Abstract
Hypertension is a public health concern. Low dose thiazide diuretics are known to effectively control blood pressure compared to that of other classes of antihypertensive drugs. In this context, we have performed an in-silico study and found that the two Sulphonamide Diuretics Hydrochlorothiazide and Indapamide bound the NADPH binding region of bacterial Dihydrofolate Reductase. Therefore, akin to Sulphonamide Antibiotics, Sulphonamide Diuretics may have antibiotic activity and thereby have the potential to modulate the gut microbiome in a way beneficial to vascular health. The in-silico experiment results were analyzed in the context of the relevant literature. We postulate that Sulphonamide Diuretics exert their antihypertensive role by modulating the gut microbiome, specifically by increasing butyrate-producing taxa in the gut. We recommend extending such work as it is plausible that Indapamide and other Sulphonamide Diuretics may be beneficial for both diabetes and hypertension.
Electronic supplementary material
The online version of this article (10.1007/s40203-020-00056-9) contains supplementary material, which is available to authorized users.
Keywords: Hypertension, Diuretics, Thiazide, Sulphonamide, Antibiotics, Gut microbiome, Diabetes mellitus
Introduction
Hypertension is a silent killer of humans (Arima et al. 2011). It precipitates coronary artery disease, cerebrovascular accident, and chronic renal failure, to name a few (Escobar 2002). Decades of research have led to the development of several classes of antihypertensive medications with different targets, which are used alone or in combination. Among the antihypertensive drugs, Sulphonamide Diuretics are a class of their own. These are commonly used medications for treating hypertension and offer an extra benefit to patients in low doses. They are more effective than other classes of antihypertensive agents (Roush and Sica 2016). Hydrochlorothiazide is perhaps one of the most commonly used Diuretic. It is often used orally as a first-line antihypertensive medication (Roush and Sica 2016). Indapamide is also a thiazide-like Diuretic. Some authorities believe that Indapamide controls systolic blood pressure better than Hydrochlorothiazide (Roush and Sica 2016).
Several Sulphonamides are known to have antimicrobial activity by binding to bacterial Dihydrofolate Reductase (DHFR) enzyme (Reeves et al. 1978; Wood 1942). DHFR is a critical enzyme involved in the folate synthesis pathway of bacteria. It catalyzes the reduction of dihydrofolate to tetrahydrofolate through the transfer of hydride from NADPH. The Met20 loop of DHFR consists of the amino-acid sequence VDRVIGMENAMPWNL and is involved in NADPH binding (Sawaya and Kraut 1997). During binding of the enzyme in its closed conformation, NADP + interacts with Ile14 of the Met20 loop (Sawaya and Kraut 1997). Gut microbial DHFR inhibition is believed to expand butyrate-producing taxa of the gut microbiome, which in turn supports good health (Maniar et al. 2017; Vallianou et al. 2019).
Sulphonamide Diuretics have basic Sulphonamide backbones similar to Sulphonamide Antibiotics (Fig. 1). However, to the best of our knowledge, the antibiotic activity of Sulphonamide Diuretics has not been investigated. Furthermore, there is currently no information available regarding whether Sulphonamide Diuretics inhibit bacterial DHFR or affect the human gut microbiome.
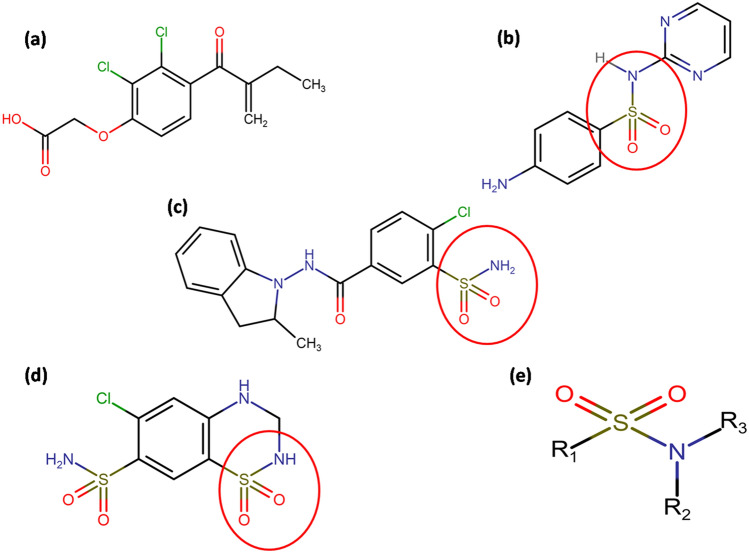
Fig. 1.
Line diagrams of the drugs considered: a Ethacrynic acid, b Sulphadiazine, c Indapamide, d Hydrochlorothiazide and e basic structure of Sulphonamide. The basic Sulphonamide moiety is circled in figures (b–d)
Vascular tone is seen as a function of the gut microbiome, and gut dysbiosis is among the theories which explain the hypertensive state (Yang et al. 2015). Brain-gut axis is a term becoming popular to describe the effect of gut microbiota on regulatory functions of the brain and vice versa to maintain overall homeostasis. Changes in the microbial community of the gastrointestinal tract affect blood pressure in experimental animals (Adnan et al. 2017). Antibiotic administration is also reported to affect the blood pressure of experimental animals. Furthermore, faecal transplantation affects blood pressure. Resistant hypertension may be controlled after antibiotic administration, and probiotic administration controls the hypertensive state (Qi et al. 2015). All these pieces of evidence prove that the gut microbiome is vital for the maintenance of blood pressure.
We believe that modification of the gut microbiome of patients may be associated with the extra benefits of these hypertensive medicines. The inhibition of bacterial DHFR affects the gut microbiome (Maniar et al. 2017; Vallianou et al. 2019). Sulphonamide Antibiotics inhibit bacterial DHFR; however, whether Sulphonamide Diuretics also interact with and inhibit bacterial DHFR is unknown. In this context, we reviewed the existing literature and explored this subject using tools of computational biology to generate in-silico data regarding the potential of Sulphonamide Diuretics to modulate the gut microbiome via the interaction with bacterial DHFR. In this paper, we present step by step the in-silico data supporting this theory.
Materials and methods
In silico studies
Three different categories of drugs were considered in this study (a) Sulphonamide Diuretics- Indapamide (Pubchem ID-3702) and Hydrochlorothiazide (3639), (b) a Sulphonamide Antibiotic- Sulphadiazine (5215), and (c) a non-sulphonamide diuretic-Ethacrynic acid (Pubchem ID-3278). The non-Sulphonamide Diuretic was considered the control. To evaluate the interactions of the drugs with the bacterial DHFR enzyme [Protein Data Bank (PDB) ID 1RX7], the docking tool AUTODOCK 4.2 (Morris et al. 2009) and Molecular Dynamic (MD) simulation tool GROMACS 2018.4 (Abraham et al. 2018) were used. The CD1 atom of Ile14 was chosen as the grid centre for docking, and docking was performed six times for each drug. For each docking, ten bound confirmations of the drug were obtained. Based on the binding energies obtained while running Autodock, the mean and standard deviations (SD) values of binding energy were calculated and listed in Table 1. The docked structures exhibiting the highest negative binding energy were considered for MD simulation. Drug topology files were generated using the SwissParam tool (Zoete et al. 2011), and CHARMM force-field (MacKerell et al. 1998) was used throughout the simulation. The DHFR-drug complexes were solvated in 1.2 nm cubic boxes of water using the TIP3P water model, and neutralization was performed using 11 sodium ions (Na+). Energy minimization was then performed for 50,000 steps using the steepest descent method. The system was then equilibrated using NVT (isothermal-isochoric ensemble) and NPT (isothermal-isobaric ensemble) protocols for each 50,000 steps. Then the entire system was considered for MD simulation of 10 ns runs at 300K temperature and 1 bar pressure and results were visualized using Pymol (DeLano 2002). Xmgrace plotting tool (Turner 2005) was used to analyze the graphs of potential energy, root mean square deviation (RMSD), the radius of gyration, hydrogen bonds, and other parameters as discussed in the results. If any two polar types of atoms, such as oxygen and nitrogen of a drug and the protein were visually located within a 3.5 Å distance, they were considered probable for hydrogen-bond formation. Superposition was performed using Pymol (DeLano 2002). The line diagrams of the drugs shown in Fig. 1 were prepared using Chemspace, which was available at https://chem-space.com/real-space. The two-dimensional (2D) representations of the DHFR-drug interactions at the NADPH binding site were generated using Poseview, which was available at https://proteins.plus/2ozr#poseview.
Table 1.
Mean binding energy and interaction energy values after docking and molecular dynamic (MD) run of protein with ligands
| Drug | Binding energy at Ile14 (mean ± SD) (KJ/mol) | Interaction energy after MD run of 10 ns (KJ/mol) |
|---|---|---|
| Ethacrynic acid (control) | – 21.4074 ± 0.36978 | – 35.5336 |
| Sulphadiazine (Antibiotic) | – 27.8354 ± 0.567008 | – 150.338 |
| Indapamide (Diuretic) | – 34.9712 ± 0.43396 | – 134.441 |
| Hydrochlorothiazide (Diuretic) | – 26.8201 ± 0.278589 | – 101.446 |
The mean values of binding energy of various drugs—Ethacrynic acid, Sulphadiazine, Indapamide, Hydrochlorothiazide along with SD values obtained after docking with respect to grid centre at CD1 of isoleucine14 of DHFR
In-vitro studies
If the tested diuretics inhibit bacterial Dihydrofolate Reductase, then the drug is expected to inhibit bacterial growth. Hydrochlorothiazide is a more popular drug compared to Indapamide. So we have checked the effect of Hydrochlorothiazide on E. coli MTCC 1687 growth. The strain is collected from the stool of healthy individuals. The bacterium was cultured overnight at 37 °C in Nutrient Broth.
Further serial dilution was done using this primary culture. 10− 5 dilution was used further for experimental purpose. On the other hand, 1 mg/ml stock solution of Hydrochlorothiazide (Aquazide, Sun Pharma, India) was prepared after dissolving in 1% ammonical distilled water. Next, the experiment was set in different aliquots. The test sets contained bacteria in nutrient broth and different concentrations of drug (6–14 µg/ml) in separate aliquots. The control sets had bacteria and appropriate control. The blank set contained only nutrient broth.
200 µl of each test, control and blank was added to 96 well ELISA plate, six times and placed in Multimode reader (TECAN). An overnight cycle of 45 reads was set up in Multimode Reader by providing the temperature of 37 °C for bacterial growth. The machine was set to take the OD of each sample after every 10,000 s at 620 nm. The effect of different concentrations of the drug on bacterial growth is plotted by subtracting the absorbance of the test from the control at 620 nm.
Results
It has been reported that Ile14 is one of the critical residues in theM20 loop of bacterial DHFR proteins involved in NADPH binding (Sawaya and Kraut 1997; Cao et al. 2018). Therefore, we considered the CD1 atom of Ile14 as the grid centre for the docking of drugs at the NADPH binding site of DHFR. Binding orientations of the drug molecules after docking are shown in Fig. 2. In all the DHFR-drug complexes, at least one hydrogen bond is observed. In addition to the hydrogen bond(s), non-bonded interactions were also seen. The binding energies of Ethacrynic acid, Sulphadiazine, Indapamide, and Hydrochlorothiazide with DHFR are shown in Table 1. Compared to the DHFR-ethacrynic acid complex, DHFR-sulphadiazine, DHFR-indapamide, and DHFR-hydrochlorothiazide complexes had higher negative binding energies, indicating more excellent stability of the complexes. The 2D interaction figures demonstrated the presence of hydrogen bonds within the complexes. Hydrophobic and pi-pi interactions were also observed (Fig. 3a, b, d, and f).
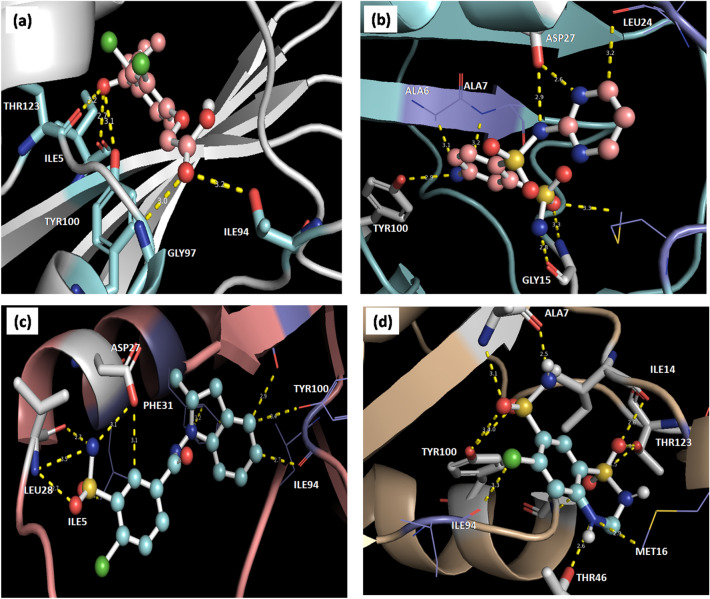
Fig. 2.
Interactions of different drugs with Dihydrofolate Reductase (DHFR) after docking with a ethacrynic acid, b Sulphadiazine, c indapamide and d Hydrochlorothiazide are represented. In ball and stick—drug, cartoon—DHFR, stick –residue of DHFR interacting with drug, yellow thick dotted line—hydrogen bond and yellow thin line—non bonded interactions are represented
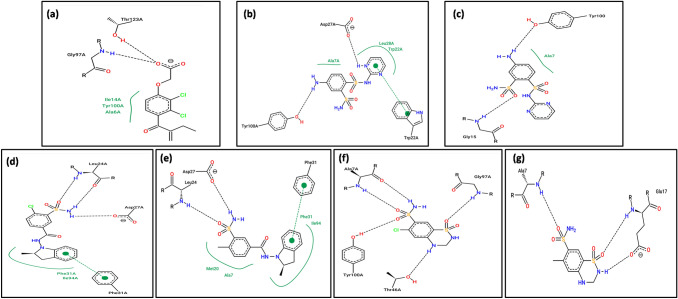
Fig. 3.
2D representation of interactions between residues of DHFR and drugs. a Ethacrynic acid, b, c Sulphadiazine, d, e Indapamide and f, g Hydrochlorothiazide. In (a, b, d and f) interactions after docking while in (c, e and g) interactions after 10 ns MD are represented. The arc represents the hydrophobic interaction. The pi-pi interaction if any, is also represented
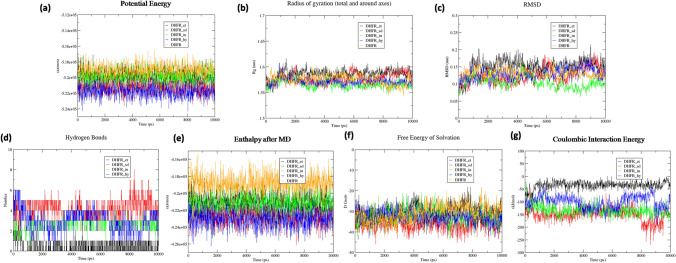
The DHFR-drug complexes and free DHFR obtained from the protein database were considered for MD simulation to compare the stabilities of the DHFR-drug complexes. 2D representations of the DHFR-drug interactions after MD simulation are shown in Fig. 3c, e, and g. Some of the hydrogen bonds observed during docking were also present after MD simulation. Even the pi–pi stacking in the DHFR-indapamide complex remained. The DHFR residues identified in drug binding after docking and MD simulation are included in Table 2. A series of parameters were analyzed after MD simulations and are represented in Fig. 4 and supplementary Figure S1. Potential energy curves indicated the stability of the DHFR-drug complexes compared to that of free DHFR and the DHFR-ethacrynic acid complex (Fig. 4a). The radius of gyration analysis revealed movement around an average value of 1.575 nm, except for the DHFR-ethacrynic acid complex (Fig. 4b). The radius of gyration was calculated with respect to the structure obtained after energy minimization. The RMSD curve reflected the same observation that fluctuation was less in the DHFR-hydrochlorothiazide and DHFR-indapamide complexes and higher in the DHFR-ethacrynic complex (Fig. 4c). All three graphs indicated the stability of the respective complexes. There were 3–5, 1–3, and 2–4 hydrogen bonds observed for Sulphadiazine, Indapamide and Hydrochlorothiazide, respectively. In the DHFR-ethacrynic acid complex, an average of one hydrogen bond was found throughout the run (Fig. 4d).
Table 2.
Residues of the bacterial Dihydrofolate Reductase (DHFR) observed in close contact with the drugs by hydrogen bonds and non bonded interactions after docking and Molecular Dynamic (MD) simulation are given
| Drug | Interactions | Residues interacting after Autodock | Residues interacting after MD simulation |
|---|---|---|---|
| Ethacrynic acid | Hydrogen bonds | Ile 5 | None |
| Ile 94 | None | ||
| Gly 97 | None | ||
| Tyr 100 | None | ||
| Thr 123 | None | ||
| Sulphadiazine | Hydrogen bonds | Gly 15 | Gly 15 |
| Glu 17 | |||
| Asp 27 | Asp 27 | ||
| Tyr 100 | Tyr 100 | ||
| Non bonded | Ala 6 | ||
| Ala 7 | Met 16 | ||
| Leu 24 | Leu 24 | ||
| Indapamide | Hydrogen bonds | Leu 24 | Glu 17 |
| Asn 23 | |||
| Asp 27 | Leu 24 | ||
| Asp 27 | |||
| Non bonded | Ile 5 | None | |
| Leu 28 | None | ||
| Phe 31 | None | ||
| Ile94 | None | ||
| Tyr 100 | None | ||
| Hydrochlorothiazide | Hydrogen bonds | Ala 7 | |
| Ile14 | Ala 7 | ||
| Thr 46 | Glu 17 | ||
| Gly 97 | Thr 46 | ||
| Tyr 100 | |||
| Thr 123 | |||
| Non bonded |
Met 16 Ile 94 |
Glu 17 His 45 |
Fig. 4.
Molecular dynamics (MD) simulation results of the DHFR-drug complexes after 10 ns run are presented. The codes of different drugs used in the graphs are et Ethacrynic acid, sd Sulphadiazine, in Indapamide and hy Hydrochlorothiazide
Enthalpy analysis demonstrated the DHFR-drug complexes were more stable compared to that of free DHFR (Fig. 4e). The enthalpy figure revealed a similar order of stability like potential energy. The free energy of Solvation curves of free DHFR and the DHFR-ethacrynic acid complex was identical throughout the run time. Free energy of Solvation of DHFR-sulphadiazine was most significant, while those for DHFR-hydrochlorothiazide and DHFR-indapamide were similar throughout the run time (Fig. 4f). Coulombic interaction energy of DHFR-hydrochlorothiazide, DHFR-indapamide and DHFR-sulphadiazine showed more favourable interactions than that of the DHFR-ethacrynic acid complex. It was observed that the Coulombic interaction energy of DHFR-hydrochlorothiazide and DHFR-sulphadiazine complexes fluctuated around − 150 KJ/mol while for the DHFR-Indapamide complex, the values fluctuated around − 100 KJ/mol (Fig. 4g). Moreover, we evaluated the enthalpy curve after equilibration using the NPT protocol and observed the fluctuation patterns were similar to the patterns observed after the complete simulation run (Figure S1a).
Solvent accessible surface areas were greater for all the drug-bound DHFR complexes compared to that of free DHFR (Figure S1b). As Ile14 of DHFR is an important residue for NADPH binding, we considered this residue as the grid centre for docking. The accessible surface area for ligand binding and root mean square fluctuation (RMSF) of Ile14 were separately observed throughout the trajectory (Figures S1c, d). For the DHFR-hydrochlorothiazide complex, both parameters were more significant compared to those for the DHFR-sulphadiazine and DHFR-Indapamide complexes, which exhibited similar trends. Superposed conformations of the DHFR-drug complexes after docking, and MD simulation are shown in Figure S2. For the DHFR-ethacrynic acid complex, the RMSD value was higher, while in the DHFR-sulphadiazine, DHFR-Indapamide, and DHFR-hydrochlorothiazide complexes, the values were lower (Table 3). This indicated that there were no significant changes in the conformation of DHFR-drug complexes other than the control DHFR-ethacrynic complex.
Table 3.
Root Mean Square Deviation (RMSD) values of superposed structures obtained after docking and Molecular dynamic (MD) simulation
| Drugs | RMSD values of superposed structures |
|---|---|
| Ethacrynic acid (Control) | 1.185 |
| Sulphadiazine (Antibiotic) | 0.853 |
| Indapamide (Diuretic) | 0.886 |
| Hydrochlorothiazide (Diuretic) | 1.051 |
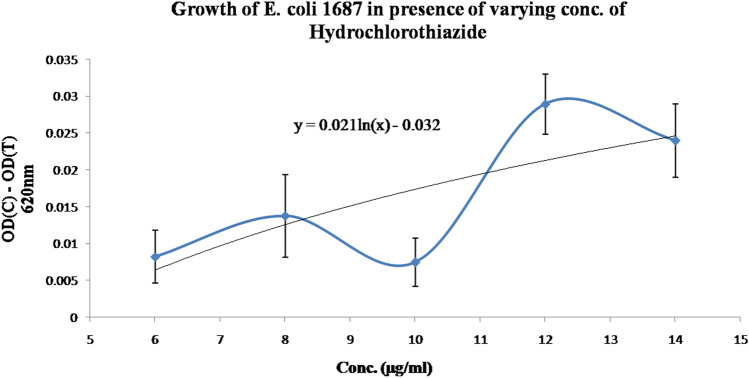
The results of the in-vitro experiments supported the conclusion of the in-silico study. It is observed that there is a dose-dependent increment of the difference of the absorbance of the control aliquot and test aliquot at 620 nm when increasing Hydrochlorothiazide doses are considered ranging from 6 to 14 µg/ml (Fig. 5). This indicates that there is a dose-dependent inhibition of bacterial growth in the presence of Hydrochlorothiazide. This is possible when the drug binds at the bacterial DHFR akin to Sulphonamide Antibiotics.
Fig. 5.
Effect of different doses of Hydrochlorothiazide on the growth of E. coli MTCC 1687 follows a logarithmic trend. The logarithmic equation for the above is shown in the graph
Discussion
Sulphonamide Antibiotics are drugs known to bind with DHFR (Reeves et al. 1978; Wood 1942). The interaction energies are shown in Table 2, and Fig. 4d, f, and g are consistent with this view. However, there are no previous reports that Diuretics, such as Hydrochlorothiazide and Indapamide, bind to DHFR. According to our current results, Sulphonamide Diuretics Hydrochlorothiazide and Indapamide did bind with DHFR, similar to the Sulphonamide Antibiotic Sulphadiazine (Table 2; Figs. 2, 3, and 4). The potential energy and enthalpy curves throughout the trajectory of Sulphadiazine and Hydrochlorothiazide had overlapping patterns, indicating the complexes had almost the same energies (Fig. 4a, e). Therefore, these Diuretics may act similar to Antibiotics from the perspective of binding with DHFR. We believe this observation is crucial for explaining the fact that low doses of thiazide and other Sulphonamide Diuretics produce superior antihypertensive effects compared to that of other classes of antihypertensive drugs. It may be noted that the non-sulphonamide diuretic Ethacrynic acid requires a higher dose for controlling blood pressure compared to thiazide diuretics, although it has 100% bioavailability (Scriabine 2007). For Ethacrynic acid, only one hydrogen bond was observed throughout the run (Fig. 4d), and the Coulombic interaction energy fluctuated around − 25 KJ/mol (Fig. 4g). The contention that Sulphonamide Diuretics can act as antibiotics gets support from the results of the in-vitro experiment. It is observed that Hydrochlorothiazide reduces the growth of E. coli MTCC 1687 in a dose-dependent manner (Fig. 5). Therefore, Hydrochlorothiazide and the other Sulphonamide Diuretics may act as an antibiotic.
It is worth noting that the antibiotic actions of various drugs used to treat chronic conditions like diabetes mellitus are beginning to be revealed. Among the oral antidiabetic drugs, Metformin is known to exhibit antibiotic action by inhibiting DHFR (Maniar et al. 2017; Gabel et al. 2017). Also, it has been reported that the antibiotic effect of Metformin expands the butyrate-producing taxa of the gut microbiome (Maniar et al. 2017; Vallianou et al. 2019). Butyrate is a known vasodilator, which, therefore, would be predicted to oppose hypertension (Mortensen et al. 1990). As expected, Metformin produces hypotensive effects in experimental animals, diabetic patients, and other human subjects (Muntzel et al. 1999; Landin-Wilhelmsen 1992; Majithiya and Balaraman 2006). However, contradictory evidence also exists in the literature. For instance, Metformin has also been shown to attenuate the reduction of in oral glucose-induced hypotension (Wulffelé et al. 2005; Borg et al. 2019).
Based on our observations, it was clear that the Sulphonamide Diuretics interacted with bacterial DHFR, akin to Sulphonamide Antibiotics. Therefore, it is highly probable that Sulphonamide Diuretics will inhibit the gut bacterial flora expressing DHFR and expand the butyrate-producing taxa, similar to that of Metformin. Since Sulphonamide Diuretics and Metformin are consumed long-term, unlike Sulphonamide Antibiotics, these drugs may act as gut microbiome modifiers. Thus, butyrate-induced hypotension would be expected in persons consuming Hydrochlorothiazide or Indapamide. However, not all cases of persons taking Metformin report clinical hypotension (Wulffelé et al. 2005). This suggests that other mechanism, in addition to butyrate, must be involved in the better antibiotic effect of these Diuretics. We hypothesize that the diuretic action, coupled with the antibiotic activity, was the reason for the extra benefit observed for the Sulphonamide Diuretics in treating hypertension (Fig. 6). On the other hand, it is not known whether Sulphonamide Antibiotics, if consumed for a prolonged period, would cause hypotension. We believe that the Sulphonamide Antibiotics with diuretic effects, if any, have the potential to control blood pressure.
Fig. 6.
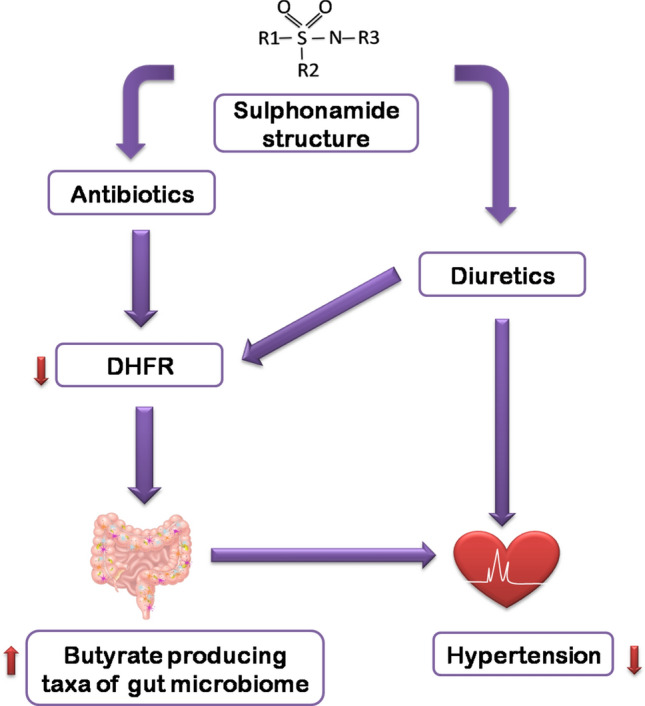
The proposed hypothesis—Sulphonamide core structure contains the sulfonyl group attached to an amine group, where the R1, R2 and R3 are respective side groups. Both Sulphonamide Antibiotics and Diuretics contain this moiety. How Sulphonamide Diuretics will control hypertension is illustrated
These findings should also be considered from another angle. If Sulphonamide Diuretics inhibit DHFR and modulate the gut microbiome to increase the butyrate-producing taxa, then similar to Metformin, they should also be beneficial to patients with diabetes mellitus. However, thiazides are known to increase the risk of diabetes (Zhang and Zhao 2016), which appears to be contradictory to our hypothesis. Thiazides are a class of drugs that increase uric acid in vivo (Raja et al. 2019). Increased uric acid is known to cause systemic inflammation; which further increases the risk of diabetes (McAdams et al. 2012). Therefore, the risk of diabetes mellitus is increased with the use of thiazides. This appears to be a thiazide-specific effect. All Sulphonamide Diuretics are not known to increase the risk of diabetes mellitus. Indapamide is recommended among the other class of Diuretics for patients with diabetes (Kuo et al. 2003). In the case of Indapamide, uric acid production is less compared to that of thiazides (Elliott et al. 1991), and it reduces blood pressure when administered at a low dose on alternate days (Inaba et al. 2004).
We believe that in the case of Indapamide and other Diuretics with Sulphonamide backbones, it is essential to know whether they increase butyrate-producing taxa of the gut microbiome. There are preliminary pieces of evidence suggesting that Indapamide improves glycemic and lipemic health, in addition to effectively controlling blood pressure (Sharabi et al. 1996). We believe this is due to its gut microbiome modifying actions, acting through the proliferation of the butyrate-producing taxa of gut microbiota (Maniar et al. 2017; Pluznick 2017). Only a focused clinical study for this purpose can verify these assertions. Designing and carrying out such a clinical trial is essential as both diabetes and hypertension remain silent killers. If a trial bears out the expected results, we will catch two birds (diabetes and hypertension) with a single stone (Sulphonamide Diuretics like Indapamide).In the future, an in-vitro study of the DHFR–Sulphonamide Diuretic interaction should be performed to validate our in-silico observations.
Conclusion
We hypothesize (Fig. 6) that Sulphonamide Diuretics are able to bind to bacterial DHFR, similar to Sulphonamide antibiotics. This binding should cause the Sulphonamide Diuretics to exhibit antibiotic property. Therefore, long term consumption of Sulphonamide can affect the gut microbiome to influence vascular health and blood pressure. Keeping into consideration the widespread occurrence of hypertension focussed research to investigate these issues are highly warranted.
Electronic supplementary material
Below is the link to the electronic supplementary material.
S1. Molecular Dynamics (MD) simulation results of the DHFR-Drug complexes after 10 ns are presented. The codes of different drugs used in the graphs are et- Ethacrynic acid, sd- Sulphadiazine, in- Indapamide and hy- Hydrochlorothiazide
S2. The superposed drug-DHFR complexes ((a)-Ethacrynic, (b)-Sulphadiazine, (c)-Indapamide and (d)-Hydrochlorothiazide) after docking and Molecular Dynamic (MD) simulation are shown. After docking in cyan cartoon – DHFR and red stick – drug, while after MD simulation in silver cartoon – DHFR and blue stick – drug are shown
Acknowledgements
SK and DB acknowledge PGIMER, Chandigarh for providing financial assistance vide Endst No. PGI/MERC/2020/4655-58 dated 7.10.2019 (Collectively) and No. 71/2-Edu-16/415 dated 23.1.2019.
Compliance with ethical standards
Conflict of interest
The authors declares that they have no competing interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Rajasri Bhattacharyya, Email: bdr.rajasri@yahoo.in.
Dibyajyoti Banerjee, Email: dibyajyoti5200@yahoo.co.in.
References
- Abraham MJ, van der Spoel D, Lindahl E, Hess B, and the GROMACS development team (2018) GROMACS User Manual version 2018.4. http://www.gromacs.org/. Accessed 4 Nov 2020
- Adnan S, Nelson JW, Ajami NJ, et al. Alterations in the gut microbiota can elicit hypertension in rats. Physiol Genom. 2017;49:96–104. doi: 10.1152/physiolgenomics.00081.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arima H, Barzi F, Chalmers J. Mortality patterns in hypertension. J Hypertens. 2011;29:S3–S7. doi: 10.1097/01.hjh.0000410246.59221.b1. [DOI] [PubMed] [Google Scholar]
- Borg MJ, Jones KL, Sun Z, et al. Metformin attenuates the postprandial fall in blood pressure in type 2 diabetes. Diabetes Obes Metab. 2019;21:1251–1254. doi: 10.1111/dom.13632. [DOI] [PubMed] [Google Scholar]
- Cao H, Gao M, Zhou H, et al. The crystal structure of a tetrahydrofolate-bound dihydrofolate reductase reveals the origin of slow product release. Commun Biol. 2018;1:226. doi: 10.1038/s42003-018-0236-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delano WL (2020) The pymol molecular graphics system. http://www.pymol.org [Internet]. [cited 2020 Sep 21]. https://ci.nii.ac.jp/naid/10020095229/. Accessed 4 Nov 2020
- Elliott WJ, Weber RR, Murphy MB. A double-blind, randomized, placebo-controlled comparison of the metabolic effects of low-dose hydrochlorothiazide and indapamide. J Clin Pharmacol. 1991;31:751–757. doi: 10.1002/j.1552-4604.1991.tb03772.x. [DOI] [PubMed] [Google Scholar]
- Escobar E. Hypertension and coronary heart disease. J Hum Hypertens. 2002;16:S61–S63. doi: 10.1038/sj.jhh.1001345. [DOI] [PubMed] [Google Scholar]
- Gabel SA, Duff MR, Pedersen LC, DeRose EF, Krahn JM, Howell EE et al (2017) A structural basis for biguanide activity. Biochemistry. Sep 12;56(36):4786–98 [DOI] [PMC free article] [PubMed]
- Inaba M, Noguchi Y, Yamamoto T, et al. Effects of a low dose of indapamide, a diuretic, given daily or every-other-day on blood pressure and metabolic parameters. Hypertens Res. 2004;27:141–145. doi: 10.1291/hypres.27.141. [DOI] [PubMed] [Google Scholar]
- Kuo S-W, Pei-Dee, Hung Y-J, et al. Effect of indapamide SR in the treatment of hypertensive patients with type 2 diabetes. Am J Hypertens. 2003;16:623–628. doi: 10.1016/S0895-7061(03)00896-3. [DOI] [PubMed] [Google Scholar]
- Landin-Wilhelmsen K. Metformin and blood pressure. J Clin Pharm Ther. 1992;17:75–79. doi: 10.1111/j.1365-2710.1992.tb01271.x. [DOI] [PubMed] [Google Scholar]
- MacKerell AD, Jr, Brooks B, Brooks CL, III, Nilsson L, Roux B, Won Y, Karplus M. CHARMM: the energy function and its parameterization. In: Schleyer PVR, Allinger NL, Clark T, Gasteiger J, Kollman PA, Schaefer HF III, Schreiner PR, editors. Encyclopedia of computational chemistry. Chichester: Wiley; 1998. pp. 271–277. [Google Scholar]
- Maniar K, Moideen A, Mittal A, et al. A story of metformin-butyrate synergism to control various pathological conditions as a consequence of gut microbiome modification: genesis of a wonder drug? Pharmacol Res. 2017;117:103–128. doi: 10.1016/j.phrs.2016.12.003. [DOI] [PubMed] [Google Scholar]
- Majithiya JB, Balaraman R. Metformin reduces blood pressure and restores endothelial function in aorta of streptozotocin-induced diabetic rats. Life Sci. 2006;78:2615–2624. doi: 10.1016/j.lfs.2005.10.020. [DOI] [PubMed] [Google Scholar]
- McAdams DeMarco MA, Maynard JW, Baer AN, et al. Diuretic use, increased serum urate levels, and risk of incident gout in a population-based study of adults with hypertension: the Atherosclerosis Risk in Communities cohort study. Arthritis Rheum. 2012;64:121–129. doi: 10.1002/art.33315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris GM, Huey R, Lindstrom W, Sanner MF, Belew RK, Goodsell DS, Olson AJ. Autodock4 and AutoDockTools4: automated docking with selective receptor flexiblity. J Comput Chem. 2009;16:2785–91. doi: 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortensen FV, Nielsen H, Mulvany MJ, et al. Short chain fatty acids dilate isolated human colonic resistance arteries. Gut. 1990;31:1391–1394. doi: 10.1136/gut.31.12.1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muntzel MS, Hamidou I, Barrett S. Metformin attenuates salt-induced hypertension in spontaneously hypertensive rats. Hypertension. 1999;33:1135–1140. doi: 10.1161/01.HYP.33.5.1135. [DOI] [PubMed] [Google Scholar]
- Pluznick JL. Microbial short chain fatty acids and blood pressure regulation. Curr Hypertens Rep. 2017;19:25. doi: 10.1007/s11906-017-0722-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi Y, Aranda JM, Rodriguez V, et al. Impact of antibiotics on arterial blood pressure in a patient with resistant hypertension—a case report. Int J Cardiol. 2015;201:157–158. doi: 10.1016/j.ijcard.2015.07.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raja R, Kavita F, Amreek F, et al. Hyperuricemia associated with thiazide diuretics in hypertensive adults. Cureus. 2019;11(8):e5457. doi: 10.7759/cureus.5457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roush GC, Sica DA. Diuretics for hypertension: a review and update. AJHYPE. 2016;29:1130–1137. doi: 10.1093/ajh/hpw030. [DOI] [PubMed] [Google Scholar]
- Reeves DS, Bint AJ, Bullock DW. Use of antibiotics. Sulphonamides, co-trimoxazole, and tetracyclines. BMJ. 1978;2:410–413. doi: 10.1136/bmj.2.6134.410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawaya MR, Kraut J. Loop and subdomain movements in the mechanism of Escherichia coli dihydrofolate reductase: crystallographic evidence. Biochemistry. 1997;36:586–603. doi: 10.1021/bi962337c. [DOI] [PubMed] [Google Scholar]
- Scriabine A. 6.32—hypertension. In: Taylor JB, Triggle DJ, editors. Comprehensive medicinal chemistry II. Oxford: Elsevier; 2007. pp. 705–728. [Google Scholar]
- Sharabi Y, Grossman E, Nussinovitch N, et al. Indapamide—a substitute diuretic for hypertensives with hyperglycemia and/or dyslipidemia. Harefuah. 1996;131:233–236. [PubMed] [Google Scholar]
- Turner PJ, Grace XM. Version 5.1.19. Center for coastal and land-margin research. Beaverton: Oregon Graduate Institute of Science and Technology; 2005. [Google Scholar]
- Vallianou NG, Stratigou T, Tsagarakis S. Metformin and gut microbiota: their interactions and their impact on diabetes. Hormones (Athens) 2019;18:141–144. doi: 10.1007/s42000-019-00093-w. [DOI] [PubMed] [Google Scholar]
- Wood WB. Studies on the antibacterial action of the sulfonamide drugs. J Exp Med. 1942;75:369–381. doi: 10.1084/jem.75.4.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wulffelé MG, Kooy A, Lehert P, et al. Does metformin decrease blood pressure in patients with Type 2 diabetes intensively treated with insulin? Diabetics Med. 2005;22:907–913. doi: 10.1111/j.1464-5491.2005.01554.x. [DOI] [PubMed] [Google Scholar]
- Yang T, Santisteban MM, Rodriguez V, et al. Gut dysbiosis is linked to hypertension. Hypertension. 2015;65:1331–1340. doi: 10.1161/HYPERTENSIONAHA.115.05315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Zhao Q. Association of thiazide-type diuretics with glycemic changes inhypertensive patients: a systematic review and meta-analysis of randomizedcontrolled clinical trials. J Clin Hypertens (Greenwich) 2016;18:342–351. doi: 10.1111/jch.12679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoete V, Cuendet MA, Grosdidier A, Michielin O. SwissParam, a fast force field generation tool for small organic molecules. J Comput Chem. 2011;32(11):2359–68. doi: 10.1002/jcc.21816. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
S1. Molecular Dynamics (MD) simulation results of the DHFR-Drug complexes after 10 ns are presented. The codes of different drugs used in the graphs are et- Ethacrynic acid, sd- Sulphadiazine, in- Indapamide and hy- Hydrochlorothiazide
S2. The superposed drug-DHFR complexes ((a)-Ethacrynic, (b)-Sulphadiazine, (c)-Indapamide and (d)-Hydrochlorothiazide) after docking and Molecular Dynamic (MD) simulation are shown. After docking in cyan cartoon – DHFR and red stick – drug, while after MD simulation in silver cartoon – DHFR and blue stick – drug are shown