Abstract
Background
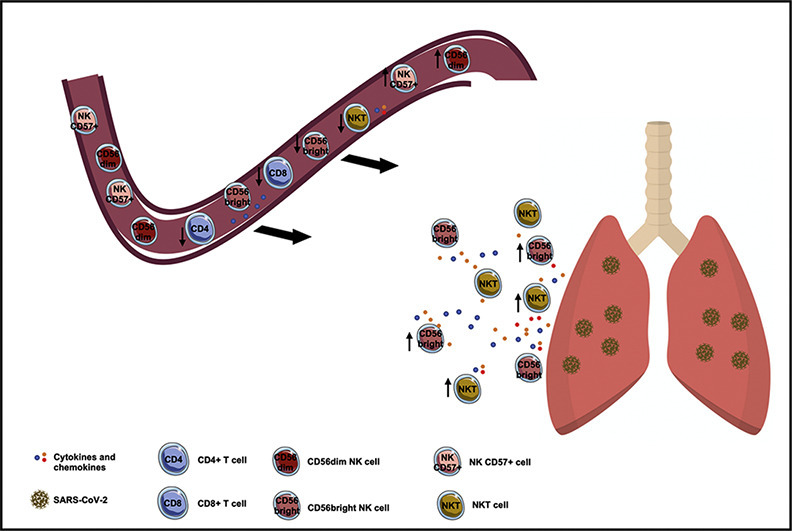
NK cells seem to be mainly involved in COVID-19 pneumonia. Little is known about NKT cells which represent a bridge between innate and adaptive immunity.
Methods
We characterized peripheral blood T, NK and NKT cells in 45 patients with COVID-19 pneumonia (COVID-19 subjects) and 19 healthy donors (HDs). According to the severity of the disease, we stratified COVID-19 subjects into severe and non-severe groups.
Results
Compared to HDs, COVID-19 subjects showed higher percentages of NK CD57+ and CD56dim NK cells and lower percentages of NKT and CD56bright cells. In the severe group we found a significantly lower percentage of NKT cells. In a multiple logistic regression analysis, NKT cell was independently associated with the severity of the disease.
Conclusions
The low percentage of NKT cells in peripheral blood of COVID-19 subjects and the independent association with the severity of the disease suggests a potential role of this subset.
Keywords: Flow Cytometry, SARS-CoV-2, Immunophenotyping analysis, CD56bright, CD56dim
Graphical abstract
Highlights
-
•
High percentages of NK CD57+ cells and CD56dimNK cells in COVID-19 subjects
-
•
Low percentages of CD56bright and NKT cells in COVID-19 subjects
-
•
Severe COVID-19 pneumonia and NKT cell reduction were independently associated
1. Introduction
The pathogenesis of coronavirus disease-2019 (COVID-19) pneumonia related to the novel coronavirus, severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2), is still unknown. Although virological, epidemiological, clinical, routine laboratory, imaging, and management outcome features of patients with COVID-19 pneumonia have been rapidly defined [[1], [2], [3], [4], [5], [6], [7]], the immune system response is not well understood.
Beside dyspnea, hypoxemia and acute respiratory distress, lymphopenia and cytokine release syndrome are also important clinical features in patients with severe COVID-19 pneumonia [4] suggesting that homeostasis of the immune system plays an important role in the development of COVID-19 pneumonia. The increase of neutrophils/lymphocytes ratio and the reduction of lymphocytes were correlated with the severity of the disease and death [[8], [9], [10]]. Furthermore, exhaustion markers, such as the inhibitory NK group 2 member A (NKG2A) receptors on NK cells and CD8+ T cells, are up-regulated in patients with COVID-19 pneumonia and these cytotoxic cells reduced perforin and granzyme content, suggesting an impaired cytotoxic immune response in COVID-19 pneumonia [11].
De Biasi et al., have recently reported an increase of CD57 expression on CD8+ T cells [13]. CD57 is considered a key marker of in vitro replicative senescence and is associated either to human aging or prolonged chronic infection [14]. Cell surface markers, mainly absence of CD28 or expression of CD57, have been used to identify T lymphocytes as senescent in vitro. In the immune system, immunosenescence includes a shift towards less functional T cells [15,16]. However, CD57 expression is reported to be also a marker of mature NK cells. Functionally, CD57 is associated with NK cell adhesion and homing to inflamed tissue [17].
Apart T and NK cells, NKT cells represent a unique subset that shares some characteristics with both NK and T cells and are particularly interesting due to their powerful role in the immune system. This population seems to be involved in tissue damage after acute myocardial infarction and chronic pulmonary diseases [18,19].
To better understand the mechanisms underlying functional immune impairment in COVID-19 pneumonia, we investigated the characteristics of peripheral blood T, NK and NKT cells in patients with COVID-19 pneumonia correlating these obtained findings with clinical parameters.
2. Methods
2.1. Study population
This study was approved by ethics committee of Policlinico Umberto I, Sapienza University of Rome (protocol number 298/2020) and written informed consent was obtained for each subject.
A case-control design was used. Cases were patients with COVID-19 pneumonia (COVID-19 subjects) admitted to the Policlinico Umberto I Hospital, Sapienza University of Rome. COVID-19 pneumonia was initially diagnosed based on the clinical symptoms and later confirmed by detecting SARS-CoV-2 RNA in nasal and pharyngeal swab specimens using a SARS-CoV-2 nucleic acid detection kit according to the manufacturer's protocol (RealStar® SARS-CoV-2 Altona Diagnostic, Germany). For each patient with COVID-19 pneumonia enrolled, the following data was extracted from electronic medical records: age, sex, medical history, symptoms, laboratory findings, chest CT or radiology findings.
As the control group, healthy donors (HDs) with similar age and sex, absence of symptoms, negative swab for SARS-CoV-2 RNA detection and negative serostatus for SARS-CoV-2 were enrolled.
Based on the severity disease, COVID-19 subjects were stratified into two groups: severe and non-severe. A non-severe case was defined as a confirmed case with fever, respiratory symptoms and radiographic evidence of pneumonia, while a severe case was defined with dyspnea or respiratory failure. Specifically, severe illness was defined according to the following criteria at the time of the admission: breathing rate ≥ 30 times/min, pulse oximeter oxygen saturation (SpO2) ≤93% at rest and ratio of partial pressure of arterial oxygen (PaO2) to fraction of inspired oxygen (FiO2) ≤300 mmHg.
2.2. Flow cytometry antibody staining
At the enrollment, peripheral whole blood samples were collected in heparin tube for each subject and tested within three hours withdrawal. Using multiparameter flow cytometry, the subset distribution and immunophenotype of polymorphonuclear (PMN) cells, T cells, CD3 + CD8- cells, CD3 + CD8+ cells, NK cells, CD56dim NK cells, CD56bright NK cells, NKT cells were investigated in peripheral whole blood samples from COVID-19 subjects and HDs. Specifically, Pacific Blue-conjugated anti-CD3 and APC/Cy7-conjugated anti-CD56 antibodies were used to defined T (CD3 + CD56-), NK (CD3-CD56+) and NKT (CD3 + CD56+) cells.
For T cells, using the APC-conjugated anti-CD8 antibody, two populations were defined: CD3 + CD8- (CD3 + CD56-CD8-) and CD3 + CD8+ (CD3 + CD56-CD8+) and their relative percentage of immunosenescence as lack of CD28- and expression of CD57+ was evaluated.
For NK cells, the APC/Cy7-conjugated anti-CD56 and PE/Cy7-conjugated anti-CD16 antibodies were used to identify CD56dim (CD3-CD56dimCD16+/−) and CD56bright (CD3-CD56brightCD16+/−). The expression of the PE-conjugated anti-CD57 was evaluated on total NK cells (CD3-CD56 + CD57+).
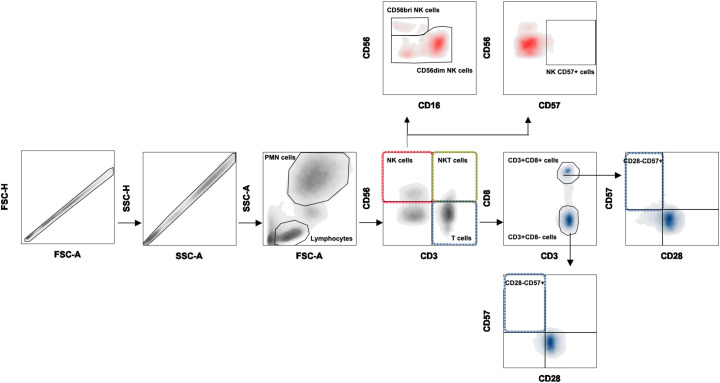
For NKT cells were defined as the co-expression of the Pacific Blue-conjugated anti-CD3 and APC/Cy7-conjugated anti-CD56 antibodies. Gating strategy is shown in Fig. 1 .
Fig. 1.
Flow cytometric gating strategy.
Using forward scatter (FSC) and side scatter (SSC), the PMN and lymphocyte populations were gated. After gating lymphocytes population, T cells were defined as CD3 + CD56- and then characterized into CD3 + CD8- (CD3 + CD56-CD8-) and CD3 + CD8+ (CD3 + CD56-CD8+) cells and their relative frequency of immunosenescence (CD28-CD57+) was evaluated. The NK cell population was identified as CD3-CD56+ cells and the expression of CD57 was evaluated. Further NK cells were categorized into different subsets according to the expression of CD16 and CD56 molecules as CD56bright (CD3-CD56brightCD16+/−), CD56dim (CD3-CD56dimCD16+/−). Finally, the NKT cell population was defined as CD3 + CD56 + .
Briefly, for each panel, 50 μl of whole blood was stained with the relative mix of monoclonal antibodies (all from BioLegend). The mixture was incubated in darkness at 4 °C for 20 min. Following direct immunofluorescence staining of peripheral blood cells with monoclonal antibodies, lyse red blood cells was performed by incubating in dark at room temperature for 10 min (BD Biosciences). The cells were washed twice in phosphate-buffered saline (PBS) containing 1% of Fetal Calf Serum (FCS). Next, cells were fixed in PBS containing 0.5% of formaldehyde (Sigma-Aldrich) before analysis. The stained blood was acquired using MACSQuant (Miltenyi Biotec, Germany) and analyzed using FlowJo™ v10.6.2 software.
2.3. Statistical analysis
All data were reported as median with IQR (25th-75th percentile). Flow cytometry data were described as percentages. Nonparametric comparative Mann-Whitney test was used to compare medians between the two groups. The 2-tailed χ 2 test or Fisher's exact test was used for comparing proportions of categorical variables. Spearman correlation coefficient was calculated for assessing the correlation between quantitative variables. Moreover, a linear regression model was built for all peripheral blood cell subsets as dependent variables, while the independent variable of interest was being a COVID-19 subject. The analyses were conducted using age and gender as possible confounders. The goodness of fit was assessed using the R2 value. The results are reported as Beta coefficients (p value).
Finally, considering only cases, a multivariate logistic regression analysis was used to evaluate the effects of the severity of the disease of different peripheral blood cell subsets, using age, gender and presence of comorbidities as potential confounders. Results are expressed as OR with 95% CI.
All statistical analyses were performed using GraphPad Prism v.8.4.1 software and p ≤ 0.05 was considered statistically significant. Linear and logistic regression analyses were performed with the statistical software SPSS, release 25.0.
3. Results
3.1. Characteristics of study population
Between March 2020 and April 2020, a total of 45 patients with laboratory confirmed COVID-19 pneumonia (COVID-19 subjects) hospitalized at Policlinico Umberto I, Sapienza University of Rome, Italy, were included in the study. In addition, 19 healthy donors (HDs) with similar age and sex were unrolled as the control group. Demographic and clinical characteristics of study population are reported in Table 1 .
Table 1.
Demographics and clinical characteristics of study population.
| HDs (n = 19) | COVID-19 subjects (n = 45) | Severe (n = 14) | Non-severe (n = 31) | p value | |
|---|---|---|---|---|---|
| Characteristics | |||||
| Age, median (IRQ) years | 61 (55.5–66.0) | 62 (54.3–75.0) | 71 (62.3–85.5) | 58 (52.0–66.0) | 0.017a |
| Male/Female | 7/12 | 17/28 | 8/6 | 9/22 | 0.072a |
| Smoking | 5 | 4 | 1 | 3 | nsa |
| Incubation period (days) | – | 9 (5–10.5) | 7 (6.3–10) | 9 (5–11) | nsa |
| Comorbid condition | |||||
| Any | – | 26 | 12 | 14 | 0.021a |
| Chronical obstructive pulmonary disease | – | 4 | 3 | 1 | |
| Hypertension | – | 16 | 7 | 9 | |
| Cardiovascular disease | – | 7 | 4 | 3 | |
| Diabetes | – | 7 | 3 | 4 | |
| Malignant tumor | – | 7 | 3 | 4 | |
| Dyslipidemia | – | 6 | 3 | 3 | |
| Obesity | – | 5 | 2 | 3 | |
| Signs and symptoms | |||||
| Fever | – | 35 | 12 | 23 | |
| Dry cough | – | 20 | 4 | 16 | |
| Shortness of breath | – | 9 | 5 | 4 | |
| Confusion | – | 1 | 1 | 0 | |
| Anosmia and ageusia | – | 3 | 2 | 1 | |
| Laboratory findings | |||||
| WBC (x 109/L) | (4.4–11.3)c | 4.8 (3.5–5.8) | 5.4 (4.6–8.0) | 4.2 (3.3–5.7) | 0.061b |
| Lymphocytes (x 109/L) | (1.0–4.8)c | 1.0 (0.8–1.3) | 0.9 (0.6–1.2) | 1.0 (0.8–1.3) | nsb |
| CRP (mg/dl) | (0.0–0.6)c | 3.5 (1.7–10.6) | 9.6 (4.2–16.6) | 2.2 (0.8–5.2) | 0.002b |
| P/F ratio | >400c | 329 (248–438) | 177 (143–248) | 383 (315.3–470) | <0.0001b |
| AST (UI/L) | (9-45)c | 21 (17–31) | 31 (21.8–53) | 18.5 (14.8–24.5) | 0.0006b |
| Ferritin (ng/ml) | (12−300)c | 473 (235.3–816) | 871 (268–3044) | 466 (227–583) | nsb |
| LDH (U/L) | (135-225)c | 257 (204–310) | 404 (269.5–438) | 241 (185.5–272.5) | <0.0001b |
| D-dimer (μg/ml) | (50-420)c | 932 (486–1733) | 1612 (1331–2382) | 598.5 (433.3–1012) | 0.001b |
HDs: healthy donors, IQR: interquartile range, CRP: C-reactive protein, WBC: white blood cells, P/F: PaO2/FiO2, AST: aspartate aminotransferase, LDH: lactate dehydrogenase.
Bold indicates p value significant.
The 2-tailed χ2 test or Fisher's exact test was used for comparing proportions between severe and non-severe groups.
The nonparametric comparative Mann-Whitney test was used to compare medians between severe and non-severe groups.
Normal range of laboratory findings in healthy donors.
Among COVID-19 subjects, 57.8% had at least one comorbidity (chronic obstructive pulmonary disease, hypertension, cardiovascular disease, diabetes, malignant tumor, dyslipidemia, obesity) and the most common symptoms were fever (77.8%), dry cough (44.4%), shortness of breath (20%) and anosmia and ageusia (6.7%). According to CT or X-ray findings, all COVID-19 subjects showed bilateral pneumonia. At the admission, in blood test, laboratory findings showed increase of CRP, ferritin, lactate dehydrogenase (LDH) and D-dimer levels compared to normal range. Otherwise, a reduction of PaO2/FiO2 (P/F) ratio was observed. Aspartate aminotransferase (AST) levels were in the normal range (Table 1).
Among all COVID-19 subjects, 31.1% was clinically diagnosed as severe COVID-19 pneumonia (severe group) and 68.9% as non-severe COVID-19 pneumonia (non-severe group) (Table 1). In the severe group, COVID-19 subjects were older and showed a higher prevalence of comorbidities (85.7%) compared to the non-severe group (45.1%). In the severe group, laboratory findings showed a significant higher level of CRP, P/F ratio, AST, LDH, and D-dimer compared to the non-severe group (Table 1).
3.2. Immunophenotyping analysis findings in COVID-19 group compared to HDs
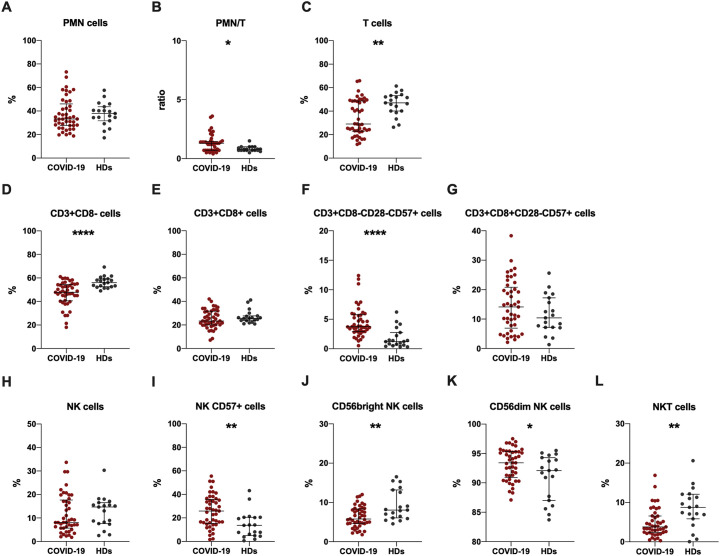
First, the evaluation of PMN cell percentage/T cell percentage (PMN/T) ratio showed a significantly higher ratio in COVID-19 subjects compared to HDs (Fig. 2B).
Fig. 2.
Immunophenotyping analysis performed in COVID-19 subjects and HDs.
Data are shown as median (lines) and interquartile ranges (whiskers). PMN: polymorphonuclear cell percentage, PMN/T: PMN cell percentage/T cell percentage. HDs: healthy donors. *p < 0.05, **0.01 < p < 0.001, **** p < 0.0001 (Mann-Whitney test).
Total T cells were immunophenotyped and subsequentially sub-divided according to the CD8 expression in two subsets: CD3 + CD8- and CD3 + CD8+ cells. For each of these subsets immunesenescence percentages (CD28-CD57+) were evaluated. Compared to HDs, COVID-19 subjects showed a significantly lower percentages of total T cells (Fig. 2C) and CD3 + CD8- cells (Fig. 2D). Otherwise, COVID-19 subjects showed a significantly higher percentage of CD3 + CD8-CD28-CD57+ cells compared to HDs (Fig. 2F). Moreover, we observed higher percentages of CD3 + CD8 + CD28-CD57+ cells in COVID-19 subjects compared to HDs, although not statistically significant (Fig. 2G).
Total NK cells were defined as CD3-CD56+ and CD57 expression was evaluated. COVID-19 subjects showed lower percetanges of total NK cells compared to HDs, although not statistically significant (Fig. 2H). However, a higher percentage of NK CD57+ cells in COVID-19 subjects compared to HDs was observed (Fig. 2I).
NK cells were divided into a CD56bright and CD56dim population. COVID-19 subjects showed a significantly lower percentage of CD56bright NK cells (Fig. 1J) and a significantly higher percentage of CD56dim NK cells (Fig. 2K).
Apart from NK and T cells, peripheral blood comprises other leucocyte subsets. For instance, co-expression of CD3 and CD56 is used to identify NKT cells. Compared to HDs, COVID-19 subjects showed a significantly lower percentage of NKT cells (Fig. 2L). All results are reported in Table 2 .
Table 2.
Immunophenotyping analysis data on study population.
| HDs (n = 19) | COVID-19 subjects (n = 45) | p valuea | Severe (n = 14) | Non-severe (n = 31) | p valueb | |
|---|---|---|---|---|---|---|
| PMN cells (%) | 37.9 (31.7–43.6) | 33.2 (27.7–46.0) | ns | 29.5 (21.6–35.8) | 34.0 (29.0–54.0) | 0.032 |
| PMN/T ratio | 0.8 (0.7–1.0) | 1.3 (0.7–1.5) | 0.024 | 1.6 (1.3–2.3) | 0.7 (0.7–1.3) | <0.0001 |
| T cells (%) | 47 (40.1–53.2) | 29.0 (23.0–48.7) | 0.025 | 23.0 (17.4–25.9) | 42.2 (25.3–50.0) | 0.0004 |
| CD3 + CD8- cells (%) | 56 (52.5–59.2) | 47.8 (40.6–54.4) | <0.0001 | 40 (30.3–54.2) | 49.9 (45.1–54.6) | 0.049 |
| CD3 + CD8+ cells (%) | 25.6 (23.7–27.7) | 23.4 (20.0–31-3) | ns | 20.9 (18.1–24) | 26.4 (20.8–32.9) | 0.018 |
| CD3 + CD8-CD28-CD57+ cells (%) | 1.2 (0.6–2.8) | 3.7 (2.9–5.8) | ns | 3.9 (3.2–5.9) | 3.7 (2.8–5.7) | ns |
| CD3 + CD8 + CD28-CD57+ cells (%) | 10.4 (7.2–17.2) | 14.1 (6.9–20.8) | <0.0001 | 16.3 (10.2–22.6) | 11.6 (4.9–19.7) | ns |
| NK cells (%) | 14.7 (8.2–16.5) | 8.1 (5.7–17.7) | ns | 9.0 (5.9–18.5) | 8.1 (5.7–18.0) | ns |
| NK CD57+ cells (%) | 13.7 (5.0–20.5) | 25.9 (15.3–35.8) | 0.001 | 24.1 (12.9–35.3) | 26.5 (15.9–36.4) | ns |
| CD56bright cells (%) | 8.1 (6.1–13.2) | 5.8 (4.6–8.3) | 0.005 | 5.1 (3.2–7.8) | 6.7 (4.8–8.5) | ns |
| CD56dim cells (%) | 92.1 (87–94.3) | 93.4 (90.9–95.4) | 0.034 | 94.9 (92–96.4) | 92.7 (90.4–95.1) | ns |
| NKT cells (%) | 8.8 (5.8–12.1) | 3.8 (2.4–7.0) | 0.002 | 2.1 (0.8–4.1) | 4.7 (3.0–8.6) | 0.001 |
The nonparametric comparative Mann-Whitney test was used to compare medians between HDs and COVID-19 subjects
The nonparametric comparative Mann-Whitney test was used to compare medians between severe and non-severe groups. HDs: healthy donors, PMN/T: polymorphonuclear cell percentage/T cell percentage, ns: not significant.
3.3. Immunophenotyping analysis findings and COVID-19 severity
COVID-19 subjects were stratified according to the severity of the disease into two groups: severe and non-severe (Table 2).
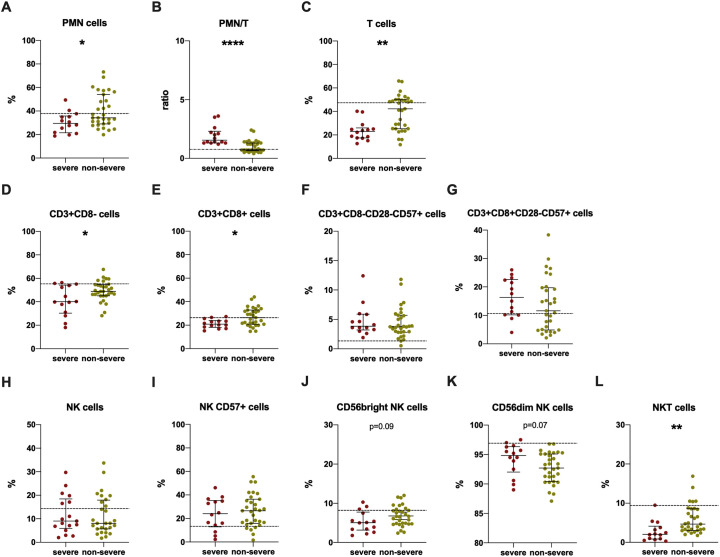
The severe group showed a significantly higher PMN/T ratio (Fig. 3B) and a significantly lower percentage of total T cells (Fig. 3C), CD3 + CD8- cells (Fig. 3D) and CD3 + CD8+ cells (Fig. 3E) compared to the non-severe group. A higher percentage of CD3 + CD8 + CD28-CD57+ cells in severe group compared to non-severe one was found, although not statistically significant (Fig. 3G).
Fig. 3.
Immunophenotyping analysis performed in severe and non-severe groups.
Data are shown as median (lines) and interquartile ranges (whiskers). Dotted grey lines represent median values for healthy donors. PMN/T: PMN cell percentage/T cell percentage. *p < 0.05, **0.01 < p < 0.001, **** p < 0.0001 (Mann-Whitney test).
The evaluation of NK subsets showed a lower percentage of CD56bright (Fig. 3J) and a higher percentage of CD56dim in the severe group compared to the non-severe one, although not statistically significant (Fig. 3K).
Finally, the severe group showed a statistically lower percentage of NKT cells compared to the non-severe group (Fig. 3L).
3.4. Multiple linear and logistic regression analysis and correlation between immunophenotyping subsets and clinical data
Multiple linear regression analysis adjusted for gender and ages was performed. The higher PMN/T ratio and the percentage of CD56dim NK cells as well as the lower percentages of T cells, CD56bright NK cells and NKT cells resulted independently associated to COVID-19 pneumonia (Table 3 ).
Table 3.
Multiple linear regression analysis. Dependent variables: peripheral blood cell subsets; Independent variable: being a COVID-19 subject.
| ß | p value | R2 of the Model | |
|---|---|---|---|
| PMN/T ratio | 0.441 | 0.008 | 0.244 |
| T cells (%) | −11.327 | 0.002 | 0.265 |
| CD3 + CD8- cells (%) | −9.253 | 0.000 | 0.295 |
| CD3 + CD8+ cells (%) | −1.972 | 0.329 | 0.023 |
| CD56bright NK cells (%) | −2.850 | 0.001 | 0.251 |
| CD56dim NK cells (%) | 2.115 | 0.012 | 0.184 |
| NKT cells (%) | −3.808 | 0.001 | 0.158 |
PMN/T: polymorphonuclear cell percentage/T cell percentage, ß: coefficient.
Simple and multiple logistic regression analysis was performed in COVID-19 subjects. The increase of PMN/T ratio and the reduction in the percentages of T cells, CD3 + CD8+ cells and NKT cells resulted independently associated to the severity of the disease in patients with COVID-19 pneumonia (Table 4 ).
Table 4.
Simple and multiple logistic regression results for the severity of the disease.
| Odd ratio crude (95% CI) | p value | Odd ratio adjusted⁎ (95% CI) | p value | |
|---|---|---|---|---|
| PMN/T ratio | 12.14 (2.37–62.25) | 0.003 | 11.45 (1.87–70.34) | 0.008 |
| T cells (%) | 0.90 (0.84–0.97) | 0.004 | 0.91 (0.84–0.98) | 0.018 |
| CD3 + CD8+ cells (%) | 0.85 (0.751–0.96) | 0.014 | 0.70 (0.55–0.89) | 0.003 |
| NKT cells (%) | 0.52 (0.32–0.85) | 0.008 | 0.25 (0.83–0.73) | 0.011 |
for gender, comorbidities and ages. PMN/T: PMN cell percentage/T cell percentage, NKT: natural killer T, ß: coefficient.
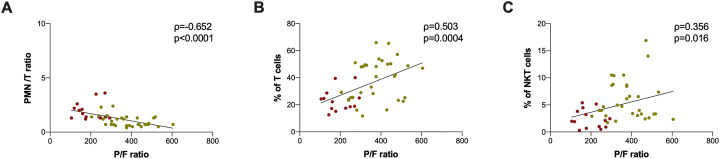
A negative correlation between P/F ratio and PMN/T ratio was observed (Fig. 4A). Otherwise, a positive correlation between P/F ratio and the percentage of T cells (Fig. 4B) as well as between P/F ratio and the percentage of NKT cells was found (Fig. 4C).
Fig. 4.
Correlation between laboratory findings and immunophenotyping data.
In red are reported severe COVID-19 subjects, in yellow non-severe COVID-19 subjects. PMN/T: PMN cell percentage/T cell percentage, P/F: PaO2/FiO2. Correlation was performed using Spearman test (Spearman coefficient [ρ] and statistical significance [p] are reported in the graphics). Linear correlation was evaluated by using the regression test, for A) R2 = 0.311, p < 0.0001, for B) R2 = 0.2411, p = 0.0006, for C) R2 = 0.1083, p = 0.0273. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
4. Discussion
Here, we reported immunophenotyping analysis of peripheral blood cells in a cohort of patients with laboratory confirmed COVID-19 pneumonia in Rome, Italy.
To the best of our knowledge, this is the first study that characterized NK populations according to CD56 expression and that investigated NKT cells in patients with COVID-19 pneumonia correlating these subsets to clinical parameters and severity of the disease.
The most relevant findings are that in COVID-19 subjects we observed significantly a higher percentage of NK CD57+ cells and CD56dim NK cells compared to HDs. On the other hand, in COVID-19 subjects we found a significantly lower percentage of CD56bright and NKT cells compared to HDs. Moreover, the low percentage of NKT cells resulted independently associated to the severity of the disease and positively correlated to P/F ratio.
Consistent with previous studies [5,20,21], at admission almost half of patients with COVID-19 pneumonia in our cohort had at least one underlying disorder (i.e. hypertension, diabetes, chronic obstructive pulmonary disease) and the laboratory abnormalities predominantly included elevated inflammatory markers, such as CRP, LDH and D-dimer among others. When all patients with COVID-19 pneumonia were stratified according to the severity of the disease, those in the severe group were older and showed a higher prevalence of comorbidities (i.e. hypertension and cardiovascular disease) compare to patients in the non-severe group. This evidence underlines that the presence of chronic comorbidities affects weaker immune functions likely making it more vulnerable to severe COVID-19 pneumonia. Moreover, the laboratory abnormalities observed were more noticeable in the severe group compared to the non-severe one.
Zheng et al. reported that among the lymphocyte populations, NK cells could be mainly involved in the COVID-19 response [11]. In line with previous report [12], in COVID-19 subjects we observed a lower percentage of NK cells compared to HDs as well as in severe group compared to non-severe one, although not statistically significant. However, in COVID-19 subjects we found a significantly higher percentage of NK CD57+ cells compared to HDs. CD57 expression is reported to be a marker of mature NK cells. Functionally, CD57 is associated with cell adhesion and homing to inflamed tissue [17]. Therefore, the increase of CD57 expression on NK cells that we observed in COVID-19 subjects might be a result of increased migration from the periphery into tissues. This evidence is in line with a previous report in which Liao et al. compared single-cell RNA sequencing analysis of bronchoalveolar lavage fluid samples from patients with COVID-19 with healthy donors and found higher proportions of NK cells in COVID-19 patients, suggesting trafficking of NK cells to the lungs [22]. Moreover, considering that CD57 expression on human lymphocytes indicates an inability to proliferate, these cells display high cytotoxic potential, and NK CD57+ cells exhibit both memory-like features and potent effector functions. Thus, the increased expression of CD57 on NK cells that we observed is in line with a previous report by Jiang et al. in which they showed an increase of cytotoxic potential of NK cells in COVID-19 subjects [12].
In our study we characterized two NK cell subsets: the cytotoxic CD56dim NK subset and the regulatory CD56bright NK subset [23,24]. In COVID-19 subjects we observed a significantly lower percentage of CD56bright NK cells and a higher percentage of CD56dim NK cells compared to HDs. The same trend was observed in the severe group compared to the non-severe one. The lower percentage of CD56bright NK cells observed in COVID-19 subjects may be attributed to a migration in tissue and secondary lymphoid organs where they exert their function in response to inflammation caused by invading pathogens [25,26]. On the other hand, the higher percentage of CD56dim observed in COVID-19 subjects, suggest that upon well-established SARS-CoV-2 infection, NK cells remaining in the periphery may have an exhausted phenotype.
Apart from NK and T cells, peripheral blood comprises other leucocyte subsets such as NKT cells which are the only cell line to express both T cell and NK receptors on their surface membranes [27].
Like NK cells, NKT cells possess cytotoxic capabilities, but are primarily considered to have an important regulatory function via the secretion of large amounts of pro- or anti-inflammatory cytokines upon activation, thereby resulting in amplification or dampening of the immune response [28]. Despite of the low proportion of NKT cells among circulating mononuclear cells, defects in NKT cell frequency and cytokine production have been reported in several infectious and autoimmune diseases [29].
As reported by Mazzoni et al. [30], in COVID-19 subjects we observed a significant lower percentage of NKT cells compared to HDs. We reported also the same finding in the severe group compared to the non-severe one. Ths evidence is in line with previous reports in chronic obstructive pulmonary disease (COPD) in which a reduction of NKT cells in peripheral blood [31] and an increased number of NKT cells in the lungs were observed [32]. Moreover, in acute viral infection pulmonary NKT cells contribute to pathogen clearance, but this might be at the expense of uncontrolled inflammation [33]. This role in immune surveillance against viruses is partly based on their capacity to mature dendritic cells (DCs) and subsequently activate potent cytotoxic NK cells and CD8 + T cells [34]. The natural role of NKT cells has been examined during experimental influenza infection [35]. NKT deficiency resulted in worse inflammation, albeit through different mechanisms according to experimental conditions [[36], [37], [38], [39]]. As suggest by De Santo et al. [37], using a H1N1 strain the increased pathology observed in NKT-deficient animals was due to an enhanced virus spread. The authors proposed that NKT cells control influenza A virus (IAV) replication by blunting the immunosuppressive activity of myeloid-derived suppressive cells, a population that prevents specific CD8+ T-cell response.
In COVID-19 subjects we observed significantly higher PMN/T ratio and percentage of CD3 + CD8-CD28-CD57+ cells as well as significantly lower percentages of total T cells and CD3 + CD8- cells compared to HDs. Stratifying COVID-19 subjects according to the severity of the disease, we observed a significantly higher PMN/T ratio and a significantly lower percentage of T cells in the severe group compared to the non-severe one underlining that the increase of neutrophil/lymphocytes ratio and lymphopenia are potential risk factors for severity of COVID-19 pneumonia. Interestingly, in the severe group beside a significantly lower percentage of CD3 + CD8+ cells we found a higher percentage of CD3 + CD8 + CD28-CD57+ cells compared to the non-severe one, although not statistically significant. However, this finding suggests a higher functional exhaustion of this subset as already shown by other authors [13].
In a multiple linear regression analysis adjusted for gender and ages, the higher PMN/T ratio and CD56dim NK cell percentage as well as the lower percentage of T cells, CD56bright NK cells and NKT cells resulted independently associated to COVID-19 pneumonia.
Furthermore, a multivariate analysis regression identified the high PMN/T ratio and the low percentages of total T cells, CD3 + CD8+ cells and NKT cells independently associated to the severity of the disease, excluding confounding factors as gender, age and presence of comorbidities.
Finally, a negative correlation between P/F ratio and PMN/T ratio while a positive correlation between P/F ratio and T cell percentages were found, highlighting the important role of T cells in COVID-19 progression. Intriguingly, among all cell subsets investigated in our study, only NKT cells resulted positively correlated to P/F ratio.
The major limitation of this study is the sample size relatively small, and most patients included in this study had non-severe COVID-19 pneumonia. However, we believe that these preliminary data could stimulate additional researches on larger cohorts of patients with COVID-19 pneumonia to elucidate lymphocyte subsets in the immunological mechanisms of COVID-19.
In summary, the present study showed a reduction of regulatory cell subsets in peripheral blood of patients with COVID-19 pneumonia suggesting that these subsets may play a crucial role in the COVID-19 pneumonia. Specifically, the reduction of CD56bright NK and NKT cells observed in COVID-19 subjects may be explained by their potential recruitment into infected tissues (i.e. lung) leading to an exaggerated or not controlled immune responses. Finally, the independent association between the severe COVID-19 pneumonia and low percentage of NKT cells as well as the positive correlation between NKT cells and P/F ratio that we found suggest a potential role of this subset as biomarker of the severity of the disease. Large-scale multicenter researches are needed to confirm our hypothesis.
Declaration of Competing Interest
The authors declare that they have no conflict of interest.
Acknowledgments
The authors would like to acknowledge Dr. Giorgio Maria Masci (Sapienza University of Rome, Italy) for his writing assistance; Dr. Serena Vita (National Institute for Infectious Diseases Lazzaro Spallanzani IRCCS, Rome, Italy) and Dr. Marco Iannetta (Tor Vergata, University of Rome, Italy) for fruitful discussions. This work was supported by Sapienza, University of Rome.
References
- 1.Chan J.F.-W., Kok K.-H., Zhu Z., Chu H., K. K.-W. To, Yuan S., Yuen K.-Y. Genomic characterization of the 2019 novel human-pathogenic coronavirus isolated from a patient with atypical pneumonia after visiting Wuhan. Emerg Microbes Infect. 2020;9:221–236. doi: 10.1080/22221751.2020.1719902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chan J.F.-W., Yuan S., Kok K.-H., K. K.-W. To, Chu H., Yang J., Xing F., Liu J., Yip C.C.-Y., Poon R.W.-S., Tsoi H.-W., Lo S.K.-F., Chan K.-H., Poon V.K.-M., Chan W.-M., Ip J.D., Cai J.-P., Cheng V.C.-C., Chen H., Hui C.K.-M., Yuen K.-Y. A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: a study of a family cluster. Lancet. 2020;395:514–523. doi: 10.1016/S0140-6736(20)30154-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Epidemiology Working Group for NCIP Epidemic Response, Chinese Center for Disease Control and Prevention The epidemiological characteristics of an outbreak of 2019 novel coronavirus diseases (COVID-19) in China. Zhonghua Liu Xing Bing Xue Za Zhi. 2020;41:145–151. doi: 10.3760/cma.j.issn.0254-6450.2020.02.003. [DOI] [PubMed] [Google Scholar]
- 4.Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X., Cheng Z., Yu T., Xia J., Wei Y., Wu W., Xie X., Yin W., Li H., Liu M., Xiao Y., Gao H., Guo L., Xie J., Wang G., Jiang R., Gao Z., Jin Q., Wang J., Cao B. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang D., Hu B., Hu C., Zhu F., Liu X., Zhang J., Wang B., Xiang H., Cheng Z., Xiong Y., Zhao Y., Li Y., Wang X., Peng Z. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus-infected pneumonia in Wuhan, China. JAMA. 2020 Mar 17;323(11):1061–1069. doi: 10.1001/jama.2020.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wu F., Zhao S., Yu B., Chen Y.-M., Wang W., Song Z.-G., Hu Y., Tao Z.-W., Tian J.-H., Pei Y.-Y., Yuan M.-L., Zhang Y.-L., Dai F.-H., Liu Y., Wang Q.-M., Zheng J.-J., Xu L., Holmes E.C., Zhang Y.-Z. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhou P., Yang X.-L., Wang X.-G., Hu B., Zhang L., Zhang W., Si H.-R., Zhu Y., Li B., Huang C.-L., Chen H.-D., Chen J., Luo Y., Guo H., Jiang R.-D., Liu M.-Q., Chen Y., Shen X.-R., Wang X., Zheng X.-S., Zhao K., Chen Q.-J., Deng F., Liu L.-L., Yan B., Zhan F.-X., Wang Y.-Y., Xiao G.-F., Shi Z.-L. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Qin C., Zhou L., Hu Z., Zhang S., Yang S., Tao Y., Xie C., Ma K., Shang K., Wang W., Tian D.-S. Dysregulation of immune response in patients with COVID-19 in Wuhan, China. Clin. Infect. Dis. 2020 Jul 28;71(15):762–768. doi: 10.1093/cid/ciaa248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu J., Liu Y., Xiang P., Pu L., Xiong H., Li C., Zhang M., Tan J., Xu Y., Song R., Song M., Wang L., Zhang W., Han B., Yang L., Wang X., Zhou G., Zhang T., Li B., Wang Y., Chen Z., Wang X. Neutrophil-to-lymphocyte ratio predicts critical illness patients with 2019 coronavirus disease in the early stage. J. Transl. Med. 2020;18:206. doi: 10.1186/s12967-020-02374-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang A.-P., Liu J.-P., Tao W.-Q., Li H.-M. The diagnostic and predictive role of NLR, d-NLR and PLR in COVID-19 patients. Int. Immunopharmacol. 2020;84 doi: 10.1016/j.intimp.2020.106504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zheng M., Gao Y., Wang G., Song G., Liu S., Sun D., Xu Y., Tian Z. Functional exhaustion of antiviral lymphocytes in COVID-19 patients. Cell. Mol. Immunol. 2020;17:533–535. doi: 10.1038/s41423-020-0402-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jiang Y., Wei X., Guan J., Qin S., Wang Z., Lu H., Qian J., Wu L., Chen Y., Chen Y., Lin X. COVID-19 pneumonia: CD8+ T and NK cells are decreased in number but compensatory increased in cytotoxic potential. Clin. Immunol. 2020;108516 doi: 10.1016/j.clim.2020.108516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.De Biasi S., Meschiari M., Gibellini L., Bellinazzi C., Borella R., Fidanza L., Gozzi L., Iannone A., Lo Tartaro D., Mattioli M., Paolini A., Menozzi M., Milić J., Franceschi G., Fantini R., Tonelli R., Sita M., Sarti M., Trenti T., Brugioni L., Cicchetti L., Facchinetti F., Pietrangelo A., Clini E., Girardis M., Guaraldi G., Mussini C., Cossarizza A. Marked T cell activation, senescence, exhaustion and skewing towards TH17 in patients with COVID-19 pneumonia. Nat. Commun. 2020;11:3434. doi: 10.1038/s41467-020-17292-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pinti M., Appay V., Campisi J., Frasca D., Fülöp T., Sauce D., Larbi A., Weinberger B., Cossarizza A. Aging of the immune system: focus on inflammation and vaccination. Eur. J. Immunol. 2016;46:2286–2301. doi: 10.1002/eji.201546178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brenchley J.M., Karandikar N.J., Betts M.R., Ambrozak D.R., Hill B.J., Crotty L.E., Casazza J.P., Kuruppu J., Migueles S.A., Connors M., Roederer M., Douek D.C., Koup R.A. Expression of CD57 defines replicative senescence and antigen-induced apoptotic death of CD8+ T cells. Blood. 2003;101:2711–2720. doi: 10.1182/blood-2002-07-2103. [DOI] [PubMed] [Google Scholar]
- 16.Effros R.B. Loss of CD28 expression on T lymphocytes: a marker of replicative senescence. Dev. Comp. Immunol. 1997;21:471–478. doi: 10.1016/s0145-305x(97)00027-x. [DOI] [PubMed] [Google Scholar]
- 17.Osterburg A.R., Lach L., Panos R.J., Borchers M.T. Unique natural killer cell subpopulations are associated with exacerbation risk in chronic obstructive pulmonary disease. Sci. Rep. 2020;10 doi: 10.1038/s41598-020-58326-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tang, Y., X. Li, M. Wang, Q. Zou, S. Zhao, B. Sun, L. Xu, and Y. Jiang. 2013. Increased numbers of NK cells, NKT-like cells, and NK inhibitory receptors in peripheral blood of patients with chronic obstructive pulmonary disease. Clin. Dev. Immunol. 2013: 721782. [DOI] [PMC free article] [PubMed]
- 19.Papakosta D., Manika K., Gounari E., Kyriazis G., Kontakiotis T., Spyropoulos G., Kontakioti E., Zarogoulidis K. Bronchoalveolar lavage fluid and blood natural killer and natural killer T-like cells in cryptogenic organizing pneumonia. Respirology. 2014;19:748–754. doi: 10.1111/resp.12305. [DOI] [PubMed] [Google Scholar]
- 20.Zhang J.-J., Dong X., Cao Y.-Y., Yuan Y.-D., Yang Y.-B., Yan Y.-Q., Akdis C.A., Gao Y.-D. Clinical characteristics of 140 patients infected with SARS-CoV-2 in Wuhan, China. Allergy. 2020 Jul;75(7):1730–1741. doi: 10.1111/all.14238. Epub 2020 Feb 27. [DOI] [PubMed] [Google Scholar]
- 21.Tian S., Hu N., Lou J., Chen K., Kang X., Xiang Z., Chen H., Wang D., Liu N., Liu D., Chen G., Zhang Y., Li D., Li J., Lian H., Niu S., Zhang L., Zhang J. Characteristics of COVID-19 infection in Beijing. J. Inf. Secur. 2020;80:401–406. doi: 10.1016/j.jinf.2020.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liao M., Liu Y., Yuan J., Wen Y., Xu G., Zhao J., Cheng L., Li J., Wang X., Wang F., Liu L., Amit I., Zhang S., Zhang Z. Single-cell landscape of bronchoalveolar immune cells in patients with COVID-19. Nat. Med. 2020 Jun;26(6):842–844. doi: 10.1038/s41591-020-0901-9. Epub 2020 May 12. [DOI] [PubMed] [Google Scholar]
- 23.Ferlazzo G., Münz C. NK cell compartments and their activation by dendritic cells. J. Immunol. 2004;172:1333–1339. doi: 10.4049/jimmunol.172.3.1333. [DOI] [PubMed] [Google Scholar]
- 24.Caligiuri M.A. Human natural killer cells. Blood. 2008;112:461–469. doi: 10.1182/blood-2007-09-077438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moretta A., Marcenaro E., Sivori S., Della Chiesa M., Vitale M., Moretta L. Early liaisons between cells of the innate immune system in inflamed peripheral tissues. Trends Immunol. 2005;26:668–675. doi: 10.1016/j.it.2005.09.008. [DOI] [PubMed] [Google Scholar]
- 26.De Maria A., Bozzano F., Cantoni C., Moretta L. Revisiting human natural killer cell subset function revealed cytolytic CD56(dim)CD16+ NK cells as rapid producers of abundant IFN-gamma on activation. Proc. Natl. Acad. Sci. U. S. A. 2011;108:728–732. doi: 10.1073/pnas.1012356108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu L., Van Kaer L. Natural killer T cells in health and disease. Front Biosci (Schol Ed) 2011;3:236–251. doi: 10.2741/s148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Krijgsman D., Hokland M., Kuppen P.J.K. The role of natural killer T cells in cancer-a Phenotypical and functional approach. Front. Immunol. 2018;9:367. doi: 10.3389/fimmu.2018.00367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Godfrey D.I., Hammond K.J., Poulton L.D., Smyth M.J., Baxter A.G. NKT cells: facts, functions and fallacies. Immunol. Today. 2000;21:573–583. doi: 10.1016/s0167-5699(00)01735-7. [DOI] [PubMed] [Google Scholar]
- 30.Mazzoni A., Salvati L., Maggi L., Capone M., Vanni A., Spinicci M., Mencarini J., Caporale R., Peruzzi B., Antonelli A., Trotta M., Zammarchi L., Ciani L., Gori L., Lazzeri C., Matucci A., Vultaggio A., Rossi O., Almerigogna F., Parronchi P., Fontanari P., Lavorini F., Peris A., Rossolini G.M., Bartoloni A., Romagnani S., Liotta F., Annunziato F., Cosmi L. Impaired immune cell cytotoxicity in severe COVID-19 is IL-6 dependent. J. Clin. Invest. 2020 Sep 1;130(9):4694–4703. doi: 10.1172/JCI138554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Urbanowicz R.A., Lamb J.R., Todd I., Corne J.M., Fairclough L.C. Altered effector function of peripheral cytotoxic cells in COPD. Respir. Res. 2009;10:53. doi: 10.1186/1465-9921-10-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim E.Y., Battaile J.T., Patel A.C., You Y., Agapov E., Grayson M.H., Benoit L.A., Byers D.E., Alevy Y., Tucker J., Swanson S., Tidwell R., Tyner J.W., Morton J.D., Castro M., Polineni D., Patterson G.A., Schwendener R.A., Allard J.D., Peltz G., Holtzman M.J. Persistent activation of an innate immune response translates respiratory viral infection into chronic lung disease. Nat. Med. 2008;14:633–640. doi: 10.1038/nm1770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tessmer M.S., Fatima A., Paget C., Trottein F., Brossay L. NKT cell immune responses to viral infection. Expert Opin. Ther. Targets. 2009;13:153–162. doi: 10.1517/14712590802653601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Johnson T.R., Hong S., Van Kaer L., Koezuka Y., Graham B.S. NK T cells contribute to expansion of CD8(+) T cells and amplification of antiviral immune responses to respiratory syncytial virus. J. Virol. 2002;76:4294–4303. doi: 10.1128/JVI.76.9.4294-4303.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Paget C., Trottein F. Role of type 1 natural killer T cells in pulmonary immunity. Mucosal Immunol. 2013;6:1054–1067. doi: 10.1038/mi.2013.59. [DOI] [PubMed] [Google Scholar]
- 36.Kok W.L., Denney L., Benam K., Cole S., Clelland C., McMichael A.J., Ho L.-P. Pivotal advance: invariant NKT cells reduce accumulation of inflammatory monocytes in the lungs and decrease immune-pathology during severe influenza a virus infection. J. Leukoc. Biol. 2012;91:357–368. doi: 10.1189/jlb.0411184. [DOI] [PubMed] [Google Scholar]
- 37.De Santo C., Salio M., Masri S.H., Lee L.Y.-H., Dong T., Speak A.O., Porubsky S., Booth S., Veerapen N., Besra G.S., Gröne H.-J., Platt F.M., Zambon M., Cerundolo V. Invariant NKT cells reduce the immunosuppressive activity of influenza a virus-induced myeloid-derived suppressor cells in mice and humans. J. Clin. Invest. 2008;118:4036–4048. doi: 10.1172/JCI36264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Maazi H., Singh A.K., Speak A.O., Lombardi V., Lam J., Khoo B., Inn K.S., Sharpe A.H., Jung J.U., Akbari O. Lack of PD-L1 expression by iNKT cells improves the course of influenza a infection. PLoS One. 2013;8:e59599. doi: 10.1371/journal.pone.0059599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Paget C., Ivanov S., Fontaine J., Blanc F., Pichavant M., Renneson J., Bialecki E., Pothlichet J., Vendeville C., Barba-Spaeth G., Barba-Speath G., Huerre M.-R., Faveeuw C., Si-Tahar M., Trottein F. Potential role of invariant NKT cells in the control of pulmonary inflammation and CD8+ T cell response during acute influenza a virus H3N2 pneumonia. J. Immunol. 2011;186:5590–5602. doi: 10.4049/jimmunol.1002348. [DOI] [PubMed] [Google Scholar]