Abstract
COVID-19 pandemic caused by SARS-CoV-2 has already claimed millions of lives worldwide due to the absence of a suitable anti-viral therapy. The CoV envelope (E) protein, which has not received much attention so far, is a 75 amino acid long integral membrane protein involved in assembly and release of the virus inside the host. Here we have used artificial intelligence (AI) and pattern recognition techniques for initial screening of FDA approved pharmaceuticals and nutraceuticals to target this E protein. Subsequently, molecular docking simulations have been performed between the ligands and target protein to screen a set of 9 ligand molecules. Finally, we have provided detailed insight into their mechanisms of action related to the varied symptoms of infected patients.
Keywords: SARS-CoV-2 infection, E protein, Drug designing, Nutraceuticals, AI based screening, Drug repurposing, Pattern recognition, Molecular docking simulations
Abbreviations: ABCA1, ATP-binding cassette transporter 1; AC, Acetyl Cyclase; Ang, Angiotensin; AT1R, Angiotensin1 Receptor; ATP, Adenosine Tri Phosphate; CAP, Community acquired Pneumonia; ARDS, Acute Respiratory Distress Syndrome; CETP, Cholesteryl ester transfer protein; COX, Cyclooxygenase; DAG, Diacylglycerol; HDL, high Density Lipoprotein; IL, Interleukin; IP3, Inositol Tri Phosphate; LCAT, Lecithin–cholesterol acyltransferase; LDL, Low Density Lipoprtein; MLCK, Myosin Light Chain Kinase; PG, Prostaglandin; PIP2, Phosphatidylinositol4,5-bisphosphate; PLC, PhospholipaseC; PK, Protein Kinase; PPAR, Peroxisome proliferator-activated receptor; ROS, Reactive Oxygen Species; TNF, Tumor necrosis Factor; B, Predicted mode of action of the repurposed FDA approved pharmaceuticals and nutraceuticals against E-protein to combat SARS-CoV-2 mediated disease symptoms
1. Introduction
SARS-CoV-2, as named by the World Health Organization (WHO) to be responsible for causing Corona Virus Disease 2019 (COVID-19), was declared as pandemic on March 2020 [1]. Till date there has been more than 17 million confirmed cases worldwide claiming lives of more than 650,000 people [2]. The causative agent of the pandemic is a positive stranded enveloped virus having a genome size of ~30 Kb and belonging to genus β-CoVs of Coronaviridae family [3]. The virus shows the highest sequence similarity as well as disease symptoms with SARS-COV [4], which emerged as a disease of zoonotic origin in 2003, reporting Acute Respiratory Distress Syndrome (ARDS) and pneumonia among patients [5]. However, SARS-CoV-2 has higher transmission rate as compared to SARS-CoV and thus emerged as the cause of the worldwide pandemic [6].
SARS-CoV-2 is made up of 14 ORFs together encoding for 27 proteins [7]. These include four major structural proteins, viz., the spike (S) protein, nucleocapsid (N) protein, membrane (M) protein, and the envelope (E) protein) [8].
Apart from the structural make up, involvement of these structural proteins in various aspects of viral life cycle and the role of the non-structural proteins in pathogenesis have garnered attention [[9], [10], [11]]. Drug repurposing against these SARS-CoV-2 targets is a promising approach to combat the viral infection [[12], [13], [14], [15]]. Majority of the drug designing efforts have been directed towards the viral Spike glycoprotein to disrupt host ACE2 receptor recognition and binding so as to prevent viral entry inside the human host [[16], [17], [18], [19]].
On the contrary, among the other structural proteins of SARS-CoV-2 which have been less investigated is the E protein which is the smallest but also the most intriguing one due to its highest sequence conservation with that of SARS-CoV [8]. We therefore have attempted to target the viral E-protein, as a therapeutic intervention against the disease.
The E protein of SARS-CoV-2 shares high amino acid conservation with that of SARS-CoV. The E protein of SARS-CoV is a short, integral membrane protein of 76–109 amino acid residues with its size varying between 8.4 and 12 kDa [20]. It has a short, hydrophilic amino terminus consisting of 7–12 amino acids preceding a large hydrophobic transmembrane domain (TMD) of 25 amino acids [21]. The presence of two neutral, non-polar amino acids, valine and leucine in the TMD domain makes it strongly hydrophobic [22]. The protein terminates with a long, hydrophilic carboxyl terminus [23]. TMD is also found to oligomerize forming a pentameric structure by self-interaction which demarcates an ion conducting hydrophilic pore viroporin [24,25]. The major portion of the protein is believed to get incorporated in the Golgi, ER and ERGIC membranes in their pentameric form where positively charged, basic amino acids (viz. lysine or arginine) of the protein affix the pore to the negatively charged membrane phospholipids through electrostatic interactions [26].
It is also suggested that E protein aids in viral assembly, morphogenesis, budding and pathogenesis [27]. Deletion of E protein exhibits formation of significantly crippled viruses though the replication efficiency of the virus is not hampered [28]. Moreover, its role in pathogenesis is implicated through various modes of interactions with the host proteins [29]. Redistribution of syntenin from nucleus to cytoplasm by E protein results in the activation of p38 MAPK, triggering an over-production of pro-inflammatory cytokines like IL-1β, resulting in edema and other characteristic symptoms of ARDS [30]. Interaction between E and Bcl-xL protein is suspected to induce SARS characteristic lymphopenia [31].
Knocking down viroporin activity showed reduction in IL-1β production and edema accumulation, restoration of lung epithelial wall integrity and proper functioning of Na+/K+ ATPase [32].
The point mutations in TMD of E-protein are N15A and V25F, could suppress viroporin activity. Particularly V25F results in disruption of pentameric structure which is a crucial evidence for its role in oligomerisation [33]. Interestingly, in a mice model experiment by Nieto-Torres et al., N15A and V25F mutant viruses incorporated compensatory mutations either in these two positions or in close proximities within the TMD domain to render the ion channel active. Those mutations are N15A ➔ A15D and V25F ➔ F25C, L19A, F20 L, F26 L, L27S, T30I, L37R. Only those mice were able to survive and recover which did not incorporate these mutations [32].
This put forward the immense importance of these amino acids spanning region between 15th to 37th position of E-protein in regulating ion channel activity. Thus, we hypothesize that target inhibition of TMD of E-protein, particularly blocking the amino acids playing the roles in viroporin oligomerization could inhibit the pentamer formation and associated pathogenic reactions.
The name of several drugs and phytochemicals have come forward to target against the pentameric structure of E-protein, though very few of them are FDA approved [34]. Here, we aim to combat the spreadability of the virus at the early stage of viral E-Protein synthesis in its monomeric form, before oligomerization and initiation of viroporin pathogenesis.
In this work, we have adopted artificial intelligence (AI) based deep learning and pattern recognition techniques to screen out FDA approved drugs and food supplements which can target E-protein. Our main focus is to target the protein region between 15th to 37th residue, responsible for modulating channel activity. Computational molecular docking based virtual screening was performed thereafter for final selection of the drugs. Further, we have provided detailed insight into their mechanisms of action that could serve as an easy remedial reference for developing therapeutic measures against SARS-CoV-2.
2. Materials and methods
2.1. Sequence analysis of E protein
Human SARS CoV E protein sequence has been downloaded from UniProt(ID:P59637) [35]. It is 76 amino acid residues spanning 26,117 bps to 26,347 bps of the SARS-CoV genome. Human SARS-CoV-2 E protein sequence of 75 amino acids (265245–26,472, 228 bases) has been downloaded from NCBI protein database (ID: YP_009724392.1) [36]. Protein sequence of the three other structural proteins i.e. Matrix (M), Nucleocapsid (N) and Spike (S) proteins of SARS-CoV and SARS-CoV-2 have been downloaded from UniProt and NCBI protein bank respectively. Sequence alignment has been performed with ClustalW in Jalview [37]. In order to check for sequence conservation among SARS-CoV-2 E proteins, 12,087 SARS-CoV-2 genome sequences have been downloaded from GSAID portal(https://www.gisaid.org/) till 26.04.2020 [38]. Custom scripts have been used to analyze sequence conservation after removing partially sequenced genomes.
2.2. Structure of E protein
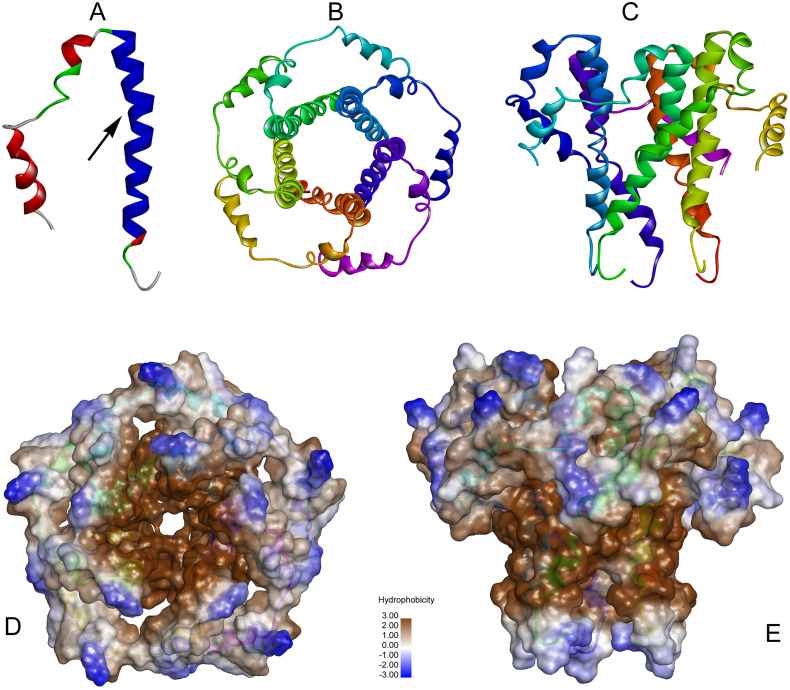
The three-dimensional structure of human SARS CoV Envelop (E) protein has been obtained from the Swiss-Model [39] repository, (https://swissmodel.expasy.org/repository/uniprot/P0DTC4) on 14.04.2020. The structure of the E protein obtained from Swiss-Model repository is a homopentamer. As it is a membrane protein, it has a patch of hydrophobic region. In this pentamer, each monomer consists of Coil (18.97%), Turn (20.68%) and Helix (60.35%) (Fig. 1 ). In this work, we have attempted to target the monomeric structure of the E-protein so as to prevent the subsequent formation of the active pentameric structure of the E-protein. This would in turn hamper the viral activity within human host. From the pentameric structure (obtained from Swiss-Model repository), the structure of the monomeric unit of E-protein was extracted using Discovery Studio 2.5. The monomeric structure of E protein is shown in Fig. 1A. In this figure, the blue region, marked with an arrow, is our zone of interest. This area lies between Asn15 to Leu37. This zone is highly responsible for the protein-protein interactions (PPI) during the pentameric formation of E protein. Fig. 1B and C are showing the top view and the side view of the pentamer respectively. Fig. 1D and E showing the hydrophobic surface view of the pentamer from top and side respectively.
Fig. 1.
Structures of human SARS-CoV E-protein.
A) Monomeric structure of E-protein, the arrow showing the zone of interest involved in protein-protein interactions.
B) Pentameric structure of E-protein, top view.
C) Pentameric structure of E-protein, side view.
D) Hydrophobic surface view of the pentamer from the top.
E) Hydrophobic surface view of the pentamer from the side.
2.3. Drug dataset and pre-processing
Multiple open source drug databases have been considered for this work. Primary drug database includes ChEMBL v.26 [40,41](https://www.ebi.ac.uk/chembl/). Besides this we have utilized Enamine Bio reference Compounds (https://www.enaminestore.com/products/bioreference-compounds) and Chemoinformatic tools and database (https://chemoinfo.ipmc.cnrs.fr/TMP/tmp.32396/e-Drug3D_1930_v3.sdf) as secondary databases. These sdf files are converted into canonical SMILES format using RDKit tools (https://www.rdkit.org/). After removal of duplicate drugs, 12,715 drugs were retained to be processed further.
2.4. In-silico screening of selected drugs
Artificial Intelligence (AI) based deep learning and pattern recognition techniques were used for initial screening and selection of suitable ligands to target the monomeric structure of the viral E-protein. The aforementioned techniques require identification of some specific patterns in the ligands derived from the sequence and structural features of the target E-protein.
(a) AI based in-silico screening method: For this, we first examined the different features of the viral E-protein and extracted conserved characteristic features from it. Such feature extraction was greatly facilitated by the absolute amino acid sequence conservation of the E-protein among the different viral strains from all over the globe.
We considered the following features of the E-protein:
-
1.
The amino acid sequence length- The monomeric structure of the E-protein is 75 amino acid residues long. Hence, the targeting ligand should be commensurate with this size of the protein.
-
2.
The domains of the E-protein- The protein has trans-membrane domain spanning the amino acid residues running between position 15 to 37. Therefore, the targeting ligands must have sufficient hydrophobic patches in order to cover this region.
Using these criteria, we downloaded the ligand datasets from ChEMBL v.26, Enamine Bio reference Compounds and Chemoinformatic tools & database in sdf format and converted them to canonical SMILES format by RDKit. The propensities of the ligands to interact with the target were analyzed by AI based deep learning models IVS2vec [42].
IVS2vec model takes 2 types of input in forms of two individual numeric vectors. One input is the protein molecule and the other one is the ligand molecule. Mol2vec [43] (which works on the principle of Word2vec [44] model) converts the SMILES formatted molecules into 300-dimensional numeric vectors. Pre-trained IVS2vec model performs the prediction task upon 600 dimensional vectors of targeted protein and molecular ligands. The structure of this deep learning based model consists of Dense Fully Connected Neural Network (DFCNN) that is already trained with PDBbind [45] database. Prediction task is executed on 2 regions of monomeric structure of the E-protein. One region considered all the amino acids constituting the monomeric structure and another one consisted of the amino acids from 15th up to 37th position of the E-protein, as it's known for its transmembrane domain which is one of the main functional sites of the targeted protein. The set of drugs having score of binding > 85% for both these regions of the protein have been selected.
(b) Pattern recognition based in-silico screening technique: Beside AI, we have also considered pattern recognition techniques for filtering important drugs. In order to select a suitable drug candidate to target this monomeric structure of E-protein, we used the following two structural features:
-
1.
Since this protein has a membrane spanning region, the candidate drug molecule must contain a long hydrophobic tail to target the membrane spanning region of the E-protein. Based upon this property we have searched for the candidate ligands having long aliphatic carbon chains of ≥16 carbon atoms which have the greater probability to bind to the membrane spanning region [46].
-
2.
The region of the protein spanning the amino acid residues 15 to 37 is known to be involved in oligomerization of the protein to make a viable and active pentameric structure necessary for exerting its activity in the host cell. The aforementioned region contains 3 aromatic Phe residues (at position 20, 23 and 26). Therefore, the candidate drug molecule would also need to have aromatic ring structures in order to fit into the three-dimensional space occupied by the said region of E-protein in such a way that oligomerization process would no longer take place. Thus, we have searched for drug molecules containing 2 aromatic rings.
The set of drugs obtained after imposing screening techniques (a) and (b) are further screened based on their (i) existing functions (i.e. to check how far its existing functionality correlates with the symptoms of SARS-CoV-2 specially in the context of the function of E-protein); (ii) FDA approval status and (iii) severity of the reported side effects of these drugs.
The set of drugs obtained after all these screening phases have been considered for subsequent molecular docking.
2.5. Molecular docking based virtual screening
The screened ligands obtained above were then subjected to computational molecular docking based virtual screening. From the SMILES information of the ligands, their three-dimensional structures have been built and then the ligand structure optimization has been performed using Discovery Studio 2.5. We used Autodock VINA [47] for this computational molecular docking. In order to get an unbiased docking result, blind docking has been performed to check the affinity of these ligands towards the zone of protein-protein interactions (PPI).
Here one of the important aspects we have taken care of, while looking into the binding affinity between the ligand and the target protein is the correlation between the binding affinity and docking score. It has been observed that binding affinity between the targeted protein and ligand cannot be judged solely by docking score [48] as they are poorly correlated [49,50]. Orientation of binding [48], site of binding [51] and type of binding energy [50] are also important aspects which need to be considered to judge binding affinity. On the contrary, docking score is highly correlated with the molecular weight of both the ligand and target protein [48]. Hence, higher the molecular weight of the ligand and target protein more will be the docking score and vice-versa.
In this work, we have targeted monomeric structure of E-protein for SARS-CoV-2 constituting 75 amino acids only. For strong interaction with small protein, ligands with length ≤ 100 in terms of SMILES symbol are considered for docking [52,53] as large molecules may fail to invade the target biologically. As specified above, interaction potential between protein and ligand can be inferred efficiently by considering structure based features instead of ligand based features [54]. Further, Guedes et al. [51], specified that binding affinity depends on the site of binding. In this case, TMD of the E-protein is mostly crucial and responsible for the virulence factor of SARS-CoV-2. We have implemented blind docking, and drugs inhibiting that specific TMD site would reduce the pathogenicity of the virus. The entire workflow is provided in Fig. 2 .
Fig. 2.
Workflow showing step wise screening of potential drugs and nutraceuticals targeting SARS-CoV2 E-Protein.
+: shows the adding up of molecules screened by multiple pattern recognition approaches.
U: shows the union of molecules screened both by AI based and Pattern recognition approaches.
3. Results
3.1. Sequence conservation of E-protein at amino acid level and nucleotide
The pairwise nucleotide sequence alignment between SARS-CoV-2 and SARS-CoV has been represented in Fig. 3 along with the consensus sequence. Protein sequence conservation score of all four structural proteins are provided in Table 1. We observed that E-protein shares the highest sequence similarity among all structural proteins when compared between SARS-CoV-2 and SARS-CoV. The details of pairwise alignment of all the structural proteins between the two strains have been provided in Supplementary File SF1.
Fig. 3.
Amino acid sequence alignment and conservation pattern of SARS-CoV and SARS-CoV-2.
Transmembrane (TMD) domain between position15 to position 37 has been highlighted in red. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Table 1.
Similarity between SARS-CoV andSARS-CoV-2 structural proteins at protein level.
| Structural Protein | NCBI protein bank accession No. SARS-CoV-2 | Length SARS-CoV-2 | UniProt Accession No. SARS-CoV | Length SARS-CoV | Percentage similarity between SARS-CoV-2 and SARS-CoV protein |
|---|---|---|---|---|---|
| E (Envelope) | P59637 | 75 | YP_009724392.1 | 76 | 94.74 |
| N (Nucleoprotein) | P59595 | 419 | QIH45060.1 | 422 | 90.52 |
| M (Matrix) | P59596 | 222 | QII57163.1 | 223 | 90.45 |
| S (Spike) | P59594 | 1273 | YP_009724390.1 | 1255 | 76.27 |
Such highly conserved nature of E-protein between the previous and current strains of the virus instigated us to target this protein while looking for the possible therapeutic interventions against the virus.
Our next step was to look into the degree of similarity between the SARS-CoV-2 nucleotide sequences collected from different patient samples at different geographical locations so as to estimate its conserved nature in due course of the spread of the virus to different geographical locations across the globe. Among 11,943 sequences (obtained from GSAID) that have been considered after removing partially sequenced sequences, 97.22% of them showed 100% conservation of E-protein at nucleotide level. Among the remaining sequences with E-protein mutations, those having sequencing errors with NNNN reads were not considered. 44% of the mutated E-protein sequences showed conservations among the entire 69 bases of interest; spanning amino acids from 15 to 37th position of the TMD domain. Sequence conservation information has been provided in Table 2 .
Table 2.
Conservation among SARS-CoV-2 E-protein sequences at nucleotide level obtained from different patients worldwide.
| Sequence downloaded from GSAID | 12,087 |
| Partial sequences removed | 144 |
| Sequence analyzed | 11,943 |
| E-protein conservation | 11,612 (97.23%) |
| Total Mutated E sequences | 331 (2.77%) |
| Sequences discarded due to sequencing errors | 111 |
| Mutated sequences considered | 220 |
| Conservation of core 69 bases from position 15 to 37 | 97 (44%) |
| Mutation in the core 69 bases from position 15 to 37 | 123 (56%) |
3.2. Screening of drugs using computational methods
As mentioned in Materials and Methods section, various computational techniques like AI based deep learning and pattern recognition techniques have been implemented to screen a set of drugs from the initial list of drugs obtained from ChEMBL v.26 (https://www.ebi.ac.uk/chembl/), Enamine Bio reference Compounds (https://www.enaminestore.com/products/bioreference-compounds) and Chemoinformatic tools and database (https://chemoinfo.ipmc.cnrs.fr/TMP/tmp.32396/e-Drug3D_1930_v3.sdf). These set of drugs are further screened based on their existing functions, FDA approval and severity of their side effects. Table 3 enlist the full set of drugs which were sent out for docking.
Table 3.
List of drugs selected for molecular docking.
| Screening approach | Drugs Name |
|---|---|
| Artificial Intelligence | Cinametic Acid |
| Dodecanoic Acid/ Lauric Acid | |
| Salmeterol | |
| Guaifenesin | |
| Mexiletine | |
| Midorine | |
| Benzyl Benzoate | |
| Ricinoleic Acid | |
| Capsaicin | |
| Pterostilbene | |
| Artificial Intelligence and Pattern Recognition on Aromatic Carbon | Nabumetone |
| Naproxen | |
| Artificial Intelligence and Pattern Recognition on Long Aliphatic Carbon Chain | Palmidrol |
| Ascorbyl Palmitate | |
| Cetalkonium | |
| Pattern Recognition on Aromatic Carbon | Nafcillin |
| Pattern Recognition on Long Aliphatic Carbon Chain | Octacosanol |
| Vitamin A Palmitate |
3.2.1. Screening of drugs using AI based Machine Learning technique
The initial list consisted of 12,715 drugs. Among those, 395 were screened as they fulfilled 2 properties (i) Their length is ≤100 in terms of SMILES string format and (ii) Their binding score with all alpha carbon atoms as well as with carbon atoms of E-protein spanning between 15-37th position is >85% using IVS2vec model. Subsequent to AI based screening, the drugs are further filtered, based on existing functions correlating with the symptoms of SARS-CoV-2, FDA approval status and severity of the reported side effects of these drugs. Finally, 18 drugs have been screened by this method (Table 3).
3.2.2. Screening of drugs based on pattern recognition
3.2.2.1. Long carbon chain based pattern recognition
E-protein consists of membrane spanning region with long hydrophobic tail. The suitable ligand which can bind to this membrane spanning region needs to have a hydrophobic part. Drugs having the necessary hydrophobic zone were screened.
Work of Duangjit et al. showed that the hydrophobicity of long carbon chain is greater than short carbon chains and the threshold length of it could be fixed to 16 [46]. This led us to filter those drug molecules (with hydrophobic zone) having long aliphatic carbon chain of length ≥ 16.
Further we checked their existing functions, FDA approval status and reported side effects. Based on all these, we filtered out 6 drug molecules shown in Table 3. These are Docosanol, Vitamin A Palmitate, Ascorbyl Palmitate, Cetalkonium, Octacosanol and Palmidrol. Among them Docosanol have been rejected as it is a topical medication. Ascorbyl Palmitate, Cetalkonium and Palmidrol have also been screened through AI based screening. Octacosanol and Vitamin A Palmitate are the exclusive set of drugs which got screened through this pattern recognition based criterion.
3.2.2.2. Aromatic carbon ring based pattern recognition
As TMD of E-protein containing 3 aromatic Phe residues (at position 20, 23 and 26); so the drugs possessing aromatic carbon ring has got the higher probability to bind with Phe amino acid. Based on this property we have selected list of drugs which has 2 or more aromatic carbon rings. Further, imposing the criterion of functionality, FDA approval status and reported side effects lead to the selection of only 1 drug Nafcillin by this procedure.
Table 3 summarizes the set of ligands which has made up to the final stage for molecular docking.
3.3. Molecular docking based virtual screening results
From the molecular docking based virtual screening we have found that from the entire list provided in Tables 3, 9 ligands could bind with the E-protein in three distinct clusters (Fig. 4 ). Cluster 1 consists of 7 ligand molecules; cluster 2 consists of 1 ligand molecule and cluster 3 consists of 1 ligand molecules.
Fig. 4.
Binding of the ligand molecules with E-protein.
Cluster 1 contains most of the ligands. Both cluster 1 and cluster2 were found to cover the region of protein-protein interaction site (indicated in blue). In other words, the aforementioned ligands in the cluster 1 and 2 could bind to the amino acid residues necessary for protein-protein interactions to make the active pentameric structure of the E-protein. The ligands present in the Cluster 3 were found to cover the outer surface of the E-protein in such a way that the flexibility of the protein necessary for interacting with other partners would somehow be hampered. The aromatic ring like structures present in the ligands would help them to stack properly with the Phe residues present in the oligomerization domain (amino acid residues 15 to 37) of E-protein. This would further enable the ligands to fix the aforementioned part of the protein in a compact static state in such a way that the protein-protein interactions necessary for oligomerization would no longer take place. Table 4 depicts the cluster-wise list of the ligand molecules along with their docking score. Table 5 denotes the detailed function and mode of action of these 9 drug molecules.
Table 4.
Cluster-wise ligand list with their docking score.
| Cluster-1 |
Cluster-2 |
Cluster-3 |
|||
|---|---|---|---|---|---|
| Molecule | Docking score (kcal/mol) | Molecule | Docking score (kcal/mol) | Molecule | Docking score (kcal/mol) |
| Nafcillin | −6.1 | Ascorbyl palmitate | −4.2 | Guaifenesin | −3.8 |
| Nabumetone | −5.1 | ||||
| Cinametic acid | −3.9 | ||||
| Octacosanol | −3.8 | ||||
| Dodecanoic acid | −3.7 | ||||
| Palmidrol | −3.5 | ||||
| Salmeterol | −3.4 | ||||
Table 5.
Mode of action of the selected drugs to be repurposed for COVID-19 and their application in existing diseases.
| Drug | Nature | Mode of Action | Prescribed in Diseases |
|---|---|---|---|
| Nafcillin | Pharmaceutical | beta lactamase resistant antibiotic. Bacterial cell wall synthesis inhibitor | Nonspecific lower respiratory tract infection as well as community-acquired pneumonia (CAP) |
| Nabumetone | Pharmaceutical | Anti-inflammatory cyclooxygenase inhibitor drug. Inhibits prostaglandin synthesis. | osteoarthritis and rheumatoid arthritis |
| Octacosanol | Nutraceutical | Lowers total cholesterol and break down LDL (low density lipoproteins) or “bad” cholesterol. | High cholesterol, atherosclerosis |
| Cinametic acid | Nutraceutical | ACE(angiotensin converting enzyme) inhibitor. | |
| Lauric acid | Nutraceutical | A medium chain fatty acid which can disrupt viral envelope. Also increases HDL (High density lipoprotein) or “good” cholesterol. | Influenza |
| Ascorbyl Palmitrate | Nutraceutical | Anti-oxidant. Protects the body against oxidative damage by free radicals. | Vitamin C deficiency |
| Palmidrol | Nutraceutical | Cannabinoid receptor agonist. Lowers the release of inflammatory cytokines and histamine. | Influenza |
| Salmeterol | Pharmaceutical | Beta2 adrenergic receptor agonist. Helps in the dilation of bronchioles and prevent air passage narrowing induced by mast cell released histamine. | Asthma and COPD (Chronic obstructive pulmonary disease) |
| Guaifenesin | Pharmaceutical | Increase the volume of trachea and bronchi. Aids in the clearance of mucous. | Cough and common cold |
We then tried to identify the residue specificity of the ligands in order to identify the amino acid residues of the E-protein binding most strongly with the ligand atoms. The docking scores of all the 9 ligands are presented in Table 4. The binding interactions of all the ligands are presented in Supplementary File SF2.
The sets of ligands which are found to be important one in the context of Envelope protein are Nafcillin, Nabumetone, Octacosanol, Cinametic Acid, Lauric Acid, Salmeterol, Palmidrol, Ascorbyl Palmitate and Guaifenesin. Their molecular structures generated by RDKit tool are provided in the Supplementary Fig. 1.
The following are the supplementary data related to this article.Supplementary Fig. 1.
Molecular structure of the final set of drugs sent for docking.
3.4. Looking into the mechanism of action of the selected drugs and nutraceuticals based on the varied symptoms of COVID infected patients
The known mechanism of action of the screened ligands are discussed below in the perspective of its actions against the main threatening outcome in COVID-19 patients i.e. Acute Respiratory Distress Syndrome (ARDS) as well as other (mainly) age related risk factors like hypertension, cholesterol etc. which has made aged people more vulnerable towards COVID-19 attack. Their detailed mode of action is depicted in Fig. 5A.
Fig. 5.
A. Existing mechanism of action of the repurposed FDA approved pharmaceuticals and nutraceuticals within human Abbreviations: ABCA1- ATP-binding cassette transporter 1 AC- Acetyl Cyclase Ang- Angiotensin AT1R- Angiotensin1 Receptor ATP- Adenosine Tri Phosphate CAP- Community acquired Pneumonia ARDS- Acute Respiratory Distress Syndrome CETP- Cholesteryl ester transfer protein COX- Cyclooxygenase DAG –Diacylglycerol HDL- high Density Lipoprotein IL- Interleukin IP3- Inositol Tri Phosphate LCAT- Lecithin–cholesterol acyltransferase LDL- Low Density Lipoprotein MLCK- Myosin Light Chain Kinase PG- Prostaglandin PIP2-Phosphatidylinositol4,5-bisphosphate PLC- Phospholipase C PK- Protein Kinase PPAR - Peroxisome proliferator-activated receptor ROS- Reactive Oxygen Species TNF- Tumor necrosis Factor.
B. Predicted mode of action of the repurposed FDA approved pharmaceuticals and nutraceuticals against E-protein to combat SARS-CoV-2 mediated disease symptoms.
Nafcillin is a semi synthetic, narrow spectrum antibiotic which is a beta lactamase resistant penicillin [55]. The bactericidal action of penicillin is to inhibit cell wall synthesis due to the presence of beta lactam ring. [56]. However, certain bacteria develop resistance against the beta lactam ring by synthesizing beta lactam inhibitors i.e. beta-lactamase or penicillinase [57]. To combat this resistance, penicillinase resistance drugs are introduced in the market [58]. Currently, Nafcillin is being used in the treatment of penicillinase producing staphylococcal species particularly methicillin sensitive Staphylococcus aureus (MSSA) [59]. Nafcillin is also being used to treat non specific lower respiratory tract infection as well as community-acquired pneumonia (CAP) [60]. Nafcillin is not known to cause life threatening adverse side effects and in our analysis, it shows the highest binding affinity with the TMD domain of monomeric E-protein. Thus, Nafcillin can be taken into consideration for redirecting its purpose for the treatment of SARS-CoV-2 infection as it could also combat bacterial co-infection in a COVID patient which produce the same symptoms as seen in SARS-CoV-2 infection (Fig. 5A).
Nabumetone is an FDA approved non selective anti inflammatory drug (NSAID) which is being used for its anti inflammatory and anti pyretic effects [61]. It is a prodrug which goes though biotransformation within the liver to produce the active component, 6-methoxy-2-naphthylacetic acid (6MNA) [62], that inhibits the synthesis of prostaglandins by acting on Cyclooxygenase (COX) I and II [63]. Prostaglandins are responsible for initiating fever by signalling the hypothalamus to increase body temperature [64]. Prostaglandin also acts as inflammatory mediator acting on blood vessels to promote inflammatory response [65,66]. NSAIDs mediate anti-inflammatory effect by preventing vasodilation, reducing capillary permeability and cytokine release from endothelial cells [67]. Altogether, these effects impede the migration of immunocompetent cells the site of injury thereby preventing uncontrolled immune system activation and inflammation [68] (Fig. 5A).
During the initial phase of COVID-19, we see the activation of uncontrolled immune response where leukocytes are recruited all over the body [69]. This is followed by an immunosuppression state with death of T and B cells further exacerbating the situation [70]. Permeability of the blood capillaries, lining the lung alveolar wall increases, causing fluids to leak inside, resulting in pulmonary edema and ARDS [71]. Literature evidences suggest that such situation is triggered by the viral E-protein [32]. Hence, anti-inflammatory agent Nabumetone, by targeting on the monomeric E-protein can hamper its formation of functional pentameric form which in turn could be beneficial in lowering the state of uncontrolled immune response. Although reported to cause less nephrotoxicity and Gastro intestinal (GI) toxicity than other NSAIDs, it still produce considerable side effects and is prescribed to be used by caution particularly for people with heart or kidney condition, high blood pressure and complications arising from thrombosis [72,73].
Octacosanol is the main component of plant extracted natural wax and is a low–molecular-weight primary aliphatic alcohol [74]. Its role is mainly investigated for the treatment of Parkinson's disease [75]. It is approved as Nutraceuticals by FDA and is marketed as the main component of Policosanol (PC), a generic term for natural mixture of primary alcohols isolated originally from sugarcane wax [76]. The first PC supplement produced by Dalmer Laboratories in La Habana, Cuba [77] has been approved as a medication for lowering total cholesterol (TC) and breaking down low density lipoprotein cholesterol (LDL-C) in more than 25 countries including India(Trade name: Zetanol) [78]. It is known to lower cholesterol levels by inducing phosphorylation of AMPK and reducing the activity of HMG-CoA reductase, which is one of the key component of cholesterol biosynthesis [79]. It is also said to increase efflux of tissue LDL by High Density Lipoprotein(HDL) via reverse cholesterol transport pathway and resulting in bigger HDL particles by inhibiting the activity cholesteryl ester transfer protein(CETP) [80,81]. Bigger HDL particles are postulated to exert more protective function in comparison to total HDL particles [82].
COVID-19 is known to severely affect the elderly population. People with atherosclerosis and higher cholesterol levels are more vulnerable to severe symptoms [83]. Cholesterol is an essential component for cell membrane structural makeup [84]. Synthesized by the liver, cholesterol is transported via bloodstreams packed within lipoprotein particles to various organs including lungs [85,86]. LDL is the major blood cholesterol carrier [87]. When LDL levels are high, organs could no longer take up the excess LDLs. They are then oxidised and engulfed by macrophages, get deposited along the arterial wall resulting in plaque formation (atherosclerosis) which increases the risk of stroke and heart attacks [88]. Moreover excess cholesterol in the lung is transported out via ABCA1 efflux pump back to the liver for recycling [89]. This mechanism of effluxion is inhibited during chronic inflammation and reduced with age causing macrophage rich cholesterol to deposit around tissue periphery resulting in increase tissue cholesterol [90,91]. SARS-CoV-2 uses Spike(S) protein to enter within the host cell cytosol by binding to surface receptor angiotensin-converting enzyme II (ACE2) followed by cleavage of S protein with host proteases [92]. A recent work by Wang et al. pointed that increased tissue cholesterol deposits with age can provide more viral entry points by trafficking and concentrating ACE2 as well as host proteases within lipid raft [93]. Hence, Octacosanol can be utilized both as a tissue cholesterol lowering agent to reduce risk of viral entry beside its repurposed role to inhibit E-protein oligomerization.
Cinametic acid is an FDA approved food additive, mainly obtained from oil of cinnamon and other plant sources [94]. Among the many therapeutic functions of cinnamic acid, one of its role has also been linked to inhibit angiotensin-converting enzyme (ACE) [95]. ACE converts Angiotensin (Ang) I to Ang II [96]. Ang II is responsible for constricting blood vessels and increase blood pressure or hypertension, one of the risk factors for COVID-19 via binding to Angiotensin1 Receptor(AT1R) and activating a cascade of signalling pathways [97,98](shown in Fig. 5A). Role of Cinametic acid in inhibiting ACE will hamper conversion of Ang I to Ang II which can reduce hypertension. Further, Ang II gets converted to Ang-(I-VII) by ACE2 in the absence of ACE [99]. Ang(I-VII) has a protective role in the system as it binds to Mas1R receptor and helps in attenuating inflammation and fibrosis related lung injury. Moreover, this engagement of ACE2 in AngII conversion could in turn decrease the number of free tissue ACE2 for binding of SARS-CoV-2 spike proteins proposed by South et al. [100].
Lauric acid/Dodecanoic acid, major source of which is coconut, can also be found in other plants [101]. It is a medium chain saturated fatty acid [102]. It can also be found in cow, goat and human breast milk [103]. Inside the body, lauric acid gets converted to the biologically active compound Monolaurin which is known for exerting antimicrobial activity [104], has been recognized as safe (GRAS) by the U.S. Food and Drug Administration (FDA) and are available as capsules. Particularly medium chain fatty acids are known to exert antiviral action in high doses by disrupting viral envelope and making the virus susceptible to immune attack. In this respect also, Lauric acid is an indispensable choice for SARS-CoV-2 inhibition as breakdown of viral coating (Fig. 5A) would result in easy clearance of virus from the system without giving rise to medical [105] complications. Further its inhibiting action on the viral E-protein oligomerization increases its potential as a therapeutic agent against the viral attack.
Ascorbyl palmitate is an FDA approved small molecule. Mainly, it is a fat soluble form of Vitamin C formed by the ester of ascorbic acid and palmitic acid [106]. Being an amphipathic molecule, it has an advantage of being more stable and easily enters into cell membranes [107]. Within human red blood cells, ascorbyl palmitate can exhibit protective property against oxidative damage by free radicals [108] (Fig. 5A). The pathophysiological processes involved in COVID-19 can be linked to oxidative stress which increases with age [109]. Particularly, deprivation of antioxidant mechanisms is pivotal for viral replication and proliferation [110]. Generation of high level of Reactive Oxygen species during viral infection can lead to oxidative damage of cells as a result of increased inflammation [111]. The role of E-protein in activating inflammasome pathway has already been discussed in the introduction. Hence, ascorbyl palmitate which targets E-protein can be a great boost to the immune system not only by reducing oxidative stress but also by enhancing phagocytosis, chemocytosis and interferon production.
Palmidrol / palmitoylethanolamide(PEA) has been initially identified in egg yolk. PEA is a natural fatty acid amide synthesized locally in animal and human tissues from the most common fatty acid source palmitic acid [112]. PEA is known to act locally and is produced when needed thus maintaining tight balance between production and breakdown [113]. PEA is known to be involved in the endocannabinoid system due to its affinity for the novel orphan cannabinoid receptors GPR55 and GPR119 [114,115]. In addition to its anti cancer, anti epileptic and neuroprotective properties, PEA is widely known to relieve inflammation associated with respiratory infections such as influenza [116]. Influenza is characterized by the production of pro inflammatory cytokines such as IL-6, IL-10 and TNF(Tumor Necrosis Factor)-α [117,118]. PEA is known to lessen the production of inflammatory cytokines by acting as the agonist of PPAR-α receptor and reduces the expression of NF-κβ [119] (Fig. 5A). It is also responsible for shifting the activated mast cells to the resting state by preventing mast cell migration and release of mast cell mediators like histamine through degranulation [120]. Thus, PEA can combat inflammation through various modes of action and has been found advantageous for treating respiratory viral infections for its minimal side effects and promising efficacy. Currently, PEA is available as a FDA approved nutraceutical under trade name PeaPure and is marketed under the trade name Nomast in Spain and Italy for special medical purposes [121]. These properties along with its capability to inhibit E protein oligomerization makes it a potential nutraceutical to fight against COVID 19.
Salmeterol is a long-acting beta2-adrenergic agonist approved by FDA for treatment of asthma and COPD or high altitude pulmonary edema by stimulating beta2-adrenoceptors predominant in the bronchial smooth muscles [122]. beta2 receptor coupled to the Gs protein, when activated, catalyse the formation of cyclic adenosine monophosphate (cAMP) and activate protein kinase A(PKA), thus inactivating myosin light-chain kinase [123] (Fig. 5A). This results in dilation of bronchioles and proper passage of air through lung airways which gets obstructed and narrowed due to the accumulation of pus, swelling of alveolar wall linings muscle spasms [124]. These would otherwise lead to shortness of breath, difficulty in breathing, cough and congestion [125]. Such manifestations are also observed among COVID-19 patients which is caused by E-protein and thus Salmeterol which can target E-protein have a high potential to be used for anti viral therapy for COVID-19. Salmeterol can also prevent air passage narrowing induced by mast cell released histamine [126]. However, beta2-adrenergic receptors are also found to be present in heart to some extent and thus salmeterol is advised to be administered with caution to patients which could otherwise lead up to undesirable cardiac effects [127].
Guaifenesin is an FDA approved over the counter(OTC) or non prescription expectorant for treatment of cough and common cold [128]. It aids in the clearance of mucous and other respiratory tract secretion by increasing the volume of trachea and bronchi and reducing mucus viscosity which would otherwise lead to congestion, chronic bronchitis and COPD which are commonly seen in ARDS. Hereby, the action of Guaifenesin result in more productive cough [129], thus combating the condition of ARDS. This is also expected to happen if administered to COVID-19 patients as it has the potential to disrupt the formation of the pentameric structure of E-protein which causes ARDS. It is a relatively safe drug to use though not recommended for children below the age of six [130].
The overall discussion above regarding the known mechanism of action of the ligands (shown in Fig. 5A) show that most of them are able to combat the various symptoms of Acute Respiratory Distress Syndrome (ARDS), the signature indicator in COVID-19 patients. Other drugs can alleviate major COVID-19 risk factors such as hypertension, age induced tissue cholesterol etc.
On the other hand, we have shown that these ligands can target the monomeric structure of viral E-protein, thus inhibiting the formation of pentameric form of E-protein. As mentioned in the introduction, functional E-protein triggers an over-expression of inflammatory cytokines like IL-1β, resulting in edema and other characteristic symptoms of ARDS [32]. Hence, these ligands being capable of inhibiting the formation of functional E-protein has the potential of subduing ARDS related complications (Fig. 5B). Thus, their repurposed role can aid in overall patient recovery by reverting SARS-CoV-2 induced conditions of ARDS.
4. Conclusion
The viroporin channel activity of SARS-CoV E-protein in its pentameric form plays a significant role in viral pathogenesis and is producing the characteristics symptoms associated with ARDS. After synthesis of monomeric E-protein following replication, they make their way to the ERGIC compartment, where most of them get inserted into the membranes in its oligomerized form and carry out their pathogenic activity by interacting with several host proteins. The TMD domain of E-protein is responsible for this oligomerization and mutating the hydrophobic residue valine 25 (V25) to phenylalanine (V25F) can abolish the oligomeric structure completely whereas mutation of asparagine 15 (N15) to alanine (N15A) can reduce the chance of pentamer formation to some extent. It has also been reported that the loss of viroporin activity could result in significant relief from edema and inflammation. To cope up with such changes to oligomerize again, the virus introduces certain mutations from position 15 to 37 of the TMD region. Hence, the importance of this region in maintaining the oligomeric form and carrying out pathogenesis seems to be immense. SARS-CoV-2 E-protein sequence shares 94.74% similarity with that of E-protein SARS-CoV and the amino acid sequences in the region 15 to 37 were found to be totally conserved among SARS-CoV and SARS-CoV-2. In this study, we aim to target the monomeric E-protein so as to prevent its oligomerization and subsequent pathogenic activities. The loss of oligomerization would result in formation of defective viral particles.
We have obtained drug information from various available sources and have utilized AI based deep learning for the first phase screening of drugs based on their binding affinity for E-protein. The small size of the E-protein, along with the membrane spanning nature and amino acid properties of the concerned region have also been considered during the process of drug screening. Thus, pattern based searching was also employed to screen suitable candidates in the first phase of screening. Next, our focus was to concentrate only on FDA approved commercial drugs which are already available easily along with FDA approved Nutraceuticals which have immense health benefits as dietary supplements. Our concern was also to check the severity of the adverse side effects of these drugs. Hence, second phase of screening was done based on these criteria and selected drugs were utilized for blind docking with the SARS-CoV-2 E-protein to obtain unbiased results in the third phase. Although there exists lack of scoring function which justifies the binding affinity between the ligand and the target protein universally [131], we have considered orientation of binding, site of binding and type of binding energy along with docking score to judge the binding affinity between them. Hereby, we have come up finally with 9 ligand molecules which has been found to be grouped into three distinct clusters, where ligands in cluster 1 and 2 interacts with the concerned amino acid residues in the zone of interest and those in cluster 3 covers the outer surface of the E-protein in a way that could hinder the flexibility of the protein necessary for interacting with other partners. Finally, we have checked the mechanism of action of these drugs and nutraceuticals emphasizing the mode in which they could combat the diverse complications associated with COVID-19 such as cytokine induced inflammation, COPD and ARDS, ACE induced hypertension, tissue cholesterol induced complications among others. Recommendation of the drugs for each patient based on their medical history as well as the symptoms experienced by them, would lead to better result and could avoid unwanted clinical complications. Overall, validation of the action of these repurposed pharmaceuticals and nutraceuticals in clinical settings will strengthen our conclusion and thus could portray the COVID-19 therapeutic roadmap which will serve as a ready remedy to stop this rapidly evolving pandemic.
Multiple sequence alignment between Envelope(E), Matrix(M), Nucleocaspid(N) and Spike(S) protein of SARS-CoV and SARS-CoV-2.
Interaction of the final set of 9 ligand molecules with SARS-CoV-2 E-protein.
Acknowledgments
Acknowledgement
We are grateful to Council of Scientific and Industrial Research (CSIR) and Department of Science and Technology (DST) for financial support. We also thank University of Kalyani for their constant support to carry out this research work.
References
- 1.Peters A., et al. Understanding the emerging coronavirus: what it means for health security and infection prevention. J. Hosp. Infect. 2020;104(4):440–448. doi: 10.1016/j.jhin.2020.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Worldometers.info 2020. Worldometers.info/coronavirus
- 3.Wu D., et al. The SARS-CoV-2 outbreak: what we know. Int. J. Infect. Dis. 2020;94:44–48. doi: 10.1016/j.ijid.2020.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zheng J. SARS-CoV-2: an emerging coronavirus that causes a global threat. Int. J. Biol. Sci. 2020;16(10):1678–1685. doi: 10.7150/ijbs.45053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cheng V.C., et al. Severe acute respiratory syndrome coronavirus as an agent of emerging and reemerging infection. Clin. Microbiol. Rev. 2007;20(4):660–694. doi: 10.1128/CMR.00023-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wolfel R., et al. Virological assessment of hospitalized patients with COVID-2019. Nature. 2020;581(7809):465–469. doi: 10.1038/s41586-020-2196-x. [DOI] [PubMed] [Google Scholar]
- 7.Satija N., Lal S.K. The molecular biology of SARS coronavirus. Ann. N. Y. Acad. Sci. 2007;1102:26–38. doi: 10.1196/annals.1408.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wu A., et al. Genome composition and divergence of the novel coronavirus (2019-nCoV) originating in China. Cell Host Microbe. 2020;27(3):325–328. doi: 10.1016/j.chom.2020.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Glowacka I., et al. Evidence that TMPRSS2 activates the severe acute respiratory syndrome coronavirus spike protein for membrane fusion and reduces viral control by the humoral immune response. J. Virol. 2011;85(9):4122–4134. doi: 10.1128/JVI.02232-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McBride R., van Zyl M., Fielding B.C. The coronavirus nucleocapsid is a multifunctional protein. Viruses. 2014;6(8):2991–3018. doi: 10.3390/v6082991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mousavizadeh L., Ghasemi S. Genotype and phenotype of COVID-19: their roles in pathogenesis. J. Microbiol. Immunol. Infect. 2020 doi: 10.1016/j.jmii.2020.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wu C., et al. Analysis of therapeutic targets for SARS-CoV-2 and discovery of potential drugs by computational methods. Acta Pharm. Sin. B. 2020;10(5):766–788. doi: 10.1016/j.apsb.2020.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Saxena A. Drug targets for COVID-19 therapeutics: ongoing global efforts. J. Biosci. 2020;45 doi: 10.1007/s12038-020-00067-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shah B., Modi P., Sagar S.R. In silico studies on therapeutic agents for COVID-19: drug repurposing approach. Life Sci. 2020;252:117652. doi: 10.1016/j.lfs.2020.117652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sinha S.K., et al. An in-silico evaluation of different Saikosaponins for their potency against SARS-CoV-2 using NSP15 and fusion spike glycoprotein as targets. J. Biomol. Struct. Dyn. 2020:1–12. doi: 10.1080/07391102.2020.1762741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.de Oliveira O.V., et al. Repurposing approved drugs as inhibitors of SARS-CoV-2 S-protein from molecular modeling and virtual screening. J. Biomol. Struct. Dyn. 2020:1–10. doi: 10.1080/07391102.2020.1772885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Choudhary S., Malik Y.S., Tomar S. Identification of SARS-CoV-2 cell entry inhibitors by drug repurposing using in silico structure-based virtual screening approach. Front. Immunol. 2020;11:1664. doi: 10.3389/fimmu.2020.01664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pandey P., et al. Targeting SARS-CoV-2 spike protein of COVID-19 with naturally occurring phytochemicals: an in silico study for drug development. J. Biomol. Struct. Dyn. 2020:1–11. doi: 10.1080/07391102.2020.1796811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kulkarni S.A., et al. Computational evaluation of major components from plant essential oils as potent inhibitors of SARS-CoV-2 spike protein. J. Mol. Struct. 2020;1221:128823. doi: 10.1016/j.molstruc.2020.128823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kuo L., Hurst K.R., Masters P.S. Exceptional flexibility in the sequence requirements for coronavirus small envelope protein function. J. Virol. 2007;81(5):2249–2262. doi: 10.1128/JVI.01577-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Torres J., et al. Conductance and amantadine binding of a pore formed by a lysine-flanked transmembrane domain of SARS coronavirus envelope protein. Protein Sci. 2007;16(9):2065–2071. doi: 10.1110/ps.062730007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu Q., et al. The E protein is a multifunctional membrane protein of SARS-CoV. Genom. Proteom. Bioinforma. 2003;1(2):131–144. doi: 10.1016/S1672-0229(03)01017-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li Y., et al. Structure of a conserved Golgi complex-targeting signal in coronavirus envelope proteins. J. Biol. Chem. 2014;289(18):12535–12549. doi: 10.1074/jbc.M114.560094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Verdia-Baguena C., et al. Coronavirus E protein forms ion channels with functionally and structurally-involved membrane lipids. Virology. 2012;432(2):485–494. doi: 10.1016/j.virol.2012.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pervushin K., et al. Structure and inhibition of the SARS coronavirus envelope protein ion channel. PLoS Pathog. 2009;5(7) doi: 10.1371/journal.ppat.1000511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nieto-Torres J.L., et al. Severe acute respiratory syndrome coronavirus envelope protein Ion Channel activity promotes virus fitness and pathogenesis. PLoS Pathog. 2014;10(5) doi: 10.1371/journal.ppat.1004077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schoeman D., Fielding B.C. Coronavirus envelope protein: current knowledge. Virol. J. 2019;16(1):69. doi: 10.1186/s12985-019-1182-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ortego J., et al. Generation of a replication-competent, propagation-deficient virus vector based on the transmissible gastroenteritis coronavirus genome. J. Virol. 2002;76(22):11518–11529. doi: 10.1128/JVI.76.22.11518-11529.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Javier R.T., Rice A.P. Emerging theme: cellular PDZ proteins as common targets of pathogenic viruses. J. Virol. 2011;85(22):11544–11556. doi: 10.1128/JVI.05410-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jimenez-Guardeno J.M., et al. The PDZ-binding motif of severe acute respiratory syndrome coronavirus envelope protein is a determinant of viral pathogenesis. PLoS Pathog. 2014;10(8) doi: 10.1371/journal.ppat.1004320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang Y., et al. Bcl-xL inhibits T-cell apoptosis induced by expression of SARS coronavirus E protein in the absence of growth factors. Biochem. J. 2005;392(Pt 1):135–143. doi: 10.1042/BJ20050698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nieto-Torres J.L., et al. Severe acute respiratory syndrome coronavirus envelope protein ion channel activity promotes virus fitness and pathogenesis. PLoS Pathog. 2014;10(5) doi: 10.1371/journal.ppat.1004077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Torres J., et al. Protein-protein interactions of Viroporins in coronaviruses and Paramyxoviruses: new targets for antivirals? Viruses. 2015;7(6):2858–2883. doi: 10.3390/v7062750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gupta M.K., et al. In-silico approaches to detect inhibitors of the human severe acute respiratory syndrome coronavirus envelope protein ion channel. J. Biomol. Struct. Dyn. 2020:1–11. doi: 10.1080/07391102.2020.1751300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.The UniProt, C UniProt: the universal protein knowledgebase. Nucleic Acids Res. 2017;45(D1):D158–D169. doi: 10.1093/nar/gkw1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Coordinators N.R. Database resources of the National Center for biotechnology information. Nucleic Acids Res. 2016;44(D1):D7–19. doi: 10.1093/nar/gkv1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Clamp M., et al. The Jalview Java alignment editor. Bioinformatics. 2004;20(3):426–427. doi: 10.1093/bioinformatics/btg430. [DOI] [PubMed] [Google Scholar]
- 38.Elbe S., Buckland-Merrett G. Data, disease and diplomacy: GISAID's innovative contribution to global health. Global Chall. 2017;1(1):33–46. doi: 10.1002/gch2.1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schwede T., et al. SWISS-MODEL: an automated protein homology-modeling server. Nucleic Acids Res. 2003;31(13):3381–3385. doi: 10.1093/nar/gkg520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gaulton A., et al. ChEMBL: a large-scale bioactivity database for drug discovery. Nucleic Acids Res. 2012;40(Database issue):D1100–D1107. doi: 10.1093/nar/gkr777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gaulton A., et al. The ChEMBL database in 2017. Nucleic Acids Res. 2017;45(D1):D945–D954. doi: 10.1093/nar/gkw1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang H., et al. IVS2vec: a tool of inverse virtual screening based on word2vec and deep learning techniques. Methods. 2019;166:57–65. doi: 10.1016/j.ymeth.2019.03.012. [DOI] [PubMed] [Google Scholar]
- 43.Jaeger S., Fulle S., Turk S. Mol2vec: unsupervised machine learning approach with chemical intuition. J. Chem. Inf. Model. 2018;58(1):27–35. doi: 10.1021/acs.jcim.7b00616. [DOI] [PubMed] [Google Scholar]
- 44.Mikolov T., et al. Efficient estimation of word representations in vector space. ICLR: Proceeding of the International Conference on Learning Representations Workshop Track, Arizona, USA. 2013:1301–3781. [Google Scholar]
- 45.Wang R., et al. The PDBbind database: collection of binding affinities for protein-ligand complexes with known three-dimensional structures. J. Med. Chem. 2004;47(12):2977–2980. doi: 10.1021/jm030580l. [DOI] [PubMed] [Google Scholar]
- 46.Duangjit S., et al. Role of the charge, carbon chain length, and content of surfactant on the skin penetration of meloxicam-loaded liposomes. Int. J. Nanomedicine. 2014;9:2005–2017. doi: 10.2147/IJN.S60674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Trott O., Olson A.J. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010;31(2):455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ramirez D., Caballero J. Is it reliable to take the molecular docking top scoring position as the best solution without considering available structural data? Molecules. 2018;23(5) doi: 10.3390/molecules23051038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Warren G.L., et al. A critical assessment of docking programs and scoring functions. J. Med. Chem. 2006;49(20):5912–5931. doi: 10.1021/jm050362n. [DOI] [PubMed] [Google Scholar]
- 50.Kim R., Skolnick J. Assessment of programs for ligand binding affinity prediction. J. Comput. Chem. 2008;29(8):1316–1331. doi: 10.1002/jcc.20893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Guedes I.A., de Magalhaes C.S., Dardenne L.E. Receptor-ligand molecular docking. Biophys. Rev. 2014;6(1):75–87. doi: 10.1007/s12551-013-0130-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Groner B., Weber A., Mack L. Increasing the range of drug targets: interacting peptides provide leads for the development of oncoprotein inhibitors. Bioengineered. 2012;3(6):320–325. doi: 10.4161/bioe.21272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lagorce D., et al. Computational analysis of calculated physicochemical and ADMET properties of protein-protein interaction inhibitors. Sci. Rep. 2017;7:46277. doi: 10.1038/srep46277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schneider M., et al. Towards accurate high-throughput ligand affinity prediction by exploiting structural ensembles, docking metrics and ligand similarity. Bioinformatics. 2020;36(1):160–168. doi: 10.1093/bioinformatics/btz538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sidell S., et al. New antistaphylococcal antibiotics. Comparative in vitro and in vivo activity of cephalothin, nafcillin, cloxacillin, oxacillin, and methicillin. Arch. Intern. Med. 1963;112:21–28. doi: 10.1001/archinte.1963.03860010067005. [DOI] [PubMed] [Google Scholar]
- 56.Donowitz G.R., Mandell G.L. Beta-lactam antibiotics (1) N. Engl. J. Med. 1988;318(7):419–426. doi: 10.1056/NEJM198802183180706. [DOI] [PubMed] [Google Scholar]
- 57.Herzberg O., Moult J. Bacterial resistance to beta-lactam antibiotics: crystal structure of beta-lactamase from Staphylococcus aureus PC1 at 2.5 a resolution. Science. 1987;236(4802):694–701. doi: 10.1126/science.3107125. [DOI] [PubMed] [Google Scholar]
- 58.McDougal L.K., Thornsberry C. The role of beta-lactamase in staphylococcal resistance to penicillinase-resistant penicillins and cephalosporins. J. Clin. Microbiol. 1986;23(5):832–839. doi: 10.1128/jcm.23.5.832-839.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schweizer M.L., et al. Comparative effectiveness of nafcillin or cefazolin versus vancomycin in methicillin-susceptible Staphylococcus aureus bacteremia. BMC Infect. Dis. 2011;11(1):279. doi: 10.1186/1471-2334-11-279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wynn M., et al. Evaluation of the efficacy and safety of outpatient parenteral antimicrobial therapy for infections with methicillin-sensitive Staphylococcus aureus. South. Med. J. 2005;98(6):590–595. doi: 10.1097/01.SMJ.0000145300.28736.BB. [DOI] [PubMed] [Google Scholar]
- 61.Friedel H.A., Langtry H.D., Buckley M.M. Nabumetone. A reappraisal of its pharmacology and therapeutic use in rheumatic diseases. Drugs. 1993;45(1):131–156. doi: 10.2165/00003495-199345010-00009. [DOI] [PubMed] [Google Scholar]
- 62.Mikami E., et al. Simultaneous analysis of naproxen, nabumetone and its major metabolite 6-methoxy-2-naphthylacetic acid in pharmaceuticals and human urine by high-performance liquid chromatography. J. Pharm. Biomed. Anal. 2000;23(5):917–925. doi: 10.1016/s0731-7085(00)00365-4. [DOI] [PubMed] [Google Scholar]
- 63.Matsunaga K., et al. Inhibitory effects of nabumetone, a cyclooxygenase-2 inhibitor, and esculetin, a lipoxygenase inhibitor, on N-methyl-N-nitrosourea-induced mammary carcinogenesis in rats. Jpn. J. Cancer Res. 1998;89(5):496–501. doi: 10.1111/j.1349-7006.1998.tb03289.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bernheim H.A., Gilbert T.M., Stitt J.T. Prostaglandin E levels in third ventricular cerebrospinal fluid of rabbits during fever and changes in body temperature. J. Physiol. 1980;301:69–78. doi: 10.1113/jphysiol.1980.sp013189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kuehl F.A., Jr. And R.W. Egan, Prostaglandins, arachidonic acid, and inflammation. Science. 1980;210(4473):978–984. doi: 10.1126/science.6254151. [DOI] [PubMed] [Google Scholar]
- 66.Rubanyi G.M., Vanhoutte P.M. Hypoxia releases a vasoconstrictor substance from the canine vascular endothelium. J. Physiol. 1985;364:45–56. doi: 10.1113/jphysiol.1985.sp015728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Fiorucci S., Santucci L., Distrutti E. NSAIDs, coxibs, CINOD and H2S-releasing NSAIDs: what lies beyond the horizon. Dig. Liver Dis. 2007;39(12):1043–1051. doi: 10.1016/j.dld.2007.09.001. [DOI] [PubMed] [Google Scholar]
- 68.Ghlichloo I., Gerriets V. 2020. Nonsteroidal Anti-inflammatory Drugs (NSAIDs), in StatPearls. (Treasure Island (FL)) [PubMed] [Google Scholar]
- 69.Georgiev T. Coronavirus disease 2019 (COVID-19) and anti-rheumatic drugs. Rheumatol. Int. 2020;40(5):825–826. doi: 10.1007/s00296-020-04570-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Cao C., Yu M., Chai Y. Pathological alteration and therapeutic implications of sepsis-induced immune cell apoptosis. Cell Death Dis. 2019;10(10):782. doi: 10.1038/s41419-019-2015-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Matthay M.A., Aldrich J.M., Gotts J.E. Treatment for severe acute respiratory distress syndrome from COVID-19. Lancet Respir. Med. 2020;8(5):433–434. doi: 10.1016/S2213-2600(20)30127-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Arai Y., et al. Low direct cytotoxicity of nabumetone on gastric mucosal cells. Dig. Dis. Sci. 2005;50(9):1641–1646. doi: 10.1007/s10620-005-2909-x. [DOI] [PubMed] [Google Scholar]
- 73.Bjarnason I., et al. Mechanisms of damage to the gastrointestinal tract from nonsteroidal anti-inflammatory drugs. Gastroenterology. 2018;154(3):500–514. doi: 10.1053/j.gastro.2017.10.049. [DOI] [PubMed] [Google Scholar]
- 74.Kabir Y. In: Health Benefits of Octacosanol and Other Long-Chain Aliphatic Fatty Alcohols from Plants, in Herbal Medicine in India: Indigenous Knowledge, Practice, Innovation and its Value. Sen S., Chakraborty R., editors. Springer Singapore; Singapore: 2020. pp. 413–425. [Google Scholar]
- 75.Wang T., et al. Anti-parkinsonian effects of octacosanol in 1-methyl-4-phenyl-1,2,3,6 tetrahydropyridine-treated mice. Neural Regen. Res. 2012;7(14):1080–1087. doi: 10.3969/j.issn.1673-5374.2012.14.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Irmak S., Dunford N.T. Policosanol contents and compositions of wheat varieties. J. Agric. Food Chem. 2005;53(14):5583–5586. doi: 10.1021/jf050508r. [DOI] [PubMed] [Google Scholar]
- 77.Janikula M. Policosanol: a new treatment for cardiovascular disease? Altern. Med. Rev. 2002;7(3):203–217. [PubMed] [Google Scholar]
- 78.Stockigt J. In: Clinical Strategies in the Testing of Thyroid Function. Endotext L.J. De Groot, et al., editors. 2000. (South Dartmouth (MA)) [Google Scholar]
- 79.Nam D.E., et al. Policosanol attenuates cholesterol synthesis via AMPK activation in Hypercholesterolemic rats. J. Med. Food. 2019;22(11):1110–1117. doi: 10.1089/jmf.2019.4491. [DOI] [PubMed] [Google Scholar]
- 80.Cho K.-H. Springer Singapore; Singapore: 2019. Enhancement of HDL by Policosanol, in High-Density Lipoproteins as Biomarkers and Therapeutic Tools: Volume 2. Improvement and Enhancement of HDL and Clinical Applications; pp. 115–261. [Google Scholar]
- 81.Shen J., Luo F., Lin Q. Policosanol: extraction and biological functions. J. Funct. Foods. 2019;57:351–360. [Google Scholar]
- 82.Kwiterovich P.O., Jr. The metabolic pathways of high-density lipoprotein, low-density lipoprotein, and triglycerides: a current review. Am. J. Cardiol. 2000;86(12A):5L–10L. doi: 10.1016/s0002-9149(00)01461-2. [DOI] [PubMed] [Google Scholar]
- 83.Vuorio A., Watts G.F., Kovanen P.T. Familial hypercholesterolaemia and COVID-19: triggering of increased sustained cardiovascular risk. J. Intern. Med. 2020;287(6):746–747. doi: 10.1111/joim.13070. [DOI] [PubMed] [Google Scholar]
- 84.Ayuyan A.G., Cohen F.S. The chemical potential of plasma membrane cholesterol: implications for cell biology. Biophys. J. 2018;114(4):904–918. doi: 10.1016/j.bpj.2017.12.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Patton, K.T.T., Gary A., Anatomy and Physiology (7 ed.). ISBN 978-9996057762. RetrievedMosby/Elsevier. 20 March 2019:2010. [Google Scholar]
- 86.Mahley R.W. Apolipoprotein E: cholesterol transport protein with expanding role in cell biology. Science. 1988;240(4852):622–630. doi: 10.1126/science.3283935. [DOI] [PubMed] [Google Scholar]
- 87.Pfisterer S.G., Peranen J., Ikonen E. LDL-cholesterol transport to the endoplasmic reticulum: current concepts. Curr. Opin. Lipidol. 2016;27(3):282–287. doi: 10.1097/MOL.0000000000000292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Chistiakov D.A., Bobryshev Y.V., Orekhov A.N. Macrophage-mediated cholesterol handling in atherosclerosis. J. Cell. Mol. Med. 2016;20(1):17–28. doi: 10.1111/jcmm.12689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Yvan-Charvet L., Wang N., Tall A.R. Role of HDL, ABCA1, and ABCG1 transporters in cholesterol efflux and immune responses. Arterioscler. Thromb. Vasc. Biol. 2010;30(2):139–143. doi: 10.1161/ATVBAHA.108.179283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.van Dam M.J., et al. Association between increased arterial-wall thickness and impairment in ABCA1-driven cholesterol efflux: an observational study. Lancet. 2002;359(9300):37–42. doi: 10.1016/S0140-6736(02)07277-X. [DOI] [PubMed] [Google Scholar]
- 91.Clee S.M., et al. Age and residual cholesterol efflux affect HDL cholesterol levels and coronary artery disease in ABCA1 heterozygotes. J. Clin. Invest. 2000;106(10):1263–1270. doi: 10.1172/JCI10727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Hoffmann M., Kleine-Weber H., Pohlmann S. A multibasic cleavage site in the spike protein of SARS-CoV-2 is essential for infection of human lung cells. Mol. Cell. 2020;78(4):779–784. doi: 10.1016/j.molcel.2020.04.022. (e5) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Wang H., et al. The role of high cholesterol in age-related COVID19 lethality. bioRxiv. 2020 [Google Scholar]
- 94.Sova M. Antioxidant and antimicrobial activities of cinnamic acid derivatives. Mini-Rev. Med. Chem. 2012;12(8):749–767. doi: 10.2174/138955712801264792. [DOI] [PubMed] [Google Scholar]
- 95.Mnafgui K., et al. Anti-obesity and cardioprotective effects of cinnamic acid in high fat diet- induced obese rats. J. Food Sci. Technol. 2015;52(7):4369–4377. doi: 10.1007/s13197-014-1488-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Hamdi H.K., Castellon R. A genetic variant of ACE increases cell survival: a new paradigm for biology and disease. Biochem. Biophys. Res. Commun. 2004;318(1):187–191. doi: 10.1016/j.bbrc.2004.04.004. [DOI] [PubMed] [Google Scholar]
- 97.Sugimoto K., et al. Association between angiotensin II type 1 receptor gene polymorphism and essential hypertension: the Ohasama study. Hypertens. Res. 2004;27(8):551–556. doi: 10.1291/hypres.27.551. [DOI] [PubMed] [Google Scholar]
- 98.Turu G., Balla A., Hunyady L. The role of beta-Arrestin proteins in Organization of Signaling and Regulation of the AT1 angiotensin receptor. Front. Endocrinol. (Lausanne) 2019;10:519. doi: 10.3389/fendo.2019.00519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Perrotta F., et al. COVID-19 and the elderly: insights into pathogenesis and clinical decision-making. Aging Clin. Exp. Res. 2020;32(8):1599–1608. doi: 10.1007/s40520-020-01631-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.South A.M., et al. Controversies of renin-angiotensin system inhibition during the COVID-19 pandemic. Nat. Rev. Nephrol. 2020;16(6):305–307. doi: 10.1038/s41581-020-0279-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Dayrit F.M. The properties of Lauric acid and their significance in coconut oil. J. Am. Oil Chem. Soc. 2015;92(1):1–15. [Google Scholar]
- 102.Dayrit F.M. Vol. 143. 2014. Lauric Acid is a Medium-Chain Fatty Acid, Coconut Oil is a Medium-Chain Triglyceride; pp. 157–166. [Google Scholar]
- 103.Beijers R.J., Schaafsma A. Long-chain polyunsaturated fatty acid content in Dutch preterm breast milk; differences in the concentrations of docosahexaenoic acid and arachidonic acid due to length of gestation. Early Hum. Dev. 1996;44(3):215–223. doi: 10.1016/0378-3782(95)01710-0. [DOI] [PubMed] [Google Scholar]
- 104.Hierholzer J.C., Kabara J.J. In vitro effects of Monolaurin compounds on enveloped Rna and DNA viruses. J. Food Saf. 1982;4(1):1–12. doi: 10.1111/j.1745-4565.1982.tb00429.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Thormar H., et al. Inactivation of enveloped viruses and killing of cells by fatty acids and monoglycerides. Antimicrob. Agents Chemother. 1987;31(1):27–31. doi: 10.1128/aac.31.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Spiclin P., Gasperlin M., Kmetec V. Stability of ascorbyl palmitate in topical microemulsions. Int. J. Pharm. 2001;222(2):271–279. doi: 10.1016/s0378-5173(01)00715-3. [DOI] [PubMed] [Google Scholar]
- 107.Jurkovic P., et al. Skin protection against ultraviolet induced free radicals with ascorbyl palmitate in microemulsions. Eur. J. Pharm. Biopharm. 2003;56(1):59–66. doi: 10.1016/s0939-6411(03)00062-6. [DOI] [PubMed] [Google Scholar]
- 108.Ross D., et al. Ascorbate 6-palmitate protects human erythrocytes from oxidative damage. Free Radic. Biol. Med. 1999;26(1–2):81–89. doi: 10.1016/s0891-5849(98)00198-1. [DOI] [PubMed] [Google Scholar]
- 109.Delgado-Roche L., Mesta F. Oxidative stress as key player in severe acute respiratory syndrome coronavirus (SARS-CoV) infection. Arch. Med. Res. 2020;51(5):384–387. doi: 10.1016/j.arcmed.2020.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Khomich O.A., et al. Redox biology of respiratory viral infections. Viruses. 2018:10(8). doi: 10.3390/v10080392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Komaravelli N., Casola A. Respiratory viral infections and subversion of cellular antioxidant defenses. J. Pharmacogenom. Pharmacoproteom. 2014;5(4) doi: 10.4172/2153-0645.1000141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Kuehl F.A., et al. The identification of n-(2-hydroxyethyl)-palmitamide as a naturally occurring anti-inflammatory agent. J. Am. Chem. Soc. 1957;79(20):5577–5578. [Google Scholar]
- 113.Bisogno T. Endogenous cannabinoids: structure and metabolism. J. Neuroendocrinol. 2008;20(s1):1–9. doi: 10.1111/j.1365-2826.2008.01676.x. [DOI] [PubMed] [Google Scholar]
- 114.LoVerme J., et al. The search for the palmitoylethanolamide receptor. Life Sci. 2005;77(14):1685–1698. doi: 10.1016/j.lfs.2005.05.012. [DOI] [PubMed] [Google Scholar]
- 115.Godlewski G., et al. Receptors for acylethanolamides-GPR55 and GPR119. Prostaglandins Other Lipid Mediat. 2009;89(3–4):105–111. doi: 10.1016/j.prostaglandins.2009.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Keppel Hesselink J.M., de Boer T., Witkamp R.F. Palmitoylethanolamide: a natural body-own anti-inflammatory agent, effective and safe against influenza and common cold. Int. J. Inflamm. 2013;2013:151028. doi: 10.1155/2013/151028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Yu X., et al. Intensive cytokine induction in pandemic H1N1 influenza virus infection accompanied by robust production of IL-10 and IL-6. PLoS One. 2011;6(12) doi: 10.1371/journal.pone.0028680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Mogensen T.H., Paludan S.R. Molecular pathways in virus-induced cytokine production. Microbiol. Mol. Biol. Rev. 2001;65(1):131–150. doi: 10.1128/MMBR.65.1.131-150.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Dubrac S., Schmuth M. PPAR-alpha in cutaneous inflammation. Dermato-Endocrinology. 2011;3(1):23–26. doi: 10.4161/derm.3.1.14615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Cerrato S., et al. Effects of palmitoylethanolamide on immunologically induced histamine, PGD2 and TNFalpha release from canine skin mast cells. Vet. Immunol. Immunopathol. 2010;133(1):9–15. doi: 10.1016/j.vetimm.2009.06.011. [DOI] [PubMed] [Google Scholar]
- 121.Keppel Hesselink J.M., de Boer T., Witkamp R.F. Palmitoylethanolamide: a natural body-own anti-inflammatory agent, effective and safe against influenza and common cold. Int. J. Inflamm. 2013;2013:151028. doi: 10.1155/2013/151028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Roberts J.A., et al. The long-acting beta2-agonist salmeterol xinafoate: effects on airway inflammation in asthma. Eur. Respir. J. 1999;14(2):275–282. doi: 10.1034/j.1399-3003.1999.14b07.x. [DOI] [PubMed] [Google Scholar]
- 123.Deshpande D.A., Penn R.B. Targeting G protein-coupled receptor signaling in asthma. Cell. Signal. 2006;18(12):2105–2120. doi: 10.1016/j.cellsig.2006.04.008. [DOI] [PubMed] [Google Scholar]
- 124.Storms W., et al. A comparison of the effects of oral montelukast and inhaled salmeterol on response to rescue bronchodilation after challenge. Respir. Med. 2004;98(11):1051–1062. doi: 10.1016/j.rmed.2004.05.008. [DOI] [PubMed] [Google Scholar]
- 125.Widdicombe J.G. The activity of pulmonary stretch receptors during bronchoconstriction, pulmonary oedema, atelectasis and breathing against a resistance. J. Physiol. 1961;159:436–450. doi: 10.1113/jphysiol.1961.sp006819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Gentilini G., et al. Salmeterol inhibits anaphylactic histamine release from Guinea-pig isolated mast cells. J. Pharm. Pharmacol. 1994;46(1):76–77. doi: 10.1111/j.2042-7158.1994.tb03725.x. [DOI] [PubMed] [Google Scholar]
- 127.Cazzola M., et al. Cardiac effects of formoterol and salmeterol in patients suffering from COPD with preexisting cardiac arrhythmias and hypoxemia. Chest. 1998;114(2):411–415. doi: 10.1378/chest.114.2.411. [DOI] [PubMed] [Google Scholar]
- 128.Dicpinigaitis P.V., et al. Inhibition of cough-reflex sensitivity by benzonatate and guaifenesin in acute viral cough. Respir. Med. 2009;103(6):902–906. doi: 10.1016/j.rmed.2008.12.008. [DOI] [PubMed] [Google Scholar]
- 129.Albrecht H.H., Dicpinigaitis P.V., Guenin E.P. Role of guaifenesin in the management of chronic bronchitis and upper respiratory tract infections. Multidiscip. Respir. Med. 2017;12:31. doi: 10.1186/s40248-017-0113-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.The National Cancer Policy Summit: Opportunities and Challenges in Cancer Research and Care. 2011. (Washington (DC)) [PubMed] [Google Scholar]
- 131.Plewczynski D., et al. Can we trust docking results? Evaluation of seven commonly used programs on PDBbind database. J. Comput. Chem. 2011;32(4):742–755. doi: 10.1002/jcc.21643. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Multiple sequence alignment between Envelope(E), Matrix(M), Nucleocaspid(N) and Spike(S) protein of SARS-CoV and SARS-CoV-2.
Interaction of the final set of 9 ligand molecules with SARS-CoV-2 E-protein.