Abstract
Background
Infections caused by triazole drug-resistant Aspergillus fumigatus are an increasing problem. The sensitivity of standard culture is poor, abrogating susceptibility testing. Early detection of resistance can improve patient outcomes, yet tools for this purpose are limited.
Objectives
To develop and validate a pyrosequencing technique to detect resistance-conferring cyp51A polymorphisms from clinical respiratory specimens and A. fumigatus isolates.
Methods
Method validation was performed by Sanger sequencing and pyrosequencing of 50 A. fumigatus isolates with a spectrum of triazole susceptibility patterns. Then, 326 Aspergillus quantitative PCR (qPCR)-positive respiratory samples collected over a 27 month period (January 2017–March 2019) from 160 patients at the UK National Aspergillosis Centre were assessed by cyp51A pyrosequencing. The Sanger sequencing and pyrosequencing results were compared with those from high-volume culture and standard susceptibility testing.
Results
The cyp51A genotypes of the 50 isolates analysed by pyrosequencing and Sanger sequencing matched. Of the 326 Aspergillus qPCR-positive respiratory specimens, 71.2% were reported with no A. fumigatus growth. Of these, 56.9% (132/232) demonstrated a WT cyp51A genotype and 31.5% (73/232) a resistant genotype by pyrosequencing. Pyrosequencing identified the environmental TR34/L98H mutation in 18.7% (61/326) of the samples in contrast to 6.4% (21/326) pan-azole resistance detected by culture. Importantly, pyrosequencing detected resistance earlier than culture in 23.3% of specimens.
Conclusions
The pyrosequencing assay described could detect a wide range of cyp51A polymorphisms associated with triazole resistance, including those not identified by commercial assays. This method allowed prompt recognition of resistance and the selection of appropriate antifungal treatment when culture was negative.
Introduction
Aspergillus fumigatus is the most common filamentous fungus associated with acute and chronic pulmonary infections and fungal asthma.1,2 The various forms of pulmonary aspergillosis affect many millions of people annually.3,4 Acute invasive pulmonary aspergillosis (IA) is estimated to develop in 200000 patients annually and has a mortality rate of approximately 50%5 if not diagnosed and treated promptly. In contrast to rapidly progressing IA, chronic pulmonary aspergillosis (CPA) and allergic bronchopulmonary aspergillosis (ABPA) are insidious, progressive conditions leading to the gradual destruction of the airways.1,6 The estimated global burden of CPA is over 200000 in Europe alone, with a 5 year mortality of 50%–85% despite treatment.7,8 The estimated global burden of ABPA is higher (1062000 cases in Europe), but the mortality rate is lower.9
Triazole-class antifungal drugs are used as first-line treatment for pulmonary aspergillosis due to their good efficacy and tolerability. The broadly active, but generally less well-tolerated, amphotericin B is the second-line treatment choice.10 However, the rising trend of aspergillosis caused by triazole-drug-resistant A. fumigatus provides an increasing challenge to the management of aspergillosis patients.11–15 The current incidence of A. fumigatus triazole resistance is largely unknown due to continued reliance on culture-based susceptibility testing and poor sensitivity of standard culture methods. The SCARE-network study demonstrated a prevalence of 3.2% across 19 countries.16 Compounding the spectre of resistance is the paucity of antifungal agents available for clinical use.7,17
The development of triazole resistance is frequently associated with polymorphisms in the promoter region and ORF of the A. fumigatus cyp51A gene, which encodes lanosterol 14α-demethylase, the target of triazole drugs.18 Azole exposure is the main driver for resistance, either in the patient during long-term triazole therapy or in the environment as a response to the agricultural use of azoles.19 The former is associated with specific cyp51A point mutations, with codons 54, 138, 220 and 448 implicated predominantly.20,21 Patients treated for CPA or ABPA with long courses (months to years) of triazole antifungals are particularly susceptible to this mechanism of resistance.22,23 Acquisition of an azole-resistant environmental strain of A. fumigatus, associated with a tandem repeat (TR) within the promoter region (TR34, TR46 and TR53) combined with specific cyp51A point mutations, is more frequently seen in IA patients, but also in CPA and ABPA patients.19,24,25
Rapid detection of triazole resistance can provide critical guidance in early, effective treatment and reduces the mortality of aspergillosis.26 Culture is still the mainstay of detecting resistance, although 85% of cultures of respiratory specimens from patients with aspergillosis are negative for A. fumigatus.27–29 When culture is positive, susceptibility testing may only be available at reference centres, whereby the total turnaround time, from sampling to an MIC result, is typically 1–2 weeks.30,31 Direct screening of the cyp51A region via molecular methods can overcome these issues and is becoming increasingly accessible,32,33 with the availability of commercial quantitative PCR (qPCR) platforms, such as Pathonostics AsperGenius, Ademtech MycoGENIE and Bruker Fungiplex® Aspergillus Azole-R IVD. These assays combine detection of A. fumigatus and, if the signal strength is high enough, some cyp51A polymorphisms.27,34–36 However, this confined scope of detectable mutations limits their ability to determine some key resistance mutations associated with long-term azole therapy. Sanger sequencing provides a well-established method for gene characterization, yet there are difficulties associated with insensitivity, laboratory workflow and therefore turnaround time.37 A more accessible and efficient sequencing technology is required for this purpose.
We report the design and validation of a pyrosequencing method to determine resistance-conferring polymorphisms within the cyp51A region of A. fumigatus38 from isolates and clinical respiratory samples. This assay was then used to screen respiratory samples from patients attending the UK National Aspergillosis Centre (NAC) to determine its utility to detect triazole resistance compared with high-volume culture (HVC)29 and routine susceptibility testing.
Materials and methods
Study design and ethical considerations
A pyrosequencing method was developed to detect polymorphisms within the cyp51A region of A. fumigatus. This method was validated against Sanger sequencing using 50 A. fumigatus isolates obtained from the Mycology Reference Centre Manchester (MRCM) culture collection (Table 1). These isolates originated from respiratory samples collected from 24 patients with CPA or ABPA as part of their standard care at the NAC. A spectrum of isolates with different triazole resistance patterns were collected from patients who demonstrated signs of clinical failure despite good serum antifungal drug levels. Then, 326 Aspergillus qPCR-positive respiratory samples from 160 patients who attended the NAC over a 27 month period (January 2017–March 2019) were screened for cyp51A polymorphisms by pyrosequencing. The pyrosequencing results were compared with those from HVC and standard susceptibility testing to determine resistance patterns and assay utility. This study was assessed through the NHS Health Research Authority system (HRA)39 and was found to meet the UK NHS definition of a retrospective service evaluation for which formal ethical review was therefore not required.
Table 1.
Comparative cyp51A sequencing and MIC values for A. fumigatus species complex clinical isolates
| Isolate |
cyp51A genotype |
Isolate MIC (mg/L) |
|||||
|---|---|---|---|---|---|---|---|
| Sanger sequencing | pyrosequencing | ITC | AMB | VRC | POS | ISA | |
| 1 | no polymorphisms | no polymorphisms | 0.5 | 0.5 | 0.25 | 0.25 | NAa |
| 2 | no polymorphisms | no polymorphisms | 0.5 | 1 | 0.5 | 0.125 | NA |
| 3 | no polymorphisms | no polymorphisms | 0.25 | 0.5 | 1 | 0.125 | NA |
| 4 | no polymorphisms | no polymorphisms | 0.25 | 0.5 | 1 | 0.125 | 1 |
| 5 | no polymorphisms | no polymorphisms | 0.5 | 1 | 0.5 | 0.125 | 1 |
| 6 | no polymorphisms | no polymorphisms | 0.125 | 0.25 | 0.25 | 0.03 | 0.5 |
| 7 | no polymorphisms | no polymorphisms | 0.5 | 0.5 | 0.25 | 0.125 | NA |
| 8b | no polymorphisms | no polymorphisms | >8 | 0.25 | 2 | 1 | >8 |
| 9 | no polymorphisms | no polymorphisms | 0.25 | 2 | 1 | 0.06 | NA |
| 10 | D262Yc | no polymorphisms | 0.25 | 0.5 | 0.5 | 0.06 | NA |
| 11 | no polymorphisms | no polymorphisms | 0.5 | 0.5 | 0.5 | 0.125 | NA |
| 12 | G448S | G448S | >8 | 0.5 | 8 | 0.5 | 8 |
| 13 | A284Tc | no polymorphisms | >8 | 0.5 | >8 | >8 | >8 |
| 14 | TR34/L98H | TR34/L98H | >8 | 1 | 4 | 0.5 | 8 |
| 15b | no polymorphisms | no polymorphisms | >8 | 1 | 4 | 1 | 8 |
| 16 | F219V | F219V | >8 | 0.5 | 0.5 | 1 | NA |
| 17 | G54W | G54W | >8 | 0.5 | 1 | >8 | NA |
| 18 | G54W | G54W | >8 | 0.5 | 1 | >8 | NA |
| 19 | no polymorphisms | no polymorphisms | 0.5 | 0.5 | 1 | 0.25 | NA |
| 20 | P216H | P216H | >8 | 0.25 | 0.5 | 0.125 | 1 |
| 21d | G448S | G448S | >8 | 2 | >8 | >8 | >8 |
| 22b | no polymorphisms | no polymorphisms | >8 | 0.5 | 0.5 | 1 | NA |
| 23 | no polymorphisms | no polymorphisms | 1 | 0.5 | 1 | 0.25 | NA |
| 24 | M220T | M220T | >8 | 0.25 | 1 | 0.5 | 2 |
| 25 b,d | no polymorphisms | no polymorphisms | >8 | 1 | 4 | 2 | 4 |
| 26 | A284Tc | no polymorphisms | >8 | 0.5 | 8 | >8 | >8 |
| 27d | no polymorphisms | no polymorphisms | 0.5 | 1 | 0.25 | 0.125 | NA |
| 28 | TR34/L98H | TR34/L98H | >8 | 1 | 2 | 0.5 | 8 |
| 29 | TR34/L98H | TR34/L98H | >8 | 0.5 | 4 | 1 | 8 |
| 30 | no polymorphisms | no polymorphisms | 0.5 | 0.5 | 2 | 0.125 | NA |
| 31e | no polymorphisms | no polymorphisms | NA | NA | NA | NA | NA |
| 32e | no polymorphisms | no polymorphisms | NA | NA | NA | NA | NA |
| 33f | TR34/L98H | TR34/L98H | >8 | 0.5 | >8 | 1 | NA |
| 34f | M220T | M220T | >8 | 1 | 0.5 | 0.25 | NA |
| 35f | M220K | M220K | >8 | 2 | 2 | 1 | 2 |
| 36f | G54E | G54E | >8 | 2 | 0.5 | >8 | 0.5 |
| 37f | G54R | G54R | >8 | 1 | 0.25 | 2 | NA |
| 38f | G54R | G54R | >8 | 2 | 0.25 | 1 | NA |
| 39f | M220T | M220T | >8 | 1 | 2 | 0.5 | 2 |
| 40f | G54V | G54V | >8 | 0.5 | 0.5 | 1 | NA |
| 41f | TR34/L98H | TR34/L98H | >8 | 0.25 | 4 | 0.5 | NA |
| 42f | G54E | G54E | >8 | 1 | 0.25 | 1 | NA |
| 43 | F219I | F219I | >8 | 0.5 | 0.5 | 1 | 2 |
| 44 | TR34/L98H | TR34/L98H | >8 | 0.25 | 4 | 1 | 8 |
| 45 | G448S | G448S | 2 | 1 | 4 | 0.25 | >8 |
| 46 d,g | TR46/Y121F/T289A | TR46/Y121F/T289A | >8 | 1 | >8 | 1 | NA |
| 47 d,g | TR46/Y121F/T289A | TR46/Y121F/T289A | >8 | 1 | >8 | 1 | NA |
| 48 b,d | no polymorphisms | no polymorphisms | >8 | 0.5 | 8 | 2 | >8 |
| 49 | TR34/L98H | TR34/L98H | >8 | 0.5 | 4 | 1 | 8 |
| 50d | TR34/L98H | TR34/L98H | >8 | 1 | 4 | 0.5 | 4 |
ITC, itraconazole; AMB, amphotericin B; VRC, voriconazole; POS, posaconazole; ISA, isavuconazole.
Susceptibility results are not available.
Isolates that demonstrate no cyp51A polymorphisms, but demonstrate phenotypic triazole resistance.
Pyrosequencing is not yet available for this position.
Isolate confirmed as A. fumigatus by ITS, bt2 and cam Sanger sequencing.
Reference A. fumigatus isolates ATCC 46645 and AF293, respectively.
Sanger sequencing result from a previous publication; isolates confirmed as A. fumigatus by microsatellite typing;22 also pyrosequenced in forward and reverse directions.
Sanger sequencing result from a previous publication.59
Isolates and susceptibility testing
Aspergillus isolates and reference strains (ATCC 46645 and AF293) were obtained from the MRCM depository. A. fumigatus species complex isolates with a spectrum of macro- and micro-morphological characteristics and susceptibility profiles were selected. The identification was confirmed by internal transcribed spacer (ITS), β-tubulin (bt2) and calmodulin (cam) Sanger sequencing or microsatellite typing22 of 17/50 isolates (Table 1). Isolates were grown on Sabouraud chloramphenicol dextrose agar (Oxoid, Basingstoke, UK) at 37°C for 3 days. Spore suspensions were made in PBS with 0.5% Tween 20 (Sigma–Aldrich) for DNA extraction. Respiratory samples, comprising sputa, bronchial washes and bronchoalveolar lavages, were processed using HVC as described previously.29 Susceptibilities were determined using the EUCAST broth microdilution method.40 Routine isavuconazole susceptibility testing was initiated at the MRCM in July 2015 and therefore was not performed for the older isolates tested in this study.
Detection of Aspergillus DNA
DNA was extracted from fungal isolates using the PrepMan Ultra Sample Preparation Reagent (Applied Biosystems). Bead beating was performed with a MagNA Lyser instrument (Roche Life Sciences). Extraction of DNA from respiratory samples (minimum 1.0 mL) was performed using a pre-lysis step with the EXTRAblood Prelysis Kit (ELITechGroup); sputum, but not bronchoalveolar lavage, samples were liquefied prior to this step using dithiothreitol. Automated extraction following liquefaction was performed with the ELITe STAR instrument and ELITe STAR 200 Extraction Kit (ELITechGroup) according to the manufacturer’s instructions. The presence of Aspergillus spp. was confirmed by qPCR using the Aspergillus spp. ELITe MGB Kit (ELITechGroup) on the 7500 Fast Dx Real-Time PCR instrument (Applied Biosystems) as per the manufacturer’s instructions.
Design of the pyrosequencing assay
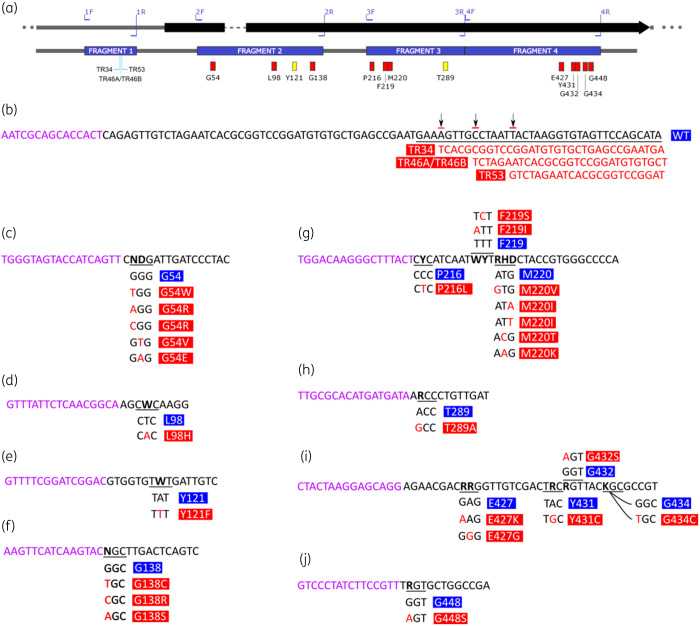
The pyrosequencing method was designed to encompass the 5′ untranslated region and codons of interest within the cyp51A locus and all polymorphisms known to date (Figure 1) using the published reference genomes of A. fumigatus strains AF293 and A1163 (CBS144.89).41 The method consists of two steps: amplification of the four fragments (Figure 1a) using biotinylated primers (Table S1, available as Supplementary data at JAC Online) and pyrosequencing of each fragment separately (Figure 1b–j) using sense or anti-sense sequencing primers (Table S2).
Figure 1.
Overview of the cyp51A gene and the targeted regions and reads. (a) Diagram of A. fumigatus cyp51A demonstrating all the sites monitored by pyrosequencing. Upper diagram shows PCR priming sites. TR sites are shown beneath fragment 1. SNPs associated with resistance are indicated by red rectangles. SNPs associated with TR46-mediated resistance are indicated by yellow rectangles. (b) Expected sense-direction pyrosequencing read for the TR region shown (black text) preceded by the pyrosequencing primer (purple text). TR insertion sites are indicated by arrows, with the expected alternative sequences aligned below (red text). (c to j) Expected sense-direction pyrosequencing reads for amino acid substitution ‘hotspots’ associated with triazole resistance. WT nucleotide sequences are indicated in black, polymorphism(s) are indicated in red and pyrosequencing primers are indicated in purple: (c) glycine 54 (G54); (d) leucine 98 (L98); (e) tyrosine 121 (Y121); (f) glycine 138 (G138); (g) phenylalanine 216 (P216) to methionine 220 (M220); (h) threonine 289 (T289); (i) aspartic acid 427 (E427) to glycine 434 (G434); and (j) glycine 448 (G448). Underlined text indicates the codon target of interest, with variations shown below. Bold or red text indicates a nucleotide position associated with resistance polymorphisms.
Amplification
Amplification was performed on extracts with more than 460 copies of 18S ribosomal DNA (rDNA), confirmed by Aspergillus spp. ELITe MGB qPCR, using a Q-Sat 96 thermal cycler (Hain Lifescience, now part of Bruker Corporation) and the PyroMark PCR Kit (QIAGEN). The number of polymorphisms screened per fragment dictated the volume of master mix required, with a minimum of 25 μL required per assay. For each 25 μL reaction volume, 12.5 μL of PyroMark PCR Master Mix, 0.2 μM forward and reverse primer (one of the pair was biotinylated), 2.5 μL of CoralLoad dye, 5 μL of QIAGEN Q solution and 4 μL of template DNA were used. Thermal cycling was performed with a hot-start step of 95°C for 15 min, followed by a touch-down phase consisting of 10 cycles of denaturation at 95°C for 30 s, annealing between 58 and 56°C (0.2°C decrease with each cycle) for 30 s and elongation at 72°C for 45 s. A subsequent 40 cycles of 30 s at 95°C, 30 s at 56°C and 45 s at 72°C followed, before a 10 min extension step at 72°C. PCR products were analysed by 2% agarose gel electrophoresis.
Pyrosequencing
Amplicons were prepared for pyrosequencing on the PyroMark Q24 Advanced platform (QIAGEN) using PyroMark Q24 Advanced Reagents as per the manufacturer’s instructions. Codons for glycine 54 (Gly-54), leucine 98 (Leu-98), tyrosine 121 (Tyr-121), proline 216 (Pro-216), phenylalanine 219 (Phe-219), methionine 220 (Met-220), threonine 289 (Thr-289) and the region comprising promoter-associated TRs were analysed for polymorphisms (Figure 1) from January 2017. Codons glycine 138 (Gly-138), aspartic acid 427 (Asp-427), tyrosine 431 (Tyr-431), glycine 432 (Gly-432), glycine 434 (Gly-434) and glycine 448 (Gly-448) were screened from mid-September 2018. Individual assays were designed using the QIAGEN Assay Design software (QIAGEN) to span each polymorphism by de novo sequencing (SEQ) or allele quantification (AQ) depending on the multiplicity of the polymorphisms. SEQ assays were used for positions where multiple nucleotide polymorphisms were possible within one codon, whereas semi-quantitative AQ assays were designed for codons with multiple alleles at a single base-pair position (Figure 1 and Table S2). Both sense and anti-sense primers were used for pyrosequencing of 10 isolates during the validation phase, with results compared against Sanger sequencing of the same region.
Sanger sequencing
DNA extracts from isolates and respiratory samples were prepared as above for Sanger sequencing, except that amplification was performed with HotStarTaq Plus Master Mix (QIAGEN) using overlapping sequencing primers (Table S1). Purification of amplicons was performed with the QIAGEN PCR Purification Kit (QIAGEN) according to the manufacturer’s instructions. Sanger sequencing (of both sense and anti-sense strands) was performed by Eurofins Genomics Germany GmbH (Ebersberg, Germany).
Data analysis
The pyrosequencing and Sanger sequencing methods were assessed by comparing the amplification efficiency, which was calculated as the percentage of the actual amplicons produced divided by the total number of amplicons expected. Furthermore, the readability of both sequencing techniques was assessed by calculating the percentage number of reads yielding sequence data divided by the total number of reads. Pyrosequencing data (exemplified in Figure S1) was analysed with use of PyroMark Q24 Advanced software version 3.0.0 (QIAGEN) and compared against known sequences of strain A1163.41 Sequence quality was checked with use of the software’s quality grading system in addition to manual interpretation. Triazole susceptibility was inferred from the sequencing results by comparison with a reference table of A. fumigatus cyp51A polymorphisms.42
Results
Pyrosequencing validation
The pyrosequencing method was as effective as Sanger sequencing for producing readable sequences. A comparison of both sequencing methods revealed that the amplification efficiency was greater for Sanger sequencing than for pyrosequencing (99% versus 96%, respectively). Similarly, the readability of sequences was 99% effective for Sanger sequencing versus 94% for pyrosequencing.
The two sequencing methods produced matching cyp51A genotypes for all the isolates analysed (Table 1), with the exception of three isolates that harboured non-synonymous polymorphisms not currently screened by pyrosequencing: two isolates with alanine 284 to threonine (A284T) and one isolate with glutamic acid 262 to tyrosine (D262Y). A284T has been described to confer reduced susceptibility to itraconazole, voriconazole and posaconazole,43 confirmed in this study along with isavuconazole resistance. D262Y is clinically insignificant,44 also confirmed here.
When the genotypic profiles obtained by pyrosequencing were compared with phenotypic susceptibilities, 16 (32%) isolates had no detectable cyp51A polymorphisms associated with triazole resistance, which corresponded to their phenotypic susceptibility profiles (Table 1). However, five (10%) isolates, which had demonstrated triazole-resistant phenotypes, were found to have no cyp51A polymorphisms by either sequencing method. Twenty-seven (54%) isolates harboured a cyp51A genotype associated with resistance, confirming the phenotypic susceptibility patterns (Table 1). The remaining two isolates contained new polymorphisms, proline 216 to histidine (P216H) and phenylalanine 219 to valine (F219V), both of which coincided with itraconazole-resistant phenotypes. The most numerous polymorphisms were seen at codons glycine 54 (14%), methionine 220 (8%), leucine 98 (16%, combined with the TR34 insertion) and glycine 448 (6%). For 10 isolates, bi-directional pyrosequencing confirmed the reproducibility of forward and reverse reads (Table 1).
Screening human respiratory specimens
A total of 335 respiratory samples were analysed during the study period. Of these samples, 40 were selected randomly for method validation and showed consistent agreement between both sequencing techniques. Pyrosequencing was more sensitive than Sanger sequencing for detecting single, as well as mixed, polymorphisms and for producing complete resistance genotypes (Table S3). Moreover, Sanger sequencing failed to detect one instance each of TR34/L98H and glycine 54 to arginine (G54R) polymorphisms, which were detected by pyrosequencing. Conversely, Sanger sequencing detected one instance of methionine 220 to lysine (M220K) not detected by pyrosequencing. Nine of 335 samples (2.7%) were culture positive for other Aspergillus spp. (Table 2) and were excluded from further analysis as pyrosequencing is limited to A. fumigatus species complex (Figures S2 and S3). Notably, pyrosequencing results were obtained in eight of these specimens, including one case of a glycine 54 to tryptophan (G54W) polymorphism in a background of Aspergillus niger growth (Table S4), indicating that genotyping was possible when A. fumigatus growth was not detected.
Table 2.
Summary of the results of cyp51A pyrosequencing and susceptibility data obtained from respiratory specimens
| Respiratory specimensa | n | % |
|---|---|---|
| Patients | 160 | |
| resistant genotypes in | ||
| CPA/CCPA | 47 | |
| APBA/CPA | 10 | |
| ABPA | 12 | |
| Aspergillus bronchitis with/out fungal sensitization | 4 | |
| resistant phenotypes in | ||
| CPA/CCPA | 21 | |
| APBA/CPA | 5 | |
| ABPA | 6 | |
| Aspergillus bronchitis with/out fungal sensitization | 0 | |
| Samples | 335 | |
| A. fumigatus species complex | 326 | |
| confirmed non-A. fumigatus | 9 | 2.7 |
| HVC results | ||
| positive samples | 94 | 28.8 |
| negative samples | 232 | 71.2 |
| resistant A fumigatus (of positive cultures) | 53 | 56.4 |
| resistant A fumigatus (of all samples) | 16.3 | |
| susceptible A fumigatus (of positive cultures) | 41 | 43.6 |
| susceptible A fumigatus (of all samples) | 12.6 | |
| WT susceptibility inferred by pyrosequencing (of all samples) | 132 | 40.5 |
| Pan-azole resistance | ||
| found by HVC, but not pyrosequencing | 21 | 6.4 |
| found by pyrosequencing, but not HVC | 61 | 18.7 |
| found by pyrosequencing or HVC | 82 | 25.1 |
| Resistance to at least one azole | ||
| found by HVC, but not pyrosequencing | 23 | 7.1 |
| found by pyrosequencing, but not HVC | 73 | 22.4 |
| found by pyrosequencing or HVC | 96 | 29.4 |
| Resistance matched by culture and pyrosequencing (of HVC positives, n = 94) | 25 | 26.6 |
| Agreement of all phenotypes between culture and pyrosequencing | 53 | 56.4 |
| No susceptibility results by pyrosequencing or HVC | 43 | 13.2 |
| Resistance results obtained by pyrosequencing before HVC results | 76 | 23.3 |
| Polymorphisms found by pyrosequencing: | ||
| G54E, R, V, W | 28 | 8.6 |
| TR34/L98H | 59 | 18.1 |
| M220I, K | 4 | 1.2 |
| P216L | 2 | 0.6 |
| F219I | 2 | 0.6 |
| WT | 99 | 30.4 |
| partial WTb | 77 | 23.6 |
| mixedc | 5 | 1.5 |
| Success rate with 460 copies (n = 326)d | 275 | 84.4 |
| Success rate with 1000 copies (n = 229)d | 208 | 90.8 |
Respiratory specimens consisted of 331 sputa, 3 bronchial washes and 1 bronchoalveolar lavage.
Pyrosequencing results were obtained from at least one of four fragments, together representing 90% coverage of the cyp51A gene length.
Polymorphisms found in the same sample: G54R and M220V, I from two CPA patients; M220V, K and L98H from two other CPA patients; and G54W and L98H in an ABPA patient.
Pertains to the pyrosequencing amplification success rate when the minimum yield of Aspergillus spp. qPCR is 460 or 1000 18S rRNA copies, respectively.
Of all the respiratory specimens analysed (Tables S3 and S4), only 28.8% (94/326) reported A. fumigatus growth after 14 days despite the use of higher culture volumes (Table 2). Pyrosequencing demonstrated a WT cyp51A genotype in 40.5% (132/326) and resistant genotypes in 22.4% (73/326) of samples. In contrast, HVC showed WT phenotypic susceptibility in 12.6% (41/326) and resistance in 16.3% (53/326) of samples (Table 2). A wide range of genotypes were identified, including multiple resistant genotypes in five specimens (Tables 2, S3 and S4). Agreement between inferred resistance patterns deduced by pyrosequencing and susceptibility testing was only 56.4% (53/94). Intriguingly, 11 respiratory samples were found to have a pan-azole-resistant or resistant genotype, but demonstrated susceptible MICs (Tables S3 and S4), suggesting phenotypic testing missed the cyp51A resistance detected by pyrosequencing. In contrast, 23 samples demonstrated a resistant phenotype, which pyrosequencing failed to detect, either outright (11/23) or due to paucity of pyrosequencing results, because amplicons could not be generated for the whole cyp51A region (12/23). Nevertheless, resistance was demonstrated by pyrosequencing before HVC results were available in 23.3% (76/326) of cases (Table 2).
The respiratory specimens analysed in this study came from 160 patients, of which 95 (59.4%) were diagnosed with CPA (including chronic cavitary pulmonary aspergillosis, CCPA), 28 (17.5%) were diagnosed with ABPA, 20 (12.5%) were diagnosed with both ABPA and CPA, 2 (1.3%) had sub-acute invasive aspergillosis, 7 (4.4%) had Aspergillus bronchitis, 4 (2.5%) had severe asthma with fungal sensitization, 2 (1.3%) had Aspergillus bronchitis with or without fungal sensitization and 2 (1.3%) had cystic fibrosis. In pyrosequenced samples, resistance was found most often in CPA patient specimens (Table 2), with 29.5% (28/95) demonstrating the pan-azole-resistant genotype and 14.5% (14/95) the glycine 54 polymorphisms, and secondly in ABPA patient specimens, with 28.6% (8/28) and 14.3% (4/28) demonstrating the pan-azole-resistant genotype and the glycine 54 polymorphisms, respectively. Resistance determined by culture was significantly lower in all patient groups (Tables 2, S3 and S4) and no resistance was found, by either pyrosequencing or culture, in patients with sub-acute invasive aspergillosis, severe asthma with fungal sensitization or cystic fibrosis. Mixed resistant genotypes determined by pyrosequencing were found in five patients: four with CPA and one with ABPA (Table 2); for none of these cases were there cultures available for susceptibility testing. Finally, concomitant presence of WT and resistant genotypes occurred in 33 samples, predominantly from CPA patients, with ABPA/CPA and APBA patient specimens demonstrating similar patterns in 9 and 8 specimens, respectively.
Discussion
The rising incidence of aspergillosis caused by triazole-resistant A. fumigatus is an alarming and significant global issue. Mortality and morbidity from both invasive and chronic forms of aspergillosis could be greatly reduced by rapid detection of triazole resistance and prompt revision of antifungal treatment.1,3,6 As triazole resistance in A. fumigatus is frequently associated with polymorphisms in the cyp51A gene, molecular methods targeting this region offer a new approach to diagnostic antifungal stewardship. Screening for cyp51A genotype prevalence can also provide essential epidemiological data for local empirical guideline development.26 The pyrosequencing assay described here was shown to be able to detect a wide range of A. fumigatus cyp51A polymorphisms associated with triazole resistance, including those not identified by commercial assays. This method allowed prompt recognition of resistance and the selection of appropriate antifungal treatment when culture was negative.
Pyrosequencing, unlike Sanger sequencing, was able to detect both susceptible and resistant genotypes when concomitantly present in a clinical sample. Polyclonal colonization is particularly common in patients with chronic or allergic conditions, but is seen in patients with invasive disease as well. In patients on antifungal therapy, this polyclonal colonization is likely to lead to dynamic ratios of the various A. fumigatus genotypes.17,22,36,45 The PyroMark platform excels in detecting the characteristics of mixed populations when assays are designed appropriately.46 In our study, where possible, AQ assays were used to allow the detection and semi-quantification of mixed templates (Figure S1), although detection (but not quantification) is also possible using SEQ assays. The AQ assay could theoretically be used to track the advancement of a resistant A. fumigatus population when performed regularly on serial samples. In practice, the emergence of a resistant cyp51A genotype could be monitored before phenotypic resistance was detected, as evidenced in over 20% of the samples analysed. Consequently, the ability to detect subtleties in mixed samples could permit modification of patient treatment before a resistant phenotype became dominant within an infected patient. The advantage of this technique is the duality of its application. The presence of a resistant genotype(s) in an IA patient can be confirmed rapidly and direct urgent therapy. In contrast, pyrosequencing is ideal for long-term monitoring for CPA and ABPA patients whose mycobiomes may evolve more slowly and for whom, in particular, being able to monitor WT/resistant organisms is key for therapeutic success.
Pyrosequencing was significantly more effective than susceptibility testing for assessing the presence of triazole resistance, because of the low rate of culture positivity, even when using HVC. Yet, when culture was available, resistance was detected by phenotypic means more frequently than by pyrosequencing. This paradox may be due to the fact that selection of isolates for susceptibility testing is based on their typical morphological phenotype rather than on molecular confirmation of A. fumigatus. It is well known that isolates from patients with a long history of antifungal therapy and antifungal resistance are often phenotypically atypical and appear similar to cryptic species. Consequently, a closely related species, such as Aspergillus fischeri or Aspergillus lentulus, with some level of intrinsic resistance to triazoles, may have been reported as azole-resistant A. fumigatus,47 but they would not yield any polymorphisms by pyrosequencing, because the resistance mechanism is not cyp51A based. Also, as pyrosequencing is specific to A. fumigatus, it would fail on cryptic species. Moreover, phenotypic resistance may have originated from efflux, overexpression of cyp51B,48,49 uncharacterized cyp51A mutations or polymorphisms in gene(s) other than cyp51A,50–52 none of which this pyrosequencing assay can identify.
The findings in our study highlight the significant flexibility of pyrosequencing compared with qPCR-based commercial assays. For example, pyrosequencing identified the A284T polymorphism43 together with an undescribed P216H polymorphism associated with itraconazole resistance in pan-azole-resistant isolates recovered from a single patient. Other than the insertion and ORF polymorphisms associated with the environmental pan-azole resistance mutations, and recently evidence for glycine 54 and methionine 220 polymorphisms,27 detection of other cyp51A polymorphisms is lacking in commercial assays. Moreover, pyrosequencing yields cyp51 sequence data, which can distinguish other members of the species complex from A. fumigatus, which is not possible using qPCR assays. In addition, assays for detecting novel mechanisms can be incorporated into the Q24 Advanced PyroMark workflow with relative ease. For example, either the tyrosine 433 to asparagine (Y433N) in cyp51A53 or the lysine 363 to arginine (K363R) erg5 polymorphism52 may explain the pan-triazole and amphotericin B resistance found in the isolates in this study lacking cyp51A polymorphisms. The advantage of pyrosequencing over newer sequencing approaches is the determination of definitive sequence information rather than a reliance on the restriction pattern differences generated by surveyor nucleases and detected by gel electrophoresis.54
The main limitation of this method is the challenge of amplifying low numbers of the single-copy cyp51A gene, as evidenced by an 84% amplification success rate from respiratory samples and 96% when using isolates. In addition, fragment amplification efficiencies varied; for example, fragment 3 (which contains hotspot amino acids proline 216 to methionine 220) is the most successful, while the promoter region (fragment 1) proved the most difficult to amplify, regardless of the iterations of primer sequences assayed during method development. These differences could be explained by varying amplification efficiencies or primer complementarity, amplification insensitivity, PCR inhibition and inferior DNA quality due to the lengthy extraction process required for respiratory samples55 compared with fungal cultures.29 As a consequence of these limitations, we determined that there should be greater than 1000 Aspergillus spp. rDNA copies in a respiratory sample to improve downstream pyrosequencing. Considering the 18S rDNA:single gene ratio in A. fumigatus varies from 38:1 to 91:1,56 this cut-off translates to a limit of detection of 11–26 genomes, in line with probe-based qPCR resistance assays.34,57,58 Consequently, a 1000 copy cut-off improved the pyrosequencing success rate from 84.4% to 90.8%. Although use of an A. fumigatus-specific probe to further discriminate non-Aspergillus templates would not completely rule out those respiratory specimens containing templates of members of A. fumigatus species complex (see Figure S2), it would reduce testing of samples containing other Aspergillus species (such as Aspergillus flavus and A. niger; Figure S3) and improve the success rate of the method even further.
In our centre, cyp51A findings via this method are routinely discussed with clinicians and compared against other laboratory findings to inform decisions on revision of antifungal treatment. A significant level of resistance was not unexpected in this group of patients, consisting mainly of ABPA and CPA patients, who are known to acquire A. fumigatus cyp51A polymorphisms over the course of long-term triazole therapy. Our cyp51A pyrosequencing screening approach is an effective means of deciphering resistance mechanisms from a range of clinically relevant samples containing A. fumigatus and is particularly effective when phenotypic testing is not possible. This technique is easily adaptable, requires low computing power and can be implemented easily within a molecular-based clinical laboratory workflow.
Supplementary Material
Acknowledgements
Part of this data was presented at the Ninth Trends in Medical Mycology Conference, Nice, France, 2019 (Poster 161).
We thank the members of the NHS MRCM and the NAC for their continued support. We thank Marcin Walczak for assembling the NAC database, which was used to confirm patient data and diagnoses, and Maisie Palmer for generating A. fumigatus species complex data. We also thank Lottie Brown for presenting part of this data at the Ninth Trends in Medical Mycology Conference.
Funding
This research was co-funded by the National Institutes of Health Research Manchester Biomedical Research Centre (BRC-1215-20007) and the National Health Service England.
Transparency declarations
L.N.-F. has received travel grants from and has been paid for talks by Gilead Sciences. C.B.M. has received grant support from the Fungal Research Trust and Pfizer, has received travel grants from Astellas and Gilead, and has been paid for lectures on behalf of Pfizer. M.D.R. has received grant support from Gilead Sciences, Pfizer and MSD, acts as a consultant and speaker for Gilead Sciences and Pfizer, is co-founder of Richardson Bio-Tech (Guangzhou) Limited and is a long-standing member of the European Society for Clinical Microbiology and Infectious Diseases Aspergillosis Guidelines, Mucormycosis and Rare Moulds writing group. D.W.D. holds founder shares in F2G Ltd, a University of Manchester spin-out antifungal discovery company, acts or has recently acted as a consultant to Scynexis, Pulmatrix, Pulmocide, Zambon, iCo Therapeutics, Roivant, Mayne Pharma, Biosergen and Fujifilm, has been paid for talks on behalf of Hikma, Gilead, Mylan and Pfizer (in the last 3 years), was co-applicant of the original University of South Manchester NHS Foundation Trust/NHS England business case that secured funding for this project and is a long-standing member of the Infectious Diseases Society of America Aspergillosis Guidelines group, the European Society for Clinical Microbiology and Infectious Diseases Aspergillosis Guidelines group and the British Society for Medical Mycology Standards of Care committee. R.R.-R. has been paid for lectures on behalf of Astellas, Gilead, Pfizer, MSD and Basilea. All other authors: none to declare.
Author contributions
L.N.-F. conceived the project. L.N.-F., S.P.A.-H. and D.H. designed the validation study. S.P.A.-H., D.H. and R.M. performed the experiments. L.N.-F. compiled the audit data. L.N.-F., S.P.A.-H. and D.H. analysed the results. S.P.A.-H. and L.N.-F. co-wrote the manuscript. C.B.M., M.D.R., R.R.-R. and D.W.D. gave clinical and conceptual advice and contributed to writing the paper.
References
- 1. Kosmidis C, Denning DW. The clinical spectrum of pulmonary aspergillosis. Thorax 2015; 70: 270–7. [DOI] [PubMed] [Google Scholar]
- 2. Sugui JA, Kwon-Chung KJ, Juvvadi PR et al. Aspergillus fumigatus and related species. Cold Spring Harb Perspect Med 2014; 5: a019786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Brown GD, Denning DW, Gow NAR et al. Hidden killers: human fungal infections. Sci Transl Med 2012; 4: 165rv13. [DOI] [PubMed] [Google Scholar]
- 4. Denning DW, Pashley C, Hartl D et al. Fungal allergy in asthma—state of the art and research needs. Clin Transl Allergy 2014; 4: 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kosmidis C, Denning DW. Chapter 189: Opportunistic and systemic fungi In: Cohen J, Powderly WG, Opal SM, eds. Infectious Diseases (Fourth Edition) Volume 2. Elsevier, 2017; 1681–709. [Google Scholar]
- 6. Taccone FS, Van den Abeele A-M, Bulpa P et al. Epidemiology of invasive aspergillosis in critically ill patients: clinical presentation, underlying conditions, and outcomes. Crit Care 2015; 19: 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Denning DW, Cadranel J, Beigelman-Aubry C et al. Chronic pulmonary aspergillosis: rationale and clinical guidelines for diagnosis and management. Eur Respir J 2016; 47: 45–68. [DOI] [PubMed] [Google Scholar]
- 8. Lowes D, Al-Shair K, Newton PJ et al. Predictors of mortality in chronic pulmonary aspergillosis. Eur Respir J 2017; 49: 1601062. [DOI] [PubMed] [Google Scholar]
- 9. Denning DW, Pleuvry A, Cole DC. Global burden of allergic bronchopulmonary aspergillosis with asthma and its complication chronic pulmonary aspergillosis in adults. Med Mycol 2013; 51: 361–70. [DOI] [PubMed] [Google Scholar]
- 10. Ullmann AJ, Aguado JM, Arikan-Akdagli S et al. Diagnosis and management of Aspergillus diseases: executive summary of the 2017 ESCMID-ECMM-ERS guideline. Clin Microbiol Infect 2018; 24: e1–38. [DOI] [PubMed] [Google Scholar]
- 11. Abdolrasouli A, Petrou MA, Park H et al. Surveillance for azole-resistant Aspergillus fumigatus in a centralized diagnostic mycology service, London, United Kingdom, 1998-2017. Front Microbiol 2018; 9: 2234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chen Y, Lu Z, Zhao J et al. Epidemiology and molecular characterizations of azole resistance in clinical and environmental Aspergillus fumigatus isolates from China. Antimicrob Agents Chemother 2016; 60: 5878–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chowdhary A, Sharma C, Meis JF. Azole-resistant aspergillosis: epidemiology, molecular mechanisms, and treatment. J Infect Dis 2017; 216 Suppl 3: S436–44. [DOI] [PubMed] [Google Scholar]
- 14. Hurst SF, Berkow EL, Stevenson KL et al. Isolation of azole-resistant Aspergillus fumigatus from the environment in the south-eastern USA. J Antimicrob Chemother 2017; 72: 2443–6. [DOI] [PubMed] [Google Scholar]
- 15. Sewell TR, Zhang Y, Brackin AP et al. Elevated prevalence of azole-resistant Aspergillus fumigatus in urban versus rural environments in the United Kingdom. Antimicrob Agents Chemother 2019; 63: e00548–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. van der Linden JWM, Arendrup MC, Warris A et al. Prospective multicenter international surveillance of azole resistance in Aspergillus fumigatus. Emerg Infect Dis 2015; 21: 1041–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Camps SMT, van der Linden JWM, Li Y et al. Rapid induction of multiple resistance mechanisms in Aspergillus fumigatus during azole therapy: a case study and review of the literature. Antimicrob Agents Chemother 2012; 56: 10–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hagiwara D, Watanabe A, Kamei K et al. Epidemiological and genomic landscape of azole resistance mechanisms in Aspergillus fungi. Front Microbiol 2016; 7: 1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Resendiz Sharpe A, Lagrou K, Meis JF et al. Triazole resistance surveillance in Aspergillus fumigatus. Med Mycol 2018; 56 Suppl 1: S83–92. [DOI] [PubMed] [Google Scholar]
- 20. Garcia-Rubio R, Alcazar-Fuoli L, Monteiro MC et al. Insight into the significance of Aspergillus fumigatus cyp51A polymorphisms. Antimicrob Agents Chemother 2018; 62: e00241–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Verweij PE, Howard SJ, Melchers WJG et al. Azole-resistance in Aspergillus: proposed nomenclature and breakpoints. Drug Resist Updat 2009; 12: 141–7. [DOI] [PubMed] [Google Scholar]
- 22. Howard SJ, Cerar D, Anderson MJ et al. Frequency and evolution of azole resistance in Aspergillus fumigatus associated with treatment failure. Emerg Infect Dis 2009; 15: 1068–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Howard SJ, Pasqualotto AC, Denning DW. Azole resistance in allergic bronchopulmonary aspergillosis and Aspergillus bronchitis. Clin Microbiol Infect 2010; 16: 683–8. [DOI] [PubMed] [Google Scholar]
- 24. Fuhren J, Voskuil WS, Boel CHE et al. High prevalence of azole resistance in Aspergillus fumigatus isolates from high-risk patients. J Antimicrob Chemother 2015; 70: 2894–8. [DOI] [PubMed] [Google Scholar]
- 25. Snelders E, Karawajczyk A, Verhoeven RJA et al. The structure–function relationship of the Aspergillus fumigatus cyp51A L98H conversion by site-directed mutagenesis: the mechanism of L98H azole resistance. Fungal Genet Biol 2011; 48: 1062–70. [DOI] [PubMed] [Google Scholar]
- 26. Verweij PE, Ananda-Rajah M, Andes D et al. International expert opinion on the management of infection caused by azole-resistant Aspergillus fumigatus. Drug Resist Updat 2015; 21-22: 30–40. [DOI] [PubMed] [Google Scholar]
- 27. Singh A, Sharma B, Mahto KK et al. High-frequency direct detection of triazole resistance in Aspergillus fumigatus from patients with chronic pulmonary fungal diseases in India. J Fungi 2020; 6: 67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Takazono T, Izumikawa K. Recent advances in diagnosing chronic pulmonary aspergillosis. Front Microbiol 2018; 9: 1810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Vergidis P, Moore CB, Novak-Frazer L et al. High-volume culture and quantitative real-time PCR for the detection of Aspergillus in sputum. Clin Microbiol Infect 2020; 26: 935–40. [DOI] [PubMed] [Google Scholar]
- 30. Rodriguez-Tudela JL, Alcazar-Fuoli L, Alastruey-Izquierdo A et al. Time of incubation for antifungal susceptibility testing of Aspergillus fumigatus: can MIC values be obtained at 24 hours? Antimicrob Agents Chemother 2007; 51: 4502–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Guinea J, Verweij PE, Meletiadis J et al. How to: EUCAST recommendations on the screening procedure E.Def 10.1 for the detection of azole resistance in Aspergillus fumigatus isolates using four-well azole-containing agar plates. Clin Microbiol Infect 2019; 25: 681–7. [DOI] [PubMed] [Google Scholar]
- 32. Buil JB, Zoll J, Verweij PE et al. Molecular detection of azole-resistant Aspergillus fumigatus in clinical samples. Front Microbiol 2018; 9: 515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Dudakova A, Spiess B, Tangwattanachuleeporn M et al. Molecular tools for the detection and deduction of azole antifungal drug resistance phenotypes in Aspergillus species. Clin Microbiol Rev 2017; 30: 1065–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Dannaoui E, Gabriel F, Gaboyard M et al. Molecular diagnosis of invasive aspergillosis and detection of azole resistance by a newly commercialized PCR kit. J Clin Microbiol 2017; 55: 3210–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Postina P, Skladny J, Boch T et al. Comparison of two molecular assays for detection and characterization of Aspergillus fumigatus triazole resistance and cyp51A mutations in clinical isolates and primary clinical samples of immunocompromised patients. Front Microbiol 2018; 9: 555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Schauwvlieghe AFAD, Vonk AG, Buddingh EP et al. Detection of azole-susceptible and azole-resistant Aspergillus coinfection by cyp51A PCR amplicon melting curve analysis. J Antimicrob Chemother 2017; 72: 3047–50. [DOI] [PubMed] [Google Scholar]
- 37. Wickes BL, Wiederhold NP. Molecular diagnostics in medical mycology. Nat Commun 2018; 9: 5135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. van der Torre MH, Novak-Frazer L, Rautemaa-Richardson R. Detecting azole-antifungal resistance in Aspergillus fumigatus by pyrosequencing. J Fungi 2020; 6: 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.HRA Decision Tool. http://www.hra-decisiontools.org.uk/research/.
- 40.EUCAST. EUCAST Method for Susceptibility Testing of Moulds, Version 9.3.1, January 2017. http://www.eucast.org/astoffungi/methodsinantifungalsusceptibilitytesting/susceptibility_testing_of_moulds/.
- 41. O’Leary NA, Wright MW, Brister JR et al. Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res 2015; 44: D733–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Stensvold CR, Jørgensen LN, Arendrup MC. Azole-resistant invasive aspergillosis: relationship to agriculture. Curr Fungal Infect Rep 2012; 6: 178–91. [Google Scholar]
- 43. Bueid A, Howard SJ, Moore CB et al. Azole antifungal resistance in Aspergillus fumigatus: 2008 and 2009. J Antimicrob Chemother 2010; 65: 2116–18. [DOI] [PubMed] [Google Scholar]
- 44. Escribano P, Recio S, Peláez T et al. In vitro acquisition of secondary azole resistance in Aspergillus fumigatus isolates after prolonged exposure to itraconazole: presence of heteroresistant populations. Antimicrob Agents Chemother 2012; 56: 174–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Alvarez-Perez S, Garcia ME, Bouza E et al. Characterization of multiple isolates of Aspergillus fumigatus from patients: genotype, mating type and invasiveness. Med Mycol 2009; 47: 601–8. [DOI] [PubMed] [Google Scholar]
- 46. Mello FCA, Lago BV, Lewis-Ximenez LL et al. Detection of mixed populations of wild-type and YMDD hepatitis B variants by pyrosequencing in acutely and chronically infected patients. BMC Microbiol 2012; 12: 96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Lamoth F. Aspergillus fumigatus-related species in clinical practice. Front Microbiol 2016; 7: 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Buied A, Moore CB, Denning DW et al. High-level expression of cyp51B in azole-resistant clinical Aspergillus fumigatus isolates. J Antimicrob Chemother 2012; 68: 512–14. [DOI] [PubMed] [Google Scholar]
- 49. Fraczek MG, Bromley M, Buied A et al. The cdr1B efflux transporter is associated with non-cyp51A-mediated itraconazole resistance in Aspergillus fumigatus. J Antimicrob Chemother 2013; 68: 1486–96. [DOI] [PubMed] [Google Scholar]
- 50. Camps SMT, Dutilh BE, Arendrup MC et al. Discovery of a hapE mutation that causes azole resistance in Aspergillus fumigatus through whole genome sequencing and sexual crossing. PLoS One 2012; 7: e50034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Hagiwara D, Arai T, Takahashi H et al. Non-cyp51A azole-resistant Aspergillus fumigatus isolates with mutation in HMG-CoA reductase. Emerg Infect Dis 2018; 24: 1889–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Rybak JM, Ge W, Wiederhold NP et al. Mutations in hmg1, challenging the paradigm of clinical triazole resistance in Aspergillus fumigatus. mBio 2019; 10: e00437–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Chen P, Liu M, Zeng Q et al. Uncovering new mutations conferring azole resistance in the Aspergillus fumigatus cyp51A gene. Front Microbiol 2020; 10: 3127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Arai T, Majima H, Watanabe A et al. A simple method to detect point mutations in Aspergillus fumigatus cyp51A gene using a Surveyor nuclease assay. Antimicrob Agents Chemother 2020; 64: e02271–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. White PL, Barnes RA. Aspergillus PCR – platforms, strengths and weaknesses. Med Mycol 2006; 44 Suppl 1: S191–8. [DOI] [PubMed] [Google Scholar]
- 56. Herrera ML, Vallor AC, Gelfond JA et al. Strain-dependent variation in 18S ribosomal DNA copy numbers in Aspergillus fumigatus. J Clin Microbiol 2009; 47: 1325–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. White PL, Posso RB, Barnes RA. Analytical and clinical evaluation of the PathoNostics AsperGenius assay for detection of invasive aspergillosis and resistance to azole antifungal drugs during testing of serum samples. J Clin Microbiol 2015; 53: 2115–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. White PL, Posso RB, Barnes RA. Analytical and clinical evaluation of the PathoNostics AsperGenius assay for detection of invasive aspergillosis and resistance to azole antifungal drugs directly from plasma samples. J Clin Microbiol 2017; 55: 2356–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Moore CB, Novak-Frazer L, Muldoon E et al. First isolation of the pan-azole-resistant Aspergillus fumigatus cyp51A TR46/Y121F/T289A mutant in a UK patient. Int J Antimicrob Agents 2017; 49: 512–14. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.