Abstract
The tumor mutational burden (TMB) has been reported as a predictive marker of the response to immune checkpoint inhibition (ICI) therapy in previous melanoma clinical trials. However, the TMB alone is not sufficient to accurately predict immunotherapy benefit. Additional biomarkers are needed for better stratification of immunotherapy-sensitive patients. In the present study, mutation data and survival information of patients with melanoma were collected from several immunotherapy studies, and tumor heterogeneity was estimated using mutant-allele tumor heterogeneity (MATH). The benefit score was defined as the ratio between the TMB and tumor heterogeneity, and optimal critical values were selected to group patients and evaluate their response to ICI treatment. The benefit score significantly improved the performance of stratifying the overall survival of patients compared with the TMB alone as a predictor in two independent cohorts (p = 0.0068 vs. p = 0.1 and p = 0.045 vs. p = 0.13), in which patients were treated with Ipilimumab and Nivolumab, respectively. In another cohort of patients with melanoma receiving mixed ICI treatment, the benefit score was also positively associated with higher overall survival (p = 0.022) and outperformed the TMB alone, with a significance of p = 0.089. The benefit score showed a positive correlation with clonal TMB, a reported immunotherapy marker, and exceeded it in immunotherapy response prediction. Besides, a high benefit score was found to be associated with higher proportions of natural killer cells, lower proportions of M2 macrophages and elevated CD8 T cells, all of which favor ICI therapy. In summary, tumor heterogeneity combined with the TMB showed superior efficacy in predicting the response to ICI therapy. This might further help to delineate the mechanisms of immunotherapy in patients with melanoma.
Keywords: tumor mutational burden, tumor heterogeneity, immune checkpoint inhibition, melanoma, benefit score
Introduction
In recent years, immune checkpoint inhibition (ICI) therapy has benefited patients with advanced cancers, as exemplified for lung cancer (1) and melanoma (2). The identification of predictive markers and the mechanisms of resistance to immunotherapy have been subjects of intense research, because only a minority of patients are responsive to checkpoint blockers. Several markers have been proposed, including tumor-infiltrating lymphocytes (3), programmed cell death protein 1 (PD-1) and PD-1 ligand (PD-L1) expression (4), or mutational load (5). A difference in intratumoral PD-L1 expression was observed to contribute to discrepancies in ICI therapy trials and was approved by the FDA as a companion diagnostic for clinical use (6). The tumor mutational burden (TMB) was reported recently as a predictor of pan-cancer survival (7). However, none of these markers has been fully validated to perfectly predict the response to ICI (8, 9).
Among all identified markers, the TMB has proven its potential to predict the response of ICI in melanoma, based on the hypothesis that an elevated TMB corresponds to more neoantigens (10–12). However, it was also found that the effectiveness of the TMB was confounded by disease subtype, whereas a selective response was noticed between different genomic features (13). Putative biomarkers, such as tumor heterogeneity, have been reported to contribute to this variation in the immune response (14). Ultraviolet beta (UVB)-induced heterogeneity was confirmed to be able to diminish the immune response in melanoma, which contributes to loss of efficacy of checkpoint blockade (15), suggesting the potential of tumor heterogeneity as a candidate marker to achieve better performance in ICI response prediction, together with the TMB (16).
In the present study, the TMB and tumor heterogeneity were estimated using somatic mutation data collected from several studies, in which patients with melanoma were treated with checkpoint inhibitors. Next, we defined the benefit score as a metric to integrate the TMB and tumor heterogeneity, and evaluated its predictive performance for the response to ICI therapy.
Materials and Methods
Data Collection
After excluding studies without sufficient data, three melanoma cohorts that employed ICI therapy were brought into the analysis: The Van Allen cohort (12), the Miao cohort (17), and the Riaz cohort (18). Genomic profiling based on whole exome sequencing (WES) and clinical characteristics, such as age, sex, tumor stage, and survival information were gathered from publicly accessible resources. Somatic mutations and copy number segmentation data of 206 patients with melanoma profiled by The Cancer Genome Atlas (TCGA) were obtained from a previous pan-cancer report (19).
The Benefit Score
We defined the benefit score as the ratio between the TMB and tumor heterogeneity, where the TMB denoted the number of nonsynonymous somatic mutations and heterogeneity was determined using mutant-allele tumor heterogeneity (MATH), as calculated using the R package, maftools (20). For the convenience of calculation, MATH was set to 1 when its value was equal to zero. Mutations with allelic fractions less than 0.05 or a coverage of no more than 30 were excluded, as described previously (12).
Clonal Tumor Mutation Load
The clonal TMB, representing the number of clonal nonsynonymous somatic mutations, is a measure used to evaluate the clonal tumor mutation load of patients. In the Riaz cohort, cancer cell fraction (CCF) of mutations was estimated using PyClone (21), and mutations with a lower confidential interval of CCF exceeded 95% were defined as clonal mutations. In the Miao cohort, mutational clonality was assessed using ABSOLUTE (22), and mutations with a CCF of 1, or those whose probability of being clonal exceeded the probability of being subclonal, were identified clonal. Clonal mutations in the TCGA dataset were determined using the same method as in the Miao cohort.
Statistical Analysis
Correlation analysis between the TMB and MATH, as well as the benefit score and the clonal TMB, was implemented using the Spearman method. Patients in each cohort were stratified into two groups based on the critical value of the benefit score inferred using the function surv_cutpoint in the R package survival, using the maximally selected log-rank statistic; the log-rank test was also performed. The prognostic significance of the benefit score was examined using a Cox multivariate regression model that incorporated clinical and molecular features, and the hazard ratios (HRs) were calculated. Fraction data of each tumor immune cell type in the Riaz cohort were downloaded from The Cancer Imaging Archive (TCIA) (https://tcia.at/home) (8). Cell fractions before treatment and fraction changes during treatment were compared between the high and low benefit score sets using a two-tailed Wilcoxon test.
Results
The Benefit Score as a Metric for Integration of the TMB and Tumor Heterogeneity
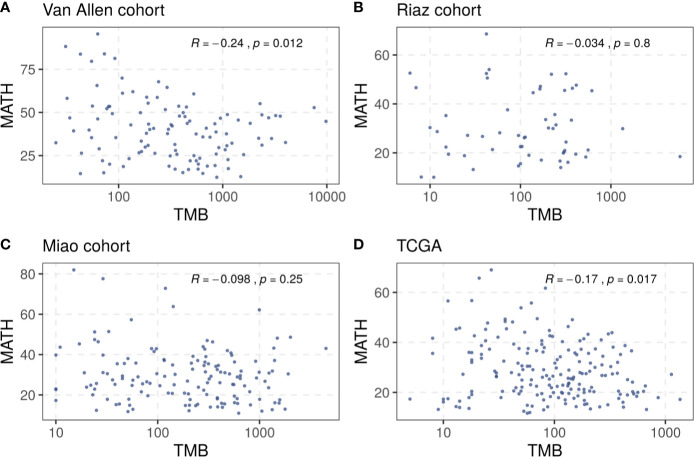
A total of 321 patients with melanoma from three cohorts, the Van Allen cohort, the Riaz cohort, and the Miao cohort, were included in our investigation ( Supplementary Table S1 ). The TMB and tumor heterogeneity were measured independently using the data collected from each cohort. A small number of samples, four in the Riaz cohort and nine in the Miao cohort, were excluded from subsequent analysis because the tumor heterogeneity could not be estimated due to an inadequate number of mutations. The TMB did not correlate clearly with the MATH value in these cohorts and a similar result was observed in the TCGA melanoma dataset ( Figure 1 ), rationalizing the integration of these two factors to merge their prediction effects. Consequently, we defined the benefit score as the TMB divided by the MATH value as a proxy for tumor heterogeneity.
Figure 1.
Correlation between TMB and tumor heterogeneity in (A) the Van Allen cohort, (B) the Riaz cohort, (C) the Miao cohort, and (D) the TCGA dataset. Spearman correlation coefficients and statistical significance are labeled at the top right in each part.
The Association Between the Benefit Score and the Clonal TMB
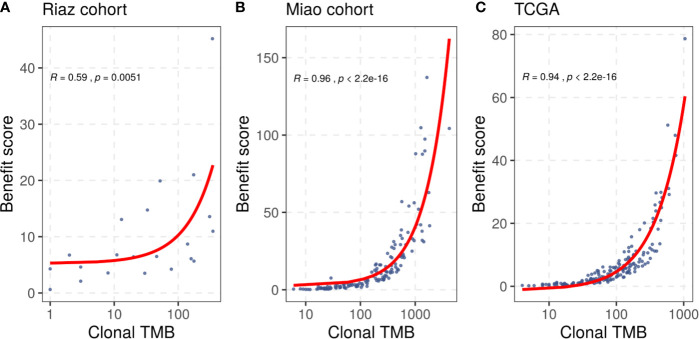
In addition to the benefit score, the clonal TMB is also simultaneously related to the TMB and tumor heterogeneity, and was reported to be associated with the response of patients treated with checkpoint blockers. We compared the benefit score with the clonal TMB based on the clonal results collected from the included studies and the correlation result is shown in Figure 2 . We found that the benefit score correlated positively with the clonal TMB in the Riaz cohort (R = 0.59, p = 0.0051) and the Miao cohort (R = 0.96, p < 0.001). A similar trend was observed in the TCGA melanoma dataset (R = 0.94, p < 0.001). The diversity in the significance of the correlation between the Riaz cohort and the other two cohorts might have been caused by the number of patients and approach used to estimate clonality.
Figure 2.
Correlation between the benefit score and clonal TMB in (A) the Riaz cohort, (B) the Miao cohort, and (C) the TCGA dataset. Spearman correlation coefficients and statistical significance are labeled at the top left in each part.
The Benefit Score Yields a Better ICI Response Prediction Performance
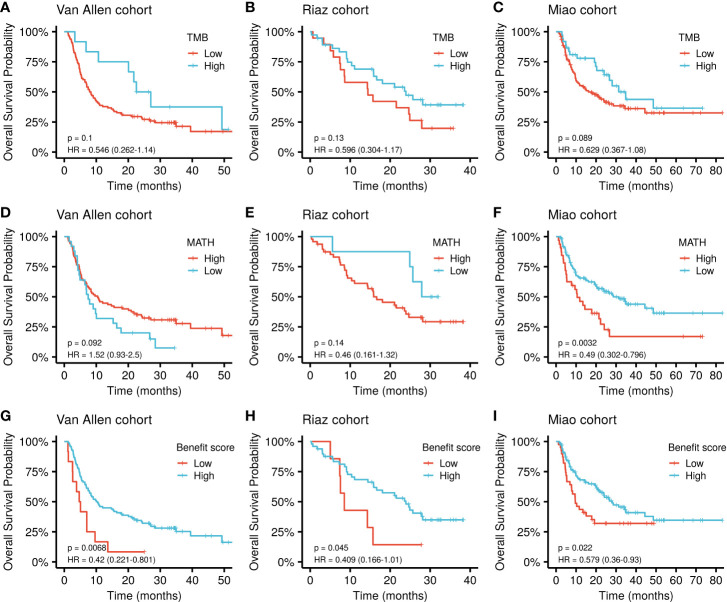
The cut-off value for each marker was determined based on the principle of comparing their best performance to stratify the patients ( Supplementary Table S2 ). The benefit score showed a significantly improved performance in stratifying the overall survival (OS) of patients receiving ICI treatment compared with that of the TMB alone as a predictor in the Van Allen cohort (p = 0.0068, HR = 0.42 vs. p = 0.1, HR = 0.546; Figure 3A ). The same results were obtained using the Riaz cohort (p = 0.045, HR = 0.409 vs. p = 0.13, HR = 0.596; Figure 3B ) and the Miao cohort (p = 0.022, HR = 0.579 vs. p = 0.089, HR = 0.629; Figure 3C ). Meanwhile, the benefit score also exceeded the ability of MATH to predicting higher OS in the Van Allen cohort (p = 0.0068, HR = 0.42 vs. p = 0.092, HR = 1.52; Figure 3D ) and the Riaz cohort (p = 0.045, HR = 0.409 vs. p = 0.14, HR = 0.46; Figure 3E ). Only in the Miao cohort, MATH was superior to the benefit score (p = 0.0032, HR = 0.49 vs. p = 0.022, HR = 0.579; Figure 3F ). Survival analysis result of the benefit score was is shown in Figures 3G–I . In addition, the superiority of the benefit score over the clonal TMB was demonstrated in the Riaz cohort (p = 0.045, HR = 0.409 vs. p = 0.33, HR = 0.664; Supplementary Figure S1A ) and the Miao cohort (p = 0.022, HR = 0.579 vs. p = 0.037, HR = 0.624; Supplementary Figure S1B ). A slight difference, without statistical significance, was observed when simultaneously considering the clonal TMB and the benefit score, indicating synergistic effects might exist between these two features ( Supplementary Figures S1C, D ).
Figure 3.
Kaplan-Meier curve of overall survival by TMB, MATH, and benefit scores in different datasets. (A–C) Survival curves for the TMB in each cohort. (D–F) Survival curves for MATH in each cohort. (G–I) Survival curves for the benefit score in each cohort. P values of the log-rank test are shown at the top right for each curve.
Cox multivariate regression was implemented to verify the benefit score’s prediction of ICI therapy response, independently of other clinical or molecular factors. Relevant covariates including age, sex, tumor stage, previous treatment, and driver gene mutations, together with the benefit score, were incorporated into the model. The result demonstrated that a low benefit score was independently associated with shorter OS, after adjusting for age, sex, and tumor stage in the Van Allen cohort (HR = 2.16, p = 0.02, Table 1 ). Despite that fact that a similar result was not significant in the other two cohorts, a lower benefit score still had a higher hazard ratio when correcting for age and sex in the Miao cohort (HR = 1.30, p = 0.308, Table 1 ), and while controlling for BRAF, NF1, and RAS mutations, and previous Ipilimumab treatment, in the Riaz cohort (HR = 2.31, p = 0.085, Table 1 ). The benefit score outperformed the TMB, with more significantly higher hazard ratios, as estimated by multivariate regression, in all cohorts ( Supplementary Table S3 ).
Table 1.
Multivariate Cox regression analysis of overall survival with the benefit score and other covariates in different cohorts.
| Cohort | Covariates | HR | 95% CI | P-value |
|---|---|---|---|---|
| Van Allen cohort | Age | 1.00 | 0.99–1.02 | 0.629 |
| Sex | ||||
| Male | 0.73 | 0.45–1.18 | 0.200 | |
| Stage | ||||
| IV | 4.20 | 1.31–13.5 | 0.016* | |
| Benefit score | ||||
| Low | 2.16 | 1.13–4.12 | 0.020* | |
| Riaz cohort | Mutation Subtype | |||
| BRAF | 0.90 | 0.40–2.04 | 0.802 | |
| NF1 | 1.16 | 0.32–4.16 | 0.822 | |
| RAS | 0.68 | 0.27–1.73 | 0.420 | |
| Prior treatment | ||||
| Yes | 0.89 | 0.44–1.78 | 0.738 | |
| Benefit score | ||||
| Low | 2.31 | 0.89–6.01 | 0.085 | |
| Miao cohort | Age | 1.01 | 1.00–1.03 | 0.135 |
| Sex | ||||
| Male | 0.77 | 0.45–1.32 | 0.344 | |
| Benefit score | ||||
| Low | 1.30 | 0.79–2.14 | 0.308 |
HR, Hazard Ratio; CI, Confidence Interval; *indicates statistical significance.
The Benefit Score and the Tumor Microenvironment
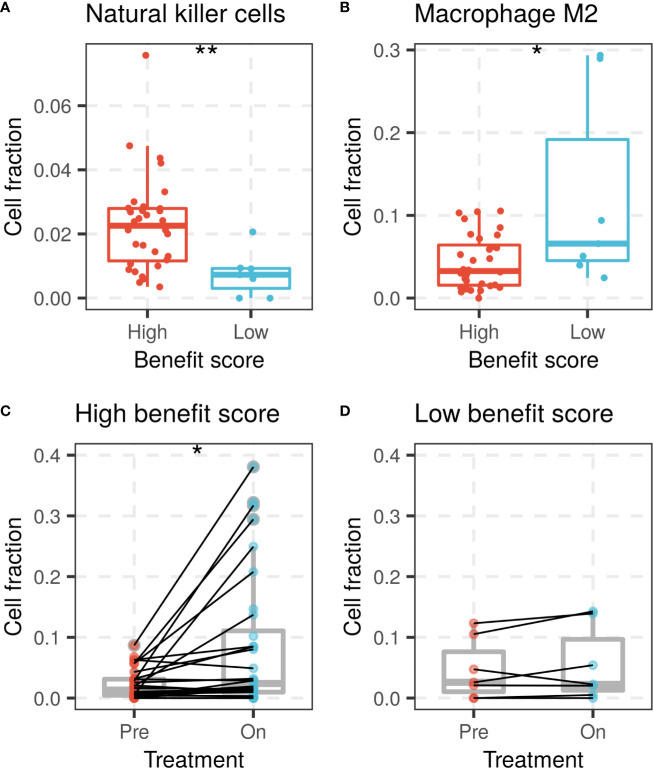
The benefit score was also investigated for its association with variation in the tumor microenvironment, mainly the immune cell composition. Immune cell proportions were calculated from gene expression profiles and compared between the high and low benefit score groups in the Riaz cohort. A high benefit score tumor was significantly associated with a higher proportion of activated natural killer (NK) cells and a lower proportion of M2 macrophages compared with those in the low benefit score group (p = 0.0041 and p = 0.049, respectively, Figures 4A, B ). A significant elevation of the CD8 T cell fraction during treatment was found in the high benefit score group, but not in the low benefit score group (p = 0.014 and p = 0.29, respectively, Figures 4C, D ). The observation that higher proportions of NK cells, lower proportions of M2 macrophages and elevated CD8 T cells are favorable factors for immunotherapy supports the result that the benefit score is a remarkable predictor.
Figure 4.
NK cells and macrophage M2 fractions of patients between high and low benefit score groups in the Riaz cohort. (A) NK cell fraction of patients grouped by benefit score is shown by a box plot at left, while (B) the macrophage M2 fraction is shown on the right side. CD8 T cell fraction change during ICI treatment in (C) high benefit score group and (D) low benefit score group. (*p < 0.05, **p < 0.01).
Discussion
The ability of the TMB to predict the response to immunotherapy response has been reported to be slightly restrictive in recent research (13, 23). In the present study, we combined the TMB with tumor heterogeneity to form the benefit score as a new indicator to achieve a better predictive effect compared with that of the TMB alone. The observed prognostic significance was consistent with previous reports, proving that integration of the TMB and tumor heterogeneity forms a superior predictive marker for the response to ICI treatment.
A high TMB is a relative measure of a tumor being more immunogenic by producing more neoantigens. However, the effectiveness of the antitumor immune response would be greatly reduced if the presented antigens were subclone-specific, because immune surveillance is promoted by clonal neoantigens (14). The chance of a neoantigen being clonal cannot be reflected by the TMB and high neoantigen heterogeneity usually originates from a more complicated tumor evolutionary structure. Our benefit score covers both the neoantigen burden and clonality, without direct clonal neoantigen measurement. A high benefit score corresponds to either a high mutational load, low genetic heterogeneity, or both, i.e., it reflects a high neoantigen load and the avoidance of immune evasion mediated by tumor heterogeneity, which would likely be more sensitive to immunotherapy. Similar to our benefit score, the clonal TMB considers clonality and the neoantigen burden simultaneously. It also strongly correlates with the benefit score, except that the clonal TMB showed a poorer performance than the benefit score to predict the ICI response, because the immune response induced by tumor-specific epitopes generated from subclones are neglected by the clonal TMB.
It has been reported that NK cells contribute to PD-1/PD-L1 antibody-mediated immunotherapy by expressing PD-L1 as a cytolytic effector (24, 25), while M2 macrophages help to generate an immune-evasive microenvironment in evolved cancers (26, 27). A high benefit score was found to be associated with a higher proportion of NK cells, a lower proportion of M2 macrophages and elevated CD8 T cells in the Riaz cohort, in which patients were treated with Nivolumab, in accordance with its favorable survival prediction.
Despite of significant results achieved in the present study, a disadvantage common in most cases involving heterogeneity evaluation was that genetic diversity might not be adequately estimated using in silico methods in a single tumor region sample, which could lead to unstable results. Such a limitation, together with the small cohort, might have interfered with the robustness of the multivariate analysis, resulting in an insignificant outcome. Analysis of multiple regions or single cell sequencing in a larger cohort would be good methods to validate the prognostic value of our findings in the future.
In summary, our study suggested that integration of the tumor mutational load and heterogeneity provides a better predictive marker of the response to ICI treatment for patients with melanoma. The potential significance of the benefit score could be further investigated and applied using more sophisticated approaches in other types of cancer.
Data Availability Statement
The original contributions presented in the study are included in the article/ Supplementary Material . Further inquiries can be directed to the corresponding authors.
Author Contributions
YG, CY, YY, and GZ conceived and designed the study. YG, CY, NH, YY, and JW conducted data collection, analysis, and interpretation. YG and CY wrote the manuscript. All authors contributed to the article and approved the submitted version.
Conflict of Interest
Authors CY, NH, GZ, JW, and YY were employed by the company GloriousMed Clinical Laboratory (Shanghai) Co., Ltd.
The remaining author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The handling editor declared a past co-authorship with one of the authors YY.
Acknowledgments
The authors would like to thank the cBioPortal database and others involved research for their publicly available data.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2020.571545/full#supplementary-material
References
- 1. Borghaei H, Paz-Ares L, Horn L, Spigel DR, Steins M, Ready NE, et al. Nivolumab versus Docetaxel in Advanced Nonsquamous Non-Small-Cell Lung Cancer. N Engl J Med (2015) 373(17):1627–39. 10.1056/NEJMoa1507643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Hodi FS, O’Day SJ, McDermott DF, Weber RW, Sosman JA, Haanen JB, et al. Improved survival with ipilimumab in patients with metastatic melanoma. N Engl J Med (2010) 363(8):711–23. 10.1056/NEJMoa1003466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Tang H, Wang Y, Chlewicki LK, Zhang Y, Guo J, Liang W, et al. Facilitating T Cell Infiltration in Tumor Microenvironment Overcomes Resistance to PD-L1 Blockade. Cancer Cell (2016) 30(3):500. 10.1016/j.ccell.2016.08.011 [DOI] [PubMed] [Google Scholar]
- 4. Patel SP, Kurzrock R. PD-L1 Expression as a Predictive Biomarker in Cancer Immunotherapy. Mol Cancer Ther (2015) 14(4):847–56. 10.1158/1535-7163.MCT-14-0983 [DOI] [PubMed] [Google Scholar]
- 5. Rizvi NA, Hellmann MD, Snyder A, Kvistborg P, Makarov V, Havel JJ, et al. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science (2015) 348(6230):124–8. 10.1126/science.aaa1348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Garon EB, Rizvi NA, Hui R, Leighl N, Balmanoukian AS, Eder JP, et al. Pembrolizumab for the Treatment of Non–Small-Cell Lung Cancer. N Engl J Med (2015) 372(21):2018–28. 10.1056/NEJMoa1501824 [DOI] [PubMed] [Google Scholar]
- 7. Samstein RM, Lee CH, Shoushtari AN, Hellmann MD, Shen R, Janjigian YY, et al. Tumor mutational load predicts survival after immunotherapy across multiple cancer types. Nat Genet (2019) 51(2):202–6. 10.1038/s41588-018-0312-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Charoentong P, Finotello F, Angelova M, Mayer C, Efremova M, Rieder D, et al. Pan-cancer Immunogenomic Analyses Reveal Genotype-Immunophenotype Relationships and Predictors of Response to Checkpoint Blockade. Cell Rep (2017) 18(1):248–62. 10.1016/j.celrep.2016.12.019 [DOI] [PubMed] [Google Scholar]
- 9. Spencer KR, Wang J, Silk AW, Ganesan S, Kaufman HL, Mehnert JM. Biomarkers for Immunotherapy: Current Developments and Challenges. Am Soc Clin Oncol Educ Book (2016) 35:e493–503. 10.1200/EDBK_160766 [DOI] [PubMed] [Google Scholar]
- 10. Eroglu Z, Zaretsky JM, Hu-Lieskovan S, Kim DW, Algazi A, Johnson DB, et al. High response rate to PD-1 blockade in desmoplastic melanomas. Nature (2018) 553(7688):347–50. 10.1038/nature25187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Snyder A, Makarov V, Merghoub T, Yuan J, Zaretsky JM, Desrichard A, et al. Genetic basis for clinical response to CTLA-4 blockade in melanoma. N Engl J Med (2014) 371(23):2189–99. 10.1056/NEJMoa1406498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Van Allen EM, Miao D, Schilling B, Shukla SA, Blank C, Zimmer L, et al. Genomic correlates of response to CTLA-4 blockade in metastatic melanoma. Science (2015) 350(6257):207–11. 10.1126/science.aad0095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Liu D, Schilling B, Liu D, Sucker A, Livingstone E, Jerby-Amon L, et al. Integrative molecular and clinical modeling of clinical outcomes to PD1 blockade in patients with metastatic melanoma. Nat Med (2019) 25(12):1916–27. 10.1038/s41591-019-0654-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. McGranahan N, Furness AJ, Rosenthal R, Ramskov S, Lyngaa R, Saini SK, et al. Clonal neoantigens elicit T cell immunoreactivity and sensitivity to immune checkpoint blockade. Science (2016) 351(6280):1463–9. 10.1126/science.aaf1490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wolf Y, Bartok O, Patkar S, Eli GB, Cohen S, Litchfield K, et al. UVB-Induced Tumor Heterogeneity Diminishes Immune Response in Melanoma. Cell (2019) 179(1):219–35 e21. 10.1016/j.cell.2019.08.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kim K, Kim HS, Kim JY, Jung H, Sun JM, Ahn JS, et al. Predicting clinical benefit of immunotherapy by antigenic or functional mutations affecting tumour immunogenicity. Nat Commun (2020) 11(1):951. 10.1038/s41467-020-14562-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Miao D, Margolis CA, Vokes NI, Liu D, Taylor-Weiner A, Wankowicz SM, et al. Genomic correlates of response to immune checkpoint blockade in microsatellite-stable solid tumors. Nat Genet (2018) 50(9):1271–81. 10.1038/s41588-018-0200-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Riaz N, Havel JJ, Makarov V, Desrichard A, Urba WJ, Sims JS, et al. Tumor and Microenvironment Evolution during Immunotherapy with Nivolumab. Cell (2017) 171: (4):934–49 e16. 10.1016/j.cell.2017.09.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Raynaud F, Mina M, Tavernari D, Ciriello G. Pan-cancer inference of intra-tumor heterogeneity reveals associations with different forms of genomic instability. PLoS Genet (2018) 14(9):e1007669. 10.1371/journal.pgen.1007669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Mayakonda A, Lin DC, Assenov Y, Plass C, Koeffler HP. Maftools: efficient and comprehensive analysis of somatic variants in cancer. Genome Res (2018) 28(11):1747–56. 10.1101/gr.239244.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Roth A, Khattra J, Yap D, Wan A, Laks E, Biele J, et al. PyClone: statistical inference of clonal population structure in cancer. Nat Methods (2014) 11(4):396–8. 10.1038/nmeth.2883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Carter SL, Cibulskis K, Helman E, McKenna A, Shen H, Zack T, et al. Absolute quantification of somatic DNA alterations in human cancer. Nat Biotechnol (2012) 30(5):413–21. 10.1038/nbt.2203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hellmann MD, Ciuleanu TE, Pluzanski A, Lee JS, Otterson GA, Audigier-Valette C, et al. Nivolumab plus Ipilimumab in Lung Cancer with a High Tumor Mutational Burden. N Engl J Med (2018) 378(22):2093–104. 10.1056/NEJMoa1801946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hsu J, Hodgins JJ, Marathe M, Nicolai CJ, Bourgeois-Daigneault MC, Trevino TN, et al. Contribution of NK cells to immunotherapy mediated by PD-1/PD-L1 blockade. J Clin Invest (2018) 128(10):4654–68. 10.1172/JCI99317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Dong W, Wu X, Ma S, Wang Y, Nalin AP, Zhu Z, et al. The Mechanism of Anti-PD-L1 Antibody Efficacy against PD-L1-Negative Tumors Identifies NK Cells Expressing PD-L1 as a Cytolytic Effector. Cancer Discov (2019) 9(10):1422–37. 10.1158/2159-8290.CD-18-1259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Qian BZ, Pollard JW. Macrophage diversity enhances tumor progression and metastasis. Cell (2010) 141(1):39–51. 10.1016/j.cell.2010.03.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Mantovani A, Marchesi F, Malesci A, Laghi L, Allavena P. Tumour-associated macrophages as treatment targets in oncology. Nat Rev Clin Oncol (2017) 14(7):399–416. 10.1038/nrclinonc.2016.217 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The original contributions presented in the study are included in the article/ Supplementary Material . Further inquiries can be directed to the corresponding authors.