Abstract
Background
Prevotella species are commonly isolated from the reproductive tract of women with obstetric/gynecologic health complications. However, contributions of this genus to changes in local microenvironment are not well characterized. Our objective was to evaluate species-specific effects of Prevotella on the human endometrial epithelium.
Methods
Thirteen Prevotella strains, originally isolated from the human oral cavity, amniotic fluid, endometrium, or vagina (including women with bacterial vaginosis), were obtained from BEI and ATCC resources. Bacteria were evaluated in silico and in vitro using human endometrial epithelial cells (EEC) grown as monolayers or a 3-dimensional (3D) model.
Results
Genomic characterization illustrated metabolic and phylogenetic diversity of Prevotella genus. Among tested species, P. disiens exhibited cytotoxicity. Scanning electron microscopy analysis of the 3D EEC model revealed species-specific colonization patterns and alterations of ultracellular structures. Infection with sialidase-producing P. timonensis resulted in elongated microvilli, and increased MUC3 and MUC4 expression. Infections with Prevotella species, including P. bivia, did not result in significant proinflammatory activation of EEC.
Conclusions
Collectively, findings indicate that Prevotella species are metabolically diverse and overall not cytotoxic or overtly inflammatory in EEC; however, these bacteria can form biofilms, alter barrier properties of the endometrial epithelium, and ultimately impact colonization of secondary colonizers.
Keywords: ascending infection, bacterial vaginosis, biofilm, colonizer, genital inflammation, membrane-associated mucin, epithelial layer, pathway prediction, sialidase, upper reproductive tract infection
Phylogenetically and metabolically diverse Prevotella interacted with human 3D endometrial epithelial cells (EEC) in a species-specific manner. P. disiens was cytotoxic to the EEC. P. timonensis exhibited high sialidase activity altered membrane-associated mucins more than P. bivia and other species.
Prevotella species are integral members of biofilm communities at various mucosal sites, such as the gut [1], oral cavity [2], and respiratory [3] and reproductive tracts [4]. These microorganisms may play a dual role in host health and their impact can be site-specific. In the gut, Prevotella abundance has been positively associated with high fiber consumption [5], but also with greater risk for obesity and type 2 diabetes [1]. In the oral cavity, Prevotella species contribute to periodontitis by facilitating biofilm formation and neutralizing the pH to support pathogenic growth [2].
In the female reproductive tract (FRT), Prevotella is commonly isolated from women with bacterial vaginosis (BV) [6]. Furthermore, abundance of Prevotella in the vagina has been associated with obesity and inflammation [7], increased risk of acquisition of sexually transmitted infections [8–10], preterm birth [11], and premature rupture of membranes [11, 12]. However, the role of Prevotella species in the etiology and manifestation of BV and other gynecologic and reproductive sequelae is still controversial. While some clinical reports strongly associate Prevotella species with elevated levels of proinflammatory cytokines in the lower FRT [4, 7], other reports suggest that Prevotella species are not inflammatory alone, but act as early colonizers in FRT biofilms and enhance the growth of other BV-associated bacteria including Gardnerella vaginalis [13]. For example, P. bivia has been shown to stimulate G. vaginalis growth in vitro by providing ammonia [14]. Additionally, sialidase produced by Prevotella and Gardnerella species has been hypothesized to enhance colonization of the FRT with other BV-associated microorganisms [13].
The ascension of vaginal bacteria to the upper FRT is associated with adverse gynecologic, obstetric, and reproductive health outcomes [15]. Prevotella species, such as P. bivia and/or P. disiens, have been detected in endometrial or Fallopian tube samples collected from woman with BV [15, 16], pelvic inflammatory disease [17], and pyometra [18]. These clinical findings indicate that Prevotella can transcend the cervical mucus plug and ascend from the vagina to the upper FRT. Recently, a study demonstrated the ascension of P. bivia into the uterus of mice, which was facilitated by G. vaginalis [19]. Although, Prevotella species are consistently found in the upper FRT and associated with inflammatory diseases [4], the mechanistic impact of Prevotella on endometrial epithelium, contributing to gynecologic and obstetric sequalae, is unknown.
The majority of the studies that focus on pathogenicity of Prevotella species in the FRT rely on 16S rRNA gene sequencing. These association-based studies lack strain-specific genetic and metabolic differences among Prevotella species, which may lead to misinterpretation of the potential role of these strains in the etiology of the FRT diseases. Additionally, previous studies mainly focused on P. bivia because it is the most commonly isolated Prevotella species in women with BV [20]. However, emerging studies demonstrate that other Prevotella species, such as P. amnii and P. timonensis, are also found in the FRT [21]. In fact, P. timonensis and P. disiens have been found primarily in women with BV compared to healthy women [22].
Previously, our group has established a physiologically relevant and robust human 3-dimensional (3D) endometrial epithelial cell (EEC) model [23]. Herein, we utilized this model and evaluated 13 Prevotella strains, originally isolated from the endometrium, vagina, amniotic fluid, and oral cavity, to characterize the physiological and host defense interactions between the endometrial epithelium and Prevotella species.
METHODS
In Silico Analysis of Bacterial Genomes
16S rRNA gene sequences of Prevotella strains (listed in Table 1) were downloaded from the Silva database [24] (https://www.arb-silva.de). The sequences were aligned using Multiple Alignment using Fast Fourier Transform (MAFFT) version 7 pipeline, the multiple alignment program for nucleotide sequences [25]. The tree was constructed using unweighted pair group method with arithmetic mean clustering method. The tree was visualized using interactive tree of life [26] (https://itol.embl.de) display tool. Metabolic pathway and enzyme analyses were performed using MetaCyc database [27] (https://metacyc.org) (Supplementary Methods).
Table 1.
Bacterial Strains Used in the Study
| Species | Strain | Isolation Site | Health Status | Source | Sequence ID, Silva Database |
|---|---|---|---|---|---|
| P. amnii | CRIS 21A-A | Amniotic fluid | Unknown | BEI | ADFQ01000002 |
| P. bivia | VPI6822 | Endometrium | Unknown | ATTC | AJVZ01000014 |
| P. bivia | DNF00188 | Vagina | BV | BEI | JRNF01000130 |
| P. bivia | DNF00650 | Vagina | BV | BEI | JRNM01000057 |
| P. bivia | GED7880 | Vagina | BV | BEI | LTAG01000045 |
| P. bivia | GED7760C | Vagina | BV | BEI | LRQF02000029 |
| P. buccae | D17 | Oral cavity | Unknown | BEI | ACRB01000001 |
| P. corporis | MJR7716 | Vagina | Pregnancy | BEI | LRQG01000067 |
| P. denticola | Oral Taxon 291 | Oral cavity | Unknown | BEI | CP002589 |
| P. denticola | DNF00960 | Vagina | BV | BEI | JRNO01000111 |
| P. disiens | DNF00882 | Vagina | BV | BEI | JRNR01000204 |
| P. histicola | F0411 | Oral cavity | Dental plaque | BEI | AFXP01000013 |
| P. timonensis | CRIS 5C-B1 | Vagina | Unknown | BEI | ADEF01000012 |
| G. vaginalis | JCP8151B | Vagina | BV | BEI | Not included |
Abbreviations: ATTC, American Type Culture Collection; BEI, Biodefense and Emerging Infections Research Resources Repository; BV, bacterial vaginosis; G., Gardnerella; ID, identity; P., Prevotella.
Bacterial Strains and Growth Conditions
All Prevotella strains and the G. vaginalis strain JCP8151B (Table 1) were obtained from Biodefense and Emerging Infections Research Resources Repository (https://www.beiresources.org) or the American Type Culture Collection (ATTC). Bacteria were grown on tryptic soy agar (Becton Dickinson) supplemented with 5% defibrinated sheep blood (Quad Five) at 37 °C under anaerobic conditions generated using GasPak EZ Anaerobe Container System.
Human Endometrial Epithelial Cell Culture and Generation of 3-Dimensional Model
Human endometrial adenocarcinoma cell line (HEC-1A; ATCC HTB-112) was cultured in supplemented modified McCoy 5A medium at 37°C in 5% carbon dioxide atmosphere. Human 3D EEC model was generated as previously described [23]. Briefly, cells were grown on collagen-coated dextran microcarrier beads (Cytodex-3; Sigma-Aldrich) in a rotating wall vessel bioreactor (Synthecon) for 21 days. Following differentiation, 3D aggregates were transferred into 24-well plates (approximately 2.5 × 106 cells/mL) for infection assays.
Infection Assays
Prevotella and Gardnerella strains were grown for 20 hours as described above and resuspended in sterile Dulbecco phosphate-buffered saline (PBS). Monolayer and 3D EEC were infected at a multiplicity of infection (MOI) of 10 for 24 hours under anaerobic conditions generated using the GasPak EZ Anaerobe Container System. Cytotoxicity was evaluated via trypan blue exclusion or crystal violet staining, as described previously [28]. The quantification of the cell density was performed using Image J software [29] (Supplementary Methods).
Scanning Electron Microscopy
Samples were fixed and processed for scanning electron microscopy (SEM) as described previously [30] and imaged using a JSM-6300 JEOL microscope. Images were acquired using an IXRF model 500 digital processor (IXRF Systems) and pseudocolored using Adobe Photoshop CS5.1.
Gene Expression Assays
The 3D EEC model was infected with the Prevotella strains and G. vaginalis or treated with microbial products, poly(I:C) (100 μg/mL) and flagellin (0.5 μg/mL), as positive controls for 24 hours. The 3D aggregates were harvested and pellets were stored at −80°C until RNA extraction. Total RNA was extracted from 3D EEC aggregates using Quick-RNA Miniprep Kit (Zymo Research) according to the manufacturer’s instructions. cDNA was synthesized from 1 µg of total RNA using iScript Reverse Transcription Supermix (Bio-Rad). Gene expression was analyzed by quantitative real-time polymerase chain reaction (PCR) assays using custom primers (Supplementary Table 1) and iTaq Universal SYBR Green Supermix (Bio-Rad) as described previously [23] (Supplementary Methods).
Sialidase Assay
Sialidase activity was determined in overnight Prevotella cultures using Amplex Red Neuroaminidase (Sialidase) Assay Kit (A-22178) according to the manufacturer’s instructions.
Statistical Methods
All experiments were performed at least in triplicate. Statistical analyses were performed using Prism v8 software (GraphPad). Analysis of variance (ANOVA) with Tukey adjustment for multiple comparison was used to determine the differences in the means. An adjusted P value less than .05 was considered significant. Hierarchical clustering analysis of the cytokines was performed using ClustVis tool [31].
RESULTS
Members of Prevotella Genus Are Phylogenetically Diverse and Exhibit Variable Metabolic Capabilities
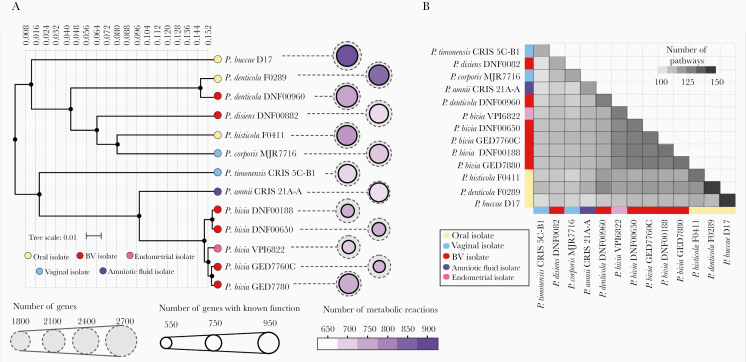
In order to reveal evolutionary relatedness and genomic differences among Prevotella strains, we constructed a phylogenetic tree based on the variations in the 16S rRNA gene sequences. The strains formed 2 main clusters primarily driven by oral and FRT strains (Figure 1A). The FRT cluster included vaginal P. bivia, which shares a common ancestor with P. amnii and P. timonensis. The second cluster was dominated by oral strains (P. buccae, P. denticola, and P. histicola) but also contained vaginal strains (P. corporis, P. denticola, and P. disiens). Similarities between the vaginal and oral isolates indicate the interrelatedness of the species in these 2 ecosystems.
Figure 1.
Prevotella isolates are genetically and metabolically diverse and share genetic elements based on isolation source. A, Phylogenetic tree constructed based on 16S rRNA gene sequences of the Prevotella strains using unweighted pair group method with arithmetic mean algorithm. The 16S rRNA amplicons were downloaded from the Silva database (https://www.arb-silva.de) and the sequences were aligned using the Multiple Alignment using Fast Fourier Transform (MAFFT) version 7 program (https://mafft.cbrc.jp/alignment/server/) [25]. Number of genes and number of metabolic reactions were extracted from the MetaCyc database (https://metacyc.org) [27]. The outer grey circles represent number of genes detected in each strain. The size of the inner circle represents number of genes with known functions. The color intensity of the inner circle represents number of metabolic reactions. P. bivia strains were phylogenetically more similar to P. amnii and P. timonensis than the other vaginal strains in the analysis. P. disiens and P. denticola strains were more metabolically diverse and clustered with the oral strains. P. bivia species had relatively smaller genomes, number of genes with known functions, and metabolic reactions in comparison to other Prevotella species isolated from female reproductive tract or oral cavity. B, Number of metabolic pathways and shared metabolic pathways among the Prevotella species based on predictions from the MetaCyc database. Oral isolates had the highest number of pathways whereas P. bivia strain had the greatest number of shared pathways. Abbreviation: BV, bacterial vaginosis.
Using the MetaCyc pipeline, we compared the genomes of the strains to highlight their predicted metabolic and enzymatic relatedness. Oral strains, P. buccae D17 and P. denticola F0289, had the greatest number of genes, genes with known functions, predicted number of metabolic reactions (Figure 1A), and metabolic pathways (Figure 1B), particularly amino acid biosynthesis and carbohydrate degradation (Supplementary Table 2). P. bivia strains, besides P. bivia GED7880, had the lowest number of genes. The genome of P. timonensis CRIS 5C-B1 encoded the smallest number of pathways among the analyzed strains followed by P. disiens DNF00882 and P. corporis MJR7716. All strains evaluated have the metabolic capacity to synthesize many amino acids, whereas only a few have the capacity to synthesize valine, tryptophan, and lysine. Similarly, all strains evaluated exhibit serine, threonine, glutamine, and glutamate degradation pathways. Notably, P. disiens had the fewest number of carbohydrate degradation pathways, but harbored genes encoding specific hydrolases, an oxidoreductase, and isomerase that were not present in other strains (Supplementary Table 3). This in silico analysis demonstrated genetic and predicted metabolic versatility of Prevotella strains that may be instrumental in their interactions with the endometrial epithelium.
Prevotella disiens Exhibits Cytotoxicity in 3-Dimensional Endometrial Epithelial Model
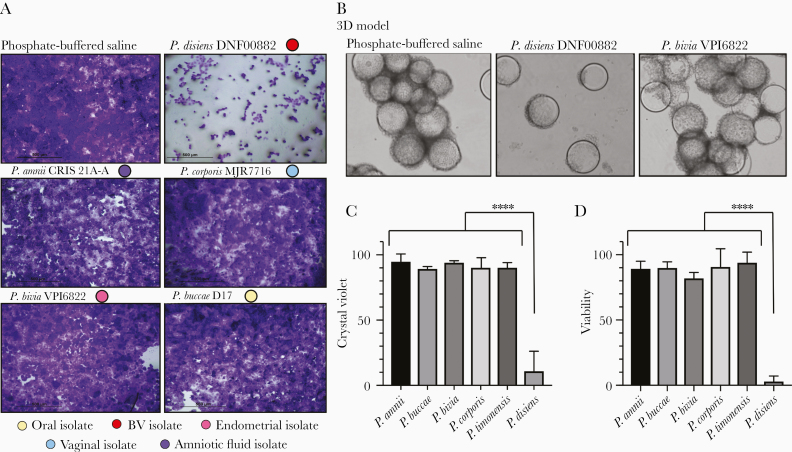
To assess the cytotoxic effect of Prevotella species on endometrial epithelium, we performed an initial screening by infecting EEC monolayers (Supplementary Figure 1 and Figure 2A) using 13 Prevotella strains (Table 1) at a MOI of 10 for 24 hours followed by crystal violet staining. Among strains tested, P. disiens DNF00882 was the only strain that showed dramatic cytotoxicity at this MOI both on monolayers and the 3D ECC model (Figure 2B). Based on crystal violet staining and quantitative densitometry of the images, about 90% of cells were estimated to be cleared upon P. disiens infection (Figure 2C). As the 3D EEC model is more resilient to cytotoxicity than conventional monolayers [30], we further characterized the impact of selected strains using the trypan blue exclusion method in a bioreactor-derived human 3D EEC model (Figure 2D and Supplementary Figure 2). We observed less than 3% viable cells following infection with P. disiens DNF00882 at a MOI of 10 for 24 hours. The other Prevotella strains included in the study at this MOI did not induce significant cytotoxicity (viability > 90%), confirming that the majority of the Prevotella species, including the most commonly isolated P. bivia, are not cytotoxic to EEC grown as monolayers or in the 3D model at MOI of 10.
Figure 2.
Among the Prevotella strains tested, P. disiens was dramatically cytotoxic to the endometrial epithelial cells grown as monolayers as well as in the 3D model. Human EEC models (monolayers and 3D) were incubated with Prevotella strains for 24 hours at MOI 10. Monolayer cultures were used to screen 13 Prevotella strains (Supplementary Figure 1) and 3D model was incubated with selected Prevotella strains for more robust evaluation of cytotoxicity. A, Crystal violet staining of the endometrial epithelial cells grown as monolayers and (B) light microscopy images of the 3D EEC aggregates visualized the damage on endometrial epithelial cells due to P. disiens infection (× 100). Light microscopy images were converted to grayscale using the same brightness and contrast filters. C, Crystal violet staining densities of the cells in the monolayer model were quantified using Fiji (ImageJ) software. The comparisons were performed relative to PBS control. All the incubations were performed under the same conditions. D, Viabilities of the endometrial epithelial cells were significantly lower when the 3D EEC aggregates were incubated with P. disiens compared to other Prevotella species. Viability calculations were based on trypan blue exclusion in comparison to PBS control. Student t test **** P < .001. Data are representative of at least 3 independent experiments. C and D, Error bars represent standard deviation from the mean. Abbreviations: 3D, 3-dimensional; BV, bacterial vaginosis; EEC, endometrial epithelial cell; MOI, multiplicity of infection; PBS, phosphate-buffered saline.
Prevotella Species Exhibit Distinct Colonization Patterns in Human 3-Dimensional Endometrial Epithelial Cell Model
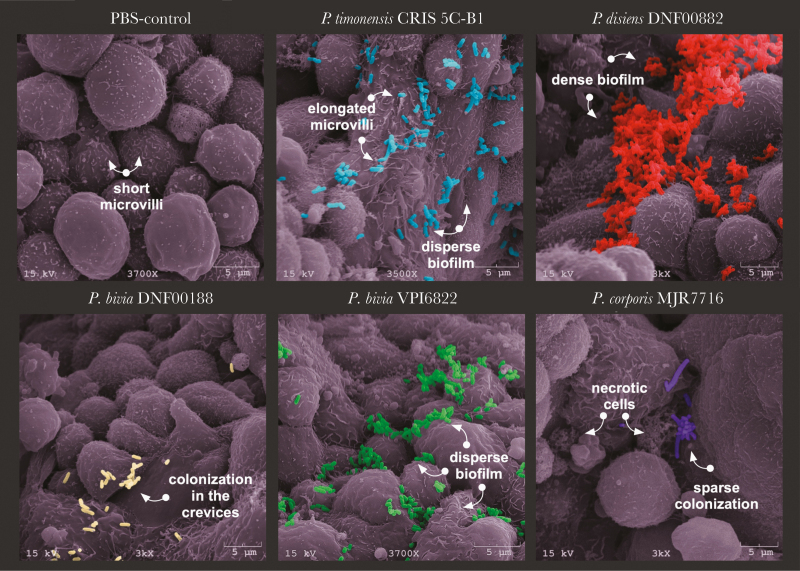
The human 3D EEC model recapitulates physiologically relevant properties of endometrial epithelium including microvilli, extracellular secretions, and cellular junctions [23]. SEM images of the 3D model infected with various Prevotella species highlighted key differences in colonization patterns, localization, and ultrastructural changes to the endometrial epithelium (Figure 3). Control PBS-treated cells exhibited varying levels of microvilli density and extracellular secretions. Following infection of the 3D model with P. timonensis CRIS 5C-B1, we observed rearrangement of the microvilli. P. timonensis formed a sparse biofilm on the surface of the 3D model and the bacteria were entrapped within a “web” of elongated microvilli. In contrast, P. disiens DNF00882 formed dense biofilms, mostly in the crevices between the cells. Because the 3D model was infected for only 4 hours, there was no apparent evidence of cytotoxicity on the cells. Similarly, 2 P. bivia strains (DNF00188 and VPI6822) colonized mainly the crevices; however, instead of dense biofilms, these strains formed scattered clusters. P. corporis MJR7716 were also visualized in the deep crevices among the cells, indicating Prevotella strains mainly colonize the intercellular spaces, potentially to strengthen their attachment and resistance from dissociation and simultaneously interact with multiple cells. Additionally, Prevotella sp. colonized the 3D ECC at different rates (Supplementary Figure 3). Distinct colonization patterns of the Prevotella species on the 3D EEC model may impact epithelial gene expression and barrier function.
Figure 3.
Prevotella isolates colonize human 3D endometrial epithelial cell model in a species-specific manner. The 3D EEC aggregates were incubated with Prevotella species at MOI 10 for 4 hours under anaerobic conditions. The SEM images depict PBS treatment and colonization with P. timonensis CRIS 5C-B1, P. disiens DNF00882, P. bivia DNF00188, P. bivia VPI6822, and P. corporis MJR7716. SEM images also demonstrated viability of Prevotella strains. Prevotella species induced species-specific microvilli remodeling and cellular secretion patterns. The images were pseudocolored using Adobe Photoshop. The images presented were selected from a minimum of 3 representative SEM images. Abbreviations: 3D, 3-dimensional; EEC, endometrial epithelial cell; MOI, multiplicity of infection; PBS, phosphate-buffered saline; SEM, scanning electron microscopy.
P. timonensis Alters Host Membrane-Associated Mucins and Has High Sialidase Activity
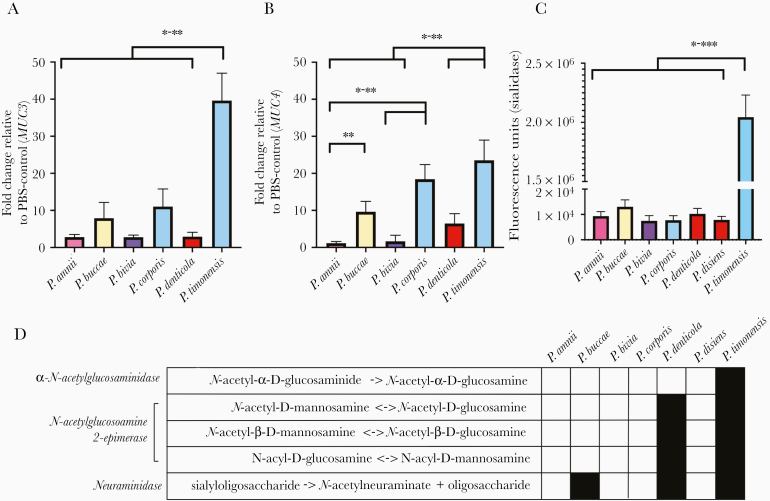
Mucin expression in the 3D EEC model was previously profiled [23] and shown to express membrane-associated mucins (MAMs). We measured the gene expression of relatively abundant MAMs, MUC1, MUC3, MUC4, and MUC20, following infection with Prevotella strains. We did not observe a significant induction of MUC1 and MUC20 expression by any of Prevotella strains tested. However, P. timonensis, P. buccae, and P. corporis induced the expression of MUC3 and MUC4 (Figure 4A and 4B). These strains induced MUC4 expression more than 10-fold and significantly higher compared to the other strains tested (P < .05). Furthermore, the expression levels of MUC3 were drastically higher following infection with P. timonensis compared to other Prevotella species (P < .05).
Figure 4.
The molecular interactions between P. timonensis and human 3-dimensional endometrial epithelial cell model rely on mucin metabolism. Relative expression levels of membrane-associated mucins (A) MUC3 and (B) MUC4 in comparison to phosphate-buffered saline (PBS) treated cells. P. timonensis, P. buccae, and P. corporis induced significantly higher levels of MUC3 and MUC4. C, Sialidase activity of the selected Prevotella strains. P. timonensis had more than 1000-times greater sialidase activity in comparison to the representative Prevotella strains. P. bivia VPI6822 was selected to represent Prevotella strains. Data are representative of at least 3 independent experiments. D, Predictions of enzymatic reactions involved in mucin degradation or metabolism based on Prevotella genomes in MetaCyc database. P. denticola and P. timonensis strains are predicted to contain genes that can metabolize mucins. P. timonensis harbors another gene that encodes transformation of mucins. Strain designations: P. amnii CRIS 21A-A, P. buccae D17, P. bivia VPI6822, P. corporis MJR7716, P. denticola DNF00960, and P. timonensis CRIS 5C-B1. Tukey’s multiple comparisons results: *P < 0.05, **P < 0.01, and ***P < 0.001.
Prevotella species can alter mucin metabolism not only by stimulating their production, but also their degradation. Bacterial sialidases initiate degradation of mucins by catalyzing the cleavage of sialic acids [32]. The majority of the strains screened had similar levels of sialidase activity (Figure 4C). Strikingly, P. timonensis had significantly greater sialidase activity (P < .0001) and induced relatively greater expression of the MAMs in 3D EEC model compared to other strains. Interestingly, P. timonensis exhibited the greatest number of mucin degradation pathways (Figure 4D).
Prevotella Isolates Do Not Induce a Prominent Proinflammatory Response
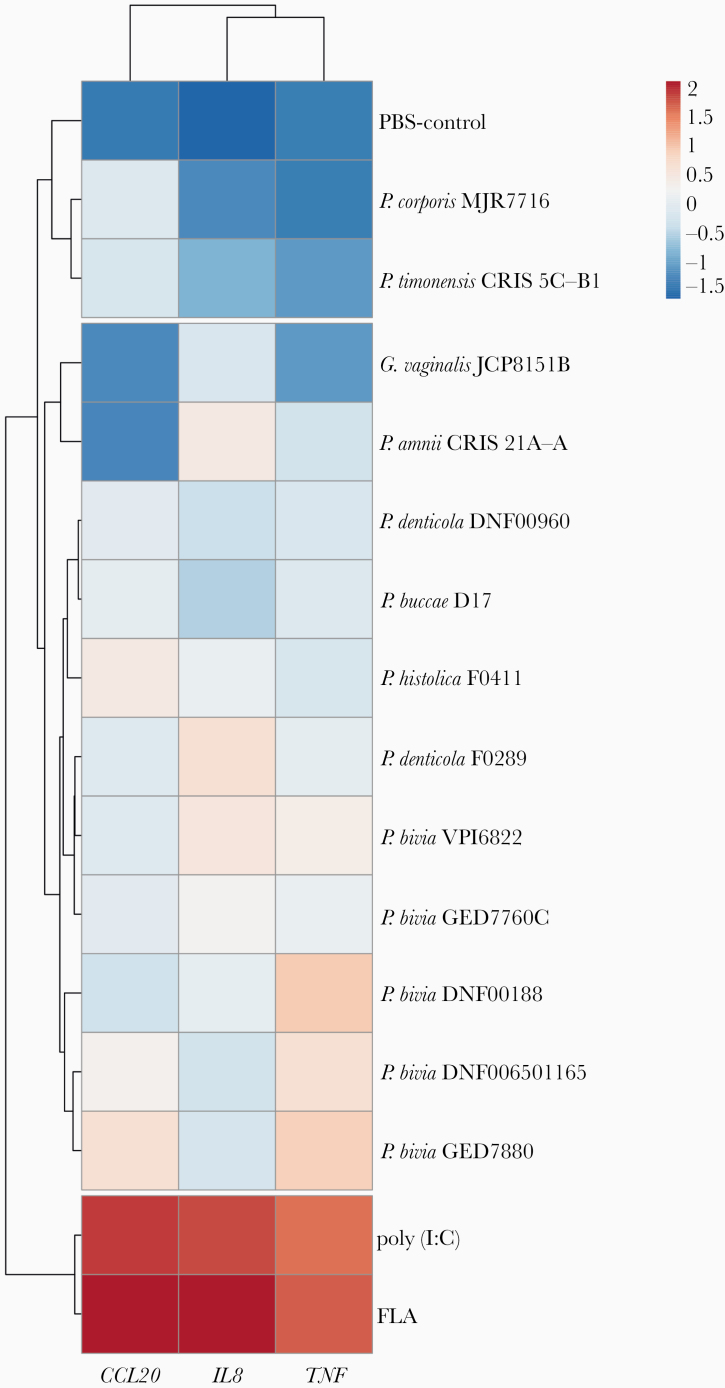
Because Prevotella species have been frequently isolated from individuals with inflammatory diseases of the upper FRT [15–18], we investigated whether Prevotella species directly induce the expression of proinflammatory cytokines in the 3D EEC model (Figure 5). P. disiens was excluded from this analysis due to its cytotoxic effect on EEC. We quantified expression levels of CCL20, interleukin-8 (IL8), and tumor necrosis factor (TNF) genes relative to glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and subsequently performed hierarchical clustering analysis on gene expression data. We compared the expression levels to positive (flagellin and poly(I:C)) and negative (PBS) controls. All Prevotella strains and G. vaginalis evaluated did not significantly induced the expression of the tested proinflammatory mediators in comparison to positive controls (Figure 5 and Supplementary Figure 4). However, variations were detected in the expression levels of the targeted genes depending on the strain. P. timonensis and P. corporis strains clustered with PBS-treated aggregates implying a relatively low level of proinflammatory epithelial activation compared to infections with other Prevotella species. In contrast, P. bivia strains induced the expression of IL8, TNF, and CCL20 at higher magnitude compared to other strains, but not significantly. Overall, Prevotella species did not overtly induce proinflammatory responses in the EEC consistent with our findings in the lower FRT [33, 34].
Figure 5.
Prevotella strains do not robustly induce an inflammatory response in the human 3D endometrial epithelial cell model. The expression of proinflammatory genes in the human 3D EEC following infection with Prevotella species for 24 hours. Expression levels of IL8, TNF, and CCL20 genes were normalized to expression levels of GAPDH gene and log10-transformed prior to the hierarchal clustering analysis. The clustering was performed based on Euclidean distances. FLA and poly(I:C) were used as positive controls for induction of proinflammatory genes and Gardnerella vaginalis infections were used to compare the response from Prevotella infections. Data are representative of at least 3 independent experiments. Abbreviations: 3D, 3-dimensional; EEC, endometrial epithelial cell; FLA, flagellin; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; IL8, interleukin-8; PBS, phosphate-buffered saline; TNF, tumor necrosis factor.
DISCUSSION
Endometrial epithelium is the first line of host defense in the uterus. Prevotella spp. are known to ascend to the upper FRT; however, the mechanistic impact of Prevotella spp. on the endometrial epithelium has not been extensively studied. Prevotella species are commonly detected in women with diverse anaerobic vaginal bacterial consortia with or without BV [35]. However, lack of strain/species-level resolution obscures a complete understanding of their virulence in the FRT. Herein, we investigated the impact of phylogenetically and metabolically diverse Prevotella strains on pathophysiological changes in the upper FRT using a human 3D EEC model.
We demonstrated that Prevotella is a ubiquitous and diverse genus with niche-specific genomic and metabolic capabilities, further validating the need for host response characterization to infection with different species/strains. The greater genetic and metabolic diversity observed in oral isolates in comparison to FRT isolates is possibly due to the adaptation to a diverse niche that requires versatility. Among the isolates, P. bivia had relatively smaller genomes and less metabolic diversity than the oral or other FRT isolates. P. bivia isolates had the lowest number of genes with known functions, which may play a role in microbe-microbe and microbe-host interactions. Notably, microorganisms with smaller genome sizes are predicted to rely on other microorganisms for cellular functions [36]. Studies that further characterize genomes of Prevotella isolates would provide insights into the role of these species in women’s health. Using this phylogenetic analysis to inform our experiments, we investigated species/strain-specific interactions of Prevotella isolates with a human 3D EEC model.
Cytotoxicity or damage to the epithelium is a virulence mechanism that pathogens employ to invade host tissues. The majority of Prevotella species, including P. bivia strains, were not cytotoxic to the 3D model. A lack of cytotoxicity of P. bivia was consistent with our previous findings in the lower FRT [33, 34]. However, P. disiens exhibited high cytotoxic activity, confirming that the Prevotella genus contains species with high pathogenic potential for the upper FRT. Interestingly, in a cohort of pregnant women, abundance of P. disiens in the vagina differentiated symptomatic women with dilated cervix and exposed fetal membranes from women with normal pregnancy [37]. Prevotella species in the gut are known ammonia producers and some species, such as P. intermedia associated with lung disease, are known to produce toxins [38]. Therefore, it is possible that P. disiens damages endometrial epithelial aggregates via these pathophysiological mechanisms [39]. However, additional studies with P. disiens are needed to better understand the mechanisms of cytotoxicity and to determine its role in pathogenesis of FRT diseases.
Employing SEM, we observed species-specific colonization patterns and alterations in the ultracellular endometrial epithelial architecture. SEM demonstrated that 3D EEC exhibited healthy cellular morphology under anaerobic conditions and the ability of Prevotella spp. to colonize the model. P. timonensis formed sparse biofilms and induced ultrastructural reorganization of the epithelial surface resulting in elongated microvilli. Previously, it has been shown that elongation of microvilli enhances adhesion of bacteria, including pathogens such as Vibrio haemolyticus [40] to the gut epithelium. Therefore, the microvilli alteration following P. timonensis infection might influence adhesion of other secondary colonizers and/or bacterial ascension.
We also investigated the impact of the Prevotella strains on epithelial barrier properties, including mucin expression. MAMs can be expressed in response to microorganisms or microbial products and play a dual role in pathogen adherence/clearance [41]. In an intestinal epithelial cell model, adherence of commensal Lactobacillus has been shown to increase MUC3 production, which in turn decreased adherence of a pathogenic E. coli [42]. Herein, we observed induction of MUC3 and MUC4 expression by specific Prevotella strains, including vaginal P. timonensis, P. corporis, and oral P. buccae strains. An increase in the expression of MAMs can promote bacterial clearance by allowing epithelia to produce a protective barrier that entraps pathogens [43]. Notably, the abundance of Prevotella [44] and expression of MUC4 [45] have been associated with endometrial polyps and endometriosis, respectively.
Prevotella species can alter mucus metabolism not only by stimulating mucin production but also contributing to degradation of these highly glycosylated proteins [19]. Microorganisms secrete sialidase enzymes that cleave sialic acid residues from host glycoproteins to harvest energy [46]. P. bivia has been known to exhibit sialidase activity [19]; however, our experiments showed that P. timonensis exhibited 1000-times higher sialidase activity than the Prevotella species tested. Interestingly, our in silico analysis found that genes encoding α-N-acetylglucosaminidase, an enzyme capable of cleaving O-glycans of type 3 mucins [47], was only detected in P. timonensis genomes among the analyzed strains. In the FRT, bacterial sialidase activity has been linked to BV, acquisition of sexually transmitted infections [48], and preterm labor [49]. G. vaginalis has been shown to stimulate ascension of P. bivia to the upper FRT due to its sialidase activity [19]. Similarly, the high sialidase activity of P. timonensis could also compromise the cervical mucus plug and thereby uterine health by allowing vaginal bacteria to ascend to the upper FRT. Our genomic and in vitro analyses on P. timonensis and mucins indicate a potential pathogenic role of this species in the FRT.
Prevotella species are often associated with inflammatory diseases [7]; however, the extent to which these microorganisms directly impact epithelial inflammation in the uterine microenvironment has not been investigated previously. Our findings demonstrated that Prevotella strains, including those isolated from BV-positive individuals, induced minimal proinflammatory epithelial cell activation in the 3D EEC model. Previous reports from our group have shown that P. bivia did not induce a significant inflammatory response in vaginal or cervical epithelial models [33, 34]. Using the 3D EEC model, we extended these findings to the upper FRT. Nevertheless, it is important to note that the Prevotella genus contains species, for example P. copri and P. intermedia, that are inflammatory at other mucosal sites [4]. Notably, Prevotella sp. may contribute to inflammation in the upper FRT indirectly through its mutual interactions with other secondary colonizers and inflammatory microorganisms [13]. Overall, Prevotella spp. may play a role in biofilm formation in the FRT and support colonization of other bacteria, including BV-associated microorganisms or sexually transmitted pathogens [13], that can lead to dysbiosis and subsequently adverse gynecological and reproductive outcomes.
Our study evaluated species/strain-specific host responses to Prevotella infections in a 3D EEC model, which recapitulates many characteristics of human endometrial tissue. However, this model has some limitations, including use of an adenocarcinoma cell line and varied Toll-like receptor expression [23]. However, this cell line has been widely used to study endometrial physiological/pathophysiological mechanisms [50]. Future studies employing primary in vitro systems are needed to extend these findings.
In summary, Prevotella resulted in minimal proinflammatory epithelial activation and induced changes to the cellular physiology and integrity of the epithelium in a species-specific fashion. Cytotoxicity of P. disiens indicated that species with high pathogenic potential exists within the Prevotella genus. Sialidase activity and increased mucin expression by specific Prevotella species indicated their role in endometrial epithelial barrier function and the metabolic microenvironment. The human 3D EEC model in combination with in silico analyses allowed us to provide mechanistic insights into the role of Prevotella isolates in the context of the endometrial epithelium that can be used in contextualizing sequencing findings related to the upper FRT and gynecologic health.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Notes
Acknowledgments. The authors thank David Lowry, Arizona State University Electron Microscope Laboratory, for technical support of SEM sample preparation and analyses and Mary Salliss for her assistance with generating the pseudocolored SEM images. The bacterial strains were obtained through Biodefense and Emerging Infections Research Resources, National Institute of Allergy and Infectious Diseases, National Institutes of Health as part of the Human Microbiome Project.
Financial support. This work was supported by the Alternatives Research and Development Foundation; and the National Cancer Institute grant supplement from the Office of Research on Women’s Health (grant number P30CA023074).
Potential conflicts of interest. M. M. H.-K. has been a consultant for Lupin Pharmaceuticals and Becton Dickinson. All other authors report no potential conflicts. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
Presented in part: Global Health Keystone Symposia, December 8-12, 2019, Cape Town, South Africa; and 58th American Society for Microbiology Annual Branch Meeting, April 13, 2019, Flagstaff, AZ.
References
- 1. Larsen N, Vogensen FK, van den Berg FW, et al. Gut microbiota in human adults with type 2 diabetes differs from non-diabetic adults. PLoS One 2010; 5:e9085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Takahashi N. Microbial ecosystem in the oral cavity: metabolic diversity in an ecological niche and its relationship with oral diseases. Int Congr Ser 2005; 1284:103–12. [Google Scholar]
- 3. Di Cicco M, Pistello M, Jacinto T, et al. Does lung microbiome play a causal or casual role in asthma? Pediatr Pulmonol 2018; 53:1340–5. [DOI] [PubMed] [Google Scholar]
- 4. Larsen JM. The immune response to Prevotella bacteria in chronic inflammatory disease. Immunology 2017; 151:363–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. De Filippo C, Cavalieri D, Di Paola M, et al. Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc Natl Acad Sci U S A 2010; 107:14691–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hillier SL, Krohn MA, Rabe LK, Klebanoff SJ, Eschenbach DA. The normal vaginal flora, H2O2-producing lactobacilli, and bacterial vaginosis in pregnant women. Clin Infect Dis 1993; 16(suppl 4);S273–81. [DOI] [PubMed] [Google Scholar]
- 7. Si J, You HJ, Yu J, Sung J, Ko GP. Prevotella as a hub for vaginal microbiota under the influence of host genetics and their association with obesity. Cell Host Microbe 2017; 21:97–105. [DOI] [PubMed] [Google Scholar]
- 8. Hashemi FB, Ghassemi M, Faro S, Aroutcheva A, Spear GT. Induction of human immunodeficiency virus type 1 expression by anaerobes associated with bacterial vaginosis. J Infect Dis 2000; 181:1574–80. [DOI] [PubMed] [Google Scholar]
- 9. Balle C, Lennard K, Dabee S, et al. Endocervical and vaginal microbiota in South African adolescents with asymptomatic Chlamydia trachomatis infection. Sci Rep 2018; 8:11109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Masson L, Barnabas S, Deese J, et al. Inflammatory cytokine biomarkers of asymptomatic sexually transmitted infections and vaginal dysbiosis: a multicentre validation study. Sex Transm Infect 2019; 95:5–12. [DOI] [PubMed] [Google Scholar]
- 11. Mikamo H, Sato Y, Hayasaki Y, Kawazoe K, Hua YX, Tamaya T. Bacterial isolates from patients with preterm labor with and without preterm rupture of the fetal membranes. Infect Dis Obstet Gynecol 1999; 7:190–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Baldwin EA, Walther-Antonio M, MacLean AM, et al. Persistent microbial dysbiosis in preterm premature rupture of membranes from onset until delivery. PeerJ 2015; 3:e1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Muzny CA, Łaniewski P, Schwebke JR, Herbst-Kralovetz MM. Host-vaginal microbiota interactions in the pathogenesis of bacterial vaginosis. Curr Opin Infect Dis 2020; 33:59–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Pybus V, Onderdonk AB. Evidence for a commensal, symbiotic relationship between Gardnerella vaginalis and Prevotella bivia involving ammonia: potential significance for bacterial vaginosis. J Infect Dis 1997; 175:406–13. [DOI] [PubMed] [Google Scholar]
- 15. Mitchell CM, Haick A, Nkwopara E, et al. Colonization of the upper genital tract by vaginal bacterial species in nonpregnant women. Am J Obstet Gynecol 2015; 212:611.e1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Swidsinski A, Verstraelen H, Loening-Baucke V, Swidsinski S, Mendling W, Halwani Z. Presence of a polymicrobial endometrial biofilm in patients with bacterial vaginosis. PLoS One 2013; 8:e53997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Sweet RL. Role of bacterial vaginosis in pelvic inflammatory disease. Clin infect Dis 1995; 20(suppl 2):S271–5. [DOI] [PubMed] [Google Scholar]
- 18. Mikamo H, Kawazoe K, Izumi K, Sato Y, Tamaya T. Studies on the clinical implications of anaerobes, especially Prevotella bivia, in obstetrics and gynecology. J Infect Chemother 1998; 4:177–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gilbert NM, Lewis WG, Li G, Sojka DK, Lubin JB, Lewis AL. Gardnerella vaginalis and Prevotella bivia trigger distinct and overlapping phenotypes in a mouse model of bacterial vaginosis. J Infect Dis 2019; 220:1099–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Muzny CA, Blanchard E, Taylor CM, et al. Identification of key bacteria involved in the induction of incident bacterial vaginosis: a prospective study. J Infect Dis 2018; 218:966–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Elwood C, Albert AYK, McClymont E, et al. Different and diverse anaerobic microbiota were seen in women living with HIV with unsuppressed HIV viral load and in women with recurrent bacterial vaginosis: a cohort study. BJOG 2020; 127:250–9. [DOI] [PubMed] [Google Scholar]
- 22. Margolis E, Fredricks DN. Bacterial vaginosis-associated bacteria. In: Tang Y-W, Sussman M, Liu D, et al. Molecular medical microbiology. 2nd ed.San Diego, CA: Elsevier, 2015:1487–96. [Google Scholar]
- 23. Laniewski P, Gomez A, Hire G, So M, Herbst-Kralovetz MM. Human three-dimensional endometrial epithelial cell model to study host interactions with vaginal bacteria and Neisseria gonorrhoeae. Infect Immun 2017; 85:e01049-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Quast C, Pruesse E, Yilmaz P, et al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res 2013; 41:D590–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Katoh K, Misawa K, Kuma K, Miyata T. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res 2002; 30:3059–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Letunic I, Bork P. Interactive tree of life (iTOL): an online tool for phylogenetic tree display and annotation. Bioinformatics 2007; 23:127–8. [DOI] [PubMed] [Google Scholar]
- 27. Caspi R, Foerster H, Fulcher CA, et al. MetaCyc: a multiorganism database of metabolic pathways and enzymes. Nucleic Acids Res 2006; 34:D511–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wilkinson EM, Łaniewski P, Herbst-Kralovetz MM, Brotman RM. Personal and clinical vaginal lubricants: impact on local vaginal microenvironment and implications for epithelial cell host response and barrier function. J Infect Dis 2019; 220:2009–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Abràmoff MD, Magalhães PJ, Ram SJ. Image processing with imageJ. Biophotonics Int 2004; 11:36–41. [Google Scholar]
- 30. Hjelm BE, Berta AN, Nickerson CA, Arntzen CJ, Herbst-Kralovetz MM. Development and characterization of a three-dimensional organotypic human vaginal epithelial cell model. Biol Reprod 2010; 82:617–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Metsalu T, Vilo J. ClustVis: a web tool for visualizing clustering of multivariate data using principal component analysis and heatmap. Nucleic Acids Res 2015; 43:W566–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Juge N, Tailford L, Owen CD. Sialidases from gut bacteria: a mini-review. Biochem Soc Trans 2016; 44:166–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Gardner JK, Łaniewski P, Knight A, Haddad LB, Swaims-Kohlmeier A, Herbst-Kralovetz MM. Interleukin-36γ is elevated in cervicovaginal epithelial cells in women with bacterial vaginosis and in vitro after infection with microbes associated with bacterial vaginosis. J Infect Dis 2020; 221:983–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Doerflinger SY, Throop AL, Herbst-Kralovetz MM. Bacteria in the vaginal microbiome alter the innate immune response and barrier properties of the human vaginal epithelia in a species-specific manner. J Infect Dis 2014; 209:1989–99. [DOI] [PubMed] [Google Scholar]
- 35. Ravel J, Gajer P, Abdo Z, et al. Vaginal microbiome of reproductive-age women. Proc Natl Acad Sci U S A 2011; 108(suppl 1):4680–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Morris JJ, Lenski RE, Zinser ER. The black queen hypothesis: evolution of dependencies through adaptive gene loss. MBio 2012; 3:e00036-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Brown RG, Chan D, Terzidou V, et al. Prospective observational study of vaginal microbiota pre- and post-rescue cervical cerclage. BJOG 2019; 126:916–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ulrich M, Beer I, Braitmaier P, et al. Relative contribution of Prevotella intermedia and Pseudomonas aeruginosa to lung pathology in airways of patients with cystic fibrosis. Thorax 2010; 65:978–84. [DOI] [PubMed] [Google Scholar]
- 39. Mégraud F, Neman-Simha V, Brügmann D. Further evidence of the toxic effect of ammonia produced by Helicobacter pylori urease on human epithelial cells. Infect Immun 1992; 60:1858–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zhou X, Massol RH, Nakamura F, et al. Remodeling of the intestinal brush border underlies adhesion and virulence of an enteric pathogen. MBio 2014; 5:e01639-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Gipson IK, Spurr-Michaud S, Tisdale A, Menon BB. Comparison of the transmembrane mucins MUC1 and MUC16 in epithelial barrier function. PLoS One 2014; 9:e100393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Mack DR, Ahrne S, Hyde L, Wei S, Hollingsworth MA. Extracellular MUC3 mucin secretion follows adherence of Lactobacillus strains to intestinal epithelial cells in vitro. Gut 2003; 52:827–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Linden SK, Sutton P, Karlsson NG, Korolik V, McGuckin MA. Mucins in the mucosal barrier to infection. Mucosal Immunol 2008; 1:183–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Fang RL, Chen LX, Shu WS, Yao SZ, Wang SW, Chen YQ. Barcoded sequencing reveals diverse intrauterine microbiomes in patients suffering with endometrial polyps. Am J Transl Res 2016; 8:1581–92. [PMC free article] [PubMed] [Google Scholar]
- 45. Chang CY, Chang HW, Chen CM, et al. MUC4 gene polymorphisms associate with endometriosis development and endometriosis-related infertility. BMC Med 2011; 9:19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Wiggins R, Hicks SJ, Soothill PW, Millar MR, Corfield AP. Mucinases and sialidases: their role in the pathogenesis of sexually transmitted infections in the female genital tract. Sex Transm Infect 2001; 77:402–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Fujita M, Tsuchida A, Hirata A, et al. Glycoside hydrolase family 89 alpha-N-acetylglucosaminidase from Clostridium perfringens specifically acts on GlcNAc alpha1,4Gal beta1R at the non-reducing terminus of O-glycans in gastric mucin. J Biol Chem 2011; 286:6479–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ketterer MR, Rice PA, Gulati S, et al. Desialylation of Neisseria gonorrhoeae lipooligosaccharide by cervicovaginal microbiome sialidases: the potential for enhancing infectivity in men. J Infect Dis 2016; 214:1621–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Cauci S, Culhane JF. High sialidase levels increase preterm birth risk among women who are bacterial vaginosis-positive in early gestation. Am J Obstet Gynecol 2011; 204:142.e1-9. [DOI] [PubMed] [Google Scholar]
- 50. Kuramoto H, Tamura S, Notake Y. Establishment of a cell line of human endometrial adenocarcinoma in vitro. Am J Obstet Gynecol 1972; 114:1012–9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.