Abstract
Leaves are the major organ for photosynthesis in most land plants, and leaf structure is optimized for the maximum capture of sunlight and gas exchange. Three polarity axes, the adaxial–abaxial axis, the proximal-distal axis, and the medial-lateral axis are established during leaf development to give rise to a flattened lamina with a large area for photosynthesis and blades that are extended on petioles for maximum sunlight. Adaxial cells are elongated, tightly packed cells with many chloroplasts, and their fate is specified by HD-ZIP III and related factors. Abaxial cells are rounder and loosely packed cells and their fate is established and maintained by YABBY family and KANADI family proteins. The activities of adaxial and abaxial regulators are coordinated by ASYMMETRIC LEAVES2 and auxin. Establishment of the proximodistal axis involves the BTB/POZ domain proteins BLADE-ON-PETIOLE1 and 2, whereas homeobox genes PRESSED FLOWER and WUSCHEL-RELATED HOMEOBOX1 mediate leaf development along the mediolateral axis. This review summarizes recent advances in leaf polarity establishment with a focus on the regulatory networks involved.
Keywords: adaxial–abaxial polarity, proximodistal polarity, mediolateral polarity, leaf, lamina outgrowth
Introduction
Leaves are the major organ for photosynthesis and are specified for maximum capture of sunlight and minimum loss of water. Leaf morphology and development differ in the two main groups of angiosperms: parallel veined monocots and reticulate veined eudicots (Conklin et al., 2018). Proper leaf morphology is essential for light interception, transpiration rates, carbon fixation, photosynthesis, and crop yield and is established through numerous genetic pathways (Xu et al., 2018). In monocots, such as rice and maize, leaves form as strap-shaped structures with a sheathing leaf base around the shoot apical meristem (SAM), whereas eudicot leaves are initiated as bumps around the periphery of the SAM and ultimately form a flat structure composed of a lamina and petiole (Conklin et al., 2018; Xu et al., 2018). Arabidopsis thaliana (Arabidopsis) will be used here as representative of dicot leaf development (Christenhusz and Byng, 2016). Leaves are generated from a group of stem cells located at the tip of the shoot called the SAM. Approximately 500 cells constitute the Arabidopsis SAM and are divided into three functional and cytohistological zones: the central zone, the peripheral zone, and the rib zone (reviewed in Dodsworth, 2009; Barton, 2010; Du et al., 2018). The number of cells in the central zone is relatively constant, and cell division in this zone gives rise not only to stem cells that maintain the central zone but also to daughter cells that become incorporated in the peripheral zone. Cell division in the peripheral zone gives rise to lateral organs, including leaves and flowers. The rib zone cells are underneath the central zone and peripheral zones. After leaf primordia are initiated from the flanks of the peripheral zone, three polarities, the adaxial–abaxial polarity, the proximal–distal polarity, and the medial–lateral polarity, are established (Figure 1A). Compared to the deep understanding of the mechanisms of adaxial–abaxial polarity establishment, much less is known about how proximal–distal polarity and medial–lateral polarities are established. Several previous reviews have focused on adaxial–abaxial polarity (Byrne, 2006; Szakonyi et al., 2010; Liu et al., 2012; Du et al., 2018). Here, we focus on gene regulatory network regulating the establishment of all three axes during leaf development with an emphasis on recent advances in order to provide a basis for future studies.
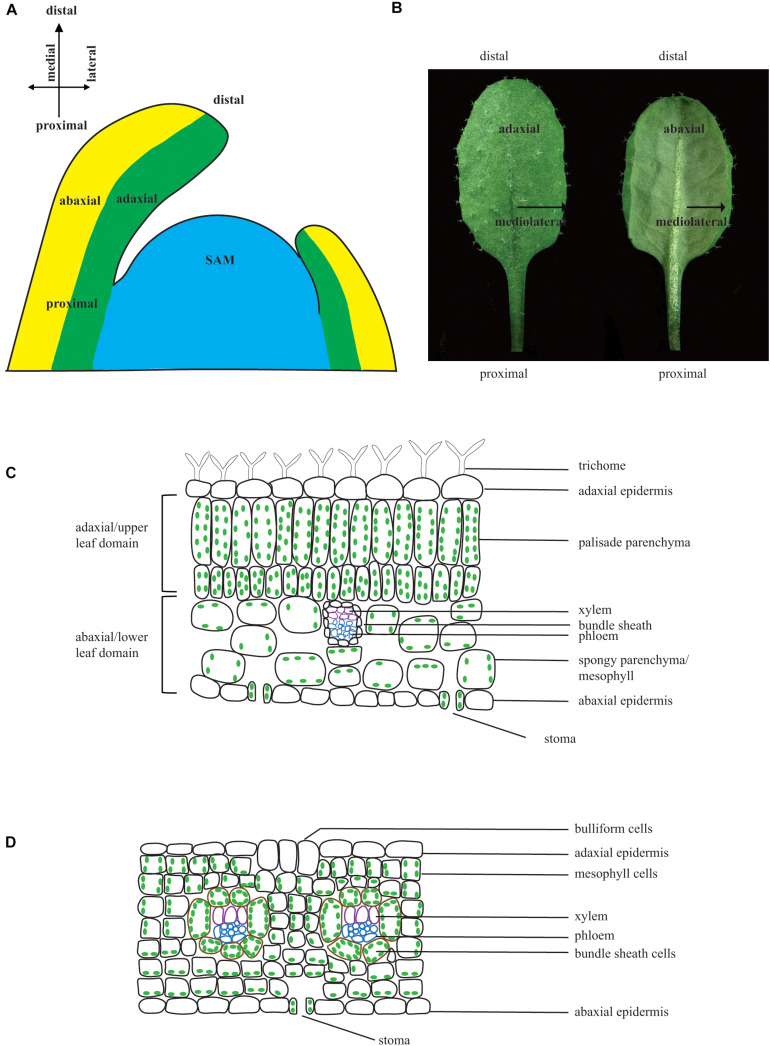
FIGURE 1.
Leaf polarities. (A) Schematic diagram of a longitudinal section of a SAM showing the adaxial–abaxial domains, proximal–distal domains, and medial-lateral domains. (B) Arabidopsis leaf with adaxial side (left) and abaxial side (right). (C) Schematic diagram of cross section through a typical dicot C3 plant leaf. A leaf is enclosed by the upper (adaxial) epidermis and lower (abaxial) epidermis. The upper epidermis has trichomes. The upper mesophyll cells are tightly organized and enriched in chloroplast, whereas the lower mesophyll cells are loosely organized with fewer chloroplasts. Vascular bundles exhibit polarity with adaxially localized xylem and abaxially localized phloem. Stoma are abundant on the lower epidermis and trichome may or may not be present on the lower epidermis. (D) Schematic diagram of cross section through a monocot C4 plant leaf. Bulliform cells are present on the adaxial surface, and bundle sheath cells enclosing the vascular bundle are large and have chloroplasts.
Establishment of Adaxial–Abaxial Polarity
Adaxial–abaxial polarity is comparable to dorsiventral polarity in animals. When leaf primordia are initiated and separated from the SAM, the portion that is closest to the SAM is the adaxial domain, whereas the portion that is away from the SAM is the abaxial domain. Initially, leaf primordia fold over the SAM and later bend away from the SAM because of faster growth on the adaxial domain than the abaxial domain. As a result, the adaxial side becomes the upper side of the leaf, and the abaxial side becomes the lower side of the leaf (Figure 1B). As leaves are normally flattened during lamina outgrowth for maximum sunlight capture, the abaxial and adaxial side cells become morphologically different. In Arabidopsis leaves, cells on the adaxial (upper) domain are relatively elongated and tightly compacted, with many chloroplasts to optimize light capture and photosynthesis. Cells on the abaxial (bottom) domain are rounder and have large air spaces between them, with many stomata on this side to promote gas exchange (Figure 1C). Trichomes are always present on the adaxial surface of true leaves; however, they are not present on the abaxial surface until the plant is in the adult phase (Eshed et al., 2001; Kerstetter et al., 2001; Poethig, 2003; Liu et al., 2012). The leaf anatomy in the monocot C4 plant maize differs from that of Arabidopsis, as it uses different mechanisms for photosynthesis. Cells on the adaxial and abaxial domain are similar in size, and there are some bulliform cells on the adaxial surface, which may play a role in leaf rolling in response to water deficit stress (Xu et al., 2018). The bundle sheath cells in C3 plants are normally similar in size to the vascular cells and do not contain chloroplast, but the bundle sheath cells in C4 plants are larger than the vascular cells and normally contain chloroplast such that they can photosynthesize (Figure 1D). Although the morphological difference between adaxial and abaxial cells becomes apparent at later stages of leaf development, the expression of adaxial–abaxial identity genes is observed in globular-stage cotyledons and leaf primordia, suggesting that the establishment of adaxial–abaxial polarity occurs very early in leaf development (Eshed et al., 2001; Kerstetter et al., 2001; Emery et al., 2003). Molecular and genetic studies have shown that adaxial identity is established and maintained by a group of plant-specific homeodomain/leucine zipper (HD-ZIP) transcription factors, whereas abaxial identity is established and maintained by KANADI (KAN) family (GARP domain transcription factors) and YABBY (YAB) family transcription factors. Other factors are involved in the establishment of adaxial–abaxial identity by regulating or interacting with the above three groups of proteins (Figure 2A) (Talbert et al., 1995; Eshed et al., 2001, 2004; Kerstetter et al., 2001; McConnell et al., 2001; Emery et al., 2003; Prigge et al., 2005; Wenkel et al., 2007).
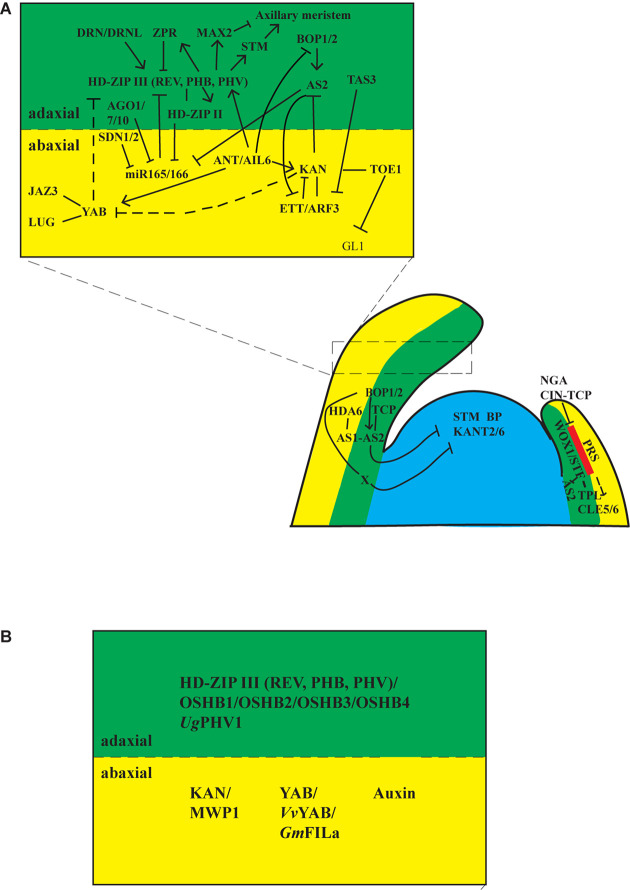
FIGURE 2.
Regulatory networks for establishing leaf polarities. (A) Gene regulatory networks in Arabidopsis. Upper, Gene regulatory networks for adaxial/abaxial polarity. Adaxial identity is established by HD-ZIP III transcription factors, whereas the abaxial identity is established by KAN and YAB transcription factors. The activities of HD-ZIP III are repressed by ZRP proteins and miR165/166. HD-ZIP III proteins activate and interact with HD-ZIP II transcription factors to repress expression of miR165/166; thus, a feedback loop is formed between miR165/166-HD-ZIP III-HD-ZIP II. miR165/166 are processed and sequestered by AGO1, AGL7, AGO10, and other miRNA processing proteins. SDN1 and SDN2 proteins mediate the degradation of miR165/166. A second feedback loop functions between AS2-KAN-ETT. ETT is repressed by TAS3, ETT physically interacts with KAN to repress AS2 and KAN, and AS2 represses ETT. AS2 also represses miR165/166. Thus, AS2 is a hub between these two feedback loops. KAN interacts directly with TOE1 to form a repressive loop at the GL1 locus to suppress abaxial trichome formation in juvenile leaves. DRN and DRNL bind to HD-ZIPIII proteins and assist them in activating STM and MAX2 to activate or suppress axillary meristem formation on the adaxial side of leaves. Lower, Gene regulatory networks for proximodistal and mediolateral polarities. Proximodistal polarity established by BOP1/2 and AS1/2 complexes. BOP1/2 proteins activate AS2 directly, and AS2 physically interacts with AS1 and TCP transcription factors. AS1 associates with HDA6 to restrict class I KNOX genes (STM, BP, KNAT2, and KNAT6) in the SAM to suppress meristematic activities in leaves. BOP1/2 proteins function through AS2-dependent and AS2-independent pathways to repress class I KNOX genes. WOX1 and PRS are expressed at the boundary between adaxial and abaxial domains. WOX1 and PRS are restricted from the tip of the leaf by NGS and CIN-TCP transcription factors. TPL physically interacts with STF (WOX1) to repress AS2 and promote lateral expansion of the lamina. CLE5 and CLE6 act downstream of WOX1 and PRS. Red area indicates the expression domain of WOX1 and PRS. Filled lines represent direct interaction and dashed lines indicate indirect or genetic interaction. (B) Conserved gene regulatory network among plants. Adaxial polarity is established by HD-ZIP III (REV, PHB, PHV) transcription factors. Orthologs of the HD-ZIP III transcription factors, OSHB1/2/3/4 in rice, and UgPHV1 in Utricularia have similar roles as HD-ZIP III transcription factors in Arabidopsis. Abaxial polarity is established by YAB, KAN, and auxin pathways in Arabidopsis. The KAN1 ortholog MWP1 in maize, the YAB ortholog VvYAB in grapevine, and GmFILa in soybean have conserved roles as the YAB proteins in Arabidopsis. How auxin is involved in establishing adaxial–abaxial polarity is complicated. Nevertheless, it is required for polarity establishment in Arabidopsis and Chinese cabbage.
Establishment of Adaxial Identity by HD-ZIP III and Related Factors
HD-ZIP III Transcription Factors Specify Adaxial Identity
The first protein that was reported to mediate adaxial–abaxial polarity was an MYB domain transcription factor PHANTASTICA (PHAN) in Antirrhinum majus. Mutations in PHAN converted flat leaves into needle-like structures (Waites and Hudson, 1995). In Arabidopsis, plant-specific class III HD-ZIP family proteins, including PHABULOSA (PHB)/ATHB14, PHAVOLULA (PHV)/ATHB9, REVOLUTA (REV)/ATHB8, and INCURVATA4/CORONA/ATHB15, establish adaxial identity (Emery et al., 2003; Prigge et al., 2005). Expression analysis showed that PHB, PHV, and REV are restricted to the adaxial domain of developing leaf primordia (Eshed et al., 2001; McConnell et al., 2001; Otsuga et al., 2001; Emery et al., 2003). Semidominant gain-of-function phb, phv, and rev mutants showed loss of abaxial identity and adaxialization of leaves (McConnell et al., 2001; Emery et al., 2003). Although recessive single loss-of-function mutants in these genes do not cause obvious defects in adaxial identity, the phb phv rev triple mutant exhibits severe phenotypes that include needle-like leaves without blades and absence of the SAM (Emery et al., 2003). Genetic studies showed that ATHB8 and ATHB15 act partially redundantly with PHB, PHV, and REV (Prigge et al., 2005). Specification of adaxial polarity is not only important for the generation of flat leaves for maximizing sunlight capture, but it is also important for the development of axillary meristems, which arise from the adaxial side of leaves. Consistent with this, axillary meristem activity was absent in the phb phv rev triple mutant (Emery et al., 2003; Prigge et al., 2005). The protein MORE AXILLARY BRANCHES2 (MAX2) is required for the production and perception of the strigolactone signal that suppresses axillary meristems. REV and MAX2 were expressed in overlapping regions in leaf veins and petioles, and REV activates MAX2 directly by binding to the MAX2 promoter (Brandt et al., 2012; Hong et al., 2020). Consistent with MAX2 acting downstream of REV, radialization of vascular elements in rev10D-dominant mutants was suppressed by max2 (Hong et al., 2020). In addition, REV directly activates SHOOT MERISTEMLESS (STM) in leaf axils to promote axillary meristem formation (Shi et al., 2016). This regulation appears to be promoted by DORNROSCHEN (DRN) and DORNROSCHEN-LIKE (DRNL), two AP2/ERF transcription factors that physically interact with REV and bind to a region near the transcriptional start site of STM (Zhang et al., 2018). The interaction of DRN/DRNL with REV promotes REV binding to STM regulatory regions (Shi et al., 2016; Zhang et al., 2018). The AP2 domain of DRN/DRNL interacts with the PAS domain of REV (Zhang et al., 2018). LITTLE ZIPPER3 (ZPR3) destabilizes the DRN/DRNL-REV interaction by competing for binding to REV (Zhang et al., 2018). Together, molecular and genetic studies have demonstrated that plant-specific class III HD-ZIP proteins establish adaxial identity and activate STM expression to promote axillary meristem formation (Figure 2A).
Regulation of HD-ZIP IIIs by miR165/166
Defects in adaxial/abaxial identity are also observed in plants lacking ARGONAUTE1 (AGO1), a key member of the AGO family that binds with regulatory small RNAs to form the RNA induced silencing complex (reviewed in Chen, 2009; Wang J. et al., 2019). Mutations in AGO1 result in trumpet-like or radialized leaves, mimicking semidominant phb-d or phv-d leaves. miR165/166 are expressed in the abaxial domain of the leaf, complementary to that of PHB and PHV. Overexpression of a PHB construct containing sequence changes that reduce the complementary of the mRNA to miR165/166 but do not alter the amino acid sequence resulted in a phb-d phenotype (Mallory et al., 2004). Together, these lines of evidence indicate that PHB and PHV are directly regulated by miR165/166. miR165/166 also functions non-cell-autonomously to repress HD-ZIP III transcription factors. MIR165A, MIR166A, and MIR166B are expressed in the abaxial epidermis of leaves (Yao et al., 2009). Mature miR165 is detected in the abaxial epidermis and one to two cell layers underneath the epidermis (Kidner and Martienssen, 2004), suggesting that miR165 and possibly also miR166 migrate to neighboring cells and create an miR165/166 gradient. Consistent with this, confocal analysis on PHB-GFP protein showed that it is not evenly distributed in the adaxial domain of leaves (Miyashima et al., 2011; Tatematsu et al., 2015). A pair of artificially designed versions of miR165 and PHB were used to analyze their interaction. A mutated form of PHB (PHBm, which does not respond to endogenous miR165/166) was expressed in most of the leaf primordia. Introducing a mutated form of miR165 (miR165mu) complementary to PHBm gradually repressed the PHBm protein in the abaxial cells of leaves, indicating non-cell-autonomous function of endogenous miR165/166 (Miyashima et al., 2011; Tatematsu et al., 2015).
Proteins involved in transcriptional regulation of miR165/166, such as ASYMMETRIC LEAVES1 (AS1) and AS2, or in biogenesis of miR165/miR166, such as HASTY (HST), AGO1, AGO7, AGO10, and RNA-dependent RNA polymerase 6 (RDR 6), also function in modification of adaxial cell fate in leaves (Bollman et al., 2003; Li et al., 2005; Xu et al., 2006; Liu et al., 2009; Zhu et al., 2011). AGO10 is expressed at the adaxial side of leaf primordia and in the provasculature underneath the SAM to restrict the expression of miR165/166 to the abaxial side of leaves and to maintain stem cell fate in the SAM (Lynn et al., 1999). AGO10 has higher binding affinity to miR165/166 than AGO1 (Zhu et al., 2011), such that levels of miR165/166 are maintained to certain levels to restrict the expression of HD-ZIPIII to the adaxial side. The balance of miR165/166 is also maintained by the degradation activities of SMALL RNA DEGRADING NUCLEASE (SDN1) and SDN2 (Yu et al., 2017). AGO10 associated miR165/166 is more susceptible to SDN1 and SDN2 mediated trimming. The trimmed and unmethylated miR165/166 are then tailed by the nucleotidyl-transferase HESO1 (Yu et al., 2017). AGO7 is also expressed on the adaxial side of leaf, and it acts synergistically with AS2 in promoting adaxial cell fate (Xu et al., 2006; Hoyer et al., 2019). Yeast-one-hybrid analysis showed that TEOSINTE BRANCHED/CYCLOIDEA/PCF (TCP) transcription factors bind to the “TGGTCC” motif of AGO7, and SQUAMOSA PROMOTER BINDING PROTEIN-LIKE (SPL) binds to the “GTCA” motif of AGO7. However, truncated AGO7 promoter activity analysis showed that the SPL binding site is not required for the polar localization of AGO7. Additionally, the SPL and TCP binding sites are not strictly required for the function of AGO7 in leaves (Hoyer et al., 2019). It will be interesting to know what controls polar localization of AGO7 and AGO10 to understand how HD-ZIPIII transcription factors are regulated.
HD-ZIP III-ZPR Feedback Loop
The DNA-binding activities of HD-ZIP transcription factors require homodimerization through their leucine zipper domain (Tron et al., 2001, 2004, 2005). A group of LITTLE ZIPPER (ZPR) proteins ZPR1, ZPR2, ZPR3, and ZPR4 have a leucine zipper domain similar to that of HD-ZIP III. Transcripts of ZPR are localized to the adaxial portion of leaf primordia, like the HD-ZIP III transcription factors but at much lower levels. Plants ectopically expressing ZPR3 had rod-like and downward curling leaves, indicating abaxialization. This could be explained by the activities of the ZPR proteins, in that the zipper domain of the ZPR proteins forms heterodimers with HD-ZIP III to prevent the homodimerization of HD-ZIP III proteins. Thus, the activities of HD-ZIP III proteins are reduced in 35S:ZPR3 (Wenkel et al., 2007; Kim et al., 2008; Magnani and Barton, 2011). ZPR3 and ZPR4 are induced by dexamethasone in the dexamethasone-inducible REV fusion plant, and ZPRs were also identified as direct targets of REV, thus forming a feedback loop that modulates adaxial identity protein activity (Figure 2A) (Wenkel et al., 2007; Kim et al., 2008).
HD-ZIP II Transcription Factors
Like class III HD-ZIP genes, class II HD-ZIP genes HOMEOBOX ARABIDOPSIS THALIANA 2 (HAT2), HAT3, and ARABIDOPSIS THALIANA HOMEOBOX 4 (ATHB4) are expressed in the adaxial domain of leaf primordia (Brandt et al., 2012; Turchi et al., 2013). HD-ZIP II genes are regulated by a red/far-red light ratio that induces the shade avoidance response (Ruberti et al., 2012). These proteins are directly activated by REV and physically interact with REV to directly repress miR165/166, thus maintaining distinct adaxial and abaxial domains (Figure 2A) (Bou-Torrent et al., 2012; Brandt et al., 2012; Turchi et al., 2013; Merelo et al., 2016).
Establishment of Abaxial Identity by KANADI and YABBY Proteins
KANADI Proteins
The flattened leaf lamina results from the parallel development of adaxial and abaxial cells. Abaxial identity is established by members of two gene families, the KANADIs and the YABBYs (Eshed et al., 2001, 2004; Emery et al., 2003; Kerstetter et al., 2001; Prigge et al., 2005; Izhaki and Bowman, 2007). The KAN family shares a common GARP domain of MYB transcription factors, and three members of this family, KAN1, KAN2, and KAN3, are expressed in the abaxial portion of leaf primordia (Kerstetter et al., 2001; Eshed et al., 2004). None of the kan1, kan2, kan3, and kan4 single mutants exhibited a dramatic loss of abaxial polarity phenotype; however, the leaves of the kan1/2 double mutant are narrow and have ectopic outgrowths on their abaxial side. The kan1/2/3 triple mutant looked like the kan1/2 double mutant, whereas kan1/2/4 and kan1/2/3/4 mutants produced radialized rosette leaves or radialized leaf-like structures on the hypocotyl (Eshed et al., 2001, 2004; Izhaki and Bowman, 2007). Overexpression of KAN1, KAN2, or KAN3 resulted in radialized leaves without blades, and the plants lacked an SAM (Eshed et al., 2001, 2004; Emery et al., 2003; Kerstetter et al., 2001; Izhaki and Bowman, 2007). Thus, KAN1, KAN2, KAN3, and KAN4 function redundantly in specifying leaf abaxial identity.
KAN1 is an abaxial identity gene, but the kan1 mutant was first identified in screens for juvenile-to-adult phase change mutants (Kerstetter et al., 2001). Trichomes are polarly located on the adaxial side of juvenile leaves, whereas trichomes are present on both sides of adult leaves. The production of trichomes on the abaxial side of leaves is a hallmark for adult leaves and is promoted by a group of miR156-targeted SPL transcription factors (Wu et al., 2009). SPL9 and SPL15 directly activate the expression of miR172b, a precursor for mature miR172 (Wu et al., 2009; Hyun et al., 2016). Mutations in miR172-targeted AP2 genes, such as target of eat1 (toe1) and toe2, slightly accelerate abaxial trichome production (Wu et al., 2009), whereas the toe1 toe2, toe1 toe2 ap2, and the toes toe2 smz snz mutants are gradually more accelerated in abaxial trichome production (Wang L. et al., 2019). Abaxial trichome production is accelerated in kan1 (Kerstetter et al., 2001; Wang L. et al., 2019), and KAN1 is found to associate with TOE1 in yeast two-hybrid screens. The N-terminal domain of KAN1/2 binds to miR172-targeted AP2 proteins, whereas KAN3/4 do not bind to them. ChIP and EMSA assays showed that both KAN1 and TOE1 bind to the 3′ end of a trichome initiation gene GLABRA1 (GL1), and mutations in this KAN1/TOE1-binding site resulted in precocious abaxial trichome production (Wang L. et al., 2019; Xu et al., 2019). Since KAN1 and TOE1 bind to the 3′ end of GL1, how do they repress the transcription of GL1? A chromosome conformation capture (3C) assay showed that the 3′ end of GL1 was brought to its 5′ end by the KAN1–TOE1 complex. In a 3C assay, chromatin is crossed linked, digested with restriction enzymes, and ligated. The ligation product is then subjected to quantitative polymerase chain reactions by primers spanning the 5′ end and 3′ end to give 100- to 150-bp amplicons. Truncation of the 3′ end significantly reduced the amount of 5′–3′ amplicons, whereas insertion of an 8-kb fragment between the 5′ end and 3′ end did not interfere with the activity of KAN1–TOE1 complex, indicating that cis-elements at the 3′ end are required for the formation of the KAN1–TOE1 repressing loop and cis-elements in the middle are not critical. Therefore, the KAN1–TOE1 complex is involved with the temporal and spatial production of abaxial trichomes. In juvenile leaves where miR156 levels are high, levels of TOE1 are also high, and TOE1 directly interacts with KAN1 on the abaxial side of the leaf to form a repressive loop at GLABRA1 (GL1). In adult leaves, when levels of miR156 are low, TOE1 activity is also low, which deactivates the KAN1–TOE1 repressive loop and derepresses GL1 (Figure 2A) (Wang L. et al., 2019; Xu et al., 2019).
YABBY Proteins
The YABBY gene family encodes HMG-like proteins, and members of this family in Arabidopsis are initially expressed in the whole leaf primordium and later restricted to the abaxial domain as the primordium elongates (Sawa et al., 1999; Siegfried et al., 1999). The fil/yab1, yab2, yab3, and yab4 single mutants do not have visible defects in leaf polarity; the yab1/3 double mutant, however, exhibits some loss of abaxial cell identity, while the lamina looks normal compared to wild type. The yab1/2/3/5 quadruple mutant shows more severe defects with fully radialized leaves, indicating functional redundancy among members of this gene family (Siegfried et al., 1999; Sarojam et al., 2010). The failure of leaf laminar outgrowth in loss-of-function yab mutants (yab1/2/3/5 and pANT > > miR-YAB1) is associated with loss of leaf margin cells and discontinuous vein patterning, which might be caused by lower auxin levels in their leaves (Goldshmidt et al., 2008; Sarojam et al., 2010). WUSCHEL (WUS), a gene required for SAM maintenance, is restricted to the SAM in WT but reactivated in pANT > > miRYAB1 leaf primordia, suggesting that YAB proteins suppress meristematic activity in leaf primordia to promote proper leaf development (Sarojam et al., 2010). Ectopic expression of YAB1 or YAB3 under the 35S promoter resulted in the abaxialization of leaves and loss of the SAM (Siegfried et al., 1999). Interestingly, the yab1/3/5 triple mutation suppresses the abaxial outgrowths seen on kan1/2/4 leaves. The leaves of the kan1/2/4 yab1/3/5 sextuple mutant are rod-like (Izhaki and Bowman, 2007), indicating a synergistic interaction between YAB and KAN families in establishing abaxial cell fate. Additionally, mutations in the auxin biosynthetic YUCCA (YUC) genes can also suppress the outgrowths in the kan1/2 double mutant, and the yuc1/2/4 triple mutation enhances the loss of abaxial identity in a kan1/2 background (Wang et al., 2011).
It is not known if the synergistic interaction between YAB family proteins and KAN family proteins is attributable to a physical YAB–KAN interaction. FIL/YAB1 is ectopically expressed in kan1/2 leaf outgrowths, suggesting that YAB might be repressed by KAN (Eshed et al., 2004). YAB proteins physically interact with the GRO-TUP1–like proteins LEUNIG (LUG) and LEUNIG-HOMOLOG (LUH) to repress PHB (Stahle et al., 2009). On the other hand, the activities of YAB proteins are attenuated by JASMONATE-ZIM DOMAIN protein JAZ3 (Boter et al., 2015). AINTEGUMENTA (ANT), which encodes an APETALA2/ETHYLENE RESPONSE FACTOR transcription factor, functions redundantly with three other AINTEGUMENTA-LIKE (AIL) family members, AIL5, AIL6, and AIL7, in floral organogenesis and leaf development (Krizek, 2009, 2015; Mudunkothge and Krizek, 2012). ANT, AIL5, and AIL6 were initially expressed throughout incipient leaf primordia (Long and Barton, 2000; Prasad et al., 2011). The ant ail6 ail7 triple mutant produces fewer rosette leaves, which are narrower than those in ant single mutants or ant ail6 double mutants (Mudunkothge and Krizek, 2012; Krizek, 2015). An ant mutation enhances defects in leaf polarity in fil and fil yab3 mutant backgrounds through synergistic regulation of PHB (Nole-Wilson and Krizek, 2006). RNA-seq analysis in whole inflorescence and ChIP-Seq analysis in stage 6/7 flowers revealed that ANT binds directly to the adaxial polarity genes PHB and BLADE-ON-PETIOLE1 (BOP1) and to the abaxial polarity genes KAN2 and YAB3 (Figure 2A) (Krizek et al., 2020), suggesting that ANT is involved in regulating organ polarity. Together, the studies of ANT and AIL proteins in Arabidopsis suggest that they have a role in establishing adaxial/abaxial polarity in lateral organs, but how they are involved in this process remains to be resolved.
AS2 Functions as a Hub for Juxtaposition of Adaxial–Abaxial Cell Fate
The adaxial and abaxial cell fates are specified by the interaction of distinct transcription factor families, with an LOB domain transcription factor ASYMMETRIC LEAVES2 (AS2) seeming to function as a hub (Figure 2A). AS2 is expressed in the adaxial domain of leaves (Iwakawa et al., 2007; Wu et al., 2008; Jun et al., 2010; Husbands et al., 2015) and directly represses miR166 to restrict expression of miR166 to the abaxial domain, resulting in the expression of HD ZIP III genes on the adaxial domain (Eshed et al., 2001; McConnell et al., 2001; Otsuga et al., 2001; Emery et al., 2003; Wu et al., 2008; Jun et al., 2010; Husbands et al., 2015). KAN1, which is expressed on the abaxial domain, represses AS2 transcription directly to restrict its expression to the adaxial domain (Wu et al., 2008). AS2 also functions synergistically with RDR6 in adaxial patterning (Li et al., 2005). RDR6 is involved in trans-acting siRNA (TAS1–TAS4) biogenesis (Yoshikawa et al., 2005; Chen, 2009). TAS3 is expressed in the adaxial domain of leaves and represses the expression of the AUXIN RESPONSE FACTORS (ARFs) ETTIN (ETT, also termed ARF3) and ARF4 (Fahlgren et al., 2006; Garcia et al., 2006; Hunter et al., 2006). ARF3 is expressed in the abaxial domain and physically interacts with KAN proteins to facilitate KAN activity (Pekker et al., 2005; Garcia et al., 2006; Kelley et al., 2012). AS2 represses ETT directly, and AS2 activity is required for the DNA methylation of ETT on its sixth exon (Iwasaki et al., 2013). Two nucleolus localized proteins, NUCLEOLINI and RNA HELICASE10 are associated with the AS2-dependent DNA methylation of ETT (Vial-Pradel et al., 2018). It is also reported that replication factor C subunit 3 and DNA polymerase subunit INCURVATA2 function as modifiers of AS2 in repressing ETT and ARF4 (Luong et al., 2018). Therefore, a feedback loop among ETT-KAN-AS2 acts to spatially define adaxial–abaxial cell fates.
Other factors, such as AS1, TCP, and BLADE-ON-PETIOLE 1 and 2 (BOP1/2) transcription factors, and histone acetylase/deacetylase epigenetic factors also interact with AS2 directly or indirectly to specify adaxial–abaxial polarity. AS1 (the ortholog of PHAN in Arabidopsis) and miR319-targted TCP transcription factors physically interact with AS2 in adaxial–abaxial patterning (Xu et al., 2003; Guo et al., 2008; Li et al., 2012). The BTB/POZ domain protein BOP1, however, binds to the AS2 promoter directly to activate AS2 expression (Ha et al., 2007; Jun et al., 2010). At least three lines of evidence indicated that AS1/AS2 interacts with epigenetic factors in establishing adaxial cell fate. First, treating an as2 mutant with trichostatin A (a histone deacetylase inhibitor) resulted in radialized leaves, indicating enhanced loss of adaxial polarity (Ueno et al., 2007). Second, AS1 interacted with histone deacetylase HDA6 in vitro and in vivo, and the AS1–HDA complex was recruited to repress the KNOX I genes in leaf patterning (Luo et al., 2012). Third, a histone acetylase ELONGATE was reported to function redundantly with AS1 and AS2 in establishing adaxial cell fate (Kojima et al., 2011).
Mutant screens in as2 backgrounds led to the discovery of ribosomal proteins (r-proteins), whose mutation enhances leaf abaxial polarity in as1 or as2 backgrounds (Horiguchi et al., 2011). AS2 is localized to the nucleus and forms AS2 bodies around the peripheral region of the nucleolus. Truncation analysis on AS2 motifs revealed that ICG and LZL regions are required for the nuclear localization of AS2. The zinc-finger DNA-binding motif of AS2 is strongly conserved among AS2/LOB protein family, and the sequences within and close to this motif are essential for the formation of AS2 bodies, which in turn is essential for the function of AS2 (Luo et al., 2020). Florescence in situ hybridization analysis showed that AS2 bodies are close to 45S rDNA, and they both are close to chromocenters. Therefore, the synergistic interaction between AS2 and ribosomal proteins might be caused by overlapping actions of AS2 bodies and ribosomal DNA in the chromocenter, which might be important for the epigenetic repression of its targets (Luo et al., 2020). Other two ribosome-related mutants, oligocellula2 (oli2) and g-patch domain protein (gdp1), were also found in the sensitized as2 mutant screens. OLI2 and GDP1 function synergistically to promote cell proliferation, and mutations in them caused narrow leaves. However, they do not strongly enhance the polarity defect of as2 (Kojima et al., 2018), indicating that leaf polarity induced by r-proteins function in a different pathway to OLI2 and GDP1.
Role of Auxin in Specifying Adaxial–Abaxial Identity
The plant hormone auxin is required for lateral organ initiation and many other aspects of plant development (Reinhardt et al., 2000; Benkova et al., 2003; Friml et al., 2003; Simonini et al., 2016, 2017, 2018). Several lines of evidence suggest that auxin is also involved in leaf patterning along the adaxial–abaxial axis. It was reported in 2007 that PIN-FORMED1 (PIN1), an auxin efflux carrier, and the auxin reporter DR5 are abnormally localized in kan1/2/4 and rev phb phv triple mutants (Izhaki and Bowman, 2007). DR5 and PIN1 were also abnormally distributed in the yab1/2/3/5 quadruple mutant in several organs, such as embryo, leaf primordium, and leaf vein, consistent with their roles in cotyledon development, leaf adaxial–abaxial differentiation, and leaf margin patterning (Sarojam et al., 2010). Auxin seems localized on the adaxial domain of leaf primordia as indicated by the distribution of DII (an auxin sensor) and PIN1 (Qi et al., 2014; Shi et al., 2017). However, other studies argue that auxin is not polarly localized because maximum auxin levels are found between the REV and KAN1 domains, and quantitative analysis on the auxin sensor R2D2 showed that there is no polar localization of R2D2 in lateral organ primordia, although there is transient asymmetric distribution of mDII (Caggiano et al., 2017; Bhatia et al., 2019). How auxin could be involved in lateral organ development (including leaf development) has been discussed by Heisler and Byrne (2020), and whether auxin is asymmetrically distributed in leaf primordia remains to be resolved. REV directly activates two auxin biosynthesis genes, TAA1 and YUC5 (Brandt et al., 2012). KAN1, a transcriptional repressor, regulates biosynthesis, transport, and signaling of auxin. The auxin response factor ARF3/ETT physically interacts with KAN proteins on the abaxial domain of leaves (Kelley et al., 2012; Huang et al., 2014). RNA-seq analysis of cells sorted by different fluorescence markers (pREV:REV-2YPET and pKAN1:KAN1–2GFP) revealed that auxin-related genes are regulated by both REV and KAN1. REV activates auxin-related genes initially but represses them at a later time point. KAN1 functions antagonistically to REV in regulating a common group of targets (Ram et al., 2020). Auxin acts through binding to TIR1/AFB F-box proteins, leading to the release of Aux/IAA from ARFs. There are 23 ARFs in the Arabidopsis genome. Among them, ARF3, ARF4, and ARF5 seem to have roles in adaxial–abaxial patterning (Pekker et al., 2005; Garcia et al., 2006; Guan et al., 2017). ARF3/ETT is atypical as it does not interact with Aux/IAA (Simonini et al., 2016). Sequence analysis of ETT combined with yeast two hybrid assays showed that a 27-kDa ETT variant ETT_388-594, encompassing the entire predicted ETT-specific domain, contains auxin responsive elements (Simonini et al., 2018). RNA-seq analysis on ett mutants and ChIP-seq analysis on ETT-GFP transgenic plants revealed that ETT regulates a wide range of genes, which include ethylene, cytokinin, and auxin biosynthesis, response, and signaling genes. ETT also binds to a bHLH proteins that interact with ETT. Among the polarity genes, KAN3 is a direct target of ETT and is upregulated in ett (Simonini et al., 2017). KAN proteins also interact with ETT physically, and their interactions have a role in ovule development (Kelley et al., 2012). The RNA-seq, ChIP-seq, and genetic interactions between ETT and KAN were analyzed in reproductive tissues; therefore, mechanisms of how auxin is involved in adaxial–abaxial cell fate determination in leaves remain to be investigated by molecular and genetic studies.
Adaxial–Abaxial Polarity in Plants Other Than Arabidopsis
Studies in species other than Arabidopsis showed that the mechanisms for establishment and maintenance of adaxial–abaxial polarity are conserved among some plants. The HD-ZIP III transcription factors in rice are encoded by OSHB genes. Similar to HD-ZIP III genes in Arabidopsis, OSHB1/OSHB2/OSHB3/OSHB4 are expressed in the adaxial domain of rice leaves (Itoh et al., 2008). Ectopic expression of these OSHBm genes resulted in rod-like leaf structures, similar to the semidominant rev, phv, and phb mutations in Arabidopsis (Itoh et al., 2008). A calpain-like cysteine protease ADAXIALIZED LEAF 1 (ADL1) is required for the maintenance rather than establishment of adaxial polarity as the expression of OSHB is increased in mature adl1 leaves, but not in immature adl1 leaves. As a result, the adl1 leaf is adaxialized (Hibara et al., 2009). Rolling of leaves in rice indicates loss of adaxial–abaxial identity. Several proteins in rice, including HD-ZIP IV family member Roc8, a KAN ortholog SHALLOT-LIKE1 (SLL1), and a CHD3/Mi-2 chromatin remodeling factor ROLLED FINE STRIPED (RFS), have been discovered to play essential roles in controlling rolling of leaves in rice (Zhang et al., 2009; Cho et al., 2018; Sun et al., 2020). The milkweed pod1 (mwp1) gene encodes a KAN ortholog in maize. Molecular and genetic analysis of mwp1 showed that it is required for maintenance of the abaxial identity of the maize leaf (Candela et al., 2008). Evolutionary studies on KAN family genes in ferns and lycophytes revealed that KAN family genes are expressed in the abaxial domain in ferns but not in lycophytes, suggesting that KAN family genes are conserved within ferns and some angiosperms, but not in lycophytes (Zumajo-Cardona et al., 2019). Similar results were also observed for the YAB gene family in that the grapevine VvYABBY gene and the soybean YAB family gene GmFILa are expressed in the abaxial domain of leaves, and overexpression of those genes in tomato or Arabidopsis resulted in abaxialization of leaves (Xiang et al., 2013; Yang et al., 2019; Zhang S. et al., 2019). Together, these studies suggest conserved roles of KAN and YAB genes in some angiosperms (Figure 2B).
Chinese cabbage (Brassica rapa ssp. pekinensis) produces flattened leaves early in the rosette stage, and the leaves become curved inward and folded at the heading stage, indicating adaxialization of leaves. External application of auxin to Chinese cabbage and RNA-seq analysis on the heading mutant flat growth-1 (fg-1) revealed that auxin plays an essential role in establishing adaxial–abaxial polarity (heading) in Chinese cabbage (He et al., 2000; Li et al., 2019), indicating a conserved role of auxin in establishing adaxial–abaxial polarity. Although the molecular nature of FG is still unknown, six adaxial identity genes BrREV, BrHB8.1, BrHB8.2, BrHB9, BrHB14.1, and BrHB14.2 and six abaxial identity genes BrKAN1, BrBOP2, BrYAB1.1, BrYAB1.2, BrYAB2, and BrYAB3 were differently expressed in fg-1 mutants, suggesting that heading of Chinese cabbage is caused by ectopic expression of adaxial/abaxial polarity genes.
Utricularia gibba produces both planar and non-planar (needle-like or cup-shaped) leaves. Planar leaves are formed when the expression of UgPHV1 is restricted at the adaxial domain, and needle-like leaves are formed when UgPHV1 is ectopically expressed, similar to the role of PHV in Arabidopsis (Whitewoods et al., 2020). Computational modeling on U. gibba planar leaves and non-planar leaves, together with molecular and genetic studies, revealed that UgPHV1 patterns the leaf via regulating growth rate along orthoplanar (adaxial–abaxial), proximodistal, and perpendicular to both (mediolateral) axes. Planar leaves are formed when growth rates along the orthoplanar and perpendicular axes are higher than the growth rate along the proximodistal axis. Non-planar leaves are formed when the growth rate along the orthoplanar axis is higher than the growth rates along the proximodistal and perpendicular axes (Whitewoods et al., 2020). These results suggest that adaxial–abaxial polarity is formed by restricting growth rate along the proximodistal axis and that the adaxial identity gene PHV1 has a role in restricting growth along the proximodistal axis. This work indicates that polarities are the results of oriented growth, rather than the cause.
Establishment of Proximal-Distal Polarity
Proximal–distal polarity is established as the primordium grows away from the SAM. The domain close to the SAM is the proximal part, which gives rise to the petiole. The domain farthest from the SAM is the distal part, which gives rise to the blade. The best-known genes that control proximal–distal polarity are BOP1/2, as the bop1 bop2 double mutant exhibits blade growth on the petiole, diminishing the petiole domain (proximal domain). BOP1/2 genes are expressed at organ boundaries and have a wide range of roles in Arabidopsis development, including leaf patterning, floral organ patterning, floral meristem identity, floral organ abscission, and pedicel patterning (Hepworth et al., 2005; Norberg et al., 2005; Ha et al., 2007; Jun et al., 2010). BOP1/2 genes are expressed at the proximal and adaxial side of leaf primordia, and their activities in the adaxial domain activate the expression of another adaxial gene AS2 (Ha et al., 2007; Jun et al., 2010). AS2 is expressed throughout initiating leaf primordia but becomes localized to the adaxial L1 layer in older leaf primordia (Iwakawa et al., 2007; Wu et al., 2008; Jun et al., 2010). Mutations in AS2 resulted in an upward curling petiole and sometimes a small leaflet on the petiole (Lin et al., 2003; Xu et al., 2003; Iwakawa et al., 2007). Both the bop1 bop2 and as2 leaf phenotypes are associated with ectopic expression of meristem class I homeobox KNOX genes, including SHOOT MERISTEMLESS (STM), BREVIPEDICELLUS (BP), KNAT2, and KNAT6 (Semiarti et al., 2001; Byrne et al., 2002; Iwakawa et al., 2007; Ikezaki et al., 2010; Jun et al., 2010). The AS1–AS2 complex directly represses class I KNOX genes to suppress ectopic growth on the petiole (Guo et al., 2008). This is supported by genetic analysis showing that mutations in class I KNOX genes suppress the ectopic growth on bop1 bop2 and as2 petioles (Ha et al., 2010; Ikezaki et al., 2010). BOP1/2 proteins not only directly activate AS2 but also function redundantly with AS1 and AS2 to pattern the proximodistal axis, as growth on bop1 bop2 as1 and bop1 bop2 as2 petioles is much more dramatic than on the bop1 bop2 double mutant or on as1 or as2 single mutants (Figure 2A) (Ha et al., 2007).
Leaves of maize and rice do not have petioles. Instead, they have a sheath that wraps around the shoot in place of a petiole. Loss-of-function studies in OsBOP1/2/3 mutants revealed that these genes promote the growth of the distal blade. Leaf 1 of wild-type cultivated rice has only a sheath, whereas leaf 1 of the osbop triple mutant produces blades without a sheath. Overexpression of OsBOP1 in rice does not affect the growth of the sheath; however, the length of the blade is shorter (Toriba et al., 2019). Thus, the sheath to blade ratio is affected by loss-of-function and gain-of-function mutations in OsBOP. Rhizomes are modified stems (without any blade) growing underground. Rhizomes of Oryza longistaminata have much higher levels of BOP1 than the leaves above ground, suggesting that BOP1 suppresses blade growth in rhizomes (Toriba et al., 2020). The growth of the sheath is also correlated with high levels of miR156, and OsSPL14 represses the expression of BOP1/2/3 genes (Toriba et al., 2019), suggesting the role of miR156-SPL in establishing the proximal–distal polarity.
Establishment of Medial-Lateral Polarity
The blade of a leaf expands laterally to increase its surface area for maximum capacity of photosynthesis. Two WUSCHEL-RELATED HOMEOBOX (WOX) genes, WOX1 and PRESSED FLOWER (PRS) (also known as WOX3), were found to be involved in establishing mediolateral polarity. PRS encodes a homeodomain protein that shares 68% identity with WUS. WOX1, PRS, and the PRS ortholog MAEWEST (MAW) in Petunia are expressed at the boundary between adaxial–abaxial domains at the P2 stage and at the leaf margin at later stages. The wox1 prs double mutant and maw single mutant have narrower lamina than wild type, as well as an abnormal abaxial cell fate in lamina (Matsumoto and Okada, 2001; Vandenbussche et al., 2009; Nakata et al., 2012; Caggiano et al., 2017). Domain localization of expression of the maize orthologs of PRS, NARROW SHEATH1 (NS1) and NS2 is similar to that of PRS, and leaves of a ns1 ns2 double mutant are much narrower than wild type (Nardmann et al., 2004). The WOX1 orthologs in tobacco (LAMINA1), petunia (MAEWEST), Medicago truncatula (STENOFOLIA, STF), and barley (NARROW LEAFED DWARF1) have conserved roles in promoting lamina expansion along the mediolateral axis (Vandenbussche et al., 2009; Tadege et al., 2011a,b; Yoshikawa et al., 2016). In M. truncatula, STF recruits TOPLESS (TPL) and requires the activity of TPL to promote cell proliferation at the adaxial–abaxial boundary (Zhang et al., 2014). ANGUSTIFOLIA3 (AN3) physically interacts with LEUNIG (LUG), functioning redundantly with WOX1 and PRS to promote lamina lateral growth (Zhang F. et al., 2019). Two CLAVATA3/ESR-RELATED peptide ligand gene family members, CLE5 and CLE6, were activated by WOX1 and PRS. However, the cle5 and cle6 single or double mutants did not show any defect in leaf development, probably due to the redundancy within the CLE family (DiGennaro et al., 2018). In most plants, the lamina margin is determinant and will not give rise to any new structures. The meristem activity in leaves is restricted by several NGATHA (NGA) and CINCINNATA-class-TCP (CIN-TCP) transcription factors, which act redundantly to restrict the activities of PRS in the leaf margin (Alvarez et al., 2016). Together, studies in several species suggest that leaf development along the mediolateral axis is promoted by PRS and WOX1. NGA and CIN-TCP transcription factors restrict the expression of PRS, whereas CLE5 and CLE6 are activated by PRS and WOX1. TPL, AN3, and LUG act redundantly with PRS and WOX1 to restrict the expression of AS2 at lateral ends to promote the lateral growth of lamina (Figure 2A).
Future Perspectives
Genetic studies in Arabidopsis showed that plants that cannot specify an adaxial fate, such as the phb phv rev triple mutant and 35S:KAN1 ectopic expression construct, or plants with loss of both adaxial and abaxial identities, such as the kan1/kan2/kan4/phb/phv/rev sextuple mutant, do not produce SAMs, suggesting that specification of polarity might be a prerequisite for SAM formation (Talbert et al., 1995; Kerstetter et al., 2001; Otsuga et al., 2001; Emery et al., 2003). However, none of these polarity genes are expressed in the central zone of the SAM, which is essential for maintenance of the SAM. How these adaxial–abaxial identity genes affect SAM formation is uncertain. One hypothesis is that a mobile signal moves from leaf primordia to the SAM and regulates the transcription or activity of meristematic genes. WUS is expressed in a small group of central cells in and underneath the L3 layer of the SAM, and it is required for maintenance of the SAM (Mayer et al., 1998). Although YAB1 does not move, the expression of WUS is expanded in yab1/3 double mutants, indicating that downstream targets of YAB might be mobile and mediate the change of WUS expression (Goldshmidt et al., 2008). But what these downstream mobile targets are and how they maintain the SAM remain unknown. In 1954, Sussex proposed that signals from the SAM determine adaxial–abaxial polarity, suggesting a feedback loop between SAM identity genes and leaf adaxial–abaxial identity genes. Auxin and a lipophilic signal were proposed to be candidates for the “Sussex signal,” but it remains to be determined (Kuhlemeier and Timmermans, 2016). Moreover, the establishment of adaxial–abaxial polarity, proximodistal polarity, and mediolateral polarity seems to be coordinated rather than independent of each other. BOP1/2 are involved in both adaxial–abaxial polarity and proximal–distal polarity, and the leaves of a phb phv rev triple mutant are rod-like without blade outgrowth, indicating the loss of adaxial polarity, distal polarity, and lateral polarity at the same time. Ectopic expression of ARF5/MP resulted in loss of normal adaxial–abaxial and mediolateral development at the same time (Guan et al., 2017; Shi et al., 2017). Mechanisms of how these three polarities (adaxial–abaxial, proximal–distal, and mediolateral) are coestablished remain to be discovered. Computational modeling on U. gibba indicated that leaf shape variation (sheet-like or needle like) is correlated with different growth rates along the growth axes. A combination of computational modeling with molecular and genetic approaches could enable us to uncover how polarity regulators promote or restrict growth rates in certain areas that eventually shape leaves along the polarity axes. Studies on the development of unifacial leaves suggest strikingly different mechanisms in leaf patterning. In bifacial leaves, loss of adaxial identity resulted in rod-like leaves, whereas in unifacial leaves, loss of adaxial identity resulted in either cylindrical leaves (Juncus wallichianus) or flattened leaves (Juncus prismatocarpus). Molecular studies showed that DROOPING LEAF (DL), a YABBY paralog, is expressed in the middle of flattened leaves (J. prismatocarpus), like the expression of PRS in Arabidopsis. However, a similar expression pattern of DL was not observed in cylindrical leaves (J. wallichianus). Consistently, PRS is detected in the flattened leaf margin while not in the cylindrical leaf margin (Yamaguchi et al., 2010). Further analysis on cell division direction revealed that there is more thickening cell division on the adaxial domain than on the abaxial domain, which gives rise to flattened leaves like bifacial leaves (Yin and Tsukaya, 2019). Further investigation on unifacial leaf development may provide new insight into how leaves are flattened. Compared to the mechanisms in adaxial–abaxial polarity, much less is known about leaf development along the proximodistal polarity and/or mediolateral polarity axes, due to the lack of mutants. Other strategies, such as conditional screening of mutants combined with RNA-seq or studying plant species that have natural variation in proximodistal polarity and/or mediolateral polarity, may lead us to uncover new or redundant genetic pathways that pattern the leaf along the three polarity axes. Leaf shape diversity sometimes is associated with developmental phase transitions and stress responses (Wang et al., 2009; Wu et al., 2009; Kumar and Wigge, 2010; Xu et al., 2016; Leichty and Poethig, 2019). Understanding the mechanisms in leaf shape patterning may provide vital insight into developmental phase transitions and environmental plasticity.
Author Contributions
MX wrote the first draft of the manuscript and drew the figures. Both authors revised the manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Dr. Beth Krizek and Dr. Gail Pruss for critical reading of the manuscript.
Footnotes
Funding. This work was supported by the National Science Foundation (NSF) grant IOS 1947274.
References
- Alvarez J. P., Furumizu C., Efroni I., Eshed Y., Bowman J. L. (2016). Active suppression of a leaf meristem orchestrates determinate leaf growth. Elife 5:e15023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton M. K. (2010). Twenty years on: the inner workings of the shoot apical meristem, a developmental dynamo. Dev. Biol. 341 95–113. 10.1016/j.ydbio.2009.11.029 [DOI] [PubMed] [Google Scholar]
- Benkova E., Michniewicz M., Sauer M., Teichmann T., Seifertova D., Jurgens G., et al. (2003). Local, efflux-dependent auxin gradients as a common module for plant organ formation. Cell 115 591–602. 10.1016/s0092-8674(03)00924-3 [DOI] [PubMed] [Google Scholar]
- Bhatia N., Ahl H., Jönsson H., Heisler M. G. (2019). Quantitative analysis of auxin sensing in leaf primordia argues against proposed role in regulating leaf dorsoventrality. Elife 8:e39298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bollman K. M., Aukerman M. J., Park M. Y., Hunter C., Berardini T. Z., Poethig R. S. (2003). HASTY, the Arabidopsis ortholog of exportin 5/MSN5, regulates phase change and morphogenesis. Development 130 1493–1504. 10.1242/dev.00362 [DOI] [PubMed] [Google Scholar]
- Boter M., Golz J. F., Gimenez-Ibanez S., Fernandez-Barbero G., Franco-Zorrilla J. M., Solano R. (2015). FILAMENTOUS FLOWER is a direct target of JAZ3 and modulates response to Jasmonate. Plant Cell 27 3160–3174. 10.1105/tpc.15.00220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bou-Torrent J., Salla-Martret M., Brandt R., Musielak T., Palauqui J. C., Martinez-Garcia J. F., et al. (2012). ATHB4 and HAT3, two class II HD_ZIP transcription factors, control leaf development in Arabidopsis. Plant Signal Behav. 7 1382–1387. 10.4161/psb.21824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandt R., Salla-Martret M., Bou-Torrent J., Musielak T., Stahl M., Lnz C., et al. (2012). Genome-wide binding-site analysis of REVOLUTA reveals a link between leaf patterning and light-mediated growth response. Plant J. 72 31–42. 10.1111/j.1365-313x.2012.05049.x [DOI] [PubMed] [Google Scholar]
- Byrne M. E. (2006). Shoot meristem function and leaf polarity: the role of class III HD-ZIP genes. PLoS Genet. 2:e89. 10.1371/journal.pgen.0020089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrne M. E., Simorowski J., Martienssen R. A. (2002). ASYMMETRIC LEAVES1 reveals knox gene redundancy in Arabidopsis. Development 129 1957–1965. [DOI] [PubMed] [Google Scholar]
- Caggiano M. P., Yu X., Bhatia N., et al. (2017). Cell type boundaries organize plant development. Elife 6:e27421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Candela H., Johnston R., Gerhold A., Foster T., Hake S. (2008). The milkweed pod1 gene encodes a KANADI protein that is required for abaxial/adaxial patterning in maize leaves. Plant Cell 20 2073–2087. 10.1105/tpc.108.059709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X. (2009). Small RNAs and their roles in plant development. Annu. Rev. Cell Dev. Biol. 25 21–44. 10.1146/annurev.cellbio.042308.113417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho S.-H., Lee C.-H., Gi E., Yim Y., Koh H.-J., Kang K., et al. (2018). The Rice Rolled Fine Striped (RFS) CHD3/Mi-2 chromatin remodeling factor epigenetically regulates genes involved in oxidative stress responses during leaf development. Front. Plant Sci. 9:364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christenhusz M. J., Byng J. W. (2016). The number of known plants species in the world and its annual increase. Phytotaxa 261:201. 10.11646/phytotaxa.261.3.1 24943631 [DOI] [Google Scholar]
- Conklin P. A., Strable J., Li S., Scanlon M. J. (2018). On the mechanisms of development in monocot and eudicot leaves. New Phytologist. 221 706–724. 10.1111/nph.15371 [DOI] [PubMed] [Google Scholar]
- DiGennaro P., Grienenberger E., Dao T. Q., Jun J. H., Fletcher J. C. (2018). Peptide signaling molecules CLE5 and CLE6 affect Arabidopsis leaf shape downstream of leaf patterning transcription factors and auxin. Plant Direct. 2:e00103. 10.1002/pld3.103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodsworth S. (2009). A diverse and intricate signaling network regulates stem cell fate in the shoot apical meristem. Dev. Biol. 336 1–9. 10.1016/j.ydbio.2009.09.031 [DOI] [PubMed] [Google Scholar]
- Du F., Guan C., Jiao Y. (2018). Molecular mechanisms of leaf morphogenesis. Mol. Plant 11 1117–1134. 10.1016/j.molp.2018.06.006 [DOI] [PubMed] [Google Scholar]
- Emery J. F., Floyd S. K., Alvarez J., Shed Y., Hawker N. P., Izhaki A., et al. (2003). Radial patterning of Arabidopsis shoots by Class III HD-ZIP and KANADI genes. Curr. Biol. 13 1768–1774. 10.1016/j.cub.2003.09.035 [DOI] [PubMed] [Google Scholar]
- Eshed Y., Baum S., Bowman J. (1999). Distinct mechanisms promote polarity establishment in carpels of Arabidopsis. Cell 99 199–209. 10.1016/s0092-8674(00)81651-7 [DOI] [PubMed] [Google Scholar]
- Eshed Y., Baum S. F., Perea J. V., Bowman J. L. (2001). Establishment of polarity in lateral organs of plants. Curr. Biol 11 1251–1260. 10.1016/s0960-9822(01)00392-x [DOI] [PubMed] [Google Scholar]
- Eshed Y., Izhaki A., Baum S. F., Floyd S. K., Bowman J. L. (2004). Asymmetric leaf development and blade expansion in Arabidopsis mediated by KANADI and YABBY activities. Development 137 767–774. [DOI] [PubMed] [Google Scholar]
- Fahlgren N., Montgomery T. A., Howell M. D., Allen E., Dvorak S. K., Alexander A. L., et al. (2006). Regulation of AUXIN RESPONSE FACTOR by TAS3 ta-siRNA affects developmental timing and patterning in Arabidopsis. Curr. Biol. 16 939–944. 10.1016/j.cub.2006.03.065 [DOI] [PubMed] [Google Scholar]
- Fletcher J. C. (2018). The CLV-WUS Stem cell signaling pathway: a road map to crop yield optimization. Plants (Basel) 7:87. 10.3390/plants7040087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friml J., Vieten A., Sauer M., Weijers D., Schwarz H., Hamann T., et al. (2003). Efflux-dependent auxin gradients establish the apical-basal axis of Arabidopsis. Nature 426 147–153. 10.1038/nature02085 [DOI] [PubMed] [Google Scholar]
- Garcia D., Collier S. A., Byrne M. E., Martienssen R. A. (2006). Specification of leaf polarity in Arabidopsis via the trans-acting siRNA pathway. Curr. Biol. 16 933–938. 10.1016/j.cub.2006.03.064 [DOI] [PubMed] [Google Scholar]
- Goldshmidt A., Alvarez J. P., Bowman J. L., Eshed Y. (2008). Signals derived from YABBY gene activities in organ primordia regulate growth and partitioning of Arabidopsis shoot apical meristems. Plant Cell 20 1217–1230. 10.1105/tpc.107.057877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan C., Wu B., Yu T., Wang Q., Krogan N. T., Liu X., et al. (2017). Spatial auxin signaling controls leaf flattening in Arabidopsis. Curr. Biol. 27 2940–2950. 10.1016/j.cub.2017.08.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo M., Thomas J., Collins G., Timmermans M. C. P. (2008). Direct repression of KNOX1 loci by the ASYMMETRIC LEAVES1 complex of Arabidopsis. Plant Cell 20 48–58. 10.1105/tpc.107.056127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha C. M., Jun J. H., Fletcher J. C. (2010). Control of Arabidopsis leaf morphogenesis through regulationof the YABBY and KNOX families of transcription factors. Genetics 186 197–206. 10.1534/genetics.110.118703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha C. M., Jun J. H., Nam H. G., Fletcher J. C. (2007). BALDE-ON-PETIOLE 1 and 2 control Arabidopsis lateral organ fate through regulation of LOB domain and adaxial-abaxial polarity genes. Plant Cell 19 1809–1825. 10.1105/tpc.107.051938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han H., Krizek B. A. (2016). AINTEGUMENTA-LIKE6 can functionally replace AINTEGUMENTA but alters Arabidopsis flower development when misexpressed at high levels. Plant Mol. Biol. 92 597–612. 10.1007/s11103-016-0535-y [DOI] [PubMed] [Google Scholar]
- Hay A., Tsiantis M. (2006). The genetic basis for differences in leaf form between Arabidopsis thaliana and its wild relative Cardamine hirsuta. Nat. Genet. 38 942–947. 10.1038/ng1835 [DOI] [PubMed] [Google Scholar]
- Hay A., Tsiantis M. (2016). Cardamine hirsuta: a comparative view. Curr. Opin. Genet. Dev. 39 1–7. 10.1016/j.gde.2016.05.005 [DOI] [PubMed] [Google Scholar]
- He Y. K., Xue W. X., Sun Y. D., Yu X. H., Liu P. L. (2000). Leafy head formation of the progenies of transgenic plants of Chinese cabbage with exogenous auxin genes. Cell Res. 10 151–160. 10.1038/sj.cr.7290044 [DOI] [PubMed] [Google Scholar]
- Heisler M. G., Byrne M. E. (2020). Progress in understanding the role of auxin in lateral organ development in plants. Curr. Opin. Plant Biol. 53 73–79. 10.1016/j.pbi.2019.10.007 [DOI] [PubMed] [Google Scholar]
- Hepworth S. R., Zhang Y., McKim S., Li X., Haugh G. (2005). BLADE-ON-PETIOLE-dependent signaling controls leaf and floral patterning in Arabidopsis. Plant Cell 17 1434–1448. 10.1105/tpc.104.030536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hibara K., Obara M., Hayashida E., et al. (2009). The ADAXIALIZED LEAF1 gene functions in leaf and embryonic pattern formation in rice. Dev. Biol. 334 345–354. 10.1016/j.ydbio.2009.07.042 [DOI] [PubMed] [Google Scholar]
- Hong S. Y., Botterweg-Paredes E., Doll J., et al. (2020). Multi-level analysis of the interactions between REVOLUTA and MORE AXILLARY BRANCHES 2 in controlling plant development reveals parallel, independent and antagonistic functions. Development 147:dev183681. 10.1242/dev.183681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horiguchi G., Mola-Morales A., Perez-Perez J. M., Kojima K., Robles P., Ponce M. R., et al. (2011). Differential contributions of ribosomal protein genes to Arabidopsis thaliana leaf development. Plant J. 65 724–736. 10.1111/j.1365-313x.2010.04457.x [DOI] [PubMed] [Google Scholar]
- Hoyer J. S., Pruneda-Paz J. L., Breton G., Hassert M. A., Holcomb E. E., Fowler H., et al. (2019). Functional dissection of the ARGONAUTE7 promoter. Plant Direct 3:e00102. 10.1002/pld3.102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang T., Harrar Y., Lin C., Reinhart B., Newell N. R., Talavera-Rauh F., et al. (2014). Arabidopsis KANADI 1 acts as a transcriptional repressor by interacting with a specific cis-element and regulates auxin biosynthesis, transport, and signaling in opposition to HD-ZIP III factors. Plant Cell 26 246–262. 10.1105/tpc.113.111526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter C., Sun H., Poethig R. S. (2003). The Arabidopsis heterochronic gene ZIPPY is an ARGONAUTE family member. Curr. Biol. 13 1734–1739. 10.1016/j.cub.2003.09.004 [DOI] [PubMed] [Google Scholar]
- Hunter C., Willmann M. R., Wu G., Yoshikawa M., De La Luz Gutierreznava M., Poethig R. S. (2006). Trans-acting siRNA-mediated repression of ETTIN and ARF4 regulated heteroblasty in Arabidopsis. Development 133 2973–2981. 10.1242/dev.02491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Husbands A. Y., Enkovics A. H., Nogueira F. T., Lodha M., Timmermans M. C. (2015). The ASYMMETRIC LEAVES complex employs multiple modes of regulation to affect adaxial-abaxial patterning and leaf complexity. Plant Cell 27 3321–3335. 10.1105/tpc.15.00454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyun Y., Richter R., Vincent C., Martinez-Gallegos R., Porri A., Coupland G. (2016). Multi-layered regulation of SPL15 and cooperation with SOC1 integrate endogenous flowering pathways at the Arabidopsis shoot meristem. Dev Cell 37 254–266. 10.1016/j.devcel.2016.04.001 [DOI] [PubMed] [Google Scholar]
- Ikezaki M., Kojima M., Sakakibara H., Kojima S., Ueno Y., Machida C., et al. (2010). Genetic networks regulated by ASYMMETRIC LEAVES1 (AS1). and AS2 in leaf development in Arabidopsis thaliana: KNOX genes control five morphological events. Plant J. 61 70–82. 10.1111/j.1365-313x.2009.04033.x [DOI] [PubMed] [Google Scholar]
- Itoh J., Hibara K., Sato Y., Nagato Y. (2008). Developmental role and auxin responsiveness of class III homeodomain leucine zipper gene family members in rice. Plant Physiol. 147 1960–1975. 10.1104/pp.108.118679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwakawa H., Iwasaki M., Kojima S., Ueno Y., Soma T., Tanaka H., et al. (2007). Expression of the ASYMMETRIC LEAVES2 gene in the adaxial domain of Arabidopsis leaves represses cell proliferation in this domain and is critical for the development of properly expanded leaves. Plant J. 51 173–184. 10.1111/j.1365-313x.2007.03132.x [DOI] [PubMed] [Google Scholar]
- Iwasaki M., Takahashi H., Iwakawa H., Nakagawa A., Ishikawa T., Tanaka H., et al. (2013). Dural regulation of ETTIN (ARF3). gene expression by AS1-AS2, which maintains the DNA methylation level, is involved in stabilization of leaf adaxial-abaxial partitioning in Arabidopsis. Development 140 1958–1969. 10.1242/dev.085365 [DOI] [PubMed] [Google Scholar]
- Izhaki A., Alvares J. P., Cinnamon Y., Genin O., Liberman-Aloni R., Eyal Y. (2018). The tomato BLADE-ON-PETIOLE and TERMINAING FLOWER regulate leaf axil patterning along the proximal-distal axes. Front. Plant Sci. 9:1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izhaki A., Bowman J. L. (2007). KANDI and class III HD-Zip gene families regulate embryo patterning and modulate auxin flow during embryogenesis in Arabidopsis. Plant Cell 19 495–508. 10.1105/tpc.106.047472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jun J. H., Ha C. M., Fletcher J. C. (2010). BLADE-ON-PETIOLE1 coordinate organ determinacy and axial polarity in Arabidopsis by directly activating ASYMMETRIC LEAVES2. Plant Cell 22 62–76. 10.1105/tpc.109.070763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley D. R., Arreola A., Gallagher T. L., Gasser C. S. (2012). ETTIN (ARF3) physically interacts with KANADI proteins to form a function complex essential for integument development and polarity determination in Arabidopsis. Development 139 1105–1109. 10.1242/dev.067918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerstetter R. A., Bollman K., Taylor R. A., Bomblies K., Poethig R. S. (2001). KANADI regulates organ polarity in Arabidopsis. Nature 411 706–709. 10.1038/35079629 [DOI] [PubMed] [Google Scholar]
- Kidner C. A., Martienssen R. A. (2004). Spatially restricted microRNA directs leaf polarity through ARGONAUTE1. Nature 428 81–84. 10.1038/nature02366 [DOI] [PubMed] [Google Scholar]
- Kierzkowski D., Runions A., Vuolo F., et al. (2019). A growth-based framework for leaf shape development and diversity. Cell 177 1405–1418. 10.1016/j.cell.2019.05.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y. S., Kim S. G., Lee M., Lee I., Park H. Y., Seo P. J., et al. (2008). HD-ZIP III activity is modulated by competitive inhibitors via a feedback loop in Arabidopsis shoot apical meristem development. Plant Cell 20 920–933. 10.1105/tpc.107.057448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojima K., Tamura J., Chiba H., Fukada K., Tsukaya H., Horiguchi G. (2018). Two nucleolar proteins, GDP1 and OLI2, function as ribosome biogenesis factors and are preferentially involved in promotion of leaf cell proliferation without strongly affecting leaf adaxial-abaxial patterning in Arabidopsis thaliana. Front. Plant Sci. 8:2240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojima S., Iwasaki M., Rakahashi H., Imai T., Matsumura Y., Fleury D., et al. (2011). Asymmetric leaves2 and Elongator, a histone acetyltransferase complex, mediate the establishment of polarity in leaves of Arabidopsis thaliana. Plant Cell Physiol. 52 1259–1273. 10.1093/pcp/pcr083 [DOI] [PubMed] [Google Scholar]
- Krizek B. A. (2009). AINTEGUMENTA and AINTEGUMENTA-LIKE6 act redundantly to regulate Arabidopsis floral growth and patterning. Plant Physiol. 150 1916–1929. 10.1104/pp.109.141119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krizek B. A. (2015). AINTEGUMENTA-LIKE genes have partly overlapping functions with AINTEGUMENTA but make distinct contributions to Arabidopsis thaliana flower development. J. Exp. Bot. 66 4537–4549. 10.1093/jxb/erv224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krizek B. A., Blakley I. C., Ho Y. Y., Freese N., Loraine A. E. (2020). The Arabidopsis transcription factor AINTEGUMENTA orchestrates patterning genes and auxin signaling in the establishment of floral growth and form. Plant J. [Epub ahead of print]. 10.1111/tpj.14769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhlemeier C., Timmermans M. C. (2016). The Sussex signal: insights into leaf dorsiventrality. Development 143 3230–3237. 10.1242/dev.131888 [DOI] [PubMed] [Google Scholar]
- Kumar S. V., Wigge P. A. (2010). H2A.Z-containing nucleosomes mediate the thermosensory response in Arabidopsis. Cell 140 136–147. 10.1016/j.cell.2009.11.006 [DOI] [PubMed] [Google Scholar]
- Leichty A. R., Poethig R. S. (2019). Development and evolution of age-dependent defense in ant-acacias. Proc. Natl. Acad. Sci. U.S.A. 116 15596–15601. 10.1073/pnas.1900644116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Wang H., Yuan Z., Cao Z., Yang Z., Xu Y., et al. (2005). The putative RNA-dependent polymerase RDR6 acts synergistically with ASYMMETRIC LEAVES1 and 2 to repress BREVIPEDICELLUS and Micro165/166 in Arabidopsis leaf development. Plant Cell 17 2157–2171. 10.1105/tpc.105.033449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Zhang X., Lu Y., Feng D., Gu A., Wang S., et al. (2019). Characterization of non-heading mutation in heading chinese cabbage (Brassica rapa L. ssp. pekinensis). Front. Plant Sci. 10:112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Li B., Shen W. H., Huang H., Dong A. (2012). TCP transcription factors interact with AS2 in the repression of class I KNOX genes in Arabidopsis thaliana. Plant J. 71 99–107. 10.1111/j.1365-313x.2012.04973.x [DOI] [PubMed] [Google Scholar]
- Lin W. C., Shuai B., Springer P. S. (2003). The Arabidopsis LATERAL ORGAN BOUNDARIES-domain gene ASYMMETRIC LEAVES2 functions in the repression of KNOX gene expression and in adaxial-abaxial patterning. Plant Cell 15 2241–2252. 10.1105/tpc.014969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q., Yao X., Pi L., Wang H., Cui X., Huang H. (2009). The ARGONAUTE10 gene modulates shoot apical meristem maintenance and establishment of leaf polarity by repressing miR165/166 in Arabidopsis. Plant J. 58 27–40. 10.1111/j.1365-313x.2008.03757.x [DOI] [PubMed] [Google Scholar]
- Liu T., Reinhart B. J., Manani E., Huang T., Kerstetter R., Barton M. K. (2012). Of blades and branches: understanding and expanding the Arabidopsis Ad/Abaxial regulatory network through target gene identification. Cold Spring Harb. Symp. Auant. Biol. 77 31–45. 10.1101/sqb.2013.77.014480 [DOI] [PubMed] [Google Scholar]
- Long J., Barton M. K. (2000). Initiation of axillary and floral meristems in Arabidopsis. Dev. Biol. 218 341–353. 10.1006/dbio.1999.9572 [DOI] [PubMed] [Google Scholar]
- Luo L., Ando S., Sakamoto Y., Suzuki T., Takahashi H., Ishibashi N., et al. (2020). The formation of perinucleolar bodies is important for normal leaf development and requires the zinc-finger DNA-binding motif in Arabidopsis ASYMMETRIC LEAVES2. Plant J. 101 1118–1134. 10.1111/tpj.14579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo M., Yu C. W., Chen F. F., Zhao L., Tian G., Liu X., et al. (2012). Histone deacetylase HDA6 is functionally associated with AS1 in repression of KNOX genes in Arabidopsis. PLos Genet. 8:e1003114. 10.1371/journal.pgen.1003114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luong T. Q., Keta S., Asai T., Kojima S., Nakagawa A., Micol J. L., et al. (2018). A genetic link between epigenetic repressor AS1-AS2 and DNA replication factors in establishment of adaxial-abaxial leaf polarity of Arabidopsis. Plant Biotechnol. (Tokyo) 35 39–49. 10.5511/plantbiotechnology.18.0129b [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynn K., Fernandez A., Aida M., Sedbrook J., Tasaka M., Masson P., et al. (1999). The PINHEAD/ZWILLE gene acts pleiotropically in Arabidopsis development and has overlapping functions with the ARGONAUTE1 gene. Development 126 469–481. [DOI] [PubMed] [Google Scholar]
- Shi B., Zhang C., Tian C., Wang J., Wang Q., Xu T., et al. (2016). Two-step regulation of a meristematic cell population acting in shoot branching in Arabidopsis. PLoS Genet. 12:e1006168. 10.1371/journal.pgen.1006168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnani E., Barton M. K. (2011). A per-ARNT-sim-like sensor domain uniquely regulates the activity of the homeodomain leucine zipper transcription factor REVOLUTA in Arabidopsis. Plant Cell 23 567–582. 10.1105/tpc.110.080754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magne K., George J., Berbel Tornero A., Broquet B., Madueno F., Andersen S. U., et al. (2018). Lotus japonicus NOOT-BOP-COCH-LIKE1 is essential for nodule, nectary, leaf and flower development. Plant J. 94 880–894. 10.1111/tpj.13905 [DOI] [PubMed] [Google Scholar]
- Mallory A. C., Reinhart B. J., Jones-Rhaodes M. W., Tang G., Zamore P. D., Barton M. K., et al. (2004). MicroRNA control PHABULOSA in leaf development: importance of pairing to the microRNA 5’region. EMBO J. 23 2256–3364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto N., Okada K. (2001). A homeobox gene, PRESSED FLOWER, regulates lateral axis-dependent development of Arabidopsis flowers. Genes Dev. 15 3355–3364. 10.1101/gad.931001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer K. F., Schoof H., Haecker A., Lenhard M., Jürgens G., Laux T. (1998). Role of WUSCHEL in regulating stem cell fate in the Arabidopsis shoot meristem. Cell 95 805–815. 10.1016/s0092-8674(00)81703-1 [DOI] [PubMed] [Google Scholar]
- McConnell J. R., Barton M. K. (1998). Leaf polarity and meristem formation in Arabidopsis. Development 125 2935–2942. [DOI] [PubMed] [Google Scholar]
- McConnell J. R., Emery J., Eshed Y., Bao N., Bowman J., Barton M. K. (2001). Role of PHABULOSA and PHAVOLUTA in determining radial pattering in shoots. Nature 411 709–713. 10.1038/35079635 [DOI] [PubMed] [Google Scholar]
- Merelo P., Ram H., Pia, Caggiano M., Ohno C., Ott F., et al. (2016). Regulation of MIR165/166 by class II and class II homeodomain leucine zipper proteins establishes leaf polarity. Proc. Natl. Acad. Sci. U.S.A. 113 11973–11978. 10.1073/pnas.1516110113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyashima S., Koi S., Hashimoto T., Nakajima K. (2011). Non-cell-autonomous microRNA165 acts in a dose-dependent manner to regulate multiple differentiation status in the Arabidopsis root. Development 138 2303–2313. 10.1242/dev.060491 [DOI] [PubMed] [Google Scholar]
- Mudunkothge J. S., Krizek B. A. (2012). Three Arabidopsis AIL/PLT genes act in combination to regulate shoot apical meristem function. Plant J. 71 108–121. 10.1111/j.1365-313x.2012.04975.x [DOI] [PubMed] [Google Scholar]
- Muszynski M. G., Moss-Taylor L., Chudalayandi S., Cahill J., Del Valle-Echevarria A. R., Alvarez-Castro I., et al. (2020). The maize hairy sheath frayed1 (Hsf1) mutation alters leaf patterning through increased cytokinin signaling. Plant Cell 32 1501–1518. 10.1105/tpc.19.00677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakata M., Matsumoto N., Tsugeki R., Rikirsch E., Laux T., Okada K. (2012). Roles of the middle domain-specific WUSCHEL-RELATED HOMEOBOX genes in early development of leaves in Arabidopsis. Plant Cell 24 519–535. 10.1105/tpc.111.092858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nardmann J., Ji J., Werr W., Scanlon M. J. (2004). The maize duplicate genes narrow sheath1 and narrow sheath2 encode a conserved homeobox gene function in a lateral domain of shoot apical meristems. Development 131 2827–2839. 10.1242/dev.01164 [DOI] [PubMed] [Google Scholar]
- Nole-Wilson S., Krizek B. A. (2006). AINTEGUMENTA contributes to organ polarity and regulates growth of lateral organs in combination with YABBY genes. Plant Physiol. 141 977–987. 10.1104/pp.106.076604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norberg M., Holmlund M., Nilsson O. (2005). The BLADE-ON-PETIOLE genes act redundantly to control the growth and development of lateral organs. Development 132 2203–2213. 10.1242/dev.01815 [DOI] [PubMed] [Google Scholar]
- Otsuga D., DeGuzman B., Prigge M. J., Drews G. N., Clark S. E. (2001). REVOLUTA regulates meristem initiation at lateral positions. Plant J. 25 223–236. 10.1111/j.1365-313x.2001.00959.x [DOI] [PubMed] [Google Scholar]
- Pekker I., Alvarez J. P., Eshed Y. (2005). Auxin response factors mediate Arabidopsis organ asymmetry via modulation of KANADI activity. Plant Cell 17 2899–2910. 10.1105/tpc.105.034876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Perez J. M., Candela H., Robles P., Lopez-Torrejon G., del Pozo J. C., Micol J. L. (2010). A role for AUXIN RESISTANT3 in the coordination of leaf growth. Plant Cell Physiol. 51 1661–1673. 10.1093/pcp/pcq123 [DOI] [PubMed] [Google Scholar]
- Poethig R. S. (2003). Phase change and the regulation of developmental timing in plants. Science 301 334–336. 10.1126/science.1085328 [DOI] [PubMed] [Google Scholar]
- Prasad K., Grigg S. P., Barkoulas M., Yadav R. K., Sanchez-Perez G. F., Pinon V., et al. (2011). Arabidopsis PLETHORA transcription factors control phyllotaxis. Curr. Biol. 21 1123–1128. 10.1016/j.cub.2011.05.009 [DOI] [PubMed] [Google Scholar]
- Prigge M. J., Otsuga D., Alonso J. M., Echker J. R., Drews G. N., Clark S. E. (2005). Class III homeodomain-leucin zipper gene family members have overlapping, antagonistic, and distinct roles in Arabidopsis development. Plant Cell 17 61–76. 10.1105/tpc.104.026161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi J., Wang Y., Yu T., Cunha A., Wu B., Vernoux T., et al. (2014). Auxin depletion from leaf primordia contributes to organ patterning. Proc. Natl. Acad. Sci. U.S.A. 111 18769–18774. 10.1073/pnas.1421878112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ram H., Sahadevan S., Gale N., et al. (2020). An integrated analysis of cell-type specific gene expression reveals genes regulated by REVOLUTA and KANADI1 in the Arabidopsis shoot apical meristem. PLoS Genet. 16:e1008661. 10.1371/journal.pgen.1008661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rast-Somssich M. I., Broholm S., Jenkins H., et al. (2015). Alternate wiring of a KNOXI genetic network underlies differences in leaf development of A. thaliana and C. hirsuta. Genes Dev. 29 2391–2404. 10.1101/gad.269050.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhardt D., Mandel T., Kuhlemeier C. (2000). Auxin regulates the initiation and radial position of plant lateral organs. Plant Cell 12 507–518. 10.2307/3871065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruberti I., Sessa G., Ciolfi A., Possenti M., Carabelli M., Morelli G. (2012). Plant adaption to dynamically changing environment: the shade avoidance response. Biotechnol. Adv. 30 1047–1058. 10.1016/j.biotechadv.2011.08.014 [DOI] [PubMed] [Google Scholar]
- Sarojam R., Sappl P. G., Goldshmidt A., Efroni I., Floyd S. K., Eshed Y., et al. (2010). Differentiating Arabidopsis shoots from leaves by combined YABBY activities. Plant Cell 22 2113–2130. 10.1105/tpc.110.075853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawa S., Watanabe K., Goto K., Liu Y. G., Shibata D., Kanaya E., et al. (1999). FILAMRNAYLOUS FLOWER, a meristem and organ identity gene of Arabidopsis, encodes a protein with a zinc finger and HMG-related domains. Gene Dev. 13 1079–1088. 10.1101/gad.13.9.1079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semiarti E., Ueno Y., Tsukaya H., Iwakawa H., Machida C., Machida Y. (2001). The ASYMMETRIC LEAVES2 gene of Arabidopsis thaliana regulates formation of a symmetric lamina, establishment of venation and repression of meristem-related homeobox genes in leaves. Development 128 1771–1783. [DOI] [PubMed] [Google Scholar]
- Shi J., Dong J., Xie J., Wang H., Yang Z., Jiao Y., et al. (2017). Model for the role of auxin polar transport in patterning of the leaf adaxial-abaxial axis. Plant J. 92 469–480. 10.1111/tpj.13670 [DOI] [PubMed] [Google Scholar]
- Siegfried K. R., Eshed Y., Baum S. F., Otsuga D., Drewa G. N., Bowman J. L. (1999). Members of the YABBY gene family specify abaxial cell fate in Arabidopsis. Development 126 4117–4128. [DOI] [PubMed] [Google Scholar]
- Simonini S., Bencivenga S., Trick M., Østergaard L. (2017). Auxin-induced modulation of ETTIN activity orchestrates gene expression in Arabidopsis. Plant Cell 29 1864–1882. 10.1105/tpc.17.00389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonini S., Deb J., Moubayidin L., Stephenson P., Valluru M., Freire-Rios A., et al. (2016). A noncanonical auxin-sensing mechanism is required for organ morphogenesis in Arabidopsis. Genes Dev. 30 2286–2296. 10.1101/gad.285361.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonini S., Mas P. J., Mas C., Østergaard L., Hart D. J. (2018). Auxin sensing is a property of an unstructured domain in the Auxin Response Factor ETTIN of Arabidopsis thaliana. Sci. Rep. 8:13563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahle M., I, Kuehlich J., Staron L., Von Arnim A. G., Golz J. F. (2009). YABBY and the transcriptional corepressors LEUNIG and LEUNIG HOMOLOG maintain leaf polarity and meristem activity in Arabidopsis. Plant Cell 21 3105–3118. 10.1105/tpc.109.070458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J., Cui X., Teng S., Zhao K., Wang Y., Chen Z., et al. (2020). HD-ZIP IV gene Roc8 regulates the size of bulliform cells and lignin content in rice. Plant Biotechnol. J. 10.1111/pbi.13435 [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sussex I. M. (1954). Experiment on the cause of dorsiventrality in leaves. Nature 174 351–352. 10.1038/174351a0 [DOI] [PubMed] [Google Scholar]
- Szakonyi D., Moschopoulos A., Byrne M. E. (2010). Perspectives on leaf dorsoventral polarity. J. Plant Res. 123 281–290. 10.1007/s10265-010-0336-3 [DOI] [PubMed] [Google Scholar]
- Tadege M., Lin H., Bedair M., Berbel A., Wen J., Rojas C. M., et al. (2011a). STENOFOLIA regulates blade outgrowth and leaf vascular patterning in Medicago truncatula and Nicotiana sylvestris. Plant Cell 23 2125–2142. 10.1105/tpc.111.085340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tadege M., Lin H., Niu L., Mysore K. S. (2011b). Control of dicot leaf blade expansion by a WOX gene, STF. Plant Signal. Behav. 6 1861–1864. 10.4161/psb.6.11.17761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talbert P. B., Alder H. T., Parks D. W., Comai L. (1995). The REVOLATA gene is necessary for apical meristem development and for limiting cell division in the leaves and stems of Arabidopsis thaliana. Development 121 2723–2735. [DOI] [PubMed] [Google Scholar]
- Tatematsu K., Toyokura K., Okada K. (2015). Requirement of MIR165A primary transcript sequence for its activity pattern in Arabidopsis leaf primordia. Plant Signal Behav. 10:e1055432. 10.1080/15592324.2015.1055432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toriba T., Okunaga H., Nagasawa K., Nie F., Yoshida A., Kyozuka J. (2020). Suppression of leaf blade by BLADE-ON-PETIOLE orthologs is a common strategy for underground rhizome growth. Curr. Biol. 30 509–516. 10.1016/j.cub.2019.11.055 [DOI] [PubMed] [Google Scholar]
- Toriba T., Tokunaga H., Shiga T., Nie F., Naramoto S., Honda E., et al. (2019). BLADE-ON-PETIOLE genes temporally and developmentally regulate the sheath to blade ratio of rice leaves. Nat. Commun. 10:619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tron A. E., Bertoncini C. W., Palena C. M., Chan R. L., Gonzalez D. H. (2001). Combinational interactions of two amino acids with a single base pair define target specificity in plant dimeric homeodomain proteins. Nucleic Acids Res. 29 4866–4872. 10.1093/nar/29.23.4866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tron A. E., Comelli R. N., Gonzalez D. H. (2005). Structure of homeodomain-leucine zipper/DNA complex studies using hydroxyl radical cleavage of DNA and methylation interference. Biochemistry 44 16796–16803. 10.1021/bi0513150 [DOI] [PubMed] [Google Scholar]
- Tron A. E., Welchen E., Gonzalez D. H. (2004). Engineering the loop region of homeodomain protein promotes efficient binding to a monomeric binding site. Biochemistry 43 15845–15851. 10.1021/bi048254a [DOI] [PubMed] [Google Scholar]
- Turchi L., Carabelli M., Rizza V., Possenti M., Sassi M., Penalosa A., et al. (2013). Arabidopsis HD-ZIP II transcription factors control apical embryo development and meristem function. Development 140 2118–2129. 10.1242/dev.092833 [DOI] [PubMed] [Google Scholar]
- Ueno Y., Ishikawa T., Watanabe K., Terakura S., Iwakawa H., Okada K., et al. (2007). Histone deacetylases and ASYMMETRIC LEAVES2 are involved in the establishment of polarity in leaves of Arabidopsis. Plant Cell 19 445–457. 10.1105/tpc.106.042325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandenbussche M., Horstman A., Zethof J., Koes R., Rijpkema A. S., Gerats T. (2009). Differential recruitment of WOX transcription factors for lateral development and organ fusion in Petunia and Arabidopsis. Plant Cell 21 2269–2283. 10.1105/tpc.109.065862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vial-Pradel S., Keta S., Nomoto M., Luo L., Takahashi H., Suzuki M., et al. (2018). Arabidopsis zinc-finger-like protein ASYMMETRIC LEAVES2 (AS2) and two nucleolar proteins maintain gene body DNA methylation in the leaf polarity gene ETTIN (ARF3). Plant Cell Physiol. 59 1385–1397. [DOI] [PubMed] [Google Scholar]
- Vlad D., Kierzkowski D., Rast M. I., et al. (2014). Leaf shape evolution through duplication, regulatory diversification, and loss of a homeobox gene. Science 343 780–783. 10.1126/science.1248384 [DOI] [PubMed] [Google Scholar]
- Waites R., Hudson A. (1995). Phantastica: a gene required for dorsiventrality of leaves in Antirrhinum majus. Development 121 2143–2154. [Google Scholar]
- Wang H., Chen J., Wen J., et al. (2008). Control of compound leaf development by FLORICAULA/LEAFY ortholog SINGLE LEAFLET1 in Medicago truncatula. Plant Physiol. 146 1759–1772. 10.1104/pp.108.117044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Mei J., Ren G. (2019). Plant microRNAs: biogenesis, homeostasis, and degradation. Front. Plant Sci. 10:360. 10.3389/fpls.2019.00360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J. W., Czech B., Weigel D. (2009). miR156-rgulated SPL transcription factors define an endogenous flowering pathway in Arabidopsis thaliana. Cell 138 738–749. 10.1016/j.cell.2009.06.014 [DOI] [PubMed] [Google Scholar]
- Wang L., Zhou C. M., Mai Y. X., Li L. Z., Gao J., Shang G. D., et al. (2019). A spatiotemporally regulated transcriptional complex underlies heteroblastic development of leaf hairs in Arabidopsis thaliana. EMBO J. 38:e100063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Xu B., Wang H., Li J., Huang H., Xu L. (2011). YUCCA genes are expressed in response to leaf adaxial-abaxial juxtaposition and are required for leaf margin development. Plant Physiol. 157 1805–1819. 10.1104/pp.111.186395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenkel S., Emery J., Hou B. H., Evans M. M. S., Barton M. K. (2007). A feedback regulatory module formed by LITTLE ZIPPER and HD-ZIP III genes. Plant Cell 19 3379–3390. 10.1105/tpc.107.055772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitewoods C. D., Gonçalves B., Cheng J., Cui M., Kennaway R., Lee K., et al. (2020). Evolution of carnivorous traps from planar leaves through simple shifts in gene expression. Science 367 91–96. 10.1126/science.aay5433 [DOI] [PubMed] [Google Scholar]
- Wu G., Huang T., Poethig R. S., Springer P. S., Kerstetter R. A. (2008). KANADI 1 regulated adaxial-abaxial polarity in Arabidopsis by directly repressing the transcription of ASYMMETRIC LEAVES2. Proc. Natl. Acad. Sci. U.S.A. 105 16392–16397. 10.1073/pnas.0803997105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G., Park M. Y., Corway S. R., Wang J. W., Weigel D., Poethig R. S. (2009). The sequential action of miR156 and miR172 regulates developmental timing in Arabidopsis. Cell 138 750–759. 10.1016/j.cell.2009.06.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang J., Liu R. Q., Li T. M., Han L. J., Zou Y., Xu T. F., et al. (2013). Isolation and characterization of two VpYABBY genes from wild Chinese Vitis pseudoreticulata. Protoplasma 250 1315–2135. 10.1007/s00709-013-0514-y [DOI] [PubMed] [Google Scholar]
- Xu L., Xu Y., Dong A., Sun Y., Pi L., Xu Y., et al. (2003). Novel as1 and as2 defects in leaf adaxial-adaxial polarity reveal the requirement for ASYMMETRIC LEAVES1 and 2 and ERECTA functions in specifying lead adaxial identity. Development 130 4097–4107. 10.1242/dev.00622 [DOI] [PubMed] [Google Scholar]
- Xu L., Yang L., Pi L., Liu Q., Ling Q., Wang H., et al. (2006). Genetic interaction between the AS1-AS2 and RDR6-SGS3-AGO7 pathways for leaf morphogenesis. Plant Cell Physiol. 47 853–863. 10.1093/pcp/pcj057 [DOI] [PubMed] [Google Scholar]
- Xu M., Hu T., Smith M. R., Poethig R. S. (2016). Epigenetic regulation of vegetative phase change in Arabidopsis. Plant Cell 28 28–41. 10.1105/tpc.15.00854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu P., Ali A., Han B., Wu X. (2018). Current advances in molecular basis and mechanisms regulating leaf morphology in rice. Front. Plant Sci. 9:1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y., Qian Z., Zhou B., Wu G. (2019). Age-dependent heteroblastic development of leaf hairs in Arabidopsis. New Phytol. 224 741–748. 10.1111/nph.16054 [DOI] [PubMed] [Google Scholar]
- Yamaguchi T., Yano S., Tsukaya H. (2010). Genetic framework for flattened leaf blade formation in unifacial leaves of Juncus prismatocarpus. Plant Cell 22 2141–2155. 10.1105/tpc.110.076927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H., Shi G., Li X., Hu D., Cui Y., Hou J., et al. (2019). Overexpression of a soybean YABBY gene, GmFILa, causes leaf curling in Arabidopsis thaliana. BMC Plant Biol. 19:234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao X., Wang H., Li H., Yuan Z., Li F., Yang L., et al. (2009). Two types of cis-acting elements control the abaxial epidermis-specific transcription of the MIR165a and MIR166a genes. FEBS Lett. 583 3711–3717. 10.1016/j.febslet.2009.10.076 [DOI] [PubMed] [Google Scholar]
- Yin X., Tsukaya H. (2019). Morphogenesis of flattened unifacial leaves in Juncus prismatocarpus (Juncaceae). New Phytol. 222 1101–1111. 10.1111/nph.15649 [DOI] [PubMed] [Google Scholar]
- Yoshikawa M., Peragine A., Park M. Y., Poethig R. S. (2005). A pathway for the biogenesis of trans-acting siRNAs in Arabidopsis. Genes Dev. 19 2164–2175. 10.1101/gad.1352605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshikawa T., Tanaka S. Y., Masumoto Y., Nobori N., Ishii H., Hibara K., et al. (2016). Barley NARROW LEAFED DWARF1 encoding a WUSCHEL-RELATED HOMEOBOX 3 (WOX3) regulates the marginal development of lateral organs. Breed Sci. 66 416–424. 10.1270/jsbbs.16019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y., Ji L., Le B. H., Zhai J., Chen J., Luscher E., et al. (2017). ARGONAUTE10 promotes the degradation of miR165/6 through the SDN1 and SDN2 exonucleases in Arabidopsis. PLoS Biol. 15:e2001272. 10.1371/journal.pbio.2001272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C., Wang J., Wenkel S., Chandler J. W., Werr W., Jiao Y. (2018). Spatiotemporal control of axillary meristem formation by interacting transcriptional regulators. Development 145:dev158352. 10.1242/dev.158352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F., Wang H., Kalve S., Wolabu T. W., Nakashima J., Golz J. F., et al. (2019). Control of leaf blade outgrowth and floral organ development by LEUNIG, ANGUSTIFOLIA3 and WOX transcriptional regulators. New Phytol. 223 2024–2038. 10.1111/nph.15921 [DOI] [PubMed] [Google Scholar]
- Zhang F., Wang Y., Li G., Tang Y., Kramer E. M., Tadege M. (2014). STENOFOLIA recruits TOPLESS to repress ASYMMETRIC LEAVES2 at the leaf margin and promote leaf blade outgrowth in Medicago truncatula. Plant Cell 26 650–664. 10.1105/tpc.113.121947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G. H., Xu Q., Zhu X. D., Qian Q., Xue H. W. (2009). SHALLOT-LIKE1 is a KANADI transcription factor that modulates rice leaf rolling by regulating leaf abaxial cell development. Plant Cell 21 719–735. 10.1105/tpc.108.061457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S., Wang L., Sun X., Li Y., Yao J., van Nocker S., et al. (2019). Genome-wide analysis of the YABBY gene family in grapevine and functional characterization of VvYABBY4. Front. Plant Sci. 10:1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B., Holmlund M., Lorrain S., Norberg M., Bako L., Fankhauser C., et al. (2017). Blade-on-petiole proteins act in an E3 ubiquitin ligase complex to regulate phytochrome interacting factoR 4 abundance. Elife 6:e26759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou C., Han L., Zhao Y., Wang H., Nakashima J., Tong J., et al. (2019). Transforming compound leaf patterning by manipulating REVOLUTA in Medicago truncatula. Plant J. 100 562–571. 10.1111/tpj.14469 [DOI] [PubMed] [Google Scholar]
- Zhu H., Wang R., Zhou X., Sze S.-H., Liou L. W., Barefoot A., et al. (2011). ARGONAUTE 10 specifically sequesters miR166/miR165 to regulate shot apical meristem. Cell 145 242–256. 10.1016/j.cell.2011.03.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zumajo-Cardona C., Vasco A., Ambrose B. A. (2019). The evolution of the KANADI gene family and leaf development in lycophytes and ferns. Plants 8:313. 10.3390/plants8090313 [DOI] [PMC free article] [PubMed] [Google Scholar]