Abstract
The premise of Open Science is that research and medical management will progress faster if data and knowledge are openly shared. The value of Open Science is nowhere more important and appreciated than in the rare disease (RD) community. Research into RDs has been limited by insufficient patient data and resources, a paucity of trained disease experts, and lack of therapeutics, leading to long delays in diagnosis and treatment. These issues can be ameliorated by following the principles and practices of sharing that are intrinsic to Open Science. Here, we describe how the RD community has adopted the core pillars of Open Science, adding new initiatives to promote care and research for RD patients and, ultimately, for all of medicine. We also present recommendations that can advance Open Science more globally.
Keywords: open science, ontology, FAIR data, common data elements, rare disease patients, data standards
INTRODUCTION
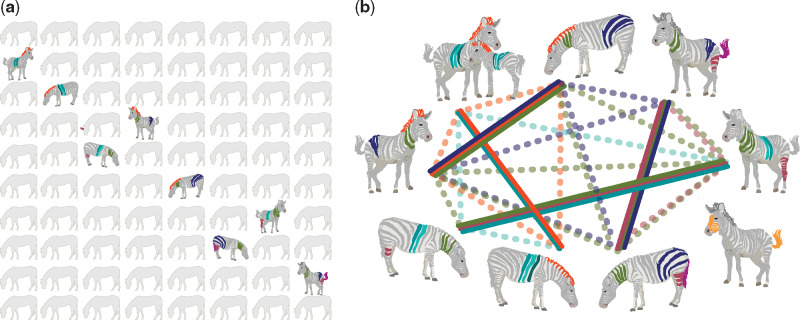
In the United States, a rare disease (RD) is defined as one that affects fewer than 200,000 persons; for Japan, it is fewer than 50,000; and for South Korea, fewer than 20,000. In contrast, Europe and Australia define rare as 1 in 2000 individuals.1,2 Taken together, RDs represent a public health problem; ∼10% of people eventually present with an RD.2–4 Roughly 5000–8000 RDs have been described, but the number of RDs is estimated to exceed 10,000.5 Most RDs are severe and chronic and some are life-threatening. RDs, which are often inherited, frequently present in childhood and can have deleterious long-term effects. Patients with RDs often face diagnostic delays; it can take 7 years or more to reach an accurate diagnosis.6,7 Delayed or inaccurate diagnoses hinder the development of effective treatment plans, preclude prognoses and genetic counseling, create skepticism among relatives, colleagues, and physicians, and exclude patients from a community of individuals with similar experiences. Appropriate information and medical expertise on RDs are often insufficient, and access to care is difficult. Because many RDs affect multiple organ systems, care can be fragmented across several specialties. Electronic health records (EHRs) are not well suited for recording and sharing information about RDs; it remains difficult to stratify patients into useful classifications and to identify individuals with specific RDs8,9 (Figure 1).
Figure 1.
Rare diseases. (A) RDs are individually rare but collectively impact ∼10% of the population. Here, RDs are represented in the classic aphorism, “When you hear hoofbeats, think of horses, not zebras”—in other words, look for the most common disease that matches the symptoms, not the rarest one. It was originally used by Theodore Woodward, professor at the University of Maryland School of Medicine in the 1940s. (B) Defining RDs requires carefully matching a patient’s spectrum of phenotypes with the phenotypic profile of candidate diseases, here represented by a single color-feature. Each zebra (patient) has a constellation of phenotypes that may match none, some (dashed lines), or all (solid lines) of the phenotypes of other zebras. The diagnosis of RDs often involves recognition of phenotypic patterns and is aided by computational phenotype analysis.
Programs have been established to accelerate the diagnosis of very RDs, identify new RDs, and provide improved RD patient care. One such program was the NIH Undiagnosed Diseases Program (UDP),10–13 which expanded to the Undiagnosed Disease Network (UDN). This NIH-funded consortium includes 12 clinical sites and analytical cores around the United States14,15; both the UDN and the UDP provide multidisciplinary clinical evaluations, research collaborations, and translational validations for RD patients. The UDN uses many hundreds of open data resources that have helped inform many diagnoses, illustrating the success of Open Science for diagnosing RD patients. Similar RD diagnostic initiatives in other countries have been instantiated in Japan in 2015 (Initiative on Rare and Undiagnosed Disease [IRUD]16) in Western Australia in 2013 (Rare and Undiagnosed Diseases Diagnostic Service, RUDDS), and in other countries. The Undiagnosed Diseases Network International (UDNI), established in 2014, is dedicated to discovering new diseases and defining standards for sharing data and best practices in RD programs throughout the world.11 With the Cross-Border Healthcare Directive (2011/24/EU), the European Union established a mandatory framework to foster cooperation addressed to RDs within European Reference Networks.17 Despite these laudatory efforts to coordinate internationally, there are not enough programs worldwide to provide the care needed for the many RDs patients. In addition, RD patients often lack a supporting community that shares the same disease, despite the many support groups such as the National Organization for Rare Disorders (NORD), European Organization for Rare Diseases (EURORDIS), and Coordination of Rare Diseases at Sanford (CoRDS).
OPEN SCIENCE AND MAKING DATA FAIR
Individually, RDs are rare, and so any one physician, researcher, or institution will not accrue sufficient experience, data, or knowledge to effectively treat or research RDs. Therefore, progress in diagnosing, treating, and understanding a particular RD requires the synthesis of all available data from multiple institutions.
To facilitate this exchange of data, the field has started to embrace the principles of Open Science. The premise of Open Science is that research will progress faster if data and knowledge are openly shared with proper data safety measures and ethical frameworks.18 Open Science, an umbrella term for a wide range of activities from basic biological research to clinical research, makes it easier for scientists and clinicians to share and access knowledge, resources, tools, and data. Open Science considers scientific knowledge a product of social collaboration that belongs to the community; hence, the public should have access to it at little or no cost.
In a very real sense, Open Science means open data. To be open, the data need to be FAIR, that is, Findable, Accessible, Interoperable, and Reusable (FAIR) for humans and machines.19 These FAIR Guiding Principles,20 adopted in 2014, are followed by many organizations world-wide, including the G20, NIH, and IRDiRC (the International Rare Disease Research Consortium). Many projects, such as the European Joint Programme on Rare Diseases, are now working on the implementation of FAIR. Germany, France, and The Netherlands decided to support communities in organizing Global Open FAIR implementation networks. The RDs GO FAIR Network was established to foster implementation in the RD domain.21 Also important are factors specifically related to data reusability, such as traceability (eg, provenance and attribution), data licensing, and connectedness of the data.22–24
FAIR data stewardship is challenging, because it requires a wide range of expertise: knowledge of the domain, local IT systems, local and cloud storage systems, local and global data access policies, machine-readable formats for data and knowledge, and software for communication between FAIR resources. Making data FAIR should be considered a team effort. There is no comprehensive suite of tools for a stakeholder to make data FAIR; ELIXIR’s “service bundles” may provide that in the future, but teams of experts are needed.
The FAIR principles require data to be prepared for reuse. Moreover, for diseases with low prevalence, sparsity of data necessitates that data are prepared for analysis across multiple sources. Current lack of interoperability is an obstacle for Open Science.25 Data scientists must go through a laborious and error-prone process of finding data, assuring access and permissions, and making data compatible and optimally reusable. By experience, this post hoc data preparation may take up a substantial part of their time,26 and inevitably leads to an inability to address certain research questions. Open Science needs international collaboration, infrastructure, and good data stewardship to address the costly inefficiency caused by data that are not prepared for reuse.
Sharing data can be problematic in general, but particularly in the RD domain, because of (1) ethical and legal constraints that can differ among institutes, regions, and countries, (2) the scale of the distribution of RD data, and (3) hesitation of scientists to share data that are precious to their careers. The FAIR principles can provide an alternative approach to centralizing data, especially clinical data, from multiple sources for analysis. When data are FAIR “at source,” distributed analysis can be effectively performed, with only the result of the analysis leaving the source and the data secure and private. In principle, all source data are available, enabling analyses ranging from counting how many patients show certain conditions to distributed machine learning to predict treatment outcomes. Some computer algorithms will be too demanding for distributed analysis, but even in that case, application of the FAIR principles will prepare data for efficient analyses.
Another significant challenge is data licensing. Integrative analytical platforms aimed at facilitating RD research and mechanism and drug discovery, such as the Monarch Initiative,27 the NCATS Biomedical Data Translator,28–30 and the Gabriella Miller Kids First Data Resource Portal,31 rely on the ability to integrate and redistribute data from other third-party public knowledge sources. The more FAIR-ready these sources are, the more the integrated data may be effectively applied for RDs. However, a recent study evaluating more than 50 data sources suggested that current licensing terms may significantly impede the use, reuse, and redistribution of data. The lack of legal data redistribution is a fundamental problem for RDs, for which maximal utility must be garnered from all possible knowledge sources. Custom licenses constitute the largest single class of licenses found in these data resources, suggesting that the providers either did not know about standard licenses or believed that standard licenses did not meet their needs.22,23 The (Re)usable Data Project32 aims to help data providers evaluate the impact of their licensing terms on downstream users, and is already assisting RD data providers to improve their reusability.
Despite these challenges, the benefits of FAIR outweigh the cost of implementation. Theoretically, the additional time to make data compatible for multi-source analysis by a data analyst is zero when data are already FAIR.33 Considering that RD data sets are precious and reused often, the efficiency gain multiplies quickly. The RD community was the first community in Europe to embrace the concept of a “Bring Your Own Data” workshop (BYOD) aimed at learning how to make data interoperable. BYODs for RD registry managers have been organized by the Istituto Superiore di Sanitá since 2015,34 and are planned to continue as part of an annual summer school at least until 2023 with support from the European Joint Program on Rare Diseases (EJPRD). Inspired by the feedback from these BYODs, “RDs GO FAIR” was created to foster adoption of FAIR principles toward a critical mass of FAIR data resources.17 Through interdisciplinary collaboration fostered by RDs GO FAIR and others, and activities of ELIXIR, BBMRI (Better Biology Makes Reality Interesting), the NIH, the EJPRD, NORD, and EURORDIS, we expect gradual maturation of guidelines, supporting tools, FAIR data stewardship (including in patient organizations), and for-profit and not-for-profit service providers. A FAIR ecosystem thus brings about an Open Science environment where new analysis possibilities can be explored under well-defined and transparent conditions for sensitive data.
OPEN SCIENCE IN THE RARE DISEASE FIELD
RD patient empowerment and resources
Patients, families, and their advocates are key stakeholders that have not always been sufficiently engaged in many biomedical research initiatives.35 Engaging patients as partners in product development is important to better understand the patient perspectives and the pathogenesis of the disease. Patients and caregivers are often the best advocates for raising awareness and describing the clinical manifestations and the daily progress of the disease and treatments.36 Engagement of patients and other stakeholders (such as caregivers, advocacy organizations, and clinicians) in clinical research can help to ensure that research efforts address relevant clinical questions and patient-centered health outcomes.37 Numerous RD programs and organizations exist, including the NIH Rare Disease Clinical Research Network (RDCRN),38 the EURORDIS-Rare Diseases Europe,39 Patient-Centered Outcomes Research Institute (PCORI),40 the Genetic Alliance,41 NORD,42 and the Innovative Medicine Initiative (IMI).43
Many patients and their families look for ways to improve dissemination of their data and help catalyze research in their RD in a hope for faster and better diagnosis and treatment. There are many inspiring examples of individual patient or a parent who with little resources but with much determination, they established a foundation for their RD, shared their data and created a successful collaboration between scientific researchers and patient organizations. A few to mention are: Syndromes Without A Name USA (SWAN),44–47 Ngly1.org foundation,48,49 the Chordoma Foundation,50 the Castleman Disease Collaborative Network,51 the Joshua Frase Foundation,52 the Cystic Fibrosis Foundation,53 and the PXE International.54 In all of these cases, there has been not only a patient or parent creating research programs and collaborations but also data sharing and data reutilization to support diagnosis and discovery. These foundational patient-scientist collaborations are a clear window into what will become the de facto standard, that is, Open Science, international collaborations involving patients, clinicians, researchers, and data technologists in a global venue.
The diversity of the aforementioned activities has contributed to the mention of the importance of patient engagement in RD clinical trials in the US Food and Drug Administration (FDA)’s 2019 draft guidance document for industry.35 The role of patients’ and parents’ support groups is growing beyond the boundaries of individual national initiatives aimed at raising public awareness and promoting medical care and social benefits.
Common data elements
Open access to data is not sufficient to make the data useful to science; data must also be structured, documented, interoperable, and curated. The magnitude of this task has led to the development of programs and software that helps automate data curation, data integration, and data mining; it has also underscored the need for machine learning and language processing.8,55–58 Health data comes from many different sources, and many different people produce, curate, and use the data. Integration is obstructed when systems and studies use different words to describe the same objects or concepts, use the same words intending different meanings, or use different data formats or structures.
Common data elements (CDEs) are a universal language that describes the data collected in a study. CDEs make data meaningful by structuring and defining commonly used, community shaped, recommended measures, and assessment instruments. Using CDEs when first collecting biomedical data makes it easier to develop meaningful analyses and research projects. When data is associated with CDEs, they can be more readily analyzed and reused to accelerate research into disease pathogenesis and therapeutic development. Although some CDEs were originally developed to address the needs of a specific research domain or clinical application, many CDEs address universal concepts of interest to a wide variety of domains for a variety of data collection purposes, such as demographic characteristics of research participants. In many cases, CDEs related to RDs may be broadly applicable for collecting data about other diseases, or for rapidly pivoting to collecting well-defined data critical for research related to emerging diseases, such as lung function measures that might have been developed for people with cystic fibrosis, and might be leveraged for use with patients with COVID-19. Identifying and reusing existing CDEs paves the way for smoothly finding, interpreting, and exchanging data. Unambiguous definitions are critical. For comparability among sources, CDEs should describe not only the data to be collected, but also rich metadata, that is the manner in which the data are collected and how the data are recorded. CDEs should define the parameter space for the data point and, instead of using natural language, they should encourage the use of standardized terminologies and ontologies. While consistency of data collection and the use of CDEs within an individual study are essential for maintaining data quality and enabling analysis, consistency of data collection across multiple studies brings additional value by promoting data sharing.59
Nevertheless, despite potential benefits and the extensive use of CDEs across clinical research studies, there are some challenges. There may be differences across studies in the interpretation and implementation of the data elements; researchers must ensure that CDEs are valid in different populations recruited for a study (eg, participants may have different cultural and linguistic backgrounds). Adoption of CDEs can be inhibited by existing research practices and legacy data systems. Conversely, use of clinical research data beyond the original purpose for which it was collected requires that researchers ensure that the collected data and its use is consistent with the informed consent and research ethics.
Data collection and annotation with a well-defined, controlled vocabulary and terms allow describing the meaning of data in a human and machine-readable way, enable data harmonization and meta-analyses, and enhance data sharing. Lack of standardization hinders data sharing and interoperability, so the use of CDEs is particularly critical for research and clinical care for people with RDs. The National Institutes of Health (NIH) Common Data Element Repository (CDE-R), developed and hosted by the US National Library of Medicine (NLM), is a platform for identifying related data elements in use across diverse areas, for harmonizing data elements, and for linking CDEs to other existing standards and terminologies.60,61 NLM and others across NIH work to ensure that formal vocabularies used to describe people, health problems, and health care processes are sufficiently robust to encompass the full range of health and disease across all populations and all communities.62–75 The CDE-R contains many CDEs developed for and by the RD research community, the Global Rare Diseases Registry Data Repository (GRDR).76,77 The PhenX toolkit is a catalog of measurement protocols, developed with a robust community consensus protocol.78 PhenX notes that its protocols can be used to combine studies to increase statistical power, enable comparisons of studies to validate results, and increase the impact of individual studies. PhenX has been used for the application of standardized measures in many clinical research studies, many of which are submitted to dbGaP. PhenX contains a collection of measures for Rare Genetic Conditions79 that, while very useful, would require significant expansion beyond their current remit of 10 per domain to be relevant to the 10 000 RDs that exist. PhenX also allows the creation of clinical data collection forms in standardized tools such as REDCap,80 which prospectively is a great advantage in standardizing data. All of the aforementioned efforts help support improved interoperability of clinical data across studies; they are critically important for RDs, for which data from one study or a different RD may help inform others.
Many RDs lack consistent identifiable terms, limiting literature searches, registry interoperability, and comparability in clinical information systems. Despite the advances in the creation of CDEs, many RDs lack a comprehensive set of disease definitions, associated phenotypes, genetic variations, treatments, prognoses, and other disease characteristics. However, CDE-development efforts that involve multidisciplinary collaboration, including informatics expertise, can address some of these challenges by identifying synonymy, clearly defining terms, and achieving consensus of key stakeholders for adoption of the CDEs. For example, this process was used to develop CDEs and guidance for health information exchange of newborn screening orders and results for lysosomal storage disorders. We now detail ongoing efforts to address this gap; the next steps would be to implement such components into CDEs, clinical systems such as EHRs, clinical decision support tools, and RD registries.
Data collected for RD research typically includes laboratory measurements, clinical observations, imaging, genomics and other 'omics data, as well as patient-reported outcomes (PROs). However, one of the biggest challenges for RD diagnosis is that RDs are not well-represented in terminologies typically used within EHRs, diagnostic settings, or other clinical information systems. The aforementioned CDEs for RD are intended to address this issue, but standardized ontologies are still lacking for use in those CDEs and clinical systems. Ontologies provide precise definitions of terms and relationships between different terms, which makes it possible to provide better quality checks, remove ambiguity, and provide much greater computability and utility in diagnostic or other algorithms. Precision medicine would greatly benefit from improved logical representation of clinical terminologies for classifying patients9; simply put, RD diagnostics requires it.
The Human Phenotype Ontology
The Human Phenotype Ontology (HPO) provides a structured, comprehensive, and well-defined set of terms that describe phenotypic abnormalities seen in human disease. It also provides a collection of disease-phenotype annotations, that is, computational assertions that a disease is associated with a given phenotypic abnormality. The HPO was created to enable “deep phenotyping,” that is, capture of symptoms and phenotypic findings using a logically constructed hierarchy of phenotypic terms.81,82 The HPO is a flagship project of the Monarch Initiative, an international consortium dedicated to developing integrative semantic technologies for disease diagnosis and mechanism discovery.27,83–85 The HPO allows algorithms to match sets of patient phenotype profiles in a “fuzzy” non-exact manner to gold standard RD profiles, other patients, and model organisms, greatly facilitating diagnosis.86–88 The HPO has therefore become the de facto standard for representing clinical phenotype data for diagnosis for rare genetic diseases by the 100 000 Genomes Project,89 the UDP,13,90 and Undiagnosed Diseases Network (UDN), as well as thousands of other clinics, laboratories, tools, and databases59,91,92; it is also a IRDiRC (International Rare Diseases Research Consortium) Recognized Resource.93
Although the focus of the HPO has, to date, been on RDs, it has been extended to provide a computational foundation for phenotype-driven analysis of genomes and other translational research on complex human disease.91 For example, many of the laboratory data recorded in EHRs for RD patients are expressed in an exact manner, such as measurements captured using the Logical Observation Identifiers Names and Codes (LOINC) standard for identifying medical laboratory observations. Recent efforts have been made to support interoperability between HPO and LOINC, such that direct measurements can be converted into HPO codes and used for diagnostic purposes.94
Deep phenotyping can be time-consuming and may miss key phenotypic features because they are not assessed (eg, phenotypes in internal organs that are only observable if a CT is performed) or not reported (eg, an inconsolable child or heavy snoring may not be documented in a clinical setting). Patients could therefore provide informative contributions to their computable phenotype profiles; however, the “terminology gap” between medical professionals and patients can limit patient participation both in research studies and in clinical phenotyping. Current patient vocabularies provide broad consumer equivalents for clinical findings, medical procedures and equipment but are not well integrated with research terminologies. For undiagnosed patients and those with RDs, affected individuals themselves are an especially critical source of phenotyping information. These patients accumulate a clear, firsthand knowledge about their condition, first from observing how the condition progresses daily, but also from multiple clinician evaluations and from other families and patients with similar conditions. In some cases, patients’ self-phenotyping combined with additional investigations has led to clinical diagnoses.95
To address these issues, the HPO was further developed to allow capture of patient-generated phenotypic profiles for use in diagnostic and patient community settings (registries, forums, clinics, and patient websites). To achieve this, a patient-centered lexicon of relevant terms was developed and added to the HPO.59,91,92 These terms are frequently referred to in plain language but can also include clinical terms (eg, Myopia [HP: 0000545] has a lay synonym “Near-sightedness”). Since the lay translation of the HPO uses the same logical infrastructure as the HPO itself, patient-generated phenotyping data can be readily combined with clinical phenotyping data to prioritize variants, improve diagnostic rates, and examine expressivity, penetrance and disease progression. Formal evaluation of the diagnostic capabilities of the lay HPO is in progress, and includes an informatics comparison against the gold-standard HPO disease annotations used in genomic diagnostics for patients with RDs. The lay HPO is expected to serve as a resource that will allow patients and families to become more effective partners in translational research, empowering families to achieve an accurate diagnosis and enabling people to improve the lives of others with RDs by increasing medical knowledge through their personal perspectives. The lay HPO should also enable RD patients to share their phenotyping profiles openly on the web using standards such as Phenopackets (see below), which allows the use of informatics to support open querying for similar patients to improve diagnosis.
Databases to share rare disease knowledge
While the HPO81 has become a global ontological standard for representing phenotypic attributes of RDs, community coordination of RD disease terminology is still emerging. Different terminological and database resources have been developed that describe RDs. The Online Mendelian Inheritance in Man (OMIM) began in the early 1960s by Dr. Victor A. McKusick as a catalog of Mendelian traits and disorders and has since become a global standard for documentation of Mendelian diseases.96,97 OMIM provides highly curated knowledge on genes and genetic diseases, phenotypes, and the relationships between them. Each disease listed in OMIM has a current summary of information based on expert review of the biomedical literature. Orphanet was established in France by the INSERM (French National Institute for Health and Medical Research) in 1997, and provides the community information and nomenclature on RDs; it is focused on improving the visibility of RDs in health and research information systems, particularly in Europe. The ORDO98 Orphanet rare disease terminology, an IRDiRC Recognized Resource,93 has been successfully used in the RD-Connect Sample Catalogue,99,100 which is an open data repository with information about biological samples from RD patients that are available to scientists for (re-)use. Disease infoSearch (diseaseinfosearch.org) is a crowdsourced database of thousands of diseases that helps patients find resources and studies, and integrates information from numerous sources, such as the NIH Genetic and Rare Diseases Information Center (https://rarediseases.info.nih.gov/). These RD-spanning databases are complemented by gene-, disease-, and/or locus-specific databases. For example, both the Human Genome Variation Society (HGVS)101 and the Leiden Open Variation Database (LOVD) list approximately 1500 expert-curated locus-specific mutation databases.102
Approaches to discovering the genetic basis of disease include linkage studies, genome-wide association studies, and a variety of designs involving next-generation sequencing including whole-exome and whole-genome sequencing. The majority of software used for these analyses are open access, greatly facilitating the pace of discovery. The results of many studies are also readily available. For example, the National Human Genome Research Institute (NHGRI)-EBI GWAS catalog reports over 70,000 variant-trait associations from >5000 studies.103 GenBank has freely released DNA sequence data since 1982.104 ClinVar is a public archive of reports of the relationships among human variations and phenotypes, with supporting evidence.105 The Genome Aggregation Database (gnomAD) is an aggregation and harmonization of exome and genome sequencing data from a variety of large-scale sequencing projects. Summary data are available for the wider scientific community based on genomic sequences of over 140,000 individuals. Also, the Database of Genotypes and Phenotypes (dbGaP) at NIH and the European Genome-phenome Archive (EGA) at EBI.106 These are examples of resources that are facilitating major progress in the discovery of genes and their functional characterization, leading to progress toward improved diagnosis and treatment.
Recently, major knowledge sources on RDs such as Orphanet, OMIM, ClinGen, MedGen, GARD, NCI Thesaurus, and others have been working together to harmonize disease definitions in a new ontology called “Mondo”,107,108 meaning “world”. While Mondo is still in development, the new ontology already provides a computational framework for defining RDs based upon logical representation of a variety of attributes such as phenotypes, genetic variants, treatment, onset, frequency, etc. Algorithmic and manual curation efforts have been used to align these RD terminologies, yielding preliminary estimates that the total number of RDs may exceed 10,000, that is, many more than the ∼7000 estimated during the inception of the Orphan Drug Act.109 More than half of these RDs can be found in three or more resources, whereas ∼4 K are unique to a given source. This preliminary analysis suggests that there could be a substantially higher number of RDs than currently assumed, with obvious implications for diagnostics, drug discovery and treatment. However, it should be emphasized that much more rigorous analysis is needed to establish the accuracy of this estimate.
Because RD patient presentations are heterogeneous and may not perfectly match existing disease definitions based on very small populations, it is critically important to share patients’ phenotypic information to support diagnosis, matchmaking, patient registries, communities, and target drug development. Further, despite the substantial improvements in exome analysis that have revealed numerous new rare Mendelian disease genes, the specific causal gene cannot be identified for more than half of patients.109 For these patients, evidence for causality depends on identifying other affected individuals with a similar phenotype and functionally impactful variants in the same candidate gene. In order to support this n-of-1 patient matching, the Global Alliance for Genomics and Health (GA4GH) initiated the Matchmaker Exchange (MME).110 MME is a federated network connecting different patient databases containing genomic and phenotypic data using a common application programming interface and allowing data exchange among them. MME has helped diagnose thousands of patients globally, by connecting these regional resources in a data sharing network that preserves privacy and maintains clinical review of potential matches and subsequent diagnoses.
While the MME has significantly advanced diagnostic potential for very RD patients, it does depend upon a patient being registered within a participating MME database. To increase computability of the phenotype data and to maximize potential open data sharing of patient phenotype information, the GA4GH created Phenopackets. Phenopackets is a standard file format for sharing phenotypic information that enables structured data sharing of information about a participant’s phenotype, such as clinical diagnosis, age of onset, results from lab tests, and disease severity.111 It can link to separate files containing a patient’s genetic sequence and pedigree, if available. Phenopackets are expected to standardize phenotypic data exchange within medical and scientific settings. This will allow phenotypic data to flow among clinics, databases, clinical labs, journals, and patient registries in ways that are currently feasible only for more quantifiable data, like sequence data. As more Phenopackets for RD patients are shared, clinicians, biologists, RD registries, and disease and drug researchers will build more complete models of disease and match similar patients (Figure 2). In addition, the use of Phenopackets to better represent and share the heterogeneity of RD presentation will lend itself well to drug repurposing. However, repurposing drugs similarly relies on sharing knowledge that has already been generated but may otherwise be difficult to access for those trying to repurpose.112 Monarch’s RD diagnosis tool Exomiser113,114 now takes Phenopackets as input, and Phenopackets are being adopted for projects such as the Japanese Agency for Medical Research and Development’s BioBank Network (biobank-search.megabank.tohoku.ac.jp) as well as SOLVE-RD (solve-rd.eu), the RD project of the European Commission.
Figure 2.
Phenopackets provide a mechanism for structured, de-identified, patient-level phenotype data sharing for computational use across the globe and in different information systems. Image credit: GA4GH Communications Team.
RD registries
Registries are considered key instruments for developing RD clinical research, enhancing patient care and health planning, and improving social, economic, and quality-of-life outcomes115,116 for the analysis of the natural history of RDs.117 Traditionally, registries have been either population-based or hospital-based. The former aim to capture all cases from a specific population and are focused on incident cases, seeking to describe the natural history of diseases.118 The latter provide responses to different clinical questions, serving as a source of patients for clinical trials and identifying and analyzing biomarkers as clinical prognosis factors.119 Both strategies are valid and are complementary because each can control for different types of biases.
Defining a standardized set of data elements is a key function and a key challenge for all registries120; the process of standardization is closely linked to the original sources of information used. The primary source of information is the patient and/or the physician collecting information directly from the patient; these sources have been used for centuries. However, standardizing the phenotype is not simple because we want the data collected to represent the patient’s clinical course. Standards such as CDEs, PROs,121 and ontologies such as HPO82 are not used by most registries; those that use them often do so in an ad hoc manner. Therefore, the main challenge for capture and reuse of registry data is transforming the physician’s free text or bespoke encoding into a standardized form. Specifically, how can the reliability between observers and within observers be guaranteed in an RD registry.122 Is the phenotype collected at a single point enough to define the full natural history of the disease? How long should be the follow-up period for each specific RD? How can a registry help in the analysis of natural and temporal variability of diseases? In fact, the only way to provide valid health outcomes is to guarantee the quality of all procedures included in the registry123; the use of ontologies instead of classical registry-specific standardization provides added value. Such standardization uses strict definitions, controls all parameters for each data element, and provides a high level of certainty about the data already collected and saved. Conversely, ontologies allow clinicians a certain level of confidentiality and flexibility because the terms are probabilistically linked. Ontologies and related standards facilitate data sharing among registries and improve interoperability between clinical and research systems.
Other secondary sources of information such as EHRs124 can provide some structured information are usually well standardized. EHRs can provide some information for certain types of registries, but since they have been built for other purposes with different criteria, they are not always appropriate for the aims of registries (EHRs typically contain a problem list functionality, while standardized and structured capture of symptoms is almost never available). RD registries have the capacity to reveal new disease genes, modifier variants, and new or very rare phenotypes, as well as the assessment of biomarkers, new treatments, and the impact of the implementation of health measurements. However, maximizing a registry’s ability to address unmet needs of RD patients requires data sharing and phenotypic and omics data, by researchers. Well designed and managed registries are regularly used for these purposes, but they must adapt their methods by collecting data directly from the EHR to identify the phenotypes instead of searching and recording specific data elements.
At early stages of registry planning, patient groups can provide support both as advisors and as partners. Patient groups can propose the creation of registries to healthcare institutions and work in a partnership. These outstanding, emerging possibilities should be carefully considered.125 As a recent example, several patient associations have contacted the Italian National Health Institute to establish and maintain disease-specific registries, and formal agreements have been signed between each association and the Institute. Further, the use of standardized registry software such as NORD’s Natural Histories Patient Registry Platform, RDconnect’s Registry Finder126 Coordination of Rare Diseases at Sanford (CoRDS), and the Program for Engaging Everyone Responsibly (PEER), are all examples of improved interoperability and data sharing and evolution with the standards over time. Ideally, such platforms will eventually robustly support both patient-generated individual content and synchronization with EHR data—something that is likely to improve clinical trial efficacy, recruitment, and engagement.
In general, early engagement of patient groups can substantially contribute to the success of the registry. The patient’s general contribution will assure that the Registry meets the patient’s needs and priorities, as well as their own data sharing wishes More specifically, patient engagement supports recruitment, relevance to patient healthcare, and the transparency of the process.127 Nevertheless, robust guidance on this issue is still insufficient and approaches to meet the challenge should be refined. Methods of engagement may vary based on the registry’s aim and many other factors. However, direct participation of the Registry governance at several levels is suitable for engaging patient partners in decision-making. Patient engagement in registries is an evolving field that presents both opportunities and challenges. Early engagement in the planning phase, consistent engagement throughout the registry functioning, relevance to patient needs, empowerment of each team component as well as transparency will create a tool that will both serve the patients and society and provide novel and integrated know-how. Simply put, RD registries are key to maximizing data sharing, patient communication across the globe for RD communities, delineating disease mechanisms, and promoting drug discovery; however, they are challenged in interoperability, maintenance, multi-model data types and sources, and governance.
Facial imaging, an artificial intelligence technology for RD research and diagnostics
The ability of artificial intelligence (AI) technologies to integrate and analyze data from different sources can be used to overcome some of the RDs’ challenges.128,129 In recent years, there have been significant advances in disease diagnosis as a result of new technologies for collecting and analyzing data. Researchers and clinicians are using these technologies to diagnose rare genetic diseases by scanning a person’s face or a photograph. AI can also be applied to speech structure and patient movement.129
The eagerness of the RD patients and their advocates to share their data and collaborate, despite the many privacy concerns, has facilitated the implementation of state of the art technologies in diagnosis and improving quality of life. Such new technologies include the ability to diagnose rare genetic diseases by scanning a person’s face or a photograph. Many RDs are manifested in a distinctive and recognizable facial phenotype, such as Noonan syndrome and Cornelia de Lange syndrome.130,131 Algorithms that analyze facial images have matured in recent years so that they predict several hundred RDs with a high degree of accuracy.20 Three-dimensional facial analysis (3DFA), an evolving deep phenotyping application, provides detailed representation and analysis of the RD phenotypes that can generate biological insights. In the RD domain, 3DFA is increasingly being implemented primarily for diagnostic purposes132–134 but also for monitoring existing and trial therapies.133,135,136
Advanced facial analysis platforms such as Cliniface,137 FACE2GENE,138 FaceBase,139 and DeepGestalt130 can point doctors in the direction of specific disorders or genes that could be responsible for the patient’s symptoms, potentially reducing the number of diagnostic tests needed to confirm the diagnosis. Facial analysis can also offer greater diagnostic certainty when the genetic causation remains undetermined or when molecular testing is unavailable, for example, in resource poor environments. AI and other analytic approaches provide objective analysis of phenotypes and the association of phenotype and genotype to streamline diagnostics, including genomic sequence interpretation.129,140 The application of facial analysis to RD diagnosis and care will require open source approaches as well as platforms that facilitate pre-competitive tools and partnerships, and that can be integrated with multi-omics initiatives.
An example is the Cliniface 3D facial analysis platform.137 Cliniface 3D tools have been shared for integration in multi-omics platforms for RD research, including through the Personalized Medicine Center for Children at the Telethon Kids Institute, and it is being prepared for partnership with the National Rare Diseases Registry System of China.141 Cliniface has been implemented across multiple research and clinical environments, including state-wide for the Western Australian Health Department, and is being increasingly integrated with the Patient Archive knowledge management platform142 which is connected to MME.110 Cliniface converts 3D facial images to text-based descriptions, specifically HPO terms. Converting face-to-text reduces the risk of individual identification, mitigating against the inherently identifying nature of facial data. These text-based descriptions can be shared through MME or Phenopackets, and they can be incorporated into text-based diagnostic support algorithms.
One of the most promising resources for facial data sharing is the Minerva Initiative.143 While it was originally launched for 2-D data sharing, the underlying principles are intentionally extensible to 3-D data. The initiative includes a research data resource (Minerva Image Resource—MIR) and an open research consortium (Minerva Consortium—MC) which allows the sharing of identifiable patient data, such as facial photographs and collaborative research projects on RD. It operates in the spirit of Open Science to enable precision public health. The Minerva Initiative has the following objectives: to build a community of researchers and clinicians, to continue to develop ethical structures and provisions for working on identifiable clinical images, and to deliver secure data sharing among consortium members. It has been constructed to align with the goals and objectives of the GA4GH.144 The Minerva Consortium (MC) is an international network of clinicians and researchers, from both public and private organizations. The public website Minerva&Me allows anyone around the world to participate directly in the Minerva Initiative.145 Initiatives such as the Minerva Initiative are poised to lead the way in terms of not only amassing data but also using integrative technologies for accessing and using data at the point of care.
While 3DFA was originally developed for RD diagnostic applications, it can also be applied to treatment monitoring for both rare and common diseases, as demonstrated in a new project traversing specialties at the Perth Children’s Hospital and Western Australia’s premier clinical trials facility, Linear Clinical Research. In addition, while 3DFA is yielding translational insights into innovations for diagnosis, treatment, and monitoring in the RD domain, it also examines the overlap between rare and more common diseases and, therefore, mechanistic research. Notably, population-level studies demonstrated that common genetic variations (polymorphisms) were associated with discrete patterns of facial variation. Notably, these facial signatures recapitulated the characteristic facies of the respective genetic syndrome due to rare genetic variation (pathogenic variants).146 An example of a common disease that is poised for 3D facial translational research is obstructive sleep apnea (OSA). OSA is a condition seen in RDs such as mucopolysaccharidoses, where it regularly has an earlier onset than in the general population. These findings highlight the overlap between common and rare phenotypes, with implications for possible reciprocal (rare-common) insights.147
Data acquisition, analysis, and sharing mechanisms for identifiable facial data are key to RD diagnosis and research, but specialized approaches are required to simultaneously facilitate more Open Science while respecting patient privacy.
Telemedicine
Developments in modern communication technology such as telemedicine have created new opportunities for the delivery of health services to remote areas and unprivileged communities. Telemedicine refers to communication tools for medical care delivery at a distance, including telephones, smart phones, interactive televideo, “store-and-forward” images and medical record transmission via personal computers, and remote monitoring.148 High-speed telecommunications systems, in addition to the invention of devices capable of capturing and transmitting images and other data in digital form, have facilitated better sharing, collaboration, and efficiency in telemedicine. As a result, health professionals can communicate faster, more widely, and more directly with other clinicians and patients regardless of location.
Access to medical care is a major concern for RD patients and their families not only in rural areas and developing countries. Among the main issues are a lack of physicians specialized in RD treatment; concerns about sharing personal information and the security of personal information, few programs and resources to support low socioeconomic families with travel accommodation, as well as loss of income associated with obtaining care from specialists at long distances.149
RD and undiagnosed patients are usually dispersed over a large geographical area, yet they require multidisciplinary experts. As a result, a correct diagnosis may be delayed, and ready access to ongoing care is limited. Thus, telemedicine can profoundly change patient care for individuals with RD and directly address challenges of geography, travel burden, and access to experts; it can provide open access and global data sharing. Telemedicine can increase patient access to health care services otherwise unavailable149 as well as for patients in developing countries and rural/remote area.150 If utilized to its potential, telemedicine may open the way for more equitable distribution of knowledge and medical care throughout the world.149,150 In 2020, the Mayo Clinic plans to serve 200 million patients, many of them from outside the United State and most of them remotely.149
Telemedicine can revolutionize the way in which healthcare is delivered and allow the home to become a preferred place of care. The advantages of this approach are patient satisfaction, reduced travel requirements to health care providers, clinics and hospitals, early intervention for disease progression, support for caregivers, and economic benefits associated with reduced hospitalization rates.151
In addition to the increased connectivity between providers and patients, telemedicine also provides a means for researchers to connect to potential participants. Mobile and wearable medical devices enable patients to share and transmit a wealth of digital health data to databases contributing to patient registries, natural history studies, and clinical trials. Telemedicine has already been used and proven its value for chronic non-RDs, such as congestive heart failure and chronic obstructive pulmonary disease152 as well as some RDs, such as mesothelioma,153,154 cystic fibrosis,155 diabetes,156–159 Prader–Willi syndrome,160 and juvenile idiopathic arthritis.161 As promising as telemedicine sounds, it cannot be a replacement for in-person examination. There are significant limitations and barriers that need to be addressed and overcome, including quality of patient-clinician interaction, insurance coverage, reimbursement for services, privacy and legal issues of state licensure laws and liability concerns.
Providing care through telemedicine technology may not work for every organization. However, with the move toward personalized medicine, incorporating telemedicine into the health system can offer benefits to physicians and patients.162 Examples for reductions in use of services are hospital admissions/re-admissions, length of hospital stay, and emergency department visits that translate into reduced mortality.152 To increase the uses and implementation of telemedicine, more resources and studies are needed to evaluate the net value, visibility, and access for patients and the health care providers.
Telemedicine can emerge as an important component of the health care delivery system that relies on sharing medical information, knowledge and collaboration, which are the building blocks necessary to facilitate Open Science. RD patients and their families seem to more enthusiastically share personal information and collaborate because they desperately want to find the correct diagnosis, experts, and treatment.
In the context of global health, telemedicine is beginning to have an important impact on many aspects of healthcare, especially in developing countries and in rural areas, opening the way for distribution of knowledge and medical care throughout the world.150 Although Open Science can aid the RD community, the RD community can be instrumental for Open Science and aid to further the development of Open Science by adopting and incorporating telemedicine and new technologies into health care delivery.
Ethical and legal considerations
We have demonstrated the significant need for Open Science practice to share data and collaborate in support of RD diagnosis, research, and patient care. Open Science creates some dilemmas and opposing forces with regards to privacy and ethical concerns. Experience with the RD patients and their families has demonstrated that their eagerness for adequate diagnosis and treatment override the privacy concerns. Nevertheless, as new technologies, and systems are developed and implemented, the ethical and legal challenges increase.163,164
Global data sharing creates significant challenges for the responsible stewardship of the growing number of large and complex datasets, including oversight, accountability, and data management. Ethical and legal frameworks are required to protect the rights of affected individuals, while still sharing data appropriately to promote progress in RD research and health care. For example, before increasing the availability and dissemination of RD patient data, scientists must consider participant protections and appropriate data use, consent, and participant understanding of data sharing, ownership, reuse, analysis and the generation of new or derived data, among other concerns.
A number of international organizations165 have devoted considerable attention and resources to developing regulatory frameworks for open data production, dissemination and use. The frameworks have resulted in national policies on Open Science, and documents such as the international accord Open Data in a Big Data World,166 the Open Science policy by the European Commission,167 and the National Academies of Science “Open Science by Design: Realizing a Vision for 21st Century Research”,168 which recommends that data be made FAIR based on legal and ethical considerations. The Biobanking and BioMolecular Resources Research Infrastructure- European Research Infrastructure Consortium (BBMRI ERIC) is building an international Code of Conduct for health research, with the aim of contributing to the proper application of the regulation, taking into account the specific features of processing personal data in the area of health research in order to clarify and specify certain rules of the General Data Protection Regulation (GDPR) for those who process personal data for scientific research in the area of health.169 Similarly, the NIH Genomic Data Sharing Policy170 includes provisions for the sharing of large-scale genomic data while taking into account participant protections and limitations on data use based on the consent of the study participants. All of these efforts emphasize the importance of good data practices in the sharing, dissemination, and re-use of biomedical data, particularly considering issues of privacy, confidentiality, intellectual property and security.165 Clinical trial data sharing has been particularly challenging when it involves the pharmaceutical industry or other entities with IP interests. With the extremely small RD cohorts, it is especially important to coordinate and to share results globally. The Vivli (https://vivli.org/) program aims to support sharing and reuse of clinical research data, including individual participant-level data from completed clinical trials globally. Medical journals generally require clinical trials to be posted on clinicaltrials.gov, and FDAAA requirements include submission of the data of both positive and negative studies. However, for RDs, it is especially important to share trial information as trials are being designed and launched, so that different studies can be aligned and patients can be recruited from around the world. PAGs for each RD have been successful in doing so, and approaches such as those attempted in OpenTrials (https://opentrials.net/) are laudatory but are not yet established enough to support the RD community.
Approaching ethical challenges from an international standpoint is central to the promise of Open Science.171
Addressing Indigenous rights and interests in genomic and other data sharing is critical for equitable scientific translation. While Indigenous experiences with genetic research have been shaped by a series of negative interactions, there is increasing recognition that equitable benefits can only be realized through greater participation of Indigenous communities. Issues of trust, accountability, return of benefit and equity will need to be addressed. In this context, it is notable that the Research Data Alliance International Indigenous Data Sovereignty Interest Group172 developed the CARE Principles for Indigenous Data Governance. These principles identify Collective benefit, Authority to control, Responsibility, and Ethics to be used alongside other data centric principles.
While critically important and relevant to all health care domains, endeavors such as these do not specifically take into account the special Open Science needs of RD patients and caregivers. RD advocacy and methods for robust and informed data sharing must be developed alongside policies and secure infrastructure that are specifically designed for sharing data about RD patients—who by their very rarity have a much greater likelihood of re-identification or even a desire to share identified data. Toward the end of supporting genomic health ethics for all types of genetic diseases, the GA4GH has created a “Framework for Responsible Sharing of Genomic and Health-Related Data”. It contains foundational principles and core elements for responsible data sharing and is guided by concern for human rights, including the right to benefit from the progress of science, as well as privacy, non-discrimination, and procedural fairness. This is pivotal for RD patients, since traditional medical privacy laws around the world may not adequately support the Open Science strategies that RD diagnosis and research necessitates.
The best practices and ethical-legal considerations for FAIR data sharing in the context of RDs are still evolving. A necessary improvement in the management of data in a FAIR-er direction is the annotation of patient data with Ethical Legal Social Issue (ELSI) requirements and choices as determined at the time of collection. This could constitute a great addition to the quality of data that could be transferred along with the data itself. This means that if a certain dataset was collected under the condition of a specific use (eg, cancer research only in Europe) this information should travel with the data, ensuring sustainable and ELSI reusability of the data. While the promises of the Open Science paradigm and the FAIRification of data are key to effective research, especially in RD, compliance with existing regulatory requirements and ethical norms is necessary to ensure long-term sustainability of data stewardship.
New perspectives, understanding and challenges introduced by rapidly developing machine learning approaches increase the necessity of open data sharing to realize the public good, but simultaneously can give rise to new ethical and legal dilemmas. Among the challenges already becoming apparent are the potential risks for re-identification, incidental/secondary findings, and biases for equitable access to algorithmically assisted decision making. Particularly in the context of RD, implications of new machine learning will influence the best practices and acceptable frameworks for FAIR data sharing in the coming years.
A ROADMAP FOR OPEN SCIENCE IN RARE DISEASES
To accelerate the diagnosis and care of RD patients, we propose a set of recommendations to advance Open Science:
Create shared RD definitions, models, and governance.
Consider how to realize the FAIR principles in all aspects of the RD data lifecycle for any given RD, clinical system, or research initiative.
Create metrics for successful compliance with RD-GO FAIR.
Support RD tools that enable patients to share their own data in a well-informed manner and establish standards for consistent representation of phenotype data (eg, Phenopackets and HPO) as well as genotype and pedigree data.
Adopt new standards for registries to support interoperability and data sharing internationally.
Develop methods to create “proxy” data to share representations or subsets of personally identifiable data (such as facial images) in a deidentified manner.
Establish networks of controlled-access data that can be searched using diagnostic algorithms for research on RDs.
Increase centers specializing in RDs, train more clinicians in diagnosing and treating RDs, and create improved clinical decision-making guidelines related to RDs.
Create opportunities for patients to be better informed and encourage patient engagement with the scientific community to increase openness and data sharing.
Welcome and attribute openly-developed novel technologies and interventions in RD clinical settings.
A fundamental component of addressing the RD public health challenge involves improvements in ethical Open Science, whose core principles are data sharing and collaboration. RD families and their advocates, as well as RD physicians and scientists, have led the way toward openness, data sharing, and collaboration to find diagnoses, treatments, and improved quality of life. Despite privacy concerns, institutional policies, and technological barriers, the RD community has demonstrated that they are thought leaders in Open Science, forging the way forward for the world.
FUNDING
This work was supported by the U.S. Department of Health and Human Services National Institutes of Health (5r24od011883), National Institutes of Health (NIH) Office of the Director (OD); the Monarch Initiative (1R24OD011883) and the Eunice Kennedy Shriver National Institute of Child Health and Human Development (U54 HD079123).
AUTHOR CONTRIBUTIONS
All authors contributed texts and ideas, participated in critical revision of the article, and approved the final version. WAG, RMG, MAH, PNR, and YRR, participated in the initial discussion and the conception/design of the article. MAH, PNR, and YRR supervised the conception, design, and revision of the manuscript.
ACKNOWLEDGMENTS
This research was supported in part by the Office of Strategic Initiatives, Office of the Director, and the Intramural Research Program at the National Library of Medicine (NLM) at the National Institutes of Health (NIH). The findings and conclusions in this article are those of the authors and do not necessarily represent the official position of HRSA, NIH, NLM, or the Department of Health and Human Services.
The authors would like to thank Dr. Mike Huerta (the Director of the Office of Strategic Initiatives, Associate Director of the National Library of Medicine, NIH) for his enthusiastic support to initiate this article and his fruitful and valuable advice throughout the writing of the manuscript.
We would like to dedicate this article to Dr. Stephen Groft, for his great mentorship and exemplary caring and kindness toward others. Dr. Groft devoted his career to the Rare Disease community, providing hope and voice for rare disease patients and their families. This article is also dedicated to the rare disease patients, their families, the caregivers, the medical practitioners and the research community, for their commitment to sharing data and Open Science.
CONFLICT OF INTEREST
MAH and JAM have a conflict of interest. Both are the founders of Pryzm Health.
REFERENCES
- 1. Taruscio D, Floridia G, Salvatore M, Groft SC, Gahl WA. Undiagnosed diseases: Italy-US collaboration and international efforts to tackle rare and common diseases lacking a diagnosis. Adv Exp Med Biol 2017; 1031: 25–38. [DOI] [PubMed] [Google Scholar]
- 2. Richter T, Nestler-Parr S, Babela R, et al. Rare disease terminology and definitions-A systematic global review: report of the ISPOR Rare Disease Special Interest Group. Value Health 2015; 18 (6): 906–14. [DOI] [PubMed] [Google Scholar]
- 3. Forrest CB, Bartek RJ, Rubinstein Y, Groft SC. The case for a global rare-diseases registry. Lancet 2011; 377 (9771): 1057–9. [DOI] [PubMed] [Google Scholar]
- 4. Khosla N, Valdez R. A compilation of national plans, policies and government actions for rare diseases in 23 countries. Intractable Rare Dis Res 2018; 7 (4): 213–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Haendel M, Vasilevsky N, Unni D, et al. How many rare diseases are there? Nat Rev Drug Discov 2020; 19: 77–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Evans WR. Dare to think rare: diagnostic delay and rare diseases. Br J Gen Pract 2018; 68: 224–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Vandeborne L, van Overbeeke E, Dooms M, De Beleyr B, Huys I. Information needs of physicians regarding the diagnosis of rare diseases: a questionnaire-based study in Belgium. Orphanet J Rare Dis 2019; 14 (1): 99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Colbaugh R, Glass K, Rudolf C, Tremblay Volv Global Lausanne Switzerland M. Learning to identify rare disease patients from electronic health records. AMIA Annu Symp Proc 2018; 2018: 340–7. [PMC free article] [PubMed] [Google Scholar]
- 9. Haendel MA, Chute CG, Robinson PN. Classification, ontology, and precision medicine. N Engl J Med 2018; 379 (15): 1452–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gahl WA, Markello TC, Toro C, et al. The National Institutes of Health Undiagnosed Diseases Program: insights into rare diseases. Genet Med 2012; 14 (1): 51–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gahl WA, Boerkoel CF, Boehm M. The NIH Undiagnosed Diseases Program: bonding scientists and clinicians. Dis Model Mech 2012; 5 (1): 3–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gahl WA, Tifft CJ. The NIH Undiagnosed Diseases Program: lessons learned. JAMA 2011; 305 (18): 1904–5. [DOI] [PubMed] [Google Scholar]
- 13. Gall T, Valkanas E, Bello C, et al. Defining disease, diagnosis, and translational medicine within a homeostatic perturbation paradigm: the national institutes of health undiagnosed diseases program experience. Front Med (Lausanne) 2017; 4: 62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Taruscio D, Groft SC, Cederroth H, et al. Undiagnosed Diseases Network International (UDNI): White paper for global actions to meet patient needs. Mol Genet Metab 2015; 116 (4): 223–5. [DOI] [PubMed] [Google Scholar]
- 15.UDNI. UDNI. UDNI—Undiagnosed Diseases Network International. 2019. http://www.udninternational.org/ Accessed June 26, 2019.
- 16.Initiative on Rare and Undiagnosed Diseases (IRUD) | Japan Agency for Medical Research and Development. https://www.amed.go.jp/en/program/IRUD/ Accessed December 24, 2019.
- 17.GO FAIR. Implementation Networks—GO FAIR. GO FAIR; 2019. https://www.go-fair.org/implementation-networks/ Accessed July 22, 2019.
- 18. Burgelman J-C, Pascu C, Szkuta K, et al. Open science, open data, and open scholarship: european policies to make science fit for the twenty-first century. Front Big Data 2019; 2: 43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wilkinson MD, Dumontier M, Sansone S-A, et al. Evaluating FAIR maturity through a scalable, automated, community-governed framework. Sci Data 2019; 6 (1): 174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Molster C, Youngs L, Hammond E, Dawkins H; National Rare Diseases Coordinating Committee, National Rare Diseases Working Group. Key outcomes from stakeholder workshops at a symposium to inform the development of an Australian national plan for rare diseases. Orphanet J Rare Dis 2012; 7 (1): 50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rare Diseases—GO FAIR. GO FAIR. https://www.go-fair.org/implementation-networks/overview/rare-diseases/ Accessed December 24, 2019.
- 22. Carbon S, Champieux R, McMurry JA, Winfree L, Wyatt LR, Haendel MA. An analysis and metric of reusable data licensing practices for biomedical resources. PLoS One 2019; 14 (3): e0213090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Haendel. Socio-legal Barriers to Data Reuse. NLM Musings from the Mezzanine. 2019. https://nlmdirector.nlm.nih.gov/2019/06/11/socio-legal-barriers-to-data-reuse/ Accessed July 22, 2019.
- 24. Haendel M, Su A, McMurry J. FAIR-TLC: Metrics to Assess Value of Biomedical Digital Repositories: Response to RFI NOT-OD-16-133. 2016. https://zenodo.org/record/203295.
- 25.B2SHARE. https://b2share.eudat.eu/records/6ceeed13eb6340fcb132bcb5b5e3d69a Accessed December 24, 2019.
- 26. Donoho D. 50 years of data science. J Comput Graph Stat 2017; 26 (4): 745–66. [Google Scholar]
- 27. Shefchek KA, Harris NL, Gargano M, et al. The Monarch Initiative in 2019: an integrative data and analytic platform connecting phenotypes to genotypes across species. Nucleic Acids Res 2020; 48 (D1): D704–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Biomedical Data Translator. National Center for Advancing Translational Sciences; 2017. https://ncats.nih.gov/translator Accessed December 24, 2019.
- 29.Biomedical Data Translator Consortium. Toward a universal biomedical data translator. Clin Transl Sci 2019; 12: 86–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Biomedical Data Translator Consortium. The Biomedical Data Translator Program: conception, culture, and community. Clin Transl Sci 2019; 12: 91–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Guo Y, Heath AP, Raman P, et al. Gabriella Miller Kids First Data Resource Center: Harmonizing genomic and clinical information to support childhood cancer and structural birth defect research. Eur J Hum Genet 2019; 27: 1174–813. [Google Scholar]
- 32.(Re)usable Data Project. http://www.reusabledata.org Accessed December 24, 2019.
- 33.OpenPHACTS. Open PHACTS. Open PHACTS. 2019. https://www.openphactsfoundation.org/ Accessed July 23, 2019.
- 34.CNMR. Centro Nazionale Malattie Rare. Centro Nazionale Malattie Rare; 2019. http://old.iss.it/cnmr/? lang=1&id=2662&tipo=3 Accessed June 26, 2019.
- 35. Vernon M, Marsh K. Patient engagement in clinical trial protocol design and recruitment strategies: what does it mean for orphan drug manufacturers? Evidera 2019: 1–4. https://www.evidera.com/patient-engagement-in-clinical-trial-protocol-design-and-recruitment-strategies-what-does-it-mean-for-orphan-drug-manufacturers/. [Google Scholar]
- 36. Lapteva L, Vatsan R, Purohit-Sheth T. Regenerative medicine therapies for rare diseases. Transl Sci Rare Dis 2018; 3 (3–4): 121–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Schliebner S, Pruitt B. How Technology is Reshaping the Rare Disease Landscape. 2018. https://prahs.com/resources/whitepapers/Technology%20Reshaping%20Rare%20Disease_PRA%20White%20Paper.pdf Accessed December 15, 2019.
- 38.NCTS. About the RDCRN | The Rare Diseases Clinical Research Network. National Center for Advancing Translational Sciences; 2015. https://ncats.nih.gov/rdcrn/about Accessed July 23, 2019.
- 39.EURODIS. EURORDIS Rare Diseases Europe. EURORDIS; 2019. https://www.eurordis.org/ Accessed July 23, 2019.
- 40.PCORI. Rare Diseases Topic Spotlight. Patient Centered Outcomes Research Institute; 2018. https://www.pcori.org/research-results/topics/rare-diseases Accessed July 23, 2019. [Google Scholar]
- 41.Genetic Alliance. Genetic Alliance. Genetic Alliance; 2019. http://www.geneticalliance.org/ Accessed July 23, 2019.
- 42.NORD. National Organization for Rare Disorders. National Organization for Rare Disorders; 2019. https://rarediseases.org/ Accessed July 23, 2019.
- 43.IMI Innovative Medicines Initiative | PARADIGM | Patients active in research and dialogues for an improved generation of medicines: advancing meaningful patient engagement in the life cycle of medicines for better health outcomes. IMI Innovative Medicines Initiative. https://www.imi.europa.eu/projects-results/project-factsheets/paradigm Accessed May 20, 2020.
- 44.Swan USA. Swan USA. http://swanusa.org/ Accessed Decembe 24, 2019.
- 45. Genes G. Global Genes Partners with SWAN USA to Help Undiagnosed Rare Disease Patients Seek a Medical Diagnosis through Free Whole Exome Sequencing Program. PR Newswire; 2014. https://www.prnewswire.com/news-releases/global-genes-partners-with-swan-usa-to-help-undiagnosed-rare-disease-patients-seek-a-medical-diagnosis-through-free-whole-exome-sequencing-program-243552661.html Accessed Decembe 24, 2019.
- 46.Home | SWAN UK. SWAN UK. https://www.undiagnosed.org.uk/ Accessed Decembe 24, 2019.
- 47.GeneDx, OPKO Health, Inc. OPKO’s GeneDx Recognizes International Rare Disease Day with Donation of Free Exome Tests to Syndromes without a Name. PR Newswire; 2016. https://www.prnewswire.com/news-releases/opkos-genedx-recognizes-international-rare-disease-day-with-donation-of-free-exome-tests-to-syndromes-without-a-name-300227519.html Accessed Decembe 24, 2019.
- 48.NGLY1.org—N-Glycanase Deficiency—NGLY1. NGLY1. http://Ngly1.org Accessed Decembe 24, 2019.
- 49. Weintraub K. A Battle Plan for a War on Rare Diseases. The New York Times. 2018. https://www.nytimes.com/2018/09/10/health/matthew-might-rare-diseases.html Accessed Decembe 24, 2019.
- 50.Chordoma Foundation. Chordoma Foundation. Chordoma Foundation; 2019. https://www.chordomafoundation.org/ Accessed July 23, 2019.
- 51.CDCN. Castleman Disease Collaborative Network. Castleman Disease Collaborative Network; 2019. https://www.cdcn.org/ Accessed July 23, 2019.
- 52. Frase J. The Joshua Frase Foundation Supports Research for Myotubular Myopathy. The Joshua Frase Foundation Supports Research for Myotubular Myopathy; 2019. https://www.joshuafrase.org/ Accessed July 23, 2019.
- 53.CFF. Cystic Fibrosis Foundation. Cystic Fibrosis Foundation; 2019. https://www.cff.org/ Accessed July 23, 2019.
- 54.PXE International | PXE International. https://www.pxe.org/ Accessed May 20, 2020.
- 55.NIH. NIH Releases Strategic Plan for Data Science. NIH News & Events; 2018. https://www.nih.gov/news-events/news-releases/nih-releases-strategic-plan-data-science Accessed July 17, 2019.
- 56.NLM. NLM Launches 2017-2027 Strategic Plan. NLM News & Events. U.S. National Library of Medicine; 2018. https://www.nlm.nih.gov/news/NLM_Launches_2017_to_2027_Strategic_Plan.html Accessed July 17, 2019.
- 57. Shen F, Zhao Y, Wang L, et al. Rare disease knowledge enrichment through a data-driven approach. BMC Med Inform Decis Mak 2019; 19 (1): 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Arbabi A, Adams DR, Fidler S, Brudno M. Identifying clinical terms in medical text using ontology-guided machine learning. JMIR Med Inform 2019; 7 (2): e12596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Sheehan J, Hirschfeld S, Foster E, et al. Improving the value of clinical research through the use of common data elements. Clin Trials 2016; 13 (6): 671–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.NIH Common Data Elements (CDE) Repository. https://cde.nlm.nih.gov/ Accessed February 6, 2020.
- 61.NIH Common Data Element (CDE) Resource Portal. NIH Common Data Element (CDE) Resource Portal. U.S. National Library of Medicine; 2012. https://www.nlm.nih.gov/cde/index.html Accessed February 6, 2020.
- 62. Goetz KE, Reeves MJ, Tumminia SJ, Brooks BP. eyeGENE(R): a novel approach to combine clinical testing and researching genetic ocular disease. Curr Opin Ophthalmol 2012; 23 (5): 355–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Redeker NS, Anderson R, Bakken S, et al. Advancing symptom science through use of common data elements. J Nurs Scholarsh 2015; 47 (5): 379–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Moore SM, Schiffman R, Waldrop-Valverde D, et al. Recommendations of common data elements to advance the science of self-management of chronic conditions. J Nurs Scholarsh 2016; 48 (5): 437–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Corwin EJ, Moore SM, Plotsky A, et al. Feasibility of combining common data elements across studies to test a hypothesis. J Nurs Scholarsh 2017; 49 (3): 249–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Knisely MR, Maserati M, Heinsberg LW, et al. Symptom science: advocating for inclusion of functional genetic polymorphisms. Biol Res Nurs 2019; 21 (4): 349–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Corwin EJ, Berg JA, Armstrong TS, et al. Envisioning the future in symptom science. Nurs Outlook 2014; 62 (5): 346–51. [DOI] [PubMed] [Google Scholar]
- 68. Page GG, Corwin EJ, Dorsey SG, et al. Biomarkers as common data elements for symptom and self-management science. J Nurs Scholarsh 2018; 50 (3): 276–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Menon DK, Schwab K, Wright DW, Maas AI. Demographics and clinical assessment working group of the international and interagency initiative toward common data elements for research on traumatic brain injury and psychological health. Position statement: definition of traumatic brain injury. Arch Phys Med Rehabil 2010; 91 (11): 1637–40.21044706 [Google Scholar]
- 70. Roos M, López Martin E, Wilkinson MD. Preparing data at the source to foster interoperability across rare disease resources. Adv Exp Med Biol 2017; 1031: 165–79. [DOI] [PubMed] [Google Scholar]
- 71. Nelson LD, Ranson J, Ferguson AR, et al. Validating multidimensional outcome assessment using the TBI common data elements: an analysis of the TRACK-TBI pilot sample. J Neurotrauma 2017; 34 (22): 3158–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Duhaime A-C, Gean AD, Haacke EM, et al. Common data elements in radiologic imaging of traumatic brain injury. Arch Phys Med Rehabil 2010; 91 (11): 1661–6. [DOI] [PubMed] [Google Scholar]
- 73. Cheadle C, Cao H, Kalinin A, Hodgkinson J. Advanced literature analysis in a big data world. Ann NY Acad Sci 2017; 1387 (1): 25–33. [DOI] [PubMed] [Google Scholar]
- 74. Abhyankar S, Goodwin RM, Sontag M, Yusuf C, Ojodu J, McDonald CJ. An update on the use of health information technology in newborn screening. Semin Perinatol 2015; 39 (3): 188–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Abhyankar S, Lloyd-Puryear MA, Goodwin R, et al. Standardizing newborn screening results for health information exchange. AMIA Annu Symp Proc 2010; 2010: 1–5. [PMC free article] [PubMed] [Google Scholar]
- 76. Hendershot T, Pan H, Haines J, et al. Using the PhenX toolkit to add standard measures to a study. Curr Protoc Hum Genet 2015; 86: 1.21.1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Rubinstein YR, McInnes P. NIH/NCATS/GRDR® common data elements: a leading force for standardized data collection. Contemp Clin Trials 2015; 42: 78–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.PhenX Toolkit. http://phenxtoolkit.org Accessed December 23, 2019.
- 79.PhenX Toolkit: Domains. http://phenxtoolkit.org/domains/view/220000 Accessed December 23, 2019.
- 80. Harris PA, Taylor R, Minor BL, et al. The REDCap consortium: building an international community of software platform partners. J Biomed Inform 2019; 95: 103208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Köhler S, Carmody L, Vasilevsky N, et al. Expansion of the human phenotype ontology (HPO) knowledge base and resources. Nucleic Acids Res 2019; 47 (D1): D1018–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Robinson PN, Köhler S, Bauer S, Seelow D, Horn D, Mundlos S. The human phenotype ontology: a tool for annotating and analyzing human hereditary disease. Am J Hum Genet 2008; 83 (5): 610–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Mungall CJ, McMurry JA, Köhler S, et al. The Monarch Initiative: an integrative data and analytic platform connecting phenotypes to genotypes across species. Nucleic Acids Res 2017; 45 (D1): D712–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Mungall CJ, Washington NL, Nguyen-Xuan J, et al. Use of model organism and disease databases to support matchmaking for human disease gene discovery. Hum Mutat 2015; 36 (10): 979–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. McMurry JA, Köhler S, Washington NL, et al. Navigating the phenotype Frontier: the Monarch initiative. Genetics 2016; 203 (4): 1491–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Köhler S, Schulz MH, Krawitz P, et al. Clinical diagnostics in human genetics with semantic similarity searches in ontologies. Am J Hum Genet 2009; 85 (4): 457–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Bauer S, Köhler S, Schulz MH, Robinson PN. Bayesian ontology querying for accurate and noise-tolerant semantic searches. Bioinformatics 2012; 28 (19): 2502–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Schulz MH, Köhler S, Bauer S, Vingron M, Robinson PN. Exact score distribution computation for ontological similarity searches. BMC Bioinformatics 2011; 12: 441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Chief Medical Officer annual report 2016: Generation Genome. GOV.UK. https://www.gov.uk/government/publications/chief-medical-officer-annual-report-2016-generation-genome Accessed June 30, 2018.
- 90. Bone WP, Washington NL, Buske OJ, et al. Computational evaluation of exome sequence data using human and model organism phenotypes improves diagnostic efficiency. Genet Med 2016; 18 (6): 608–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Groza T, Köhler S, Moldenhauer D, et al. The human phenotype ontology: semantic unification of common and rare disease. Am J Hum Genet 2015; 97 (1): 111–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Vasilevsky NA, Foster ED, Engelstad ME, et al. Plain-language medical vocabulary for precision diagnosis. Nat Genet 2018; 50 (4): 474–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.IRDiRC. IRDiRC Recognized Resources. IRDiRC; 2018. http://www.irdirc.org/research/irdirc-recognized-resources/current-irdirc-recognized-resources/ Accessed July 17, 2019.
- 94. Zhang XA, Yates A, Vasilevsky N, et al. Semantic integration of clinical laboratory tests from electronic health records for deep phenotyping and biomarker discovery. NPJ Digit Med 2019; 2: 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Konopka BM. Biomedical ontologies—a review. Biocybern Biomed Eng 2015; 35 (2): 75–86. [Google Scholar]
- 96. Amberger JS, Bocchini CA, Schiettecatte F, Scott AF, Hamosh A. OMIM.org: Online Mendelian Inheritance in Man (OMIM®), an online catalog of human genes and genetic disorders. Nucleic Acids Res 2015; 43 (D1): D789–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Amberger JS, Bocchini CA, Scott AF, Hamosh A. OMIM.org: leveraging knowledge across phenotype-gene relationships. Nucleic Acids Res 2019; 47 (D1): D1038–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Vasant D, Chanas L, Malone J, et al. Orphanet Rare Disease ontology (ORDO): An Ontology Connecting Rare Disease, Epidemiology and Genetic Data. https://www.orpha.net/consor/cgi-bin/index.php.
- 99.RD-Connect. RD-Connect Sample Catalogue. RD-Connect Sample Catalogue; 2019. https://samples.rd-connect.eu/ Accessed July 17, 2019.
- 100. van Enkenvort D. Molgenis-Rdconnect-Report. Github; 2018. https://github.com/djvanenckevort/molgenis-rdconnect-report Accessed June 26, 2019.
- 101. Horaitis O, Talbot CC Jr, Phommarinh M, Phillips KM, Cotton R. A database of locus-specific databases. Nat Genet 2007; 39 (4): 425. [DOI] [PubMed] [Google Scholar]
- 102. Fokkema I, Taschner PEM, Schaafsma GCP, Celli J, Laros JFJ, den Dunnen JT. LOVD v.2.0: the next generation in gene variant databases. Hum Mutat 2011; 32 (5): 557–63. [DOI] [PubMed] [Google Scholar]
- 103. MacArthur J, Bowler E, Cerezo M, et al. The new NHGRI-EBI Catalog of published genome-wide association studies (GWAS Catalog). Nucleic Acids Res 2017; 45 (D1): D896–D901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.NCBI Resource Coordinators. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res 2016; 44: D7–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Landrum MJ, Lee JM, Benson M, et al. ClinVar: public archive of interpretations of clinically relevant variants. Nucleic Acids Res 2016; 44 (D1): D862–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Buniello A, MacArthur JAL, Cerezo M, et al. The NHGRI-EBI GWAS Catalog of published genome-wide association studies, targeted arrays and summary statistics 2019. Nucleic Acids Res 2019; 47 (D1): D1005–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Wg OT. Mondo Disease Ontology. http://obofoundry.org/ontology/mondo.html Accessed January 30, 2020.
- 108. Mungall CJ, Koehler S, Robinson P, Holmes I, Haendel M. k-BOOM: a Bayesian approach to ontology structure inference, with applications in disease ontology construction. bioRxiv 2019: 048843. https://www.biorxiv.org/content/10.1101/048843v3 Accessed December 14, 2019.
- 109. Wright CF, FitzPatrick DR, Firth HV. Paediatric genomics: diagnosing rare disease in children. Nat Rev Genet 2018; 19 (5): 325–325. [DOI] [PubMed] [Google Scholar]
- 110. Philippakis AA, Azzariti DR, Beltran S, Brookes AJ, et al. The Matchmaker Exchange: a platform for rare disease gene discovery. Hum Mutat 2015; 36 (10): 915–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.phenopacket-schema. Github. https://github.com/phenopackets/phenopacket-schema Accessed December 24, 2019.
- 112. Southall NT, Natarajan M, Lau LPL, et al. ; on behalf of the IRDiRC Data Mining and Repurposing Task Force. The use or generation of biomedical data and existing medicines to discover and establish new treatments for patients with rare diseases—recommendations of the IRDiRC Data Mining and Repurposing Task Force. Orphanet J Rare Dis 2019; 14 (1): 225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Robinson PN, Köhler S, Oellrich A, Wang K, Mungall CJ, et al. ; Sanger Mouse Genetics Project. Improved exome prioritization of disease genes through cross-species phenotype comparison. Genome Res 2014; 24 (2): 340–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Exomiser. Github. https://github.com/exomiser/Exomiser Accessed December 23, 2019.
- 115. Taruscio D, Gainotti S, Mollo E, et al. The current situation and needs of rare disease registries in Europe. Public Health Genomics 2013; 16 (6): 288–98. [DOI] [PubMed] [Google Scholar]
- 116.European Medicines Agency. European Medicines Agency; 2020. https://www.ema.europa.eu/en?cookies=disabled Accessed May 20, 2020.
- 117. Alonso-Ferreira V, Sánchez-Díaz G, Villaverde-Hueso A, Posada de la Paz M, Bermejo-Sánchez E. A Nationwide Registry-based study on mortality due to rare congenital anomalies. Int J Environ Res Public Health 2018; 15 (8): 1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Mazzucato M, Visonà Dalla Pozza L, Minichiello C, et al. The epidemiology of transition into adulthood of rare diseases patients: results from a population-based registry. Int J Environ Res Public Health 2018; 15 (10): 2212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Jansen-van der Weide MC, Gaasterland CMW, Roes KCB, et al. Rare disease registries: potential applications towards impact on development of new drug treatments. Orphanet J Rare Dis 2018; 13 (1): 154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Taruscio D, Mollo E, Gainotti S, Posada de la Paz M, Bianchi F, Vittozzi L. The EPIRARE proposal of a set of indicators and common data elements for the European platform for rare disease registration. Arch Public Health 2014; 72 (1): 35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Weldring T, Smith S. Patient-reported outcomes (PROs) and patient-reported outcome measures (PROMs). Health Serv Insights 2013; 6: 61–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Maaroufi M, Landais P, Messiaen C, Jaulent M-C, Choquet R. Federating patients identities: the case of rare diseases. Orphanet J Rare Dis 2018; 13 (1): 199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Coi A, Santoro M, Villaverde-Hueso A, et al. The quality of rare disease registries: evaluation and characterization. Public Health Genomics 2016; 19 (2): 108–15. [DOI] [PubMed] [Google Scholar]
- 124. Ambinder EP. Electronic health records. JOP 2005; 1 (2): 57–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Timotijevic L, Barnett J, Brown K, Raats MM, Shepherd R. Scientific decision-making and stakeholder consultations: the case of salt recommendations. Soc Sci Med 2013; 85: 79–86. [DOI] [PubMed] [Google Scholar]
- 126.Registry & Biobank Finder for Registries—RD-Connect. https://rd-connect.eu/what-we-do/phenotypic-data/rb-finder-for-registries/ Accessed December 24, 2019.
- 127. Santanello N, Largent J, Myers E, Smalley JB. Engaging Patients as Partners throughout the Registry Life Cycle. Rockville, MD: Agency for Healthcare Research and Quality (US); 2018. [Google Scholar]
- 128. Brasil S, Pascoal C, Francisco R, Dos Reis Ferreira V, Videira PA, Valadão AG. Artificial intelligence (AI) in rare diseases: is the future brighter? Genes 2019; 10 (12): 978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Hsieh T-C, Mensah MA, Pantel JT, et al. PEDIA: prioritization of exome data by image analysis. Genet Med 2019; 21 (12): 2807–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Gurovich Y, Hanani Y, Bar O, et al. Identifying facial phenotypes of genetic disorders using deep learning. Nat Med 2019; 25 (1): 60–4. [DOI] [PubMed] [Google Scholar]
- 131. Kline AD, Moss JF, Selicorni A, et al. Diagnosis and management of Cornelia de Lange syndrome: first international consensus statement. Nat Rev Genet 2018; 19 (10): 649–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Hammond P, Suttie M, Hennekam RC, Allanson J, Shore EM, Kaplan FS. The face signature of fibrodysplasia ossificans progressiva. Am J Med Genet A 2012; 158A (6): 1368–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Kung S, Walters M, Claes P, Goldblatt J, Le Souef P, Baynam G. A dysmorphometric analysis to investigate facial phenotypic signatures as a foundation for non-invasive monitoring of lysosomal storage disorders. JIMD Rep 2013; 8: 31–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Hu H, Haas SA, Chelly J, et al. X-exome sequencing of 405 unresolved families identifies seven novel intellectual disability genes. Mol Psychiatry 2016; 21 (1): 133–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Baynam GS, Walters M, Dawkins H, Bellgard M, Halbert AR, Claes P. Objective monitoring of mTOR inhibitor therapy by three-dimensional facial analysis. Twin Res Hum Genet 2013; 16 (4): 840–4. [DOI] [PubMed] [Google Scholar]
- 136. Kung S, Walters M, Claes P, et al. Monitoring of therapy for mucopolysaccharidosis type I using dysmorphometric facial phenotypic signatures. JIMD Rep 2015; 22: 99–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Cliniface. Cliniface. Cliniface; 2019. https://cliniface.org/ Accessed July 23, 2019.
- 138.Face2Gene. Face2Gene. Face2Gene; 2019. https://www.face2gene.com/ Accessed July 23, 2019.
- 139.FaceBase. FaceBase. FaceBase; 2019. https://www.facebase.org/ Accessed July 23, 2019.
- 140. Ramesh AN, Kambhampati C, Monson JRT, Drew PJ. Artificial intelligence in medicine. Ann R Coll Surg Engl 2004; 86 (5): 334–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Feng S, Liu S, Zhu C, Gong M, Zhu Y, Zhang S. National rare diseases registry system of china and related cohort studies: vision and roadmap. Hum Gene Ther 2018; 29 (2): 128–35. [DOI] [PubMed] [Google Scholar]
- 142.Patient Archive. Patient Archive. Patient Archive; 2019. http://patientarchive.org/#/home Accessed July 23, 2019.
- 143. Nellåker C, Alkuraya F, Baynam G, et al. ; Minerva Consortium. Enabling global clinical collaborations on identifiable patient data: the Minerva Initiative. Front Genet 2019; 10: 611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Austin CP, Cutillo CM, Lau LPL, et al. ; on behalf of the International Rare Diseases Research Consortium (IRDiRC). Future of rare diseases research 2017-2027: an IRDiRC perspective. Clin Transl Sci 2018; 11 (1): 21–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Minerva & Me. Minerva and Me—Help Rare Disease Research. Minerva and Me; 2019. https://www.minervaandme.com/ Accessed July 23, 2019 [Google Scholar]
- 146. Claes P, Liberton DK, Daniels K, et al. Modeling 3D facial shape from DNA. PLoS Genet 2014; 10 (3): e1004224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Gahl WA. The battlefield of rare diseases: where uncommon insights are common. Sci Transl Med 2012; 4 (154): 154ed7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148. Strode SW, Gustke S, Allen A. Technical and clinical progress in telemedicine. JAMA 1999; 281 (12): 1066–8. [DOI] [PubMed] [Google Scholar]
- 149.HIMSS. HIMSS Analytics. HIMSS Analytics; 2019. https://www.himssanalytics.org/ Accessed July 17, 2019
- 150. Edworthy SM. Telemedicine in developing countries. BMJ 2001; 323 (7312): 524–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Augustine EF, Dorsey ER, Saltonstall PL. The care continuum: an evolving model for care and research in rare diseases. Pediatrics 2017; 140 (3): e20170108. [DOI] [PubMed] [Google Scholar]
- 152. Darkins A, Sanders JH. Remote patient monitoring in home healthcare: lessons learned from advanced users. J Manage Market Healthcare 2009; 2 (3): 238–52. [Google Scholar]
- 153. Hinsch N, Rauofi R, Stauch G. Benign cystic mesothelioma of the peritoneum in a 12-year-old boy, diagnosed via telepathology. BMJ Case Rep 2015; 2015: bcr2015211419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154. Siegert CJ, Fisichella PM, Moseley JM, Shoni M, Lebenthal A. Open access phone triage for veterans with suspected malignant pleural mesothelioma. J Surg Res 2017; 207: 108–14. [DOI] [PubMed] [Google Scholar]
- 155. Tagliente I, Trieste L, Solvoll T, Murgia F, Bella S. Telemonitoring in cystic fibrosis: a 4-year assessment and simulation for the next 6 years. Interact J Med Res 2016; 5 (2): e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156. Sun C, Sun L, Xi S, et al. Mobile phone-based telemedicine practice in older Chinese patients with type 2 diabetes mellitus: randomized controlled trial. JMIR Mhealth Uhealth 2019; 7 (1): e10664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157. Sasso FC, Pafundi PC, Gelso A, et al. ; on behalf of NO BLIND Study Group. Telemedicine for screening diabetic retinopathy: the NO BLIND Italian multicenter study. Diabetes Metab Res Rev 2018; 35: e3113. [DOI] [PubMed] [Google Scholar]
- 158. Vujosevic S, Pucci P, Casciano M, et al. A decade-long telemedicine screening program for diabetic retinopathy in the north-east of Italy. J Diabetes Complications 2017; 31 (8): 1348–53. [DOI] [PubMed] [Google Scholar]
- 159. Ting DSW, Tan G. Telemedicine for diabetic retinopathy screening. JAMA Ophthalmol 2017; 135 (7): 722–3. [DOI] [PubMed] [Google Scholar]
- 160. Duis J, van Wattum PJ, Scheimann A, et al. A multidisciplinary approach to the clinical management of Prader-Willi syndrome. Mol Genet Genomic Med 2019; 7 (3): e514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161. Strickler AS, Palma J, Charris R, et al. Contribution of the use of basic telemedicine tools to the care of children and adolescents with juvenile idiopathic arthritis at the Puerto Montt Hospital, Chile. Rev Chil Pediatr 2018; 89 (1): 59–66. [DOI] [PubMed] [Google Scholar]
- 162. Bashshur RL, Shannon GW, Smith BR, et al. The empirical foundations of telemedicine interventions for chronic disease management. Telemed J E Health 2014; 20 (9): 769–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163. Baker DB, Kaye J, Terry SF. Governance through privacy, fairness, and respect for individuals. eGEMs 2016; 4 (2): 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164. Fecher B, Friesike S. Open Science: one term, five schools of thought In: Bartling S, Friesike S, eds. Opening Science: The Evolving Guide on How the Internet is Changing Research, Collaboration and Scholarly Publishing. Cham: Springer International Publishing; 2014: 17–47. [Google Scholar]
- 165. Leonelli S. Locating ethics in data science: responsibility and accountability in global and distributed knowledge production systems. Philos Trans A Math Phys Eng Sci 2016; 374: 20160122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Science International. Open Data in a Big Data World. 2015. https://council.science/publications/open-data-in-a-big-data-world Accessed December 10, 2019.
- 167.European Union. Open innovation, open science, open to the world: a vision for Europe. Moedas C, ed. Luxembourg: Publications Office of the European Union; 2015. [Google Scholar]
- 168.National Academies of Sciences, Engineering, and Medicine, Policy and Global Affairs, Board on Research Data and Information, Committee on Toward an Open Science Enterprise. In: Open Science by Design: Realizing a Vision for 21st Century Research Washington, DC: National Academies Press; 2018. [PubMed]
- 169.EU. A Code of Conduct for Health Research. EU General Data Protection Regulation; 2018. http://code-of-conduct-for-health-research.eu/.
- 170.National Institutes of Health. Final NIH Genomic Data Sharing Policy. Federal Register; 2014: 51345–54. https://www.federalregister.gov/d/2014-20385.
- 171. Kass N. On the Ethics of Open Science. Sage Bionetworks; 2019. https://sagebionetworks.org/in-the-news/on-the-ethics-of-open-science-2/ Accessed July 23, 2019.
- 172.International Indigenous Data Sovereignty IG. RDA; 2017. https://www.rd-alliance.org/groups/international-indigenous-data-sovereignty-ig Accessed December 30, 2019.