Abstract
Peptidyl-tRNA hydrolase 2 (PTRH2; Bit-1; Bit1) is an underappreciated regulator of adhesion signals and Bcl2 expression. Its key roles in muscle differentiation and integrin-mediated signaling are central to the pathology of a recently identified patient syndrome caused by a cluster of Ptrh2 gene mutations. These loss-of-function mutations were identified in patients presenting with severe deleterious phenotypes of the skeletal muscle, endocrine, and nervous systems resulting in a syndrome called Infantile-onset Multisystem Nervous, Endocrine, and Pancreatic Disease (IMNEPD). In contrast, in cancer PTRH2 is a potential oncogene that promotes malignancy and metastasis. PTRH2 modulates PI3K/AKT and ERK signaling in addition to Bcl2 expression and thereby regulates key cellular processes in response to adhesion including cell survival, growth, and differentiation. In this Review, we discuss the state of the science on this important cell survival, anoikis and differentiation regulator, and opportunities for further investigation and translation. We begin with a brief overview of the structure, regulation, and subcellular localization of PTRH2. We discuss the cluster of gene mutations thus far identified which cause developmental delays and multisystem disease. We then discuss the role of PTRH2 and adhesion in breast, lung, and esophageal cancers focusing on signaling pathways involved in cell survival, cell growth, and cell differentiation.
Subject terms: Apoptosis, Tumour-suppressor proteins, Growth disorders
Facts
PTRH2 is a bi-functional protein with pro-survival and anoikis functions.
PTRH2 regulates adhesion mediated pro-survival signaling by upregulation of Bcl2 transcription.
Loss of adhesion promotes PTRH2-mediated anoikis by promoting PTRH2 translocation to the cytoplasm and interaction with the Gro/Tle transcriptional co-repressor.
Loss of PTRH2 induces premature skeletal muscle differentiation through increased caspase-3 activation.
Congenital Ptrh2 gene mutations cause multisystem disease including a congenital myopathy.
Open questions
Is PTRH2 the executioner for adhesion-dependent regulation of anoikis?
How does cellular localization impact PTRH2 bi-functional roles?
Is the enzymatic activity of PTRH2 required for any of these functions?
In cancer, what is the context that determines whether PTRH2 is pro-survival or anti-metastatic?
Introduction
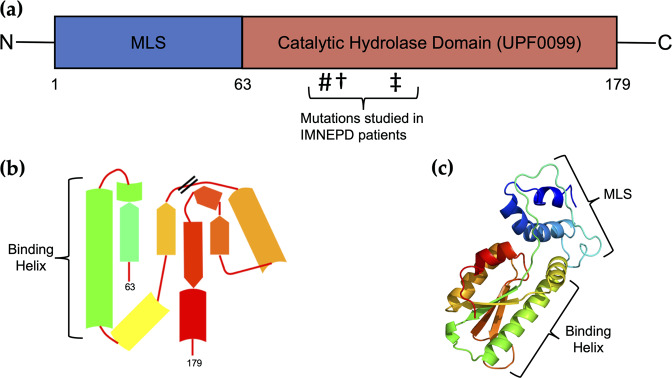
Peptidyl-tRNA Hydrolase (PTRH2; Bit1) is a nuclear-encoded, 19 kDa protein of 179 residues located at the plasma membrane, mitochondria, endoplasmic reticulum, and Golgi apparatus in eukaryotic cells1–4. The relevance of PTRH2 in human disease was identified after its homologs were characterized in microorganisms. Bacterial and archaeal peptidyl-tRNA hydrolases are biologically essential as they maintain the efficiency of translation turnover by hydrolyzing aberrant peptidyl-tRNA. This catalytic activity is conserved in yeast, bacteria, and eukaryotes, although it is nonessential in yeast5. In humans, the PTRH2 gene is on chromosome 17 and is expressed in all tissue types6,7. The 3D structure of PTRH2, as determined by 2 Å resolution crystallography, is an α/β domain consisting of a five-strand beta-sheet flanked by two alpha-helices on either side (Fig. 1)1. PTRH2’s crystallographic asymmetrical structure suggests it may bind to proteins via the largest alpha-helix located within its catalytic domain1. Indeed, PTRH2 mutations have been identified in patients suffering from Infantile-onset Multisystem Nervous, Endocrine, and Pancreatic Disease (IMNPED) that are clustered within this catalytic domain8–12. PTRH2 regulates cell functions including cell survival and death, muscle differentiation, and cancer cell metastasis.
Fig. 1. PTRH2 structure.
a PTRH2 is 179 residues in length and contains a mitochondrial localization sequence (MLS) and a hydrolase domain (UPF0099). #Missense glutamine to proline mutation of the 85th residue (p.Q85P). †Frameshift mutation at the 90th residue (p.Ala90fs). ‡Nonsense mutation at the 108th residue (p.W108*). b PTRH2 catalytic domain folds into an α/β structure with a binding helix. c 3D structure of PTRH2 generated by X-ray crystal diffraction and visualized in Phyre286.
The study of PTRH2 loss of function mutations in IMNEPD patients and in a global knockout mouse has revealed its essential roles in the regulation of pro-survival and anoikis signaling in addition to cellular stress responses, cell size, and myoblast differentiation. PTRH2’s functions are determined by its cellular localization, binding partners, phosphorylation state, and integrin-mediated attachment. In cells attached to the extracellular matrix (ECM), PTRH2 mediates stress resistance through an integrin-Bcl2 pro-survival signaling pathway3,13,14. Conversely, in detached cells, PTRH2 is localized to the cytoplasm where it binds to the Amino-terminal Enhancer of Split (AES; also known as Transducin-Like Enhancer family member 5; TLE5) to initiate anoikis2. Anoikis is programmed cell death (apoptosis) that occurs when cells lose integrin-mediated attachment to the ECM15. Anoikis is important in normal embryonic development and is suppressed in detached cancer cells16. It occurs in cells and tissues where specific ECM adhesion is required for cell survival and where the loss of this adhesion inactivates cell survival pathways and instead activates a signal transduction cascade leading to activation of caspases and cell death. How cell attachment suppresses anoikis is not completely understood; however, anti-apoptotic Bcl2 family members block anoikis17,18. In vitro, functional analysis of PTRH2 found the catalytic domain region increases cell survival whereas a peptide fragment within the first 62 amino acids of the mitochondrial localization sequence (MLS) promotes cell death due to anoikis19. Because of its ability to initiate anoikis in detached cells, in part by inhibiting transcription via an unknown mechanism, PTRH2 is also referred to as Bcl2 inhibitor of transcription (Bit-1; Bit1)2.
Thus, the function of PTRH2/Bit1 in regulating cell survival or anoikis is context-dependent and related to the adhesion state of the cell and may further relate to the specific cell type. These roles are central to PTRH2 function in IMNEPD and cancer. Due to its context-dependent effects on cell survival and cell death, the ablation of PTRH2 in different cancers may either enhance or suppress malignancy. By summarizing current knowledge of PTRH2 and its essential roles in development, homeostasis, and metastasis, we hope to inspire further research of its potential as a therapeutic target in the treatment of congenital diseases and cancer.
IMNEPD syndrome reveals the significance of PTRH2 in development and homeostasis
Analysis of exome and genome data from the Genome Aggregation Database version 2 (gnomAD v.2.1.1) indicates deleterious mutations of PTRH2 are extremely rare and selected against with allele frequencies on the order of 0.0004%20. Due to PTRH2’s recessive inheritance pattern, it is no surprise that IMNEPD is extremely rare in normal populations and has only been documented in children born to consanguineous mutant allele carriers8–12. Despite its rarity, studies of this disease and the individuals affected by it are important because they identify multisystemic manifestations caused by PTRH2 gene mutations indicating important roles for the protein and potential therapeutic applications.
Three PTRH2 loss of function gene mutations manifest as disease phenotypes affecting multiple tissues
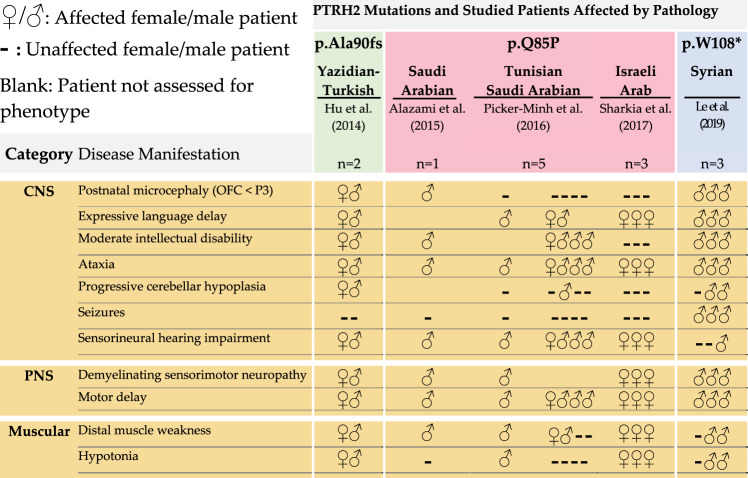
Whole-exome sequencing of consanguineous families and screening for deleterious genes was instrumental in the discovery of recessive PTRH2-null mutations that cause IMNEPD8,9,11. This method is highly effective for the identification of mutations that are rare in human populations due to significant negative selection. Hu et al. utilized whole-exome sequencing to identify a mutant PTRH2 allele in two children of consanguineous heterozygous carriers of Yazidian–Turkish descent8. This mutation (p.Ala90fs; Tables 1 and 2) consists of a CT deletion at positions 269 and 270, respectively. The resultant frameshift mutation yields a truncated 102-residue PTRH2 variant that lacks a catalytic domain, resulting in a non-functional protein. While this mutation does not alter PTRH2 mRNA transcript levels, there is decreased total PTRH2 protein in fibroblasts isolated from affected patients compared to age-matched controls8. Le et al.12 identified three patients born to first cousins of Syrian descent. These patients harbored another truncated PTRH2 variant due to a point-nonsense mutation (c.324G>A) that creates a stop codon at residue 108 (p.W108*; Tables 1, 2).
Table 1.
IMNEPD: neuromuscular pathology.
Table 2.
IMNEPD: physiological and developmental pathology.
Whole-exome sequencing was later used to screen the autozygome of 143 consanguineous families in which multiple members presented with similar neurogenetic phenotypes. Relevant to IMNEPD, intellectual disability was the most prevalent phenotype; however, other relevant phenotypes including ataxia and motor delay were also documented. A third mutant variant of PTRH2 was identified among a list of 68 other recessive genes associated with the neurogenetic disease. This PTRH2 variant results from a missense, glutamine to proline substitution that eliminates a hydrogen bond and introduces a kink within an alpha-helix (p.Q85P; Tables 1 and2). This mutation is proposed to destabilize the structure and function of the catalytic domain9. Later studies have reported this mutation and its phenotypes in eight other children from three different consanguineous families. Similar to the truncated variant, isolated patient fibroblasts with this missense mutation displayed significantly decreased PTRH2 protein levels compared to controls; yet no differences in mRNA levels10,11. These findings reinforce the loss of function nature of PTRH2 in the etiology of IMNEPD.
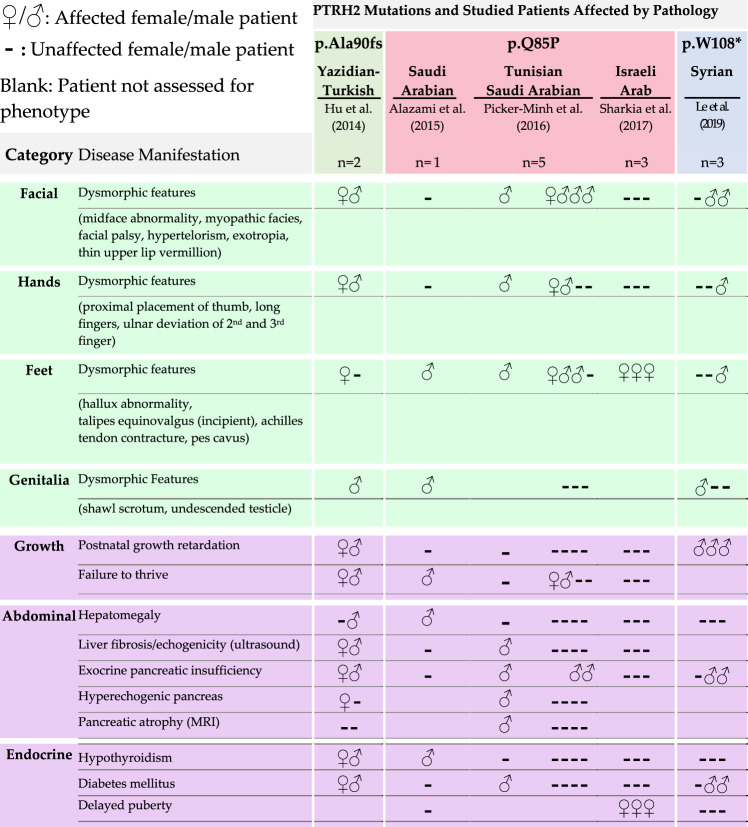
All three PTRH2 mutations manifest postnatally with similar disease phenotypes despite variations in expression and severity (Table 2). Patients harboring these PTRH2 gene mutations experienced no complications at birth and were born within the normal height and weight ranges. However, the 102-residue p.Ala90fs mutant yielded the most severe phenotypes, manifesting congenitally as distinctive dysmorphisms of the face and limbs, EEG abnormalities, microcephaly, neonatal hypotonia, stunted postnatal growth, and failure to thrive8. The patients harboring the nonsense mutation p.W108* shared some of these phenotypes with the addition of seizures12. Ultrasound revealed increased echogenicity of the liver and fibrosis of skeletal muscle while Magnetic Resonance Imaging showed cerebellar atrophy and microencephaly. Furthermore, analysis of patient blood samples indicated metabolic disorders in the form of hypothyroidism, mild diabetes mellitus, and exocrine pancreas insufficiency8,10.
While these phenotypes were absent or scarce in the patients with the missense mutation p.Q85P, progressive phenotypes common to PTRH2 mutations include developmental delays of speech and motor ability, demyelinating sensorimotor neuropathies, ataxia, distal muscle weakness, sensorineural hearing loss, and intellectual disability (Table 1). Importantly, motor and speech delays and signs of demyelinating sensorimotor neuropathy were reported in all patients presenting with a PTRH2 gene mutation8–12.
Ptrh2-null mouse model recapitulates IMNEPD and provides new functional insight
The study of IMNEPD cases is thus far limited to a small patient cohort of 13 patients from five families. In studying children of consanguineous families, it is possible that some of the reported phenotypes may have been caused by the homozygous inheritance of other recessive disease-associated genes. This is a potential explanation of the variation of phenotypic appearance and severity reported among patients. Additionally, the young age range of the patients (3–27 years of age) at the time of study prevents investigators from making conclusions about phenotypes that may manifest later in life.
Similar to human patients, Ptrh2-null mice are born equal in size to age-matched wild type littermates but develop disease phenotypes postnatally. At seven days post-birth, Ptrh2-null mice are significantly smaller than age-matched littermate controls and display ataxia and muscle weakness and die within 2 weeks after birth. Similar to patients, Ptrh2 knockout mice present with atrophy and hypoplasia of the cerebellum along with underdeveloped pancreas and liver parenchyma8. They have smaller glomeruli and muscle fibers compared to age-matched littermate controls and increased ERK activity21. Further analysis of these mice will enhance our understanding of the mechanisms by which PTRH2 causes IMNEPD.
PTRH2 functions at the axis of cell survival and cell death
PTRH2 confers stress resistance protection through pro-survival signaling in ECM-attached cells
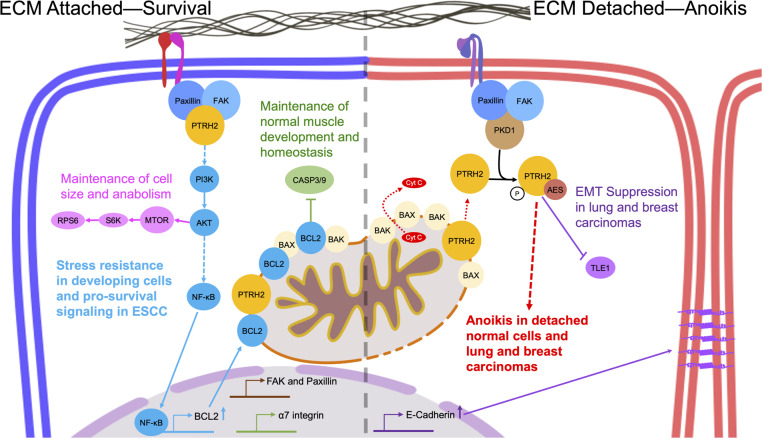
PTRH2 was identified in an expression cloning screen to alter the transcription and protein levels of the apoptosis regulator B-cell lymphoma-2 (Bcl2) in response to integrin-mediated adhesion2,3,22. Griffiths et al. demonstrated PTRH2 regulates pro-survival signals in cells bound to the ECM by integrins. They reported knockdown of endogenous PTRH2 increased staurosporine-induced mitochondrial apoptosis in a variety of cell types and PTRH2 re-expression rescued these cells3. Upon integrin ligation, FAK is recruited to focal adhesions to induce signaling pathways that include activation of PI3K and ERK Kinase. Integrin recruitment and activation of FAK blocks apoptosis in a number of cell types23. FAK mediates integrin-activated bcl2 transcription13. Griffiths and colleagues mapped the signal transduction pathway activated by PTRH2. They found that expression of FRNK (a dominant-negative form of FAK that blocks integrin-mediated activation of FAK) abrogated the activation of Bcl2 by PTRH2. Moreover, Bcl2 upregulation by PTRH2 requires PI3K as cells expressing activated PI3K (p110-CAAX) demonstrated significantly increased Bcl2 transcription and protein expression whereas expression of dominant-negative PI3K (p85D) blocks PTRH2-induced Bcl2 transcription. Furthermore, knockdown of PTRH2 prevents activation of AKT and Bcl2 expression in adherent cells by inhibiting. NFkB translocation to the nucleus where it activates NFkB-dependent transcription of genes including Bcl224. Indeed, in cells attached to the ECM, PTRH2 expression increased PI3K-AKT and subsequent Bcl2 transcription. Taken together, PTRH2 promotes integrin-mediated cell survival through a PTRH2-FAK-PI3K-AKT-NFkB pathway that upregulates Bcl2 transcription and protein expression (Fig. 2)3,13,14.
Fig. 2. PTRH2 is involved in diverse signaling pathways depending on its cellular localization.
In cells attached to the ECM by integrins, PTRH2 complexes with FAK at the cell membrane to initiate a PI3K signaling pathway to regulate homeostasis, development, stress resistance, and promote cell survival. In cells not bound to the ECM by integrins, PTRH2 is released from mitochondria into the cytosol where it complexes with AES/TLE to induce anoikis and suppress EMT.
The loss of this PTRH2 pro-survival pathway is a key characteristic in cells isolated from IMNEPD patients. Indeed, patient fibroblasts are more susceptible to staurosporine-induced mitochondrial apoptosis compared to age-matched healthy controls. In these patient fibroblasts re-expression of wild type PTRH2 restores activation of a FAK-PI3K-AKT-NFκB-Bcl2 pro-survival pathway8.
PTRH2 initiates cell death by interacting with cytoplasmic GRO/TLE
Under detached conditions, PTRH2 is released from mitochondria and presumably focal adhesions into the cytosol where it forms a complex with AES/TLE5, a dominant-negative member of the Groucho/TLE family of transcriptional corepressors, to induce cell death (Fig. 2). In adherent cells when PTRH2 is artificially targeted to the cytoplasm it binds to AES to induce cell death2,25. As a transcriptional co-repressor, AES binds Nuclear Factor Kappa B (NF-κB) in the nucleus to turn off transcription of NF-κB-regulated genes26. Although the molecular mechanism of PTRH2-mediated cell death has not been elucidated, Jenning and colleagues hypothesize that PTRH2 retains AES/TLE1 in the cytoplasm keeping TLE1 out of the nucleus and turning off AES and TLE1’s pro-survival activity27. Indeed, artificially overexpressing cytoplasmic PTRH2 retains TLE1 in the cytoplasm in an AES-dependent manner. The authors further speculate that this is due to a PTRH2/AES/TLE1 tri-complex; although that has not yet been demonstrated. Additionally, TLE family member 1 (TLE1) functions as a co-repressor with other proteins of the TLE family to regulate transcription and these may also be affected28,29. Nuclear TLE1 expression inhibits PTRH2-mediated anoikis by reducing the formation of a cytoplasmic PTRH2-AES complex by keeping AES in the nucleus. Complexing and sequestration of AES by TLE1 in the nucleus, in addition to integrin ligation, are the only known mechanisms that counteract anoikis induced by cytoplasmic PTRH230.
Localization of PTRH2 to the cytoplasm is regulated by protein kinase D
Signaling activated by ECM-detached integrins upstream of PTRH2 remains poorly understood. However, detachment of integrins leads to PTRH2 cytoplasmic localization and activation of anoikis. There, PTRH2 is phosphorylated at two serines (5 and 87) by activated serine/threonine-protein kinase D (PKD1)25. Activated PKD1 is enriched in cells detached from ECM. It is unknown if this phosphorylation of cytosolic-PTRH2 enhances anoikis signaling, however, the authors hypothesize that PTRH2 phosphorylation at serine 5, within the MLS, may block its mitochondrial recruitment keeping PTRH2 in the cytosol25. In a similar fashion, the mitochondrial translocation of Cofilin or 2′,3′-cyclic nucleotide-3′-phosphodiesterase (CNP2) is blocked by phosphorylation of serine residues within their MLS sequences31,32. This may be due to the conversion of serine to a more acidic phosphoserine, which disrupts mitochondrial translocation33,34.
PTRH2 functions in stress responses via a secretory pathway
PTRH2 can be localized to the endoplasmic reticulum (ER) and Golgi apparatus4. In the Golgi, PTRH2 regulates ERK signaling in detergent-resistant microdomains, which are sphingomyelin and cholesterol-enriched regions of intracellular membranes35. In these microdomains signaling complexes assemble and signal at internal membrane structures as they would at the plasma membrane36. Similar to the mitochondrial-cytosolic model of PTRH2-mediated anoikis, PTRH2 in the Golgi coincides with dephosphorylated ERK1, thereby reducing ERK survival signals and decreasing cell viability. However, if PTRH2 is trapped at the ER and prohibited from progressing to the Golgi, ERK survival signaling is activated due to specific enrichment of phosphorylated, active MAPK pathway components such as BRAF, MEK1/2, and ERK1/2. Cells with PTRH2 constitutively localized to the ER showed significantly increased adhesion-independent viability4. PTRH2 exists as an SDS resistant 28 kD dimer at the ER and a signaling-active 16 kD monomer at the Golgi. PTRH2 is also suspected to be associated with the membrane of the ER and Golgi with its C-terminal catalytic domain-oriented in the cytoplasm. This allows PTRH2 to complex with ERK1 localized to the exterior of the Golgi where it disrupts ERK signaling. Furthermore, when ER stress is induced by tunicamycin treatment, this ERK1-PTRH2 complex dissociates and expression of PTRH2 is induced4. This pathway is functionally relevant in the organogenesis of the lens and retina. PTRH2 and PKD1 are both expressed differentially in lens fibroblasts and astrocytes during eye development. Retinal astrocytes undergo anoikis during the remodeling of the developing eye. This is mediated, in part, by Golgi-localized PTRH2 inhibiting ERK signaling. In rats with a mutation of a gene encoding βA3/A1-crystallin, eye organogenesis is dysregulated due to aberrant lens fibroblasts and retinal astrocyte signaling. In lens fibroblasts, this mutation prevents normal de-nucleation required for a transparent lens37. In astrocytes, it blocks PTRH2 from trafficking to the Golgi, thereby upregulating survival signaling and hindering essential anoikis38.
The only eye defects reported in IMNEPD patients were myopia in three sisters with the PTRH2 missense mutation (p.Q85P). Cerebellar atrophy was reported in IMNEPD patients with the PTRH2 truncation mutation (p.Ala90fs and p.W108*) and ataxia and cerebellar hypoplasia was observed in the Ptrh2-null mice8,11. Potentially, these neurological phenotypes may be caused by deregulation of anoikis or loss of pro-survival signaling in cerebellar astrocytes harboring a PTRH2 mutation because anoikis in astrocytes is essential in the developing cerebellum39.
PTRH2 has regulatory roles in diverse cellular processes
In addition to its well-studied role in pro-survival and anoikis signaling, there is evidence that IMNEPD phenotypes are also caused, in part, by deregulation of cell size, myogenesis, dystrophin complex signaling, and epithelial-mesenchymal transition (EMT; Fig. 2).
PTRH2 regulates cell size via the mTOR pathway
Histology on tissues isolated from Ptrh2-null mice demonstrates significantly smaller cross-sectional areas in neurons, myocytes, exocrine pancreatic acini, and hepatocytes compared to age-matched littermate controls8. These findings correspond to cerebellar atrophy, skeletal myopathy, exocrine pancreas insufficiency, and liver pathologies reported in IMNEPD patients. Additionally, ribosomal protein S6 (RPS6) is decreased in Ptrh2-null mouse brain and IMNEPD patient fibroblasts, suggesting that abnormal cell size may be due to decreased activity of the mechanistic target of rapamycin (mTOR) pathway8. It may be that PTRH2 regulates mTOR through its activation of the FAK-PI3K-AKT pathway because, in the context of cell size, mTOR is activated by way of PI3K/AKT40–43.
PTRH2 regulates myogenic differentiation and maintains skeletal muscle homeostasis
Distal muscle weakness is one of the most prevalent phenotypes documented in IMNEPD patients and in the Ptrh2-null mice. Most of the patients suffer from walking gait abnormalities due to severe joint contractures and are wheelchair dependent8,10,12. In Ptrh2-null mice, severe muscle wasting and skeletal myopathy occur44,45. Indeed, Ptrh2 loss of function severely deregulates myogenic differentiation and disrupts essential integrin signaling in skeletal muscle3,8,44,45. PTRH2 levels progressively increase with skeletal muscle differentiation44. In myoblasts isolated from the gastrocnemius of Ptrh2-null mice, differentiation occurs prematurely due, in part, to a decrease of Bcl2 protein expression, thus significantly increasing the activity of caspases 3 and 9. Notably, during myoblast differentiation, these pro-apoptotic caspases have a non-death function where activated caspase-3 proteolytically cleaves kinases such as serine/threonine kinase 4 (STK4; MST1) or homeodomain interacting protein kinase 2 (HIPK2)46–48.
Ptrh2-null myoblasts exhibit earlier and higher expression levels of troponin T, myosin-2, and myogenin compared to age-matched controls, suggesting premature differentiation occurs when PTRH2 protein is absent. Re-expression of PTRH2 in null myoblasts rescues early differentiation and decreases caspase activity similar to littermate age-matched controls44.
Doe et al.45 demonstrated that Ptrh2-null myofibers are not only hypotrophic but also exhibit pathological features of congenital muscular dystrophy. Muscular dystrophies are caused by mutations in proteins that comprise the dystrophin protein complex (DPC) that anchors the cytoskeleton underlying the sarcolemma to the ECM and scaffolds signaling proteins. Constant deregulation of signaling at the DPC and its mechanical support of the sarcolemma leads to oxidative stress, which increases myofiber permeability and damages the sarcolemma. The damaged myofiber is lost to necrosis and is replaced via regeneration by satellite cells49–53. Ptrh2-null mice suffer myofiber damage, which was shown by increased creatine kinase activity and permeability to Evans blue dye. Active regeneration of Ptrh2-null muscle fibers was also identifiable by a significant increase in central nuclei, which indicates muscle damage. These mice displayed significant increases in eMyHC expression and endomysial fibrosis; hallmarks of muscular dystrophy45,53–58.
The α7β1 integrin is required for healthy muscle development and loss of α7β1 integrin causes Duchenne’s muscular dystrophy in humans and mice59. This integrin binds to laminin within the ECM and functions in the regulation of mechanotransduction, hypertrophy, and survival by signaling through AKT, mTOR, and ERK60–62. PTRH2 and α7β1 integrin form a complex at the sarcolemma of myofibrils. Interestingly, the ablation of PTRH2 results in a decrease of α7 integrin expression suggesting a feedback loop. Ptrh2- and α7-null muscle have decreased PI3K-AKT activation indicative of diminished survival signaling via a FAK-PI3K-AKT pathway downstream of α7β1 integrin45.
Muscular dystrophy is studied in three different mouse models each representing different deficiencies of DPC components: α7-deficient, laminin-α2 dyW null, and dystrophin-deficient mdx. PTRH2 expression and AKT signaling has been found to be decreased in α7-deficient and laminin-α2 dyW null mice but increased in dystrophin-deficient mdx mice45. In mice, the ablation of PTRH2 alone produces a more severe and lethal myopathy similar to the muscular dystrophy phenotype found in the double knockout of dystrophin and integrin α7β161. This suggests a pivotal role for PTRH2 in skeletal muscle development.
PTRH2 regulates epithelial-to-mesenchymal transition (EMT) in lens epithelial cells
Following cataract surgery, lens epithelial cells (LECs) at the anterior capsule of the lens differentiate into a fibroblast-like morphology. This process has been shown in vitro and in vivo to be caused by transforming growth factor-beta (TGFβ) and fibroblast growth factor 2 (FGF2). LECs post-surgery display increased migration and deposit an excess of ECM, which occludes the pre-operative lens63,64. This process is mediated in, part, by PTRH2 that is expressed at high levels in the cytoplasm of human LECs. When PTRH2 protein levels are knocked down in LECs, EMT is inhibited despite treatment with TGFβ2 as shown by decreased alpha-smooth muscle actin and migration. TGFβ2 increases PTRH2 mRNA and protein levels in human LECs65. Myopia was documented in three patients with the PTRH2 missense mutation (p.Q85P)11. Moreover, EMT is an essential process in neural development that is severely dysregulated in IMNEPD patients66,67.
PTRH2 in cancer
PTRH2 has multiple pro-oncogenic and metastasis suppressing functions due to its bi-functional roles in pro-survival signaling, anoikis, and EMT. Due to the heterogeneity and genetic instability of cancers, multifunctional proteins such as PTRH2 may act differently on the available pathways68,69. For example, while the loss of PTRH2 expression in cancers of the lung and breast suppresses anoikis, it is upregulated in late-stage esophageal squamous cell carcinomas, perhaps because of its potent cell survival function30,70–72,57,58,73. This section will highlight the outcomes of PTRH2 signaling in specific cancers.
PTRH2 is a regulator of metastasis and EMT in lung and breast cancers
Metastasis is the process by which a primary tumor cell disseminates to distant sites in the body through the lymphatic or circulatory systems74. This requires suppression of anoikis pathways and activation of pro-survival pathways, independent of ECM attachment75. EMT is a hallmark of metastasis whereby tumor cells become more motile, invasive, and fibroblastic in shape70,76,77.
In the context of cancer, PTRH2 has been identified as a potential metastasis suppressor via its regulation of anoikis and EMT. In breast cancer, higher-grade breast carcinomas positive for lymph node metastases stain the least for PTRH2 compared to less invasive breast cancers or normal breast tissue. Additionally, knockdown of PTRH2 protein expression in the breast cancer cell line, MCF7 decreased anoikis sensitivity, increased cell adhesion, and decreased ERK phosphorylation. In vivo, xenograft tumors from MCF7 cells with decreased PTRH2 protein expression did not differ in size or growth rate compared to control tumors. However, metastasis to the lungs increased in the MCF7 tumors with knockdown PTRH2. These tumors contained higher levels of phosphorylated ERK1/278. These results suggest that loss of PTRH2 expression confers a metastatic advantage in invasive breast cancers.
In lung cancer, a signaling axis containing PTRH2, ERK, and TLE1 prevents EMT. The depletion of PTRH2 protein levels in lung cancer cells (A549 and BEAS-2B) increases migration and vimentin expression but decreases E-cadherin expression thereby promoting increased metastasis in vivo. Overexpression of PTRH2 increases E-cadherin expression and decreases motility and metastasis in vivo. These changes occurred in cells where cytosolic-localized PTRH2 protein was overexpressed; suggesting that cytosolic-PTRH2 regulates EMT. In agreement with previous findings in breast cancer, overexpression of cytosolic-PTRH2 promotes the degradation of TLE1. In detached conditions, cytoplasmic PTRH2 may direct TLE1 to be degraded by the proteasome by an unknown mechanism30. This would prevent TLE1 from being recruited to a repressor complex to inhibit E-cadherin expression57,73. Furthermore, the regulation of E-cadherin by PTRH2 and TLE1 also involves ERK. In cells with knockdown of PTRH2 protein and treated with an ERK inhibitor, TLE1’s ability to repress E-cadherin transcription was inhibited compared to controls. This suggests that in normal lung tissue, PTRH2 maintains E-cadherin expression by inhibiting ERK. However, in cancer cells with knockdown of PTRH2 protein expression, EMT is driven by active ERK inducing TLE1 repression of E-cadherin transcription79.
PTRH2 is downstream of estrogen receptor and signals through a PI3K/AKT pathway in ovarian cancer
In ovarian serous papillary adenocarcinomas, low PTRH2 protein expression significantly correlates with higher histologic grade and poorer clinical prognosis. In vitro, cytosolic-PTRH2 protein overexpression in Caov-3 cells decreases cell viability80. PTRH2 is also detected in sera of patients with serous papillary adenocarcinomas81.
As previously described, PTRH2 functions on an integrin-PI3K/AKT pro-survival pathway8. In suspended ovarian cancer cells, estrogen (E2) binding to its estrogen (E2) binding estrogen receptor alpha (ERα) blocks the anoikis-initiating release of PTRH2 from the mitochondria and FAs to the cytoplasm through a PI3K/AKT pathway. Cells co-treated with E2 and an AKT inhibitor (CCT128930) have higher cytoplasmic levels of PTRH2 compared to cells with intact PI3K/AKT signaling. This suggests that PTRH2’s translocation to the cytoplasm is inhibited by signals from active AKT82. This points to a potential feedback mechanism in which AKT, activated downstream of pro-survival PTRH2-FAK-PI3K signaling, blocks FA and mitochondrial PTRH2 translocation to the cytoplasm.
PTRH2 supports metastasis in esophageal squamous cell carcinomas (ESCC)
Up until this point, PTRH2 has been discussed for its role in metastasis suppression. However, based on its function in adhesion mediated pro-survival signaling, PTRH2 also acts as an oncogene in specific cancers58. Fan et al. analyzed patient-derived esophageal squamous cell carcinomas (ESCC) tissues for differential expression of PTRH2, AIF, and Bcl2 with respect to patient gender, age, and TNM classification, which assesses ESCC extent based on primary tumor size (T) and metastasis to lymphatics (N) or distant sites (M). PTRH2 protein expression is significantly upregulated in ESCC tissues compared to dysplastic and normal tissues. Specifically, higher PTRH2 protein expression correlates with invasive ESCCs, which are poorly differentiated and lymph node metastasis-positive (TMN stages III-IV).
PTRH2 protein expression also correlated with significantly increased Bcl2 and AIF protein levels72. Upon the shRNA knockdown of PTRH2 protein in ESCC cell lines (EC9706 and TE1), cell proliferation, migration, and invasion were significantly downregulated compared to controls. Supporting its pro-survival role, knockdown of PTRH2 protein levels increased anoikis. In an in vivo xenograft mouse model, knockdown of PTRH2 protein expression in EC9706 cells decreased tumor size, Bcl2 and matrix metalloproteinase-2 (MMP-2) expression, and increased anoikis58.
The oncogenic role of PTRH2 in ESCC may be partly explained by PTRH2’s activation of Bcl2 expression downstream of integrin ligation and PTRH2’s interaction with and activation of FAK3 and its ability to activate a FAK-paxillin pathway58. As an adaptor protein of FAK, paxillin is required for FAK activation and localization to FAs. Paxillin allows FAK to engage in pro-survival signaling and regulate migration83,84. Griffiths and colleagues showed PTRH2 is in a complex with FAK at FAs and PTRH2 may have a role in maintaining FAK-paxillin signaling at the transcriptional level3,58. In this ESCC study, microarray data revealed a multitude of differentially expressed genes between PTRH2 knockdown cells and controls. Many of these genes regulated functions where PTRH2 is involved namely cell adhesion, interactions with integrins and myogenesis including Wnt, TLE1, Talin, FAK, and Paxillin58.
A peptide derived from PTRH2 as potential cancer therapeutic
Chen et al. identified a small peptide fragment from the MLS region of PTRH2 that has potential as cancer therapeutic by promoting cell death. The authors called this peptide the PTRH2-Cell Death Domain (PTRH2-CDD). PTRH2-CDD inactivates PTRH2’s pro-survival function and promotes cell death in a caspase-independent manner. It decreased breast cancer cells (MCF-10CA1a) viability in vitro. MCF-10CA1a is an aggressive breast cancer cell line containing a PIK3CA H1047R activating mutation that consistently develops tumors in vivo85.
Moreover, intra-tumoral injections of iRGD-PTRH2-CDD significantly reduced weight and volume of MCF-10CA1a xenograft tumors and PTRH2-CDD was found in the tumor niche19. The PTRH2-CDD peptide contains a species-conserved transmembrane sequence between residues 14 to 333,4. The ablation of this sequence increased PTRH2’s localization to the perinuclear region of the cytoplasm. On the other hand, expression of the PTRH2 catalytic domain alone significantly increased cell survival in HEK 293T cells compared to the expression of full-length PTRH219. IMNEPD patient cells with catalytic domain mutations display reduced cell viability that is rescued by the expression of wild type PTRH28. These findings point to the catalytic domain as a pro-survival region of PTRH2; however, it remains to be determined if the enzymatic activity of this domain is required. Moreover, the PTRH2-CDD peptide exemplifies the bi-functional role of PTRH2. PTRH2-CDD is unable to localize at the OMM and is retained in the cytoplasm where it presumably complexes with GRO/TLE to sequester TLE and thereby promote cell death. Conversely, the PTRH2 catalytic domain alone has a pro-survival effect potentially through an unknown complex at the OMM that may block PTRH2 release into the cytoplasm and subsequent anoikis. The molecular mechanism of how PTRH2 functions as both a pro-survival and a pro-anoikis regulator is an area of an ongoing investigation. It will be helpful to dissect the role of PTRH2 at the OMM to determine how its function at the mitochondria promotes cell survival and blocks anoikis-induction.
Conclusions
PTRH2 has a crucial role in pro-survival and anoikis signaling decisions. It is an essential participant in integrin-mediated pro-survival signaling pathways in response to adhesion, which allows the cell to make life and death decisions based on specific environmental cues. In cancer, PTRH2 promotes cell survival and metastasis or cell death depending on cancer and signaling context. In human development, PTRH2 provides a cell survival role and is an important regulator of muscle differentiation whereby loss of PTRH2 function causes congenital IMNEPD due to loss of pro-survival signals. PTRH2, therefore, has great potential to be developed as a biomarker and therapeutic target for developmental disorders and specific cancers. To facilitate this, further work is needed to better define the mechanisms that regulate the localization of PTRH2, how PTRH2 activates PI3K and regulates TLE function, and whether enzymatic activity of the catalytic domain is essential to its function.
Acknowledgements
This work is funded by the NIH R01HD091162 (to M.L.M.) and the B.H. and Alice C. Beams Endowed Professorship in Cancer Research (to J.W.R.).
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Edited by Inna Lavrik
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Pereda JMde, et al. Crystal structure of a human peptidyl-tRNA hydrolase reveals a new fold and suggests basis for a bifunctional activity. J. Biol. Chem. 2004;279:8111–8115. doi: 10.1074/jbc.M311449200. [DOI] [PubMed] [Google Scholar]
- 2.Jan Y, et al. A mitochondrial protein, Bit1, mediates apoptosis regulated by integrins and Groucho/TLE corepressors. Cell. 2004;116:751–762. doi: 10.1016/S0092-8674(04)00204-1. [DOI] [PubMed] [Google Scholar]
- 3.Griffiths GS, et al. Bit-1 mediates integrin-dependent cell survival through activation of the NFκB pathway. J. Biol. Chem. 2011;286:14713–14723. doi: 10.1074/jbc.M111.228387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yi P, et al. MAPK scaffolding by BIT1 in the Golgi complex modulates stress resistance. J. Cell Sci. 2010;123:1060–1072. doi: 10.1242/jcs.059717. [DOI] [PubMed] [Google Scholar]
- 5.Rosas-Sandoval G, et al. Orthologs of a novel archaeal and of the bacterial peptidyl–tRNA hydrolase are nonessential in yeast. Proc. Natl Acad. Sci. USA. 2002;99:16707–16712. doi: 10.1073/pnas.222659199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Uhlen, M. et al. A pathology atlas of the human cancer transcriptome. Science357, eaan2507 (2017). [DOI] [PubMed]
- 7.Regev, A. et al. Human Cell Atlas. eLife. 6, e27041 (2017). 10.7554/eLife.27041.
- 8.Hu H, et al. Mutations in PTRH2 cause novel infantile-onset multisystem disease with intellectual disability, microcephaly, progressive ataxia, and muscle weakness. Ann. Clin. Transl. Neurol. 2014;1:1024–1035. doi: 10.1002/acn3.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Alazami AM, et al. Accelerating novel candidate gene discovery in neurogenetic disorders via whole-exome sequencing of prescreened multiplex consanguineous families. Cell Rep. 2015;10:148–161. doi: 10.1016/j.celrep.2014.12.015. [DOI] [PubMed] [Google Scholar]
- 10.Picker-Minh S, et al. Phenotype variability of infantile-onset multisystem neurologic, endocrine, and pancreatic disease IMNEPD. Orphanet. J. Rare Dis. 2016;11:52. doi: 10.1186/s13023-016-0433-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sharkia R, et al. Homozygous mutation in PTRH2 gene causes progressive sensorineural deafness and peripheral neuropathy. Am. J. Med. Genet. A. 2017;173:1051–1055. doi: 10.1002/ajmg.a.38140. [DOI] [PubMed] [Google Scholar]
- 12.Le C, et al. Infantile-onset multisystem neurologic, endocrine, and pancreatic disease: case and review. Can. J. Neurol. Sci. J. Can. Sci. Neurol. 2019;46:459–463. doi: 10.1017/cjn.2019.35. [DOI] [PubMed] [Google Scholar]
- 13.Matter ML, Ruoslahti E. A signaling pathway from the alpha5beta1 and alpha(v)beta3 integrins that elevates bcl-2 transcription. J. Biol. Chem. 2001;276:27757–27763. doi: 10.1074/jbc.M102014200. [DOI] [PubMed] [Google Scholar]
- 14.Matter ML, Zhang Z, Nordstedt C, Ruoslahti E. The α5β1 integrin mediates elimination of amyloid-β peptide and protects against apoptosis. J. Cell Biol. 1998;141:1019–1030. doi: 10.1083/jcb.141.4.1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gilmore AP. Anoikis. Cell Death Differ. 2005;12:1473–1477. doi: 10.1038/sj.cdd.4401723. [DOI] [PubMed] [Google Scholar]
- 16.Douma S, et al. Suppression of anoikis and induction of metastasis by the neurotrophic receptor TrkB. Nature. 2004;430:1034–1039. doi: 10.1038/nature02765. [DOI] [PubMed] [Google Scholar]
- 17.Frisch SM, Vuori K, Kelaita D, Sicks S. A role for Jun-N-terminal kinase in anoikis; suppression by bcl-2 and crmA. J. Cell Biol. 1996;135:1377–1382. doi: 10.1083/jcb.135.5.1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ilić D, et al. Extracellular matrix survival signals transduced by focal adhesion kinase suppress p53-mediated apoptosis. J. Cell Biol. 1998;143:547–560. doi: 10.1083/jcb.143.2.547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen R, et al. Application of a proapoptotic peptide to intratumorally spreading cancer therapy. Cancer Res. 2013;73:1352–1361. doi: 10.1158/0008-5472.CAN-12-1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Karczewski, K. J. et al. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature581, 434–443 (2020). 10.1038/s41586-020-2308-7. [DOI] [PMC free article] [PubMed]
- 21.Kairouz-Wahbe R, et al. Anoikis effector Bit1 negatively regulates Erk activity. Proc. Natl Acad. Sci. USA. 2008;105:1528–1532. doi: 10.1073/pnas.0711357105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Matter, M. L. & Ramos, J. W. in Cell-Cell Interactions: Methods and Protocols (ed. Colgan, S. P.) 155–165 (Humana Press, 2006). 10.1385/1-59745-113-4:155.
- 23.Cohen LA, Guan J-L. Mechanisms of focal adhesion kinase regulation. Curr. Cancer Drug Targets. 2005;5:629–643. doi: 10.2174/156800905774932798. [DOI] [PubMed] [Google Scholar]
- 24.Kurland JF, et al. NF-κB1 (p50) homodimers contribute to transcription of the bcl-2 oncogene. J. Biol. Chem. 2001;276:45380–45386. doi: 10.1074/jbc.M108294200. [DOI] [PubMed] [Google Scholar]
- 25.Biliran H, Jan Y, Chen R, Pasquale EB, Ruoslahti E. Protein kinase D is a positive regulator of Bit1 apoptotic function. J. Biol. Chem. 2008;283:28029–28037. doi: 10.1074/jbc.M803139200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tetsuka T, et al. Inhibition of nuclear factor-κB-mediated transcription by association with the amino-terminal enhancer of split, a groucho-related protein lacking WD40 repeats. J. Biol. Chem. 2000;275:4383–4390. doi: 10.1074/jbc.275.6.4383. [DOI] [PubMed] [Google Scholar]
- 27.Jenning S, Pham T, Ireland SK, Ruoslahti E, Biliran H. Bit1 in anoikis resistance and tumor metastasis. Cancer Lett. 2013;333:147–151. doi: 10.1016/j.canlet.2013.01.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Palaparti A, Baratz A, Stifani S. The Groucho/Transducin-like enhancer of split transcriptional repressors interact with the genetically defined amino-terminal silencing domain of histone H3. J. Biol. Chem. 1997;272:26604–26610. doi: 10.1074/jbc.272.42.26604. [DOI] [PubMed] [Google Scholar]
- 29.Chen G, Courey AJ. Groucho/TLE family proteins and transcriptional repression. Gene. 2000;249:1–16. doi: 10.1016/S0378-1119(00)00161-X. [DOI] [PubMed] [Google Scholar]
- 30.Brunquell C, et al. TLE1 is an Anoikis regulator and is downregulated by Bit1 in breast cancer cells. Mol. Cancer Res. 2012;10:1482–1495. doi: 10.1158/1541-7786.MCR-12-0144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chua BT, et al. Mitochondrial translocation of cofilin is an early step in apoptosis induction. Nat. Cell Biol. 2003;5:1083–1089. doi: 10.1038/ncb1070. [DOI] [PubMed] [Google Scholar]
- 32.Lee J, O’Neill RC, Park MW, Gravel M, Braun PE. Mitochondrial localization of CNP2 is regulated by phosphorylation of the N-terminal targeting signal by PKC: implications of a mitochondrial function for CNP2 in glial and non-glial cells. Mol. Cell. Neurosci. 2006;31:446–462. doi: 10.1016/j.mcn.2005.10.017. [DOI] [PubMed] [Google Scholar]
- 33.von Heijne G. Mitochondrial targeting sequences may form amphiphilic helices. EMBO J. 1986;5:1335–1342. doi: 10.1002/j.1460-2075.1986.tb04364.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bedwell DM, Strobel SA, Yun K, Jongeward GD, Emr SD. Sequence and structural requirements of a mitochondrial protein import signal defined by saturation cassette mutagenesis. Mol. Cell. Biol. 1989;9:1014–1025. doi: 10.1128/MCB.9.3.1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gkantiragas I, et al. Sphingomyelin-enriched microdomains at the Golgi Complex. Mol. Biol. Cell. 2001;12:1819–1833. doi: 10.1091/mbc.12.6.1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chiu VK, et al. Ras signalling on the endoplasmic reticulum and the Golgi. Nat. Cell Biol. 2002;4:343–350. doi: 10.1038/ncb783. [DOI] [PubMed] [Google Scholar]
- 37.Sinha D, et al. A spontaneous mutation affects programmed cell death during development of the rat eye. Exp. Eye Res. 2005;80:323–335. doi: 10.1016/j.exer.2004.09.014. [DOI] [PubMed] [Google Scholar]
- 38.Ma B, et al. βA3/A1-Crystallin controls anoikis-mediated cell death in astrocytes by modulating PI3K/AKT/mTOR and ERK survival pathways through the PKD/Bit1-signaling axis. Cell Death Dis. 2011;2:e217. doi: 10.1038/cddis.2011.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Krueger BK, Burne JF, Raff MC. Evidence for large-scale astrocyte death in the developing cerebellum. J. Neurosci. J. Soc. Neurosci. 1995;15:3366–3374. doi: 10.1523/JNEUROSCI.15-05-03366.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kim D-H, et al. mTOR interacts with Raptor to form a nutrient-sensitive complex that signals to the cell growth machinery. Cell. 2002;110:163–175. doi: 10.1016/S0092-8674(02)00808-5. [DOI] [PubMed] [Google Scholar]
- 41.Sarbassov DD, et al. Rictor, a novel binding partner of mTOR, defines a rapamycin-insensitive and raptor-independent pathway that regulates the cytoskeleton. Curr. Biol. 2004;14:1296–1302. doi: 10.1016/j.cub.2004.06.054. [DOI] [PubMed] [Google Scholar]
- 42.Saxton RA, Sabatini DM. mTOR signaling in growth, metabolism, and disease. Cell. 2017;168:960–976. doi: 10.1016/j.cell.2017.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rommel C, et al. Mediation of IGF-1-induced skeletal myotube hypertrophy by PI(3)K/Akt/mTOR and PI(3)K/Akt/GSK3 pathways. Nat. Cell Biol. 2001;3:1009–1013. doi: 10.1038/ncb1101-1009. [DOI] [PubMed] [Google Scholar]
- 44.Griffiths GS, et al. Bit‐1 is an essential regulator of myogenic differentiation. J. Cell Sci. 2015;128:1707–1717. doi: 10.1242/jcs.158964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Doe J, et al. PTRH2 gene mutation causes progressive congenital skeletal muscle pathology. Hum. Mol. Genet. 2017;26:1458–1464. doi: 10.1093/hmg/ddx048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fernando P, Kelly JF, Balazsi K, Slack RS, Megeney LA. Caspase 3 activity is required for skeletal muscle differentiation. Proc. Natl Acad. Sci. USA. 2002;99:11025–11030. doi: 10.1073/pnas.162172899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.de la Vega L, Hornung J, Kremmer E, Milanovic M, Schmitz ML. Homeodomain-interacting protein kinase 2-dependent repression of myogenic differentiation is relieved by its caspase-mediated cleavage. Nucleic Acids Res. 2013;41:5731–5745. doi: 10.1093/nar/gkt262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bell, R., Al-Khalaf, M. & A. Megeney, L. The beneficial role of proteolysis in skeletal muscle growth and stress adaptation. Skelet. Muscle6, 16 (2016). [DOI] [PMC free article] [PubMed]
- 49.Disatnik MH, et al. Evidence of oxidative stress in mdx mouse muscle: studies of the pre-necrotic state. J. Neurol. Sci. 1998;161:77–84. doi: 10.1016/S0022-510X(98)00258-5. [DOI] [PubMed] [Google Scholar]
- 50.Renault V, et al. Skeletal muscle regeneration and the mitotic clock. Exp. Gerontol. 2000;35:711–719. doi: 10.1016/S0531-5565(00)00151-0. [DOI] [PubMed] [Google Scholar]
- 51.Martins AS, Shkryl VM, Nowycky MC, Shirokova N. Reactive oxygen species contribute to Ca2+ signals produced by osmotic stress in mouse skeletal muscle fibres. J. Physiol. 2008;586:197–210. doi: 10.1113/jphysiol.2007.146571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Whitehead NP, Yeung EW, Froehner SC, Allen DG. Skeletal muscle NADPH oxidase is increased and triggers stretch-induced damage in the mdx mouse. PLoS ONE. 2010;5:e15354. doi: 10.1371/journal.pone.0015354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Allen DG, Whitehead NP, Froehner SC. Absence of dystrophin disrupts skeletal muscle signaling: roles of Ca2+, reactive oxygen species, and nitric oxide in the development of muscular dystrophy. Physiol. Rev. 2016;96:253–305. doi: 10.1152/physrev.00007.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Düsterhöft S, Yablonka-Reuveni Z, Pette D. Characterization of myosin isoforms in satellite cell cultures from adult rat diaphragm, soleus and tibialis anterior muscles. Differ. Res. Biol. Divers. 1990;45:185–191. doi: 10.1111/j.1432-0436.1990.tb00472.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bernasconi P, et al. Expression of transforming growth factor-beta 1 in dystrophic patient muscles correlates with fibrosis. Pathogenetic role of a fibrogenic cytokine. J. Clin. Invest. 1995;96:1137–1144. doi: 10.1172/JCI118101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bernasconi P, et al. Transforming growth factor-β1 and fibrosis in congenital muscular dystrophies. Neuromuscul. Disord. 1999;9:28–33. doi: 10.1016/S0960-8966(98)00093-5. [DOI] [PubMed] [Google Scholar]
- 57.Yao X, et al. The Anoikis effector Bit1 inhibits EMT through attenuation of TLE1-mediated repression of E-cadherin in lung cancer cells. PLoS ONE. 2016;11:e0163228. doi: 10.1371/journal.pone.0163228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fan T, et al. Bit1 knockdown contributes to growth suppression as well as the decreases of migration and invasion abilities in esophageal squamous cell carcinoma via suppressing FAK-paxillin pathway. Mol. Cancer. 2016;15:23. doi: 10.1186/s12943-016-0507-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Guo C, et al. Absence of α7 integrin in dystrophin-deficient mice causes a myopathy similar to Duchenne muscular dystrophy. Hum. Mol. Genet. 2006;15:989–998. doi: 10.1093/hmg/ddl018. [DOI] [PubMed] [Google Scholar]
- 60.Burkin DJ, et al. Transgenic expression of {alpha}7{beta}1 integrin maintains muscle integrity, increases regenerative capacity, promotes hypertrophy, and reduces cardiomyopathy in dystrophic mice. Am. J. Pathol. 2005;166:253–263. doi: 10.1016/S0002-9440(10)62249-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rooney JE, et al. Severe muscular dystrophy in mice that lack dystrophin and alpha7 integrin. J. Cell Sci. 2006;119:2185–2195. doi: 10.1242/jcs.02952. [DOI] [PubMed] [Google Scholar]
- 62.Boppart MD, Burkin DJ, Kaufman SJ. Alpha7beta1-integrin regulates mechanotransduction and prevents skeletal muscle injury. Am. J. Physiol. Cell Physiol. 2006;290:C1660–C1665. doi: 10.1152/ajpcell.00317.2005. [DOI] [PubMed] [Google Scholar]
- 63.Marcantonio JM, Vrensen GFJM. Cell biology of posterior capsular opacification. Eye. 1999;13:484. doi: 10.1038/eye.1999.126. [DOI] [PubMed] [Google Scholar]
- 64.Meacock WR, Spalton DJ, Stanford MR. Role of cytokines in the pathogenesis of posterior capsule opacification. Br. J. Ophthalmol. 2000;84:332–336. doi: 10.1136/bjo.84.3.332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wu X, Ruan J, Ma B, Luo M. Bit1—a potential positive regulator of epithelial–mesenchymal transition in lens epithelial cells. Graefes Arch. Clin. Exp. Ophthalmol. 2016;254:1311–1318. doi: 10.1007/s00417-016-3357-3. [DOI] [PubMed] [Google Scholar]
- 66.Duband JL, Monier F, Delannet M, Newgreen D. Epithelium-mesenchyme transition during neural crest development. Cells Tissues Organs. 1995;154:63–78. doi: 10.1159/000147752. [DOI] [PubMed] [Google Scholar]
- 67.Kerosuo L, Bronner-Fraser M. What is bad in cancer is good in the embryo: Importance of EMT in neural crest development. Semin. Cell Dev. Biol. 2012;23:320–332. doi: 10.1016/j.semcdb.2012.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Calin GA, Vasilescu C, Negrini M, Barbanti-Brodano G. Genetic chaos and antichaos in human cancers. Med. Hypotheses. 2003;60:258–262. doi: 10.1016/S0306-9877(02)00383-3. [DOI] [PubMed] [Google Scholar]
- 69.Giam M, Rancati G. Aneuploidy and chromosomal instability in cancer: a jackpot to chaos. Cell Div. 2015;10:3. doi: 10.1186/s13008-015-0009-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Fuchs IB, et al. The prognostic significance of epithelial-mesenchymal transition in breast cancer. Anticancer Res. 2002;22:3415–3419. [PubMed] [Google Scholar]
- 71.Fan T, et al. Down-regulation of the apoptosis-inducing factor or Bcl-2 inhibitor of transcription by RNA interference can alleviate TAp63γ-induced apoptosis in esophageal squamous carcinoma EC9706 cells. Int. J. Oncol. 2009;35:359–367. [PubMed] [Google Scholar]
- 72.Fan T, et al. Implications of Bit1 and AIF overexpressions in esophageal squamous cell carcinoma. Tumor Biol. 2014;35:519–527. doi: 10.1007/s13277-013-1073-8. [DOI] [PubMed] [Google Scholar]
- 73.Yao X, et al. TLE1 inhibits anoikis and promotes tumorigenicity in human lung cancer cells through ZEB1-mediated E-cadherin repression. Oncotarget. 2017;8:72235–72249. doi: 10.18632/oncotarget.19703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Liotta LA, Steeg PS, Stetler-Stevenson WG. Cancer metastasis and angiogenesis: An imbalance of positive and negative regulation. Cell. 1991;64:327–336. doi: 10.1016/0092-8674(91)90642-C. [DOI] [PubMed] [Google Scholar]
- 75.Frisch SM, Francis H. Disruption of epithelial cell-matrix interactions induces apoptosis. J. Cell Biol. 1994;124:619–626. doi: 10.1083/jcb.124.4.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Cano A, et al. The transcription factor Snail controls epithelial–mesenchymal transitions by repressing E-cadherin expression. Nat. Cell Biol. 2000;2:76–83. doi: 10.1038/35000025. [DOI] [PubMed] [Google Scholar]
- 77.Boyer B, Roche S, Denoyelle M, Paul Thiery J. Src and Ras are involved in separate pathways in epithelial cell scattering. EMBO J. 1997;16:5904–5913. doi: 10.1093/emboj/16.19.5904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Karmali PP, et al. Metastasis of tumor cells is enhanced by downregulation of Bit1. PLoS ONE. 2011;6:e23840. doi: 10.1371/journal.pone.0023840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Yao X, et al. Downregulation of Bit1 expression promotes growth, anoikis resistance, and transformation of immortalized human bronchial epithelial cells via Erk activation-dependent suppression of E-cadherin. Biochem. Biophys. Res. Commun. 2018;495:1240–1248. doi: 10.1016/j.bbrc.2017.11.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hua W, Miao S, Zou W, Yang H, Chen BL. Pathological implication and function of Bcl2-inhibitor of transcription in ovarian serous papillary adenocarcinomas. Neoplasma. 2013;60:143–150. doi: 10.4149/neo_2013_019. [DOI] [PubMed] [Google Scholar]
- 81.Hua W, et al. Monoclonal antibodies against human Bit1, an apoptosis-associated mitochondrial protein. Hybridoma. 2009;28:167–171. doi: 10.1089/hyb.2008.0095. [DOI] [PubMed] [Google Scholar]
- 82.Zheng J, et al. Estrogen decreases anoikis of ovarian cancer cell line Caov-3 through reducing release of Bit1. DNA Cell Biol. 2014;33:847–853. doi: 10.1089/dna.2014.2453. [DOI] [PubMed] [Google Scholar]
- 83.Hagel M, et al. The adaptor protein paxillin is essential for normal development in the mouse and is a critical transducer of fibronectin signaling. Mol. Cell. Biol. 2002;22:901–915. doi: 10.1128/MCB.22.3.901-915.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zouq NK, et al. FAK engages multiple pathways to maintain survival of fibroblasts and epithelia - differential roles for paxillin and p130Cas. J. Cell Sci. 2009;122:357–367. doi: 10.1242/jcs.030478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Bessette DC, et al. Using the MCF10A/MCF10CA1a breast cancer progression cell line model to investigate the effect of active, mutant forms of EGFR in breast cancer development and treatment using gefitinib. PLoS ONE. 2015;10:e0125232. doi: 10.1371/journal.pone.0125232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJE. The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 2015;10:845. doi: 10.1038/nprot.2015.053. [DOI] [PMC free article] [PubMed] [Google Scholar]