Abstract
Background
Mitogen-activated protein kinase 14 (MAPK14) acts as an integration point for multiple biochemical signal pathways. High expressions of MAPK14 have been found in a variety of tumors. Runt‑related transcription factor 2 (RUNX2) is related to many tumors, especially in tumor invasion and metastasis. However, the mechanism of these two genes in bladder cancer remains unclear.
Methods
TCGA database and Western blot were used to analyze the mRNA and protein levels of the target gene in bladder cancer tissues and adjacent tissues. The proliferation ability of bladder cancer cells was tested by colony forming and EdU assay. The migration ability of cells was detected by transwell assay. Immunoprecipitation was utilized to detect protein–protein interaction. Cycloheximide chase assay was used to measure the half-life of RUNX2 protein.
Results
Phosphorylated mitogen-activated protein kinase 14 (P-MAPK14, Thr180/Tyr182) was highly expressed in bladder cancer tissues and bladder cancer cell lines. Accordingly, P-MAPK14 could be combined with RUNX2 and maintain its protein stability and promote the proliferation and migration of bladder cancer cells. In addition, the functional degradation caused by the downregulation of MAPK14 and P-MAPK14 could be partially compensated by the overexpression of RUNX2.
Conclusion
These results suggest that P-MAPK14 might play an important role in the development of bladder cancer and in the regulation of RUNX2 protein expression. P-MAPK14 might become a potential target for the treatment of bladder cancer.
Keywords: MAPK14, RUNX2, P-MAPK14, bladder cancer, ubiquitination
Introduction
Bladder cancer, the ninth most common cancer worldwide, is a highly heterogeneous disease and an important cause of cancer metastasis-related death.1 In the United States, it has been estimated that there would be about 80,470 new bladder cancer cases and 17,670 deaths in 2019.2 The incidence of bladder cancer is higher in males and 70% of patients with bladder cancer are initially diagnosed as non-muscular invasive diseases. There are several risk factors related to bladder cancer, such as smoking, aging and exposure to the products of chemical industries.1,3 In recent years, despite the progress in clinical diagnosis and treatment of bladder cancer, it remains to be a perplexing problem in clinical management due to its high metastasis rate.4 Therefore, a comprehensive understanding of the pathogenetic mechanism of bladder cancer would improve the prognosis of patients.
P38 mitogen-activated protein kinase (MAPK) contains four isoforms (α, β, γ, δ), which are encoded by four different genes, sharing a sequence with high homology.5,6 MAPK14 is a member of the P38 MAPK family, also called P38α.7 It has been reported that MAPK14 plays an important role in coordinating DNA damage response and limiting chromosomal instability during breast cancer progression. Moreover, reduced MAPK14 level leads to DNA damage and increased chromosomal instability in breast cancer cells, ultimately resulting in cancer cell death and tumor regression.8 However, MAPK14 plays a dual role in colon tumors. MAPK14 protects intestinal epithelial cells from colon cancer-related colitis by regulating the function of the intestinal epithelial barrier, but it also contributes to the maintenance of colon tumors.9 In bladder cancer, although some studies have pointed out that MAPK14 signaling pathway is involved in promoting or inhibiting the proliferation and migration of tumor cells, most of the studies are not directed at MAPK14.10–12 Therefore, the mechanism of MAPK14 gene in bladder cancer remains to be clarified, and further study is necessary.
As a key factor of osteoblast differentiation, transcription factor Runt-related transcription factor 2 (RUNX2) plays a crucial role in osteoblast differentiation.13,14 RUNX2 is a member of the mammalian RUNX family of transcription factors, and it has been shown to be involved in tumor development.15 Phosphorylated RUNX2 is tightly related to the metastasis of prostate cancer.16 Overexpression of RUNX2 could increase the cancer cell proliferation in mantle cell lymphoma.17 In addition, it has been reported that RUNX2 and p53 may be functionally related to bladder cancer, and may be associated with the development and invasion of bladder tumors.18 However, the expression pattern and the role of RUNX2 in bladder cancer require to be elucidated.
In this study, the transcriptional level of MAPK14 in bladder urothelial carcinoma from the TCGA database was analyzed, and it was shown that MAPK14 was poorly expressed in bladder cancer tissues, both in the overall analysis and in the analysis of 19 paired samples. Then, protein levels of MAPK14 and phosphorylated MAPK14 (P-MAPK14) in bladder cancer tissues and bladder cancer cell lines were investigated. It was found that P-MAPK14 was highly expressed in both bladder cancer tissues and bladder cancer cell lines. After MAPK14 protein was knocked down by small interfering RNA (siRNA), the level of P-MAPK14 protein declined, and the clonal formation, proliferation and migration ability of bladder cancer cells decreased. Downregulation of P-MAPK14 protein did not reduce the RUNX2 mRNA level but resulted in the decrease of RUNX2 protein abundance. Co-immunoprecipitation assay indicated that P-MAPK14 might interact with RUNX2. Further studies revealed that P-MAPK14 might preserve the stability of RUNX2 protein by reducing its ubiquitination degradation pathway. This study suggested that P-MAPK14 might promote bladder cancer cell proliferation and migration by maintaining the stability of RUNX2 protein.
Materials and Methods
The Cancer Genome Atlas (TCGA) Data
TCGA is a program for retrieving and processing data from the open access GDC data portal. From pre-treated bladder urothelial carcinoma, 414 cancer samples were selected, including 19 pairs of matched bladder urothelial carcinoma tissues and adjacent normal tissues, and the target gene MAPK14 was analyzed.
Tissue Samples
Thirty-six cases of bladder cancer with adjacent normal tissues were collected. The patients did not receive preoperative chemoradiotherapy, and the adjacent normal tissues obtained were at least 5 cm away from the cancer tissues. The tissue was stored in the refrigerator at −80°C before use. Informed consent was signed by all patients and the study was approved by the Ethics Committee of the First Hospital of China Medical University.
Cell Culture and Treatment
Human bladder cancer cells 5637, UMUC3, T24, J82 and immortalized human urothelial cells SV-HUC were cultured in RPMI-1640 (HyClone, USA) medium. The medium was supplemented with 10% fetal bovine serum (HyClone, USA). All cell lines were obtained from the Type Culture Collection of the Chinese Academy of Sciences (Shanghai, China) and were cultured under humidified air containing 5% CO2 at 37°C. When the confluence of cells reached at 80%, the cells were digested with trypsin for subculture.
VX-702, cycloheximide (CHX) and MG-132 were purchased from MCE (MedChemExpress, USA). Dimethyl sulfoxide (DMSO) was purchased from Sigma-Aldrich (St Louis, MO, USA). The cells were treated with VX-702 (20 nmol/L) for 24 hours or 48 hours before the protein was extracted for analysis. To explore the degradation pathway of RUNX2, MG-132 (20 μmol/L) was added to stabilize the cell lines with shMAPK14 for 4 hours, and then the proteins were collected for Western blot analysis. The method for determining the half-life of RUNX2 protein by cyclohexylamine was described earlier.19 After 48 hours of transfection, cells were treated with CHX (100 μg/mL) for the indicated time.
siRNAs and Plasmids Transfection
The siRNAs of MAPK14 were synthesized by JTSBIO Co (China). The MAPK14 siRNA and negative control siRNA (siNC) sequences were used as follows (5ʹ‑3ʹ): siMAPK14#1 sense, CCAGACCAUUUCAGUCCAUTT and anti-sense, AUGGACUGAAAUGGUCUGGTT; siMAPK14#2 sense, CCUUGCACAUGCCUACUUUTT and anti-sense, AAAGUAGGCAUGUGCAAGGTT; siNC sense, UUCUCCGAACGUGUCACGUTT and anti-sense, ACGUGACACGUUCGGAGAATT. The plasmids for empty vector, shMAPK14 and OE-RUNX2 were purchased from GeneChem (Shanghai, China). Lipofectamine®3000 Transfection Kit (Invitrogen) was used for transfection and the specific dose was determined according to the manufacturer’s instructions. RNA or protein was extracted when the transfection time reached 24 or 48 hours. In addition, cell lines stably downregulating MAPK14 (shMAPK14) were screened with puromycin (2 μg/mL) for 4 weeks.
Western Blot
Total protein of tissues and cells lines was extracted with RIPA lysates (containing protease and phosphatase inhibitors). Protein concentration was detected using a BCA Protein Assay Kit (Beyotime, China). The equal amount of protein was separated by 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Then, proteins were transferred onto polyvinylidene fluoride membranes (BIO-RAD, USA). After blocked by 5% skim milk for 1 hour, the proteins were then tested with the following antibodies: anti-MAPK14 (1:1000, 9218, CST), anti-P-MAPK14 (1:1000, 4511, CST), anti-RUNX2 (1:1000, 12,556, CST), anti-Tubulin (1:1000, 2128S, CST), anti-GAPDH (1:1000, 5174, CST) at 4°C overnight. The membrane was subsequently incubated with the secondary anti-rabbit antibody at 37°C for 1 hour. Finally, the luminescence system (Bio-Rad, CA, USA) and ECL luminescence reagent were used for detection (Absin Biotechnology, Shanghai, China). ImageJ software (USA) was used for quantitative analysis. Protein abundance was normalized with tubulin or GAPDH.
RNA Extraction and qPCR
Total RNA of bladder cancer cells was extracted with TRIzol reagent (Invitrogen) according to the manufacturer’s instructions. Reverse transcription was performed using PrimeScriptTM RT reagent Kit (Takara, Japan) following the manufacturer’s instructions. QPCR analyses were performed using TB Green® Premix Ex TaqTM II (Takara, Japan). GAPDH was used as the internal reference. The 2−ΔΔCT method was used for detection of the relative expression level. Primer sequences designed for target genes were as follows: MAPK14 sense, CCCGAGCGTTACCAGAACC and anti-sense, TCGCATGAATGATGGACTGAAAT; RUNX2 sense, GCGCATTCCTCATCCCAGTA and anti-sense, GGCTCAGGTAGGAGGGGTAA; GAPDH sense, ACAACTTTGGTATCGTGGAAGG and anti-sense, GCCATCACGCCACAGTTTC.
Co-Immunoprecipitation
The cells were washed with precooled 1×PBS when the confluence of cells reached at 90%, and 0.5 mL of cold 1× RIPA lysates was added into the culture dish. After centrifugation at 4°C, 14,000 g for 30 min, the supernatant was transferred to the new tube. The primary antibody was added into the cell lysate, and then the lysate containing the antibody was rotated overnight at 4°C. The magnetic beads were then added to the pyrolysis solution and the mixture was rotated at 4°C for 4 hours. The magnetic beads were extracted with a magnetic rack, and the denaturation was expected to continue with the Western blot procedure. Negative control IgG was acquired from Cell Signaling Technology (1:20, 5873S, CST).
Cell Colony Forming
The transfected cells were collected and counted, and then redeposited into the six-well plates to reach 500 cells per well. After 10 days of culture at 37°C with 5%CO2, the cells in the six-well plate were stained with crystal violet for 10 minutes. The cells were photographed and counted to evaluate the viability and proliferation of individual cells.
EdU Assay
The cells were planted in 24-well plates. After 48 h transfection, EdU (1:1000) reagent was added (BeyoClick™, EDU-488, China). Culture was continued for another 2 hours, and experiments were conducted according to the manufacturer’s instructions. Finally, the number of proliferating cells was counted by taking photos under a fluorescence microscope (Olympus Corporation, Japan).
Cell Migration Assay
Cell migration ability was analyzed using transwell chamber (Corning, USA). Cells were collected and counted after transfection for 48 hours. The transwell chamber was placed in advance in 600 μL of serum medium, and 10,000 cells were seeded with 200 μL of serum‐free medium into the upper chamber. After 12 hours of culture in the incubator, the chamber was taken out and stained with crystal violet for 10 minutes. Then, cells in the upper chamber were gently wiped off with cotton swabs. Images were taken under a microscope and analyzed with ImageJ.
Statistical Analysis
The data from at least three independent experiments were expressed as mean ± standard deviation (SD). Differences between groups were analyzed by Student’s t-test. Statistical analysis was conducted with GraphPad Prism version 7.0 (La Jolla, CA, USA). P < 0.05 was considered statistically significant (*P < 0.05, **P < 0.01, ***P < 0.001, nsP > 0.05).
Results
Phosphorylated MAPK14 is Highly Expressed in Bladder Cancer Tissues and Bladder Cancer Cell Lines
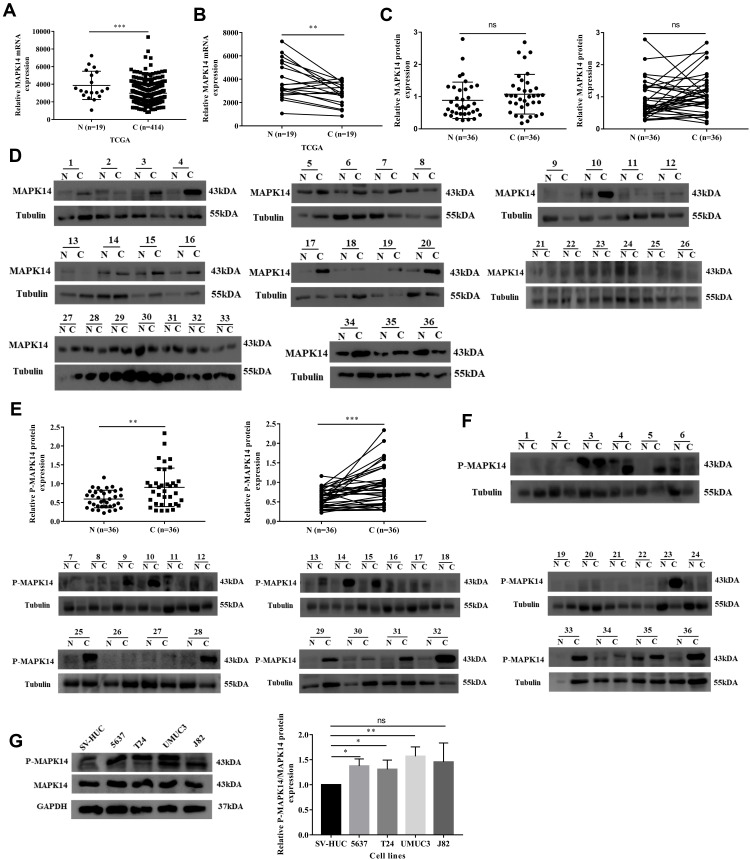
In the TCGA database, the transcriptional expression level of MAPK14 was analyzed, and it was found that mRNA expression of MAPK14 was low in bladder cancer tissues in either the overall analysis or the paired analysis (Figure 1A and B). Considering that P-MAPK14 is the main activation form of MAPK14, the expression levels of MAPK14 and P-MAPK14 in 36 pairs of bladder cancer specimens and matched adjacent non-cancerous bladder tissues were analyzed by immunoblotting. The protein level of MAPK14 in bladder cancer samples was not statistically significant with that in adjacent non-cancerous tissues, while P-MAPK14 in bladder cancer samples was significantly higher than that in adjacent tissues (Figure 1C–F). Moreover, compared with SV-HUC (immortalized human urothelial cells), high protein expression of P-MAPK14 was also observed in 5637, T24, and UMUC3 cell lines (Figure 1G). These findings suggest that P-MAPK14 was overexpressed in both human bladder cancer cell lines and bladder cancer tissues.
Figure 1.
P-MAPK14 protein was upregulated in bladder cancer samples and bladder cancer cells lines. (A and B) In the TCGA database, the transcriptional expression level of MAPK14 in bladder cancer tissues was low in either the overall analysis or the paired analysis; (C and D) MAPK14 protein levels in 36 pairs of bladder cancer tissues (C) and adjacent non-cancerous bladder tissues (N); (E and F) P-MAPK14 protein levels in 36 pairs of bladder cancer tissues (C) and adjacent non-cancerous bladder tissues (N); (G) P-MAPK14 and MAPK14 protein levels were detected in bladder cancer cells lines and SV-HUC cells by Western blot assay. (*P < 0.05, **P < 0.01, ***P < 0.001, nsP > 0.05).
Downregulation of MAPK14/P-MAPK14 Protein Can Inhibit Proliferation and Migration of Bladder Cancer Cells
The role of MAPK14 in promoting cell proliferation and migration in many tumors has been ascertained by many studies. To reveal the function of P-MAPK14 in bladder cancer cells, the biological function of P-MAPK14 in bladder cancer cells was studied. As showed by Western blot, the protein levels of MAPK14 and P-MAPK14 were significantly reduced by MAPK14 siRNA (Figure 2A). Colony formation assay showed that the T24 and UMUC3 cells colony formation ability were impeded by depletion of MAPK14 (Figure 2B). Similarly, EdU assay showed that the proliferation ability of bladder cancer cells T24 and UMUC3 were decreased significantly after 48 hours of cell transfection with MAPK14 siRNA (Figure 2C). To investigate the effect of P-MAPK14 in bladder cancer cell migration, cell lines T24 and UMUC3 with high metastatic ability were used. Consistently, migration assay showed that cells migration ability was reduced by the silencing of MAPK14 and P-MAPK14 (Figure 2D). These results suggest that MAPK14 and P-MAPK14 might be involved in cell clonal formation, proliferation, and migration.
Figure 2.
Downregulation of MAPK14 results in decrease expression of P-MAPK14 and inhibit bladder cancer cell colony formation, proliferation and migration (A) After transfected with MAPK14 siRNAs in T24 and UMUC3, protein levels of MAPK14 and P-MAPK14 were detected by Western blot; (B) Downregulation of MAPK14 suppressed bladder cancer cells colony formation ability; (C) EdU assay was used to examine the proliferation ability of T24 and UMUC3 cells (magnification × 200); (D) Transwell assay was used to examine the migration ability of T24 and UMUC3 cells (magnification × 20). (*P < 0.05, **P < 0.01, ***P < 0.001).
P-MAPK14 Regulates the RUNX2 Protein Expression in Bladder Cancer Cells
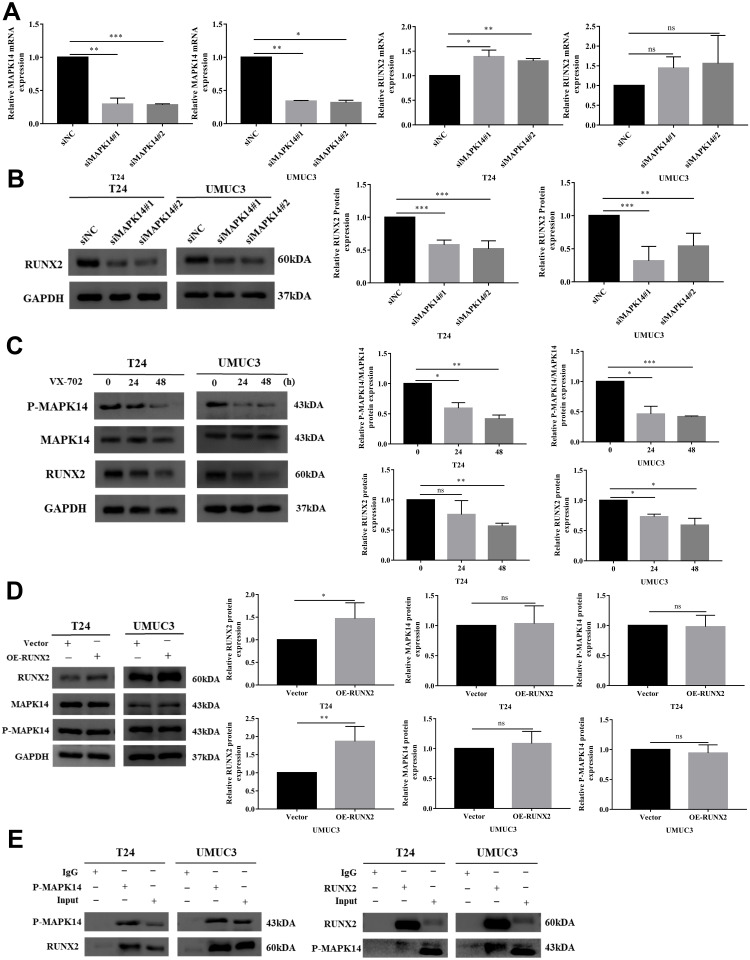
It has been shown that lack of MAPK14 can lead to the decrease in RUNX2 protein level.20 To study whether P-MAPK14 can regulate the RUNX2 protein in bladder cancer cells, endogenous MAPK14 was knocked down with two independent siRNAs in T24 and UMUC3 cells. The MAPK14 mRNA level was significantly decreased, but the RUNX2 mRNA level was increased in T24 cells. Similarly, RUNX2 was an upregulated trend in UMUC3 cell lines, whereas it was not statistically significant (Figure 3A). Western blot was used to explore whether the translation level of RUNX2 can be changed by the downregulation of MAPK14 and P-MAPK14 proteins. In surprise, in the absence of MAPK14 and P-MAPK14 proteins, RUNX2 protein level declined and was inconsistent with the transcriptional level (Figure 3B). Since the siRNA of MAPK14 could simultaneously reduce the protein levels of MAPK14 and P-MAPK14, in order to explore whether the decrease of RUNX2 protein was caused by the decrease of P-MAPK14, the inhibitor VX-702 of P-MAPK14 was utilized in this study, which can reduce the phosphorylation level of MAPK14 without the degradation of MAPK14 protein. Western blot result showed that RUNX2 protein abundance was decreased significantly with the decrease of P-MAPK14 protein level (Figure 3C). In order to investigate whether RUNX2 can affect the stability of MAPK14 and P-MAPK14 protein, bladder cancer cell lines overexpressing RUNX2 protein was constructed. Western blot showed that RUNX2 overexpression had no effect on MAPK14 and P-MAPK14 protein levels compared with empty vector group (Figure 3D). To investigate the regulatory relationship between P-MAPK14 and RUNX2, immunoprecipitation assay was performed in two bladder cancer cell lines T24 and UMUC3, Western blot result showed that P-MAPK14 and RUNX2 might be bind to each other in the two cell lines (Figure 3E). In summary, the results mainly show that P-MAPK14 regulates RUNX2 protein levels.
Figure 3.
P-MAPK14 regulates the RUNX2 protein expression in bladder cancer cells (A) After transfected with MAPK14 siRNAs in T24 and UMUC3, MAPK14 and RUNX2 mRNA levels were determined by RT-PCR; (B) RUNX2 protein levels in T24 and UMUC3 cells were measured by Western blot; (C) After T24 and UMUC3 cells were treated with VX-702, protein levels of MAPK14, P-MAPK14 and RUNX2 were detected by Western blot; (D) MAPK14 and P-MAPK14 protein expression was detected by Western blot after overexpression of RUNX2 in T24 and UMUC3 cells; (E) The interaction between P-MAPK14 and RUNX2 was detected by Co-Immunoprecipitation assay in T24 and UMUC3 cells. (*P < 0.05, **P < 0.01, ***P < 0.001, nsP > 0.05).
P-MAPK14 Maintains the Stability of the RUNX2 Protein
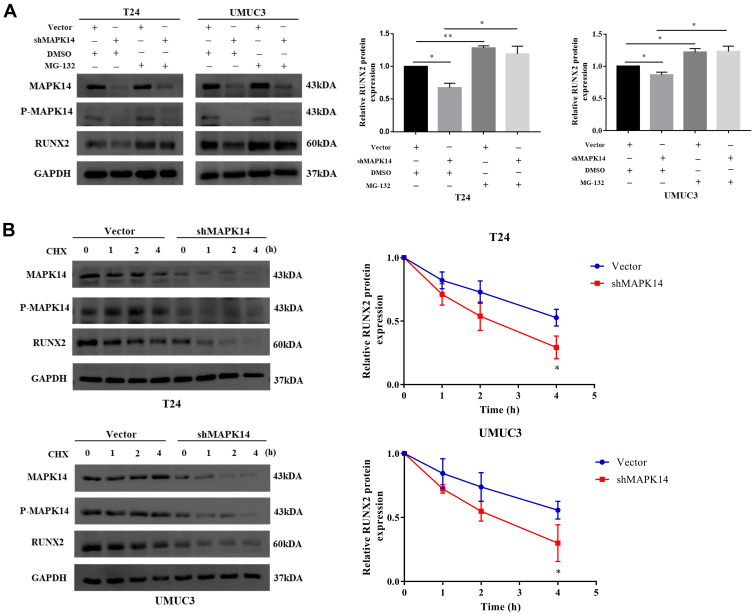
To determine the effect of P-MAPK14 on the stability of RUNX2 protein, T24 and UMUC3 cells stably expressing empty vector or shMAPK14 were treated with DMSO or MG‐132 (20 μmol/L), a proteasome inhibitor, for 4 hours. Western blot analysis showed that RUNX2 protein level was decreased due to the absence of MAPK14. When cells treated with MG-132, RUNX2 protein was no significant change (Figure 4A). Next, the half-life of the RUNX2 protein was analyzed by the cycloheximide chase assay. To further determine the effect of P-MAPK14 on RUNX2 protein degradation, T24 and UMUC3 cells were transfected with empty vector and shMAPK14 plasmid, respectively. After transfection for 48 hours, cells were treated with CHX (100 μg/mL) for the indicated time, then harvested the cell protein for immunoblotting. The half-life of the RUNX2 protein was reduced after MAPK14/P-MAPK14 depletion (Figure 4B). These results suggest that MAPK14/P-MAPK14 might regulate the stability of the RUNX2 protein, probably by inhibiting its ubiquitination degradation pathway.
Figure 4.
P-MAPK14 maintains the stability of the RUNX2 protein (A) Cells stably knockdown of MAPK14 (shMAPK14) and control group (Vector) were treated with DMSO or MG-132, followed by Western blot assay; (B) Cells stably knockdown of MAPK14 (shMAPK14) and control group (Vector) were treated with 100 μg/mL cycloheximide for the indicate times. Total cellular lysates were subjected to Western blot assay. (*P < 0.05, **P < 0.01).
Overexpression of RUNX2 Partially Reverses the Reduction in Cell Proliferation and Migration Caused by Reduced MAPK14/P-MAPK14 Expression
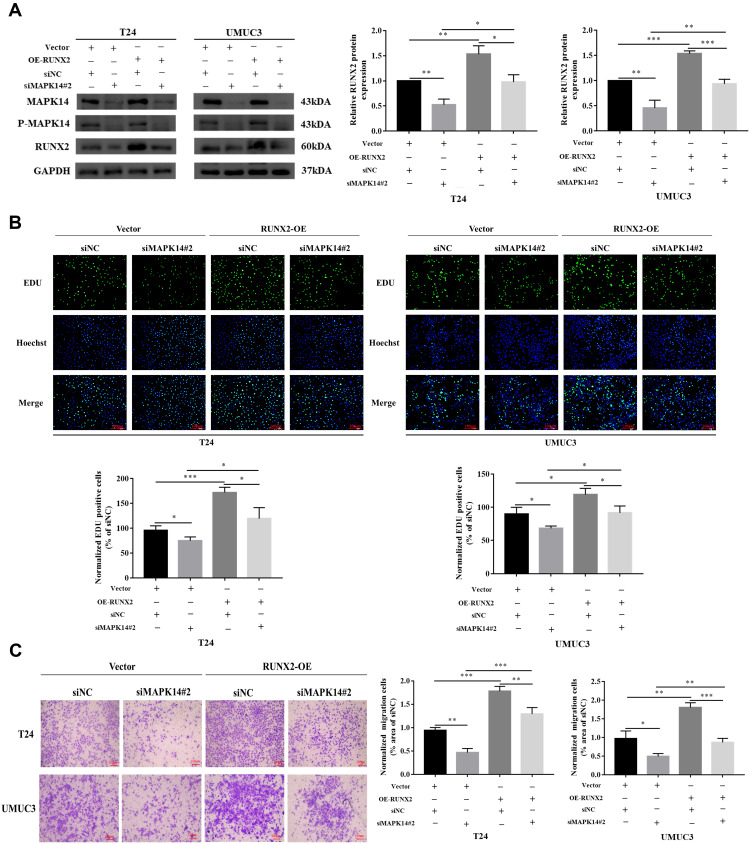
The MAPK14 protein level and RUNX2 protein level were significantly reduced by siMAPK14#2. When RUNX2 was overexpressed, the RUNX2 protein level induced by MAPK14 siRNA was partially restored (Figure 5A). EdU analysis showed that the proliferation ability of T24 and UMUC3 cells was inhibited when deficiency of MAPK14 protein, while the function of cell proliferation was partially restored after overexpression of RUNX2 (Figure 5B). Cell migration test showed that MAPK14 downregulation diminished the migration of bladder cancer cells, while the reintroduction of RUNX2 partially restored the activity of MAPK14 downregulation cells (Figure 5C). These data suggest that P-MAPK14 might promote the progression of bladder cancer partially by stabilizing RUNX2 protein.
Figure 5.
Overexpression of RUNX2 could partially restore the reduction in proliferation and migration caused by decreased MAPK14/P-MAPK14 levels (A) Cells overexpression of RUNX2 protein and control group (Vector) were transfected with MAPK14 siRNA or NC, the MAPK14 and RUNX2 proteins were detected by Western blot; (B) Proliferation ability of T24 and UMUC3 cells was determined by EdU assay (magnification × 200); (C) Transwell assay was used to examine the migration ability of T24 and UMUC3 cells (magnification × 20). (*P < 0.05, **P < 0.01, ***P < 0.001).
Discussion
It has been estimated that P38 MAPK can phosphorylate a variety of proteins and may have about 200 to 300 substrates, including many transcription factors. Some targets are downstream kinases that could be activated by phosphorylation. MAPK14, one of the four P38 MAPK, plays an important role in the cellular cascade caused by extracellular stimuli, such as pro-inflammatory cytokines or physiological stress.21–23 As a crucial member of the P38 MAPK family, MAPK14 is highly expressed in a variety of cells and the main form of activation is phosphorylated MAPK14 (P-MAPK14, Thr180/Tyr182). Our previous in vivo and in vitro studies have shown that knockdown of MAPK14 can inhibit the proliferation and migration of renal cell clear cell carcinoma.24
Although the transcription level of MAPK14 analyzed in the TCGA database was low in bladder urothelial carcinoma. This result was likely due to the limited number of normal tissue samples. Our results identified that P-MAPK14 plays an important role in the proliferation and migration of bladder cancer cells. In the present study, P-MAPK14 protein expression was detected in bladder cancer tissues and adjacent normal tissues, four bladder cancer cell lines, and one normal urinary epithelial cell line by Western blot assay. The results showed that P-MAPK14 was highly expressed in both bladder cancer tissues and bladder cancer cell lines. It has been reported that benzethonium chloride can inhibit the proliferation and promote apoptosis of lung cancer cells by activating P38 MAPK.25 In addition, the role of P-MAPK14 in bladder cancer cells in T24 and UMUC3 was explored in this study. It was found that after the abundance of P-MAPK14 protein was reduced by MAPK14 siRNA, the clone formation ability and proliferation and migration ability of cells were significantly decreased. Considering that its main functional form is P-MAPK14, and the protein was highly expressed in both bladder cancer tissues and bladder cancer cell lines. It could be hypothesized that P-MAPK14 might promote the development of bladder cancer.
RUNX2 is the most critical transcription factor when bone marrow mesenchymal stem cells (BMSCS) differentiate and mature into osteoblasts in the process of bone development.26 Previous studies have shown that RUNX2 is a major regulator of tumorigenesis and is associated with tumor invasion.27,28 In urothelial carcinoma of the bladder, it has been reported that microRNA-154 and MicroRNA-217 can directly bind to RUNX2 mRNA, then inhibit the expression of RUNX2 protein, resulting in the suppression of bladder cancer progression.15,29 In recent years, many studies have reported a correlation between MAPKs and RUNX2 in tumors.30,31 Further study has shown that P38 MAPK can phosphorylate and activate RUNX2.32 In this study, surprisingly, it was found that the transcription level of RUNX2 was increased by the downregulation of MAPK14 and P-MAPK14 proteins. However, RUNX2 protein level was not consistent with the transcriptional level. It could be inferred that the increased transcriptional level of RUNX2 might be provoked by the internal homeostasis due to the decreased RUNX2 protein level. Since the RUNX2 protein level decreased after MAPK14 was downregulated, the inhibitor of P-MAPK14 was used to further investigate the reasons for the downregulation of RUNX2 protein. It was noted that RUNX2 protein level was significantly reduced, indicating that P-MAPK14 was likely involved in regulating RUNX2 protein stability. At the same time, it was also found that P-MAPK14 and RUNX2 could be bind to each other, which is consistent with the results of bone formation and bone microenvironment studies in mice.33
RUNX2 protein degradation pathway has been reported and most studies indicate that it is degraded by ubiquitination pathway.34,35 In order to detect whether the protein degradation of RUNX2 was mediated by proteasome, proteasome inhibitors MG-132 was used in this study.36 It was found that MG-132 could effectively increase the expression of RUNX2 protein, in addition, the protein level of RUNX2 in the absence of MAPK14 protein could be recovered to some extent. It has been reported that MAPK14 could also be degraded by ubiquitination,37 but this phenomenon was not found in our study. After bladder cancer cells were incubated with MG-132, there was no significant difference in protein level of MAPK14 as compared with the control group. Moreover, cycloheximide tracing experiment showed that the degradation rate of RUNX2 protein was significantly accelerated in the absence of MAPK14 protein. In summary, P-MAPK14 might activate and maintain the stability of the RUNX2 protein by binding and phosphorylating RUNX2. Finally, it was found that the ability of bladder cancer cells proliferation and migration was partially restored by protein rescue experiment.
In conclusion, P-MAPK14 is highly expressed in bladder cancer tissues, and P-MAPK14 can promote cell proliferation and migration. Our results also show that RUNX2 is a substrate of P-MAPK14, and P-MAPK14 can prevent RUNX2 degradation by binding to it. However, our data do not confirm that downregulation of MAPK14 and P-MAPK14 lead to the increase of RUNX2 transcription and the mechanism of P-MAPK14 regulating RUNX2 ubiquitination pathway degradation need further study. Considering that inhibitors of P-MAPK14 have been identified, these findings might provide a theoretical basis for the treatment of bladder cancer.
Funding Statement
This study was supported by Shenyang Plan Project of Science and Technology (Grant No. F19-112-4-098), China Medical University’s 2017 discipline promotion program (Grant No. 3110117040), and 2017 National Key R&D Program Key Projects of Precision Medical Research (2017YFC0908000).
Data Sharing Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Ethics Approval and Consent to Participate
The Ethics Committee of the First Hospital of China Medical University (Shenyang, China) approved the use of human tissue samples for experiments. All participants provided written informed consent for the entire study.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Disclosure
The authors declare that they have no conflict of interests.
References
- 1.Albarakati N, Khayyat D, Dallol A, Al-Maghrabi J, Nedjadi T. The prognostic impact of GSTM1/GSTP1 genetic variants in bladder cancer. BMC Cancer. 2019;19:991. doi: 10.1186/s12885-019-6244-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019;69(1):7–34. doi: 10.3322/caac.21551 [DOI] [PubMed] [Google Scholar]
- 3.National Institute for Health and Care Excellence. Bladder cancer: diagnosis and management of bladder cancer: © NICE (2015) bladder cancer: diagnosis and management of bladder cancer. BJU Int. 2017;120(6):755–765. doi: 10.1111/bju.14045 [DOI] [PubMed] [Google Scholar]
- 4.Babjuk M, Burger M, Compérat EM, et al. European association of urology guidelines on non-muscle-invasive bladder cancer (tat1 and carcinoma in situ) - 2019 update. Eur Urol. 2019;76(5):639–657. [DOI] [PubMed] [Google Scholar]
- 5.Mai L, Zhu X, Huang F, He H, Fan W. p38 mitogen-activated protein kinase and pain. Life Sci. 2020;256:117885. doi: 10.1016/j.lfs.2020.117885 [DOI] [PubMed] [Google Scholar]
- 6.Han J, Wu J, Silke J. An overview of mammalian p38 mitogen-activated protein kinases, central regulators of cell stress and receptor signaling. F1000Res. 2020;9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zheng T, Zhang B, Chen C, et al. Protein kinase p38α signaling in dendritic cells regulates colon inflammation and tumorigenesis. Proc Natl Acad Sci U S A. 2018;115(52):E12313–12313 E12322. doi: 10.1073/pnas.1814705115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cánovas B, Igea A, Sartori AA, et al. Targeting p38α increases DNA damage, chromosome instability, and the anti-tumoral response to taxanes in breast cancer cells. Cancer Cell. 2018;33(6):1094–1110.e8. doi: 10.1016/j.ccell.2018.04.010 [DOI] [PubMed] [Google Scholar]
- 9.Gupta J, Del Barco Barrantes I, Igea A, et al. Dual function of p38α MAPK in colon cancer: suppression of colitis-associated tumor initiation but requirement for cancer cell survival. Cancer Cell. 2014;25(4):484–500. doi: 10.1016/j.ccr.2014.02.019 [DOI] [PubMed] [Google Scholar]
- 10.Hendrickx N, Dewaele M, Buytaert E, et al. Targeted inhibition of p38alpha MAPK suppresses tumor-associated endothelial cell migration in response to hypericin-based photodynamic therapy. Biochem Biophys Res Commun. 2005;337(3):928–935. doi: 10.1016/j.bbrc.2005.09.135 [DOI] [PubMed] [Google Scholar]
- 11.Wu YJ, Su TR, Dai GF, Su JH, Liu CI. Flaccidoxide-13-acetate-induced apoptosis in human bladder cancer cells is through activation of p38/JNK, mitochondrial dysfunction, and endoplasmic reticulum stress regulated pathway. Mar Drugs. 2019;17:5. doi: 10.3390/md17050287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shin SS, Ko MC, Park YJ, et al. Hydrangenol inhibits the proliferation, migration, and invasion of EJ bladder cancer cells via p21WAF1-mediated G1-phase cell cycle arrest, p38 MAPK activation, and reduction in Sp-1-induced MMP-9 expression. EXCLI J. 2018;17:531–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Terauchi K, Kobayashi H, Yatabe K, et al. The NAD-dependent deacetylase sirtuin-1 regulates the expression of osteogenic transcriptional activator runt-related transcription factor 2 (runx2) and production of matrix metalloproteinase (MMP)-13 in chondrocytes in osteoarthritis. Int J Mol Sci. 2016;17:7. doi: 10.3390/ijms17071019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lin X, Chen Q, Xiao Y, et al. Phosphate glass fibers facilitate proliferation and osteogenesis through Runx2 transcription in murine osteoblastic cells. J Biomed Mater Res A. 2020;108(2):316–326. doi: 10.1002/jbm.a.36818 [DOI] [PubMed] [Google Scholar]
- 15.Zhao X, Ji Z, Xie Y, Liu G, Li H. MicroRNA-154 as a prognostic factor in bladder cancer inhibits cellular malignancy by targeting RSF1 and RUNX2. Oncol Rep. 2017;38(5):2727–2734. doi: 10.3892/or.2017.5992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ge C, Zhao G, Li Y, et al. Role of Runx2 phosphorylation in prostate cancer and association with metastatic disease. Oncogene. 2016;35(3):366–376. doi: 10.1038/onc.2015.91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wen S, Zeng M, Li Y, et al. Downregulation of MANCR inhibits cancer cell proliferation in mantle cell lymphoma possibly by interacting with RUNX2. Acta Biochim Biophys Sin. 2019;51(11):1142–1147. [DOI] [PubMed] [Google Scholar]
- 18.Abdelzaher E, Kotb AF. High coexpression of runt-related transcription factor 2 (RUNX2) and p53 independently predicts early tumor recurrence in bladder urothelial carcinoma patients. Appl Immunohistochem Mol Morphol. 2016;24(5):345–354. doi: 10.1097/PAI.0000000000000193 [DOI] [PubMed] [Google Scholar]
- 19.Zhang FL, Cao JL, Xie HY, et al. Cancer-associated MORC2-mutant M276I regulates an hnRNPM-mediated CD44 splicing switch to promote invasion and metastasis in triple-negative breast cancer. Cancer Res. 2018;78(20):5780–5792. [DOI] [PubMed] [Google Scholar]
- 20.Zhang W, Guo H, Jing H, et al. Lactoferrin stimulates osteoblast differentiation through PKA and p38 pathways independent of lactoferrin’s receptor LRP1. J Bone Miner Res. 2014;29(5):1232–1243. [DOI] [PubMed] [Google Scholar]
- 21.Gills JJ, Castillo SS, Zhang C, et al. Phosphatidylinositol ether lipid analogues that inhibit AKT also independently activate the stress kinase, p38alpha, through MKK3/6-independent and -dependent mechanisms. J Biol Chem. 2007;282(37):27020–27029. doi: 10.1074/jbc.M701108200 [DOI] [PubMed] [Google Scholar]
- 22.Igea A, Nebreda AR. The stress kinase p38α as a target for cancer therapy. Cancer Res. 2015;75(19):3997–4002. doi: 10.1158/0008-5472.CAN-15-0173 [DOI] [PubMed] [Google Scholar]
- 23.Campbell RM, Anderson BD, Brooks NA, et al. Characterization of LY2228820 dimesylate, a potent and selective inhibitor of p38 MAPK with antitumor activity. Mol Cancer Ther. 2014;13(2):364–374. doi: 10.1158/1535-7163.MCT-13-0513 [DOI] [PubMed] [Google Scholar]
- 24.Liu J, Yu X, Yu H, et al. Knockdown of MAPK14 inhibits the proliferation and migration of clear cell renal cell carcinoma by downregulating the expression of CDC25B. Cancer Med. 2020;9(3):1183–1195. doi: 10.1002/cam4.2795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang XH, Wang Y, Hong P, et al. Benzethonium chloride suppresses lung cancer tumorigenesis through inducing p38-mediated cyclin D1 degradation. Am J Cancer Res. 2019;9(11):2397–2412. [PMC free article] [PubMed] [Google Scholar]
- 26.Yang JX, Xie P, Li YS, Wen T, Yang XC. Osteoclast-derived miR-23a-5p-containing exosomes inhibit osteogenic differentiation by regulating Runx2. Cell Signal. 2020;70:109504. doi: 10.1016/j.cellsig.2019.109504 [DOI] [PubMed] [Google Scholar]
- 27.Wang ZQ, Keita M, Bachvarova M, et al. Inhibition of RUNX2 transcriptional activity blocks the proliferation, migration and invasion of epithelial ovarian carcinoma cells. PLoS One. 2013;8(10):e74384. doi: 10.1371/journal.pone.0074384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Niu DF, Kondo T, Nakazawa T, et al. Transcription factor Runx2 is a regulator of epithelial-mesenchymal transition and invasion in thyroid carcinomas. Lab Invest. 2012;92(8):1181–1190. doi: 10.1038/labinvest.2012.84 [DOI] [PubMed] [Google Scholar]
- 29.Huang W, Lu Y, Wang F, Huang X, Yu Z. Downregulation of circular RNA hsa_circ_0000144 inhibits bladder cancer progression via stimulating miR-217 and suppressing RUNX2 expression. Gene. 2018;678:337–342. doi: 10.1016/j.gene.2018.08.036 [DOI] [PubMed] [Google Scholar]
- 30.Xin BC, Wu QS, Jin S, Luo AH, Sun DG, Wang F. Berberine promotes osteogenic differentiation of human dental pulp stem cells through activating EGFR-MAPK-runx2 pathways. Pathol Oncol Res. 2020;26(3):1677–1685. doi: 10.1007/s12253-019-00746-6 [DOI] [PubMed] [Google Scholar]
- 31.Artigas N, Ureña C, Rodríguez-Carballo E, Rosa JL, Ventura F. Mitogen-activated protein kinase (MAPK)-regulated interactions between Osterix and Runx2 are critical for the transcriptional osteogenic program. J Biol Chem. 2014;289(39):27105–27117. doi: 10.1074/jbc.M114.576793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Greenblatt MB, Shim JH, Glimcher LH. Mitogen-activated protein kinase pathways in osteoblasts. Annu Rev Cell Dev Biol. 2013;29:63–79. doi: 10.1146/annurev-cellbio-101512-122347 [DOI] [PubMed] [Google Scholar]
- 33.Greenblatt MB, Shim JH, Zou W, et al. The p38 MAPK pathway is essential for skeletogenesis and bone homeostasis in mice. J Clin Invest. 2010;120(7):2457–2473. doi: 10.1172/JCI42285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chava S, Chennakesavulu S, Gayatri BM, Reddy A. A novel phosphorylation by AMP-activated kinase regulates RUNX2 from ubiquitination in osteogenesis over adipogenesis. Cell Death Dis. 2018;9(7):754. doi: 10.1038/s41419-018-0791-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhu W, He X, Hua Y, Li Q, Wang J, Gan X. The E3 ubiquitin ligase WWP2 facilitates RUNX2 protein transactivation in a mono-ubiquitination manner during osteogenic differentiation. J Biol Chem. 2017;292(27):11178–11188. doi: 10.1074/jbc.M116.772277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lu Q, Lu D, Shao ZM, Li DQ. Deubiquitinase ubiquitin-specific protease 9X regulates the stability and function of E3 ubiquitin ligase ring finger protein 115 in breast cancer cells. Cancer Sci. 2019;110(4):1268–1278. doi: 10.1111/cas.13953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu Q, Zhang S, Chen G, Zhou H. E3 ubiquitin ligase Nedd4 inhibits AP-1 activity and TNF-α production through targeting p38α for polyubiquitination and subsequent degradation. Sci Rep. 2017;7(1):4521. doi: 10.1038/s41598-017-04072-2 [DOI] [PMC free article] [PubMed] [Google Scholar]