Abstract
The taxonomic positions of two novel aerobic, Gram-stain-positive Actinobacteria, designated RB20T and RB56T, were determined using a polyphasic approach. Both were isolated from the fungus-farming termite Macrotermes natalensis. Results of 16S rRNA gene sequence analysis revealed that both strains are members of the genus Nocardia with the closest phylogenetic neighbours Nocardia miyunensis JCM12860T (98.9 %) and Nocardia nova DSM44481T (98.5 %) for RB20T and Nocardia takedensis DSM 44801T (98.3 %), Nocardia pseudobrasiliensis DSM 44290T (98.3 %) and Nocardia rayongensis JCM 19832T (98.2 %) for RB56T. Digital DNA–DNA hybridization (DDH) between RB20T and N. miyunensis JCM12860T and N. nova DSM 44481T resulted in similarity values of 33.9 and 22.0 %, respectively. DDH between RB56T and N. takedensis DSM44801T and N. pseudobrasiliensis DSM44290T showed similarity values of 20.7 and 22.3 %, respectively. In addition, wet-lab DDH between RB56T and N. rayongensis JCM19832T resulted in 10.2 % (14.5 %) similarity. Both strains showed morphological and chemotaxonomic features typical for the genus Nocardia , such as the presence of meso-diaminopimelic acid (A2pm) within the cell wall, arabinose and galactose as major sugar components within whole cell-wall hydrolysates, the presence of mycolic acids and major phospholipids (diphosphatidylglycerol, phosphatidylethanolamine, phosphatidylinositol), and the predominant menaquinone MK-8 (H4, ω-cyclo). The main fatty acids for both strains were hexadecanoic acid (C16 : 0), 10-methyloctadecanoic acid (10-methyl C18 : 0) and cis-9-octadecenoic acid (C18 : 1 ω9c). We propose two novel species within the genus Nocardia : Nocardia macrotermitis sp. nov. with the type strain RB20T (=VKM Ac-2841T=NRRL B65541T) and Nocardia aurantia sp. nov. with the type strain RB56T (=VKM Ac-2842T=NRRL B65542T).
Keywords: Macrotermes natalensis, Nocardia, termite gut
Introduction
Members of the genus Nocardia are characterized as Gram-positive, non-motile, aerobic bacteria that form a branched mycelium which is easily fragmented forming rod to coccoid-like structures [1]. The genus was established by Trevisan in 1889 [2]. They form a distinct clade within the class Actinobacteria together with Corynebacteriaceae and Mycobacteriaceae due to the presence of mycolic acids in the cell membrane [3].
Like members of these two families, strains of Nocardia have been mostly recognized as pathogens of humans, plants and animals [4–7]. Nonetheless, they were also isolated from soil [8] and as symbionts of plants and marine sponges [9, 10]. In light of these studies, biochemistry- and pharmacology-driven studies have shown that Nocardia species harbour an enormous biosynthetic potential to produce structurally unique natural products with antiviral, antifungal, antibacterial and immunosuppressive functions [11–14].
We have recently focused on the phylogenetic and chemical characterization of Actinobacteria associated with fungus-growing termites [15], which are terrestrial eusocial invertebrates that occupy most available habitats in (sub)tropical regions where they contribute up to 20 % of carbon mineralization in savannah ecosystems [16–19]. Microbial profiling studies of fungus-growing termite species showed that the core community of the termite gut was distinct from those of the lower and higher non-fungus-growing termites, which suggested an adaptation to different nutritional environments in the host gut [20]. Building on microbial profiling studies, we pursued in parallel a cultivation-based approach to analyse the microbial diversity of fungus-growing termite systems [15]. Here, we describe the isolation of two new Nocardia species isolated from the gut of fungus-growing termite Macrotermes natalensis.
Isolation and ecology
In February 2015, termite workers of the genus Macrotermes natalensis were collected from a termite colony Mn160 (25° 44′ 34.7″ S 28° 15′ 38.7″ E, Pretoria, South Africa) and actinobacterial strains RB20T and RB56T were isolated from termite guts as previously described [15]. Chitin agar plates supplemented with 0.05 g l−1 cycloheximide were incubated aerobically for 21 days at 30 °C and checked daily for the appearance of colonies. Single colonies were transferred onto International Streptomyces Project (ISP) 2 medium. The isolated pure cultures of RB20T and RB56T were maintained on ISP2 at 30 °C and as glycerol suspensions (25%, v/v) at −80 °C.
16S rRNA gene phylogeny
Genomic DNA extraction, genome sequencing, PCR amplification and sequencing of the 16S rRNA genes of RB20T and RB56T were carried out as previously described [20]. Additionally, sequences of the 16S rRNA genes of RB20T and RB56T were extracted from whole genome data (accession no. WEGK00000000, WEGI00000000.1) using Artemis [21]. blastn analysis was determined using the NCBI database and the results indicated that strains RB20T and RB56T were members of the genus Nocardia . The 16S rRNA gene sequences of selected Nocardia reference strains were downloaded from the LPSN database (date of access: 2 March 2020) [22] and pairwise sequence similarities were calculated as recommended by Meier-Kolthoff et al. [23] on the GGDC web server [24, 25]. The sina sequence alignment service was used to generate 16S rRNA gene sequence alignments [26]. Phylogenetic trees were reconstructed with mega version 7.0.26 [27] using the neighbour-joining (NJ) [28] and maximum likelihood (ML) [29] algorithms. The evolutionary distance model of Tamura [30] was used to generate evolutionary distance matrices for the algorithms with deletion of complete gaps and missing data. For the ML algorithm, discrete Gamma distribution was used (+G) and the rate variation model allowed for some sites to be evolutionarily invariable (+I). For the NJ algorithm, rate variation among sites was modelled with a gamma distribution. The reliability of the tree topology was evaluated by bootstrap analysis with 1000 resamplings [31].
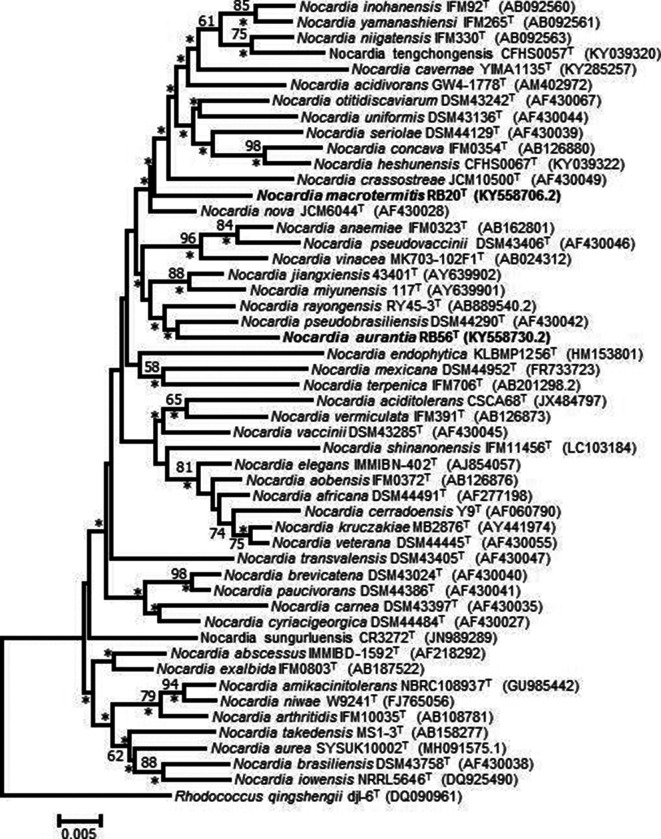
Strain RB20T shared highest 16S rRNA gene similarity with Nocardia miyunensis 117T (=JCM12860T; 98.9 %) [32], Nocardia nova DSM44481T (=JCM6044T; 98.5 %) [33], Nocardia niigatensis IFM330T (=NBRC100131T; 98.4 %) and Nocardia pseudobrasiliensis DSM44290T (=NBRC108224T; 98.3 %) [34]. Strain RB56T shared highest 16S rRNA similarity with Nocardia takedensis DSM44801T (=MS1-3T=NBRC 100417T; 98.3 %) [35], Nocardia pseudobrasiliensis DSM44290T (=NBRC108224T; 98.3 %) and Nocardia rayongensis JCM19832T (=RY45-3T; 98.2 %) [36]. Lower levels of 16S rRNA gene sequence similarity (<98.2 %) were found to all other type strains of Nocardia species (Table S1 and S2, available in the online version of this article).
Phylogenetic analysis using ML and NJ trees indicated that strain RB20T formed a cluster with a larger clade containing N. nova JCM6044T. Strain RB56T clustered with N. rayongensis RY45-3T and N. pseudobrasiliensis DSM44290T (Figs 1 and S1). However, the bootstrap support for the topology of this cluster was very low. Based on the analyses of the 16S rRNA gene sequence similarities and phylogenetic trees, N. miyunensis 117T, N. nova DSM44481T, N. pseudobrasiliensis DSM44290T and N. rayongensis JCM19832T were selected as reference strains.
Fig. 1.
Neighbour-joining tree based on almost-complete 16S rRNA gene sequences showing the relationship between strain RB20T and RB56T and species of the genus Nocardia. Rhodococcus qingshengii djl-6T was used to root the tree. Asterisks donate branches that were also recovered in the maximum-likelihood tree (Fig. S1). Only bootstrap values above 50 % (1000 pseudoreplications) are shown. Bar, 0.005 substitutions per nucleotide position.
Genome features
The DNA G+C content of the genomic DNA was determined from the whole genome sequences [23, 37]. DNA–DNA hybridization (DDH) was performed by the Deutsche Sammlung von Mikroorganismen und Zellkulturen (DSMZ) identification service as a classical wet-lab experiment. The required DNA was obtained as described by Cashion et al. [38] and the DDH experiments were performed in duplicate according to the methods of De Ley et al. [39] and Huss et al. [40]. Furthermore, DDH analysis was performed in silico using genomes deposited at public databases (Tables S3 and S4) [41].
It was recommended by Stackebrandt and Ebers [42] that a 16S rRNA gene sequence similarity range above 98.7–99.0 % requires additional genome analysis to prove the genomic uniqueness of novel isolates. To confirm the novel species status, we then compared DNA–DNA similarities of the closest type strains of the closest species of the genus Nocardia ( N. miyunensis JCM12860T [32], N. nova DSM44481T [33], N. takedensis DSM44801T [35], N. pseudobrasiliensis DSM44290T [34], N. rayongensis JCM19832T [36] and our isolates. First, digital DDH (dDDH) values were determined for RB20T and the closest relatives, N. miyunensis JCM12860T and N. nova DSM44481T, resulting in 33.9 and 22.0 %, respectively (Table S3). For strain RB56T and the closest relatives N. takedensis DSM44801T and N. pseudobrasiliensis DSM44290T the dDDH values were 20.7 and 22.3 %, respectively (Table S4). Wet-lab DDH was performed for RB56T and N. rayongensis JCM19832T due to the lack of whole genome sequence data for N. rayongensis JCM19832T and resulted in a DDH value of 14.5 % (10.2 %). In both cases, the obtained values are below the threshold value of 70 % for the definition of bacterial species recommended by Wayne et al. [43].
Genome analysis of RB20T and RB56T showed that both strains had a similar genome size of approximately 8.6 Mb with 60 contigs for RB20T and 67 contigs for RB56T (Table S5) N50 size of RB20T is 425 626 bp and for RB56T 451 059 bp. Total CDS were 7454 and 7605 for RB20T and RB56T and the genomes had a completeness of 98.9 and 99.7 %. The G+C content was 67.2 % for RB20T and 69.4 % for RB56T, which is typical for this genus (64–72 %) [1].
Physiology and chemotaxonomy
For chemotaxonomic analyses, freeze-dried cells were obtained from culture grown in ISP2 for 3 days at 28 °C on a rotary shaker at 180 r.p.m. The diagnostic diamino acid of the cell wall was determined in whole-cell hydrolysates by paper chromatography according to Hasegawa [44]. Whole-cell sugars were examined according to Schumann [45]. The occurrence of free mycolic acids was determined by TLC as described by Minnikin [46]. Respiratory quinones of the strains were extracted and separated as described by Collins et al. [47] and identified as described by Wink et al. [48]. To verify the occurrence of the menaquinone MK-8 (H4, ω-cyclo) type strains of N. asterioides (IMET 7547T) [49, 50] and N. carnea (IMET 7504T) producing this menaquinone were analyzed in parallel. Polar lipids were extracted by the method described by Minnikin [51] and identified by two-dimensional thin-layer chromatography as described by Collins and Jones [52]. Extraction and analysis of fatty acids was done by the DSMZ Identification service by described standard methods [53]. The glycolysation of the muramic acid of the peptidoglycan was analyzed as described by Schumann [45]. The reference strain investigated in parallel was Rhodococcus rhodochrous IMET 7374T containing glycolyl and Nocardoides albus IMET 7807T containing acetyl muramic acid. Gram-staining was performed as described by Kamlage et.al. [54]. Acid fastness was tested by the methods described by Rohde [55]. Decomposition of purines, tyrosine and organic acids was tested using the method described by Gordon et al. [56]. Antibiotic susceptibility tests were performed with yeast malt agar using the method described previously [57]. Antibiotics were purchased from Bio-Rad, bioMérieux, Difco, BD and BBL.
Morphological characteristics of the strains were determined on cultures grown for 5–14 days on ISP2 agar (ISP2 containing additional 20 g l−1 agar) at 30 °C using light microscope (Imager M2, Carl Zeiss) and a field emission scanning electron microscope. Scanning electron microscopy was performed as described by Groth et al. [58]. Culture characteristics were determined on various ISP media for up to 18 days according to Shirling and Gottlieb [59] and similar to the approach described by Wink et al. [60]. Anaerobic and microaerophilic growth was tested by cultivating the strains at 28 °C in chambers with anaerobic or microaerophilic atmosphere generated by GENbox anaerob or GENbox microaer (bioMérieux cat. nos. 96124 and 96125). Colony colour was determined using Baumann’s Farbatlas 1 (Paul Baumann/Aue). Carbohydrate utilization was determined using ISP9 (carbon utilization medium) supplemented with 1 % sole carbon source. Melanoid pigment production was examined on peptone–yeast extract iron agar (ISP6), tyrosine agar (ISP7) and a synthetic medium from Suter [61] with and without tyrosine (1 g l−1). Sodium chloride tolerance was tested on ISP2 by changing sodium chloride concentrations from 1–15 %. The pH tolerance (pH range 4–10) was tested in ISP2 broth using a buffer system described by Xu et al. [62].
Whole-cell hydrolysates of RB20T and RB56T contained meso-diaminopimelic acid and the carbohydrates arabinose, galactose and traces of glucose. Free mycolic acids were present. The muramic acid of the peptidoglycan of both strains was glycosylated. Both strains were acid fast.
The predominant menaquinone MK-8 (H4, ω-cyclo) and small amounts of menaquinone MK-9(H2) were detected in both strains (Table 1).
Table 1.
Physiological properties that separate the isolates from the type strains of phylogenetically close Nocardia species
Strains: 1, RB20T; 2, Nocardia miyunensis JCM 12860T; 3, Nocardia nova DSM 44481T; 4, RB56T, 5, Nocardia takedensis DSM 44801T; 6, Nocardia pseudobrasiliensis DSM 44290T; 7, Nocardia rayongensis JCM 19832T. Data were taken from this study and previous studies [32–36]. Utilization tests were analyzed as followed: ++, grows better than positive control (basal medium with glucose); +, grows like positive control (basal medium with glucose); (+), better than negative control but not like positive control; −, not better than negative control (basal medium with water). All strains were positive for utilization of d-glucose and negative for utilization of raffinose and cellulose. Decomposition of purines, tyrosine and organic acids: −, no decomposition; (+), weak decomposition; +, decomposition; ++, very good decomposition.
|
Characteristics |
1 |
2 |
3 |
4 |
5 |
6 |
7 |
|---|---|---|---|---|---|---|---|
|
DNA G+C content (mol%) |
67.2 |
67.0 |
67.3 |
69.4 |
68.6 |
67.1 |
71.0 |
|
Chemotaxonomic |
|
|
|
|
|
|
|
|
Major menaquinone* |
MK-8 (H4, ω-cyclo) |
MK-8 (H6, ω-cyclo)† |
MK-8 (H4, ω-cyclo)‡ |
MK-8* (H4, ω-cyclo) |
MK-8 (H4, ω-cyclo)§ |
MK-8 (H4, ω-cyclo)$ |
MK-8 (H4, ω-cyclo)¶ |
|
Major fatty acids |
C16 : 0, 10-methyl C18 : 0 |
C16 : 0, 10-methyl C18 : 0 |
C16 : 0, C18 : 1 ω9c |
C16 : 0, C18 : 1 ω9c |
C16 : 0, C18 : 1 ω9c |
C16 : 0, C18 : 1 ω9c |
C16 : 0, C18 : 1 ω9c |
|
pH tolerance range for growth |
5–7 |
4–8 |
4–8 |
5–7 |
6–8 |
5–9 |
4–7 |
|
Optimum pH for growth |
6–7 |
6–7 |
6–7 |
6–7 |
6–7 |
6–7 |
6–7 |
|
Temperature growth range (°C) |
15–37 |
15–37 |
15–37 |
15–37 |
15–37 |
15–45 |
15–37 |
|
Optimum temperature for growth (°C) |
28 |
28 |
28 |
28 |
28 |
28 |
28 |
|
Anaerobic growth |
− |
− |
− |
− |
− |
− |
− |
|
Microaerophilic growth |
+ |
+ |
+ |
(+) |
+ |
+ |
+ |
|
Growth at NaCl concentration (% w/v) |
0–3 |
0–3 |
0–7 |
0–1 |
0–3 |
0–9 |
0–3 |
|
Utilization of sole carbon sources |
|
|
|
|
|
|
|
|
Sucrose |
(+) |
+ |
(+) |
(+) |
− |
− |
− |
|
d-Arabinose |
+ |
+ |
+ |
+ |
− |
− |
− |
|
d-Xylose |
+ |
+ |
(+) |
+ |
− |
− |
+ |
|
Inositol |
+ |
− |
− |
− |
− |
+ |
− |
|
d-Mannitol |
+ |
++ |
− |
− |
− |
+ |
(+) |
|
d-Fructose |
+ |
++ |
− |
(+) |
− |
+ |
+ |
|
l-Rhamnose |
+ |
− |
− |
+ |
− |
− |
− |
|
Decomposition of purines, tyrosine and organic acids |
|
|
|
|
|
|
|
|
Citrate |
− |
(+) |
− |
− |
− |
+ |
(+) |
|
Lactate |
− |
− |
− |
− |
(+) |
− |
− |
|
Acetate |
− |
(+) |
+ |
(+) |
− |
(+) |
(+) |
|
Propionate |
+ |
+ |
+ |
(+) |
(+) |
+ |
(+) |
|
Malate |
++ |
+ |
+ |
+ |
− |
+ |
+ |
|
Pyruvate |
(+) |
+ |
− |
(+) |
+ |
+ |
(+) |
|
Tyrosine |
− |
− |
(+) |
− |
− |
(+) |
− |
|
Adenine |
− |
− |
+ |
− |
− |
+ |
− |
|
Hypoxanthine |
(+) |
+ |
+ |
(+) |
+ |
+ |
+ |
|
Xanthine |
− |
− |
− |
− |
− |
− |
− |
Strains RB20T and RB56T both exhibited similar polar lipid profiles with the major compounds diphosphatidylglycerol, phosphatidylethanolamine, phosphatidylinositol, phosphatidylinositol mannoside, two phospholipids (PL1, PL2) and two glycolipids (GL1, GL2). Strain RB20T contained additional two unpolar lipids (L1, L2) and more polar lipid L3, which were not present in RB56T. In contrast, RB56T revealed a third phospholipid (PL3) as well as two polar lipids (L4, L5), which is different from the lipids of RB20T (Fig. S5).
The overall cellular fatty acid profiles of RB20T and RB56T were consistent with those of the genus Nocardia (Tables 1 and S7). The fatty acid profile of strain RB20T was composed of the major fatty acids C16 : 0 (39.6 %), C18 : 0 10-methyl (19.0 %), C18 : 1 ω9c (13.4 %) and C18 : 0 (9.9 %). The closest-related species, N. miyunensis JCM12860T and N. nova DSM44481T, had similar fatty acid profiles and contained predominant amounts of C16 : 0 (39.5 and 38.3 %), C18 : 0 10-methyl (18.8 and 14.7 %) and C18 : 1 ω9c (13.9 and 16.6 %).
In comparison, the fatty acid profile of strain RB56T exhibited the major fatty acids C16 : 0 (42.8 %), C18 : 1 ω9c (16.1 %) and C18 : 0 10-methyl (12.6 %) and minor amounts of C14 : 0 (6.9 %). The closest relative N. takedensis DSM44801T was characterized by a relatively high amount of C18 : 1ω9c (27.4 %) and the presence of C18 : 0 (4.3 %) and C20 : 1ω9c (4.4 %) (Tables 1 and S7).
The following morphological and phenotypic characteristics were documented for strains RB20T and RB56T, respectively (Table 1).
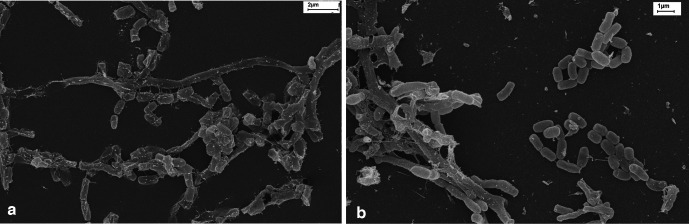
Strain RB20T grew at a pH range from pH 5 to 7 (optimum, pH 7) and at 0–3 % (w/v) NaCl (optimum, 0–1 %). Strain RB20T tolerated a temperature range from 15 to 37 °C, but with only weak growth at 15 and 37 °C, and an optimal growth temperature of 28 °C. RB20T showed fragmenting hyphae into short rod or coccoid forms (Fig. 2a).
Fig. 2.
Scanning electron micrograph images of strain RB20T (left) and strain RB56T (right) cultivated at 28 °C on ISP3 agar for 14 days. Bars, 2 µm for RB20T and 1 µm for RB56T.
Strain RB20T grew well on ISP1–ISP7 and Suter medium. White aerial mycelium was well developed on ISP1–5 and ISP7. The substrate mycelium was beige-white on ISP1, ISP4 and ISP5, beige-orange on ISP2 and ISP3, and greyish ISP6 and Suter medium (Fig. S2). Overall, strain RB20T exhibited different phenotypic characteristics compared to the reference strains N. miyunensis JCM12860T, N. nova DSM44481T and N. pseudobrasiliensis DSM44290T (Fig. S2, Table S6) with the following major differences: While RB20T showed good growth on ISP2, ISP6 and Suter medium (+Tyr) and white-beige aerial and substrate mycelium, the closest relative N. miyuensis JCM 12860T showed only very weak growth and ochre to orange aerial and substrate mycelium on ISP2 and ISP6, and good growth but ochre aerial and substrate mycelium on Suter medium (+Tyr). Similarly, RB20T exhibited white-beige aerial and substrate mycelium on ISP7, while N. nova DSM 44481T produced orange aerial and substrate mycelium.
Strain RB56T grew at pH range 5–7 (optimum, pH 7.0) and at 0–1 % (w/v) NaCl (optimum, 0 %). Strain RB56T tolerated a temperature range of 15–37 °C, with only weak growth at 15, 37 and 45 °C. The optimal growth temperature was 28 °C. On ISP2 medium, strain RB56T formed short, round and ellipsoidal cells (Fig. 2b).
Strain RB56T showed good growth on ISP2, ISP5 and ISP7, moderate growth on ISP1, ISP3, ISP4, and weak growth on ISP6 and Suter medium (Figs S3 and S4, Table S6). The substrate mycelium was orange on ISP1, ISP2, ISP6, ISP7 and Suter medium, white on ISP3, yellowish-white on ISP4 and orange-yellow on ISP5. White aerial mycelium developed on ISP3–5, white yellowish aerial mycelium on ISP7 and very poor orange aerial mycelium on ISP1 and ISP2. A soluble reddish pigment was observed on ISP7.
Overall, strain RB56T exhibited different phenotypic characteristics to the reference strains N. takedensis DSM44801T, N. pseudobrasiliensis DSM44290T and N. rayongensis JCM19832T with the following major differences: While RB56T showed good growth on ISP3 and white aerial and substrate mycelium, N. takedensis DSM 44801T showed only weak growth and orange yellow substrate mycelium. Similarly, N. pseudobrasiliensis DSM 44290T and N. rayongensis JCM 19832T showed good growth on ISP4 with white aerial and substrate mycelium, whilst RB56T and N. takedensis DSM 44801T grew only moderately to weakly with yellowish to orange substrate mycelium. Finally, growth of RB56T was only weak on Suter medium (with/without tyrosine), whilst all reference strains grew well showing orange to brown soluble pigmentation.
Both strains, RB20T and RB56T, were resistant to oxytetracycline, azlocillin, lincomycin, trimethoprim, carbenicillin, piperacillin, cefoxitin, mezlocillin, penicillin G, cephalothin and chlortetracycline.
Strain RB20T was furthermore resistant against tetracycline and novobiocin, whereas RB56T exhibited resistance against polymyxin B and erythromycin (Table S8).
The morphological, physiological, genetic and chemotaxonomic data support the delineation of RB20T and RB56T as two novel species of the genus Nocardia .
Description of Nocardia macrotermitis sp. nov.
Nocardia macrotermitis (ma.cro.ter´mi.tis. N.L. gen. n. macrotermitis, of the termite Macrotermes, from where the organism was first isolated).
Cells are Gram-stain-positive, aerobic and acid-fast. Colonies form branched vegetative mycelium that fragment into short rod and coccoid forms. Good growth occurs on all media tested within 12 days. Aerial mycelium is formed on ISP2, ISP3, ISP5, ISP7 and Suter and poorly on ISP1, ISP4 and ISP6.
The pH range for growth is pH 5–7. Growth temperature range is between 15–45 °C with optimum growth at 28 °C. The maximum concentration of NaCl for growth is 3 % (w/v). The strain grows on the following sole carbon sources: glucose, arabinose, xylose, inositol, mannitol, fructose and rhamnose; but does not grow with raffinose and cellulose.
The strain is able to utilize the organic acids propionate, malate and decomposes pyruvate and hypoxanthine weakly. Resistant to oxytetracycline, azlocillin, lincomycin, tetracycline, chlortetracycline, trimethoprim, carbenicillin, piperacillin, cefoxitin, mezlocillin, penicillin G, cephalothin and novobiocin.
The diagnostic diamino acid of the cell wall is meso-diaminopimelic acid (meso-A2pm). Mycolic acids and N-glycosylmuramic acid in the glycan part of the peptidoglycan are present.
The sugars in whole-cell hydrolysates are arabinose, galactose and traces of glucose. Major polar lipids are diphosphatidylglycerol, phosphatidylethanolamine, phosphatidylinositol, phosphatidylinositol mannoside, two phospholipids (PL1, PL2) and two glycolipids (GL1, GL2). Furthermore, two unpolar lipids (L1, L2) and a polar lipid (L3) are present. The predominant menaquinone is MK-8(H4, ω-cyclo). The major fatty acids are hexadecanoic acid (C16 : 0), 10-methyl octadecanoic acid (10-methyl C18 : 0), cis-9-octadecenoic acid (C18 : 1 ω9c) and octadecanoic acid (C18 : 0). The DNA G+C content of strain RB20T is 67.2 mol%.
The type strain, RB20T (=VKM Ac-2841T=NRRL-B65541T), was isolated from the gut of the termite Macrotermes natalensis (major worker). The strain has been deposited in the All-Russian Collection of Microorganisms (=VKM Ac-2841T) and the Agricultural Research Service Culture Collection (=NRRL B65541T). The GenBank/EMBL accession number for the partial 16S rRNA gene sequence is KY558706.2. This Whole Genome Shotgun project has been deposited at DDBJ/ENA/GenBank under the accession WEGK00000000 (Bio project PRJNA545686, Biosample SAMN11902338). The version described in this paper is version WEGK01000000.
Description of Nocardia aurantia sp. nov.
Nocardia aurantia (au.ran'ti.a. N.L. fem. adj. aurantia, orange-coloured, referring to the gold-coloured substrate mycelium).
Cells are Gram-stain-positive, aerobic and acid-fast. Colonies form branched vegetative mycelium that fragment into short rod and coccoid forms. Good growth occurs on ISP2, ISP5 and ISP7, moderate growth on ISP1, ISP3 and ISP4, and poor growth on ISP6 and Suter medium. Aerial mycelium is formed on ISP3, ISP4 and ISP7 media, very poorly on ISP1, ISP2 and ISP5, but not at all on ISP6 or Suter medium. Short, round, ellipsoidal spores are formed. A reddish pigment is produced on ISP7.
The pH range for growth is pH 5–7. The growth temperature range is 15–37 °C with optimal growth at 28 °C. The maximum concentration of NaCl for growth is 1 % (w/v). The strain grows on the following sole carbon sources: glucose, arabinose, xylose and rhamnose; but not with raffinose, cellulose, inositol or mannitol.
The strain is able to weakly utilize the organic acids acetate, propionate, pyruvate and hypoxanthine, and decomposes malate. Resistant to oxytetracycline, azlocillin, lincomycin, trimethoprim, carbenicillin, piperacillin, cefoxitin, mezlocillin penicillin G, cephalothin, chlortetracycline, polymyxin B and erythromycin.
The diagnostic diamino acid of the cell wall is meso-diaminopimelic acid (meso-A2pm). Mycolic acids and N-glycosylmuramic acid in the glycan part of the peptidoglycan are present.
Whole-cell hydrolysates contain arabinose, galactose and traces of glucose. Major polar lipids are diphosphatidylglycerol, phosphatidylethanolamine, phosphatidylinositol, phosphatidylinositol mannoside, three unidentified phospholipids (PL1, PL2 and PL3), two unknown lipids and two glycolipids (GL1, GL2). The predominant menaquinone is MK-8(H4, ω-cyclo). The major fatty acids are hexadecaonic acid (C16 : 0), cis-9-octadecenoic acid (C18 : 1 cis-9) and 10-methyl octadecanoic acid (10-methyl C18 : 0). The DNA G+C content of strain RB56T is 69.4 mol%.
The type strain, RB56T (=VKM Ac-2842T=NRRL-B65542T), was isolated from the gut of the termite Macrotermes natalensis (major worker). The strain has been deposited in the All-Russian Collection of Microorganisms (=VKM Ac-2842T) and the Agricultural Research Service Culture Collection (=NRRL B65542T). The GenBank/EMBL accession number for the partial 16S rRNA gene sequence is KY558730.2. This Whole Genome Shotgun project has been deposited at DDBJ/ENA/GenBank under the accession WEGI00000000 (Bio project PRJNA545686, Biosample SAMN11902338). The version described in this paper is version WEGI01000000.
Supplementary Data
Funding information
J.S and R.B. were funded by the International Leibniz Research School for Microbial and Biomolecular Interactions (ILRS). Furthermore R.B was funded by the Jena School for Microbial Communication (JSMC, DFG). Financial support from the Boehringer Ingelheim Foundation, the Daimler Benz foundation and the German Research Foundation (CRC 1127 (ChemBioSys, A6) and BE-4799/3–1) to C.B. and the Villum Kann Rasmussen foundation for a Young Investigator Fellowship (VKR10101) to M.P. is greatly acknowledged.
Acknowledgements
We thank the Oerlemans family (Mookgophong) for permission to sample colonies at their farm. We thank Susanne Linde (Elektronenmikroskopisches Zentrum FSU Jena) for electron microscopy pictures.
Author contributions
Authors contributed equally to the manuscript
Conflicts of interest
The authors declare that there are no conflicts of interest.
Footnotes
Abbreviations: DDH, DNA–DNA hybridization; ISP, International Streptomyces Project; ML, maximum-likelihood; NJ, neighbour-joining.
Five supplementary figures and eight supplementary tables are available with the online version of this article.
The GenBank/EMBL accession numbers for the partial 16S rRNA gene sequences of strains RB20T and RB56T are KY558706.2 and KY558730.2, respectively. This Whole Genome Shotgun project has been deposited at DDBJ/ENA/GenBank under the accessions WEGK00000000.1 and WEGI00000000.1, respectively. The versions described in this paper are version WEGK01000000.1 and WEGI01000000.1, respectively.
References
- 1.Goodfellow M, Maldonado LA. The families Dietziaceae, Gordoniaceae, Nocardiaceae and Tsukamurellaceae. In: Dworkin M, Falkow S, Rosenberg E, Schleifer KH, Stackebrandt E, editors. The Prokaryotes. USA: Springer-Verlag; 2006. pp. 843–888. [Google Scholar]
- 2.Trevisan V. I generi E Le specie delle battieriacee. 1889; Milan
- 3.Embley TM, Stackebrandt E. The molecular phylogeny and systematics of the actinomycetes. Annu Rev Microbiol. 1994;48:257–289. doi: 10.1146/annurev.mi.48.100194.001353. [DOI] [PubMed] [Google Scholar]
- 4.Demaree JB, Smith NR. Nocardia vaccinii n. sp. causing galls on Blue-berry plants. Phytopathology. 1952;42:249–252. [Google Scholar]
- 5.Kudo T, Hatai K, Seino A. Nocardia seriolae sp. nov. causing nocardiosis of cultured fish. Int J Syst Bacteriol. 1988;38:173–178. doi: 10.1099/00207713-38-2-173. [DOI] [Google Scholar]
- 6.Friedman CS, Beaman BL, Chun J, Goodfellow M, Gee A, et al. Nocardia crassostreae sp. nov., the causal agent of nocardiosis in Pacific oysters. Int J Syst Bacteriol. 1998;48:237–246. doi: 10.1099/00207713-48-1-237. [DOI] [PubMed] [Google Scholar]
- 7.McNeil MM, Brown JM, Magruder CH, Shearlock KT, Saul RA, et al. Disseminated Nocardia transvalensis infection: an unusual opportunistic pathogen in severely immunocompromised patients. J Infect Dis. 1992;165:175–178. doi: 10.1093/infdis/165.1.175. [DOI] [PubMed] [Google Scholar]
- 8.Cross T, Rowbotham EN, Mishustin E, Tepper Z, Antoine-Prtaels F, et al. Ecology of nocardioform actinomycetes. Academic Press; 1976. pp. 337–371. [Google Scholar]
- 9.Thawai C, Rungjindamai N, Klanbut K, Tanasupawat S. Nocardia xestospongiae sp. nov., isolated from a marine sponge in the Andaman sea. Int J Syst Evol Microbiol. 2017;67:1451–1456. doi: 10.1099/ijsem.0.001736. [DOI] [PubMed] [Google Scholar]
- 10.Xing K, Qin S, Fei S-M, Lin Q, Bian G-K, et al. Nocardia endophytica sp. nov., an endophytic actinomycete isolated from the oil-seed plant Jatropha curcas L . Int J Syst Evol Microbiol. 2011;61:1854–1858. doi: 10.1099/ijs.0.027391-0. [DOI] [PubMed] [Google Scholar]
- 11.Männle D, McKinnie SMK, Mantri SS, Steinke K, Lu Z, et al. Comparative Genomics and Metabolomics in the Genus Nocardia . mSystems. 2020;5:e00125–20. doi: 10.1128/mSystems.00125-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mukai A, Fukai T, Matsumoto Y, Ishikawa J, Hoshino Y, et al. Transvalencin Z, a new antimicrobial compound with salicylic acid residue from Nocardia transvalensis IFM 10065. J Antibiot. 2006;59:366–369. doi: 10.1038/ja.2006.53. [DOI] [PubMed] [Google Scholar]
- 13.Komatsu K, Tsuda M, Shiro M, Tanaka Y, Mikami Y, et al. Brasilicardins B-D, new tricyclic terpenoids [correction of terpernoids] from actinomycete Nocardia brasiliensis . Bioorg Med Chem. 2004;12:5545–5551. doi: 10.1016/j.bmc.2004.08.007. [DOI] [PubMed] [Google Scholar]
- 14.Kunimoto T, Sawa T, Wakashiro T, Hori M, Umezawa H. Biosynthesis of the formycin family. J Antibiot. 1971;24:253–258. doi: 10.7164/antibiotics.24.253. [DOI] [PubMed] [Google Scholar]
- 15.Benndorf R, Guo H, Sommerwerk E, Weigel C, Garcia-Altares M, et al. Natural products from Actinobacteria associated with fungus-growing termites. Antibiotics. 2018;7:E83. doi: 10.3390/antibiotics7030083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ramadhar TR, Beemelmanns C, Currie CR, Clardy J. Bacterial symbionts in agricultural systems provide a strategic source for antibiotic discovery. J Antibiot. 2014;67:53–58. doi: 10.1038/ja.2013.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Aanen DK, Eggleton P, Rouland-Lefevre C, Guldberg-Froslev T, Rosendahl S, et al. The evolution of fungus-growing termites and their mutualistic fungal symbionts. Proc Natl Acad Sci U S A. 2002;99:14887–14892. doi: 10.1073/pnas.222313099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.da Costa RR, Hu H, Li H, Poulsen M. Symbiotic plant biomass decomposition in fungus-growing termites. Insects. 2019;10:87. doi: 10.3390/insects10040087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Poulsen M. Towards an integrated understanding of the consequences of fungus domestication on the fungus-growing termite gut microbiota, Environ . Microbiol. 2015;17:2562–2572. doi: 10.1111/1462-2920.12765. [DOI] [PubMed] [Google Scholar]
- 20.Otani S, Mikaelyan A, Nobre T, Hansen LH, Koné N'Golo A, Koné NA, et al. Identifying the core microbial community in the gut of fungus-growing termites. Mol Ecol. 2014;23:4631–4644. doi: 10.1111/mec.12874. [DOI] [PubMed] [Google Scholar]
- 21.Rutherford K, Parkhill J, Crook J, Horsnell T, Rice P, et al. Artemis: sequence visualization and annotation. Bioinformatics. 2000;16:944–945. doi: 10.1093/bioinformatics/16.10.944. [DOI] [PubMed] [Google Scholar]
- 22.Parte AC. LPSN--list of prokaryotic names with standing in nomenclature. Nucleic Acids Res. 2014;42:D613–D616. doi: 10.1093/nar/gkt1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Meier-Kolthoff JP, Göker M, Spröer C, Klenk H-P. When should a DDH experiment be mandatory in microbial taxonomy? Arch Microbiol. 2013;195:413–418. doi: 10.1007/s00203-013-0888-4. [DOI] [PubMed] [Google Scholar]
- 24.Meier-Kolthoff JP, Auch AF, Klenk H-P, Göker M. Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinformatics. 2013;14:60. doi: 10.1186/1471-2105-14-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Available from http://ggdc.dsmz.de/
- 26.Pruesse E, Peplies J, Glöckner FO. SINA: accurate high-throughput multiple sequence alignment of ribosomal RNA genes. Bioinformatics. 2012;28:1823–1829. doi: 10.1093/bioinformatics/bts252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kumar S, Stecher G, Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 29.Felsenstein J. Evolutionary trees from DNA sequences: a maximum likelihood approach. J Mol Evol. 1981;17:368–376. doi: 10.1007/BF01734359. [DOI] [PubMed] [Google Scholar]
- 30.Tamura K. Estimation of the number of nucleotide substitutions when there are strong transition-transversion and G+C-content biases. Mol Biol Evol. 1992;9:678–687. doi: 10.1093/oxfordjournals.molbev.a040752. [DOI] [PubMed] [Google Scholar]
- 31.Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 32.Cui Q, Wang L, Huang Y, Liu Z, Goodfellow M. Nocardia jiangxiensis sp. nov. and Nocardia miyunensis sp. nov., isolated from acidic soils. Int J Syst Evol Microbiol. 2005;55:1921–1925. doi: 10.1099/ijs.0.63644-0. [DOI] [PubMed] [Google Scholar]
- 33.Tamura T, Ohji S, Ichikawa N, Hosoyama A, Yamazoe A, et al. Reclassification of Nocardia species based on whole genome sequence and associated phenotypic data. J Antibiot. 2018;71:633–641. doi: 10.1038/s41429-018-0043-1. [DOI] [PubMed] [Google Scholar]
- 34.Ruimy R, Riegel P, Carlotti A, Boiron P, Bernardin G, et al. Nocardia pseudobrasiliensis sp. nov., a new species of Nocardia which groups bacterial strains previously identified as Nocardia brasiliensis and associated with invasive diseases. Int J Syst Bacteriol. 1996;46:259–264. doi: 10.1099/00207713-46-1-259. [DOI] [PubMed] [Google Scholar]
- 35.Yamamura H, Hayakawa M, Nakagawa Y, Tamura T, Kohno T, et al. Nocardia takedensis sp. nov., isolated from moat sediment and scumming activated sludge. Int J Syst Evol Microbiol. 2005;55:433–436. doi: 10.1099/ijs.0.63189-0. [DOI] [PubMed] [Google Scholar]
- 36.Tanasupawat S, Phongsopitanun W, Suwanborirux K, Ohkuma M, Kudo T. Nocardia rayongensis sp. nov., isolated from Thai peat swamp forest soil. Int J Syst Evol Microbiol. 2016;66:1950–1955. doi: 10.1099/ijsem.0.000971. [DOI] [PubMed] [Google Scholar]
- 37.Auch AF, Klenk H-P, Göker M. Standard operating procedure for calculating genome-to-genome distances based on high-scoring segment pairs. Stand Genomic Sci. 2010;2:142–148. doi: 10.4056/sigs.541628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cashion P, Holder-Franklin MA, McCully J, Franklin M. A rapid method for the base ratio determination of bacterial DNA. Anal Biochem. 1977;81:461–466. doi: 10.1016/0003-2697(77)90720-5. [DOI] [PubMed] [Google Scholar]
- 39.De Ley J, Cattoir H, Reynaerts A. The quantitative measurement of DNA hybridization from renaturation rates. Eur J Biochem. 1970;12:133–142. doi: 10.1111/j.1432-1033.1970.tb00830.x. [DOI] [PubMed] [Google Scholar]
- 40.Huss VA, Festl H, Schleifer KH. Studies on the spectrophotometric determination of DNA hybridization from renaturation rates. Syst Appl Microbiol. 1983;4:184–192. doi: 10.1016/S0723-2020(83)80048-4. [DOI] [PubMed] [Google Scholar]
- 41.Goris J, Konstantinidis KT, Klappenbach JA, Coenye T, Vandamme P, et al. DNA-DNA hybridization values and their relationship to whole-genome sequence similarities. Int J Syst Evol Microbiol. 2007;57:81–91. doi: 10.1099/ijs.0.64483-0. [DOI] [PubMed] [Google Scholar]
- 42.Stackebrandt E, Ebers J. Taxonomic parameters revisited: tarnished gold standards. Microbiol Today. 2006;33:152–155. [Google Scholar]
- 43.Wayne LG, Moore WEC, Stackebrandt E, Kandler O, Colwell RR, et al. Report of the ad hoc committee on reconciliation of approaches to bacterial systematics. Int J Syst Evol Microbiol. 1987;37:463–464. doi: 10.1099/00207713-37-4-463. [DOI] [Google Scholar]
- 44.Hasegawa T, Takizawa M, Tanida S. A rapid analysis for chemical grouping of aerobic actinomycetes. J Gen Appl Microbiol. 1983;29:319–322. doi: 10.2323/jgam.29.319. [DOI] [Google Scholar]
- 45.Schumann P. Peptidoglycan structure. Meth Microbiol. 2011;38:101–129. [Google Scholar]
- 46.Minnikin DE, Alshamaony L, Goodfellow M. Differentiation of Mycobacterium, Nocardia, and related taxa by thin-layer chromatographic analysis of whole-organism methanolysates. J Gen Microbiol. 1975;88:200–204. doi: 10.1099/00221287-88-1-200. [DOI] [PubMed] [Google Scholar]
- 47.Collins MD, Pirouz T, Goodfellow M, Minnikin DE. Distribution of menaquinones in actinomycetes and corynebacteria. J Gen Microbiol. 1977;100:221–230. doi: 10.1099/00221287-100-2-221. [DOI] [PubMed] [Google Scholar]
- 48.Wink J, Schumann P, Atasayar E, Klenk H-P, Zaburannyi N, et al. ‘Streptomyces caelicus’, an antibiotic-producing species of the genus Streptomyces, and Streptomyces canchipurensis Li et al. 2015 are later heterotypic synonyms of Streptomyces muensis Ningthoujam et al. 2014. Int J Syst Evol Microbiol. 2017;67:548–556. doi: 10.1099/ijsem.0.001612. [DOI] [PubMed] [Google Scholar]
- 49.Gordon RE, Mihm JM. A comparison of Nocardia asteroides and Nocardia brasiliensis . J Gen Microbiol. 1959;20:129–135. doi: 10.1099/00221287-20-1-129. [DOI] [PubMed] [Google Scholar]
- 50.Roth A, Andrees S, Kroppenstedt RM, Harmsen D, Mauch H. Phylogeny of the genus Nocardia based on reassessed 16S rRNA gene sequences reveals underspeciation and division of strains classified as Nocardia asteroides into three established species and two unnamed taxons. J Clin Microbiol. 2003;41:851–856. doi: 10.1128/JCM.41.2.851-856.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Minnikin DE, Collins MD, Goodfellow M. Fatty acid and polar lipid composition in the classification of Cellulomonas, Oerskovia and related taxa. J Appl Bacteriol. 1979;47:87–95. doi: 10.1111/j.1365-2672.1979.tb01172.x. [DOI] [PubMed] [Google Scholar]
- 52.Collins MD, Jones D. Lipids in the classification and identification of coryneform bacteria containing peptidoglycans based on 2, 4-diaminobutyric acid. J Appl Bacteriol. 1980;48:459–470. doi: 10.1111/j.1365-2672.1980.tb01036.x. [DOI] [Google Scholar]
- 53.Fatty Acid Analysis Available from. https://www.dsmz.de/services/services-microorganisms/identification/analysis-of-cellular-fatty-acids.html
- 54.Kamlage B. Methods for general and molecular bacteriology. In: Gerhardt P, Murray RGE, Wood WA, Krieg NR, editors. 791 pages, numerous figures and tables. Washington, DC: American Society for Microbiology; 1994. [Google Scholar]
- 55.Rohde M. Microscopy. In: Rainey F, Oren A, editors. Methods in Microbiology. 1st ed. Vol. 38. Oxford, UK: Academic Press; 2011. pp. 61–100. [Google Scholar]
- 56.Gordon RE, Barnett DA, Handerhan JE, Pang CHN. Nocardia coeliaca, Nocardia autotrophica, and the Nocardin strain. Int J Syst Bacteriol. 1974;24:54–63. doi: 10.1099/00207713-24-1-54. [DOI] [Google Scholar]
- 57.Groth I, Schütze B, Boettcher T, Pullen CB, Rodriguez C, et al. Kitasatospora putterlickiae sp. nov., isolated from rhizosphere soil, transfer of Streptomyces kifunensis to the genus Kitasatospora as Kitasatospora kifunensis comb. nov., and emended description of Streptomyces aureofaciens Duggar 1948. Int J Syst Evol Microbiol. 2003;53:2033–2040. doi: 10.1099/ijs.0.02674-0. [DOI] [PubMed] [Google Scholar]
- 58.Groth I, Schumann P, Rajney FA, Martin K, Schuetze B, et al. Bogoriella caseilytica gen. nov., sp. nov., a new alkaliphilic actinomycete from a soda lake in Africa. Int J Syst Bacteriol. 1997;47:788–794. doi: 10.1099/00207713-47-3-788. [DOI] [PubMed] [Google Scholar]
- 59.Shirling EB, Gottlieb D. Methods for characterization of Streptomyces species. Int J Syst Bacteriol. 1966;16:313–340. doi: 10.1099/00207713-16-3-313. [DOI] [Google Scholar]
- 60.Wink JM. Compendium of actinobacteria methods for the taxonomic description of the actinobacteria. Available at: https://www.dsmz.de/collection/catalogue/microorganisms/special-groups-of-organisms/compendium-of-actinobacteria
- 61.Suter MA. Isolierung und Charakterisierung von Melanin-negativen Mutanten aus Streptomyces glaucescens . Diss. Naturwiss. ETH Zürich, Nr. 6276, 0000. 1978 [Google Scholar]
- 62.Xu P, Li W-J, Tang S-K, Zhang Y-Q, Chen G-Z, et al. Naxibacter alkalitolerans gen. nov., sp. nov., a novel member of the family 'Oxalobacteraceae' isolated from China. Int J Syst Evol Microbiol. 2005;55:1149–1153. doi: 10.1099/ijs.0.63407-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.