Abstract
Objective
Colon cancer (CC) is the third most common cancer with a high rate of incidence and mortality. Therefore, it is highly necessary to explore novel targets of CC.
Methods
The miRNA-seq and RNA-seq data of CC were accessed from the TCGA database. Differential analysis was performed using the “edgeR” package to identify differentially expressed miRNAs (DE_miRNAs). The downstream target genes of the target miRNA were then predicted by miRNA target prediction databases to identify the target mRNA. Normal colon cell line CCD-18Co and CC cell lines HCT-116, HT-29, SW620 and SW480 were chosen, and qRT-PCR was conducted to detect miR-141 expression in these cell lines. qRT-PCR and Western blot were carried out to determine PHLPP2 mRNA and protein expression, respectively. Dual-luciferase reporter gene assay was performed to verify the targeting relationship between miR-141 and PHLPP2 3ʹUTR. CCK-8 assay and colony formation assay were carried out to detect cell proliferation. Meanwhile, tumor xenograft model in nude mice was constructed to assess CC cell tumorigenic ability in vivo.
Results
miR-141 was markedly up-regulated in CC tissue. CC cell proliferation and in vivo tumorigenic ability were suppressed by miR-141 silencing but promoted by miR-141 over-expression. PHLPP2 was significantly down-regulated in cancer tissue. Dual-luciferase reporter gene assay indicated that miR-141 could bind to PHLPP2 3ʹUTR. PHLPP2 expression was noticeably elevated upon miR-141 deficiency but significantly inhibited upon miR-141 over-expression. CCK-8 and colony formation assay suggested that miR-141 facilitated CC cell proliferation by silencing PHLPP2.
Conclusion
miR-141 promotes CC cell proliferation by targeted silencing PHLPP2.
Keywords: miR-141, PHLPP2, colon cancer, proliferation
Introduction
Colon cancer (CC) is one of the major causes leading to tumor-related deaths and its incidence rate is rising in many countries.1 According to Surveillance, Epidemiology, and End Results Program of the USA, it was estimated that CC made up 8.3% of the total cancer cases and 8.4% of the total cancer-related deaths in 2019 (Available at: https://seer.cancer.gov/statfacts/html/colorect.html; cited 5 July 2019). Although the overall rate of incidence and mortality has been reduced by the progress made in screening and treatment methods, CC remains a burden and it is predicted that CC patients will increase by 60% worldwide by 2030.2 As a result, it is highly necessary to find novel molecular markers.
MicroRNAs (miRNAs) are evolutionarily conservative endogenous non-coding RNA molecules about 22 nucleotides in length3 that regulate gene expression by binding to mRNAs at the 3ʹ-untranslated region (3ʹUTR).4 Research suggests that miRNAs play a crucial role in various biological processes, such as cell proliferation,5 differentiation,6 invasion,7 apoptosis8 and metabolism.4 Besides, another evidence demonstrates that dysregulation of miRNAs is related to the occurrence and development of various human cancers.9 For example, miR-101-5p has ectopic expression in breast cancer and can weaken the aggressive phenotypes of BrCa cells, such as proliferation, migration and invasion.10 While miR-302a is poorly expressed in colorectal cancer (CRC) and suppresses CRC metastasis and cetuximab resistance.11 miR-141 is a member of miR-200 family reported to be differentially expressed in multiple human malignant tumors.12 For instance, miR-141 is up-regulated in ovarian cancer13 and down-regulated in hepatocellular carcinoma (HCC) and prostate cancer,14,15 but the specific regulatory mechanism of miR-141 in various cancers remains unclear.
PHLPP2 (PH domain leucine-rich-repeats protein phosphatase 2) is able to regulate the activity of AGC kinase including Akt.4. It is indispensable to keep PHLPP2 expression in balance for pathological prevention as the change of PHLPP2 steady-state level is connected with various diseases, including diabetes, hepatic steatosis and cancers.16,17 Recently, many studies have unveiled that PHLPP2 is ubiquitously lost in various cancers and plays an important role in multiple biological processes, such as cancer cell proliferation, metastasis, autophagy and apoptosis.18–23 Nonetheless, the diagnostic and prognostic significance of PHLPP2 in CC have not been fully researched.
In this study, we investigated miR-141 expression in CC cells and validated its effect on CC cell proliferation and tumorigenic ability. Additionally, we also studied the regulatory mechanism of miR-141 and PHLPP2 in CC cells. Our study will shed light on the mechanism of CC tumorigenesis and verify that miR-141 is a potential therapeutic target for CC treatment.
Materials and Methods
Bioinformatics Analysis
The miRNA-seq and RNA-seq data of CC were accessed from the TCGA-COAD dataset (https://portal.gdc.cancer.gov/) and subjected to differential analysis using the “edgeR” package. |logFC|>2 and p value<0.05 were set as the threshold to screen out differentially expressed genes (DEGs). TargetScan (http://www.targetscan.org/vert_72/), miRDB (http://mirdb.org/) and miRTarBase (http://mirtarbase.mbc.nctu.edu.tw/php/index.php) databases were employed for target prediction to validate the candidate mRNAs, which were then intersected with the differentially expressed mRNAs (DE_mRNAs) in the TCGA-COAD dataset. Subsequently, the mRNA which showed a marked effect on patient’s prognosis was chosen.
Cell Culture
Normal colon cell line CCD-18Co (ATCC® CRL-1459™) and CC cell lines HCT-116 (ATCC® CCL-247), HT-29 (ATCC® HTB-38), SW620 (ATCC® CCL-227™) and SW480 (ATCC® CCL-228™) were all purchased from the American Type Culture Collection (ATCC; Rockville, MD, USA). All the cells were kept in a humidified atmosphere of 5% CO2 at 37°C, among which CCD-18Co cell line was cultured in Eagle’s Minimum Essential Medium (EMEM; Catalog No. 30–2003) containing 10% fetal bovine serum (FBS; Gibco, Grand Island, NY, USA), HCT-116 and HT-29 cell lines were grown in McCoy’s 5a Medium Modified (Catalog No. 30–2007) containing 10% FBS, while SW620 and SW480 cell lines were maintained in Leibovitz’s L-15 Medium (Catalog No. 30–2008) containing 10% FBS. The mediums were replaced every two or three days depending on cell growth until cells were cultured to be 80–90% confluent. Thereafter, cells were used for subculture and then harvested when reaching the logarithmic phase.
Cell Transfection
Inhibitor NC, miR-141 inhibitor, mimic NC, miR-141 mimic, oe-NC and oe-PHLPP2 ordered from GenePharma (Shanghai, China) were transfected into cells (1×106 cells per well in a 6-well plate) by Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific, Inc.) following the manufacturer’s protocol. After transfection, cells were cultured for additional 6 h at 37°C and 5% CO2, after which the mediums were changed. All the cells were collected after 48 h of transfection.
Real-Time Quantitative PCR (qRT-PCR)
Total RNA was extracted from CC tissue and cells using Trizol reagent (16,096,020; Thermo Fisher Scientific, NY, USA) according to the manufacturer’s instructions. Then, 5 µg RNA was transcribed into complementary DNA (cDNA) by using the cDNA Reverse Transcription Kit (K1622; Fermentas Inc., Ontario, CA, USA). qRT-PCR was performed with the TaqMan MicroRNA Assay and TaqMan® Universal PCR Master Mix under the following thermocycler conditions: 95°C for 2 min, and 45 cycles of 95°C for 15 s and 60°C for 45 s, followed by 72°C for 45 s. Each measurement was normalized to U6 to ensure comparable amounts of miRNA in all wells.
According to the TaqMan Gene Expression Assays protocol (Applied Biosystems, Foster City, CA, USA), qRT-PCR was conducted to assess mRNA under the following thermocycler conditions: 95°C for 10 min, and 35 cycles of 95°C for 15 s and 60°C for 30 s, followed by 72°C for 45 s. GAPDH was used as an internal reference. Three repeated wells were set for each treatment. The primers were ordered from TAKARA (Beijing, China) and listed in Table 1. The quantitative value was expressed using the 2-ΔΔCt method: ΔΔCt=ΔCt (experiment group) -ΔCt (control group), where ΔCt = Ct (target gene) -Ct (internal reference).24
Table 1.
Primers for qRT-PCR
| Genes | Forward (5ʹ→3ʹ) | Reverse (5ʹ→3ʹ) |
|---|---|---|
| miR-141 | GTCCATCTCCATCAGTACAGGTTG | AGCCATCTTACTCACAGAGTGTG |
| U6 | GCTTGCTTCACACACACATA | AAAAAACATCGACTCADG |
| PHLPP2 | AGGTTCCTGAGCATCTCTTC | GTTCAGGCCCTTCAGTTGAG |
| GAPDH | TGCACCACCAACTGCTTAGC | GGCATGCACTGTGGTCATGAG |
Western Blot
Total proteins were extracted from cells after transfection and the concentration of proteins was measured by bicinchoninic acid (BCA) protein assay kit (Thermo Fisher Scientific, USA). Protein samples were separated on 10% sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred onto nitrocellulose membranes (ZY-160FP, SHZYSW, Shanghai, China). After being blocked with 5% non-fat milk at room temperature for 1 h, the membranes were oscillated in 3% BSA in an oscillator for 1 h. The membranes were washed with Tris-buffered saline with Tween (TBST; Solarbio, Beijing, China) in triplicate with 10 min each time and then incubated with diluted primary rabbit polyclonal antibodies PHLPP2 (ab71973, 1:2000, abcam, UK) and GAPDH (ab9485, 1:2500, abcam, UK) overnight at 4°C. The membranes were washed with TBST in triplicate with 10 min each time. Subsequently, goat anti-rabbit immunoglobulin G (IgG) H&L (Alexa Fluor® 790) secondary antibody (ab205718, 1:5000, abcam, UK) was added onto the membranes and incubated for 1 h. The membranes were washed with TBST in triplicate with 10 min each time, followed by development with electrochemiluminescence (ECL) reagent (ECL808-25, Biomiga, USA) for 1 min at room temperature. Images of the protein bands were observed and captured under an optical luminometer (GE, USA). Subsequently, analysis of relative protein expression was performed with ImageProPlus6.0 (MediaCybernetics, USA) gray scale scanning software. GAPDH was used as an internal reference. The experiment was repeated three times.
Cell Counting Kit-8 (CCK-8) Assay
HCT-116 and SW480 cells were digested and resuspended after transfection and were then seeded in 96-well plates at 1×105 cells/mL (100 μL/well) and cultured overnight. Subsequently, at 0 h, 24 h, 48 h, 72 h and 96 h, each well was added with 10 μL CCK-8 solution (Beyotime, Shanghai, China) and cells were incubated for additional 4 h. At last, the absorbance of each well at 450 nm was detected by using a microplate reader.
Colony Formation Assay in vitro
Soft agar plate was made in 6-well plates, with the bottom layer being 0.5% agar mixed with serum-free medium. Firstly, cells were seeded in 35-mm tissue cultivation dishes for 24 h and then transfected with different reagents. After cells were digested with trypsin, 1×103 cells and 0.35% agar were mixed in the cell medium containing 10% FBS and seeded into the agar plate as the top agar layer. Subsequently, cells were cultured in an incubator at 37°C for 3 weeks. The qualified colonies were stained with 0.1% crystal violet, washed extensively with phosphate-buffered saline (PBS) and counted.
Dual-Luciferase Reporter Gene Assay
The wild-type (WT) PHLPP2 3ʹUTR that could bind with miR-141 and the mutant (MUT) PHLPP2 3ʹUTR formed by site-directed mutagenesis method were, respectively, cloned into psiCheck2 luciferase vector, contributing to the establishment of PHLPP2-WT and PHLPP2-MUT vectors. PHLPP2-WT/PHLPP2-MUT vectors were co-transfected with mimic NC/miR-141 mimic into HCT-116 and SW480 cells. The luciferase activities were assessed using the Dual-Luciferase Reporter Assay Kit (Promega, Madison, WI, USA). After 48 h of incubation, the cells were lysed using 1×Passive lysis buffer and Dual-Luciferase Reporter Assay Kit (Promega) was used to evaluate the firefly luciferase activities. Firefly luciferase activity was normalized to Renilla luciferase activity. The experiment was performed in triplicate.
Tumor Formation in Nude Mice
Twelve male BALB/C nude mice (4–6 weeks old, weighing 18–24 g) derived from Hunan SJA Laboratory Animal (Hunan, China) were fed in a specific pathogen-free (SPF) environment. The mice were randomly divided into the antagomir NC group and the miR-141 antagomir group with 6 mice in each group. Lipofectamine 2000 reagent (Invitrogen) was employed to transfect antagomir NC and miR-141 antagomir into CC cell line SW480. The stably transfected SW480 cells were obtained to prepare cell suspension at 1×106 cells/mL, which was subcutaneously injected into nude mice. Tumor volume was monitored once a week. The long diameter (A) and short diameter (B) of the xenograft tumor were measured by a vernier caliper each week. Tumor volume was calculated with a formula of: Volume= (A×B2)/2. At the end of the 5th week, mice were given a euthanasia with tumors being isolated and then fixed in 4% paraformaldehyde. Tumor tissues were frozen and stored in liquid nitrogen for further study 4 weeks later. All the animals being experimented were used for medical research and this experiment was approved by the Ethics Committee of Laboratory Animal.
Statistical Analysis
All statistical analyses were performed using SPSS 21.0 (SPSS, Inc., Chicago, IL, USA) software. All the data were expressed as mean ± standard deviation (SD), and differences between two groups were analyzed by Student’s t-test. p<0.05 was considered statistically significant and p<0.01 was considered highly statistically significant.
Results
miR-141 is Highly Expressed in CC Cells
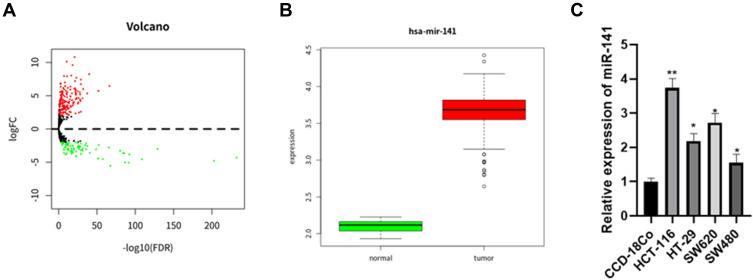
Normal samples (n=8) and CC tissue samples (n=445) were used for analysis of differentially expressed miRNAs (DE_miRNAs), and 271 DE_miRNAs in total were obtained (Figure 1A), among which miR-141 was markedly up-regulated in cancer tissue (Figure 1B). A study validated that miR-141 is able to serve as an oncogene,25 and it is abnormally expressed in CC and involved in various cell processes, including epithelial-mesenchymal transition (EMT), proliferation, migration, invasion and drug resistance.26 We chose normal colon cell line CCD-18Co and CC cell lines HCT-116, HT-29, SW620 and SW480, in which miR-141 expression was detected via qRT-PCR. The results suggested that compared with CCD-18Co, miR-141 was highly expressed in CC cells (Figure 1C). HCT-116 and SW480 cell lines with the highest and the lowest miR-141 expression were chosen for follow-up experiments.
Figure 1.
miR-141 is highly expressed in CC cells. (A) Volcano plot of DE_miRNAs in the TCGA-COAD dataset. Red and green represent markedly up-regulated and down-regulated DE_miRNAs, respectively; (B) Relative expression of miR-141 in normal and tumor samples in the TCGA-COAD dataset; (C) miR-141 expression in normal colon cell line and CC cell lines were detected using qRT-PCR; * p<0.05, ** p<0.01. The experiment was repeated in triplicate.
CC Cell Proliferation is Suppressed by miR-141 Silencing but Promoted by miR-141 Over-Expression
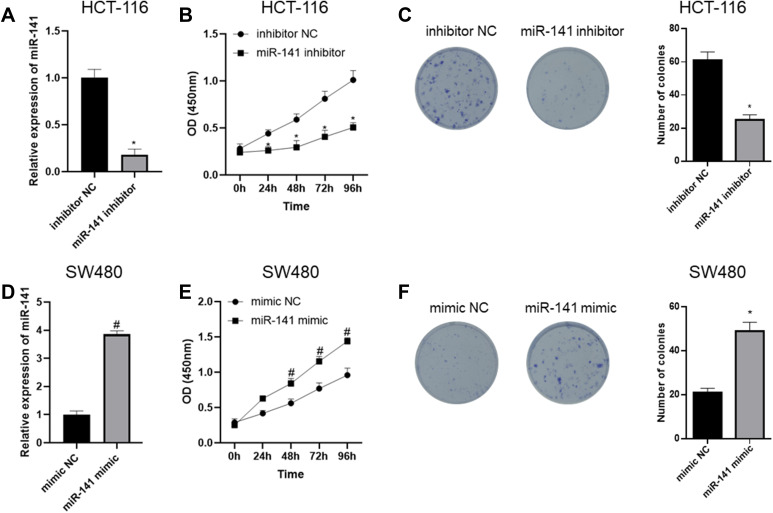
In order to further verify the mechanism by which miR-141 regulates CC cell proliferation, we firstly silenced miR-141 in HCT-116 cell line and detected miR-141 expression via qRT-PCR. The results showed that compared with the inhibitor NC group, miR-141 expression in the miR-141 inhibitor group was significantly reduced (Figure 2A). Cell proliferation was assessed by CCK-8 assay and colony formation assay, the results of which indicated that compared with the inhibitor NC group, cell proliferation in the miR-141 inhibitor group was noticeably decreased (Figure 2B and C). Meanwhile, we over-expressed miR-141 in SW480 cell line and determined the expression of miR-141 through qRT-PCR and found that compared with the mimic NC group, miR-141 expression in the miR-141 mimic group was markedly increased (Figure 2D). We further detected cell proliferation by CCK-8 and colony formation assay, discovering that compared with the mimic NC group, cell proliferation in the miR-141 mimic group was prominently raised (Figure 2E and F). Collectively, the above results demonstrated that CC cell proliferation was inhibited by miR-141 silencing but fostered by miR-141 over-expression.
Figure 2.
CC cell proliferation is suppressed by miR-141 silencing but promoted by miR-141 over-expression. (A) qRT-PCR was done to detect miR-141 expression after miR-141 was silenced in HCT-116 cells; (B) CCK-8 assay was performed to detect cell viability upon miR-141 silencing; (C) colony formation assay was carried out to detect cell proliferation upon miR-141 silencing; (D) qRT-PCR was done to detect miR-141 expression after miR-141 was overexpressed in SW480 cells; (E) CCK-8 assay was performed to detect cell viability upon miR-141 overexpression; (F) colony formation assay in vitro was carried out to detect cell proliferation upon miR-141 overexpression; *Represents comparison with inhibitor NC, p<0.05, #Represents comparison with mimic NC, p<0.05. The experiment was repeated in triplicate.
miR-141 Represses Tumor Formation of CC Cells
After stably transfected cell lines were constructed, they were subcutaneously injected into nude mice to construct xenograft tumor model in nude mice so as to verify the effect of miR-141 on the in vivo tumorigenic ability of CC cells. Firstly, qRT-PCR was carried out to detect miR-141 expression in transfected cells, which revealed that compared with the antagomir NC group, the expression of miR-141 in the miR-141 antagomir group was markedly reduced (Figure 3A). The detection of cell tumor growth speed each week out of five weeks after subcutaneous injection indicated that compared with the antagomir NC group, tumor formation speed in the miR-141 antagomir was significantly decreased (Figure 3B). All mice were sacrificed five weeks later, and the tumors were taken out and weighed, which unveiled that compared with the antagomir NC group, tumor weight in the miR-141 antagomir was noticeably reduced (Figure 3C and D). Taken together, miR-141 suppressed CC cell tumor formation in vivo.
Figure 3.
miR-141 suppresses CC cell tumor formation in vivo. (A) qRT-PCR was carried out to detect miR-141 expression in transfected cells; (B) detection of cell tumor growth speed each week out of five weeks after subcutaneous injection; (C) picture of xenograft tumors; (D) statistics of tumor weight; *p<0.05. N=6 nude mice.
miR-141 Targeted Silences PHLPP2 in CC Cells
In order to further detect the functional mechanism by which miR-141 regulates CC cell proliferation, firstly we performed differential analysis of mRNAs by using 41 normal samples and 462 cancer samples and obtained 2042 DE_mRNAs (Figure 4A). Next, TargetScan, miRDB and miRTarBase databases were used to predict the target genes of miR-141, which were then intersected with down-regulated DE_mRNAs in the TCGA-COAD dataset, and 2 DE_mRNAs which had the binding sites with miR-141 were obtained (Figure 4B), among which PHLPP2 was markedly down-regulated in cancer tissues (Figure 4C) and led to a poor prognosis in patients (Figure 4D). A study showed that low PHLPP2 expression promotes cell proliferation.18 Consequently, we speculated that miR-141 was likely to regulate CC cell proliferation by targeting PHLPP2.
Figure 4.
miR-141 targeted silences PHLPP2 in CC cells. (A) Volcano plot of DE_mRNAs in the TCGA-COAD dataset. Red and green represent markedly up-regulated and down-regulated DE_mRNAs, respectively; (B) Venn diagram of down-regulated DE_mRNAs and the predicted target mRNAs of miR-141; (C) relative expression of PHLPP2 in normal and tumor samples in the TCGA-COAD dataset; (D) survival analysis of PHLPP2 in the TCGA-COAD dataset; (E) qRT-PCR and (F) Western blot were conducted to detect PHLPP2 mRNA and protein expression in normal colon cell line and CC cell lines; (G, H) miR-141 was silenced in CC cell line HCT-116 and PHLPP2 mRNA and protein expression were detected by (G) qRT-PCR and (H) Western blot; (I, J) miR-141 was over-expressed in CC cell line SW480 and PHLPP2 mRNA and protein expression were detected by (I) qRT-PCR and (J) Western blot; (K) miR-141 was predicted to bind with PHLPP2 3ʹUTR through TargetScan website; (L, M) dual-luciferase reporter gene assay was performed; miR-141 was silenced and over-expressed in CC cell line HCT-116 and SW480, respectively; * in (I) represents comparison with CCD-18Co, p<0.05, ** p<0.01; * in (G) and (L) represents comparison with the inhibitor NC group, p<0.05; #in (I) and (M) represents comparison with the mimic NC group, p<0.05. The experiment was repeated in triplicate.
To validate our speculation, we conducted qRT-PCR and Western blot to detect PHLPP2 mRNA and protein expression in normal colon cell line CCD-18Co and CC cell lines HCT-116, HT-29, SW620 and SW480, which unveiled that compared with CCD-18Co, PHLPP2 was poorly expressed in CC cells (Figure 4E and F). miR-141 was silenced and over-expressed in CC cell line HCT-116 and SW480, respectively, and PHLPP2 mRNA and protein expression were detected by qRT-PCR and Western blot. The results suggested that compared with the inhibitor NC group, PHLPP2 expression in the miR-141 inhibitor group was markedly increased (Figure 4G and H); compared with the mimic NC group, PHLPP2 expression in the miR-141 mimic group was significantly decreased (Figure 4I and J). miR-141 was predicted to bind with PHLPP2 3ʹUTR through TargetScan website (Figure 4K). Dual-luciferase reporter gene assay demonstrated that the luciferase activity of PHLPP2-WT could be markedly enhanced by silencing miR-141 (Figure 4L) but weakened by over-expressing miR-141 (Figure 4M), whereas PHLPP2-MUT was not noticeably affected by silencing or over-expressing miR-141. Collectively, miR-141 targeted to down-regulate PHLPP2 in CC cells.
miR-141 Facilitates CC Cell Proliferation by Silencing PHLPP2
In order to clarify the functional mechanism by which miR-141 regulates CC cell proliferation by targeting PHLPP2, we simultaneously over-expressed miR-141and PHLPP2in CC cell line SW480. Firstly, qRT-PCR and Western blot were carried out to detect the expression of miR-141 and PHLPP2, which indicated that compared with the mimic NC+oe-NC group, miR-141 expression was markedly increased while PHLPP2 expression was significantly reduced in the miR-141 mimic+oe-NC group, and PHLPP2 expression was remarkably raised in the mimic NC+oe-PHLPP2 group; whereas compared with the miR-141 mimic+oe-NC group, PHLPP2 expression was noticeably elevated in the miR-141 mimic+oe-PHLPP2 group (Figure 5A and B). CCK-8 and colony formation assay were performed to detect cell proliferation, which revealed that compared with the mimic NC+oe-NC group, cell proliferation was markedly increased in the miR-141 mimic+oe-NC group but decreased in the mimic NC+oe-PHLPP2 group; whereas compared with the miR-141 mimic+oe-NC group, cell proliferation was remarkably reduced in the miR-141 mimic+oe-PHLPP2 group (Figure 5C and D). Taken together, miR-141 fostered CC cell proliferation by silencing PHLPP2.
Figure 5.
miR-141 facilitates CC cell proliferation by silencing PHLPP2. (A) qRT-PCR was conducted to detect miR-141 and PHLPP2 mRNA expression; (B) Western blot was performed to detect PHLPP2 protein expression; (C) CCK-8 assay was carried out to assess cell viability; (D) colony formation assay was conducted to evaluate cell proliferation; *Represents comparison with the mimic NC+oe-NC group, p<0.05; #Represents comparison with the miR-141 mimic+oe-NC group, p<0.05. The experiment was repeated in triplicate.
Conclusion
miR-141 is a member of miR-200 family and has been reported to be associated with various cancers.27 For example, miR-141 is down-regulated in HCC,14 pancreatic ductal adenocarcinoma28 and renal cell carcinoma,29 in which it plays a role as a potential tumor suppressor gene. However, miR-141 has been found to act as an oncogene in nasopharyngeal carcinoma, suggesting that it plays dual effects on different cancers.3 Earlier Liu et al30 validated that miR-141 expression is higher than that of normal tissue in non-small cell lung cancer. However, there have been no further reports on the functional mechanism of miR-141 in CC. In this study, we found that miR-141 was highly expressed in CC tissue by bioinformatics analysis, which was further verified by the detection of miR-141 expression in CC cells and normal colon cells using qRT-PCR. Additionally, by conducting CCK-8 assay, colony formation assay and tumor formation in nude mice experiment, we validated that miR-141 could promote CC tumor growth and unveiled that it played a carcinogenic role in CC.
PHLPP2 has been found to be down-regulated in various cancers31,32 and is able to suppress the invasion of bladder cancer by facilitating the autophagy and degradation of MMP2 protein.33 Besides, PHLPP1 is capable of inhibiting the metastasis of melanoma by repressing AKT2 activation.22 However, the regulatory function of PHLPP2 in CC needs further research. In this study, we further investigated the molecular mechanism by which miR-141 promoted CC cell proliferation. We discovered that PHLPP2 was a theoretical target gene of miR-141 through prediction using publicly available bioinformatics websites, which was subsequently validated by dual-luciferase reporter assay. Meanwhile, we also found that PHLPP2 could reverse miR-141-induced CC cell proliferation as revealed by CCK-8 and colony formation assay. Collectively, miR-141 negatively regulated PHLPP2 expression in CC cells by directly targeting PHLPP2 3ʹUTR so as to foster CC cell proliferation.
In all, this study verified that miR-141 was up-regulated in CC and facilitated CC cell proliferation by targeted inhibiting the expression of PHLPP2. Therefore, miR-141 is likely to be a potential therapeutic target of CC treatment.
Funding Statement
There is no funding to report.
Highlights
miR-141 targeted to regulate PHLPP2 in colon cancer;
Colon cancer cell proliferation and in vivo tumorigenic ability could be suppressed by miR-141 down-regulation but promoted by miR-141 over-expression;
miR-141 fostered colon cancer cell proliferation by silencing PHLPP2.
Ethics approval
This study was conducted in accordance with Guide for the care and use of laboratory animals and was approved by the Institutional Review Boards of Zhejiang Jinhua Guangfu Hospital.
Disclosure
The authors declare that they have no potential conflicts of interest for this work.
References
- 1.Krajewska JB, Fichna J, Mosinska P. One step ahead: miRNA-34 in colon cancer-future diagnostic and therapeutic tool? Crit Rev Oncol Hematol. 2018;132:1–8. doi: 10.1016/j.critrevonc.2018.09.006 [DOI] [PubMed] [Google Scholar]
- 2.Arnold M, Sierra MS, Laversanne M, et al. Global patterns and trends in colorectal cancer incidence and mortality. Gut. 2017;66(4):683–691. doi: 10.1136/gutjnl-2015-310912 [DOI] [PubMed] [Google Scholar]
- 3.Mei Z, He Y, Feng J, et al. MicroRNA-141 promotes the proliferation of non-small cell lung cancer cells by regulating expression of PHLPP1 and PHLPP2. FEBS Lett. 2014;588(17):3055–3061. doi: 10.1016/j.febslet.2014.06.020 [DOI] [PubMed] [Google Scholar]
- 4.Ambros V. The functions of animal microRNAs. Nature. 2004;431(7006):350–355. doi: 10.1038/nature02871 [DOI] [PubMed] [Google Scholar]
- 5.Gao H, Ran M, Luo H, et al. miR-499 promotes immature porcine Sertoli cell growth by the PI3K/AKT pathway by targeting the PTEN gene. Reproduction. 2019. doi: 10.1530/REP-19-0303 [DOI] [PubMed] [Google Scholar]
- 6.Guan D, Li Y, Cui Y, et al. Down-regulated miR-374c and Hsp70 promote Th17 cell differentiation by inducing Fas expression in experimental autoimmune encephalomyelitis. Int J Biol Macromol. 2019. doi: 10.1016/j.ijbiomac.2019.11.147 [DOI] [PubMed] [Google Scholar]
- 7.Bao Q, Liao X, Li R, Ding N. KCNQ1OT1 promotes migration and inhibits apoptosis by modulating miR-185-5p/Rab14 axis in oral squamous cell carcinoma. Dev Growth Differ. 2019;61(9):466–474. doi: 10.1111/dgd.12638 [DOI] [PubMed] [Google Scholar]
- 8.Jin Y, Zhan X, Zhang B, et al. Polydatin exerts an antitumor effect through regulating the miR-382/PD-L1 axis in colorectal cancer. Cancer Biother Radiopharm. 2020;35(2):83–91. doi: 10.1089/cbr.2019.2999 [DOI] [PubMed] [Google Scholar]
- 9.Garzon R, Calin GA, Croce CM. MicroRNAs in cancer. Annu Rev Med. 2009;60(1):167–179. doi: 10.1146/annurev.med.59.053006.104707 [DOI] [PubMed] [Google Scholar]
- 10.Toda H, Seki N, Kurozumi S, et al. RNA-sequence-based microRNA expression signature in breast cancer: tumor-suppressive miR-101-5p regulates molecular pathogenesis. Mol Oncol. 2020;14(2):426–446. doi: 10.1002/1878-0261.12602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sun L, Fang Y, Wang X, et al. miR-302a inhibits metastasis and cetuximab resistance in colorectal cancer by targeting NFIB and CD44. Theranostics. 2019;9:8409–8425. doi: 10.7150/thno.36605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Saleeb R, Kim SS, Ding Q, et al. The miR-200 family as prognostic markers in clear cell renal cell carcinoma. Urol Oncol. 2019;37(12):955–963. doi: 10.1016/j.urolonc.2019.08.008 [DOI] [PubMed] [Google Scholar]
- 13.Iorio MV, Visone R, Di Leva G, et al. MicroRNA signatures in human ovarian cancer. Cancer Res. 2007;67(18):8699–8707. doi: 10.1158/0008-5472.CAN-07-1936 [DOI] [PubMed] [Google Scholar]
- 14.Liu Y, Ding Y, Huang J, et al. MiR-141 suppresses the migration and invasion of HCC cells by targeting Tiam1. PLoS One. 2014;9(2):e88393. doi: 10.1371/journal.pone.0088393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Porkka KP, Pfeiffer MJ, Waltering KK, et al. MicroRNA expression profiling in prostate cancer. Cancer Res. 2007;67(13):6130–6135. doi: 10.1158/0008-5472.CAN-07-0533 [DOI] [PubMed] [Google Scholar]
- 16.Kim K, Ryu D, Dongiovanni P, et al. Degradation of PHLPP2 by KCTD17, via a glucagon-dependent pathway, promotes hepatic steatosis. Gastroenterology. 2017;153(6):1568–1580e1510. doi: 10.1053/j.gastro.2017.08.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yeh ST, Zambrano CM, Koch WJ, Purcell NH. PH domain leucine-rich repeat protein phosphatase 2 (PHLPP2) regulates G-protein–coupled receptor kinase 5 (GRK5)-induced cardiac hypertrophy in vitro. J Biol Chem. 2018;293(21):8056–8064. doi: 10.1074/jbc.M117.809913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li CF, Li YC, Jin JP, Yan ZK, Li D. miR-938 promotes colorectal cancer cell proliferation via targeting tumor suppressor PHLPP2. Eur J Pharmacol. 2017;807:168–173. doi: 10.1016/j.ejphar.2017.04.023 [DOI] [PubMed] [Google Scholar]
- 19.Strotbek M, Schmid S, Sánchez-González I, et al. miR-181 elevates Akt signaling by co-targeting PHLPP2 and INPP4B phosphatases in luminal breast cancer. Int J Cancer. 2017;140(10):2310–2320. doi: 10.1002/ijc.30661 [DOI] [PubMed] [Google Scholar]
- 20.Liao Y, Deng Y, Liu J, et al. MiR-760 overexpression promotes proliferation in ovarian cancer by downregulation of PHLPP2 expression. Gynecol Oncol. 2016;143(3):655–663. doi: 10.1016/j.ygyno.2016.09.010 [DOI] [PubMed] [Google Scholar]
- 21.Molina JR, Agarwal NK, Morales FC, et al. PTEN, NHERF1 and PHLPP form a tumor suppressor network that is disabled in glioblastoma. Oncogene. 2012;31(10):1264–1274. doi: 10.1038/onc.2011.324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yu Y, Dai M, Lu A, Yu E, Merlino G. PHLPP1 mediates melanoma metastasis suppression through repressing AKT2 activation. Oncogene. 2018;37(17):2225–2236. doi: 10.1038/s41388-017-0061-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Toivanen R, Furic L. A balancing act: PHLPP2 fine tunes AKT activity and MYC stability in prostate cancer. J Cell Biol. 2019;218(6):1771–1772. doi: 10.1083/jcb.201904119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xie M, Ma T, Xue J, et al. The long intergenic non-protein coding RNA 707 promotes proliferation and metastasis of gastric cancer by interacting with mRNA stabilizing protein HuR. Cancer Lett. 2019;443:67–79. doi: 10.1016/j.canlet.2018.11.032 [DOI] [PubMed] [Google Scholar]
- 25.Li J-H, Zhang Z, Du M-Z, et al. microRNA-141-3p fosters the growth, invasion, and tumorigenesis of cervical cancer cells by targeting FOXA2. Arch Biochem Biophys. 2018;657:23–30. doi: 10.1016/j.abb.2018.09.008 [DOI] [PubMed] [Google Scholar]
- 26.Gao Y, Feng B, Han S, et al. The roles of MicroRNA-141 in human cancers: from diagnosis to treatment. Cell Physiol Biochem. 2016;38(2):427–448. doi: 10.1159/000438641 [DOI] [PubMed] [Google Scholar]
- 27.Bendoraite A, Knouf EC, Garg KS, et al. Regulation of miR-200 family microRNAs and ZEB transcription factors in ovarian cancer: evidence supporting a mesothelial-to-epithelial transition. Gynecol Oncol. 2010;116(1):117–125. doi: 10.1016/j.ygyno.2009.08.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhu Z-M, Xu Y-F, Su Q-J, et al. Prognostic significance of microRNA-141 expression and its tumor suppressor function in human pancreatic ductal adenocarcinoma. Mol Cell Biochem. 2014;388(1–2):39–49. doi: 10.1007/s11010-013-1897-y [DOI] [PubMed] [Google Scholar]
- 29.Chen X, Wang X, Ruan A, et al. miR-141 is a key regulator of renal cell carcinoma proliferation and metastasis by controlling EphA2 expression. Clin Cancer Res. 2014;20(10):2617–2630. doi: 10.1158/1078-0432.CCR-13-3224 [DOI] [PubMed] [Google Scholar]
- 30.Liu XG, Zhu W-Y, Huang -Y-Y, et al. High expression of serum miR-21 and tumor miR-200c associated with poor prognosis in patients with lung cancer. Med Oncol. 2012;29(2):618–626. doi: 10.1007/s12032-011-9923-y [DOI] [PubMed] [Google Scholar]
- 31.Cai J, Fang L, Huang Y, et al. miR-205 targets PTEN and PHLPP2 to augment AKT signaling and drive malignant phenotypes in non-small cell lung cancer. Cancer Res. 2013;73(17):5402–5415. doi: 10.1158/0008-5472.CAN-13-0297 [DOI] [PubMed] [Google Scholar]
- 32.Gao T, Furnari F, Newton AC. PHLPP: a phosphatase that directly dephosphorylates Akt, promotes apoptosis, and suppresses tumor growth. Mol Cell. 2005;18(1):13–24. doi: 10.1016/j.molcel.2005.03.008 [DOI] [PubMed] [Google Scholar]
- 33.Peng M, Wang J, Zhang D, et al. PHLPP2 stabilization by p27 mediates its inhibition of bladder cancer invasion by promoting autophagic degradation of MMP2 protein. Oncogene. 2018;37(43):5735–5748. doi: 10.1038/s41388-018-0374-1 [DOI] [PMC free article] [PubMed] [Google Scholar]