Abstract
Background
Increasing first-line treatment failures in low- and middle-income countries (LMICs) have led to increased use of integrase strand transfer inhibitors (INSTIs) such as dolutegravir. However, HIV-1 susceptibility to INSTIs in LMICs, especially with previous raltegravir exposure, is poorly understood due to infrequent reporting of INSTI failures and testing for INSTI drug resistance mutations (DRMs).
Methods
A total of 51 non-subtype B HIV-1 infected patients failing third-line (raltegravir-based) therapy in Uganda were initially selected for the study. DRMs were detected using Sanger and deep sequencing. HIV integrase genes of 13 patients were cloned and replication capacities (RCs) and phenotypic susceptibilities to dolutegravir, raltegravir and elvitegravir were determined with TZM-bl cells. Spearman’s correlation coefficient was used to determine cross-resistance between INSTIs.
Results
INSTI DRMs were detected in 47% of patients. HIV integrase-recombinant virus carrying one primary INSTI DRM (N155H or Y143R/S) was susceptible to dolutegravir but highly resistant to raltegravir and elvitegravir (>50-fold change). Two patients, one with E138A/G140A/Q148R/G163R and one with E138K/G140A/S147G/Q148K, displayed the highest reported resistance to raltegravir, elvitegravir and even dolutegravir. The former multi-DRM virus had WT RC whereas the latter had lower RCs than WT.
Conclusions
In HIV-1 subtype A- and D-infected patients failing raltegravir and harbouring INSTI DRMs, there is high-level resistance to elvitegravir and raltegravir. More routine monitoring of INSTI treatment may be advised in LMICs, considering that multiple INSTI DRMs may have accumulated during prolonged exposure to raltegravir during virological failure, leading to high-level INSTI resistance, including dolutegravir resistance.
Introduction
HIV-1 integrase strand transfer inhibitors (INSTIs), the latest class of ART, have changed the HIV-1 treatment landscape around the world. The second-generation INSTI dolutegravir has a higher genetic barrier to resistance and has proved to be an effective INSTI when used in combination therapy for treatment-naive patients as well as individuals harbouring viruses resistant to raltegravir or elvitegravir.1,2 Under 2018 WHO guidelines, a dolutegravir-based regimen is preferred for first-line ART for all adults and adolescents except for those women of childbearing age who wish to become pregnant. Dolutegravir is also the preferred second-line drug of choice for those failing NNRTI-containing treatment regimens.3 For children >4 weeks of age, dolutegravir is favoured for second-line treatments following failure of NNRTIs or PI-containing ART.3 Following FDA approval in 2013, and prior to 2014, dolutegravir was rarely administered in low- and middle-income countries (LMICs) but with the roll-out of tenofovir disoproxil fumarate/lamivudine/dolutegravir (TDF/3TC/DTG) at a cost of US $75 annually per patient-year,4 over 4 million patients are now receiving a dolutegravir-containing regimen,5 eclipsing the number of patients receiving dolutegravir in high-income countries (HICs). In Uganda, the focus site of this study, generic dolutegravir is now part of the preferred first-line regimen (TDF/3TC/DTG) in all adults.6
Raltegravir performs a pivotal role as salvage and paediatric treatment and elvitegravir can be used in ART-naive patients in Uganda and other LMICs. However, raltegravir and elvitegravir resistance has been reported but mostly in HIV subtype B due to early use of INSTIs.7 In addition, there is infrequent reporting of INSTI failures and testing for INSTI drug resistance mutations (DRMs) in non-B subtypes, especially subtypes A and D. Switching patients to salvage treatment in Uganda is done under the discretion of a committee by the Ministry of Health, which relies on resistance predictions from HIV drug resistance databases.6 However, there appears to be subtype specificity associated with resistance to INSTIs with some mutations. For example, substitutions at Q148 of HIV integrase (IN), which confer resistance to raltegravir, elvitegravir and cross-resistance to dolutegravir, appear more frequently in subtype B than in non-B subtypes.8 Thus, it becomes imperative to assess the impact of raltegravir and elvitegravir-associated mutations in vitro to further elucidate the impact of these mutations in subtype A and D INs.
Despite the rapid and large scale roll-out of dolutegravir in LMICs, there are very limited data on the susceptibility of non-subtype B HIV-1 to INSTIs,8,9 including cross-resistance to dolutegravir conferred by raltegravir-resistant HIV-1 variants emerging in non-B HIV-1 subtype-infected patients failing raltegravir-based regimens.8 In addition, increasing spread of HIV-1 non-B subtypes in regions where subtype B previously predominated, such as Europe and America,10–12 calls for the urgent need for more genotypic drug susceptibility testing for INSTI resistance.
Even with the use of generic dolutegravir for first-line ART in LMICs, treatment failures with the NNRTI-based and subsequent boosted PI-based regimens are still followed by raltegravir-based ART. Failure of this third-line treatment in LMICs leaves patients with few options since entry inhibitors, i.e. enfuvirtide and maraviroc, are not readily available. In our previous study, we found that over 50% of patients failing raltegravir-based therapy in Uganda harboured raltegravir-associated DRMs.13
In this study, non-B HIV-1 IN chimeric viruses derived from 11 Ugandan patients failing a raltegravir-based third-line regimen showed significant resistance to raltegravir and elvitegravir but remained susceptible to dolutegravir when the ‘common’ raltegravir DRMs were present: N155H and Y143R/S. However, viruses harbouring three or four of E138K/A, G140A, S147G, Q148K/R and G163R DRMs showed high-level resistance to all INSTIs and only one had significantly impaired replicative rates.
Materials and methods
Samples for the study
Samples were collected from WHO, the College of American Pathologists and the NIH Virology Quality Assurance-accredited Center for AIDS Research (CFAR) laboratory of the Joint Clinical Research Centre (JCRC) in Kampala, Uganda. The patient database in the CFAR laboratory was used to retrieve the patient demographic, medical and treatment histories. A total of 60 plasma samples were collected from patients failing raltegravir-based third-line regimens. Virological failure was defined as a viral load >1000 copies/mL and immunological failure was defined as a CD4+ T cell count <250 cells/mm3. Written consent was obtained from all patients prior to sample storage. Ethical clearance was obtained from the Institutional Review Boards at the JCRC and University Hospitals Cleveland Medical Center/Case Western Reserve University (EM-10-07 and 10-05-35).
RNA extraction and PCR amplification
HIV-1 viral RNA was extracted from plasma using a QIAamp viral RNA Mini Kit (QIAGEN) and the HIV-1 IN-coding region was amplified using a Superscript III single RT–PCR system (Thermo Fisher Scientific) as previously described13 (Table S1, available as Supplementary data at JAC Online).
Sanger sequencing and sequence analysis
The HIV-1 IN-coding regions were amplified and analysed using Sanger sequencing as previously described.13 Briefly, a quantified and purified PCR product was sequenced to cover the full length of the HIV-1 IN (1–288 amino acids) (Table S1). Sequences were exported and analysed in RECall (beta v3.02) as recommended by WHO.14 The Stanford HIV-1db Sierra web service algorithm v8.3 was used to predict resistance phenotypes.15
Library preparation and deep sequencing
An amplicon-based deep sequencing method was used to detect variants and confirm the presence of DRMs originally identified by Sanger sequencing (Table S1). Briefly, two overlapping PCR products spanning the full length of HIV-1 IN were purified using Agencourt AMPure XP (Beckman Coulter) and quantified using a Quant-iT PicoGreen dsDNA Assay Kit (Thermo Fisher Scientific). The barcodes were added using a Nextera XT Index Kit v2 (Illumina) (Table S1) and paired-end sequencing done on a MiSeq instrument (Illumina). Analysis was performed with MiSeq Reporter analysis software v2.6 (Illumina) and drug resistance interpretation was done using Stanford HIV-1db Sierra web service algorithm v8.3.15 Samples confirmed by deep sequencing to harbour INSTI-associated DRMs (n = 11) or having no known INSTI-associated DRMs (n = 2) were selected for phenotypic testing for dolutegravir, raltegravir and elvitegravir (Table 1 and Table S2). HIV-1 subtype classification and detection of recombination forms was done using SCUEAL.16
Table 1.
Virological and clinical characteristics of patients with INSTI-associated mutations in the study
| Sample ID | cART | Viral load (copies/mL) | Predicted subtype | DRMs detected |
|---|---|---|---|---|
| UG1179 | LPVr/RAL | ND | A | N155H |
| UG11 | ATVr/RAL | ND | D | N155H |
| UG537 | LPVr/RAL | 255 641 | A/D | N155H |
| UG138 | DRVr/RAL | 275 059 | A | N155H, T97A, L74I |
| UG42 | LPVr/RAL | 155 982 | D | N155H, E157Q, G163R, M50L, L74I, V151I |
| UG35 | DRVr/RAL | 2 293 840 | A | Y143R, T97A, M50I, L74IM |
| UG1044 | TDF/3TC/DRVr/RAL | 29 200 | A | Y143S, T97A |
| UG481 | TDF/FTC/DRVr/RAL | 3914 | A | Y143R, TA97AT, G163R |
| UG23 | LPVr/RAL | 850 | A | E138A, T97A, V151A |
| UG1059 | TDF/3TC/LPVr/RAL | 14 200 | A | E138A, G140A, Q148R, G163R |
| UG206 | LPVr/RAL | 1 515 000 | D | E138K, G140A, Q148K, S147G |
| UG14 | ART naive | 3008 | A | none |
| UG98 | ART naive | ND | D | none |
The HIV subtype was predicted using the SCUEAL subtype classification algorithm. Viral loads were assayed using Abbott m2000sp/rt or Roche COBAS Amplicor Monitor ultrasensitive tests, v1.5.
Major INSTI DRMs are shown in bold.
cART, combined ART; ND, not determined; ATVr, atazanavir/ritonavir; DRVr, darunavir/ritonavir; LPVr, lopinavir/ritonavir; RAL, raltegravir; FTC, emtricitabine.
Cells and antiviral compounds
TZM-bl, U87.CD4.CXCR4 and HEK293T cell lines were obtained through the NIH AIDS Research and Reference Reagent Program (Division of AIDS, NIAID, NIH). TZM-bl and HEK293T cells were maintained in DMEM supplemented with 10% FBS and 100 mg/L penicillin/streptomycin. U87.CD4.CXCR4 cells were maintained in DMEM supplemented with 10% FBS, 100 mg/L penicillin/streptomycin, 300 mg/L G418 and 1 mg/L puromycin (Invitrogen, Carlsbad, CA, USA). All cell lines were subcultured every 3–4 days at 37°C under 5% CO2. The TZM-bl cells had reporter luciferase and β-galactosidase reporter genes that can be activated by expression of HIV-1 Tat. Elvitegravir was obtained from Gilead Sciences Inc. (Foster City, CA, USA) and raltegravir and dolutegravir from NIH.
Construction of HIV-1 in chimeric viruses
HIV-1 full-length IN PCR products were recombined into a near full-length HIV-1 (pREC_NFL_IN/URA3) vector using transfected Saccharomyces cerevisiae MYA-906 cells (ATCC) based on the yeast homologous recombination/gap repair system.17 Plasmids were extracted from the yeast cells using phenol/chloroform (Thermo Fisher Scientific) and transformed into electrocompetent Escherichia coli Stbl4 cells (Thermo Fisher Scientific). Plasmids were extracted from the bacteria using QIAGEN Miniprep Kits (Hilden, Germany). The presence of mutation(s) in the generated plasmid was confirmed by Sanger sequencing. pREC_NFL_INT plasmids were co-transfected into recombination/gap repair system HEK 293T cells (3 × 104 cells/well) along with the complementing plasmid pCMV_cplt using Fugene 6 reagent (Promega, Madison, WI, USA).17 The produced heterodiploid virus particles containing one copy of the pREC_NFL_INT and cplt HIV-1 RNAs were further propagated in U87.CD4.CXCR4 cells to produce a complete HIV-1 genome. Following virus propagation, viral RNA was resequenced to confirm the presence of the various DRMs.
INSTI resistance assays in TZM-bl cells
The susceptibility of patient-derived viruses to dolutegravir, raltegravir and elvitegravir was determined using short-term resistance assays with TZM-bl cells. Briefly, 20 000 cells were seeded into each well of a 96-well plate and infected with the controls NL4-3, UG14, UG98 and mutant viruses in the presence of 10-fold dilutions of dolutegravir, raltegravir or elvitegravir (100 μM to 10−8 μM) and diethylaminoethyl (DEAE)-dextran (1 mg/mL). The amount of virus added to each well was normalized to 0.01 moi based on infectious titre. After 48 h incubation at 37°C and 5% CO2, the infectivity of viruses was quantified using X-gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) as previously described.18 The stained colonies were counted using a Cytation 5 plate reader (BioTek, USA) and confirmed by manual counting using a fluorescence microscope. Drug susceptibility curves were generated using non-linear regression curve-fitting features of GraphPad Prism 8.0 software (GraphPad Software Inc., San Diego, CA, USA). Drug resistance was expressed as fold change (FC) in EC50 between controls and mutant viruses based on at least two sets of experiments, each performed in quadruplicate.
X-gal staining assay
The X-gal (40 mg/mL) substrate was used to detect β-galactosidase enzyme expression from infected cells.18 The TZM-bl cells infected for 48 h were fixed with glutaraldehyde (0.2%) and formaldehyde (0.8%). The cells were sequentially stained with potassium ferrocyanide (0.2 M), potassium ferricyanide (0.2 M), magnesium chloride (2 M) and X-gal substrate, and then incubated for 2 h at 37°C.
Infectivity assay in TZM-bl cells
Infectivity of HIV-1 IN chimeric mutant viruses was determined using a short-term infectivity assay in TZM-bl cells. Briefly, 20 000 TZM-bl cells in the presence of DEAE-dextran (1 mg/mL) were infected with increasing concentrations of either mutants or controls. Cells were fixed and stained, and β-galactosidase expression was measured using the X-gal protocol as described above. The fold decreases in infectivity were expressed as a percentage of relative decrease in AUC, the amount of virus needed for TZM-bl cells to produce the maximal level of β- galactosidase in an infection.
Statistical analyses
Statistical analyses were performed using non-linear regression in GraphPad Prism 8.1.2. The level of cross-resistance was analysed using Spearman’s rank order test.
Results
Samples and patient demographics
Samples were obtained from 60 patients failing a third-line raltegravir-based regimen in Uganda. Of them, 51 (85%) were successfully amplified and sequenced by Sanger and deep sequencing.13INSTI-associated DRMs were detected in 24/51 (47%) patient samples. Eleven samples with INSTI DRMs were subsequently selected for phenotypic assays. Of the 11 samples selected for phenotypic studies, 8 were obtained from individuals on dual therapy (PI + raltegravir) while the other 3 were treated with triple therapy (i.e. NRTI + PI + raltegravir). The average viral load count was 454 669 copies/mL (range 850–2 293 840) (Table 1). Seven of 11 (64%) viruses were classified as HIV-1 subtype A, 27% subtype D (3/11) and 1 sample had a subtype A/D recombinant (Table 1 and Table S2). Two patient INs with no INSTI-associated DRMs [UG98 (subtype D) and UG14 (subtype A)] and NL4-3 HIV-1 (subtype B) were included in the phenotypic assays as reference strains (Table 1).
As previously published,13 nearly 4% (2/51) of the patients failing a raltegravir-containing therapy harboured a virus with multiple DRMs to INSTIs, i.e. the subtype A sample UG1059 with E138A/G140A/G163R/Q148R and the subtype D sample UG206 with E138K/G140A/S147G/Q148K. These were selected for cloning and phenotypic assays along with the other nine samples with INSTI DRMs. Whereas Q148H only emerged in the context of G140S mutants in subtype B, the Q148R/K mutation appeared to be associated with G140A in non-B subtypes. Overall, N155H was the predominant INSTI DRM, found in 9 of 51 patients. For this study, three IN regions with N155H only (UG1179, UG11 and UG537) and two patients’ INs containing N155H plus the secondary mutations L74I/T97A or M50L/L74I/V151I/E157Q/G163R (UG138 and UG42) were cloned for phenotypic analyses (Table 1 and Table S2). Another three had Y143R plus secondary mutations M50I/L74IM/T97A (UG35) or T97AT/G163R (UG481) or Y143S plus T97A (UG1044). One patient (UG23) had E138A, T97A and V15IA mutations (Table 1 and Table S2).
N155H and Y143R/S emergence during raltegravir treatment in subtype A and D confer high-level resistance to raltegravir and elvitegravir but not to dolutegravir
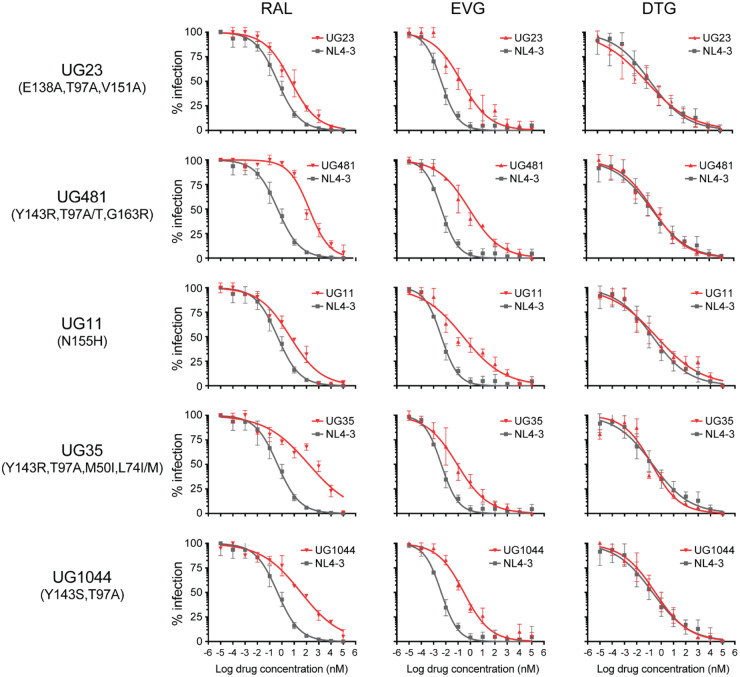
The N155H mutation in HIV-1 subtype B confers a significant level of resistance to both raltegravir and elvitegravir. In drug susceptibility assays, the three subtype D and circulating recombinant form A/D patient-derived recombinant viruses (UG11, UG537 and UG1179) carrying only N155H had a 10- to 19-fold decrease in susceptibility to raltegravir and 63- to 78-fold to elvitegravir, but only a 1.3-fold change in dolutegravir susceptibility when compared with the susceptibility of WT NL4-3 (Figures 1 and 2 and Figure S1). Slightly higher levels of resistance were observed with these N155H viruses when compared with the WT subtype D or A IN cloned into NL4-3. When L74I and T97A was found in addition to N155H in a subtype A IN, there was no effect on susceptibility to dolutegravir (FC 1.57) but increased resistance to raltegravir (FC 39) and elvitegravir (FC 48) was evident (when compared with the WT NL4-3). Presence of the secondary mutations E157Q, G163R, M50L, L74I and V151I in addition to N155H in the subtype D IN resulted in a low 2.8-fold resistance to dolutegravir but high-level resistance to elvitegravir (FC 70) and raltegravir (FC 56).
Figure 1.
The susceptibility of viruses with INSTI DRMs. The log values of drug concentrations were plotted against percentage of infections in TZM-bl cells detected by the X-gal assay. The colonies were counted using ELISpot and they were from 2–3 independent experiments, each run in quadruplicate. The change in EC50 relative to WT (NL4-3) is shown. RAL, raltegravir; EVG, elvitegravir; DTG, dolutegravir. This figure appears in colour in the online version of JAC and in black and white in the print version of JAC.
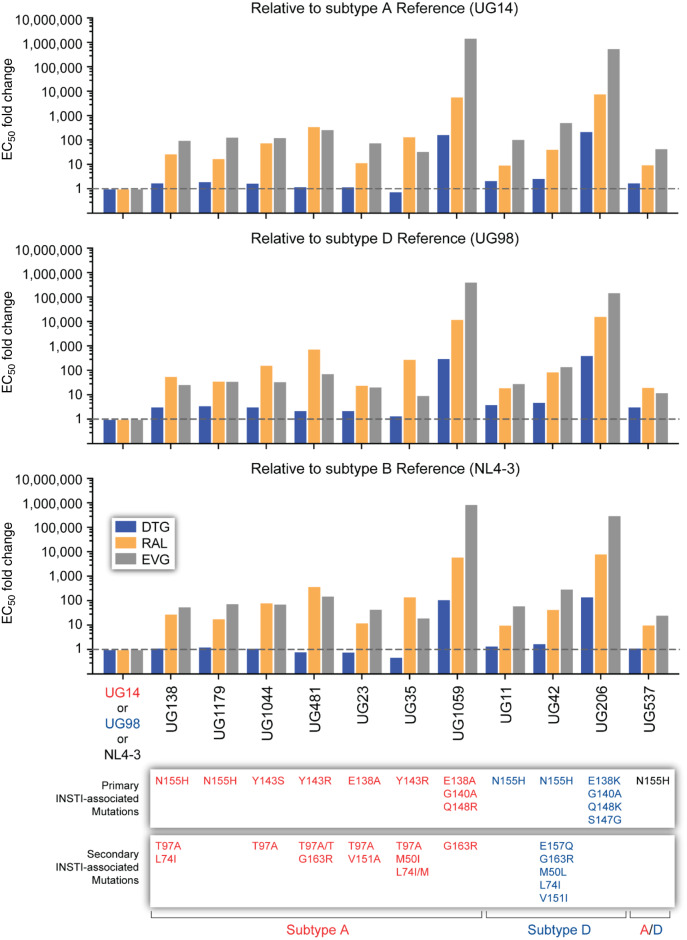
Figure 2.
The susceptibility of mutants by subtype. The FC in EC50 values of dolutegravir, raltegravir and elvitegravir relative to each subtype was determined by short-term infection assay in TZM-bl cells. Each value represents the mean FC of EC50 from 2–3 independent experiments, each done in quadruplicate. DTG, dolutegravir; RAL, raltegravir; EVG, elvitegravir. This figure appears in colour in the online version of JAC and in black and white in the print version of JAC.
The Y143R/S mutation only emerged in HIV-1 subtype A-infected patients and conferred slightly higher levels of resistance to raltegravir than to elvitegravir in subtype A HIV-1. The T97A, M50I and L74I/M secondary mutations had minimal or no impact on INSTI resistance. Susceptibility to dolutegravir was not affected by Y143R/S with the other secondary mutations (Figures 1 and 2 and Figure S1).
Multiple INSTI DRMs with secondary mutations confer high-level resistance to dolutegravir, raltegravir and elvitegravir
Q148H/K/R alone typically has minimal effect on dolutegravir susceptibility. In patients failing a raltegravir-based treatment in Uganda, the Q148R mutation in subtype A HIV-1 was found in combination with the primary INSTI DRMs E138A and G140A and, in subtype D, a Q148K mutation with E138K, G140A and S147G. With the IN from both patients, the chimeric viruses were highly resistant to elvitegravir, raltegravir (both >1000-fold) and dolutegravir (>100-fold) (Figure 2, Table 2 and Figure S1).
Table 2.
The mean EC50 of different HIV INSTIs for recombinant viruses in the study
| DRMs | DTG |
RAL |
EVG |
|||
|---|---|---|---|---|---|---|
| EC50 (nM) | 95% CI (nM) | EC50 (nM) | 95% CI (nM) | EC50 (nM) | 95% CI (nM) | |
| WT | 0.34 | 0.11–0.34 | 0.48 | 0.32–0.59 | 0.00 | 0.003–0.005 |
| N155H, T97A, L74I | 0.40 | 0.17–0.54 | 13.8 | 6.42–43.5 | 0.27 | 0.003–0.008 |
| N155H | 0.44 | 0.14–1.38 | 8.91 | 4.93–28.9 | 0.37 | 0.08–0.92 |
| N155H | 0.40 | 0.16–0.55 | 4.96 | 2.70–5.49 | 0.12 | 0.09–0.23 |
| N155H | 0.49 | 0.18–0.87 | 4.84 | 2.76–7.93 | 0.30 | 0.12–0.53 |
| Y143S, T97A | 0.39 | 0.19–0.54 | 39.6 | 17.7–71.01 | 0.35 | 0.15–0.64 |
| Y143R, T97AT, G163R | 0.28 | 0.14–0.43 | 183.7 | 99.8–262.4 | 0.75 | 0.48–1.28 |
| E138A, T97A, V151A | 0.27 | 0.03–0.22 | 6.02 | 3.60–8.12 | 0.22 | 0.09–0.34 |
| Y143R, T97A, M50I, L74IM | 0.17 | 0.08–0.33 | 69.3 | 30.09–100.3 | 0.10 | 0.05–0.13 |
| N155H, E157Q, G163R, M50L, L74I, V151I | 0.60 | 0.18–1.62 | 21.2 | 5.33–57.9 | 1.45 | 0.37–2.69 |
| E138A, G140A, Q148R, G163R | 37.8 | 11.88–120.5 | 2996.03 | 2699–17863 | 4180.00 | 3085–5257 |
| E138K, G140A, S147G, Q148K | 49.6 | 27.6–87.5 | 4002.00 | 2624–6060 | 1550.9 | 1107–4491 |
| None (UG14) | 0.22 | 0.11–0.32 | 0.50 | 0.34–5.19 | 0.00 | 0.001–0.006 |
| None (UG98) | 0.08 | 0.04–0.17 | 0.13 | 0.05–0.29 | 0.03 | 0.009–0.035 |
DTG, dolutegravir; RAL, raltegravir; EVG, elvitegravir.
Finally, the chimeric virus from an HIV-1 subtype A-infected patient with the secondary mutations T97A, V151A and E138A had resistance to elvitegravir (45-fold) and low-level resistance to raltegravir (12-fold) but WT susceptibility to dolutegravir (Figures 1 and 2 and Figure S1).
For all the chimeric viruses studied for drug susceptibility, the IN genes of HIV-1 subtype A- and D-infected patients containing INSTI DRMs were cloned into a subtype B backbone. Despite the concerns with complementation compatibility, the same level of resistance with these IN chimeric viruses was observed when compared with WT NL4-3 or the NL4-3 containing the WT subtype A and D IN-coding regions (Figure S2).
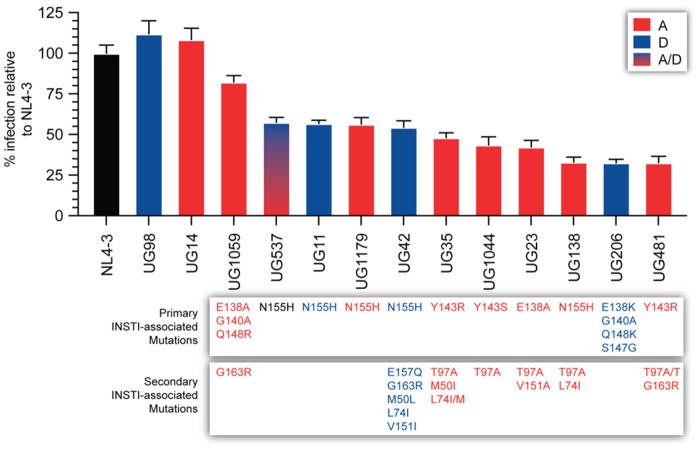
INSTI DRMs can impair RC regardless of subtype
Using TZM-bl cells, the RCs of these IN chimeric HIV-1 (all with an NL4-3 backbone) were compared with WT NL4-3, UG14 and UG98 references. All the IN chimeric HIV-1 with INSTI DRMs had significantly reduced RC compared with the reference WT strains. Reduced RC exhibited by the viruses was not simply due to poor complementation, considering the IN of subtype A and D in a NL4-3 backbone had slightly higher replication rates than WT NL4-3. Viruses UG138-A (N155H; secondary mutations, T97A/L74I) and UG481-A (Y143R; secondary mutations, T97A/G163R) had the lowest RCs at ∼30% of NL4-3 and <30% of the subtype A and D references. UG206-D, with four primary DRMs (E138K, G140A, S147G and Q148K) also had a low RC (29%). However, there was no clear pattern of reduced replication rates based on any of the primary or secondary DRMs (Figure 3). UG23-A, UG1044-A and UG35-A replicated at 40% to 45% of the references, while UG1179-A, UG537-A/D, UG42-D and UG11-D were at 50% to 60% of WT HIV-1. The other chimeric HIV-1 with three primary DRMs in IN (E138A, G140A and Q148R) and G163R as a secondary mutation had the highest RC (>80% of WT).
Figure 3.
Relative viral infectivity of recombinant viruses. The viral infectivity of resistant mutants compared with controls and WT was determined using short-term infection assay in TZM-bl cells. AUC was used to measure the relative decrease in infectivity. The data shown represent means and SD from independent experiments performed in triplicate. This figure appears in colour in the online version of JAC and in black and white in the print version of JAC.
Cross-resistance observed between dolutegravir and elvitegravir
Correlation coefficients of FC values for dolutegravir, raltegravir and elvitegravir were assessed to determine the level of cross-resistance between the INSTIs. Dolutegravir FC values for resistance were found to correlate with elvitegravir FC values for resistance (P = 0.0073, R = 0.8), but not with FC values for raltegravir resistance (P = 0.54, R = 0.21).
Discussion
Resistance to INSTIs is well characterized in HICs with a 12, 7 and 6 year history of employing raltegravir, elvitegravir and dolutegravir, respectively. The four typical major pathways of INSTI resistance involve N155H, Q148H/K/R and Y143R/H/C for raltegravir and elvitegravir and R263K for dolutegravir and bictegravir as primary INSTI DRMs. Both in vitro and in vivo selection of dolutegravir resistance is conferred by either an accumulation of INSTI DRMs or the emergence of R263K. Both pathways confer low-level dolutegravir resistance and significant reductions in RC, and often require the emergence of compensatory mutations, e.g. H51Y with R263K.19
In this study, we produced 11 chimeric viruses harbouring the IN genes of Ugandan patients failing third-line raltegravir-based treatment and with known DRMs. Chimeric IN HIV-1 produced with single DRMs resulted in significant resistance to elvitegravir and raltegravir and a 43% to 68% decrease in RC. Even these single DRMs with compensatory/secondary mutations emerging in subtype A and D IN do not confer cross-resistance to dolutegravir. HIV-1 containing the IN of two patients, UG1059 and UG206, with three and four DRMs, respectively, showed high-level resistance to elvitegravir and raltegravir and >100-fold cross-resistance to dolutegravir. The levels of INSTI resistance shown by these two patient-derived viruses are among the highest ever recorded with phenotypic tests. Although the subtype D UG206 had a 70% reduction in RC, the RC for UG1059 was not significantly different from WT.
Few studies have assessed phenotypic INSTI resistance using the IN derived from INSTI failures in Uganda or in sub-Saharan Africa. One study describes resistance to INSTIs, even dolutegravir, in subtype A, B, C, D, F, G, CRF01, CRF02 and other circulating recombinant forms (CRFs) related to the Q148H/K/R resistance pathway but these Q148H/K/R mutations were infrequent in this cohort when compared with INSTI failures in subtype B-infected patients.8,20,21 There are, however, several genotypic studies describing common INSTI DRMs in untreated and in INSTI-treated patients in sub-Saharan Africa. For nearly 10 years, raltegravir-based treatment has been recommended by WHO for third-line regimens22 and as such is rarely used in treatment within LMICs. In just 4 years, generic dolutegravir and TDF/3TC/DTG has become accessible to 3.9 million HIV-infected individuals in sub-Saharan Africa and other developing countries.3,5 This important roll-out of dolutegravir was briefly interrupted with a report suggesting a teratogenic effect in babies born to mothers who start dolutegravir treatment from the time of conception.23 Nonetheless, there has been no extensive screen for susceptibility of African non-subtype B HIV-1 to dolutegravir and especially not with HIV-1 derived from those failing a raltegravir-based regimen. This study suggests that 5% of raltegravir failures may harbour dolutegravir-resistant HIV-1. Even this frequency of dolutegravir cross-resistance in raltegravir failures in subtype A- and D-infected individuals could be a cause of concern. Furthermore, a recent study by our team suggests that 28% of raltegravir failures harbour raltegravir- and elvitegravir-resistant viruses with previously uncharacterized INSTI DRMs related to subtype A and D (M. Avino, E. Ndashimye, D. J. Lizotte, A. S. Olabode, R. M. Gibson, A. A. Meadows, C. M. Kityo, E. Nabulime, F. Kyeyune, I. Nankya, M. E. Quiñones-Mateu, E. J. Arts and A. F. Y. Poon, unpublished data). Half of these new INSTI-resistant genotypes in subtype A and D HIV-1 displayed cross-resistance to dolutegravir.
HIV-1 IN mutation N155H is selected early under raltegravir pressure.24 We have previously shown predominant selection of N155H (17.6%) in HIV-1 patients failing third-line raltegravir-based regimens in Uganda13 and thus, understanding the susceptibility of viruses with a single N155H mutation is important in assessing whether dolutegravir could be utilized in a fourth-line regimen given the limited availability of other salvage drugs such as enfuvirtide or maraviroc in Uganda. In our study, three viruses with a single N155H mutation were susceptible to dolutegravir (FC 1.6–2.4); two of these are comparable to N155H in subtype B HIV-1 (FC 1.2).25 Given the low-level phenotypic resistance associated with dolutegravir failure in subtype B-infected individuals, even our modest 2.4-fold decreased susceptibility to dolutegravir with N155H may predict potential treatment failure with dolutegravir-containing ART use as a fourth-line treatment in Uganda.
Prolonged exposure to INSTIs following treatment failure can lead to an accumulation of primary DRMs and compensatory secondary mutations to increase INSTI resistance and/or restore viral fitness.26 Achieving viral suppression with dolutegravir in patients failing raltegravir or elvitegravir is reduced with each added primary INSTI DRM (G140H/A/S, E138A/K/T) in association with substitutions at Q148, as seen in the VIKING-3 study.1 Viral suppression with dolutegravir was also reduced with the INSTI mutations E138K, G140A, S147G, Q148R and T97A in a subtype C-infected, raltegravir-experienced patient from Botswana.27 However, this accumulation of INSTI DRMs is extremely rare in HICs, due in part to frequent patient visits, viral load monitoring and drug resistance testing. In Uganda, like many other LMICs, less frequent clinic visits and viral load testing, intermittent adherence and limited drug resistance testing could contribute to multi-DRM viruses with high-level INSTI resistance. Of 18 patients with raltegravir resistance in this Ugandan cohort, 1 had E138A, G140A, Q148R and G163R in a subtype A virus and 1 had E138K, G140A, S147G and Q148K in subtype D. To our knowledge, the presence of these four primary INSTI DRMs within IN has never been observed in subtype B-infected patients naive to or failing INSTIs.28 A combination of two primary INSTI DRMs (Q148H and G140S) with secondary mutations (T97A and L74M) conferred resistance to dolutegravir in subtype B viruses.29 The loss of RC observed with UG206 could also be compensated by the emergence of secondary mutations to support active infection in the presence of drug. Thus, the frequent appearance of multi-DRM HIV-1 in subtype A (as with UG1059) and subtype D (with UG206) upon INSTI treatment failure could have devastating consequences for the future roll-out of dolutegravir. Hopefully, an 11% frequency of multiple DRMs will not persist with larger cohorts on raltegravir-based treatment in sub-Saharan Africa.
Conclusions
In this study, we show that INSTI-resistant viruses in the majority of patients in Uganda failing raltegravir-based third-line treatment remain susceptible to dolutegravir and have impaired RC. However, accumulation of primary INSTI DRMs leads to high-level resistance to all INSTIs currently available in LMICs.
Supplementary Material
Acknowledgements
This work was partially presented at the 28th International Workshop on HIV Drug Resistance and Treatment Strategies, Johannesburg, South Africa, 16–18 October 2019 (Abstract number 8).
We thank the JCRC HIV drug resistance working team, which includes Francis Ssali, Helen Musana, Joselyne Nansimbe, Aggrey Bukuru and William Tamale (all JCRC, Kampala, Uganda) and all the staff of JCRC, Kampala, Uganda.
Appreciation also goes to patients receiving treatment and care at JCRC, Kampala, Uganda, for consent to provide clinical samples for the study, and feedback from other members of the Arts laboratory and colleagues at the Department of Microbiology and Immunology at Western University, London, Ontario, Canada.
Funding
This work was supported by National Institutes of Health (AI-49170) and Gilead to E.J.A; CWRU/UH Center for AIDS Research (P30 AI036219) and University of Otago, Webster Family Chair in Viral Pathogenesis to M.E.Q.-M.; Government of Canada through Genome Canada and the Ontario Genomics Institute (OGI-131) to M.A.; and Queen Elizabeth II Diamond Jubilee scholarship through Western University (DLI O19375892122) and Ontario Graduate scholarship international to E.N.
Transparency declarations
None to declare.
Author contributions
E.N., Y.L., A.M., C.T. and P.S.R. performed drug resistance assays and experimentation in this article and E.N., M.A., A.F.Y.P., A.S.O., R.M.G., M.E.Q.-M. performed data analyses. I.N., F.K. and C.M.K. recruited all the patients for this study. E.J.A. and M.E.Q.-M. procured the funding, and E.J.A designed, supervised and guided the overall direction of the study as the principal investigator.
References
- 1. Castagna A, Maggiolo F, Penco G et al. Dolutegravir in antiretroviral-experienced patients with raltegravir- and/or elvitegravir-resistant HIV-1: 24-week results of the phase III VIKING-3 study. J Infect Dis 2014; 210: 354–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Raffi F, Rachlis A, Stellbrink H-J et al. Once-daily dolutegravir versus raltegravir in antiretroviral-naive adults with HIV-1 infection: 48 week results from the randomised, double-blind, non-inferiority SPRING-2 study. Lancet 2013; 381: 735–43. [DOI] [PubMed] [Google Scholar]
- 3.Update of Recommendations on First-and Second-Line Antiretroviral Regimes, 2019. https://apps.who.int/iris/bitstream/handle/10665/325892/WHO-CDS-HIV-19.15-eng.pdf?ua=1.
- 4.ARV Market Report: The State of the Antiretroviral Drug Market in Low- And Middle-Income Countries, 2016-2021. https://clintonhealthaccess.org/wp-content/uploads/2017/09/2017-ARV-Market-Report_Final-2.pdf.
- 5.Medicines Patent Pool. Press Release: Five Years On, 3.9 Million People in the Developing World Have Access to HIV Treatment Dolutegravir, Thanks to Access-Oriented Voluntary Licensing Agreements. https://medicinespatentpool.org/news-publications-post/five-years-on-3-9-million-people-in-the-developing-world-have-access-to-hiv-treatment-dolutegravir-thanks-to-access-oriented-voluntary-licensing-agreements/.
- 6.Consolidated Guidelines for the Prevention and Treatment of HIV and AIDS in Uganda. https://elearning.idi.co.ug/pluginfile.php/5675/mod_page/content/19/Uganda%20HIV%20%20Guidelines%20-%20September2018.pdf.
- 7. Arts EJ, Hazuda DJ. HIV-1 antiretroviral drug therapy. Cold Spring Harb Perspect Med 2012; 2: a007161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Doyle T, Dunn DT, Ceccherini-Silberstein F et al. Integrase inhibitor (INI) genotypic resistance in treatment-naive and raltegravir-experienced patients infected with diverse HIV-1 clades. J Antimicrob Chemother 2015; 70: 3080–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Inzaule SC, Hamers RL, Noguera-Julian M et al. Primary resistance to integrase strand transfer inhibitors in patients infected with diverse HIV-1 subtypes in sub-Saharan Africa. J Antimicrob Chemother 2018; 73: 1167–72. [DOI] [PubMed] [Google Scholar]
- 10. Snoeck J, Kantor R, Shafer RW et al. Discordances between interpretation algorithms for genotypic resistance to protease and reverse transcriptase inhibitors of human immunodeficiency virus are subtype dependent. Antimicrob Agents Chemother 2006; 50: 694–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Vermund SH, Leigh-Brown AJ. The HIV epidemic: high-income countries. Cold Spring Harb Perspect Med 2012; 2: a007195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Yerly S, Vora S, Rizzardi P et al. Acute HIV infection: impact on the spread of HIV and transmission of drug resistance. AIDS 2001; 15: 2287–92. [DOI] [PubMed] [Google Scholar]
- 13. Ndashimye E, Avino M, Kyeyune F et al. Absence of HIV-1 drug resistance mutations supports the use of dolutegravir in Uganda. AIDS Res Hum Retroviruses 2018; 34: 404–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Woods CK, Brumme CJ, Liu TF et al. Automating HIV drug resistance genotyping with RECall, a freely accessible sequence analysis tool. J Clin Microbiol 2012; 50: 1936–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Tang MW, Liu TF, Shafer RW. The HIVdb system for HIV-1 genotypic resistance interpretation. Intervirology 2012; 55: 98–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kosakovsky Pond SL, Posada D, Stawiski E et al. An evolutionary model-based algorithm for accurate phylogenetic breakpoint mapping and subtype prediction in HIV-1. PLoS Comput Biol 2009; 5: e1000581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Dudley DM, Gao Y, Nelson KN et al. A novel yeast-based recombination method to clone and propagate diverse HIV-1 isolates. Biotechniques 2009; 46: 458–67. [DOI] [PubMed] [Google Scholar]
- 18. Kimpton J, Emerman M. Detection of replication-competent and pseudotyped human immunodeficiency virus with a sensitive cell line on the basis of activation of an integrated β-galactosidase gene. J Virol 1992; 66: 2232–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Mesplède T, Quashie PK, Osman N et al. Viral fitness cost prevents HIV-1 from evading dolutegravir drug pressure. Retrovirology 2013; 10: 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Fourati S, Charpentier C, Amiel C et al. Cross-resistance to elvitegravir and dolutegravir in 502 patients failing on raltegravir: a French national study of raltegravir-experienced HIV-1-infected patients. J Antimicrob Chemother 2015; 70: 1507–12. [DOI] [PubMed] [Google Scholar]
- 21. Boyd MA, Moore CL, Molina J-M et al. Baseline HIV-1 resistance, virological outcomes, and emergent resistance in the SECOND-LINE trial: an exploratory analysis. Lancet HIV 2015; 2: e42–51. [DOI] [PubMed] [Google Scholar]
- 22.WHO. Antiretroviral Therapy for HIV Infection in Adults and Adolescents. https://www.who.int/hiv/pub/arv/adult2010/en/.
- 23. Zash R, Makhema J, Shapiro RL. Neural-tube defects with dolutegravir treatment from the time of conception. N Engl J Med 2018; 379: 979–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Baldanti F, Paolucci S, Gulminetti R et al. Early emergence of raltegravir resistance mutations in patients receiving HAART salvage regimens. J Med Virol 2010; 82: 116–22. [DOI] [PubMed] [Google Scholar]
- 25. Kobayashi M, Yoshinaga T, Seki T et al. In vitro antiretroviral properties of S/GSK1349572, a next-generation HIV integrase inhibitor. Antimicrob Agents Chemother 2011; 55: 813–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hurt CB, Sebastian J, Hicks CB et al. Resistance to HIV integrase strand transfer inhibitors among clinical specimens in the United States, 2009-2012. Clin Infect Dis 2014; 58: 423–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Seatla KK, Avalos A, Moyo S et al. Four-class drug-resistant HIV-1 subtype C in a treatment experienced individual on dolutegravir-based antiretroviral therapy in Botswana. AIDS 2018; 32: 1899–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Anstett K, Brenner B, Mesplede T et al. HIV drug resistance against strand transfer integrase inhibitors. Retrovirology 2017; 14: 36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zhang WW, Cheung PK, Oliveira N et al. Accumulation of multiple mutations in vivo confers cross-resistance to new and existing integrase inhibitors. J Infect Dis 2018; 218: 1773–6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.