Abstract
Background
Advanced liver disease due to hepatitis C virus (HCV) is a leading cause of human immunodeficiency virus (HIV)-related morbidity and mortality. There remains a need to develop noninvasive predictors of clinical outcomes in persons with HIV/HCV coinfection.
Methods
We conducted a nested case-control study in 126 patients with HIV/HCV and utilized multiple quantitative metabolomic assays to identify a prognostic profile that predicts end-stage liver disease (ESLD) events including ascites, hepatic encephalopathy, hepatocellular carcinoma, esophageal variceal bleed, and spontaneous bacterial peritonitis. Each analyte class was included in predictive modeling, and area under the receiver operator characteristic curves (AUC) and accuracy were determined.
Results
The baseline model including demographic and clinical data had an AUC of 0.79. Three models (baseline plus amino acids, lipid metabolites, or all combined metabolites) had very good accuracy (AUC, 0.84–0.89) in differentiating patients at risk of developing an ESLD complication up to 2 years in advance. The all combined metabolites model had sensitivity 0.70, specificity 0.85, positive likelihood ratio 4.78, and negative likelihood ratio 0.35.
Conclusions
We report that quantification of a novel set of metabolites may allow earlier identification of patients with HIV/HCV who have the greatest risk of developing ESLD clinical events.
Keywords: biomarker, end-stage liver disease, metabolomic, hepatic decompensation, accuracy
Advanced liver disease is a leading cause of human immunodeficiency virus (HIV)-related morbidity and mortality, accounting for 13% of all deaths in the large, prospective, multinational D:A:D (data collection on adverse events of anti-HIV drugs) observational cohort [1, 2]. In the United States, chronic hepatitis C virus (HCV) infection is the leading cause of liver disease and related mortality in HIV-infected individuals and has a synergistic effect on liver disease pathogenesis [1]. In a large, well-designed study in the Veterans Health Administration, compared with HCV monoinfected patients, HIV/HCV coinfected patients had a higher rate of hepatic decompensation (hazard ratio, 1.56; confidence interval, 1.31–1.86), and this risk occurred despite maintaining excellent HIV control [3]. The recent introduction of direct acting antiviral (DAA) therapies is expected to improve liver-related outcomes for all persons living with HCV infection, including those with HIV infection [4]. However, recent studies suggest that HIV-infected patients may continue to suffer adverse consequences of liver disease, with the majority experiencing events after HCV eradication if they have moderate (Metavir F2) fibrosis or greater at the time of treatment [5].
There remains a need to develop noninvasive means of assessment of liver disease severity and progression in persons living with HIV. The currently available noninvasive assessments for liver fibrosis are based on serum markers or imaging elastography, but have poor diagnostic performance characteristics for disease progression, and have not been validated for predicting end-stage liver disease (ESLD)-related outcomes in patients with HIV/HCV coinfection [6, 7]. Although these noninvasive tools are frequently used in clinical practice for all chronic liver diseases, they were not designed to provide insight into specific disease pathogenesis, such as HIV-HCV coinfection [8]. This is of particular importance in patients with HIV infection due to the multiple etiologies of liver injury, including viral hepatitis coinfections, direct and indirect HIV effects, and antiretroviral-related toxicity.
Lipids play multiple important roles in biological systems and have a vast repertoire in pathogenesis of human diseases, including cancer, atherosclerosis, and insulin resistance [9]. Multiple bioactive lipids and metabolites have been implicated in liver disease pathogenesis, including sphingolipids [10–15] and oxidized polyunsaturated fatty acids (PUFAs) [16–20]. Sphingolipids, in particular ceramides, are reported to modulate the hepatocellular susceptibility to stress and death ligands-induced cell death via the tumor necrosis factor (TNF) signaling pathway [13, 21, 22]. Oxylipins are some of the key metabolites of PUFA oxidation, and they are important effectors of inflammation, coagulation, and vascular regulation. In particular, cytochrome P450-dependent arachidonic acid metabolites have been targeted for study in cirrhosis and portal hypertension because the liver has the highest total cytochrome P450 content and contains the highest levels of individual cytochrome enzymes involved in the metabolism of PUFAs [17, 18]. In animal models of cirrhosis, metabolites of both the ω and ω-1 hydrolase and epoxygenases, which include the oxylipins hydroxyeicosatetraenoic acids and epoxyeicosatrienoic acids, are associated with increased portal resistance [17, 18]. In addition, epoxyeicosatrienoic acids have been reported to contribute to liver regeneration [23] and are hypothesized to contribute to regenerative nodules in patients with cirrhosis.
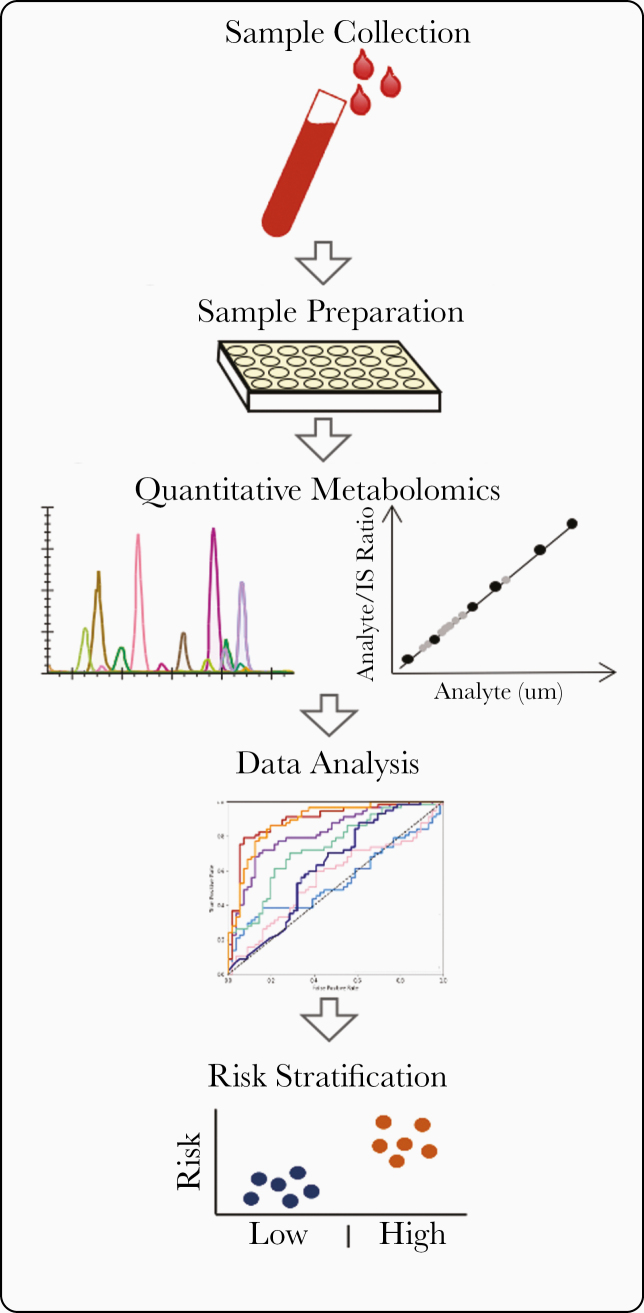
The aim of the present study was to utilize a series of targeted metabolomic assays in a broad-spectrum discovery approach to identify a metabolic profile that predicts ESLD events in a well characterized cohort of persons with HIV/HCV coinfection (Figure 1). To accomplish this, we utilized a long-standing research biorepository of patients with HIV/HCV coinfection to (1) identify samples preceding ESLD events and (2) develop a case-control study to assess differences in metabolite expression between patients who develop incident ESLD events and those who do not experience ESLD-related complications, and (3) assess the impact of adding metabolomic biomarkers to predictive clinical models of incident ESLD to improve prognostic performance characteristics.
Figure 1.
Schematic of approach to comprehensive metabolomic profiling to improve risk stratification of patients with liver disease.
METHODS
This is a retrospective, nested case-control study in which adult patients (aged ≥ 18 years) with HIV and HCV coinfection were identified from the Duke HIV Research Database and Biorepository, which has >60 000 longitudinal plasma samples stored at −80°C from patients engaged in HIV care at the Duke Infectious Diseases clinic since 1998. All subjects provided informed consent for use of their personal health information and plasma samples for future research purposes. The research project was approved by the Duke Institutional Review Board.
A potential case was defined as any patient with HIV/HCV coinfection and with a plasma sample available in the biorepository 1–5 years prior to the date of development of an incident ESLD event. Exclusion criteria included: a prior history of ESLD (decompensation) event, autoimmune disease, recent (past 12 months) HIV or HCV infection, solid organ transplantation, active malignancy, chronic hepatitis B virus infection (positive HBsAg), failed HCV treatment within 6 months of index sample collection or successful HCV treatment at any time, other chronic liver disease, use of systemic immunosuppressants, and/or daily use of a nonsteroidal anti-inflammatory drug (except low-dose aspirin). Incident ESLD events were defined as the first diagnosis or presentation of ascites, hepatic encephalopathy, esophageal variceal bleed, spontaneous bacterial peritonitis, or hepatocellular carcinoma (HCC). Cases were identified through DEDUCE, a web-based clinical research query tool that allows for searching the Duke Health data set, which currently covers over 3.4 million patients, spanning 37 years of care. A total of 565 patients with HIV/HCV coinfection were queried for ICD9/10 codes (Supplementary Table 1) for ESLD and the date of the first encounter with an ESLD ICD9/10 code. Encounters were from outpatient, emergency department, or inpatient visits. When an encounter included multiple ESLD diagnoses, the primary diagnosis was used as the defining incident event. All case charts were manually reviewed by a single individual (M. M.) to confirm the ESLD event was the first presentation. When needed, cases were discussed and reviewed independently by 2 physicians (S. N. and K. P.). Controls were matched 1:1 to cases by age (within 5 years of age), sex, race/ethnicity, and HIV RNA (suppression defined as <200 copies/mL versus viremia) at the time of the sample collection. Controls were defined as patients with HIV/HCV coinfection but without ESLD events and the same exclusion criteria were applied. Control charts were manually reviewed by M. M. to confirm there was no history of ESLD events. Subjects did not have to have cirrhosis for inclusion. Potential predictive variables including: body mass index, tobacco use, alcohol use, antiretroviral medications, comorbidities including diabetes mellitus, use of cholesterol-lowering medications, and routine laboratory values (aminotransferases, bilirubin, albumin, creatinine, HIV viral load, CD4, and platelet count) were extracted from the research database. Alcohol use was characterized as none, mild (occasional or social drinking), moderate (3–7 weekly standard drinks for women and 5–14 for men), or heavy (≥8 weekly drinks for women and ≥15 for men). The Centers for Disease Control and Prevention definition of 1 standard drink = 14 g of alcohol was used for quantification of intake. A plasma sample was identified for cases 1–5 years prior to the date of the incident ESLD event and for controls, a plasma sample was identified for the year of the age used for matching. When more than 1 plasma sample was available for a case during the 1–5 year period, choice of sample was determined by availability of a paired control, prioritizing samples 2–3 years in advance of the ESLD event and in the order 2, 3, 4, 5, and then 1 year prior to ESLD event.
Targeted Metabolomics Analysis
The Duke Proteomics and Metabolomics Shared Resource performed metabolite quantification with 4 separate mass spectrometry-based methods quantifying >500 metabolites in plasma including amino acids, amines, lipids, oxylipins, free fatty acids, and bile acids. AbsoluteIDQ p400HR and Bile Acids kits (Biocrates AG) were used for the majority of these analyte classes, while custom targeted assays were developed for oxylipins (n = 109) and free fatty acids (n = 17). AbsoluteIDQ p400HR targets over 400 metabolites from 11 analyte groups: amino acids, biogenic amines, acylcarnitines, monosaccharides (hexose), diglycerides, triglycerides, lysophposphatidylcholines, phosphatidylcholines, sphingomyelins, ceramides, and cholesteryl esters. The Biocrates Bile Acids assay quantifies 20 bile acids, 16 of which are normally detected in human plasma. Full method details as well as analytes targeted in each of these panels are in the Supplementary Material. Samples for cases and controls were deidentified and randomly distributed on the plates and the laboratory was blinded to case-control status and sample position on the plates.
Statistical Analysis
Our initial sample size calculation was based on pilot data of oxylipin profiling in patients with HIV/HCV coinfection. Based on our cohort of 565 patients, we originally assumed we would have at least 90 patients meeting case definition, in which case we estimated we would be able to detect effect sizes as small as 0.42 with 80% power and type I error rate 0.05. Our final case number was lower than expected (n = 62); however, with top effect sizes of 0.96, 1.02, and 1.35 in the pilot study we were still sufficiently powered to identify multiple lipid metabolites associated with ESLD complications.
Routine laboratory values were log2 transformed and means were imputed for missing values. Values were standardized by subtracting the mean and dividing by the standard deviation. Analyte expression levels were log2 transformed and lower limit of detection (LOD) values were imputed for missing and <LOD values. LASSO regression was used to predict scores for each patient. In the absence of an independent validation dataset, predictive accuracy was assessed by c-statistic using leave-one-out cross-validation to control for overfitting [24]. Multiple models were built to predict the composite of all incident ESLD events. First a clinical baseline model was built using demographic (age, sex, and race) and clinical variables (FIB-4 index, bilirubin, albumin, creatinine, HIV viral load, CD4, alcohol use, diabetes, cholesterol medications, aspirin, and antiretroviral medication) from the time of sample collection. Additional models were built from the baseline model for each metabolite panel and an overall model built of baseline plus all metabolite variables. Area under the receiver operator characteristic curves (AUCs) were created and compared for each model. Optimal performance characteristics for each model are also reported as sensitivity, specificity, and positive and negative likelihood ratios. We used the Youden index to determine the optimal performance characteristics for the models. The Youden index is the sum of sensitivity and specificity (technically, Youden = sensitivity + specificity − 1) at a given threshold, and the optimal threshold it determined by the largest value of this sum, which effectively maximizes both sensitivity and specificity (while giving both an equal weight).
Top predictors were also sought to identify the most predictive permutations of combinations of 3 or 4 analytes. Only analytes with individual AUC ≥ 0.65 were included. Exploratory models were also built for the most common individual ESLD, that is ascites and hepatic encephalopathy.
RESULTS
Patient Characteristics
Overall, 124 patients with HIV/HCV coinfection were included in this case-control study (62 cases and 62 controls). All patients had compensated liver disease at baseline. Overall, the cohort was 37% female, 77% black race, and median age was 49 years (SD 7.8) at the time of the sample collection (Table 1). Incident liver events incurred by cases during this study period included ascites (50%), bleeding esophageal varices (3.2%), hepatic encephalopathy (33.9%), hepatocellular carcinoma (11.3%), and spontaneous bacterial peritonitis (1.6%). Samples used for the metabolomics profiling preceded the incident liver event by a mean of 2 years (SD 1.1). Cases with ESLD events were numerically more likely to report alcohol (40.3% vs 33.9%; P = .57) and tobacco use (71% vs 56.5%; P = .13), and were more likely to have a diagnosis of diabetes (14.5% vs 8.1%; P = .40) (Table 1). Cases were more likely to have an FIB-4 index consistent with severe fibrosis (>3.25) than controls (61% vs 23%). The median model for end-stage liver disease (MELD) score at baseline for cases was 9.9 (interquartile range [IQR], 5.7–16.9) and for controls 6.3 (IQR, 2.9–8.0) (P = <.01). Overall, 86% of patients were on antiretrovirals and 53% had HIV RNA <200 copies/mL at the time of sample collection. The median CD4 count was 358 cells/mm3.
Table 1.
Baseline Demographics and Clinical Characteristics of Study Population
| Characteristics | Cases (n = 62) | Controls (n = 62) | Overall (n = 124) |
|---|---|---|---|
| Female, No. (%) | 23 (37.1) | 23 (37.1) | 46 (37.1) |
| Race, No. (%) | |||
| Black | 48 (77.4) | 47 (75.8) | 95 (76.6) |
| White | 11 (17.7) | 12 (19.4) | 23 (18.5) |
| Other | 3 (4.8) | 3 (4.8) | 6 (4.8) |
| Ethnicity | |||
| Hispanic or Latino, No. (%) | 2 (3.2) | 2 (3.2) | 4 (3.2) |
| Age at sample, y, median (Q1, Q3) | 48 (45, 54) | 48 (44, 53) | 48 (44, 54) |
| Height, cm, median (Q1, Q3) | 170 (165, 178) | 170 (165, 177) | 170 (165, 178) |
| Weight, kg, median (Q1, Q3) | 75 (64, 91) | 78 (67, 88) | 76 (65, 88) |
| BMI, m2/kg, median (Q1, Q3) | 26 (22, 29) | 26 (23, 30) | 26 (22, 29) |
| Comorbidities, No. (%) | |||
| Diabetic | 9 (14.5) | 5 (8.1) | 14 (11.3) |
| Alcohol | 25 (40.3) | 21 (33.9) | 46 (37.1) |
| Tobacco | 44 (71.0) | 35 (56.5) | 79 (63.7) |
| Liver event, No. (%) | |||
| Ascites | 31 (50.0) | 0 (0.0) | 31 (25.0) |
| Esophageal varices | 2 (3.2) | 0 (0.0) | 2 (1.6) |
| Hepatic encephalopathy | 21 (33.9) | 0 (0.0) | 21 (16.9) |
| Hepatocellular carcinoma | 7 (11.3) | 0 (0.0) | 7 (5.6) |
| Spontaneous bacterial peritonitis | 1 (1.6) | 0 (0.0) | 1 (0.8) |
| None | 0 (0.0) | 62 (100.0) | 62 (50.0) |
| Laboratory results | |||
| HIV RNA < 200 copies/mL, No. (%) | 31 (50.0) | 35 (56.5) | 66 (53.2) |
| CD4, cells/µL, median (Q1, Q3) | 344 (196, 513) | 375 (228, 680) | 358 (200, 589) |
| Albumin, g/dL, median (Q1, Q3) | 3.2 (2.7, 3.7) | 3.8 (3.3, 4.0) | 3.4 (3.0, 3.9) |
| Bilirubin, mg/dL, median (Q1, Q3) | 0.8 (0.6, 1.5) | 0.6 (0.5, 1.1) | 0.7 (0.5, 1.3) |
| Creatinine, mg/dL, median (Q1, Q3) | 1.0 (0.8, 1.7) | 0.9 (0.8, 1.1) | 1.0 (0.8, 1.2) |
| INR, median (Q1, Q3) | 1.1 (1.0, 1.3) | 1.1 (1.0, 1.1) | 1.1 (1.0, 1.3) |
| MELD, median (Q1, Q3) | 9.9 (5.7, 16.9) | 6.3 (2.9, 8.0) | 9.4 (5.2, 16.6) |
| AST, U/L baseline, median (Q1, Q3) | 78.5 (50.3, 118.0) | 53 (36.5, 73.8) | 65.5 (42.0, 93.3) |
| ALT, U/L baseline, median (Q1, Q3) | 47.0 (27.3, 68.8) | 49.0 (33.3, 63.8) | 48.5 (31.0, 65.3) |
| Platelets, 109/L, baseline, median (Q1, Q3) | 145 (102, 204) | 199 (148, 247) | 183 (112, 225) |
| FIB-4, baseline, median (Q1, Q3) | 4.0 (2.2, 7.3) | 1.9 (1.3, 3.1) | 2.6 (1.6, 5.4) |
| FIB-4 > 1.45, No. (%) | 57 (91.9) | 43 (69.4) | 100 (80.6) |
| FIB-4 > 3.25, No. (%) | 38 (61.3) | 14 (22.6) | 52 (42.0) |
| Medications, No. (%) | |||
| Aspirin | 9 (14.5) | 13 (21.0) | 22 (17.7) |
| Cholesterol | 3 (4.8) | 3 (4.8) | 6 (4.8) |
| Entry inhibitors | 1 (1.6) | 1 (1.6) | 2 (1.6) |
| Integrase inhibitors | 4 (6.5) | 9 (14.5) | 13 (10.5) |
| NNRTI | 14 (22.6) | 10 (16.1) | 24 (19.4) |
| NRTI | 56 (90.3) | 62 (100.0) | 118 (95.2) |
| PI | 32 (51.6) | 28 (45.2) | 60 (48.4) |
Abbreviations: ALT, alanine transaminase; AST, aspartate transaminase; BMI, body mass index; HIV, human immunodeficiency virus; INR, international normalized ratio; MELD, model for end-stage liver disease; NNRTI, nonnucleoside reverse transcriptase inhibitor; NRTI, nucleoside reverse transcriptase inhibitor; PI, protease inhibitor.
Lipid Metabolite Models for All Clinical Outcomes
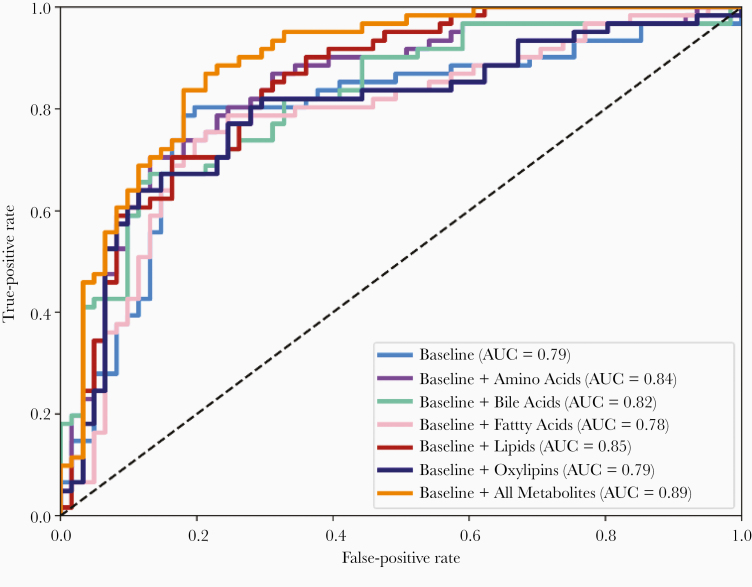
The clinical variables retained in the baseline model included: body mass index, laboratory values at the time of sample collection (albumin, total bilirubin, creatinine, international normalized ratio [INR], and CD4 absolute count), FIB-4 index, and tobacco use (AUC = 0.79). Metabolites were then added to the baseline model with the following results: baseline + amino acids (AUC = 0.84), baseline + bile acids (AUC = 0.82), baseline + fatty acids (AUC = 0.78), baseline + lipids (AUC = 0.85), baseline + oxylipins (AUC = 0.79), and baseline + all metabolites (AUC = 0.89) (Figure 2). The top predictive variables/metabolites for each model are presented in Table 2. Three models had AUC > 0.84 and the highest accuracy was from the all-metabolites model. The all-metabolites model included top predictive metabolites from amino acids, bile acids, and lipids including valine (a branched-chain amino acid), deoxycholic acid (DCA) and taurochenodeoxycholic acid, and multiple lipids including lysophosphatidylcholine (LPC), phosphatidylcholine (PC), and sphingomyelin. The all-metabolites model had a sensitivity of 0.70, specificity of 0.85, positive likelihood ratio of 4.78, and negative likelihood ratio of 0.35. The amino acid model had the best test characteristics with a sensitivity of 0.70, specificity of 0.86, positive likelihood ratio of 5.38, and negative likelihood ratio of 0.34. Test characteristics for all models are provided in Table 3.
Figure 2.
Receiver operating characteristic curves for predictive models of composite end-stage liver disease events. Light blue, baseline clinical variables model; purple, baseline + amino acid metabolites model; green, baseline + bile acid metabolites model; pink, baseline + free fatty acid metabolites model; red, baseline + lipid metabolites model; dark blue, baseline + oxylipin metabolites model; orange, baseline + all-metabolites model. Abbreviation: AUC, area under the curve.
Table 2.
Composite End-Stage Liver Disease Models Variables
| Baseline | Baseline + Amino Acids | Baseline + Bile Acids | Baseline + Fatty Acids | Baseline + Lipids | Baseline + Oxylipins | Baseline + All Metabolites |
|---|---|---|---|---|---|---|
| (AUC = 0.79) | (AUC = 0.84) | (AUC = 0.82) | (AUC = 0.78) | (AUC = 0.85) | (AUC = 0.79) | (AUC = 0.89) |
| INR | INR | INR | INR | ALBUMIN | INR | CREAT |
| ALBUMIN | CREAT | ALBUMIN | ALBUMIN | CREAT | ALBUMIN | DCA |
| CREAT | FIB-4 | CREAT | CREAT | FIB-4 | CREAT | TCDCA |
| CD4 | Arg | CD4 | CD4 | AC(3:0) | FIB-4 | 12S-HHTrE |
| FIB-4 | His | FIB-4 | FIB-4 | AC(14:2-OH) | 12S-HHTrE | AC(3:0) |
| Phe | CDCA | FA 16:1 (palmitoleic) | LPC(15:0) | 5(S)-HEPE | LPC(17:0) | |
| Val | DCA | FA 20:4 omega-6 (AA) | LPC(17:0) | 14-HDoHE | LPC-O(16:1) | |
| Met-SO | TCDCA | FA 22:5 omega-6 (Osbond) | LPC(20:4) | PC(42:7) | ||
| TUDCA | LPC-O(16:1) | PC-O(36:1) | ||||
| PC(32:0) | PC-O(40:8) | |||||
| PC(42:7) | SM(35:1) | |||||
| PC-O(36:1) | DG(39:0) | |||||
| PC-O(40:8) | His | |||||
| SM(35:1) | Val | |||||
| SM(36:2) | Met-SO | |||||
| CE(22:5) | SDMA | |||||
| DG(39:0) |
Abbreviations: AC, acylcarnitine; AUC, area under curve; CDCA, chenodesoxycholic acid; CE, cholesteryl ester; CREAT, creatinine; DCA, deoxycholic acid; DG, diacylglycerol; FA, fatty acid; LPC, lysophosphatidylcholine; HDoHE, hydroxy-docosahexaenoic acid; HEPE, hydroxyeicosapentaenoic acid; HHTrE, hydroxyheptadecatrenoic acid; INR, international normalized ratio; Met-SO, methionine sulfoxide; PC, phosphatidylcholine; SDMA, symmetric dimethylarginine; SM, sphingomyelin; TCDCA, taurochenodeoxycholic acid.
Table 3.
Classification Performance Metrics for Composite End-Stage Liver Disease Models
| Model | AUC | Sensitivity, % | Specificity, % | Positive Likelihood Ratio | Negative Likelihood Ratio |
|---|---|---|---|---|---|
| Baseline | 0.79 | 57 | 85 | 3.89 | 0.50 |
| Baseline + amino acids | 0.84 | 70 | 86 | 5.38 | 0.34 |
| Baseline + bile acids | 0.82 | 67 | 80 | 3.42 | 0.41 |
| Baseline + fatty acids | 0.78 | 68 | 81 | 3.82 | 0.38 |
| Baseline + lipids | 0.85 | 70 | 81 | 3.91 | 0.36 |
| Baseline + oxylipins | 0.79 | 67 | 83 | 4.10 | 0.39 |
| Baseline + all metabolites | 0.89 | 70 | 85 | 4.78 | 0.35 |
Abbreviation: AUC, area under the curve.
Lipid Metabolite Models for Individual Liver-Related Events
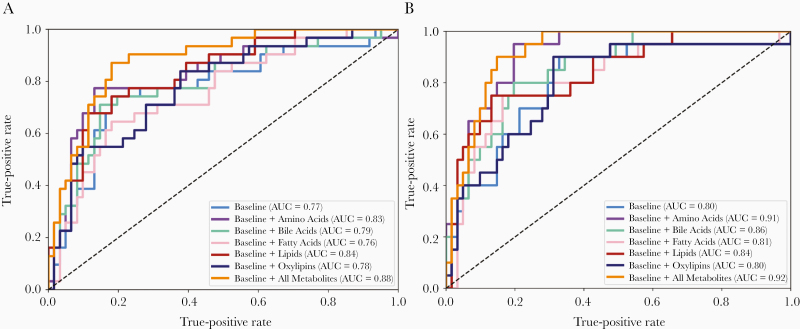
Exploratory models of the most commonly observed individual ESLD events were also built for ascites and hepatic encephalopathy. These event-targeted models resulted in variations in the AUC (Figure 3A and B). For the model of ascites (Figure 3A), the all-metabolites model had an AUC of 0.88 with top metabolites including valine, acylcarnitine, and 5 lipid metabolites (Supplementary Table 2). Eight of these metabolites were also in the composite outcome all-metabolites model. For the model of hepatic encephalopathy (Supplemental Figure 1B), the amino acid model AUC increased to 0.91 and the all-metabolites model AUC increased to 0.92. The top amino acids included arginine, methionine, histamine, valine, and methionine sulfoxide, which were also in the composite outcome amino acid model (Supplementary Table 3).
Figure 3.
Receiver operating characteristic curves for predictive models of (A) ascites and (B) hepatic encephalopathy. Light blue, baseline clinical variables model; purple, baseline + amino acid metabolites model; green, baseline + bile acid metabolites model; pink, baseline + free fatty acid metabolites model; red, baseline + lipid metabolites model; dark blue, baseline + oxylipin metabolites model; orange, baseline + all-metabolites model. Abbreviation: AUC, area under the curve.
Top Predictors of ESLD
With the goal of identifying a limited set of metabolites that optimize the predictive power of the model while increasing the feasibility that the biosignature could be applied in a clinical setting, combinations of 3 or 4 optimal predictors of the composite outcome of incident ESLD events were evaluated. Multiple permutations of metabolites with and without baseline variables were completed. Multiple models were identified with AUC = 0.84, with a predominance of the predictive model AUCs resulting from lipid metabolite only combinations (Table 4).
Table 4.
Optimization Model Performance of Top Predictors of Composite End-Stage Liver Disease
| Top Predictor Metabolites | AUC | Sensitivity, % | Specificity, % | Positive Likelihood Ratio | Negative Likelihood Ratio |
|---|---|---|---|---|---|
| SM(37:1), PC(32:0), LPC(16:0) | 0.84 | 63 | 83 | 3.9 | 0.43 |
| LPC(15:0), PC(32:0), Cer(41:1) | 0.84 | 55 | 85 | 3.78 | 0.52 |
| LPC(15:0), SM(37:1), PC(32:0) | 0.84 | 65 | 88 | 5.71 | 0.39 |
| TCA, PC(32:0), LPC(16:0) | 0.84 | 68 | 80 | 3.5 | 0.39 |
| PC(32:0), LPC-O(16:1), DG(39:0) | 0.84 | 65 | 83 | 4 | 0.41 |
| LPC(20:4), PC(32:0), Val | 0.84 | 62 | 90 | 6.33 | 0.42 |
| LPC(15:0), PC(32:0), Cer(43:1) | 0.84 | 63 | 83 | 3.9 | 0.43 |
| LPC(15:0), PC(32:0), DG(39:0) | 0.84 | 68 | 81 | 3.82 | 0.38 |
| LPC(15:0), PC(32:0), SM(39:1) | 0.84 | 59 | 83 | 3.6 | 0.49 |
| LPC(15:0), TCA, Val | 0.84 | 68 | 83 | 4.2 | 0.37 |
Abbreviations: AUC, area under the curve; Cer, ceramide; DG, diacylglycerol; LPC, lysophosphatidylcholine; PC, phosphatidylcholine; SM, sphingomyelin; TCA, taurocholic acid.
DISCUSSION
As more patients access DAAs and achieve cure of chronic HCV infection, the need for a clinical prediction tool or biomarker to differentiate patients with increased risk of liver-related complications is increasingly important. This is the first study to report that quantification of a novel set of lipid metabolites may allow earlier identification of patients with HIV/HCV coinfection at greatest risk of developing clinically significant ESLD events. Our composite liver-events models accurately identified patients at the greatest risk of hepatic decompensation or HCC over the subsequent 2 years, which could allow for individualizing clinical care pathways for these highest-risk patients, including the potential for closer surveillance and individualized education on risk reduction.
Multiple lipid pathways have been implicated in liver disease pathogenesis including sphingolipids [10–15] and oxidized PUFAs and their metabolites [16–20]. In addition, bile acids and branched chain amino acids have also been investigated in liver disease and metabolic end-organ complications [25–27]. One of the strengths of our study is that we utilized established and validated multitargeted metabolomic discovery platforms to identify a metabolic risk profile that predicts ESLD events, in a well-characterized single-center cohort of persons with HIV/HCV coinfection. Three models, baseline + amino acids, baseline + lipid metabolites, and baseline + all metabolites, had excellent accuracy (AUC 0.84, 0.85, and 0.89, respectively) in differentiating patients with HIV/HCV coinfection who are at risk of developing an ESLD complication. The accuracy of our metabolite models are better than other published predictors of liver-related events, liver biopsy [28], and FIB-4 index [29] in people with HIV/HCV coinfection. The accuracy was similar to that of transient elastography, which is a diagnostic not available in many clinical practices [30].
Due to the number of metabolites remaining in the top performing models we also looked for permutations of limited (3 or 4) metabolite combinations, which improve the feasibility (throughput time and potential cost) of the clinical development of a metabolite biosignature. Multiple permutations, many of which shared common lipid metabolites, LPC(15:0) and PC(32:0), had good accuracy (AUC 0.84) in differentiating patients with HIV/HCV coinfection at risk of ESLD complications.
Earlier identification of patients at the greatest risk of developing ESLD outcomes could allow for a more patient tailored approach to clinical management and education, as well as the potential to limit the cost of unnecessary testing that may otherwise be done in those patients identified as lowest risk. Patients stratified as high risk may benefit from more frequent clinical follow-up, more aggressive education and counseling on other liver injuries such as alcohol and tobacco use, metabolic risk for fatty liver disease, and potentially may benefit from higher-resolution imaging or more frequent imaging for HCC screening. In addition, patients stratified as low risk could be provided reassurance regarding their liver health and surveillance strategies modified based on comorbid risk factors for disease progression.
In addition to improving the prognostic accuracy of ESLD complications in people with HIV/HCV coinfection, discovery of metabolomic predictors of outcomes may also provide insight on the mechanisms of ongoing liver fibrogenesis and/or impaired fibrosis regression pathways resulting in the increased risk. For example, sphingolipids such as ceramides are reported to modulate the hepatocellular susceptibility to stress and death ligands-induced cell death via the TNF signaling pathway [13, 21, 22]. In both our lipid and all-metabolite models, lysophosphatidylcholines, phosphatidylcholines, and sphingomyelins (including ceramides) were highly predictive of clinical events, and have been previously associated with risk of hepatocellular carcinoma [31] and cirrhosis [10]. Valine, a branched chain amino acid, was retained in multiple models, and has been previously identified as a biomarker of obesity-associated conditions such as insulin resistance, type 2 diabetes, and other cardiometabolic diseases such as nonalcoholic fatty liver disease [25]. While total bile acids have been long-standing biomarkers of liver injury and function, the investigation of individual bile acids to help differentiate types of liver injury remains novel, particularly in cohorts with HIV. Although not independently predictive of clinical events, several bile acids were retained in the composite all-metabolite model as well as others, including DCA and taurochenodeoxycholic acid, metabolites of cholic acid and chenodeoxycholic acid, respectively, the 2 primary bile acids secreted by the liver [27]. DCA is an unconjugated form of cholic acid and is hydrophobic. Accumulation of DCA is thought to result in oxidative stress via mitochondrial damage, disruption of cell membranes, and production of reactive oxygen species, leading to apoptosis and necrosis [27].
The primary limitation of our study is the retrospective design that required identification of ESLD events via encounter diagnoses. We attempted to limit this weakness by doing manual chart review of every patient identified as having an ESLD event and reaching a consensus to ensure that this was a true incident event. In addition, due to the retrospective nature of the study, reporting of alcohol and tobacco use was not uniformly or systematically collected, and was thus obtained following extensive manual chart review of medical records. Cases and controls were not matched by stage of liver disease due to a lack of consistent liver disease staging available in the medical records. Without an independent cohort for validation, we relied on the leave-one-out cross-validation resampling approach to reduce overfitting bias and provide generalizable estimates of accuracy. Further validation will be required in additional cohorts including a cohort with reliable staging consistent with cirrhosis. Lastly, we were not able to include data on noninvasive tools such as imaging elastography (magnetic resonance elastography or vibration-controlled transient elastography), as these modalities were not routinely available at our center at the time of baseline sample collection. In spite of these limitations, the work presented here is novel and promising. Our study provides a potential noninvasive biomarker panel that can predict liver disease events up to 2 years in advance, providing an opportunity for individualized care to patients with HIV/HCV infection.
In conclusion, we report that quantification of a novel set of metabolites may allow earlier identification of patients with HIV/HCV coinfection who have the greatest risk of developing clinically significant ESLD events over the next approximately 2 years, including incident ascites, hepatic encephalopathy, hepatocellular carcinoma, esophageal variceal bleed, and spontaneous bacterial peritonitis. Patients stratified as high risk may have ongoing liver injury and fibrogenesis, as suggested by differences in expression of sphingolipids including ceramide, amino acids, and cholic acid pathway bile acid metabolites. Further validation of our findings are needed in cohorts of persons with HIV infection, including an independent cohort of active HCV coinfection and cohorts with lower overall prevalence of events, and importantly, for patients after DAA therapy to determine if this prognostic biosignature persists after HCV eradication.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Notes
Financial support. This work was supported by the National Institute of Diabetes and Digestive and Kidney Diseases (grant number R01DK112295 to S. N.); and Duke Center for AIDS Research (grant number AI064518).
Potential conflicts of interest. S. N. reports research support to her institution from AbbVie, Gilead, Tacere, and Janssen; consulting relationship with BioMarin and Vir for which she has stock options with Vir; and Event Adjudication Committee for PRA and FHI 360. K. P. reports research support to his institution from Gilead; advisory board membership with Gilead and Merck; and consulting relationship with Gilead. All other authors report no potential conflicts. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
Presented in part: American Association for the Study of Liver Diseases Meeting, San Francisco, CA, 9–13 November 2018.
References
- 1. Konerman MA, Mehta SH, Sutcliffe CG, et al. Fibrosis progression in human immunodeficiency virus/hepatitis C virus coinfected adults: prospective analysis of 435 liver biopsy pairs. Hepatology 2014; 59:767–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Smith CJ, Ryom L, Weber R, et al. ; D:A:D Study Group Trends in underlying causes of death in people with HIV from 1999 to 2011 (D:A:D): a multicohort collaboration. Lancet 2014; 384:241–8. [DOI] [PubMed] [Google Scholar]
- 3. Lo Re V 3rd, Kallan MJ, Tate JP, et al. Hepatic decompensation in antiretroviral-treated patients co-infected with HIV and hepatitis C virus compared with hepatitis C virus-monoinfected patients: a cohort study. Ann Intern Med 2014; 160:369–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Naggie S, Cooper C, Saag M, et al. ; ION-4 Investigators Ledipasvir and sofosbuvir for HCV in patients coinfected with HIV-1. N Engl J Med 2015; 373:705–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Zahnd C, Salazar-Vizcaya L, Dufour JF, et al. ; Swiss HIV; Swiss Hepatitis C Cohort Studies Modelling the impact of deferring HCV treatment on liver-related complications in HIV coinfected men who have sex with men. J Hepatol 2016; 65:26–32. [DOI] [PubMed] [Google Scholar]
- 6. Shaheen AA, Myers RP. Systematic review and meta-analysis of the diagnostic accuracy of fibrosis marker panels in patients with HIV/hepatitis C coinfection. HIV Clin Trials 2008; 9:43–51. [DOI] [PubMed] [Google Scholar]
- 7. Vergara S, Macías J, Rivero A, et al. ; Grupo para el Estudio de las Hepatitis Viricas de la SAEI The use of transient elastometry for assessing liver fibrosis in patients with HIV and hepatitis C virus coinfection. Clin Infect Dis 2007; 45:969–74. [DOI] [PubMed] [Google Scholar]
- 8. Peters MG, Bacchetti P, Boylan R, et al. Enhanced liver fibrosis marker as a noninvasive predictor of mortality in HIV/hepatitis C virus-coinfected women from a multicenter study of women with or at risk for HIV. AIDS 2016; 30:723–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kolter T. A view on sphingolipids and disease. Chem Phys Lipids 2011; 164:590–606. [DOI] [PubMed] [Google Scholar]
- 10. Grammatikos G, Ferreiros N, Bon D, et al. Variations in serum sphingolipid levels associate with liver fibrosis progression and poor treatment outcome in hepatitis C virus but not hepatitis B virus infection. Hepatology 2015; 61:812–22. [DOI] [PubMed] [Google Scholar]
- 11. Grammatikos G, Mühle C, Ferreiros N, et al. Serum acid sphingomyelinase is upregulated in chronic hepatitis C infection and nonalcoholic fatty liver disease. Biochim Biophys Acta 2014; 1841:1012–20. [DOI] [PubMed] [Google Scholar]
- 12. Li JF, Qu F, Zheng SJ, et al. Plasma sphingolipids: potential biomarkers for severe hepatic fibrosis in chronic hepatitis C. Mol Med Rep 2015; 12:323–30. [DOI] [PubMed] [Google Scholar]
- 13. Marí M, Colell A, Morales A, et al. Acidic sphingomyelinase downregulates the liver-specific methionine adenosyltransferase 1A, contributing to tumor necrosis factor-induced lethal hepatitis. J Clin Invest 2004; 113:895–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Marí M, Fernández-Checa JC. Sphingolipid signalling and liver diseases. Liver Int 2007; 27:440–50. [DOI] [PubMed] [Google Scholar]
- 15. Moles A, Tarrats N, Morales A, et al. Acidic sphingomyelinase controls hepatic stellate cell activation and in vivo liver fibrogenesis. Am J Pathol 2010; 177:1214–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Loomba R, Quehenberger O, Armando A, Dennis EA. Polyunsaturated fatty acid metabolites as novel lipidomic biomarkers for noninvasive diagnosis of nonalcoholic steatohepatitis. J Lipid Res 2015; 56:185–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Sacerdoti D, Gatta A, McGiff JC. Role of cytochrome P450-dependent arachidonic acid metabolites in liver physiology and pathophysiology. Prostaglandins Other Lipid Mediat 2003; 72:51–71. [DOI] [PubMed] [Google Scholar]
- 18. Sacerdoti D, Jiang H, Gaiani S, McGiff JC, Gatta A, Bolognesi M. 11,12-EET increases porto-sinusoidal resistance and may play a role in endothelial dysfunction of portal hypertension. Prostaglandins Other Lipid Mediat 2011; 96:72–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Feldstein AE, Werneburg NW, Canbay A, et al. Free fatty acids promote hepatic lipotoxicity by stimulating TNF-alpha expression via a lysosomal pathway. Hepatology 2004; 40:185–94. [DOI] [PubMed] [Google Scholar]
- 20. Puri P, Baillie RA, Wiest MM, et al. A lipidomic analysis of nonalcoholic fatty liver disease. Hepatology 2007; 46:1081–90. [DOI] [PubMed] [Google Scholar]
- 21. García-Ruiz C, Colell A, Marí M, et al. Defective TNF-alpha-mediated hepatocellular apoptosis and liver damage in acidic sphingomyelinase knockout mice. J Clin Invest 2003; 111:197–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Pagadala M, Kasumov T, McCullough AJ, Zein NN, Kirwan JP. Role of ceramides in nonalcoholic fatty liver disease. Trends Endocrinol Metab 2012; 23:365–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sacerdoti D, Pesce P, Di Pascoli M, Brocco S, Cecchetto L, Bolognesi M. Arachidonic acid metabolites and endothelial dysfunction of portal hypertension. Prostaglandins Other Lipid Mediat 2015; 120:80–90. [DOI] [PubMed] [Google Scholar]
- 24. Molinaro AM, Simon R, Pfeiffer RM. Prediction error estimation: a comparison of resampling methods. Bioinformatics 2005; 21:3301–7. [DOI] [PubMed] [Google Scholar]
- 25. White PJ, Newgard CB. Branched-chain amino acids in disease. Science 2019; 363:582–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Luo L, Aubrecht J, Li D, et al. Assessment of serum bile acid profiles as biomarkers of liver injury and liver disease in humans. PLoS One 2018; 13:e0193824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Chiang JYL. Bile acid metabolism and signaling in liver disease and therapy. Liver Res 2017; 1:3–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Berenguer J, Zamora FX, Aldámiz-Echevarría T, et al. ; Grupo de Estudio del SIDA (GESIDA) HIV/HCV Cohort Study Group Comparison of the prognostic value of liver biopsy and FIB-4 index in patients coinfected with HIV and hepatitis C virus. Clin Infect Dis 2015; 60:950–8. [DOI] [PubMed] [Google Scholar]
- 29. Lo Re V 3rd, Kallan MJ, Tate JP, et al. Predicting risk of end-stage liver disease in antiretroviral-treated human immunodeficiency virus/hepatitis C virus-coinfected patients. Open Forum Infect Dis 2015; 2:ofv109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Pérez-Latorre L, Sánchez-Conde M, Rincón D, et al. Prediction of liver complications in patients with hepatitis C virus-related cirrhosis with and without HIV coinfection: comparison of hepatic venous pressure gradient and transient elastography. Clin Infect Dis 2014; 58:713–8. [DOI] [PubMed] [Google Scholar]
- 31. Cotte AK, Cottet V, Aires V, et al. Phospholipid profiles and hepatocellular carcinoma risk and prognosis in cirrhotic patients. Oncotarget 2019; 10:2161–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.