Abstract
Symbiotic microorganisms associated with insects can produce a wide array of metabolic products, which provide an opportunity for the discovery of useful natural products. Selective isolation of bacterial strains associated with the dung beetle, Onthophagus lenzii, identified two strains, of which the antibiotic-producing Brevibacillus sp. PTH23 inhibited the growth of Bacillus sp. CCARM 9248, which is most closely related to the well-known entomopathogen, Bacillus thuringiensis. A comprehensive chemical investigation based on antibiotic activity discovered two new antibiotics, named lenzimycins A and B (1-2), which inhibited growth of Bacillus sp. CCARM 9248. The 1H and 13C NMR, MS, MS/MS, and IR analyses elucidated the structures of 1 and 2, which comprised a novel combination of fatty acid (12-methyltetradecanoic acid), glycerol, sulfate, and N-methyl ethanolamine. Furthermore, the acid hydrolysis of 1 revealed the absolute configuration of 12-methyltetradecanoic acid as 12S by comparing its optical rotation value with authentic (R)- and (S)-12-methyltetradecanoic acid. In addition to inhibition of Bacillus sp. CCARM 9248, lenzimycins A and B were found to inhibit the growth of some human pathogenic bacteria, including Enterococcus faecium and certain strains of Enterococcus faecalis. Furthermore, the present study elucidated that lenzimycins A and B activated a reporter system designed to detect the bacterial cell envelope stress, thereby indicating an activity against the integrity of the bacterial cell wall.
Keywords: antibiotic, Brevibacillus, Bacillus, dung beetle, lenzimycin, natural product
Introduction
Insects harbor symbiotic or pathogenic microorganisms; therefore, they have gained increasing interest as a reservoir for biotechnologically and pharmaceutically important microbes (Bode, 2010). In the past 20 years, several studies on insect-microbe interactions have reported on the role of bacterial natural products as molecular mediators in symbiotic insect systems (Cantley and Clardy, 2015; Pishchany and Kolter, 2020). There is an accumulating body of research which reports the insect symbiosis as biotechnological resources. For instance, Oh et al. (2009) analyzed the mutualistic associations in fungus-growing ant, Apterostigma dentigerum at the molecular level and found that the symbiotic bacterium, Pseudonocardia, produces dentigerumycin, a cyclic depsipeptide with highly modified amino acids, to selectively inhibit the associated parasitic fungus (Escovopsis sp.). Similarly, chemical ecological investigation of attine ants, Acromyrmex rugosus and their symbionts revealed a combination of antifungal compounds that inhibited the specialized fungal pathogen, Escovopsis. These small bacterial molecules were also evaluated as inhibitors against the human protozoan parasite Leishmania donovani (Ortega et al., 2019). It has also been reported that the symbiotic Streptomyces bacteria, located in specialized antennal glands, excrete antibiotics to protect wasp larvae from pathogenic fungal infestation (Kaltenpoth et al., 2005; Kroiss et al., 2010). Furthermore, studies have shown that the stingless bees, Melipona scutellaris, engage in defensive symbiosis with actinobacteria that produce antibacterial small molecules including the lobophorin, anthracycline, and cyclic hexadepsipeptide families, helping to protect their colonies against the entomopathogenic Paenibacillus larvae (Rodríguez-Hernández et al., 2019; Menegatti et al., 2020).
Dung beetles (Coleoptera: Scarabaeidae) are interesting Coleopteran insects, which feed on herbivores’ feces and make brood balls with feces for larval development. Although the ecology has not been studied much in most of the dung beetle species, their life cycle is closely linked with feces. Therefore, they may possess symbiotic relationships with chemically prolific bacteria, some of which may aid in control of the microbes in their ecosystems. In our previous studies, we have isolated a new tricyclic macrocyclic lactam, tripartilactam from the Streptomyces sp. found in the brood ball of a dung beetle species, Copris tripartitus (Park et al., 2012; Hwang et al., 2020). Furthermore, Kim et al. (2013) have isolated a Streptomyces strain producing tripartin, a novel dichlorinated indanone from the larva of C. tripartitus. Additionally, new alkenyl cinnamic acid-bearing cyclic peptides, coprisamides A and B (Um et al., 2015), and naphthoquinone-oxindole alkaloids, coprisidins A and B (Um et al., 2016), have also been discovered as natural products of bacterial isolates obtained from the gut of dung beetles.
Given the abundant species diversity of dung beetles and their ecological functions, it is expected to serve as a promising source in discovery of chemical compounds with potential pharmaceutical applications. In this study, we focused on a dung beetle species, Onthophagus lenzii (Yasuda, 1986), which is phylogenetically distant from C. tripartitus. The dung beetle O. lenzii (specimen shown in Figure 1) was collected from the city of Osan-si located on the outskirts of Seoul in South Korea. During the selective cultivation of bacteria using the excretion of the dung beetles, a Brevibacillus sp. PTH23 with an ability to inhibit a co-isolate, Bacillus sp. CCARM9247, which is a close relative of the well-known entomopathogen Bacillus thuringiensis, was isolated and further investigated to identify its antibiotic metabolites. Subsequent large-scale cultivation of PTH23 strain and chromatographic purification combined with a series of bioassay tests discovered two novel sulfated amino lipid antibiotics, lenzimycins A and B (Figure 2), which were also effective against some nosocomial human pathogenic bacteria such as Staphylococcus aureus, Enterococcus faecium, and Enterococcus faecalis. Furthermore, we also elucidated the structures and evaluated the biological functions of lenzimycins A and B (1-2).
FIGURE 1.
A specimen Onthophagus lenzii.
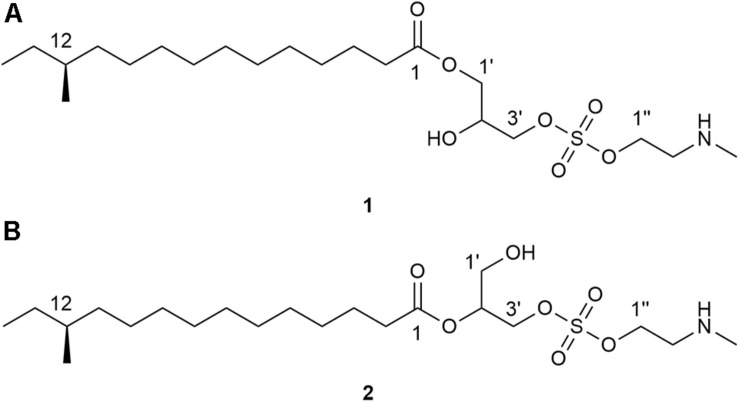
FIGURE 2.
Structures of lenzimycins A and B (1-2).
Materials and Methods
General Experimental Procedures
Optical rotations were measured using a JASCO P-2000 polarimeter with a 1-cm cell. The 1H NMR spectra were measured with the Bruker Avance III HD 850 MHz spectrometer at the National Center for Inter-University Research Facilities (NCIRF) at Seoul National University; the 13C and 2D NMR spectra were recorded on the Bruker-800 MHz and JEOL JNM-ECA-600 MHz NMR spectrometers, respectively, at the College of Pharmacy, Seoul National University. Chemical shifts of all NMR spectra were referenced to the residual protonated solvent peaks for CDCl3 (δH/δC, 7.26/77.0). Low-resolution electrospray ionization (ESI) LC/MS data were acquired on an Agilent Technologies 6130 quadrupole mass spectrometer coupled with an Agilent Technologies 1200-series HPLC. High-resolution fast atom bombardment (HR-FAB) mass spectra were obtained using a JEOL JMS-600W high-resolution mass spectrometer at NCIRF. High-resolution electrospray ionization (ESI) LC/MS and ESI MS/MS data were obtained using an AB SCIEX Q-TOF 5600 high-resolution mass spectrometer at the National Instrumentation Center for Environmental Management (NICEM, Seoul, South Korea).
Collection of Onthophagus lenzii (Harold)
A specimen of O. lenzii used in this study was collected from the entrance of the Doksanseong Fortress, Osan-si, Gyeonggi-do Province, South Korea, in March 2016. The adult dung beetle was secured inside a container using a tweezer and immediately transferred to the laboratory at Seoul National University. The collected specimen had a body length 6–12 mm and a glossy black body color, based on which it was identified as the dung beetle, Onthophagus (Strandius) lenzii Harold, 1874 (Coleoptera: Scarabaeidae). O. lenzii, widely distributed throughout Asia including Korea, Japan, and China, is the most dominant species among the genus Onthophagus (Coleoptera: Scarabaeidae: Scarabaeinae) in Korea (Kim, 2012). The prevailing appearance period of adults occurs annually from April to October, as they emerge from underground after an overwintering adult stage. These are the environmental cleanup insects, which eat and decompose the feces of cows, horses, and other large mammals (Yasuda, 1986). Moreover, the females of O. lenzii (Harold) make brood rooms and lay eggs under cow dung pats.
Isolation of Bacterial Strains From the Dung Beetle
The regurgitated dark red excretion from the mouth of the dung beetle was scrapped by using a sterilized loop and a needle and diluted in 40 mL distilled water. The suspension was heated in a water bath at 55°C for 5 min. For bacterial isolation, 1 mL of the bacterial suspension was spread on each isolation agar medium (ISP1, ISP2, ISP4, CD, K, Chitin, and actinomycete isolation medium with cycloheximide) (Supplementary Table S1) and incubated at 30°C for 2 weeks. Based on their initial phenotypic observations, two types of colonies were selected and isolated from actinomycete isolation medium, and they were designated as PTH23 and CCARM924 (Supplementary Figure S15).
Identification and Phylogenetic Analysis of Bacterial Isolates
The genomic DNAs of the isolated strains, PTH23 and CCARM9248, were extracted from the cell suspension cultures. The 16S rRNA gene was amplified by PCR with the bacterial universal forward primer 27F and reverse primer 1525R (Xu et al., 2003), and sequenced. Subsequently, similarity searches for the 16S rRNA gene sequences of the isolates were performed by the EzBioCloud server1 (Yoon et al., 2017) (GenBank accession number for PTH23: MT947196, and GenBank accession number for CCARM9248: MT947187), and the phylogenetic analysis was performed by the neighbor-joining method (Saitou and Nei, 1987) in the MEGA 7.0 program package (Kumar et al., 2016). Kimura’s two-parameter model was used to calculate the evolutionary distances (Kimura, 1980), and the bootstrap values were determined based on 1000 replications (Felsenstein, 1985).
Cultivation and Extraction of PTH23
Based on the inhibitory zones, the strain PTH23 was presumed to have inhibitory effects on the growth of CCARM9248. Therefore, the PTH23 strain was further cultured on a large scale. First, the PTH23 strain was cultivated in 50 mL of ISP2 medium (4 g yeast extract, 10 g malt extract, and 4 g glucose in 1 L distilled water) in a 125-mL flask, and incubated for 2 days on a rotary shaker at 180 rpm at 30°C. Five milliliters of the broth culture was transferred to 200 mL of ISP2 medium in a 500-mL flask for initial scale up and incubated for 2 days on a rotary shaker at 160 rpm at 25°C. Then for the large-scale culture, 25 mL of the broth culture (obtained through initial scale up) was inoculated to 1 L of ISP2 medium in 2.8-L flasks (60 ea × 1 L, total volume 60 L) and incubated for 3 days on a rotator shaker at 160 rpm at 25°C. Further, the 60-L culture broth of the PTH23 strain was extracted with 90 L of ethyl acetate using a separation funnel. The residual water in the collected ethyl acetate layer was removed by adding anhydrous sodium sulfate. The extract was concentrated to dryness in a rotary evaporator in vacuo to obtain the final concentrated extract of 15 g.
Purification of the PTH23 Extract
The extract of PTH23 was absorbed onto Celite, then loaded on a 2 g Sep-Pak C18 cartrige and fractionated with a step gradient solvent system (five fractions eluted with 100 mL each of MeOH-H2O from 20 to 100%). This process was repeated ten times to fractionate all the extract. After fractionation, lenzimycins A and B (1-2) were detected in the 80% MeOH-H2O fraction. The fraction eluted with 80% MeOH-H2O was filtered with a syringe filter (Advantec, 25HP045AN) and purified by semi-preparative reversed-phase HPLC column (YMC triart C18: 250 mm × 10 mm, 5 μm) with an isocratic solvent system [90% MeOH-H2O, refractive index (RI) detection, flow rate: 2 mL/min]. The antibiotics lenzimycins A (1) (3 mg) and B (6 mg) (2) were eluted at 18 min and 16 min, respectively.
Lenzimycin A (1)
Colorless powder; [α]−5.2 (c 0.50, MeOH) IR (neat) νmax 3274, 2924, 2853, 1735, 1633, 1464 cm–1; 1H and 13C NMR data, see Table 1; HRFABMS [M+H]+ m/z 454.2832 (calcd. for C21H44NO7S, 454.2838, error: −1.4 ppm).
TABLE 1.
1H and 13C NMR spectral data for lenzimycins A and B (1-2) in CDCl3.
| Position |
1a |
2b |
||||||
| δC | Type | δH, | mult (J in Hz) | δC | Type | δH, | mult (J in Hz) | |
| 1 | 173.8 | C | 173.3 | C | ||||
| 2 | 34.1 | CH2 | 2.30 | t (7.7) | 34.3 | CH2 | 2.30 | t (7.0) |
| 3 | 24.9 | CH2 | 1.60 | m | 24.9 | CH2 | 1.60 | m |
| 4–10 | 30.0-27.1 | CH2 | 1.31-1.23 | m | 30.0-27.1 | CH2 | 1.31-1.23 | m |
| 11 | 36.6 | CH2 | 1.27, 1.07 | m | 36.6 | CH2 | 1.27, 1.07 | m |
| 12 | 34.4 | CH | 1.28 | m | 34.4 | CH | 1.28 | m |
| 12-Me | 19.2 | CH3 | 0.83 | t (6.5) | 19.2 | CH3 | 0.84 | t (6.5) |
| 13 | 29.5 | CH2 | 1.25, 1.12 | m | 29.5 | CH2 | 1.25, 1.12 | m |
| 14 | 11.4 | CH3 | 0.84 | t (7.2) | 11.4 | CH3 | 0.85 | t (7.2) |
| 1′ | 64.5 | CH2 | 4.11 | m | 59.8 | CH2 | 3.73 | d (5.3) |
| 2′ | 69.1 | CH | 3.98 | m | 72.5 | CH | 4.97 | m |
| 3′ | 68.1 | CH2 | 4.00, 3.90 | m | 63.4 | CH2 | 4.08 | m |
| 1″″ | 61.6 | CH2 | 4.19 | m | 61.2 | CH2 | 4.20 | m |
| 2″ | 49.7 | CH2 | 3.15 | m | 49.9 | CH2 | 3.14 | m |
| 2″-NH | 9.64 | br s | 9.73 | br s | ||||
| 2″-N-Me | 33.6 | CH3 | 2.70 | s | 33.7 | CH3 | 2.71 | s |
a1H and 13C NMR were recorded at 800 MHz and 200 MHz, respectively.b1H and 13C NMR were recorded at 850 MHz and 212.5 MHz, respectively.
Lenzimycin B (2)
Colorless powder; [α]−1.0 (c 0.95, MeOH) IR (neat) νmax 3398, 2925, 2853, 1733, 1643, 1466 cm–1; 1H and 13C NMR data, see Table 1; HRFABMS [M+H]+ m/z 454.2833 (calcd. for C21H44NO7S, 454.2838, error: −1.3 ppm).
Acid Hydrolysis of Lenzimycin A (1)
To elucidate the absolute configurations of the fatty acid component, 1 (2.5 mg) was hydrolyzed in 0.5 mL of 6 N HCl and stirred at 120°C for 1 h. Then the reaction vial was cooled in an ice bath for 3 min. After the evaporation of the solvent in vacuo, the obtained dry material was dissolved in 0.5 mL of water and evaporated in vacuo for three times to remove residual HCl. The hydrolysate was purified by semi-preparative reversed-phase HPLC column (YMC triart C18: 250 mm × 10 mm, 5 μm) with an isocratic solvent system (90% MeOH-H2O, RI detection, flow rate: 2 mL/min). The desired product eluted at 31 min and confirmed as (S)-12-methyltetradecanoic acid (3, 0.5 mg) using 1H NMR and ESI-HRMS analyses.
(S)-12-Methyltetradecanoic Acid (3)
Colorless oil; [α] +5.6 (c 0.33, CHCl3) ESI-HRMS [M−H]– at m/z 241.2167 (calcd for C15H29O2, 241.2162, error: 2.1 ppm) 1H NMR (600 MHz, CDCl3), δH 2.35 (2H, t, J = 7.4), 1.64 (2H, q, J = 7.4), 1.26 (17H, m), 1.12 (1H, m), 1.07 (1H, m), 0.86 (3H, t, J = 7.4), 0.84 (3H, d, J = 6.4).
Minimum Inhibitory Concentration Assays Using Lenzimycins and Other Antibiotics
To investigate the antimicrobial activity of 1 and 2, a series of minimal inhibitory concentration (MIC) tests was performed against various microbial strains, mostly the well-known nosocomial pathogens (Table 2). The MIC evaluation for the strains listed in Table 2A was carried out using a plate media dilution method (dilution range from 0.25 to 128 μg/mL) following the guidelines of Clinical Laboratory Standard Institute (Clinical and Laboratory Standards Institute [CLSI], 2018) and by using Mueller-Hinton I medium (BBL, Sparks, MD, United States). The MIC testing of the panel of four E. faecalis strains shown in Table 2B was performed in liquid culture using Brain Heart Infusion Broth (FLUKA). Several clinically important antibiotics, such as ampicillin, norfloxacin, vancomycin, teicoplanin, and bacitracin (Sigma-Aldrich) were used for comparative analysis of the antimicrobial activities of 1 and 2.
TABLE 2A.
Antibacterial activity of lenzimycins A and B (1-2) against a panel of bacterial species.
| Number | Strain | Accession number | Remarks |
MIC (μg/mL) |
|||
| 1a | 2b | AMPc | NORd | ||||
| 1 | Bacillus sp. | CCARM 9248 | 64 | 8 | 16 | 0.12 | |
| 2 | Bacillus cereus | CCARM 0002 | 128 | 128 | 8 | 1 | |
| 3 | Enterococcus faecalis | CCARM 5171 | susceptible | >128 | >128 | 2 | 1 |
| 4 | Enterococcus faecalis | CCARM 5025 | VREe (vanB) | >128 | >128 | 2 | 2 |
| 5 | Enterococcus faecium | CCARM 5024 | VREe (vanA) | 0.5 | 1 | 32 | 4 |
| 6 | Staphylococcus aureus | CCARM 0205 | susceptible | 32 | 64 | 2 | 1 |
| 7 | Staphylococcus aureus | CCARM 3855 | susceptible | >128 | >128 | 0.25 | 1 |
| 8 | Staphylococcus aureus | CCARM 3089 | MDRf | >128 | >128 | 64 | 128 |
| 9 | Escherichia coli | CCARM 0230 | >128 | >128 | 2 | 0.03 | |
| 10 | Escherichia coli | CCARM 0237 | >128 | >128 | 8 | 0.5 | |
| 11 | Escherichia coli | CCARM 0235 | >128 | >128 | 4 | 0.06 | |
| 12 | Pseudomonas aeruginosa | CCARM 0219 | >128 | >128 | >128 | 1 | |
| 13 | Pseudomonas aeruginosa | CCARM 0223 | >128 | >128 | >128 | 0.5 | |
| 14 | Pseudomonas aeruginosa | CCARM 0225 | >128 | >128 | 0.5 | 0.25 | |
| 15 | Salmonella Typhimurium | CCARM 0240 | >128 | >128 | 1 | 0.06 | |
| 16 | Enterobacter cloacae | CCARM 0252 | >128 | >128 | >128 | 0.06 | |
| 17 | Enterobacter cloacae | CCARM 0253 | >128 | >128 | 1 | 0.03 | |
| 18 | Klebsiella oxytoca | CCARM 0248 | >128 | >128 | >128 | 0.03 | |
| 19 | Klebsiella aerogenes | CCARM 0249 | >128 | >128 | 2 | 0.06 | |
| 20 | Escherichia coli | ATCC 25922 | QCg strain | >128 | >128 | 4 | 0.06 |
| 21 | Enterococcus faecalis | ATCC 29212 | QCg strain | >128 | >128 | 0.5 | 2 ∼ 8 |
| 22 | Enterococcus faecium | ATCC 19434 | QCg strain | 16 | 8 | 0.5 | – |
| 23 | Pseudomonas aeruginosa | ATCC 27853 | QCg strain | >128 | >128 | >128 | 1 ∼ 4h |
| 24 | Staphylococcus aureus | ATCC 29213 | QCg strain | >128 | >128 | 0.13 | 0.5 ∼ 2h |
| 25 | Salmonella enteric | ATCC 14028 | QCg strain | 128 | 128 | 0.13 | – |
| 26 | Klebsiella pneumoniae | ATCC 10031 | QCg strain | >128 | >128 | 32 | – |
a lenzimycin A, blenzimycin B, cAmpicillin, dNorfloxacin, eVancomycin-resistant Enterococci, fMultiple drug resistance, gQuality control, and hAcceptable range.
TABLE 2B.
Antibacterial activity of lenzimycins A and B (1-2) against additional clinical isolates of Enterococci.
| Strain | Remarks |
MIC (μg/mL) |
||||
| 1a | 2b | Vc | Td | Be | ||
| Enterococcus faecalis JH2-2 | Parent strain, vancomycin sensitive | 3.1 | 1.6 | 25 | 3.1 | 25 |
| Enterococcus faecalis BM4316 | JH2-2::Tn1546, inducible resistance, VanA-type VREf | 4.2 | 10.4 | >100 | >100 | 83 |
| Enterococcus faecalis JH2-2::I | JH2-2::Tn1549, inducible resistance, VanB-type VREf | 6.3 | 6.3 | >100 | 50 | 83 |
| Enterococcus faecalis JH2-2::I C1 | point mutation in vanS, constitutive resistance | 4.2 | 6.3 | 100 | 83 | 83 |
a lenzimycin A, blenzimycin B, cVancomycin, dTeicoplanin, eBacitracin, and fVancomycin-resistant Enterococci.
Bioassays for Activity Against the Bacterial Cell Wall
To elucidate the mechanism of action of 1 and 2, the cell wall stress bioassay was performed. For the assay, approximately 107 spores of the Streptomyces coelicolor reporter strain harboring plasmid pIJ6880 (Hong et al., 2002) were spread onto MMCGT agar medium supplemented with 80 μg/mL kanamycin; then, lenzimycins A and B dissolved in DMSO were applied to paper disks, and incubated for 2–4 days at 30°C. The antibiotics vancomycin (10 μg) and bacitracin (50 μg) were used as positive controls, whereas novobiocin (50 μg), that acts by targeting the DNA gyrase enzyme, was used as a negative control. DMSO (5 μL) was included as a solvent blank.
Additionally, we also performed the vancomycin synergy assay. The vancomycin-resistant strain S. coelicolor wild type M600 was spread onto MMCGT agar containing either 0 μg/mL or 10 μg/mL vancomycin, and the antibiotics were applied to paper disks. The results were scored after 2–4 days of incubation at 30°C. Any change in diameter of the dark zone of inhibition around the white paper disks was assessed by comparing the results on the plates containing and lacking vancomycin. Bacitracin (B, 50 μg) was used as a positive control, whereas teicoplanin (T, 5 μg) was used as a control for antagonism. Kanamycin (K, 5 μg), which acts through inhibition of protein synthesis by targeting the 30S subunit of rRNA, was used as a negative control.
Results and Discussion
Isolation of Brevibacillus sp. PTH23 From the Dung Beetle, Onthophagus lenzii
Two different types of isolates, PTH23, and CCARM9248, were identified from the excretion of the O. lenzii. As shown in Supplementary Figure S15, the colonies pertaining to PTH23 (middle of the clear circular zone of growth inhibition on the agar) inhibited the growth of CCARM9248, (growing around the zone of inhibition) on the isolation plate. Sequencing of the 16S rRNA genes and phylogenetic analysis revealed that the strains PTH23 and CCARM9248 shared high similarity with Brevibacillus brevis NBRC 15304T (100%) and B. thuringiensis ATCC 10792T (99.8%), respectively (Supplementary Figures S16, S17). We therefore designated them as Brevibacillus sp. PTH23 and Bacillus sp. CCARM9248. The zone of inhibition on the agar could be due to the production of antibiotic metabolites from Brevibacillus sp. PTH23 which inhibited the growth of the co-isolate, Bacillus sp. CCARM9248.
Identification and Structure Elucidation of Key Metabolic Inhibitors of Bacillus sp. CCARM9248, Lenzimycins, From the Brevibacillus sp. PTH23 Extract
The Brevibacillus sp. PTH23 extract was fractionated by C18 reversed-phase column chromatography, and the fraction (80% MeOH-H2O), which displayed antibacterial activity, was analyzed by LC/MS. The chemical profile of the fraction revealed the presence of two metabolites, which had the molecular ions [M+H]+ commonly at m/z 454 in low-resolution ESI-MS analysis. Then, these compounds were further purified and designated as lenzimycins A and B (1-2) (Figure 2). Lenzimycin A (1) was obtained as a colorless oil. Its molecular formula C21H43NO7S, corresponding to one degree of unsaturation number, was deduced by the high resolution fast-atom bombardment mass spectroscopy (HR-FAB-MS) and NMR data (Table 1). The 1H NMR spectra of 1 (in CDCl3 at 800 MHz) identified one exchangeable proton (δH 9.64), seven protons attached to oxygenated carbons [δH 4.19 (2H overlapped), 4.11 (2H overlapped), 4.00, 3.98, and 3.90], five protons attached to a nitrogenous carbon [δH 3.15 (2H overlapped), 2.70 (3H)], twenty-three aliphatic protons (δH 2.30-1.12), and two aliphatic methyl groups (δH 0.84 and 0.83). The 13C and HSQC NMR data of 1 revealed the presence of one carbonyl carbon (δC 173.8), one oxymethine carbon (δC/δH 69.1/3.98), three oxymethylene carbons (δC/δH 68.1/4.00 and 3.90, 64.5/4.11, and 61.6/4.19), one methylene carbon bearing a nitrogen (δC/δH 49.7/3.15), twelve sp3 methylene carbons (δC/δH 36.6-24.9/2.30-1.23), one N-methyl group (δC/δH 33.6/2.70), and two methyl carbons (δC/δH 19.2/0.83 and 11.4/0.84) (Table 1).
Based on the COSY correlation analyses, three partial structures were elucidated. An array of COSY correlations from H2-2 (δH 2.30) to the terminal methyl protons H3-14 (δH 0.84) assembled a long lipophilic chain. The branch methyl group 12-Me (δC 19.2) was connected to C-12 (δC 34.4) by COSY correlation between H3-12-Me (δH 0.83) and H-12 (δH 1.28). This connectivity was further confirmed by HMBC correlations from H3-12-Me to C-11 (δC 36.6), C-12, and C-13 (δC 29.5). The HMBC correlation between H2-2/C-1 (δC 173.8) finally identified this substructure as 12-methyltetradecanoic acid. Consecutive COSY correlations from H2-1′ (δH 4.11) to H2-3′ (δH 4.00 and 3.90) through H-2′ (δH 3.98) established the three-carbon spin system from C-1′ (δC 64.5) to C-3′ (δC 68.1). Based on the 13C chemical shifts of C-1′, C-2′ (δC 69.1), and C-3′, these carbons were deduced to bear oxygen, thus confirming the second partial structure as glycerol. COSY correlation between H2-1″ (δH 4.19) and H2-2″ (δH 3.15) established the connectivity between C-1″ (δC 61.6) and C-2″ (δC 49.7). A broad singlet NH proton (δH 9.64) showed COSY correlations with H2-2″ and N-methyl protons (δH 2.70), thereby elucidating N-methyl ethanolamine which was supported by HMBC correlation from N-methyl protons to C-2″ (Figure 3A).
FIGURE 3.
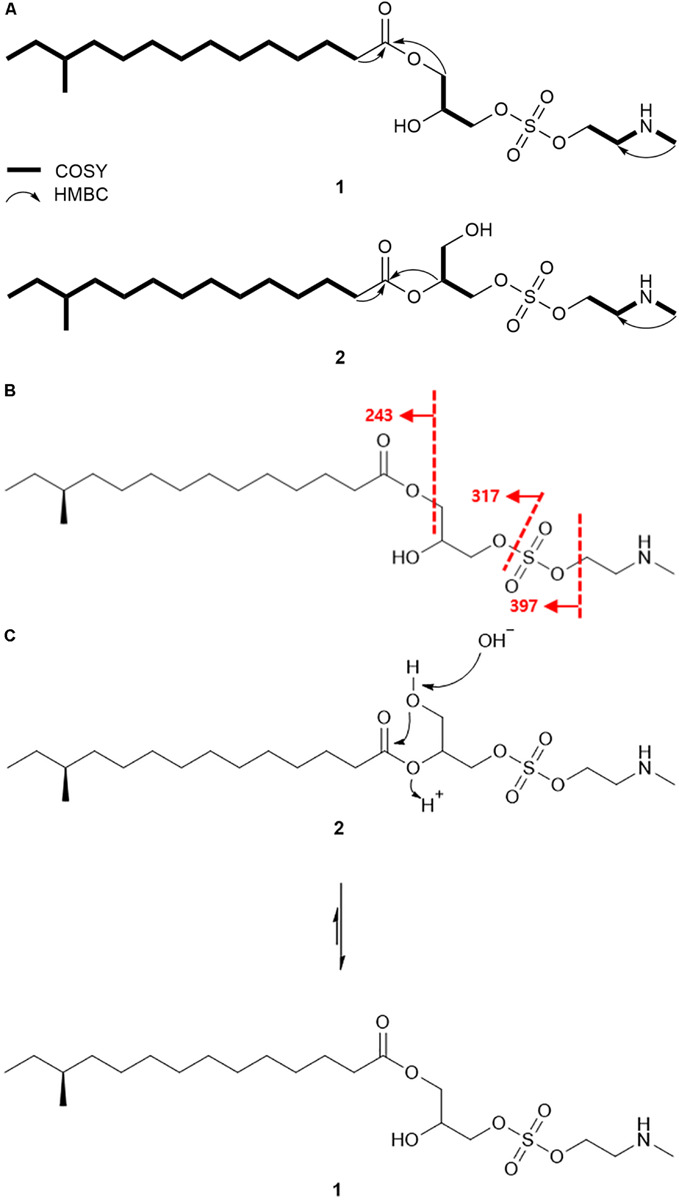
Detailed analysis of planar structures of lenzimycins. (A) Key 1H-1H COSY and HMBC correlations of lenzimycins A and B (1-2). (B) ESI-HR-MS/MS fragmentation of lenzimycin A (1). (C) Rearrangement between lenzimycins A and B (1-2).
HMBC correlations further assembled these partial structures. The H2-1′/C-1 HMBC correlation secured the connectivity between 12-methyltetradecanoic acid and glycerol through an ester linkage at the C-1′ position, and explained the molecular structure with C18H35O4, which in combination with N-methyl ethanolamine (C3H8NO) accounted for C21H43NO5 of 1. Furthermore, the IR data showed a strong absorption at 1464 cm–1 (Pretsch et al., 2008), thereby indicating the presence of a sulfate group, which was speculated to fit between the oxygenated terminal at C-3′ and C-2″ by placing the sulfur and two oxygen atoms. This addition connected glycerol and N-methyl ethanolamine thus completed the planar structure of 1 (Figure 3A).
The NMR-based structure of lenzimycin A (1) was confirmed by HR-ESI-MS/MS analysis. C-O at C-3′ and S-O bond connected at C-1″ were fragmented to yield the ion at m/z 397 by losing N-methyl ethanolamine; subsequently, it identified an ion at m/z 317 which represented the loss of the sulfate group along with N-methyl ethanolamine. Moreover, the mass difference between m/z 397 and 317 also confirmed the identity and location of a sulfate group, whereas, 12-methyltetradecanoic acid was represented by the ion at m/z 243 (Figure 3B).
Lenzimycin B (2) was obtained as a colorless oil, and its molecular formula was deduced to be C21H43NO7S, [identical to lenzimycin A (1)] through HR-FAB-MS data. The 1H and 13C NMR spectra of 2 displayed high similarity with those of 1 but the chemical shifts of the glycerol substructure (C-1′-C-3′) were noticeably different between these. Lenzimycin B (2) showed 1H signals at δH 4.97 (1H), 4.08 (2H), and 3.73 (2H) and 13C resonances at δC 72.5, 63.4, and 59.8 whereas lenzimycin A (1) displayed the corresponding protons at δH 4.11 (2H), 4.00 (1H), 3.98 (1H), and 3.90 (1H) and the carbons at δC 69.1, 68.1, and 64.5. Analysis of the HSQC NMR spectrum assigned all the C-H one bond correlations. Subsequent analysis of COSY and HMBC correlations assigned the three partial structures, 12-methyltetradecanoic acid, glycerol, and N-methyl ethanolamine, identical to 1. The IR absorption at 1466 cm–1 (Pretsch et al., 2008) also indicated the same sulfate group as lenzimycin A (1). Although mostly similar HMBC correlations were observed for the entire molecule of 2, 12-methyltetradecanoic acid was connected to C-2′ through an ester bond based on the crucial H-2′ (δH 4.97) /C-1 (δC 173.3) HMBC correlation, which finally determined its structure as a regioisomer of 1 with a different substitution pattern in the glycerol moiety (Figure 3A).
Lenzimycins A and B (1-2) have two stereogenic centers in their structures. The absolute configuration of C-12 in 12-methyltetradecanoic acid was determined by acid hydrolysis of lenzimycin A (1) followed by measurement of the optical rotation. After acid hydrolysis, 12-methyltetradecanoic acid (3) was obtained and structurally confirmed by 1H NMR and ESI-HR-MS analysis. The optical rotation value ([α] +5.6) was consistent with that of (S)-12-methyltetradecanoic acid ([α] +5.4) but opposite to the value of (R)-12-methyltetradecanoic acid ([α]−5.8) (Hansen et al., 1953), clearly assigning a 12S configuration, whereas the other chiral center at C-2′ remained unassigned. During purification and structure elucidation of the lenzimycins, it was observed that lenzimycin B (2) was converted to lenzimycin A (1) (Figure 3C). This could be due to the rearrangement of the fatty acid chain from the C-2′ to the C-1′ position as reported in other fatty acid glycerides (Biswas and Ganguly, 1960).
Lenzimycins A and B (1-2) were structurally novel as evidenced by the unprecedented combination of fatty acid, glycerol, sulfate, and N-methyl ethanolamine, which has not been reported either as natural products or synthetic compounds. The partial combination of 12-methyltetradecanoic acid and glycerol in 1 reported as aggreceride A (Supplementary Figure S19), showing inhibitory activity against platelet aggregation, has been identified from a Streptomyces strain (Ǒmura et al., 1986). However, to the best of our knowledge, fatty acid monoglycerides coupled with a sulfate group have not been reported as natural products, thereby indicating the novelty of the lenzimycins.
Antibiotic Activity of Lenzimycins
Even though the ecology of the oriental dung beetle Onthophagus species, including O. lenzii, is not well understood, it is clear that this dung beetle species is exposed to various entomopathogens in their habitat as they utilize microbe-rich feces of herbivores. B. thuringiensis is a representative entomopathogenic bacterium that is ubiquitously found in insect ecosystems. From the excretion of O. lenzii dung beetle, we isolated a potential symbiont of Brevibacillus sp. PTH23 and this bacterial strain seem to produce metabolites that inhibit the growth of entomopathogenic Bacillus species. Some metabolic agents isolated from Brevibacillus sp. are useful as a broad-spectrum treatment for Gram-positive bacteria, while some are even effective against Gram-negative bacterial infections (Hassi et al., 2012). We investigated the potential antibiotic activity of lenzimycins A and B against the Bacillus sp. CCARM9248. The estimated MIC values were 64 μg/mL and 8 μg/mL, respectively. Further, a panel of Gram-positive and Gram-negative bacteria that are mostly well-known nosocomial pathogens, including E. faecalis, E. faecium, S. aureus, Escherichia coli, Pseudomonas aeruginosa, Klebsiella oxytoca, Klebsiella aerogenes, Klebsiella pneumoniae, Enterobacter cloacae, Salmonella enterica, and Bacillus cereus were also tested to elucidate the broad range applicability of 1 and 2. The analyses showed markedly narrow-spectrum antibiotic effects as they were effective against only a few Enterococcus strains such as vancomycin-resistant E. faecium (VREF: vanA) with MIC values of 0.5 ∼ 1.0 μg/mL, E. faecium ATCC 19434 with MIC values of 8.0 ∼ 16 μg/mL, and an S. aureus strain CCARM 0205 with MIC values of 32 ∼ 64 μg/mL (Table 2A).
To further define the anti-enterococcal activity of the lenzimycins, additional MIC tests were performed using a different set of four well-studied clinical isolates of E. faecalis that respond differently to the glycopeptide antibiotic vancomycin (Foucault et al., 2010). E. faecalis JH2-2 is a clinical isolate which possesses no vancomycin resistance system and is sensitive to glycopeptide antibiotics. E. faecalis JH2-2::I is a derivative of JH2-2 containing a transposon, Tn1549, which confers an inducible, VanB-type resistance to glycopeptide antibiotics. The related strain, E. faecalis JH2-2::I C1, exhibits a constitutive resistance to glycopeptide antibiotics due to a point mutation in the vanS gene of the parent JH2-2::I strain. E. faecalis BM4316 is another clinical isolate containing transposon Tn1546 that confers an inducible, VanA-type glycopeptide resistance. VanA- or -B type VRE are distinguished by their susceptibility toward the two glycopeptide antibiotics, vancomycin and teicoplanin. Both VanA- and B-type VREs show high level, inducible resistance to vancomycin, but only the VanA-type is also highly resistant to teicoplanin, while the VanB-type shows teicoplanin sensitivity. This is believed to be due to the differences in how VanS senses the two different glycopeptide structures. VanS in VanA-type strains is activated in the presence of vancomycin, teicoplanin and other structurally unrelated cell wall-targeting antibiotics like moenomycin or bacitracin, while VanS in VanB-type strains is activated only by vancomycin and closely structurally related glycopeptides. The lenzimycins were found to be most active against the vancomycin-sensitive strain JH2-2 but also showed activity against both the inducible VanA- and VanB-type resistant strains, and the constitutively resistant JH2-2::I C1 strain (Table 2B). The growth of the other bacterial strains tested was not significantly affected except those of B. cereus CCARM 0002 and S. enterica ATCC 14028 which showed a weak inhibition (MIC = 128 μg/mL) (Table 2A).
Lenzimycins Induce Bacterial Cell Wall Stress
To investigate the activity of lenzimycins against the bacterial cell wall, a disk diffusion assay using a S. coelicolor reporter strain was performed. The reporter strain harbors a plasmid pIJ6880 that contains a sigEp-neo gene fusion construct designed to express the neo kanamycin resistance gene from a sigE promoter, which is inducible by cell wall stress (Hong et al., 2002). Growth in the presence of kanamycin, therefore, indicates cell wall stress, and this system has previously been used to provide a simple but effective bioassay screening tool to identify compounds which compromise the integrity of the bacterial cell envelope (Truman et al., 2014). In the disk diffusion assay, both lenzimycins A and B (1-2) stimulated the growth of the reporter strains, thereby indicating the bioactivity via weakening of the bacterial cell envelope (Supplementary Figure S18A). Interestingly, this activity was synergistic with the activity of the peptidoglycan biosynthesis inhibitor vancomycin in S. coelicolor which possesses an inducible VanB-type vancomycin resistance gene, vanJ, which is natively under the control of a vancomycin inducible promoter (Novotna et al., 2012; Supplementary Figure S18B). The findings suggested a different mode of action for the lenzimycins than that for vancomycin.
Conclusion
Chemical investigation of a Brevibacillus strain associated with the dung beetle, O. lenzii, revealed new antibiotic compounds, lenzimycins A and B (1-2). As the symbiotic interactions in the O. lenzii ecosystem have not been studied, it remains unclear whether the lenzimycins are the small molecules bridging microbial interactions in the dung beetle ecosystem. However, based on the antibiotic activity of the lenzimycins against Bacillus sp. CCARM 9248, most closely related to the entomopathogen B. thuringiensis, these compounds were shown to potentially have a protective role against general entomopathogenic bacteria that threaten the beetle’s health. Structural elucidation of the lenzimycins A and B (1-2) revealed that these unique antibiotics comprised unprecedented combination of a fatty acid, a glycerol, a sulfate, and an N-methyl ethanolamine moiety, which has not been reported in any natural and/or synthetic chemical compounds. Additionally, these new natural products exhibit antibiotic activity against some nosocomial human pathogenic bacteria including certain strains of Enterococcus that are resistant to the front-line glycopeptide antibiotic, vancomycin. Collectively, these results suggested the pharmaceutical potential of the lenzimycins. Furthermore, this study implicated that the chemical studies of insect-associated bacteria could be an effective research direction in the search for sources of novel natural antibiotics.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Author Contributions
D-CO conceptualized the study. JA, S-HH, and SK cultured and isolated the compound. B-YK analyzed the phylogeny of the bacterial strains. DW collected the dung beetle specimen. S-HH, DW, JA, JS, and D-CO performed spectroscopic analysis. H-JH, ES, K-BO, and JL designed and performed the microbial activity assays. S-IJ identified the beetle specimen. JA, S-IJ, JS, H-JH, and D-CO wrote and edited the text. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors submitted a patent application for this work. B-YK was employed by the company ChunLab, Inc. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors appreciate the significant help of the Culture Collection of Antimicrobial Resistant Microbes for antimicrobial assays against antibiotic resistant pathogens.
Funding. This work was supported by the National Research Foundation of Korea (NRF) grant fund by the Korean Government (Ministry of Science and ICT) (2018R1A4A1021703).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.599911/full#supplementary-material
References
- Biswas A. K., Ganguly D. (1960). Esterification of fatty acids with glycerol. Nature 188 56–57. 10.1038/188056b0 [DOI] [Google Scholar]
- Bode H. B. (2010). “Insect-associated microorganisms as a source for novel secondary metabolites with therapeutic potential,” in Insect Biotechnology, ed. Vilcinskas A. (Dordrecht: Springer; ), 77–93. 10.1007/978-90-481-9641-8_5 [DOI] [Google Scholar]
- Cantley A. M., Clardy J. (2015). Animals in a bacterial world: opportunities for chemical ecology. Nat. Prod. Rep. 32 888–892. 10.1039/c4np00141a [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clinical and Laboratory Standards Institute [CLSI] (2018). Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria that Grow Aerobically, 7th Edn Wayne, PA: Clinical and Laboratory Standards Institute. [Google Scholar]
- Felsenstein J. (1985). Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39 783–791. 10.2307/2408678 [DOI] [PubMed] [Google Scholar]
- Foucault M. L., Depardieu F., Courvalin P., Grillot-Courvalin C. (2010). Inducible expression eliminates the fitness cost of vancomycin resistance in enterococci. Proc. Natl. Acad. Sci. U.S.A. 107 16964–16969. 10.1073/pnas.1006855107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen R. P., Shorland F. B., Cooke N. J. (1953). The branched-chain fatty acids of Mutton Fat. II. The isolation of (+)-12-methyltetradecanoic acid and of 13-methyltetradecanoic acid. Biochem. J. 53 373–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassi M., Guendouzi S. E., Haggoud A., David S., Ibnsouda S., Houari A., et al. (2012). Antimycobacterial activity of a Brevibacillus laterosporus strain isolated from a moroccan soil. Braz. J. Microbiol. 43 1516–1522. 10.1590/s1517-83822012000400036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong H., Paget M., Buttner M. (2002). A signal transduction system in Streptomyces coelicolor that activates expression of a putative cell wall glycan operon in response to vancomycin and other cell wall-specific antibiotics. Mol. Microbiol. 44 1199–1211. 10.1046/j.1365-2958.2002.02960.x [DOI] [PubMed] [Google Scholar]
- Hwang S., Kim E., Lee J., Shin J., Yoon Y. J., Oh D.-C. (2020). Structure revision and the biosynthetic pathway of tripartilactam. J. Nat. Prod. 83 578–583. 10.1021/acs.jnatprod.9b00819 [DOI] [PubMed] [Google Scholar]
- Kaltenpoth M., Göttler W., Herzner G., Strohm E. (2005). Symbiotic bacteria protect wasp larvae from fungal infestation. Curr. Biol. 15 475–479. 10.1016/j.cub.2004.12.084 [DOI] [PubMed] [Google Scholar]
- Kim J. I. (ed.) (2012). “Laparosticti (Arthropoda: Insecta: Coleoptera: Scarabaeoidea),” in Insect Fauna of Korea, (Incheon: National Institute of Biological Resources; ), 78–79. [Google Scholar]
- Kim S.-H., Kwon S. H., Park S.-H., Lee J. K., Bang H.-S., Nam S.-J., et al. (2013). Tripartin, a histone demethylase inhibitor from a bacterium associated with a dung beetle larva. Org. Lett. 15 1834–1837. 10.1021/ol4004417 [DOI] [PubMed] [Google Scholar]
- Kimura M. (1980). A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 16 111–120. 10.1007/bf01731581 [DOI] [PubMed] [Google Scholar]
- Kroiss J., Kaltenpoth M., Schneider B., Schwinger M.-G., Hertweck C., Maddula R. K., et al. (2010). Symbiotic streptomycetes provide antibiotic combination prophylaxis for wasp offspring. Nat. Chem. Biol. 6 261–263. 10.1038/nchembio.331 [DOI] [PubMed] [Google Scholar]
- Kumar S., Stecher G., Tamura K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33 1870–1874. 10.1093/molbev/msw054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menegatti C., Lourenzon V. B., Rodríguez-Hernández D., da Paixao Melo W. G., Ferreira L. L. G., Andricopulo A. D., et al. (2020). Meliponamycins: antimicrobials from stingless bee-associated Streptomyces sp. J. Nat. Prod. 83 610–616. 10.1021/acs.jnatprod.9b01011 [DOI] [PubMed] [Google Scholar]
- Novotna G., Hill C., Vincent K., Liu C., Hong H.-J. (2012). A novel membrane protein, VanJ, conferring resistance to teicoplanin. Antimicrob. Agents Chemother. 56 1784–1796. 10.1128/aac.05869-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh D.-C., Poulsen M., Currie C. R., Clardy J. (2009). Dentigerumycin: a bacterial mediator of an ant-fungus symbiosis. Nat. Chem. Biol. 5 391–393. 10.1038/nchembio.159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ǒmura S., Nakagawa A., Fukamachi N., Otoguro K., Kobayashi B. (1986). Aggreceride, a new platelet aggregation inhibitor from Streptomyces. J. Antibiot. 39 1180–1181. 10.7164/antibiotics.39.1180 [DOI] [PubMed] [Google Scholar]
- Ortega H. E., Ferreira L. L., Melo W. G., Olivera A. L. L., Alvarenga R. F. R., Lopes N. P., et al. (2019). Antifungal compounds from Streptomyces associated with attine ants also inhibit Leishmania donovani. PLoS Negl. Trop. Dis. 13:e0007643. 10.1371/journal.pntd.0007643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S.-H., Moon K., Bang H.-S., Kim S.-H., Kim D.-G., Oh K.-B., et al. (2012). Tripartilactam, a cyclobutane-bearing tricyclic lactam from a Streptomyces sp. in a dung beetle’s brood ball. Org. Lett. 14 1258–1261. 10.1021/ol300108z [DOI] [PubMed] [Google Scholar]
- Pishchany G., Kolter R. (2020). On the possible ecological roles of antimicrobials. Mol. Microbial. 113 580–587. 10.1111/mmi.14471 [DOI] [PubMed] [Google Scholar]
- Pretsch E., Buhlmann P., Affolter C. (2008). “IR spectroscopy,” in Structure Determination of Organic Compounds: Tables of Spectral Data, 4th Edn, ed. Pretsch E. (Berlin: Springer; ), 304–307. [Google Scholar]
- Rodríguez-Hernández D., Melo W. G. P., Menegatti C., Lourenzon V. B., Nascimento F. S., Pupo M. T. (2019). Actinobacteria associated with stingless bees biosynthesize bioactive polyketides against bacterial pathogens. New J. Chem. 43 10109–10117. 10.1039/c9nj01619h [DOI] [Google Scholar]
- Saitou N., Nei M. (1987). The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4 406–425. [DOI] [PubMed] [Google Scholar]
- Truman A. W., Kwun M. J., Cheng J., Yang S. H., Suh J.-W., Hong H.-J. (2014). Antibiotic resistance mechanisms inform discovery: identification and characterization of a novel amycolatopsis strain producing ristocetin. Antimicrob. Agents Chemother. 58 5687–5695. 10.1128/aac.03349-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Um S., Bach D.-H., Shin B., Ahn C.-H., Kim S.-H., Bang H.-S., et al. (2016). Naphthoquinone–oxindole alkaloids, coprisidins A and B, from a gut-associated bacterium in the dung beetle, Copris tripartitus. Org. Lett. 18 5792–5795. 10.1021/acs.orglett.6b02555 [DOI] [PubMed] [Google Scholar]
- Um S., Park S. H., Kim J., Park H. J., Ko K., Bang H.-S., et al. (2015). Coprisamides A and B, new branched cyclic peptides from a gut bacterium of the dung beetle Copris tripartitus. Org. Lett. 17 1272–1275. 10.1021/acs.orglett.5b00249 [DOI] [PubMed] [Google Scholar]
- Xu P., Li W. J., Xu L. H., Jiang C. L. (2003). A microwave-based method for genomic DNA extraction from actinomycetes. Microbiology 30 82–84. [Google Scholar]
- Yasuda H. (1986). Fecundity of two dung beetle species, Onthophagus lenzii Harold and Liatongus phanaeoides Westwood (Coleoptera: Scarabacidae). Appl. Ent. Zool. 21 177–179. 10.1303/aez.21.177 [DOI] [Google Scholar]
- Yoon S.-H., Ha S.-M., Kwon S., Lim J., Kim Y., Seo H., et al. (2017). Introducing EzBioCloud: a taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int. J. Syst. Evol. Microbiol. 67 1613–1617. 10.1099/ijsem.0.001755 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.