Abstract
Methane-oxidizing bacteria (methanotrophs) play a vital role in reducing atmospheric methane emissions, and hence mitigating their potent global warming effects. A significant proportion of the methane released is thermogenic natural gas, containing associated short-chain alkanes as well as methane. It was one hundred years following the description of methanotrophs that facultative strains were discovered and validly described. These can use some multi-carbon compounds in addition to methane, often small organic acids, such as acetate, or ethanol, although Methylocella strains can also use short-chain alkanes, presumably deriving a competitive advantage from this metabolic versatility. Here, we review the diversity and molecular ecology of facultative methanotrophs. We discuss the genetic potential of the known strains and outline the consequent benefits they may obtain. Finally, we review the biotechnological promise of these fascinating microbes.
Keywords: biogeochemical cycling, facultative methanotrophs, Methane, Methane monooxygenase, Methylocella, Methylocystis, Methylocapsa
The global methane budget and its significance for climate
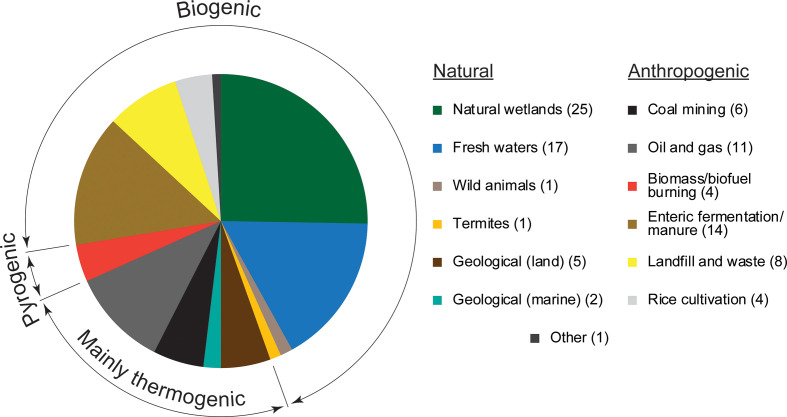
Methane, the most abundant hydrocarbon in the atmosphere and a potent greenhouse gas, is one of the most significant contributors to climate change. The atmospheric concentration of methane increased to over 1800 ppb by 2012, 2.5 times the pre-industrial value [1]. Moreover, methane is a much more effective greenhouse gas than carbon dioxide, with a global warming potential 28 times that of CO2 (per unit mass) over 100 years. The short lifetime in the atmosphere (approximately 9 years [2]) and relatively minor source/sink imbalance suggests that reduction in methane emissions would have rapid and relatively achievable benefits for climate [3], but clearly prediction and mitigation of methane emissions requires a comprehensive understanding of global and regional-scale budgets, and of how the sources and sinks respond to changing conditions. Globally, 540–884 Tg of methane are emitted annually from various natural and anthropogenic sources [1] (Fig. 1). Apart from a minor abiotic (chemical) source [4], methane arises from the biological degradation of organic matter, either by the activity of methanogenic archaea, or by the heat and pressure-mediated breakdown of subterranean organic material over geological timescales, or by the incomplete burning of biomass (termed biogenic, thermogenic or pyrogenic methane, respectively).
Fig. 1.
Summary of natural and anthropogenic methane sources to the atmosphere. A proportion of coal bed methane is of biogenic origin [179]. The magnitude of each source as a percentage of the total (736 Tg CH4 y−1) is shown in parentheses. Data from reference [1].
Biogenic methane is produced by methanogenic Archaea under anaerobic conditions, mainly in wetlands, landfill sites, rice paddies, the rumen of cattle and the hindgut of termites. Thermogenic methane, the other major source, is of geological origin, produced from the chemical decay of buried sedimentary organic material. Thermogenic methane emissions derive from both anthropogenic fossil fuel extraction and distribution (114–133 Tg y−1), and natural sources. The natural sources include macro- and micro-seeps, mud volcanoes, geothermal areas, volcanoes and submarine seeps (33–75 Tg y−1) [1]. This thermogenic ‘natural gas’ contains substantial amounts of other climate-active gases, mainly ethane (a photochemical pollutant) and propane (an ozone precursor), 2–4 and 1–2.4 Tg y−1 from natural sources, respectively [5, 6]. Visible seepage (macro-seeps) and diffuse but pervasive micro-seepage occurs over a considerable proportion of the Earth’s surface, including much of Northern Europe and Russia, and many regions in the USA, including the Appalachian Basin [7], where the gas contains, in addition to methane, up to 35 vol% ethane and propane [8, 9].
A large proportion (over 50% [10]) of biogenic and thermogenic methane is subsequently consumed in both anoxic and oxic zones by methane-oxidizing microbes (methanotrophs) before its release to the atmosphere. The major sink for atmospheric methane (90 %) is photochemical oxidation by hydroxyl radicals, predominately in the troposphere but also in the stratosphere [11], although soil-dwelling methanotrophs draw down approximately 30 Tg y−1 of atmospheric methane [12, 13].
Microbial growth on methane and short-chain alkanes
Methanotrophs, which are widespread in freshwater, marine and terrestrial environments, are bacteria able to grow on methane as their sole source of carbon and energy and are a subset of methylotrophs, micro-organisms that grow on one-carbon compounds such as methanol and methylated amines. Anaerobic oxidation of methane plays an important role in mediating emissions in anoxic zones, principally in marine but also in freshwater environments [14, 15], but here we consider only the aerobes. All aerobic methanotrophs use a methane monooxygenase (MMO) to oxidize methane to methanol, which is further oxidized to formaldehyde by methanol dehydrogenase (MDH). There are two forms of MMO, a membrane-associated copper-containing enzyme (particulate methane monooxygenase, pMMO) and a cytoplasmic enzyme (soluble methane monooxygenase, sMMO). The sMMOs form one group of a large family of soluble diiron centre monooxygenases (SDIMOs), which bacteria use to grow on a wide range of hydrocarbons and which, based on DNA sequence, gene layout, subunit composition and substrate specificity, can be assigned to one of six major groups [16, 17]. The growth substrates of SDIMO groups 1 and 2 are aromatic compounds or alkenes, group 3 comprises the sMMOs, group 4 contains alkene monooxygenase and groups 5 and 6 contain mainly propane monooxygenases. Nearly all methanotrophs possess the pMMO, and a minority also possess the sMMO. Methylocella spp. and two strains from the genera Methyloferula and Methyloceanibacter do not contain the pMMO and thus rely solely on the sMMO to oxidize methane. In strains that use both enzymes, the expression and activity of these enzymes is controlled by copper (the ‘copper switch’), reviewed by Semrau et al. [18].
Microbes growing on other short-chain alkanes (e.g. ethane, propane or butane) have also been characterized. Many of these are Gram-positive Actinobacteria, including Rhodococcus , Nocardioides and Mycobacterium but also Proteobacteria, for example Pseudomonas [19, 20]. The initial oxidation of short-chain alkanes is usually catalysed by a monooxygenase, frequently an SDIMO related to the group 3 methane monooxygenases, but which is instead from group 5 or group 6 of the SDIMO family [21], although the butane monooxygenase of Thauera butanivorans is more closely related to the sMMO [22]. These microbes are metabolically versatile compared to methanotrophs, and generally grow on a range of multicarbon compounds, but not methane [20].
Methanotrophs from approximately two dozen genera are in cultivation, and taxonomically they fall into the classes Alphaproteobacteria and Gammaproteobacteria, and the phylum Verrucomicrobia. In addition, members of the candidate phylum NC10 possess methane monooxygenase and oxidize methane coupled with oxygenic denitrification in anoxic conditions [15]. Historically, the proteobacterial methanotrophs were subdivided into type I and type II, based on physiological traits, including the arrangement of intracytoplasmic membranes, which also corresponds with their taxonomy (Gammaproteobacteria or Alphaproteobacteria) and assimilation of carbon via the RuMP pathway or the serine cycle, respectively [23]. Subsequently, with the discovery of more strains including methanotrophs that assimilate carbon autotrophically via the Calvin–Benson–Bassham (CBB) cycle [24, 25], these categories had to be adjusted and additional subdivisions were added, making this distinction less clear-cut [26].
Facultative methanotrophs
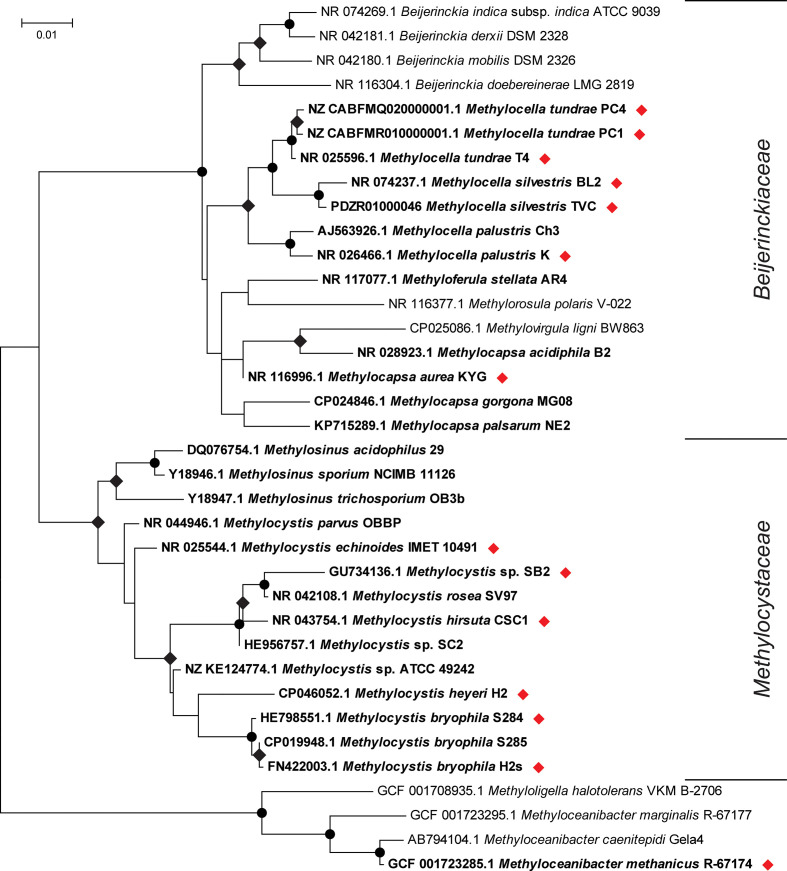
Although in some cases methanotrophs were shown to assimilate small amounts of carbon from multi-carbon compounds, including carboxylic and amino acids, supplemental to their primary metabolism while growing on methane [27–31], until recently methanotrophy was considered to be an obligate trait; despite several ultimately unconfirmed reports, by the turn of the last century no cultivated examples were known to grow on multi-carbon compounds (containing C–C bonds) as the sole source of carbon and energy, in the absence of methane [32]. In the past two decades, however, using novel media formulations and innovative techniques coupled with improved molecular methods for verification, several facultative methanotrophs have been isolated [33, 34]. These belong to the Alphaproteobacteria; Methylocystis ( Methylocystaceae ), Methyloceanibacter (Rhizobia incertae sedis), or Methylocapsa and Methylocella ( Beijerinckiaceae ) (Table 1, Fig. 2) [35–39]. Crenothrix polyspora , a gammaproteobacterium, was reported to grow on glucose and acetate [40], although no example exists in pure culture. The first of these facultative methanotrophs to be discovered were the Methylocella strains, M. palustris , M. silvestris and M. tundrae [41–43]. Although initially described as only growing on C1 compounds, they were later shown to use a wide variety of C2 – C6 multi-carbon compounds including alcohols and organic acids [38, 44, 45] and also, for several of the strains at least, ethane and propane, typical components of thermogenic natural gas. Subsequently, additional facultative strains of Beijerinckia and Methylocystis were isolated (or identified as facultative), originating from forest, peat or aquifer ecosystems in northern Europe, Russia or the USA (Table 1) as well as a single marine strain, Methyloceanibacter methanicus R-67174. In contrast to the Methylocella strains, these are much more limited in their substrate utilization, able to use only acetate or ethanol. Their growth rates on these substrates are low compared to on methane, with published rates on acetate or ethanol in the range 7–42 % of the corresponding rate on methane [35–37, 39, 46], in contrast with Methylocella , which grows more rapidly on many multi-carbon compounds than on methane [38, 44, 45].
Table 1.
Facultative methanotrophs
|
Taxon |
Strain |
Family |
Environment |
Location |
Multi-C substrates |
pH optimum |
Ref. |
|---|---|---|---|---|---|---|---|
|
H2s |
Sphagnum peat |
Germany |
Acetate |
6.0–6.5 |
[39] |
||
|
S284 |
Sphagnum peat bog |
European N. Russia |
Acetate |
6.0–6.5 |
[35] |
||
|
IMET 10491 |
Sewage sludge |
Germany |
Acetate |
nr |
[180] |
||
|
H2 |
Sphagnum peat |
Germany |
Acetate |
5.8–6.2 |
[181] |
||
|
CSC1 |
Uncontaminated aquifer |
CA, USA |
Acetate |
7.0 |
[182] |
||
|
Methylocystis sp. SB2 |
SB2 |
Spring bog |
MI, USA |
Acetate, ethanol |
6.8 |
[37] |
|
|
KYG |
Forest soil |
Germany |
Acetate |
6.0–6.2 |
[36] |
||
|
BL2 |
Forest soil |
Germany |
Organic acids, alcohols, ethane, propane |
5.5 |
[43] |
||
|
K |
Sphagnum peat bog |
W.Siberia |
Organic acids, alcohols |
5.5 |
[42] |
||
|
T4 |
Sphagnum peat |
N.Russia |
Organic acids, alcohols |
5.5–6.0 |
[41] |
||
|
TVC |
Tundra soil |
N.Canada |
Organic acids, ethanol, propane |
5.8 |
[47] |
||
|
PC1 |
Stream water and sediment |
NY, USA |
Organic acids, alcohols, ethane, propane |
5.8 |
[45] |
||
|
PC4 |
Stream water and sediment |
NY, USA |
Organic acids, alcohols, ethane, propane |
5.8 |
[45] |
||
|
R-67174 |
Rhizobiales incertae sedis |
Marine sediment |
North Sea |
Acetate |
7.3 |
[49] |
nr, not reported.
Fig. 2.
Phylogeny, based on 16S rRNA genes, of alphaproteobacterial methanotrophs (in bold) together with other closely related non-methanotrophic representatives. Facultative strains are identified with red diamonds. The tree was drawn using the maximum-likelihood method in mega7 [183], with bootstrap values (500 replications) greater than 90 or 50 % shown as circles or diamonds, respectively, at the nodes. The tree is drawn to scale and the scale bar indicates substitutions per site. There were a total of 1524 positions in the final dataset.
Recent efforts to obtain additional Methylocella strains resulted in isolation of Methylocella silvestris strain TVC, originating from stream sediment/soil from a permafrost location in N. Canada [47]. Next, the observation that Methylocella silvestris could grow on propane and methane concurrently [44], prompted sampling from environments where these gases co-occur, specifically natural gas seeps at sites including streams in northern New York State, where ethane and propane together comprise up to 35 % v/v [9, 48]. Two isolates, Methylocella tundrae strains PC1 and PC4, were obtained from these environments [45].
Methane and alkane monooxygenases of facultative methanotrophs
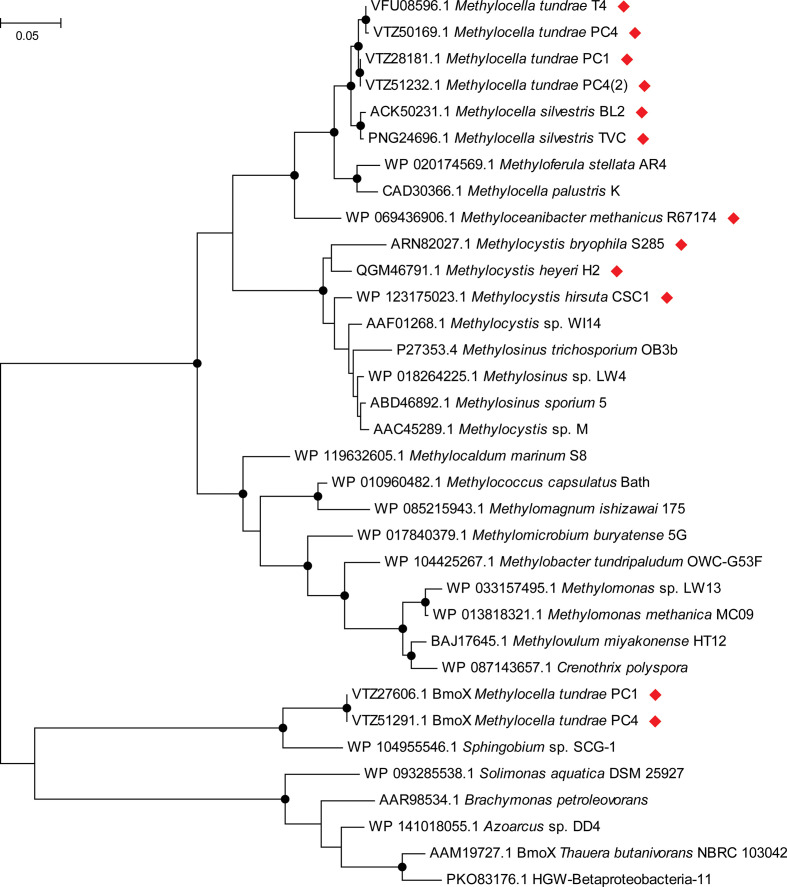
So far, all known Methylocella strains differ from almost all other methanotrophs in possessing only the sMMO rather than the copper-dependent membrane-associated pMMO, the only other sMMO-only methanotrophs described being Methyloferula stellata AR4 and Methyloceanibacter methanicus R-67174. Interestingly, the latter strain is facultative and grows to a low density on acetate, although it is the only methane-utilizing member of this genus, which otherwise contains facultative methylotrophs able to grow on several multi-carbon compounds [49–51]. Of the other facultative methanotroph strains, Methylocapsa aurea and Methylocystis sp. SB2 contain only the pMMO, whereas the rest contain both forms of MMO (Fig. 3). M. tundrae PC4 contains two similar but not identical sMMO operons (96 % nucleotide identity between the operons mmoXYBZDCRG, 88–99% amino acid identity between polypeptides). Although many methanotrophs contain multiple copies of the pmoCAB operon, additional copies of the sMMO genes are hitherto unknown [52]. M. tundrae strains PC1 and PC4 also contain an additional SDIMO gene cluster with identical subunit layout to the sMMO [45]. These MmoX-like sequences form a small subgroup (described as BmoX in Figs 3 and 4), distinct from the MmoX sequences of characterized sMMOs, clustering with a sequence from Sphingobium sp. SCG-1, isolated from soil associated with a major natural gas leak [53], and more distantly with BmoX from Thauera butanivorans , which grows on butane (but not methane) as the sole source of carbon and energy [54] (Fig. 4). However, neither of these Methylocella strains can grow on butane [45], so the role of these genes remains unclear.
Fig. 3.
Genetic potential of facultative methanotrophs with determined genome sequences. The presence of a gene or genes encoding a reaction or pathway is shown as black squares. Whole-genome nucleotide sequences were searched with representative protein query sequences using TBLASTN [184]. BL2, Methylocella silvestris BL2; TVC, Methylocella silvestris TVC; PC1, Methylocella tundrae PC1; PC4, Methylocella tundrae PC4; T4, Methylocella tundrae T4; H2, Methylocystis heyeri H2; S285, Methylocystis bryophila S285; SB2, Methylocystis sp. SB2; CSC1, Methylocystis hirsuta CSC1; KYG, Methylocapsa aurea KYG; R-67164, Methyloceanibacter methanicus R-67174. MmoX, soluble methane monooxygenase; PmoA, particulate methane monooxygenase; BmoX, butane monooxygenase; PrmA, propane monooxygenase; MxaF, Ca-dependent methanol dehydrogenase; XoxF, lanthanide-dependent methanol dehydrogenase; ExaF, lanthanide-dependent ethanol dehydrogenase; MauA, methylamine dehydrogenase; NMG, N-methylglutamate pathway; Serine, serine cycle; RuMP, ribulose monophosphate pathway; CBB, Calvin–Benson–Bassham pathway; ActP, acetate-specific permease; ICL, isocitrate lyase; MS, malate synthase; ECM, ethylmalonyl-CoA pathway; NarGHJI, respiratory nitrate reductase; NapAB, periplasmic nitrate reductase; NirK, copper-containing nitrite reductase; NirS, multi-haem nitrite reductase; cNorB, cytochrome c-dependent nitric oxide reductase; NosZ, nitrous oxide reductase; Nif, nitrogenase; PufLM, photosynthetic reaction centre; Hhy-5, high-affinity group 5 hydrogenase; CODH, carbon monoxide dehydrogenase.
Fig. 4.
Relationship of the α-subunits of the sMMOs and sMMO-like proteins from facultative methanotrophs (indicated with red diamonds) and other representative strains. The sequences at the bottom of the figure, which form a group with BmoX (butane monooxygenase) of Thauera butanivorans, are from non-methanotrophs, except for those from Methylocella tundrae strains PC1 and PC4. The tree was drawn using the maximum-likelihood method in mega7 [183], with bootstrap values (500 replications) greater than 75 % shown as solid circles at the nodes. The tree is drawn to scale and the scale bar indicates substitutions per site. There were a total of 540 amino acid residues in the final dataset.
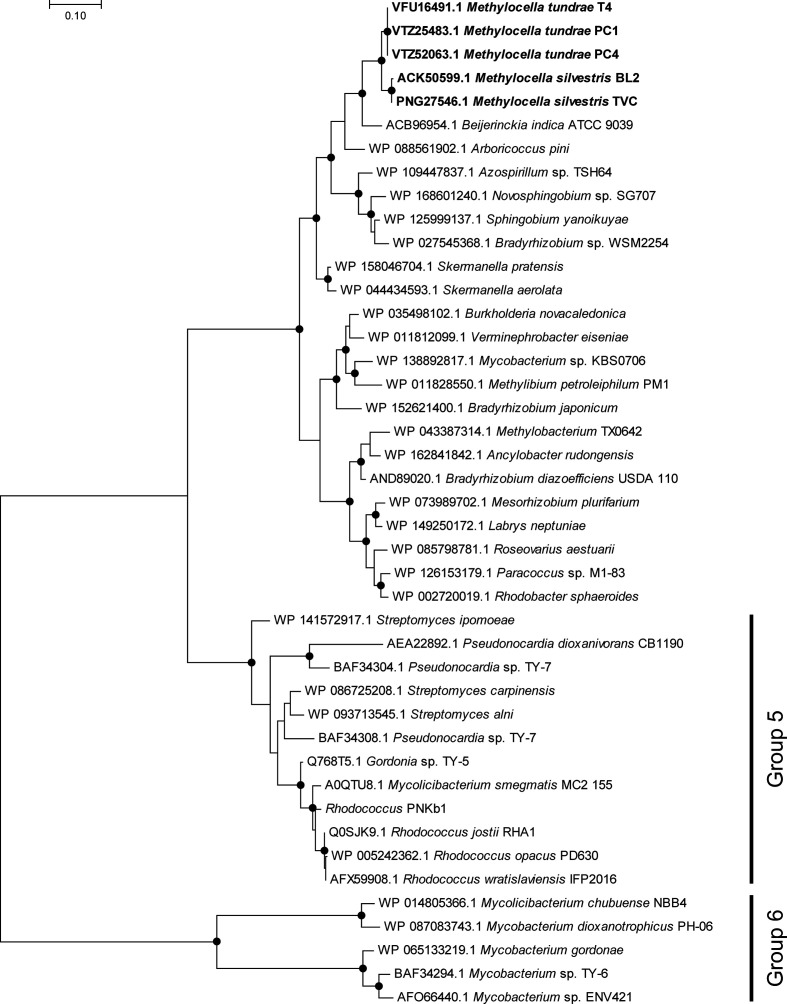
All the Methylocella strains contain a propane monooxygenase-like (PrMO) sequence belonging to SDIMO group 5 [17], and several of them have been tested and can grow on propane [44, 45, 47, 55]. The hydroxylase alpha subunits (encoded by prmA) form a distinct cluster within group 5, distinct from the group 5 and 6 enzymes of characterized propanotrophs such as Gordonia sp. TY-5, Rhodococcus sp. RHA1 or Mycobacterium sp. TY-6, and instead cluster with sequences from a diverse range of organisms not known for growth on short-chain alkanes (Fig. 5). Interestingly, this prm gene cluster, together with genes encoding propionyl-CoA carboxylase and methylmalonyl-CoA mutase and epimerase, involved in the subsequent metabolism of 1-propanol (the product of terminal propane oxidation), are located on a megaplasmid in M. tundrae T4, but chromosomally encoded in M. silvestris [55, 56]. In Methylocella silvestris BL2, while prmA was required for growth on propane at high concentrations (20 % v/v), it was not essential for growth at the lower concentrations typically found in the natural environment [44]. However, the propane monooxygenase was implicated in growth on 2-propanol and acetone, as it was in other bacteria [57, 58], suggesting that the environmental role of this enzyme is not restricted to oxidation of short-chain alkanes.
Fig. 5.
The Methylocella PrmA (propane monooxygenase α-subunit) sequences (shown in bold), group with those of diverse strains not known for propane oxidation, distinct from the propanotrophs in SDIMO groups 5 and 6. The tree was drawn using the maximum-likelihood method in mega7 [183], with bootstrap values (500 replications) greater than 75 % shown as solid circles at the nodes. The tree is drawn to scale and the scale bar indicates substitutions per site. There were a total of 440 amino acid residues in the final dataset.
Alcohol oxidation and one-carbon assimilation
Methanol dehydrogenase (MDH), the second essential enzyme for methane metabolism, catalyses the conversion of methanol to formaldehyde. The classical MDH is a soluble periplasmic pyrroloquinoline quinone (PQQ)-containing enzyme, with an α2β2 structure, consisting of two large subunits (MxaF) and two small subunits (MxaI) and containing a Ca2+ ion at the active site [59]. MDH passes electrons to a specific cytochrome c L (MxaG), and the mxa operon also encodes several genes responsible for Ca insertion and enzyme maturation, including periplasmic solute-binding protein MxaJ, of unknown function. Relatively recently a homologous MDH (Xox-MDH) dependent on a lanthanide (Ln) rare-earth element, rather than calcium, was identified [60]. Although xoxF genes (encoding the lanthanide-dependent subunit) were detected in methanotrophs many years ago, their function was not established until the discovery of the role of lanthanides as co-factors [61]. Many methylotrophs and methanotrophs contain both forms of MDH and recently several studies have shown that lanthanides regulate their relative expression and activity [62–64]. The fact that rare-earth elements are not actually rare, but occur in the Earth’s crust in similar amounts to other metals with biological importance (e.g. copper or zinc [65]), and that Xox genes and enzymes are highly active and abundant, suggests that Xox-MDHs may be at least as important as Mxa-MDHs [66, 67]. The xox operons are less complex than mxa operons; Xox seems not to require the accessory genes required by the calcium-containing MDH and, in addition, most characterized examples lack the small subunit. Xox-type MDHs can be subdivided into five clades, XoxF1–XoxF5, with additional lanthanide-dependent homologues with non-methanol substrates [68]. Of the Xox-MDHs, clade XoxF5 appears to be the most abundant, except that XoxF4 is present to the exclusion of XoxF5 in the order Methylophilales. The structures of examples from clade 2 and clade 5 have been determined [61, 69–71] and several enzymes have been purified [68]. The Ln-dependent enzymes can be identified by the presence of an Ln-coordinating Asp residue two positions from the catalytic Asp residue (highly conserved in both MxaF and XoxF proteins), which is occupied by Ala in MxaF proteins [68, 71, 72]. Virtually all methanotrophs that contain Mxa-MDH also contain Xox-MDH, often more than one copy and sometimes from different clades [73, 74]. The advantage to a methanotroph of this methanol-oxidizing flexibility is unclear, but it may be important as a response to changing environmental conditions or for controlling symbiotic transfer of metabolites (e.g. methanol) to other members of the microbial community [75, 76].
All the facultative methanotroph strains except for Methyloceanibacter (which encodes XoxF1) encode one or two copies of XoxF5 (Fig. 3). The Methylocella strains also encode one or both of clade 1 and clade 3 enzymes. All also encode an Mxa-MDH, except for M. tundrae strains PC1 and PC4, which contain only Xox-MDHs [45]. These two strains were unable to grow on methanol in the absence of a rare-earth element, in common with a relatively small number of other methylotrophs [61, 77–79], which require these elements for methylotrophic growth. Subsequent to the isolation of these Xox-only Methylocella strains, additional methylotrophic (not methanotrophic) Xox-only members of the Beijerinckiaceae have been discovered [80]. Some of the facultative methanotrophs can grow on ethane or ethanol and so the identity of the enzyme(s) responsible for ethanol oxidation is of interest. Methylorubrum extorquens and Pseudomonas putida express lanthanide- and PQQ-dependent dehydrogenases with good ethanol compared to methanol-oxidizing activity, termed ExaF or PedH, respectively [81, 82]. However, in M. extorquens both forms of MDH can also efficiently use ethanol as substrate [81, 83], and it required deletion of all three MDHs as well as ExaF to prevent growth on either methanol or ethanol [81]. In contrast, analysis of the Methylocystis sp. SB2 transcriptome (grown without added lanthanide) showed that expression of Mxa-MDH was downregulated 50-fold during growth on ethanol in comparison to growth on methane, and expression of an NAD(P)-dependent short-chain dehydrogenase was fivefold upregulated [84] suggesting that the MDH is not primarily responsible for ethanol oxidation under these conditions in this strain. Since, of the facultative methanotrophs, only Methylocella tundrae PC4 encodes a PQQ-dependent ethanol dehydrogenase homologous to ExaF/PedH (Fig. 3) and since Methylocella spp. can grow on a range of alcohols [44, 45, 47], more research is needed to identify the enzymes responsible.
Methylated amines are present in many soils and aquatic environments [85] and many bacteria have evolved to metabolize these compounds, using one of a number of pathways [86]. Methylocella silvestris BL2 can grow on mono-, di- or tri-methylamine using the N-methylglutamate pathway [87–89] and the required genes (gmas and associated genes) are present in all the Methylocella isolates, but not in the other facultative strains, although Methyloceanibacter methanicus R-67174 appears to contain remnants of this gene cluster, but did not grow on methylamine [49]. Interestingly, none of the facultative methanotroph strains encode the key enzyme of the alternative proteobacterial pathway, methylamine dehydrogenase (MauAB), suggesting that the Methylocystis and Methylocapsa strains cannot utilize this source of carbon and, perhaps equally important in oligotrophic habitats, nitrogen, thus emphasizing the metabolic versatility of Methylocella.
All of the facultative methanotrophs assimilate carbon using the serine cycle (Fig. 3); none of the isolates possesses genes for the key enzymes of the RuMP cycle, 3-hexulose-6-phosphate synthase or 6-phospho-3-hexuloisomerase. However, the Methylocella and Methylocapsa strains possess CBB cycle genes, of which the deduced RubisCO large subunits (CbbL) share 88–90% amino acid identity with the form I enzyme from Bradyrhizobium diazoefficiens [90] and 92–94% identity with CbbL from closely related Beijerinckia mobilis , which was reported to grow autotrophically on methanol [91], raising the possibility that the CBB cycle may contribute to carbon fixation in some facultative strains under as-yet undefined conditions.
Growth on multi-carbon compounds
As mentioned above, the ability to grow on acetate and/or ethanol (and, in the case of Methylocella , a range of other multi-C compounds) in addition to methane is the defining feature of all so-far discovered facultative methanotrophs. However, Methylocella grows much better on two-carbon compounds than the other strains and it has been suggested that the inability of obligate methanotrophs to grow on multi-carbon compounds, such as acetate, is due to the lack of appropriate membrane transporter mechanisms [34, 92, 93]. In this context, it is interesting to note that whereas all the facultative strains contain a number of relatively uncharacterized membrane transporters, which possibly allow organic acids to enter the cell, only the Methylocella strains possess close homologues of actP, an acetate-specific permease (67–71% amino acid identity with ActP characterized in Escherichia coli [94] and 68–70% identity with META1p2533 from Methylorubrum extorquens AM1 [95, 96]).
When two-carbon compounds enter the cell, their assimilation presents another difficulty, since the TCA cycle oxidizes acetyl-CoA to two molecules of CO2, generating energy but not supplying carbon for assimilation. For many years, assimilation of acetate was considered to require the activity of the two enzymes of the glyoxylate shunt, isocitrate lyase (ICL) and malate synthase (MS), which together bypass the decarboxylation reactions of the TCA cycle, forming four-carbon molecules from two acetyl-CoA [97]. During one-carbon growth of serine cycle methylotrophs, the formation of glyoxylate (the substrate of the essential serine cycle enzyme serine-glyoxylate aminotransferase), from acetyl-CoA, can be catalysed by ICL together with non-decarboxylating enzymes of the TCA cycle. However, the lack of ICL genes and activity in many methylotrophs prompted researchers to seek an alternative pathway, which was finally resolved with the discovery of the ethylmalonyl-CoA (EMC) pathway (reviewed by Anthony [98]). Interestingly, whereas the Methylocystis and Methyloceanibacter strains encode the EMC pathway genes, all the Methylocella strains and Methylocapsa aurea KYG encode the glyoxylate pathway enzymes (Fig. 3), which are comparatively uncommon in serine cycle methanotrophs. Deletion of ICL or MS in M. silvestris BL2 resulted in severe growth defects on C1 and C2 compounds [99, 100], confirming the operation of the glyoxylate shunt in Methylocella .
Additional metabolic capabilities
Methanotrophs use a variety of survival strategies to persist in conditions of varying substrate availability and under the influence of environmental stress. Where resources are scarce, these strategies may include energy supplementation by oxidation of other soil or atmospheric trace gases, or by mechanisms such as phototrophy [101–104]. Utilization of hydrogen by methanotrophs has previously been observed, together with the ability to use this energy source to provide the reducing power required for methane oxidation [105, 106]. For example, Methylocystis sp. SC2, grown in batch culture, was able to oxidize comparatively high concentrations of supplied hydrogen under moderately limiting methane and oxygen concentrations, using a low affinity group 1d hydrogenase (also encoded by Methylocystis strains H2 and 285), achieving increased biomass yield from methane and depleting hydrogen to below the limits of detection [107]. Methylocystis strains 285 and SB2 and Methylocapsa aurea KYG encode a high-affinity group 5 NiFe uptake hydrogenase [108], similar to that used by other methanotrophs such as Verrucomicrobia to grow on low concentrations of hydrogen [101, 109], potentially allowing these strains to obtain energy from hydrogen in the atmosphere (<1 ppmv) under oligotrophic conditions [110]. Methylocella silvestris strains BL2 and TVC and Methylocapsa aurea KYG encode a group 2a enzyme, perhaps involved in recycling hydrogen produced as a bi-product of nitrogen fixation, although an enzyme from the same group can oxidize atmospheric hydrogen in Mycolicobacterium smegmatis (which also expresses a group 5 enzyme) [111]. Interestingly, the Methylocella tundrae strains appear to lack hydrogenases, despite possessing nitrogen fixation genes, suggesting that they are unable to use exogenous or internally produced hydrogen. Moreover, none of the facultative methanotrophs encode a carbon monoxide dehydrogenase, an enzyme used by many soil bacteria to enhance survival in oligotrophic conditions [112].
Aerobic anoxygenic phototrophs are abundant in aquatic ecosystems where the additional energy derived from light may give them a competitive advantage [113–115]. Some phototrophs are methylotrophs, and it is interesting to note that the Methylocella silvestris and Methylocystis sp. SB2 genomes contain the required pufLM and bacteriochlorophyll biosynthesis genes [116, 117] (Fig. 3), although the significance of this is currently unknown.
Where methane is abundant and oxygen may be limiting, some methanotrophs thrive at low-oxygen tensions. At the extreme, Candidatus Methylomirabilis oxyfera uses nitric oxide dismutation to generate its own molecular oxygen for use by methane monooxygenase, while other methanotrophs economize on oxygen consumption by denitrification [118–121], iron reduction [122, 123], or by fermentation [124, 125], maintaining a supply of molecular oxygen for methane activation. While the denitrification enzymes are also important for detoxification [126], the use of nitrogen compounds as electron acceptor may be significant in methane oxidation [127]. Of the facultative methanotrophs, only the Methylocella strains encode enzymes of the denitrification pathway, with M. silvestris BL2 containing all genes required for denitrification as far as nitrous oxide, but lacking nosZ, encoding the enzyme catalysing the final reduction step to dinitrogen. This gene is present in Methylocella tundrae T4, together with a copy of norB, (nitric oxide reductase) although this latter gene contains an internal stop codon, suggesting that it is not functional in M. tundrae T4. In common with many methanotrophs, the facultative isolates all contain the genetic requirements for nitrogen fixation (Fig. 3).
Environmental occurrence and ecology
Methanotrophs from the Methylocystaceae ( Methylocystis and Methylosinus ) and Beijerinckiaceae ( Methylocapsa and Methylocella ) have mostly been isolated from wetlands and forest soils in the Northern hemisphere [128] and in these and similar environments they are frequently among the most abundant methanotrophs [129–136]. Methylocystis and, to a lesser extent Methylocella , are also frequently found in rice paddies [137–139], another major source of methane to the atmosphere. In fact, Methylocella-related 16S rRNA gene sequences have been found in many diverse environments, ranging from moderately acidic to alkaline (summarized by Rahman et. al. [140]), although we should bear in mind that whereas all members of the Methylocystaceae (and of the gammaproteobacterial methanotroph families) are methanotrophs, the same is not true for the Methylocella family, Beijerinckiaceae , which contains methanotrophs, methylotrophs and heterotrophs, nor for the genus Methyloceanibacter , which contains non-methane-oxidizing facultative methylotrophs [51, 92]. The difficulty of distinguishing methanotrophs of the Beijerinckiaceae from heterotrophic members of this family is compounded since, before the discovery of Methylocella , the pMMO was thought to be diagnostic for all methanotrophs and so gene probes were developed using this gene. These have been extensively used to characterize methanotroph communities [128] but cannot detect Methylocella or Methyloceanibacter . Therefore, the identification of Methylocella-like DNA sequences in environmental DNA does not prove methanotrophy, and further confirmation is required. This can be obtained by methods which identify the community, active in response to methane, such as transcriptomics, proteomics or DNA-stable isotope probing. These methods have identified active Methylocella in methane-exposed material from, for example, peatlands, forest soil, landfill cover soil, alkaline coal mine, warm springs, acidic aquifers and natural gas seeps [45, 131, 141–147]. However, a consequence of the frequent reliance on pmoA gene probes is that Methylocella and other sMMO-only methanotrophs must frequently have been (and still are) overlooked in cultivation-independent studies.
Multi-carbon compounds, such as aliphatic, cyclic and aromatic organic acids including acetate, are frequently detectable in the oxic soil horizons and surface sediments which comprise the habitats of methanotrophs, sometimes reaching millimolar concentrations [148–150]. The impact of alternative carbon sources on methanotrophic activity is not clear; some reports have suggested that organic acids inhibit methane oxidation [151–153] while others showed methane oxidation still occurring in the presence of alternative carbon sources [39, 154, 155], or that acetate stimulated the activity of methanotrophs [156–158]. It was suggested that use of acetate by facultative methanotrophs might be a survival strategy when the supply of methane is intermittent, and reports have shown that transcription of the pMMO genes continues under these conditions [39, 153–155]. In Methylocella silvestris , acetate repressed sMMO gene transcription, at least at the concentration tested (5 mM) [159, 160], although this was not the case for propane [44]. Recently, with the availability of additional genome sequences, Farhan Ul Haque et al. [161] designed improved Methylocella -specific mmoX primers, which could be used to quantify Methylocella -like sMMO gene sequences in environmental samples. When environments exposed to short-chain-alkane-containing natural gas were examined, Methylocella were found to be extraordinarily abundant and active, in some cases making up 85 % of the methanotroph community, or 12 % of the total bacterial population [45, 161]. In these environments Methylocella may have benefited from the additional ethane and propane, and/or obligate methanotrophs might have been inhibited by these gases or by the products of their co-oxidation [162].
Biotechnological potential of facultative methanotrophs
Methanotrophs, which are ubiquitous, have potential for bioremediation of polluted sites. For example, it was shown that introduction of methane enhanced aerobic degradation of halogenated hydrocarbons (reviewed by Semrau [163]). Both forms of the MMO can transform these halogenated compounds, although the data suggested that despite its slower degradation rate, the pMMO was ultimately the more effective system. This being the case, the use of facultative Methylocystis strains is attractive. In these strains, the pMMO is expressed in the presence of acetate or ethanol [39, 154], which could be used to provide the reductant required by the MMO. This would both be easier to introduce into polluted sites than methane and would also avoid competition for binding to the monooxygenase. As another example of bioremediation, Methylocella , was among bacteria associated with degradation of plastics in landfill lysimeters [164, 165].
Obligate methanotrophs such as Methylococcus capsulatus, which can grow relatively quickly and to high cell densities, have been exploited for production of single-cell protein [166]. While Methylocella , which grows more slowly, may not offer the same potential for the production of low value, bulk chemicals, it can still be grown to high cell densities in fermenter culture [44, 159] and due to its metabolic versatility it should be examined further in this respect. Large-scale production of methanol from methane is an attractive proposition and promising results have been obtained in several studies [167]. For example, co-cultures of Methylomonas methanica with Methylocella tundrae , immobilized in silica gel, were fed with simulated biogas. Interestingly, methanol production was enhanced (nearly 100 %) by addition of hydrogen, achieving approximately 0.32 g l−1 and 66 % conversion efficiency [168]. An obstacle to the use of the MMO to produce methanol is the requirement to supply a costly electron donor (formate) to enable methane oxidation, but facultative methanotrophs may offer an attractive solution since they can instead use compounds, for example acetate, frequently present in the waste stream [169].
The biocatalytic potential of Methylocella spp. has not so far been exploited, despite their metabolic versatility when compared to obligate methanotrophs. The sMMO has long been regarded as a highly versatile biocatalyst, catalysing the oxidation of a wide range of alkanes, alkenes and even aromatic compounds as large as naphthalene [163, 166, 170]. It is possible to use whole cells of methanotrophs such as Methylococcus capsulatus to produce chemicals such as propylene oxide (from propylene), although the toxic nature of this product requires a recycling system to regenerate the whole-cell biocatalyst [171, 172]. If Methylocella is less susceptible to the toxic effects of propylene epoxide, it could offer an advantage over M. capsulatus for production of this chemical since it could be supplied with alternative energy sources to drive the oxidation of propylene by sMMO. In addition to the metabolic versatility of Methylocella , these strains have an advantage over obligate methanotrophs since expression of the sMMO is not repressed by copper [173]. The prospect of using the sMMO as a biocatalyst while using a multi-carbon compound such as succinate or acetate, as carbon and energy source, suggests that Methylocella might be useful as a cell platform for production of high value commodities, for example chiral alcohols and epoxides.
Final conclusions
It was nearly one hundred years after the first description of a methanotroph [174] before facultative methanotrophs were isolated, which highlights the difficulty of identifying this trait in environmental samples [33] but suggests that more facultative strains await discovery. As mentioned above, phylogeny alone cannot identify facultative methanotrophs from any of the currently identified genera, and it is not easy to devise experiments to identify facultative methanotrophs in the environment with methods used, to good effect, for obligate strains, for example using labelled substrates. Where identifiable genetic markers exist for specific metabolic traits, this can be used to screen (meta)genomic libraries derived from labelled nucleic acids (for example in DNA- or RNA-SIP or single-cell labelling experiments [175–177]), followed by targeted isolation techniques [45], but such traits may not always be easy to identify in genomes. This underlines the crucial importance of isolating and characterizing strains in the laboratory, now possible using several innovative techniques [178]. Facultative methanotrophs are clearly competitive in several environments, and research effort should be directed towards describing their behaviour in synthetic and natural communities and identifying the environmental conditions in which their role is important to the wider methane-oxidizing community. For example, is their facultative ability important in competition with obligate methanotrophs in methane-rich environments and how does oxygen availability affect this? What are the conditions where a denitrifying capability is important, and when is the potential to extract energy from alternative sources useful? In biotechnology and bioremediation their unique metabolic potential also justifies investigation.
Funding information
Funding was supplied by the Leverhulme Trust Research project (RPG2016-050) to JCM and Leverhulme Trust Early Career Fellowship (ECF2016-626) to ATC.
Acknowledgements
We thank Marcela Hernandez Garcia for useful comments on the manuscript.
Conflicts of interest
The authors declare that there are no conflicts of interest.
Footnotes
Abbreviations: EMC, ethylmalonyl CoA; ICL, isocitrate lyase; MDH, methanol dehydrogenase; MMO, methane monooxygenase; MS, malate synthase; pMMO, particulate methane monooxygenase; PrMO, propane monooxygenase; SDIMO, soluble diiron centre monooxygenase; sMMO, soluble methane monooxygenase; TCA, tricarboxylic acid.
References
- 1.Saunois M, Bousquet P, Poulter B, Peregon A, Ciais P, et al. The global methane budget 2000–2012. Earth Syst Sci Data. 2016;8:697–751. doi: 10.5194/essd-8-697-2016. [DOI] [Google Scholar]
- 2.Prather MJ, Holmes CD, Hsu J. Reactive greenhouse gas scenarios: systematic exploration of uncertainties and the role of atmospheric chemistry. Geophys Res Lett. 2012;39:L09803. doi: 10.1029/2012GL051440. [DOI] [Google Scholar]
- 3.Hansen J, Sato M, Ruedy R, Lacis A, Oinas V. Global warming in the twenty-first century: an alternative scenario. Proc Natl Acad Sci U S A. 2000;97:9875–9880. doi: 10.1073/pnas.170278997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Etiope G, Sherwood Lollar B. Abiotic methane on earth. Rev Geophys. 2013;51:276–299. doi: 10.1002/rog.20011. [DOI] [Google Scholar]
- 5.Etiope G, Ciccioli P. Earth's degassing: a missing ethane and propane source. Science. 2009;323:478. doi: 10.1126/science.1165904. [DOI] [PubMed] [Google Scholar]
- 6.Dalsøren SB, Myhre G, Hodnebrog Øivind, Myhre CL, Stohl A, et al. Discrepancy between simulated and observed ethane and propane levels explained by underestimated fossil emissions. Nat Geosci. 2018;11:178–184. doi: 10.1038/s41561-018-0073-0. [DOI] [Google Scholar]
- 7.Etiope G. Natural Gas Seepage: the Earth’s Hydrocarbon Degassing. Cham, Switzerland: Springer International Publishing; 2015. [Google Scholar]
- 8.Etiope G, Klusman RW. Microseepage in drylands: flux and implications in the global atmospheric source/sink budget of methane. Glob Planet Change. 2010;72:265–274. doi: 10.1016/j.gloplacha.2010.01.002. [DOI] [Google Scholar]
- 9.Schimmelmann A, Ensminger SA, Drobniak A, Mastalerz M, Etiope G, et al. Natural geological seepage of hydrocarbon gas in the Appalachian Basin and Midwest USA in relation to shale tectonic fracturing and past industrial hydrocarbon production. Sci Total Environ. 2018;644:982–993. doi: 10.1016/j.scitotenv.2018.06.374. [DOI] [PubMed] [Google Scholar]
- 10.Reeburgh WS. Global methane biogeochemistry. In: Holland HD, Turekian KK, editors. Treatise on Geochemistry. Amsterdam: Elsevier; 2007. pp. 1–32. [Google Scholar]
- 11.Lelieveld J, Crutzen PJ, Dentener FJ. Changing concentration, lifetime and climate forcing of atmospheric methane. Tellus B Chem Phys Meteorol. 1998;50:128–150. doi: 10.3402/tellusb.v50i2.16030. [DOI] [Google Scholar]
- 12.Curry CL. Modeling the soil consumption of atmospheric methane at the global scale. Global Biogeochem Cycles. 2007;21:GB4012. doi: 10.1029/2006GB002818. [DOI] [Google Scholar]
- 13.Dunfield PF. The soil methane sink. In: Reay DS, Hewitt N, Smith K, Grace J, editors. Greenhouse Gas Sinks. Wallingford, UK: CAB International; 2007. pp. 152–179. [Google Scholar]
- 14.Knittel K, Boetius A. Anaerobic oxidation of methane: progress with an unknown process. Annu Rev Microbiol. 2009;63:311–334. doi: 10.1146/annurev.micro.61.080706.093130. [DOI] [PubMed] [Google Scholar]
- 15.Welte CU, Rasigraf O, Vaksmaa A, Versantvoort W, Arshad A, et al. Nitrate- and nitrite-dependent anaerobic oxidation of methane. Environ Microbiol Rep. 2016;8:941–955. doi: 10.1111/1758-2229.12487. [DOI] [PubMed] [Google Scholar]
- 16.Leahy JG, Batchelor PJ, Morcomb SM. Evolution of the soluble diiron monooxygenases. FEMS Microbiol Rev. 2003;27:449–479. doi: 10.1016/S0168-6445(03)00023-8. [DOI] [PubMed] [Google Scholar]
- 17.Coleman NV, Bui NB, Holmes AJ. Soluble di-iron monooxygenase gene diversity in soils, sediments and ethene enrichments. Environ Microbiol. 2006;8:1228–1239. doi: 10.1111/j.1462-2920.2006.01015.x. [DOI] [PubMed] [Google Scholar]
- 18.Semrau JD, DiSpirito AA, Yoon S. Methanotrophs and copper. FEMS Microbiol Rev. 2010;34:496–531. doi: 10.1111/j.1574-6976.2010.00212.x. [DOI] [PubMed] [Google Scholar]
- 19.Rojo F. Degradation of alkanes by bacteria. Environ Microbiol. 2009;11:2477–2490. doi: 10.1111/j.1462-2920.2009.01948.x. [DOI] [PubMed] [Google Scholar]
- 20.Shennan JL. Utilisation of C2–C4 gaseous hydrocarbons and isoprene by microorganisms. J Chem Technol Biotechnol. 2006;81:237–256. [Google Scholar]
- 21.Holmes AJ, Coleman NV. Evolutionary ecology and multidisciplinary approaches to prospecting for monooxygenases as biocatalysts. Antonie van Leeuwenhoek. 2008;94:75–84. doi: 10.1007/s10482-008-9227-1. [DOI] [PubMed] [Google Scholar]
- 22.Osborne CD, Haritos VS. Beneath the surface: evolution of methane activity in the bacterial multicomponent monooxygenases. Mol Phylogenet Evol. 2019;139:106527. doi: 10.1016/j.ympev.2019.106527. [DOI] [PubMed] [Google Scholar]
- 23.Hanson RS, Hanson TE. Methanotrophic bacteria. Microbiol Rev. 1996;60:439–471. doi: 10.1128/MMBR.60.2.439-471.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.van Teeseling MCF, Pol A, Harhangi HR, van der Zwart S, Jetten MSM, et al. Expanding the verrucomicrobial methanotrophic world: description of three novel species of Methylacidimicrobium gen. nov. Appl Environ Microbiol. 2014;80:6782–6791. doi: 10.1128/AEM.01838-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rasigraf O, Kool DM, Jetten MSM, Sinninghe Damsté JS, Ettwig KF. Autotrophic carbon dioxide fixation via the Calvin-Benson-Bassham cycle by the denitrifying methanotroph “Candidatus Methylomirabilis oxyfera”. Appl Environ Microbiol. 2014;80:2451–2460. doi: 10.1128/AEM.04199-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Op den Camp HJM, Islam T, Stott MB, Harhangi HR, Hynes A, et al. Environmental, genomic and taxonomic perspectives on methanotrophic Verrucomicrobia . Environ Microbiol Rep. 2009;1:293–306. doi: 10.1111/j.1758-2229.2009.00022.x. [DOI] [PubMed] [Google Scholar]
- 27.Patel RN, Hoare SL, Hoare DS, Taylor BF. [14C]Acetate assimilation by a type I obligate methylotroph, Methylococcus capsulatus . Appl Environ Microb. 1977;34:607–610. doi: 10.1128/AEM.34.5.607-610.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Eccleston M, Kelly DP. Assimilation and toxicity of some exogenous C1 compounds, alcohols, sugars and acetate in the methane-oxidizing bacterium Methylococcus capsulatus . J Gen Microbiol. 1973;75:211–221. doi: 10.1099/00221287-75-1-211. [DOI] [PubMed] [Google Scholar]
- 29.Shishkina VN, Trotsenko YA. Multiple enzymic lesions in obligate methanotrophic bacteria. FEMS Microbiol Lett. 1982;13:237–242. doi: 10.1111/j.1574-6968.1982.tb08264.x. [DOI] [Google Scholar]
- 30.Wadzinski AM, Ribbons DW. Utilization of acetate by Methanomonas methanooxidans . J Bacteriol. 1975;123:380–381. doi: 10.1128/JB.123.1.380-381.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xing X-H, Wu H, Luo M-F, Wang B-P. Effects of organic chemicals on growth of Methylosinus trichosporium OB3b. Biochem Eng J. 2006;31:113–117. doi: 10.1016/j.bej.2006.06.001. [DOI] [Google Scholar]
- 32.Semrau JD, DiSpirito AA, Vuilleumier S. Facultative methanotrophy: false leads, true results, and suggestions for future research. FEMS Microbiol Lett. 2011;323:1–12. doi: 10.1111/j.1574-6968.2011.02315.x. [DOI] [PubMed] [Google Scholar]
- 33.Dedysh SN, Dunfield PF. Facultative and obligate methanotrophs how to identify and differentiate them. Methods Enzymol. 2011;495:32–61. doi: 10.1016/B978-0-12-386905-0.00003-6. [DOI] [PubMed] [Google Scholar]
- 34.Dedysh SN, Dunfield PF. Facultative methane oxidizers. In: McGenity TJ, editor. Taxonomy, Genomics and Ecophysiology of Hydrocarbon-Degrading Microbes. Cham: Springer International Publishing; 2018. pp. 1–20. editor. [Google Scholar]
- 35.Belova SE, Kulichevskaya IS, Bodelier PLE, Dedysh SN. Methylocystis bryophila sp. nov., a facultatively methanotrophic bacterium from acidic Sphagnum peat, and emended description of the genus Methylocystis (ex Whittenbury et al. 1970) Bowman et al. 1993. Int J Syst Evol Microbiol. 2013;63:1096–1104. doi: 10.1099/ijs.0.043505-0. [DOI] [PubMed] [Google Scholar]
- 36.Dunfield PF, Belova SE, Vorob'ev AV, Cornish SL, Dedysh SN. Methylocapsa aurea sp. nov., a facultative methanotroph possessing a particulate methane monooxygenase, and emended description of the genus Methylocapsa . Int J Syst Evol Microbiol. 2010;60:2659–2664. doi: 10.1099/ijs.0.020149-0. [DOI] [PubMed] [Google Scholar]
- 37.Im J, Lee SW, Yoon S, DiSpirito AA, Semrau JD. Characterization of a novel facultative Methylocystis species capable of growth on methane, acetate and ethanol. Environ Microbiol Rep. 2011;3:174–181. doi: 10.1111/j.1758-2229.2010.00204.x. [DOI] [PubMed] [Google Scholar]
- 38.Dedysh SN, Knief C, Dunfield PF. Methylocella species are facultatively methanotrophic. J Bacteriol. 2005;187:4665–4670. doi: 10.1128/JB.187.13.4665-4670.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Belova SE, Baani M, Suzina NE, Bodelier PLE, Liesack W, et al. Acetate utilization as a survival strategy of peat-inhabiting Methylocystis spp. Environ Microbiol Rep. 2011;3:36–46. doi: 10.1111/j.1758-2229.2010.00180.x. [DOI] [PubMed] [Google Scholar]
- 40.Stoecker K, Bendinger B, Schöning B, Nielsen PH, Nielsen JL, et al. Cohn’s Crenothrix is a filamentous methane oxidizer with an unusual methane monooxygenase. Proc Natl Acad Sci U S A. 2006;103:2363–2367. doi: 10.1073/pnas.0506361103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dedysh SN, Berestovskaya YY, Vasylieva LV, Belova SE, Khmelenina VN, et al. Methylocella tundrae sp. nov., a novel methanotrophic bacterium from acidic tundra peatlands. Int J Syst Evol Microbiol. 2004;54:151–156. doi: 10.1099/ijs.0.02805-0. [DOI] [PubMed] [Google Scholar]
- 42.Dedysh SN, Liesack W, Khmelenina VN, Suzina NE, Trotsenko YA, et al. Methylocella palustris gen. nov., sp. nov., a new methane-oxidizing acidophilic bacterium from peat bogs, representing a novel subtype of serine-pathway methanotrophs. Int J Syst Evol Microbiol. 2000;50:955–969. doi: 10.1099/00207713-50-3-955. [DOI] [PubMed] [Google Scholar]
- 43.Dunfield PF, Khmelenina VN, Suzina NE, Trotsenko YA, Dedysh SN. Methylocella silvestris sp. nov., a novel methanotroph isolated from an acidic forest cambisol. Int J Syst Evol Microbiol. 2003;53:1231–1239. doi: 10.1099/ijs.0.02481-0. [DOI] [PubMed] [Google Scholar]
- 44.Crombie AT, Murrell JC. Trace-gas metabolic versatility of the facultative methanotroph Methylocella silvestris . Nature. 2014;510:148–151. doi: 10.1038/nature13192. [DOI] [PubMed] [Google Scholar]
- 45.Farhan Ul Haque M, Crombie AT, Murrell JC. Novel facultative Methylocella strains are active methane consumers at terrestrial natural gas seeps. Microbiome. 2019;7:134. doi: 10.1186/s40168-019-0741-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bordel S, Rodríguez Y, Hakobyan A, Rodríguez E, Lebrero R, et al. Genome scale metabolic modeling reveals the metabolic potential of three Type II methanotrophs of the genus Methylocystis . Metab Eng. 2019;54:191–199. doi: 10.1016/j.ymben.2019.04.001. [DOI] [PubMed] [Google Scholar]
- 47.Wang J, Geng K, Farhan Ul Haque M, Crombie A, Street LE, et al. Draft genome sequence of Methylocella silvestris TVC, a facultative methanotroph isolated from permafrost. Genome Announc. 2018;6:e00040–00018. doi: 10.1128/genomeA.00040-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Etiope G, Drobniak A, Schimmelmann A. Natural seepage of shale gas and the origin of “eternal flames” in the Northern Appalachian Basin, USA. Mar Petrol Geol. 2013;43:178–186. doi: 10.1016/j.marpetgeo.2013.02.009. [DOI] [Google Scholar]
- 49.Vekeman B, Kerckhof F-M, Cremers G, de Vos P, Vandamme P, et al. New Methyloceanibacter diversity from North Sea sediments includes methanotroph containing solely the soluble methane monooxygenase. Environ Microbiol. 2016;18:4523–4536. doi: 10.1111/1462-2920.13485. [DOI] [PubMed] [Google Scholar]
- 50.Vorobev AV, Baani M, Doronina NV, Brady AL, Liesack W, et al. Methyloferula stellata gen. nov., sp. nov., an acidophilic, obligately methanotrophic bacterium that possesses only a soluble methane monooxygenase. Int J Syst Evol Microbiol. 2011;61:2456–2463. doi: 10.1099/ijs.0.028118-0. [DOI] [PubMed] [Google Scholar]
- 51.Takeuchi M, Katayama T, Yamagishi T, Hanada S, Tamaki H, et al. Methyloceanibacter caenitepidi gen. nov., sp. nov., a facultatively methylotrophic bacterium isolated from marine sediments near a hydrothermal vent. Int J Syst Evol Microbiol. 2014;64:462–468. doi: 10.1099/ijs.0.053397-0. [DOI] [PubMed] [Google Scholar]
- 52.Kang CS, Dunfield PF, Semrau JD. The origin of aerobic methanotrophy within the Proteobacteria. FEMS Microbiol Lett. 2019;366:fnz096. doi: 10.1093/femsle/fnz096. [DOI] [PubMed] [Google Scholar]
- 53.Conley S, Franco G, Faloona I, Blake DR, Peischl J, et al. Methane emissions from the 2015 Aliso Canyon blowout in Los Angeles, CA. Science. 2016;351:1317–1320. doi: 10.1126/science.aaf2348. [DOI] [PubMed] [Google Scholar]
- 54.Arp DJ. Butane metabolism by butane-grown ‘Pseudomonas butanovora’. Microbiology. 1999;145:1173–1180. doi: 10.1099/13500872-145-5-1173. [DOI] [PubMed] [Google Scholar]
- 55.Kox MAR, Farhan Ul Haque M, van Alen TA, Crombie AT, Jetten MSM, et al. Complete genome sequence of the aerobic facultative methanotroph Methylocella tundrae strain T4. Microbiol Resour Announc. 2019;8:e00286–00219. doi: 10.1128/MRA.00286-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chen Y, Crombie A, Rahman MT, Dedysh SN, Liesack W, et al. Complete genome sequence of the aerobic facultative methanotroph Methylocella silvestris BL2. J Bacteriol. 2010;192:3840–3841. doi: 10.1128/JB.00506-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kotani T, Yamamoto T, Yurimoto H, Sakai Y, Kato N. Propane monooxygenase and NAD+-dependent secondary alcohol dehydrogenase in propane metabolism by Gordonia sp. strain TY-5. J Bacteriol. 2003;185:7120–7128. doi: 10.1128/JB.185.24.7120-7128.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Furuya T, Nakao T, Kino K. Catalytic function of the mycobacterial binuclear iron monooxygenase in acetone metabolism. FEMS Microbiol Lett. 2015;362:fnv136. doi: 10.1093/femsle/fnv136. [DOI] [PubMed] [Google Scholar]
- 59.Anthony C. The quinoprotein dehydrogenases for methanol and glucose. Arch Biochem Biophys. 2004;428:2–9. doi: 10.1016/j.abb.2004.03.038. [DOI] [PubMed] [Google Scholar]
- 60.Picone N, Op den Camp HJ. Role of rare earth elements in methanol oxidation. Curr Opin Chem Biol. 2019;49:39–44. doi: 10.1016/j.cbpa.2018.09.019. [DOI] [PubMed] [Google Scholar]
- 61.Pol A, Barends TRM, Dietl A, Khadem AF, Eygensteyn J, et al. Rare earth metals are essential for methanotrophic life in volcanic mudpots. Environ Microbiol. 2014;16:255–264. doi: 10.1111/1462-2920.12249. [DOI] [PubMed] [Google Scholar]
- 62.Chistoserdova L. New pieces to the lanthanide puzzle. Mol Microbiol. 2019;111:1127–1131. doi: 10.1111/mmi.14210. [DOI] [PubMed] [Google Scholar]
- 63.Skovran E, Martinez-Gomez NC. Just add lanthanides. Science. 2015;348:862–863. doi: 10.1126/science.aaa9091. [DOI] [PubMed] [Google Scholar]
- 64.Farhan Ul Haque M, Kalidass B, Bandow N, Turpin EA, DiSpirito AA, et al. Cerium regulates expression of alternative methanol dehydrogenases in Methylosinus trichosporium OB3b. Appl Environ Microbiol. 2015;81:7546–7552. doi: 10.1128/AEM.02542-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tyler G. Rare earth elements in soil and plant systems - A review. Plant Soil. 2004;267:191–206. doi: 10.1007/s11104-005-4888-2. [DOI] [Google Scholar]
- 66.Chistoserdova L. Lanthanides: new life metals? World J Microbiol Biot. 2016;32:1–7. doi: 10.1007/s11274-016-2088-2. [DOI] [PubMed] [Google Scholar]
- 67.Huang J, Yu Z, Groom J, Cheng J-F, Tarver A, et al. Rare earth element alcohol dehydrogenases widely occur among globally distributed, numerically abundant and environmentally important microbes. Isme J. 2019;13:2005–2017. doi: 10.1038/s41396-019-0414-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Keltjens JT, Pol A, Reimann J, Op den Camp HM. PQQ-dependent methanol dehydrogenases: rare-earth elements make a difference. Appl Microbiol Biotechnol. 2014;98:6163–6183. doi: 10.1007/s00253-014-5766-8. [DOI] [PubMed] [Google Scholar]
- 69.Jahn B, Pol A, Lumpe H, Barends T, Dietl A, et al. Similar but not the same: first kinetic and structural analyses of a methanol dehydrogenase containing a europium ion in the active site. ChemBioChem. 2018;19:1147–1153. doi: 10.1002/cbic.201800130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Deng YW, Ro SY, Rosenzweig AC. Structure and function of the lanthanide-dependent methanol dehydrogenase XoxF from the methanotroph Methylomicrobium buryatense 5GB1C. J Biol Inorg Chem. 2018;23:1037–1047. doi: 10.1007/s00775-018-1604-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Good NM, Fellner M, Demirer K, Hu J, Hausinger RP, et al. Lanthanide-dependent alcohol dehydrogenases require an essential aspartate residue for metal coordination and enzymatic function. J Biol Chem. 2020;295:8272–8284. doi: 10.1074/jbc.RA120.013227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Anthony C, Williams P. The structure and mechanism of methanol dehydrogenase. Biochim Biophys Acta. 2003;1647:18–23. doi: 10.1016/S1570-9639(03)00042-6. [DOI] [PubMed] [Google Scholar]
- 73.Semrau JD, DiSpirito AA, Gu W, Yoon S. Metals and methanotrophy. Appl Environ Microbiol. 2018;84:e02289-17. doi: 10.1128/AEM.02289-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Taubert M, Grob C, Howat AM, Burns OJ, Dixon JL, et al. xoxF encoding an alternative methanol dehydrogenase is widespread in coastal marine environments. Environ Microbiol. 2015;17:3937–3948. doi: 10.1111/1462-2920.12896. [DOI] [PubMed] [Google Scholar]
- 75.Krause SMB, Johnson T, Samadhi Karunaratne Y, Fu Y, Beck DAC, et al. Lanthanide-dependent cross-feeding of methane-derived carbon is linked by microbial community interactions. Proc Natl Acad Sci U S A. 2017;114:358–363. doi: 10.1073/pnas.1619871114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yu Z, Beck DAC, Chistoserdova L. Natural selection in synthetic communities highlights the roles of Methylococcaceae and Methylophilaceae and suggests differential roles for alternative methanol dehydrogenases in methane consumption. Front Microbiol. 2017;8:2392. doi: 10.3389/fmicb.2017.02392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wang L, Suganuma S, Hibino A, Mitsui R, Tani A, et al. Lanthanide-dependent methanol dehydrogenase from the legume symbiotic nitrogen-fixing bacterium Bradyrhizobium diazoefficiens strain USDA110. Enzyme Microb Technol. 2019;130:109371. doi: 10.1016/j.enzmictec.2019.109371. [DOI] [PubMed] [Google Scholar]
- 78.Wilson SM, Gleisten MP, Donohue TJ. Identification of proteins involved in formaldehyde metabolism by Rhodobacter sphaeroides . Microbiology. 2008;154:296–305. doi: 10.1099/mic.0.2007/011346-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kato S, Takashino M, Igarashi K, Kitagawa W. Isolation and genomic characterization of a proteobacterial methanotroph requiring lanthanides. Microbes Environ. 2020;35:n/a.:ME19128. doi: 10.1264/jsme2.ME19128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wegner C-E, Gorniak L, Riedel S, Westermann M, Küsel K. Lanthanide-dependent methylotrophs of the family Beijerinckiaceae: physiological and genomic insights. Appl Environ Microbiol. 2019;86:e01830-19. doi: 10.1128/AEM.01830-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Good NM, Vu HN, Suriano CJ, Subuyuj GA, Skovran E, et al. Pyrroloquinoline quinone-containing ethanol dehydrogenase in Methylobacterium extorquens AM1 extends lanthanide-dependent metabolism to multi-carbon substrates. J Bacteriol. 2016;198:3109–3118. doi: 10.1128/JB.00478-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wehrmann M, Billard P, Martin-Meriadec A, Zegeye A, Klebensberger J. Functional role of lanthanides in enzymatic activity and transcriptional regulation of pyrroloquinoline quinone-dependent alcohol dehydrogenases in Pseudomonas putida KT2440. MBio. 2017;8:e00570-17. doi: 10.1128/mBio.00570-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Dunstan PM, Anthony C, Drabble WT. Microbial metabolism of C1 and C2 compounds. The involvement of glycollate in the metabolism of ethanol and of acetate by Pseudomonas AM1. Biochem J. 1972;128:99–106. doi: 10.1042/bj1280099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Vorobev A, Jagadevan S, Jain S, Anantharaman K, Dick GJ, et al. Genomic and transcriptomic analyses of the facultative methanotroph Methylocystis sp. strain SB2 grown on methane or ethanol. Appl Environ Microbiol. 2014;80:3044–3052. doi: 10.1128/AEM.00218-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Yu Z, Zhang Q, Kraus TEC, Dahlgren RA, Anastasio C, et al. Contribution of amino compounds to dissolved organic nitrogen in forest soils. Biogeochemistry. 2002;61:173–198. doi: 10.1023/A:1020221528515. [DOI] [Google Scholar]
- 86.Anthony C. Biochemistry of Methylotrophs. London: Academic Press; 1982. [Google Scholar]
- 87.Chen Y, Patel NA, Crombie A, Scrivens JH, Murrell JC. Bacterial flavin-containing monooxygenase is trimethylamine monooxygenase. Proc Natl Acad Sci U S A. 2011;108:17791–17796. doi: 10.1073/pnas.1112928108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Chen Y, Scanlan J, Song L, Crombie A, Rahman MT, et al. γ-Glutamylmethylamide is an essential intermediate in the metabolism of methylamine by Methylocella silvestris . Appl Environ Microbiol. 2010;76:4530–4537. doi: 10.1128/AEM.00739-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zhu Y, Jameson E, Parslow RA, Lidbury I, Fu T, et al. Identification and characterization of trimethylamine N -oxide (TMAO) demethylase and TMAO permease in Methylocella silvestris BL2. Environ Microbiol. 2014;16:3318–3330. doi: 10.1111/1462-2920.12585. [DOI] [PubMed] [Google Scholar]
- 90.Horken KM, Tabita FR. Closely related form I ribulose bisphosphate carboxylase/oxygenase molecules that possess different CO2/O2 substrate specificities. Arch Biochem Biophys. 1999;361:183–194. doi: 10.1006/abbi.1998.0979. [DOI] [PubMed] [Google Scholar]
- 91.Dedysh SN, Smirnova KV, Khmelenina VN, Suzina NE, Liesack W, et al. Methylotrophic autotrophy in Beijerinckia mobilis . J Bacteriol. 2005;187:3884–3888. doi: 10.1128/JB.187.11.3884-3888.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Tamas I, Smirnova AV, He Z, Dunfield PF. The (d)evolution of methanotrophy in the Beijerinckiaceae--a comparative genomics analysis. Isme J. 2014;8:369–382. doi: 10.1038/ismej.2013.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ward N, Larsen Øivind, Sakwa J, Bruseth L, Khouri H, et al. Genomic insights into methanotrophy: the complete genome sequence of Methylococcus capsulatus (Bath) PLoS Biol. 2004;2:e303. doi: 10.1371/journal.pbio.0020303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Gimenez R, Nuñez María Felisa, Badia J, Aguilar J, Baldoma L. The gene yjcG, cotranscribed with the gene acs, encodes an acetate permease in Escherichia coli . J Bacteriol. 2003;185:6448–6455. doi: 10.1128/JB.185.21.6448-6455.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Schneider K, Peyraud R, Kiefer P, Christen P, Delmotte N, et al. The ethylmalonyl-CoA pathway is used in place of the glyoxylate cycle by Methylobacterium extorquens AM1 during growth on acetate. J Biol Chem. 2012;287:757–766. doi: 10.1074/jbc.M111.305219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Han D, Dedysh SN, Liesack W. Unusual genomic traits suggest Methylocystis bryophila S285 to be well adapted for life in peatlands. Genome Biol Evol. 2018;10:623–628. doi: 10.1093/gbe/evy025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Kornberg HL, Krebs HA. Synthesis of cell constituents from C2-units by a modified tricarboxylic acid cycle. Nature. 1957;179:988–991. doi: 10.1038/179988a0. [DOI] [PubMed] [Google Scholar]
- 98.Anthony C. How half a century of research was required to understand bacterial growth on C1 and C2 compounds; the story of the serine cycle and the ethylmalonyl-CoA pathway. Sci Prog. 2011;94:109–137. doi: 10.3184/003685011X13044430633960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Crombie A, Murrell JC. Development of a system for genetic manipulation of the facultative methanotroph Methylocella silvestris BL2. Methods Enzymol. 2011;495:119–133. doi: 10.1016/B978-0-12-386905-0.00008-5. [DOI] [PubMed] [Google Scholar]
- 100.Bordel S, Crombie AT, Muñoz R, Murrell JC. Genome scale metabolic model of the versatile methanotroph Methylocella silvestris . Microb Cell Fact. 2020;19:144. doi: 10.1186/s12934-020-01395-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Carere CR, Hards K, Houghton KM, Power JF, McDonald B, et al. Mixotrophy drives niche expansion of verrucomicrobial methanotrophs. ISME J. 2017;11:2599–2610. doi: 10.1038/ismej.2017.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Ward L, Shih PM, Hemp J, Kakegawa T, Fischer WW, et al. Phototrophic methane oxidation in a member of the Chloroflexi phylum. bioRxiv. 2019;531582 [Google Scholar]
- 103.Ji M, Greening C, Vanwonterghem I, Carere CR, Bay SK, et al. Atmospheric trace gases support primary production in Antarctic desert surface soil. Nature. 2017;552:400–403. doi: 10.1038/nature25014. [DOI] [PubMed] [Google Scholar]
- 104.Conrad R. Soil microorganisms as controllers of atmospheric trace gases (H2, CO, CH4, OCS, N2O, and NO) Microbiol Rev. 1996;60:609–640. doi: 10.1128/MMBR.60.4.609-640.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Hanczár T, Csáki R, Bodrossy L, Murrell CJ, Kovács KL. Detection and localization of two hydrogenases in Methylococcus capsulatus (Bath) and their potential role in methane metabolism. Arch Microbiol. 2002;177:167–172. doi: 10.1007/s00203-001-0372-4. [DOI] [PubMed] [Google Scholar]
- 106.Chen YP, Yoch DC. Regulation of two nickel-requiring (inducible and constitutive) hydrogenases and their coupling to nitrogenase in Methylosinus trichosporium OB3b. J Bacteriol. 1987;169:4778–4783. doi: 10.1128/JB.169.10.4778-4783.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Hakobyan A, Zhu J, Glatter T, Paczia N, Liesack W. Hydrogen utilization by Methylocystis sp. strain SC2 expands the known metabolic versatility of type IIa methanotrophs. Metab Eng. 2020;61:181–196. doi: 10.1016/j.ymben.2020.05.003. [DOI] [PubMed] [Google Scholar]
- 108.Piché-Choquette S, Constant P. Molecular hydrogen, a neglected key driver of soil biogeochemical processes. Appl Environ Microbiol. 2019;85:e02418-18. doi: 10.1128/AEM.02418-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Mohammadi SS, Schmitz RA, Pol A, Berben T, Jetten MSM, et al. The acidophilic methanotroph Methylacidimicrobium tartarophylax 4AC grows as autotroph on H2under microoxic conditions. Front Microbiol. 2019;10:2352. doi: 10.3389/fmicb.2019.02352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Greening C, Constant P, Hards K, Morales SE, Oakeshott JG, et al. Atmospheric hydrogen scavenging: from enzymes to ecosystems. Appl Environ Microbiol. 2015;81:1190–1199. doi: 10.1128/AEM.03364-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Greening C, Berney M, Hards K, Cook GM, Conrad R. A soil actinobacterium scavenges atmospheric H2 using two membrane-associated, oxygen-dependent [NiFe] hydrogenases. Proc Natl Acad Sci U S A. 2014;111:4257–4261. doi: 10.1073/pnas.1320586111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Cordero PRF, Bayly K, Man Leung P, Huang C, Islam ZF, et al. Atmospheric carbon monoxide oxidation is a widespread mechanism supporting microbial survival. ISME J. 2019;13:2868–2881. doi: 10.1038/s41396-019-0479-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Garcia-Chaves MC, Cottrell MT, Kirchman DL, Ruiz-González C, Del Giorgio PA. Single-cell activity of freshwater aerobic anoxygenic phototrophic bacteria and their contribution to biomass production. ISME J. 2016;10:1579–1588. doi: 10.1038/ismej.2015.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Yurkov VV, Beatty JT. Aerobic anoxygenic phototrophic bacteria. Microbiol Mol Biol Rev. 1998;62:695–724. doi: 10.1128/MMBR.62.3.695-724.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Koblížek M. Ecology of aerobic anoxygenic phototrophs in aquatic environments. FEMS Microbiol Rev. 2015;39:854–870. doi: 10.1093/femsre/fuv032. [DOI] [PubMed] [Google Scholar]
- 116.Miroshnikov KK, Belova SE, Dedysh SN. Genomic determinants of phototrophy in methanotrophic Alphaproteobacteria. Microbiology. 2019;88:548–555. doi: 10.1134/S0026261719050102. [DOI] [Google Scholar]
- 117.Oshkin IY, Miroshnikov KK, Grouzdev DS, Dedysh SN. Pan-genome-based analysis as a framework for demarcating two closely related methanotroph genera Methylocystis and Methylosinus . Microorganisms. 2020;8:768. doi: 10.3390/microorganisms8050768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Ettwig KF, Butler MK, Le Paslier D, Pelletier E, Mangenot S, et al. Nitrite-driven anaerobic methane oxidation by oxygenic bacteria. Nature. 2010;464:543–548. doi: 10.1038/nature08883. [DOI] [PubMed] [Google Scholar]
- 119.Kits KD, Klotz MG, Stein LY. Methane oxidation coupled to nitrate reduction under hypoxia by the Gammaproteobacterium Methylomonas denitrificans, sp. nov. type strain FJG1. Environ Microbiol. 2015;17:3219–3232. doi: 10.1111/1462-2920.12772. [DOI] [PubMed] [Google Scholar]
- 120.Dam B, Dam S, Blom J, Liesack W. Genome analysis coupled with physiological studies reveals a diverse nitrogen metabolism in Methylocystis sp. strain SC2. PLoS One. 2013;8:e74767. doi: 10.1371/journal.pone.0074767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Kits KD, Campbell DJ, Rosana AR, Stein LY. Diverse electron sources support denitrification under hypoxia in the obligate methanotroph Methylomicrobium album strain BG8. Front Microbiol. 2015;6:1072. doi: 10.3389/fmicb.2015.01072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Tanaka K, Yokoe S, Igarashi K, Takashino M, Ishikawa M, et al. Extracellular electron transfer via outer membrane cytochromes in a methanotrophic bacterium Methylococcus capsulatus (Bath) Front Microbiol. 2018;9:2905. doi: 10.3389/fmicb.2018.02905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Zheng Y, Wang H, Liu Y, Zhu B, Li J, et al. Methane-dependent mineral reduction by aerobic methanotrophs under hypoxia. Environ Sci Tech Let. 2020;7:606–612. doi: 10.1021/acs.estlett.0c00436. [DOI] [Google Scholar]
- 124.Kalyuzhnaya MG, Yang S, Rozova ON, Smalley NE, Clubb J, et al. Highly efficient methane biocatalysis revealed in a methanotrophic bacterium. Nat Commun. 2013;4:2785. doi: 10.1038/ncomms3785. [DOI] [PubMed] [Google Scholar]
- 125.Gilman A, Fu Y, Hendershott M, Chu F, Puri AW, et al. Oxygen-limited metabolism in the methanotroph Methylomicrobium buryatense 5GB1C. PeerJ. 2017;5:e3945. doi: 10.7717/peerj.3945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Stein LY, Klotz MG. Nitrifying and denitrifying pathways of methanotrophic bacteria. Biochem Soc Trans. 2011;39:1826–1831. doi: 10.1042/BST20110712. [DOI] [PubMed] [Google Scholar]
- 127.Stein LY. The long-term relationship between microbial metabolism and greenhouse gases. Trends Microbiol. 2020;28:500–511. doi: 10.1016/j.tim.2020.01.006. [DOI] [PubMed] [Google Scholar]
- 128.Knief C. Diversity and habitat preferences of cultivated and uncultivated aerobic methanotrophic bacteria evaluated based on pmoA as molecular marker. Front Microbiol. 2015;6:1346. doi: 10.3389/fmicb.2015.01346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Dedysh SN. Exploring methanotroph diversity in acidic northern wetlands: molecular and cultivation-based studies. Microbiology. 2009;78:655–669. doi: 10.1134/S0026261709060010. [DOI] [Google Scholar]
- 130.Dedysh SN, Pankratov TA, Belova SE, Kulichevskaya IS, Liesack W. Phylogenetic analysis and in situ identification of bacteria community composition in an acidic Sphagnum peat bog. Appl Environ Microbiol. 2006;72:2110–2117. doi: 10.1128/AEM.72.3.2110-2117.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Radajewski S, Webster G, Reay DS, Morris SA, Ineson P, et al. Identification of active methylotroph populations in an acidic forest soil by stable-isotope probing. Microbiology. 2002;148:2331–2342. doi: 10.1099/00221287-148-8-2331. [DOI] [PubMed] [Google Scholar]
- 132.Chen Y, Dumont MG, McNamara NP, Chamberlain PM, Bodrossy L, et al. Diversity of the active methanotrophic community in acidic peatlands as assessed by mRNA and SIP-PLFA analyses. Environ Microbiol. 2008;10:446–459. doi: 10.1111/j.1462-2920.2007.01466.x. [DOI] [PubMed] [Google Scholar]
- 133.Kip N, Dutilh BE, Pan Y, Bodrossy L, Neveling K, et al. Ultra-deep pyrosequencing of pmoA amplicons confirms the prevalence of Methylomonas and Methylocystis in Sphagnum mosses from a Dutch peat bog. Environ Microbiol Rep. 2011;3:667–673. doi: 10.1111/j.1758-2229.2011.00260.x. [DOI] [PubMed] [Google Scholar]
- 134.Kip N, Fritz C, Langelaan ES, Pan Y, Bodrossy L, et al. Methanotrophic activity and diversity in different Sphagnum magellanicum dominated habitats in the southernmost peat bogs of Patagonia. Biogeosciences. 2012;9:47–55. doi: 10.5194/bg-9-47-2012. [DOI] [Google Scholar]
- 135.Dedysh SN, Derakshani M, Liesack W. Detection and enumeration of methanotrophs in acidic Sphagnum peat by 16S rRNA fluorescence in situ hybridization, including the use of newly developed oligonucleotide probes for Methylocella palustris . Appl Environ Microbiol. 2001;67:4850–4857. doi: 10.1128/AEM.67.10.4850-4857.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Kolb S, Horn MA. Microbial CH4 and N2O consumption in acidic wetlands. Front Microbiol. 2012;3:78. doi: 10.3389/fmicb.2012.00078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Henckel T, Friedrich M, Conrad R. Molecular analyses of the methane-oxidizing microbial community in rice field soil by targeting the genes of the 16S rRNA, particulate methane monooxygenase, and methanol dehydrogenase. Appl Environ Microbiol. 1999;65:1980–1990. doi: 10.1128/AEM.65.5.1980-1990.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Bao Z, Okubo T, Kubota K, Kasahara Y, Tsurumaru H, et al. Metaproteomic identification of diazotrophic methanotrophs and their localization in root tissues of field-grown rice plants. Appl Environ Microbiol. 2014;80:5043–5052. doi: 10.1128/AEM.00969-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Horz HP, Yimga MT, Liesack W. Detection of methanotroph diversity on roots of submerged rice plants by molecular retrieval of pmoA, mmoX, mxaF, and 16S rRNA and ribosomal DNA, including pmoA-based terminal restriction fragment length polymorphism profiling. Appl Environ Microbiol. 2001;67:4177–4185. doi: 10.1128/AEM.67.9.4177-4185.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Rahman MT, Crombie A, Chen Y, Stralis-Pavese N, Bodrossy L, et al. Environmental distribution and abundance of the facultative methanotroph Methylocella . ISME J. 2011;5:1061–1066. doi: 10.1038/ismej.2010.190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Cébron A, Bodrossy L, Chen Y, Singer AC, Thompson IP, et al. Identity of active methanotrophs in landfill cover soil as revealed by DNA-stable isotope probing. FEMS Microbiol Ecol. 2007;62:12–23. doi: 10.1111/j.1574-6941.2007.00368.x. [DOI] [PubMed] [Google Scholar]
- 142.Han B, Chen Y, Abell G, Jiang H, Bodrossy L, et al. Diversity and activity of methanotrophs in alkaline soil from a Chinese coal mine. FEMS Microbiol Ecol. 2009;70:196–207. doi: 10.1111/j.1574-6941.2009.00707.x. [DOI] [PubMed] [Google Scholar]
- 143.Putkinen A, Larmola T, Tuomivirta T, Siljanen HMP, Bodrossy L, et al. Peatland succession induces a shift in the community composition of Sphagnum-associated active methanotrophs. FEMS Microbiol Ecol. 2014;88:596–611. doi: 10.1111/1574-6941.12327. [DOI] [PubMed] [Google Scholar]
- 144.Gupta V, Smemo KA, Yavitt JB, Basiliko N. Active methanotrophs in two contrasting North American peatland ecosystems revealed using DNA-SIP. Microb Ecol. 2012;63:438–445. doi: 10.1007/s00248-011-9902-z. [DOI] [PubMed] [Google Scholar]
- 145.Sharp CE, Martínez-Lorenzo A, Brady AL, Grasby SE, Dunfield PF. Methanotrophic bacteria in warm geothermal spring sediments identified using stable-isotope probing. FEMS Microbiol Ecol. 2014;90:92–102. doi: 10.1111/1574-6941.12375. [DOI] [PubMed] [Google Scholar]
- 146.Shao Y, Hatzinger PB, Streger SH, Rezes RT, Chu KH. Evaluation of methanotrophic bacterial communities capable of biodegrading trichloroethene (TCE) in acidic aquifers. Biodegradation. 2019;30:173–190. doi: 10.1007/s10532-019-09875-w. [DOI] [PubMed] [Google Scholar]
- 147.Morris SA, Radajewski S, Willison TW, Murrell JC. Identification of the functionally active methanotroph population in a peat soil microcosm by stable-isotope probing. Appl Environ Microbiol. 2002;68:1446–1453. doi: 10.1128/AEM.68.3.1446-1453.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Strobel BW. Influence of vegetation on low-molecular-weight carboxylic acids in soil solution—a review. Geoderma. 2001;99:169–198. doi: 10.1016/S0016-7061(00)00102-6. [DOI] [Google Scholar]
- 149.Vranova V, Rejsek K, Formanek P, Aliphatic FP. Aliphatic, cyclic, and aromatic organic acids, vitamins, and carbohydrates in soil: a review. Sci World J. 2013:524239. doi: 10.1155/2013/524239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Hines ME, Duddleston KN, Kiene RP. Carbon flow to acetate and C1 compounds in northern wetlands. Geophys Res Lett. 2001;28:4251–4254. doi: 10.1029/2001GL012901. [DOI] [Google Scholar]
- 151.Wieczorek AS, Drake HL, Kolb S. Organic acids and ethanol inhibit the oxidation of methane by mire methanotrophs. FEMS Microbiol Ecol. 2011;77:28–39. doi: 10.1111/j.1574-6941.2011.01080.x. [DOI] [PubMed] [Google Scholar]
- 152.Rahman MT, Crombie A, Moussard H, Chen Y, Murrell JC. Acetate repression of methane oxidation by supplemental Methylocella silvestris in a peat soil microcosm. Appl Environ Microbiol. 2011;77:4234–4236. doi: 10.1128/AEM.02902-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Leng L, Chang J, Geng K, Lu Y, Ma K. Uncultivated Methylocystis species in paddy soil include facultative methanotrophs that utilize acetate. Microb Ecol. 2015;70:88–96. doi: 10.1007/s00248-014-0540-0. [DOI] [PubMed] [Google Scholar]
- 154.Yoon S, Im J, Bandow N, Dispirito AA, Semrau JD. Constitutive expression of pMMO by Methylocystis strain SB2 when grown on multi-carbon substrates: implications for biodegradation of chlorinated ethenes. Environ Microbiol Rep. 2011;3:182–188. doi: 10.1111/j.1758-2229.2010.00205.x. [DOI] [PubMed] [Google Scholar]
- 155.Pratscher J, Dumont MG, Conrad R. Assimilation of acetate by the putative atmospheric methane oxidizers belonging to the USCα clade. Environ Microbiol. 2011;13:2692–2701. doi: 10.1111/j.1462-2920.2011.02537.x. [DOI] [PubMed] [Google Scholar]
- 156.West AE, Schmidt SK. Acetate stimulates atmospheric CH4 oxidation by an alpine tundra soil. Soil Biol Biochem. 1999;31:1649–1655. doi: 10.1016/S0038-0717(99)00076-0. [DOI] [Google Scholar]
- 157.Sullivan BW, Selmants PC, Hart SC. Does dissolved organic carbon regulate biological methane oxidation in semiarid soils? Glob Chang Biol. 2013;19:2149–2157. doi: 10.1111/gcb.12201. [DOI] [PubMed] [Google Scholar]
- 158.Sullivan BW, Selmants PC, Hart SC. What is the relationship between soil methane oxidation and other C compounds? Glob Chang Biol. 2014;20:2381–2382. doi: 10.1111/gcb.12533. [DOI] [PubMed] [Google Scholar]
- 159.Theisen AR, Ali MH, Radajewski S, Dumont MG, Dunfield PF, et al. Regulation of methane oxidation in the facultative methanotroph Methylocella silvestris BL2. Mol Microbiol. 2005;58:682–692. doi: 10.1111/j.1365-2958.2005.04861.x. [DOI] [PubMed] [Google Scholar]
- 160.Smirnova AV, Dunfield PF. Differential transcriptional activation of genes encoding soluble methane monooxygenase in a facultative versus an obligate methanotroph. Microorganisms. 2018;6:20. doi: 10.3390/microorganisms6010020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Farhan Ul Haque M, Crombie AT, Ensminger SA, Baciu C, Murrell JC. Facultative methanotrophs are abundant at terrestrial natural gas seeps. Microbiome. 2018;6:118. doi: 10.1186/s40168-018-0500-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Dalton H, Stirling DI, Quayle JR, Higgins IJ, Bull AT. Co-metabolism. Philos Trans R Soc Lond B Biol Sci. 1982;297:481–496. doi: 10.1098/rstb.1982.0056. [DOI] [PubMed] [Google Scholar]
- 163.Semrau JD. Bioremediation via methanotrophy: overview of recent findings and suggestions for future research. Front Microbiol. 2011;2:209. doi: 10.3389/fmicb.2011.00209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Muenmee S, Chiemchaisri W, Chiemchaisri C. Enhancement of biodegradation of plastic wastes via methane oxidation in semi-aerobic landfill. Int Biodeterior Biodegradation. 2016;113:244–255. doi: 10.1016/j.ibiod.2016.03.016. [DOI] [Google Scholar]
- 165.Muenmee S, Chiemchaisri W, Chiemchaisri C. Microbial consortium involving biological methane oxidation in relation to the biodegradation of waste plastics in a solid waste disposal open dump site. Int Biodeterior Biodegradation. 2015;102:172–181. doi: 10.1016/j.ibiod.2015.03.015. [DOI] [Google Scholar]
- 166.Smith TJ, Dalton H, Vazquez-Duhalt R, Quintero-Ramirez R. Petroleum Biotechnology - Developments and Perspectives. Amsterdam: Elsevier; 2004. Biocatalysis by methane monooxygenase and its implications for the petroleum industry; pp. 177–192. [Google Scholar]
- 167.Baba T, Miyaji A. Catalysis and the Mechanism of Methane Conversion to Chemicals. Singapore: Springer Singapore; 2020. Application of biocatalysts for the production of methanol from methane; pp. 73–101. [Google Scholar]
- 168.Patel SKS, Kumar V, Mardina P, Li J, Lestari R, et al. Methanol production from simulated biogas mixtures by co-immobilized Methylomonas methanica and Methylocella tundrae . Bioresour Technol. 2018;263:25–32. doi: 10.1016/j.biortech.2018.04.096. [DOI] [PubMed] [Google Scholar]
- 169.Ge X, Yang L, Sheets JP, Yu Z, Li Y. Biological conversion of methane to liquid fuels: status and opportunities. Biotechnol Adv. 2014;32:1460–1475. doi: 10.1016/j.biotechadv.2014.09.004. [DOI] [PubMed] [Google Scholar]
- 170.Jiang H, Chen Y, Jiang P, Zhang C, Smith TJ, et al. Methanotrophs: multifunctional bacteria with promising applications in environmental bioengineering. Biochem Eng J. 2010;49:277–288. doi: 10.1016/j.bej.2010.01.003. [DOI] [Google Scholar]
- 171.Stanley SH, Dalton H. The biotransformation of propylene to propylene oxide by Methylococcus capsulatus (Bath): 1. Biocatalysis. 1992;6:163–175. doi: 10.3109/10242429209014893. [DOI] [Google Scholar]
- 172.Stanley SH, Richards Anthony O'L, Suzuki M, Dalton H. The biotransformation of propylene to propylene oxide by Methylococcus capsulatus (Bath): 2. A study of the biocatalyst stability. Biocatalysis. 1992;6:177–190. doi: 10.3109/10242429209014894. [DOI] [Google Scholar]
- 173.Nielsen AK, Gerdes K, Murrell JC. Copper-dependent reciprocal transcriptional regulation of methane monooxygenase genes in Methylococcus capsulatus and Methylosinus trichosporium . Mol Microbiol. 1997;25:399–409. doi: 10.1046/j.1365-2958.1997.4801846.x. [DOI] [PubMed] [Google Scholar]
- 174.Söhngen N. Über bakterien, welche methan ALS kohlenstoffnahrung und energiequelle gebrauchen. Zentrabl Bakteriol Parasitenk Infektionskr. 1906;15:513–517. [Google Scholar]
- 175.Berry D, Mader E, Lee TK, Woebken D, Wang Y, et al. Tracking heavy water (D2O) incorporation for identifying and sorting active microbial cells. Proc Natl Acad Sci U S A. 2015;112:E194–E203. doi: 10.1073/pnas.1420406112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Huang WE, Stoecker K, Griffiths R, Newbold L, Daims H, et al. Raman-FISH: combining stable-isotope Raman spectroscopy and fluorescence in situ hybridization for the single cell analysis of identity and function. Environ Microbiol. 2007;9:1878–1889. doi: 10.1111/j.1462-2920.2007.01352.x. [DOI] [PubMed] [Google Scholar]
- 177.Dumont MG, Hernández García M, editors. Stable Isotope Probing; Methods and Protocols. Totowa, NJ US: Humana Press; 2019. [Google Scholar]
- 178.Vartoukian SR, Palmer RM, Wade WG. Strategies for culture of ‘unculturable’ bacteria. FEMS Microbiol Lett. 2010;309:1–7. doi: 10.1111/j.1574-6968.2010.02000.x. [DOI] [PubMed] [Google Scholar]
- 179.Gao L, Mastalerz M, Schimmelmann A, Rodvelt G, et al. The origin of coalbed methane. In: Thakur P, Schatzel SJ, Aminian K, Mosser MH, editors. Coal Bed Methane. 2nd ed. Amsterdam: Elsevier; 2020. pp. 3–34. [Google Scholar]
- 180.Meyer J, Haubold R, Heyer J, Böckel W. Contribution to the taxonomy of methanotrophic bacteria: correlation between membrane type and GC-value. J Basic Microbiol. 1986;26:155–160. doi: 10.1002/jobm.3620260305. [DOI] [Google Scholar]
- 181.Dedysh SN, Belova SE, Bodelier PLE, Smirnova KV, Khmelenina VN, et al. Methylocystis heyeri sp. nov., a novel type II methanotrophic bacterium possessing ‘signature’ fatty acids of type I methanotrophs. Int J Syst Evol Microbiol. 2007;57:472–479. doi: 10.1099/ijs.0.64623-0. [DOI] [PubMed] [Google Scholar]
- 182.Lindner AS, Pacheco A, Aldrich HC, Costello Staniec A, Uz I, et al. Methylocystis hirsuta sp. nov., a novel methanotroph isolated from a groundwater aquifer. Int J Syst Evol Microbiol. 2007;57:1891–1900. doi: 10.1099/ijs.0.64541-0. [DOI] [PubMed] [Google Scholar]
- 183.Kumar S, Stecher G, Tamura K. mega7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Gertz EM, Yu Y-K, Agarwala R, Schäffer AA, Altschul SF. Composition-based statistics and translated nucleotide searches: improving the TBLASTN module of blast . BMC Biol. 2006;4:41. doi: 10.1186/1741-7007-4-41. [DOI] [PMC free article] [PubMed] [Google Scholar]