Abstract
Lateral gene transfer (LGT) has impacted prokaryotic genome evolution, yet the extent to which LGT compromises vertical evolution across individual genes and individual phyla is unknown, as are the factors that govern LGT frequency across genes. Estimating LGT frequency from tree comparisons is problematic when thousands of genomes are compared, because LGT becomes difficult to distinguish from phylogenetic artefacts. Here we report quantitative estimates for verticality across all genes and genomes, leveraging a well-known property of phylogenetic inference: phylogeny works best at the tips of trees. From terminal (tip) phylum level relationships, we calculate the verticality for 19,050,992 genes from 101,422 clusters in 5,655 prokaryotic genomes and rank them by their verticality. Among functional classes, translation, followed by nucleotide and cofactor biosynthesis, and DNA replication and repair are the most vertical. The most vertically evolving lineages are those rich in ecological specialists such as Acidithiobacilli, Chlamydiae, Chlorobi and Methanococcales. Lineages most affected by LGT are the α-, β-, γ-, and δ- classes of Proteobacteria and the Firmicutes. The 2,587 eukaryotic clusters in our sample having prokaryotic homologues fail to reject eukaryotic monophyly using the likelihood ratio test. The low verticality of α-proteobacterial and cyanobacterial genomes requires only three partners—an archaeal host, a mitochondrial symbiont, and a plastid ancestor—each with mosaic chromosomes, to directly account for the prokaryotic origin of eukaryotic genes. In terms of phylogeny, the 100 most vertically evolving prokaryotic genes are neither representative nor predictive for the remaining 97% of an average genome. In search of factors that govern LGT frequency, we find a simple but natural principle: Verticality correlates strongly with gene distribution density, LGT being least likely for intruding genes that must replace a preexisting homologue in recipient chromosomes. LGT is most likely for novel genetic material, intruding genes that encounter no competing copy.
Author summary
Because multicellular life is a latecomer in Earth history, most of evolutionary history is microbial evolution. Scientists investigate microbial evolution by studying the evolution of genes. One of the main surprises of the genomic era is the amount of lateral gene transfer that has gone on in prokaryote genome evolution. Gene transfer clouds evolutionary history, but by how much: How lateral and how vertical is the microbial evolutionary process across genes, genomes and lineages? We introduce measures of verticality in genome evolution that permit a ranking of genes and lineages according to their degree of verticality. We show that genes already present in genomes are less likely to be replaced by a newly introduced copy than genes that offer new evolutionary opportunities for the recipient, providing a simple and natural mechanism that limits and promotes lateral gene transfer frequency. Only a very small minority of prokaryotic genes evolve vertically. While the 100 genes that are most widely used to describe the phylogenetic relationships of microbes are indeed the most vertical, they are not at all representative for the evolution of other genes. These findings have broad implications for how we understand the evolutionary process as inferred from gene trees.
Introduction
Prokaryotes undergo recombination that is facilitated by the mechanisms of lateral gene transfer (LGT) [1,2]—transformation, conjugation, transduction, and gene transfer agents [3]. These mechanisms introduce DNA into the cell for recombination and do not obey taxonomic boundaries, species or otherwise. Over time they generate pangenomes [4,5] that are superimposed upon vertical evolution of a conserved core. About 30 genes are present in all genomes [6–9], a few more are nearly universal [10], many are found only in strains of one species [5], but the vast majority of genes are distributed between those extremes according to a power law [11]. Previous work has shown that LGT is subject to natural barriers [12,13], that LGT affects core metabolism less than it affects peripheral metabolism [14] and that LGT is affected by regulatory interaction networks [15]. LGT generates collections of genes in each genome that are of different evolutionary age [16], transferred genes are non-randomly associated [17,18], and major events of gene flux have occurred during evolution [9,19]. In principle, each gene should be transferable, because the mechanisms that introduce DNA into the cell are not selective with regard to the nature of sequences introduced, notwithstanding the CRISPR activity associated with phage defense [20]. If all genes are transferrable, what determines verticality?
At the level of strains or species, gene distributions within rapidly evolving pangenomes have been well-studied [21–25]. Less well understood are the factors that govern the distribution of genes across prokaryotic genomes at higher taxonomic levels. These reflect processes that occurred in deep evolutionary time and, in some cases, underpin the physiological identity of major prokaryotic clades. Though LGT impacts prokaryotic evolution, it does not obscure lineage identity, because despite the abundance of LGT, biologists 100 years ago were able to recognize the identity of many higher level taxa, for example Cyanobacteria and Spirochaetes [26], that we still recognize today. Hence there must exist a spectrum of verticality in prokaryote lineage evolution. It follows that a natural spectrum of verticality across prokaryotic genes should exist as well. Here we rank 101,422 gene families from 5,655 prokaryotic genomes according to conservative estimates of verticality and report how this attribute affects phylogenetic inference in microbial evolution in general and as it impacts inference of eukaryote origin in particular.
Results
The verticality of genes
The two main parameters influencing reconstruction of gene evolution across prokaryotes are sequence conservation and phylogenetic distribution, both of which are easy to estimate from clustering methods based on pairwise sequence comparisons. The degree of congruence among trees for overlapping leaf sets is, by contrast, determined by two unknowns: the accuracy of phylogenetic inference relative to the true gene trees, and the relative amount of LGT that has, or has not, occurred in the evolution of each gene (verticality V). There are many methods of tree comparison, but not for measures of gene verticality. If a gene occurs in many lineages, one invariably observes discordance between the branching pattern generated by the gene and that generated by some standard such as rRNA, yet whether such discordance is due to LGT or to technical issues involving alignment and phylogeny [27] is virtually impossible to determine, because knowledge of the amino acid substitution process underlying sequence divergence in real alignments is irretrievable from real data [28]. That problem is exacerbated in trees having thousands of leaves, where random phylogenetic differences are inevitable. For example, there are 3 · 1080 possible topologies for a tree with 52 leaves, and there are about 1080 protons in the universe [29]. A comparison of two trees, each with 52 (or 520, or 5,200) leaves for an alignment of 400 amino acid sites, evaluates many branches that are not better than random.
Earlier surveys of lateral gene transfer across 116 prokaryotic genomes using nucleotide frequency comparisons were reported over a decade ago [30]. In the era of computers that can calculate all trees for all genes, we sought a measure of verticality that is based on phylogenetic principles but independent of the problems inherent to topological comparisons of large trees. To obtain such an estimate, we leveraged two simple but robust assumptions. First, we assume that the higher order taxa of prokaryotes (referred to here as phyla) that microbiologists have traditionally recognized based on morphological, physiological and rRNA sequence criteria are real and constitute monophyletic groups. On that premise, the null hypothesis for phylogenetic behavior of a given gene in a given prokaryotic phylum is vertical evolution (phylum monophyly). Our second assumption for estimating verticality is that molecular phylogeny works most reliably at the tips of trees, the terminal branches. This assumption is the basis of Neighbor Joining [31], almost all alignment programs [32], and maximum likelihood methods, which typically start the topology search from an NJ tree [33]. By reading the trees only at the tips, we disregard phyletic patterns in deeper branches, where pairwise sequence similarity fades and the processes underlying sequence differences, alignments, and branching pattern differences become more numerous, more poorly constrained and more prone to inference errors.
To estimate V, we read the information contained in each tree solely with regard to the branching patterns of phyla by posing the following recursive set of questions: 1) For each phylum that exists in our data, do sequences from the phylum occur in the tree? 2) If so, do they form a monophyletic group (a clade) or are they singletons? 3) How many clades do they form in that tree? 4) For each clade for tree i and phylum j, what is the phylogenetic composition of the sister group? That set of questions is repeated for all phyla in tree i, the results are tabulated, and the procedure repeated for the next tree. The resulting data contains information both about the verticality of all genes (how often phyla appeared monophyletic for each gene) and about the verticality of genome evolution in all phyla (how often phyla were monophyletic across all genes in the phylum). In a world without LGT and perfect data that reconstructs the true tree from the alignment, all phyla would be monophyletic, all genes from the same phylum would have the same sister phylum and each gene would appear to be inherited vertically. In real data, LGT exists and the data are not perfect, but by looking only at the tips we can estimate verticality without random effects among deeper branches. Note that the true branching order of phyla relative to one another has no bearing upon our estimate of V, nor does the relative branching of lower order taxa within each phylum. For a given gene, we calculate V as follows. For each tree, phyla that are not monophyletic are given a score of zero, the number of genomes present in the tree for each monophyletic phylum is divided by the number of genomes from that phylum among the 5,655 genomes in the data; that proportion is summed across all monophyletic phyla in the tree, that sum is V for that tree or cluster. For n phyla, V obtains a value between 0 and n.
This measure scores the verticality of a gene across all phyla in which it occurs and gives a higher rank to genes that recover phylum monophyly in a tree sampling many phyla than to those with a more narrow distribution, where the opportunity to observe LGT in tree tips is reduced. Note that an accurate taxonomic assignment for each gene is important for estimating V, for which reason we do not include metagenomic data, where binning can lead to assemblies of genes from different lineages. Clustering all 19,050,992 genes yielded 448,821 clusters with genes spanning at least two sequenced genomes, with 261,058 clusters spanning at least three genomes for tree reconstruction with an average of 66.4 genomes and 68.7 sequences each. Removing trees that contained sequences from only one phylum left 101,422 trees containing on average 138.8 genomes and 146.7 sequences (median 18 for both).
The first question we asked was whether gene duplications are frequent, as they might emulate LGT and thus mask verticality. For smaller data sets it is known that gene duplications in prokaryotes are generally rare as compared to eukaryotes [34] and that genome sizes constrain the number of duplicates (or transfers) that a genome can accommodate [11]. Estimating ancient duplications for this data set is not possible as duplications and transfers would be indistinguishable, but recent duplications can be quantified. We found 32,277 cases in which the sister of a terminal leaf (gene) occurred within the same prokaryotic genome. For 5,655 prokaryotic genomes this yields 5.7 genome specific duplications per genome. For comparison, 150 eukaryote genomes [35] harbor 109,056 genome specific duplications corresponding to 727 genome specific duplications per genome. Thus, based upon the values for recent duplications in the present sample, gene duplications per genome are 134-fold less frequent in prokaryotes than in eukaryotes. We also plotted the fraction of terminal duplicates normalized for genome size and compared the distribution in eukaryotes versus prokaryotes using all genomes. The cumulative distribution function (S1A Fig) shows that a eukaryotic genome has, on average, 4% recent duplications while prokaryotes have 0.2%. This is not an effect of unequal sample size, because the average 20:1 ratio is robust for 100 random samples of 150 prokaryotic genomes (S1B Fig). That duplications are 20–134 fold less frequent in prokaryotes than in eukaryotes in this sample of 5,655 genomes corresponds well with the earlier estimate from six groups of closely related bacteria that ~98% of gene families in prokaryotes result from LGT, not duplication [34]. It suggests that in prokaryotic genomes, duplication (paralogy) does not impact estimates of V in prokaryotic genomes to an appreciable extent, a caveat for methods that allow and infer roughly equal probabilities of LGT and duplication, both for prokaryotes and for eukaryotes [36].
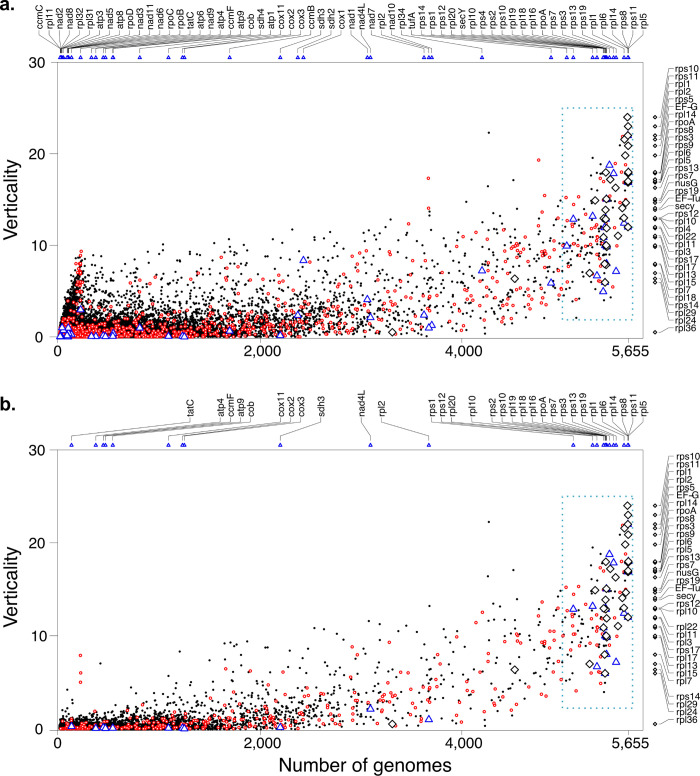
The values of V obtained for all genes allows us to rank them by their relative degree of verticality or LGT, as one prefers. What governs LGT? Few factors have been suggested to govern the rate of LGT that genes undergo. It has been suggested that LGT is limited by the number of intermolecular interactions in which a molecule in involved [37]. Although many genes with high values of V encode ribosomal proteins, which have many interactions, many ribosomal proteins have modest values of V. We found that the majority of highly vertical genes are soluble proteins as opposed to being components of macromolecular complexes, and that verticality V strongly correlates with the gene’s distribution frequency across genomes, as shown in Fig 1, where the value of V estimated for each gene is plotted against the number of genomes in which it occurs. Fig 1A shows the verticality and distribution of all 101,422 clusters that generate trees. Fig 1B displays the verticality the 8,547 clusters that contain more conserved sequences, that is, those that have an average branch length ≤ 0.1 substitutions per site. The spike of sequences at the left of Fig 1A represents sequences that tend to be vertically inherited within closely related lineages but whose clusters span only a few genomes because they are not well conserved, for which reason the spike, which encompasses 836 clusters (0.8%; see S1 Table), is not present in Fig 1B.
Fig 1.
Comparison of estimated verticality and number of genomes in a protein cluster for a. all clusters (n = 101,422) and b. all conserved clusters (average branch length ≥ 0.1; n = 8,547). Unrooted trees were analyzed if at least two taxonomic groups were present. Verticality was calculated as the sum of monophyletic taxonomic groups in a cluster adjusted by the fraction of a taxonomic group represented in the cluster. The procedure for determining verticality on the basis of an example is shown in S3 Fig. This value correlates with the number of genomes, an approximation of universality, which is even more apparent when clusters of high evolutionary rate were filtered out (a.: p < 10−300, Pearson´s R2 = 0.726; b.: p < 10−300, R2 = 0.829). In both plots clusters of special interest were marked: The eukaryotic-prokaryotic clusters (EPCs) are highlighted in red and the clusters that correspond to a gene from the mitochondrial genome of Reclinomonas americana [45] are displayed in blue triangles along the abscissa of the plot and in the graph. For the latter, the gene identifier was noted above each plot. Ribosomal proteins are indicated by the black diamond on the right of each plot and in the graph [6]. Notably, the ribosomal protein clusters show a steep gradient of verticality among conserved clusters with similarly wide distribution.
The value of V as calculated has desirable properties because it takes distribution into account. In order to see whether verticality is correlated with distribution, we also calculated values of verticality that are independent of distribution, using the number of monophyletic phyla per tree multiplied by the average root-to-tip distance [38] (weighted verticality, Vw; S10 Table) instead of dividing by the number of phyla in which the gene is present. The correlation between gene distribution frequency and weighted verticality Vw as inferred independent of distribution frequency was significant at p < 10−300 (S2 Fig, S2 Table). From that one obtains a very general observation about verticality and gene distribution: The most densely distributed genes tend to have the highest verticality, that is, the lowest frequency of recent LGT as determined by phylogenetic criteria.
Why should the most densely distributed genes tend to be most resistant to LGT? We suggest that the reason is simple: If a well-regulated, codon-bias adapted [2] resident copy of a gene already exists in the genome, it would have to be displaced by the intruding copy. Selection in prokaryotes can be intense, as evidenced by codon bias itself: synonymous substitutions that impair codon bias for highly expressed genes are tenaciously counter selected in nature [2]. The existence of a preexisting copy of a gene in the genome reduces the probability of LGT in a highly significant manner (R2 = 0.726; Fig 1B). This is all the more noteworthy because the genes that most frequently enter a recipient cell via LGT in nature will be those that are themselves the most widespread genes in nature—that is, the most common genes will be introduced into recipients with the highest frequency. Prokaryotic genes thus have a home field advantage relative to intruders.
The mechanisms of LGT (transduction, transformation, conjugation, gene transfer agents) operate constantly across all prokaryotic genomes in the wild. All things being equal, new coding sequences enter the prokaryotic genome as a random sample of genes available in the environment [39,24], producing natural variation in gene content upon which selection and drift [40] can act to prolong or curtail the gene’s lifespan, or residence time, in the descendant clonal lineage. Genes that interfere with the workings of the cell [13] are eliminated quickly from the accessory genome and therefore have a short residence time. Neutral genes that merely constitute functionless ballast can persist in the pangenome longer before loss, while genes that are of value under circumstances encountered by the recipient can become fixed [23,24], in which case they start to shift from the accessory genome to the core genome, thereby defining new genomic lineages of vertical core descent.
The gene families that we observe to be the most vertical (Fig 1, S1 Fig) are those that are most widely distributed among genomes and hence the most prevalent in nature. This would be puzzling were it not for an inhibitory effect that presence of a preexisting copy exerts on the success rate of LGT. Transposases constitute a special case. They are likely the most common genes in nature [41], but there are different classes of transposases [41], hence they do not fall into one cluster. The fate of transposases is not governed by selection and drift, as they self-amplify within genomes, increasing their copy number by virtue of their ability to do so [42], not by virtue of selection and drift.
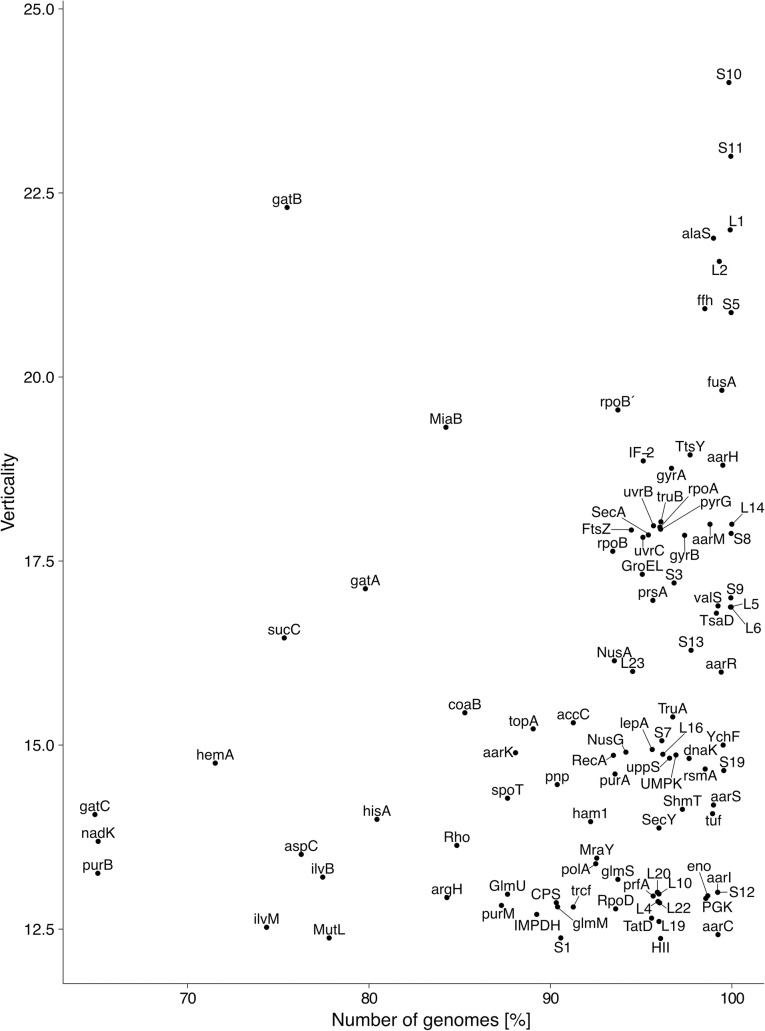
The verticality of genes has practical importance for prokaryotic phylogeny, because modern approaches to prokaryotic systematics typically aim to increase the amount of information per lineage beyond that provided by ribosomal RNA. Since 1997, phylogenetic studies of prokaryotic genomes have typically concatenated dozens of sequences into longer alignments [6,43,44]. However, it is not enough to just combine sequences into longer alignments, the sequences ideally need to share the same evolutionary history. V provides a measure for how vertically a gene tends to evolve over evolutionary time spans. Ranking all genes by their verticality (Fig 1; S1 Table) provides criteria for inclusion of genes for phylogenetic studies. For orientation, in Fig 1 we have labelled along the ordinate the genes in current use for phylogenetic studies of archaeal lineages and their relationship to the host that acquired the mitochondrion at eukaryote origin [45]. They differ in their degree of verticality. A number of sequences that are not widely used for phylogeny exhibit higher verticality; these are shown in Fig 2 and listed in S6 Table. Similarly, genes encoded in mitochondrial DNA are typically used to investigate the relationship of mitochondria to bacterial lineages [46]. Those genes are a subset of the genes found in Reclinomonas americana mitochondrial DNA [47], which are indicated along the abscissa in Fig 1.
Fig 2. Comparison of estimated verticality and number of genomes [%] for the 100 most vertical clusters.
Identity and Annotation of clusters can be found in S6 Table. This is a representation of some of the clusters shown in the blue rectangle of Fig 1A.
From the standpoint of phylogenetics, the main message of Fig 1 is twofold. First, the genes most commonly used as markers in broad scale prokaryotic phylogenetic studies are, in terms of their distribution and their verticality, not representative for the genome as a whole. Worse, without the comparative information from Fig 1 they could even be positively misleading, because without measures to compare verticality across genes, one might assume that the tendency of the most widely distributed genes to be vertically inherited is representative for the phylogenetic behavior of all genes. But that is not the case. Widely distributed genes tend to be vertically inherited but they are not a representative sample for the phylogenetic behavior of the genome as a whole. The vast majority of prokaryotic genes are not inherited vertically, hence the small vertically inherited sample is not a good proxy for the phylogenetic behavior of prokaryotic genes. Vertically inherited genes in prokaryotes are not a random sample, they are a biased sample. This is also known as the tree of 1% [9] and is most clearly seen in Fig 1B, where the more conservatively evolving, hence phylogenetically more useful genes are shown. The vast majority of genes that occur in two or more phyla in prokaryotes fail to recover phylum monophyly to any appreciable extent, also for estimates of V that are independent of distribution (S2 Fig), and most of them are present in only very few phyla to begin with. The mean and median values of V in Fig 1A are 0.27 and 0.04, in Fig 1B 0.70 and 0.06, respectively. The second main message of Fig 1 concerns the relationship of eukaryotic clusters to prokaryotic clusters. We mapped these prokaryotic clusters to eukaryotic clusters (see Methods) as indicated by red circles in Fig 1. Their significance will be discussed in a later section.
The most vertical and lateral genes and categories
Table 1 lists the 20 most vertically and 20 least vertically inherited genes in sequenced prokaryotic genomes, both for the complete sample and for the conserved fraction of genes. Among the most vertical are the ribosomal proteins, ribosomal protein S10 currently being the most vertical protein in genomes, followed by other proteins involved in information processing. The least vertically inherited genes by our conservative tip-based approach, comprise various categories (Table 1), the complete lists of genes ranked by verticality is given in S1 Table.
Table 1. Maximum likelihood trees from 19,050,992 protein sequences from 5,433 bacterial and 212 archaeal species were calculated for clusters obtained by MCL, yielding 101,422 trees with at least four sequences and two taxonomic groups present. Each of the 101,422 trees were assigned a protein label according to the NCBI sequence header that was represented the most. On the left panel all trees were annotated and sorted according to their verticality score for the genes (Vg).
The number of organisms in the respective cluster is stated as Nspec. On the right panel the same values are stated only for conserved protein families–determined by average branch length ≤ 0.1.
| All 101,422 protein families | The 8,547 most conserved protein families | |||||
|---|---|---|---|---|---|---|
| Vg | Protein family | Nspec | V | Protein family | Nspec | |
| Most vertical | ||||||
| 24.00 | 30S ribosomal protein S10 | 5,646 | 24.00 | 30S ribosomal protein S10 | 5,646 | |
| 23.00 | 30S ribosomal protein S11 | 5,652 | 23.00 | 30S ribosomal protein S11 | 5,652 | |
| 22.30 | Asp/glu–tRNA amidotransferase subunit B | 4,269 | 22.30 | Asp/glu–tRNA amidotransferase subunit B | 4,269 | |
| 22.00 | 50S ribosomal protein L1 | 5,650 | 22.00 | 50S ribosomal protein L1 | 5,650 | |
| 21.89 | Alanine–tRNA ligase | 5,598 | 21.89 | Alanine–tRNA ligase | 5,598 | |
| 21.57 | 50S ribosomal protein L2 | 5,616 | 21.57 | 50S ribosomal protein L2 | 5,616 | |
| 20.93 | Sec family type I SRPa protein | 5,571 | 20.93 | Sec family type I SRPa protein | 5,571 | |
| 20.88 | 30S ribosomal protein S5 | 5,653 | 20.88 | 30S ribosomal protein S5 | 5,653 | |
| 19.82 | Translation elongation factor G | 5,624 | 19.82 | Translation elongation factor G | 5,624 | |
| 19.55 | DNA-directed RNA polymerase subunit beta | 5,300 | 19.55 | DNA-directed RNA polymerase subunit beta | 5,300 | |
| 19.32 | tRNA methylthiotransferase MiaB | 4,764 | 18.86 | Translation initiation factor IF-2 | 5,379 | |
| 18.94 | Signal recognition particle-docking protein FtsY | 5,525 | 18.80 | Histidine–tRNA ligase | 5,627 | |
| 18.86 | Translation initiation factor IF-2 | 5,379 | 18.76 | DNA gyrase subunit A | 5,467 | |
| 18.80 | Histidine–tRNA ligase | 5,627 | 18.00 | 50S ribosomal protein L14 | 5,655 | |
| 18.76 | DNA gyrase subunit A | 5,467 | 18.00 | Methionine–tRNA ligase | 5,587 | |
| 18.03 | tRNA pseudouridine synthase B | 5,434 | 17.98 | Excinuclease ABC subunit B | 5,411 | |
| 18.00 | 50S ribosomal protein L14 | 5,655 | 17.96 | DNA-directed RNA polymerase subunit alpha | 5,431 | |
| 18.00 | Methionine–tRNA ligase | 5,587 | 17.93 | CTP synthetase | 5,433 | |
| 17.98 | Excinuclease ABC subunit B | 5,411 | 17.88 | 30S ribosomal protein S8 | 5,653 | |
| 17.96 | DNA-directed RNA polymerase subunit alpha | 5,431 | 17.85 | Preprotein translocase subunit SecA | 5,395 | |
| Most lateral | ||||||
| 0 | Heavy metal-responsive transcriptional regulator | 2,392 | 0 | SDH cyt b556 large subunit | 2,344 | |
| 0 | SDH cyt b556 large subunit | 2,344 | 0 | RnfH family protein | 2,004 | |
| 0 | Anaerobic ribo.-triPb reductase activating protein | 2,078 | 0 | Hypothetical protein | 1,964 | |
| 0 | Thiol:disulfide interchange protein DsbC | 1,952 | 0 | Amino acid ABC transporter permease | 1,666 | |
| 0 | RnfH family protein | 2,004 | 0 | Succinate dehydrogenase, HMc anchor protein | 1,800 | |
| 0 | Disulfide bond formation protein B 1 | 1,808 | 0 | LysR family transcriptional regulator | 1,267 | |
| 0 | Hypothetical protein | 1,964 | 0 | Hypothetical protein | 1,688 | |
| 0 | Amino acid ABC transporter permease | 1,666 | 0 | Maleylacetoacetate isomerase | 1,430 | |
| 0 | LysR family transcriptional regulator | 1,431 | 0 | Sigma-E factor regulatory protein RseB | 1,599 | |
| 0 | Succinate dehydrogenase, HMc anchor protein | 1,800 | 0 | tRNA synthase TrmP | 1,567 | |
| 0 | LysR family transcriptional regulator | 1,267 | 0 | tRNA 5-methoxyuridine(34) synthase CmoB | 1,525 | |
| 0 | Hypothetical protein | 1,688 | 0 | Chemotaxis phosphatase CheZ family protein | 1,483 | |
| 0 | Maleylacetoacetate isomerase | 1,430 | 0 | Hypothetical protein | 1,505 | |
| 0 | Sigma-E factor regulatory protein RseB | 1,599 | 0 | Hypothetical protein | 1,345 | |
| 0 | tRNA synthase TrmP | 1,567 | 0 | Outer membrane protein assembly protein | 1,301 | |
| 0 | tRNA 5-methoxyuridine(34) synthase CmoB | 1,525 | 0 | Deoxyribonuclease I | 1,269 | |
| 0 | Chemotaxis phosphatase CheZ family protein | 1,483 | 0 | Formate dehydrogenase accessory protein FdhE | 1,241 | |
| 0 | Hypothetical protein | 1,505 | 0 | Flagellar export protein FliJ | 1,208 | |
| 0 | Hypothetical protein | 1,345 | 0 | Hypothetical protein | 1,200 | |
| 0 | Hypothetical protein | 1,325 | 0 | Hypothetical protein | 1,179 | |
Notes
a SRP protein–general secretory pathway protein signal recognition particle protein
b ribo.-triP–ribonucleoside-triphosphate
c HM–hydrophobic membrane
Although we have no estimate of V for rRNA, as its sequence in part defines phyla, the tendency we see for widely distributed protein coding genes to resist LGT would also explain why rRNA is itself so refractory to transfer [13,48], the rRNA genes that are present in a recipient genome are difficult to improve upon or match in functional efficiency, and the rRNA gene product can comprise up to 20% of the cell’s dry weight [49]. Genes for rRNA thereby carry great inertia against LGT and are therefore difficult to displace by intruding copies. The rank of functional categories (Table 2) with respect to verticality reveals that the clusters functionally associated with translation rank highest, followed by nucleotide metabolism (many proteins without intermolecular interactions), replication, folding and vitamin synthesis. Genes for vitamin synthesis are not highly expressed but are widely distributed and are highly vertical. The least vertical categories comprise drug resistance and community interactions. Cognoscenti might surmise that there are no real surprises in the ranking of functional categories with respect to V, an indication that our measure of V is recovering meaningful information about gene evolution.
Table 2. Assignment of KEGG level B functional annotations.
On the left panel all prokaryotic maximum likelihood trees were annotated and sorted according to their average verticality score (Vavg). The number of clusters employed for this analysis are indicated (Nclust). The same procedure was performed on the right panel only for conserved protein families–determined by average branch length ≤ 0.1.
| All 101,422 protein families | The 8,547 most conserved protein families | ||||
|---|---|---|---|---|---|
| Function | Vavg | Nclust | Function | Vavg | Nclust |
| Translation | 5.31 | 2,428 | Translation | 14.82 | 284 |
| Metabolism of cofactors and vitamins | 4.86 | 2,443 | Nucleotide metabolism | 10.21 | 160 |
| Nucleotide metabolism | 4.28 | 1,419 | Metabolism of cofactors and vitamins | 7.95 | 199 |
| Amino acid metabolism | 3.83 | 3,777 | Carbohydrate metabolism | 7.23 | 534 |
| Carbohydrate metabolism | 3.63 | 4,836 | Replication and repair | 7.11 | 187 |
| Biosynthesis of other secondary metabolites | 3.62 | 507 | Energy metabolism | 7.07 | 208 |
| Glycan biosynthesis and metabolism | 3.42 | 3,349 | Amino acid metabolism | 7.06 | 438 |
| Metabolism | 3.31 | 4,260 | Folding, sorting and degradation | 6.77 | 118 |
| Energy metabolism | 3.28 | 2,705 | Metabolism of other amino acids | 5.87 | 81 |
| Xenobiotics biodegradation and metabolism | 3.26 | 1,606 | Metabolism | 5.67 | 337 |
| Replication and repair | 3.14 | 3,502 | Enzyme families | 5.53 | 164 |
| Transport and catabolism | 3.02 | 2,843 | Biosynthesis of other secondary metabolites | 5.50 | 25 |
| Metabolism of terpenoids and polyketides | 2.97 | 1,473 | Xenobiotics biodegradation and metabolism | 5.36 | 103 |
| Metabolism of other amino acids | 2.95 | 745 | Glycan biosynthesis and metabolism | 5.33 | 158 |
| Transcription | 2.84 | 7,245 | Signal transduction | 5.10 | 240 |
| Folding, sorting and degradation | 2.79 | 1,873 | Membrane transport | 4.69 | 1,431 |
| Lipid metabolism | 2.65 | 2,864 | Cell motility | 4.37 | 124 |
| Enzyme families | 2.59 | 3,735 | Metabolism of terpenoids and polyketides | 4.31 | 85 |
| Cellular processes and signaling | 2.49 | 3,905 | Transport and catabolism | 4.31 | 143 |
| Signal transduction | 2.48 | 6,712 | Lipid metabolism | 4.20 | 215 |
| Membrane transport | 2.46 | 19,992 | Transcription | 4.12 | 409 |
| Genetic information processing | 2.31 | 4,838 | Cellular processes and signaling | 3.75 | 257 |
| Cellular community prokaryotes | 2.21 | 3,986 | Cellular community prokaryotes | 3.55 | 172 |
| Drug resistance | 2.15 | 1,754 | Genetic information processing | 3.23 | 269 |
| Cell motility | 1.94 | 3,620 | Drug resistance | 3.10 | 88 |
| Poorly characterized | 1.41 | 178,665 | Poorly characterized | 1.68 | 2,970 |
The verticality of phyla
By averaging the verticality of all genes that occur in a given phylum, we can also estimate the verticality of phyla and rank them accordingly. This is shown in Table 3, for bacteria and archaea separately, where Pmono indicates the proportion of trees in which the given phylum was monophyletic. No phyla were always monophyletic, with values of Pmono topping out at about 0.8, meaning that the phylum was monophyletic in 80% of the trees in which its sequences occurred. At the level of phyla, for all genes and for the conserved genes, Acidithiobacilli emerge as the most vertically evolving bacteria, while the Thermococcales and Methanococcales emerge as the most vertically evolving archaea. The most laterally evolving bacteria are the Erysipelotrichia, a group of firmicutes related to Clostridia, and the Clostridia themselves for all genes, while for the conserved genes, the Gammaproteobacteria finish last when it comes to avoiding LGT. The archaea most riddled by LGT are the halophiles, which are methanogens that acquired their respiratory chain and aerobic lifestyle from bacteria [19]. Though not strict, there is a clear tendency for bacteria with a specialist lifestyle to resist LGT, and a tendency for generalists like the divisions of the proteobacteria to harbor less vertically evolving chromosomes, that, is to undergo LGT.
Table 3. Verticality of prokaryotic taxa across protein families with at least two taxonomic groups.
The list of bacterial (top) and archaeal (bottom) taxa occurring in all trees (right) and only trees that were filtered for conservation (average branch length in the tree ≤ 0.1) (left). Archaeal and bacterial phyla with less than 5 representative species in the dataset were excluded. Pmono refers the proportion of monophyletic trees. Nmono indicates the number of trees in which this taxon is monophyletic whereas Ntrees shows the number of occurrences of the phyla in the respective dataset.
| All trees– 101,423 | Conserved trees– 8,547 | |||||||
|---|---|---|---|---|---|---|---|---|
| Taxon | Pmono | Nmono | Ntrees | Pmono | Nmono | Ntrees | ||
| Bacteria | ||||||||
| Acidithiobacillia | 0.81 | 1,677 | 2,067 | 0.91 | 629 | 688 | ||
| Chlamydiae | 0.74 | 1,378 | 1,867 | 0.75 | 482 | 642 | ||
| Tenericutes | 0.68 | 2,770 | 4,076 | 0.50 | 391 | 776 | ||
| Actinobacteria | 0.60 | 30,050 | 49,958 | 0.37 | 1,214 | 3,293 | ||
| Bacilli | 0.59 | 24,365 | 41,526 | 0.25 | 1,017 | 3,997 | ||
| Chlorobi | 0.59 | 1,728 | 2,946 | 0.80 | 494 | 619 | ||
| Thermotogae | 0.57 | 2,252 | 3,937 | 0.65 | 495 | 764 | ||
| Cyanobacteria | 0.56 | 8,655 | 15,446 | 0.64 | 843 | 1,319 | ||
| Deinococcus-Thermus | 0.54 | 3,156 | 5,858 | 0.63 | 705 | 1,113 | ||
| Synergistetes | 0.53 | 1,001 | 1,872 | 0.70 | 484 | 692 | ||
| Epsilonproteobacteria | 0.52 | 3,815 | 7,270 | 0.37 | 513 | 1,397 | ||
| Fusobacteria | 0.51 | 1,805 | 3,516 | 0.60 | 717 | 1,194 | ||
| Spirochaetes | 0.50 | 5,063 | 10,130 | 0.44 | 683 | 1,564 | ||
| Bacteroidetes | 0.49 | 11,677 | 23,755 | 0.40 | 759 | 1,879 | ||
| Gammaproteobacteria | 0.48 | 29,439 | 61,803 | 0.18 | 1,078 | 5,874 | ||
| Negativicutes | 0.45 | 1,892 | 4,170 | 0.59 | 804 | 1,371 | ||
| Nitrospirae | 0.43 | 1,377 | 3,180 | 0.47 | 359 | 762 | ||
| Alphaproteobacteria | 0.43 | 18,086 | 41,953 | 0.35 | 1,312 | 3,735 | ||
| Aquificae | 0.43 | 1,210 | 2,826 | 0.43 | 290 | 672 | ||
| Planctomycetes | 0.40 | 1,755 | 4,399 | 0.55 | 533 | 961 | ||
| Chloroflexi | 0.39 | 2,349 | 6,003 | 0.46 | 521 | 1,141 | ||
| Acidobacteria | 0.38 | 1,789 | 4,666 | 0.58 | 625 | 1,077 | ||
| Betaproteobacteria | 0.38 | 14,203 | 37,225 | 0.34 | 1,601 | 4,775 | ||
| Deltaproteobacteria | 0.37 | 8,512 | 23,013 | 0.38 | 1,005 | 2,618 | ||
| Verrucomicrobia | 0.36 | 1,146 | 3,152 | 0.56 | 601 | 1,067 | ||
| Clostridia | 0.32 | 7,481 | 23,638 | 0.34 | 1,084 | 3,196 | ||
| Erysipelotrichia | 0.17 | 344 | 2,001 | 0.43 | 451 | 1,058 | ||
| Archaea | ||||||||
| Thermococcales | 0.73 | 2,482 | 3,380 | 0.79 | 271 | 341 | ||
| Methanococcales | 0.73 | 1,612 | 2,220 | 0.83 | 236 | 283 | ||
| Methanobacteriales | 0.68 | 1,949 | 2,857 | 0.79 | 282 | 356 | ||
| Sulfolobales | 0.66 | 2,223 | 3,387 | 0.75 | 280 | 374 | ||
| Archaeoglobales | 0.62 | 1,415 | 2,286 | 0.79 | 252 | 318 | ||
| Methanomicrobiales | 0.60 | 1,616 | 2,693 | 0.74 | 301 | 406 | ||
| Methanosarcinales | 0.60 | 3,392 | 5,654 | 0.63 | 318 | 503 | ||
| Thermoproteales | 0.55 | 1,537 | 2,775 | 0.61 | 257 | 420 | ||
| Thermoplasmatales | 0.49 | 662 | 1,364 | 0.58 | 212 | 366 | ||
| Desulfurococcales | 0.41 | 852 | 2,072 | 0.44 | 130 | 298 | ||
| Natrialbales | 0.32 | 1,459 | 4,503 | 0.42 | 246 | 588 | ||
| Haloferacales | 0.27 | 980 | 3,593 | 0.40 | 205 | 513 | ||
| Halobacteriales | 0.20 | 1,024 | 5,057 | 0.30 | 178 | 591 | ||
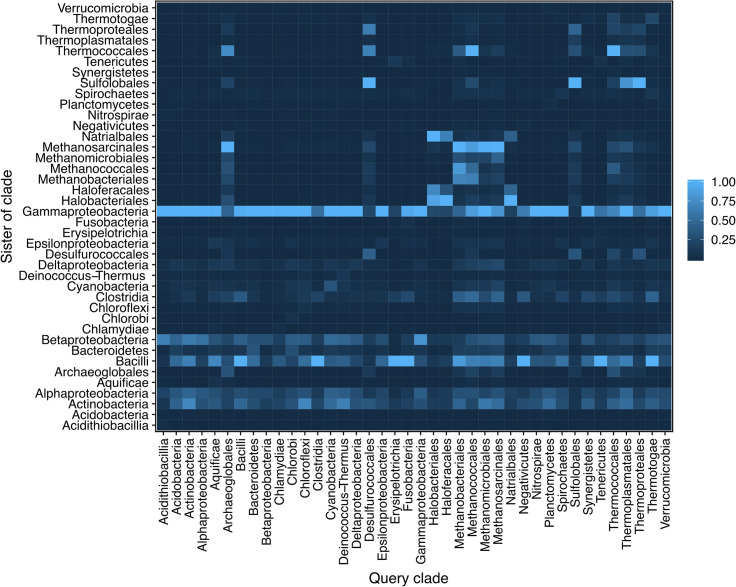
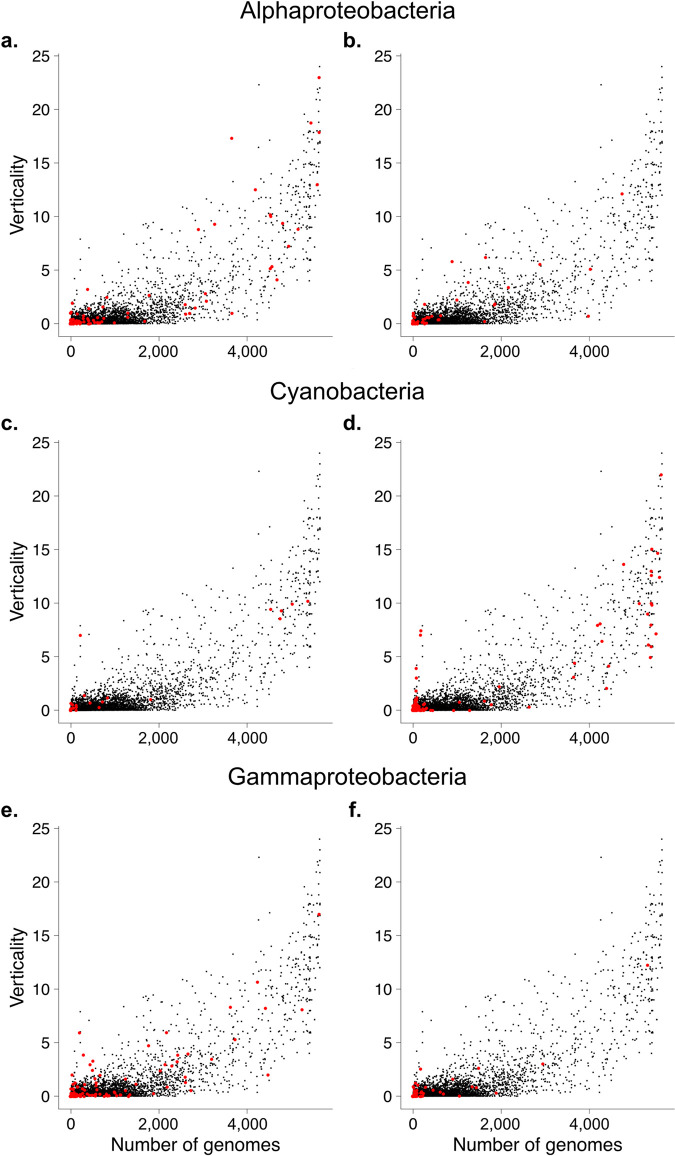
The Gammaproteobacteria were the worst offenders when it came to LGT among the 8,547 conserved gene trees, showing gammaproteobacterial monophyly in less than 20% of trees that contained the phylum. Of course, it is possible that verticality is violated by recurrent exchanges among specific pairs of taxa or by phylogenetic artefact involving true neighbors, which for Gammaproteobacteria would be the Betaproteobacteria in traditional schemes. In order to check for such effects, each time we scored a tip-resident clade in our trees, we also scored the phylogenetic membership within its sister group. A sister group can either itself be monophyletic, containing sequences from only one phylum, or it can be mixed, containing sequences from two or more different phyla. The summary is shown in Fig 3, where the frequencies of phyla in the sister group are shown. Note that a phylum can appear as its own sister group when its monophyletic clade is broken by recent LGT to a member of a different phylum: the gene tree does not change, but the taxon label of the donated gene does, leaving sequences of the donor phylum that branch below the recent export in the sister group. This is illustrated in S3d Fig. While methanogens and halophilic archaea tend to interleave, as do archaea as a whole, the dominant signal in the sister group plot is that Gammaproteobacteria tend to be the sister of virtually every phylum, meaning that they are the recipient of genes from all phyla in our sample. The tendency to undergo recent LGT—recent because we are only scoring terminal branches—is also clearly manifest in Bacilli, Betaproteobacteria, Alphaproteobacteria and Actinobacteria, all of which harbor lineages with large genomes, large pangenomes, and diverse generalist lifestyles.
Fig 3. Relative occurrence of a taxonomic group as the sister group of each clade in the unrooted trees.
For each taxonomic group in a cluster the sister was determined and counted. Multiple occurrences of different groups in the sister were accounted for by their relative occurrence. If the taxonomic group was paraphyletic, each monophyletic subgroup was determined and the sister of these were noted. The values of these subgroups were added up by multiplying the individual values of the sister by the fraction of the subgroup of the whole taxonomic group. To compare, the final values of each taxonomic group were normalized by dividing by the highest count a possible sister has gotten. It is apparent that Gammaproteobacteria are always overrepresented. It is not clear if the observed effects are due to overrepresentation of certain taxa in the data set or due to relative abundance of LGT.
The verticality of individual genomes
Averaging the value of verticality across all genes in a genome gives an estimate for the verticality of the genome, Vg. The verticality of all genomes investigated here is given in S4 Table. The most vertical genomes are those with the highest proportion of genes involved in translation. This is because the process of reductive genome evolution [50] always hones in on the ribosome, translation and information processing, as these functions are prerequisite to gene expression. The widely distributed genes involved in information processing are the most vertical (Table 1), such that the gammaproteobacterial endosymbiont Carsonella ruddii [51] which possesses only 166 protein coding genes, is the most vertical genome in our sample with Vg = 9.44. Conversely, the least vertical genomes are the largest, with the actinobacterium Amycolatopsis mediterranei (Vg = 0.84) having a genome over 10 Mb coming in last. Among the archaea, the most vertical genomes were those of H2 dependent autotrophs (S4 Table). The most vertical genome was that of the highly reduced free living archaeon, Ignicoccus hospitalis [52] (Vg = 4.10) an extreme specialist that grows only on H2 + S0, followed by nine H2 dependent methanogens, starting with the thermophilic methanogen Methanothermus fervidus (Vg = 4.09), with a genome of 1.2 Mb [53]. The most lateral archaeal genome was that of the halophile Haloterrigena turkmenica (Vg = 1.66).
Eukaryotes
In an ideal world of vertically inherited genes and infallible phylogeny, all genes would produce the same tree and all eukaryotic genes would trace to the same alphaproteobacterium (the mitochondrion) and the same are archaeon (host), plus the same cyanobacterium in the case of eukaryotes with plastids. But the real data from real genomes reveals that only a small minority of prokaryotic genes, much less than 1%, tend to be inherited vertically. How does the non-verticality of prokaryotic genes, genomes, and phyla impact our ability to infer the origin of eukaryotic genes? For all 3,420,731 protein coding genes from 150 eukaryotic genomes, we constructed clusters, merged them with their cognate prokaryotic clusters to generate eukaryote-prokaryote clusters (EPCs), constructed alignments and ML trees (see Methods). The red circles in Fig 1 mark the prokaryotic clusters that were merged with their unique cognate eukaryotic clusters. The first question concerned eukaryote monophyly. There are many claims in the literature for LGT from prokaryotes to eukaryotes, but few are supported by prokaryotic reference samples that reflect the availability of genome data and fewer still, if any, are supported by systematic tests for eukaryote monophyly. Therefore, we looked closely at the possibility of LGT vs. eukaryote monophyly in our sample.
Among the 2,575 maximum-likelihood (ML) trees reconstructed from the merged eukaryote-prokaryote clusters, only 475 of the best trees found (18.4%) failed to recover eukaryotes as monophyletic. Does that finding represent evidence for LGT to eukaryotes in 18% of these trees, that is, is the best tree identified significantly better than the case of eukaryote monophyly? To test whether the lack of eukaryote monophyly in those 475 trees is due to reconstruction errors or due to prokaryote-eukaryote LGT, we compared the ML trees against trees with constrained eukaryote monophyly using likelihood tests. We employed the Kishino-Hasegawa test (KH), the approximately-unbiased test (AU) and the Shimodaira-Hasegawa test (SH) (see Methods for details). At the 5% significance level (p-value ≤ 0.05), the KH test rejected eukaryote monophyly for 6% of the trees (30 out of 475), that is, the null hypothesis (eukaryote monophyly) was rejected at a rate very close to that expected by chance. The AU test rejected eukaryote monophyly for 3 trees while the SH test did not reject eukaryote monophyly for any tree at the p-value of ≤ 0.05 (S4 Fig and S5 Table). Thus, the absence of a pure eukaryotic clade in some of the best trees found by ML trees results from challenges in distinguishing alternative trees that are statistically identical to the true tree, or to trees recovering eukaryote monophyly, in terms of their likelihood values, a problem that becomes more acute for phylogenetic inference using large samples because the tree space for the ML method to search grows exponentially. In terms of traits, eukaryotes are the strongest monophylum in nature, a status corroborated by the lack of any evidence that would support a case for the non-monophyly (LGT) of eukaryotic genes.
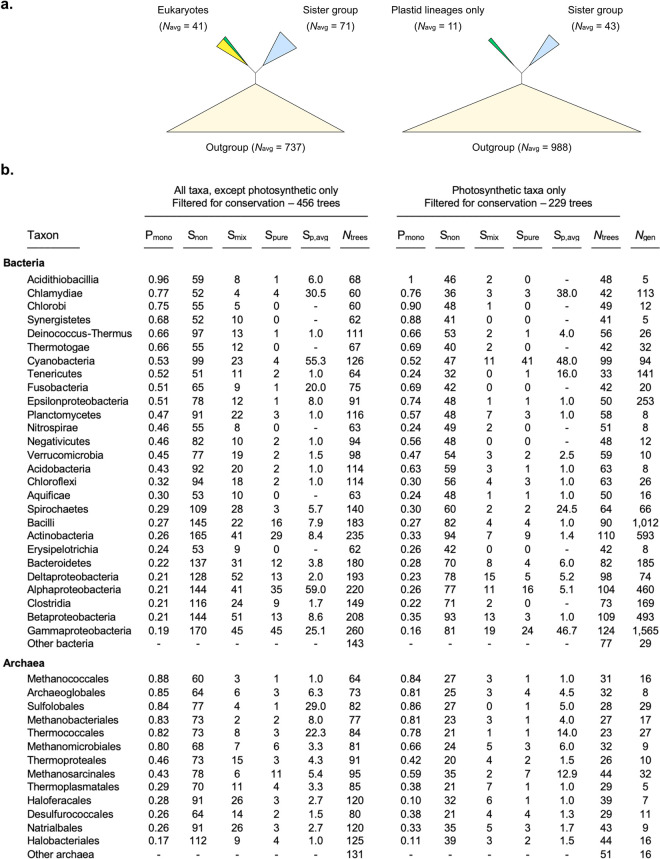
What do trees say about the origin of eukaryotic genes? In the following, to avoid the effects of trees for poorly conserved genes (Fig 1A), we consider only those 685 trees in which the eukaryotic cluster mapped to one of the conserved prokaryotic clusters in Fig 1B. For each tree, we determined the prokaryotic sister group to the eukaryotic clade, and scored whether it was a pure sister containing sequences from only one prokaryotic phylum or a mixed sister group containing a mixed sister group from two or more phyla. The results are summarized in Fig 4B.
Fig 4. Identification of the prokaryotic sister group to the eukaryotes in 2,575 eukaryotic-prokaryotic unrooted gene trees (EPC).
a. shows the average clade sizes for eukaryotes, the sister group to eukaryotes and the outgroup in the analyzed trees for (right) the 229 trees with only plastid derived lineages and (left) for the 456 EPCs containing all taxa except photosynthetic lineages. b. details the list of bacterial (top) and archaeal (bottom) phyla occurring in the trees with only plant lineages (right) and all other trees (left) that were filtered for conservation (average branch length of the tree ≤ 0.1). Archaeal and bacterial phyla with less than 5 representative species in the dataset were collapsed into ‘other archaea’ and ‘other bacteria’ groups. Pmono refers to the proportion of trees with a branch (split) separating the species of the respective phylum from all the others in the tree; Snon refers to the number of occurrence of the phylum only in the outgroup clade; Smix refers to the number of occurrences of the phylum as a mixed sister (more than one phylum in the clade); Spure refers to the number of occurrences of the phylum as pure sister (as the single phylum); Sp,avg shows the average size of the sister clade when the phylum occurs as a pure sister clade. Ntrees show the number of occurrences of the phyla across the trees and Ngen indicates the number of species in each taxon included in the complete dataset.
By the measure of phylogenetic inference, every prokaryotic phylum sampled in our study appears as a donor of genes to the eukaryote common ancestor, either by presence in a mixed sister group or as a pure sister (Fig 4B). This is true not only for bacteria, which would be expected to trace mitochondrial ancestry, but also for archaea, which since their discovery have been linked to the origin of the host. Can we naïvely interpret such observations at face value? Is it reasonable to believe that every phylum sampled here donated a gene, or several, to eukaryotes at their origin? If we break the data down to families, genera, or species, the number of donors grows accordingly (all prokaryotic organisms employed in this study were in the sister group to eukaryotes at least once), such that each gene in eukaryotes would correspond to an individual donation, as some would argue [54]. But that logic leads straight to the erroneous conclusion that ancestral plastid and mitochondrial genomes were assembled by acquisition one gene at a time [55] the converse of what they are in plain sight, namely reduced genomes of single bacterial endosymbionts [50] that underwent reductive evolution by transferring genes to the nucleus. Worse yet, the same problem ensues at the origin of plastids (Fig 4B, right column), because for photosynthetic eukaryotes again all phyla, including the archaea, appear as donors. Many genes that are germane to photosynthesis in eukaryotes trace to the plant common ancestor (plants being monophyletic) but only a minority of them trace specifically to Cyanobacteria, and those that do, do not trace to the same cyanobacterium [56,57].
If we only consider pure sister groups to eukaryotes, the most common apparent gene donor was Gammaproteobacteria, followed by Alphaproteobacteria, Actinobacteria and Bacilli. There is at least one theory in the literature invoking the participation of those groups at eukaryote origin [58]. However, a similar pattern recurs for plastids, which have the strongest pure sister signal from Cyanobacteria followed again by Gammaproteobacteria (for which there is no plastid origin theory) and Alphaproteobacteria. The problem of inferring symbionts from gene trees becomes more evident when we consider apparent archaeal contributions to the origins of plastids (Fig 4B), because there are no archaea that synthesize chlorophyll. We are confronted with a conflict. Blind inference of symbionts from trees cannot account for the origin of organelle genomes, the strongest form of evidence for the origin of organelles in the first place. The ‘one tree one donor’ logic carries a weighty premise that is never spelled out by its proponents, namely that the donated genes never underwent LGT among free living prokaryotes in the 1.5 billion years since organelle origin. If we approach the problem from the standpoint of theory testing in the presence of prior knowledge about the underlying process, namely one symbiont 1.5 billion years ago (as evidenced by the single origin of plastids and mitochondria respectively), what would look like many donors if we were to assume that prokaryotic gene evolution is vertical, is clearly the result of LGT among free-living prokaryotes, where, in real data, gene evolution is lateral.
For example, were the gammaproteobacterial signal in heterotrophic eukaryotes a result of gene acquisitions from donors with gammaproteobacterial rRNA, then that same signal would reflect a gammaproteobacterial origin of plastids (Fig 4B), which seems unlikely and is not covered by any theory. If on the other hand it were due to the low verticality of Gammaproteobacteria as a phylum, then Gammaproteobacteria should appear as the sister to many different groups of prokaryotes, which is precisely the observation (Fig 3). We asked whether there is a non-random signal across all genes that singles out Cyanobacteria (plastids) and Alphaproteobacteria (mitochondria) specifically as donors. This is shown in Fig 5, where we have plotted the distribution of trees that identify Alphaproteobacteria, Cyanobacteria or Gammaproteobacteria as pure sisters to (donors of) eukaryotic genes. Though Gammaproteobacteria appear as the pure sister in many trees (Fig 4B), the genes that do so are primarily of low verticality. Only the Alphaproteobacteria have a significant enrichment of vertical genes as sisters relative to the sample (Fig 5A), but the significance is marginal (p < 0.01). The Cyanobacteria are not significantly enriched in high verticality sisters, because of a large number of low verticality cases (Fig 5C and 5D). The majority of the gammaproteobacterial sister cases are low verticality genes (Fig 5E and 5F).
Fig 5.
Mapping of EPCs to prokaryotic clusters. The EPCs were separated according to the pure sister group of eukaryotes in the unrooted trees: a. and b. Alphaproteobacteria, c. and d. Cyanobacteria, e. and f. Gammaproteobacteria. The left panel shows EPCs that may include all eukaryotic supergroups but no groups that include only photosynthetic lineages, the right panel shows only EPCs that only include photosynthetic eukaryotes (lineages from SAR, Hacrobia and Archaeplastida). Meaning the latter are indicative of plastid endosymbiosis. Plots for all taxa see S5 Fig.
Throughout this discussion, we recall that the ancestor of mitochondria was not a phylum of proteobacteria, it was a single proteobacterium that engaged in a singular symbiosis. The same is true for plastids, whose origin was not the result of a symbiosis with the cyanobacterial phylum, it was a symbiosis with a single cyanobacterium. The genes that trace to those organelle origin events are, however, like almost all prokaryotic genes, of low verticality within prokaryotes.
A critic might ask whether eukaryotes, if their genes are of monophyletic origin relative to prokaryotes, score higher than all prokaryotes in terms of a comparable measure of verticality (supergroups instead of phyla). The problem there is a different one, namely paralogy. The underlying theme of eukaryotic genome evolution is recurrent gene and genome duplication [59,60], massive paralogy impairs eukaryote gene monophyly although gene duplications carry phylogenetic information in their own right [35]. The genes that have remained in plastid and mitochondrial genomes encode proteins involved in the electron transport chain of the bioenergetic membrane supporting photosynthesis and respiration, respectively, and the ribosomal proteins [61] involved in synthesizing those proteins in the organelle [62]. Why do those ribosomal proteins reflect an alphaproteobacterial [46] and cyanobacterial [56] origin of the organelle more clearly than non-ribosomal genes? It is not because non-ribosomal genes were acquired from different biological donors. Rather, it is because the prokaryotic reference set of ribosomal proteins is inherited in a vertical manner among free living prokaryotes; all other prokaryotic genes are inherited more laterally (Fig 1), evoking the illusion of many different donors to eukaryotes in phylogenetic analyses (Fig 4B). Yet that illusion rests upon the tacit assumption that prokaryotes inherit their genes vertically, which is however untrue [2,34,63,64,65].
Discussion
Even though gene evolution in prokaryotes has substantial lateral components, rRNA-based investigations and some protein phylogenetic studies tend to recover groups that microbiologists recognized long before molecular systematics. Hence the groups are in some cases real and there must be a vertical component to prokaryote evolution. The vertical component has, however, been difficult to quantify across lineages. Equally elusive have been estimates for verticality itself, yet suitable methods to quantify that component have been obscure, as have means to quantify verticality across prokaryotic genes. Quantification of discordance in tree comparisons represents one approach [66] to estimate LGT or lack thereof, but its utility is limited when large genome samples are involved, because the number of possible trees exceeds the number that a computer can examine by hundreds of orders of magnitude for trees containing 60 leaves or more. By exploiting the common wisdom that phylogeny works better at the tips of trees than at their deeper branches, we have obtained robust estimates of verticality.
Though many genes that are currently used in molecular systematic studies based on their widespread occurrence have low verticality, across all genes V does increase with distribution density. We suggest that this is so because the displacement of a well-regulated preexisting copy is less likely than the transient and rarely permanent, in some cases lineage founding [67], acquisition of novel traits. That most genes in prokaryotes have both restricted distribution and low verticality underscores the need to identify genes that are inherited vertically across large data sets for the purpose of higher-level broad scale phylogenetic analyses. We found no genes among the 101,422 total clusters and 8,547 conserved clusters that recovered monophyly of all 40 phyla. At the same time all phyla were disguised as gene donors to eukaryotes both at the origin of mitochondria and at the origin of plastids because of LGT among the prokaryotic reference set.
The spectrum of verticality across genes observed here precludes the need to propose, based on trees that implicate non-alphaproteobacterial or non-cyanobacterial gene donors, genetic contributors at the origin of eukaryotes beyond the host, the mitochondrion and, later, the cyanobacterial antecedent of plastids, because LGT among prokaryotes can account for the same gene-tree based observations, more directly and with fewer corollary assumptions, while simultaneously accounting for a larger set of observations among the prokaryotic reference set. The criterion of verticality can furthermore be of practical use in the selection of genes for molecular systematic studies.
Methods
Prokaryotic dataset
Protein sequences for 5,655 prokaryotic genomes were downloaded from NCBI [68] (version September 2016; see S3 Table for detailed species composition). We performed all-vs-all BLAST [69] searches (BlastP version 2.5.0 with default parameters) and selected all reciprocal best hits with e-value ≤ 10−10. The protein pairs were aligned with the Needleman-Wunsch algorithm [70] (EMBOSS needle) and the pairs with global identity values < 25% were discarded. The retained global identity pairs were used for clustering using Markov clustering algorithm [71] (MCL) version 12–068, changing default parameters for pruning (-P 180000, -S 19800, -R 25200). Clusters distributed in at least 4 genomes spanning 2 prokaryotic phyla were retained, resulting in 101,422 used clusters in total. Sequence alignments for each cluster were generated using MAFFT [72], with the iterative refinement method that incorporates local pairwise alignment information (L-INS-i; version 7.130). The resulting alignments were used to reconstruct maximum-likelihood trees with RAxML version 8.2.8 [73] (parameters: -m PROTCATWAG -p 12345) (S9 Table). The trees were rooted with the Minimal Ancestor Deviation method (MAD) [74].
Eukaryotic dataset
Protein sequences for 150 eukaryotic genomes were downloaded from NCBI, Ensembl Protists and JGI (see S7 Table for detailed species composition). To construct gene families, we performed an all-vs-all BLAST [66] of the eukaryotic proteins (BlastP version 2.5.0 with default parameters) and selected the reciprocal best BLAST hits with e-value ≤ 10−10. The protein pairs were aligned with the Needleman-Wunsch algorithm (EMBOSS needle) [70] and the pairs with global identity values < 25% were discarded. The retained global identity pairs were used to construct gene families with the MCL algorithm [71] (version 12–068) with default parameters. We considered only the gene families with multiple gene copies in at least two eukaryotic genomes. Protein-sequence alignments for the multi-copy gene families were generated using MAFFT [72], with the iterative refinement method that incorporates local pairwise alignment information (L-INS-i, version 7.130). The alignments were used to reconstruct maximum likelihood trees with IQ-tree [75], applying the parameters ‘-bb 1000’ and ‘-alrt 1000’ (version 1.6.5), with subsequent rooting with MAD [74].
Eukaryotic-prokaryotic dataset
To assemble a dataset of conserved genes for phylogenies linking prokaryotes and eukaryotes, eukaryotic, archaeal and bacterial protein sequences were first clustered separately before homologous clusters between eukaryotes and prokaryotes were identified. Eukaryotic protein sequences from 150 genomes (S7 Table) were clustered with MCL [71] using global identities from best reciprocal BLAST hits for protein pairs with e-value ≤ 10−10 and global identity ≥ 40%. The clusters with genes distributed in at least two eukaryotic genomes were retained. Similarly, prokaryotic protein sequences from 5,655 genomes were clustered using the best reciprocal BLAST for protein pairs with e-value ≤ 10−10 and global identity ≥ 25% (for archaea and bacteria, separately). The resulting clusters with gene copies in at least 5 prokaryotic genomes were retained. Eukaryotic and prokaryotic clusters were merged using the reciprocal best cluster procedure [57]. We merged a eukaryotic cluster with a prokaryotic cluster if ≥ 50% of the eukaryotic sequences in the cluster have their best reciprocal BLAST hit in the same prokaryotic cluster and vice-versa (cut-offs: e-value ≤ 10−10 and local identity ≥ 30%) yielding 2,587 eukaryotic-prokaryotic clusters (EPCs). EPCs with ambiguous cluster assignment were discarded. Protein-sequence alignments for 2,575 EPCs were generated using MAFFT (L-INS-i, version 7.130); for twelve clusters, the alignment did not compute as sequence quality was low. The alignments were used to reconstruct maximum-likelihood trees with IQ-tree (version 1.6.5) employing the parameters ‘-bb 1000’ and ‘-alrt 1000’ (S5 Table).
Verticality
The verticality measure for each gene was defined as the sum of monophyly scores for all monophyletic taxa present in the unrooted trees. Only for the calculation of the average root-to-tip measurements (S2 Fig) rooted trees were necessary. This supplementary analysis was then performed with MAD rooted trees. Our species set contains 42 taxa corresponding mostly to phyla level, except for Proteobacteria, Firmicutes and Achaea (see S8 Table). For a given tree, the monophyly score Sa for taxon a was defined as:
where na is the number of species in the tree affiliated to a and Na is the total number of species from a among the 5,655 genomes in our set. The verticality measure Vg for a gene was then defined as:
The analyses were conducted with custom scripts using NewickUtilities [76] and ETE [77]. Taxon and genome verticality were defined as the average gene verticality across all gene trees where the taxon (or genome) were present. In addition, weighted taxon verticality for each taxon was defined as the weighted average across all gene trees where the phylum appears, weighted meaning here that values of monophyletic clusters were summed up while values of paraphyletic clusters were subtracted.
Functional annotation
Two annotation strategies were performed for each protein cluster. First, protein annotation information according to the BRITE (Biomolecular Reaction pathways for Information Transfer and Expression) hierarchy was downloaded from the Kyoto Encyclopedia of Genes and Genomes (KEGG v. September 2017) website [78], including protein sequences and their assigned function according to the KO numbers. The protein sequences of the 5,655 organisms were mapped to the KEGG database using local alignments with ‘blastp’. Only the best BLAST hit of the given protein with an e-value ≤ 10−10 and alignment coverage of 80% was selected. After assigning a function based on the KO numbers of KEGG for each protein in the clusters, the majority rule was applied to identify the function for each cluster. The occurrence of the function of each protein in the cluster was added and the most prevalent function was assigned for each cluster.
The second annotation used the NCBI headers. For this, the appearance of a word among all sequence headers of a cluster was counted. Then, each header was given a score based on the sum of how often its words appeared among all headers. The header with the highest score was then chosen as the cluster annotation.
Tests for eukaryote monophyly
For 475 gene trees where eukaryotes were not recovered as monophyletic, we conducted the Kishino-Hasegawa (KH) test [79], the Shimodaira-Hasegawa (SH) test [80] and the approximately-unbiased (AU) test [81] to assess whether the observed non-monophyly was statistically significant. We reconstructed trees constraining eukaryotic sequences to be monophyletic, but not imposing any other topological constraint, using FastTree [82] (version 2.1.10 SSE3) and recording all trees explored during the tree search with the ‘-log’ parameter. The sample of monophyletic trees were used as input in IQ-tree (version 2.0.3; parameter: ‘-zb 100000 -au’) to perform the KH, SH and AU tests against the unconstrained tree (non-monophyletic). If the best constrained tree did not show significant difference relative to the unconstrained tree (p-value ≤ 0.05), then we considered that eukaryotic monophyly cannot be rejected.
Sister analyses
Prokaryotes
The sister for each prokaryotic taxon was defined as the clade with the smallest branch to the query clade. Two cases had to be differentiated: Mono- or paraphyletic taxonomic groups in a tree. Monophyly was tested as described above with NewickUtilities. For these taxonomic groups, the sister groups could also be directly obtained by using NewickUtilities (nw_clade -s). Finally, all different taxonomic groups in the sister groups were given a score equal to their proportion in the sister group. For paraphyly of a taxonomic group (main group), the monophyletic subgroups were determined with the python package ETE 3 [77]. Each of these subgroups was handled as an individual group in the cluster and the sister clades were determined. Again, if several taxonomic groups were present in a sister group, then these were given a score equal to their proportion in the sister. To get from the scores of each subgroup to the total score of the main group, each subgroup´s scores was multiplied by the proportion of genomes the subgroup has of the main group. Subsequently, the score of a potential sister group to the main group was calculated by summing up its adjusted score over all subgroups. For each taxonomic group, sister scores were normalized by dividing each score through the highest sister score and then plotted as a heatmap.
Eukaryotes
To infer the prokaryotic sisters to eukaryotes we used 2,575 EPC trees. The majority of the EPC trees (2,100) support eukaryotic monophyly. For 475 trees for which eukaryotes did not branch together we recalculated trees constraining eukaryotic monophyly because the Shimodaira-Hasegawa tests failed to reject eukaryotic monophyly for all the 475 trees (see Methods section ‘tests for eukaryote monophyly’ and main text). Note that in unrooted trees for which eukaryotes are monophyletic, the prokaryotic side of the tree is bisected by one internal node into two prokaryotic subclades, each subclade being the potential sister to eukaryotes (Fig 4A). We considered the prokaryotic subclade with the smallest number of leaves for our inferences of sister-relations.
Terminal gene duplications
Terminal gene duplications were inferred using the rooted gene trees as pairs of genes sampled from the same genome that appeared as reciprocal sisters in the tree. Gene trees with ambiguous MAD roots were discarded.
Statistical tests
To test the correlations of variables, the Pearson´s correlation test was used [83]. The test results of various combinations for example Number of genomes and number of phyla, that are not mentioned in the text are given in S2 Table.
Supporting information
(XLSX)
(TXT)
(XLSX)
(XLSX)
(XLSX)
(TXT)
(TXT)
(DOCX)
(XLSX)
a. Shows the cumulative frequency of the proportion of duplications of all 5,655 prokaryotic organisms (red) compared to the 150 eukaryotes (blue) in our dataset. b. Shows the cumulative frequency of 100 random sample sets of 150 prokaryotic organisms each (red) versus the 150 eukaryotic organisms (blue) in the dataset.
(TIF)
Comparison of verticality, normalized by multiplying raw monophyly count by their average root to leave distance of each tree, and number of genomes in a protein cluster for a. all clusters (n = 101,422) and b. all conserved clusters (average branch length ≥ 0.1; n = 8,547). The plot is created analogous to Fig 1 in the main text and this alternative verticality calculation also correlates to number of genomes (A: p < 10–300, Pearson´s R2 = 0.571; B: p < 10–300, R2 = 0.686). The correlation is more consistent when comparing verticality to number of phyla represented in a cluster (a: p < 10–300, Pearson´s R2 = 0.754; b: p < 10–300, R2 = 0.767, see S2 Table for more details). The eukaryotic-prokaryotic clusters (EPCs) are highlighted in red and the clusters that correspond to a gene from the mitochondrial genome of Reclinomonas americana [45] are displayed in blue triangles along the abscissa of the plot and in the graph. For the latter, the gene identifier was noted above each plot. Ribosomal proteins are indicated by the black diamond on the right of each plot and in the graph [6]. Notably, these clusters show a steep decline in clusters with lower verticality among the conserved clusters.
(TIF)
Each phylum is indicated by one color as depicted in the legend of the table. If the phylum is monophyletic in the tree, the number of genomes in the tree are divided by the number of genomes of this phylum present in the dataset of 5,655 organisms–phyla e and f in the panels a. and b. of the figure. If the phylum is paraphyletic, the verticality is set to '0'–phyla g and h in panels c. and d. of the figure. This number represents the verticality for each phylum. The sum of all verticality scores for the phyla in the tree is then the verticality for the tree and conversely, for the gene.
(TIF)
The Kishino-Hasegawa (KH) test, Shimodaira-Hasegawa (SH) test and the Approximately-Unbiased (AU) test were performed for 475 prokaryote-eukaryote genes for which eukaryotes were not recovered monophyletic in the ML trees. The histogram shows the distribution of p-values (horizontal axis) for the tests of the unconstrained ML trees against ML trees with constrained eukaryote monophyly. A test was considered significant (eukaryote monophyly was rejected) if p-value ≤ 0.05.
(TIF)
Mapping of EPCs to prokaryotic clusters. The EPCs were separated according to the pure sister group of eukaryotes in the trees and plotted in the same way as in Fig 4 of the main text. The left panel shows EPCs that may include all eukaryotic supergroups, the right panel shows only EPCs that include archaeplastidal eukaryotes. Meaning the latter are indicative of plastid endosymbiosis. For a better overview a headline is included in each plot that lists the taxonomic group represented, if it shows EPCs linked to the mitochondrial (‘P and O’, left panel) or to the plastidal endosymbiosis event (‘Plant only’, right panel), and the number of EPCs that are shown as red dots.
(GZ)
Acknowledgments
We thank the central computing unit, ZIM, at the University of Düsseldorf for providing the computational platform for these analyses.
Data Availability
Files for S9 Table are available on the repository of our university: http://dx.doi.org/10.25838/d5p-12. The other supplementary files are provided with the manuscript and Supporting Information.
Funding Statement
This study was supported by the European Research Council (666053), the Volkswagen Foundation (93 046), and the Moore-Simons Project on the Origin of the Eukaryotic Cell (9743) which were awarded to WFM. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.McDaniel LD, Young E, Delaney J, Ruhnau F, Ritchie KB, Paul JH. High frequency of horizontal gene transfer in the oceans. Science 2010;330(6000):50 10.1126/science.1192243 [DOI] [PubMed] [Google Scholar]
- 2.Ochman H, Lawrence JG, Groisman EA. Lateral gene transfer and the nature of bacterial innovation. Nature 2000;405(6784):299–304. 10.1038/35012500 [DOI] [PubMed] [Google Scholar]
- 3.Popa O, Dagan T. Trends and barriers to lateral gene transfer in prokaryotes. Curr Opin Microbiol 2011;14(5):615–623. 10.1016/j.mib.2011.07.027 [DOI] [PubMed] [Google Scholar]
- 4.Rasko DA, Rosovitz MJ, Myers GSA, Mongodin EF, Fricke WF, Gajer P, et al. The pangenome structure of Escherichia coli: comparative genomic analysis of E. coli commensal and pathogenic isolates. J Bacteriol 2008;190(20):6881–6893. 10.1128/JB.00619-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lukjancenko O, Wassenaar TM, Ussery DW. Comparison of 61 sequenced Escherichia coli genomes. Microb Ecol 2010;60(4):708–720. 10.1007/s00248-010-9717-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hansmann S, Martin W. Phylogeny of 33 ribosomal and six other proteins encoded in an ancient gene cluster that is conserved across prokaryotic genomes: influence of excluding poorly alignable sites from analysis. Int J Syst Evol Microbiol 2000;50 Pt 4:1655–1663 10.1099/00207713-50-4-1655 [DOI] [PubMed] [Google Scholar]
- 7.Charlebois RL, Doolittle WF. Computing prokaryotic gene ubiquity: rescuing the core from extinction. Genome Res 2004;14(12):2469–2477. 10.1101/gr.3024704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ciccarelli FD, Doerks T, von Mering C, Creevey CJ, Snel B, Bork P. Toward automatic reconstruction of a highly resolved tree of life. Science 2006;311(5765):1283–1287. 10.1126/science.1123061 [DOI] [PubMed] [Google Scholar]
- 9.Dagan T, Martin W. The tree of one percent. Genome Biol 2006;7(10):118 10.1186/gb-2006-7-10-118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Koonin EV, Wolf YI, Puigbò P. The phylogenetic forest and the quest for the elusive tree of life. Cold Spring Harb Symp Quant Biol 2009;74:205–213. 10.1101/sqb.2009.74.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dagan T, Artzy-Randrup Y, Martin W. Modular networks and cumulative impact of lateral transfer in prokaryote genome evolution. Proc Natl Acad Sci U S A 2008;105(29):10039–10044. 10.1073/pnas.0800679105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ku C, Martin WF. A natural barrier to lateral gene transfer from prokaryotes to eukaryotes revealed from genomes: the 70% rule. BMC Biol 2016;14(1):89 10.1186/s12915-016-0315-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sorek R, Zhu Y, Creevey CJ, Francino MP, Bork P, Rubin EM. Genome-wide experimental determination of barriers to horizontal gene transfer. Science 2007;318(5855):1449–1452. 10.1126/science.1147112 [DOI] [PubMed] [Google Scholar]
- 14.Pál C, Papp B, Lercher MJ. Adaptive evolution of bacterial metabolic networks by horizontal gene transfer. Nat Genet 2005;37(12):1372–1375. 10.1038/ng1686 [DOI] [PubMed] [Google Scholar]
- 15.Lercher MJ, Pál C. Integration of horizontally transferred genes into regulatory interaction networks takes many million years. Mol Biol Evol 2008;25(3):559–567. 10.1093/molbev/msm283 [DOI] [PubMed] [Google Scholar]
- 16.Chen W-H, Trachana K, Lercher MJ, Bork P. Younger genes are less likely to be essential than older genes, and duplicates are less likely to be essential than singletons of the same age. Mol Biol Evol 2012;29(7):1703–1706. 10.1093/molbev/mss014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dilthey A, Lercher MJ. Horizontally transferred genes cluster spatially and metabolically. Biol Direct 2015;10:72 10.1186/s13062-015-0102-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grassi L, Caselle M, Lercher MJ, Lagomarsino MC. Horizontal gene transfers as metagenomic gene duplications. Mol Biosyst 2012;8(3):790–795. 10.1039/c2mb05330f [DOI] [PubMed] [Google Scholar]
- 19.Nelson-Sathi S, Dagan T, Landan G, Janssen A, Steel M, McInerney JO, et al. Acquisition of 1,000 eubacterial genes physiologically transformed a methanogen at the origin of Haloarchaea. Proc Natl Acad Sci U S A 2012;109(50):20537–20542. 10.1073/pnas.1209119109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineau S, et al. CRISPR provides acquired resistance against viruses in prokaryotes. Science 2007;315(5819):1709–1712. 10.1126/science.1138140 [DOI] [PubMed] [Google Scholar]
- 21.Holt KE, Wertheim H, Zadoks RN, Baker S, Whitehouse CA, Dance D, et al. Genomic analysis of diversity, population structure, virulence, and antimicrobial resistance in Klebsiella pneumoniae, an urgent threat to public health. Proc Natl Acad Sci U S A 2015;112(27):E3574–E3581. 10.1073/pnas.1501049112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brockhurst MA, Harrison E, Hall JPJ, Richards T, McNally A, MacLean C. The ecology and evolution of pangenomes. Curr Biol 2019;29(20):R1094–R1103. 10.1016/j.cub.2019.08.012 [DOI] [PubMed] [Google Scholar]
- 23.Croll D, McDonald BA. The accessory genome as a cradle for adaptive evolution in pathogens. PLoS Pathog 2012;8(4):e1002608 10.1371/journal.ppat.1002608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McInerney JO, McNally A, O’Connell MJ. Why prokaryotes have pangenomes. Nat Microbiol 2017;2:17040 10.1038/nmicrobiol.2017.40 [DOI] [PubMed] [Google Scholar]
- 25.Vernikos G, Medini D, Riley DR, Tettelin H. Ten years of pan-genome analyses. Curr Opin Microbiol 2015;23:148–154. 10.1016/j.mib.2014.11.016 [DOI] [PubMed] [Google Scholar]
- 26.Chatton E. Pansporella perplexa. Amoebien a spores protégées parasite des daphnies. Réflexions sur la biologie et la phylogénie des protozoaires. Ann Sci Nat Zool 1925;8:5–85. [Google Scholar]
- 27.Creevey CJ, Fitzpatrick DA, Philip GK, Kinsella RJ, O’Connell MJ, Pentony MM, et al. Does a tree-like phylogeny only exist at the tips in the prokaryotes? Proc Biol Sci 2004;271(1557):2551–2558. 10.1098/rspb.2004.2864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Semple C, Steel MA. Phylogenetics Reprinted. Oxford: Oxford Univ. Press; 2009. (Oxford lecture series in mathematics and its applications; vol 24). [Google Scholar]
- 29.McPherson RA. The Numbers Universe: An outline of the dirac/eddington numbers as scaling factors for fractal, black hole universes. Electronic Journal of Theoretical Physics 2008;5(18). [Google Scholar]
- 30.Nakamura Y, Itoh T, Matsuda H, Gojobori T. Biased biological functions of horizontally transferred genes in prokaryotic genomes. Nat Genet 2004;36(7):760–766. 10.1038/ng1381 [DOI] [PubMed] [Google Scholar]
- 31.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 1987;4(4):406–425. 10.1093/oxfordjournals.molbev.a040454 [DOI] [PubMed] [Google Scholar]
- 32.Landan G, Graur D. Heads or tails: a simple reliability check for multiple sequence alignments. Mol Biol Evol 2007;24(6):1380–1383. 10.1093/molbev/msm060 [DOI] [PubMed] [Google Scholar]
- 33.Criscuolo A. morePhyML: improving the phylogenetic tree space exploration with PhyML 3. Mol Phylogenet Evol 2011;61(3):944–948. 10.1016/j.ympev.2011.08.029 [DOI] [PubMed] [Google Scholar]
- 34.Treangen TJ, Rocha EPC. Horizontal transfer, not duplication, drives the expansion of protein families in prokaryotes. PLoS Genet 2011;7(1):e1001284 10.1371/journal.pgen.1001284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tria FDK, Brückner J, Skejo J, Xavier JC, Zimorski V, Gould SB, et al. Gene duplications trace mitochondria to the onset of eukaryote complexity; 2019. (vol 176) bioRxiv. 10.1101/781211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Szöllősi GJ, Davín AA, Tannier E, Daubin V, Boussau B. Genome-scale phylogenetic analysis finds extensive gene transfer among fungi. Philos Trans R Soc Lond B, Biol Sci 2015;370(1678):20140335 10.1098/rstb.2014.0335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jain R, Rivera MC, Lake JA. Horizontal gene transfer among genomes: the complexity hypothesis. Proc Natl Acad Sci U S A 1999;96(7):3801–3806. 10.1073/pnas.96.7.3801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rambaut A, Lam TT, Max Carvalho L, Pybus OG. Exploring the temporal structure of heterochronous sequences using TempEst (formerly Path-O-Gen). Virus Evol 2016;2(1):vew007 10.1093/ve/vew007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Niehus R, Mitri S, Fletcher AG, Foster KR. Migration and horizontal gene transfer divide microbial genomes into multiple niches. Nat Commun 2015;6:8924 10.1038/ncomms9924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nei M. Molecular evolutionary genetics. New York: Columbia University Press; 1987. [Google Scholar]
- 41.Aziz RK, Breitbart M, Edwards RA. Transposases are the most abundant, most ubiquitous genes in nature. Nucleic Acids Res 2010;38(13):4207–4217. 10.1093/nar/gkq140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nevers P, Saedler H. Transposable genetic elements as agents of gene instability and chromosomal rearrangements. Nature 1977;268(5616):109–115. 10.1038/268109a0 [DOI] [PubMed] [Google Scholar]
- 43.Goremykin VV, Hansmann S, Martin WF. Evolutionary analysis of 58 proteins encoded in six completely sequenced chloroplast genomes: Revised molecular estimates of two seed plant divergence times. Pl Syst Evol 1997;206(1–4):337–351. [Google Scholar]
- 44.Martin W, Stoebe B, Goremykin V, Hapsmann S, Hasegawa M, Kowallik KV. Gene transfer to the nucleus and the evolution of chloroplasts. Nature 1998;393(6681):162–165. 10.1038/30234 [DOI] [PubMed] [Google Scholar]
- 45.Imachi H, Nobu MK, Nakahara N, Morono Y, Ogawara M, Takaki Y, et al. Isolation of an archaeon at the prokaryote-eukaryote interface. Nature 2020;577(7791):519–525. 10.1038/s41586-019-1916-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fan L, Wu D, Goremykin V, Xiao J, Xu Y, Garg S, et al. Phylogenetic analyses with systematic taxon sampling show that mitochondria branch within alphaproteobacteria. Nat Ecol Evol 2020. 10.1038/s41559-020-1239-x [DOI] [PubMed] [Google Scholar]
- 47.Lang BF, Burger G, O’Kelly CJ, Cedergren R, Golding GB, Lemieux C, et al. An ancestral mitochondrial DNA resembling a eubacterial genome in miniature. Nature 1997;387(6632):493–497. 10.1038/387493a0 [DOI] [PubMed] [Google Scholar]
- 48.Tian R-M, Cai L, Zhang W-P, Cao H-L, Qian P-Y. Rare Events of Intragenus and Intraspecies Horizontal Transfer of the 16S rRNA Gene. Genome Biol Evol 2015;7(8):2310–2320. 10.1093/gbe/evv143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schönheit P, Buckel W, Martin WF. On the origin of heterotrophy. Trends Microbiol 2016;24(1):12–25. 10.1016/j.tim.2015.10.003 [DOI] [PubMed] [Google Scholar]
- 50.Husnik F, Keeling PJ. The fate of obligate endosymbionts: reduction, integration, or extinction. Curr Opin Genet Dev 2019;58–59:1–8. 10.1016/j.gde.2019.07.014 [DOI] [PubMed] [Google Scholar]
- 51.Tamames J, Gil R, Latorre A, Peretó J, Silva FJ, Moya A. The frontier between cell and organelle: genome analysis of Candidatus Carsonella ruddii. BMC Evol Biol 2007;7:181 10.1186/1471-2148-7-181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Podar M, Anderson I, Makarova KS, Elkins JG, Ivanova N, Wall MA, et al. A genomic analysis of the archaeal system Ignicoccus hospitalis-Nanoarchaeum equitans. Genome Biol 2008;9(11):R158 10.1186/gb-2008-9-11-r158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Anderson I, Djao ODN, Misra M, Chertkov O, Nolan M, Lucas S, et al. Complete genome sequence of Methanothermus fervidus type strain (V24S). Stand Genomic Sci 2010;3(3):315–324. 10.4056/sigs.1283367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gabaldón T. Relative timing of mitochondrial endosymbiosis and the “pre-mitochondrial symbioses” hypothesis. IUBMB Life 2018;70(12):1188–1196. 10.1002/iub.1950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kapust N, Nelson-Sathi S, Schönfeld B, Hazkani-Covo E, Bryant D, Lockhart PJ, et al. Failure to recover major events of gene flux in real biological data due to method misapplication. Genome Biol Evol 2018;10(5):1198–1209. 10.1093/gbe/evy080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Martin W, Rujan T, Richly E, Hansen A, Cornelsen S, Lins T, et al. Evolutionary analysis of Arabidopsis, cyanobacterial, and chloroplast genomes reveals plastid phylogeny and thousands of cyanobacterial genes in the nucleus. Proc Natl Acad Sci U S A 2002;99(19):12246–12251. 10.1073/pnas.182432999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ku C, Nelson-Sathi S, Roettger M, Garg S, Hazkani-Covo E, Martin WF. Endosymbiotic gene transfer from prokaryotic pangenomes: Inherited chimerism in eukaryotes. Proc Natl Acad Sci U S A 2015;112(33):10139–10146. 10.1073/pnas.1421385112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Martin WF, Garg S, Zimorski V. Endosymbiotic theories for eukaryote origin. Philos Trans R Soc Lond B, Biol Sci 2015;370(1678):20140330 10.1098/rstb.2014.0330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hittinger CT, Carroll SB. Gene duplication and the adaptive evolution of a classic genetic switch. Nature 2007;449(7163):677–681. 10.1038/nature06151 [DOI] [PubMed] [Google Scholar]
- 60.van de Peer Y, Maere S, Meyer A. The evolutionary significance of ancient genome duplications. Nat Rev Genet 2009;10(10):725–732. 10.1038/nrg2600 [DOI] [PubMed] [Google Scholar]
- 61.Maier U-G, Zauner S, Woehle C, Bolte K, Hempel F, Allen JF, et al. Massively convergent evolution for ribosomal protein gene content in plastid and mitochondrial genomes. Genome Biol Evol 2013;5(12):2318–2329. 10.1093/gbe/evt181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Allen JF, Martin WF. Why have organelles retained genomes? Cell Syst 2016;2(2):70–72. 10.1016/j.cels.2016.02.007 [DOI] [PubMed] [Google Scholar]
- 63.Vos M, Hesselman MC, Te Beek TA, van Passel MWJ, Eyre-Walker A. Rates of lateral gene transfer in prokaryotes: High but why? Trends Microbiol 2015; 23(10):598–605. 10.1016/j.tim.2015.07.006 [DOI] [PubMed] [Google Scholar]
- 64.Sela I, Wolf YI, Koonin EV. Theory of prokaryotic genome evolution. Proc Natl Acad Sci U S A 2016;113(41):11399–11407. 10.1073/pnas.1614083113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Martin W. Mosaic bacterial chromosomes: a challenge en route to a tree of genomes. Bioessays 1999;21(2):99–104. [DOI] [PubMed] [Google Scholar]
- 66.Puigbò P, Wolf YI, Koonin EV. Genome-wide comparative analysis of phylogenetic trees: The prokaryotic forest of life. Methods Mol Biol 2019;1910:241–269. 10.1007/978-1-4939-9074-0_8 [DOI] [PubMed] [Google Scholar]
- 67.Wright ES, Baum DA. Exclusivity offers a sound yet practical species criterion for bacteria despite abundant gene flow. BMC Genomics 2018;19(1):724 10.1186/s12864-018-5099-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.O’Leary NA, Wright MW, Brister JR, Ciufo S, Haddad D, McVeigh R, et al. Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res 2016;44(D1):D733–D745 10.1093/nar/gkv1189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. Journal of Molecular Biology 1990;215(3):403–10. 10.1016/S0022-2836(05)80360-2 [DOI] [PubMed] [Google Scholar]
- 70.Rice P, Longden I, Bleasby A. EMBOSS: the European molecular biology open software suite. Trends Genet. 2000;(16):276–277. 10.1016/s0168-9525(00)02024-2 [DOI] [PubMed] [Google Scholar]
- 71.Enright AJ, van Dongen S, Ouzounis CA. An efficient algorithm for large-scale detection of protein families. Nucleic Acids Res 2002;30(7):1575–1584. 10.1093/nar/30.7.1575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 2013;30(4):772–780. 10.1093/molbev/mst010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014;30(9):1312–1313. 10.1093/bioinformatics/btu033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Tria FDK, Landan G, Dagan T. Phylogenetic rooting using minimal ancestor deviation. Nat Ecol Evol 2017;1:193 10.1038/s41559-017-0193 [DOI] [PubMed] [Google Scholar]
- 75.Nguyen L-T, Schmidt HA, von Haeseler A, Minh BQ. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol 2015;32(1):268–274. 10.1093/molbev/msu300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Junier T, Zdobnov EM. The Newick utilities: high-throughput phylogenetic tree processing in the UNIX shell. Bioinformatics 2010;26(13):1669–1670. 10.1093/bioinformatics/btq243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Huerta-Cepas J, Serra F, Bork P. ETE 3: Reconstruction, analysis, and visualization of phylogenomic data. Mol Biol Evol 2016;33(6):1635–1638. 10.1093/molbev/msw046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kanehisa M, Sato Y, Kawashima M, Furumichi M, Tanabe M. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res 2016;44(D1):D457–D462 10.1093/nar/gkv1070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kishino H, Hasegawa M. Evaluation of the maximum likelihood estimate of the evolutionary tree topologies from DNA sequence data, and the branching order in hominoidea. Journal of molecular evolution 1989;29(2):170–9. 10.1007/BF02100115 [DOI] [PubMed] [Google Scholar]
- 80.Shimodaira H, Hasegawa M. Multiple comparisons of log-likelihoods with applications to phylogenetic inference. Mol Biol Evol 1999;16(8):1114–1116 10.1093/oxfordjournals.molbev.a026201 [DOI] [Google Scholar]
- 81.Shimodaira H. An approximately unbiased test of phylogenetic tree selection. Systematic biology 2002;51(3):492–508. 10.1080/10635150290069913 [DOI] [PubMed] [Google Scholar]
- 82.Price MN, Dehal PS, Arkin AP. FastTree 2–approximately maximum-likelihood trees for large alignments. PLoS ONE 2010;5(3):e9490 10.1371/journal.pone.0009490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Havlicek LL, Peterson NL. Robustness of the pearson correlation against violations of assumptions. Percept Mot Skills 1976;43(3_suppl):1319–1334 10.2466/pms.1976.43.3f.1319 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(XLSX)
(TXT)
(XLSX)
(XLSX)
(XLSX)
(TXT)
(TXT)
(DOCX)
(XLSX)
a. Shows the cumulative frequency of the proportion of duplications of all 5,655 prokaryotic organisms (red) compared to the 150 eukaryotes (blue) in our dataset. b. Shows the cumulative frequency of 100 random sample sets of 150 prokaryotic organisms each (red) versus the 150 eukaryotic organisms (blue) in the dataset.
(TIF)
Comparison of verticality, normalized by multiplying raw monophyly count by their average root to leave distance of each tree, and number of genomes in a protein cluster for a. all clusters (n = 101,422) and b. all conserved clusters (average branch length ≥ 0.1; n = 8,547). The plot is created analogous to Fig 1 in the main text and this alternative verticality calculation also correlates to number of genomes (A: p < 10–300, Pearson´s R2 = 0.571; B: p < 10–300, R2 = 0.686). The correlation is more consistent when comparing verticality to number of phyla represented in a cluster (a: p < 10–300, Pearson´s R2 = 0.754; b: p < 10–300, R2 = 0.767, see S2 Table for more details). The eukaryotic-prokaryotic clusters (EPCs) are highlighted in red and the clusters that correspond to a gene from the mitochondrial genome of Reclinomonas americana [45] are displayed in blue triangles along the abscissa of the plot and in the graph. For the latter, the gene identifier was noted above each plot. Ribosomal proteins are indicated by the black diamond on the right of each plot and in the graph [6]. Notably, these clusters show a steep decline in clusters with lower verticality among the conserved clusters.
(TIF)
Each phylum is indicated by one color as depicted in the legend of the table. If the phylum is monophyletic in the tree, the number of genomes in the tree are divided by the number of genomes of this phylum present in the dataset of 5,655 organisms–phyla e and f in the panels a. and b. of the figure. If the phylum is paraphyletic, the verticality is set to '0'–phyla g and h in panels c. and d. of the figure. This number represents the verticality for each phylum. The sum of all verticality scores for the phyla in the tree is then the verticality for the tree and conversely, for the gene.
(TIF)
The Kishino-Hasegawa (KH) test, Shimodaira-Hasegawa (SH) test and the Approximately-Unbiased (AU) test were performed for 475 prokaryote-eukaryote genes for which eukaryotes were not recovered monophyletic in the ML trees. The histogram shows the distribution of p-values (horizontal axis) for the tests of the unconstrained ML trees against ML trees with constrained eukaryote monophyly. A test was considered significant (eukaryote monophyly was rejected) if p-value ≤ 0.05.
(TIF)
Mapping of EPCs to prokaryotic clusters. The EPCs were separated according to the pure sister group of eukaryotes in the trees and plotted in the same way as in Fig 4 of the main text. The left panel shows EPCs that may include all eukaryotic supergroups, the right panel shows only EPCs that include archaeplastidal eukaryotes. Meaning the latter are indicative of plastid endosymbiosis. For a better overview a headline is included in each plot that lists the taxonomic group represented, if it shows EPCs linked to the mitochondrial (‘P and O’, left panel) or to the plastidal endosymbiosis event (‘Plant only’, right panel), and the number of EPCs that are shown as red dots.
(GZ)
Data Availability Statement
Files for S9 Table are available on the repository of our university: http://dx.doi.org/10.25838/d5p-12. The other supplementary files are provided with the manuscript and Supporting Information.