Abstract
BACKGROUND
The correct interpretation of the ECG is pivotal for the accurate diagnosis of many cardiac abnormalities, and conventional computerized interpretation has not been able to reach physician‐level accuracy in detecting (acute) cardiac abnormalities. This study aims to develop and validate a deep neural network for comprehensive automated ECG triage in daily practice.
METHODS AND RESULTS
We developed a 37‐layer convolutional residual deep neural network on a data set of free‐text physician‐annotated 12‐lead ECGs. The deep neural network was trained on a data set with 336.835 recordings from 142.040 patients and validated on an independent validation data set (n=984), annotated by a panel of 5 cardiologists electrophysiologists. The 12‐lead ECGs were acquired in all noncardiology departments of the University Medical Center Utrecht. The algorithm learned to classify these ECGs into the following 4 triage categories: normal, abnormal not acute, subacute, and acute. Discriminative performance is presented with overall and category‐specific concordance statistics, polytomous discrimination indexes, sensitivities, specificities, and positive and negative predictive values. The patients in the validation data set had a mean age of 60.4 years and 54.3% were men. The deep neural network showed excellent overall discrimination with an overall concordance statistic of 0.93 (95% CI, 0.92–0.95) and a polytomous discriminatory index of 0.83 (95% CI, 0.79–0.87).
CONCLUSIONS
This study demonstrates that an end‐to‐end deep neural network can be accurately trained on unstructured free‐text physician annotations and used to consistently triage 12‐lead ECGs. When further fine‐tuned with other clinical outcomes and externally validated in clinical practice, the demonstrated deep learning–based ECG interpretation can potentially improve time to treatment and decrease healthcare burden.
Keywords: deep learning, deep neural networks, electrocardiography, triage
Subject Categories: Arrhythmias, Electrophysiology, Ventricular Fibrillation, Epidemiology, Electrocardiology (ECG)
Nonstandard Abbreviations and Acronyms
- CIE
computerized interpretation of the ECG
- DNN
deep neural network
- PDI
polytomous discriminatory index
Clinical Perspective
What Is New?
Our findings indicate that a deep neural network may be used to support the physician in ECG triage and reduce the clinical workload with an improved prioritization of ECGs for interpretation by the cardiologist.
The study shows that, in comparison with earlier studies that combined ECG recordings with other imaging modalities or laboratory findings, it is also feasible to use the less structured and noisy physician labels to successfully train a deep neural network for comprehensive ECG triaging.
Moreover, this is one of the first studies to visualize regions in the ECG important for the decisions of the deep neural network.
What Are the Clinical Implications?
The proposed end‐to‐end deep neural network can triage 12‐lead ECGs into normal, abnormal, and acute with high discrimination across all categories with an overall concordance statistic of 0.93 (95% CI, 0.92–0.95).
In clinical practice, this could lead to improved time to treatment for acute cardiac disorders and decreased and better‐balanced workloads for clinicians.
Further improvement with other clinical outcomes, prospective validation in other populations, and implementation studies are needed before implementation in clinical practice is possible.
With more than 300 million ECGs obtained annually worldwide, the ECG is a fundamental tool in the everyday practice of clinical medicine.1 The correct interpretation of the ECG is pivotal for the accurate diagnosis of a wide spectrum of cardiac abnormalities and requires the expertise of an experienced cardiologist. The life‐threatening nature of a suspected acute coronary syndrome and ventricular arrhythmias requires not only accurate but also timely ECG interpretation and places a heavy logistic burden on clinical practice.
Automated triage of ECGs in categories that need acute, nonacute, or no attention may therefore be of great support in daily practice. Accurately prioritizing different ECGs could lead to improvements in time to treatment and possibly decrease healthcare costs.2 Especially in prehospital care and noncardiology departments, expert knowledge to interpret ECGs might not always be readily available.3, 4, 5 However, a consistent and fast automated algorithm that supports the physician in comprehensive triage of the ECG remains lacking.
Computerized interpretation of the ECG (CIE) was introduced more than 50 years ago and became increasingly important in aiding the physician interpretation in many clinical settings. However, current CIE algorithms have not been able to reach physician‐level accuracy in diagnosing cardiac abnormalities.5 The accurate interpretation of arrythmias and ST‐segment abnormalities remains the most problematic, and many algorithms suffer from high amounts of false positives for these disorders.4, 5, 6, 7, 8, 9 Overdiagnosis and a failure to correct the erroneous interpretation by an overreading physician has shown to lead to unnecessary interventions and medication use.10, 11
With the development of algorithms that can benefit from the large‐scale processing of raw data without the need for hand‐crafted feature extraction, a substantial improvement of CIE is forthcoming. Several of these techniques, deep neural networks (DNN) in particular, have shown to be highly effective in similar applications as speech recognition and image classification.12, 13, 14 DNNs are computer algorithms based on the structure and function of the human brain. Their hidden layers of neurons can be trained to discover complex patterns in signals such as the ECG.15 In comparison with conventional CIE algorithms, DNNs have the advantage that they jointly optimize both pattern discovery and classification in an end‐to‐end approach that only needs the raw waveforms as input. In medicine, deep learning showed promising results when applied to arrhythmia detection in single‐lead ECG recordings and to early detection of atrial fibrillation in normal sinus rhythm ECGs.16, 17 When combined with ultrasound or laboratory findings, deep‐learning algorithms were able to detect reduced ejection fraction and hyperkalemia in 12‐lead ECGs.18, 19
This study aims to develop and validate a DNN for comprehensive automated ECG triage that could support daily clinical practice.
METHODS
Study Participants
The data set contained all 12‐lead ECGs from patients aged between 18 and 85 years, recorded in the University Medical Center Utrecht from January 2000 to August 2019, and obtained at noncardiology departments. All extracted data were deidentified in accordance with the EU General Data Protection Regulation and written informed consent was therefore not required by the University Medical Center Utrecht ethical committee.
Training Data Acquisition and Annotation
All ECGs were recorded on a General Electric MAC 5500 (GE Healthcare, Chicago, IL). We extracted raw 10‐second 12‐lead ECG data waveforms from the MUSE ECG system (MUSE version 8; GE Healthcare). All recordings in the University Medical Center Utrecht acquired in noncardiology departments were systematically annotated by a physician as part of the regular clinical workflow. These physicians were all trained to interpret and annotate an ECG as part of their cardiology residency. During the annotation, the physicians had access to the name, sex, and age of the patient; the computer‐calculated conduction intervals; the previous ECG recordings; and the full patient records. The ECGs were divided into the following 4 triage categories based on how quickly a cardiologist has to be consulted: (1) normal, (2) not acute abnormal (consultation with low priority), (3) subacute abnormal (consultation with moderate priority), and (4) acute abnormal (consultation with high priority).
The free‐text physician ECG annotations were labeled into 1 of the 4 triage categories using a text mining–based approach. First, the annotations were tokenized and all frequent (ie, occurring >20 times) terms and multiword collocations were extracted. These terms, such as “STEMI,” and collocations, such as “first degree AV‐block” and “1st degree AV block,” contained multiple variations of diagnostic ECG statements. Therefore, they were mapped to the standardized statements of the American Heart Association's Electrocardiography Diagnostic Statement List.20 Second, a panel of 3 electrophysiologists defined the triage category for every standardized diagnostic statement. The used diagnostic statements and their corresponding triage category are provided in Figure 1. Third, a final triage category was assigned to every ECG. When multiple statements were given, the final triage category was the maximum category. All text‐mining steps were performed with the quanteda package for R (version 3.5; R Foundation for Statistical Computing, Vienna, Austria).21 An overview of the text‐mining steps can be found in Figure 2.
Figure 1. ECG diagnoses with their corresponding triage categories.
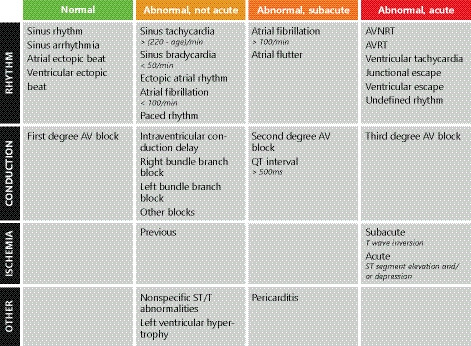
Triage categories as defined by the panel of electrophysiologists, with (1) normal, (2) not acute abnormal (consultation without priority), (3) subacute abnormal (consultation with some priority), and (4) acute abnormal (consult immediately). The ECG diagnoses derived from the text‐mining algorithm were used to categorize the training data using these rules. When multiple diagnoses were given, the final triage category was the maximum category. AV indicates atrioventricular; AVNRT, atrioventricular nodal reentrant tachycardia; and AVRT, atrioventricular reentrant tachycardia.
Figure 2. Overview of the labeling into triage categories in the training and validation data sets.
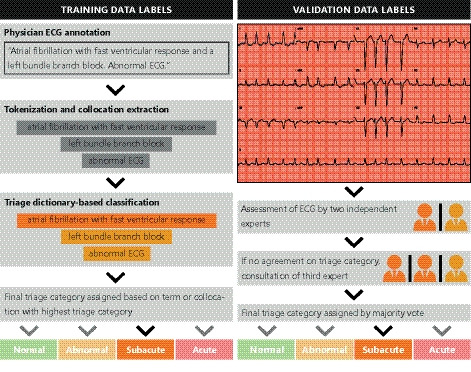
The training labels (left), used for training, are derived from the free‐text annotation given to the ECG by a single physician in daily practice. The ECG diagnoses are mapped to triage categories using the rules defined by a panel of electrophysiologists (Figure 1). The validation labels (right), used for validation of the deep neural network, are given by the expert panel based on visual inspection of a 12‐lead ECG.
Validation Data Annotation
For the validation of the DNN, a data set with higher annotation reliability was required. Therefore, an independent data set was annotated and triaged by the reference standard, a panel of 5 practicing senior electrophysiologists or cardiologists. All records were annotated by 2 independent annotators who were blinded to the other annotation. In case of disagreement in the triage category, a third annotator was consulted, and the majority vote was used as the final label. The recordings with 3 discordant votes were discussed in a joint panel meeting, and the recordings of insufficient quality were excluded. Annotation was performed using an online tool, where the expert had access to the 12‐lead ECG, computer‐calculated conduction intervals, and age and sex of the patient. The experts were instructed to classify the ECGs into 1 of the 4 triage categories based on the rules in Figure 1. The input and annotation steps in the validation data set are schematically shown in Figure 2.
As manual annotation by a panel is time intensive, a sample‐size calculation was performed to achieve adequate precision of the validation performance measures. For this, a minimum of 50 cases per category was needed.22 As the smallest triage category in the training data set has a prevalence of approximately 5%, the validation data set consisted of 1000 recordings from unique patients. All ECGs of these patients were excluded from the training data set.
Algorithm Development
As leads III, aVR, aVL and aVF are derivatives of the other leads and contain no new information, we only used the raw 10‐second, 8‐channel waveforms (I, II, and V1–V6), sampled at 500 Hz, as the input for our DNN. We applied an architecture similar to the Inception ResNet network by combining blocks of convolutional layers in parallel with residual connections.23, 24
This network is built with layers of identical blocks with a preactivation design consisting of 2 one‐dimensional convolutional layers, preceded by batch normalization, rectified linear unit activation, and dropout.25, 26, 27 Every residual block consists of 3 parallel branches: 1 with a normal convolutional layer, 1 with a dilated convolutional layer, and 1 with a shortcut connection, where the input to the block is added to the output unadjusted.28 This enables the network to determine the features in 2 different time dimensions, where the dilated convolution covers a complete heartbeat. The output of the last block was flattened and used as input to a fully connected layer with a rectified linear unit nonlinearity, followed by dropout. The output layer consisted of 4 nodes, 1 for every triage category, and a softmax function was used to produce a probability distribution over all triage categories. A similar auxiliary output was added in the middle of the network, and its loss was added to the total loss during training.
After hyperparameter and architecture optimizations, the final selected network consisted of 16 residual blocks with 2 one‐dimensional convolutional layers with filter size 5 and a dilation of 100 (Figure S1). Every other block downsampled the input using a strided convolution, and the number of filters was doubled every fourth block. Dropout was performed with a probability of 30%. The fully connected layer consisted of 256 nodes. This resulted in a final network with 37 layers.
This network was trained using the Adam optimizer with a learning rate of 0.0005 and a mini batch size of 128.29 Weighted focal loss was used to counteract the category imbalance in the data set and to minimize the number of false negatives.30 Training was terminated when the loss stopped decreasing in the 5% subset of the training data set. Network training was performed using the PyTorch package (version 1.3) on a Titan Xp GPU (NVIDIA Corporation, Santa Clara, CA).31
The different network architectures and hyperparameters were chosen using a combination of manual tuning and random grid search. The network with the lowest loss in a 5% randomly sampled subset of the training data set was chosen. When multiple architectures showed similar performance, the simplest architecture was selected. The following hyperparameters were assessed: the use of dilated convolutions, residual connections, max pooling, an auxiliary loss and/or fully connected layers, the number of layers, the size and number of convolutional filters, the dropout rate, the learning rate, and the weights of the loss. We also experimented with an ordinal loss method instead of a multinomial loss method and with adding age and sex to the flattened layer, but this did not result in increased performance.32, 33
Visualization of the DNN
To improve understanding of the decisions of the DNN, guided gradient‐weighted class activation mapping, a technique for visual explanations in convolutional neural networks, was adjusted for use in 1‐dimensional data.34 Guided gradient‐weighted class activation mapping is a combination between the fine‐grained guided backpropagation and gradient‐weighted class activation mapping, which produces a coarse class‐discriminative heatmap based on the final convolutional layer.34, 35 The heatmap is superimposed over the ECG recording and shows the regions in the ECG important to the DNN for predicting a specific triage category.
Statistical Analysis
Interobserver agreement was quantified using squared‐weighted Cohen's κ for 2 reviewers or tests and ordinal Krippendorff α for more than 2 reviewers.36, 37 Considering the imbalance in category frequencies, overall algorithm discriminatory performance was assessed with the unweighted mean of all pairwise concordance (or c) statistics (also known as area under the receiver operating curve) and the polytomous discriminatory index (PDI).38, 39, 40 The first metric estimates the probability to correctly distinguish between all pairs of 2 patients from different categories, where a value of 0.5 denotes random performance and 1 perfect performance. The second assesses the discrimination between all categories simultaneously in a set approach. It estimates the probability to correctly identify a specific patient in a set of patients from every category, where 0.25 denotes random and 1 perfect performance with 4 categories.38, 39, 41 As a second step, category‐specific performance is assessed with the c‐statistic, PDI, sensitivity, specificity, and positive and negative predictive values. All category‐specific measures, except the PDI, were applied in a 1‐versus‐other approach.
All statistical analyses were performed using R version 3.5 (R Foundation for Statistical Computing). The Transparent Reporting of a Multivariable Prediction Model for Individual Prognosis or Diagnosis Statement for the reporting of diagnostic models was followed, where appropriate.42 All data are presented as mean±SD or median with interquartile range. The 95% CIs around the performance measures were obtained using 2000 bootstrap samples.
RESULTS
The total training data set consisted of 336 835 recordings of 142 040 patients. The distribution of triage categories was unbalanced with the most recordings in category 2 (45.5%) and the least in category 4 (4.8%). In the validation data set, there was consensus between the 2 experts in 736 cases (73.6%). After consultation of a third tie‐breaker expert (248 cases, 24.8%), the panel meeting (29 cases, 2.9%), and the exclusion of recordings of insufficient quality, 984 validation cases were used for the analysis. There was good interobserver agreement, with a Krippendorff α of 0.72. Conflicts in the first expert annotation round occurred the most between categories 1 and 2 (162/255, 64%), between categories 2 and 3 (30/255, 12%), and between categories 2 and 4 (24/255, 9.5%). Disagreement between categories 1 and 2 was mostly attributed to different assessments on the presence of nonspecific ST‐segment or T‐wave abnormalities. For categories 2 and 3 and categories 3 and 4, the most common difference was the interpretation of ST‐segment elevation or depression. Table 1 summarizes the patient demographics and triage category distributions of the recordings in the training and validation data sets.
Table 1.
Patient Demographics and Distribution of Triage Categories in the Training and Validation Data Sets
| Training (n=336.835) | Validation (n=984) | |
|---|---|---|
| Male sex, n (%) | 188 858 (56.1) | 402 (54.3) |
| Age, mean (SD), y | 60.8 (15.5) | 60.4 (15.3) |
| Location, n (%) | ||
| Emergency department | 92 532 (27.5) | 310 (31.5) |
| Intensive care unit | 20 045 (6.0) | 63 (6.4) |
| Noncardiology outpatient clinic | 73 170 (21.7) | 161 (16.4) |
| Noncardiology ward | 86 630 (25.7) | 263 (26.7) |
| Preoperative screening | 6300 (1.9) | 8 (0.8) |
| Recovery ward | 53 994 (16.0) | 163 (16.6) |
| Other | 4164 (1.2) | 16 (1.6) |
| Triage category, n (%) | ||
| Normal | 142 456 (42.3)a | 418 (42.5)b |
| Abnormal, not acute | 153 360 (45.5)a | 410 (41.7)b |
| Abnormal, subacute | 24 731 (7.3)a | 76 (7.7)b |
| Abnormal, acute | 16 288 (4.8)a | 80 (8.1)b |
A 5% randomly sampled subset of the training data set was used for model tuning and internal validation. The validation data set is independent from the training data set.
Distribution based on text‐mining categorization of annotations by physician in daily practice.
Distribution based on the expert consensus panel annotations.
The overall discriminations, as measured by the unweighted mean of pairwise c‐statistics and the PDI, of the DNNs demonstrated in this article were 0.93 (95% CI, 0.92–0.95) and 0.83 (95% CI, 0.79–0.87), respectively. The c‐statistics, PDIs, sensitivities, specificities, positive predictive values, and negative predictive values per triage category in a 1‐versus‐other approach are shown in Table 2, whereas the confusion matrix is provided in Figure 3. Visualizations of the regions in the ECG important for the DNN to predict a specific category are shown in Figure 4. The full 12‐lead ECGs can be found in Figures S2 through S6.
Table 2.
Diagnostic Performance Measures per Triage Category for the Deep Neural Network in the Panel‐Annotated Validation Data Set
| Normal | Abnormal, Not Acute | Abnormal, Subacute | Abnormal, Acute | |
|---|---|---|---|---|
| C‐statistic (95% CI) | 0.95 (0.94 to 0.96) | 0.91 (0.89–0.93) | 0.94 (0.90–0.97) | 0.94 (0.90–0.96) |
| PDI (95% CI) | 0.91 (0.87–0.93) | 0.80 (0.75–0.84) | 0.82 (0.75–0.88) | 0.80 (0.73–0.87) |
| Sensitivity | 0.87 | 0.76 | 0.64 | 0.79 |
| Specificity | 0.88 | 0.89 | 0.98 | 0.94 |
| Positive predictive value | 0.85 | 0.83 | 0.78 | 0.55 |
| Negative predictive value | 0.90 | 0.84 | 0.97 | 0.98 |
The c‐statistics, sensitivities, specificities, positive and negative predictive values are calculated in a 1‐vs‐other approach and compare the category with the highest probability to the reference standard. The PDI estimates the probability that a patient from that category is correctly identified from a set of cases from every category. C‐statistic indicates concordance statistic, equivalent to area under the receiver operating characteristic curve; and PDI, polytomous discriminatory index.
Figure 3. Confusion matrix for the deep neural network. Rows represent the categories given by the reference standard (expert panel), and columns represent the categories predicted by the deep neural network.
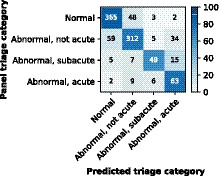
The color map is normalized per row and represents the percentage in the true triage category.
Figure 4.
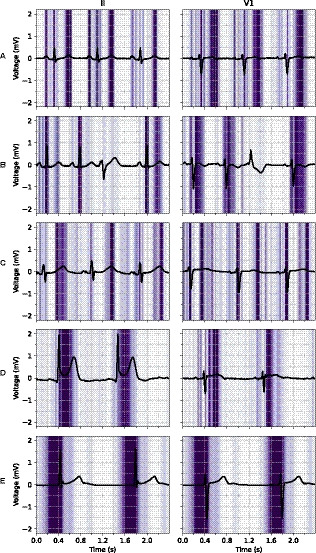
Examples of ECG leads II and V1 with a superimposed guided gradient‐weighted class activation mapping visualization showing regions important for the deep neural network to predict a certain triage category. A, Normal ECG with focus on the P‐wave, QRS‐complex, and T‐wave. B, Normal ECG with a single ignored premature ventricular complex. C, Subacute ECG with a long QT interval and a focus on the beginning and end of the QT‐segment. D, Acute ECG with an inferior ST‐segment–elevation myocardial infarction and a focus on the ST‐segment and J‐point. E, Acute ECG with a junctional escape rhythm and a focus on the pre‐QRS‐segment, where the P‐wave is missing. The full 12‐lead ECGs are available in Figures S2 through S6.
The DNN predicted a lower triage category than the true category (undertriage) in 88 (8.9%) and a higher category (overtriage) in 107 (11%) of the recordings in the validation data set. Most undertriage (59/88, 67%) occurred between categories 1 and 2, and these undertriaged recordings were categorized as 2 by the panel based on nonspecific ST‐segment abnormalities (26/59, 44%), old ischemia (12/59, 20%), left ventricular hypertrophy (7/59, 12%) or other reasons (14/59, 24%). All 9 acute category 4 recordings triaged as category 2 contained ST‐depression or T‐wave inversion and no ST‐elevation. In the category 2 recordings overtriaged as 4, the panel did mention nonspecific ST‐segment abnormalities in 20/34 (59%) recordings and old ischemia in 8/34 (24%).
As the labeling procedures for the training and validation data sets differ (Figure 2), the performance of the DNN could be dependent on errors in 2 steps in the training labeling procedure. First, the interobserver agreement between manual categorization of the free‐text physician ECG annotations into triage categories and the text mining–based categorization was excellent in the validation data set, with a weighted Cohen's α of 0.96. Second, agreement between the text mining–based triage categories and the reference standard was good (Cohen's α, 0.74). The overall c‐statistic and PDI for predicting the reference standard triage category with the text mining–based categories were 0.86 (95% CI, 0.85–0.88) and 0.48 (95% CI, 0.43–0.53), respectively.
DISCUSSION
This study is among the first to apply DNNs to a large data set of 12‐lead ECGs for automatic interpretation. We demonstrated that a deep‐learning approach performs well in detecting abnormalities for triage of 12‐lead ECGs. Our DNN has an excellent c‐statistic of 0.93 (95% CI, 0.92–0.95) and a good PDI of 0.83 (95% CI, 0.79–0.87), with high positive and negative predictive values across all triage categories. These findings indicate that a deep‐learning approach may be used to support the physician in ECG triage and reduce clinical workload with an improved prioritization of ECGs for interpretation by the cardiologist.
Interpretation of the ECG requires extensive knowledge of the wide variety of electrical manifestations of heart disease and a good understanding of normal variety. This has been a challenge for both manual and computerized interpretations and has led to a collection of definitions, measurements, and criteria to aid clinical decision making.5, 20 This challenge is extensively described in earlier studies, but comparisons are difficult, as the studies demonstrate wide variations in diagnostic measures, and an international accepted standard for the validation of ECG diagnoses is still missing.4, 5 For comprehensive ECG interpretations, noncardiologist physicians correctly identified 36% to 96% of the diagnoses, with significant differences between physicians and increasing performance for more experienced physicians.4, 43, 44, 45 Most studies focused on particular aspects of ECG interpretation, such as normal–abnormal differentiation, arrythmia classification, and detection of ST‐segment–elevation myocardial infarction. Overall, for these aspects physicians have higher false negative rates, whereas computerized algorithms have higher false positive rates when compared with expert panels.4, 5, 7, 8, 9, 43, 44, 45 The DNN could improve both the high false positive and negative rates while producing consistent results not dependent on external factors, such as physician experience.
Conventional CIE uses manually derived features, which only capture a fraction of the available information for any manifestation of heart disease in the obtained raw signal. This is one of the reasons that could explain the excellent performance of our algorithm and DNNs in general, as their integrated feature discovery and classification incorporates the whole raw input signal. In addition, conventional CIE algorithms are tuned to produce complete interpretations of the ECG and are less focused on one of their most important uses, quick triage. By training on a large physician‐annotated 12‐lead ECG data set, where the labels are mapped to predefined triage categories, we focus on a single task and are able to achieve high accuracy. The large size of the data set makes that the network has seen a wide variety of ECGs and should therefore be well generalizable.
Although the DNN does not use any manually selected features of the signal, visualizations show that the network bases its decisions on the same regions in the ECG as would experts. As shown in Figure 3, the network correctly identifies a normal ECG, a long QT‐segment, ST‐segment–elevation myocardial infarction, and a junctional escape rhythm in sensible regions and correctly ignores a premature ventricular complex in a normal ECG. Furthermore, inspection of the misclassifications of the DNN shows a similar pattern to the disagreement between the experts in the panel. The correct interpretation of ST‐segment and T‐wave abnormalities is apparently challenging for both the cardiologist and the DNN.
The DNN is trained on triage category labels that were automatically derived using text mining on free‐text annotations by a single physician in daily practice. Disagreement measures show that the text‐mining categories do not completely agree with the labels given by the expert panel. Most of this disagreement is caused by disagreements between the expert panel labels and the automatically categorized single‐physician labels (Cohen's α, 0.74). Considering this substantial disagreement between training and validation labels, we might expect that the DNN cannot outperform the performance measures for prediction with only the text mining–based triage categories. However, the DNN exceeds both the overall c‐statistic and PDI of the text mining–based triage categories and shows to be robust against considerable training label noise. This is in line with previous research that showed that the DNN can handle label noise quite well.46
Other research demonstrated the value of DNN for ECG interpretation for similar problems, where a single‐lead ECG was used for arrhythmia classification and a 12‐lead ECG for early detection of atrial fibrillation, contractile dysfunction, and hyperkalemia.16, 17, 18, 19 Our study shows that, in comparison with combining ECG recordings with other imaging modalities or laboratory findings, it is also feasible to use the less structured and noisy physician labels to successfully train a DNN for comprehensive ECG triaging. Moreover, this is one of the first studies to visualize regions in the ECG important for the decisions of the DNN.34
Triage is the process of classifying according to the severity of the case to determine how quickly action is needed. Careful triage is needed to prioritize those cases where timely action reduces morbidity and mortality among patients. For a triage algorithm to be effective, it is important that undertriage (eg, failure to detect patients with acute disease) and overtriage (eg, false alarms) are minimized. Our DNN shows very high negative predictive values for the highest categories, subacute and acute (Table 2). This can potentially reduce time to treatment for patients with acute cardiac disorders as the algorithm is able to provide triage advice immediately after the ECG is acquired and before the ECG is assessed by a physician with sufficient expertise. However, the sensitivities for the subacute and acute categories of 64% and 79% are partly attributed to undertriage (Figure 3) and therefore need further improvement before clinical implementation is possible. The algorithm shows relatively high positive predictive values, which will decrease the amount of false alarms in otherwise normal ECGs. Because most hospital‐acquired ECGs fall into this category, a modest improvement can already significantly decrease the workload for physicians.
This study has several limitations to address. Although a reasonably large training data set was used, the acute categories remained relatively small. This is customary to an unselected real‐world data set but entails a chance of underprediction. We made use of the focal loss method, used in computer‐vision DNN algorithms, to counteract this problem.26 In the validation data set, the triage category distribution was similar, but CIs showed adequate precision in the smaller categories as well. The representative sampling of the validation is also a strength, making it possible to derive positive and negative predictive values, which are most important to the patient. We believe that the panel‐annotated validation data set in this study provides a good measure of generalizability to hospital populations comparable to ours. It has been shown that ethnicity influences the ECG and could be taken into account to improve automated interpretation.47 External validation is therefore needed when used with different recording machines and in different populations, such as patients in the general practice or populations with different ethnical compositions. This is beyond the scope of this study and will most likely require (re)training on a such a data set.
Both manual and computerized ECG interpretations are hard to standardize, as can be seen by the high disagreement rates between the experts (Krippendorff α, 0.72). This number is comparable with earlier studies on the interobserver agreement between experts on ECG interpretation.4, 6, 48 The panel‐annotated validation data set used in this study is the current best reference standard available, but in clinical practice, many other diagnostic tests are used to interpret the ECG findings. Therefore, we suspect the diagnostic accuracy of our algorithm could be further optimized with hard clinical outcome data, such as a diagnosis and localization of myocardial infarction with coronary artery angiography, cardiac enzymes, and electrolyte disorders from laboratory data and even mortality. Both optimization with clinical outcome data and external validation are necessary before clinical implementation is possible.
Another future perspective of the DNN is the capability to continuously improve and learn by adding new cases. Traditionally, neural networks did not provide uncertainty around their predictions, but this was changed because of new insights from several different bayesian methods.49 When combining uncertainty around predictions with active learning, it becomes possible to let uncertain cases be annotated by a cardiologist and improve the algorithm, whereas easier cases can be classified automatically.50 Moreover, to determine the most important ECG leads, the algorithm could be trained and evaluated with fewer input channels. This could make the use of a similar algorithm with home‐monitoring devices with less leads possible.
In conclusion, our end‐to‐end DNN can triage 12‐lead ECGs into normal, abnormal, and acute with high discrimination across all categories. In clinical practice, this could lead to improved time to treatment for acute cardiac disorders and decreased and better‐balanced workloads for clinicians. Further improvement with other clinical outcomes, prospective validation in other populations, and implementation studies are needed before implementation in clinical practice is possible.
Sources of Funding
None.
Disclosures
None.
Supporting information
Figures S1–S6
Acknowledgments
We gratefully acknowledge the support of NVIDIA Corporation with the donation of the Titan Xp GPU used for this research. We thank Ronald Groenemeijer for his technical and logistic support in acquiring the data.
(J Am Heart Assoc. 2020;9:e015138 DOI: 10.1161/JAHA.119.015138.)
For Sources of Funding and Disclosures, see page 10.
Data Availability
The anonymized expert panel–annotated validation data set used in this study is available from the corresponding author upon request. The training data and analytic methods are not available to other researchers.
References
- 1. Holst H, Ohlsson M, Peterson C, Edenbrandt L. A confident decision support system for interpreting electrocardiograms. Clin Physiol. 1999;19:410–418. [DOI] [PubMed] [Google Scholar]
- 2. Diercks DB, Kontos MC, Chen AY, Pollack CV, Wiviott SD, Rumsfeld JS, Magid DJ, Gibler WB, Cannon CP, Peterson ED, et al. Utilization and impact of pre‐hospital electrocardiograms for patients with acute ST‐segment elevation myocardial infarction. J Am Coll Cardiol. 2009;53:161–166. [DOI] [PubMed] [Google Scholar]
- 3. Eslava D, Dhillon S, Berger J, Homel P, Bergmann S. Interpretation of electrocardiograms by first‐year residents: the need for change. J Electrocardiol. 2009;42:693–697. [DOI] [PubMed] [Google Scholar]
- 4. Salerno SM, Alguire PC, Waxman HS. Competency in interpretation of 12‐lead electrocardiograms: a summary and appraisal of published evidence. Ann Intern Med. 2003;138:751. [DOI] [PubMed] [Google Scholar]
- 5. Schläpfer J, Wellens HJ. Computer‐interpreted electrocardiograms. J Am Coll Cardiol. 2017;70:1183–1192. [DOI] [PubMed] [Google Scholar]
- 6. Holmvang L, Hasbak P, Clemmensen P, Wagner G, Grande P. Differences between local investigator and core laboratory interpretation of the admission electrocardiogram in patients with unstable angina pectoris or non‐Q‐wave myocardial infarction (a thrombin inhibition in myocardial ischemia [TRIM] substudy). Am J Cardiol. 1998;82:54–60. [DOI] [PubMed] [Google Scholar]
- 7. Berge HM, Steine K, Andersen TE, Solberg EE, Gjesdal K. Visual or computer‐based measurements: important for interpretation of athletes’ ECG. Br J Sports Med. 2014;48:761–767. [DOI] [PubMed] [Google Scholar]
- 8. Garvey JL, Zegre‐Hemsey J, Gregg R, Studnek JR. Electrocardiographic diagnosis of ST segment elevation myocardial infarction: an evaluation of three automated interpretation algorithms. J Electrocardiol. 2016;49:728–732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Shah AP, Rubin SA. Errors in the computerized electrocardiogram interpretation of cardiac rhythm. J Electrocardiol. 2007;40:385–390. [DOI] [PubMed] [Google Scholar]
- 10. Bogun F, Anh D, Kalahasty G, Wissner E, Serhal CB, Bazzi R, Weaver WD, Schuger C. Misdiagnosis of atrial fibrillation and its clinical consequences. Am J Med. 2004;117:636–642. [DOI] [PubMed] [Google Scholar]
- 11. Southern WN, Arnsten JH. The effect of erroneous computer interpretation of ECGs on resident decision making. Med Decis Mak. 2009;29:372–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Graves A, Mohamed A, Hinton G. Speech recognition with deep recurrent neural networks 2013:6645–6649. [Google Scholar]
- 13. Gulshan V, Peng L, Coram M, Stumpe MC, Wu D, Narayanaswamy A, Venugopalan S, Widner K, Madams T, Cuadros J, et al. Development and validation of a deep learning algorithm for detection of diabetic retinopathy in retinal fundus photographs. J Am Med Assoc. 2016;316:2402–2410. [DOI] [PubMed] [Google Scholar]
- 14. Tschandl P, Rosendahl C, Akay BN, Argenziano G, Blum A, Braun RP, Cabo H, Gourhant J‐Y, Kreusch J, Lallas A, et al. Expert‐level diagnosis of nonpigmented skin cancer by combined convolutional neural networks. JAMA Dermatol. 2018;155:58–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Goodfellow I, Bengio Y, Courville A. Deep Learning. Cambridge, MA: The MIT Press; 2016. [Google Scholar]
- 16. Hannun AY, Rajpurkar P, Haghpanahi M, Tison GH, Bourn C, Turakhia MP, Ng AY. Cardiologist‐level arrhythmia detection and classification in ambulatory electrocardiograms using a deep neural network. Nat Med. 2019;25:65–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Attia ZI, Noseworthy PA, Lopez‐Jimenez F, Asirvatham SJ, Deshmukh AJ, Gersh BJ, Carter RE, Yao X, Rabinstein AA, Erickson BJ, et al. An artificial intelligence‐enabled ECG algorithm for the identification of patients with atrial fibrillation during sinus rhythm: a retrospective analysis of outcome prediction. Lancet. 2019;6736:1–7. [DOI] [PubMed] [Google Scholar]
- 18. Galloway CD, Valys AV, Shreibati JB, Treiman DL, Petterson FL, Gundotra VP, Albert DE, Attia ZI, Carter RE, Asirvatham SJ, et al. Development and validation of a deep‐learning model to screen for hyperkalemia from the electrocardiogram. JAMA Cardiol. 2019;55905:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Attia ZI, Kapa S, Lopez‐Jimenez F, McKie PM, Ladewig DJ, Satam G, Pellikka PA, Enriquez‐Sarano M, Noseworthy PA, Munger TM, et al. Screening for cardiac contractile dysfunction using an artificial intelligence–enabled electrocardiogram. Nat Med. 2019;25:70–74. [DOI] [PubMed] [Google Scholar]
- 20. Mason JW, Hancock EW, Gettes LS. Recommendations for the standardization and interpretation of the electrocardiogram: Part II: electrocardiography diagnostic statement list: a scientific statement from the American Heart Association Electrocardiography and Arrhythmias Committee, Council. Circulation. 2007;115:1325–1332. [DOI] [PubMed] [Google Scholar]
- 21. Benoit K, Watanabe K, Wang H, Nulty P, Obeng A, Müller S, Matsuo A. Quanteda: an R package for the quantitative analysis of textual data. J Open Source Softw. 2018;3:774. [Google Scholar]
- 22. Collins GS, Ogundimu EO, Altman DG. Sample size considerations for the external validation of a multivariable prognostic model: a resampling study. Stat Med. 2016;35:214–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. He K, Zhang X, Ren S, Sun J. Deep residual learning for image recognition. Available at: http://arxiv.org/abs/1512.03385. Published December 10, 2015. Accessed February 5, 2019.
- 24. Szegedy C, Ioffe S, Vanhoucke V, Alemi A. Inception‐v4, inception‐ResNet and the impact of residual connections on learning. Published 2016. Available at: http://arxiv.org/abs/1602.07261. Accessed April 26, 2019.
- 25. Ioffe S, Szegedy C. Batch normalization: accelerating deep network training by reducing internal covariate shift In: Bach F, Blei D, eds. Proceedings of the 32nd International Conference on Machine Learning. Lille, France: Proceedings of Machine Learning Research; 2015:448–456. [Google Scholar]
- 26. Srivastava N, Hinton G, Krizhevsky A, Sutskever I, Salakhutdinov R. Dropout: a simple way to prevent neural networks from overfitting. J Mach Learn Res. 2014;15:1929–1958. [Google Scholar]
- 27. Glorot X, Bordes A, Bengio Y. Deep sparse rectifier neural networks xavier. In: Proceedings from the 14th International Conference on Artificial Intelligence and Statistics; Fort Lauderdale, USA, 2011; April 11–13, 2011.
- 28. Yu F, Koltun V. Multi‐scale context aggregation by dilated convolutions. Available at: http://arxiv.org/abs/1511.07122. Published 2015. Accessed June 10, 2019.
- 29. Kingma DP, Ba J. Adam: a method for stochastic optimization. AIP Conf Proc. 2014;1631:58–62. [Google Scholar]
- 30. Lin TY, Goyal P, Girshick R, He K, Dollar P. Focal loss for dense object detection. In: Proceedings of the IEEE International Conference on Computer Vision Venice, Italy; 2017:2999–3007. Accessed October 22–29, 2017. [Google Scholar]
- 31. Paszke A, Gross S, Massa F, Lerer A, Bradbury J, Chanan G, Killeen T, Lin Z, Gimelshein N, Antiga L, et al. PyTorch: An Imperative Style, High‐Performance Deep Learning Library. In: Advances in Neural Information Processing Systems. Vancouver, Canada: December 8–14, 2019;8024–8035. [Google Scholar]
- 32. Cheng J, Wang Z, Pollastri G. A neural network approach to ordinal regression. In: Proceedings of the International Joint Conference on Neural Networks Hong Kong, China: 2008;1279–1284. [Google Scholar]
- 33. van Leeuwen KG, Sun H, Tabaeizadeh M, Struck AF, van Putten MJA, Westover MB. Detecting abnormal electroencephalograms using deep convolutional networks. Clin Neurophysiol. 2018;130:77–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Selvaraju RR, Cogswell M, Das A, Vedantam R, Parikh D, Batra D. Grad‐CAM: visual explanations from deep networks via gradient‐based localization. In: Proceedings of the IEEE International Conference on Computer Vision Venice, Italy; 2017:618–626. [Google Scholar]
- 35. Springenberg JT, Dosovitskiy A, Brox T, Riedmiller M. Striving for simplicity: the all convolutional net. San Diego, United States: International Conference on Learning Representations. 2015:1–14. [Google Scholar]
- 36. Krippendorff K, Hayes AF. Answering the call for a standard reliability measure for coding data. Commun Methods Meas. 2007;1:77–89. [Google Scholar]
- 37. Fleiss JL, Cohen J. The equivalence of weighted kappa and the intraclass correlation coefficient as measures of reliability. Educ Psychol Meas. 1973;33:613–619. [Google Scholar]
- 38. Van Calster B, Vergouwe Y, Looman CWN, Van Belle V, Timmerman D, Steyerberg EW. Assessing the discriminative ability of risk models for more than two outcome categories. Eur J Epidemiol. 2012;27:761–770. [DOI] [PubMed] [Google Scholar]
- 39. Van Calster B, Van Belle V, Vergouwe Y, Timmerman D, Van Huffel S, Steyerberg EW. Extending the c‐statistic to nominal polytomous outcomes: the Polytomous Discrimination Index. Stat Med. 2012;31:2610–2626. [DOI] [PubMed] [Google Scholar]
- 40. Hand DJ, Till RJ. A simple generalisation of the area under the ROC curve for multiple class classification problems. Mach Learn. 2001;45:171–186. [Google Scholar]
- 41. Li J, Gao M, D'Agostino R. Evaluating classification accuracy for modern learning approaches. Stat Med. 2019;38:2477–2503. [DOI] [PubMed] [Google Scholar]
- 42. Collins GS, Reitsma JB, Altman DG, Moons KGM. Transparent reporting of a multivariable prediction model for Individual Prognosis Or Diagnosis (TRIPOD): the TRIPOD statement. Ann Intern Med. 2015;162:55. [DOI] [PubMed] [Google Scholar]
- 43. Goy JJ, Schlaepfer J, Stauffer JC. Competency in interpretation of the 12‐lead electrocardiogram among swiss doctors. Swiss Med Wkly. 2013;143:8–10. [DOI] [PubMed] [Google Scholar]
- 44. Veronese G, Germini F, Ingrassia S, Cutuli O, Donati V, Bonacchini L, Marcucci M, Fabbri A. Emergency physician accuracy in interpreting electrocardiograms with potential ST‐segment elevation myocardial infarction: is it enough? Acute Card Care. 2016;18:7–10. [DOI] [PubMed] [Google Scholar]
- 45. McCabe JM, Armstrong EJ, Ku I, Kulkarni A, Hoffmayer KS, Bhave PD, Waldo SW, Hsue P, Stein JC, Marcus GM, et al. Physician accuracy in interpreting potential ST‐segment elevation myocardial infarction electrocardiograms. J Am Heart Assoc. 2013;2:000268 DOI: 10.1161/JAHA.113.000268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Rolnick D, Veit A, Belongie S, Shavit N. Deep learning is robust to massive label noise. ArXiv. Available at: http://arxiv.org/abs/1705.10694. Published 2017. Accessed April 10, 2019. [Google Scholar]
- 47. MacFarlane PW, Katibi IA, Hamde ST, Singh D, Clark E, Devine B, Francq BG, Lloyd S, Kumar V. Racial differences in the ECG—selected aspects. J Electrocardiol. 2014;47:809–814. [DOI] [PubMed] [Google Scholar]
- 48. Brosnan M, La Gerche A, Kumar S, Lo W, Kalman J, Prior D. Modest agreement in ECG interpretation limits the application of ECG screening in young athletes. Heart Rhythm. 2015;12:130–136. [DOI] [PubMed] [Google Scholar]
- 49. Blundell C, Cornebise J, Kavukcuoglu K, Wierstra D. Weight uncertainty in neural networks. Available at: http://arxiv.org/abs/1505.05424. Published 2015. Accessed April 26, 2019.
- 50. Gal Y, Islam R, Ghahramani Z. Deep bayesian active learning with image data. Available at: http://arxiv.org/abs/1703.02910. Published 2017. Accessed April 26, 2019.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figures S1–S6
Data Availability Statement
The anonymized expert panel–annotated validation data set used in this study is available from the corresponding author upon request. The training data and analytic methods are not available to other researchers.


