Abstract
Thirty-three Yersinia strains previously characterized by the French Yersinia National Reference Laboratory (YNRL) and isolated from humans and animals were suspected to belong to six novel species by a recently described core genome multilocus sequence typing scheme. These strains and five additional strains from the YNRL were characterized using a polyphasic taxonomic approach including a phylogenetic analysis based on 500 core genes, determination of average nucleotide identity (ANI), determination of DNA G+C content and identification of phenotypic features. Phylogenetic analysis confirmed that the 38 studied strains formed six well-demarcated clades. ANI values between these clades and their closest relatives were <94.7 % and ANI values within each putative novel species were >97.5 %. Distinctive biochemical characteristics were identified in five out of the six novel species. All of these data demonstrated that the 38 strains belong to six novel species of the genus Yersinia : Yersinia artesiana sp. nov., type strain IP42281T (=CIP 111845T=DSM 110725T); Yersinia proxima sp. nov., type strain IP37424T (=CIP 111847T=DSM 110727T); Yersinia alsatica sp. nov., type strain IP38850T (=CIP 111848T=DSM 110726T); Yersinia vastinensis sp. nov., type strain IP38594T (=CIP 111844T=DSM 110738T); Yersinia thracica sp. nov., type strain IP34646T (=CIP 111842T=DSM 110736T); and Yersinia occitanica sp. nov., type strain IP35638T (=CIP 111843T=DSM 110739T).
Keywords: cgMLST, species, Yersinia, Yersiniaceae
The genus Yersinia is a member of the order Enterobacteriales and belongs to the family Yersiniaceae [1]. The genus Yersinia includes three prominent human and animal pathogens: Yersinia pestis , the causative agent of plague [2], Yersinia pseudotuberculosis and Yersinia enterocolitica (biotypes 1B, 2, 3, 4 and 5), responsible for enteric yersiniosis [3], and the putatively pathogenic species Yersinia wautersii [4]. Two species are pathogenic only for animals: Yersinia ruckeri is the causative agent of enteric redmouth disease in salmonid fish [5] and Yersinia entomophaga is known to cause disease in larvae of the New Zealand grass grub [6]. In addition, 14 non-pathogenic species have been described since the 1980s: Yersinia aldovae [7], Yersinia aleksiciae [8], Yersinia bercovieri [9], Yersinia frederiksenii [10], Yersinia intermedia [11], Yersinia kristensenii [12], Yersinia massiliensis [13], Yersinia mollaretii [9], Yersinia nurmii [14], Yersinia pekkanenii [15], Yersinia rohdei [16], Yersinia similis [17] and the most recently described Yersinia hibernica [18] and Yersinia canariae [19].
Seven putative novel species were identified from 1348 Yersinia genomes during the recent investigation of the genetic diversity and population structure of the genus Yersinia [20]. The seven clades were named Yersinia frederiksenii 2, Yersinia frederiksenii 3, NEW 2, Yersinia kristensenii 2, Yersinia kristensenii 3, NEW 3 and NEW 4 [20]. NEW 2 corresponds to Yersinia canariae , the species recently described by Nguyen et al. [19]. The present study aimed to determine the taxonomic status of the six remaining putative novel species, using a polyphasic approach.
In order to increase the number of strains included in each clade, we added three, one and one strains to NEW 3, Y. kristensenii 2 and Y. frederiksenii 3 clades, respectively. A total of 38 strains belonging to these six putative novel species were studied (Table 1). All strains belong to the collection of the French Yersinia National Reference Laboratory (YNRL) for plague and other yersiniosis. They were isolated in France from clinical samples and in Germany, Bulgaria and Italy from animals. They were initially identified as Y. enterocolitica biotype 1A (strains of NEW 3 and NEW 4), Y. frederiksenii (strains of Y. frederiksenii 2 and 3) and Y. kristensenii (strains of Y. kristensenii 2 and 3) by phenotypic characterization and considered as new taxa after applying the recently described Yersinia -core genome multilocus sequence typing (cgMLST) [20] (Table 1).
Table 1.
Strains belonging to the six novel species
|
cgMLST cladea |
Proposed novel species |
Strain |
Phenotypic characterization |
Assembly accession number |
Isolation |
|||||
|---|---|---|---|---|---|---|---|---|---|---|
|
Species |
Serotype |
Source |
Material |
Country |
Area |
Year |
||||
|
NEW 3 |
IP39904 |
Yersinia enterocolitica biotype 1A |
Not typeable |
GCA_902170805.1 |
Human |
Stool |
France |
Halennes les Haubourdin |
2018 |
|
|
IP41384 |
Not typeable |
GCA_902726525b |
Human |
Stool |
France |
Le Haillan |
2019 |
|||
|
IP42281T |
Not typeable |
GCA_902726545b |
Human |
Stool |
France |
Barlin |
2019 |
|||
|
IP42750 |
Not typeable |
GCA_902726535b |
Human |
Stool |
France |
Volckerinckhove |
2019 |
|||
|
NEW 4 |
IP37424T |
Yersinia enterocolitica biotype 1A |
O:10-34 |
GCA_902170785.1 |
Human |
Stool |
France |
Saint Laurent de la Salanque |
2016 |
|
|
IP37838 |
O:10-34 |
GCA_902170665.1 |
Human |
Stool |
France |
Chalon sur Saône |
2016 |
|||
|
IP38046 |
O:10-34 |
GCA_902171005.1 |
Human |
Stool |
France |
Limoges |
2016 |
|||
|
IP38191 |
O:10 |
GCA_902170985.1 |
Human |
Stool |
France |
Besançon |
2016 |
|||
|
IP38663 |
O:10-34 |
GCA_902170895.1 |
Human |
Stool |
France |
Toulouse |
2017 |
|||
|
IP38819 |
O:10 |
GCA_902170975.1 |
Human |
Stool |
France |
Cholet |
2017 |
|||
|
IP38868 |
O:34 |
GCA_902170815.1 |
Human |
Stool |
France |
Levallois Perret |
2017 |
|||
|
IP38950 |
O:10-34 |
GCA_902170945.1 |
Human |
Stool |
France |
Levallois Perret |
2017 |
|||
|
IP39432 |
O:6,31 |
GCA_902170955.1 |
Human |
Stool |
France |
Angoulême |
2017 |
|||
|
IP39924 |
O:10-34 |
GCA_902170885.1 |
Human |
Stool |
France |
Metz |
2016 |
|||
|
IP35553 |
O:52-52,53 |
GCA_902170365.1 |
Human |
Stool |
France |
Le Puy en Velay |
2013 |
|||
|
IP37124 |
O:16-16,29 |
GCA_902170395.1 |
Human |
Stool |
France |
Levallois Perret |
2015 |
|||
|
IP37802 |
O:16-16,29 |
GCA_902170375.1 |
Human |
Stool |
France |
Strasbourg |
2016 |
|||
|
IP38166 |
Not typeable |
GCA_902170345.1 |
Human |
Stool |
France |
Levallois Perret |
2016 |
|||
|
IP38403 |
Not typeable |
GCA_902170385.1 |
Human |
Stool |
France |
Strasbourg |
2016 |
|||
|
IP38767 |
Not typeable |
GCA_902170275.1 |
Human |
Stool |
France |
Nancy |
2017 |
|||
|
IP38850T |
O:40 |
GCA_902170305.1 |
Human |
Stool |
France |
Strasbourg |
2017 |
|||
|
IP39458 |
Not typeable |
GCA_902170285.1 |
Human |
Stool |
France |
Brumath |
2017 |
|||
|
IP39797 |
Not typeable |
GCA_902170325.1 |
Human |
Stool |
France |
Strasbourg |
2017 |
|||
|
IP37831 |
O:16-16,29 |
GCA_902170405.1 |
Human |
Stool |
France |
Romilly sur Seine |
2016 |
|||
|
IP38006 |
O:16-16,29 |
GCA_902170245.1 |
Human |
Stool |
France |
Rodez |
2016 |
|||
|
IP38178 |
O:16-16,29 |
GCA_902170255.1 |
Human |
Stool |
France |
Brumath |
2016 |
|||
|
IP38594T |
O:16-16,29 |
GCA_902726565b |
Human |
Stool |
France |
Nemours |
2017 |
|||
|
IP38831 |
O:16-16,29 |
GCA_902170295.1 |
Human |
Stool |
France |
Rethel |
2017 |
|||
|
IP6945 |
O:16 |
GCA_001123825.1 |
Pig |
Stool |
Germany |
Unknown |
1977 |
|||
|
IP34646T |
O:16-16,29 |
GCA_902170565.1 |
Fish |
Unknown |
Bulgaria |
Sofia |
2012 |
|||
|
IP35448 |
O:16-16,29 |
GCA_902170455.1 |
Bird |
Unknown |
Bulgaria |
Sofia |
2013 |
|||
|
IP42199 |
Not typeable |
GCA_902726555b |
Wild boar |
Stool |
Italy |
Parma |
2019 |
|||
|
IP28581 |
O:12,25-12,26 |
GCA_902170535.1 |
Unknown |
Unknown |
Unknown |
Unknown |
2005 |
|||
|
IP35638T |
O:12,25-12,26 |
GCA_902170605.1 |
Human |
Stool |
France |
Rodez |
2014 |
|||
|
IP37484 |
O:12,25-12,26 |
GCA_902170505.1 |
Human |
Stool |
France |
Draguignan |
2014 |
|||
|
IP38487 |
O:12,25-12,26 |
GCA_902170595.1 |
Human |
Stool |
France |
Saint Mandé |
2016 |
|||
|
IP38810 |
O:12,25-12,26 |
GCA_902170465.1 |
Human |
Stool |
France |
Romilly sur Seine |
2017 |
|||
|
IP38921 |
O:12,25-12,26 |
GCA_902170475.1 |
Human |
Stool |
France |
Saumur |
2017 |
|||
a, According to Savin et al. [20]; b, this study.
Genomic features and 16S rRNA gene
Genome sequences of strains were determined as previously described by Savin et al. [20]. All genomes were sequenced using a NextSeq 500 instrument (Illumina) and sequencing libraries were prepared using a Nextera XT DNA library preparation kit (Illumina). Paired-end reads of 150 nucleotides were obtained using the Mid Output or High Output kits (Illumina). A de novo assembly was performed using SPAdes version 3.12.0 [21]. A minimum sequencing depth of 50× was obtained for each genome. On average, genomes of the 38 studied strains were assembled into 116 contigs (min, 20; max, 419) with a total size of 4.57 Mb (min, 4.12; max, 5.25) and with an N50 value of 149 309 (min, 29 468; max, 490 559; Table S1, available in the online version of this article).The ENA accession numbers of the nucleotide sequences are listed in Table 1. DNA G+C content was determined from the whole genome sequence (Table S1). The presence of plasmid in the genomes was investigated in silico using PlasmidSeeker [22].
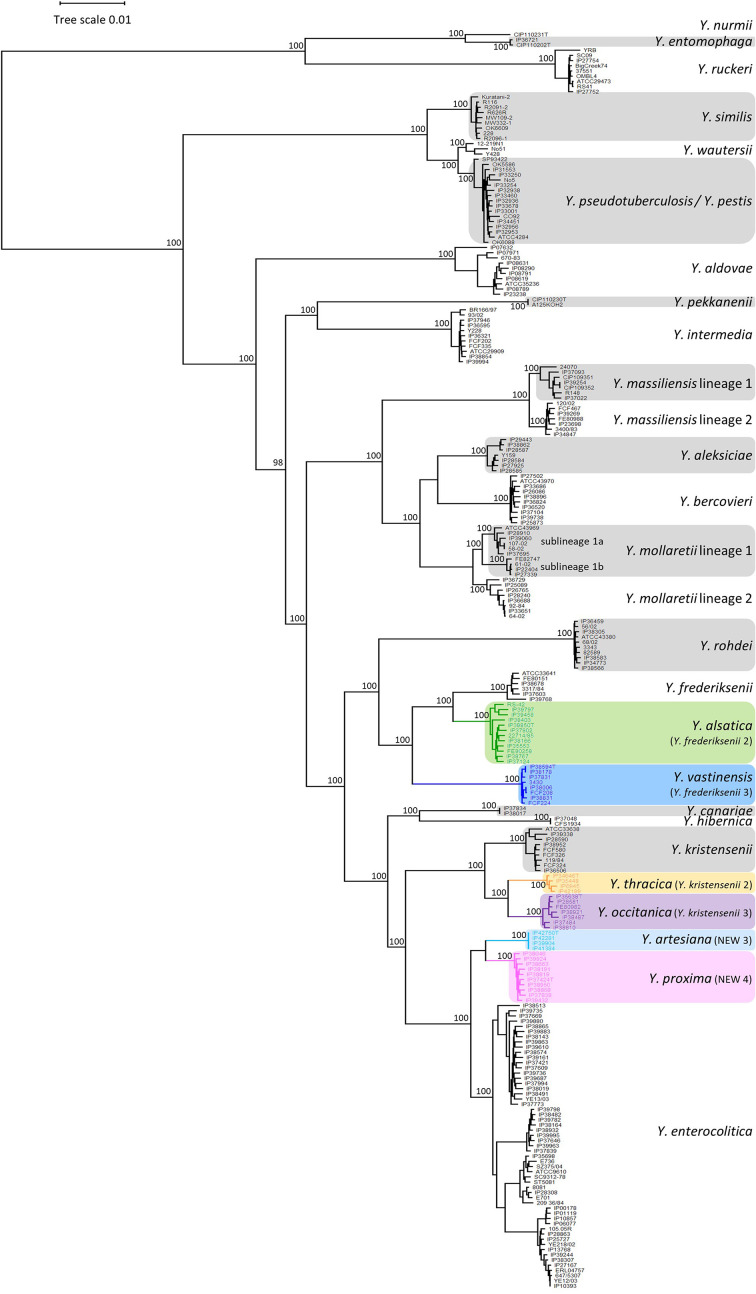
The phylogenetic analysis included the concatenated amino acid sequences of the 500 core genes selected for the Yersinia cgMLST scheme described by Savin et al. in 2019 [20]. The concatenated sequences of the five strains sequenced for this study together with the Y. hibernica type strain (CFS1934T) were compared to those of the 236 strains from the Savin et al. study leading to a total of 242 strains belonging to the 20 described species and the six undescribed taxa. A maximum-likelihood phylogenetic reconstruction was performed using RAxML 8.2.8 [23] (Fig. 1). The two strains IP37834 and IP38017 falling into to the NEW 2 clade were named Y. canariae as they clustered with the type strain in the phylogenetic tree generated by Nguyen et al. [19]. The phylogenetic analysis confirmed that the six undescribed clades are strongly demarcated from the already known species. The 38 strains studied here fell into those six clades: NEW 3 (IP39904, IP41384, IP42281 and IP42750); NEW 4 (IP37424, IP37838, IP38046, IP38191, IP38663, IP38819, IP38868, IP38950, IP39432 and IP39924); Y. frederiksenii 2 (IP35553, IP37124, IP37802, IP38166, IP38403, IP38767, IP38850, IP39458 and IP39797); Y. frederiksenii 3 (IP37831, IP38006, IP38178, IP38594 and IP38831); Y. kristensenii 2 (IP6945, IP34646, IP35448 and IP42199); and Y. kristensenii 3 (IP28581, IP35638, IP37484, IP38487, IP38810 and IP38921).
Fig. 1.
Maximum-likelihood phylogenetic tree of the genus Yersinia (242 strains) based on 500 concatenated multiple sequence alignments. Boostrap support values are shown close to the branches. Bar, 0.01 amino acid substitutions per character.
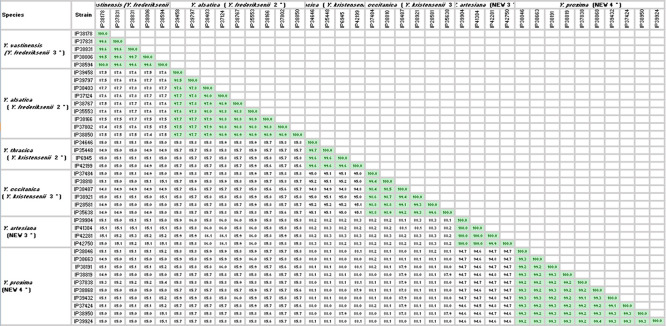
The average nucleotide identity (ANI) values were calculated using fastANI version 1.1 [24]. ANI values between the six groups and their closest relatives were determined by Savin et al. in 2019 and were <94.7 % [20]. ANI values obtained within each clade in our study by adding the genome of the five sequenced strains were all >97.5 % (Table 2). These observations are concordant with the species status of these six groups according to the proposed delineation cut-off of 95–96 % [25].
Table 2.
Average nucleotide identity (ANI) matrix for the 38 Yersinia genomes: nine Y. frederiksenii 2 strains; five Y. frederiksenii 3 strains; four Y. kristensenii 2 strains; six Y. kristensenii 3 strains; four NEW 3 strains, 10 NEW 4 strains
|
*Clade name according to Savin et al. [20].
The 16S rRNA gene sequences of six strains (IP42281, IP37424, IP38850, IP38594, IP34646 and IP35638) were amplified using primers designed by Janvier and Grimont [26]. Amplicons were purified by QIAquick PCR purification kit (Qiagen) and sequenced by Eurofins, resulting in 1477–1491 bp products. Nucleotide sequences of 16S rRNA gene for the seven strains were deposited in GenBank. Accession numbers for IP42281T, IP34646T, IP35638T, IP37424T, IP38594T and IP38850T are LR745664, LR745665, LR745666, LR745667, LR745669 and LR745670, respectively. Sequences were compared to the sequences extracted from the whole genome assembly. Similarity between the product sequences and the extracted sequences was 100 % for the six type strains, ensuring the authenticity of the genome data.
Phenotypic characteristics
Cells were cultured aerobically for 48 h at 28 °C on TSA (trypticase soy agar) and CIN (cefsulodin–irgasan–novobiocin) agar plates. Gram staining of cells was carried out from colonies on TSA and cell morphology was observed using an optical microscope. Motility and nitrate reductase were tested on mannitol motility nitrate (MMN) semisolid medium (Biorad). Catalase activity was determined by observing bubble production in a 3 % (v/v) hydrogen peroxide solution. Oxidase activity was evaluated using discs impregnated with tetramethyl-p-phenylendiamine (Biorad). Tween esterase activity was detected using agar medium containing Tween 80, a positive reaction indicated by the production of aggregates in the media [27]. Pyrazinamidase activity was evaluated using agar medium with pyrazinamide, a positive reaction indicated by a brownish colour in the presence of ferrous salts [28]. Other biochemical characteristics were obtained using API 20E kits (bioMérieux): β-galactosidase, arginine dihydrolase, lysine decarboxylase, ornithine decarboxylase, citrate utilization, H2S production, urease, tryptophan deaminase, indole production, acetoin production (Voges–Proskauer) and gelatinase, and using API 50CH strips (bioMérieux) for testing sugar fermentation. Tests were performed at 28 °C. Strains were serotyped with a set of 47 O:antigen-specific rabbit antisera. The characteristics of the six novel species are listed below in the species description.
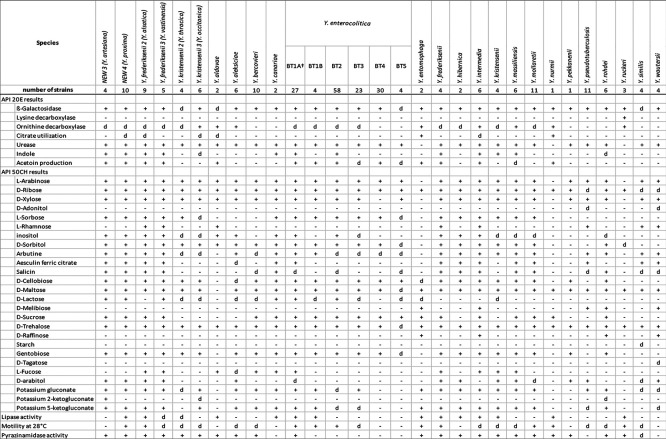
Distinctive phenotypic features between the six putative novel species and the other described Yersinia species and subspecies, except Y. pestis, were obtained by comparison to type strains and other strains characterized at the YNRL by the phenotypic method. Strains are listed in Table S2. The results are shown in Table 3. Strains from clades NEW 3, NEW 4, Y. kristensenii 2 and 3 can be distinguished from their closest relatives by the following biochemical tests: indole and acetoin production, l-fucose, d-arabitol and potassium 2- and 5-ketogluconate fermentation and lipase activity. In contrast, strains belonging to the clades Y. frederiksenii 2 and 3 cannot be distinguished from their closest relative Y. frederiksenii by the biochemical tests.
Table 3.
Distinctive biochemical characteristics between the six novel species and the other Yersinia species and subspecies (except Y. pestis )
BT, biotype; +, 90 % or more strains positive; −, 90 % or more strains negative; d, 11–89 % of strains positive.
|
Three strains from each clade were also characterized using MALDI-ToF MS. The protein patterns of all the strains tested matched with patterns of already known Yersinia species with high score values above 2.3 (database version 8.0.0.0-7311-7854; RUO), threshold of reliable species identification. Therefore, these strains cannot be distinguished from their genetically closest relatives using this technique.
On the basis of the genetic and phenotypic results presented here, we concluded that the 38 isolates should be assigned to the following six novel species: NEW 3 clade as Yersinia artesiana sp. nov. (type strain IP42281T,) NEW 4 clade as Yersinia proxima sp. nov. (type strain IP37424T), Y. frederiksenii 2 clade as Yersinia alsatica sp. nov. (type strain IP38850T), Y. frederiksenii 3 clade as Yersinia vastinensis sp. nov. (type strain IP38594T), Y. kristensenii 2 as Yersinia thracica sp. nov. (type strain IP34646T) and Y. kristensenii 3 as Yersinia occitanica sp. nov. (type strain IP35638T).
Description of Yersinia artesiana sp. nov.
Yersinia artesiana (ar.te.si.a’na. M.L. fem. adj. artesiana, pertaining to Artois county in France where the type strain IP42281T was isolated).
Cells are short Gram-stain-negative rods. Colonies on CIN agar are small, circular, 3.5 mm diameter and have a deep-red centre surrounded by a transparent pale border. Colonies on TSA are beige, smooth and convex. The four strains are not motile on MMN semisolid medium and reduce nitrate. Oxidase is negative. Catalase is positive. Tween esterase is negative and pyrazinamidase is positive. In API20E tests, all strains are positive for β-galactosidase, urease, indole and acetoin production (Voges–Proskauer); and negative for arginine dihydrolase, lysine decarboxylase, citrate utilization, H2S production, tryptophan deaminase and gelatinase for all strains. Three out of the four strains, including the type strain, are negative for ornithine decarboxylase. In API 50CH tests, acid production from glycerol, l-arabinose, ribose, d-xylose, galactose, d-glucose, d-fructose, d-mannose, l-sorbose, inositol, mannitol, sorbitol, N-acetylglucosamine, arbutin, aesculin, salicin, cellobiose, maltose, lactose, sucrose, trehalose, gentiobiose, d-arabitol, gluconate, potassium 2-ketogluconate and potassium 5-ketogluconate is positive; no acid production from erythritol, d-arabinose, l-xylose, adonitol, methyl β-d-xylopyranoside, rhamnose, dulcitol, methyl α-d-mannoside, methyl α-d-glucoside, amygdalin, melibiose, inulin, melezitose, d-raffinose, starch, glycogen, xylitol, turanose, d-lyxose, d-tagatose, d-fucose, l-fucose or l-arabitol. All strains are not typeable for the O:antigen.
The type strain, IP42281T (=CIP 111845T=DSM 110725T), as well as strains IP39904, IP41384 and IP42750, were isolated from human stool. The complete genome of IP42281T has been deposited into ENA (accession number GCA_902726545). The DNA G+C content of the type strain is 47.8 mol%.
Description of Yersinia proxima sp. nov.
Yersinia proxima (pro’xi.ma. L. fem. adj. proxima closest, referring to its genetic closeness to Y. enterocolitica ).
Cells are short Gram-stain-negative rods. Colonies on CIN agar are small, circular, 3.5 mm diameter and have a deep-red centre surrounded by a transparent pale border. Colonies on TSA are beige, smooth and convex. The ten strains are motile on MMN semisolid medium and reduce nitrate. Oxidase is negative. Catalase is positive. Tween esterase and pyrazinamidase are positive. In API20E tests, all strains are positive for β-galactosidase, urease, indole and acetoin production (Voges–Proskauer); negative for arginine dihydrolase, lysine decarboxylase, H2S production, tryptophan deaminase and gelatinase. Seven out of the ten strains, including the type strain, are negative for ornithine decarboxylase and five strains, including the type strain, are positive for citrate. In API 50CH tests, acid production from glycerol, l-arabinose, ribose, d-xylose, galactose, d-glucose, d-fructose, d-mannose, l-sorbose, inositol, mannitol, sorbitol, N-acetylglucosamine, arbutin, aesculin, salicin, cellobiose, maltose, lactose, sucrose, trehalose, gentiobiose, d-arabitol, gluconate and potassium 5-ketogluconate is positive; no acid production from erythritol, d-arabinose, l-xylose, adonitol, methyl β-d-xylopyranoside, rhamnose, dulcitol, methyl α-d-mannoside, methyl α-d-glucoside, amygdalin, melibiose, inulin, melezitose, d-raffinose, starch, glycogen, xylitol, turanose, d-lyxose, d-tagatose, d-fucose, l-fucose, l-arabitol and potassium 2-ketogluconate. Nine out of the ten strains, including the type strain, belong to the O:10–34 serotype.
The type strain, IP37424T (=CIP 111847T=DSM 110727T), and strains IP37838, IP38046, IP38191, IP38663, IP38819, IP38868, IP38950, IP39432 and IP39924 were isolated from human stool. The complete genome of IP37424T has been deposited into ENA (accession number GCA_902170785). The DNA G+C content of the type strain is 47 mol%.
Description of Yersinia alsatica sp. nov.
Yersinia alsatica (al.sa’ti.ca. M.L. fem. adj. alsatica from Alsace, the region in the eastern part of France where the type strain IP38850T was isolated).
Cells are short Gram-stain-negative rods. Colonies on CIN agar are small, circular, 5 mm diameter and have a deep-red centre surrounded by a transparent pale border. Colonies on TSA are beige, smooth and convex. The nine strains are motile on MMN semisolid medium. Only eight strains, including the type strain, reduce nitrate. Oxidase is negative. Catalase is positive. Tween esterase and pyrazinamidase are positive. In API20E tests, all strains are positive for β-galactosidase, urease, indole and acetoin production (Voges–Proskauer); negative for arginine dihydrolase, lysine decarboxylase, H2S production, tryptophan deaminase and gelatinase. Five out of the nine strains, including the type strain, are positive for ornithine decarboxylase; and four strains, including the type strain, are positive for citrate. In API 50CH tests, all strains are positive for acid production from glycerol, l-arabinose, ribose, d-xylose, galactose, d-glucose, d-fructose, d-mannose, l-sorbose, rhamnose, inositol, mannitol, sorbitol, N-acetylglucosamine, arbutin, aesculin, salicin, cellobiose, maltose, sucrose, trehalose, gentiobiose, l-fucose, d-arabitol, gluconate and potassium 5-ketogluconate; and negative for acid production from erythritol, d-arabinose, l-xylose, adonitol, methyl β-d-xylopyranoside, dulcitol, methyl α-d-mannoside, methyl α-d-glucoside, amygdalin, lactose, melibiose, inulin, melezitose, d-raffinose, starch, glycogen, xylitol, turanose, d-lyxose, d-tagatose, d-fucose, l-arabitol and potassium 2-ketogluconate. The type strain belongs to the O:40 serotype.
The type strain, IP38850T (=CIP 111848T=DSM 110726T), as well as strains IP35553, IP37124, IP37802, IP38166, IP38403, IP38767, IP39458, and IP39797, were isolated from human stool. The complete genome of IP38850T has been deposited into ENA (accession number GCA_902170305). The DNA G+C content of the type strain is 47.6 mol%.
Description of Yersinia vastinensis sp. nov.
Yersinia vastinensis (vas.ti.nen’sis. M.L. fem. adj. vastinensis pertaining to Gatinais county in France where the type strain IP38594T was isolated).
Cells are short Gram-stain-negative rods. Colonies on CIN agar are small, circular, 2.5 mm diameter and have a deep-red centre surrounded by a transparent pale border. Colonies on TSA are beige, smooth and convex. Two out of the five strains, including the type strain, are not motile on MMN semisolid medium and all strains reduce nitrate. Oxidase is negative. Catalase is positive. Tween esterase is negative in four strains, including the type strain and pyrazinamidase is positive in all strains. In API20E tests, all strains are positive for β-galactosidase, urease, indole and acetoin production (Voges–Proskauer) and negative for arginine dihydrolase, lysine decarboxylase, citrate, H2S production, tryptophan deaminase and gelatinase. Four out of the five strains, including the type strain, are positive for ornithine decarboxylase. In API 50CH tests, acid production from glycerol, l-arabinose, ribose, d-xylose, galactose, d-glucose, d-fructose, d-mannose, l-sorbose, rhamnose, inositol, mannitol, sorbitol, N-acetylglucosamine, arbutin, aesculin, salicin, cellobiose, maltose, lactose, sucrose, trehalose, gentiobiose, l-fucose, d-arabitol, gluconate and potassium 5-ketogluconate is positive; no acid production from erythritol, d-arabinose, l-xylose, adonitol, methyl β-d-xylopyranoside, dulcitol, methyl α-d-mannoside, methyl α-d-glucoside, amygdalin, melibiose, inulin, melezitose, d-raffinose, starch, glycogen, xylitol, turanose, d-lyxose, d-tagatose, d-fucose, l-arabitol and potassium 2-ketogluconate. All strains belong to the O:16–16,29 serotype.
The type strain, IP38594T (=CIP 111844T=DSM 110738T), as well as strains IP37831, IP38006, IP38178 and IP38831, were isolated from human stool. The complete genome of IP38594T has been deposited into ENA (accession number GCA_902726565). The DNA G+C content of the type strain is 46.9 mol%.
Description of Yersinia thracica sp. nov.
Yersinia thracica (thra’ci.ca. L. fem. adj. thracica referring to Thrace, the ancient province including Bulgaria where the type strain IP34646T was isolated).
Cells are short Gram-stain-negative rods. Colonies on CIN agar are small, circular, 2.5 mm diameter and have a deep-red centre surrounded by a transparent pale border. Colonies on TSA are beige, smooth and convex. Two out of the four strains, including the type strain, are not motile on MMN semisolid medium. All strains reduce nitrate. Oxidase is negative. Catalase and pyrazinamidase are positive. Tween esterase is negative in three strains including the type strain. In API20E tests, all strains are positive for urease; and negative for arginine dihydrolase, lysine decarboxylase, citrate, H2S production, tryptophan deaminase, indole and acetoin production (Voges–Proskauer), and gelatinase. Three out of the four strains, including the type strain, are positive for β-galactosidase and ornithine decarboxylase. In API 50CH tests, acid production from glycerol, l-arabinose, ribose, d-xylose, galactose, d-glucose, d-fructose, d-mannose, l-sorbose, mannitol, sorbitol, N-acetylglucosamine, cellobiose, maltose, trehalose and gentiobiose is positive; no acid production from erythritol, d-arabinose, l-xylose, adonitol, methyl β-d-xylopyranoside, rhamnose, dulcitol, methyl α-d-mannoside, methyl α-d-glucoside, amygdalin, aesculin, salicin, melibiose, sucrose, inulin, melezitose, d-raffinose, starch, glycogen, xylitol, turanose, d-lyxose, d-tagatose, d-fucose, l-fucose, d-arabitol, l-arabitol, potassium 2-ketogluconate and potassium 5-ketogluconate. Only the type strain is negative for inositol. Three out of the four strains, including the type strain, are positive for arbutin and negative for lactose and gluconate. Three out of the four strains, including the type strain, belong to the O:16 serotype and one strain is not typeable.
Strains of this novel species were isolated only in animals. The type strain, IP34646T (=CIP 111842T=DSM 110736T) was isolated from diseased rainbow trouts (Onchorhynchus mykiss) in Bulgaria. Strains IP6945, IP35448 and IP42199 were isolated from pig stool, bird and wild boar, respectively. The complete genome of IP34646T has been deposited into ENA (accession number GCA_902170565). The DNA G+C content of the type strain is 47.4 mol%.
Description of Yersinia occitanica sp. nov.
Yersinia occitanica (oc.ci.ta’ni.ca. M.L. fem. adj. occitanica, referring to Occitanie province in France where the type strain IP35638T was isolated).
Cells are short Gram-stain-negative rods. Colonies on CIN agar are small, circular, 5 mm diameter and have a deep-red centre surrounded by a transparent pale border. Colonies on TSA are beige, smooth and convex. Four out of the six strains, including the type strain, are motile on MMN semisolid medium and all strains reduce nitrate. Oxidase is negative. Catalase is positive. Tween esterase is negative and pyrazinamidase is positive. In API20E tests, all strains are positive for β-galactosidase, ornithine decarboxylase, urease; negative for arginine dihydrolase, lysine decarboxylase, H2S production, tryptophan deaminase, acetoin production (Voges–Proskauer) and gelatinase. Four out of the six strains, including the type strain, are negative for citrate and five strains, including the type strain, produce indole. In API 50CH tests, acid production from glycerol, l-arabinose, ribose, d-xylose, galactose, d-glucose, d-fructose, d-mannose, mannitol, sorbitol, N-acetylglucosamine, cellobiose, maltose, trehalose, gentiobiose, gluconate and potassium 5-ketogluconate is positive; no acid production from erythritol, d-arabinose, l-xylose, adonitol, methyl β-d-xylopyranoside, rhamnose, dulcitol, methyl α-d-mannoside, methyl α-d-glucoside, amygdalin, aesculin, salicin, melibiose, sucrose, inulin, melezitose, raffinose, starch, glycogen, xylitol, turanose, d-lyxose, d-tagatose, d-fucose, l-fucose, d-arabitol and l-arabitol. Five out of the six strains, including the type strain, are l-sorbose-positive; 50 % of the strains, including the type strain are negative for inositol, arbutin and lactose fermentation and four strains, including the type strain are negative for potassium 2-ketogluconate. All strains belong to the O:12,25–12,26 serotype.
The type strain, IP35638T (=CIP 111843T=DSM 110739T), as well as strains IP37484, IP38487, IP38810 and IP38921, were isolated from human stool. The complete genome of IP35638T has been deposited into ENA (accession number GCA_902170605). The DNA G+C content of the type strain is 47.8 mol%.
IP38810 hosts a plasmid similar to the pKpN01-COL plasmid, with 99.97 % identity and 100 % coverage.
Supplementary Data
Funding information
The Yersinia Research Unit is a member of the LabEX IBEID (ANR LBX-62 IBEID). This work was financially supported by the Institut Pasteur (all authors), the European Programme for Public Health Microbiology (EUPHEM) of the European Centre for Disease Prevention and Control (H.A.) and Santé publique France (A.-S.L.G., C.S.).
Acknowledgements
We thank the French microbiologists who sent their Yersinia strains to the NRL for characterization including Françoise Hamida for providing IP35638, Marie-France Aran for providing IP37424, Aline Salabelle for providing IP38594, Thomas Gueudet for providing IP38850 and Anne-Sophie Calippe for providing IP42281. We also acknowledge Silvia Herrera-Léon as a EUPHEM coordinator for reviewing the manuscript.
Conflicts of interest
The authors declare that there are no conflicts of interest.
Footnotes
Abbreviations: ANI, average nucleotide identity; cgMLST, core genome multilocus sequence typing; CIN, cefsulodin–irgasan–novobiocine; MMN, mannitol motility nitrate; TSA, trypticase soy agar; YNRL, Yersinia National Reference Laboratory.
Two supplementary tables are available with the online version of this article.
The GenBank/ENA accession numbers for 16S rRNA gene sequences of strains IP42281T, IP34646T, IP35638T, IP37424T, IP38594T and IP38850T are LR745664, LR745665, LR745666, LR745667, LR745669 and LR745670, respectively. The GenBank/ENA accession numbers for whole genome sequences of the type strains IP42281T, IP34646T, IP35638T, IP37424T, IP38594T and IP38850T are GCA_902726545, GCA_902170565.1, GCA_902170605.1, GCA_902170785.1, GCA_902726565 and GCA_902170305.1, respectively. The Genbank/ENA accession numbers for whole genome sequences of strains IP41384, IP42750 and IP42199, are GCA_902726525, GCA_902726535 and GCA_902726555, respectively.
References
- 1.Adeolu M, Alnajar S, Naushad S, S Gupta R. Genome-based phylogeny and taxonomy of the 'Enterobacteriales': proposal for Enterobacterales ord. nov. divided into the families Enterobacteriaceae, Erwiniaceae fam. nov., Pectobacteriaceae fam. nov., Yersiniaceae fam. nov., Hafniaceae fam. nov., Morganellaceae fam. nov., and Budviciaceae fam. nov. Int J Syst Evol Microbiol. 2016;66:5575–5599. doi: 10.1099/ijsem.0.001485. [DOI] [PubMed] [Google Scholar]
- 2.Demeure C, Dussurget O, Fiol GM, Le Guern A-S, Savin C, et al. Yersinia pestis and plague: an updated view on evolution, virulence determinants, immune subversion, vaccination and diagnostics. Microbes Infect. 2019;21:202–212. doi: 10.1016/j.micinf.2019.06.007. [DOI] [PubMed] [Google Scholar]
- 3.Carniel E, Autenrieth I, Cornelis G, Fukushima H, Guinet F, et al. Y. enterocolitica and Y. pseudotuberculosis. In: Dworkin MM, Falkow S, Rosenberg E, Schleifer KH, Stackebrandt E, et al., editors. The Prokaryotes, a Handbook on the Biology of Bacteria. 3rd ed. Vol. 6. Springer; 2006. pp. 270–398. [Google Scholar]
- 4.Savin C, Martin L, Bouchier C, Filali S, Chenau J, et al. The Yersinia pseudotuberculosis complex: characterization and delineation of a new species, Yersinia wautersii . Int J Med Microbiol. 2014;304:452–463. doi: 10.1016/j.ijmm.2014.02.002. [DOI] [PubMed] [Google Scholar]
- 5.Kumar G, Menanteau-Ledouble S, Saleh M, El-Matbouli M. Yersinia ruckeri, the causative agent of enteric redmouth disease in fish. Vet Res. 2015;46:103. doi: 10.1186/s13567-015-0238-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hurst MRH, Beattie AK, Jones SA, Hsu P-C, Calder J, et al. Temperature-dependent Galleria mellonella mortality as a result of Yersinia entomophaga infection. Appl Environ Microbiol. 2015;81:6404–6414. doi: 10.1128/AEM.00790-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bercovier H, Steigerwalt AG, Guiyoule A, Huntley-Carter G, Brenner DJ. Yersinia aldovae (formerly Yersinia enterocolitica-like group X2): a new species of Enterobacteriaceae isolated from aquatic ecosystems. Int J Syst Bacteriol. 1984;34:166–172. doi: 10.1099/00207713-34-2-166. [DOI] [Google Scholar]
- 8.Sprague LD, Neubauer H. Yersinia aleksiciae sp. nov. Int J Syst Evol Microbiol. 2005;55:831–835. doi: 10.1099/ijs.0.63220-0. [DOI] [PubMed] [Google Scholar]
- 9.Wauters G, Janssens M, Steigerwalt AG, Brenner DJ. Yersinia mollaretii sp. nov. and Yersinia bercovieri sp. nov., formerly called Yersinia enterocolitica biogroups 3A and 3B. Int J Syst Bacteriol. 1988;38:424–429. doi: 10.1099/00207713-38-4-424. [DOI] [Google Scholar]
- 10.Ursing J, Don Brennert J, Bercovier H, Richard Fanning G, Steigerwalt AG, et al. Yersinia frederiksenii: a new species of Enterobacteriaceae composed of rhamnose-positive strains (formerly called atypical Yersinia enterocolitica or Yersinia enterocolitica-like) Curr Microbiol. 1980;4:213–217. doi: 10.1007/BF02605859. [DOI] [Google Scholar]
- 11.Brenner DJ, Bercovier H, Ursing J, Alonso JM, Steigerwalt AG, et al. Yersinia intermedia: a new species of Enterobacteriaceae composed of rhamnose-positive, melibiose-positive, raffinose-positive strains (formerly called Yersinia enterocolitica or Yersinia enterocolitica-like) Curr Microbiol. 1980;4:207–212. doi: 10.1007/BF02605858. [DOI] [Google Scholar]
- 12.Bercovier H, Ursing J, Don Brenner J, Steigerwalt AG, Fanning GR, et al. Yersinia kristensenii: a new species of Enterobacteriaceae composed of sucrose-negative strains (formerly called atypical Yersinia enterocolitica or Yersinia enterocolitica-like) Curr Microbiol. 1980;4:219–224. doi: 10.1007/BF02605860. [DOI] [Google Scholar]
- 13.Merhej V, Adékambi T, Pagnier I, Raoult D, Drancourt M. Yersinia massiliensis sp. nov., isolated from fresh water. Int J Syst Evol Microbiol. 2008;58:779–784. doi: 10.1099/ijs.0.65219-0. [DOI] [PubMed] [Google Scholar]
- 14.Murros-Kontiainen A, Fredriksson-Ahomaa M, Korkeala H, Johansson P, Rahkila R, et al. Yersinia nurmii sp. nov. Int J Syst Evol Microbiol. 2011;61:2368–2372. doi: 10.1099/ijs.0.024836-0. [DOI] [PubMed] [Google Scholar]
- 15.Murros-Kontiainen A, Johansson P, Niskanen T, Fredriksson-Ahomaa M, Korkeala H, et al. Yersinia pekkanenii sp. nov. Int J Syst Evol Microbiol. 2011;61:2363–2367. doi: 10.1099/ijs.0.019984-0. [DOI] [PubMed] [Google Scholar]
- 16.Aleksic S, Steigerwalt AG, Bockemuhl J, Huntleycarter GP, Brenner DJ. Yersinia-rohdei sp. nov., isolated from human and dog feces and Surface-Water. Int J Syst Bacteriol. 1987;37:327–332. [Google Scholar]
- 17.Sprague LD, Scholz HC, Amann S, Busse H-J, Neubauer H. Yersinia similis sp. nov. Int J Syst Evol Microbiol. 2008;58:952–958. doi: 10.1099/ijs.0.65417-0. [DOI] [PubMed] [Google Scholar]
- 18.Nguyen SV, Muthappa DM, Hurley D, Donoghue O, McCabe E, et al. Yersinia hibernica sp. nov., isolated from pig-production environments. Int J Syst Evol Microbiol. 2019;69:2023–2027. doi: 10.1099/ijsem.0.003422. [DOI] [PubMed] [Google Scholar]
- 19.Nguyen SV, Greig DR, Hurley D, Donoghue O, Cao Y, et al. Yersinia canariae sp. nov., isolated from a human yersiniosis case. Int J Syst Evol Microbiol. 2020;70:2382–2387. doi: 10.1099/ijsem.0.004047. [DOI] [PubMed] [Google Scholar]
- 20.Savin C, Criscuolo A, Guglielmini J, Le Guern A-S, Carniel E, et al. Genus-wide Yersinia core-genome multilocus sequence typing for species identification and strain characterization. Microb Genom. 2019;5 doi: 10.1099/mgen.0.000301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012;19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Roosaare M, Puustusmaa M, Möls M, Vaher M, Remm M. PlasmidSeeker: identification of known plasmids from bacterial whole genome sequencing reads. PeerJ. 2018;6:e4588. doi: 10.7717/peerj.4588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jain C, Rodriguez-R LM, Phillippy AM, Konstantinidis KT, Aluru S. High throughput ANI analysis of 90K prokaryotic genomes reveals clear species boundaries. Nat Commun. 2018;9:5114. doi: 10.1038/s41467-018-07641-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Konstantinidis KT, Tiedje JM. Genomic insights that advance the species definition for prokaryotes. Proc Natl Acad Sci U S A. 2005;102:2567–2572. doi: 10.1073/pnas.0409727102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Janvier M, Grimont PA. The genus Methylophaga, a new line of descent within phylogenetic branch gamma of Proteobacteria. Res Microbiol. 1995;146:543–550. doi: 10.1016/0923-2508(96)80560-2. [DOI] [PubMed] [Google Scholar]
- 27.Lee WH. Two plating media modified with tween 80 for isolating Yersinia enterocolitica . Appl Environ Microbiol. 1977;33:215–216. doi: 10.1128/AEM.33.1.215-216.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kandolo K, Wauters G. Pyrazinamidase activity in Yersinia enterocolitica and related organisms. J Clin Microbiol. 1985;21:980–982. doi: 10.1128/JCM.21.6.980-982.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.