Abstract
Background
To characterize the effects of mutation subtypes and concomitant pathogenic mutations on progression-free survival (PFS) and overall survival (OS) in advanced non-small cell lung cancer (NSCLC) patients with epidermal growth factor receptor (EGFR) exon 20 insertion (ex20ins) mutations treated with chemotherapy.
Methods
We retrospectively found that patients who underwent genomic analysis from January 2017 to December 2019, and 101 patients with advanced EGFR ex20ins NSCLC were found. Binary logistic regression and Cox regression were used to determine how EGFR ex20ins mutation subtypes and concomitant mutations are associated with PFS and OS.
Results
A total of 8,348 patients were screened and 101 advanced EGFR ex20ins NSCLC patients were detected. Fifty-five patients who received chemotherapy (n=49) or TKIs (n=6) as first-line treatment were recorded for PFS and OS. PFS and OS were significantly longer in the platinum-based chemotherapy group (median PFS: 7.6 versus 5.6 months; P=0.001; median OS: 19.9 versus 7.4 months; P=0.027) than in the TKI group. Common mutations include Ala767_Val769dupAlaSerVal (A767_V769dupASV), Ser768_Asp770dupSerValAsp (S768_D770dupSVD) and Ala763_Tyr764insPheGlnGluAla (A763_Y764insFQEA). On binary logistic regression, common mutations (OR =17.04, 95% CI: 1.39–209.56; P=0.027) and number of concomitant mutations ≤1 (OR =34.67, 95% CI: 2.02–595.48; P=0.015) is significantly associated with durable clinical benefit (DCB). On multivariable analysis, common mutations (HR =0.26, 95% CI: 0.0.10–0.63; P=0.003) and the number of concomitant mutations ≤1 (HR =0.33, 95% CI: 0.15–0.73; P=0.006) were significantly associated with longer PFS.
Conclusions
Common mutations and the number of concomitant mutations ≤1 correlate with a biomarker that predicts benefit from chemotherapy and confers excellent prognosis for advanced patients with advanced EGFR ex20ins NSCLC. Patients with common mutations and with only one concomitant mutation had the greatest PFS and patients with uncommon mutations, and with over one concomitant mutation had the worst prognosis.
Keywords: EGFR exon 20 insertion, non-small cell lung cancer (NSCLC), chemotherapy, concomitant mutation
Introduction
Among all cancer types, lung cancer is still the leading cause of cancer-related death (1,2). The main drug treatments for advanced non-small cell lung cancer (NSCLC) include chemotherapy, targeted therapy and immunotherapy. For patients with specific genomic mutations, targeted therapy can significantly improve their prognosis and quality of life. Immunotherapy showed a survival advantage to NSCLC, but only 20% of patients can benefit from it (3,4). Chemotherapy is still a standard choice for NSCLC patients with genomic mutations, which are not sensitive to targeted therapy or patients without genomic mutations. Epidermal growth factor receptor (EGFR) mutations were found in 40% to 50% of Asian patients and 10% to 15% of white patients (5,6). Among these patients with EGFR mutations, the single point mutation leucine-858 to arginine (L858R) in exon 21 and variable deletions of at least three amino acid residues in exon 19 account for 85% to 90%, which are regarded as the best predictors for targeted therapy. These mutations are called classic EGFR mutations and are more common in women, Asians, never smokers, and those with adenocarcinoma (7,8). EGFR ex20ins mutations define a unique molecular subtype of NSCLC, associated with insensitivity to tyrosine kinase inhibitors (TKIs), and can be detected in 4–10% of EGFR mutation patients, especially in adenocarcinoma and female patients (9-12). Several previous studies have reported that besides the A763_Y764insFQEA mutant, the insensitivity to TKIs may be related to the active kinase conformation with an unaltered adenosine triphosphate-binding pocket, and the inserted residues form a wedge at the end of the C helix. The heterogeneity of insertion length and position leads to different types of insertion mutations with different therapeutic responses (13). Chemotherapy is the dominant treatment for advanced NSCLC with EGFR ex20ins.
To date, many studies have proved that chemotherapy is associated with better progression-free survival (PFS) compared with approved EGFR TKIs as first or second line treatment of EGFR ex20ins mutations (14,15). However, not all patients with EGFR ex20ins have a long-term response to chemotherapy. The fraction of patients who derive clinical benefit from chemotherapy is still undefined. An urgent need exists to find clinically practical information to find a subset of patients. In this study, we summarized clinical data for patients with EGFR ex20ins mutations and investigated potential factors that may predict the efficacy of chemotherapy from clinical characteristics and molecular features. We present the following article in accordance with the REMARK reporting checklist (available at http://dx.doi.org/10.21037/atm-20-6172).
Methods
Patients and data collection
The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The institutional review board approved our study of Shanghai Chest Hospital, and all the patients supplied written consent before invasive procedures and chemotherapy treatment. This study included patients with pathologically confirmed lung cancer who were diagnosed at Shanghai Chest Hospital between January 2017 and December 2019. All patients with EGFR mutations were found from our medical record system. The inclusion criteria were: (I) stage III/IV lung adenocarcinoma patients; (II) patients with EGFR exon20ins mutation. Exclusion criteria included the following: (I) without complete medical records or follow-up information; (II) patients who received surgery only; (III) patients who received chemotherapy combined with immunotherapy or TKI combined with chemotherapy; (IV) Overall survival (OS) ≤3 months. Finally, fifty-five patients were included in our study. Therapeutic and prognostic information was retrospectively collected. The clinical or pathological stage was determined on the seventh edition of the TNM classification for NSCLC (16).
EGFR mutation detection
Specimens used for EGFR mutation detection were obtained by surgery, fine-needle small biopsy guided by ultrasound or computed tomography, transbronchial biopsy, or malignant effusion cell blocks. Polymerase chain reaction (PCR) testing and next generation sequencing (NGS, 68 genes, Table S1) were used for EGFR mutation detection in this study. PCR can only detect specific mutations and cannot fully reflect the concomitant mutations. The kit for PCR was provided by Amoy Diagnostics Co., Ltd. (AmoyDx). Most patients diagnosed in 2017 used PCR testing. However, NGS can sequence gene mutations in the entire genome, and we used it for some analysis in this study.
Table S1. Genes covered under the next generation sequencing (NGS).
| AKT1 | BRCA1 | DDR2 | FGF4 | JAK1 | NF1 | PTCH1 | TOP2A |
| ALK | BRCA2 | DPYD | FGFR1 | JAK2 | NOTCH1 | PTEN | TP53 |
| APC | CCND1 | EGFR | FGFR2 | KDR | NRAS | RAF1 | TSC1 |
| AR | CD74 | ERBB2 | FGFR3 | KIT | NRG1 | RB1 | TSC2 |
| ARAF | CDK4 | ERBB3 | FLT3 | KRAS | NTRK1 | RET | UGT1A1 |
| ATM | CDK6 | ERBB4 | HRAS | MAP2K1 | NTRK2 | ROS1 | |
| AXL | CDKN2A | ESR1 | IDH1 | MET | NTRK3 | SMAD4 | |
| BCL2L11 | CTNNB1 | FGF19 | IDH2 | MTOR | PDGFRA | SMO | |
| BRAF | CYP2D6 | FGF3 | IGF1R | MYC | PIK3CA | STK11 |
Treatment and clinical assessment
Fifty-five patients with advanced NSCLC EGFR ex20ins received systemic chemotherapy or EGFR TKIs (icotinib, gefitinib, or osimertinib). Chemotherapy regimens include pemetrexed, docetaxel, and gemcitabine, combined with platinum (cisplatin, carboplatin). Tumor assessment was performed using Response Evaluation Criteria in Solid Tumors version 1.1 (17) by computed tomography (CT) scans, nuclear magnetic resonance imaging (MRI), or abdominal ultrasound at every 4–8 weeks during treatment. Evaluation of the response includes complete response (CR), partial response (PR), stable disease (SD), or progressive disease (PD). The PFS was calculated from the date the patient received treatment to disease progression or last follow-up visit. OS was defined from the same date until death or last follow-up visit. Also, durable clinical benefit (DCB: CR/PR/SD that lasted >6 months) and no durable benefit (NDB: PD or SD that lasted ≤6 months) can react efficacy (18). The cutoff date was May 30, 2020.
Statistical analysis
Percentages and absolute numbers are used to describe categorical variables. Logistic regression models were developed to estimate the adjusted odds ratios (ORs) for DCB. Cox proportional hazard regression models were used to examine the univariable and multivariable analysis of relevant variables and estimated hazard ratios (HRs) for PFS. The median PFS and OS were estimated using the Kaplan-Meier method, and the log-rank test was used for comparison. Differences in common mutations and uncommon mutations were examined by using the Mann-Whitney U test for two-group comparisons. The quantitative relationship between the mutation type and the number of concomitant mutations was measured using linear regression analysis measured. All p values were two-sided, and a P value <0.05 was considered statistically significant. All statistical analysis were performed using SPSS (version 23.0, IBM, Armonk, NY).
Results
Patient characteristics
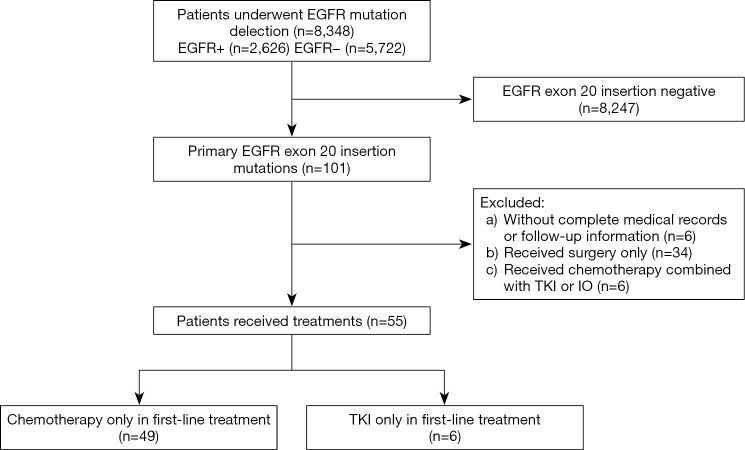
Among 8,348 patients who underwent genomic analysis, 101 had positive EGFR ex20ins mutations, accounting for 4.2% (101/2,426) of the patients with positive EGFR mutations. Among these 101 patients, six patients had incomplete medical records or follow-up information. Thirty-four patients only received surgery, and six advanced patients received combination therapy. Therefore, 55 patients who received platinum-based chemotherapy (n=49) or TKIs (n=6) as first-line therapy were included in our analysis (Figure 1). The baseline clinicopathological characteristics of the 55 patients are listed in Table 1.
Figure 1.
A flow diagram of the patient selection steps from 8,348 individuals. EGFR, epidermal growth factor receptor; TKI, tyrosine kinase inhibitor; IO, immunotherapy.
Table 1. Baseline characteristics of patients received chemotherapy or TKI (n=55).
| Variables | Number (%) |
|---|---|
| Age (median, range) | 59 [35–75] |
| Gender | |
| Male | 27 (56.4) |
| Female | 28 (54.4) |
| Clinical stage | |
| IIIB | 5 (9.1) |
| IV | 50 (90.9) |
| Site of metastasis | |
| Bone | 24 (43.6) |
| Pleura | 25 (45.5) |
| Lung | 22 (40.0) |
| Brain | 12 (21.8) |
| Liver | 4 (7.3) |
| Specimens | |
| Cell blocks | 7 (12.7) |
| Small biopsy | 23 (46.0) |
| Operation | 8 (14.5) |
| Sequencing technique | |
| Polymerase chain reaction | 16 (29.1) |
| Next generation sequencing | 39 (70.9) |
| First-line treatment | |
| Chemotherapy | 49 (89.1) |
| Tyrosine kinase inhibitor | 6 (10.9) |
| *Mutation type | |
| Common mutations | 24 (61.5) |
| Uncommon mutations | 15 (38.5) |
| *Number of concomitant mutations | |
| ≤1 | 18 (46.2) |
| >1 | 21 (53.8) |
*, patients received next generation sequencing (n=39). TKI, tyrosine kinase inhibitor.
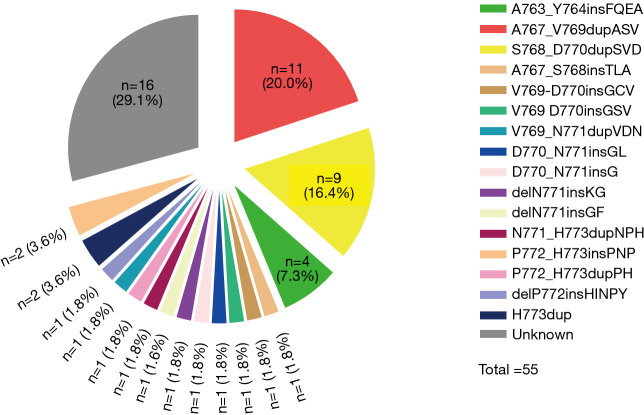
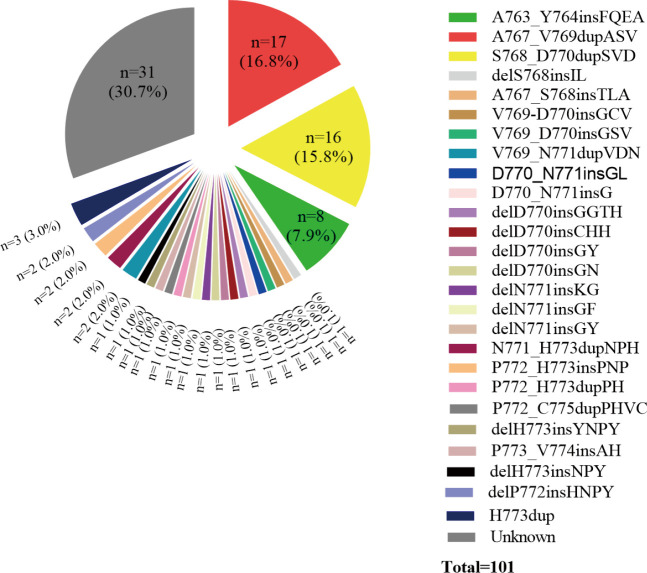
Among the 55 patients, 37 (70.9%) patients had specific and data of amino acid sequence changes by NGS testing. Sixteen different subtypes of insertion variants were recorded (Figure 2). The most common subtype of EGFR ex20ins was Ala767_Val769dupAlaSerVal (A767_V769dupASV) (n=11). The second and third most common variants were Ser768_Asp770dupSerValAsp (S768_D770dupSVD) (n=9) and Ala763_Tyr764insPheGlnGluAla (A763_Y764insFQEA) (n=4). These three types of mutations each accounted for over 5% of all patients. Therefore, in this article, we refer to these three molecular subtypes as common mutations and the others as uncommon mutations. We also recorded the percentage of molecular subtypes of the 101 positive EGFR ex20ins patients in Figure S1. Common mutations and uncommon mutations accounted for 58.6% and 41.4%, respectively (41/70 vs. 29/70).
Figure 2.
Percentage of individual EGFR ex20ins molecular subtypes among patients received first-line chemotherapy or TKI. EGFR ex20ins, epidermal growth factor receptor exon 20 insertion.
Figure S1.
Percentage of the individual EGFR ex20ins molecular subtypes among all positive patients.
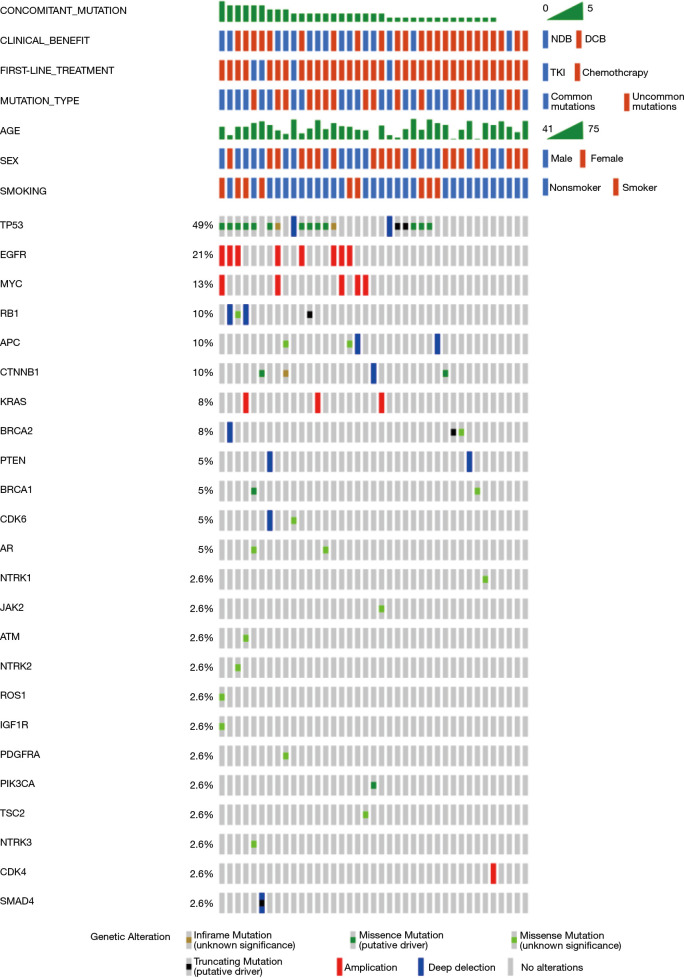
Thirty-five target patients (35/39, 89.7%) harbored concomitant mutations. Most cases have one to five concomitant mutations. Among these patients, 54% had over one concomitant mutation, and 46% had one or none. TP53 alteration and EGFR amplification were the two most common genetic alterations and were detected in 49% and 21% of the samples, respectively. Expanded lists of genes and clinical elements of target patients and all positive EGFR ex20ins patients are shown in Figures 3 and S2.
Figure 3.
Genomic landscape of advanced non-small cell lung cancer with EGFR ex20ins in this study. EGFR ex20ins, epidermal growth factor receptor exon 20 insertions.
Figure S2.
The genomic landscape of all positive advanced NSCLC with EGFR ex20ins with an expanded list of genes and clinical elements. Comparison of median PFS and OS between first-line chemotherapy and first-line EGFR TKIs. PFS, progression-free survival; OS, overall survival; EGFR, epidermal growth factor receptor; TKI, tyrosine kinase inhibitor.
Survival analysis
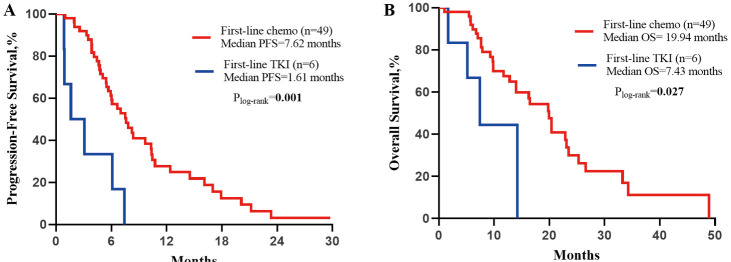
We finally collected 49 advanced EGFR ex20ins NSCLC patients receiving platinum-based chemotherapy as first-line treatment and six patients receiving TKIs for PFS and OS. Details of the chemotherapy regimens and those of the EGFR TKIs are shown in Table S2. The median (range) follow-up duration for the platinum-based chemotherapy group was 29.0 (17.9–41.9) months, and for the TKI group was 14 (2.3–25.3) months. At the time of the last follow-up, 41 of 49 patients (83.7%) in the platinum-based chemotherapy group. All patients in the TKI group had progressed. Thirty-two of 49 (65.3%) patients died in the chemotherapy group, and 4 of 6 (66.7%) patients died in the TKI group. The median PFS for the chemotherapy group was 7.6 months (95% CI: 5.7–9.6 months), which was longer than the median PFS for the TKI group (5.6 months, 95% CI: 0.0–4.2 months; HR =0.23, 95% CI: 0.09–0.57; P=0.001; Figure S3A). The median OS for the chemotherapy group was 19.9 months (95% CI: 15.9–24.0 months), which was longer than the median OS for the TKI group (7.4 months, 95% CI: 3.0–11.9 months; HR =0.31, 95% CI: 0.1–0.9; P=0.027; Figure S3B).
Table S2. Details of first-line chemotherapy regimens and EGFR TKIs.
| Treatment | Number |
|---|---|
| Pemetrexed + Carboplatin/Cisplatin | 45 |
| Paclitaxed + Carboplatin | 2 |
| Gemcitabine + Carboplatin | 1 |
| Docetaxel + Carboplatin | 1 |
| Icotinib | 3 |
| Gefitinib | 1 |
| Afatinib | 1 |
| Osimertinib | 1 |
TKIs, tyrosine kinase inhibitors.
As the efficacy of chemotherapy was better than that of TKIs in advanced EGFR ex20ins NSCLC patients, we were interested in learning who is the most suitable population for chemotherapy. To evaluate the influence of various factors on DCB, we found that common mutations (OR =5.333, 95% CI: 1.162–24.469; P=0.031) and the number of concomitant mutations ≤1 (OR =23.571, 95% CI: 2.522–220.326; P=0.006) were associated with DCB, whereas MYC amplification was independently associated with NDB (OR =0.095, 95% CI: 0.009–0.986; P=0.049; Table 2). After adjusting for potential confounders, we confirmed that common mutation and concomitant mutation number ≤1 were also significantly associated with DCB. The genomic landscape that depicts number of concomitant mutations >1/≤1 (Figure S4A) or common mutations/uncommon mutations (Figure S4B) of advanced NSCLC EGFR ex20ins in this study were showed.
Table 2. Binary logistic regression analysis of DCB.
| Variables | Univariate analysis | Multivariate analysis | |||
|---|---|---|---|---|---|
| OR (95% CI) | P | OR (95% CI) | P | ||
| Age: ≤60 vs. >60 | 0.593 (0.149–2.365) | 0.459 | |||
| Gender: male vs. female | 0.972 (0.246–3.849) | 0.968 | |||
| Smoking: smoker vs. non-smoker | 2.062 (0.350–12.168) | 0.424 | |||
| Site of metastasis | |||||
| Bone | 1.108 (0.272–4.509) | 0.886 | |||
| Pleura | 0.593 (0.149–2.365) | 0.459 | |||
| Lung | 2.308 (0.492–10.818) | 0.289 | |||
| *Mutation type: common mutation vs. uncommon mutation | 5.333 (1.162–24.469) | 0.031 | 17.042 (1.386–209.561) | 0.027 | |
| *Concomitant mutation number: ≤1 vs. >1 | 23.571 (2.522–220.326) | 0.006 | 34.666 (2.018–595.480) | 0.015 | |
| *Concomitant mutations | |||||
| TP53 | 0.692 (0.168–2.850) | 0.611 | |||
| EGFR amplification | 0.221 (0.041–1.178) | 0.077 | |||
| MYC amplification | 0.095 (0.009–0.986) | 0.049 | 0.165 (0.010–2.702) | 0.207 | |
*, patients received next generation sequencing (n=35). Italic P values indicate the higher correlations. NDB, durable clinical benefit; HR, odds ratio; CI, confidence intervals.
Univariate analysis identified that common mutations (HR =0.307, 95% CI: 0.128–0.739; P=0.008) and the number of concomitant mutations ≤1 (HR =0.396, 95% CI: 0.183–0.853; P=0.018; Table 3) were associated with longer PFS. On multivariate analysis, common mutations (HR =0.255, 95% CI: 0.104–0.625; P=0.003) and the number of concomitant mutations so (HR =0.334, 95% CI: 0.153–0.732; P=0.006) remained significantly associated with longer PFS. There was no statistical significance between the mutation type and the number of concomitant mutations (VIF =1.018; P=0.770; Figure S5), which were two independent factors.
Table 3. Univariable and multivariable analyses for progression-free survival.
| Variables | Univariate analysis | Multivariate analysis | |||
|---|---|---|---|---|---|
| HR (95% CI) | P | HR (95% CI) | P | ||
| Age: ≤60 vs. >60 | 1.377 (0.743–2.554) | 0.310 | |||
| Gender: male vs. female | 0.923 (0.488–1.746) | 0.806 | |||
| Smoking: smoker vs. non-smoker | 0.594 (0.286–1.236) | 0.164 | |||
| Clinical stage: III vs. IV | 0.670 (0.205–2.187) | 0.507 | |||
| Site of metastasis | |||||
| Bone | 1.284 (0.687–2.400) | 0.434 | |||
| Pleura | 1.378 (0.734–2.587) | 0.318 | |||
| Lung | 0.902 (0.479–1.699) | 0.749 | |||
| *Mutation type: common mutation vs. uncommon mutation | 0.307 (0.128–0.739) | 0.008 | 0.255 (0.104–0.625) | 0.003 | |
| *Concomitant mutation number: ≤1 vs. >1 | 0.396 (0.183–0.853) | 0.018 | 0.334 (0.153–0.732) | 0.006 | |
| *Concomitant mutation | |||||
| TP53 | 0.919 (0.417–2.027) | 0.834 | |||
| EGFR amplification | 1.959 (0.804–4.769) | 0.139 | |||
| MYC amplification | 2.187 (0.810–5.906) | 0.123 | |||
*, patients received next generation sequencing (n=35). Italic P values indicate the higher correlations. NGS, next generation sequencing; HR, hazard ratio; CI, confidence intervals.
Figure S5.
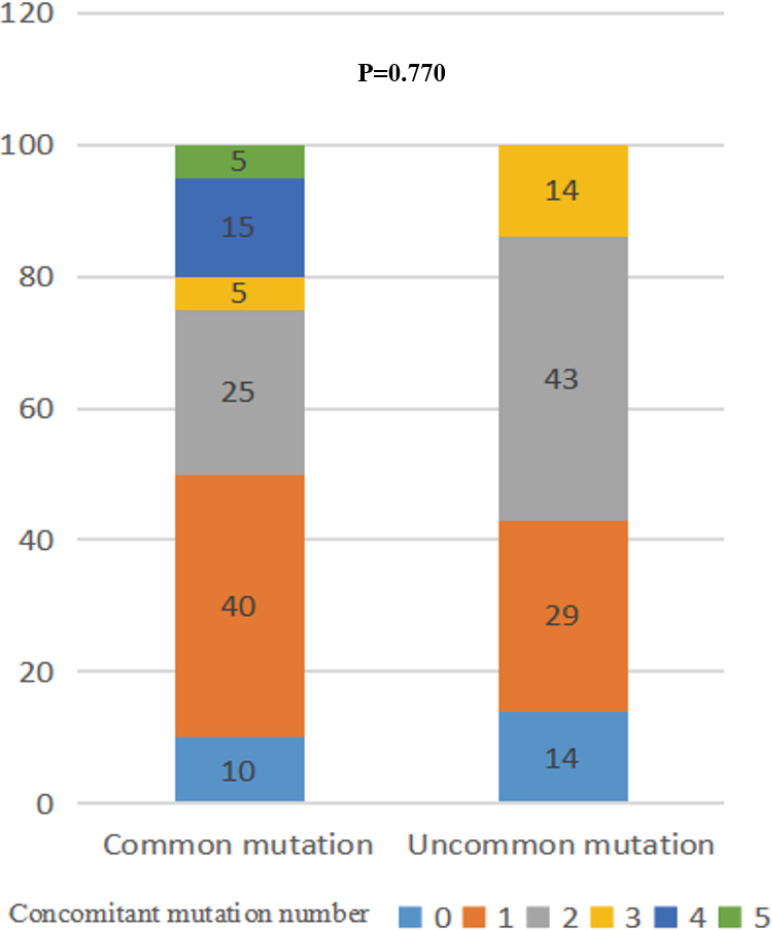
Percentage of number of concomitant mutations between common and uncommon mutations.
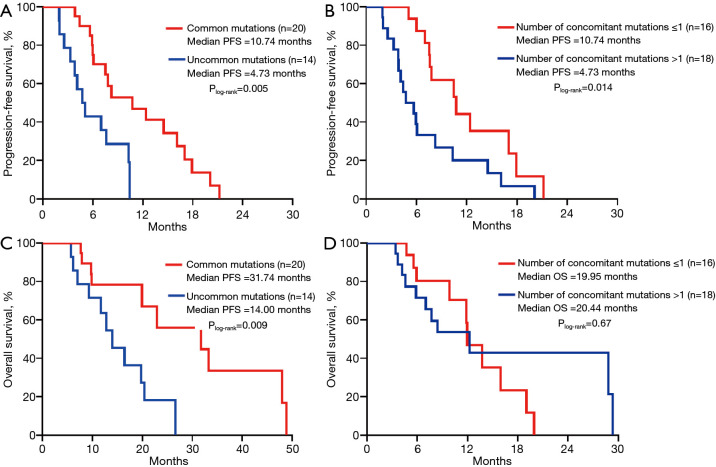
On Kaplan-Meier analysis, common mutations (mPFS =10.74 months, 95% CI: 4.8–16.7 months; P=0.005; mOS =31.74 months, 95% CI: 7.51–55.97 months) had longer PFS and OS than the group of uncommon mutations (Figure 4A,B). Also, the median PFS for the number of concomitant mutations ≤1 was 10.74 months (95% CI: 6.3–15.2; P=0.014), which was substantially longer than the number of concomitant mutations >1 (Figure 4C). However, the groups of concomitant mutation numbers were not associated with differences in OS (mOS 19.95 vs. 20.44 months; P=0.67; Figure 4D).
Figure 4.
Comparison of median PFS and OS between (A,B) common/uncommon mutations (C,D) number of concomitant mutations ≤1/>1. PFS, progression-free survival; OS, overall survival.
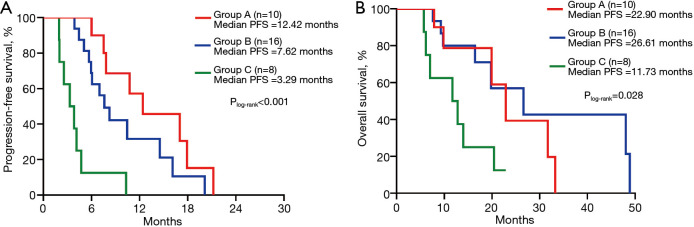
According to the above results, we divided patients into three groups: group A, patients with common mutation and the number of concomitant mutations ≤1; group B, patients with uncommon mutation and the number of concomitant mutations ≤1 or with common mutation and the number of concomitant mutations >1; and group C, patients with uncommon mutation and the number of concomitant mutations >1. On Kaplan-Meier analysis, patients in group A had the longest PFS (12.42 months, 95% CI: 4.75–20.08 months) compared with patients in group B (7.62 months, 95% CI: 5.41–9.83 months; P=0.122), and group C (3.29 months, 95% CI: 1.51–5.06; P<0.001). The difference in the PFS was found between group B and group C (P<0.001; Figure 5A). Although there was no significance in the OS between group A and group B, or group A and group C, group B had a significant association with group C (P=0.028; Figure 5B).
Figure 5.
Comparison of median PFS and OS between Group A (common mutations + number of concomitant mutations ≤1), Group B (uncommon mutations + number of concomitant mutations ≤1 or common mutations + number of concomitant mutations >1), Group C (uncommon mutations + number of concomitant mutations >1). PFS, progression-free survival; OS, overall survival.
Responses to chemotherapy between the three groups
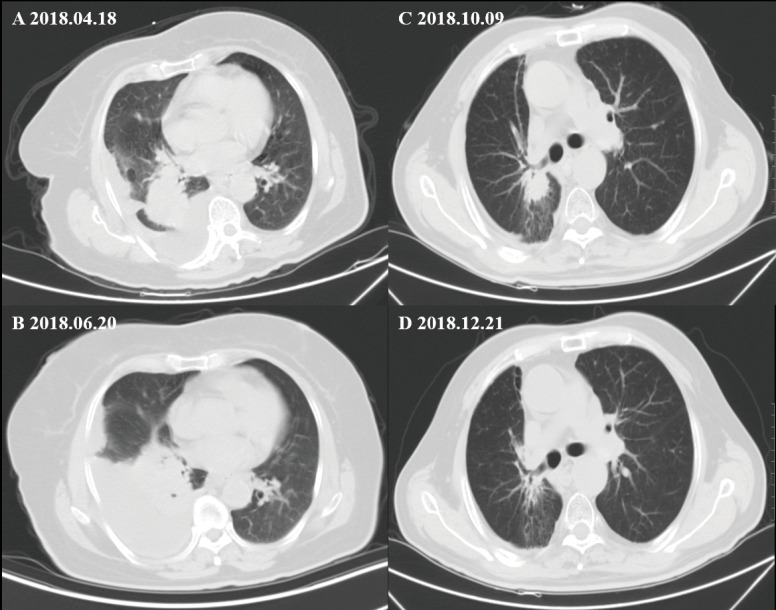
Among these three groups, all the 34 patients had enough data for their greatest response during treatment to be evaluated. Among these patients, eight patients had a PR, 23 showed an SD, and the remaining three were evaluated as having progressive disease. The ORR (CR + PR) was 23.5%, and the disease control rate (CR + PR + SD) was 91.2%. The ORRs in the three groups (group A, group B, group C) were 30%, 25%, and 12.5% (Figure S6). The ORR or DCR of group A was higher than group B or C, but it had no statistical significance (ORR, P=0.769; DCR, P=0.206). To reflet the efficiency of chemotherapy between different groups more directly, we showed the computed tomography images of two patients in group A and group C respectively (Figure S7).
Figure S6.
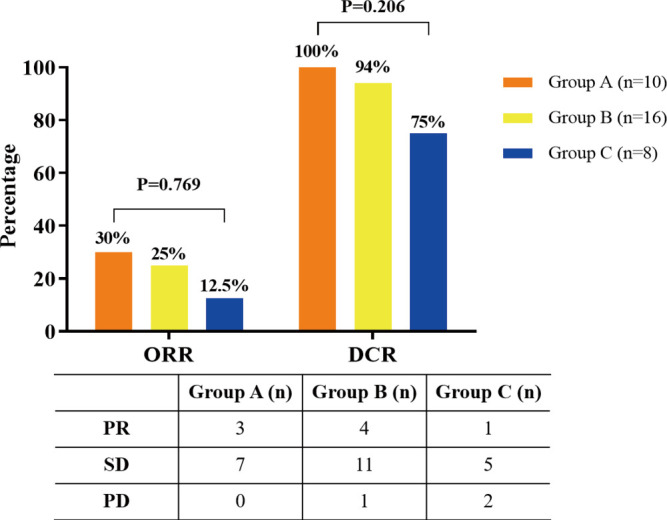
Objective response rate and disease control rate of Group A (common mutations + number of concomitant mutations ≤1), Group B (uncommon mutations + number of concomitant mutations ≤1 or common mutations + number of concomitant mutations >1), Group C (uncommon mutations + number of concomitant mutations >1).
Figure S7.
The computed tomography images exhibit two NSCLC patients with EGFR ex20ins who showed different response to chemotherapy. (A,B) One patient in group C achieved progressive disease after two cycles of chemotherapy. (C,D) One patient in group A achieved partial response after two cycles of chemotherapy.
Discussion
To our knowledge, this is the first report to evaluate the role of molecular features derived from targeted NGS in determining response or resistance to chemotherapy in patients with advanced EGFR ex20ins, and we identify that common mutation, including A767_V769dupASV, S768_D770dupSVD, A763_Y764insFQEA, correlate with a biomarker that predicts benefit from chemotherapy. Patients with only one concomitant mutation may receive help more from chemotherapy than those with over one concomitant mutation. Finally, our findings showed that patients with common mutations and the number of concomitant mutations o1 could benefit the most from chemotherapy, and the patients with uncommon mutations and the number of concomitant mutations >1 benefited the least.
We collected 101 EGFR ex20ins NSCLC, which were mainly patients in China. Wu et al. (14) identified that the three most common molecular subtypes of EGFR ex20ins are A767_V769dupASV, S768_D770dupSVD, A763_Y764insFQEA, consistent with our conclusion while studies in Caucasian or Hispanic patients showed that more unique molecular subtypes were found in Caucasian patients and H773insH was the most common mutation in Hispanic patients (19). Because of distinct races, the molecular subtypes are also different.
As we have known, increases in tumor mutation burden (TMB) are driven by several factors, including DNA replication errors mediated by defective tumor suppressor genes, deficient DNA mismatch repair (dMMR) mechanisms, and exposure to mutagens (20,21). Fewer concomitant mutations may be associated with less defective tumor suppressor genes, including TP53, which could lead to DNA replication errors. Therefore, the number of concomitant mutations may be associated with the overall tumor burden. Several studies have proved that low TMB was associated with a survival advantage in patients with NSCLC treated with chemotherapy (22-24). In this study, we found that the number of concomitant mutations was negatively associated with the clinical benefit of chemotherapy. Our results may provide the clinical support for earlier reported patients with NSCLC with low TMB might derive a better prognosis from chemotherapy (22,23,25).
Yasuda et al. found that A763_Y764insFQEA can shift the register of the C-helix toward its N-terminus and alter the length of the b3-aC loop, which had a high affinity for gefitinib in vitro and was highly sensitive to erlotinib in an engineered cell line model (13). A763_Y764insFQEA is a particular molecular subtype of EGFR ex20ins, which may be sensitive to TKIs like classical NSCLC EGFR mutations (10,26-28). Interestingly, one case with A763_Y764insFQEA had PD after a one-month treatment with afatinib in our study. The potential reason could be because of the concomitant mutation of TP53, defective tumor suppressor genes, which may reduce responsiveness to TKIs and was a poor prognosis in EGFR-mutated NSCLC patients (29,30). In our study, we concluded that common mutations could receive help from platinum-based chemotherapy. Therefore, chemotherapy could be an excellent possibility when advanced NSCLC patients with EGFR ex20ins FQEA cannot receive help from target treatment.
The retrospective design limits our study. Also, 16 patients (29.1%) in this study were detected by PCR testing, and we did not know the specific subtypes of insertion among these patients.
In conclusion, this study proves that patients with advanced EGFR ex20ins NSCLC can receive help from platinum-based chemotherapy. Common mutations and the number of concomitant mutations ≤1 is associated with positive prognosis and may be predictive of benefit with chemotherapy. Additionally, patients with common mutations and no more than one concomitant mutation had the greatest PFS comparatively with patients with other mutation types, and they may have helped more from chemotherapy. Patients with uncommon mutations and more than one concomitant mutation have the worst prognosis. This study has increased our knowledge of the advanced EGFR ex20ins NSCLC and should aid clinicians in developing more tailored therapies.
Supplementary
The article’s supplementary files as
Figure S3.
Comparison of median PFS and OS between first-line chemotherapy and first-line EGFR TKIs. PFS, progression-free survival; OS, overall survival; EGFR, epidermal growth factor receptor; TKI, tyrosine kinase inhibitor.
Figure S4.
The genomic landscape that depicts (A) number of concomitant mutations >1/≤1 or (B) common mutations/uncommon mutations of advanced NSCLC EGFR ex20ins in this study.
Acknowledgments
The authors much appreciate all patients who contributed to this study.
Funding: This work was supported by the National Key Research and Development Program of China (Project No. 2017YFC0112703), and Shanghai Chest Hospital-Collaborative Innovation Project (Project No. YJXT20190204).
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The institutional review board approved our study of Shanghai Chest Hospital, and all the patients supplied written consent before invasive procedures and chemotherapy treatment.
Reporting Checklist: The authors have completed the REMARK reporting checklist. Available at http://dx.doi.org/10.21037/atm-20-6172
Data Sharing Statement: Available at http://dx.doi.org/10.21037/atm-20-6172
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/atm-20-6172). The authors have no conflicts of interest to declare.
(English Language Editor: J. Chapnick)
References
- 1.Ferlay J, Soerjomataram I, Dikshit R, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer 2015;136:E359-86. 10.1002/ijc.29210 [DOI] [PubMed] [Google Scholar]
- 2.Rivera MP, Henderson LM. Lung cancer screening and shared decision making in cancer survivors: the long and winding road. Transl Lung Cancer Res 2019;8:119-23. 10.21037/tlcr.2018.12.14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rittmeyer A, Barlesi F, Waterkamp D, et al. Atezolizumab versus docetaxel in patients with previously treated non-small-cell lung cancer (OAK): a phase 3, open-label, multicentre randomised controlled trial. Lancet 2017;389:255-65. 10.1016/S0140-6736(16)32517-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tsiara A, Liontos M, Kaparelou M, et al. Implementation of immunotherapy in the treatment of advanced non-small cell lung cancer (NSCLC). Ann Transl Med 2018;6:144. 10.21037/atm.2018.02.07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rosell R, Moran T, Queralt C, et al. Screening for epidermal growth factor receptor mutations in lung cancer. N Engl J Med 2009;361:958-67. 10.1056/NEJMoa0904554 [DOI] [PubMed] [Google Scholar]
- 6.Mitsudomi T, Kosaka T, Endoh H, et al. Mutations of the epidermal growth factor receptor gene predict prolonged survival after gefitinib treatment in patients with non-small-cell lung cancer with postoperative recurrence. J Clin Oncol 2005;23:2513-20. 10.1200/JCO.2005.00.992 [DOI] [PubMed] [Google Scholar]
- 7.Tsao AS, Tang XM, Sabloff B, et al. Clinicopathologic characteristics of the EGFR gene mutation in non-small cell lung cancer. J Thorac Oncol 2006;1:231-9. 10.1016/S1556-0864(15)31573-2 [DOI] [PubMed] [Google Scholar]
- 8.Huang A, Shen Q, Yu X, et al. Efficacy, safety and prognostic factors analysis of first-line icotinib treatment in advanced non-small cell lung cancer patients with mutated EGFR. Transl Cancer Res 2018;7:600-8. 10.21037/tcr.2018.05.30 [DOI] [Google Scholar]
- 9.Greulich H, Chen TH, Feng W, et al. Oncogenic transformation by inhibitor sensitive and resistant EGFR mutants. PLoS Med 2005;2:e313. 10.1371/journal.pmed.0020313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yasuda H, Kobayashi S, Costa DB. EGFR exon 20 insertion mutations in non-small-cell lung cancer: preclinical data and clinical implications. Lancet Oncol 2012;13:e23-31. 10.1016/S1470-2045(11)70129-2 [DOI] [PubMed] [Google Scholar]
- 11.Oxnard GR, Lo PC, Nishino M, et al. Natural history and molecular characteristics of lung cancers harboring EGFR exon 20 insertions. J Thorac Oncol 2013;8:179-84. 10.1097/JTO.0b013e3182779d18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Arcila ME, Nafa K, Chaft JE, et al. EGFR exon 20 insertion mutations in lung adenocarcinomas: prevalence, molecular heterogeneity, and clinicopathologic characteristics. Mol Cancer Ther 2013;12:220-9. 10.1158/1535-7163.MCT-12-0620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yasuda H, Park E, Yun CH, et al. Structural, biochemical, and clinical characterization of epidermal growth factor receptor (EGFR) exon 20 insertion mutations in lung cancer. Sci Transl Med 2013;5:216ra177. 10.1126/scitranslmed.3007205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wu JY, Yu CJ, Shih JY. Effectiveness of Treatments for Advanced Non–Small-Cell Lung Cancer With Exon 20 Insertion Epidermal Growth Factor Receptor Mutations. Clin Lung Cancer 2019;20:e620-30. 10.1016/j.cllc.2019.06.018 [DOI] [PubMed] [Google Scholar]
- 15.Yang G, Li J, Xu H, et al. GFR exon 20 insertion mutations in Chinese advanced non-small cell lung cancer patients: molecular heterogeneity and treatment outcome from nationwide real-world study. Lung Cancer 2020;145:186-94. 10.1016/j.lungcan.2020.03.014 [DOI] [PubMed] [Google Scholar]
- 16.Rami-Porta R, Ball D, Crowley J, et al. The IASLC Lung Cancer Staging Project: proposals for the revision of the T descriptors in the forthcoming (seventh) edition of the TNM classification for lung cancer. J Thorac Oncol 2007;2:593-602. 10.1097/JTO.0b013e31807a2f81 [DOI] [PubMed] [Google Scholar]
- 17.Therasse P, Arbuck SG, Eisenhauer EA, et al. New guidelines to evaluate the response to treatment in solid tumors. European Organization for Research and Treatment of Cancer, National Cancer Institute of the United States, National Cancer Institute of Canada. J Natl Cancer Inst 2000;92:205-16. 10.1093/jnci/92.3.205 [DOI] [PubMed] [Google Scholar]
- 18.Rizvi NA, Hellmann MD, Snyder A, et al. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science 2015;348:124-8. 10.1126/science.aaa1348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cardona AF, Rojas L, Zatarain-Barrón ZL, et al. EGFR exon 20 insertion in lung adenocarcinomas among Hispanics (geno1.2-CLICaP). Lung Cancer 2018;125:265-72. 10.1016/j.lungcan.2018.10.007 [DOI] [PubMed] [Google Scholar]
- 20.Penault-Llorca F, Radosevic-Robin N. Tumor mutational burden in non-small cell lung cancer-the pathologist’s point of view. Transl Lung Cancer Res 2018;7:716-21. 10.21037/tlcr.2018.09.26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chalmers ZR, Connelly CF, Fabrizio D, et al. Analysis of 100,000 human cancer genomes reveals the landscape of tumor mutational burden. Genome Med 2017;9:34. 10.1186/s13073-017-0424-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Carbone DP, Reck M, Paz-Ares L, et al. First-line nivolumab in stage IV or recurrent non-small-cell lung cancer. N Engl J Med 2017;376:2415-26. 10.1056/NEJMoa1613493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Devarakonda S, Rotolo F, Tsao MS, et al. Tumor mutation burden as a biomarker in resected non-small-cell lung cancer. J Clin Oncol 2018;36:2995-3006. 10.1200/JCO.2018.78.1963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Willis C, Fiander M, Tran D, et al. Tumor Mutational Burden in Lung Cancer: A Systematic Literature. Oncotarget 2019;10:6604-22. 10.18632/oncotarget.27287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nie W, Qian J, Xu MD, et al. Prognostic and Predictive Value of Blood Tumor Mutational Burden in Patients With Lung Cancer Treated With Docetaxel. J Natl Compr Canc Netw 2020;18:582-9. 10.6004/jnccn.2019.7383 [DOI] [PubMed] [Google Scholar]
- 26.Riess JW, Gandara DR, Frampton GM, et al. Diverse EGFR Exon 20 Insertions and Co-Occurring Molecular Alterations Identified by Comprehensive Genomic Profiling of NSCLC. J Thorac Oncol 2018;13:1560-8. 10.1016/j.jtho.2018.06.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Arcila ME, Nafa K, Chaft JE, et al. EGFR exon 20 insertion mutations in lung adenocarcinomas: prevalence, molecular heterogeneity, and clinicopathologic characteristics. Mol Cancer Ther 2013;12:220-9. 10.1158/1535-7163.MCT-12-0620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Voon PJ, Tsui DWY, Rosenfeld N, et al. Letter to Editor: EGFR Exon 20 Insertion A763-Y764insFQEA and response to Erlotinib. Mol Cancer Ther 2013;12:20-5. [DOI] [PubMed] [Google Scholar]
- 29.Canale M, Petracci E, Delmonte A, et al. Impact of TP53 Mutations on Outcome in EGFR-Mutated Patients Treated With First-Line Tyrosine Kinase Inhibitors. Clin Cancer Res 2017;23:2195-202. 10.1158/1078-0432.CCR-16-0966 [DOI] [PubMed] [Google Scholar]
- 30.Xu Y, Tong X, Yan J, et al. Short-Term Responders of Non-Small Cell Lung Cancer Patients to EGFR Tyrosine Kinase Inhibitors Display High Prevalence of TP53 Mutations and Primary Resistance Mechanisms. Transl Oncol 2018;11:1364-9. 10.1016/j.tranon.2018.08.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The article’s supplementary files as