Abstract
Background
The prevalence and types of fibroblast growth factor receptor (FGFR) mutations vary significantly among different ethnic groups. The optimal application of FGFR inhibitors depends on these variations being comprehensively understood. However, such an analysis has yet to be conducted in Chinese patients.
Methods
We retrospectively screened the genomic profiling results of 10,582 Chinese cancer patients across 16 cancer types to investigate the frequency and distribution of FGFR aberrations.
Results
FGFR aberrations were identified in 745 patients, equating to an overall prevalence of 7.0%. A majority of the aberrations occurred on FGFR1 (56.8%), which was followed by FGFR3 (17.7%), FGFR2 (14.4%), and FGFR4 (2.8%). Further, 8.5% of patients had aberrations of more than 1 FGFR gene. The most common types of aberrations were amplification (53.7%), other mutations (38.8%), and fusions (5.6%). FGFR fusion and amplification occurred concurrently in 1.9% of the patients. FGFR aberrations were detected in 12 of the 16 cancers, with the highest prevalence belonging to colorectal cancer (CRC) (31%). Other FGFR-aberrant cancer types included stomach (16.8%), breast (14.3%), and esophageal (12.7%) cancer. Breast tumors were also more likely than other cancer types to have concurrent FGFR rearrangements and amplifications (P<0.001). In comparison with the public dataset, our cohort had a significantly higher number of FGFR aberrations in colorectal (P<0.001) and breast cancer (P=0.05).
Conclusions
Among the Chinese cancer patients in our study, the overall prevalence of FGFR aberrations was 7.0%. FGFR1 amplification was the most common genetic alteration in CRC, breast cancer, and lung cancer; while FGFR2 amplification was more commonly observed in gastric cancer than in other cancers in our cohort. Our study advances the understanding of the distribution of FGFR aberrations in various cancer types in the Chinese population, which will facilitate the further development of FGFR inhibitors.
Keywords: Fibroblast growth factor receptors (FGFRs), FGFR amplifications, FGFR mutations, FGFR fusions, genomic profiling
Introduction
Fibroblast growth factor receptors (FGFRs) are a family of four homologous, highly conserved transmembrane tyrosine kinase receptors (FGFR1-4) (1). While FGFRs are widely distributed throughout the body, they are not constitutively active in nonmalignant cells. FGFRs bind to fibroblast growth factor (FGF) ligands, leading to FGFR dimerization followed by phosphorylation of tyrosine residues, which triggers a series of intracellular events that activate major signaling pathways including the RAS/MAPK, PI3K/AKT, and JAK/STAT pathways (2,3). FGFR signaling plays a role in various biological processes including cellular proliferation, migration, anti-apoptosis, angiogenesis, wound healing, and tissue regeneration (4,5). The constitutive activation of FGFR signaling results in dysregulated proliferation and angiogenesis, the development of drug resistance, and immune evasion (5-8). FGFR aberrations, including gene amplification, chromosomal translocation, and/or mutations, have been reported in a broad range of cancers, including breast (9), urothelial (10), gastric (11), lung (8), and prostate cancer, as well as multiple myeloma (12).
There is significant variation in the frequency of different types of FGFR aberrations across different cancers (7). For instance, FGFR1 has been reported to be amplified in as many as 19% of squamous cell lung carcinomas (13), 6% of small cell lung carcinomas, and 1% of lung adenocarcinomas (14). FGFR1 amplification is also prevalent in breast cancer, with 15% of hormone receptor-positive breast cancers and 5% triple-negative breast cancers (TNBC) have been found to harbor FGFR1 amplification (15-17). FGFR2 amplification, which occurs less frequently than FGFR1 amplification, is present in 4–9% of gastric cancers, especially in diffuse-type gastric cancer (18), and is often associated with poor prognosis (19). FGFR2 amplification has also been found in 4% of TNBCs. Mutations in FGFR3 are extremely common in non-muscle invasive bladder cancers (75%) and are also found in 15% of high-grade invasive bladder cancers (20-22).
In recent years, oncogenic fusions in FGFRs have been discovered in a number of cancers; to date, more than 40 different FGFR fusion proteins have been detected (3,20,23). FGFR1 rearrangements are rare compared to FGFR2 or FGFR3 rearrangements. FGFR2 fusions have been identified in approximately 13.6% of intrahepatic cholangiocarcinomas (24-26), while FGFR3 fusions are commonly observed in glioblastomas and bladder cancers (27). Transforming acidic coiled-coil containing protein 3 (TACC3) is the most common fusion partner of FGFR3 (28,29). The identification of oncogenic aberrations of FGFR family members and their potential as therapeutic targets have encouraged the development of multiple FGFR inhibitors, which are currently the focus of clinical studies at different phases in various cancers. Erdafitinib (JNJ-42756493), a potent tyrosine kinase inhibitor of FGFR1-4, has been approved by the US Food and Drug Administration (FDA) as the first targeted therapy for previously-treated patients with locally advanced and metastatic urothelial cancer harboring FGFR alterations (30). Selective FGFR1-3 tyrosine kinase inhibitors, pemigatinib (INCB05482) and infigratinib (BGJ398) have been granted accelerated approval and fast-track designation, respectively, by the US FDA for the treatment of intrahepatic cholangiocarcinoma with FGFR2 rearrangements (31-33). Several trials have shed light on the specific patient population who would benefit from FGFR-targeted drugs (30-39); however, studies that have investigated the distribution of FGFR aberrations in different cancers have focused primarily on Caucasians (20). A few studies investigated FGFR mutations in Chinese squamous non-small cell lung cancer patients and their associated clinical significance (40,41). However, there lacks a study that interrogates the FGFR mutation spectrum in Chinese cancer patients. In the present study, we examined the frequency of FGFR aberrations including amplification, fusions and all non-silent mutations, as well as the distribution of these mutation types across 16 different cancers in Chinese patients. We present the following article in accordance with the MDAR reporting checklist (available at http://dx.doi.org/10.21037/atm-20-5118).
Methods
Patients
Patients who were diagnosed with any of 16 cancer types in the 3 participating hospitals: Xiangya Hospital, Hainan General Hospital, and The Seventh Affiliated Hospital, Sun Yat-sen University between September, 2015, and April, 2018, were retrospectively screened for FGFR aberrations based on their genomic profiling results from plasma or formalin-fixed paraffin-embedded (FFPE) tumor samples. All of the patients had submitted samples for sequencing to Burning Rock Biotech. The cohort included 10,582 patients across the following 16 cancer types: lung cancer (8,922 patients), breast cancer (750 patients), gastric cancer (149 patients), hepatobiliary cancer (101 patients), pancreatic cancer (87 patients), soft tissue sarcoma (STS, 76 patients), esophageal cancer (71 patients), ovarian cancer (59 patients), colorectal cancer (CRC, 58 patients), head and neck cancer (37 patients), renal carcinoma (32 patients), endometrial cancer (21 patients), osteogenic sarcoma (21 patients), cervical cancer (16 patients), melanoma (10 patients), and lymphoma (10 patients). All procedures performed in this study involving human participants were in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Institutional Review Board of Xiangya Hospital (number: 2018121148). Due to the study’s retrospective nature, written informed consent was waived.
DNA extraction
Sample processing, NGS library construction, and subsequent sequencing analysis were performed in Burning Rock Biotech, a College of American Pathologists (CAP)-accredited and Clinical Laboratory Improvement Amendments (CLIA)-certified clinical laboratory. Briefly, circulating cell-free DNA (cfDNA) and tumor DNA were extracted from plasma and FFPE tumor samples using the QIAamp Circulating Nucleic Acid Kit (Qiagen, Hilden, Germany) and the QIAamp DNA FFPE Tissue Kit (Qiagen, UK), respectively, according to the manufacturer’s instructions. The DNA concentration was quantified with the Qubit 2.0 Fluorometer and the Qubit dsDNA HS Assay Kit (Life Technologies, Carlsbad, USA).
Library construction and sequencing
DNA shearing was performed on tissue DNA using the M220 Focused-ultrasonicator (Covaris, Woburn, MA, USA), followed by end repair, phosphorylation, and adaptor ligation. Fragments in the range of 200–400 bp were size selected by Agencourt AMPure XP beads (Beckman Coulter, Brea, CA, USA) followed by hybridization with capture probe baits, hybrid selection with magnetic beads, and PCR amplification. Target capture was performed using commercially-available panels consisting of either 168 genes (Lung Plasma), 295 genes (OncoScreen), or 520 genes (OncoScreen Plus), spanning 0.273, 1.44, and 1.64 megabases (Mb) of the human genome, respectively. Finally, a high-sensitivity DNA assay was performed using Bioanalyzer 2100 (Agilent, CA, USA) to assess the quality and size of the fragments and indexed samples were sequenced on Illumina NextSeq 500 paired-end system (Illumina, Inc., Hayward, CA, USA).
Sequencing coverage for FGFR 1-4
Each of the gene panels used for genomic profiling contained capture probes that interrogated the same regions for FGFR1-4, including all of the exons for FGFR1, FGFR2, FGFR3, and FGFR4, introns 3-4 for FGFR1, the intron 17 and 3' untranslated regions (UTR) for FGFR2, and introns 16-17 and 3' UTR for FGFR3.
Sequence data analysis
Burrows-Wheeler aligner v.0.7.10 was used for mapping the paired-end reads to the human genome (hg19) (42). The Genome Analysis Toolkit (GATK) v.3.2 (43) and VarScan v.2.4.3 (44) were used to perform local alignment optimization, variant calling, and annotation. DNA translocation analysis was performed using Factera v.1.4.3 (45). The variants were filtered using the VarScan filter pipeline, and loci with depths of less than 100 were filtered out. Germline mutations were also filtered out by sequencing matched white blood cells from the patients. At least two and eight supporting reads were needed for calling insertion-deletions (INDELs) and single nucleotide variations (SNVs) in plasma samples, respectively. Variants with population frequencies of over 0.1% on the Exome Aggregation Consortium (ExAC), 1,000 Genomes, dbSNP, and ESP6500SI-V2 databases were grouped as single-nucleotide polymorphisms (SNPs) and excluded from further analysis. The remaining variants were annotated with ANNOVAR (2016-02-01 release) (46) and SnpEff v.3.6 (47). Copy number variation (CNV) was detected using in-house analysis scripts based on the depth of coverage data of capture intervals. Coverage data were corrected against sequencing bias stemming from GC content and probe design. CNV was defined as the coverage data of the gene region that were quantitatively and statistically significantly different from the corresponding reference control. The limit of detection for CNVs was 1.5 for deletions and 2.64 for amplifications. FGFR aberrations were considered novel if they were previously unreported in the somatic variation databases Catalogue of Somatic Mutations in Cancer (COSMIC), cBioPortal for Cancer Genomics (48), or the Atlas of Genetics and Cytogenetics in Oncology and Hematology (49).
Statistical analysis
Continuous variables were presented as either the mean ± standard deviation (SD) or the median with interquartile ranges, and categorical variables were presented as frequencies. Continuous variables were compared with unpaired Wilcoxon signed-rank tests, and two-sided Fisher’s exact tests were used to compare categorical data, as appropriate. P<0.05 was considered statistically significant. All bioinformatics analyses were performed with R (v.3.5.3, the R Foundation for Statistical Computing, Vienna, Austria).
Results
Somatic aberrations of FGFRs
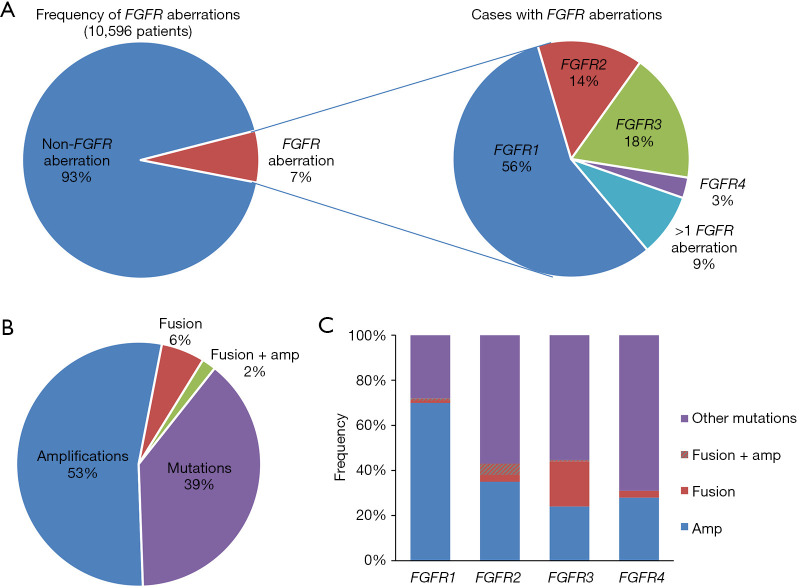
Of the 10,582 patients screened, FGFR aberrations were detected in 745 patients, showing an overall prevalence of 7.0%. FGFR amplification was harbored by 3.8% of the screened population, 0.5% had fusions, and the remaining 2.7% had other mutations. Aberrations of FGFR1, FGFR2, FGFR3, and FGFR4 were detected in 56.8%, 14.2%, 17.7%, and 2.8% of the 745 patients, respectively, and 8.5% of them had concurrent FGFR aberrations (>1 FGFR aberration, Figure 1A). Amplification was the predominant mutation type and was detected in 53.7% of the patients. Rearrangements were observed in 5.6% of patients. Non-amplification, non-fusion mutations including insertions or deletions, single base substitutions, multiple-nucleotide substitutions, and copy number deletions, which hereafter are referred to collectively as mutations or other mutations, were detected in 38.8% of the patients. Furthermore, 1.9% of patients had concurrent FGFR fusion and amplification (Figure 1B). The mutation types were distributed significantly differently across the four FGFRs (P<0.01). Of the FGFR1 aberrations, 70% were amplifications, 28% were mutations, 1% were fusions, and 1% were concurrent fusions and amplifications. Amplifications were more likely to be detected in FGFR1 than in any of the other FGFR genes (P<0.001). Of the FGFR2 aberrations, 57% were mutations, 35% were amplifications, 3% were fusions, and 5% were concurrent fusions and amplifications. FGFR2 had significantly more concurrent fusions and amplifications than the other genes (P=0.003). Of the FGFR3 aberrations, 56% were mutations, 24% were amplifications, 19% were fusions, and 1% were concurrent fusions and amplifications. FGFR3 fusions occurred more frequently than fusions in the other 3 FGFR genes (P<0.001). The predominant aberration in FGFR4 was mutations (69%), followed by amplifications (28%), and fusions (3%) (Figure 1C). Moreover, 86 novel non-amplification, non-fusion mutations were identified from our cohort; these are listed in Table S1. A total of 24, 25, 25, and 12 novel mutations were identified in FGFR1, FGFR2, FGFR3, and FGFR4, respectively. Taken together, these data revealed an overall FGFR aberration prevalence of 7.0% in Chinese patients across 16 different cancers. Also, distinct mutation frequencies and distributions of mutation types were observed in the four FGFR genes.
Figure 1.
FGFR aberrations in 10,582 cancer patients across 16 cancer types. (A) The prevalence of FGFR aberrations among all cancer patients (left), the percentages of patients with aberrations in FGFR1, FGFR2, FGFR3, and FGFR4, and the percentages of patients with aberrations in more than one FGFR gene (right); (B) the distribution of mutation types in patients with FGFR alterations; (C) the distribution of mutation types for each FGFR gene. Different types of alterations are denoted in different colors.
Table S1. Novel FGFR mutations identified in our cohort.
| Gene | Mutation type | Exon position | Description | Chromosomal position | Reference allele | Mutant allele | Frequency |
|---|---|---|---|---|---|---|---|
| FGFR1 | splice_region_variant | 1 | c.-88-3563A>T | 38318615 | T | A | 1 |
| FGFR1 | splice_acceptor_variant | 1 | c.-88-1G>T | 38315053 | C | A | 1 |
| FGFR1 | missense_variant | 2 | c.-71G>T | 38315035 | C | A | 1 |
| FGFR1 | missense_variant | 3 | p.R54C | 38287398 | G | A | 1 |
| FGFR1 | missense_variant | 3 | p.V92L | 38287284 | C | A | 2 |
| FGFR1 | missense_variant | 5 | p.E196D | 38285472 | T | A | 1 |
| FGFR1 | missense_variant | 5 | p.K163N | 38285571 | C | A | 1 |
| FGFR1 | missense_variant | 6 | p.A211G | 38283753 | G | C | 1 |
| FGFR1 | missense_variant | 6 | p.W213C | 38283746 | C | A | 1 |
| FGFR1 | missense_variant | 6 | p.I215N | 38283741 | A | T | 1 |
| FGFR1 | stop_gained | 9 | p.E363* | 38277248 | C | A | 2 |
| FGFR1 | missense_variant | 10 | p.E467G | 38275776 | T | C | 1 |
| FGFR1 | missense_variant | 10 | p.P446Q | 38275839 | G | T | 1 |
| FGFR1 | missense_variant | 11 | p.V492E | 38275465 | A | T | 2 |
| FGFR1 | missense_variant | 12 | p.G549V | 38274841 | C | A | 1 |
| FGFR1 | missense_variant | 13 | p.P579T | 38273505 | TGG | GGT | 1 |
| FGFR1 | missense_variant | 17 | p.K748N | 38271484 | C | G | 1 |
| FGFR1 | stop_gained | 17 | p.W737* | 38271517 | C | T | 1 |
| FGFR1 | missense_variant | 18 | p.M771I | 38271302 | C | T | 1 |
| FGFR1 | missense_variant | 18 | p.D782N | 38271271 | C | T | 1 |
| FGFR1 | missense_variant | 18 | p.R787C | 38271462 | G | A | 1 |
| FGFR1 | missense_variant | 18 | p.E792K | 38271241 | C | T | 1 |
| FGFR1 | missense_variant | 18 | p.G818E | 38271162 | C | T | 1 |
| FGFR1 | missense_variant | 18 | p.R821H | 38271153 | C | T | 1 |
| FGFR2 | start_lost | 2 | p.M1? | 123353329 | C | A | 1 |
| FGFR2 | missense_variant | 3 | p.G55W | 123325165 | C | A | 2 |
| FGFR2 | frameshift_variant | 3 | p.N82fs | 123325087 | GC | G | 1 |
| FGFR2 | missense_variant | 3 | p.I94V | 123325048 | T | C | 1 |
| FGFR2 | missense_variant | 3 | p.S110A | 123324999 | CTG | GCT | 1 |
| FGFR2 | missense_variant | 3 | p.D114V | 123324987 | T | A | 1 |
| FGFR2 | missense_variant | 4 | p.K151N | 123324017 | C | A | 1 |
| FGFR2 | missense_variant | 5 | p.M162I | 123310942 | C | T | 1 |
| FGFR2 | missense_variant | 7 | c.939+1207_939+1208delinsAT | 123278285 | GC | AT | 1 |
| FGFR2 | missense_variant | 7 | p.T268P | 123279630 | T | G | 1 |
| FGFR2 | splice_region_variant | 7 | p.K313= | 123279493 | C | T | 1 |
| FGFR2 | missense_variant | 9 | p.A371T | 123274807 | C | T | 1 |
| FGFR2 | missense_variant | 9 | p.P373S | 123274801 | G | A | 1 |
| FGFR2 | missense_variant | 9 | p.L376P | 123274791 | A | G | 1 |
| FGFR2 | missense_variant | 10 | p.S436F | 123263436 | G | A | 1 |
| FGFR2 | missense_variant | 11 | p.E499D | 123260404 | T | A | 2 |
| FGFR2 | missense_variant | 12 | p.E525K | 123258108 | C | T | 1 |
| FGFR2 | missense_variant | 13 | p.L560V | 123256231 | G | C | 1 |
| FGFR2 | missense_variant | 16 | p.P705R | 123244990 | G | C | 1 |
| FGFR2 | missense_variant | 16 | p.R721T | 123244942 | C | G | 1 |
| FGFR2 | missense_variant | 17 | p.M736I | 123243305 | C | T | 1 |
| FGFR2 | missense_variant | 17 | p.L761F | 123243232 | G | A | 1 |
| FGFR2 | missense_variant | 17 | p.L763F | 123243226 | G | A | 1 |
| FGFR2 | missense_variant | 18 | p.Y783F | 123239489 | T | A | 1 |
| FGFR2 | missense_variant | 18 | p.Q778P | 123239504 | T | G | 1 |
| FGFR3 | missense_variant | 3 | p.M69L | 1801076 | A | T | 1 |
| FGFR3 | missense_variant | 3 | p.K76R | 1801098 | A | G | 1 |
| FGFR3 | missense_variant | 6 | p.R207W | 1803350 | C | T | 2 |
| FGFR3 | missense_variant | 7 | p.G258A | 1803595 | G | C | 1 |
| FGFR3 | splice_region_variant | 8 | c.931-777C>T | 1804642 | C | T | 1 |
| FGFR3 | missense_variant | 8 | c.931-747_931-746inv | 1804672 | TG | CA | 1 |
| FGFR3 | missense_variant | 8 | p.E320D | 1805448 | G | C | 1 |
| FGFR3 | missense_variant | 8 | p.Y337C | 1805498 | A | G | 1 |
| FGFR3 | missense_variant | 9 | p.P401A | 1806182 | C | G | 1 |
| FGFR3 | missense_variant | 11 | p.D495G | 1807153 | A | G | 1 |
| FGFR3 | missense_variant | 13 | p.R564W | 1807521 | C | T | 1 |
| FGFR3 | missense_variant | 13 | p.V601M | 1807632 | G | A | 1 |
| FGFR3 | missense_variant | 14 | p.F636L | 1807849 | C | G | 1 |
| FGFR3 | missense_variant | 16 | p.P696T | 1808328 | C | A | 1 |
| FGFR3 | missense_variant | 16 | p.E701K | 1808343 | G | A | 1 |
| FGFR3 | missense_variant | 16 | p.A717G | 1808392 | C | G | 1 |
| FGFR3 | missense_variant | 17 | p.M725T | 1808561 | T | C | 1 |
| FGFR3 | missense_variant | 17 | p.R728L | 1808570 | G | T | 1 |
| FGFR3 | missense_variant | 17 | p.A734S | 1808587 | G | T | 2 |
| FGFR3 | missense_variant | 17 | p.K742M | 1808612 | A | T | 1 |
| FGFR3 | missense_variant | 18 | p.G774S | 1808888 | G | A | 1 |
| FGFR3 | missense_variant | 18 | p.S779C | 1808903 | A | T | 1 |
| FGFR3 | frameshift_variant | 18 | p.P796fs | 1808950 | GC | G | 1 |
| FGFR3 | frameshift_variant | 18 | p.G803fs | 1808971 | TG | T | 1 |
| FGFR3 | frameshift_variant | 18 | p.Q775fs | 1808891 | CAG | C | 1 |
| FGFR4 | missense_variant | 5 | p.R171P | 176518014 | G | C | 1 |
| FGFR4 | missense_variant | 7 | p.P279L | 176519430 | C | T | 1 |
| FGFR4 | missense_variant | 9 | p.D369N | 176520186 | G | A | 1 |
| FGFR4 | splice_region_variant | 10 | c.1397+5G>T | 176520557 | G | T | 2 |
| FGFR4 | stop_gained | 10 | p.E421* | 176520416 | G | T | 1 |
| FGFR4 | missense_variant | 13 | p.R559W | 176522578 | C | T | 1 |
| FGFR4 | missense_variant | 13 | p.R598L | 176522696 | G | T | 1 |
| FGFR4 | missense_variant | 14 | p.R635P | 176523140 | G | C | 1 |
| FGFR4 | missense_variant | 15 | p.H668N | 176523345 | C | A | 1 |
| FGFR4 | missense_variant | 16 | p.I682M | 176523635 | C | G | 1 |
| FGFR4 | missense_variant | 16 | p.S688F | 176523652 | C | T | 1 |
| FGFR4 | missense_variant | 18 | p.T774N | 176524589 | C | A | 1 |
Somatic aberrations of FGFRs across the 16 cancer types
Next, we investigated the mutation frequencies and distribution of mutation types of the four FGFR genes based on cancer type. Our cohort comprised of patients with 16 different types of cancer. In the following four types of cancer, no FGFR aberrations were identified: head and neck cancer, osteogenic sarcoma, renal carcinoma, and lymphoma. Detailed mutation frequencies and the distribution of mutation types of the 4 FGFRs in the other 12 cancers are summarized in Table 1.
Table 1. Frequencies and distributions of FGFR aberrations detected in our cohort.
| Cancer type | N | Frequency of aberrations per cancer type (%) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| All | Distribution per gene | Distribution per mutation type | ||||||||||
| FGFR1 | FGFR2 | FGFR3 | FGFR4 | Multiple FGFRs | Amp | Mutation | Fusion | Fusion + amp | ||||
| Lung | 8,922 | 6.19 | 3.49 | 0.67 | 1.33 | 0.18 | 0.52 | 3.08 | 2.71 | 0.36 | 0.03 | |
| Breast | 750 | 14.27 | 10.8 | 2.53 | 0.13 | 0.27 | 0.53 | 11.07 | 2.0 | 0.13 | 1.07 | |
| Gastric | 149 | 16.78 | 2.01 | 10.74 | 2.01 | 0 | 2.01 | 10.07 | 5.37 | 0 | 1.34 | |
| Colorectal | 58 | 31.03 | 20.69 | 1.72 | 3.45 | 1.72 | 3.45 | 17.24 | 13.79 | 0 | 0 | |
| Soft tissue sarcoma | 76 | 5.26 | 1.32 | 3.95 | 0 | 0 | 0 | 2.63 | 1.32 | 1.32 | 0 | |
| Ovarian | 59 | 11.86 | 3.39 | 1.69 | 1.69 | 1.69 | 3.39 | 6.78 | 3.39 | 1.69 | 0 | |
| Esophageal | 71 | 12.68 | 2.82 | 0 | 5.63 | 0 | 4.23 | 2.82 | 7.04 | 2.82 | 0 | |
| Pancreatic | 87 | 4.60 | 2.3 | 1.15 | 0 | 1.15 | 0 | 2.3 | 2.3 | 0 | 0 | |
| Hepatobiliary | 101 | 6.93 | 2.97 | 1.98 | 0 | 0 | 0 | 0.99 | 1.98 | 1.98 | 0 | |
| Melanoma | 10 | 20 | 20 | 0 | 0 | 0 | 0 | 10 | 0 | 10 | 0 | |
| Endometrial | 21 | 4.76 | 4.76 | 0 | 0 | 0 | 0 | 4.76 | 0 | 0 | 0 | |
| Cervical | 16 | 12.5 | 0 | 0 | 12.5 | 0 | 0 | 0 | 0 | 12.5 | 0 | |
Mutation refers to all non-amplification, non-fusion mutations including insertions or deletions, single base substitutions, multiple-nucleotide substitutions, and copy number deletion. N, screened population; amp, amplification.
Among the cancer types analyzed in this study, CRC (18/58, 31.0%) had the highest frequency of FGFR aberrations, with the majority of these aberrations identified in FGFR1 (12/18, 8 amplifications and 4 other mutations). Mutations in the other FGFR genes included FGFR2 amplification (n=1), FGFR3 amplification (n=1), FGFR3 mutation (n=1), FGFR4 mutation (n=1), and concurrent mutations in multiple FGFR genes (n=2).
Of the 10 patients with melanoma in our cohort, 2 patients harbored FGFR1 aberrations, amplification, and a fusion (FGFR1 Exon10-ADAM9 Intron 11). However, due to the extremely limited number of patients with melanoma included in our cohort, this prevalence (20.0%, 2/10) may not accurately reflect the prevalence of FGFR1 aberrations in the broader population of Chinese melanoma patients.
Meanwhile, of the 149 patients with gastric cancer, 16.8% (25/149) had FGFR aberrations, with a majority of the patients harboring mutations in FGFR2 (64.0%, 16/25). Amplification was the most frequently observed mutation type, occurring in 68.0% (17/25) of the gastric cancer patients.
Of the 750 breast cancer patients included in the cohort, 14.7% (107/750) had FGFR alterations, and the majority harbored mutations in FGFR1 (75.7%, 81/107), followed by FGFR2 (17.8%, 19/107). Only 1 patient had a mutation in FGFR3, and 2 patients had aberrations of FGFR4. Amplification was also the predominant mutation type among breast cancer patients, with 84.1% of the patients harboring amplifications (84.1%, 90/107).
The prevalence rates of FGFR aberrations between esophageal (9/71, 12.7%), cervical (2/16, 12.5%), and ovarian (7/59, 11.9%) cancer were comparable. In contrast to the other cancer types, the predominant mutation type in esophageal carcinoma was non-amplification, non-fusion mutations. Of the 16 patients with cervical cancer, 2 had FGFR3 fusions. Due to the small number of patients with cervical cancer included in our cohort, this prevalence (12.5%, 2/16) may not be representative of the actual prevalence of FGFR3 fusion among Chinese cervical cancer patients. From a screening pool of 59 patients with ovarian cancer, 7 had FGFR alterations spanning all 4 FGFR genes. Two patients had concurrent mutations in two FGFR genes.
Among the 101 patients with hepatobiliary cancer, 7 patients had FGFR mutations, equating to a prevalence of 6.9%; 3 patients had FGFR1 aberrations, 3 patients harbored FGFR2 aberrations, and 1 patient had a concurrent FGFR1/3 amplification. Of these seven patients, two had fusions, three had other mutations, and two had amplifications.
FGFR aberrations were harbored by 6.2% (552/8,922) of the patients with lung cancer in our study, with the majority of aberrations occurring in FGFR1 (59.6%, 311/552), followed by FGFR3 (119/552) and FGFR2 (60/552). FGFR4 aberrations were detected in 16 (2.9%, 16/552) patients including 7 with amplifications and 9 with other mutations. The most common mutation type among the lung cancer patients in our study was amplification.
Among the 76 patients with STS, 4 FGFR aberrations were identified, including 3 in FGFR2 and 1 in FGFR1, revealing a prevalence of 5.3%. Of the 21 patients with endometrial cancer screened, only 1 patient with an FGFR1-amplified tumor was identified (4.8%, 1/21). Of the 87 patients with pancreatic cancer, 4 (5%) had FGFR aberrations; these were FGFR1 amplification, FGFR2 amplification, FGFR1 mutation; and FGFR4 mutation, respectively.
Collectively, our analysis revealed distinct prevalence and distribution of mutation types of FGFR aberrations in 12 cancer types. FGFR1 amplification was the most common genetic alteration in CRC, breast cancer, and lung cancer, while FGFR2 amplification was more commonly observed in gastric cancer than in other cancers in our cohort.
Comparison with publicly available pan-cancer dataset
To better understand the distinct distribution of FGFR aberrations in Chinese patients with cancer, we compared our cohort with that of the Memorial Sloan Kettering Cancer Center (MSKCC) dataset obtained from cBioPortal (48,50). The coverage for the 4 FGFR genes was comparable between our panel and the panels used in the MSKCC cohort (50,51). The results are shown in Figure 2 and are summarized in Table S2. Since the MSKCC dataset does not include patients with gastric cancer, we only compared the distribution of FGFR aberrations between 11 cancers. Collectively, our cohort had a significantly higher number of FGFR aberrations in CRC (P<0.001) and breast cancer (P=0.05) than the cohort in the MSKCC dataset (Figure 2). Moreover, FGFR1 aberrations were more common in patients with CRC (20.7% vs. 3.1%; P<0.001) and melanoma (20.0% vs. 1.1%; P=0.01) in our cohort. FGFR2 aberrations were more frequently observed in patients with STS in our cohort (4.0% vs. 0.2%; P=0.01). FGFR3 aberrations occurred less frequently in patients with breast cancer (0.1% vs. 1.2%; P=0.01) and more frequently in patients with esophageal cancer (5.6% vs. 0.6%; P=0.01) in our cohort compared with the MSKCC cohort. Meanwhile, FGFR4 aberrations were significantly less common in patients with lung (0.2% vs. 1.2%; P<0.001) and breast (0.3% vs. 1.3%; P=0.03) cancer in our cohort. Concurrent FGFR aberrations were also more frequent among lung (0.5% vs. 0.1%; P=0.03), esophageal (4.2% vs. 0%; P=0.01), and ovarian (3.4% vs. 0%; P=0.045) cancer patients in our cohort in comparison with the MSKCC cohort. No significant differences were found between our data and the MSKCC dataset in the distribution of FGFR aberrations in urothelial, hepatobiliary, endometrial, or cervical cancer. Overall, our findings revealed the distinct distribution of FGFR aberrations in our cohort in certain cancer types.
Figure 2.
The prevalence and relative distributions of all FGFR aberrations (A), and in FGFR1 (B), FGFR2 (C), FGFR3 (D), and FGFR4 (E) in 11 cancer types. The frequency of FGFR aberrations is reported as the percentage of all cases screened per cancer type. The distribution of alteration types is shown as the proportion within each FGFR gene. Different FGFR genes (A) and alteration types (B,C,D,E) are denoted in different colors. The top bar reflects the data from the MSKCC data set for that particular cancer type; the bottom bar reflects the data from our cohort. (B,C,D,E) The single bars reflect only the data from our cohort due to the absence of corresponding data from the MSKCC cohort for some FGFR/cancer types. *, denotes statistical significance of P=0.05–0.01; **, denotes statistical significance of P<0.001.
Table S2. A comparison of the frequencies and distributions of FGFR aberrations in our cohort and the MSKCC dataset (Newman et al. 2014).
| Cancer type | Dataset | N | FGFR1 (%) | P value | FGFR2 (%) | P value | FGFR3 (%) | P value | FGFR4 (%) | P value | >1 FGFRs (%) | P value |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lung | MSKCC | 1,648 | 3.03 | 0.38 | 0.49 | 0.50 | 0.79 | 0.07 | 1.21 | <0.001* | 0.12 | 0.03* |
| This study | 8,922 | 3.49 | 0.67 | 1.33 | 0.18 | 0.52 | ||||||
| Breast | MSKCC | 1,234 | 12.48 | 0.28 | 1.86 | 0.34 | 1.22 | 0.01* | 1.30 | 0.03* | 0.65 | 1.00 |
| This study | 750 | 10.80 | 2.53 | 0.13 | 0.27 | 0.53 | ||||||
| Colorectal | MSKCC | 982 | 3.05 | <0.001* | 0.71 | 0.37 | 0.61 | 0.07 | 0.61 | 0.33 | 0.61 | 0.07 |
| This study | 58 | 20.69 | 1.72 | 3.45 | 1.72 | 3.45 | ||||||
| Prostate | MSKCC | 624 | 1.76 | 1.00 | 0.32 | 0.04* | 0.32 | 1.00 | 0.16 | 1.00 | 0.16 | 1.00 |
| This study | 8 | 0.00 | 12.50 | 0.00 | 0.00 | 0.00 | ||||||
| Pancreatic | MSKCC | 487 | 2.05 | 0.70 | 0.62 | 0.48 | 0.21 | 1.00 | 0.41 | 0.39 | 0.00 | 1.00 |
| This study | 87 | 2.30 | 1.15 | 0.00 | 1.15 | 0.00 | ||||||
| Soft tissue sarcoma | MSKCC | 439 | 2.51 | 1.00 | 0.23 | 0.01* | 0.68 | 1.00 | 1.37 | 0.60 | 0.00 | 1.00 |
| This study | 76 | 1.32 | 3.95 | 0.00 | 0.00 | 0.00 | ||||||
| Urothelial | MSKCC | 406 | 3.20 | 1.00 | 1.48 | 0.10 | 12.81 | 1.00 | 0.25 | 1.00 | 1.97 | 0.12 |
| This study | 6 | 0.00 | 16.67 | 0.00 | 0.00 | 16.67 | ||||||
| Melanoma | MSKCC | 350 | 1.14 | 0.01* | 3.14 | 1.00 | 0.86 | 1.00 | 2.00 | 1.00 | 1.14 | 1.00 |
| This study | 10 | 20.00 | 0.00 | 0.00 | 0.00 | 0.00 | ||||||
| Hepatobiliary | MSKCC | 346 | 1.45 | 0.39 | 1.73 | 0.43 | 2.02 | 0.36 | 0.87 | 1.00 | 0.29 | 0.40 |
| This study | 101 | 2.97 | 2.97 | 0.00 | 0.00 | 0.99 | ||||||
| Esophageal | MSKCC | 315 | 1.59 | 0.62 | 4.44 | 0.08 | 0.63 | 0.01* | 0.32 | 1.00 | 0.00 | 0.01* |
| This study | 71 | 2.82 | 0.00 | 5.63 | 0.00 | 4.23 | ||||||
| Ovarian | MSKCC | 216 | 1.85 | 0.61 | 0.93 | 0.52 | 1.39 | 1.00 | 1.85 | 1.00 | 0.00 | 0.045* |
| This study | 59 | 3.39 | 1.69 | 1.69 | 1.69 | 3.39 | ||||||
| Endometrial | MSKCC | 210 | 4.76 | 1.00 | 3.81 | 1.00 | 0.48 | 1.00 | 0.48 | 1.00 | 0.95 | 1.00 |
| This study | 21 | 4.76 | 0.00 | 0.00 | 0.00 | 0.00 | ||||||
| Cervical | MSKCC | 47 | 2.13 | 1.00 | 4.26 | 1.00 | 2.13 | 0.16 | 0.00 | 1.00 | 0.00 | 1.00 |
| This study | 16 | 0.00 | 0.00 | 12.50 | 0.00 | 0.00 |
Values in italic and P values marked with asterisks (*) are considered statistically significant. N, screened population; >1 FGFRs, detection of more than 1 FGFR aberration.
Distribution of FGFR fusion across cancer types
Next, we performed a detailed analysis of FGFR fusions. Fifty-nine FGFR fusions were distributed in 57 patients, spanning 9 of the 12 cancer types analyzed (Figure 3). Concurrent fusions were detected in two patients including one patient with STS harboring concurrent FGFR1-CREM (Intron4-intergenic region) and FGFR1-FN1 (Intron 4-Intron 20), and one patient with gastric cancer harboring concurrent FGFR2-MIR5694 (Exon 18-intergenic region) and FGFR2-PDHX (Intron 3-Exon 18). Furthermore, 13 patients had concurrent FGFR fusions and amplifications (Figures 4,S1). The majority of fusions involved FGFR3 (59.3%, 35/59), followed by fusions in FGFR1 (20.3%, 12/59), FGFR2 (18.6%, 11/59), and FGFR4 (1.7%, 1/59). Of the 59 FGFR fusions, the majority were (61.0%, 36/59) observed in patients with lung cancer; while the others were detected in patients with breast cancer (n=9), gastric cancer (n=3), hepatobiliary cancer (n=2), cervical cancer (n=2), and esophageal cancer (n=2), as well as in individual patients with melanoma, ovarian cancer, or STS. Of the cancer types with a reasonably large screening population (excluding melanoma and cervical cancer), esophageal cancer had the highest prevalence (2.8%) of FGFR fusions, followed by gastric cancer and hepatobiliary carcinoma, which each had a prevalence of 2%. In contrast, low frequencies of FGFR fusions were observed in lung and breast cancer, with fusions occurring in 0.4% and 1.2% of patients, respectively. The majority of fusions in lung cancer involved FGFR3 (80.6%, 29/36); while for breast cancer, the majority involved FGFR1 (66.7%, 6/9) (Figure 3). Meanwhile, a previously unreported rearrangement between FGFR4 and MAP1B (MAP1B intergenic region-FGFR4 Exon 7) was detected in a patient with breast cancer. Interestingly, in our analysis, FGFR-rearranged breast tumors were more likely to exist with concurrent FGFR amplifications (8/9) than other tumor types (P<0.001).
Figure 3.
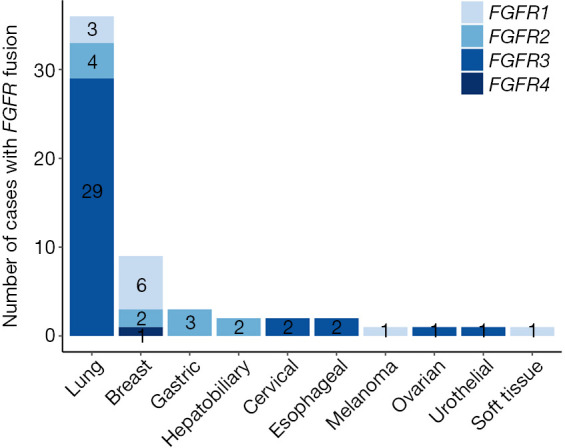
The distribution of FGFR fusions in our cohort. The frequency of FGFR fusions is reported as the number of all cases screened per cancer type. Different FGFR genes are denoted in different colors.
Figure 4.
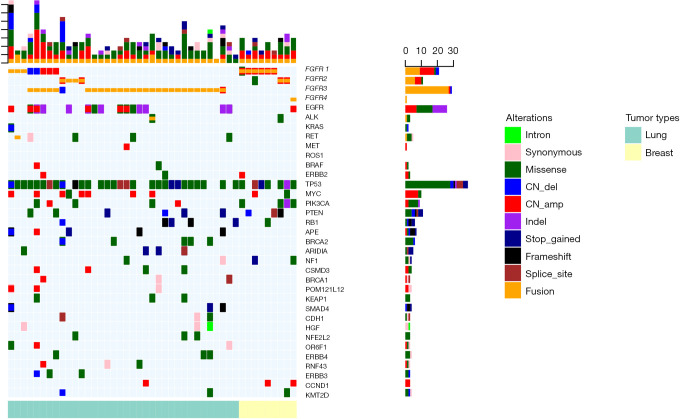
The comprehensive somatic mutation spectrum of FGFR-rearranged lung and breast cancer patients. Each column represents a patient; each row represents a gene. The number on the left represents the percentage of patients with mutations in a specific gene. The top plot represents the overall number of mutations detected in a patient. Different types of mutations are denoted in different colors. The colored annotation at the bottom of the oncoprint specifies the cancer type of the patient.
Figure S1.
Comprehensive somatic mutation spectra of patients with FGFR-rearranged cancers including gastric, hepatobiliary, cervical, esophageal, melanoma, soft tissue sarcoma, ovarian, and urothelial cancer. Each column represents a patient; each row represents a gene. The number on the left represents the percentage of patients with mutations in a specific gene. The top plot represents the overall number of mutations detected in a patient. The different colors denote different types of mutations. The colored annotation at the bottom of the oncoprint specifies the cancer type of the patient.
Overall, 31 fusion partners were identified from our cohort, including TACC3, MIR5694, and MIR1286A, which were detected in 26, 2, and 2 patients, respectively. The remaining 28 fusion partners were only detected once. TACC3-FGFR3 fusions were predominantly detected in patients with lung cancer (21/26), but were also in patients with esophageal (n=2), cervical (n=2), and ovarian (n=1) cancer. Table 2 summarizes the FGFR fusions detected in the cohort. Twenty-eight of the fusion partners detected were previously unreported. Additionally, breakpoints of fusion partners were detected in various regions including 22 in intergenic regions and 3 in the kinase domain of FGFR, while 3 had noncoding fusion partners. A further 31 breakpoints occurred outside of the abovementioned regions and thus may potentially be functional.
Table 2. List of the FGFR fusions detected in our study.
| Cancer type | FGFR family | Fusion partner | Breakpoint information for fusion partners (excluding TACC3) |
|---|---|---|---|
| Lung | FGFR1 | MIR1268A | MIR1268A intron2-FGFR1 Exon16 |
| C20orf26* | FGFR1 Intron6-C20orf26 intergenic | ||
| MIR3148* | FGFR1 Intron4-MIR3148 intergenic | ||
| FGFR2 | DPRX* | DPRX intergenic-FGFR2 Intron10 | |
| ZWINT* | FGFR2 Intron17-ZWINT intergenic | ||
| LRRC49* | FGFR2 Exon18-LRRC49 Intron8 | ||
| MIR5694 | MIR5694 intergenic-FGFR2 Intron17 | ||
| FGFR3 | PYGL* | PYGL Intron3-FGFR3 intergenic | |
| TACC3 | FAM53A Intron2-FGFR3 intergenic | ||
| FAM53A* | MUM1 intergenic-FGFR3 Exon18 | ||
| MUM1* | LOC101928201 intergenic-FGFR3 Exon3 | ||
| LOC101928201* | IDUA intergenic-FGFR3 Exon18 | ||
| IDUA* | FGFR3 Exon6-SLBP Intron3 | ||
| SLBP* | FGFR3 Intron13-LOC100507175 intergenic | ||
| LOC100507175* | |||
| Breast | FGFR1 | KCNU1* | KCNU1 intergenic-FGFR1 Exon19 |
| MIR1268A | MIR1268A Intron2-FGFR1 Exon9 | ||
| LZTS1-AS1 | LZTS1-AS1 intergenic-FGFR1 Exon6 | ||
| LINC01605* | LINC01605 Intron1-FGFR1 Exon4 | ||
| TACC1 | TACC1 Intron6-FGFR1 Exon6 | ||
| RNF5P1* | RNF5P1 intergenic-FGFR1 Intron18 | ||
| FGFR2 | EPHA6* | EPHA6 Intron5-FGFR2 Exon3 | |
| ARMT1* | ARMT1 Exon1-FGFR2 Intron11 | ||
| FGFR4 | MAP1B* | MAP1B intergenic-FGFR4 Exon7 | |
| Gastric | FGFR2 | MIR99AHG* | MIR99AHG intergenic-FGFR2 Exon18 |
| MIR56941 | FGFR2 Exon18-MIR5694 intergenic | ||
| PDHX*1 | PDHX Intron3-FGFR2 Exon18 | ||
| ATE1* | FGFR2 Intron17-ATE1 Intron10 | ||
| Hepatobiliary | FGFR2 | BICC1 | FGFR2 Intron17-BICC1 Intron2 |
| LINC00251* | FGFR2 Intron17-LINC00251 intergenic | ||
| Cervical | FGFR3 | TACC3 | |
| Esophageal | FGFR3 | TACC3 | |
| Melanoma | FGFR1 | ADAM9* | FGFR1 Exon10-ADAM9 Intron11 |
| Ovarian | FGFR3 | TACC3 | |
| Soft tissue sarcoma | FGFR1 | CREM*2 | CREM intergenic-FGFR1 Intron4 |
| FN12 | FGFR1 Intron4-FN1 Intron20 |
Asterisks (*) denote fusion partners unreported in publications and absent in somatic variation databases; 1 denotes two novel fusion partners detected from the same gastric cancer patient; 2 denotes two novel fusion partners detected from the same soft tissue sarcoma patient.
The panels we used for targeted sequencing not only interrogated critical regions of FGFR1-4, but also included various genes associated with the development and progression of cancer. Hence, we further analyzed the genomic profiles of patients with FGFR-rearranged tumors for concurrent mutations (Figures 4,S1). Since the sequencing was performed using panels comprising different numbers of genes, only the genes that were common across all of the panels were analyzed. Among the 36 patients with FGFR-rearranged lung cancer, concurrent TP53 mutations were detected in a majority (80.56%, 29/36) of the patients. Furthermore, classic lung cancer driver mutations were detected in 20 patients; these included EGFR mutations (n=15), ALK fusion (n=1), RET fusion (n=1), MET amplification (n=1), ERBB2 amplification (n=1), and KRAS Q61 mutation (n=1) (Figure 4). Among the 9 patients with FGFR-rearranged breast cancer, concurrent TP53 mutations were also detected in 7 patients (7/9). Other frequently mutated genes included PIK3CA (4/9) and PTEN (3/9). One patient had ERBB2 amplification (Figure 4). Moreover, concurrent TP53 mutations were also detected in patients with various FGFR-rearranged tumors including gastric (2/3), hepatobiliary (1/2), esophageal (2/2), ovarian (1/1), and urothelial (1/1) cancer (Figure S1). Taken together, our data revealed that the genomic profiles of patients who harbored FGFR fusions were comparable to those of patients with the same cancer type but without FGFR fusions.
Discussion
FGFR aberrations have been shown to be drivers of cancer development and progression, which makes them attractive therapeutic targets (5,7,8). The optimal application of FGFR inhibitors demands a comprehensive understanding of the prevalence and types of FGFR mutations. However, studies on the prevalence of FGFR aberrations, as well as the distribution of the mutation types across multiple cancer types, have only focused on Western populations (20), and similar research on Chinese cancer patients is limited. Herein, we conducted a pan-cancer retrospective study to investigate the prevalence and distribution of mutation types in Chinese patients with different cancers. Of the 16 cancer types included in our cohort, we revealed the overall prevalence of FGFR aberrations to be 7.0%. Consistent with the findings of a study on Caucasian cancer patients (20), in our cohort, FGFR1 and FGFR4 aberrations were found to be the most and least frequent, respectively. A majority of the FGFR aberrations in our cohort were amplifications (53.7%). In contrast, FGFR rearrangements are relatively rare, with a frequency of 7.5%, which is comparable to the prevalence reported by other studies (20,52). Furthermore, our findings revealed the distinct distribution of FGFR mutations across various cancers.
Multiple studies have investigated the prevalence of FGFR aberrations. Compared to the published data (20,26), we revealed comparable prevalence across cancers including lung, breast, STS, ovarian, and pancreatic cancer, as well as hepatobiliary carcinoma. However, we also revealed a significantly higher incidence of FGFR aberrations in Chinese patients with gastric/esophageal cancer (15.45% vs. 6.7%, P<0.001) and CRC (31.03% vs. 4.4%, P<0.001) (20). To further analyze the prevalence and the distribution of FGFR aberrations between our cohort and Caucasian patients, we compared the sequencing data from our cohort with the MSKCC dataset (51), which revealed a statistically distinct distribution of FGFR aberrations in certain cancer types between the two populations. Our cohort had a significantly higher number of FGFR aberrations in CRC (P<0.001) and breast (P=0.05) cancer than the MSKCC cohort. The prevalence of mutations in each FGFR gene also differed in some cancer types between the two populations. Interestingly, concurrent FGFR aberrations were also more frequent in Chinese patients with lung (0.52% vs. 0.12%), esophageal (4.23% vs. 0%), and ovarian (3.39% vs. 0%) cancers than in their Western counterparts.
Furthermore, based on our findings, FGFR1 amplification was the most frequently observed genomic aberration in several types of cancer, including CRC, lung, breast, and ovarian cancers. FGFR1 amplifications were detected in 13.8% (9/58) of Chinese patients with CRC in our cohort and have been reported to have a prevalence of 5.3% (24/454) in CRC primary tumors and 97.9% (92/94) of patients with CRC who have lymph node metastases (53). In our cohort, 2.4% of patients with lung cancer, 97% of whom had lung adenocarcinoma, were found to harbor FGFR1 amplifications, which is comparable to the prevalence of 1.5% reported for lung adenocarcinoma in Western population (20). The incidence of FGFR1 amplifications in lung cancer varies according to histology and tumor grade, with other studies reporting a prevalence of 20% in squamous cell carcinoma of the lung (54,55) and 6% in small cell lung carcinoma (14). In our cohort, FGFR1 amplifications were also prevalent (9.2%) among the patients with breast cancer, which is consistent with the prevalence of 9.7% reported from the Breast Cancer International Research Group (BCIRG) trials (56). FGFR2 amplifications commonly occurred in gastric cancer and breast cancer, with prevalence rates of 9.4% and 1.7%, respectively. These data were highly consistent with the rates reported in earlier studies (18,57). FGFR3 and FGFR4 amplifications were rarely observed; patients were far more likely to harbor other types of mutations. Surprisingly, in contrast to previous reports that found 10–12% of patients with endometrial cancer harbored FGFR2 mutations (excluding amplifications and fusions) and 5% of patients with cervical cancer harbored FGFR3 mutations (excluding amplifications and fusions) (20,21,58,59), we did not observe any FGFR2 and FGFR3 mutations in our patients with endometrial and cervical cancers. This discrepancy may be attributable to the limited size of our screening population in these two gynecological cancers.
FGFR fusions, which represent 8% of FGFR aberrations, have been considered as drivers of cancer development and progression (28,60,61). In our study, the majority of FGFR fusions in four cancer types (lung, cervical, esophageal, and ovarian cancer) involved FGFR3. FGFR3 fusions have also been commonly observed in bladder cancer and glioblastoma (6,62). FGFR2 fusions, the second most commonly observed fusion, were found in breast, gastric, hepatobiliary, and lung cancer. FGFR2 fusion-driven tumor cells are sensitive to FGFR inhibitors, infigratinib (BGJ398) and PD173074, thus FGFR2 fusions are recognized as a promising target (25,31-33). Despite their rarity, FGFR1 fusions were observed in breast cancer, melanoma, lung cancer, and STS in our study. We also detected a previously unreported FGFR4 fusion in a patient with breast cancer. Meanwhile, TACC3 was revealed as the most frequently observed fusion partner, which can potentially be explained by its close proximity to FGFR, and this is consistent with the results of other studies (63,64). In addition to previously reported fusion partners, including TACC3, TACC1, BICC1, and FN1 (20,23,25,27,62,63,65), we also identified numerous novel fusion partners from our cohort.
Our study has several limitations. Firstly, patients were selected for NGS-based genetic testing at the physicians’ discretion, which may have introduced sampling bias. Secondly, the size of the screening population was extremely limited for some cancer types, especially endometrial cancer, cervical cancer, and melanoma. Thirdly, due to the retrospective nature of our study, data such as detailed clinicopathological characteristics, treatment history, and survival outcomes were not available for most of the patients; hence, association analysis could not be performed. A nationwide multi-center, prospective study to evaluate the association between FGFR aberrations and the clinical outcomes of patients with various cancers is called for. Fourth, our study does not include the functional analysis of the oncogenicity of the novel FGFR fusions identified in our cohort. In vitro experiments are needed to elucidate the functionality of novel fusions. Despite these limitations, to the best of our knowledge, our study is the first to comprehensively investigate the prevalence and distribution of FGFR aberrations in a large cohort of Chinese patients spanning 16 cancer types.
Conclusions
In conclusion, our study revealed an overall prevalence of FGFR aberrations of 7.0% in Chinese patients with cancer. Our findings also demonstrate a distinct distribution of certain FGFR aberration in Chinese patients with certain types of cancer. Our study facilitates a better understanding of FGFR mutations in various cancer types in the Chinese population.
Supplementary
The article’s supplementary files as
Acknowledgments
The authors thank all of the patients who participated in this study and their families. We also thank the investigators, study coordinators, operation staff, and the whole project team who worked on this study.
Funding: This research was funded by the Guangdong Medical Science and Technology (grant number A2016295 to ZY) and Science and Technology Development Foundation Item of Nanjing Medical University (NMUB2018119 to JL). The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. All procedures involving human participants were performed in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Institutional Review Board of Xiangya Hospital (approval number: 2018121148). Due to the study’s retrospective nature, written informed consent was waived.
Footnotes
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at http://dx.doi.org/10.21037/atm-20-5118
Data Sharing Statement: Available at http://dx.doi.org/10.21037/atm-20-5118
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/atm-20-5118). Ms. JL, Dr. HHZ, Dr. AL, Dr. JX, Ms. XM, and Dr. HL report that they are employees of Burning Rock Biotech. The other authors have no conflict of interest to declare.
References
- 1.Sleeman M, Fraser J, McDonald M, et al. Identification of a new fibroblast growth factor receptor, FGFR5. Gene 2001;271:171-82. 10.1016/S0378-1119(01)00518-2 [DOI] [PubMed] [Google Scholar]
- 2.Itoh N, Ornitz DM. Fibroblast growth factors: from molecular evolution to roles in development, metabolism and disease. J Biochem 2011;149:121-30. 10.1093/jb/mvq121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gallo LH, Nelson KN, Meyer AN, et al. Functions of Fibroblast Growth Factor Receptors in cancer defined by novel translocations and mutations. Cytokine Growth Factor Rev 2015;26:425-49. 10.1016/j.cytogfr.2015.03.003 [DOI] [PubMed] [Google Scholar]
- 4.Katoh M, Nakagama H. FGF receptors: cancer biology and therapeutics. Med Res Rev 2014;34:280-300. 10.1002/med.21288 [DOI] [PubMed] [Google Scholar]
- 5.Touat M, Ileana E, Postel-Vinay S, et al. Targeting FGFR Signaling in Cancer. Clin Cancer Res 2015;21:2684-94. 10.1158/1078-0432.CCR-14-2329 [DOI] [PubMed] [Google Scholar]
- 6.Turner N, Grose R. Fibroblast growth factor signalling: from development to cancer. Nat Rev Cancer 2010;10:116-29. 10.1038/nrc2780 [DOI] [PubMed] [Google Scholar]
- 7.Dienstmann R, Rodon J, Prat A, et al. Genomic aberrations in the FGFR pathway: opportunities for targeted therapies in solid tumors. Ann Oncol 2014;25:552-63. 10.1093/annonc/mdt419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Desai A, Adjei AA. FGFR Signaling as a Target for Lung Cancer Therapy. J Thorac Oncol 2016;11:9-20. 10.1016/j.jtho.2015.08.003 [DOI] [PubMed] [Google Scholar]
- 9.Turner N, Pearson A, Sharpe R, et al. FGFR1 amplification drives endocrine therapy resistance and is a therapeutic target in breast cancer. Cancer Res 2010;70:2085-94. 10.1158/0008-5472.CAN-09-3746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tomlinson DC, Baldo O, Harnden P, et al. FGFR3 protein expression and its relationship to mutation status and prognostic variables in bladder cancer. J Pathol 2007;213:91-8. 10.1002/path.2207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jang JH, Shin KH, Park JG. Mutations in fibroblast growth factor receptor 2 and fibroblast growth factor receptor 3 genes associated with human gastric and colorectal cancers. Cancer Res 2001;61:3541-3. [PubMed] [Google Scholar]
- 12.Keats JJ, Reiman T, Belch AR, et al. Ten years and counting: so what do we know about t (4; 14)(p16; q32) multiple myeloma. Leuk Lymphoma 2006;47:2289-300. 10.1080/10428190600822128 [DOI] [PubMed] [Google Scholar]
- 13.Heist RS, Mino-Kenudson M, Sequist LV, et al. FGFR1 amplification in squamous cell carcinoma of the lung. J Thorac Oncol 2012;7:1775-80. 10.1097/JTO.0b013e31826aed28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Peifer M, Fernandez-Cuesta L, Sos ML, et al. Integrative genome analyses identify key somatic driver mutations of small-cell lung cancer. Nat Genet 2012;44:1104-10. 10.1038/ng.2396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee HJ, Seo AN, Park SY, et al. Low prognostic implication of fibroblast growth factor family activation in triple-negative breast cancer subsets. Ann Surg Oncol 2014;21:1561-8. 10.1245/s10434-013-3456-x [DOI] [PubMed] [Google Scholar]
- 16.Li W, Zheng X, Ren L, et al. Epigenetic hypomethylation and upregulation of GD3s in triple negative breast cancer. Ann Transl Med 2019;7:723. 10.21037/atm.2019.12.23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Reis-Filho JS, Simpson PT, Turner NC, et al. FGFR1 emerges as a potential therapeutic target for lobular breast carcinomas. Clin Cancer Res 2006;12:6652-62. 10.1158/1078-0432.CCR-06-1164 [DOI] [PubMed] [Google Scholar]
- 18.Matsumoto K, Arao T, Hamaguchi T, et al. FGFR2 gene amplification and clinicopathological features in gastric cancer. Br J Cancer 2012;106:727-32. 10.1038/bjc.2011.603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Su X, Zhan P, Gavine P, et al. FGFR2 amplification has prognostic significance in gastric cancer: results from a large international multicentre study. Br J Cancer 2014;110:967. 10.1038/bjc.2013.802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Helsten T, Elkin S, Arthur E, et al. The FGFR Landscape in Cancer: Analysis of 4,853 Tumors by Next-Generation Sequencing. Clin Cancer Res 2016;22:259-67. 10.1158/1078-0432.CCR-14-3212 [DOI] [PubMed] [Google Scholar]
- 21.Cappellen D, De Oliveira C, Ricol D, et al. Frequent activating mutations of FGFR3 in human bladder and cervix carcinomas. Nat Genet 1999;23:18-20. 10.1038/12615 [DOI] [PubMed] [Google Scholar]
- 22.Couffignal C, Desgrandchamps F, Mongiat-Artus P, et al. The Diagnostic and Prognostic Performance of Urinary FGFR3 Mutation Analysis in Bladder Cancer Surveillance: A Prospective Multicenter Study. Urology 2015;86:1185-90. 10.1016/j.urology.2015.07.036 [DOI] [PubMed] [Google Scholar]
- 23.Qin A, Johnson A, Ross JS, et al. Detection of Known and Novel FGFR Fusions in Non-Small Cell Lung Cancer by Comprehensive Genomic Profiling. J Thorac Oncol 2019;14:54-62. 10.1016/j.jtho.2018.09.014 [DOI] [PubMed] [Google Scholar]
- 24.Sia D, Losic B, Moeini A, et al. Massive parallel sequencing uncovers actionable FGFR2-PPHLN1 fusion and ARAF mutations in intrahepatic cholangiocarcinoma. Nat Commun 2015;6:6087. 10.1038/ncomms7087 [DOI] [PubMed] [Google Scholar]
- 25.Arai Y, Totoki Y, Hosoda F, et al. Fibroblast growth factor receptor 2 tyrosine kinase fusions define a unique molecular subtype of cholangiocarcinoma. Hepatology 2014;59:1427-34. 10.1002/hep.26890 [DOI] [PubMed] [Google Scholar]
- 26.Javle M, Bekaii-Saab T, Jain A, et al. Biliary cancer: Utility of next-generation sequencing for clinical management. Cancer 2016;122:3838-47. 10.1002/cncr.30254 [DOI] [PubMed] [Google Scholar]
- 27.Wu YM, Su F, Kalyana-Sundaram S, et al. Identification of targetable FGFR gene fusions in diverse cancers. Cancer Discov 2013;3:636-47. 10.1158/2159-8290.CD-13-0050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Carneiro BA, Elvin JA, Kamath SD, et al. FGFR3–TACC3: a novel gene fusion in cervical cancer. Gynecol Oncol Rep 2015;13:53-6. 10.1016/j.gore.2015.06.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Oliveira DM, Mirante T, Mignogna C, et al. Simultaneous identification of clinically relevant single nucleotide variants, copy number alterations and gene fusions in solid tumors by targeted next-generation sequencing. Oncotarget 2018;9:22749-68. 10.18632/oncotarget.25229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Loriot Y, Necchi A, Park SH, et al. Erdafitinib in Locally Advanced or Metastatic Urothelial Carcinoma. N Engl J Med 2019;381:338-48. 10.1056/NEJMoa1817323 [DOI] [PubMed] [Google Scholar]
- 31.Javle M, Kelley RK, Roychowdhury S, et al. Updated results from a phase II study of infigratinib (BGJ398), a selective pan-FGFR kinase inhibitor, in patients with previously treated advanced cholangiocarcinoma containing FGFR2 fusions. Ann Oncol 2018;29:viii720.
- 32.Makawita S, K, Abou-Alfa G, Roychowdhury S, et al. Infigratinib in patients with advanced cholangiocarcinoma with FGFR2 gene fusions/translocations: the PROOF 301 trial. Future Oncol 2020. [Epub ahead of print]. 10.2217/fon-2020-0299 [DOI] [PubMed] [Google Scholar]
- 33.Abou-Alfa GK, Sahai V, Hollebecque A, et al. Pemigatinib for previously treated, locally advanced or metastatic cholangiocarcinoma: a multicentre, open-label, phase 2 study. Lancet Oncol 2020;21:671-84. 10.1016/S1470-2045(20)30109-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.André F, Bachelot T, Campone M, et al. Targeting FGFR with dovitinib (TKI258): preclinical and clinical data in breast cancer. Clin Cancer Res 2013;19:3693-702. 10.1158/1078-0432.CCR-13-0190 [DOI] [PubMed] [Google Scholar]
- 35.Smyth EC, Turner NC, Peckitt C, et al. Phase II multicenter proof of concept study of AZD4547 in FGFR amplified tumours. American Society of Clinical Oncology, 2015. [Google Scholar]
- 36.Tabernero J, Bahleda R, Dienstmann R, et al. Phase I dose-escalation study of JNJ-42756493, an oral pan-fibroblast growth factor receptor inhibitor, in patients with advanced solid tumors. J Clin Oncol 2015;33:3401-8. 10.1200/JCO.2014.60.7341 [DOI] [PubMed] [Google Scholar]
- 37.Nogova L, Sequist LV, Perez Garcia JM, et al. Evaluation of BGJ398, a Fibroblast Growth Factor Receptor 1-3 Kinase Inhibitor, in Patients With Advanced Solid Tumors Harboring Genetic Alterations in Fibroblast Growth Factor Receptors: Results of a Global Phase I, Dose-Escalation and Dose-Expansion Study. J Clin Oncol 2017;35:157-65. 10.1200/JCO.2016.67.2048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Krook MA, Lenyo A, Wilberding M, et al. Efficacy of FGFR Inhibitors and Combination Therapies for Acquired Resistance in FGFR2-Fusion Cholangiocarcinoma. Mol Cancer Ther 2020;19:847-57. 10.1158/1535-7163.MCT-19-0631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Roskoski R, Jr. The role of fibroblast growth factor receptor (FGFR) protein-tyrosine kinase inhibitors in the treatment of cancers including those of the urinary bladder. Pharmacol Res 2020;151:104567. 10.1016/j.phrs.2019.104567 [DOI] [PubMed] [Google Scholar]
- 40.Li JJ, Yan S, Pan Y, et al. FGFR genes mutation is an independent prognostic factor and associated with lymph node metastasis in squamous non-small cell lung cancer. Cancer Biol Ther 2018;19:1108-16. 10.1080/15384047.2018.1480294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dong M, Li T, Chen J. Progress on the Study of Targeting FGFR in Squamous Non-small Cell Lung Cancer. Zhongguo Fei Ai Za Zhi 2018;21:116-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009;25:1754-60. 10.1093/bioinformatics/btp324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McKenna A, Hanna M, Banks E, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 2010;20:1297-303. 10.1101/gr.107524.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Koboldt DC, Zhang Q, Larson DE, et al. VarScan 2: somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res 2012;22:568-76. 10.1101/gr.129684.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Newman AM, Bratman SV, Stehr H, et al. FACTERA: a practical method for the discovery of genomic rearrangements at breakpoint resolution. Bioinformatics 2014;30:3390-3. 10.1093/bioinformatics/btu549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res 2010;38:e164. 10.1093/nar/gkq603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cingolani P, Platts A, Wang le L, et al. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin) 2012;6:80-92. 10.4161/fly.19695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gao J, Aksoy BA, Dogrusoz U, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal 2013;6:pl1. 10.1126/scisignal.2004088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Huret JL, Ahmad M, Arsaban M, et al. Atlas of genetics and cytogenetics in oncology and haematology in 2013. Nucleic Acids Res 2013;41:D920-4. 10.1093/nar/gks1082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zehir A, Benayed R, Shah RH, et al. Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nat Med 2017;23:703-13. 10.1038/nm.4333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cheng DT, Mitchell TN, Zehir A, et al. Memorial Sloan Kettering-Integrated Mutation Profiling of Actionable Cancer Targets (MSK-IMPACT): A Hybridization Capture-Based Next-Generation Sequencing Clinical Assay for Solid Tumor Molecular Oncology. J Mol Diagn 2015;17:251-64. 10.1016/j.jmoldx.2014.12.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Porta R, Borea R, Coelho A, et al. FGFR a promising druggable target in cancer: Molecular biology and new drugs. Crit Rev Oncol Hematol 2017;113:256-67. 10.1016/j.critrevonc.2017.02.018 [DOI] [PubMed] [Google Scholar]
- 53.Göke F, Göke A, von Mässenhausen A, et al. Fibroblast growth factor receptor 1 as a putative therapy target in colorectal cancer. Digestion 2013;88:172-81. 10.1159/000355018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yang W, Yao YW, Zeng JL, et al. Prognostic value of FGFR1 gene copy number in patients with non-small cell lung cancer: a meta-analysis. J Thorac Dis 2014;6:803-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Weiss J, Sos ML, Seidel D, et al. Frequent and focal FGFR1 amplification associates with therapeutically tractable FGFR1 dependency in squamous cell lung cancer. Sci Transl Med 2010;2:62ra93. 10.1126/scitranslmed.3001451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nuciforo P, Burzykowski T, Lambertini C, et al. Abstract P3-10-24: Fibroblast Growth Factor Receptor 1 Amplification and Overexpression in Breast Cancer Tissue Microarrays Using Chromogenic In Situ Hybridization and Immunohistochemistry. Cancer Res 2010;70:P3-10-24-P3-10-24.
- 57.Turner N, Lambros MB, Horlings HM, et al. Integrative molecular profiling of triple negative breast cancers identifies amplicon drivers and potential therapeutic targets. Oncogene 2010;29:2013-23. 10.1038/onc.2009.489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dutt A, Salvesen HB, Chen TH, et al. Drug-sensitive FGFR2 mutations in endometrial carcinoma. Proc Natl Acad Sci U S A 2008;105:8713-7. 10.1073/pnas.0803379105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rosty C, Aubriot MH, Cappellen D, et al. Clinical and biological characteristics of cervical neoplasias with FGFR3 mutation. Mol Cancer 2005;4:15. 10.1186/1476-4598-4-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kohler LH, Mireskandari M, Knosel T, et al. FGFR1 expression and gene copy numbers in human lung cancer. Virchows Arch 2012;461:49-57. 10.1007/s00428-012-1250-y [DOI] [PubMed] [Google Scholar]
- 61.Sabnis AJ, Bivona TG. FGFR fusions in the driver's seat. Cancer Discov 2013;3:607-9. 10.1158/2159-8290.CD-13-0185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Stransky N, Cerami E, Schalm S, et al. The landscape of kinase fusions in cancer. Nat Commun 2014;5:4846. 10.1038/ncomms5846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Williams SV, Hurst CD, Knowles MA. Oncogenic FGFR3 gene fusions in bladder cancer. Hum Mol Genet 2013;22:795-803. 10.1093/hmg/dds486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Singh D, Chan JM, Zoppoli P, et al. Transforming fusions of FGFR and TACC genes in human glioblastoma. Science 2012;337:1231-5. 10.1126/science.1220834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Petersen I, Kotb WF, Friedrich KH, et al. Core classification of lung cancer: correlating nuclear size and mitoses with ploidy and clinicopathological parameters. Lung Cancer 2009;65:312-8. 10.1016/j.lungcan.2008.12.013 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The article’s supplementary files as