Abstract
Structural studies of challenging targets such as G protein-coupled receptors (GPCRs) have accelerated during the last several years due to the development of new approaches, including small-wedge and serial crystallography. Here, we describe the deposition of seven datasets consisting of X-ray diffraction images acquired from lipidic cubic phase (LCP) grown microcrystals of two human GPCRs, Cysteinyl leukotriene receptors 1 and 2 (CysLT1R and CysLT2R), in complex with various antagonists. Five datasets were collected using small-wedge synchrotron crystallography (SWSX) at the European Synchrotron Radiation Facility with multiple crystals under cryo-conditions. Two datasets were collected using X-ray free electron laser (XFEL) serial femtosecond crystallography (SFX) at the Linac Coherent Light Source, with microcrystals delivered at room temperature into the beam within LCP matrix by a viscous media microextrusion injector. All seven datasets have been deposited in the open-access databases Zenodo and CXIDB. Here, we describe sample preparation and annotate crystallization conditions for each partial and full datasets. We also document full processing pipelines and provide wrapper scripts for SWSX and SFX data processing.
Subject terms: Data publication and archiving, Nanocrystallography
| Measurement(s) | X-ray diffraction data • protein complex • protein structure data • protein crystallization |
| Technology Type(s) | small-wedge synchrotron crystallography • x-ray crystallography assay • X-ray free electron laser serial femtosecond crystallography |
| Factor Type(s) | type of G-protein-coupled receptor • type of antagonist |
Machine-accessible metadata file describing the reported data: 10.6084/m9.figshare.13128758
Background & Summary
Cysteinyl leukotrienes, produced from arachidonic acid via the 5-lipooxygenase pathway, are pro-inflammatory mediators that modulate vascular permeability and immune response; hence, they are involved in multiple disorders including asthma, cardiovascular diseases and cancer1. Cysteinyl leukotrienes elicit their action through two G protein-coupled receptors (GPCRs), CysLT1R and CysLT2R, that share 38% sequence identity1. CysLT1R is mostly expressed in the lungs and immune cells, and its stimulation leads to allergic symptoms in the airways2. CysLT2R is found additionally in cardiovascular and brain tissues, with demonstrated involvement in ischemia and acute brain injuries3,4, however, the role of this receptor remains controversial and poorly understood. Both CysLT1R and CysLT2R have been implicated in progression of various cancers5–8, while the mutated form of CysLT2R with L129Q substitution has been associated with uveal melanoma9,10. Thus, CysLTRs are important pharmaceutical targets11, what inspired us to determine their high-resolution structures in complex with antiasthmatic drugs and other prospective antagonists.
Over the last few years, small-wedge synchrotron crystallography (SWSX) and serial femtosecond crystallography (SFX) have developed into powerful techniques, enabling high-resolution structure determination of many difficult to crystallize targets12,13. Several approaches to data processing have been developed for both SFX13 and SWSX14–20, and several papers reported deposition of raw serial crystallography data for challenging targets21–25. Many datasets can be found online on SBGrid (data.sbgrid.org)26, Zenodo (zenodo.org) or CXIDB (cxidb.org)27; the latter is used for SFX and other XFEL-related data deposition, whereas SBGrid and Zenodo host SWSX among other types of data.
Recently, we have determined crystal structures of CysLT1R28 (PDB codes 6RZ4, 6RZ5) and CysLT2R29 (PDB codes 6RZ6, 6RZ7, 6RZ8, 6RZ9). Here, we present fully-annotated SWSX and SFX datasets for these structures, as well as unpublished SFX data of a new crystal form of CysLT1R. The raw diffraction data, consisting of five SWSX and two SFX datasets, represent a wide range of resolutions (2.4–3.5 Å), SWSX miniset30 wedge sizes (3–180°), and space groups (6 different space groups). We carefully document crystallization conditions and harvesting details for each dataset, allowing one to investigate crystal non-isomorphism. Finally, we describe all data processing steps, provide supporting code and intermediate results, aiming for reproducibility of deposited data processing.
Methods
The preparation of CysLT1R and CysLT2R samples, data collection, and processing have been described previously28,29. Here, we provide a summary for each sample.
Construct engineering, expression, purification, and crystallization of CysLT1R and CysLT2R
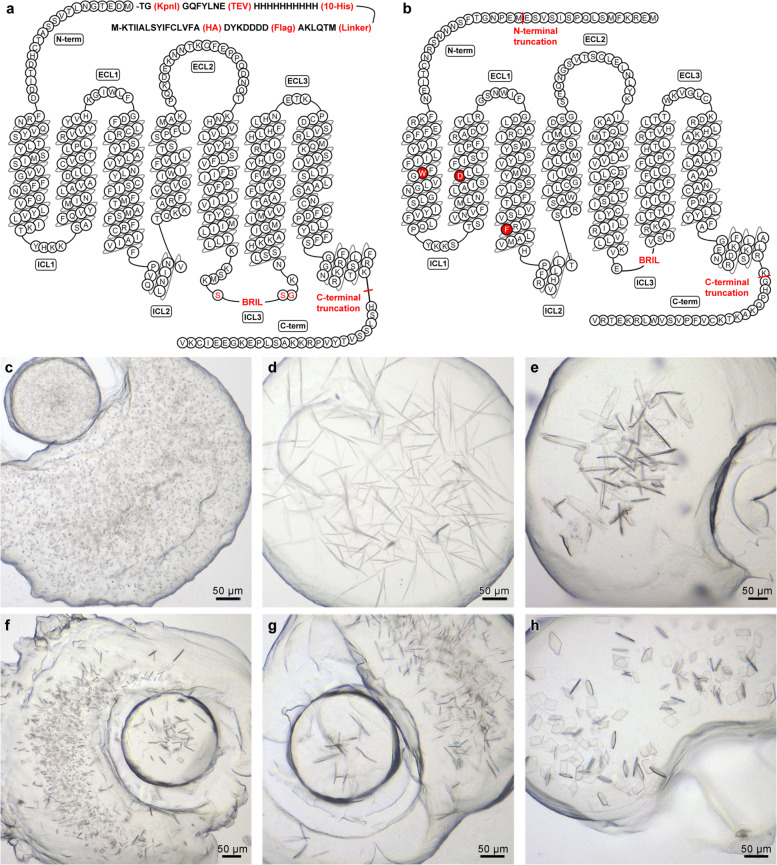
The human CysLT1R gene (UniProt ID Q9Y271) was codon-optimized for expression in Spodoptera frugiperda (Sf9) insect cell line and modified for crystallization by a C-terminal truncation at K311 and by the insertion of a fusion protein BRIL31 (thermostabilized apocytochrome b562 from Escherichia coli with mutations M7W, H102I, and R106L) in the third intracellular loop (ICL3) between K222 and K223 using the S and SG linkers on each side, respectively (Fig. 1a). For CysLT2R, the human WT gene (UniProt ID Q9NS75) was modified by truncating amino acids 1–16 from the N-terminus and 323–346 from the C-terminus and inserting BRIL into ICL3 between residues E232 and V240. Three point mutations, W511.45V, D842.50N, and F1373.51Y (superscripts refer to the generic Ballesteros-Weinstein numbering of residues in Class A GPCR32), were further introduced to improve receptor surface expression as well as its stability and yield (Fig. 1b).
Fig. 1.
Construct design and crystallization of CysLT1R and CysLT2R. (a,b) Amino acid sequence snake-plot of the CysLT1R (a) and CysLT2R (b) crystallization constructs. Protein modifications are shown in red, red background shows stabilising point mutations, red font amino acids – linkers for BRIL insertion. Initial figure drawn using GPCRdb49. The N-terminal peptide fragment (a) was added to both constructs. (c–h) microphotographs of typical crystals grown in lipidic cubic phase (LCP) for CysLT1R-zafirlukast (c), CysLT1R-pranlukast (d), and CysLT2R with antagonists: 11a (C2221 space group) (e), 11a (F222 space group) (f), 11b (g), and 11c (h) complexes51.
Each gene of interest was cloned into a modified pFastBac1 plasmid, containing a cleavable influenza hemagglutinin signal sequence (HA), a Flag tag, a AKLQTM linker, a 10 × His tag, and a Tobacco Etch Virus (TEV) protease site followed by KpnI restriction site on the N-terminal side of the inserted gene (Fig. 1c). The plasmid was then transfected into Sf9 insect cells using the bac-to-bac expression system (Invitrogen). High-titer recombinant baculovirus (>3 × 108 viral particles per ml) was obtained and used to infect Sf9 cells at a density of (2-3) × 106 cells per ml culture and a multiplicity of infection of 5–10 in the presence of a ligand: 8 µM zafirlukast (Cayman Chemical) for CysLT1R or 3 µM BayCysLT2 (Cayman Chemical) for CysLT2R. The protein surface expression and the virus titer were measured using flow cytometry. Cells were harvested 48–50 hours post infection by centrifugation at 2,000 × g and stored at −80 °C until use.
Protein purification was conducted at 4 °C. For each protein-ligand complex, the relevant ligand was added during purification. Cells were thawed and lysed by repetitive homogenization with a glass douncer followed by ultracentrifugation (30 min at 220,000 × g), 2 times in hypotonic buffer (10 mM HEPES pH 7.5, 20 mM KCl, and 10 mM MgCl2) and 3 times in high osmotic buffer (10 mM HEPES pH 7.5, 20 mM KCl, 10 mM MgCl2, and 1 M NaCl) with the addition of a protease inhibitor cocktail (500 µM 4-(2-aminoethyl)benzenesulfonyl fluoride hydrochloride (Gold Biotechnology), 1 µM E-64 (Cayman Chemical), 1 µM Leupeptin (Cayman Chemical), 150 nM Aprotinin (A.G. Scientific)).
Membranes were then incubated for 30 min in 10 mM HEPES pH 7.5, 20 mM KCl, 10 mM MgCl2, 2 mg ml−1 iodacetomide, protease inhibitor cocktail, and 25 µM ligand. Then receptors were solubilized by the addition of an equal volume of solubilisation buffer (300 mM NaCl, 2% (w/v) n-dodecyl-β-D-maltopyranoside (DDM; Avanti Polar Lipids) 0.4% (w/v) cholesteryl hemisuccinate (CHS; Sigma), 10% glycerol) and incubation for 3.5 hours. After 1-hour centrifugation (650,000 × g) to remove insolubilized material, the supernatant was incubated with a TALON IMAC resin (Clontech) overnight in the presence of 10/20 mM imidazole, 100 мМ HEPES pH 7.5 for CysLT1/2R with NaCl concentration increased to 800 mM.
The resin was then washed with 10 column volumes (CV) of wash buffer I (8 mM ATP, 100 mM HEPES pH 7.5, 10 mM MgCl2, 500 mM NaCl, 15 mM imidazole, 10 μM ligand, 10% glycerol, 0.1% DDM, 0.02% CHS), then with 5 CV of wash buffer II (25 mM HEPES pH 7.5, 250/500 mM NaCl for CysLT1/2R, 30 mM imidazole, 10 μM ligand, 10% glycerol, 0.015% DDM, 0.003% CHS), then buffer was exchanged into buffer III (25 mM HEPES pH 7.5, 250/500 mM NaCl for CysLT1/2R, 10 mM imidazole, 10 μM ligand, 10% glycerol, 0.05% DDM, 0.01% CHS) and the protein-containing resin was treated with PNGase F (Sigma) for 5 hours. Resin was further washed with 5 CV of wash buffer III and eluted with (25 mM HEPES pH 7.5, 250/500 mM NaCl for CysLT1/2R, 300 mM imidazole, 10 μM ligand, 10% glycerol, 0.05% DDM, 0.01% CHS) in several fractions. After removing imidazole using a PD10 desalting column (GE Healthcare), the protein was incubated with 50 µM ligand and a His-tagged home-made TEV protease overnight to remove the N-terminal tags. Reverse IMAC was performed on the following day and the protein was concentrated up to 50–70 mg ml−1 using a 100 kDa molecular weight cut-off centrifugal concentrator (Millipore). The protein purity was checked by SDS-PAGE, and the protein yield and monodispersity were estimated by analytical size exclusion chromatography (aSEC).
Crystals for SWSX were grown using high-throughput nanovolume LCP crystallization. The purified and concentrated protein solution was combined with a lipid mixture: 90% monoolein (Sigma), 10% cholesterol (Affymetrix) in the ratio of 2:3 v/v and homogenized using a lipid syringe mixer until a transparent gel-like LCP formed33. Crystallisation was set up in 96-well glass sandwich LCP plates (Marienfeld), with 40 nL LCP drops and 800 nL precipitant drops, which were pipetted using an NT8-LCP robot (Formulatrix). All LCP manipulations were performed at room temperature (20–23 °C), and plates were incubated and imaged at 22 °C using an automated incubator/imager RockImager 1000 (Formulatrix).
CysLT1R-pranlukast crystals had a needle shape (Fig. 1d) and gained their full size after 3-4 weeks; however, the best diffraction was obtained from samples incubated for 2 months. CysLT2R crystals grew to their full size within 1–3 weeks. Crystals of CysLT2R in complex with ligands 11a and 11b had a shape of an elongated plate with a maximal size up to 150 µm (Fig. 1e–g). In case of CysLT2R-11c complex, crystals grew as flat parallelepipeds as long as 30–50 µm in diagonal (Fig. 1h). For the full list of crystallization conditions for crystals used in the data collection see Table 1.
Table 1.
Summary of crystallization conditions for SWSX datasets.
| Dataset | Condition ID | No. minisets | Condition |
|---|---|---|---|
| CysLT1R_6RZ4 | 1 | 27 | 100 mM Tris pH8, 33% PEG400, 160 mM Sodium Nitrate, 1% v/v 1,2-Butanediol |
| 2 | 4 | 100 mM Tris pH8, 34% PEG400, 130 mM Sodium Nitrate | |
| 3 | 40 | 100 mM Tris pH8, 34% PEG400, 75 mM Sodium Nitrate | |
| 4 | 8 | 100 mM Sodium Citrate pH6, 31% PEG400, 200 mM Lithium Nitrate | |
| 5 | 23 | 100 mM Tris pH8, 33% PEG400, 160 mM Sodium Nitrate, 4% v/v 2,5-Hexanediol (mixture of isomers) | |
| 6 | 23 | 100 mM Sodium Citrate pH6, 34% PEG400, 400 mM Lithium Nitrate, 10 mM Betaine hydrochloride | |
| 7 | 13 | 100 mM Tris pH8, 34% PEG400, 266 mM Sodium Nitrate | |
| 8 | 5 | 100 mM Sodium Citrate pH6, 34% PEG400, 400 mM Lithium Nitrate, 3% w/v D-( + )-Glucose monohydrate | |
| 9 | 2 | 100 mM Sodium Citrate pH6, 34% PEG400, 400 mM Lithium Nitrate, 100 mM Cesium Chloride | |
| 10 | 2 | 100 mM Tris pH8.2, 37% PEG400, 200 mM Sodium Nitrate, 4% v/v 2,5-Hexanediol (mixture of isomers) | |
| 11 | 16 | 100 mM Sodium Citrate pH6, 34% PEG400, 400 mM Lithium Nitrate, 4% v/v 2,5-Hexanediol (mixture of isomers) | |
| 12 | 11 | 100 mM Tris pH8, 33% PEG400, 160 mM Sodium Nitrate, 10 mM Spermine tetrahydrochloride | |
| 13 | 7 | 100 mM Sodium Citrate pH6, 30% PEG400, 200 mM Lithium Nitrate | |
| 14 | 2 | 100 mM Sodium Citrate pH6, 30% PEG400, 236 mM Lithium Nitrate | |
| 15 | 1 | 100 mM Tris pH8, 33% PEG400, 160 mM Sodium Nitrate, 10 mM Spermidine | |
| 16 | 2 | 100 mM Tris pH8, 33% PEG400, 160 mM Sodium Nitrate, 0.5% w/v n-Dodecyl-b-D-maltoside | |
| CysLT2R_6RZ6 | 1 | 9 | 100 mM Ammonium Sulfate, 30% v/v PEG400, 100 mM HEPES pH7.0 |
| 2 | 6 | 130 mM Ammonium Tartrate dibasic, 30% v/v PEG400, 100 mM HEPES pH 7.0, 10 mM Spermine tetrahydrochloride | |
| 3 | 12 | 100 mM Ammonium Sulfate, 30% v/v PEG400, 100 mM HEPES pH7.0 | |
| 4 | 2 | 100 mM Lithium Sulfate monohydrate, 30% v/v PEG400, 100 mM HEPES pH7.0 | |
| 5 | 18 | 30 mM Ammonium Tartrate dibasic, 20% v/v PEG400, 100 mM HEPES pH 7.0, 10 mM Betaine hydrochloride | |
| 6 | 7 | 60 mM Ammonium Tartrate dibasic, 22% PEG400, 100 mM HEPES pH7.0 | |
| 7 | 19 | 70 mM Ammonium Tartrate dibasic, 22% PEG400, 100 mM HEPES pH7.0 | |
| 8 | 2 | 30 mM Ammonium Tartrate dibasic, 20% PEG400, 100 mM HEPES pH7.0 | |
| 9 | 7 | 100 mM Ammonium Tartrate dibasic, 30% v/v PEG400, 100 mM TRIS pH8.0 | |
| 10 | 9 | 100 mM Ammonium Tartrate dibasic, 24% PEG400, 100 mM HEPES pH7.0 | |
| 11 | 1 | 100 mM Ammonium Tartrate dibasic, 26% PEG400, 100 mM HEPES pH7.0 | |
| 12 | 1 | 120 mM Ammonium Tartrate dibasic, 26% PEG400, 100 mM HEPES pH7.0 | |
| 13 | 7 | 120 mM Ammonium Tartrate dibasic, 28% PEG400, 100 mM HEPES pH7.0 | |
| 14 | 4 | 100 mM Ammonium Tartrate dibasic, 30% v/v PEG400, 100 mM TRIS pH8.0 | |
| 15 | 17 | 100 mM Ammonium Tartrate dibasic, 30% v/v PEG400, 100 mM TRIS pH8.0 | |
| 16 | 52 | 80 mM Ammonium Tartrate dibasic, 24% PEG400, 100 mM HEPES pH7.0 | |
| 17 | 5 | 130 mM Ammonium Tartrate dibasic, 30% v/v PEG400, 100 mM HEPES pH 7.0, 10 mM Yttrium(III) Chloride hexahydrate | |
| 18 | 20 | 100 mM Ammonium Tartrate dibasic, 26% PEG400, 100 mM HEPES pH7.0 | |
| 19 | 3 | 130 mM Ammonium Tartrate dibasic, 26% PEG400, 100 mM HEPES pH7.0 | |
| 20 | 2 | 130 mM Ammonium Tartrate dibasic, 28% PEG400, 100 mM HEPES pH7.0 | |
| 21 | 1 | 30 mM Ammonium Tartrate dibasic, 20% v/v PEG400, 100 mM HEPES pH 7.0, 0.025% v/v Dichloromethane | |
| 22 | 1 | 90 mM Ammonium Tartrate dibasic, 24% PEG400, 100 mM HEPES pH7.0 | |
| 23 | 6 | 30 mM Ammonium Tartrate dibasic, 20% v/v PEG400, 100 mM HEPES pH 7.0, 1% v/v 1,2-Butanediol | |
| 24 | 9 | 30 mM Ammonium Tartrate dibasic, 20% v/v PEG400, 100 mM HEPES pH 7.0, 4% v/v 1-Propanol | |
| 25 | 11 | 130 mM Ammonium Tartrate dibasic, 27.4% PEG400, 100 mM HEPES pH7.0 | |
| 26 | 5 | 120 mM Ammonium Tartrate dibasic, 26% PEG400, 100 mM HEPES pH7.0 | |
| CysLT2R_6RZ7 | 1 | 2 | 130 mM Ammonium Tartrate dibasic, 30% v/v PEG400, 100 mM HEPES pH 7.0, 0.025% v/v Dichloromethane |
| 2 | 4 | 100 mM Ammonium Tartrate dibasic, 30% v/v PEG400, 100 mM TRIS pH8.0 | |
| 3 | 10 | 100 mM Ammonium Tartrate dibasic, 24% PEG400, 100 mM HEPES pH7.0 | |
| 4 | 9 | 100 mM Ammonium Sulfate, 30% v/v PEG400, 100 mM HEPES pH7.0 | |
| 5 | 18 | 30 mM Ammonium Tartrate dibasic, 20% v/v PEG400, 100 mM HEPES pH 7.0, 10 mM Betaine Hydrochloride | |
| 6 | 9 | 30 mM Ammonium Tartrate dibasic, 20% v/v PEG400, 100 mM HEPES pH 7.0, 4% v/v 1-Propanol | |
| 7 | 8 | 130 mM Ammonium Tartrate dibasic, 30% v/v PEG400, 100 mM HEPES pH 7.0, 10 mM Yttrium(III) Chloride hexahydrate | |
| 8 | 3 | 100 mM Lithium Sulfate monohydrate, 30% v/v PEG400, 100 mM HEPES pH7.0 | |
| 9 | 1 | 120 mM Ammonium Tartrate dibasic, 26% PEG400, 100 mM HEPES pH7.0 | |
| 10 | 6 | 110 mM Ammonium Tartrate dibasic, 26% PEG400, 100 mM HEPES pH7.0 | |
| 11 | 1 | 30 mM Ammonium Tartrate dibasic, 20% v/v PEG400, 100 mM HEPES pH 7.0, 0.025% v/v Dichloromethane | |
| 12 | 16 | 120 mM Ammonium Tartrate dibasic, 28% PEG400, 100 mM HEPES pH7.0 | |
| 13 | 14 | 130 mM Ammonium Tartrate dibasic, 28% PEG400, 100 mM HEPES pH7.0 | |
| 14 | 6 | 30 mM Ammonium Tartrate dibasic, 20% v/v PEG400, 100 mM HEPES pH 7.0, 1% v/v 1,2-Butanediol | |
| 15 | 16 | 30 mM Ammonium Tartrate dibasic, 20% PEG400, 100 mM HEPES pH7.0 | |
| 16 | 23 | 130 mM Ammonium Tartrate dibasic, 27.4% PEG400, 100 mM HEPES pH7.0 | |
| 17 | 6 | 100 mM Ammonium Tartrate dibasic, 30% v/v PEG400, 100 mM TRIS pH8.0 | |
| 18 | 6 | 130 mM Ammonium Tartrate dibasic, 30% v/v PEG400, 100 mM HEPES pH 7.0, 10 mM Spermine tetrahydrochloride | |
| CysLT2R_6RZ8 | 1 | 11 | 100 mM Potassium fluoride, 30% v/v PEG400, 100 mM TRIS pH8.0 |
| CysLT2R_6RZ9 | 1 | 18 | 210 mM Ammonium Tartrate dibasic, 30.9% PEG400, 100 mM HEPES pH7.0 |
| 2 | 4 | 100 mM Ammonium Tartrate dibasic, 30% v/v PEG400, 100 mM TRIS pH8.0 | |
| 3 | 3 | 80 mM Ammonium Tartrate dibasic, 29.14% PEG400, 100 mM HEPES pH7.0 | |
| 4 | 5 | 20 mM Ammonium Tartrate dibasic, 32.6% PEG400, 100 mM HEPES pH7.0 | |
| 6 | 8 | 250 mM Ammonium Tartrate dibasic, 32.6% PEG400, 100 mM HEPES pH7.0 | |
| 7 | 2 | 130 mM Ammonium acetate, 27.4% PEG400, 100 mM HEPES pH7.0 | |
| 8 | 2 | 50 mM Ammonium Tartrate dibasic, 34.9% PEG400, 100 mM HEPES pH7.0 | |
| 9 | 2 | 90 mM Ammonium Tartrate dibasic, 29.14% PEG400, 100 mM HEPES pH7.0 |
Microcrystals of the CysLT1R-zafirlukast complex for SFX were grown in 100 µl gas-tight Hamilton syringes as previously described34,35. Briefly, approximately 5 µl of protein-laden LCP was transferred through a coupler (Formulatrix) into a syringe, containing 50 µl of precipitant, so that LCP extends towards the plunger as a straight filament. For experiments conducted in 2016 (dataset CysLT1R_zafirlukast-P21), zafirlukast was added at 50 μM prior to the protein concentration. Crystals grew in the following precipitant conditions: 100 mM ammonium phosphate, 31–34% v/v PEG400, 100 mM HEPES pH 7.0, 1 µM zafirlukast. For experiments conducted in 2017 (dataset CysLT1R_6RZ5), zafirlukast was added at 200 μM prior to the protein concentration. Crystals grew in the following precipitant conditions: 120–200 mM sodium/potassium phosphate, 31–34% v/v PEG400, 100 mM HEPES pH 7, no zafirlukast added. Crystals grew for 1-2 weeks, reaching an average crystal size of 5 × 2 × 2 µm (Fig. 1c).
Synchrotron data collection
Crystal harvesting
Crystals were harvested directly from LCP using 50–200 µm dual thickness MicroMounts or 400–700 µm MicroMesh loops (MiTeGen) with various hole sizes and flash frozen in liquid nitrogen, as described36.
Full sets data collection
Single-crystal datasets (for CysLT1R_6RZ4 and CysLT2R_6RZ8) were collected using the following procedure. First, the best diffracting position was found using automatic X-ray centring37 with a microfocus beam, followed by characterization37 and dose estimation using BEST38 software, and further data collection as proposed by BEST. This resulted in over 90% complete datasets, however, with a relatively low resolution (>3 Å).
Partial sets data collection
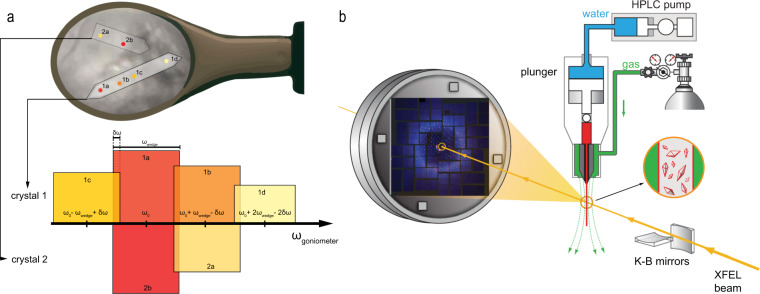
To improve resolution, SWSX partial datasets (minisets, as introduced by Basu et al.30) were collected using an updated version of the raster-scanning approach39. The process is illustrated in Fig. 2a. Each loop was first visually aligned and oriented with its plane perpendicular to the X-ray beam. Then, the whole loop was scanned with the beam to identify locations with diffracting crystals (shutterless mode was used on the ID29 and ID30b beamlines). Raster scans were performed using a minimal dose per image, which allowed for visual detection of diffraction spots, but was less than 1% of the total dose per dataset. The grid spacing was set around , where R is the beam profile radius (HWHM). The overlap between adjacent beam spots was introduced to improve accuracy in location of the best diffracting positions and to maximize the grid coverage by HWHM profiles. The grid cells showing diffraction spots were ranked by the DOZOR score37 and then manually selected for further data collection. In the case of large single crystals spanning through several grid cells, minisets were collected starting from the highest ranked location and then moving to the next best location along the crystal but skipping grid cells if they had a common edge with the cells already used for data collection to avoid collecting data from previously exposed parts of the crystal. Consecutive minisets from the same crystal were collected by ensuring 1-2° overlaps in the goniometer rotation ω angle. When the goniometer rotation angle exceeded 10° from its original orientation, a new line raster scan was performed to re-align the crystal with the beam. Each miniset was collected restricting an estimated dose per diffraction location within ∼20 MGy and using 0.1–0.2° oscillation and 3–20° total wedge size. The wedge size and the corresponding exposure time were selected based on the total number of harvested crystals from the particular condition and were adjusted by decreasing the wedge size and increasing the exposure time when preliminary data processing indicated that a complete dataset had been already collected, or in case of a weak diffraction. The beam size was chosen to match the smallest crystal dimension. A summary of miniset parameters for each SWSX entry is given in Table 2.
Fig. 2.
Synchrotron and XFEL data collection setups. (a) Schematics of the SWSX data collection process. The bar colour indicates the DOZOR score (from red – the best diffraction, to yellow – the worst). For minisets collected from the same crystal, as judged by the diffraction patterns and DOZOR score heatmap, an overlap of δω = 1–2° is introduced between consecutive sets. When the rotation angle ω exceeds ~10° from the initial orientation, as for the point 1d, an additional line scan is performed to re-align the crystal. The orientations of two different crystals 1 and 2 in the loop are assumed independent, and thus minisets from them are collected within the same ω range. (b) LCP-SFX data collection scheme. Microcrystals embedded in LCP are injected inside a vacuum chamber into the XFEL beam focus region using a viscous media microextrusion injector. A stream of sheath gas (nitrogen or helium) is used to keep the LCP stream straight. Microcrystals intersect with the XFEL beam in random orientations and diffraction patterns are collected by a CSPAD detector.
Table 2.
Summary of SWSX datasets.
| Entry | CysLT1R_6RZ4 | CysLT2R_6RZ6 | CysLT2R_6RZ7 | CysLT2R_6RZ9 | CysLT2R_6RZ8 |
|---|---|---|---|---|---|
| Reported resolution, Å | 2.7 | 2.43 | 2.43 | 2.73 | 2.7 |
| Reprocessed resolution, Å | 2.6 | 2.43 | 2.43 | 2.73 | 2.5 |
| No. collected minisets | 186 | 236 | 158 | 44 | 11 |
| No. processed minisets | 116 | 126 | 144 | 38 | 11 |
| No. minisets in the final dataset | 9 | 74 | 16 | 12 | 8 |
| Wedge size, ° | 10–180 | 3–12 | 4–20 | 10–20 | 10–20 |
| Detector/beamline | Eiger-4M/ID30a3 Pilatus3-S-6M/ID30b Pilatus-6M/ID29 | Pilatus3-S-6M/ID30b Pilatus-6M/ID29 | Eiger-4M/ID30a3 Pilatus-6M/ID23-1 Pilatus-6M/ID29 | Pilatus-6M/ID23-1 Pilatus-6M/ID29 | Pilatus-6M/ID23-1 |
XFEL data collection
Loading crystals into injector
Precipitant solutions were slowly withdrawn from 3 syringes containing microcrystals of appropriate size and density through a 22 s gauge Hamilton needle. The remaining samples of LCP with microcrystals embedded in it were consolidated from these 3 syringes into one syringe using a syringe coupler (Formulatrix). An aliquot of ~10% of 7.9 MAG lipid was added to the sample to absorb the excess of the precipitant and to avoid LCP freezing upon extrusion in the vacuum chamber40. A total sample volume of 15–20 µl was loaded into an LCP injector as described40.
LCLS data collection: 2016
An overall scheme of the data collection setup is shown in Fig. 2b. SFX data of CysLT1R_Zafirlukast-P21 were collected in August 2016 at the CXI instrument of the Linac Coherent Light Source (LCLS) at the SLAC National Accelerator Laboratory, Menlo Park, California. LCLS was operated at a wavelength of 1.305 Å (9.50 keV) delivering individual X-ray pulses of 40 fs duration and 2.6 × 1010 photons per pulse focused into a spot size of ~1.5 µm in diameter using a pair of Kirkpatrick-Baez mirrors. LCP with protein microcrystals was extruded at room temperature and at a flow rate of 0.3 μl min−1 inside a vacuum chamber into the beam focus region using an LCP injector40 with a 50-μm diameter capillary. The XFEL beam was attenuated at transmission levels of 6.1% to avoid disruptions of the LCP stream. Diffraction images were collected at an XFEL pulse repetition rate of 120 Hz using a 2.3 Megapixel Cornell-SLAC Pixel Array Detector41 (CSPAD).
A total number of 900,173 detector images were collected, of which 22,047 (2% of total) were identified as potential crystal hits with more than 15 Bragg peaks with SNR = 6.0, threshold 100 and min-pix-count 3.0 using peakfinder8 algorithm as implemented in Cheetah42. The overall time of data collection from a sample with a total volume of 27 μl was about 2 h 6 min.
LCLS data collection: 2017
SFX data of CysLT1R_6RZ5 were collected in August 2017 at the CXI instrument. LCLS was operated at a wavelength of 1.302 Å (9.52 keV) delivering individual X-ray pulses of 43 fs duration and 1.9 × 1010 photons per pulse focused into a spot size of ~1.5 µm in diameter using a pair of Kirkpatrick-Baez mirrors. LCP with protein microcrystals was extruded at room temperature and at a flow rate of 0.3 μl min−1 inside a vacuum chamber into the beam focus region using an LCP injector40 with a 50-μm diameter capillary. The XFEL beam was attenuated at transmission levels of 6.3–10% to avoid disruptions of the LCP stream. Diffraction images were collected at an XFEL pulse repetition rate of 120 Hz using a 2.3 Megapixel Cornell-SLAC Pixel Array Detector43 (CSPAD).
A total number of 390,442 detector images were collected, of which 43,417 (11% of total) were identified as potential crystal hits with more than 20 Bragg peaks with SNR = 4.0, threshold 200 and min-pix-count 3.0 using peakfinder8 algorithm as implemented in Cheetah42. The overall time of data collection from a sample with a total volume of 15 μl was about 54 min.
Data processing
All datasets, except for the SFX dataset CysLT1R_Zafirlukast-P21 (P21 space group), have been previously indexed, integrated, sorted, and merged to solve the structures of the corresponding receptor complexes by molecular replacement, as described28,29. Re-processing of the data with the same or better processing statistics as in the original manuscripts is described in the Technical validation section.
Data Records
SWSX data44–49 have been deposited to Zenodo under accession numbers provided in Table 3. Each SWSX dataset folder contains subfolders, representing each miniset collected, regardless of the angular range for data collection. Each miniset subfolder is named as XXX_YY_ZZ_NN, where XXX is the sequential number of the miniset, YY is the crystallization condition ID, ZZ is the serial number of the harvesting loop within each crystallization condition, NN – the serial number of the miniset within each loop. Each miniset subfolder contains a subfolder ‘images‘ with all diffraction images in either cbf or HDF5 format. It also contains an XDS parameter file XDS.INP with the keyword NAME_TEMPLATE_OF_DATA_FRAMES pointing to files in ‘images‘ subfolder, and other parameters as used during reprocessing (see keywords for express.py below). Also, each miniset subfolder contains all XDS-related files (including geometry correction x_geo_corr.cbf and y_geo_corr.cbf for cbf files) for this miniset (everything up to CORRECT.LP and XDS_ASCII.HKL for successfully integrated datasets, and only COLSPOT.LP for non-successful ones). A summary of all SWSX entries is shown in Table 2, and a summary of all SWSX entries crystallization conditions is present in Table 1, with a full description provided in Supplementary Table 1.
Table 3.
Data availability on Internet. Github gist, associated with the publication, has the ‘download_all.sh‘ script for Linux to download all data entries described in this publication.
| Entry | Reported resolution in PDB, Å | No. minisets (SWSX) or No. indexed lattices (SFX) | Entry page | Entry size |
|---|---|---|---|---|
| CysLT2R_6RZ6 | 2.43 | 236 (SWSX) | https://zenodo.org/record/4032836 | 48.2 GB |
| CysLT2R_6RZ7 | 2.43 | 158 (SWSX) | https://zenodo.org/record/4032837 | 88.1 GB |
| CysLT2R_6RZ9 | 2.73 | 11 (SWSX) | https://zenodo.org/record/4032841 | 18.7 GB |
| CysLT2R_ 6RZ8 | 2.70 | 44 (SWSX) | https://zenodo.org/record/4032840 | 8.9 GB |
| CysLT1R_ 6RZ4 | 2.70 | 186 (SWSX) | https://zenodo.org/record/4032826 | 90.0 GB |
| CysLT1R_ 6RZ5 | 2.53 | 43,417 (SFX) | https://www.cxidb.org/id-106.html | 134.9 GB |
| CysLT1R_Zafirlukast-P21 | N/A | 22,047 (SFX) | https://www.cxidb.org/id-107.html | 193.6 GB |
SFX data have been deposited to CXIDB as ID10650 (CysLT1R_6RZ5) and ID10751 (CysLT1R_Zafirlukast-P21). Only those images identified as crystal hits by Cheetah are included in the deposited dataset. Each SFX dataset folder contains a subfolder ‘raw_data’ with all runs as written by Cheetah, their respective cheetah.ini files and cxi files with images. Also, each SFX dataset folder contains a file ‘initial.geom’ that was used during reprocessing. A summary for all SFX entries is given in Table 4.
Table 4.
Summary of SFX datasets.
| Entry | CysLT1R_6RZ5 | CysLT1R_Zafirlukast-P21 |
|---|---|---|
| Reported resolution, Å | 2.53 | N/A |
| Reprocessed resolution, Å | 2.50 | 3.5 |
| Total No. images | 390,442 | 900,173 |
| No crystal hits | 43,417 | 22,047 |
| No. indexed images | 28,528 | 17,193 |
| No. indexed lattices | 28,900 | 17,193 |
Technical Validation
Data processing
During the preparation of this manuscript, all data were re-processed in a consistent manner. Here we present a pipeline for data processing that results in similar or better resolution values and figures of merit compared to those reported in the original papers. Data processing statistics for all datasets is shown in Table 5.
Table 5.
Crystallographic data collection statistics.
| PDB ID | CysLT1R | CysLT2R | |||||
|---|---|---|---|---|---|---|---|
| 6RZ4 | 6RZ5 | Zafirlukast-P21 | 6RZ6 | 6RZ7 | 6RZ9 | 6RZ8 | |
| Dataset type | SWSX | SFX | SFX | SWSX | SWSX | SWSX | SWSX |
| No. crystals (SFX) or No. minisets (SWSX) | 9 | 28,900 | 17,193 | 74 | 16 | 12 | 8 |
| Cell dimensions | |||||||
| a b c, Å | 59.39 45.89 87.13 | 41.58 68.56 87.37 | 43.86 159.43 73.48 | 69.34 167.95 85.44 | 81.82 142.07 171.21 | 67.67 165.01 82.66 | 78.40 78.40 171.91 |
| α β γ, ° | 90.00 95.69 90.00 | 76.33 76.69 81.19 | 90.00 95.12 90.00 | 90.00 90.00 90.00 | 90.00 90.00 90.00 | 90.00 90.00 90.00 | 90.00 90.00 90.00 |
| Space group | P21 | P1 | P21 | C2221 | F222 | I4 | C2221 |
| Resolution, Å# | 30.0-2.6 (2.67-2.60) | 30.0-2.53 (2.57-2.53) | 30.0-3.5 (3.56-3.50) | 30.0-2.3 (2.36-2.30) | 30.0-2.43 (2.49-2.43) | 30.0-2.5 (2.56-2.50) | 30.0-2.73 (2.80-2.73) |
| Completeness, %# | 99.9 (100.0) | 100.0 (100.0) | 100.0 (100.0) | 99.2 (97.6) | 98.2 (98.0) | 99.9 (100.0) | 94.7 (95.4) |
| Redundancy# | 7.24 (6.86) | 67.6 (31.0) | 128.7 (73.6) | 7.39 (5.80) | 6.15 (6.25) | 12.5 (12.8) | 3.27 (3.35) |
| Rmeas (SWSX) or Rsplit (SFX)# | 0.35 (1.05) | 0.23 (2.80) | 0.16 (2.59) | 0.30 (2.74) | 0.31 (3.33) | 0.24 (5.52) | 0.23 (3.06) |
| I/σ(I)# | 5.59 (1.93) | 2.89 (0.42) | 4.17 (0.46) | 5.75 (0.84) | 5.48 (0.79) | 8.62 (0.67) | 3.72 (0.69) |
| CC*# | 1.00 (0.57) | 0.99 (0.51) | 1.00 (0.61) | 1.00 (0.65) | 1.00 (0.63) | 1.00 (0.50) | 1.00 (0.57) |
#Values in parentheses are for highest-resolution shell.
SWSX data
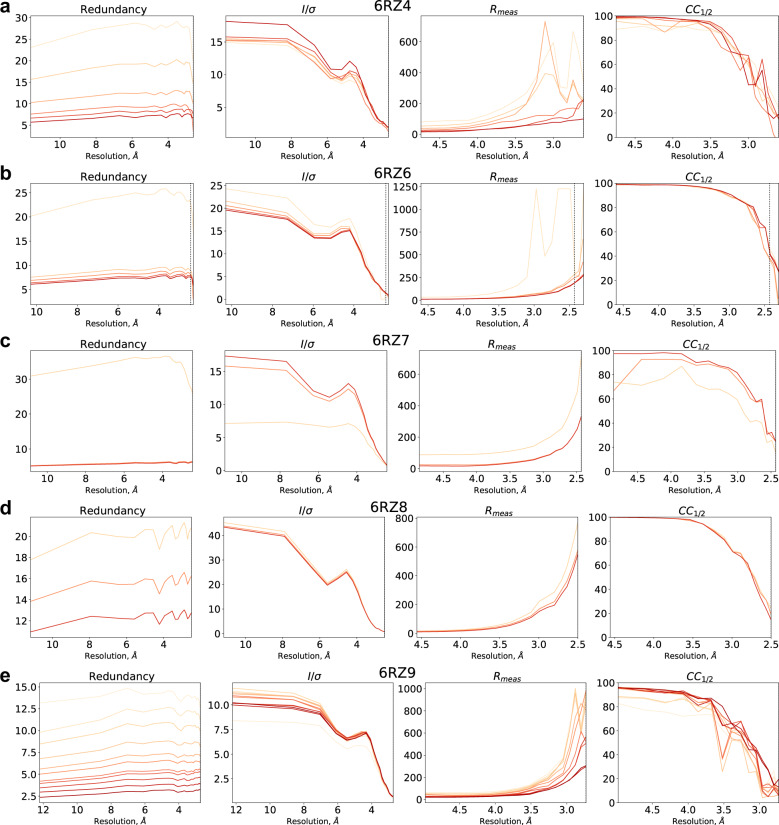
For SWSX data, the processing algorithm works as following (note that the treatment of both full datasets and minisets is the same). For each dataset, initial indexing and integration are performed by XDS within the resolution range of 40–2.5 Å using the beamline-provided XDS.INP file, without specifying the unit cell parameters and the space group identity (for Dectris images, the “neggia” library was used, as described here (https://strucbio.biologie.uni-konstanz.de/xdswiki/index.php/Eiger). Each integration runs first with the keywords “JOB = XYCORR INIT COLSPOT IDXREF DEFPIX INTEGRATE CORRECT”, then the integration parameters are updated using the output of the CORRECT step as described in the section “Final polishing: Re-INTEGRATEing with the correct spacegroup, refined geometry and fine-slicing of profiles” (https://strucbio.biologie.uni-konstanz.de/xdswiki/index.php/Optimisation#Re-INTEGRATEing_with_the_correct_spacegroup.2C_refined_geometry_and_fine-slicing_of_profiles), and integration is re-run using the keywords “JOB = DEFPIX INTEGRATE CORRECT”. After that, the algorithm attempts to scale all obtained XDS_ASCII.HKL files using XSCALE, and runs several rounds of ΔCC1/2 rejection of non-isomorphous minisets using xdscc12 subprogram as described17 (until there are no rejected minisets at the subsequent iteration). For most datasets, the miniset rejection was applied in two steps: first, using the low-resolution range (e.g. 30.0–10.0 Å, ΔCC1/2 threshold 0–2), and then using the high-resolution range (e.g. 10.0-2.5 Å, ΔCC1/2 threshold 1–5). All processing parameters are summarized in Table 6. The obtained dataset is merged and used as a REFERENCE_DATA_SET during the 2nd integration attempt of all minisets (including those rejected during the previous integration attempts). If CC1/2 in the highest resolution shell exceeds 0.15, the RESOLUTION_RANGE is increased manually for the 3rd integration. Next, another round of ΔCC1/2 rejection of non-isomorphous minisets is performed followed by merging to produce a final dataset. Improvements in the figures of merit for a dataset as a result of ΔCC1/2 rejection are shown in Fig. 3. For the CysLT2R_6RZ8 dataset (space group I4), there is an indexing ambiguity with two indexing options available for each miniset, thereby some minisets have to be re-indexed using ‘REIDX_ISET = 0 -1 0 0 -1 0 0 0 0 0 -1 0‘ keyword in XSCALE. This is done by following an iterative procedure: first, two largest minisets are merged together using two possible indexing options for the second set, and the indexing option resulting in a smaller Rmeas is chosen. Then, all other minisets are added one by one, using the indexing choice that producess smaller Rmeas for the merged dataset. For the final merged dataset, phenix.xtriage reports no significant twinning.
Table 6.
SWSX data processing parameters. For CysLT2R_6RZ6 and CysLT2R_6RZ7, only full resolution range rejection was performed.
| Entry | CysLT2R_6RZ6 | CysLT2R_6RZ7 | CysLT2R_6RZ9 | CysLT2R_6RZ8 | CysLT1R_6RZ4 | |
|---|---|---|---|---|---|---|
| Low-resolution rejection | Resolution limits, Å/No. bins | N/A | N/A | 30.0-10.0/17 | 30.0-10.0/7 | 32.0-10.0/5 |
| ΔCC1/2 threshold | N/A | N/A | 0.0 | 1.0 | 2.0 | |
| No. iterations | 0 | 0 | 5 | 5 | 4 | |
| High-resolution rejection | Resolution limits, Å/No. bins | 30.0-2.3/23 | 30.0-2.43/23 | 30.0-2.73/17 | 10.0-2.5/15 | 5.0-2.6/15 |
| ΔCC1/2 threshold | 3.0 | 3.0 | 5.0 | 1.0 | 2.0 | |
| No. iterations | 5 | 5 | 7 | 4 | 5 |
Fig. 3.
Improvements in data merging statistics for SWSX datasets during ΔCC1/2 rejection process. (a–e) Plots of redundancy, I/σ, Rmeas and CC1/2 vs resolution for each data processing iteration are shown for the following datasets: CysLT1R_6RZ4 (a), CysLT2R_6RZ6 (b), CysLT2R_6RZ7 (c), CysLT2R_6RZ8 (d), CysLT2R_6RZ9 (e). Darker curves represent latter stages of ΔCC1/2 miniset rejection.
SFX data
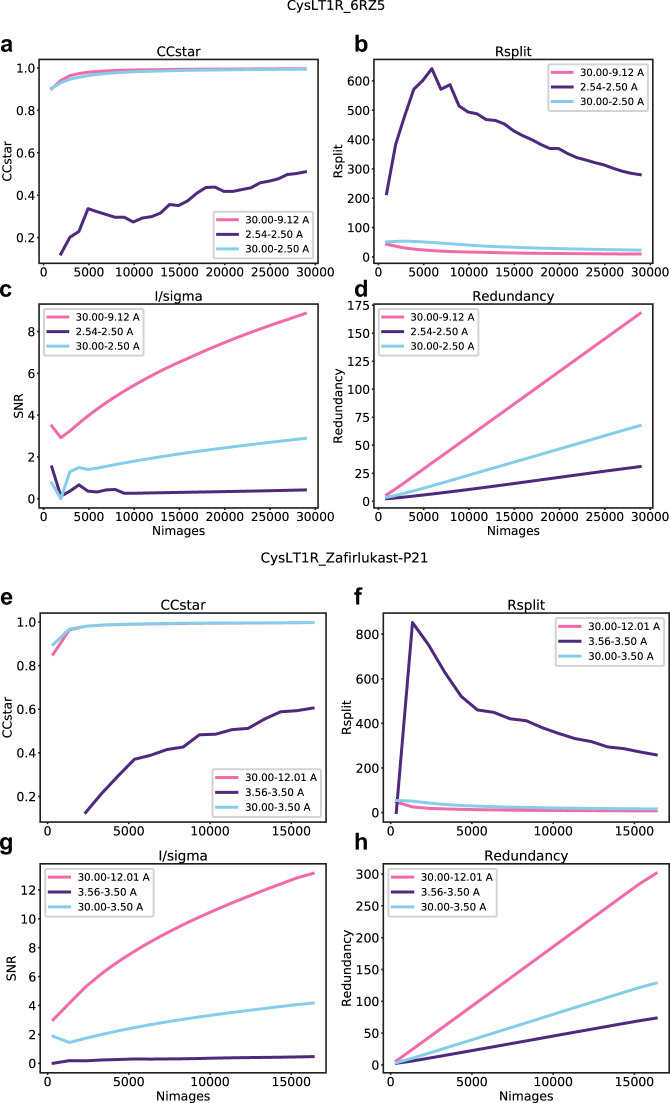
CysLT1R_6RZ5. Previously published data28 were processed using CrystFEL (v. 0.6.3 + 23ea03c7). For peak finding, peakfinder8 with min-snr = 4.5, threshold = 210 was used. For indexing, the following indexers were employed: felix, dirax, asdf, taketwo, mosflm-nolatt-cell, mosflm-nocell-latt, and xds (in that order), with–multi option enabled. Data were merged using process_hkl, with push-res = 1.8 and max-adu = 14,000. For reprocessing, the same parameters in CrystFEL (v. 0.8.0) were used. The final reprocessed dataset included 28,900 indexed lattices (67% of the frames selected by Cheetah). Among indexers, felix was the most successful one, providing 16,717 indexed lattices (57.8% of all indexed lattices). Improvements of Rsplit, CC* and I/sigma are shown in Fig. 4a–d.
Fig. 4.
Improvements in SFX data processing statistics with increasing the number of crystals. (a–d) Plots of I/σ (a), Rsplit (b), CC* (c) and redundancy (d) in three major resolution shells (low resolution, high resolution and overall) vs the number of crystals used for merging for the dataset CysLT1R_6RZ5. (e–h) The same as panels (a–d) but for the dataset CysLT1R_Zafirlukast_P21.
CysLT1R_Zafirlukast-P21
Data (previously unpublished) were processed using CrystFEL (v. 0.8.0). For peak finding, peakfinder8 with highres = 3.0, min-snr = 4.4, threshold = 20, max-res = 300 and min-res = 80 was used. For indexing, indexers dirax, taketwo, mosflm, xds, and asdf (in that order) were used. Data were merged using process_hkl, with mincc = 0.3 and push-res = 5.0. The final dataset resulted in 17,193 indexed lattices (79% of the frames selected by Cheetah). Among indexers, dirax was the most successful one, providing 14,457 indexed lattices. Improvements of Rpim, CC* and I/sigma are shown in Fig. 4e–h.
Usage Notes
Downloading data
The information about downloading data is shown in Table 3. A Linux script ‘download_all.sh‘, fetching all data using curl utility is provided on the Github gist, associated with the publication. Folder with each entry is archived in a single tar.gz file for more convenient fetching.
Data processing assistance scripts
Here, a brief description of scripts is given. Please, find a more detailed description in the github gist (https://gist.github.com/marinegor/96102c9b7ce87509a0832649d11ba927), associated with the publication.
-
create_xscale.inp.py — a simple script to include all existing XDS_ASCIIs to XSCALE.INP
Given the structure of folders as in data deposited in this publication, creates an input file for express.py in the csv format
-
express.py — the SWSX integration pipeline
Given a list of folders with XDS.INP and a path to the respective data sets, the script runs XDS for all data sets in the list, optionally adding UNIT_CELL_CONSTANTS, SPACE_GROUP_NUMBER, INCLUDE_RESOLUTION_RANGE, setting SPOT_RANGE same as DATA_RANGE, and setting REFERENCE_DATA_SET. Adds MAXIMUM_NUMBER_OF_PROCESSORS and MAXIMUM_NUMBER_OF_JOBS for processing on large clusters. Runs xscale_par afterwards.
-
xdscc.py — parsing of XDSCC.LP logfile of xdscc12 utility for rejection of minisets based on ΔCC1/2.
Analyses the output of xdscc12 utility together with the last XSCALE.INP used, providing the list of datasets with their ΔCC1/2 values. Saves list of those which have ΔCC1/2 higher than the input threshold value.
-
reject.sh — iterative “until no dataset with negative ΔCC1/2 are left” dataset rejection script
Scales XDS_ASCII.HKL files in all subfolders of the current folder. Then iteratively runs ΔCC1/2 rejection with the given resolution range and the number of cycles. Saves all intermediate XSCALE.INP-s and XSCALE.LP-s.
-
run_crystfel.sh
A wrapper for the indexamajig routine, which i) arranges all CrystFEL-related files into subfolders, ii) automatically assigns the date and time for each generated stream and respective log file, iii) links the last created stream to ‘laststream‘ link, and shuffles the input file list, so that one could quickly and reliably check the indexing rate before the indexing finishes.
analysis.sh
A wrapper for process_hkl, partialator, check_hkl, and compare_hkl routines, which produces an XSCALE.LP-like statistics table, counts images indexed with different indexers, produces a command-line visible histogram of the image resolution (for a simple estimation of the push-res parameter), and writes logs.
Supplementary information
Acknowledgements
SWSX data analysis and treatment was supported by Russian Science Foundation (project no. 19-74-00088). XFEL sample preparation and SFX data analysis was done with support of grant 18-02-40020 from the Russian Foundation for Basic Research (RFBR). A.L., A.R., V.G., A.M. and V.B. are thankful for the Ministry of Science and Higher Education of the Russian Federation (agreement # 075-00337-20-03, project FSMG-2020-0003). V.P. acknowledges support by the project Structural Dynamics of Biomolecular Systems (ELIBIO) (CZ.02.1.01/0.0/0.0/15_003/0000447) from the European Regional Development Fund and the Ministry of Education, Youth and Sports (MEYS) of the Czech Republic. Use of the Linac Coherent Light Source (LCLS), SLAC National Accelerator Laboratory, is supported by the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences under Contract No. DE-AC02-76SF00515. We acknowledge the European Synchrotron Radiation Facility for provision of beam time on ID23-1, ID30a3, ID29 and ID30b and we would like to thank structural biology group for assistance. V.C. acknowledges that the University of Southern California is his primary affiliation.
Author contributions
E.M. performed SWSX data collection, processed SFX and SWSX data, organized raw data and wrote the manuscript. A.L. produced and crystallized protein, performed SWSX data collection and wrote the manuscript. A.G. produced and crystallized protein, performed SWSX data collection and wrote the manuscript. K.K. performed SWSX data collection and processed SWSX data. S.B. deposited the data and helped with manuscript preparation. P.K. produced protein. V.P. helped with crystal harvesting and SWSX data collection. E.L. produced protein. A.R. deposited the data. V.G. supervised the project. A.M. performed SWSX data collection, helped with manuscript preparation and supervised the project. V.C. performed SFX data collection, wrote the manuscript and supervised the project. V.B. performed SWSX data collection, wrote the manuscript and supervised the project.
Code availability
The code used for data reprocessing (see usage notes in Technical validation section) is available on github gist (https://gist.github.com/marinegor/96102c9b7ce87509a0832649d11ba927). The utility xdscc12 is available through XDS-Wiki website (https://strucbio.biologie.uni-konstanz.de/xdswiki/index.php/Xdscc12). In previous publications, for SFX data processing, CrystFEL version 0.6.3 + 23ea03c7 (available on https://stash.desy.de/projects/CRYS/repos/crystfel/commits) was used. For SWSX data processing, XDS (version BUILT = 20161101 for CysLT1R_6RZ4 and BUILT = 20161205 for CysLT2R_6RZ5–9) and XSCALE (version BUILT = 20161101 for CysLT1R_6RZ4 and BUILT = 20180319 for CysLT2R_6RZ4–9) were used in the original publication, together with the “neggia” library for reading HDF5 images, as described (https://strucbio.biologie.uni-konstanz.de/xdswiki/index.php/Eiger). For data reprocessing, XDS and XSCALE version BUILT = 20190315, and CrystFEL 0.8.0, were used.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Vadim Cherezov, Email: cherezov@usc.edu.
Valentin Borshchevskiy, Email: borshchevskiy.vi@phystech.edu.
Supplementary information
is available for this paper at 10.1038/s41597-020-00729-2.
References
- 1.Bäck M, et al. International Union of Basic and Clinical Pharmacology. LXXXIV: Leukotriene Receptor Nomenclature, Distribution, and Pathophysiological Functions. Pharmacol. Rev. 2011;63:539–584. doi: 10.1124/pr.110.004184. [DOI] [PubMed] [Google Scholar]
- 2.Singh RK, Tandon R, Dastidar SG, Ray A. A review on leukotrienes and their receptors with reference to asthma. J. Asthma. 2013;50:922–931. doi: 10.3109/02770903.2013.823447. [DOI] [PubMed] [Google Scholar]
- 3.Shi Q-J, et al. Intracerebroventricular injection of HAMI 3379, a selective cysteinyl leukotriene receptor 2 antagonist, protects against acute brain injury after focal cerebral ischemia in rats. Brain Res. 2012;1484:57–67. doi: 10.1016/j.brainres.2012.09.020. [DOI] [PubMed] [Google Scholar]
- 4.Colazzo F, Gelosa P, Tremoli E, Sironi L, Castiglioni L. Role of the Cysteinyl Leukotrienes in the Pathogenesis and Progression of Cardiovascular Diseases. Mediators Inflamm. 2017;2017:1–13. doi: 10.1155/2017/2432958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Magnusson C, et al. Low expression of CysLT1R and high expression of CysLT2R mediate good prognosis in colorectal cancer. Eur. J. Cancer. 2010;46:826–835. doi: 10.1016/j.ejca.2009.12.022. [DOI] [PubMed] [Google Scholar]
- 6.Magnusson C, et al. Cysteinyl leukotriene receptor expression pattern affects migration of breast cancer cells and survival of breast cancer patients. Int. J. Cancer. 2011;129:9–22. doi: 10.1002/ijc.25648. [DOI] [PubMed] [Google Scholar]
- 7.Tsai M-J, et al. Cysteinyl Leukotriene Receptor Antagonists Decrease Cancer Risk in Asthma Patients. Sci. Rep. 2016;6:23979. doi: 10.1038/srep23979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Duah E, et al. Cysteinyl leukotriene 2 receptor promotes endothelial permeability, tumor angiogenesis, and metastasis. Proc. Natl. Acad. Sci. 2019;116:199–204. doi: 10.1073/pnas.1817325115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Moore AR, et al. Recurrent activating mutations of G-protein-coupled receptor CYSLTR2 in uveal melanoma. Nat. Genet. 2016;48:675–680. doi: 10.1038/ng.3549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ceraudo, E. et al. Uveal Melanoma Oncogene CYSLTR2 Encodes a Constitutively Active GPCR Highly Biased Toward Gq Signaling. bioRxiv 1–60, 10.1101/663153 (2019).
- 11.Yokomizo T, Nakamura M, Shimizu T, Sasaki F, Yokomizo T. Leukotriene receptors as potential therapeutic targets. J. Clin. Invest. 2018;128:2691–2701. doi: 10.1172/JCI97946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yamamoto M, et al. Protein microcrystallography using synchrotron radiation. IUCrJ. 2017;4:529–539. doi: 10.1107/S2052252517008193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mishin A, et al. An outlook on using serial femtosecond crystallography in drug discovery. Expert Opin. Drug Discov. 2019;14:933–945. doi: 10.1080/17460441.2019.1626822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zander U, et al. Merging of synchrotron serial crystallographic data by a genetic algorithm. Acta Crystallogr. Sect. D Struct. Biol. 2016;72:1026–1035. doi: 10.1107/S2059798316012079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Santoni G, Zander U, Mueller-Dieckmann C, Leonard G, Popov A. Hierarchical clustering for multiple-crystal macromolecular crystallography experiments: the ccCluster program. J. Appl. Cryst. 2017;50:1844–1851. doi: 10.1107/S1600576717015229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Foadi J, et al. Clustering procedures for the optimal selection of data sets from multiple crystals in macromolecular crystallography. Acta Crystallogr. Sect. D Biol. Crystallogr. 2013;69:1617–1632. doi: 10.1107/S0907444913012274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Assmann G, Brehm W, Diederichs K. Identification of rogue datasets in serial crystallography. J. Appl. Crystallogr. 2016;49:1021–1028. doi: 10.1107/S1600576716005471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hanson MA, et al. Crystal Structure of a Lipid G Protein-Coupled Receptor. Science (80-.). 2012;335:851–855. doi: 10.1126/science.1215904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Diederichs K. Dissecting random and systematic differences between noisy composite data sets. Acta Crystallogr. Sect. D Struct. Biol. 2017;73:286–293. doi: 10.1107/S2059798317000699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brehm W, Diederichs K. Breaking the indexing ambiguity in serial crystallography. Acta Crystallogr. Sect. D Biol. Crystallogr. 2014;70:101–109. doi: 10.1107/S1399004713025431. [DOI] [PubMed] [Google Scholar]
- 21.Asada H, et al. Crystal structure of the human angiotensin II type 2 receptor bound to an angiotensin II analog. Nat. Struct. Mol. Biol. 2018;25:570–576. doi: 10.1038/s41594-018-0079-8. [DOI] [PubMed] [Google Scholar]
- 22.White TA, et al. Serial femtosecond crystallography datasets from G-protein-coupled receptors. Sci. Data. 2016;3:160057. doi: 10.1038/sdata.2016.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Toyoda Y, et al. Ligand binding to human prostaglandin E receptor EP4 at the lipid-bilayer interface. Nat. Chem. Biol. 2019;15:18–26. doi: 10.1038/s41589-018-0131-3. [DOI] [PubMed] [Google Scholar]
- 24.Kato HE, et al. Structural mechanisms of selectivity and gating in anion channelrhodopsins. Nature. 2018;561:349–354. doi: 10.1038/s41586-018-0504-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim YS, et al. Crystal structure of the natural anion-conducting channelrhodopsin GtACR1. Nature. 2018;561:343–348. doi: 10.1038/s41586-018-0511-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Morin A, et al. Collaboration gets the most out of software. Elife. 2013;2:1–6. doi: 10.7554/eLife.01456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maia FRNC. The coherent X-ray imaging data bank. Nature Methods. 2012;9:854–855. doi: 10.1038/nmeth.2110. [DOI] [PubMed] [Google Scholar]
- 28.Luginina A, et al. Structure-based mechanism of cysteinyl leukotriene receptor inhibition by antiasthmatic drugs. Sci. Adv. 2019;5:eaax2518. doi: 10.1126/sciadv.aax2518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gusach A, et al. Structural basis of ligand selectivity and disease mutations in cysteinyl leukotriene receptors. Nat. Commun. 2019;10:5573. doi: 10.1038/s41467-019-13348-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Basu S, et al. Automated data collection and real-time data analysis suite for serial synchrotron crystallography. J. Synchrotron Radiat. 2019;26:244–252. doi: 10.1107/S1600577518016570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chun E, et al. Fusion partner toolchest for the stabilization and crystallization of G protein-coupled receptors. Structure. 2012;20:967–976. doi: 10.1016/j.str.2012.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ballesteros JA, Weinstein H. Integrated methods for the construction of three-dimensional models and computational probing of structure-function relations in G protein-coupled receptors. Methods Neurosci. 1995;25:366–428. doi: 10.1016/S1043-9471(05)80049-7. [DOI] [Google Scholar]
- 33.Caffrey M, Cherezov V, Caffrey M, Cherezov V. Crystallizing membrane proteins using lipidic mesophases. Nat. Protoc. 2009;4:706–731. doi: 10.1038/nprot.2009.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu W, Ishchenko A, Cherezov V. Preparation of microcrystals in lipidic cubic phase for serial femtosecond crystallography. Nat. Protoc. 2014;9:2123–2134. doi: 10.1038/nprot.2014.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ishchenko A, Cherezov V, Liu W. Preparation and delivery of protein microcrystals in lipidic cubic phase for serial femtosecond crystallography. J. Vis. Exp. 2016;2016:e54463. doi: 10.3791/54463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu, W. & Cherezov, V. Crystallization of Membrane Proteins in Lipidic Mesophases. J. Vis. Exp. e2501, 10.3791/2501 (2011). [DOI] [PMC free article] [PubMed]
- 37.Svensson O, Malbet-Monaco S, Popov A, Nurizzo D, Bowler MW. Fully automatic characterization and data collection from crystals of biological macromolecules. Acta Crystallogr. Sect. D Biol. Crystallogr. 2015;71:1757–1767. doi: 10.1107/S1399004715011918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Popov AN, Bourenkov GP. Choice of data-collection parameters based on statistic modelling. Acta Crystallogr. Sect. D Biol. Crystallogr. 2003;59:1145–1153. doi: 10.1107/S0907444903008163. [DOI] [PubMed] [Google Scholar]
- 39.Cherezov V, et al. Rastering strategy for screening and centring of microcrystal samples of human membrane proteins with a sub-10 $μ$m size X-ray synchrotron beam. J. R. Soc. Interface. 2009;6:587–597. doi: 10.1098/rsif.2009.0142.focus. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Weierstall U, et al. Lipidic cubic phase injector facilitates membrane protein serial femtosecond crystallography. Nat. Commun. 2014;5:3309. doi: 10.1038/ncomms4309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hart, P. et al. The CSPAD megapixel x-ray camera at LCLS. In X-Ray Free-Electron Lasers: Beam Diagnostics, Beamline Instrumentation, and Applications., 10.1117/12.930924 (2012).
- 42.Barty A, et al. Cheetah: software for high-throughput reduction and analysis of serial femtosecond X-ray diffraction data. J. Appl. Crystallogr. 2014;47:1118–1131. doi: 10.1107/S1600576714007626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Herrmann S, et al. CSPAD-140k: A versatile detector for LCLS experiments. Nucl. Instruments Methods Phys. Res. Sect. A Accel. Spectrometers, Detect. Assoc. Equip. 2013;718:550–553. doi: 10.1016/j.nima.2013.01.057. [DOI] [Google Scholar]
- 44.Marin E, 2020. CysLT1R receptor complex with Zafirlukast (P21 space group) structure (SFX@LCLS) Coherent X-ray Imaging Data Bank. [DOI]
- 45.Marin E, 2019. CysLT1R_6RZ4. Zenodo. [DOI]
- 46.Marin E, 2019. CysLT2R_6RZ6. Zenodo. [DOI]
- 47.Marin E, 2019. CysLT2R_6RZ7. Zenodo. [DOI]
- 48.Marin E, 2019. S. CysLT2R_6RZ8. Zenodo. [DOI]
- 49.Marin E, 2019. CysLT2R_6RZ9. Zenodo. [DOI]
- 50.Pándy-Szekeres G, et al. GPCRdb in 2018: adding GPCR structure models and ligands. Nucleic Acids Res. 2018;46:D440–D446. doi: 10.1093/nar/gkx1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Marin E, 2020. CysLT1R receptor complex with Zafirlukast (P1 space group) structure (SFX@LCLS) Coherent X-ray Imaging Data Bank. [DOI]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Marin E, 2020. CysLT1R receptor complex with Zafirlukast (P21 space group) structure (SFX@LCLS) Coherent X-ray Imaging Data Bank. [DOI]
- Marin E, 2019. CysLT1R_6RZ4. Zenodo. [DOI]
- Marin E, 2019. CysLT2R_6RZ6. Zenodo. [DOI]
- Marin E, 2019. CysLT2R_6RZ7. Zenodo. [DOI]
- Marin E, 2019. S. CysLT2R_6RZ8. Zenodo. [DOI]
- Marin E, 2019. CysLT2R_6RZ9. Zenodo. [DOI]
- Marin E, 2020. CysLT1R receptor complex with Zafirlukast (P1 space group) structure (SFX@LCLS) Coherent X-ray Imaging Data Bank. [DOI]
Supplementary Materials
Data Availability Statement
The code used for data reprocessing (see usage notes in Technical validation section) is available on github gist (https://gist.github.com/marinegor/96102c9b7ce87509a0832649d11ba927). The utility xdscc12 is available through XDS-Wiki website (https://strucbio.biologie.uni-konstanz.de/xdswiki/index.php/Xdscc12). In previous publications, for SFX data processing, CrystFEL version 0.6.3 + 23ea03c7 (available on https://stash.desy.de/projects/CRYS/repos/crystfel/commits) was used. For SWSX data processing, XDS (version BUILT = 20161101 for CysLT1R_6RZ4 and BUILT = 20161205 for CysLT2R_6RZ5–9) and XSCALE (version BUILT = 20161101 for CysLT1R_6RZ4 and BUILT = 20180319 for CysLT2R_6RZ4–9) were used in the original publication, together with the “neggia” library for reading HDF5 images, as described (https://strucbio.biologie.uni-konstanz.de/xdswiki/index.php/Eiger). For data reprocessing, XDS and XSCALE version BUILT = 20190315, and CrystFEL 0.8.0, were used.