Abstract
How the measurement of aging biomarkers in peripheral blood T-lymphocytes (PBTLs) is influenced by cell composition is unclear. Here, we collected peripheral blood and isolated CD3+ PBTLs from 117 healthy couples between the ages of 21 and 72. Each sample was profiled for Horvath epigenetic clock (DNAm), p16INK4a expression, cytomegalovirus (CMV) seropositivity and 74 mRNA markers of PBTL subtype, differentiation, immune checkpoints, and cytokine production. Correlations between individual aging biomarkers (DNAm or p16INK4a) and PBTL mRNAs were corrected for chronological age, sex, and couple. DNAm measurements correlated with CMV seropositivity as well as PBTL mRNAs indicative of effector function (CD8A, EOMES, TBX21, GZMB), poor proliferative capacity (KLRG1, CD57), differentiation (CD45RO, CD45RA), and immune checkpoints (PDCD1, TIGIT, LAG3, CD160, CD244). In contrast, only three PBTL mRNAs, CD28, CD244, and p14ARF, showed a significant association with p16INK4a. p16INK4a expression also showed a weaker association with immunosenescent PBTL subsets than DNAm in flow cytometry analyses. These data suggest that PBTL composition has a greater influence on DNAm than p16INK4a and link accelerated epigenetic aging to immunosenescent phenotypes.
Keywords: Aging biomarker, Senescence, T cell, Horvath clock, CMV
The composition and function of peripheral blood T-lymphocytes (PBTLs) changes with age, yet understanding of how PBTL dynamics impact blood-based measures of biological age is limited. Two commonly used measures of biological age are the Horvath epigenetic clock (DNAm) and p16INK4a. p16INK4a expression is elevated in senescent cells and increases logarithmically with age in many biological sample types, including PBTLs (1). The Horvath epigenetic clock provides an estimate of biological age based upon the status of specific DNA methylation sites throughout the genome [reviewed in Horvath and Raj (2)]. DNAm differs from other epigenetic clocks in that data from 51 different cell and tissue types were used to develop the algorithm such that values would remain consistent across multiple sample types (3). DNAm or p16INK4a levels exceeding the average trend for healthy individuals are considered clinical indicators of accelerated biologic aging and have been linked to gerontogenic exposures like smoking, dose dense chemotherapy, bone marrow transplantation, and inactivity (1).
In-depth explorations of the relationship between PBTL composition and p16INK4a or DNAm are limited, yet many clinical studies use peripheral blood due to ease of access. In a previous analysis of CD3+ PBTLs from 170 healthy donors, p16INK4a expression was not correlated with CD4:CD8 ratios or mRNA markers of memory T cells (BCL2, BCLXL, IL7Rα) suggesting that age-related changes in PBTL constituents have little impact on p16INK4a levels (4). Conversely, changes in peripheral blood constituents clearly influence epigenetic age and failure to adjust for sample composition may increase the rate of false positives (5). To address this issue, methods such as least-squares minimization are often employed to estimate the degree to which individual, cell-type specific methylation patterns contribute to each specimen. This process requires that unique DNA methylation patterns are established for each cell type within a given mixture. While epigenetic signatures are available for the major peripheral blood cell types (6), patterns specific to distinct cell subsets, like effector T cells, have yet to be incorporated into epigenetic clock algorithms. It is unclear whether such granularity is even necessary when analyzing DNAm data.
Here, we collected PBTLs from 117 healthy donors to attain a more detailed understanding of the relationship between cell composition, p16INK4a and DNAm. We profiled mRNA makers of PBTL differentiation, subset, cytokine production, and immune checkpoints using a Nanostring CodeSet containing custom probes for p16INK4a. Collection of DNA, RNA, and plasma from each donor sample allowed us to measure PBTL profiles, p16INK4a, DNAm, and cytomegalovirus (CMV) serostatus in the same donor. Together, our data reveal distinctions in how PBTL constituents influence p16INK4a and DNAm.
Methods
Study Population
We recruited 117 healthy adults, 21–72 years of age, from the local community between January and August, 2017 as part of a couples study approved by The Ohio State Institutional Review Board. Two same sex couples participated in the study (1 female/female and 1 male/male). All participants provided written informed consent. Couples were ineligible for the study if either partner was pregnant or had a medical history of malignancy, stroke, heart attack, lupus, surgery, multiple sclerosis, ulcerative colitis, inflammatory bowel disease, Crohn’s disease, Parkinson’s disease, or human immunodeficiency virus (HIV). Participants self-reported their height, weight, and smoking history. The Charlson Index was used to measure comorbidities (7,8).
Sample Collection and Processing
Peripheral blood was obtained via standard venipuncture. Serum was isolated from four 10 mL tubes of blood and the resulting supernatants were stored at −80°C until CMV serology could be performed as described (9). Flow cytometry, PBTL isolation, and nucleic acid purification protocols are detailed in the Supplementary Methods.
OSU_Senescence
OSU_Senescence is a custom Nanostring CodeSet that measures five controls and 74 mRNA markers indicative of PBTL subset, function, and senescence (Supplementary Table S1). Custom Nanostring probes were designed to differentiate between the two CDKN2A transcripts: p16INK4a and p14ARF. High quality total RNA (200 ng) from CD3+ PBTLs (Bioanalyzer RIN > 6.4) was converted to cDNA using SuperScript VILO Master Mix. CD244, CD274, CD276, IL17A, IL-6, p14ARF, and p16INK4a cDNAs were preamplified using Nanostring-supplied custom primers and Taqman PreAmp Master Mix. A total of 12 cycles of amplification were performed prior to standard nCounter analysis. Sample data were examined for linearity and normalized to the average transcript count for all five controls.
DNAm
Genomic DNA from CD3+ PBTLs was sent to Zymo Research Corporation for DNAm analysis. DNAm was determined using Simplified Whole-panel Amplification Reaction Method (SWARM) technology to simultaneously examine the DNA methylation status of >500 loci. Methylation patterns at these loci were used to determine epigenetic age based upon the Horvath algorithm (3).
Statistical Analyses
Due to the clustering of subjects within couples, linear mixed effects models were used for all analyses. In these models, outcomes were DNAm and p16INK4a, fixed effects were sex, chronological age, and individual OSU_Senescence markers (included separately). Couple was included as a random effect. R2 and partial R2 values were estimated for fixed effects as described (10). All analyses were repeated, splitting the sample into two groups: <40 years, 40+ years. The 40-year cutoff represents approximately one-half the average U.S. life span [78.6 years (11)]. The Benjamini-Hochberg procedure was used to control the false discovery rate for each set of models with the same outcome and sample (each set of 75/76 models). Similar mixed effects models were used to quantify associations between DNAm or p16INK4a and cohort characteristics, with each characteristic (sex, race) as the only fixed effect. Results with adjusted p < .05 were considered statistically significant. All analyses used R version 3.6 (12).
Results
Characteristics of the 117 study participants are summarized in Table 1. Participants were between 21 and 72 years of age and generally healthy with few reported comorbidities. Two study participants were current smokers (1.7%), 21 (18.0%) were former smokers and 94 (80.3%) were nonsmokers. Average BMI was in the overweight range (27.0 ± 5.7). The average DNAm of participant CD3+ PBTLs (27.1 ± 14.1 years) was lower than the chronological age of the cohort (39.0 ± 12.9 years), suggesting that the Horvath clock algorithm might perform differently in purified T cells. Log2-transformed p16INK4a levels ranged between 0.4 and 6.6 units, which equates to 62 years of chronological aging [1 unit equals ~10 years chronological aging (13,14)].
Table 1.
Cohort Characteristics and Summary of Measured Values
| Characteristic or Measure | n | Mean ± SD (Median, Range) or n (%) |
|---|---|---|
| Age | 117 | 39.0 ± 12.9 (35, 21–72) |
| 21–29 | 33 (28.2) | |
| 30–39 | 37 (31.6) | |
| 40–49 | 23 (19.7) | |
| 50–59 | 11 (9.4) | |
| > 60 | 13 (11.1) | |
| Race | 117 | |
| Asian | 4 (3.4) | |
| African American/black | 6 (5.1) | |
| Other | 4 (3.4) | |
| White | 103 (88.1) | |
| BMI | 117 | 27.0 ± 5.7 (26.5, 18.2–51.6) |
| <19.0 | 1 (0.9) | |
| 19.0–24.9 | 47 (40.2) | |
| 25.0–29.9 | 42 (35.9) | |
| 30.0–39.9 | 24 (20.5) | |
| 40+ | 3 (2.6) | |
| Comorbidities | 117 | |
| None | 102 (87.2) | |
| One | 13 (11.1) | |
| Two | 1 (0.9) | |
| Three | 1 (0.9) | |
| Smoking status | 117 | |
| Current smoker | 2 (1.7) | |
| Former smoker | 21 (18.0) | |
| Nonsmoker | 94 (80.3) | |
| Pack-years | 117 | 1.2 ± 3.6 (0, 0–23.5) |
| DNAm | 117 | 27.1 ± 14.1 (22.9, 9.1–65.2) |
| p16INK4a | 114 | 3.0 ± 1.0 (2.9, 0.4–6.6) |
| % CD3+ | 116 | 96.0 ± 4.9 (97.0, 60.9–98.7) |
| CMV seropositivity | 117 | |
| Yes | 45 (38.5) | |
| No | 72 (61.5) | |
| CMV antibody titer | 45 | 2.8 ± 0.4 (2.9, 2.0–3.5) |
We first looked to see whether race or sex influenced PBTL DNAm or p16INK4a in our cohort. There was no difference in the DNAm (F = 0.20, p = .66) or p16INK4a (F = 3.80, p = .054) levels of white and nonwhite study participants. Consistent with prior publications (4,15), levels of p16INK4a were independent of sex (F < 0.01, p = .99), whereas DNAm was higher in men than women (F = 16.2, p < .01).
We next examined the relationship between each aging biomarker and OSU_Senescence mRNA in the entire cohort and two smaller subgroups: participants <40 years of age (younger adults; n = 70) and participants 40+ years of age (older adults; n = 47; Figure 1). Correlations were corrected for couple, sex, and chronological age using linear mixed models. This ensured that any identified relationship between PBTL mRNAs and DNAm or p16INK4a would be representative of biologic aging. A complete listing of all R2, partial R2 and p values is provided in Supplementary Tables S2 and S3.
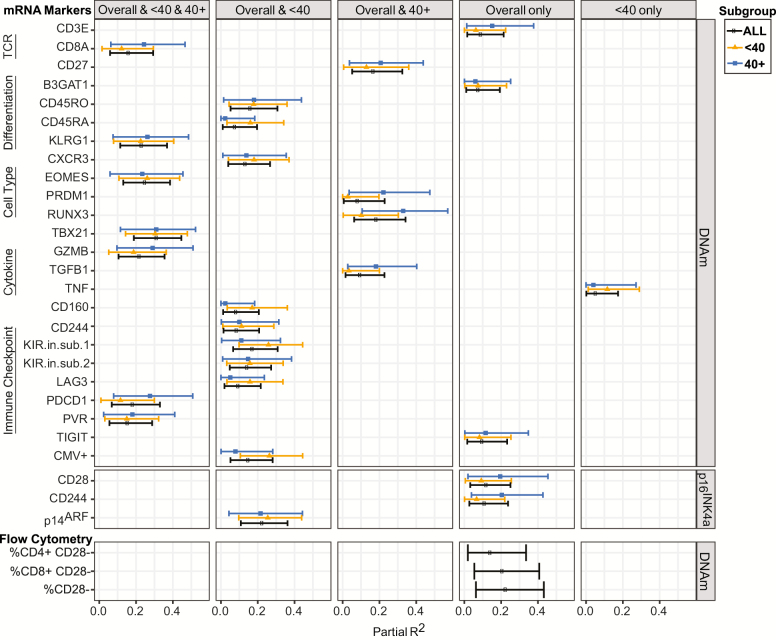
Figure 1.
Significant correlations between study measures and aging biomarkers. Linear mixed models, adjusting for sex, chronological age, and couple, were used to calculate the correlation (partial R2) between each study measure and DNAm or p16INK4a. Relationships were assessed in all study participants (overall), those under 40 years of age (<40), and those 40 years and older (40+). Values are graphed in columns according to the subgroups where statistical significance was achieved (adjusted p < .05). Points represent the estimated partial R2 with bars indicating the 95% confidence intervals. A complete listing of all study relationships appears in Supplementary Table S2.
Twenty-two of the 74 OSU_Senescence mRNAs (30%) correlated with DNAm in our cohort after adjustment for sex, chronological age, and couple (Figure 1; Supplementary Table S2). However, only seven of these mRNAs: CD8A, EOMES, GZMB, KLRG1, PDCD1, PVR, and TBX21, remained closely correlated with DNAm in both the <40 and 40+ subgroups (Figure 1). Other mRNAs had a weaker relationship with DNAm in at least one of these subgroups. For example, partial R2 values for PRDM1, RUNX3, and TGFβ1 were greater in participants over 40, whereas KIR inhibitory subunit 1, CD45RA, CD160, and TNF showed a stronger relationship with DNAm in the <40 subgroup. DNAm also correlated with CMV seropositivity, and this association was particularly robust in participants under 40 (Figure 1).
Unlike DNAm, few OSU_Senescence mRNAs strongly correlated with p16INK4a in mixed models adjusted for sex, chronological age, and couple (Figure 1; Supplementary Table S3). Two transcripts, CD28 and CD244, correlated with p16INK4a in the entire cohort, but these relationships were lost in the <40 and 40+ subgroups. p14ARF, which is transcribed from the same gene as p16INK4a, correlated with p16INK4a in the entire cohort and the <40 subgroup (Figure 1; Supplementary Table S3). p14ARF also correlated with p16INK4a in participants 40+ years of age, but this relationship did not reach statistical significance likely due to the smaller sample size. Thus, unlike DNAm, there are few robust correlations between cell composition markers and p16INK4a.
To further explore possible relationships between aging biomarkers and PBTL cell composition, we performed flow cytometry on 61 participant samples for which cryopreserved PBTLs were available. Demographic information for these 61 individuals appears in Supplementary Table S4. In each sample, we measured the percentage of CD3, CD4, and CD8 cells expressing CD28 because the loss of CD28 is considered a hallmark of immunosenescence (16). Consistent with our mRNA analysis, p16INK4a expression did not correlate with any measured PBTL subset after adjusting for sex, chronological age, and couple (Supplementary Table S5). However, the percentage of CD28+ PBTLs was negatively associated with DNAm in the CD8 and CD4 lineages (Supplementary Table S5). These data link DNAm, but not p16INK4a, to PBTL changes associated with immunosenescence.
Discussion
This is the first report to examine DNAm, p16INK4a, CMV seropositivity, and PBTL subset markers in the same patient population, sample, and cell type. We discover robust correlations between PBTL subset markers and DNAm, but not p16INK4a. These observations suggest that changes in peripheral blood constituents have a greater impact on DNAm than p16INK4a. However, distinct correlations might be observed in other populations. Only 11.1% (13/117) of our study participants were over the age of 60 and few reported any comorbidities (12.8%; 15/117) (Table 1). Most participants were also nonsmokers (80.3%; 94/117), nonobese (76.9%; 90/117) and cancer-free (100%) (Table 1).
We were unable to test whether other epigenetic clocks are correlated with PBTL subset markers due to the capture-based technical strategy chosen to measure DNAm. However, the tissue-agnostic discovery approach used to generate Horvath’s algorithm seems the most likely to provide consistency across PBTL cell types (3). Supporting this argument, a prior report found no significant difference in the DNAm of whole blood, total PBMCs, or sorted CD8+/CD4+ T cells from 4 to 6 individuals (3). Our results indicate that with increased cell granularity (74 mRNA markers) and a larger sample size (n = 117), relationships between PBTL subset markers and DNAm can be observed (Figure 1). Epigenetic profiles of these cell subsets will be needed before we can determine whether more granular cell type adjustments enhance the predictive power of DNAm.
Age-independent correlations (partial R2 values) between DNAm and individual OSU_Senescence mRNAs support the idea that PBTL epigenetic aging is marked by an expansion effector memory T cells (TEM). In all age groups, DNAm correlated with the expression of TEM markers (CD8, GZMB) and T-box transcription factors critical for TEM differentiation and function (TBX21, EOMES) (Figure 1; Supplementary Table S2). DNAm was also associated with inhibitory receptor mRNAs, including KLRG1 and PDCD1, which are highly expressed on TEM and terminally differentiated TEM cells (TEMRA) (17). Notably, corrections for sample purity (%CD3+) did not affect these relationships, suggesting that other contaminating cell types were not responsible for the correlations observed (Supplementary Table S6). Flow cytometry data also linked DNAm to the process of immunosenescence, which is associated with an expansion of terminally differentiated CD8+ PBTLs [(18); Figure 1; Supplementary Table S5]. Thus, further exploration of how age-related TEM accumulation relates to DNAm measurements in peripheral blood may provide mechanistic insight into the physiologic origins of accelerated epigenetic aging.
Our findings point to CMV as a potential cause of accelerated epigenetic aging—especially in healthy adults under 40. CMV establishes a lifelong infection in 50%–90% of world’s population (19). While most seropositive individuals are asymptomatic, epidemiological studies link CMV seropositivity to cardiovascular disease and frailty (19). Persistent efforts to control the virus can drive immune “memory inflation” characterized by the expansion of highly differentiated, CMV-specific CD8+ T cells (20). Such CMV-driven TEM expansion is consistent with our observation that DNAm correlated positively with CD45R0 (m = 0.021) and negatively with CD45RA (m = −0.028) in the under 40 subgroup. Furthermore, correlations between DNAm and receptors like CD160, CD244, LAG3, and KIRs (Figure 1; Supplementary Table S2) suggest that chronic antigen exposure plays a role in the accelerated epigenetic aging of young, healthy adults. Notably, all of these relationships were diminished in individuals over 40, hinting at a limited role for CMV in the epigenetic aging of older individuals.
Although p16INK4a expression is cell type specific (4), changes in PBTL constituents were not a major driver of p16INK4a dynamics in our study. It is interesting that p16INK4a correlated indirectly with CD28, a T cell co-stimulatory receptor lost on immunosenescent cells, and that the strength of this negative correlation appeared to increase with age (Table 1). While the relationship between CD28 surface expression and p16INK4a did not reach statistical significance in our flow cytometry data (p = .15 for CD8+CD28−; Supplementary Table S5), we cannot rule out the possibility that a relationship would be significant in a larger cohort.
In sum, this pilot study suggests that CMV and PBTL dynamics could potentially influence measurements of DNAm. Expansion of this investigation to include a greater number of older adults and additional epigenetic clocks will help clarify when and how CMV serostatus and PBTL constituents should be addressed in aging biomarker studies.
Supplementary Material
Acknowledgments
The authors thank Y. Chow (Zymo.) for technical advice. B.F.L., S.Z., C.E.B., and J.L.H. helped to prepare samples and compile data. J.P. and R.R.A. performed the statistical analyses. L.Y. and P.F. assisted with Nanostring data normalization. C.E.B. wrote the article with assistance from B.F.L., R.R.A., J.P., and J.K.K.-G. J.K.K.-G. and C.E.B. designed and oversaw the project.
Funding
This work was supported by awards from the Glenn Foundation (C.E.B.) and the National Institutes of Health (R01AG059711 to C.E.B. and R01AG057032 to J.K.K.-G.), P30CA016058 [OSUCCC Genomics Shared Resource]).
Conflict of Interest
None reported.
References
- 1. LaPak KM, Burd CE. The molecular balancing act of p16(INK4a) in cancer and aging. Mol Cancer Res. 2014;12:167–183. doi: 10.1158/1541-7786.MCR-13-0350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Horvath S, Raj K. DNA methylation-based biomarkers and the epigenetic clock theory of ageing. Nat Rev Genet. 2018;19:371–384. doi: 10.1038/s41576-018-0004-3 [DOI] [PubMed] [Google Scholar]
- 3. Horvath S. DNA methylation age of human tissues and cell types. Genome Biol. 2013;14:R115. doi: 10.1186/gb-2013-14-10-r115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Liu Y, Sanoff HK, Cho H, et al. Expression of p16(INK4a) in peripheral blood T-cells is a biomarker of human aging. Aging Cell. 2009;8:439–448. doi: 10.1111/j.1474-9726.2009.00489.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Teschendorff AE, Zheng SC. Cell-type deconvolution in epigenome-wide association studies: a review and recommendations. Epigenomics. 2017;9:757–768. doi: 10.2217/epi-2016-0153 [DOI] [PubMed] [Google Scholar]
- 6. Ziller MJ, Gu H, Müller F, et al. Charting a dynamic DNA methylation landscape of the human genome. Nature. 2013;500:477–481. doi: 10.1038/nature12433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Quan H, Li B, Couris CM, et al. Updating and validating the Charlson comorbidity index and score for risk adjustment in hospital discharge abstracts using data from 6 countries. Am J Epidemiol. 2011;173:676–682. doi: 10.1093/aje/kwq433 [DOI] [PubMed] [Google Scholar]
- 8. Hall WH, Ramachandran R, Narayan S, Jani AB, Vijayakumar S. An electronic application for rapidly calculating Charlson comorbidity score. BMC Cancer. 2004;4:94. doi: 10.1186/1471-2407-4-94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bennett JM, Glaser R, Malarkey WB, Beversdorf DQ, Peng J, Kiecolt-Glaser JK. Inflammation and reactivation of latent herpesviruses in older adults. Brain Behav Immun. 2012;26:739–746. doi: 10.1016/j.bbi.2011.11.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Edwards LJ, Muller KE, Wolfinger RD, Qaqish BF, Schabenberger O. An R2 statistic for fixed effects in the linear mixed model. Stat Med. 2008;27:6137–6157. doi: 10.1002/sim.3429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Arias E, Xu JQ. United States Life Tables, 2017. National Vital Statistics Reports. Hyattsville, MD: National Center for Health Statistics; 2019. [PubMed] [Google Scholar]
- 12. R Core Team. R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing; 2019. [Google Scholar]
- 13. Wood WA, Krishnamurthy J, Mitin N, et al. Chemotherapy and stem cell transplantation increase p16INK4a expression, a biomarker of T-cell Aging. EBioMedicine. 2016;11:227–238. doi: 10.1016/j.ebiom.2016.08.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sanoff HK, Deal AM, Krishnamurthy J, et al. Effect of cytotoxic chemotherapy on markers of molecular age in patients with breast cancer. J Natl Cancer Inst. 2014;106:dju057. doi: 10.1093/jnci/dju057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Horvath S, Gurven M, Levine ME, et al. An epigenetic clock analysis of race/ethnicity, sex, and coronary heart disease. Genome Biol. 2016;17:171. doi: 10.1186/s13059-016-1030-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Boucher N, Dufeu-Duchesne T, Vicaut E, Farge D, Effros RB, Schächter F. CD28 expression in T cell aging and human longevity. Exp Gerontol. 1998;33:267–282. doi: 10.1016/s0531-5565(97)00132-0 [DOI] [PubMed] [Google Scholar]
- 17. Legat A, Speiser DE, Pircher H, Zehn D, Fuertes Marraco SA. Inhibitory receptor expression depends more dominantly on differentiation and activation than “exhaustion” of human CD8 T cells. Front Immunol. 2013;4:455. doi: 10.3389/fimmu.2013.00455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Pawelec G. Hallmarks of human “immunosenescence”: adaptation or dysregulation? Immun Ageing. 2012;9:15. doi: 10.1186/1742-4933-9-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Nikolich-Zugich J, Cicin-Sain L, Collins-McMillen D, et al. Advances in cytomegalovirus (CMV) biology and its relationship to health, diseases, and aging. GeroScience. 2020;42:495–504. doi: 10.1007/s11357-020-00170-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Souquette A, Frere J, Smithey M, Sauce D, Thomas PG. A constant companion: immune recognition and response to cytomegalovirus with aging and implications for immune fitness. Geroscience. 2017;39:293–303. doi: 10.1007/s11357-017-9982-x [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.