
- United States Food Drug Administration: Center for Drug Evaluation and Research; Center for Veterinary Medicine, 2001. [Google Scholar]
- Caprioli R. M.; Farmer T. B.; Gile J. Molecular imaging of biological samples: localization of peptides and proteins using MALDI-TOF MS. Anal. Chem. 1997, 69, 4751–4760. 10.1021/ac970888i. [DOI] [PubMed] [Google Scholar]
- Rzagalinski I.; Volmer D. A. Quantification of low molecular weight compounds by MALDI imaging mass spectrometry - A tutorial review. Biochim. Biophys. Acta Proteins Proteom. 2017, 1865, 726–739. 10.1016/j.bbapap.2016.12.011. [DOI] [PubMed] [Google Scholar]
- Nilsson A.; Fehniger T. E.; Gustavsson L.; Andersson M.; Kenne K.; Marko-Varga G.; Andrén P. E. Fine mapping the spatial distribution and concentration of unlabeled drugs within tissue micro-compartments using imaging mass spectrometry. PLoS One 2010, 5, e11411 10.1371/journal.pone.0011411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamm G.; Bonnel D.; Legouffe R.; Pamelard F.; Delbos J. M.; Bouzom F.; Stauber J. Quantitative mass spectrometry imaging of propranolol and olanzapine using tissue extinction calculation as normalization factor. J. Proteomics 2012, 75, 4952–4961. 10.1016/j.jprot.2012.07.035. [DOI] [PubMed] [Google Scholar]
- Groseclose M. R.; Castellino S. A mimetic tissue model for the quantification of drug distributions by MALDI imaging mass spectrometry. Anal. Chem. 2013, 85, 10099–10106. 10.1021/ac400892z. [DOI] [PubMed] [Google Scholar]
- Peris-Vicente J.; Esteve-Romero J.; Carda-Broch S., In Analytical Separation Science, Vol. 5; American Cancer Society: New York, N.Y., 2015. pp. 1757−1808.
- Shah V. P.; Midha K. K.; Findlay J. W. A.; Hill H. M.; Hulse J. D.; McGilveray I. J.; McKay G.; Miller K. J.; Patnaik R. N.; Powell M. L.; Tonelli A.; Viswanathan C. T.; Yacobi A. Bioanalytical method validation--a revisit with a decade of progress. Pharm. Res. 2000, 17, 1551–1557. 10.1023/A:1007669411738. [DOI] [PubMed] [Google Scholar]
- Chumbley C. W.; Reyzer M. L.; Allen J. L.; Marriner G. A.; Via L. E.; Barry C. E. III; Caprioli R. M. Absolute Quantitative MALDI Imaging Mass Spectrometry: A Case of Rifampicin in Liver Tissues. Anal. Chem. 2016, 88, 2392–2398. 10.1021/acs.analchem.5b04409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prentice B. M.; Chumbley C. W.; Caprioli R. M. Absolute Quantification of Rifampicin by MALDI Imaging Mass Spectrometry Using Multiple TOF/TOF Events in a Single Laser Shot. J. Am. Soc. Mass Spectrom. 2017, 28, 136–144. 10.1007/s13361-016-1501-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barry J. A.; Ait-Belkacem R.; Hardesty W. M.; Benakli L.; Andonian C.; Licea-Perez H.; Stauber J.; Castellino S. Multicenter Validation Study of Quantitative Imaging Mass Spectrometry. Anal. Chem. 2019, 91, 6266–6274. 10.1021/acs.analchem.9b01016. [DOI] [PubMed] [Google Scholar]
- Pirman D. A.; Reich R. F.; Kiss A.; Heeren R. M.; Yost R. A. Quantitative MALDI tandem mass spectrometric imaging of cocaine from brain tissue with a deuterated internal standard. Anal. Chem. 2013, 85, 1081–1089. 10.1021/ac302960j. [DOI] [PubMed] [Google Scholar]
- Takai N.; Tanaka Y.; Inazawa K.; Saji H. Quantitative analysis of pharmaceutical drug distribution in multiple organs by imaging mass spectrometry. Rapid Commun. Mass Spectrom. 2012, 26, 1549–1556. 10.1002/rcm.6256. [DOI] [PubMed] [Google Scholar]
- Bezchlibnyk-Butler K.; Aleksic I.; Kennedy S. H. Citalopram--a review of pharmacological and clinical effects. J. Psychiatry Neurosci. 2000, 25, 241–254. [PMC free article] [PubMed] [Google Scholar]
- Shariatgorji M.; Nilsson A.; Fridjonsdottir E.; Vallianatou T.; Källback P.; Katan L.; Sävmarker J.; Mantas I.; Zhang X.; Bezard E.; Svenningsson P.; Odell L. R.; Andrén P. E. Comprehensive mapping of neurotransmitter networks by MALDI-MS imaging. Nat. Methods 2019, 16, 1021–1028. 10.1038/s41592-019-0551-3. [DOI] [PubMed] [Google Scholar]
- Wolf A. V.; G B. M.; Prentice P. G. In CRC Handbook of Chemistry and Physics ,65th Edition; CRC Press, Inc, 1984, p. D-241. [Google Scholar]
- Paxinos G.; Franklin K. B. J.. Paxinos and Franklin’s the mouse brain in stereotaxic coordinates, 4th ed.; Elsevier Academic Press: Amsterdam, 2013. [Google Scholar]
- Koeniger S. L.; Talaty N.; Luo Y.; Ready D.; Voorbach M.; Seifert T.; Cepa S.; Fagerland J. A.; Bouska J.; Buck W.; Johnson R. W.; Spanton S. A quantitation method for mass spectrometry imaging. Rapid Commun. Mass Spectrom. 2011, 25, 503–510. 10.1002/rcm.4891. [DOI] [PubMed] [Google Scholar]
- Källback P.; Nilsson A.; Shariatgorji M.; Andrén P. E. msIQuant--Quantitation Software for Mass Spectrometry Imaging Enabling Fast Access, Visualization, and Analysis of Large Data Sets. Anal. Chem. 2016, 88, 4346–4353. 10.1021/acs.analchem.5b04603. [DOI] [PubMed] [Google Scholar]
- Barber T. W.; Brockway J. A.; Higgins L. S. The density of tissues in and about the head. Acta Neurol. Scand. 1970, 46, 85–92. 10.1111/j.1600-0404.1970.tb05606.x. [DOI] [PubMed] [Google Scholar]
- Deininger S. O.; Cornett D. S.; Paape R.; Becker M.; Pineau C.; Rauser S.; Walch A.; Wolski E. Normalization in MALDI-TOF imaging datasets of proteins: practical considerations. Anal. Bioanal. Chem. 2011, 401, 167–181. 10.1007/s00216-011-4929-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hensler J. G. Serotonergic modulation of the limbic system. Neurosci Biobehav. Rev. 2006, 30, 203–214. 10.1016/j.neubiorev.2005.06.007. [DOI] [PubMed] [Google Scholar]
- Freund T. F.; Gulyas A. I.; Acsady L.; Gorcs T.; Toth K. Serotonergic control of the hippocampus via local inhibitory interneurons. Proc. Natl. Acad. Sci. U. S. A. 1990, 87, 8501–8505. 10.1073/pnas.87.21.8501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peris-Vicente J.; Esteve-Romero J.; Carda-Broch S. In Analytical Separation Science; American Cancer Society: New York, NY, 2015, pp. 1757–1808. [Google Scholar]
- Tiwari G.; Tiwari R. Bioanalytical method validation: An updated review. Pharm. Methods 2010, 1, 25–38. 10.4103/2229-4708.72226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green J. M. Peer Reviewed: A Practical Guide to Analytical Method Validation. Anal. Chem. 1996, 68, 305A–309A. 10.1021/ac961912f. [DOI] [Google Scholar]
- Randall E. C.; Emdal K. B.; Laramy J. K.; Kim M.; Roos A.; Calligaris D.; Regan M. S.; Gupta S. K.; Mladek A. C.; Carlson B. L.; Johnson A. J.; Lu F. K.; Xie X. S.; Joughin B. A.; Reddy R. J.; Peng S.; Abdelmoula W. M.; Jackson P. R.; Kolluri A.; Kellersberger K. A.; et al. Integrated mapping of pharmacokinetics and pharmacodynamics in a patient-derived xenograft model of glioblastoma. Nat. Commun. 2018, 9, 4904. 10.1038/s41467-018-07334-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swales J. G.; Dexter A.; Hamm G.; Nilsson A.; Strittmatter N.; Michopoulos F.; Hardy C.; Morentin-Gutierrez P.; Mellor M.; Andren P. E.; Clench M. R.; Bunch J.; Critchlow S. E.; Goodwin R. J. A. Quantitation of Endogenous Metabolites in Mouse Tumors Using Mass-Spectrometry Imaging. Anal. Chem. 2018, 90, 6051–6058. 10.1021/acs.analchem.7b05239. [DOI] [PubMed] [Google Scholar]
- Hansen H. T.; Janfelt C. Aspects of Quantitation in Mass Spectrometry Imaging Investigated on Cryo-Sections of Spiked Tissue Homogenates. Anal. Chem. 2016, 88, 11513–11520. 10.1021/acs.analchem.6b02711. [DOI] [PubMed] [Google Scholar]
- Schulz S.; Becker M.; Groseclose M. R.; Schadt S.; Hopf C. Advanced MALDI mass spectrometry imaging in pharmaceutical research and drug development. Curr. Opin. Biotechnol. 2019, 55, 51–59. 10.1016/j.copbio.2018.08.003. [DOI] [PubMed] [Google Scholar]
- Owens M. J.; Knight D. L.; Nemeroff C. B. Second-generation SSRIs: human monoamine transporter binding profile of escitalopram and R-fluoxetine. Biol. Psychiatry 2001, 50, 345–350. 10.1016/S0006-3223(01)01145-3. [DOI] [PubMed] [Google Scholar]
- Kretzschmar M.; Brust P.; Zessin J.; Cumming P.; Bergmann R.; Johannsen B. Autoradiographic imaging of the serotonin transporter in the brain of rats and pigs using S-([18F]fluoromethyl)-(+)-McN5652. Eur. Neuropsychopharmacol. 2003, 13, 387–397. 10.1016/S0924-977X(03)00039-7. [DOI] [PubMed] [Google Scholar]
- Small S. A.; Schobel S. A.; Buxton R. B.; Witter M. P.; Barnes C. A. A pathophysiological framework of hippocampal dysfunction in ageing and disease. Nat. Rev. Neurosci. 2011, 12, 585–601. 10.1038/nrn3085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dale E.; Pehrson A. L.; Jeyarajah T.; Li Y.; Leiser S. C.; Smagin G.; Olsen C. K.; Sanchez C. Effects of serotonin in the hippocampus: how SSRIs and multimodal antidepressants might regulate pyramidal cell function. CNS Spectr. 2016, 21, 143–161. 10.1017/S1092852915000425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyttel J. Pharmacological characterization of selective serotonin reuptake inhibitors (SSRIs). Int. Clin. Psychopharmacol. 1994, 9, 19–26. 10.1097/00004850-199403001-00004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.