Abstract
Objective
To advance use of real-world data (RWD) for pharmacovigilance, we sought to integrate a high-sensitivity natural language processing (NLP) pipeline for detecting potential adverse drug events (ADEs) with easily interpretable output for high-efficiency human review and adjudication of true ADEs.
Materials and methods
The adverse drug event presentation and tracking (ADEPT) system employs an open source NLP pipeline to identify in clinical notes mentions of medications and signs and symptoms potentially indicative of ADEs. ADEPT presents the output to human reviewers by highlighting these drug-event pairs within the context of the clinical note. To measure incidence of seizures associated with sildenafil, we applied ADEPT to 149 029 notes for 982 patients with pediatric pulmonary hypertension.
Results
Of 416 patients identified as taking sildenafil, NLP found 72 [17%, 95% confidence interval (CI) 14–21] with seizures as a potential ADE. Upon human review and adjudication, only 4 (0.96%, 95% CI 0.37–2.4) patients with seizures were determined to have true ADEs. Reviewers using ADEPT required a median of 89 s (interquartile range 57–142 s) per patient to review potential ADEs.
Discussion
ADEPT combines high throughput NLP to increase sensitivity of ADE detection and human review, to increase specificity by differentiating true ADEs from signs and symptoms related to comorbidities, effects of other medications, or other confounders.
Conclusion
ADEPT is a promising tool for creating gold standard, patient-level labels for advancing NLP-based pharmacovigilance. ADEPT is a potentially time savings platform for computer-assisted pharmacovigilance based on RWD.
Keywords: adverse drug event, natural language processing, software design, hypertension, pulmonary
INTRODUCTION
The 21st Century Cures Act calls for using real-world data (RWD) in the drug approval process, including in the identification of adverse drug events (ADEs).1 Generally, RWD refers to any data generated outside of clinical trials, but most often is used to designate data produced in the course of routine delivery of healthcare.2 Because patients exposed to drugs during routine care often differ from those who are not prescribed medications, causal inference—attributing a particular adverse event to a particular drug—is more challenging than it is in well-designed clinical trials. Much of the work on pharmacovigilance using RWD has focused on disproportionality statistics in structured databases.3 However, these systems are not designed for causal assessment of drug safety signals.3 For instance, disproportionality statistics do not differentiate when an apparent adverse effect of a drug may overlap with or be a manifestation or complication of the disease for which it is being used.
The preponderance of clinical data necessary for distinguishing ADEs from signs and symptoms not caused by a drug are found in clinicians’ notes.4 Natural language processing (NLP) can be used to identify terms for medications and for signs and symptoms that may suggest ADEs.4 However, most implementations of NLP for pharmacovigilance focus on analysis of signs or symptoms that clinicians have explicitly attributed in their documentation as being adverse reactions due to medications.5 Furthermore, even with explicit documentation of potential causality, current top-performing NLP systems have relatively poor performance for the end-to-end task of identifying a relationship between drugs and adverse events.6–8 Thus, differentiating in electronic health record (EHR) data when co-occurrence of drugs and signs or symptoms are true ADEs and not effects of the underlying disease, comorbidities, or other confounding medications has to date largely remained a manual task.
We developed the adverse drug event presentation and tracking (ADEPT) toolkit to assess causality of putative ADEs in EHR clinical notes. ADEPT serves as a complement to active ADE surveillance systems and ADE hypotheses generated from structured databases by partially automating review and annotation of potential ADEs identified in the unstructured clinical narrative. Manual review of notes to find relevant information is a time-consuming and laborious task, which has been made more difficult with the prevalence of copied text in EHR notes.9 ADEPT uses NLP to screen patients’ clinical notes for mentions of medications and signs or symptoms of interest,4 presents the identified terms graphically on a timeline that reviewers use to navigate relevant notes in a patient’s EHR, and allows users to review whether each patient experienced each potential ADE of interest. Thus, ADEPT is a tool for computer-assisted pharmacovigilance. Unlike most NLP annotation tasks,10–12 ADEPT was not designed for instance- or document-level annotation of entities. Instead, ADEPT was optimized for patient-level, across-document assessment of whether an identified sign or symptom was likely to be a true ADE caused directly by a medication.
In this context, temporality of events plays a crucial role. Four of ten criteria in the Naranjo Scale, an instrument for ADE evaluation, consider change over time.13 An additional criterion—whether alternate causes may explain the sign or symptom—requires knowing the patient’s clinical status and comorbidities at the time the sign or symptom occurred. Recent work in RWD-based pharmacovigilance has focused on incorporating temporality of drug and sign/symptom patterns into signal detection.14–16 However, we have identified only one such project that has incorporated visualization of temporality in ADE signal detection, and that work was based on structured data only.14 By highlighting the sequence of multiple events across a patient’s clinical timeline, ADEPT addresses the problem of determining causality from free-form, narrative clinical text. To test ADEPT, we use it to estimate the proportion of patients with incident seizures, a known but rare ADE associated with use of sildenafil to treat pediatric pulmonary hypertension (PH).
MATERIALS AND METHODS
Subjects and data processing
We previously identified a cohort of patients with PH using a computable phenotype.17 The study was approved by the institutional review board at Boston Children’s Hospital with waiver of informed consent for review of EHR data.
ADEPT used the Apache clinical Text Analysis Knowledge Extraction System (cTAKES)18 to identify mentions of medications and signs or symptoms of interest in plain-text clinical notes extracted from the local data warehouse. Because of ADEPT’s modular design, the output of any clinical NLP system capable of identifying clinical events of interest within individual notes could be substituted as input to ADEPT. We previously described the cTAKES pipeline used for extracting these potential ADEs of interest from EHR notes.4 cTAKES identified textual mentions with mappings to concept unique identifiers (CUIs) representing medications and signs or symptoms and assigned relevant attributes—negation, conditional status (eg, “if you experience headache”), and temporality—to each mention using open-source modules whose methods are described in detail elsewhere.18–20 We included only mentions that were not negated or conditional and whose temporality did not place them prior to the document creation time. We excluded certain terms that generated false-positive associations by subsuming them into a more specific concept (eg, specifying that “heparin flush” was not an instance of “flushing” as a sign or symptom), which we implemented using a custom cTAKES dictionary. As a postprocessing, rules-based step, we linked into potential relations medication and sign/symptom mentions when mentions occurred within 25 newline characters of one another. This proximity parameter was determined empirically during pipeline development to maximize F1 score.4 We excluded putative relations between medications and signs and symptoms that were temporally implausible (ie, if the sign or symptom overlapped the document creation time, but the medication term had a temporality after the document creation time, that could not be a temporally plausible relationship for an ADE). The pipeline was tuned to maximize recall. Evaluated at the document level against gold standard human annotations (interannotator agreement 0.88) for extracting temporally compatible medication and relevant sign/symptom pairs, the final pipeline had sensitivity 90%, positive predictive value 69%, and F1 score 0.78.4
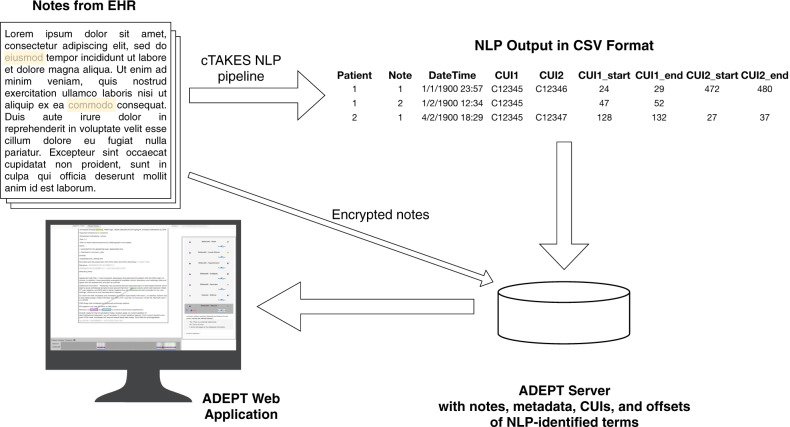
The output of cTAKES was a comma-separated file containing identifiers for each patient and note, a timestamp for the note’s creation, the CUIs identified in that note and any potential relationships among them, and the specific character spans in each note of the text relating to each CUI (Figure 1). Using the high throughput cTAKES NLP pipeline,4 ADEPT processed 149 029 notes for the 982 patients with pediatric PH. We then developed ADEPT to increase precision for the integrated task of clinical pharmacovigilance using the NLP data.
Figure 1:
Schematic of data flow in ADEPT. Clinical notes extracted from the EHR are processed using a NLP pipeline implemented in cTAKES. cTAKES identifies terms for drugs and signs/symptoms as well as relations between them representing potential ADEs. The output of cTAKES is a comma-separated values (CSV) file that includes patient and note identifiers, a timestamp of document creation time, and character spans for each CUI of interest identified. These data are uploaded into the ADEPT database alongside the encrypted raw text notes. Finally, ADEPT presents each patient’s cumulative, temporal narrative data in an easily navigable format.
ADEPT technical description
Overview and front end
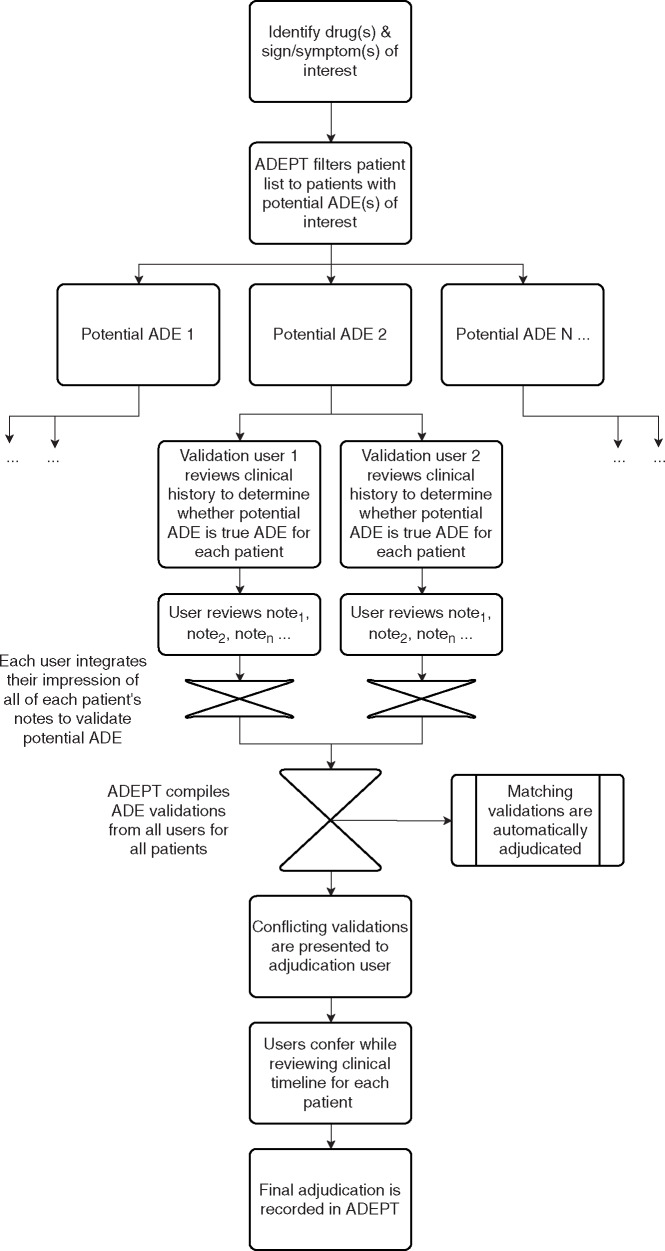
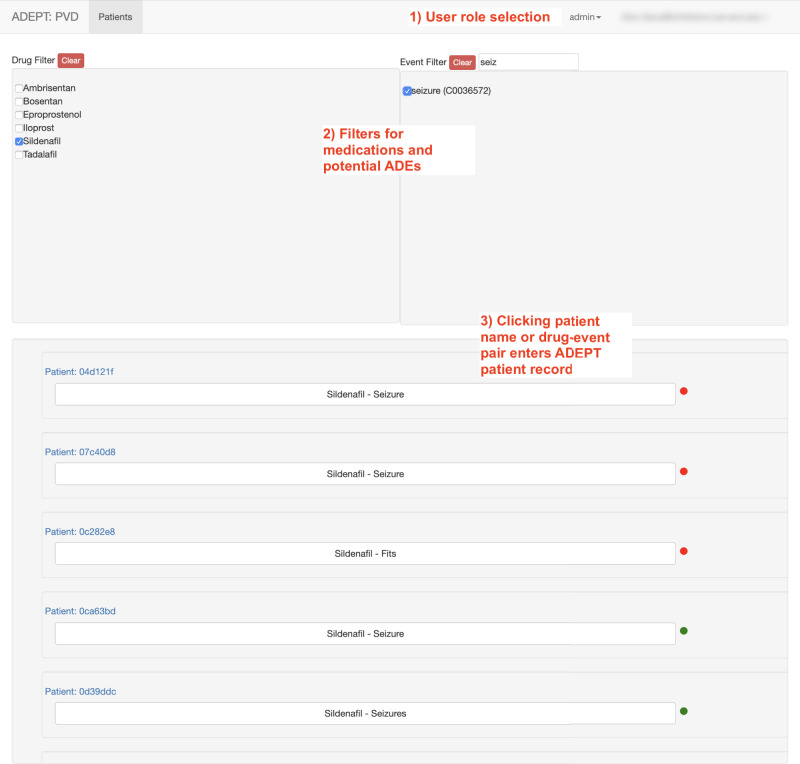
ADEPT is a Java- and JavaScript-based web application. Figure 2 presents an overview of users’ workflow within ADEPT. Users are first presented with a listing of all patients in the system and a set of filters to limit the list based on drugs or signs/symptoms representing potential ADEs (Figure 3). Applying filters limits the patient list to patients with mentions of the drugs and/or signs and symptoms identified by the NLP pipeline. Underneath each patient identifier are the (filtered) medications and potential ADEs identified for that particular patient.
Figure 2:
User workflow within ADEPT. The same workflow is repeated for each potential ADE.
Figure 3:
The patient list screen in ADEPT showing the functionality of filters to select patients with seizures as a potential ADE for sildenafil. Note that clicking on a patient number will select all potential ADEs for that patient for review, whereas clicking in the box “Sildenafil - Seizure” will select only sildenafil and seizure as the potential ADE for review.
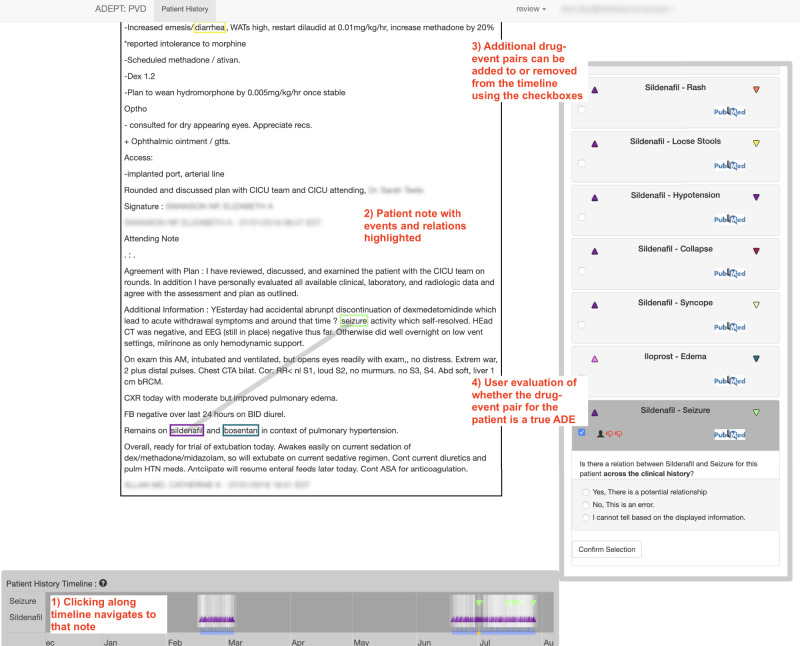
The patient-level ADE evaluation screen in ADEPT contains a large area for displaying note text, a patient history timeline with indicators for medications and signs and symptoms, and an ADE selection and annotation pane (Figure 4). The patient history timeline is used to navigate among a patient’s notes. Each note in the history is shown as a translucent-white vertical bar. Notes containing any mention of a drug or sign or symptom involved in one of the selected potential ADEs contain a color-coded triangle, where drug mentions are represented using an upward-pointed triangle and signs or symptoms are represented using a downward pointed triangle. Translucency of the colored triangles and the bars indicating notes provide an indication of note and mention density, cueing users to zoom in on times of heavy documentation. Drug-candidate ADE pairings can be selected and deselected at any time (Figure 4). Notes in which the NLP pipeline identified a potential relationship between the drug and sign or symptom are highlighted in a translucent orange color.
Figure 4:
The patient-level annotation screen in ADEPT. The selected note text is in the top-left, showing several potential terms of interest highlighted with color-coded rectangles. Note the thick line connecting “seizure” and “sildenafil,” indicating a potential plausible connection between the two terms. At the bottom-left is the Patient History Timeline navigation pane, with triangles indicating notes that contain “sildenafil” and/or “seizures.” At right is the review/adjudication pane (shown in review mode), where annotators indicate whether the potential ADE represents a true ADE.
When a user clicks on a note to navigate to it, the note displays in a large, scrollable pane in ADEPT. The visualization highlights mentions of any drugs or signs and symptoms using color-coded rectangles (Figure 4). The visualization also highlights mentions involved in potentially causal relationships identified by the NLP pipeline using a thick light-gray line. This color-coded visualization directs user attention to potential ADE descriptions within the clinical note.
The visualization of NLP output is implemented by a transparent JavaScript Canvas element overlaid on top of the note text. Using the character offsets from the cTAKES output for each note, each mention of a drug or sign or symptom identified by cTAKES is wrapped by a Hypertext Markup Language (HTML) span element. An HTML class attribute is added to the span class identifying the Unified Medical Language System (UMLS) CUI of the drug, sign, or symptom. The UMLS CUI classes are used to add the color-coded borders to the span element highlighting the mentions of interest. In addition, a Group ID is added to the HTML class attribute. The Group ID classes are used to identify mentions of drugs and signs or symptoms among which the light-gray connection bars indicating potential causal ADEs are drawn.
Once a user has completed review of the patient’s notes, the reviewer uses the right-hand pane to record whether each drug-sign/symptom pair is consistent with a true ADE for that patient (Figure 4). ADEPT supports both “review” and “adjudication” roles for users. In the review role, a user sees only his/her own annotations of whether drug-sign/symptom pairs are ADEs for each patient. In the adjudication role, a user can identify conflicting potential ADE evaluations and, after review of the evidence, make a final adjudication of whether the drug-sign/symptom pair for the patient is a true ADE.
Data structure and back end
To configure ADEPT for a particular task, the clinical note content and metadata (patient identifier, date/time of note creation, and term spans and relationships) are loaded into a Structured Query Language (SQL) database (Figure 1). The locations for NLP-identified terms are expressed in the cTAKES output as character offsets from the start of the text. Each of the NLP-identified mentions of a term are stored in the database in the CTakesHit table. Related groupings of CTakesHit entries are tied together by a many-to-one relationship with the CTakesGroup table.
Each candidate ADE identified by the NLP pipeline is stored in the Candidate table and tied to a CTakesGroup record through the candidateId column. User validations of candidate ADEs are captured in the CandidateValidation table. Candidate records which have been adjudicated are marked by the isAdjudicated column and have the adjudicator’s userId and adjudicated score recorded directly in the Candidate table.
Additional measures are taken to ensure security of the note text data, since clinical notes frequently contain personally identifiable, protected health information and other sensitive/confidential patient information. ADEPT uses Transport Layer Security (TLS)-based standards to encrypt data in transit. For encryption at rest, in addition to volume-level encryption, ADEPT encrypts the text of each note using the Java Cryptography Extension implementation of the Advanced Encryption Standard/Galois/Counter Mode/NoPadding encryption algorithm. ADEPT also utilizes typical secure coding practices, such as using stored procedures for database access, to prevent SQL injection attacks.
The only flow of execution that yields a plain-text note through ADEPT is a specific service call, which is secured via a role-based access control model in addition to user-specific rate-limiting. This provides a required human intervention to validate that the user’s token has not fallen into the wrong hands. Every action pertaining to user management, retrieval of patient data, and annotation of candidate ADEs is logged to the application log. This log is then forwarded to a log aggregation and analysis server instance. For the current project, we used Splunk Enterprise (Splunk, Inc., San Francisco, CA, USA) for tracking ADEPT interactions.
Evaluation
To test application of ADEPT to a clinical problem, we focused on sildenafil, the most commonly prescribed PH-targeted therapy in children.21 As our potential ADE of interest, we chose seizures, since preliminary work suggested that seizures were a rare ADE in patients taking sildenafil22 but a commonly recorded event in our patients’ clinical notes.4 Two clinicians (A.G. and S.F.M.) independently used ADEPT to review clinical notes of all patients identified as potentially experiencing seizures while on sildenafil.
To review potential ADEs, we initially intended to use the Naranjo scale.13 However, we noted several limitations of that assessment tool, particularly to critically ill patients,23 which describes many of the patients in the current study. Thus, we instead agreed on consensus annotation criteria that accounted for temporal plausibility of the ADE, alternative causes for the ADE, and objective evidence of the ADE, but did not assign specific scores to those components. Other components of the Naranjo scale, including readministration of the drug, placebo administration, testing for drug levels, and a dose–response relationship were rarely assessed in our patient population. For each patient, each reviewer responded to the question, “Is there a relation between sildenafil and seizure for this patient across the clinical history?” and responses (“Yes,” “No,” or “Cannot tell”) were recorded in ADEPT. Interannotator agreement for the ADE determination was calculated using Cohen’s weighted κ. Discrepant responses were resolved by discussion and consensus between the two reviewers and recorded as an adjudicated result.
Prevalence of the ADE was calculated as the percent of patients exposed to sildenafil who experienced seizures, with 95% confidence intervals calculated using the binomial approximation. The number of notes reviewed, time spent on each note, and total time spent from accessing a patient’s history to recording a response to whether the seizure was consistent with a true ADE were determined from the server log data. Times greater than 10 min [N = 3 (1.8%) of 169 actions] were considered potentially spurious and were eliminated from further analysis. Differences in number of notes and time reviewing notes were compared using the Mann–Whitney U test. A P-value < 0.05 was considered statistically significant. Data analysis was performed using R version 3.5.0 (R Foundation for Statistical Computing, Vienna, Austria).
RESULTS
The NLP pipeline identified seizures in 72 [17%, 95% confidence interval (CI) 14–21] of 416 patients taking sildenafil [age (mean ± SD) 20 ± 25 years, 46% male]. Most documents (77%) with mentions of sildenafil were written between 2012 and 2015.
While interannotator agreement was initially poor (65%, κ = 0.2), most (16 of 25, 64%) of the initially conflicting reviews were due to one of the evaluators marking that he or she could not tell whether the seizure was a true ADE and the other evaluator indicating “yes” or “no.” After adjudication, only 4 (0.96%, 95% CI 0.37–2.4) of the patients’ seizures were found to be consistent with true ADEs. In an additional 13 patients (3.1%, 95% CI 1.8–5.3), users were not able to determine whether the seizure was consistent with a true ADE. As compared to reaching consensus for discrepant annotations, single-reviewer determination of whether a sign/symptom represented an ADE required review of fewer notes and took less time in total and per note (Table 1).
Table 1. Usage statistics for ADEPT interactions
| Review | Adjudication | P-value | |
|---|---|---|---|
| Notes (N) | 8 (6–10) | 11 (9–19) | <0.001 |
| Unique notes (N) | 7 (5–9) | 10 (8–18) | <0.001 |
| Total time to result (s) | 89 (57–142) | 224 (141–317) | <0.001 |
| Time per note (s) | 9.6 (0.21–21) | 13 (3.8–24) | 0.003 |
Data shown as median (interquartile range).
DISCUSSION
ADEPT facilitates ascertainment of gold standard, human-reviewed ADEs. Our approach takes advantage the scalability of natural language processing and machine learning for high throughput to maximize sensitivity and guide reviewers, while presenting the computed data in a format that minimizes cognitive load and effort on the part of human annotators. In this sense, ADEPT is similar to systems that use preannotation to aid in annotating clinical text for other NLP tasks.10,12 ADEPT, however, differs from other NLP annotation tools, in that it facilitates clinical pharmacovigilance by providing an interface to review whether a sign or symptom represents an ADE across a patient’s entire clinical corpus, rather than in a single instance or document. This functionality facilitates developing gold standard ADE labels for ADE causality determination, in which expert clinicians review the totality of the clinical record to determine whether the incidence of a sign or symptom is caused by a medication, but does so in a way that is notably more efficient than traditional, manual review of clinical notes. We demonstrate, with a driving use case, that many potential ADEs that might be identified through a chart search actually represent signs and symptoms that cannot be attributed definitively to the patient’s being exposed to a medication, even when searching specifically for known ADEs. Such review would be laborious without the efficiency gains of ADEPT.
Given the importance of temporality in determining whether a drug-sign/symptom pair represents an ADE (exposure to the medication must precede the sign or symptom), ADEPT’s utility was heavily influenced by its ability to discriminate when a patient was exposed to a medication. Studies have shown that as many as 82% of notes may contain as much as 20% copied material from previous notes.9 Thus, although our NLP pipeline filtered out temporally impossible relationships based on temporality of terms relative to the document creation time,19 within-note temporality was not sufficient to determine the overall clinical course of the patient. In contrast, the visual representation of medication and sign or symptom mentions in ADEPT allowed annotators to find quickly first and final mentions of these terms in a patient’s clinical history, determine whether these terms truly represent initiation of a medication or only first mention within our EHR, and determine when medications were discontinued in order to assess whether signs/symptoms occurred prior to initiation or after cessation of the drug.
Determining whether a drug-sign/symptom pair is consistent with a true ADE remains a difficult task requiring some subjective assessment. Our ultimate interannotator agreement was poor (κ = 0.2), reflecting how challenging the task of determining causality in a complex, dense clinical record can be, but was similar to that reported in several other studies of ADEs that used varied scales and assessment tools.24–26 Moreover, disagreement largely stemmed from the challenge of reviewers determining definitively whether there was sufficient information in the clinical notes to rule in or out each potential ADE. Agreement was better when both reviewers found sufficient information to determine whether or not the seizure was consistent with a true ADE, although the causal relation between sildenafil and seizures was rarely explicitly recorded by the clinician. ADEPT addresses the challenge of interpretability in ADE ascertainment by highlighting the specific sentences in which drugs and signs and symptoms are mentioned and displaying that information within the full context of the patient’s clinical narrative. This contextual representation allows consideration of confounding from other medications and conditions during manual annotation of potential ADEs.
ADEPT increased efficiency of this pharmacovigilance case study by reducing the time needed to review potential ADEs. Two recent studies report times for case completion of 15–23 min,27,28 whereas using ADEPT chart review typically took annotators less than 4 min per patient to complete. This efficiency was gained despite the density of inpatient notes for a relatively ill patient population. By providing an interface that combined navigation across notes with visual indicators of the most relevant notes, annotators were able to rapidly peruse a patient’s clinical narrative.
We found that few of the potential ADEs identified by our screening NLP pipeline were adjudicated to be consistent with true ADEs. This was expected given that our NLP pipeline was tuned to maximize sensitivity, at the expense of positive predictive value, as an initial screen for ADE-related terms. In the case of seizures, the rate of confirmed ADEs in this study was similar to the rate reported in the STARTS-2 trial of sildenafil for pediatric PH.22 Of note, seizures were not a reported ADE in two other small studies of sildenafil.29,30 Using scalable, computer-assisted methods, we were able to scan the retrospective records of 416 patients taking sildenafil, examining the rate of seizures in over twice as many patients taking sildenafil compared to prior studies. These findings highlight the importance of RWD in pharmacovigilance, particularly for drugs used in children, for whom limited randomized clinical trial and postmarketing follow-up data exist.31
While many studies have used NLP for ADE detection, state-of-the-art NLP pipelines continue to have limited performance for the full, integrated task of ADE identification and depend on clinicians’ explicit assertion that a sign/symptom is related to a medication.5–8 In contrast, ADEPT is built around the idea that the connections between medications and signs/symptoms that may represent ADEs are often represented vaguely and inconsistently in the clinical narrative. In medically complex patients, in particular, many potential ADEs may represent effects of underlying or comorbid diseases or adverse effects of other treatments for those diseases. ADEPT leverages the scalability of NLP and machine learning to screen notes for potential informativeness regarding ADEs, while facilitating human review of those potential ADE mentions. This two-step process allows for the underlying NLP pipeline to be tuned to greater sensitivity—capturing implicit rather than only explicit relations—while the human review and validation allows for maintaining adequate specificity for pharmacovigilance applications. Thus, even using an NLP pipeline with relatively low positive predictive value, ADEPT facilitated efficient human screening of potential ADEs and, after adjudication to resolve disagreements between two reviewers, allowed for rapid creation of gold standard ADE labels.
This study has several limitations. Though we found that ADEPT was used effectively by one user (S.F.M.) without prior experience with the platform, further usability testing with more clinicians, epidemiologists, and researchers is needed. We were not able to directly compare ADEPT to manual chart review of potential ADEs, in which an annotators would have note-level NLP results but not the visual/navigation functions of ADEPT. However, without ADEPT, which highlighted which notes over a patient’s clinical timeline had mentions of medications alone, signs/symptoms alone, and medications and signs/symptoms in the same note with a putative relationship, annotators would have had to review an average of over 200 notes per patient with mention of either a drug or sign/symptom of interest to determine causality. In future studies, we will compare ADEPT and manual chart review to confirm the hypothesized performance gains. ADEPT is currently designed for research purposes, and further refinements to the user interface for general clinical use will be required. The NLP pipeline required significant tuning and may not perform as well at other institutions or with different patient populations. Nonetheless, the methods underlying ADEPT can be readily adapted to more robustly tuned input data. Future planned application enhancements include the ability to modify, tune, and run the NLP pipeline from within ADEPT in order to allow end-users to develop custom NLP-assisted pharmacovigilance pipelines. Finally, although the performance of the NLP pipeline was evaluated on a hold-out test set of patients and showed high sensitivity, the potential of false-negative patient-level annotations using the ADEPT system could not be evaluated since we did not have a gold standard against which to compare annotations. A strength of ADEPT is that it allowed annotators to review discrepant annotations together and record an adjudicated result.
CONCLUSION
ADEPT, a platform for NLP-based computer-assisted human annotation of potential ADEs, effectively identifies and validates rare events consistent with true ADEs in unstructured, real-world data. Future studies in NLP-based pharmacovigilance should emphasize causal inference between medications and signs or symptoms even when the signs or symptoms are not explicitly labeled as an ADE in the clinical text. Such studies should also address the relationships among signs or symptoms and medications across a patient’s clinical documents in addition to within individual documents. Systems such as ADEPT that address temporality and confounding by facilitating rapid navigation of a patient’s clinical timeline can be used to create gold standard labels and accelerate pharmacovigilance applications based on RWD. In future work, we plan to test ADEPT’s ability to facilitate validation of novel potential ADEs. The ADEPT source code is available at https://github.com/hms-dbmi/adept.
Contributors
K.D.M. obtained funding. A.G., J.P.S., and K.D.M. wrote the manuscript. A.G. and K.D.M. designed the research. A.G., J.P.S., S.F.M., G.S., and C.L. performed the research. A.G., S.F.M., P.A., and K.D.M. analyzed the data. J.P.S., C.L., G.S., and P.A. contributed new analytical tools. S.F.M., C.L., G.K.S., and P.A. critically revised the manuscript for important intellectual content. All the authors take responsibility for the final approval of the version to be published and are accountable for all aspects of the work.
FUNDING
This work was supported by the National Institutes of Health (NHLBI U01HL121518, NHLBI L40HL133929, NICHD T32HD040128, NICHD K12HD047349, NLM R01LM010090, and NCATS U01TR002623). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Conflict of interest statement. None declared.
REFERENCES
- 1. Pitts PJ, Le Louet H, Moride Y, et al. 21st century pharmacovigilance: efforts, roles, and responsibilities. Lancet Oncol 2016; 17 (11): e486–e92. [DOI] [PubMed] [Google Scholar]
- 2. Khozin S Blumenthal GM Pazdur R. Real-world data for clinical evidence generation in oncology. J Natl Cancer Inst 2017; 109 (11): djx187. [DOI] [PubMed] [Google Scholar]
- 3. Beninger P, Ibara MA. Pharmacovigilance and biomedical informatics: a model for future development. Clin Ther 2016; 38: 1–12. [DOI] [PubMed] [Google Scholar]
- 4. Geva A, Abman SH, Manzi SF, et al. Adverse drug event rates in pediatric pulmonary hypertension: a comparison of real-world data sources. J Am Med Inform Assoc 2020; 27 (2): 294–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chapman AB, Peterson KS, Alba PR, et al. Detecting adverse drug events with rapidly trained classification models. Drug Saf 2019; 42 (1): 147–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Jagannatha A, Liu F, Liu W, et al. Overview of the first natural language processing challenge for extracting medication, indication, and adverse drug events from electronic health record notes (MADE 1.0). Drug Saf 2019; 42 (1): 99–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Dandala B, Joopudi V, Devarakonda M. Adverse drug events detection in clinical notes by jointly modeling entities and relations using neural networks. Drug Saf 2019; 42 (1): 135–46. [DOI] [PubMed] [Google Scholar]
- 8. Xu D, Yadav V, Bethard S. UArizona at the MADE1.0 NLP Challenge, , . Proc Mach Learn Res 2018; 90: 57–65. [PMC free article] [PubMed] [Google Scholar]
- 9. Thornton JD, Schold JD, Venkateshaiah L, et al. Prevalence of copied information by attendings and residents in critical care progress notes. Crit Care Med 2013; 41: 382–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gobbel GT, Garvin J, Reeves R, et al. Assisted annotation of medical free text using RapTAT. J Am Med Inform Assoc 2014; 21 (5): 833–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Roberts A, Gaizauskas R, Hepple M, et al. Building a semantically annotated corpus of clinical texts. J Biomed Inform 2009; 42 (5): 950–66. [DOI] [PubMed] [Google Scholar]
- 12. Lingren T, Deleger L, Molnar K, et al. Evaluating the impact of pre-annotation on annotation speed and potential bias: natural language processing gold standard development for clinical named entity recognition in clinical trial announcements. J Am Med Inform Assoc 2014; 21 (3): 406–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Naranjo CA, Busto U, Sellers EM, et al. A method for estimating the probability of adverse drug reactions. Clin Pharmacol Ther 1981; 30 (2): 239–45. [DOI] [PubMed] [Google Scholar]
- 14. Star K, Watson S, Sandberg L, et al. Longitudinal medical records as a complement to routine drug safety signal analysis. Pharmacoepidemiol Drug Saf 2015; 24 (5): 486–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Pacurariu AC, Straus SM, Trifiro G, et al. Useful interplay between spontaneous ADR reports and electronic healthcare records in signal detection. Drug Saf 2015; 38 (12): 1201–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Harpaz R, Vilar S, Dumouchel W, et al. Combing signals from spontaneous reports and electronic health records for detection of adverse drug reactions. J Am Med Inform Assoc 2013; 20 (3): 413–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Geva A, Gronsbell JL, Cai T, et al. A computable phenotype improves cohort ascertainment in a pediatric pulmonary hypertension registry. J Pediatr 2017; 188: 224–31.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Savova GK, Masanz JJ, Ogren PV, et al. Mayo clinical Text Analysis and Knowledge Extraction System (cTAKES): architecture, component evaluation and applications. J Am Med Inform Assoc 2010; 17 (5): 507–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lin C, Dligach D, Miller TA, et al. Multilayered temporal modeling for the clinical domain. J Am Med Inform Assoc 2016; 23 (2): 387–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wu S, Miller T, Masanz J, et al. Negation's not solved: generalizability versus optimizability in clinical natural language processing. PLoS One 2014; 9 (11): e112774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Barst RJ, McGoon MD, Elliott CG, et al. Survival in childhood pulmonary arterial hypertension: insights from the registry to evaluate early and long-term pulmonary arterial hypertension disease management. Circulation 2012; 125 (1): 113–22. [DOI] [PubMed] [Google Scholar]
- 22. Barst RJ, Beghetti M, Pulido T, et al. STARTS-2: long-term survival with oral sildenafil monotherapy in treatment-naive pediatric pulmonary arterial hypertension. Circulation 2014; 129 (19): 1914–23. [DOI] [PubMed] [Google Scholar]
- 23. Seger D, Barker K, McNaughton C. Misuse of the Naranjo adverse drug reaction probability scale in toxicology. Clin Toxicol (Phila) 2013; 51 (6): 461–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Goldman JL, Chung WH, Lee BR, et al. Adverse drug reaction causality assessment tools for drug-induced Stevens-Johnson syndrome and toxic epidermal necrolysis: room for improvement. Eur J Clin Pharmacol 2019; 75 (8): 1135–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Beniwal R, Gupta LK, Khare AK, et al. Clinical profile and comparison of causality assessment tools in cutaneous adverse drug reactions. Indian Dermatol Online J 2019; 10 (1): 27–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Behera SK, Das S, Xavier AS, et al. Comparison of different methods for causality assessment of adverse drug reactions. Int J Clin Pharm 2018; 40 (4): 903–10. [DOI] [PubMed] [Google Scholar]
- 27. Ledieu T, Bouzille G, Thiessard F, et al. Timeline representation of clinical data: usability and added value for pharmacovigilance. BMC Med Inform Decis Mak 2018; 18 (1): 86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Thevelin S, Spinewine A, Beuscart JB, et al. Development of a standardized chart review method to identify drug-related hospital admissions in older people. Br J Clin Pharmacol 2018; 84 (11): 2600–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Takatsuki S, Calderbank M, Ivy DD. Initial experience with tadalafil in pediatric pulmonary arterial hypertension. Pediatr Cardiol 2012; 33 (5): 683–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Siehr SL, McCarthy EK, Ogawa MT, et al. Reported sildenafil side effects in pediatric pulmonary hypertension patients. Front Pediatr 2015; 3: 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Bourgeois FT, Hwang TJ. The pediatric research equity act moves into adolescence. JAMA 2017; 317 (3): 259–60. [DOI] [PubMed] [Google Scholar]