Abstract
Objectives
WGS-based antimicrobial susceptibility testing (AST) is as reliable as phenotypic AST for several antimicrobial/bacterial species combinations. However, routine use of WGS-based AST is hindered by the need for bioinformatics skills and knowledge of antimicrobial resistance (AMR) determinants to operate the vast majority of tools developed to date. By leveraging on ResFinder and PointFinder, two freely accessible tools that can also assist users without bioinformatics skills, we aimed at increasing their speed and providing an easily interpretable antibiogram as output.
Methods
The ResFinder code was re-written to process raw reads and use Kmer-based alignment. The existing ResFinder and PointFinder databases were revised and expanded. Additional databases were developed including a genotype-to-phenotype key associating each AMR determinant with a phenotype at the antimicrobial compound level, and species-specific panels for in silico antibiograms. ResFinder 4.0 was validated using Escherichia coli (n = 584), Salmonella spp. (n = 1081), Campylobacter jejuni (n = 239), Enterococcus faecium (n = 106), Enterococcus faecalis (n = 50) and Staphylococcus aureus (n = 163) exhibiting different AST profiles, and from different human and animal sources and geographical origins.
Results
Genotype–phenotype concordance was ≥95% for 46/51 and 25/32 of the antimicrobial/species combinations evaluated for Gram-negative and Gram-positive bacteria, respectively. When genotype–phenotype concordance was <95%, discrepancies were mainly linked to criteria for interpretation of phenotypic tests and suboptimal sequence quality, and not to ResFinder 4.0 performance.
Conclusions
WGS-based AST using ResFinder 4.0 provides in silico antibiograms as reliable as those obtained by phenotypic AST at least for the bacterial species/antimicrobial agents of major public health relevance considered.
Introduction
Antimicrobial susceptibility testing (AST) is the cornerstone of appropriate clinical use of antimicrobial agents and for surveillance programmes aiming at estimating the occurrence of antimicrobial resistance (AMR). Currently, in vitro phenotypic AST methods such as broth microdilution (BMD) and disc diffusion, which are based on principles laid down in the work by Alexander Fleming in the 1920s,1 are considered the gold standard for measuring AMR in bacteria. Despite being conceptually simple, phenotypic methods suffer from limitations that hinder reproducibility of results even when following international standards.2,3 These reproducibility issues range from the operator executing the test and reading the results, to parameters affecting bacterial growth and stability of reagents. In particular, the need for tightly controlled environmental conditions is a limiting factor for reliable AST in low- and middle-income countries where AMR claims the highest social and economic toll at present.4,5 An additional limitation of phenotypic AST is caused by incomplete agreements on panels of antimicrobial agents to test and on interpretive criteria by different organizations. It has also been argued that the laboratory conditions used for AST are poor predictors of the clinical efficacy for certain antimicrobial agent–bacterium combinations as a consequence of different bacterial growth dynamics in infection sites compared with in vitro conditions, inducible resistance genes, as well as silent genes that may revert during clinical antimicrobial treatment, among other factors.6–8 Furthermore, phenotypes can poorly inform conclusions on the epidemiology of resistance genes and bacteria, since phenotypic tests for AST have limited discriminatory power.
Genotypic approaches have been proposed as a valid alternative to phenotypic AST since the early 1990s.9 Although genotypic tests have been and are widely used for detection of AMR genes, their limited overall sensitivity hindered their application as reference AST methods.10,11 With increased accessibility and decreasing cost of next-generation sequencing (NGS), WGS has become an available methodology for routine characterization of bacteria. NGS-based methods outperform other genotypic approaches for detection of AMR since they allow detection of virtually any known AMR gene/mutation and identification of new variants of known AMR determinants.11 Furthermore, sequence data can be stored indefinitely and can be re-analysed when new AMR determinants are discovered phenotypically.11 The main caveat related to any genotypic AST method is represented by the fact that only known AMR mechanisms can be detected, whereas resistance caused by new mechanisms and/or modulation of gene expression (increased expression of efflux pumps, heteroresistance, etc.) might be overlooked.
In the last decade, at least 47 open-access bioinformatics pipelines for detecting AMR genes in NGS data have been published (reviewed in Hendriksen et al.12) and continue to be developed, but they mostly lack easy and rapidly interpretable outputs including translation of genotypes into predicted phenotypes.12–14 In 2012, we published ResFinder,15 the first online bioinformatics tool aimed at users without specialized bioinformatic skills, which provides detection of AMR genes in WGS data submitted through a web server. The ResFinder tool has been widely used, and has so far executed more than 400 000 jobs from more than 32 000 IP addresses in over 100 countries. ResFinder was expanded with PointFinder, a tool that detects chromosomal point mutations mediating resistance to selected antimicrobial agents in a few selected bacterial species16 and recently extended to include Mycobacterium tuberculosis.17
Here we describe the development and evaluation of a new and more advanced tool, ResFinder 4.0, which, in addition to the detection of AMR genes and chromosomal gene mutations, generates in silico antibiograms. ResFinder 4.0 was developed by improving and expanding the database of ResFinder and PointFinder and by rewriting the software. ResFinder 4.0 was validated using bacterial isolates of different species and genera from food, animal and human sources.
Methods
ResFinder 4.0 databases
ResFinder 4.0 contains four databases including AMR genes (ResFinder), chromosomal gene mutations mediating AMR (PointFinder), translation of genotypes into phenotypes and species-specific panels for in silico antibiograms. The databases of ResFinder15 and PointFinder16 were reviewed by experts and, when necessary, entries were removed or added. Furthermore, the PointFinder database was extended to include chromosomal gene mutations leading to ampicillin resistance in Enterococcus faecium, ciprofloxacin resistance in E. faecium and Enterococcus faecalis, and resistance to cefoxitin, chloramphenicol, ciprofloxacin, fusidic acid, linezolid, mupirocin, quinupristin–dalfopristin, rifampicin and trimethoprim in Staphylococcus aureus. The genotype-to-phenotype tables were created by experts, by using additional databases (www.bldb.eu for β-lactam resistance genes,18 http://faculty.washington.edu/marilynr/ for tetracycline as well as macrolide, lincosamide, streptogramin and oxazolidinone resistance genes) and by performing extensive literature searches. In the genotype-to-phenotype tables, the ResFinder and PointFinder entries have been associated with an AMR phenotype both at the antimicrobial class and at the antimicrobial compound level. A selection of antimicrobial compounds within each class was made to include antimicrobial agents important for clinical and surveillance purposes for the different bacterial species included (Table S1, available as Supplementary data at JAC Online). The genotype-to-phenotype tables also include: (i) the PubMed ID of relevant literature describing the respective AMR determinants and phenotypes, when available; (ii) the mechanism of resistance by which each AMR determinant functions; and (iii) notes, which may contain different information such as warnings on variable expression levels (inducible resistance, cryptic genes in some species, etc.), structural and functional information, and alternative nomenclature.
Software and interface
ResFinder 4.0 was embedded using the same web interface as previous ResFinder versions and available at the link https://cge.cbs.dtu.dk/services/ResFinder-4.0/. Importantly, in the interface, the user is prompted to specify a bacterial species, which is needed to define the specific antimicrobial panel for the in silico antibiogram (Table S2). There is the option to include all antimicrobial agents from all panels (‘Other’ option). In this case, interpretation of results must be executed carefully and knowledge on intrinsic resistance is essential because, in the ‘Other’ option, isolates intrinsically resistant to an antimicrobial agent might appear predicted as susceptible since intrinsic resistance is often mediated by structural traits (e.g. reduced permeability of the outer membrane, among others) rather than by specific genes/mutations.19
Previous versions of ResFinder were written in Perl, whereas ResFinder 4.0 was rewritten in Python 3. The ResFinder software has not previously been able to process read data (FASTQ) directly but relied on an assembly step. ResFinder 4.0 has implemented KMA,20 which aligns reads directly to the databases without the need for assembly. Like all previous versions, ResFinder 4.0 is released as open source under the Apache 2.0 license and is available at: https://bitbucket.org/genomicepidemiology/resfinder/.
Datasets for validation
ResFinder 4.0 was validated with datasets consisting of MIC values (BMD or Etest, Table 1) and WGS data (Illumina sequencing) of Escherichia coli, Salmonella spp., Campylobacter jejuni, E. faecium, E. faecalis and S. aureus of different origins (Table 1). These datasets represent a convenience sample. Phenotypic AST results were interpreted using the EUCAST epidemiological cut-off values (ECOFFs) to categorize isolates as WT (MIC ≤ECOFF) and non-WT (MIC >ECOFF) (www.eucast.org). Exceptions were: (i) one S. aureus dataset for which phenotypic AST was performed by disc diffusion and interpreted by EUCAST clinical breakpoints (Table 1); and (ii) one E. coli dataset that consisted of Illumina WGS data only and MIC values were available for the data provider but not for the ResFinder 4.0 developers, thus providing a blind test of the tool performance (Table 1). WGS data were obtained as raw reads and processed through a quality control (QC) pipeline as described here: https://bitbucket.org/genomicepidemiology/foodqcpipeline/. In brief, reads were trimmed using bbduk2 (https://jgi.doe.gov/data-and-tools/bbtools/) to a phred score of 20, reads less than 50 bp were discarded, adapters were trimmed away and a draft de novo assembly was created using SPAdes.21 From the assemblies, contigs below 500 bp were discarded. The most important parameters that were used to assess quality of sequencing data were: number of bases left after trimming, N50, number of contigs and total size of assembly. QC parameters used as guidelines were: read depth of at least 25×, N50 of >30 000 bp and a limit on the number of contigs to <500.
Table 1.
Datasets for ResFinder 4.0 validation
| Species | Isolates (n) | Observations (n) | Source | Origin | Country | Reference |
|---|---|---|---|---|---|---|
| E. coli | 95 | 1520 | animal, food | surveillance | DK | Hendriksen et al.43 |
| E. coli | 99 | 890 | animal, food | surveillance | UK | Duggett et al.44, AbuOun et al.48 |
| E. coli a | 390 | 2559 | human, animal | clinical, surveillance | DE | This study |
| Salmonella enterica | 1081 | 7489 | animal, food | surveillance | USA | McDermott et al.23 |
| C. jejuni | 239 | 1382 | animal, food | surveillance | FIN, FR, DE, LU, PL, PT | Leekitcharoenphon et al.45 |
| E. faecium | 50 | 363 | human | clinical | DE | this study |
| E. faecium b | 56 | 159 | human | clinical | BE | this study |
| E. faecalis | 50 | 235 | human and animal | clinical, surveillance | DE | Neumann et al.46, Bender et al.47 |
| S. aureus b | 63 | 504 | human | clinical, surveillance | BE | this study |
| S. aureus c | 100 | 598 | human | clinical, surveillance | DK | this study |
If not otherwise specified, phenotypic AST results were obtained by BMD.
DK, Denmark; DE, Germany; FIN, Finland; FR, France; LU, Luxembourg; PL, Poland; PT, Portugal; BE, Belgium.
Dataset for blind test of ResFinder 4.0 performance. Cefepime, chloramphenicol, ertapenem and nalidixic acid susceptibility testing were performed by Etest in a subset of isolates. All remaining AST were performed by BMD.
Phenotypic AST results were obtained by Etest.
Phenotypic AST results were obtained by disc diffusion.
WGS data (FASTQ) were used as input for ResFinder 4.0 using default parameters (≥80% identity over ≥60% of the length of the target gene) and also for SNP-based phylogenetic analysis as previously described22 to verify the genetic diversity of the validation datasets. SNP analysis was not performed for the Salmonella spp. dataset whose diversity was already described previously.23 The ResFinder 4.0 output was analysed to define AMR genotypes, i.e. patterns of resistance determinants observed for each antimicrobial, in each dataset.
Genotype–phenotype concordance was defined as presence or absence of a genetic determinant of resistance to a specific antimicrobial agent in non-WT (nWT) or WT isolates, respectively. Genotype–phenotype discordance was defined either as presence of a relevant AMR determinant in WT isolates or as absence of a relevant AMR determinant in nWT isolates. All discordances were individually analysed.
Sequence data that did not derive from previous studies (Table 1) have been deposited at NCBI (E. coli dataset from Germany: PRJNA616452; E. faecium dataset from Germany: PRJNA625631; E. faecium dataset from Belgium: PRJNA552025; S. aureus dataset from Belgium: PRJNA615176) and in the European Nucleotide Archive (S. aureus dataset from Denmark: PRJEB37586).
Results
ResFinder 4.0 databases
The ResFinder 4.0 databases validated in this study include 2690 AMR genes (ResFinder) and 266 resistance-mediating mutations in 39 selected genes (PointFinder), a genotype-to-phenotype translation database that includes 57 antimicrobial compounds (Table S1) and in silico antimicrobial panels for eight species (Table S2). Of note, a few antimicrobial agents included in the genotype-to-phenotype translation database do not appear in the in silico antimicrobial panels. Thus the database can be used for any bacterial species and is not restricted to the species having assigned in silico antimicrobial panels. Furthermore, the in silico antimicrobial panels are limited to the antimicrobial agents for which the tool can actually provide an output. Hence, for example, antimicrobial agents for which the genetic bases of resistance have not been fully elucidated (e.g. daptomycin in Enterococcus spp., among others) are not included. The most updated versions of the databases are publicly available in BitBucket (https://bitbucket.org/genomicepidemiology/resfinder_db and https://bitbucket.org/genomicepidemiology/pointfinder_db). The databases are continuously updated relying both on users’ feedback and on curators’/developers’ own research. Importantly, the BitBucket repository allows access to outdated versions of the databases, which is particularly useful in case ResFinder 4.0 is used within quality-assured procedures.
Datasets for ResFinder 4.0 validation
The SNP distances across strains used for ResFinder 4.0 validation were in the range 1–48 102 for E. coli from Denmark and the UK, 1–48 066 for E. coli from Germany, 1–20 134 for C. jejuni, 1–7131 for E. faecium 15–17 067 for E. faecalis and 1–25 424 for S. aureus (Table S3). The proportion of WT and nWT isolates in each dataset is presented in Tables 2 and 3.
Table 2.
Antimicrobial resistance phenotypes and genotype–phenotype concordance for the Gram-negative bacteria datasets using ECOFFs
| Antimicrobial |
E. coli (animal surveillance; DK) |
E. coli (animal surveillance; UK) |
E. coli (human and animal clinical and surveillance; DE) |
Salmonella sp. (human clinical, animal surveillance; USA) |
C. jejuni (animal surveillance; EU) |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| nWT | WT | concordance (%) | nWT | WT | concordance (%) | nWT | WT | concordance (%) | nWT | WT | concordance (%) | nWT | WT | concordance (%) | |
| Ampicillin | 95 | 0 | 100 | 81 | 18 | 98.9 | 202 | 0 | 100 | 249 | 822 | 98.7 | – | – | – |
| Cefepime | 78 | 17 | 71.6 | – | – | – | 137 | 0 | 100 | – | – | – | – | – | – |
| Cefotaxime | 95 | 0 | 100 | 23 | 76 | 98.9 | 370 | 0 | 98.6 | – | – | – | – | – | – |
| Cefoxitin | 46 | 49 | 97.8 | – | – | – | – | – | – | 130 | 933 | 98.9 | – | – | – |
| Ceftazidime | 94 | 1 | 98.9 | – | – | – | 282 | 0 | 99.2 | – | – | – | – | – | – |
| Chloramphenicol | 8 | 87 | 100 | – | – | – | 63 | 67 | 73.1 | 41 | 1030 | 99.7 | – | – | – |
| Ciprofloxacin | 29 | 66 | 87.3 | 64 | 35 | 95.9 | 275 | 0 | 99.2 | 22 | 1049 | 97 | 134 | 105 | 99.1 |
| Colistin | 0 | 95 | 100 | 11 | 88 | 100 | – | – | – | – | – | – | – | – | – |
| Ertapenem | 1 | 94 | 98.9 | – | – | – | 60 | 70 | 54.6 | – | – | – | – | – | – |
| Erythromycin | – | – | – | – | – | – | – | – | – | – | – | – | 3 | 236 | 99.1 |
| Gentamicin | 15 | 80 | 100 | 34 | 65 | 100 | 129 | 258 | 97.6 | 126 | 945 | 98.9 | 0 | 239 | 100 |
| Imipenem | 0 | 95 | 100 | 0 | 98 | 100 | 2 | 192 | 100 | – | – | – | – | – | – |
| Meropenem | 0 | 95 | 100 | 0 | 99 | 100 | 2 | 0 | 100 | – | – | – | – | – | – |
| Nalidixic acid | 25 | 70 | 98.9 | 39 | 60 | 90.9 | 99 | 28 | 99.2 | 10 | 1061 | 99.4 | 131 | 108 | 97.9 |
| Streptomycin | – | – | – | – | – | – | – | – | – | – | – | – | 0 | 187 | 100 |
| Sulfamethoxazole | 61 | 34 | 98.9 | – | – | – | – | – | – | – | – | – | – | – | – |
| Tetracycline | 53 | 42 | 100 | 79 | 20 | 97.9 | 138 | 33 | 98.8 | 656 | 415 | 98.6 | 130 | 109 | 99.1 |
| Tigecycline | – | – | – | – | – | – | 5 | 147 | 96.7 | – | – | – | – | – | – |
| Trimethoprim | 29 | 66 | 100 | – | – | – | – | – | – | – | – | – | – | – | – |
DK, Denmark; DE, Germany.
Table 3.
Antimicrobial resistance phenotypes and genotype–phenotype concordance for the Gram-positive bacteria datasets using ECOFFs
| Antimicrobial |
E. faecium (human clinical; DE) |
E. faecium (human clinical; BE) |
E. faecalis (human clinical; DE) |
S. aureus (human clinical and surveillance; BE) |
S. aureus (human clinical and surveillance; DK) |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| nWT | WT | concordance (%) | nWT | WT | concordance (%) | nWT | WT | concordance (%) | nWT | WT | concordance (%) | R | S | concordance (%) | |
| Ampicillin | 50 | 0 | 100 | 55 | 1 | 100 | – | – | – | – | – | – | – | – | – |
| Cefoxitin | – | – | – | – | – | – | – | – | – | 63 | 0 | 100 | 99 | 1 | 100 |
| Chloramphenicol | 0 | 50 | 64 | – | – | – | – | – | – | – | – | – | – | – | – |
| Ciprofloxacin | 50 | 0 | 100 | 3 | 1 | 100 | – | – | – | 63 | 0 | 100 | – | – | – |
| Clindamycin | – | – | – | – | – | – | – | – | – | 31 | 32 | 96.8 | 36 | 64 | 97 |
| Erythromycin | 50 | 0 | 100 | – | – | – | 39 | 11 | 96 | 32 | 31 | 95.2 | 42 | 58 | 99 |
| Gentamicin | 13 | 0 | 100 | 6 | 2 | 75 | 31 | 4 | 97.1 | 63 | 0 | 100 | 13 | 86 | 93.9 |
| Linezolid | 2 | 48 | 92 | 2 | 33 | 94.2 | 16 | 34 | 96 | 0 | 63 | 100 | 2 | 98 | 99 |
| Tetracycline | 22 | 28 | 92 | – | – | – | 43 | 7 | 98 | 46 | 17 | 76.2 | 17 | 82 | 96.9 |
| Vancomycin | 40 | 10 | 100 | 53 | 3 | 96.4 | 16 | 34 | 98 | 0 | 63 | 100 | – | – | – |
For the S. aureus DK dataset only, R, resistant and S, susceptible interpretations according to EUCAST clinical breakpoints were available.
DK, Denmark; DE, Germany; BE, Belgium.
Concordance between genotypic and phenotypic AST
E. coli
For the dataset deriving from Danish AMR surveillance in animals and meat, the overall genotype–phenotype concordance resulting from 1520 observations encompassing 16 antimicrobial agents was 97%, ranging from 71.6% for cefepime to 100% for most antimicrobial agents (Table 2).
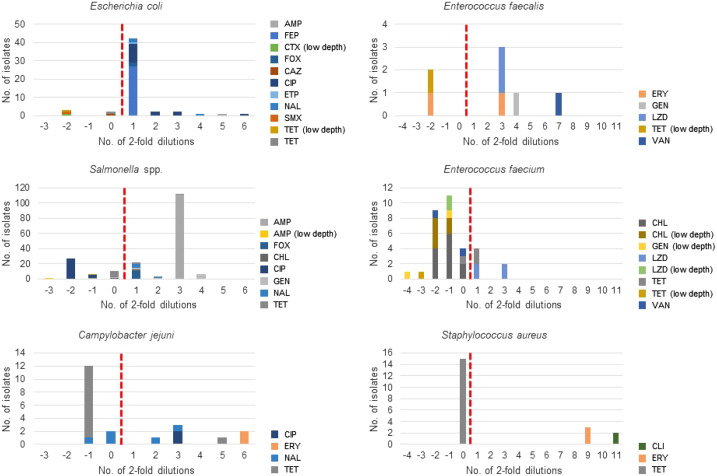
ResFinder 4.0 detected between 2 and 14 genotypes of resistance to most antimicrobial agents besides several cases in which no AMR determinant was detected (Table S4). The predicted and observed AMR phenotypes matched in 99.6% (586/588) of cases in which AMR determinants were detected and in 95.3% (889/932) of cases in which no resistance determinant was detected. Discordances were caused by detection of sul2 in an isolate with the sulfamethoxazole MIC well below the ECOFF, and blaCTX-M-1 in an isolate with the ceftazidime MIC on the ECOFF (Figure 1). Both genes were detected with a reliable read depth (see the Discussion). Additional discordances were caused by lack of detection of AMR determinants in isolates with nWT phenotype for cefepime, cefoxitin, ciprofloxacin, ertapenem and nalidixic acid. Most of these isolates had the respective MIC one step dilution above the ECOFF (Figure 1 and Table S5).
Figure 1.
Discordance between predicted (ResFinder 4.0) and observed phenotypes. The vertical dotted line divides the isolates having ‘WT phenotype with AMR determinant’ to the left and the isolates having ‘nWT phenotype without AMR determinant’ to the right. ‘Low depth’ refers to low read depth of the respective AMR determinant as explained in the Discussion. AMP, ampicillin; FEP, cefepime; CTX, cefotaxime; FOX, cefoxitin; CAZ, ceftazidime; CIP, ciprofloxacin; ETP, ertapenem; NAL, nalidixic acid; SMX, sulfamethoxazole; TET, tetracycline; ERY, erythromycin; GEN, gentamicin; LZD, linezolid; VAN, vancomycin; CHL, chloramphenicol; CLI, clindamycin.
For the dataset deriving from AMR surveillance in animals in the UK, the overall genotype–phenotype concordance for 890 observations encompassing nine antimicrobial agents was 98%, ranging from 90.9% for nalidixic acid to 100% for colistin, gentamicin, imipenem and meropenem (Table 2). ResFinder 4.0 detected between 2 and 17 genotypes of resistance to most antimicrobial agents, in addition to several cases in which no AMR determinant was detected (Table S4). The predicted and observed AMR phenotypes matched in 96.7% (325/336) of cases in which AMR determinants were detected and in 98.9% (548/554) of cases in which no resistance determinant was detected. Discordances were caused by detection of: (i) (fluoro)quinolone resistance determinants (gyrA and parC mutations and qnrB1) in eight (fluoro)quinolone WT isolates for which only interpretation of the MIC was available (Table S5); (ii) blaTEM-106 and truncated tet(M) genes in isolates with WT cefotaxime and tetracycline phenotype, respectively, and with very low read depth (see the Discussion) of these AMR genes; and (iii) detection of the tet(C) gene with reliable read depth in a tetracycline WT isolate with the MIC on the breakpoint (Figure 1). Additional discordances were caused by lack of detection of AMR determinants in isolates with nWT phenotype for ampicillin, ciprofloxacin and nalidixic acid (Figure 1). Most (60%) of the (fluoro)quinolone nWT isolates had MICs one step dilution above the ECOFF (Figure 1 and Table S5).
For the dataset used for the blind test that comprised human and animal origin isolates from clinical and surveillance samples, the overall genotype–phenotype concordance resulting from 2559 observations encompassing 13 antimicrobial agents was 95.3%, ranging from 54.6% for ertapenem to 100% for ampicillin, cefepime, imipenem and meropenem (Table 2). ResFinder 4.0 detected between 1 and 37 genotypes of resistance to most antimicrobial agents besides several cases in which no AMR determinant was detected (Table S4). The predicted and observed AMR phenotypes matched in 99.7% (1649/1654) of cases in which AMR determinants were detected and in 87.3% (790/905) of cases in which no resistance determinant was detected (Table S4). Discordances were mainly caused by lack of detection of AMR determinants in isolates with nWT phenotype for different antimicrobial agents (Table S4). Most of these discordant cases concerned chloramphenicol and ertapenem for which the phenotype was measured by Etest (Table S5).
Salmonella spp
The overall genotype–phenotype concordance for 7489 observations encompassing seven antimicrobial agents was 98.8%, ranging from 97.0% for ciprofloxacin to 99.7% for chloramphenicol (Table 2). ResFinder 4.0 detected between 3 and 25 genotypes of resistance to the different antimicrobial agents besides several cases in which no AMR determinant was detected (Table S4). The predicted and observed AMR phenotypes matched in 96.3% (1220/1266) of cases in which AMR determinants were detected and in 99.2% (6179/6223) of cases in which no resistance determinant was detected. Discordances were mainly due to detection of aac(6′)-Ib-cr with <100% identity to the database entry in ciprofloxacin WT isolates, whereas in the remaining cases the genotype–phenotype discordance was mainly observed in isolates with MIC values on the ECOFF (Figure 1). Additional discordances were mainly observed in isolates with an MIC one step dilution above the ECOFF. There were also cases in which ampicillin and gentamicin resistance determinants were not detected even though the isolates had the respective MIC well above the ECOFF (Figure 1 and Table S5).
C. jejuni
For C. jejuni, the overall genotype–phenotype concordance resulting from 1382 observations encompassing six antimicrobial agents was 99.2%, ranging from 97.9% for nalidixic acid to 100% for gentamicin and streptomycin (Table 2).
ResFinder 4.0 detected between one and eight genotypes for resistance to most antimicrobial agents besides several cases in which no AMR determinant was detected (Table S4). The predicted and observed AMR phenotypes matched in 99.0% (391/395) of cases in which AMR determinants were detected and in 99.3% (980/987) of cases in which no resistance determinant was detected. Discordances were observed in isolates with (i) gyrA mutations known to mediate (fluoro)quinolone resistance and the nalidixic acid MIC close to the ECOFF, and (ii) tet(O) with <100% identity to the ResFinder 4.0 entries and reliable read depth in an isolate with the MIC close to the breakpoint. Additional discordances were caused by lack of detection of AMR determinants in isolates with nWT phenotype for ciprofloxacin, nalidixic acid, erythromycin and tetracycline with MICs notably higher than the ECOFF (Figure 1 and Table S5).
E. faecium
Of the two E. faecium datasets, the one from Germany included 363 observations encompassing eight antimicrobial agents and the one from Belgium included 159 observations encompassing five antimicrobial agents. In the first dataset, the overall genotype–phenotype concordance was 92.8%, ranging from 64% for chloramphenicol to 100% for ampicillin, ciprofloxacin, erythromycin, gentamicin and vancomycin (Table 3). In this dataset, ResFinder 4.0 detected between 1 and 11 genotypes for resistance to all antimicrobial agents besides several cases in which no AMR determinant was detected (Table S4). The predicted and observed AMR phenotypes matched in 91.0% (223/245) of cases in which AMR determinants were detected and in 96.6% (114/118) of cases in which no resistance determinant was detected. Most discordances were due to detection of poxtA, cat and fexB with <100% identity to the database entries and often with low read depth (Figure 1). Furthermore, tet(M) was detected with reliable read depth in an isolate with the tetracycline MIC on the ECOFF (Figure 1). Additional discordances were due to lack of detection of tetracycline and linezolid resistance determinants in isolates with the respective MICs 1- and 3-fold dilution above the ECOFF, respectively (Figure 1 and Table S5).
In the second dataset, the overall genotype–phenotype concordance was 96.2%, ranging from 75% for gentamicin to 100% for ampicillin and ciprofloxacin (Table 3). In this dataset, ResFinder 4.0 detected between two and six genotypes for resistance to all antimicrobial agents besides several cases in which no AMR determinant was detected (Table S4). The predicted and observed AMR phenotypes matched in 96.6% (117/121) of cases in which AMR determinants were detected and in 94.7% (36/38) of cases in which no resistance determinant was detected. Discordances were due to detection of (i) gentamicin resistance genes in gentamicin WT isolates (n = 2), but further analysis revealed that such genes had low read depth, and (ii) vancomycin resistance genes with reliable coverage in isolates (n = 2) with different vancomycin MICs (Figure 1). Additional discordances were caused by lack of detection of AMR determinants in isolates with the linezolid MIC one step dilution above the ECOFF (Figure 1 and Table S5).
E. faecalis
For the E. faecalis dataset, the overall genotype–phenotype concordance resulting from 235 observations encompassing five antimicrobial agents was 97%, ranging from 96% for erythromycin and linezolid to 98% for tetracycline and vancomycin (Table 3).
ResFinder 4.0 detected between 1 and 17 genotypes of resistance to all antimicrobial agents besides several cases in which no AMR determinant was detected (Table S4). The predicted and observed AMR phenotypes matched in 98.6% (140/142) of cases in which AMR determinants were detected and in 94.6% (88/93) of cases in which no resistance determinant was detected. Discordances were represented by isolates having erm(B) (n = 1) and tet(M) (n = 1) with 100% and <100% identity to the ResFinder 4.0 database, respectively, and an MIC distant from the ECOFF (Figure 1). Further analysis revealed that tet(M) had very low read depth. Additional discordances were represented by lack of detection of determinants of resistance to erythromycin, gentamicin, linezolid and vancomycin in a few isolates with clear nWT phenotype (Figure 1 and Table S5).
S. aureus
Of the two S. aureus datasets, the one from Belgium included 504 observations encompassing eight antimicrobial agents, whereas the one from Denmark included 598 observations encompassing six antimicrobial agents.
For the first dataset, the overall genotype–phenotype concordance was 96%, ranging from 76.2% for tetracycline to 100% for cefoxitin, ciprofloxacin, gentamicin, linezolid and vancomycin (Table 3). In this dataset, ResFinder 4.0 detected between one and two genotypes of resistance to most antimicrobial agents besides several cases in which no AMR determinant was detected (Table S4). The predicted and observed AMR phenotypes matched in 95.1% (293/308) of cases in which AMR determinants were detected and in 97.4% (191/196) of cases in which no resistance determinant was detected. Most discordances were linked to detection of tet(M) with <100% identity to the database entry and in isolates with MICs on the ECOFF (Figure 1). Additional discordances were due to lack of detection of AMR determinants in isolates with clindamycin and erythromycin MICs notably higher than the ECOFF (Figure 1 and Table S5).
For the second dataset, the overall genotype–phenotype concordance was 97.3%, ranging from 93.9% for gentamicin to 100% for cefoxitin (Table 3). In this dataset, ResFinder 4.0 detected between one and nine genotypes of resistance to most antimicrobial agents besides several cases in which no AMR determinant was detected (Table S4). The predicted and observed AMR phenotypes matched in 94.4% (205/217) of cases in which AMR determinants were detected and in 98.9% (379/383) of cases in which no resistance determinant was detected. Discordances were due to detection of erm(A) (n = 2) and aac(6′)-aph(2″) (n = 6) with reliable read depth and to detection of tet(K) (n = 3) with low read depth (Table S4). Additional discordances were due to lack of detection of clindamycin, gentamicin, linezolid and tetracycline resistance determinants in isolates that were reported as resistant (Table S5). As phenotypic data were available only as interpretation (according to EUCAST clinical breakpoints) of disc diffusion results, we could not assess if the measured values were close to the breakpoint for resistance.
Discussion
We developed and implemented a freely accessible web server, a database and bioinformatics pipeline for detecting AMR genes and mutations and providing in silico prediction of expected phenotypes. ResFinder 4.0 was validated for six bacterial species of major public health relevance and has been evaluated for M. tuberculosis previously.17 ResFinder 4.0 can also be used to predict phenotypes for any additional bacterial species by users with profound knowledge of AMR.
Excellent concordance between genotypically predicted and phenotypically detected phenotypes was found for most antimicrobial–species combinations (Tables 2 and 3). In the vast majority of cases in which the automatically evaluated genotype–phenotype concordance was <95%, we could identify issues related to ECOFFs and/or read depth of WGS data (Figure 1), indicating that phenotypic tests and suboptimal sequence quality, respectively, were likely sources of discrepancies. Misclassifications by the phenotypic tests could have happened in the cases in which the MIC was either on or bordered the ECOFF (i.e. ± one 2-fold dilution from the ECOFF). This is due to limitation in the reproducibility of MIC results, whereby one 2-fold dilution difference across repeated tests is considered acceptable.24 Furthermore, for certain antimicrobial–bacteria combinations including chloramphenicol–enterococci, MIC reading is difficult due to trailing growth,24 and we cannot exclude that this might have also contributed to the genotype–phenotype discordances. In fact, in a previous study, we found that most discrepancies between genotype and phenotype were resolved when repeating the phenotypic test.25 Unfortunately, the isolates used in this study were not available for repeating MIC determination. Also the ECOFFs themselves might be a source of misclassification of phenotypic tests. It has been shown that the number of isolates showing genotype–phenotype concordance notably increased when adjusting the ECOFF of certain antimicrobial agents.26 Low read depth was also a likely source of some of the genotype–phenotype discordances observed (Figure 1). There is no unequivocally accepted threshold for acceptable read depth for detection of AMR genes and mutations, and our definition of ‘low read depth’ derived from a case-by-case examination. Specifically, the genes scored as ‘low read depth’ had either ‘depth <10’ or ‘depth <1/10 compared with that of additional AMR genes in the same isolate and query coverage <100%’ (which indicates an imperfect match between the query and the subject). This shows the importance of visualizing the ‘read depth’ parameter in the output. At present, such a parameter appears only in the output obtained when running ResFinder 4.0 in Unix but will also be included in the web tool output in the future. Of note, issues related to read depth depend on sequencing procedures and are not indicative of ResFinder 4.0 performance and/or feasibility of WGS-based AST. ResFinder 4.0 performed poorly in prediction of linezolid resistance. This was expected since the tool only detects linezolid resistance mediated by acquired genes, whereas resistance mediated by mutations in chromosomal genes is currently detected by LREFinder.27 This underlines the importance of becoming familiar with features of the tools used for AMR detection in WGS data to choose the most appropriate according to the user’s aims.12 Genotype–phenotype discordance might have also occurred due to inducible genes. For example, the S. aureus dataset from Denmark included erm(A)-positive isolates phenotypically classified as clindamycin susceptible (Tables S4 and S5). erm(A) can be inducible, and mutations in the translational attenuator can occur very rapidly, leading to phenotypic resistance to all lincosamides and ketolides.28 Also, occurrence of genes with <100% identity and/or coverage (in length) compared with the ResFinder 4.0 database entries might have caused some genotype–phenotype discordances since genes that underwent mutations, insertions or deletions might have become silent. In addition, there are genes for which an nWT phenotype is linked to specific sequences only and will not be expressed if a mutation appears. For example, only specific mutations in aac(6′)-Ib mediate fluoroquinolone resistance in E. coli and Salmonella,29 which explains why aac(6')-Ib-cr with <100% identity to the database entry is detected in ciprofloxacin WT Salmonella spp. isolates (Table S5). Nonetheless, in our datasets, the prediction of a resistance phenotype matched the measured nWT phenotype even in most of the cases presenting an AMR gene with <100% identity and/or coverage (in length) compared with the database entries (Tables S4 and S5). From a surveillance perspective, detection of potentially silent genes is still more informative than the reporting of a WT phenotype. As silencing mutations can be transient,30 also from a clinical perspective the finding of AMR genes may be even more relevant than the observed in vitro phenotype as it may be used as an early warning to avoid initiating inappropriate therapies.
In current evaluations of WGS as a tool for detection of AMR, most studies have used the phenotypic test as the correct answer and largely ignored the fact that phenotypic testing has important limitations in accuracy and reproducibility.31,32 In large ring trials running globally for several years, we have not been able to obtain concordances to the reference test of more than 91%–93% for Salmonella and Campylobacter.2,3 Over the years, large efforts have been put into standardizing and improving the quality of phenotypic susceptibility testing and ensuring that exactly the same methodology, growth conditions and interpretations are used. Nevertheless, even when laboratories followed the same standard operating procedure, between 0.8% and 31% of obtained MIC values were outside the acceptable range.33 Furthermore, even with exactly the same test in the same laboratory, the reading of MIC results might show inconsistencies in 5% of all results from different readers.34 In comparison, WGS data can be analysed using the same bioinformatics pipeline and with 100% reproducibility between laboratories and, even with the continued need to include novel resistance genes and additional mutations, we would argue that already today the WGS approach would be just as reliable as phenotypic testing, at least for surveillance. With further verification, WGS could potentially also be used to support clinical decision-making. We are aware that the use of WGS to guide antimicrobial therapy is often encountered with scepticism, and indeed genotype–phenotype concordance when using phenotypic results interpreted according to clinical breakpoints diminished for a few antimicrobial–species combinations (Tables S6 and S7), as also previously observed.26 However, several discordances concerned isolates that harboured AMR determinants despite being classified as phenotypically susceptible, which may lead to therapeutic failures. Thus, the current practice of using MICs to guide dosing regimens may not be as appropriate as commonly believed, as also indicated recently by others.35,36 Furthermore, the genotype-to-phenotype translation represents a valuable resource for clinical metagenomics, which is an expanding field that will greatly impact clinical medicine in the coming years.37
Implementing WGS for routine diagnostics in clinical microbiological laboratories will not be without challenges.38,39 Besides the costs associated with implementation of equipment and training staff, many microbiology laboratories have to adhere to regulations and criteria for accreditation. In addition, the current turnaround time is an issue for most technologies. However, while this is the case for established clinical microbiological laboratories, it should also be mentioned that for laboratories in low- and middle-income countries that do not already have established laboratories it could potentially be an advantage to install a workflow based on WGS.40
ResFinder 4.0 can be used with raw reads and with assembled sequences as inputs, and it performs database searches using Kmer-based mapping and BLAST, respectively. In this work, we presented the results obtained by inputting raw reads. As different processing of the same input sequences has been shown to lead to different results,41 we also evaluated genotype–phenotype concordance using assembled sequences as input (data not shown). Across 13 140 observations, which correspond to all datasets excluding the E. coli for the blind test, there were only 29 (0.2%) cases in which results differed using FASTQ (raw reads) and FASTA (assembled sequences) files of the same sequence (data not shown). In most (20/29) cases, AMR genes were detected using FASTQ but not detected using FASTA, whereas the opposite applied in the remaining cases. In 14 and 15 cases, the phenotype predicted using the FASTQ and FASTA inputs was in agreement with the observed phenotype, respectively (data not shown). Based on this, it appears that the input file format does not affect the reliability of ResFinder 4.0 results and, given that raw reads are processed considerably faster compared with assemblies, FASTQ files should be considered the preferable input for ResFinder 4.0.
The datasets available to us did not include an even distribution of WT and nWT phenotypes to all antimicrobial agents. This limitation highlights a need for making not only WGS data but also phenotypic test data as well as the bacterial isolates publicly available for those developing bioinformatics tools.42
In conclusion, we implemented and evaluated ResFinder 4.0, a bioinformatics tool whose unique feature is to provide a phenotype for each antimicrobial resistance gene detected and easily interpretable in silico antibiograms for eight bacterial species including both Gram-positive and Gram-negative bacteria of major public health relevance. The tool will be extended to allow for input of Nanopore sequence data and to include antibiograms of additional bacterial species. ResFinder 4.0 allows for rapid updates of all databases to promptly respond to users’ feedback and keep pace with improvements in knowledge on AMR. We would encourage users around the world to input additional curated databases for better and faster improvement.
Supplementary Material
Acknowledgements
We thank Kathrine Valentini Jensen for her skilful assistance in formatting the databases. We also thank Carola Fleige and Christine Günther from the Robert Koch Institute (Germany), G. Mourand from Anses Ploufragan (France), and Edyta Denis, Iwona Kania and Magdalena Staniak from the National Veterinary Research Institute (Poland) for their excellent technical assistance in laboratory analyses.
Funding
This work was supported by the European Union Horizon 2020 research and innovation programme under grant agreement 643476 to the COMPARE project (http://www.compare-europe.eu), and The Novo Nordisk Foundation (NNF16OC0021856) - Global Surveillance of Antimicrobial Resistance. The work by L. Falgenhauer and T. Chakraborty was supported by grants to the German Center of Infection Research (DZIF), and the Zoonoses Network ‘ESBL and fluoroquinolone resistance in Enterobacteriaceae (RESET)’ Consortium through the German Federal Ministry of Education and Research (BMBF; grant numbers 8000 701-3 [HZI], 01KI1313G, TI06.001, 8032808811, 8032808818 and 8032808820 to T.C.). Data for the E. faecium (dataset from Germany) and E. faecalis isolates were generated as part of a research project called InfectControl2020-IRMRESS funded by the German BMBF (grant no. 03ZZ0815A).
Transparency declarations
None to declare.
References
- 1. Wheat PF. History and development of antimicrobial susceptibility testing methodology. J Antimicrob Chemother 2001; 48 Suppl 1: 1–4. [DOI] [PubMed] [Google Scholar]
- 2. Pedersen SK, Wagenaar JA, Vigre H et al. Proficiency of WHO global foodborne infections network external quality assurance system participants in identification and susceptibility testing of thermotolerant Campylobacter spp. from 2003 to 2012. J Clin Microbiol 2018; 56: e01066–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hendriksen RS, Seyfarth AM, Jensen AB et al. Results of use of WHO Global Salm-Surv external quality assurance system for antimicrobial susceptibility testing of Salmonella isolates from 2000 to 2007. J Clin Microbiol 2009; 47: 79–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.World Health Organization. Antimicrobial Resistance: Global Report on Surveillance 2014. https://apps.who.int/iris/bitstream/handle/10665/112642/9789241564748_eng.pdf;jsessionid=845ECA593D3D087F909CF083DC999BB7?sequence=1.
- 5.World Bank Group. Resistant Infections. A Threat to Our Economic Future 2017. http://documents1.worldbank.org/curated/en/323311493396993758/pdf/final-report.pdf.
- 6. Thaker MN, Kalan L, Waglechner N et al. Vancomycin-variable enterococci can give rise to constitutive resistance during antibiotic therapy. Antimicrob Agents Chemother 2015; 59: 1405–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Acar JF, Courvalin P, Chabbert YA. Methicillin-resistant staphylococcemia: bacteriological failure of treatment with cephalosporins. Antimicrob Agents Chemother 1970; 10: 280–5. [PubMed] [Google Scholar]
- 8. Mouton JW. Soup with or without meatballs: impact of nutritional factors on the MIC, kill-rates and growth-rates. Eur J Pharm Sci 2018; 125: 23–7. [DOI] [PubMed] [Google Scholar]
- 9. Courvalin P. Genotypic approach to the study of bacterial resistance to antibiotics. Antimicrob Agents Chemother 1991; 35: 1019–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Pfaller MA. Molecular approaches to diagnosing and managing infectious diseases: practicality and costs. Emerg Infect Dis 2001; 7: 312–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Anjum MF, Zankari E, Hasman H. Molecular methods for detection of antimicrobial resistance In: Schwarz S, Cavaco LM, Shen J, eds. Antimicrobial Resistance in Bacteria from Livestock and Companion Animals. American Society for Microbiology, 2018; 33–50. [Google Scholar]
- 12. Hendriksen RS, Bortolaia V, Tate H et al. Using genomics to track global antimicrobial resistance. Front Public Health 2019; 7: 242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Rigden DJ, Fernandez XM. The 27th annual Nucleic Acids Research database issue and molecular biology database collection. Nucleic Acids Res 2020; 48: D1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Panunzi LG. sraX: a novel comprehensive resistome analysis tool. Front Microbiol 2020; 11: 52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zankari E, Hasman H, Cosentino S et al. Identification of acquired antimicrobial resistance genes. J Antimicrob Chemother 2012; 67: 2640–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zankari E, Allesøe R, Joensen KG et al. PointFinder: a novel web tool for WGS-based detection of antimicrobial resistance associated with chromosomal point mutations in bacterial pathogens. J Antimicrob Chemother 2017; 72: 2764–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Johnsen CH, Clausen P, Aarestrup FM et al. Improved resistance prediction in Mycobacterium tuberculosis by better handling of insertions and deletions, premature stop codons, and filtering of non-informative sites. Front Microbiol 2019; 10: 2464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Naas T, Oueslati S, Bonnin RA et al. β-lactamase database (BLDB)—structure and function. J Enzyme Inhib Med Chem 2017; 32: 917–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.EUCAST. Intrinsic Resistance and Exceptional Phenotypes, Expert Rules. Version 3.1 2016. https://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Expert_Rules/Expert_rules_intrinsic_exceptional_V3.1.pdf
- 20. Clausen P, Aarestrup FM, Lund O. Rapid and precise alignment of raw reads against redundant databases with KMA. BMC Bioinformatics 2018; 19: 307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bankevich A, Nurk S, Antipov D et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 2012; 19: 455–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kaas RS, Leekitcharoenphon P, Aarestrup FM et al. Solving the problem of comparing whole bacterial genomes across different sequencing platforms. PLoS One 2014; 9: e104984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. McDermott PF, Tyson GH, Kabera C et al. Whole-genome sequencing for detecting antimicrobial resistance in nontyphoidal Salmonella. Antimicrob Agents Chemother 2016; 60: 5515–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.CLSI Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria That Grow Aerobically—Eleventh Edition: M07. 2018.
- 25. Zankari E, Hasman H, Kaas RS et al. Genotyping using whole-genome sequencing is a realistic alternative to surveillance based on phenotypic antimicrobial susceptibility testing. J Antimicrob Chemother 2013; 68: 771–7. [DOI] [PubMed] [Google Scholar]
- 26. Stubberfield E, AbuOun M, Sayers E et al. Use of whole genome sequencing of commensal Escherichia coli in pigs for antimicrobial resistance surveillance, United Kingdom, 2018. Euro Surveill 2019; 24: 1900136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hasman H, Clausen P, Kaya H et al. LRE-Finder, a Web tool for detection of the 23S rRNA mutations and the optrA, cfr, cfr(B) and poxtA genes encoding linezolid resistance in enterococci from whole-genome sequences. J Antimicrob Chemother 2019; 74: 1473–6. [DOI] [PubMed] [Google Scholar]
- 28. Schmitz FJ, Petridou J, Jagusch H et al. Molecular characterization of ketolide-resistant erm(A)-carrying Staphylococcus aureus isolates selected in vitro by telithromycin, ABT-773, quinupristin and clindamycin. J Antimicrob Chemother 2002; 49: 611–17. [DOI] [PubMed] [Google Scholar]
- 29. Robicsek A, Jacoby G, Hooper DC. The worldwide emergence of plasmid-mediated quinolone resistance. Lancet Infect Dis 2006; 6: 629–40. [DOI] [PubMed] [Google Scholar]
- 30. Kime L, Randall CP, Banda FI et al. Transient silencing of antibiotic resistance by mutation represents a significant potential source of unanticipated therapeutic failure. mBio 2019; 10: e01755–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Mouton JW, Meletiadis J, Voss A et al. Variation of MIC measurements: the contribution of strain and laboratory variability to measurement precision—authors’ response. J Antimicrob Chemother 2019; 74: 1761–2. [DOI] [PubMed] [Google Scholar]
- 32. Davies TJ, Stoesser N, Sheppard AE et al. Reconciling the potentially irreconcilable? Genotypic and phenotypic amoxicillin–clavulanate resistance in Escherichia coli. Antimicrob Agents Chemother 2020; 64: e02026–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wallmann J, Böttner A, Goossens L et al. Results of an interlaboratory test on antimicrobial susceptibility testing of bacteria from animals by broth microdilution. Int J Antimicrob Agents 2006; 27: 482–90. [DOI] [PubMed] [Google Scholar]
- 34. Barry AL, Braun LE. Reader error in determining minimal inhibitory concentrations with microdilution susceptibility test panels. J Clin Microbiol 1981; 13: 228–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Mouton JW, Muller AE, Canton R et al. MIC-based dose adjustment: facts and fables. J Antimicrob Chemother 2018; 73: 564–8. [DOI] [PubMed] [Google Scholar]
- 36. Bader JC, Lakota EA, Andes DR et al. Time for precision: a world without susceptibility breakpoints. Open Forum Infect Dis 2018; 5: ofy282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ruppé E, Schrenzel J. Messages from the third International Conference on Clinical Metagenomics (ICCMg3). Microbes Infect 2019; 21: 273–7. [DOI] [PubMed] [Google Scholar]
- 38. Ellington MJ, Ekelund O, Aarestrup FM et al. The role of whole genome sequencing in antimicrobial susceptibility testing of bacteria: report from the EUCAST Subcommittee. Clin Microbiol Infect 2017; 23: 2–22. [DOI] [PubMed] [Google Scholar]
- 39. Rossen JWA, Friedrich AW, Moran-Gilad J; ESCMID Study Group for Genomic and Molecular Diagnostics (ESGMD). Practical issues in implementing whole-genome-sequencing in routine diagnostic microbiology. Clin Microbiol Infect 2018; 24: 355–60. [DOI] [PubMed] [Google Scholar]
- 40. Sonda T, Kumburu H, van Zwetselaar M et al. Molecular epidemiology of virulence and antimicrobial resistance determinants in Klebsiella pneumoniae from hospitalised patients in Kilimanjaro, Tanzania. Eur J Clin Microbiol Infect Dis 2018; 37: 1901–14. [DOI] [PubMed] [Google Scholar]
- 41. Doyle RM, O’Sullivan DM, Aller SD et al. Discordant bioinformatic predictions of antimicrobial resistance from whole-genome sequencing data of bacterial isolates: an inter-laboratory study. Microb Genom 2020; 6: e000335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Aytan-Aktug D, Clausen P, Bortolaia V et al. Prediction of acquired antimicrobial resistance for multiple bacterial species using neural networks. mSystems 2020; 5: e00774–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Hendriksen RS, Pedersen SK, Leekitcharoenphon P et al. Final Report of ENGAGE—Establishing Next-generation Sequencing Ability for Genomic Analysis in Europe https://www.efsa.europa.eu/en/supporting/pub/en-1431.
- 44. Duggett NA, Randall LP, Horton RA et al. Molecular epidemiology of isolates with multiple mcr plasmids from a pig farm in Great Britain: the effects of colistin withdrawal in the short and long term. J Antimicrob Chemother 2018; 73: 3025–33. [DOI] [PubMed] [Google Scholar]
- 45. Leekitcharoenphon P, Garcia-Graells C, Botteldoorn N et al. Comparative Genomics of Quinolone-resistant and Susceptible Campylobacter jejuni of Poultry Origin from Major Poultry Producing European countries (GENCAMP) https://www.efsa.europa.eu/en/supporting/pub/en-1398.
- 46. Neumann B, Prior K, Bender JK et al. A core genome multilocus sequence typing scheme for Enterococcus faecalis. J Clin Microbiol 2019; 57: e01686–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Bender J, Fleige C, Lange D et al. Rapid emergence of highly variable and transferable oxazolidinone and phenicol resistance gene optrA in German Enterococcus spp. clinical isolates. Int J Antimicrob Agents 2018; 52: 819–27. [DOI] [PubMed] [Google Scholar]
- 48. AbuOun M, O’Connor HM, Stubberfield EJ et al. Characterizing antimicrobial resistant Escherichia coli and associated risk factors in a cross-sectional study of pig farms in Great Britain. Front Microbiol 2020; 11: 861. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.