Abstract
The taxonomic positions of two novel aerobic, Gram-positive actinobacteria, designated strains RB29T and RB68T, were determined using a polyphasic approach. Based on 16S rRNA gene sequence analysis, the closest phylogenetic neighbours of RB29T were identified as Actinomadura rayongensis DSM 102126T (99.2 % similarity) and Actinomadura atramentaria DSM 43919T (98.7 %), and for strain RB68T was Actinomadura hibisca DSM 44148T (98.3 %). Digital DNA–DNA hybridization (dDDH) between RB29T and its closest phylogenetic neighbours, A. rayongensis DSM 102126T and A. atramentaria DSM 43919T, resulted in similarity values of 53.2 % (50.6–55.9 %) and 26.4 % (24.1–28.9 %), respectively. Additionally, the average nucleotide identity (ANI) was 93.2 % (94.0 %) for A. rayongensis DSM 102126T and 82.3 % (78.9 %) for A. atramentaria DSM 43919T. dDDH analysis between strain RB68T and A. hibisca DSM 44148T gave a similarity value of 24.5 % (22.2–27.0 %). Both strains, RB29T and RB68T, revealed morphological characteristics and chemotaxonomic features typical for the genus Actinomadura , such as the presence of meso-diaminopimelic acid in the cell wall, galactose and glucose as major sugar components within whole-cell hydrolysates and the absence of mycolic acids. The major phospholipids were diphosphatidylglycerol, phosphatidylglycerol, phosphatidylinositol and phosphatidylinositol mannoside. Predominant menaquinones were MK-9(H6) and MK-9(H8) for RB29T and MK-9(H4) and MK-9(H6) for RB68T. The main fatty acids were identified as 10-methyloctadecanoic acid (10-methyl C18:0), 14-methylpentadecanoic acid (iso-C16:0), hexadecanoic acid (C16:0) and cis-9-octadecanoic acid (C18 : 1 ω9c). Here, we propose two novel species of the genus Actinomadura : Actinomadura rubteroloni sp. nov. with the type strain RB29T (=CCUG 72668T=NRRL B-65537T) and Actinomadura macrotermitis sp. nov. with the type strain RB68T (=CCUG 72669T=NRRL B-65538T).
Keywords: Actinomadura, Macrotermes natalensis, new natural products, termite gut
Introduction
Lechevalier and Lechevalier were the first to describe the genus Actinomadura based on the type species Actinomadura madurae [1, 2]. Actinomadura belongs to the family Thermomonosporaceae together with the genera Thermomonospora , Spirillospora , Actinocorallia and Actinoallomurus [3, 4]. Members of the genus Actinomadura have a characteristic non-fragmenting, extensively branched substrate mycelium and moderate or absent aerial mycelia, which differentiate when formed into chains of spores. Species of Actinomadura are described as aerobic, Gram-positive, non-motile organisms. Characteristic chemotaxonomic markers of Actinomadura are meso-diaminopimelic acid (A2pm) as the diagnostic diaminoacid of the cell wall, and galactose, glucose, madurose, mannose and ribose as major components in cell-wall hydrolysates. Cell membranes consist dominantly of phosphatidylinositol and diphosphatylglycerol, and habour minor amounts of phosphatidylinositol and phosphatidylinositol mannoside [1, 5]. Predominant menaquinones are MK-9(H4), MK-9(H6) and MK-9(H8) [6, 7]. The fatty acid profiles of Actinomadura species are found to be mostly complex mixtures of hexadecanoic (C16:0), 14-methylpentadecanoic (iso-C16:0) and 10-methyloctadecanoic acid [4]. Actinomadura species have been isolated from diverse habitats such as soil [5–8], marine sources [9], plant tissues [10, 11] and bee hives [12], and are a known source of diverse bioactive secondary metabolites. Over the last three decades, different bioactive secondary metabolites have been reported, including polyether antibiotic [13], anthracycline [14], polyketide [15] and β-carboline [16].
Isolation and ecology
Fungus-growing termite workers of Macrotermes natalensis were collected from two colonies, Mn160 (25° 44′ 34.7″ S 28° 15′ 38.7″ E) and Mn162 (24° 40′ 30.5″ S 28° 47′ 50.4″ E) in South Africa in February 2015 and isolation procedures have been described by Benndorf et al. [17]. We focused on the isolation of actinobacteria present within the termite gut and capable of living on cellulose or chitin as a sole carbon source as these bacterial isolates are likely to be adapted to living in a lignin- and cellulose-rich environment. Actinobacteria strains RB29T and RB68T showing distinct morphotypes appeared on chitin agar supplemented with cycloheximide (0.05 g l−1) [18, 19] after aerobic incubation at 30 °C for 21 days [17]. Bacterial colonies were subcultured on yeast extract-malt extract agar (ISP2) [20] until clean isolates were obtained. Strains RB29T (alias 5-2) and RB68T were maintained on ISP2 at 30 °C and as glycerol suspensions (25 %, v/v) at −80 °C.
16S rRNA phylogeny
For phylogenetic analysis, 16S rRNA gene sequences were obtained from both strains (RB29T and RB68T) by PCR amplification [17] and additionally extracted from genomic data (vide infra Genomic Features) using Artemis [21]. blast analysis was performed using the NCBI database and 16S rRNA gene sequences of nearly all published Actinomadura reference strains downloaded from the LPSN database [22]. Pairwise sequence similarities were calculated using the method recommended by Meier-Kolthoff [23] on the GGDC web server available at http://ggdc.dsmz.de/ (Tables S1 and S2, available in the online version of this article). Sequences were aligned with the sina sequence alignment service [24]. Phylogenetic trees were reconstructed with the neighbour-joining [25] and maximum-likelihood algorithms [26] in mega software (version 7.0.26) [27]. The evolutionary distance model of Tamura and Nei [28] was used to generate evolutionary distance matrices for the maximum-likelihood and neighbour-joining algorithms with deletion of complete gaps and missing data. For the maximum-likelihood algorithm, the discrete gamma distribution was used (+G) and the rate variation model allowing for some sites to be evolutionarily invariable (+I). For the neighbour-joining algorithm, rate variation among sites was modulated with a gamma distribution (+G). The confidence values of nodes were evaluated by bootstrap analysis based on 1000 re-samplings [29].
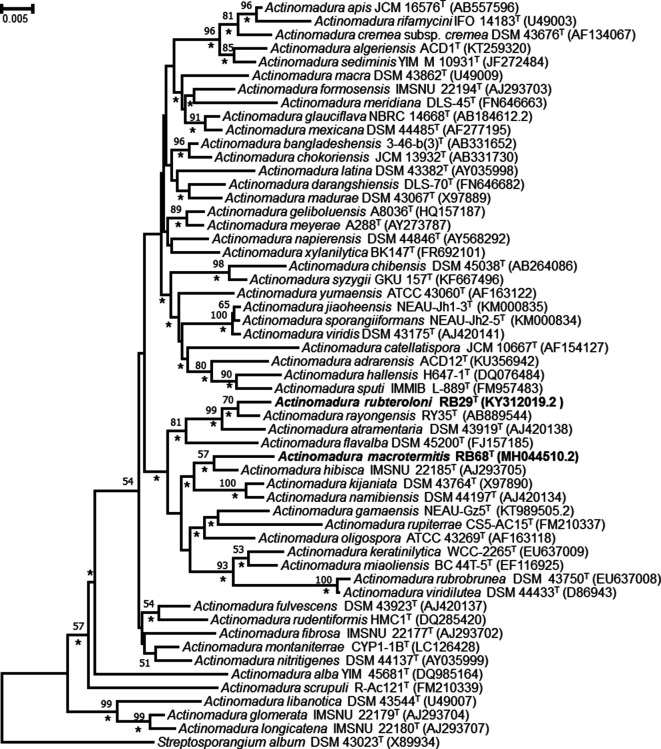
The results of our phylogenetic analysis can be summarized as follows. First, 16S rRNA gene sequence comparisons showed that strain RB29T shared highest similarity with Actinomadura rayongensis DSM 102126T (RY35-68T) (99.2 %) and Actinomadura atramentaria DSM 43919T (98.7 %). In contrast, strain RB68T shared highest 16S rRNA gene sequence similarity with Actinomadura hibisca DSM 44148T (ISMNU 22165T=HKI14) (98.3 %), Actinomadura gamaensis DSM 100815T (NEAU-Gz5T) (98.1 %), Actinomadura namibiensis DSM 44197T (98.0 %) and Actinomadura montaniterrea CYP1-1BT (98.0 %). Lower levels of 16S rRNA gene sequence similarity (<98.0 %) were found with the type strains of all other Actinomadura species (Tables S1 and S2).
Phylogenetic analysis based on maximum-likelihood and neighbour-joining trees revealed that strain RB29T formed a cluster together with A. rayongensis DSM 102126T, A. atramentaria DSM 43919T and A. flavalba DSM 45200T. Strain RB68T formed a cluster with A. hibisca DSM 44148T, A. kijaniata DSM 43764T and A. namibiensis DSM 44197T (Figs 1 and 2). These findings are supported by bootstrap values of 70 % for RB29T and A. rayongensis DSM 102126T (=RY35-68T) and 57 % for RB68T and A. hibisca DSM 44148T (Figs 1 and 2).
Fig. 1.
Neighbour-joining tree based on almost-complete 16S rRNA gene sequences showing the relationships between strains RB29T and RB68T and species of the genus Actinomadura. Streptosporangium album DSM 43023T was used as an outgroup. Asterisks indicate branches that were also recovered in the maximum-likelihood tree. Only bootstrap values above 50 % (percentages of 1000 replications) are shown. Bar, 0.005 substitutions per nucleotide position.
Fig. 2.
Whole-genome sequence tree generated with TYGS for strains RB29T and RB68T and closely related Actinomadura strains. FastME used GBDP distances calculated from genome sequences. Branch lengths are scaled in terms of GBDP distance formula d5; numbers above branches are GBDP pseudo-bootstrap support values from 100 replications. The tree was processed with mega. Thermobispora bispora DSM 43833T (CP001874) was used as an outgroup. Bar, 0.02 substitutions per nucleotide position.
Genome features
Extraction of genomic DNA, PCR amplification/sequencing of the 16S rRNA gene and genome sequencing of strains RB29T and RB68T have been described previously [17, 30] and genome data have been deposited at NCBI under NZ_MTBP00000000.1 [RB29T (alias 5-2)] and NZ_WEGH00000000.1 (RB68T). Additionally, the genome of A. rayongensis DSM102126T was sequenced (Genewiz) and genome data were deposited at NCBI (NZ_WUTW00000000.1, Table S4). The genomic DNA G+C content was determined from whole genome sequence data of strains RB29T and RB68T [31–33]. Digital DNA–DNA hybridization (dDDH) analysis was performed using the GGDC web service at http://ggdc.dsmz.de [32] and compared with other available published genomes of their corresponding genus. Additionally, average nucleotide identity (ANI) was calculated using the web services available at JSpeciesWS (http://jspecies.ribohost.com/jspeciesws) and EZBiocloud (https://www.ezbiocloud.net/tools/ani) (Tables S3 and S4) [34–36].
The results of the genomic analyses can be summarized as follows. First, whole genome sequences of strains RB29T and RB68T and of type strains of phylogenetically related Actinomadura species were used to construct a phylogenetic tree using the TYGS server (http://tygs.dsmz.de) [37]. The topology of the whole-genome sequence tree was similar to that obtained from 16S rRNA gene sequences (Figs 1 and 2).
We then determined the dDDH values (average and confidence intervals in parentheses) between strain RB29T and its closest relatives A. rayongensis DSM 102126T as 53.2 % (50.6–55.9 %) and A. atramentaria DSM 43919T as 26.4 % (24.1–28.9 %). ANI values between strain RB29T and A. rayongensis DSM 102126T and A. atramentaria DSM 43919T were 93.2 % (94.0 %) and 82.3 % (78.9 %), respectively. Overall, all calculated values were below the recommended threshold for the definition of a novel species [38]. Thus, we concluded at this step that strain RB29T represents a novel bacterial species.
The dDDH value between strain RB68T and its closest relative A. hibisca DSM 44148T was determined as 24.5 % (22.2–27.0 %) (Table S3), below the threshold value of 70 % for the delineation of bacterial species, indicating that RB68T also represents a novel bacterial species.
Finally, the GC contents of strains RB29T and RB68T were calculated to be 73.1 mol% for both genomes (Table 1).
Table 1.
Characteristic properties of strains RB29T and RB68T and related species
Strains: 1, RB29T; 2, A. rayongensis DSM 102126T; 3, A. atramentaria DSM 43919T; 4, RB68T; 5, A. hibisca DSM 44148T. All data were acquired in this study. nt, Not tested. Utilization tests were analyzed as follows; +, grows like positive control (basal medium with glucose); (+), better than negative control but not like positive control; −, not better than negative control (basal medium with water). All strains were positive for utilization of d-glucose and negative for utilization of d-arabinose, d-xylose, d-mannitol, raffinose and cellulose
|
Characteristic |
1 |
2 |
3 |
4 |
5 |
|---|---|---|---|---|---|
|
DNA G+C content (mol%) |
73.1 |
73.1 |
73.7 |
73.1 |
72.4 |
|
Chemotaxonomic |
|
|
|
|
|
|
Major menaquinone |
MK-9(H6) |
MK-9(H6) |
MK-9(H6) |
MK-9(H4) |
MK-9(H6) |
|
Major fatty acids (%) |
C18 : 0 10-methyl (22), iso-C16 : 0 (18.4) |
C16 : 0 (23.6), C18 : 1 ω9c (22.2) |
C18 : 0 10-methyl (24.3), C16 : 0 (18.2) |
C16 : 0 (30.7), C18 : 1 ω9c (19.7) |
C16 : 0 (15.7), C18 : 1ω9c (14.5) |
|
Diaminopimelic acid |
meso-A2pm |
meso-A2pm |
meso-A2pm |
meso-A2pm |
meso-A2pm |
|
Mycolic acid |
− |
− |
− |
− |
− |
|
Physiological |
|
|
|
|
|
|
Growth temperature (°C) |
20–37 |
20–45 |
15–42 |
15–37 |
18–40 |
|
Temperature optimum (°C) |
25–30 |
30 |
30 |
25–30 |
30 |
|
pH tolerance range |
5–7 |
5–8 |
5–8 |
6–8 |
6–8 |
|
Optimum pH |
6–7 |
6–7 |
6–7 |
7 |
7 |
|
NaCl tolerance (%, w/v) |
0–4 |
0–3 |
0–4 |
0–4 |
0–4 |
|
Optimum NaCl (%, w/v) |
0–1 |
0–1 |
0–1 |
0–1 |
0–1 |
|
Carbon source |
|
|
|||
|
Sucrose |
− |
− |
− |
− |
(+) |
|
Inositol |
− |
− |
− |
(+) |
+ |
|
d-Fructose |
− |
− |
− |
(+) |
(+) |
|
l-Rhamnose |
− |
− |
+ |
− |
− |
Chemotaxonomy
For chemotaxonomic analyses, freeze dried biomass was obtained from cultures grown in ISP2 broth for 3 days at 28 °C on a rotary shaker at 180 r.p.m. The diagnostic diamino acid of the cell wall was determined in whole-cell hydrolysates by paper chromatography according to Hasegawa et al. [39]. Whole-cell sugars were examined according to Schumann [40]. The occurrence of mycolic acids was determined by TLC as described by Minnikin et al. [41]. Respiratory quinones were extracted and separated as described by Collins et al. [42] and subsequently analyzed as described by Wink et al. [43]. Polar lipids were extracted using the method described by Minnikin et al. [44] and identified by two-dimensional TLC as described by Collins and Jones (Figures S8 and S9) [45]. Extraction and analysis of fatty acids was performed by the DSMZ Identification service according to standard methods [46].
In summary, we found that whole-organism hydrolysates of strains RB29T and RB68T contained meso-A2pm as the diagnostic diamino acid of the peptidoglycan, and carbohydrates composed of galactose, glucose, mannose and trace amounts of madurose (3-O-methyl-d-galactose, wall chemotype III sensu, Lechevalier et al.) [1], which are typical major carbohydrates found in members of the genus Actinomadura . Mycolic acids were not detected in the hydrolysates.
The menaquinone profile of strain RB29T consisted of the predominant menaquinone MK-9(H6) (75 %), MK-9(H8) (13 %) and minor amounts of MK-10 (7 %) and MK-9(H4) (5 %). The closest neighbour A. rayongensis DSM 102126T exhibited a similar menaquinone ratio with MK-9(H6) (71%), MK-9(H8) (15 %), MK-10 (7%) and MK-9(H4) (7 %), while A. atramentaria DSM 43919T revealed a different profile of MK-9(H6) (66 %), MK-9(H8) (16 %), MK-9(H4) (12 %) and MK-10 (6 %).
In contrast, strain RB68T had a different menaquinone profile with major amounts of MK-9(H4) (58 %), MK-9(H6) (30 %) and minor amounts of MK-9(H2) (6 %) and MK-8 (6 %). Here, the closest relative A. hibisca DSM 44148T contained MK-9(H6) (59 %), MK-9(H4) (25 %), MK-9(H8) (12 %) and MK-10 (5 %) and was thus different in its overall profile.
The major phospholipids found in strains RB29T and RB68T were diphosphatidylglycerol, phosphatidylinositol, phosphatidylinositol mannoside and phosphatidylglycerol, all of which are commonly found in most Actinomadura species (Table S7, Figs S8 and S9). RB29T additionally contained two unidentified phospholipids, two lipids and a glycolipid.
The fatty acid profile of strain RB29T (Table S7) was composed of the major fatty acids C18 : 0 10-methyl (22 %), iso-C16 : 0 (18.4 %), C17 : 0 10-methyl (8.8 %), C16 : 0 (8.4 %) and C18 : 1 ω9c (7.0 %), and minor amounts of iso-C18 : 0 (4.8 %), iso-C16 : 0 2-OH (4.5 %), C18 : 0 (3.5 %), C16 : 1 ω9c (2.8 %), C16 : 0 2-OH (2.8 %), C16 : 0 10-methyl (2.8 %), C19 : 0 cyclo C11–12 (2.6 %), C17 : 1 ω9c (2.3 %) and C15 : 0 (2.0 %).
In contrast, the closest related species A. rayongensis DSM 102126T revealed a different fatty acid composition with the following major differences: C16 : 0 (8.4 % in RB29T and 23.6 % in A. rayongensis DSM 102126T), C17 : 0 10-methyl (8.8 % and 2.6 %), C18 : 0 10-methyl (22 % and 14.1 %), C18 : 1 ω9c (7.0 % and 22.2 %) and iso-C16 : 0 (18.4 % and 7.3 %).
Analysis of strain RB68T resulted in a different fatty acid profile with C16 : 0 (30.7 %), C18 : 1 ω9c (19.7 %), C18 : 0 10-methyl (10 %) and only minor amounts of C16 : 1 ω9c (9.7 %), iso-C16 : 0 (8.4 %), C14 : 0 (5 %) and iso-C18 : 0 (1.6 %). The closest relative of RB68T, A. hibisca DSM 44148T, could be differentiated based on a more diverse profile consisting of C16 : 0 (15.7 %), C18 : 1 ω9c (14.5 %), C17 : 0 (13.9 %) and C17 : 1 ω9c (10.6 %). The latter two fatty acids were missing from strain RB68T. Additionally, A. hibisca DSM 44148T contained minor amounts of C15 : 0 (8.1 %), C18 : 0 10-methyl (6.9 %), iso-C16 : 0 (6.2 %), C16 : 1 ω9c (5.2 %), C17 : 0 10-methyl (4.5 %), C18 : 0 (3.5 %) and C14 : 0 (2.2 %).
Overall, the fatty acid profiles clearly differentiate both new isolates from their closest relatives (Table 1).
Physiology
Morphological characteristics of the strains were determined on cultures grown for 5–14 days on ISP2 agar at 30 °C using a light microscope (Imager M2; Zeiss) and a field emission scanning electron microscope. Scanning electron microscopy was performed as described by Groth et al. [47]. Growth characteristics were determined on various ISP media for up to 18 days according to Shirling and Gottlieb [20] and similar to the approach described by Wink et al. [8]. Colony colour was determined using Baumanns Farbatlas 1 (Paul Baumann/Aue). Carbohydrate utilization was determined by using ISP9 (carbon utilization medium) supplemented with 1 % of the sole carbon source. Melanoid pigment production was examined on peptone-yeast extract iron agar (ISP6), tyrosine agar (ISP7) and Suter medium [48] with and without tyrosine (1 g l–1). Tolerance to NaCl was tested on ISP2 agar at concentrations of 1–15 %. The pH tolerance (pH range 4–10) was tested in ISP2 broth using the buffer system described by Xu et al. [49], and temperature tolerance (4, 20, 28, 30, 37, 40, 45 and 60 °C) was tested on ISP2 agar.
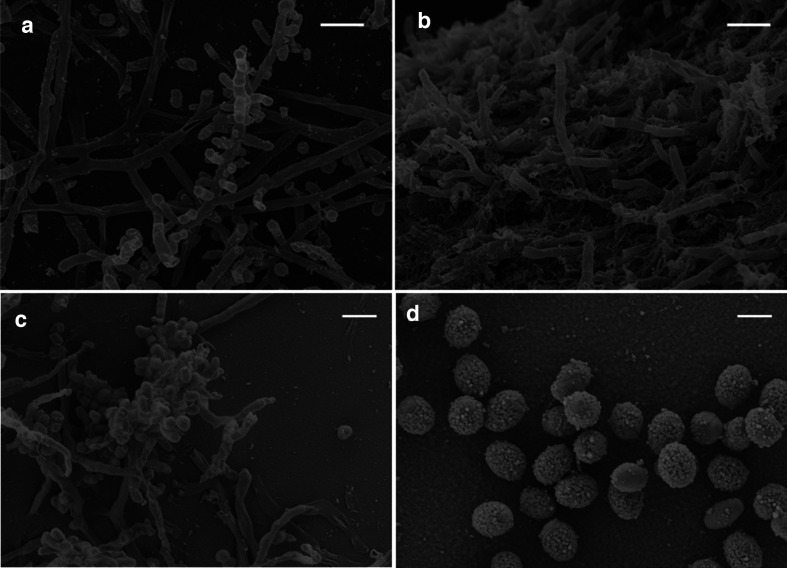
The following morphological and phenotypic characteristics were documented for strains RB29T and RB68T, respectively (Table 1). RB29T was capable of growing on all media tested, tolerated a pH range of 5.0–7.0 (optimal pH 7.0) and NaCl concentration of 0–4 % (w/v) with optimal growth between 0 and 1 % NaCl (Table 1). Note that strain RB29T tolerated only growth temperatures of up to 37 °C, while A. rayongensis DSM 102126 grew even at a temperature of 45 °C. Substrate mycelium was pale (ISP1, ISP4), pale brownish (ISP2), pale brown (Suter medium), pale pinkish (ISP3) to grey (ISP5, ISP6, ISP7). Greyish white aerial mycelium was well developed on ISP1, ISP2, ISP3 and ISP4, only very poor white aerial mycelium was developed at the colony margin on Suter medium with and without tyrosine, and no aerial mycelium occurred on ISP5, ISP6 or ISP7 (Figs S1–S3). Straight chains of two, three and rarely four cylindrical spores with a smooth shape are formed on the tip of sporulating aerial mycelium on ISP2 (Fig. 3a, c). In contrast, the closest relative A. rayongensis DSM 102126T exhibited green substrate mycelium on ISP6, light ochre substrate mycelium on ISP7 and brown substrate mycelium without aerial mycelium on Suter medium. One of the main physiological differences between RB29T and A. rayongensis DSM 102126T is the different temperature tolerance and a pronounced red pigment production in RB29T caused by the production of rubterolones [30, 50] (Figs S1–S3 and S7, Table S6). These findings were supported by additional comparative genome analyses and MS analysis showing that A. rayongensis DSM 102126T neither produces typical representatives of the rubterolone family nor harbours homologues of the responsible rbl genes (Fig. S7).
Fig. 3.
Scanning electron micrographs of cells of strain RB29T (a and c, bar 2 µm) and strain RB68T (b, bar 2 µm and d, bar 1 µm) cultivated at 28 °C on ISP2 agar for 14 days.
In summary, strain RB29T exhibited only a few distinct different phenotypic and growth characteristics compared to the closest reference type strain A. ayongensis DSM 102126T, but several distinct phenotypic characteristics compared to A. atramentaria DSM 43919T (Table S6).
For strain RB68T, phenotypic and growth characteristics can be described as follows. It was capable of growing on all media tested, tolerated a pH range of 6–8 (optimal pH 7.0) and NaCl concentration of 0–4 % (w/v) (optimal 0–1 %, Table 1). Strain RB68T exhibited well-developed substrate mycelium on all media of grey to greenish (ISP1, ISP2 and ISP6), white (ISP3 and ISP4), beige (ISP5) or bright ochre (ISP7) and light brown (Suter medium) colour (Table S6, Figs S4–S6). Sparse aerial mycelium was developed on ISP2, ISP3 and ISP4 media. A brown pigment was observed on Suter medium (supplemented with tyrosine). Straight chains of two to five cylindrical warty spores were formed on the aerial mycelium (Fig. 3b, d). Overall, strain RB68T exhibited several different phenotypic characteristics from the reference type strain A. hibisca DSM 44148T (Tables 1 and S5), which clearly supported its status as representing a novel bacterial species.
Protologue
Description of Actinomadura rubteroloni sp. nov.
Actinomadura rubteroloni (rub.ter.o.lo´ni. N.L. neut. n. rubterolonum rubterolone; N.L. gen. n. rubteroloni of rubterolone).
Aerobic, Gram-positive actinobacterium forming a branched vegetative mycelium. Good growth occurs on ISP1, ISP2, ISP3, ISP5, ISP6, ISP7 and Suter medium and moderate growth occurs on ISP4. Aerial mycelium is formed on all media within 6 days except ISP5, ISP6 and ISP7. Straight chains of two, three and rarely four cylindrical spores of smooth shape are formed on the tip of sporulating aerial mycelium on ISP2 agar. Red to pinkish pigments (identified as rubterolones) are produced on ISP1, ISP2, ISP3, ISP4, ISP5, ISP6 and Suter medium and a brown pigment on ISP7. The pH range for growth is 5–7. The maximum concentration of NaCl for growth is 4 % (w/v) supplemented on ISP2. Growth occurs at 20–37 °C on ISP2 agar. Can grow with glucose as sole carbon source, but not with d-arabinose, d-xylose, d-mannitol, raffinose, cellulose, sucrose, inositol, d-fructose or l-rhamnose. The cell-wall peptidoglycan contains meso-A2pm. The sugars in whole-cell hydrolysates are galactose, glucose, and trace madurose, mannose and ribose. The major polar lipids are diphosphatidylglycerol, phosphatidylglycerol, phosphatidylinositol and phosphatidylinositol mannoside. The predominant menaquinones are MK-9(H6) and MK-9(H8). The major fatty acids are 10-methyloctadecanoic acid (10-methyl C18 : 0), 14-methylpentadecanoic acid (iso-C16 : 0), 10-methylheptadecanoic acid (10-methyl C17 : 0), hexadecanoic acid (C16 : 0) and cis-9-octadecanoic acid (C18 : 1 ω9c).
The type strain is RB29T (=CCUG 72668T=NRRL B-65537T), which was isolated from the gut of the termite Macrotermes natalensis (major worker). The DNA G+C content of the type strain is 73.1 mol%. The GenBank/EMBL accession number for the partial 16S rRNA gene sequence of strain RB29T (alias 5-2) is KY312019.2. The whole genome shotgun (WGS) project has project accession NZ_MTBP00000000. Raw sequencing data have been deposited under accession number SRR11565158.
Description of Actinomadura macrotermitis sp. nov.
Actinomadura macrotermitis (ma.cro.ter´mi.tis. N.L. gen. n. macrotermitis of Macrotermes, of the termite genus Macrotermes).
Aerobic, Gram-positive actinobacterium forming branched vegetative mycelium. Good growth occurs on ISP1, ISP2, ISP5, ISP6 and ISP7 and moderate growth occurs on ISP3, ISP4 and Suter medium. Aerial mycelium is sparsely formed on ISP2, ISP3 and ISP4. Straight chains of two to five cylindrical warty spores are formed on aerial mycelium on ISP2, ISP3 and ISP4. An ochre pigment is produced on ISP5, a bright-ochre pigment on ISP7 and a brown pigment on Suter medium (supplemented with tyrosine). Growth occurs at 15–37 °C on ISP2 agar. The pH range for growth is 6–8. The maximum concentration of NaCl for growth is 4 % (w/v) supplemented on ISP2. Can grow with glucose, inositol or d-fructose as sole carbon source, weak growth occurs on inositol or d-fructose as sole carbon source, but not with d-arabinose, d-xylose, d-mannitol, raffinose, cellulose, sucrose or l-rhamnose. The cell-wall peptidoglycan contains meso-A2pm. The sugars in whole-cell hydrolysates are galactose, glucose, mannose, and trace madurose and ribose. The major polar lipids are diphosphatidylglycerol, phosphatidylglycerol, phosphatidylinositol and phosphatidylinositol mannoside. The predominant menaquinones are MK-9(H4) and MK-9(H6). The major fatty acids are hexadecanoic acid (C16 : 0), cis-9-octadecanoic acid (C18 : 1ω9c), 10- methyloctadecanoic acid (10-methyl C18 : 0), cis-9-hexadecanoic acid (C16 : 1 ω9c) and 14-methylpentadecanoic acid (iso-C16 : 0).
The type strain is RB68T (=CCUG 72669T=NRRL B-65538T), which was isolated from the gut of the termite Macrotermes natalensis (major worker). The DNA G+C content of the type strain is 73.1 mol%. The GenBank/EMBL accession number for the partial 16S rRNA gene sequence of strain RB68T is MH044510.2. The whole genome shotgun (WGS) project has project accession NZ_WEGH00000000. The raw sequencing data have been deposited under accession number SRR11566452.
Supplementary Data
Funding information
R.B. is supported by the International Leibniz Research School for Microbial and Biomolecular Interactions (ILRS) and Jena School for Microbial Communication (JSMC, DFG). The financial support of the Boehringer Ingelheim Foundation, the Daimler Benz foundation and the German research Foundation (CRC 1127, ChemBioSys and BE-4799/3-1) to C.B. is greatly acknowledged.
Acknowledgements
We thank the Oerlemans family (Mookgophong) for permission to sample colonies at their farm, and Susanne Linde (Elektronenmikroskopisches Zentrum FSU Jena) for electron micrographs.
Conflicts of interest
The authors declare that there are no conflicts of interest.
Footnotes
Abbreviations: ANI, average nucleotide identity; ANI, average nucleotide identity; A2pm, diaminopimelic acid; A2pm, diaminopimelic acid; dDDH, digital DNA-DNA hybridization; dDDH, digital DNA–DNA hybridization.
Seven supplementary tables and nine supplementary figures are available with the online version of this article.
The GenBank/EMBL/DDBJ accession numbers for the 16S rRNA gene sequences of strain RB29T and RB68T are KY312019.2 and MH044510.2, respectively. The Whole Genome Shotgun projects have been deposited at DDBJ/ENA/GenBank under INSDC accession numbers MTBP00000000 and WEGH00000000 respectively. The versions described in this paper are versions MTBP00000000.1 and WEGH00000000.1. The raw sequencing data have been deposited under accession numbers SRR11565158 and SRR11566452, respectively.
References
- 1.Lechevalier MP, De Bievre C, Lechevalier H. Chemotaxonomy of aerobic actinomycetes: phospholipid composition. Biochem Syst Ecol. 1977;5:249–260. doi: 10.1016/0305-1978(77)90021-7. [DOI] [Google Scholar]
- 2.Kroppenstedt RM, Goodfellow M. The family Thermomonosporaceae: Actinocorallia, Actinomadura, Spirillospora and Thermomonospora. In: Dworkin M, Falkow S, Rosenberg E, Schleifer KH, Stackebrandt E, editors. The Prokaryotes. USA: Springer-Verlag; 2006. pp. 682–724. [Google Scholar]
- 3.Zhang Z, Kudo T, Nakajima Y, Wang Y. Clarification of the relationship between the members of the family Thermomonosporaceae on the basis of 16S rDNA, 16S-23S rRNA internal transcribed spacer and 23S rDNA sequences and chemotaxonomic analyses. Int J Syst Evol Microbiol. 2001;51:373–383. doi: 10.1099/00207713-51-2-373. [DOI] [PubMed] [Google Scholar]
- 4.Tamura T, Ishida Y, Nozawa Y, Otoguro M, Suzuki K-ichiro. Transfer of Actinomadura spadix Nonomura and Ohara 1971 to Actinoallomurus spadix gen. nov., comb. nov., and description of Actinoallomurus amamiensis sp. nov., Actinoallomurus caesius sp. nov., Actinoallomurus coprocola sp. nov., Actinoallomurus fulvus sp. nov., Actinoallomurus iriomotensis sp. nov., Actinoallomurus luridus sp. nov., Actinoallomurus purpureus sp. nov. and Actinoallomurus yoronensis sp. nov. Int J Syst Evol Microbiol. 2009;59:1867–1874. doi: 10.1099/ijs.0.006858-0. [DOI] [PubMed] [Google Scholar]
- 5.Zucchi TD, Kim B-Y, Bonda ANV, Goodfellow M. Actinomadura xylanilytica sp. nov., an actinomycete isolated from soil. Int J Syst Evol Microbiol. 2013;63:576–580. doi: 10.1099/ijs.0.042325-0. [DOI] [PubMed] [Google Scholar]
- 6.Songsumanus A, Kudo T, Ohkuma M, Phongsopitanun W, Tanasupawat S. Actinomadura montaniterrae sp. nov., isolated from mountain soil. Int J Syst Evol Microbiol. 2016;66:3310–3316. doi: 10.1099/ijsem.0.001196. [DOI] [PubMed] [Google Scholar]
- 7.Miyadoh S, Amano S, Tohyama H, Shomura T. Actinomadura atramentaria, a new species of the Actinomycetales . Int J Syst Bacteriol. 1987;37:342–346. doi: 10.1099/00207713-37-4-342. [DOI] [Google Scholar]
- 8.Wink J, Kroppenstedt RM, Seibert G, Stackebrandt E. Actinomadura namibiensis sp. nov. Int J Syst Evol Microbiol. 2003;53:721–724. doi: 10.1099/ijs.0.02286-0. [DOI] [PubMed] [Google Scholar]
- 9.He J, Xu Y, Sahu MK, Tian X-P, Nie G-X, et al. Actinomadura sediminis sp. nov., a marine actinomycete isolated from mangrove sediment. Int J Syst Evol Microbiol. 2012;62:1110–1116. doi: 10.1099/ijs.0.032979-0. [DOI] [PubMed] [Google Scholar]
- 10.Qin S, Zhao G-Z, Li J, Zhu W-Y, Xu L-H, et al. Actinomadura flavalba sp. nov., an endophytic actinomycete isolated from leaves of Maytenus austroyunnanensis . Int J Syst Evol Microbiol. 2009;59:2453–2457. doi: 10.1099/ijs.0.010652-0. [DOI] [PubMed] [Google Scholar]
- 11.Rachniyom H, Matsumoto A, Indananda C, Duangmal K, Takahashi Y, et al. Actinomadura syzygii sp. nov., an endophytic actinomycete isolated from the roots of a jambolan plum tree (Syzygium cumini L. Skeels) Int J Syst Evol Microbiol. 2015;65:1946–1949. doi: 10.1099/ijs.0.000203. [DOI] [PubMed] [Google Scholar]
- 12.Promnuan Y, Kudo T, Ohkuma M, Chantawannakul P. Actinomadura apis sp. nov., isolated from a honey bee (Apis mellifera) hive, and the reclassification of Actinomadura cremea subsp. rifamycini Gauze et al. 1987 as Actinomadura rifamycini (Gauze et al. 1987) sp. nov., comb. nov. Int J Syst Evol Microbiol. 2011;61:2271–2277. doi: 10.1099/ijs.0.026633-0. [DOI] [PubMed] [Google Scholar]
- 13.Nakamura G, Isono K. A new species of Actinomadura producing a polyether antibiotic, cationomycin. J Antibiot. 1983;36:1468–1472. doi: 10.7164/antibiotics.36.1468. [DOI] [PubMed] [Google Scholar]
- 14.Oki T, Konishi M, Tomatsu K, Tomita K, Saitoh K, et al. Pradimicin, a novel class of potent antifungal antibiotics. J Antibiot. 1988;41:1701–1704. doi: 10.7164/antibiotics.41.1701. [DOI] [PubMed] [Google Scholar]
- 15.Igarashi Y, Iida T, Oku N, Watanabe H, Furihata K, et al. Nomimicin, a new spirotetronate-class polyketide from an actinomycete of the genus Actinomadura . J Antibiot. 2012;65:355–359. doi: 10.1038/ja.2012.30. [DOI] [PubMed] [Google Scholar]
- 16.Kornsakulkarn J, Saepua S, Boonruangprapa T, Suphothina S, Thongpanchang C. New β-carboline and indole alkaloids from actinomycete Actinomadura sp. bcc 24717. Phytochem Lett. 2013;6:491–494. doi: 10.1016/j.phytol.2013.06.007. [DOI] [Google Scholar]
- 17.Benndorf R, Guo H, Sommerwerk E, Weigel C, Garcia-Altares M, et al. Natural products from actinobacteria associated with fungus-growing termites. Antibiotics. 2018;7:E83. doi: 10.3390/antibiotics7030083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Visser AA, Nobre T, Currie CR, Aanen DK, Poulsen M. Exploring the potential for actinobacteria as defensive symbionts in fungus-growing termites. Microb Ecol. 2012;63:975–985. doi: 10.1007/s00248-011-9987-4. [DOI] [PubMed] [Google Scholar]
- 19.Hsu SC, Lockwood JL. Powdered chitin agar as a selective medium for enumeration of actinomycetes in water and soil. Appl Microbiol. 1975;29:422–426. doi: 10.1128/AEM.29.3.422-426.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shirling EB, Gottlieb D. Methods for characterization of Streptomyces species. Int J Syst Bacteriol. 1966;16:313–340. doi: 10.1099/00207713-16-3-313. [DOI] [Google Scholar]
- 21.Rutherford K, Parkhill J, Crook J, Horsnell T, Rice P, et al. Artemis: sequence visualization and annotation. Bioinformatics. 2000;16:944–945. doi: 10.1093/bioinformatics/16.10.944. [DOI] [PubMed] [Google Scholar]
- 22.Parte AC. LPSN--list of prokaryotic names with standing in nomenclature. Nucleic Acids Res. 2014;42:D613–D616. doi: 10.1093/nar/gkt1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Meier-Kolthoff JP, Göker M, Spröer C, Klenk H-P. When should a DDH experiment be mandatory in microbial taxonomy? Arch Microbiol. 2013b;195:413–418. doi: 10.1007/s00203-013-0888-4. [DOI] [PubMed] [Google Scholar]
- 24.Pruesse E, Peplies J, Glöckner FO. SINA: accurate high-throughput multiple sequence alignment of ribosomal RNA genes. Bioinformatics. 2012;28:1823–1829. doi: 10.1093/bioinformatics/bts252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 26.Felsenstein J. Evolutionary trees from DNA sequences: a maximum likelihood approach. J Mol Evol. 1981;17:368–376. doi: 10.1007/BF01734359. [DOI] [PubMed] [Google Scholar]
- 27.Kumar S, Stecher G, Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tamura K, Nei M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol Biol Evol. 1993;10:512–526. doi: 10.1093/oxfordjournals.molbev.a040023. [DOI] [PubMed] [Google Scholar]
- 29.Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 30.Guo H, Benndorf R, Leichnitz D, Klassen JL, Vollmers J, et al. Isolation, biosynthesis and chemical modifications of Rubterolones A-F: rare tropolone alkaloids from Actinomadura sp. 5-2. Chem Eur J. 2017;23:9338–9345. doi: 10.1002/chem.201701005. [DOI] [PubMed] [Google Scholar]
- 31.Auch AF, Klenk H-P, Göker M. Standard operating procedure for calculating genome-to-genome distances based on high-scoring segment pairs. Stand Genomic Sci. 2010;2:142–148. doi: 10.4056/sigs.541628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Meier-Kolthoff JP, Auch AF, Klenk H-P, Göker M. Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinformatics. 2013;14:60. doi: 10.1186/1471-2105-14-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Meier-Kolthoff JP, Klenk H-P, Göker M. Taxonomic use of DNA G+C content and DNA-DNA hybridization in the genomic age. Int J Syst Evol Microbiol. 2014;64:352–356. doi: 10.1099/ijs.0.056994-0. [DOI] [PubMed] [Google Scholar]
- 34.Goris J, Konstantinidis KT, Klappenbach JA, Coenye T, Vandamme P, et al. DNA-DNA hybridization values and their relationship to whole-genome sequence similarities. Int J Syst Evol Microbiol. 2007;57:81–91. doi: 10.1099/ijs.0.64483-0. [DOI] [PubMed] [Google Scholar]
- 35.Richter M, Rosselló-Móra R, Oliver Glöckner F, Peplies J. JSpeciesWS: a web server for prokaryotic species circumscription based on pairwise genome comparison. Bioinformatics. 2016;32:929–931. doi: 10.1093/bioinformatics/btv681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yoon S-H, Ha S-M, Lim J, Kwon S, Chun J. A large-scale evaluation of algorithms to calculate average nucleotide identity. Antonie van Leeuwenhoek. 2017;110:1281–1286. doi: 10.1007/s10482-017-0844-4. [DOI] [PubMed] [Google Scholar]
- 37.Meier-Kolthoff JP, Göker M. TYGS is an automated high-throughput platform for state-of-the-art genome-based taxonomy. Nat Commun. 2019;10:2182. doi: 10.1038/s41467-019-10210-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chun J, Oren A, Ventosa A, Christensen H, Arahal DR, et al. Proposed minimal standards for the use of genome data for the taxonomy of prokaryotes. Int J Syst Evol Microbiol. 2018;68:461–466. doi: 10.1099/ijsem.0.002516. [DOI] [PubMed] [Google Scholar]
- 39.Hasegawa T, Takizawa M, Tanida S. A rapid analysis for chemical grouping of aerobic actinomycetes. J Gen Appl Microbiol. 1983;29:319–322. doi: 10.2323/jgam.29.319. [DOI] [Google Scholar]
- 40.Schumann P. Peptidoglycan structure. In: Rainey F, Oren A, editors. Taxonomy of Prokaryotes, Methods in Microbiology. Vol. 38. London: Academic Press; pp. 101–129. [Google Scholar]
- 41.Minnikin DE, Alshamaony L, Goodfellow M. Differentiation of Mycobacterium, Nocardia, and related taxa by thin-layer chromatographic analysis of whole-organism methanolysates. J Gen Microbiol. 1975;88:200–204. doi: 10.1099/00221287-88-1-200. [DOI] [PubMed] [Google Scholar]
- 42.Collins MD, Pirouz T, Goodfellow M, Minnikin DE. Distribution of menaquinones in actinomycetes and corynebacteria. J Gen Microbiol. 1977;100:221–230. doi: 10.1099/00221287-100-2-221. [DOI] [PubMed] [Google Scholar]
- 43.Wink J, Schumann P, Atasayar E, Klenk H-P, Zaburannyi N, et al. ‘Streptomyces caelicus’, an antibiotic-producing species of the genus Streptomyces, and Streptomyces canchipurensis Li et al. 2015 are later heterotypic synonyms of Streptomyces muensis Ningthoujam et al. 2014. Int J Syst Evol Microbiol. 2017;67:548–556. doi: 10.1099/ijsem.0.001612. [DOI] [PubMed] [Google Scholar]
- 44.Minnikin DE, Collins MD, Goodfellow M. Fatty acid and polar lipid composition in the classification of Cellulomonas, Oerskovia and related taxa. J Appl Bacteriol. 1979;47:87–95. doi: 10.1111/j.1365-2672.1979.tb01172.x. [DOI] [PubMed] [Google Scholar]
- 45.Collins MD, Jones D. Lipids in the classification and identification of coryneform bacteria containing peptidoglycans based on 2, 4-diaminobutyric acid. J Appl Bacteriol. 1980;48:459–470. doi: 10.1111/j.1365-2672.1980.tb01036.x. [DOI] [Google Scholar]
- 46.Leibniz Institute DSMZ German Collection of Microorganisms and Cell Cultures GmbH. Available from: https://www.dsmz.de/services/services-microorganisms/identification/analysis-of-cellular-fatty-acids.html
- 47.Groth I, Schumann P, Rajney FA, Martin K, Schuetze B, et al. Bogoriella caseilytica gen. nov., sp. nov., a new alkaliphilic actinomycete from a soda lake in Africa. Int J Syst Bacteriol. 1997;47:788–794. doi: 10.1099/00207713-47-3-788. [DOI] [PubMed] [Google Scholar]
- 48.Suter MA. Isolierung und Charakterisierung von Melanin-negativen Mutanten aus Streptomyces glaucescens . ETH Zürich Doktorarbeit. 1978 [Google Scholar]
- 49.Xu P, Li W-J, Tang S-K, Zhang Y-Q, Chen G-Z, et al. Naxibacter alkalitolerans gen. nov., sp. nov., a novel member of the family 'Oxalobacteraceae' isolated from China. Int J Syst Evol Microbiol. 2005;55:1149–1153. doi: 10.1099/ijs.0.63407-0. [DOI] [PubMed] [Google Scholar]
- 50.Guo H, Benndorf R, König S, Leichnitz D, Weigel C, et al. Expanding the Rubterolone family: intrinsic reactivity and directed diversification of PKS-derived pyrans. Chem Eur J. 2018;24:11319–11324. doi: 10.1002/chem.201802066. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.